Abstract
Background:
Medulloblastoma is the most common malignant brain tumor in children, currently treated uniformly based on histopathology and clinico-radiological risk stratification leading to unpredictable relapses and therapeutic failures. Identification of molecular subgroups have thrown light on the reasons for these and now reveals clues to profile molecularly based personalized therapy against these tumors.
Methods:
Research and online contents were evaluated for pediatric medulloblastoma which included latest information on the molecular subgroups and their clinical relevance and update on efforts to translate them into clinics.
Results:
Scientific endeavors over the last decade have clearly identified four molecular variants (WNT, SHH, Group 3, and Group 4) and their demographic, genomic, and epigenetic profile. Latest revelations include significant heterogeneity within these subgroups and 12 different subtypes of MB are now identified with disparate outcomes and biology. These findings have important implications for stratification and profiling future clinical trials against these formidable tumors.
Conclusion:
With the continued outpouring of genomic/epigenomic data of these molecular subgroups and evolution of further subtypes in each subgroup, the challenge lies in comprehensive evaluation of these informations. Current and future endeavors are now needed to profile personalized therapy for each child based on the molecular risk stratification of medulloblastoma, with a hope to improve survival outcome and reduce relapses.
Keywords: Childhood medulloblastoma, targeted therapy, molecular subtypes, epigenetic machinery, newer treatment strategies, therapeutic resistance
1. INTRODUCTION
Medulloblastoma (MB) is the most common malignant pediatric brain tumor, accounting for nearly 20% of all childhood central nervous system (CNS) malignancies [1]. The major histopathological classification of these tumors consist of the classic, desmoplastic nodular (MB/N), MB with extensive nodularity (MBEN), and large cell/anaplastic (LC/A) types. Prognosis has varied between these subtypes, with MBEN having a better outcome than the LC/A tumors [2, 3]. Current treatment strategies are tailored based on clinico-radiological risk criterion. These include the standard-risk (SR) or high-risk (HR) group. Children who are older than 3 years with no evidence of metastatic disease (M0), post-surgical residual tumor less than 1.5 cm2, and histologically non-anaplastic are categorized as SR, while the remaining are considered HR [4]. Children younger than 3 years who have significant residual disease following surgery, LC/A histology and metastatic disease fare worse with poor survival outcome [5]. A multimodality therapeutic approach is used to treat children over three years, which includes maximal safe surgical resection, concurrent chemotherapy with craniospinal radiation (CSI) and a boost to the tumor bed, usually followed by adjuvant chemotherapy [6, 7]. Children younger than 3 years are treated postoperatively with high-dose chemotherapy as an irradiation-avoiding strategy or with non-high-dose chemotherapy during induction followed by a reduced dose of conformal radiotherapy (CRT) to the tumor bed [8-10], NCT00602667/SJYC07, NCT02724579.
Over the last two decades, many clinical trials have been pursued (Table 1). Though this has resulted in increased overall survival (OS), long-term side effects are concerning [11]. These include neurocognitive deficits, hearing loss, endocrine dysfunction and increased incidence of secondary malignancies [12, 13]. The uniform treatment approach based on the histopathology and risk group has been undermined by the unpredictable relapse and heterogeneity in survival outcome, and nearly one-third of patients die within 5 years of diagnosis. Four subgroups of tumors (WNT, SHH, Group 3 and Group 4) with distinctive molecular profiles and prognosis, have now been identified. These findings have been validated in multiple studies, and it is now clear that critical prognostic factors are defined not only by histology, but also by subgroup assignments and metastatic status [14]. These molecular revelations now explain the variable survival outcomes when children are treated uniformly solely based on histopathological criterion and clinical risk.
Table 1.
Clinical trials.
| S. No. |
Clinical
Trial |
Patients
(Number/Age in (Year) |
Treatment
Risk Group |
Survival Rate | Refs. |
|---|---|---|---|---|---|
| 1. | Gajjar et al, 2006 SJMB 96 |
134 3-21 yrs |
VCR, CPM, CDDP, XRT 23.4 Gy (SR) or 36.0-39.6 Gy (HR) |
5 year OS: 83% SR group 70% HR group |
[4] |
| 2. | Von Bueren et al, 2011 HIT 2000 |
45 < 4 yrs |
No XRT CPM, Mtx, Carboplatin, VP16, VCR, intraventricular Mtx M0 |
5 year OS: 80% +/- 6% | [5] |
| 3. | Rutkowski et al, 2008 HIT-SKK’87 |
29 < 3 yrs |
No XRT Procarbazine, Ifosfamide, VP16, CDDP, Ara-C, Mtx 26 pts M0/M1, 3 pts M2/M3 |
10-year OS: 58.8% ± 11.9% (complete resection), 66.7% ± 15.7% (incomplete resection) | [8] |
| 4. | Chi et al, 2004 Head Start II |
21 <10 yrs |
VCR, CDDP, VP16, CPM, Mtx HR |
3-year OS: 60% | [9] |
| 5. | Packer et al 2006 Phase III |
379 3-21 yrs |
XRT with VCR Reg A: CCNU, VCR CDDP, Reg B: CDDP, CPM, VCR SR |
5 year OS: 86% ± 9% | [15] |
| 6. | Kortmann et al, 2000 HIT’91 |
137 3-18 yrs |
XRT Neoadjuvant arm: Ifosfamide, VP16, Mtx, CDDP, Ara-C Adjuvant arm: VCR, CDDP, CCNU SR and HR |
3 year relapse-free survival: 0.70+/-0.08 |
[20] |
| 7. | Tarbell et al 2013 POG 9031 |
154 3-21 yrs |
Randomized to pre XRT CT (CT1) or post XRT CT (RT1) CDDP, VP16, Consolidation with CPM, VCR HR |
5 year OS: 73.1% CT1 arm, 76.1% RT1 arm |
[21] |
| 9. | Jakacki et al, 2011 COG Phase I/II |
161 3-21 yrs |
Carboplatin and VCR during XRT Maintenance regimen A: CPM and VCR Maintenance regimen B: CDDP, CPM, VCR HR |
5-year OS: Regimen A - 82% +/- 9% Regimen B 68% +/- 10% |
[22] |
| 10. | Von Bueren et al 2016 | 123 4-21 yrs |
Induction CT, dose escalated CSI XRT, maintenance CT HR |
5 year: EFS 62%, OS 74% for all patients 5 year OS 100% for WNT and 20% for MYCC/MYCN amplification tumors |
[23] |
| 11. | Perez-Martinez et al, 2005 | 19 2-15 yrs |
XRT for patients > 4 years Induction CT and then HDCT (busulfan-melphalan, busulfan-thiotepa, topotecan) HR/Recurrent tumors |
2 year EFS: 37.67+/-14% in all patients 57+/-15% for the HR group. |
[29] |
Abbreviations: Ara-C, cytosine arabinoside; CCNU, Lomustine; CDDP, cisplatin; CPM, cyclophosphamide; CSI, craniospinal radiation; CT, chemotherapy; EFS, event-free survival; HDCT, high dose chemotherapy; HR: high risk; Mtx- methotrexate; M0: no metastasis; OS, overall survival, SR, standard risk; XRT, radiotherapy.
2. TREATMENT STRATEGIES BASED ON RISK STRATIFICATION
For SR MB patients, following surgery, a reduced-dose radiation strategy using CSI to a total dose of 23.4 Gy plus a boost to the posterior fossa up to a total dose of 54-55.8 Gy, has been usually used along with concurrent and adjuvant chemotherapy. A large study was performed on 379 evaluable patients with SR MB who were treated with 23.4 Gy of CSI and 55.8 Gy of posterior fossa RT and randomly assigned to one of the two adjuvant chemotherapy regimens: lomustine (CCNU), cisplatin, and vincristine; or cyclophosphamide, cisplatin, and vincristine. Five-year event free survival (EFS) and overall survival (OS) for the cohort of 379 patients were 81% +/- 2.1% and 86% +/- 9%, respectively [15]. A European study (HIT-SIOP PNET4) on 340 children aged 4 to 21 years with SR MB compared conventional CSI at a dose of 23.4 Gy plus boost with hyperfractionated CSI at a dose of 36 Gy plus boost, followed by similar chemotherapy regimens for both arms. A follow-up of 4.8 years did not find a significant difference in the survival rates between the two therapeutic strategies [16]. The Children’s Oncology Group (COG) completed a study (ACNS0331) on 513 children with SR MB, evaluating and comparing limited target volume boost irradiation (tumor bed versus posterior fossa) and reduced-dose CSI (18 Gy versus standard 23.4 Gy) in a subset of patients between 3-7 years of age, with an intent to alleviate toxicity without compromising OS and EFS. The results of this study are pending, but preliminary data show no evidence of a significant difference in EFS between the two fields of radiation, and CSI cannot be safely reduced from 23.4 Gy to 18 Gy without compromising clinical outcome (NCT00085735). Currently, an ongoing study (SIOP PNET5) is evaluating the role of β-catenin status in SR MB. Patients with β-catenin-positive tumors are assigned to a low-risk (LR) treatment arm (PNET 5 MB – LR) and patients with other SR MB are assigned to a standard-risk treatment arm (PNET 5 MB – SR). The LR patients receive reduced CSI to 18 Gy without carboplatin and reduced-intensity maintenance chemotherapy (NCT02066150).
For HR MB patients, an earlier study showed that the addition of adjuvant chemotherapy was effective in improving the rate of disease-free survival in children with poor-risk MB/PNET compared to those who received radiotherapy alone [17]. Studies by the French Society of Pediatric Oncology (SFOP), COG 921, and HIT91 for HR MB did not fare well with an EFS of less than 50% [18-20]. Though these studies used multiple chemotherapy regimens, inferior survival outcome was seen, likely due to the delay in starting radiation therapy. Another study randomized 112 HR MB patients to receive chemotherapy before or after radiation followed by similar adjuvant chemotherapy. An improved 5 year EFS of 66 and 70% for the respective arms was noted, though no statistical difference in OS was achieved for patients receiving post-surgical chemotherapy before or after radiation [21]. A more recent study treated HR patients with 36 Gy CSI along with weekly vincristine and carboplatin, followed by chemotherapy with cyclophosphamide and vincristine (Reg A), and later cisplatin was added (Reg B) to the adjuvant therapy regimen. The 5-year OS was 82% and 68% for the respective arms, which showed improved survival rates over historical controls for these patients [22]. A study was performed on 123 metastatic MB patients to evaluate prognostic relevance of clinical and biologic parameters. Treatment included induction chemotherapy, dose-escalated hyperfractionated CSI, followed by maintenance chemotherapy. The 5 year OS was 74% for the entire group with a 100% OS seen in those tumors driven by WNT pathway [23].
3. TARGETED THERAPIES
Recent clinical trials have focused on using newer drugs, targeting signaling pathways and molecules which have emerged as key oncogenic drivers of these tumors. The Notch signaling pathway, which induces proliferation and survival of neural precursors, has been shown to drive MB formation [24]. Preclinical studies demonstrated overexpression and the role of vascular endothelial growth factor (VEGF), platelet derived growth factor (PDGFR), histone deacetylase (HDAC), and the cyclooxygenase 2 (COX2) proteins in inducing MB formation [25]. These events have led to clinical trials targeting these tumorigenic drivers. A completed clinical trial targeting the Notch pathway by inhibiting the γ-secretase, using a weekly dose of MK-0752 (γ-secretase inhibitor) in 10 patients with recurrent CNS malignancies (including MB), showed safety and inhibition of NOTCH expression. No objective responses were noted in any patients. Future trials using MK-0752 in combination with targeted agents (e.g., mTOR/Akt inhibitors) or chemotherapeutic agents (e.g., temozolomide) are planned to be explored in phase I trials [26]. The role of bevacizumab (VEGF inhibitor) has been explored alone and in combination with irinotecan (topoisomerase I inhibitor) and/or temozolomide (DNA alkylating agent) with good tolerability and variable response [27, 28].
4. HIGH DOSE CHEMOTHERAPY WITH AUTOLOGOUS STEM CELL RESCUE
As an irradiation-avoiding strategy, the role of high-dose chemotherapy (HDC) with autologous stem cell rescue (ASCR) has been explored for newly diagnosed and relapsed MB, with some studies showing good clinical outcome and an effective alternative therapy in younger children. Though HDC with ASCR has been shown to improve survival rates in a subset of patients, the incidence of morbidity and mortality due to veno-occlusive disease and infections have at times undermined the modest survival outcome [29-32].
