Abstract
The classical microbial strategy for depolymerization of β-mannan polysaccharides involves the synergistic action of at least two enzymes, endo-1,4-β-mannanases and β-mannosidases. In this work, we describe the first exo-β-mannanase from the GH2 family, isolated from Xanthomonas axonopodis pv. citri (XacMan2A), which can efficiently hydrolyze both manno-oligosaccharides and β-mannan into mannose. It represents a valuable process simplification in the microbial carbon uptake that could be of potential industrial interest. Biochemical assays revealed a progressive increase in the hydrolysis rates from mannobiose to mannohexaose, which distinguishes XacMan2A from the known GH2 β-mannosidases. Crystallographic analysis indicates that the active-site topology of XacMan2A underwent profound structural changes at the positive-subsite region, by the removal of the physical barrier canonically observed in GH2 β-mannosidases, generating a more open and accessible active site with additional productive positive subsites. Besides that, XacMan2A contains two residue substitutions in relation to typical GH2 β-mannosidases, Gly439 and Gly556, which alter the active site volume and are essential to its mode of action. Interestingly, the only other mechanistically characterized mannose-releasing exo-β-mannanase so far is from the GH5 family, and its mode of action was attributed to the emergence of a blocking loop at the negative-subsite region of a cleft-like active site, whereas in XacMan2A, the same activity can be explained by the removal of steric barriers at the positive-subsite region in an originally pocket-like active site. Therefore, the GH2 exo-β-mannanase represents a distinct molecular route to this rare activity, expanding our knowledge about functional convergence mechanisms in carbohydrate-active enzymes.
Keywords: glycoside hydrolase, enzyme, enzyme catalysis, protein structure, protein conformation, site-directed mutagenesis, exo-β-mannanase, GH2 family, molecular mechanism, Xanthomonas axonopodis pv. citri
Introduction
β-Mannans are polysaccharides widely distributed in nature, from constituents of glycoproteins in yeast cells (1, 2) to plant hemicellulose components (2, 3, 4). In plants, β-mannans are more abundant in softwoods (reviewed by Moreira and Filho (5)) and can correspond to up to 10% of coniferous forest residues (6). This polysaccharide comprises a backbone of β-1,4-linked d-mannopyranosyl residues (known as “β-mannan”) or a heterogeneous combination of β-1,4-d-mannosyl and β-1,4-d-glucosyl units (termed “glucomannan”), which can be further decorated with α-1,6-galactosyl side chains to yield “galactomannan” and “galactoglucomannan,” respectively (5, 7). Enzymes involved in β-mannan depolymerization have a versatile applicability in the production of fermentable sugars for advanced biofuels and other potential industrial applications (8–17).
The depolymerization of β-mannans to mannose requires a synergistic action of at least two enzymes: endo-1,4-β-mannanases, also known as 1,4-β-d-mannan mannanohydrolases (EC 3.2.1.78), and β-d-mannosidases or β-d-mannoside mannohydrolases (EC 3.2.1.25). According to the carbohydrate-active enzymes database (www.cazy.org)5 (18), β-mannanases are found in the glycoside hydrolase (GH)6 families 5, 26, 113, and 134, whereas β-mannosidases belong to GH families 1, 2, and 5. However, to hydrolyze heteromannans, in addition to β-mannanases and β-mannosidases, the presence of accessory enzymes, such as α-galactosidases, is also required to remove galactosyl side chains that restrict the capacity of β-mannanases to cleave the β-mannan backbone (19).
Beyond the classical microbial strategy for β-mannan degradation that involves the endo action of β-mannanases followed by the depolymerization of MOS by β-mannosidases, very few enzymes showed the ability to efficiently degrade both β-mannans and MOS, being named as exo-β-mannanases. These enzymes represent an instrumental simplification of the enzymatic route to obtain mannose from β-mannans. The structural basis for this interesting mode of action was unveiled for the GH5 family (20); however, there is no information about this activity within the GH2 family, which is known to classically harbor β-mannosidases. The GH5 enzymes are typically known to act on polymeric substrates, and most of characterized members are endo-acting enzymes, usually exhibiting a cleft-like catalytic interface. On the other hand, GH2 enzymes are known to act on shorter oligosaccharides or on substitutions such as arabinosyl and galactosyl side chains and, in contrast to GH5 members, feature a pocket-like active site. Thus, although similar activities are observed in these families, the molecular adaptations involved in the functional specialization are probably diverse, considering the distinct structural architecture and active site topology. Knowledge of those structural changes in the catalytic interface might be instrumental to understand their specificities and to generate relevant data for rational enzyme engineering aimed at biotechnological applications.
In this context, we discovered and characterized the first exo-β-mannanase from the GH2 family, revealing the molecular modifications in the active site that enabled this enzyme to efficiently cleave both β-mannans and MOS, producing mannose. According to crystallographic analyses, the conserved hydrophobic barrier in the pocket-like active site of GH2 mannosidases is absent in XacMan2A, generating extra productive positive subsites.
