Abstract
Scope
Bovine lactoferrin (bLF) is an ingredient of food supplements and infant formulas given its antimicrobial and antiviral properties. We modified bLF enzymatically to alter its N‐glycosylation and to isolate the glycan chains. The aims of this study include (1) to evaluate whether such derivates induce responses via pattern recognition receptors namely Toll‐like receptors (TLRs) and (2) to relate those responses to their different glycosylation profiles.
Methods and results
The unmodified and modified bLF fractions are incubated with reporter cell lines expressing pattern recognition receptors. Afterwards, we screen for TLRs and analyze for nuclear factor kappa—light‐chain enhancer of activated B cells (NF‐κB) activation. Activation of reporter cell lines show that signaling is highly dependent on TLRs. The activation pattern of bLF is reduced with the desialylated form and increased with the demannosylated form. In reporter cells for TLR, bLF activate TLR‐4 and inhibit TLR‐3. The isolated glycans from bLF inhibit TLR‐8. TLR‐2, TLR‐5, TLR‐7, and TLR‐9 are not significantly altered.
Conclusion
The profile of glycosylation is key for the biological activity of bLF. By understanding how this affects the human defense responses, the bLF glycan profile can be modified to enhance its immunomodulatory effects when used as a dietary ingredient.
Keywords: bovine lactoferrin, N‐glycosylation, NF‐κB, toll‐like receptors
1. Introduction
Milk substitutes and infant formulas play an important role in infant nutrition when breastfeeding is not possible.1 Among all the human milk components, lactoferrin (LF) is considered one of the most important elements for the newborns defense against infections and proper development and maturation of the intestinal mucosa.2 LF promotes enterocytes proliferation and differentiation.3, 4 Considering these important biological functions, LF has been incorporated in many products as a dietary ingredient to support the immune system.1, 5
LF is a cationic glycoprotein from the transferrin family.6 It is present in the secondary granules of neutrophils and in exocrine secretions such as saliva, tears, and milk.7 In humans, LF is one of the most abundant glycoproteins in breast milk and is considered to play a role in iron homeostasis and to have among others antimicrobial, antiviral, and anti‐inflammatory properties.8, 9 LF is secreted in its open apo‐form (iron‐free LF) and it binds to ferric ions (Fe3+) to become the closed holo‐form.6 Human LF is more abundant in colostrum (7 g L−1) and found at a lower concentration in mature breast milk (2–4 g L−1). For this reason, much attention is given to its functional role in human health. Therefore, in the design of infant formulas LF should closely mimic the concentration and functional aspects of LF in human breast milk.2
Human lactoferrin (hLF) and bovine lactoferrin (bLF) are not identical.10 They share a protein core with a similarity of 68–70% but a pronounced difference occurs at their glycosylation level. hLF and bLF differ both in type of glycosylation, the number of potential N‐glycosylation sites, and the glycan decoration itself.11 Glycosylation is a post‐translational modification of proteins that affects their structure, trafficking, recognition, and biological functions.12 It has been reported that glycosylation in LF protects against proteolysis,13 facilitates inter‐ or intra‐cellular signaling,14 allows proper protein folding,15 and modulates lectin N‐glycan recognition processes.16
Bovine milk glycoproteins carry N‐ and O‐linked glycans. However, bLF carries only N‐glycans with sugar moieties attached via N‐acetyl glucosamine to the asparagine residues of the protein in the specific amino acid sequence Asn‐X‐Ser/Thr, in which X can be any amino acid except proline.17 hLF has three potential sites for N‐glycosylation, that is, Asn137, Asn478, Asn623, that are always occupied, whereas bLF has five potential sites, that is, Asn 281, Asn233, Asn368, Asn476, and Asn545.11, 18 Four sites are always occupied, whereas Asn281 is glycosylated for approximately 30% in bovine colostrum, but is reduced to 15% in mature milk.19 N‐glycans from hLF differ from bLF as they are highly branched, highly sialylated, and highly fucosylated complex‐type structures and many contain Lewis (x) epitopes.20 Typically, the bLF complex‐type N‐glycans include certain epitopes, not found in hLF N‐glycans, that is, Gal(α1‐3)Gal(β1‐4)GlcNAc (αGal), GalNAc(β1‐4)GlcNAc (LacdiNAc), and N‐glycolylneuraminic acid (Neu5Gc).21, 22 In total, bLF contains 76% neutral, 9% mono‐sialylated, and 15% di‐sialylated glycan structures. From the sialic acid content, 8.5% has been reported to correspond to Neu5Gc.21 Bovine lactoferrin (bLF) glycans have been classified as 65% oligomannose type, while 35% remaining correspond to complex and hybrid type.23
Glycosylation is a tightly regulated process, considered to be programmed, temporal and sensitive to dietary regime.24, 25 Most importantly, there is growing evidence that glycoproteins play a critical role in immune recognition and that this property is linked to the structural diversity in glycosylation. Both bLF and hLF are known to modulate the immune system via Toll‐like receptors (TLRs).26 However, how this modulation via TLRs is affected by glycosylation is not yet well understood. Therefore, in the present study, we investigated the effect of modified bLF structures (desialylated, demannosylated) and the effect of N‐glycans (desialylated and demannosylated) isolated from bLF on the signaling of NF‐κB (nuclear factor kappa–light‐chain enhancer of activated B cells) via TLRs. Our study showed that these variations in glycans decorations of bLF influence signaling of TLR‐3, TLR‐4, and TLR‐8.