5. THE MOLECULAR ERA OF MEDULLOBLASTOMA
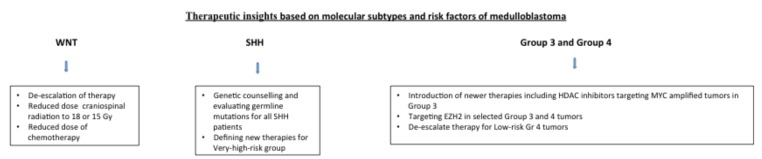
Over the last decade, various transcriptional profiling studies have identified four molecular subgroups of MB: WNT, SHH, Group 3, and Group 4 [33-39]. This has ushered a new era in the realm of MB genomics, and these subgroups are now clearly identified with distinguishing features based on demographics, histology, chromosomal profile, driver genes, and outcome (Table 2) [40-44]. Cancer genomic sequencing studies have provided insight into the drivers of these subgroups, through recurrent structural alterations, somatic mutations, and epigenetic deregulation [45-53]. Parallel research endeavors have also provided a superior understanding of important signaling pathways and mechanisms that induce therapeutic resistance. This has now enabled risk stratification of MB based on the molecular subtypes of these tumors and provide insights to newer therapies (Fig. 1).
Table 2.
Characteristics of medulloblastoma subgroups.
| WNT (10%) | SHH (30%) | Group 3 (25%) | Group 4 (35%) | |
|---|---|---|---|---|
| Clinical Features | ||||
| Age | Older children and adults (median age ~10 years) | Bimodal: < 5 years & >16 years; less common in children aged 5-16 years | Infants and young children | Children (median age ~ 9 years); can occur in all age groups |
| Gender Ratio (M:F) | ~1:1 | ~1.5:1 | ~2:1 | ~3:1 |
| Histology | Classic; rarely LC/A | Classic > desmoplastic nodular > LC/A > MBEN | Classic > LC/A | Classic; rarely LC/A |
| Proposed Cell of Origin | Lower rhombic lip progenitor cells | CGNPs of the EGL and cochlear nucleus; neural stem cells of the subventricular zone | Prominin 1+/lineage- neural stem cells; CGNPs of the EGL | Deep cerebellar nuclei in the nuclear transitory zone; upper rhombic lip progenitor cells |
| Location of Tumor | Fourth ventricle; infiltrating brainstem | Cerebellar hemispheres; rarely midline | Fourth ventricle; midline | Fourth ventricle; midline |
| Metastasis at Diagnosis | ~5-10% | ~15-20% | ~40-45% | ~35-40% |
| Prognosis (5-year survival) |
~95% | ~75% | ~50% | ~75% |
| Genomic Features | ||||
| Cytogenetics | Monosomy 6 | Loss of 9q, 10q, and 17p; Gain of 3q and 9p |
Loss of 10q, 16q, and 17p; Gain of 1q, 7, 17q, and 18 |
Loss of 8, 10, 11, and 17p; Gain of 4, 7, 17q, and 18 |
| Driver Genes (Most prevalent) |
CTNNB1; DDX3X; SMARCA4; KMT2D (MLL2); TP53 | PTCH1; TP53; KMT2D (MLL2); DDX3X; MYCN amplification; BCOR; LDB1; TCF4; GLI2 amplification | MYC amplification; PVT1 amplification; SMARCA4; OTX2 amplification; CTDNEP1; LRP1B; KMT2D (MLL2) | KDM6A; SNCAIP gain; MYCN amplification; KMT2C (MLL3); CDK6 amplification; ZMYM3 |
| Expression Signature |
WNT signaling | SHH signaling | MYC signature; photoreceptor GABAergic signature | Neuronal signature; glutamatergic signature |
Abbreviations: LC/A, large cell/anaplastic; MBEN, medulloblastoma with extensive nodularity; CGNPs, cerebellar granule neuron precursors; EGL, external granule cell layer.
Fig. (1).
Treatment strategies based on molecular subtypes and risk factors in medulloblastoma.
5.1. WNT Subgroup Medulloblastoma
WNT MBs are the least common subgroup, accounting for about 10% of all MBs [54-56]. They occur equally between males and females, with a median age of ~10 years at diagnosis [47]. The tumors rarely metastasize and are usually located in the midline of the brain, infiltrating the brainstem [57]. Most of these tumors have classic histology and the prognosis for this subgroup of patient population is excellent in comparison to the other subgroups, with a five-year survival rate of ~95% [44]. All patients in the WNT subgroup younger than 16 years at the age of diagnosis are considered to have low-risk disease (>90% survival) based on the current risk stratification consensus. This study also concluded the prognosis of children older than 16 years at diagnosis remains ill-defined [58]. Molecular-based clinical trials are currently underway to evaluate the potential of radiation therapy and chemotherapy de-escalation to reduce treatment-associated side effects, while maintaining equivalent survival rates (SJMB12, NCT01878617, PNET 5, NCT02066150, ACNS1422, and NCT02724579).
Historically, Turcot syndrome described the concurrence of a primary brain tumor, typically a MB or glioma, and multiple colorectal adenomas [59]. As the genetics of familial adenomatous polyposis (FAP) and hereditary nonpolyposis colorectal cancer (HNPCC) were elucidated, it became clear that the majority of brain tumors associated with FAP were MBs, while gliomas were more common with HNPCC. FAP is caused by inactivating, germline mutations in the APC tumor suppressor gene. Loss of function of the APC protein leads to nuclear accumulation of the WNT pathway effector molecule β-catenin. β-catenin then complexes with the T cell factor/lymphoid enhancer-binding factor (TCF/LEF) family of transcription factors and recruits other transcriptional co-activators to activate the target genes in canonical WNT/β-catenin signaling pathway—a pathway that plays a crucial role in many diverse processes governing embryonic development and disease, including cancer progression [60-62]. This led to the discovery of somatic activating point mutations in CTNNB1, the gene that encodes β-catenin [63]. More than 90% of the tumors in this subgroup have CTNNB1 mutations that cause production of a mutant β-catenin protein, that is resistant to degradation and accumulates in the nucleus to hyper-activate WNT pathway genes [46-49].
In addition to the nearly ubiquitous CTNNB1 mutations, other prevalent recurrent mutations in this subgroup include DDX3X (50%), SMARCA4 (26%), KMT2D (13%), and TP53 (13%) [44, 46-49]. Mutations in DDX3X, which encodes a DEAD-box RNA helicase, have been shown to promote cell viability and proliferation by potentiating the transactivation capacity of mutant β-catenin and also have been shown to maintain the lineage of lower rhombic lip progenitor cells, the proposed cells of origin for WNT MBs [48, 49, 57]. SMARCA4 and KMT2D (also known as MLL2) encode proteins involved in chromatin remodeling. Specifically, SMARCA4 has been identified as one of the transcriptional co-activators associated with the β-catenin-TCF/LEF transcription factor complex [64]. Mutations have also been identified in other transcriptional co-activators associated with the β-catenin-TCF/LEF complex as well (e.g., CREBBP, TRRAP, MED13) [49, 65, 66]. Though p53 mutations are frequently associated with the WNT subgroup of MB, they are not prognostic and not associated with the Li-Fraumeni syndrome, unlike in the SHH subgroup where they are related to a very bad prognosis [67]. The mutational landscape of WNT MB suggests that different subsets of tumors may be modified and potentially dependent on specific chromatin remodeling and RNA helicase complexes, affording potential opportunities for targeted therapy.
5.2. SHH Subgroup Medulloblastoma
Sonic hedgehog (SHH) pathway-driven MBs comprise approximately 30% of cases [47]. There is a bimodal age distribution in patients with this subgroup of tumors, with increased incidence among infants and young children less than 5 years of age and a second increase in frequency among older adolescents and adults greater than 16 years of age. These tumors are found in the cerebellar hemispheres, a location unique to this subtype [57, 68]. Metastatic disease at diagnosis occurs in less than 25% of cases. All MB histologies are represented among SHH MBs, with desmoplastic nodular and MBEN variants almost exclusively found in this subgroup [55, 69].
The prognosis for these patients is intermediate, with a five-year survival rate of ~75% [44]. Based on the current risk stratification consensus, patients with SHH MBs fall into the standard-risk stratum (75-90% survival)—unless they have: 1) metastatic disease and/or MYCN-amplified tumors, which constitutes high-risk disease (50-75% survival), or 2) tumors with TP53 mutations, which is considered very high risk (<50% survival) [58]. The tumors with TP53 mutations are highly enriched in children aged 3-17 and are frequently associated with LC/A morphology, constituting a higher risk group with significantly worse outcomes. In one cohort of MB, TP53 mutations accounted for 72% of deaths in children older than 5 years with SHH tumors [70].
The association of MBs with activation of the SHH signaling pathway was first identified in patients with Gorlin syndrome (nevoid basal cell carcinoma syndrome), who have a high prevalence of basal cell carcinoma and are predisposed to desmoplastic and MBEN MBs that commonly present in infancy, among other various neoplasms [71-73]. Gorlin syndrome is primarily caused by germline mutations in the PTCH1 tumor suppressor gene, which encodes patched 1 (PTC1), a cell-surface receptor for SHH and other hedgehog homologues [74,75]. In the absence of ligand binding, PTC1 inhibits the G protein-coupled receptor smoothened (SMO), thus acting as a negative regulator of the SHH signaling pathway [76]. The binding of SHH to PTC1 activates SMO, which leads to the release of GLI transcription factors from the cytoplasmic sequestration of SUFU, another negative regulator in the pathway. This results in the nuclear localization of GLI transcription factors and activation of target genes, including those involved in cellular proliferation, such as MYCN and CCND1 [77]. Thus, while germline or somatic mutations in PTCH1 are a hallmark feature in this subgroup, occurring in almost 30% of tumors, recurrent mutations and DNA structural alterations in the other SHH pathway genes acting downstream of PTCH1 (e.g., SMO, SUFU, GLI2) have also been identified [35, 46-49, 78].
Targeted therapies for SHH-driven MBs that inhibit SMO (e.g., vismodegib, GDC-0449; and sonidegib, NVP-LDE225) have entered clinical trials [79-81]. A recently completed phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma, showed that this drug was well tolerated with responses [82]. However a notable side-effect was premature closure of the growth plate, as also demonstrated in preclinical models [83]. While clinical response has been achieved in some patients with SHH MB, resistance to SMO inhibitors quickly emerges in these patients, prompting investigations into the mechanisms of acquired resistance and strategies to overcome these adaptations [84-88]. One strategy involves targeting the PI3K/Akt pathway, which is mutated in some tumors with acquired SMO inhibitor resistance, in combination with SMO antagonists [86]. Another promising combination therapy involving arsenic trioxide, which acts at the level of the GLI transcription factors, and itraconzaole, which inhibits SMO through a distinct mechanism, has been shown in pre-clinical experiments to be effective in SHH MBs that become resistant to SMO inhibitors [89]. Meanwhile, patients who did not respond to SMO inhibition in the first place tend to have mutations or amplifications in downstream SHH pathway genes (e.g., SUFU, MYCN, GLI2), causing inherent resistance to the SMO inhibitor [50, 78, 85]. These findings support that multiple levels of genomic evidence (i.e., sequencing, copy number alterations) will be imperative for the identification of potential responders and non-responders to target-based MB therapy and also for pinpointing likely candidates that mediate therapeutic resistance.
5.3. GROUP 3 Subgroup Medulloblastoma
Group 3 constitutes about 25% of MBs [47]. This subgroup carries the worst prognosis, with a five-year survival rate of ~50% [44]. While Group 3 MBs are considered to be standard risk (75-90% survival) if they do not have MYC amplification and are not metastatic, if they are metastatic at the time of diagnosis, they are classified as very high risk (<50% survival). Risk group for those tumors who are MYC amplified and non-metastatic is unknown [58, 59]. These tumors affect predominantly infants and young children (rarely teenagers) and occur twice as often in males as in females. Tumors are located in the midline/fourth ventricle [68]. Approximately 40-45% of patients already have metastatic disease at the time of diagnosis [69]. Histologically, Group 3 MBs are either classic or LC/A, with the highest prevalence of LC/A histology among the subgroups (40%).
The genetic basis for Group 3 MB is poorly understood [41-44]. No germline mutations that predispose patients to Group 3 MBs have been described. Amplification of the MYC proto-oncogene is nearly exclusive to this subgroup, but occurs in just 17% of these tumors [35, 38, 46-49, 69]. A few other recurrent somatic genomic aberrations have been reported, primarily in genes encoding chromatin remodeling proteins (e.g., SMARCA4 in 11%, KMT2D in 4%) and demethylases. However, over 50% of Group 3 tumors do not carry any of these genomic aberrations and, instead, have widespread chromosomal alterations, indicating a high level of genomic instability [42, 44].