Results
XacMan2A is an exo-β-mannanase from the GH2 family
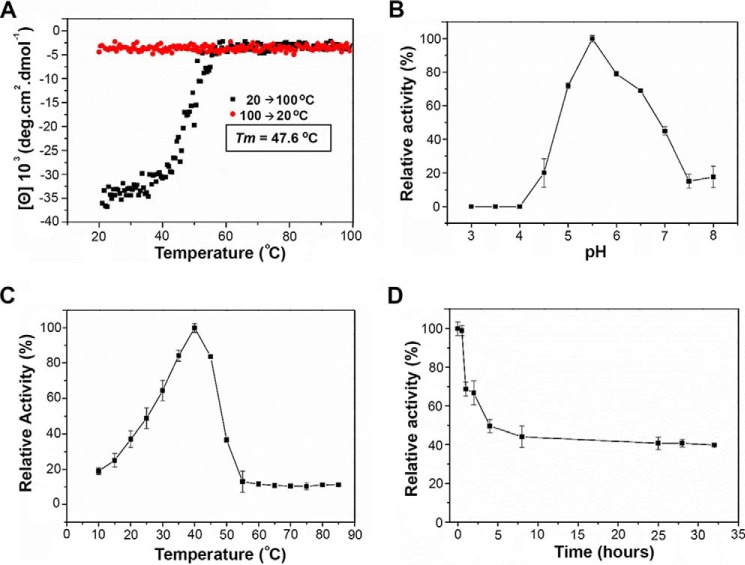
The X. axonopodis pv. citri GH2 enzyme (GenBankTM code AAM37920.1) ORF comprises 2,691 bp encoding an 886-amino acid protein with a molecular mass of 99.48 kDa. The recombinant XacMan2A was successfully overexpressed in BL21(DE3) Escherichia coli cells, and the purification procedure yielded 2.5 mg of pure and homogenous enzyme per liter of culture, despite its large size and structural complexity. CD analysis indicated a proper folded conformation with a melting temperature of 47.6 °C (Fig. 1A), which is similar to that observed in other CAZyme isolated from the same bacterium (21). XacMan2A thermal unfolding followed the canonical two-state model with a single transition (Fig. 1A), despite the five-domain organization inferred by sequence analysis and further confirmed by X-ray crystallography.
Figure 1.
Biophysical and biochemical characterization of XacMan2A. A, thermal denaturation profile of XacMan2A from 20 to 100 °C, monitored at 212 nm by CD spectroscopy. The melting temperature (Tm) was calculated from the sigmoidal fit of the denaturation curve. Shown are effects of pH (B) and temperature (C) on relative activity of XacMan2A. D, thermal stability curve. Residual activities were estimated at 40 °C in 40 mmol·liter−1 citrate-phosphate buffer, pH 5.5, using 40 mmol·liter−1 pNP-β-Man as substrate. 100% relative activity corresponded to 35.3 ± 1.2 units·mg−1 protein, estimated in the same conditions. Values shown represent means ± S.D. (error bars) from triplicate assays carried out with three separate preparations of purified recombinant XacMan2A.
Activity assays with synthetic substrates revealed that XacMan2A is only active on p-nitrophenyl-β-d-mannopyranoside (pNP-β-Man) exhibiting optimum pH and temperature of 5.5 and 40 °C, respectively, with a moderate thermotolerance (Fig. 1, B–D). Interestingly, increasing hydrolysis rates were observed from mannobiose (M2) to mannohexaose (M6), indicating the enzyme preference for longer manno-oligosaccharides (Table 1). In addition, XacMan2A was able to cleave mannose-based polysaccharides, showing high activity against β-mannan, low activity against galactomannan, and trace activity against glucomannan (Table 1).
Table 1.
Substrate specificity
| vo/[ET] (s−1) | |
|---|---|
| Synthetic (10 mmol·liter−1) | |
| pNP-β-Man | 24.5 ± 0.45 |
| pNP-α-Man | NDa |
| pNP-Glu | ND |
| pNP-Gal | ND |
| pNP-Xyl | ND |
| pNP-AraF | ND |
| Oligosaccharides (4 mmol·liter−1) | |
| Mannobiose | 0.4 ± 0.07 |
| Mannotriose | 0.8 ± 0.05 |
| Mannotetrose | 11.5 ± 0.42 |
| Mannopentaose | 27.5 ± 3.25 |
| Mannohexaose | 38.5 ± 2.86 |
| Polysaccharides (10 mg·ml−1) | |
| Mannan | 8.8 ± 0.62 |
| Galactomannan | 2.6 ± 0.27 |
| Glucomannan | 0.03 ± 0.01 |
| Arabinan | ND |
| Xylan | ND |
| CMC | ND |
| β-Glucan | ND |
a ND, activity not detected.