2. Experimental Section
2.1. Preparation of Bovine Lactoferrin Modified Structures
bLF was isolated from pooled cow milk and was obtained from Friesland Campina (Amersfoort, The Netherlands). This compound was subjected to different treatments to alter its native structure. Samples of 500 mg bLF were incubated with either sialidase (Arthrobacter ureafaciens, Sigma‐Aldrich Chemie B.V.) or α‐mannosidase (Canavalia ensiformis, Sigma‐Aldrich Chemie B.V.). Samples of bLF were dissolved at a concentration of ∼5 mg mL−1 in 50 mM sodium acetate (pH 5.0). The buffer for the mannosidase assay was supplemented with 1 mM of calcium and zinc. Either sialidase (1 mU mg−1 protein) or mannosidase (5 mU mg−1 protein) was added and incubations were performed overnight at 37 °C with continuous agitation. After 16 h, 1 mU mg−1 enzyme was added to the sialidase and incubated for 24 h. The incubation at 37 °C does not denaturalize the protein or alter its function. The heat‐induced denaturalization of bLF occurs above a temperature range of 70 °C and 90 °C and at pH lower than 5.0.27, 28, 29
The resulting products were dialyzed (SnakeSkin Dialysis Tubing, 10 kDa MWCO, 22 mm, ThermoFisher Scientific, Waltham, MA) against running filtered water for 24 h to remove cleaved monosaccharides and buffer salts. After lyophilisation, the dialyzed bLF was subjected to a monosaccharide analysis to evaluate the remaining sialic acid and mannose content. When the desired modifications were complete (>90% of sialic acid removed or a reduction of 25% of mannose), portions (50 mg) of the modified proteins were stored for the experiments together with the intact proteins.
Glycans were released from intact and modified bLF structures by incubation with PNGase F. Lyophilized bLF was dissolved at a concentration of 2.5 mg mL−1 in 100 mM sodium phosphate buffer (pH 7.5). SDS and β‐mercapto ethanol (Sigma‐Aldrich Chemie B.V.) were added to a concentration of 0.25% and 1%, respectively, and the protein denatured by heating at 85 °C for 30 min. Denatured protein was alkylated by addition of iodoacetamide to a concentration of 20 mM (55 °C; 30 min). Nonidet P‐40 (NP‐40, Sigma‐Aldrich Chemie B.V.) was added at a final concentration of 1%. PNGaseF (Flavobacterium meningosepticum, New England Biolabs, Ipswich, UK) was added at a concentration of 50 U mg−1 of bLF protein and the solution incubated overnight at 37 °C with continuous agitation. Completion of the PNGase F digestion was checked by SDS‐PAGE. The released glycans were isolated by a sequence of purification steps, including acetone precipitation, C18, and graphitized carbon solid‐phase extraction (Valk‐Weeber et al., in press). Purity of the obtained glycans was established by monosaccharide analysis and 1D 1H NMR spectroscopy (described in the Supporting Information, Materials and Methods).
2.2. Cell Culture of Reporter Cell Lines
Reagents such as selection media, Quanti‐Blue reagents, TLR agonists, and THP1 (human acute monocytic leukemia) and TLR reporter cell lines were purchased from InvivoGen (Toulouse, France). The THP1 reporter cell line express TLRs and contains a construct for secreted embryonic alkaline phosphatase (SEAP) coupled to the NF‐κB/AP‐1 promoter. This THP‐1 cell line carries extra inserts for the cosignaling molecules MD2 and CD14. The second THP‐1 cell line expresses a nonfunctional form of the TLR adaptor MyD88 (myeloid differentiation primary response protein 88). Additionally, nine human embryonic kidney HEK293‐Blue reporter cell lines (InvivoGen) containing individual constructs for TLR‐2, TLR‐3, TLR‐4, TLR‐5, TLR7, TLR‐8, and TLR‐9 were used.30, 31 All the cell lines carry a construct for SEAP coupled to the NF‐κB/AP‐1 promoter. Both THP1 cell lines were maintained in RPMI1640. The culture medium was enriched with 10% heat‐inactivated fetal bovine serum, sodium bicarbonate NaHCO3 (1.5 g L−1), l‐glutamine (2 mM Sigma‐Aldrich Chemie B.V), glucose (4.5 g L−1 Sigma‐Aldrich Chemie B.V), HEPES (10 mM Sigma‐Aldrich Chemie B.V), sodium pyruvate (1.0 mM Sigma‐Aldrich Chemie B.V), penicillin–streptomycin (50 U mL−1–50 μg mL−1, Sigma‐Aldrich Chemie B.V), and normocyn (100 μg mL−1, Sigma‐Aldrich Chemie B.V). The HEK‐Blue cell lines were maintained in DMEM (Life Technologies Europe B.V.) containing 10% heat inactivated fetal bovine serum, l‐glutamine (2.0 mM‐Sigma‐Aldrich Chemie B.V), glucose (4.5 g L−1, Sigma‐Aldrich Chemie B.V), penicillin‐streptomycin (50 U mL−1–50 μg mL−1 Sigma‐Aldrich Chemie B.V), and normocyn (100 μg mL−1 Sigma‐Aldrich Chemie B.V). HEK cells were grown to approximately 80% of confluence. After culturing for 3 passages, all reporter cell lines were maintained on selection media according to the manufacturer's protocol.
2.3. THP1 Reporter Cells Lines Stimulation
THP1 cells were centrifuged for 5 min at 300 g. The cell density per well indicated by the manufacturer (Table 1) was accomplished by appropriate dilution in culture medium. Next, in a flat bottom 96‐well plate, 100 μL of this cell suspension plus 10 μL of stimulus were added per well. The plate was incubated for 24 h at 37 °C and 5% CO2. The stimulus consisted of bLF proteins and its derivates at 2 mg mL−1, isolated bLF glycan fractions and its derivates at 1.2 mg mL−1. The culture medium and endotoxin free‐water were used as negative controls. The activity of SEAP converts the pink Quanti‐Blue substrate to blue. After 24 h, the media supernatant was mixed with the Quanti‐Blue in a ratio of 1:10 and incubated for 1 h at 37 °C and 5% CO2. The NF‐kB release was quantified at 650 nm using a Benchmark Plus Microplate Reader using Microplate Manager 5.2.1 version for data acquisition. The assays were performed with three technical repeats and each experiment was repeated three to five times.