Recently, one study shed some light on the role that these chromosomal alterations may play in the pathogenesis of Group 3 and Group 4 MB. Northcott and colleagues identified a series of atypical duplications, deletions, inversions, and complex rearrangements that converge to specifically activate the growth factor independent 1 family proto-oncogenes GFI1 and GFI1B through enhancer hijacking. While copy number alterations are not seen in the genes themselves, these intra- and inter-chromosomal structural variations repositioned GFI1 and GFI1B from transcriptionally silent regions of the genome to regions adjacent to active enhancers, including super-enhancers, thereby activating oncogene expression. Activation of GFI1 and GFI1B in a mutually exclusive manner was found in 30-40% of Group 3 MBs, such that they are now the most prevalent known oncogenic drivers in this subgroup. Currently unknown is whether GFI1 and GFI1B represent suitable druggable candidates for targeted therapy, or perhaps reveal synthetic lethal pathways that may be modulated to improve clinical outcome [52].
5.4. GROUP 4 Subgroup Medulloblastoma
Group 4 MBs are the most prevalent subgroup, accounting for nearly 35% of cases [47]. The prognosis for these patients is intermediate, with a five-year survival rate of ~75% [44]. Group 4 tumors are actually considered low-risk (>90% survival) if they are non-metastatic and have loss of chromosome 11 [58]. If tumors are non-metastatic, but do not have loss of chromosome 11, they are considered standard-risk (75-90% survival). However, if metastatic disease is present at diagnosis, they are considered high-risk (50-75% survival). These tumors affect all ages and are the most skewed in gender distribution, affecting males three times as commonly as females. Like Group 3, Group 4 tumors are located in the midline/fourth ventricle, and nearly 35-40% of patients already have metastases at diagnosis [68, 69]. Also similar to Group 3, Group 4 MBs have either classic or LC/A histology, although LC/A histology is much more infrequent.
The biology of Group 4 MBs is the least understood among the four subgroups [41-44]. Cytogenetically, the characteristic feature of this subgroup is isochromosome 17q, which is found in over 80%, and for which a driver oncogene and/or tumor suppressor gene remains unknown [35, 38, 68]. Group 4 tumors appear to share some biological similarities with Group 3 tumors as there is significant overlap of recurrent genetic mutations and copy number alterations between the two subtypes [35, 38, 46-49]. However, while MYC is amplified in Group 3, the proto-oncogenes MYCN and CDK6 are recurrently amplified in Group 4 MBs, but only in 6% and 5% of tumors, respectively. The most frequent mutation in this subgroup, occurring in 13%, is in KDM6A, a lysine demethylase regulating H3K27 methylation, a repressive chromatin mark important in cancer and stem cell biology [42, 44]. The most frequent focal somatic copy number gain is a tandem duplication of SNCAIP, a gene associated with Parkinson’s disease, found in 10% of tumors. The biology of KDM6A and SNCAIP in Group 4 MB pathogenesis remains poorly understood and is a current area of active investigation.
Though molecular subgrouping has revolutionized medulloblastoma classification, identification of significant heterogeneity within these subgroups are now emerging. 12 different subtypes of MB are now identified including two in infants with SHH MB with disparate outcomes and biology. Integrative analysis using similarity network fusion (SNF) has now further delineated group 3 from group 4 medulloblastoma. These recent revelations of MB subtypes within the four molecular subgroups determined through integrative clustering have important implications for stratification and profiling future clinical trials [90].
6. ROLE OF EPIGENETIC MODIFIERS
One theme that has emerged from the extensive sequencing studies recently published is the important role epigenetic deregulation plays in the pathogenesis of MB [91]. Histone modifiers are recurrently mutated and amplified in these genomes [44, 46-49, 92-94]. About a third of MBs across all subgroups carry mutations in histone methyltransferases (HMTs), histone demethylases (HDMs), histone acetyltransferases (HATs), histone deacetylases (HDACs), or other chromatin-associated genes (Table 3) [44].
Table 3.
Most prevalent, recurrently mutated chromatin modifiers in medulloblastoma.
| Function | Gene | Somatic |Mutation (%) | Subgroup Enrichment | ||||
|---|---|---|---|---|---|---|---|
| WNT | SHH | Group 3 | Group 4 | ||||
| H3K4 methyltransferase | KMT2D (MLL2) | 5.8 | X | X | |||
| KMT2C (MLL3) | 2.6 | X | X | ||||
| Chromatin remodeler | SMARCA4 | 5.8 | X | X | |||
| CHD7 | 1.9 | X | X | ||||
| ARID1B | 1.2 | X | X | ||||
| Histone demethylase | H3K27me2 H3K27me3 | KDM6A | 5.2 | X | |||
| H3K9me3 H3K36me3 | KDM4C | 1.0 | X | X | |||
| Transcriptional repressor | BCOR | 2.0 | X | ||||
| Transcriptional regulator | LDB1 | 1.6 | X | ||||
| Histone acetyltransferase | CREBBP | 1.3 | X | X | |||
| EP300 | 1.0 | X | |||||
| Associated with HAT complex | TRRAP | 1.0 | |||||
| Associated with HDAC complex | ZMYM3 | 1.3 | X | ||||
| Histone phosphatase | EYA4 | 1.2 | |||||
Abbreviations: HAT, histone acetyltransferase; HDAC, histone deacetylase.
Histone methylation, in particular, is deregulated in Group 3 and Group 4 tumors [49]. H3K27 trimethylation (H3K27me3) is a histone mark that maintains the undifferentiated state of stem cells by repressing lineage-specific genes [95]. H3K27me3 is written by histone methyltransferases (e.g., EZH2) and erased during differentiation by demethylases (e.g., KDM6A) [96-98]. With the removal of repressive H3K27me3 marks and coordinated deposition of H3K4me3 activating marks, chromatin remodelers (e.g., CHD7) are recruited to H3K4me3 sites to promote differentiation [99-101]. Phase I studies in children with relapsed/refractory CNS tumors showed vorinostat (HDAC inhibitor) was well-tolerated when combined with either temozolomide (NCT01076530) or bortezomib, a proteasome inhibitor (NCT00994500), highlighting the potential role epigenetic modifiers.
As discussed above, KDM6A is a lysine demethylase regulating H3K27 methylation and is the most commonly mutated gene in Group 4 MBs (13%). These tumors also harbor recurrent mutations in CHD7 and ZMYM3, which converge on the regulation of gene expression by H3K4me3 [93]. Furthermore, both Group 3 and Group 4 MBs overexpress EZH2 as a result of chromosome 7 gain—but not in those tumors carrying inactivating mutations in KDM6A, CHD7, or ZMYM3 mutations [49]. Thus, Robinson and colleagues proposed that Group 3 and Group 4 MBs preserve stem-like signatures through a variety of mechanisms: accumulation of repressive H3K27me3 marks (EZH2 overexpression), maintenance of H3K27me3 (KDM6A inactivation), or abrogation of H3K4me4-mediated differentiation (CHD7 and ZMYM3 inactivation) [49, 93].
Accordingly, there has been growing interest in investigating therapeutics that target epigenetic regulators. One novel mechanism of targeting histone modifications is by inhibiting the readers of acetylation, the BET bromodomain proteins (e.g., BRD4) [102]. Bromodomains recognize and bind to acetylation motifs on open chromatin (e.g., H3K27ac) at active enhancer and super-enhancer sites, often localizing to promoters of key transcription factors, such as MYC [103-105]. Inhibitors of bromodomains, such as JQ1, have been shown to be effective in MYC-driven, Group 3 MBs [106-108] as well as in SHH MBs with resistance to SMO inhibitors (both acquired and inherent) [109]. Inhibitors of EZH2 are also promising in targeting the subset of Group 3 and Group 4 MBs that overexpress EZH2 [110]. Additionally, Pei and colleagues recently performed a high-throughput screen that identified HDAC inhibitors as potent inhibitors of MYC-driven MB in preclinical models [111]. Furthermore, they showed that the combination of HDAC inhibitors with PI3K antagonists (specifically, panobinostat and buparlisib) act synergistically against tumor growth. Given the potentially rapid adaptation of MB cells to targeted ablation, upfront identification of effective combination therapies and potential resistance mechanisms is needed to improve therapeutic efficacy.
7. RECURRENT DISEASE AND THERAPEUTIC RESISTANCE
Recurrent MB is almost uniformly fatal, with a 2-year survival rate of 9% [19]. The mechanism(s) and genetic basis by which MB cells escape therapy and drive disease relapse is still poorly understood. Several studies were published recently that serve to enhance this understanding.
Phoenix and colleagues focused on the role of the microenvironment in determining treatment sensitivity, specifically what allows for the excellent prognosis of WNT-subtype tumors, such that they can be curable even when metastatic, compared to other subgroups [112]. They found that while other MB subgroups have normal central nervous system vasculature, WNT MBs lack a functional blood-brain barrier (BBB), which facilitates increased access of systemic chemotherapy to WNT tumors and contributes to the excellent treatment response. This disruption in the BBB is driven by the paracrine signaling of WNT antagonists secreted by the tumor, presumably produced in a negative feedback response to the constitutive mutant β-catenin activity that drives the tumor. These WNT antagonists block the normal process of paracrine WNT signaling in neighboring endothelial cells that induces angiogenesis and barriergenesis and maintains the BBB throughout life [113, 114]. Thus, the failure of chemotherapy to adequately reach its target is one mechanism by which tumor cells could escape treatment, and inhibiting WNT to disrupt the BBB and improve chemotherapy penetration in other MB subgroups is a future area of study.
Another mechanism by which disease could recur is through therapeutic resistance in a progenitor, stem cell-like population that can subsequently reconstitute the tumor. Vanner and colleagues identified such a population of Sox+ cells in a mouse model of SHH MB [115]. They showed that these cells are resistant to both anti-mitotic therapy and the targeted SMO inhibitor vismodegib, such that they become enriched after therapy. These cells also express a quiescent stem cell signature and can reconstitute the tumor hierarchy at MB relapse. Eradicating this progenitor cell population with novel therapies in addition to tumor debulking with current therapies may be an efficacious combination in order to prevent disease relapse. There is also the need to identify novel therapies targeting the very high risk group of SHH tumors with TP53 mutations as multivariable analysis has revealed that TP53 status is the most important risk factor for SHH medulloblastoma. This subset of tumors is the leading cause for therapeutic resistance and poor outcomes in the SHH subgroup [70].
Historically, when MB recurs, patients have been treated based on the premise that the recurrent tumor is biologically similar to the tumor at the time of diagnosis. This, however, has been an unsubstantiated assumption that Morrissy and Garzia et al. recently showed is untrue in the vast majority of cases [116]. The authors performed whole-genome sequencing on 33 pairs of diagnostic and post-recurrent MBs and found that the dominant clone after therapy shares only <12% of genetic events found in the dominant clone at diagnosis. Instead, it appears that the dominant clone at recurrence may arise through clonal selection of a minor clone present at the time of diagnosis. Nevertheless, despite this genetic divergence, the tumor subgroup (WNT, SHH, Group 3, or Group 4) remains stable across recurrence and metastasis [117, 118] suggesting that certain molecular aspects of MBs are maintained during tumor evolution. Thus, prior clinical trials for novel therapies designed to target tumors based on their genetic profile at the time of diagnosis may have been doomed to fail as these targets may have no longer been present at recurrence. It will be essential that future target-based clinical trials for MB ensure that the specific driver lesions are maintained at relapse if trials are to be designed in patients with recurrent MB.
CONCLUSION
The identification of the molecular subtypes of MB and ongoing research advances have incited a new era in personalized therapeutic approaches for these tumors. Future clinical trials and efforts are now looking into deescalating therapy in the WNT subgroup, which carries an excellent prognosis. Identifying methodologies to overcome drug resistance in the SHH variant is important, as downstream molecules of this pathway are emerging as key players inducing therapeutic failures and may serve as potential druggable targets. The major challenge and the focus lie in improved understanding of the elusive molecular biology of Group 3 and 4 tumors as well as judicious intensification of the therapy in these subtypes to improve clinical outcome. With the continuing surge of genomic/epigenomic data of these subgroups, the formidable task lies ahead in the comprehensive evaluation of this knowledge, and being able to deliver the most promising clinical trials, more so for the patients with high-risk and recurrent MB.
FUNDING DISCLOSURE
No funding has been received for this work from any of the following organizations: National Institutes of Health (NIH); Wellcome Trust; Howard Hughes Medical Institute (HHMI); and other(s).