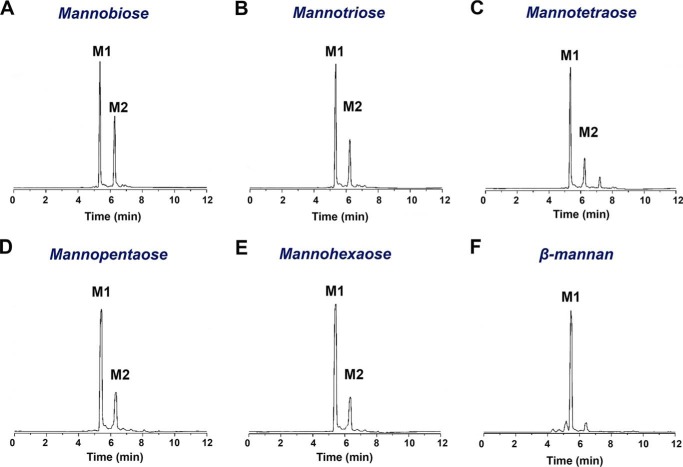
The cleavage patterns of MOS and polymeric substrates, analyzed by capillary zone electrophoresis, indicate an exo-acting mechanism with mannose as the main product (Fig. 2). The reduction of MOS (M5 and M6) with borohydride did not alter the activity, suggesting that the enzyme recognizes the nonreducing ends of the substrate.
Figure 2.
Cleavage pattern of XacMan2A. Shown is capillary zone electrophoretic analysis of M2 (A), M3 (B), M4 (C), M5 (D), M6 (E), and ivory nut mannan hydrolysis (F) by XacMan2A. For M2, M3, and M4 substrates, the hydrolysis reaction time was 240 min, whereas for M5, M6, and mannan, it was 120 min, indicating a preference for longer substrates.
Kinetic characterization with M2, M6, and β-mannan were performed (Fig. 3A); however, the saturation was not achieved with M2, supporting the adaptation of XacMan2A to cope with longer oligosaccharides and polysaccharides. On the other hand, XacMan2A efficiently cleaved both M6 and β-mannan substrates (Fig. 3, A and B). Remarkably, XacMan2A exhibited a kcat of ∼12.5 s−1 on β-mannan (Fig. 3C), which is at least 3-fold higher than that observed for the GH5 exo-β-mannanase (20), highlighting its improved capacity to act on polymeric substrates. Together, these analyses pointed out that XacMan2A is not a classic β-mannosidase but a genuine exo-β-1,4-mannanase, revealing a new activity in the GH2 family.
Figure 3.
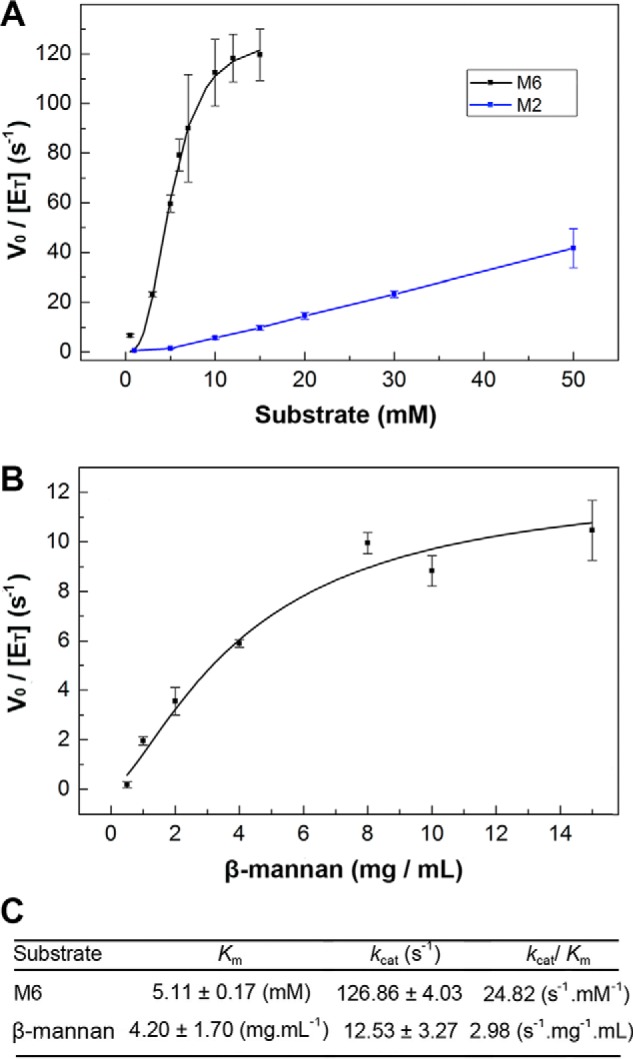
Kinetic characterization of XacMan2A. A, M2 and M6 hydrolysis, with mannose release monitored by MS. For M2, saturation was not achieved. B, ivory nut β-mannan hydrolysis, where the reducing sugar release was measured by the 3,5-dinitrosalicylic acid method. C, kinetics parameters calculated for M6 and β-mannan substrates. Error bars, S.D.