Table 1.
Summary of the agonists, agonist concentrations, and cell density applied for the stimulation experiments
| Cell line | Cell density | Agonist (positive control) | Agonist concentration |
|---|---|---|---|
| (cell mL−1) | |||
| THP1MD2CD14 | 1 × 106 | LPS‐EK | 10 μg mL−1 |
| Lipopolysaccharides from Escherichia coli K12 | |||
| THP1MyD88 def | 2 × 106 | Tri‐DAP | 100 μg mL−1 |
| L‐ala‐γ‐d‐Glu‐mDAP | |||
| HEK‐hTLR2 | 2.8 × 105 | FSL‐1 | 1 μg mL−1 |
| lipopeptide | |||
| HEK‐hTLR3 | 2.8 × 105 | Poly (I:C) LMW) | 5 μg mL−1 |
| polyinosinic–polycytidylic acid | |||
| Low molecular weight | |||
| HEK‐hTLR4 | 1.4 × 105 | LPS‐EK | 0.1 μg mL−1 |
| Lipopolysaccharides from E. coli K12 | |||
| HEK‐hTLR5 | 1.4 × 105 | Rec‐FLA‐ST | 10 μg mL−1 |
| Flagellin from Salmonella typhymurium | |||
| HEK‐hTLR7 | 2.2 × 105 | CL246 | 5 μg mL−1 |
| Adenine analog | |||
| HEK‐hTLR8 | 2.2 × 105 | ssRNA40/LyoVec | 50 μg mL−1 |
| Singled‐stranded RNA | |||
| HEK‐hTLR9 | 4.5 × 105 | ODN 2006 | 10 μM |
| Class B CpG oligonucleotide |
2.4. TLR Activation and Inhibition Assay
HEK‐Blue cell lines were detached from the bottom flask after which the cells were centrifuged and re‐suspended according to manufacturer's protocol (Table 1). Later the cells were seeded at different cell densities (Table 1) in 96‐well plates at 100 μL per well. The cells were treated with 10 μL of sample. The plates were incubated for 24 h at 37 °C and 5% CO2. After this period, 20 μL of supernatant were mixed with 180 μL of Quanti‐Blue in flat bottom 96‐well plates. The plate was incubated for 1 h at 37 °C and 5% CO2. Activation was studied by comparing the NF‐κB release of nontreated cells with the NF‐κB release from the treated cells. Inhibition of TLRs was studied by comparing the NF‐κB release of TLRs agonist with the NF‐κB release of cells treated with TLR agonist and sample. After the incubation, the analysis of SEAP was performed in the same fashion as described for the THP1 cell lines.
2.5. Statistical Analysis
Values were expressed as median with interquartile range. Normal distribution of the data sets was excluded using the Shapiro–Wilk and Kolmogorov–Smirnov tests. Statistical comparisons were performed using Mann–Whitney nonparametric U tests for unpaired observations and (two‐tailed). A p‐value < 0.05 was considered statistically significant. p values < 0.05 are denoted with *, p values < 0.01 are denoted with ** and p values < 0.001 are denoted with ***.
3. Results
3.1. Bovine Lactoferrin Induces Myd88‐Dependent Activation of THP1 MD2 CD14 Cells
To determine whether bLF has immune stimulating effects, we stimulated the THP1 MD2 CD14 cell line with bLF at a concentration of 2 mg mL−1 for 24 h. As shown in Figure 1, bLF enhanced the release of NF‐κB from 0.0044 to 0.86 (p < 0.01).
Figure 1.

bLF stimulates THP1 MD2 CD14 via TLRs. The cell lines THP1 MD2 CD14 and THP1 MD2 CD14 with a truncated MyD88 adaptor were stimulated with 2 mg mL−1 of bLF after which NF‐κB activation was measured by spectrophotometry at 650 nm. The release of NF‐κB after bLF incubation was compared with the release of NF‐κB of nontreated cells. NF‐κB release is expressed in arbitrary units. Data are represented as median with interquartile range (n = 5). Statistical differences were measured using Mann–Whitney test (**p < 0.01, ***p < 0.001).
To study whether the MyD88 protein adaptor mediates this activation, we also performed this study with THP1 MD2 CD14 with a truncated MyD88.27, 28 An activation of NF‐κB in THP1 MD2 CD14 but not in THP1 with a truncated Myd88 when incubated with bLF confirms that the activation was Myd88 and TLR dependent (Figure S1). TRIDAP is a suitable control because it signals via NOD‐1 receptors.
3.2. Consequences of Structural Modifications on bLF N‐Glycans
In order to gain insight into the effects of different glycosylation patterns in the activation of PRRs, a series of modified bLF fractions were prepared and tested on the THP1 MD2 CD14 cell lines. The iron saturation of bLF ranges from 15 to 19%.8 Therefore, under the dialysis conditions (pH 5.0), the bLF is expected to shift from the iron‐bound holo‐form to apo‐form with approximately 4–8% of iron because iron is retained in the dialysis membrane.29, 32 In order to completely remove the iron form the bLF, a pH below 2 is required but this was avoided as it might compromise function of the glycoprotein.32, 33 This dialysis step was associated with a reduction in THP1 cell activating capacity (Figure S2), possibly due to this change in form.
As shown in Figure 2A, desialylated bLF induced a statistical significantly lower activation of THP1 MD2 CD14 cells (p < 0.01). The reduction in the mannose content had the opposite effect. It enhanced the response of THP1 MD2 CD14 cell almost twofold when compared with bLF itself (Figure 2B)
Figure 2.

The absence of sialic acid has an effect on the release of NF‐κB. THP1 MD2 CD14 cells were stimulated with 2 mg mL−1 of bLF without sialic acid (A) and bLF with 25% reduced mannose on the N‐glycan chains(B). NF‐κB activation was measured by spectrophotometry at 650 nm. NF‐κB release is expressed in arbitrary units. Data are represented as median with interquartile range (n = 5). Statistical differences were measured using Mann–Whitney test (**p < 0.01).