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [http://dx.doi.org/ 10.1007/s00401-016-1545-1]. [PMID: 27157931]. [DOI] [PubMed] [Google Scholar]
- 2.Eberhart C.G., Kepner J.L., Goldthwaite P.T., Kun L.E., Duffner P.K., Friedman H.S., Strother D.R., Burger P.C. Histopathologic grading of medulloblastomas: a Pediatric Oncology Group study. Cancer. 2002;94(2):552–560. doi: 10.1002/cncr.10189. [http://dx.doi.org/10.1002/ cncr.10189]. [PMID: 11900240]. [DOI] [PubMed] [Google Scholar]
- 3.Perry A. Medulloblastomas with favorable versus unfavorable histology: how many small blue cell tumor types are there in the brain? Adv. Anat. Pathol. 2002;9(6):345–350. doi: 10.1097/00125480-200211000-00003. [http://dx.doi.org/ 10.1097/00125480-200211000-00003]. [PMID: 12409643]. [DOI] [PubMed] [Google Scholar]
- 4.Gajjar A., Chintagumpala M., Ashley D., Kellie S., Kun L.E., Merchant T.E., Woo S., Wheeler G., Ahern V., Krasin M.J., Fouladi M., Broniscer A., Krance R., Hale G.A., Stewart C.F., Dauser R., Sanford R.A., Fuller C., Lau C., Boyett J.M., Wallace D., Gilbertson R.J. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7(10):813–820. doi: 10.1016/S1470-2045(06)70867-1. [http://dx.doi.org/ 10.1016/S1470-2045(06)70867-1]. [PMID: 17012043]. [DOI] [PubMed] [Google Scholar]
- 5.von Bueren A.O., von Hoff K., Pietsch T., Gerber N.U., Warmuth-Metz M., Deinlein F., Zwiener I., Faldum A., Fleischhack G., Benesch M., Krauss J., Kuehl J., Kortmann R.D., Rutkowski S. Treatment of young children with localized medulloblastoma by chemotherapy alone: results of the prospective, multicenter trial HIT 2000 confirming the prognostic impact of histology. Neuro-oncol. 2011;13(6):669–679. doi: 10.1093/neuonc/nor025. [http://dx.doi.org/10.1093/neuonc/ nor025]. [PMID: 21636711]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massimino M., Biassoni V., Gandola L., Garrè M.L., Gatta G., Giangaspero F., Poggi G., Rutkowski S. Childhood medulloblastoma. Crit. Rev. Oncol. Hematol. 2016;105:35–51. doi: 10.1016/j.critrevonc.2016.05.012. [http://dx.doi. org/10.1016/j.critrevonc.2016.05.012]. [PMID: 27375228]. [DOI] [PubMed] [Google Scholar]
- 7.Packer R.J., Macdonald T., Vezina G., Keating R., Santi M. Medulloblastoma and primitive neuroectodermal tumors. Handb. Clin. Neurol. 2012;105:529–548. doi: 10.1016/B978-0-444-53502-3.00007-0. [http://dx.doi.org/10.1016/ B978-0-444-53502-3.00007-0]. [PMID: 22230517]. [DOI] [PubMed] [Google Scholar]
- 8.Rutkowski S., Gerber N.U., von Hoff K., Gnekow A., Bode U., Graf N., Berthold F., Henze G., Wolff J.E., Warmuth-Metz M., Soerensen N., Emser A., Ottensmeier H., Deinlein F., Schlegel P.G., Kortmann R.D., Pietsch T., Kuehl J. Treatment of early childhood medulloblastoma by postoperative chemotherapy and deferred radiotherapy. Neuro-oncol. 2009;11(2):201–210. doi: 10.1215/15228517-2008-084. [http:// dx.doi.org/10.1215/15228517-2008-084]. [PMID: 18818397]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi S.N., Gardner S.L., Levy A.S., Knopp E.A., Miller D.C., Wisoff J.H., Weiner H.L., Finlay J.L. Feasibility and response to induction chemotherapy intensified with high-dose methotrexate for young children with newly diagnosed high-risk disseminated medulloblastoma. J. Clin. Oncol. 2004;22(24):4881–4887. doi: 10.1200/JCO.2004.12.126. [http:// dx.doi.org/10.1200/JCO.2004.12.126]. [PMID: 15611503]. [DOI] [PubMed] [Google Scholar]
- 10.Ashley D.M., Merchant T.E., Strother D., Zhou T., Duffner P., Burger P.C., Miller D.C., Lyon N., Bonner M.J., Msall M., Buxton A., Geyer R., Kun L.E., Coleman L., Pollack I.F. Induction chemotherapy and conformal radiation therapy for very young children with nonmetastatic medulloblastoma: Children’s Oncology Group study P9934. J. Clin. Oncol. 2012;30(26):3181–3186. doi: 10.1200/JCO.2010.34.4341. [http://dx.doi.org/10.1200/JCO.2010.34.4341]. [PMID: 22851568]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottardo N.G., Gajjar A. Current therapy for medulloblastoma. Curr. Treat. Options Neurol. 2006;8(4):319–334. doi: 10.1007/s11940-006-0022-x. [http://dx. doi.org/10.1007/s11940-006-0022-x]. [PMID: 16942675]. [DOI] [PubMed] [Google Scholar]
- 12.Mabbott D.J., Penkman L., Witol A., Strother D., Bouffet E. Core neurocognitive functions in children treated for posterior fossa tumors. Neuropsychology. 2008;22(2):159–168. doi: 10.1037/0894-4105.22.2.159. [http://dx. doi.org/10.1037/0894-4105.22.2.159]. [PMID: 18331158]. [DOI] [PubMed] [Google Scholar]
- 13.Gudrunardottir T., Lannering B., Remke M., Taylor M.D., Wells E.M., Keating R.F., Packer R.J. Treatment developments and the unfolding of the quality of life discussion in childhood medulloblastoma: a review. Childs Nerv. Syst. 2014;30(6):979–990. doi: 10.1007/s00381-014-2388-5. [http://dx.doi.org/10.1007/s00381-014-2388-5]. [PMID: 24569911]. [DOI] [PubMed] [Google Scholar]
- 14.Khatua S., Zaky W. The biologic era of childhood medulloblastoma and clues to novel therapies. Future Oncol. 2014;10(4):637–645. doi: 10.2217/fon.13.185. [http://dx.doi.org/10.2217/fon.13.185]. [PMID: 24754593]. [DOI] [PubMed] [Google Scholar]
- 15.Packer R.J., Gajjar A., Vezina G., Rorke-Adams L., Burger P.C., Robertson P.L., Bayer L., LaFond D., Donahue B.R., Marymont M.H., Muraszko K., Langston J., Sposto R. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J. Clin. Oncol. 2006;24(25):4202–4208. doi: 10.1200/JCO.2006.06.4980. [http://dx.doi.org/10.1200/ JCO.2006.06.4980]. [PMID: 16943538]. [DOI] [PubMed] [Google Scholar]
- 16.Gatta G., Zigon G., Capocaccia R., Coebergh J.W., Desandes E., Kaatsch P., Pastore G., Peris-Bonet R., Stiller C.A., Group E.W. Survival of European children and young adults with cancer diagnosed 1995-2002. Eur. J. Cancer. 2009;45(6):992–1005. doi: 10.1016/j.ejca.2008.11.042. [http://dx.doi.org/10.1016/j.ejca.2008.11.042]. [PMID: 19231160]. [DOI] [PubMed] [Google Scholar]
- 17.Packer R.J., Siegel K.R., Sutton L.N., Evans A.E., D’Angio G., Rorke L.B., Bunin G.R., Schut L. Efficacy of adjuvant chemotherapy for patients with poor-risk medulloblastoma: a preliminary report. Ann. Neurol. 1988;24(4):503–508. doi: 10.1002/ana.410240405. [http://dx.doi.org/10. 1002/ana.410240405]. [PMID: 3239953]. [DOI] [PubMed] [Google Scholar]
- 18.Verlooy J., Mosseri V., Bracard S., Tubiana A.L., Kalifa C., Pichon F., Frappaz D., Chastagner P., Pagnier A., Bertozzi A.I., Gentet J.C., Sariban E., Rialland X., Edan C., Bours D., Zerah M., Le Gales C., Alapetite C., Doz F. Treatment of high risk medulloblastomas in children above the age of 3 years: a SFOP study. Eur. J. Cancer. 2006;42(17):3004–3014. doi: 10.1016/j.ejca.2006.02.026. [http://dx.doi.org/10. 1016/j.ejca.2006.02.026]. [PMID: 16956759]. [DOI] [PubMed] [Google Scholar]
- 19.Zeltzer P.M., Boyett J.M., Finlay J.L., Albright A.L., Rorke L.B., Milstein J.M., Allen J.C., Stevens K.R., Stanley P., Li H., Wisoff J.H., Geyer J.R., McGuire-Cullen P., Stehbens J.A., Shurin S.B., Packer R.J. Metastasis stage, adjuvant treatment, and residual tumor are prognostic factors for medulloblastoma in children: conclusions from the Children’s Cancer Group 921 randomized phase III study. J. Clin. Oncol. 1999;17(3):832–845. doi: 10.1200/JCO.1999.17.3.832. [http:// dx.doi.org/10.1200/JCO.1999.17.3.832]. [PMID: 10071274]. [DOI] [PubMed] [Google Scholar]
- 20.Kortmann R.D., Kühl J., Timmermann B., Mittler U., Urban C., Budach V., Richter E., Willich N., Flentje M., Berthold F., Slavc I., Wolff J., Meisner C., Wiestler O., Sörensen N., Warmuth-Metz M., Bamberg M. Postoperative neoadjuvant chemotherapy before radiotherapy as compared to immediate radiotherapy followed by maintenance chemotherapy in the treatment of medulloblastoma in childhood: results of the German prospective randomized trial HIT ’91. Int. J. Radiat. Oncol. Biol. Phys. 2000;46(2):269–279. doi: 10.1016/s0360-3016(99)00369-7. [http://dx.doi.org/10.1016/S0360-3016(99)00369-7]. [PMID: 10661332]. [DOI] [PubMed] [Google Scholar]
- 21.Tarbell N.J., Friedman H., Polkinghorn W.R., Yock T., Zhou T., Chen Z., Burger P., Barnes P., Kun L. High-risk medulloblastoma: a pediatric oncology group randomized trial of chemotherapy before or after radiation therapy (POG 9031). J. Clin. Oncol. 2013;31(23):2936–2941. doi: 10.1200/JCO.2012.43.9984. [http://dx.doi.org/10.1200/JCO. 2012.43.9984]. [PMID: 23857975]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakacki R.I., Burger P.C., Zhou T., Holmes E.J., Kocak M., Onar A., Goldwein J., Mehta M., Packer R.J., Tarbell N., Fitz C., Vezina G., Hilden J., Pollack I.F. Outcome of children with metastatic medulloblastoma treated with carboplatin during craniospinal radiotherapy: a Children’s Oncology Group Phase I/II study. J. Clin. Oncol. 2012;30(21):2648–2653. doi: 10.1200/JCO.2011.40.2792. [http://dx.doi.org/10. 1200/JCO.2011.40.2792]. [PMID: 22665539]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Bueren A.O., Kortmann R.D., von Hoff K., Friedrich C., Mynarek M., Müller K., Goschzik T., Zur Mühlen A., Gerber N., Warmuth-Metz M., Soerensen N., Deinlein F., Benesch M., Zwiener I., Kwiecien R., Faldum A., Bode U., Fleischhack G., Hovestadt V., Kool M., Jones D., Northcott P., Kuehl J., Pfister S., Pietsch T., Rutkowski S. Treatment of children and adolescents with metastatic medulloblastoma and prognostic relevance of clinical and biologic parameters. J. Clin. Oncol. 2016;34(34):4151–4160. doi: 10.1200/JCO.2016.67.2428. [http://dx.doi.org/10.1200/JCO.2016.67.2428]. [PMID: 27863192]. [DOI] [PubMed] [Google Scholar]
- 24.Hallahan A.R., Pritchard J.I., Hansen S., Benson M., Stoeck J., Hatton B.A., Russell T.L., Ellenbogen R.G., Bernstein I.D., Beachy P.A., Olson J.M. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64(21):7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [http://dx.doi.org/10.1158/0008-5472.CAN-04-1813]. [PMID: 15520185]. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald T.J., Aguilera D., Castellino R.C. The rationale for targeted therapies in medulloblastoma. Neuro-oncol. 2014;16(1):9–20. doi: 10.1093/neuonc/not147. [http://dx.doi.org/10.1093/neuonc/not147]. [PMID: 24305711]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman L.M., Fouladi M., Olson J., Daryani V.M., Stewart C.F., Wetmore C., Kocak M., Onar-Thomas A., Wagner L., Gururangan S., Packer R.J., Blaney S.M., Gajjar A., Kun L.E., Boyett J.M., Gilbertson R.J. Phase I trial of weekly MK-0752 in children with refractory central nervous system malignancies: a pediatric brain tumor consortium study. Childs Nerv. Syst. 2015;31(8):1283–1289. doi: 10.1007/s00381-015-2725-3. [http://dx.doi.org/10.1007/s00381-015-2725-3]. [PMID: 25930724]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguilera D., Mazewski C., Fangusaro J., MacDonald T.J., McNall-Knapp R.Y., Hayes L.L., Kim S., Castellino R.C. Response to bevacizumab, irinotecan, and temozolomide in children with relapsed medulloblastoma: a multi-institutional experience. Childs Nerv. Syst. 2013;29(4):589–596. doi: 10.1007/s00381-012-2013-4. [http://dx.doi.org/10. 1007/s00381-012-2013-4]. [PMID: 23296323]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aguilera D.G., Goldman S., Fangusaro J. Bevacizumab and irinotecan in the treatment of children with recurrent/refractory medulloblastoma. Pediatr. Blood Cancer. 2011;56(3):491–494. doi: 10.1002/pbc.22868. [http://dx.doi.org/10.1002/pbc.22868]. [PMID: 21072819]. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Martínez A., Lassaletta A., González-Vicent M., Sevilla J., Díaz M.A., Madero L. High-dose chemotherapy with autologous stem cell rescue for children with high risk and recurrent medulloblastoma and supratentorial primitive neuroectodermal tumors. J. Neurooncol. 2005;71(1):33–38. doi: 10.1007/s11060-004-4527-4. [http://dx.doi.org/10. 1007/s11060-004-4527-4]. [PMID: 15719272]. [DOI] [PubMed] [Google Scholar]