The multi-modular structural architecture, biological assembly, and negative-subsite mapping
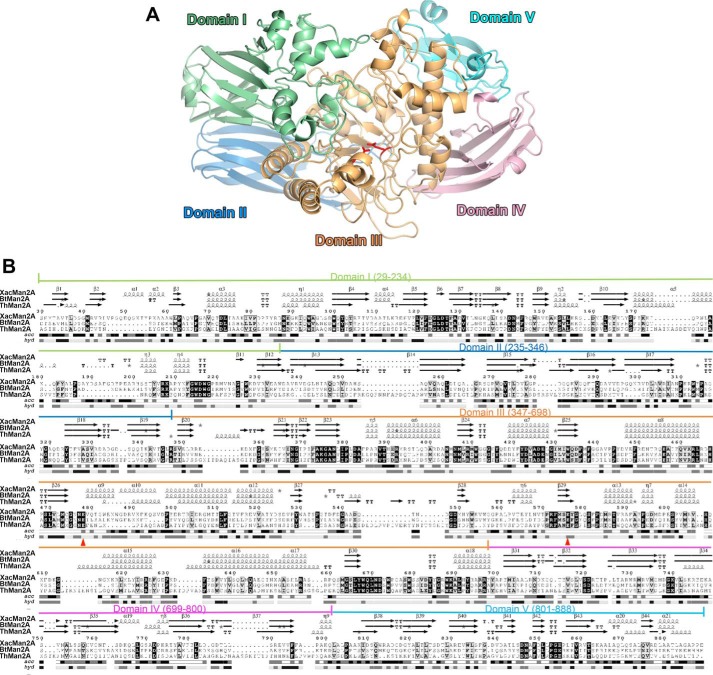
To understand the structural basis of XacMan2A mode of action, the crystallographic structure was determined in the native form and in complex with mannose (Table 2). The XacMan2A structure consists of five well-defined domains (Fig. 4, A and B) with a (β/α)8 catalytic core at the central position. This modular organization is similar to that seen in other GH2 enzymes, such as β-mannosidases, from Bacteroides thetaiotaomicron (BtMan2A) (22), Dictyoglomus thermoplilum (DtMan) (23), and Trichoderma harzianum (ThMan2A) (24), and β-galactosidases, from E. coli (25) and Arthrobacter sp. (26). XacMan2A domain I (Fig. 4, residues Ser29–Trp234) comprises two antiparallel β-sheets and an α-helical motif. Domain II (residues Asp235–Arg346) and domain IV (residues Ala699–Gln800) are structurally similar and display a β-sandwich fold, which is commonly found in carbohydrate-binding modules (reviewed by Hashimoto (27)). The catalytic domain, domain III (residues Ser347–Phe698), has a classical (β/α)8 TIM-barrel fold, commonly observed in the clan of GH-A enzymes. Domain V, located at the C terminus, is the shortest one (residues Leu801–Glu888) and is the most divergent compared with other structurally characterized GH2 enzymes.
Table 2.
Data collection and refinement statistics of XacMan2A structures
Values in parentheses are for the highest-resolution shell.
| Native | Native + Mana | E575A + Mana | E477A + Mana | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C121 | P1 | P1211 | P1211 |
| Cell dimensions | ||||
| a, b, c (Å) | 179.50, 68.57, 79.02 | 67.94, 78.97, 95.58 | 79.44, 116.05, 104.94 | 68.71, 202.13, 79.09 |
| α, β, γ (degrees) | 90, 95.16, 90 | 94.62, 103.42, 111.84 | 90, 98.17, 90 | 90, 110.3, 90 |
| Molecules/AUb | 1 | 2 | 2 | 2 |
| Resolution (Å) | 19.98–1.90 (1.96–1.90) | 38.51–2.13 (2.20–2.13) | 47.40–2.00 (2.07–2.00) | 46.57–2.20 (2.28–2.20) |
| Observed reflections | 333,657 (31,309) | 219,539 (21,464) | 556,729 (53,036) | 471,390 (33,676) |
| Unique reflections | 71,912 (6,621) | 93,216 (8,971) | 121,038 (11,289) | 97,007 (8,305) |
| CC½c | 0.989 (0.958) | 0.998 (0.967) | 0.997 (0.649) | 0.989 (0.639) |
| I/σI | 7.89 (3.52) | 14.20 (4.50) | 10.57 (1.53) | 9.08 (2.06) |
| Completeness (%) | 94.91 (88.22) | 94.11 (92.10) | 95.13 (88.90) | 94.98 (81.69) |
| Rmeas | 0.1391 | 0.05284 | 0.1199 | 0.1815 |
| Rmerge | 0.1237 (0.2736) | 0.0406 (0.1691) | 0.106 (0.9444) | 0.1616 (0.6898) |
| Multiplicity | 4.6 (4.7) | 2.4 (2.4) | 4.6 (4.7) | 4.9 (4.1) |
| Refinement | ||||
| Resolution (Å) | 19.98–1.90 | 38.51–2.13 | 43.15–2.00 | 41.97–2.20 |
| No. of reflections | 71,908 | 93,205 | 121,017 | 96,979 |
| Rwork/Rfree | 0.17/0.22 | 0.19/0.23 | 0.17/0.21 | 0.17/0.21 |
| No. of protein residues | 861 | 1722 | 1714 | 1718 |
| B-factor (Å2) | ||||
| Average | 27.10 | 37.50 | 33.10 | 28.70 |
| Macromolecules | 26.7 | 37.5 | 32.9 | 28.4 |
| Ligands | 37.1 | 60.9 | 36.1 | 66.1 |
| Solvent | 29.9 | 35.8 | 34.7 | 30.2 |
| RMSDs | ||||
| Bond lengths (Å) | 0.011 | 0.004 | 0.011 | 0.004 |
| Bond angles (°) | 1.26 | 0.84 | 1.24 | 0.86 |
| Ramachandran plot | ||||
| Favored (%) | 98 | 97 | 97 | 97 |
| Allowed (%) | 2 | 3 | 3 | 3 |
| Disallowed (%) | 0 | 0 | 0.06 | 0 |
| Molprobity clashscore | 3.37 | 3.11 | 3.45 | 2.81 |
| PDB code | 6BYC | 6BYE | 6BYG | 6BYI |
a Complex with mannose (Man).
b Asymmetric unit.
c CC½, correlation between intensities from random half-data sets (60).