Finally, to get further insight into structure–function relationships, also the effects of the isolated N‐glycan groups were tested. Fractions of the intact, desialylated, and the demannosylated bLF were treated with PGNase F to release the N‐glycans from the protein. The incubation on THP1 MD2 CD14 cells with intact N‐glycans and desialylated and demannosylated N‐glycans showed no activation (Figure S3).
3.3. Bovine Lactoferrin Effects on TLRs
The results from the THP1 cell stimulation showed that TLRs are engaged in NF‐κB signaling by bLF proteins. Next, we investigated which TLRs were involved and whether or not the intact, dialyzed, desialylated or demannosylated bLFs influence the signaling via specific TLR receptors. To this end, HEK cells expressing TLR‐2, TLR‐3, TLR‐4, TLR‐5, TLR‐7, and TLR‐9 were incubated with the different types of (un)modified bLF. HEK cells expressing TLR‐2, TLR‐5, TLR‐7, TLR‐9 were not significantly activated or inhibited by intact bLF or its counterparts with N‐glycan modifications (Figures S4, S5, S6, and S7, respectively). Additionally, when these cell lines were treated with the isolated intact, desialylated, and demannosylated N‐glycans, no effects were observed. This was different for TLR‐3, TLR‐4, and TLR‐8.
3.4. TLR‐3 Inhibition of NF‐κB Release by Modified bLF Proteins
bLF did have an inhibitory rather than an activating effect on TLR‐3 cells. When HEK cells carrying TLR‐3 were incubated with poly‐IC LMW combined with bLF, we observed almost a complete blockage of the signaling via TLR‐3. This was not dependent on the sialic acid or mannose decoration on the N‐glycans attached to the bLF: the desialylated and demannosylated forms exhibited similar, if not identical inhibitory effects on TLR‐3 (p < 0.01).
The NF‐κB released from the bLF was 10.03‐fold (p < 0.01) lower when compared to the NF‐κB released from the positive control. The desialylated bLF showed 8.05‐fold (p < 0.01) and the demannosylated bLFs 11.64‐fold (p < 0.01) lower response when compared to the Poly (I:C) activation (Figure 3).
Figure 3.
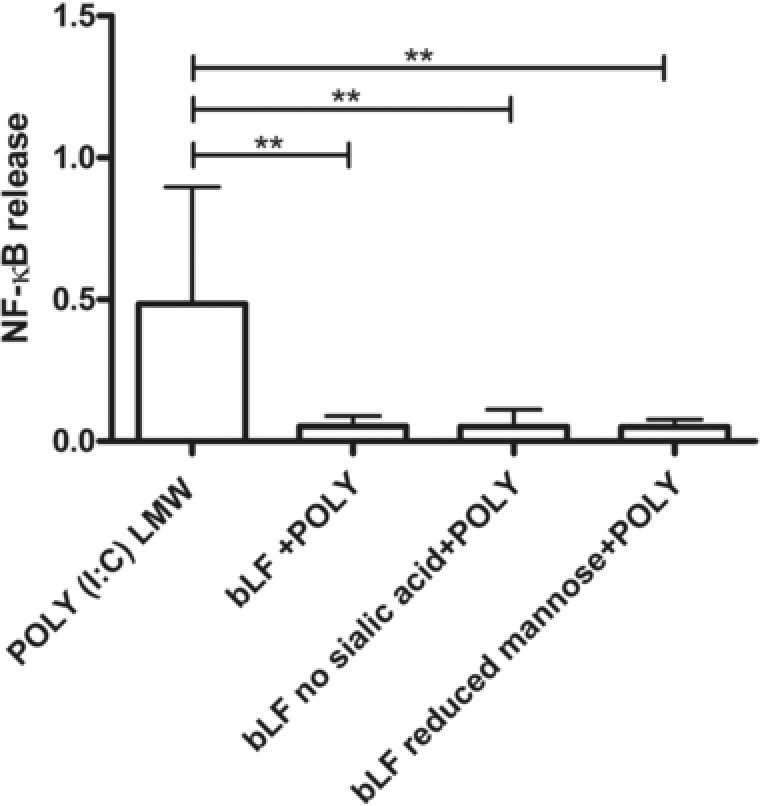
Inhibitory effects of bLF proteins on HEK‐ TLR‐3 cells. The cells were coincubated with the bLF structures and with the specific agonist for TLR‐3 Poly(I:C) LMW (5 μg mL−1). NF‐κB release was measured by spectrophotometry at 650 nm. Median and interquartile range of activation is plotted as NF‐κB release (n = 3). Statistical differences were calculated with Mann–Whitney test (**p < 0.01).
When the isolated bLF N‐glycans were incubated with this cell line, neither activation nor inhibition was observed (Figure S3). This suggest that the presence of the protein core is necessary to result in the inhibition of Poly (I:C) induced TLR‐3 activation.
3.5. Bovine Lactoferrin Proteins Induce Strong Activation of TLR‐4
As shown in Figure 4, bLF and its desialylated and demannosylated counterparts activated TLR‐4. The HEK‐Blue TLR‐4 cells were also treated with the isolated N‐glycans, but these compounds did not exert activation. This may suggest that the protein core of bLF and not the isolated N‐glycans play a role in the activation of TLR‐4. To exclude that the observed TLR‐4 activation was mediated or interfered by LPS contamination, the samples were treated with Polymyxin B.
Figure 4.

bLF glycosylation pattern influence in the release of NF‐κB in HEK‐Blue TLR‐4. Cells were incubated with bLF proteins at 2 mg mL−1 and isolated N‐glycans and desialylated and demannosylated N‐glycans at 1.2 mg mL−1. Culture medium served as negative control. NF‐κB release is expressed in arbitrary units. NF‐κB activation was measured by spectrophotometry at 650 nm. Data are represented as median with interquartile range (n = 5). Statistical differences were measured using Mann–Whitney test (***p < 0.001).