- 30.Gururangan S., Krauser J., Watral M.A., Driscoll T., Larrier N., Reardon D.A., Rich J.N., Quinn J.A., Vredenburgh J.J., Desjardins A., McLendon R.E., Fuchs H., Kurtzberg J., Friedman H.S. Efficacy of high-dose chemotherapy or standard salvage therapy in patients with recurrent medulloblastoma. Neuro-oncol. 2008;10(5):745–751. doi: 10.1215/15228517-2008-044. [http://dx.doi.org/10.1215/15228517-2008-044]. [PMID: 18755919]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butturini A.M., Jacob M., Aguajo J., Vander-Walde N.A., Villablanca J., Jubran R., Erdreich-Epstein A., Marachelian A., Dhall G., Finlay J.L. High-dose chemotherapy and autologous hematopoietic progenitor cell rescue in children with recurrent medulloblastoma and supratentorial primitive neuroectodermal tumors: the impact of prior radiotherapy on outcome. Cancer. 2009;115(13):2956–2963. doi: 10.1002/cncr.24341. [http://dx.doi.org/10.1002/cncr.24341]. [PMID: 19402050]. [DOI] [PubMed] [Google Scholar]
- 32.Ridola V., Grill J., Doz F., Gentet J.C., Frappaz D., Raquin M.A., Habrand J.L., Sainte-Rose C., Valteau-Couanet D., Kalifa C. High-dose chemotherapy with autologous stem cell rescue followed by posterior fossa irradiation for local medulloblastoma recurrence or progression after conventional chemotherapy. Cancer. 2007;110(1):156–163. doi: 10.1002/cncr.22761. [http://dx.doi.org/10.1002/cncr.22761]. [PMID: 17541945]. [DOI] [PubMed] [Google Scholar]
- 33.Pomeroy S.L., Tamayo P., Gaasenbeek M., Sturla L.M., Angelo M., McLaughlin M.E., Kim J.Y., Goumnerova L.C., Black P.M., Lau C., Allen J.C., Zagzag D., Olson J.M., Curran T., Wetmore C., Biegel J.A., Poggio T., Mukherjee S., Rifkin R., Califano A., Stolovitzky G., Louis D.N., Mesirov J.P., Lander E.S., Golub T.R. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. [http://dx.doi.org/10.1038/415436a]. [PMID: 11807556]. [DOI] [PubMed] [Google Scholar]
- 34.Al-Halabi H., Nantel A., Klekner A., Guiot M.C., Albrecht S., Hauser P., Garami M., Bognar L., Kavan P., Gerges N., Shirinian M., Roberge D., Muanza T., Jabado N. Preponderance of sonic hedgehog pathway activation characterizes adult medulloblastoma. Acta Neuropathol. 2011;121(2):229–239. doi: 10.1007/s00401-010-0780-0. [http://dx.doi.org/10.1007/s00401-010-0780-0]. [PMID: 21107850]. [DOI] [PubMed] [Google Scholar]
- 35.Cho Y.J., Tsherniak A., Tamayo P., Santagata S., Ligon A., Greulich H., Berhoukim R., Amani V., Goumnerova L., Eberhart C.G., Lau C.C., Olson J.M., Gilbertson R.J., Gajjar A., Delattre O., Kool M., Ligon K., Meyerson M., Mesirov J.P., Pomeroy S.L. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. [http://dx.doi.org/10. 1200/JCO.2010.28.5148]. [PMID: 21098324]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fattet S., Haberler C., Legoix P., Varlet P., Lellouch-Tubiana A., Lair S., Manie E., Raquin M.A., Bours D., Carpentier S., Barillot E., Grill J., Doz F., Puget S., Janoueix-Lerosey I., Delattre O. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J. Pathol. 2009;218(1):86–94. doi: 10.1002/path.2514. [http://dx.doi.org/10.1002/path.2514]. [PMID: 19197950]. [DOI] [PubMed] [Google Scholar]
- 37.Kool M., Koster J., Bunt J., Hasselt N.E., Lakeman A., van Sluis P., Troost D., Meeteren N.S., Caron H.N., Cloos J., Mrsić A., Ylstra B., Grajkowska W., Hartmann W., Pietsch T., Ellison D., Clifford S.C., Versteeg R. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3(8):e3088. doi: 10.1371/journal.pone.0003088. [http://dx.doi.org/10.1371/journal.pone.0003088]. [PMID: 18769486]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Northcott P.A., Korshunov A., Witt H., Hielscher T., Eberhart C.G., Mack S., Bouffet E., Clifford S.C., Hawkins C.E., French P., Rutka J.T., Pfister S., Taylor M.D. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [http://dx.doi.org/10.1200/JCO.2009.27.4324]. [PMID: 20823417]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson M.C., Fuller C., Hogg T.L., Dalton J., Finkelstein D., Lau C.C., Chintagumpala M., Adesina A., Ashley D.M., Kellie S.J., Taylor M.D., Curran T., Gajjar A., Gilbertson R.J. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 2006;24(12):1924–1931. doi: 10.1200/JCO.2005.04.4974. [http://dx.doi.org/10.1200/JCO.2005.04.4974]. [PMID: 16567768]. [DOI] [PubMed] [Google Scholar]
- 40.Taylor M.D., Northcott P.A., Korshunov A., Remke M., Cho Y.J., Clifford S.C., Eberhart C.G., Parsons D.W., Rutkowski S., Gajjar A., Ellison D.W., Lichter P., Gilbertson R.J., Pomeroy S.L., Kool M., Pfister S.M. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. doi: 10.1007/s00401-011-0922-z. [http://dx.doi.org/10.1007/s00401-011-0922-z]. [PMID: 22134537]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Northcott P.A., Korshunov A., Pfister S.M., Taylor M.D. The clinical implications of medulloblastoma subgroups. Nat. Rev. Neurol. 2012;8(6):340–351. doi: 10.1038/nrneurol.2012.78. [http://dx.doi.org/10.1038/nrneurol.2012. 78]. [PMID: 22565209]. [DOI] [PubMed] [Google Scholar]
- 42.Gajjar A.J., Robinson G.W. Medulloblastoma-translating discoveries from the bench to the bedside. Nat. Rev. Clin. Oncol. 2014;11(12):714–722. doi: 10.1038/nrclinonc.2014.181. [http://dx.doi.org/10.1038/nrclinonc.2014.181]. [PMID: 25348790]. [DOI] [PubMed] [Google Scholar]
- 43.Khatua S. Evolving molecular era of childhood medulloblastoma: time to revisit therapy. Future Oncol. 2016;12(1):107–117. doi: 10.2217/fon.15.284. [http://dx.doi.org/10.2217/fon.15.284]. [PMID: 26617331]. [DOI] [PubMed] [Google Scholar]
- 44.Northcott P.A., Jones D.T., Kool M., Robinson G.W., Gilbertson R.J., Cho Y.J., Pomeroy S.L., Korshunov A., Lichter P., Taylor M.D., Pfister S.M. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer. 2012;12(12):818–834. doi: 10.1038/nrc3410. [http://dx.doi. org/10.1038/nrc3410]. [PMID: 23175120]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rausch T., Jones D.T., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A., Pfaff E., Tica J., Wang Q., Massimi L., Witt H., Bender S., Pleier S., Cin H., Hawkins C., Beck C., von Deimling A., Hans V., Brors B., Eils R., Scheurlen W., Blake J., Benes V., Kulozik A.E., Witt O., Martin D., Zhang C., Porat R., Merino D.M., Wasserman J., Jabado N., Fontebasso A., Bullinger L., Rücker F.G., Döhner K., Döhner H., Koster J., Molenaar J.J., Versteeg R., Kool M., Tabori U., Malkin D., Korshunov A., Taylor M.D., Lichter P., Pfister S.M., Korbel J.O. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1-2):59–71. doi: 10.1016/j.cell.2011.12.013. [http://dx.doi.org/10.1016/j.cell.2011.12.013]. [PMID: 22265402]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones D.T., Jäger N., Kool M., Zichner T., Hutter B., Sultan M., Cho Y.J., Pugh T.J., Hovestadt V., Stütz A.M., Rausch T., Warnatz H.J., Ryzhova M., Bender S., Sturm D., Pleier S., Cin H., Pfaff E., Sieber L., Wittmann A., Remke M., Witt H., Hutter S., Tzaridis T., Weischenfeldt J., Raeder B., Avci M., Amstislavskiy V., Zapatka M., Weber U.D., Wang Q., Lasitschka B., Bartholomae C.C., Schmidt M., von Kalle C., Ast V., Lawerenz C., Eils J., Kabbe R., Benes V., van Sluis P., Koster J., Volckmann R., Shih D., Betts M.J., Russell R.B., Coco S., Tonini G.P., Schüller U., Hans V., Graf N., Kim Y.J., Monoranu C., Roggendorf W., Unterberg A., Herold-Mende C., Milde T., Kulozik A.E., von Deimling A., Witt O., Maass E., Rössler J., Ebinger M., Schuhmann M.U., Frühwald M.C., Hasselblatt M., Jabado N., Rutkowski S., von Bueren A.O., Williamson D., Clifford S.C., McCabe M.G., Collins V.P., Wolf S., Wiemann S., Lehrach H., Brors B., Scheurlen W., Felsberg J., Reifenberger G., Northcott P.A., Taylor M.D., Meyerson M., Pomeroy S.L., Yaspo M.L., Korbel J.O., Korshunov A., Eils R., Pfister S.M., Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [http://dx.doi.org/ 10. 1038/nature11284]. [PMID: 22832583]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Northcott P.A., Shih D.J., Peacock J., Garzia L., Morrissy A.S., Zichner T., Stütz A.M., Korshunov A., Reimand J., Schumacher S.E., Beroukhim R., Ellison D.W., Marshall C.R., Lionel A.C., Mack S., Dubuc A., Yao Y., Ramaswamy V., Luu B., Rolider A., Cavalli F.M., Wang X., Remke M., Wu X., Chiu R.Y., Chu A., Chuah E., Corbett R.D., Hoad G.R., Jackman S.D., Li Y., Lo A., Mungall K.L., Nip K.M., Qian J.Q., Raymond A.G., Thiessen N.T., Varhol R.J., Birol I., Moore R.A., Mungall A.J., Holt R., Kawauchi D., Roussel M.F., Kool M., Jones D.T., Witt H., Fernandez-L A., Kenney A.M., Wechsler-Reya R.J., Dirks P., Aviv T., Grajkowska W.A., Perek-Polnik M., Haberler C.C., Delattre O., Reynaud S.S., Doz F.F., Pernet-Fattet S.S., Cho B.K., Kim S.K., Wang K.C., Scheurlen W., Eberhart C.G., Fèvre-Montange M., Jouvet A., Pollack I.F., Fan X., Muraszko K.M., Gillespie G.Y., Di Rocco C., Massimi L., Michiels E.M., Kloosterhof N.K., French P.J., Kros J.M., Olson J.M., Ellenbogen R.G., Zitterbart K., Kren L., Thompson R.C., Cooper M.K., Lach B., McLendon R.E., Bigner D.D., Fontebasso A., Albrecht S., Jabado N., Lindsey J.C., Bailey S., Gupta N., Weiss W.A., Bognár L., Klekner A., Van Meter T.E., Kumabe T., Tominaga T., Elbabaa S.K., Leonard J.R., Rubin J.B., Liau L.M., Van Meir E.G., Fouladi M., Nakamura H., Cinalli G., Garami M., Hauser P., Saad A.G., Iolascon A., Jung S., Carlotti C.G., Vibhakar R., Ra Y.S., Robinson S., Zollo M., Faria C.C., Chan J.A., Levy M.L., Sorensen P.H., Meyerson M., Pomeroy S.L., Cho Y.J., Bader G.D., Tabori U., Hawkins C.E., Bouffet E., Scherer S.W., Rutka J.T., Malkin D., Clifford S.C., Jones S.J., Korbel J.O., Pfister S.M., Marra M.A., Taylor M.D. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. doi: 10.1038/nature11327. [http://dx.doi.org/10. 