Figure 4.
XacMan2A displays a penta-modular structural architecture. A, cartoon representation of the overall crystal structure of XacMan2A (PDB code 6BYC) colored by domain. Catalytic residues are shown as red sticks. Domain I is colored in pale green, domain II in light blue, domain III in light orange, domain IV in light pink, and domain V in pale cyan. B, amino acid sequence alignment of XacMan2A with GH2 β-mannosidases from B. thetaiotaomicron (BtMan2A, 39%, PDB code 2JE8 (22)) and T. harzianum (ThMan2A, 25%, PDB code 4CVU (24). The five structural domains of XacMan2A are indicated in the amino acid sequence. Acc, solvent accessibility colored from white (buried) to black (exposed). Secondary structure elements are shown above the alignment and labeled according to the XacMan2A crystal structure. Hyp, hydropathy colored in black (hydrophobic; H > 1.5), dark gray (intermediate; −1.5 ≤ H ≤ 1.5), and light gray (hydrophilic; H < 1.5). The figure was prepared using ENDscript version 2.0 (61). The catalytic residues are marked with red triangles.
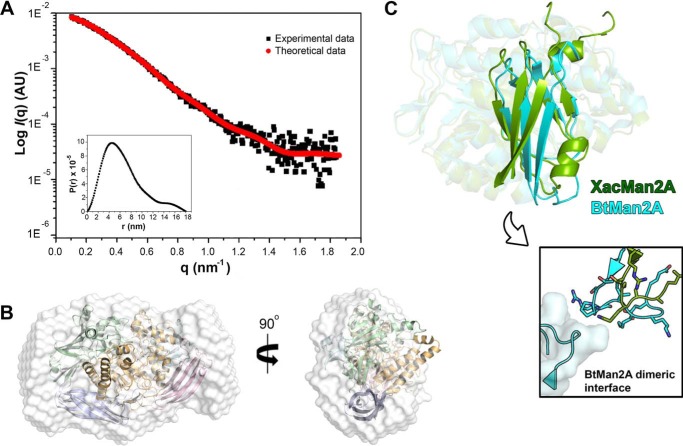
In one of the crystalline forms of XacMan2A, a dimer was found in the asymmetric unit; however, structural and energetic analyses indicate that the dimer is not stable in solution, which was further confirmed by small-angle X-ray scattering (SAXS) analysis. In solution, XacMan2A exhibited a radius of gyration (Rg) of 51.5 ± 0.1 Å, and the calculated low-resolution envelope was fully compatible with the crystallographic monomer (Fig. 5, A and B). Interestingly, the hydrodynamic behavior of XacMan2A differs from the GH2 β-mannosidase BtMan2A, which forms dimers in solution. The dimerization in BtMan2A involves extensive contacts between the domains V of each monomer (22) that are not present in the crystal packing of XacMan2A. In addition, the domain V of XacMan2A largely diverges, in structure and composition, from other GH2 enzymes (38% sequence identity with the same domain of BtMan2A), in particular at the dimerization interface (Fig. 5C), which explains the inability of XacMan2A to dimerize.
Figure 5.
XacMan2A is a monomer in solution. A, SAXS scattering and pair-distance distribution function P(r) plots. B, crystallographic structure of a monomer fitted into the SAXS envelope. C, structure superposition of catalytic domain (with transparency) and domain V (highlighted) from XacMan2A (PDB code 6BYC; green) and BtMan2A (PDB code 2JE8 (22); cyan), highlighting the pronounced differences in the relative position and conformation of domain V. In the boxed image, the dimeric interface of BtMan2A is shown, indicating the lack of conservation in relation to XacMan2A.
The catalytic residues, initially inferred by structural comparisons, were further confirmed with the mutants E477A (acid-base) and E575A (nucleophile) that were shown to be inactive against 4-nitrophenyl-p-mannopyranoside and β-mannan polysaccharide. These residues are fully conserved in the family and adopt similar conformations in BtMan2A and ThMan2A, with ∼3.5 Å of distance, which is compatible with the classical Koshland retaining mechanism (28, 29) that is expected for members of the GH-A clan.