As shown in Figure 5A, Polymyxin B does not induce the release of NF‐κB in TLR‐4. When LPS was coincubated with Polymyxin B, a complete blockade of LPS signaling was observed. In Figure 5B, the neutralized bLF showed 1.67‐fold (p < 0.001) increased NF‐κB activation compared with the control (nontreated cells). Similar to bLF, neutralized desialylated and demannosylated bLF increased 1.38‐fold (p < 0.01) and 2.85‐fold (p < 0.001), respectively, the release of NF‐κB (Figure 5C and D).
Figure 5.
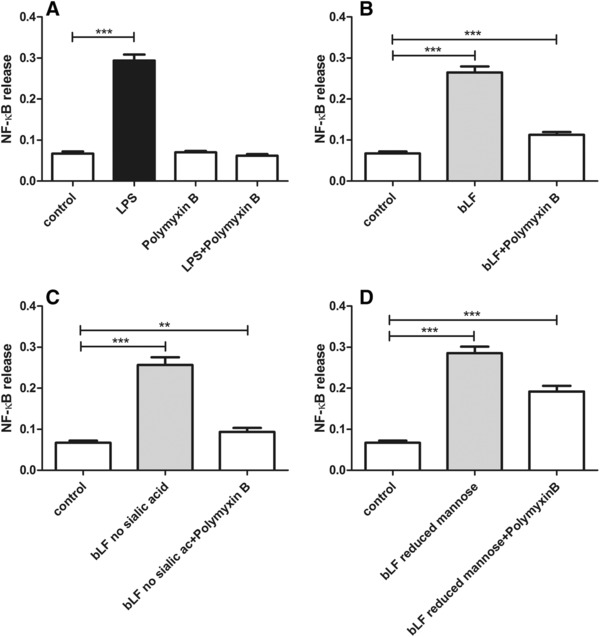
bLF proteins structural influence in the release of NF‐κB in HEK‐Blue TLR‐4. The effect of Polymyxin B blockade at 100 μg mL−1 was confirmed by coincubation of this cationic compound with LPS at 10 μg mL−1 (A). LPS neutralization was also achieved when Polymixyn B was coincubated with bLF (B), the desialylated bLF (C), and demannosylated bLF (D) at a concentration of 2 mg mL−1. Culture medium served as negative control. NF‐κB activation was measured by spectrophotometry at 650 nm. The statistical differences were measured with unpaired t‐test with (n = 4) (**p < 0.01, ***p < 0.001).
Furthermore, with the exclusion of LPS contamination, the effects of the glycosylation profiles of bLF were more evident. As shown in Figure 6, the desialylated bLF activation (0.0930, p < 0.001) was lower when compared with the activation by bLF (0.058, p < 0.001). In contrast, the demannosylated bLF activation (0.1918, p < 0.001) was higher compared with the activation exerted by bLF. The desialylated bLF showed 2.06‐fold (p < 0.001) lower NF‐κB activation compared with the demannosylated bLF.
Figure 6.

bLF proteins glycosylation profile influences the release of NF‐κB in HEK‐Blue TLR‐4. Cells were incubated with bLF proteins at 2 mg mL−1 and Polymyxin B at 100 μg mL−1. The statistical differences were measured with unpaired t‐test (n = 4) (*p < 0.05, **p < 0.01, ***p < 0.001).
3.6. TLR‐8 Inhibition of NF‐κB Release by Modified bLF Isolated N‐Glycans
TLR8 was neither activated nor inhibited by bLF (Figure S8). However, it was inhibited by the N‐glycans isolated from bLF. This was studied with the TLR agonist ssRNA40. As shown in Figure 7, the N‐glycans isolated from bLF reduced the NF‐κB release 2.12‐fold (p < 0.05). Comparable inhibitory effects were observed with the desialylated (2.47‐fold, p < 0.05) and demannosylated bLF (2.45‐fold, p < 0.05) counterparts.
Figure 7.

Inhibitory effects of isolated bLF N‐glycans on HEK‐TLR‐8 cells. The cells were coincubated with the bLF structures and with the specific agonist for TLR‐8 ssRNA40 (50 μg mL−1). NF‐κB was measured by spectrophotometry at 650 nm. Median and interquartile range of activation is plotted as NF‐κB release (n = 5). Statistical differences were calculated with Mann–Whitney test (*p < 0.05).
4. Discussion
The role of bLF in regulation of the innate immune system is of considerable interest for its nutraceutical and pharmacological potential as dietary supplement or treatment adjuvant. The immunomodulatory properties of bLF have been previously described in vitro34, 35, 36, 37 and in vivo.38, 39 However, the role of glycosylation patterns on its biological activity is not well understood. To the best of our knowledge, in the present study, we show for the first time that sialylation and mannosylation of bLF alter the immunomodulatory properties by profound differences in signaling of NF‐κB mediated via TLRs.
The innate function is guarded by pattern recognition receptors (PRRs) and involved in the regulation of host–commensal interactions.40 The most studied PRRs are TLRs that are localized in the cell surface or within endosomes and can be expressed by intestinal epithelial cells, macrophages, dendritic cells, B cells, T cells, and stromal cells.41 To determine whether bLF and its desialylated and demannosylated forms could be sensed by TLRs or other PRRs, we used the THP1 cell line, which is equipped with most of the known PRRs.42 Combined with a THP1 cell line with a truncated MyD88 adaptor, which is necessary for the signaling via TLR, the data showed that the activation of bLF and its modified counterparts was mainly TLR dependent.