1038/nature11327]. [PMID: 22832581]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pugh T.J., Weeraratne S.D., Archer T.C., Pomeranz Krummel D.A., Auclair D., Bochicchio J., Carneiro M.O., Carter S.L., Cibulskis K., Erlich R.L., Greulich H., Lawrence M.S., Lennon N.J., McKenna A., Meldrim J., Ramos A.H., Ross M.G., Russ C., Shefler E., Sivachenko A., Sogoloff B., Stojanov P., Tamayo P., Mesirov J.P., Amani V., Teider N., Sengupta S., Francois J.P., Northcott P.A., Taylor M.D., Yu F., Crabtree G.R., Kautzman A.G., Gabriel S.B., Getz G., Jäger N., Jones D.T., Lichter P., Pfister S.M., Roberts T.M., Meyerson M., Pomeroy S.L., Cho Y.J. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [http://dx.doi.org/10.1038/nature11329]. [PMID: 22820256]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson G., Parker M., Kranenburg T.A., Lu C., Chen X., Ding L., Phoenix T.N., Hedlund E., Wei L., Zhu X., Chalhoub N., Baker S.J., Huether R., Kriwacki R., Curley N., Thiruvenkatam R., Wang J., Wu G., Rusch M., Hong X., Becksfort J., Gupta P., Ma J., Easton J., Vadodaria B., Onar-Thomas A., Lin T., Li S., Pounds S., Paugh S., Zhao D., Kawauchi D., Roussel M.F., Finkelstein D., Ellison D.W., Lau C.C., Bouffet E., Hassall T., Gururangan S., Cohn R., Fulton R.S., Fulton L.L., Dooling D.J., Ochoa K., Gajjar A., Mardis E.R., Wilson R.K., Downing J.R., Zhang J., Gilbertson R.J. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [http://dx.doi.org/10.1038/nature11213]. [PMID: 22722829]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kool M., Jones D.T., Jäger N., Northcott P.A., Pugh T.J., Hovestadt V., Piro R.M., Esparza L.A., Markant S.L., Remke M., Milde T., Bourdeaut F., Ryzhova M., Sturm D., Pfaff E., Stark S., Hutter S., Seker-Cin H., Johann P., Bender S., Schmidt C., Rausch T., Shih D., Reimand J., Sieber L., Wittmann A., Linke L., Witt H., Weber U.D., Zapatka M., König R., Beroukhim R., Bergthold G., van Sluis P., Volckmann R., Koster J., Versteeg R., Schmidt S., Wolf S., Lawerenz C., Bartholomae C.C., von Kalle C., Unterberg A., Herold-Mende C., Hofer S., Kulozik A.E., von Deimling A., Scheurlen W., Felsberg J., Reifenberger G., Hasselblatt M., Crawford J.R., Grant G.A., Jabado N., Perry A., Cowdrey C., Croul S., Zadeh G., Korbel J.O., Doz F., Delattre O., Bader G.D., McCabe M.G., Collins V.P., Kieran M.W., Cho Y.J., Pomeroy S.L., Witt O., Brors B., Taylor M.D., Schüller U., Korshunov A., Eils R., Wechsler-Reya R.J., Lichter P., Pfister S.M., Project I.P.T. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25(3):393–405. doi: 10.1016/j.ccr.2014.02.004. [http://dx.doi.org/10.1016/j.ccr.2014.02.004]. [PMID: 24651015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hovestadt V., Jones D.T., Picelli S., Wang W., Kool M., Northcott P.A., Sultan M., Stachurski K., Ryzhova M., Warnatz H.J., Ralser M., Brun S., Bunt J., Jäger N., Kleinheinz K., Erkek S., Weber U.D., Bartholomae C.C., von Kalle C., Lawerenz C., Eils J., Koster J., Versteeg R., Milde T., Witt O., Schmidt S., Wolf S., Pietsch T., Rutkowski S., Scheurlen W., Taylor M.D., Brors B., Felsberg J., Reifenberger G., Borkhardt A., Lehrach H., Wechsler-Reya R.J., Eils R., Yaspo M.L., Landgraf P., Korshunov A., Zapatka M., Radlwimmer B., Pfister S.M., Lichter P. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510(7506):537–541. doi: 10.1038/nature13268. [http://dx.doi.org/10.1038/nature13268]. [PMID: 24847876]. [DOI] [PubMed] [Google Scholar]
- 52.Northcott P.A., Lee C., Zichner T., Stütz A.M., Erkek S., Kawauchi D., Shih D.J., Hovestadt V., Zapatka M., Sturm D., Jones D.T., Kool M., Remke M., Cavalli F.M., Zuyderduyn S., Bader G.D., VandenBerg S., Esparza L.A., Ryzhova M., Wang W., Wittmann A., Stark S., Sieber L., Seker-Cin H., Linke L., Kratochwil F., Jäger N., Buchhalter I., Imbusch C.D., Zipprich G., Raeder B., Schmidt S., Diessl N., Wolf S., Wiemann S., Brors B., Lawerenz C., Eils J., Warnatz H.J., Risch T., Yaspo M.L., Weber U.D., Bartholomae C.C., von Kalle C., Turányi E., Hauser P., Sanden E., Darabi A., Siesjö P., Sterba J., Zitterbart K., Sumerauer D., van Sluis P., Versteeg R., Volckmann R., Koster J., Schuhmann M.U., Ebinger M., Grimes H.L., Robinson G.W., Gajjar A., Mynarek M., von Hoff K., Rutkowski S., Pietsch T., Scheurlen W., Felsberg J., Reifenberger G., Kulozik A.E., von Deimling A., Witt O., Eils R., Gilbertson R.J., Korshunov A., Taylor M.D., Lichter P., Korbel J.O., Wechsler-Reya R.J., Pfister S.M. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–434. doi: 10.1038/nature13379. [http://dx.doi.org/10.1038/nature13379]. [PMID: 25043047]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin C.Y., Erkek S., Tong Y., Yin L., Federation A.J., Zapatka M., Haldipur P., Kawauchi D., Risch T., Warnatz H.J., Worst B.C., Ju B., Orr B.A., Zeid R., Polaski D.R., Segura-Wang M., Waszak S.M., Jones D.T., Kool M., Hovestadt V., Buchhalter I., Sieber L., Johann P., Chavez L., Gröschel S., Ryzhova M., Korshunov A., Chen W., Chizhikov V.V., Millen K.J., Amstislavskiy V., Lehrach H., Yaspo M.L., Eils R., Lichter P., Korbel J.O., Pfister S.M., Bradner J.E., Northcott P.A. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530(7588):57–62. doi: 10.1038/nature16546. [http://dx.doi.org/10.1038/ nature16546]. [PMID: 26814967]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellison D.W., Onilude O.E., Lindsey J.C., Lusher M.E., Weston C.L., Taylor R.E., Pearson A.D., Clifford S.C. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J. Clin. Oncol. 2005;23(31):7951–7957. doi: 10.1200/JCO.2005.01.5479. [http://dx. doi.org/10.1200/JCO.2005.01.5479]. [PMID: 16258095]. [DOI] [PubMed] [Google Scholar]
- 55.Ellison D.W., Dalton J., Kocak M., Nicholson S.L., Fraga C., Neale G., Kenney A.M., Brat D.J., Perry A., Yong W.H., Taylor R.E., Bailey S., Clifford S.C., Gilbertson R.J. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. doi: 10.1007/s00401-011-0800-8. [http://dx.doi.org/10.1007/s00401-011-0800-8]. [PMID: 21267586]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clifford S.C., Lusher M.E., Lindsey J.C., Langdon J.A., Gilbertson R.J., Straughton D., Ellison D.W. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5(22):2666–2670. doi: 10.4161/cc.5.22.3446. [http://dx.doi.org/10. 4161/cc.5.22.3446]. [PMID: 17172831]. [DOI] [PubMed] [Google Scholar]
- 57.Gibson P., Tong Y., Robinson G., Thompson M.C., Currle D.S., Eden C., Kranenburg T.A., Hogg T., Poppleton H., Martin J., Finkelstein D., Pounds S., Weiss A., Patay Z., Scoggins M., Ogg R., Pei Y., Yang Z.J., Brun S., Lee Y., Zindy F., Lindsey J.C., Taketo M.M., Boop F.A., Sanford R.A., Gajjar A., Clifford S.C., Roussel M.F., McKinnon P.J., Gutmann D.H., Ellison D.W., Wechsler-Reya R., Gilbertson R.J. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468(7327):1095–1099. doi: 10.1038/nature09587. [http://dx.doi.org/10.1038/nature09587]. [PMID: 21150899]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramaswamy V., Remke M., Bouffet E., Bailey S., Clifford S.C., Doz F., Kool M., Dufour C., Vassal G., Milde T., Witt O., von Hoff K., Pietsch T., Northcott P.A., Gajjar A., Robinson G.W., Padovani L., André N., Massimino M., Pizer B., Packer R., Rutkowski S., Pfister S.M., Taylor M.D., Pomeroy S.L. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131(6):821–831. doi: 10.1007/s00401-016-1569-6. [http://dx.doi.org/10.1007/s00401-016-1569-6]. [PMID: 27040285]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamilton S.R., Liu B., Parsons R.E., Papadopoulos N., Jen J., Powell S.M., Krush A.J., Berk T., Cohen Z., Tetu B. The molecular basis of Turcot’s syndrome. N. Engl. J. Med. 1995;332(13):839–847. doi: 10.1056/NEJM199503303321302. [http://dx.doi.org/10.1056/NEJM199503303321302]. [PMID: 7661930]. [DOI] [PubMed] [Google Scholar]
- 60.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [http:// dx.doi.org/10.1038/nrc3419]. [PMID: 23258168]. [DOI] [PubMed] [Google Scholar]
- 61.Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [http://dx.doi.org/10.1016/j.cell.2012.05. 012]. [PMID: 22682243]. [DOI] [PubMed] [Google Scholar]
- 62.Mosimann C., Hausmann G., Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 2009;10(4):276–286. doi: 10.1038/nrm2654. [http://dx.doi.org/10.1038/nrm2654]. [PMID: 19305417]. [DOI] [PubMed] [Google Scholar]
- 63.Zurawel R.H., Chiappa S.A., Allen C., Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58(5):896–899. [PMID: 9500446]. [PubMed] [Google Scholar]
- 64.Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20(17):4935–4943. doi: 10.1093/emboj/20.17.4935. [http://dx.doi.org/10.1093/emboj/20.17.4935]. [PMID: 11532957]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hecht A., Vleminckx K., Stemmler M.P., van Roy F., Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19(8):1839–1850. doi: 10.1093/emboj/19.8.1839. [http://dx.doi.org/10.1093/emboj/19.8.1839]. [PMID: 10775268]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carrera I., Janody F., Leeds N., Duveau F., Treisman J.E. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. USA. 2008;105(18):6644–6649. doi: 10.1073/pnas.0709749105. [http://dx.doi.org/10.1073/pnas. 0709749105]. [PMID: 18451032]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfaff E., Remke M., Sturm D., Benner A., Witt H., Milde T., von Bueren A.O., Wittmann A., Schöttler A., Jorch N., Graf N., Kulozik A.E., Witt O., Scheurlen W., von Deimling A., Rutkowski S., Taylor M.D., Tabori U., Lichter P., Korshunov A., Pfister S.M. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J. Clin. Oncol. 2010;28(35):5188–5196. doi: 10.1200/JCO.2010.31.1670. [http://dx.doi.org/10.1200/JCO.2010.31.1670]. [PMID: 21060032]. [DOI] [PubMed] [Google Scholar]
- 68.Perreault S., Ramaswamy V., Achrol A.S., Chao K., Liu T.T., Shih D., Remke M., Schubert S., Bouffet E., Fisher P.G., Partap S., Vogel H., Taylor M.D., Cho Y.J., Yeom K.W. MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am. J. Neuroradiol. 2014;35(7):1263–1269. doi: 10.3174/ajnr.A3990. [http://dx.doi.org/10.3174/ ajnr.A3990]. [PMID: 24831600]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kool M., Korshunov A., Remke M., Jones D.T., Schlanstein M., Northcott P.A., Cho Y.J., Koster J., Schouten-van Meeteren A., van Vuurden D., Clifford S.C., Pietsch T., von Bueren A.O., Rutkowski S., McCabe M., Collins V.P., Bäcklund M.L., Haberler C., Bourdeaut F., Delattre O., Doz F., Ellison D.W., Gilbertson R.J., Pomeroy S.L., Taylor M.D., Lichter P., Pfister S.M. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123(4):473–484. doi: 10.1007/s00401-012-0958-8. [http://dx.doi.org/10.1007/ s00401-012-0958-8]. [PMID: 22358457]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhukova N., Ramaswamy V., Remke M., Pfaff E., Shih D.J., Martin D.C., Castelo-Branco P., Baskin B., Ray P.N., Bouffet E., von Bueren A.O., Jones D.T., Northcott P.A., Kool M., Sturm D., Pugh T.J., Pomeroy S.L., Cho Y.J., Pietsch T., Gessi M., Rutkowski S., Bognar L., Klekner A., Cho B.K., Kim S.K., Wang K.C., Eberhart C.G., Fevre-Montange M., Fouladi M., French P.J., Kros M., Grajkowska W.A., Gupta N., Weiss W.A., Hauser P., Jabado N., Jouvet A., Jung S., Kumabe T., Lach B., Leonard J.R., Rubin J.B., Liau L.M., Massimi L., Pollack I.F., Shin Ra Y., Van Meir E.G., Zitterbart K., Schüller U., Hill R.M., Lindsey J.C., Schwalbe E.C., Bailey S., Ellison D.W., Hawkins C., Malkin D., Clifford S.C., Korshunov A., Pfister S., Taylor M.D., Tabori U. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J. Clin. Oncol. 2013;31(23):2927–2935. doi: 10.1200/JCO.2012.48.5052. [http://dx.doi.org/10.1200/JCO.2012.48. 5052]. [PMID: 23835706]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorlin R.J. Nevoid basal cell carcinoma (Gorlin) syndrome. Genet. Med. 2004;6(6):530–539. doi: 10.1097/01.gim.0000144188.15902.c4. [http://dx.doi.org/10.1097/01.GIM. 0000144188.15902.C4]. [PMID: 15545751]. [DOI] [PubMed] [Google Scholar]