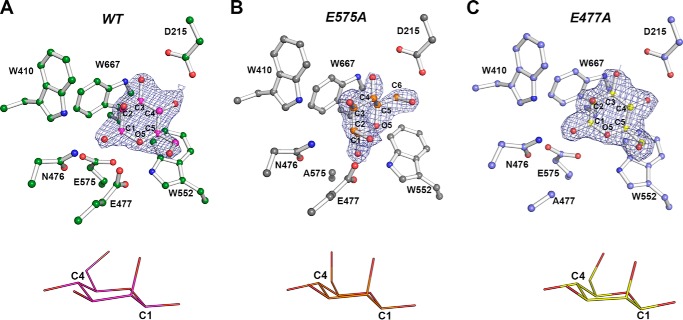
Aiming to obtain a complete description of enzyme–substrate interactions, crystals of inactive mutants were exhaustively prepared with MOS from M2 to M6 by soaking and co-crystallization experiments; however, no high quality crystallographic data were attained. Only mannose complexes were obtained for inactive and active forms of XacMan2A (Fig. 6). In all complexes, the monosaccharide was observed at the −1 subsite, which is surrounded by several tryptophan residues (Trp410, Trp552, and Trp667) and the polar residues Asp215 and Asn476 (Fig. 6) that are fully conserved among the structurally characterized GH2 β-mannosidases.
Figure 6.
Three-dimensional structure of the complexes of XacMan2A WT and mutants E575A and E477A with mannose. A–C, depiction of the 2Fo − Fc electron density maps (contoured at 1.0 σ) for mannose bound to the −1 subsite of WT XacMan2A (PDB code 6BYE) (A), E575A mutant (PBD code 6BYG) (B), and E477A mutant (PDB code 6BYI) (C). Mannose and surrounding residues are shown as ball-and-stick models. Mannose configurations are shown below the respective protein structures.
In the WT–mannose complex, the O1 atom from mannose makes polar contacts with Glu575 and Asn476, whereas the O5 atom interacts with Glu477 (Fig. 6A). In the E575A mutant structure, the O2 atom interacts with Glu477 and the O3 atom with Asn476. In addition, the residue Asp215 makes contacts with the O3 and O4 atoms in the WT protein and with the O4 and O6 atoms in the E575A mutant. The mannose displacement in the E575A structure is probably due to Glu575 side-chain absence, indicating a crucial role of the nucleophile residue in the proper positioning of substrate (Fig. 6B). The mannose molecule in the E477A mutant showed a very similar position compared with the WT complex, except for the lack of interaction between the Glu477 side chain and the O5 atom from the sugar ring (Fig. 6C). Despite the differences and the displacement found in the E575A structure, in all mannose-complexed structures (WT and mutants), the mannopyranosid ring assumes the chair geometry 4C1, which is the most stable configuration in solution (30, 31) (Fig. 6). It correlates with the mannose configuration bound to the −2 subsite of the β-mannanase from Cellvibrio japonicus (PDB code 2VX5) (32) and with the mannosyl residues from the mannotriose bound to Streptomyces sp. β-mannanase (PDB code 5JU9) (31). In GHs, the substrate saccharide unit that binds to the negative subsite generally adopts a boat- or skew-boat-type conformation before the cleavage (30), such as in retaining β-mannosidases/mannanases that often use the 1S5 to B2,5 to 0S2 (33) or the 1C4 to 3H4 to 3S1 (31) catalytic pathways. Because the XacMan2A (WT and mutants) complexes were done with the product, no such configurations in the monosaccharide were observed.
Structural adaptations in the positive-subsite region are determinant for the exo-β-mannanase activity
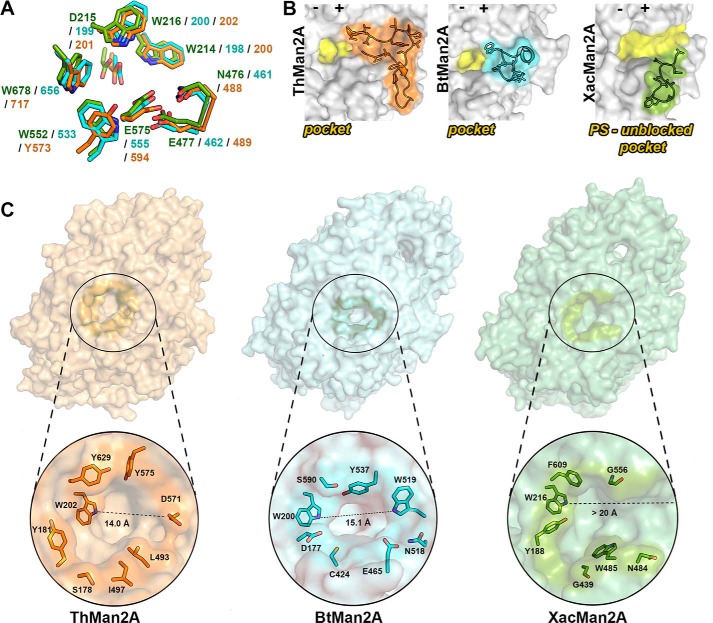
Although the catalytic acidic residues and the −1 subsite are conserved in GH2 β-mannosidases and in XacMan2A (Fig. 7A), the positive-subsite region of the latter enzyme is largely divergent in terms of composition and conformation, which might explain its particular mode of action. An important feature of the typical pocket-like active site topology in GH2 enzymes is the presence of a physical barrier in the positive-subsite region formed mainly by the loops Ala539–Asp571 in ThMan2A (24), and Pro514–Thr527 in BtMan2A (22) (Fig. 7B). However, in XacMan2A, the corresponding region (residues Gly534–Thr544) differs in length, composition, and 3D configuration, generating a continuous and more accessible active-site cleft (Fig. 7, B and C), suggesting the presence of more than two productive subsites, as expected based on MOS hydrolysis rates (Table 1). For instance, in ThMan2A (24), this loop is 32 residues long with a β-hairpin motif. In BtMan2A (22), the loop is 13 residues long and includes a tryptophan residue (Trp519), precluding the continuity of a potential positive-subsite region. In XacMan2A, this loop is the shortest one among these GH2 members with only 10 residues, and none of them represents a barrier for the positive-subsite region.