Sialylation of LF is essential for TLR signaling as the desialylated form of bLF was significantly lower compared to its sialylated counterpart. The absence or presence of sialic acid has been found to be both unfavorable and favorable for bLF structure stability and its biological activity.43 On one hand, the lack of sialic acid on bLF has been shown to reduce its ability to bind to iron up to as much as 90%.44 The iron‐binding capacity of LF is involved in the maintenance of iron homeostasis.23 On the other hand, the anti‐rotavirus activity of bLF has been observed to increase upon the removal of the sialic acid.45 De‐sialylation of LF has been assumed to favor the opening of other functional epitopes on LF that increase the interaction between the rotavirus and bLF.45 These findings are in line with studies on epitopes recognized by rotavirus V8.46 However, our data suggest that sialylation can also contribute to immune responses and thereby also contributes to the clearance of pathogens.
In contrast, the reduced mannose bLF structure has an opposing effect on immunomodulation since it had a significant higher signaling compared with the intact bLF. The type of oligomannose structures on bLF have been described to be diverse, but its effects on the modulation of the innate response are unknown.21 The antibacterial activity of bLF is partially attributed to its oligomannose type glycans that act as decoy receptors that prevent bacterial adhesion.12 Oligomannose glycans have a high affinity for Escherichia coli type 1 fimbrial lectin, thus facilitating adhesion of bacteria to bLF instead of the intestinal mucosa.47 bLF has shown to be more effective inhibitor than hLF of DC‐SIGN, a C‐type lectin that mediates the internalization of HIV‐1 virus. This occur as a consequence of the binding of the oligomannose glycans of bLF to the DC‐SIGN.48 This combined with enhanced TLR signaling as shown here might be mechanisms by which mannose glycans contribute to prevention of disease.
To the best of our knowledge, inhibitory effects of TLR‐3 by bLF have not been reported before. Nevertheless, it has been reported that the expression of TLR‐3 in mice small intestines can be downregulated by the oral administration of LF.49 TLR‐3 mediates immune responses against viral infections upon activation by its ligand double‐stranded RNA.50 However, little is known about how this receptor is regulated or how its signaling is initiated in response to its agonists.51 The in vivo injection of Poly (I:C), a ligand of TLR‐3, results in the activation of dendritic cells and natural killer cells. Uncontrolled or sustained responses via TLR‐3 have been associated with increased mortality and morbidity in infections such as Nile disease, Phlebovirus, vaccinia, and influenza A.52 A downregulation of TLR‐3 pathways by LF is a potential target for the therapeutic treatment of such diseases and can possibly be manipulated by changing the glycosylation of LF.
bLF activated TLR‐4. bLF contains a basic region close to the N‐terminus capable to bind to anionic molecules such as lipid A from LPS.36, 53 This characteristic confers LF its anti‐endotoxic effect. The work of Yong et al. suggests that LPS is even necessary for TLR‐4 signaling and even part of the immunomodulatory function35 and anti‐microbial activity of LF.34, 54, 55 To confirm that the bLF and its desialylated and demannosylated counterparts and not contaminating LPS induced release of NF‐κB, the LPS content was neutralized with Polymyxin B. Once the signaling of LPS was blocked, it was observed that the signaling of the desialylated bLF was lower compared to the demannosylated form. The change in glycosylation profile seems to affect the intensity of the signaling but not the signaling per se. These results can occur as a consequence of TLR‐4 cooperation with glycan receptors. TLR‐4 has been described to be regulated by glycan receptors such as siglecs or C‐type lectins. Siglecs are receptors recognizing specifically structures decorated with sialic acid. In particular, siglecs have been shown to act by slowing down the activation of TLRs such as TLR‐4.56 In contrast, TLR‐4 interacts with Dectin‐1 and mannose receptors upon fungal infection to induce lymphocyte proliferation.57 Although TLR‐4 is key for the antibacterial activity of bLF, our study confirms that the signaling via TLR‐4 is modulated by the composition of the glycan chains.
Finally, unlike TLR‐3 and TLR‐4, the isolated glycans from bLF and its desialylated and demannosylated forms inhibited TLR‐8. This type of interaction between carbohydrates and TLR‐8 is quite atypical because of the type of structures that endosomal receptors recognize and its localization in endosomal compartments.58 TLR‐8 senses ssRNA rich in adenylate‐uridylate present in viruses, small interfering RNAs, and imidazoquinoline compounds.59 TLR‐8 mediates the recognition of self‐RNA released from apoptotic cells.60 The inappropriate recognition of endogenous agonist like ssRNA through endosomal TLRs contribute significantly to autoimmune diseases.61 It has been described that circulating DNA or RNA complexes in patients with systemic lupus erythematosus can induce cytokine production and disease development. Therefore, the inhibition of endosomic TLRs by isolated glycans has a therapeutic potential for the treatment of autoimmune diseases.62
Our data demonstrate a pertinent role for sialylation and mannosylation of bLF in its immunomodulatory properties. We identified the PRRs TLR‐3, TLR‐4, and TLR‐8 as principle target for the N‐glycans of bLF. As these receptors are involved in many pathologies, our data do not only contribute to a better understanding of how bLF can have immunomodulatory properties.6 It also open new venues to manage disease with adapted bLF formulations.
Abbreviations
- bLF
bovine lactoferrin
- hLF
human lactoferrin
- LF
lactoferrin
- MyD88
myeloid differentiation primary response protein 88
- NF‐κB
nuclear factor kappa–light‐chain enhancer of activated B cells
- PRRs
pattern recognition receptors
- SEAP
secreted embryonic alkaline phosphatase
- THP1 cells
human acute monocytic leukemia cells
- TLR
Toll‐like receptors
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supplementary Material
Acknowledgements
RVW and SVL prepared the bLF modified structures and isolated the bLF N‐glycans. SFL performed all the cell‐based experiments and the data analysis. SFL, RVW, SVL, LB, and PDV interpreted the data. SFL and PDV wrote the paper. RVW, SVL, and LB critically reviewed the manuscript. SFL and this research project was supported by RUG‐Campus Friesland. VWR acknowledges financial support from Friesland Campina. The copyright line for this article was changed on 17 August 2018 after original online publication.