- 72.Amlashi S.F., Riffaud L., Brassier G., Morandi X. Nevoid basal cell carcinoma syndrome: relation with desmoplastic medulloblastoma in infancy. A population-based study and review of the literature. Cancer. 2003;98(3):618–624. doi: 10.1002/cncr.11537. [http://dx.doi.org/10.1002/ cncr.11537]. [PMID: 12879481]. [DOI] [PubMed] [Google Scholar]
- 73.Garrè M.L., Cama A., Bagnasco F., Morana G., Giangaspero F., Brisigotti M., Gambini C., Forni M., Rossi A., Haupt R., Nozza P., Barra S., Piatelli G., Viglizzo G., Capra V., Bruno W., Pastorino L., Massimino M., Tumolo M., Fidani P., Dallorso S., Schumacher R.F., Milanaccio C., Pietsch T. Medulloblastoma variants: age-dependent occurrence and relation to Gorlin syndrome--a new clinical perspective. Clin. Cancer Res. 2009;15(7):2463–2471. doi: 10.1158/1078-0432.CCR-08-2023. [http://dx.doi.org/10.1158/1078-0432.CCR-08-2023]. [PMID: 19276247]. [DOI] [PubMed] [Google Scholar]
- 74.Hahn H., Wicking C., Zaphiropoulous P.G., Gailani M.R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A.B., Gillies S., Negus K., Smyth I., Pressman C., Leffell D.J., Gerrard B., Goldstein A.M., Dean M., Toftgard R., Chenevix-Trench G., Wainwright B., Bale A.E. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [http://dx.doi.org/10.1016/ S0092-8674(00)81268-4]. [PMID: 8681379]. [DOI] [PubMed] [Google Scholar]
- 75.Johnson R.L., Rothman A.L., Xie J., Goodrich L.V., Bare J.W., Bonifas J.M., Quinn A.G., Myers R.M., Cox D.R., Epstein E.H., Jr, Scott M.P. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [http://dx.doi.org/10.1126/science.272.5268.1668]. [PMID: 8658145]. [DOI] [PubMed] [Google Scholar]
- 76.Mimeault M., Batra S.K. Frequent deregulations in the hedgehog signaling network and cross-talks with the epidermal growth factor receptor pathway involved in cancer progression and targeted therapies. Pharmacol. Rev. 2010;62(3):497–524. doi: 10.1124/pr.109.002329. [http://dx.doi. org/10.1124/pr.109.002329]. [PMID: 20716670]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oliver T.G., Grasfeder L.L., Carroll A.L., Kaiser C., Gillingham C.L., Lin S.M., Wickramasinghe R., Scott M.P., Wechsler-Reya R.J. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA. 2003;100(12):7331–7336. doi: 10.1073/pnas.0832317100. [http://dx. doi.org/10.1073/pnas.0832317100]. [PMID: 12777630]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor M.D., Liu L., Raffel C., Hui C.C., Mainprize T.G., Zhang X., Agatep R., Chiappa S., Gao L., Lowrance A., Hao A., Goldstein A.M., Stavrou T., Scherer S.W., Dura W.T., Wainwright B., Squire J.A., Rutka J.T., Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 2002;31(3):306–310. doi: 10.1038/ng916. [http://dx.doi.org/10.1038/ng916]. [PMID: 12068298]. [DOI] [PubMed] [Google Scholar]
- 79.Gajjar A., Stewart C.F., Ellison D.W., Kaste S., Kun L.E., Packer R.J., Goldman S., Chintagumpala M., Wallace D., Takebe N., Boyett J.M., Gilbertson R.J., Curran T. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clin. Cancer Res. 2013;19(22):6305–6312. doi: 10.1158/1078-0432.CCR-13-1425. [http://dx.doi.org/10.1158/1078-0432.CCR-13-1425]. [PMID: 24077351]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson G.W., Orr B.A., Wu G., Gururangan S., Lin T., Qaddoumi I., Packer R.J., Goldman S., Prados M.D., Desjardins A., Chintagumpala M., Takebe N., Kaste S.C., Rusch M., Allen S.J., Onar-Thomas A., Stewart C.F., Fouladi M., Boyett J.M., Gilbertson R.J., Curran T., Ellison D.W., Gajjar A. Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: Results from phase II pediatric brain tumor consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol. 2015;33(24):2646–2654. doi: 10.1200/JCO.2014.60.1591. [http://dx.doi.org/10.1200/JCO.2014.60.1591]. [PMID: 26169613]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodon J., Tawbi H.A., Thomas A.L., Stoller R.G., Turtschi C.P., Baselga J., Sarantopoulos J., Mahalingam D., Shou Y., Moles M.A., Yang L., Granvil C., Hurh E., Rose K.L., Amakye D.D., Dummer R., Mita A.C. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin. Cancer Res. 2014;20(7):1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [http://dx.doi. org/10.1158/1078-0432.CCR-13-1710]. [PMID: 24523439]. [DOI] [PubMed] [Google Scholar]
- 82.Kieran M.W., Chisholm J., Casanova M., Brandes A.A., Aerts I., Bouffet E., Bailey S., Leary S., MacDonald T.J., Mechinaud F., Cohen K.J., Riccardi R., Mason W., Hargrave D., Kalambakas S., Deshpande P., Tai F., Hurh E., Geoerger B. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro-oncol. 2017;19(11):1542–1552. doi: 10.1093/neuonc/nox109. [http://dx. doi.org/10.1093/neuonc/nox109]. [PMID: 28605510]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimura H., Ng J.M., Curran T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell. 2008;13(3):249–260. doi: 10.1016/j.ccr.2008.01.027. [http://dx.doi.org/10.1016/ j.ccr.2008.01.027]. [PMID: 18328428]. [DOI] [PubMed] [Google Scholar]
- 84.Rudin C.M., Hann C.L., Laterra J., Yauch R.L., Callahan C.A., Fu L., Holcomb T., Stinson J., Gould S.E., Coleman B., LoRusso P.M., Von Hoff D.D., de Sauvage F.J., Low J.A. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [http://dx.doi. org/10.1056/NEJMoa0902903]. [PMID: 19726761]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yauch R.L., Dijkgraaf G.J., Alicke B., Januario T., Ahn C.P., Holcomb T., Pujara K., Stinson J., Callahan C.A., Tang T., Bazan J.F., Kan Z., Seshagiri S., Hann C.L., Gould S.E., Low J.A., Rudin C.M., de Sauvage F.J. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–574. doi: 10.1126/science.1179386. [http://dx.doi.org/10.1126/ science.1179386]. [PMID: 19726788]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buonamici S., Williams J., Morrissey M., Wang A., Guo R., Vattay A., Hsiao K., Yuan J., Green J., Ospina B., Yu Q., Ostrom L., Fordjour P., Anderson D.L., Monahan J.E., Kelleher J.F., Peukert S., Pan S., Wu X., Maira S.M., García-Echeverría C., Briggs K.J., Watkins D.N., Yao Y.M., Lengauer C., Warmuth M., Sellers W.R., Dorsch M. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci. Transl. Med. 2010;2(51):51ra70. doi: 10.1126/scitranslmed.3001599. [http://dx. doi.org/10.1126/scitranslmed.3001599]. [PMID: 20881279]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dijkgraaf G.J., Alicke B., Weinmann L., Januario T., West K., Modrusan Z., Burdick D., Goldsmith R., Robarge K., Sutherlin D., Scales S.J., Gould S.E., Yauch R.L., de Sauvage F.J. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71(2):435–444. doi: 10.1158/0008-5472.CAN-10-2876. [http://dx.doi.org/10.1158/0008-5472.CAN-10-2876]. [PMID: 21123452]. [DOI] [PubMed] [Google Scholar]
- 88.Metcalfe C., de Sauvage F.J. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71(15):5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [http://dx.doi.org/10.1158/0008-5472.CAN-11-0923]. [PMID: 21771911]. [DOI] [PubMed] [Google Scholar]
- 89.Kim J., Aftab B.T., Tang J.Y., Kim D., Lee A.H., Rezaee M., Kim J., Chen B., King E.M., Borodovsky A., Riggins G.J., Epstein E.H., Jr, Beachy P.A., Rudin C.M. Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell. 2013;23(1):23–34. doi: 10.1016/j.ccr.2012.11.017. [http://dx.doi.org/10.1016/j.ccr.2012. 11.017]. [PMID: 23291299]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cavalli F.M.G., Remke M., Rampasek L., Peacock J., Shih D.J.H., Luu B., Garzia L., Torchia J., Nor C., Morrissy A.S., Agnihotri S., Thompson Y.Y., Kuzan-Fischer C.M., Farooq H., Isaev K., Daniels C., Cho B.K., Kim S.K., Wang K.C., Lee J.Y., Grajkowska W.A., Perek-Polnik M., Vasiljevic A., Faure-Conter C., Jouvet A., Giannini C., Nageswara Rao A.A., Li K.K.W., Ng H.K., Eberhart C.G., Pollack I.F., Hamilton R.L., Gillespie G.Y., Olson J.M., Leary S., Weiss W.A., Lach B., Chambless L.B., Thompson R.C., Cooper M.K., Vibhakar R., Hauser P., van Veelen M.C., Kros J.M., French P.J., Ra Y.S., Kumabe T., López-Aguilar E., Zitterbart K., Sterba J., Finocchiaro G., Massimino M., Van Meir E.G., Osuka S., Shofuda T., Klekner A., Zollo M., Leonard J.R., Rubin J.B., Jabado N., Albrecht S., Mora J., Van Meter T.E., Jung S., Moore A.S., Hallahan A.R., Chan J.A., Tirapelli D.P.C., Carlotti C.G., Fouladi M., Pimentel J., Faria C.C., Saad A.G., Massimi L., Liau L.M., Wheeler H., Nakamura H., Elbabaa S.K., Perezpeña-Diazconti M., Chico Ponce de León F., Robinson S., Zapotocky M., Lassaletta A., Huang A., Hawkins C.E., Tabori U., Bouffet E., Bartels U., Dirks P.B., Rutka J.T., Bader G.D., Reimand J., Goldenberg A., Ramaswamy V., Taylor M.D. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31(6):737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [http://dx.doi.org/10.1016/j.ccell.2017.05.005]. [PMID: 28609654]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mack S.C., Hubert C.G., Miller T.E., Taylor M.D., Rich J.N. An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 2016;19(1):10–19. doi: 10.1038/nn.4190. [http://dx.doi.org/10.1038/nn.4190]. [PMID: 26713744]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parsons D.W., Li M., Zhang X., Jones S., Leary R.J., Lin J.C., Boca S.M., Carter H., Samayoa J., Bettegowda C., Gallia G.L., Jallo G.I., Binder Z.A., Nikolsky Y., Hartigan J., Smith D.R., Gerhard D.S., Fults D.W., VandenBerg S., Berger M.S., Marie S.K., Shinjo S.M., Clara C., Phillips P.C., Minturn J.E., Biegel J.A., Judkins A.R., Resnick A.C., Storm P.B., Curran T., He Y., Rasheed B.A., Friedman H.S., Keir S.T., McLendon R., Northcott P.A., Taylor M.D., Burger P.C., Riggins G.J., Karchin R., Parmigiani G., Bigner D.D., Yan H., Papadopoulos N., Vogelstein B., Kinzler K.W., Velculescu V.E. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [http://dx.doi.org/10.1126/science.1198056]. [PMID: 21163964]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Northcott P.A., Nakahara Y., Wu X., Feuk L., Ellison D.W., Croul S., Mack S., Kongkham P.N., Peacock J., Dubuc A., Ra Y.S., Zilberberg K., McLeod J., Scherer S.W., Sunil Rao J., Eberhart C.G., Grajkowska W., Gillespie Y., Lach B., Grundy R., Pollack I.F., Hamilton R.L., Van Meter T., Carlotti C.G., Boop F., Bigner D., Gilbertson R.J., Rutka J.T., Taylor M.D. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [http://dx.doi.org/10.1038/ng.336]. [PMID: 19270706]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dubuc A.M., Remke M., Korshunov A., Northcott P.A., Zhan S.H., Mendez-Lago M., Kool M., Jones D.T., Unterberger A., Morrissy A.S., Shih D., Peacock J., Ramaswamy V., Rolider A., Wang X., Witt H., Hielscher T., Hawkins C., Vibhakar R., Croul S., Rutka J.T., Weiss W.A., Jones S.J., Eberhart C.G., Marra M.A., Pfister S.M., Taylor M.D. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125(3):373–384. doi: 10.1007/s00401-012-1070-9. [http://dx.doi.org/10.1007/s00401-012-1070-9]. [PMID: 23184418]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., Lee W., Mendenhall E., O’Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E.S., Bernstein B.E. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [http://dx.doi.org/10.1038/nature06008]. [PMID: 17603471]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [http://dx.doi.org/10.1126/science.1076997]. [PMID: 12351676]. [DOI] [PubMed] [Google Scholar]