Figure 7.
XacMan2A active-site topology differs from GH2 β-mannosidases. A, details of the conserved −1 subsite between XacMan2A (PDB code 6BYC), BtMan2A (PDB code 2JE8), and ThMan2A (PDB code 4CVU). Side chains of the active site residues are shown as sticks, and carbon atoms are displayed in green (XacMan2A), in cyan (BtMan2A), and in orange (ThMan2A). Negative and positive subsites are represented by minus and plus signs, respectively. B, representation of ThMan2A, BtMan2A, and XacMan2A active-site regions in yellow, highlighting the loops in ThMan2A (residues from Ala539 to Asp571, in orange), BtMan2A (residues from Pro514 to Thr527, in cyan), and XacMan2a (residues from Gly534 to Thr544) that contribute to determine the active-site topology (shown in yellow). C, surface representation of ThMan2A (orange), BtMan2A (cyan), and XacMan2A (green), highlighting the active-site vicinity, with some delimiting residues represented as sticks. The second Trp residue from the conserved WDW motif was used as reference for the active-site length measurements.
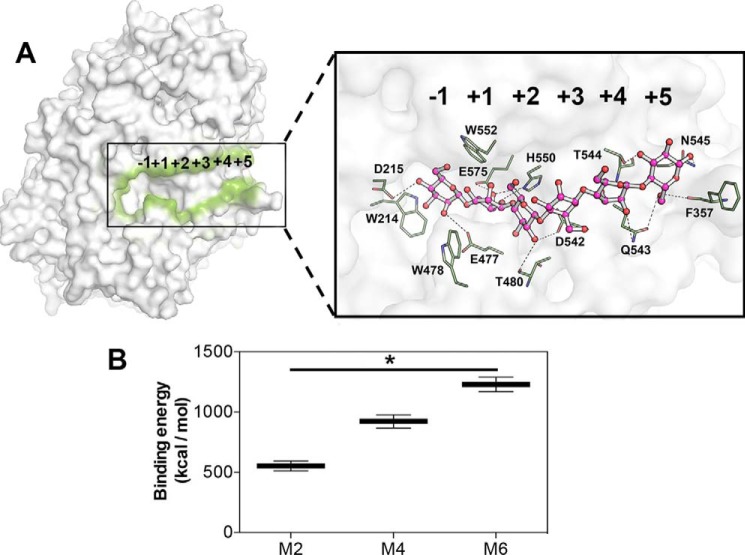
To identify those additional positive subsites, we docked different MOS (M2, M4, and M6) in XacMan2A and performed molecular dynamics simulations to obtain the relative binding energies (Fig. 8, A and B). Calculated relative binding-free energies were nearly 2-fold higher from M2 to M4, and a similar trend was observed from M4 to M6, indicating a higher affinity of XacMan2A for longer substrates (Fig. 8B). These in silico results are in accordance with biochemical data and suggest the existence of potentially six subsites ranging from −1 to +5 (Fig. 8A).
Figure 8.
Putative subsites in XacMan2A. A, representation of XacMan2A catalytic interface (contoured in green) with a docked mannohexaose (zoomed image, pink carbon atoms), indicating the putative subsites and key residues. Residues surrounding the sugar are represented as sticks with green carbon atoms. Hydrogen bonds are represented with traced lines. B, calculated relative binding free energies of XacMan2A with MOS (M2, M4, and M6) according to molecular dynamics simulations. *, p < 0.1. Error bars, S.D.
In addition to the profound changes in the positive-subsite region compared with other GH2 members, XacMan2A contains two residue substitutions that increase the active-site volumetric capacity, Gly439 and Gly556, which correspond to Asp451 and Tyr575 in ThMan2A and to Cys424 and Tyr537 in BtMan2A, respectively (Fig. 9A). To understand the functional role of these two nonconserved residues, we constructed the mutants G556Y and G439C, mimicking the BtMan2A configuration. Biochemical assays showed that both mutations abolished the enzyme activity on β-mannan polysaccharide (Fig. 9B), revealing that the decrease in volumetric capacity impaired the polymeric substrate binding. In addition, an unexpected decrease in the activity was observed on pNP-β-Man (Fig. 9C), indicating distinct structural determinants for substrate recognition in XacMan2A, since the residue Tyr537 was considered essential for BtMan2A catalysis (22).
Figure 9.