Figueroa‐Lozano S., Valk‐Weeber R. L., van Leeuwen S. S., Dijkhuizen L., de Vos P., Mol. Nutr. Food Res. 2018, 62, 1700389 10.1002/mnfr.201700389
References
- 1. Satue‐Gracia M. T., Frankel E. N., Rangavajhyala N., German J. B., J. Agric. Food Chem. 2000, 48, 4984. [DOI] [PubMed] [Google Scholar]
- 2. Aly E., Ros G., Frontela C., J. Food Res. 2013, 2, 25. [Google Scholar]
- 3. Buccigrossi V., De Marco G., Bruzzese E., Ombrato L., Bracale I., Polito G., Guarino A., Pediatr. Res. 2007, 61, 410. [DOI] [PubMed] [Google Scholar]
- 4. Liao Y. L., Jiang R. L., Lonnerdal B., Biochem. Cell Biol. Biol. Cell. 2012, 90, 476. [DOI] [PubMed] [Google Scholar]
- 5. Steijns J. M., Int. J. Dairy Technol. 2001, 54, 81. [Google Scholar]
- 6. Mayeur S., Spahis S., Pouliot Y., Levy E., Antioxid. Redox Signal. 2016, 24, 813. [DOI] [PubMed] [Google Scholar]
- 7. Nuijens J. H., van Berkel P. H., Schanbacher F. L., J. Mammary Gland Biol. Neoplasia 1996, 1, 285. [DOI] [PubMed] [Google Scholar]
- 8. Steijns J. M., van Hooijdonk A. C., Br. J. Nutr. 2000, 84(Suppl 1), S11. [DOI] [PubMed] [Google Scholar]
- 9. Roseanu A., Brock J. H., IUBMB Life 2006, 58, 235. [DOI] [PubMed] [Google Scholar]
- 10. Magnuson J. S., Henry J. F., Yip T., Hutchens W., Pediatr. Res. 1990, 28, 176. [DOI] [PubMed] [Google Scholar]
- 11. Le Parc A., Dallas D. C., Duaut S., Leonil J., Martin P., Barile D., Electrophoresis 2014, 35, 1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barboza M., Pinzon J., Wickramasinghe S., Froehlich J. W., Moeller I., Smilowitz L. T., Ruhaak L. E., Huang J., Lönnerdal B., German J. B., Medrano J. F., Weimer B. C., Lebrilla C. B., Mol. Cell. Proteomics 2012, 11, M111.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Berkel P. H., Geerts M. E., van Veen H. A., Kooiman P. M., Pieper F. R., de Boer H. A., Nuijens J. H., Biochem. J. 1995, 312, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao Y. Y., Takahashi M., Gu J. G., Miyoshi E., Matsumoto A., Kitazume S., Taniguchi N., Cancer Sci. 2008, 99, 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kleizen B., Braakman I., Curr. Opin. Cell Biol. 2004, 16, 343. [DOI] [PubMed] [Google Scholar]
- 16. Yen M. H., Wu A. M., Yang Z., Gong Y. P., Chang E. T., Biochim. Biophys. Acta ‐ Gen. Subj. 2011, 1810, 139. [DOI] [PubMed] [Google Scholar]
- 17. Aebi M., Biochim. Biophys. Acta ‐ Mol. Cell Res. 2013, 1833, 2430. [DOI] [PubMed] [Google Scholar]
- 18. O'Riordan N., Gerlach J. Q., Kilcoyne M., O'Callaghan J., Kane M., Hickey R. M., Joshi L., Food Chem. 2014, 165, 388. [DOI] [PubMed] [Google Scholar]
- 19. Van Veen H. A., Geerts M. E. J., Van Berkel P. H. C., Nuijens J. H., Eur. J. Biochem. 2004, 271, 678. [DOI] [PubMed] [Google Scholar]
- 20. Yu T., Guo C., Wang J., Hao P., Sui S., Chen X., Zhang R., Wang P., Yu G., Zhang L., Dai Y., Li N., Glycobiology 2011, 21, 206. [DOI] [PubMed] [Google Scholar]
- 21. Van Leeuwen S. S., Schoemaker R. J. W., Timmer C. J. A. M., Kamerling J. P., Dijkhuizen L., Biochim. Biophys. Acta ‐ Gen. Subj. 2012, 1820, 1444. [DOI] [PubMed] [Google Scholar]
- 22. Coddevilie B., Strecker G., Wieruszeski J., Carbohydr. Res. 1992, 236, 145. [DOI] [PubMed] [Google Scholar]
- 23. O'Riordan N., Kane M., Joshi L., Hickey R. M., Glycobiology 2014, 24, 220. [DOI] [PubMed] [Google Scholar]
- 24. Kelly D., Coutts A. G. P., Livest. Prod. Sci. 2000, 66, 161. [Google Scholar]
- 25. Zoldoš V., Novokmet M., Bečeheli I., Lauc G., Glycoconj. J. 2013, 30, 41. [DOI] [PubMed] [Google Scholar]
- 26. Actor J. K., Hwang S.‐A., Kruzel M. L., Curr. Pharm. Des. 2009, 15, 1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanchez L., Castillo H., Plirez M. D., Ena J. M., Calvo M., J. Food Sci. 1992, 57, 873. [Google Scholar]
- 28. Indyk H. E., Mcgrail I. J., Watene G. A., Filonzi E. L., Food Chem. 2007, 101, 838. [Google Scholar]
- 29. Sreedhara A., Flengsrud R., Langsrud T., Vegarud G. E., Biometals 2010, 23, 1159. [DOI] [PubMed] [Google Scholar]
- 30. Paredes‐Juarez G. A., de Haan B. J., Faas M. M., de Vos P., Materials (Basel). 2014, 7, 2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vogt L. M., Sahasrabudhe N. M., Ramasamy U., Meyer D., Pullens G., Faas M. M., Venema K., Schols H. A., de Vos P., J. Funct. Foods 2016, 22, 398. [Google Scholar]
- 32. Majka G., Śpiewak K., Kurpiewska K., Heczko P., Stochel G., Strus M., Brindell M., Anal. Bioanal. Chem. 2013, 405, 5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brisson G., Britten M., Pouliot Y., Int. Dairy J. 2007, 17, 617. [DOI] [PubMed] [Google Scholar]