- 97.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [http://dx.doi.org/10.1016/S0092-8674(02)00975-3]. [PMID: 12408863]. [DOI] [PubMed] [Google Scholar]
- 98.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [http://dx.doi.org/10.1038/nature06145]. [PMID: 17713478]. [DOI] [PubMed] [Google Scholar]
- 99.Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. [http://dx.doi.org/10.1128/MCB.01506-06]. [PMID: 17178841]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., Crawford G.E., Scacheri P.C. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19(4):590–601. doi: 10.1101/gr.086983.108. [http://dx.doi.org/10.1101/gr.086983. 108]. [PMID: 19251738]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sauvageau M., Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. doi: 10.1016/j.stem.2010.08.002. [http://dx.doi.org/10.1016/j.stem.2010.08.002]. [PMID: 20804967]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., Philpott M., Munro S., McKeown M.R., Wang Y., Christie A.L., West N., Cameron M.J., Schwartz B., Heightman T.D., La Thangue N., French C.A., Wiest O., Kung A.L., Knapp S., Bradner J.E. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [http://dx.doi.org/10.1038/nature09504]. [PMID: 20871596]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [http://dx.doi. org/10.1038/20974]. [PMID: 10365964]. [DOI] [PubMed] [Google Scholar]
- 104.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., Chesi M., Schinzel A.C., McKeown M.R., Heffernan T.P., Vakoc C.R., Bergsagel P.L., Ghobrial I.M., Richardson P.G., Young R.A., Hahn W.C., Anderson K.C., Kung A.L., Bradner J.E., Mitsiades C.S. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [http://dx. doi.org/10.1016/j.cell.2011.08.017]. [PMID: 21889194]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mertz J.A., Conery A.R., Bryant B.M., Sandy P., Balasubramanian S., Mele D.A., Bergeron L., Sims R.J., III Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [http://dx.doi. org/10.1073/pnas.1108190108]. [PMID: 21949397]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henssen A., Thor T., Odersky A., Heukamp L., El-Hindy N., Beckers A., Speleman F., Althoff K., Schäfers S., Schramm A., Sure U., Fleischhack G., Eggert A., Schulte J.H. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4(11):2080–2095. doi: 10.18632/oncotarget.1534. [http://dx.doi.org/10.18632/ oncotarget.1534]. [PMID: 24231268]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bandopadhayay P., Bergthold G., Nguyen B., Schubert S., Gholamin S., Tang Y., Bolin S., Schumacher S.E., Zeid R., Masoud S., Yu F., Vue N., Gibson W.J., Paolella B.R., Mitra S.S., Cheshier S.H., Qi J., Liu K.W., Wechsler-Reya R., Weiss W.A., Swartling F.J., Kieran M.W., Bradner J.E., Beroukhim R., Cho Y.J. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 2014;20(4):912–925. doi: 10.1158/1078-0432.CCR-13-2281. [http://dx.doi.org/ 10.1158/1078-0432.CCR-13-2281]. [PMID: 24297863]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venkataraman S., Alimova I., Balakrishnan I., Harris P., Birks D.K., Griesinger A., Amani V., Cristiano B., Remke M., Taylor M.D., Handler M., Foreman N.K., Vibhakar R. Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Oncotarget. 2014;5(9):2355–2371. doi: 10.18632/oncotarget.1659. [PMID: 24796395]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tang Y., Gholamin S., Schubert S., Willardson M.I., Lee A., Bandopadhayay P., Bergthold G., Masoud S., Nguyen B., Vue N., Balansay B., Yu F., Oh S., Woo P., Chen S., Ponnuswami A., Monje M., Atwood S.X., Whitson R.J., Mitra S., Cheshier S.H., Qi J., Beroukhim R., Tang J.Y., Wechsler-Reya R., Oro A.E., Link B.A., Bradner J.E., Cho Y.J. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat. Med. 2014;20(7):732–740. doi: 10.1038/nm.3613. [http://dx.doi. org/10.1038/nm.3613]. [PMID: 24973920]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alimova I., Venkataraman S., Harris P., Marquez V.E., Northcott P.A., Dubuc A., Taylor M.D., Foreman N.K., Vibhakar R. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int. J. Cancer. 2012;131(8):1800–1809. doi: 10.1002/ijc.27455. [http://dx.doi.org/10.1002/ ijc.27455]. [PMID: 22287205]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pei Y., Liu K.W., Wang J., Garancher A., Tao R., Esparza L.A., Maier D.L., Udaka Y.T., Murad N., Morrissy S., Seker-Cin H., Brabetz S., Qi L., Kogiso M., Schubert S., Olson J.M., Cho Y.J., Li X.N., Crawford J.R., Levy M.L., Kool M., Pfister S.M., Taylor M.D., Wechsler-Reya R.J. HDAC and PI3K Antagonists Cooperate to Inhibit Growth of MYC-Driven Medulloblastoma. Cancer Cell. 2016;29(3):311–323. doi: 10.1016/j.ccell.2016.02.011. [http://dx.doi. org/10.1016/j.ccell.2016.02.011]. [PMID: 26977882]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Phoenix T.N., Patmore D.M., Boop S., Boulos N., Jacus M.O., Patel Y.T., Roussel M.F., Finkelstein D., Goumnerova L., Perreault S., Wadhwa E., Cho Y.J., Stewart C.F., Gilbertson R.J. Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell. 2016;29(4):508–522. doi: 10.1016/j.ccell.2016.03.002. [http://dx.doi.org/10.1016/ j.ccell.2016.03.002]. [PMID: 27050100]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stenman J.M., Rajagopal J., Carroll T.J., Ishibashi M., McMahon J., McMahon A.P. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322(5905):1247–1250. doi: 10.1126/science.1164594. [http://dx.doi.org/10.1126/science. 1164594]. [PMID: 19023080]. [DOI] [PubMed] [Google Scholar]
- 114.Daneman R., Agalliu D., Zhou L., Kuhnert F., Kuo C.J., Barres B.A. Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. USA. 2009;106(2):641–646. doi: 10.1073/pnas.0805165106. [http://dx.doi.org/10.1073/pnas.0805165106]. [PMID: 19129494]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vanner R.J., Remke M., Gallo M., Selvadurai H.J., Coutinho F., Lee L., Kushida M., Head R., Morrissy S., Zhu X., Aviv T., Voisin V., Clarke I.D., Li Y., Mungall A.J., Moore R.A., Ma Y., Jones S.J., Marra M.A., Malkin D., Northcott P.A., Kool M., Pfister S.M., Bader G., Hochedlinger K., Korshunov A., Taylor M.D., Dirks P.B. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26(1):33–47. doi: 10.1016/j.ccr.2014.05.005. [http://dx.doi.org/10. 1016/j.ccr.2014.05.005]. [PMID: 24954133]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morrissy A.S., Garzia L., Shih D.J., Zuyderduyn S., Huang X., Skowron P., Remke M., Cavalli F.M., Ramaswamy V., Lindsay P.E., Jelveh S., Donovan L.K., Wang X., Luu B., Zayne K., Li Y., Mayoh C., Thiessen N., Mercier E., Mungall K.L., Ma Y., Tse K., Zeng T., Shumansky K., Roth A.J., Shah S., Farooq H., Kijima N., Holgado B.L., Lee J.J., Matan-Lithwick S., Liu J., Mack S.C., Manno A., Michealraj K.A., Nor C., Peacock J., Qin L., Reimand J., Rolider A., Thompson Y.Y., Wu X., Pugh T., Ally A., Bilenky M., Butterfield Y.S., Carlsen R., Cheng Y., Chuah E., Corbett R.D., Dhalla N., He A., Lee D., Li H.I., Long W., Mayo M., Plettner P., Qian J.Q., Schein J.E., Tam A., Wong T., Birol I., Zhao Y., Faria C.C., Pimentel J., Nunes S., Shalaby T., Grotzer M., Pollack I.F., Hamilton R.L., Li X.N., Bendel A.E., Fults D.W., Walter A.W., Kumabe T., Tominaga T., Collins V.P., Cho Y.J., Hoffman C., Lyden D., Wisoff J.H., Garvin J.H., Jr, Stearns D.S., Massimi L., Schüller U., Sterba J., Zitterbart K., Puget S., Ayrault O., Dunn S.E., Tirapelli D.P., Carlotti C.G., Wheeler H., Hallahan A.R., Ingram W., MacDonald T.J., Olson J.J., Van Meir E.G., Lee J.Y., Wang K.C., Kim S.K., Cho B.K., Pietsch T., Fleischhack G., Tippelt S., Ra Y.S., Bailey S., Lindsey J.C., Clifford S.C., Eberhart C.G., Cooper M.K., Packer R.J., Massimino M., Garre M.L., Bartels U., Tabori U., Hawkins C.E., Dirks P., Bouffet E., Rutka J.T., Wechsler-Reya R.J., Weiss W.A., Collier L.S., Dupuy A.J., Korshunov A., Jones D.T., Kool M., Northcott P.A., Pfister S.M., Largaespada D.A., Mungall A.J., Moore R.A., Jabado N., Bader G.D., Jones S.J., Malkin D., Marra M.A., Taylor M.D. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529(7586):351–357. doi: 10.1038/nature16478. [http://dx.doi.org/ 10.1038/nature16478]. [PMID: 26760213]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramaswamy V., Remke M., Bouffet E., Faria C.C., Perreault S., Cho Y.J., Shih D.J., Luu B., Dubuc A.M., Northcott P.A., Schüller U., Gururangan S., McLendon R., Bigner D., Fouladi M., Ligon K.L., Pomeroy S.L., Dunn S., Triscott J., Jabado N., Fontebasso A., Jones D.T., Kool M., Karajannis M.A., Gardner S.L., Zagzag D., Nunes S., Pimentel J., Mora J., Lipp E., Walter A.W., Ryzhova M., Zheludkova O., Kumirova E., Alshami J., Croul S.E., Rutka J.T., Hawkins C., Tabori U., Codispoti K.E., Packer R.J., Pfister S.M., Korshunov A., Taylor M.D. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14(12):1200–1207. doi: 10.1016/S1470-2045(13)70449-2. [http://dx.doi.org/10.1016/S1470-2045(13)70449-2]. [PMID: 24140199]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang X., Dubuc A.M., Ramaswamy V., Mack S., Gendoo D.M., Remke M., Wu X., Garzia L., Luu B., Cavalli F., Peacock J., López B., Skowron P., Zagzag D., Lyden D., Hoffman C., Cho Y.J., Eberhart C., MacDonald T., Li X.N., Van Meter T., Northcott P.A., Haibe-Kains B., Hawkins C., Rutka J.T., Bouffet E., Pfister S.M., Korshunov A., Taylor M.D. Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol. 2015;129(3):449–457. doi: 10.1007/s00401-015-1389-0. [http:// dx.doi.org/10.1007/s00401-015-1389-0]. [PMID: 25689980]. [DOI] [PMC free article] [PubMed] [Google Scholar]