- 34. Håversen L., Ohlsson B. G., Hahn‐Zoric M., Å. Hanson L., Mattsby‐Baltzer I., Cell. Immunol. 2002, 220, 83. [DOI] [PubMed] [Google Scholar]
- 35. Na Y. J., Han S. B., Kang J. S., Yoon Y. D., Park S. K., Yang K. H., Joe C. O., Int. Immunopharmacol. 2004, 4, 1187. [DOI] [PubMed] [Google Scholar]
- 36. Puddu P., Latorre D., Carollo M., Catizone A., Ricci G., Valenti P., Gessani S., PLoS One 2011, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anand N., Kanwar R. K., Dubey M. L., Vahishta R. K., Sehgal R., Verma A. K., Kanwar J. R., Drug Des. Devel. Ther. 2015, 9, 3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mulder A. M., Connellan P. A., Oliver C. J., Morris C. A., Stevenson L. M., Nutr. Res. 2008, 28, 583. [DOI] [PubMed] [Google Scholar]
- 39. Ramalingam S., Crawford J., Chang A., Manegold C., Perez‐Soler R., Douillard J. Y., Thatcher N., Barlesi F., Owonikoko T., Wang Y., Pultar P., Zhu J., Malik R., Giaccone G., Ann. Oncol. 2013, 24, 2875. [DOI] [PubMed] [Google Scholar]
- 40. Sotolongo J., Ruiz J. F. M., Biophys. Chem. 2005, 257, 2432. [Google Scholar]
- 41. Santaolalla R., Fukata M., Abreu M., Curr. Opin. Gastroenterol. 2011, 27, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chanput W., Mes J. J., Wichers H. J., Int. Immunopharmacol. 2014, 23, 37. [DOI] [PubMed] [Google Scholar]
- 43. Wang B., Brand‐Miller J., Eur. J. Clin. Nutr. 2003, 57, 1351. [DOI] [PubMed] [Google Scholar]
- 44. Zhenpu L., Furmanski P., Chin. J. Cancer Res. 1995, 7, 79. [Google Scholar]
- 45. Superti F., Siciliano R., Rega B., Giansanti F., Valenti P., Antonini G., Biochim. Biophys. Acta ‐ Gen. Subj. 2001, 1528, 107. [DOI] [PubMed] [Google Scholar]
- 46. Yu Y., Lasanajak Y., Song X., Hu L., Ramani S., Mickum M. L., Ashline D. J., Prasad B. V., Estes M. K., Reinhold V. N., Cummings R. D., Smith D. F., Mol. Cell. Proteomics 2014, 13, 2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teraguchi S., Shin K., Fukuwatari Y., Shimamura S., Microbiology 1996, 64, 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Groot F., Geijtenbeek T. B. H., Sanders R. W., Baldwin C. E., Sanchez‐Hernandez M., Floris R., van Kooyk Y., de Jong E. C., Berkhout B., J. Virol. 2005, 79, 3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wakabayashi H., Takakura N., Yamauchi K., Tamura Y., Clin. Vaccine Immunol. 2006, 13, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pohar J., Pirher N., Benčina M., Manček‐Keber M., Jerala R., PLoS One 2014, 9, e92391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garcia‐Cattaneo A., Gobert F.‐X., Müller M., Toscano F., Flores M., Lescure A., Del Nery E., Benaroch P., Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang T., Town T., Alexopoulou L., Anderson J. F., Fikrig E., Flavell R. A., Nat. Med. 2004, 10, 1366. [DOI] [PubMed] [Google Scholar]
- 53. Appelmelk B. J., An Y. Q., Geerts M., Thijs B. G., de Boer H. A., MacLaren D. M., de Graaff J., Nuijens J. H., Infect. Immun. 1994, 62, 2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ando K., Hasegawa K., Shindo K. I., Furusawa T., Fujino T., Kikugawa K., Nakano H., Takeuchi O., Akira S., Akiyama T., Gohda J., Inoue J., Hayakawa M., FEBS J. 2010, 277, 2051. [DOI] [PubMed] [Google Scholar]
- 55. Curran C. S., Demick K. P., Mansfield J. M., Cell. Immunol. 2006, 242, 23. [DOI] [PubMed] [Google Scholar]
- 56. Chen G. Y., Brown N. K., Wu W., Khedri Z., Yu H., Chen X., van de Vlekkert D., D'Azzo A., Zheng P., Liu Y., Elife 2014, 3, e04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loures F. V., Araújo E. F., Feriotti C., Bazan S. B., Calich V. L., Front. Microbiol. 2015, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee B. L., Barton G. M., Trends Cell Biol. 2014, 24, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T., Nat. Struct. Mol. Biol. 2015, 22, 109. [DOI] [PubMed] [Google Scholar]
- 60. Guiducci C., Gong M., Cepika A.‐M., Xu Z., Tripodo C., Bennett L., Crain C., Quartier P., Cush J. J., Pascual V., Coffman R. L., Barrat F. J., J. Exp. Med. 2013, 210, 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krieg A. M., Vollmer J., Immunol. Rev. 2007, 220, 251. [DOI] [PubMed] [Google Scholar]
- 62. Kuznik A., Bencina M., Svajger U., Jeras M., Rozman B., Jerala R., J. Immunol. 2011, 186, 4794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