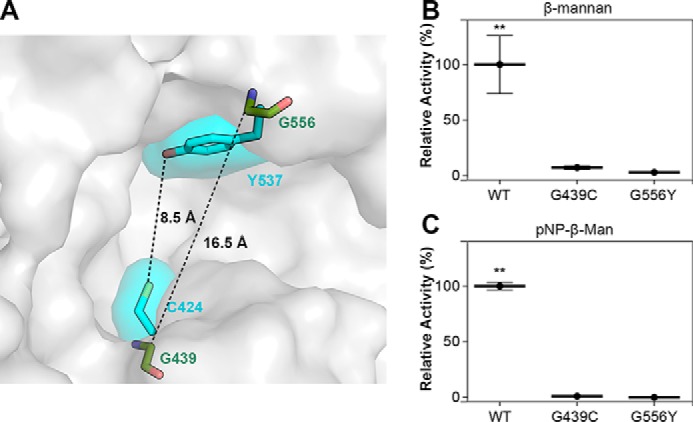
The residues Gly439 and Gly556, not conserved in GH2 β-mannosidases, are essential for XacMan2A catalytic activity. A, surface representation of XacMan2A active site with Gly439 and Gly556 residues shown as green sticks. BtMan2A residues Cys424 and Tyr537 are shown as cyan sticks and their respective surfaces. B and C, relative activities of XacMan2A and mutants against ivory nut β-mannan (15 mg·ml−1) (B) or pNP-β-Man (40 mmol·liter−1) (C). 100% corresponds to the specific activity (units·mg−1) of the WT protein on each substrate. **, p < 0.05. Error bars, S.D.
Together, our data demonstrate that the XacMan2A positive-subsite region clearly provides accessibility to the substrate with a cleft that can accommodate up to six sugar residues (Figs. 7C and 8A). These structural observations are in full agreement with the functional assays, supporting the novel mode of action of XacMan2A.
The (dis)similarities of the catalytic interface of GH2 and GH5 exo-β-mannanases
XacMan2A has the same ability of the Cellvibrio mixtus GH5 exo-β-mannanase (CmMan5A) (20) to release mannose from pNP-β-Man and from the nonreducing end of β-mannans. Structural superposition of the two catalytic domains resulted in RMSD of 2.4 Å (computed for Cα atoms) with the acid-base and nucleophile residues located in equivalent positions and oriented in a similar manner (Fig. 10A). Other conserved residues are Arg80, Asn214, Trp285, and Trp376 from the catalytic center of CmMan5A, which correspond to Arg408, Asn476, Trp552, and Trp667 in XacMan2A (Fig. 10A). These residues participate in substrate binding and also in the maintenance of the hydrogen bond network that supports catalysis. However, the conserved WDW motif adjacent to the −1 subsite of GH2 mannosidases is absent in CmMan5A. Instead, CmMan5A possesses an extended loop (Trp378–Phe412) that forms a “double” steric barrier at the negative-subsite region, preventing an endo-acting mode (20) (Fig. 10B).
Figure 10.
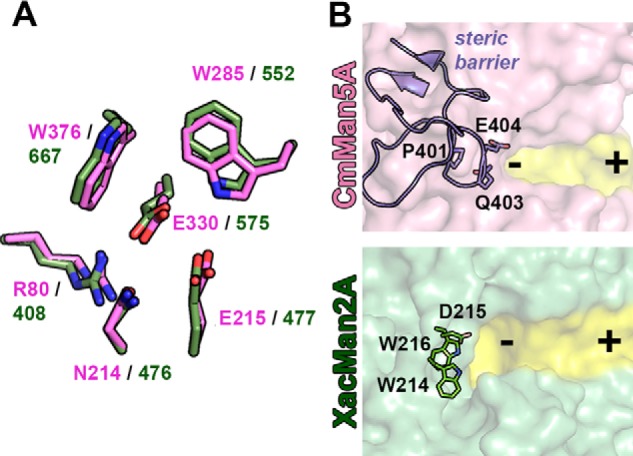
Structural comparison between GH2 and GH5 exo-mannanases. A, residues of the catalytic interface of CmMan5A (PDB code 1UUQ) and XacMan2A (PDB code 6BYC) are superposed and shown in pink and green, respectively. The catalytic residues, the acid base (Glu477 in XacMan2A and the corresponding residue in CmMan5A, Glu215) and the nucleophile (Glu575 in XacMan2A and the corresponding residue in CmMan5A, Glu330), are located in equivalent positions in the three-dimensional structures of both enzymes. Arg80, Asn214, Trp285, and Trp376 residues from the catalytic center of CmMan5A and the corresponding residues (Arg408, Asn476, Trp552, and Trp667) in XacMan2A are also conserved in very similar orientations. B, comparison between active-site topologies (highlighted in yellow) of CmMan5A and XacMan2A. The extended loop in the CmMan5A structure (Trp378–Phe412) forms a “double” steric barrier at the negative-subsite region and is represented as purple cartoon and sticks. This loop is responsible for the mode of action of CmMan5A and, in XacMan2A, the corresponding region is formed by the conserved motif WDW (residues Trp214, Asp215, and Trp216) in the GH2 family. Negative and positive subsites are represented by minus and plus signs, respectively.
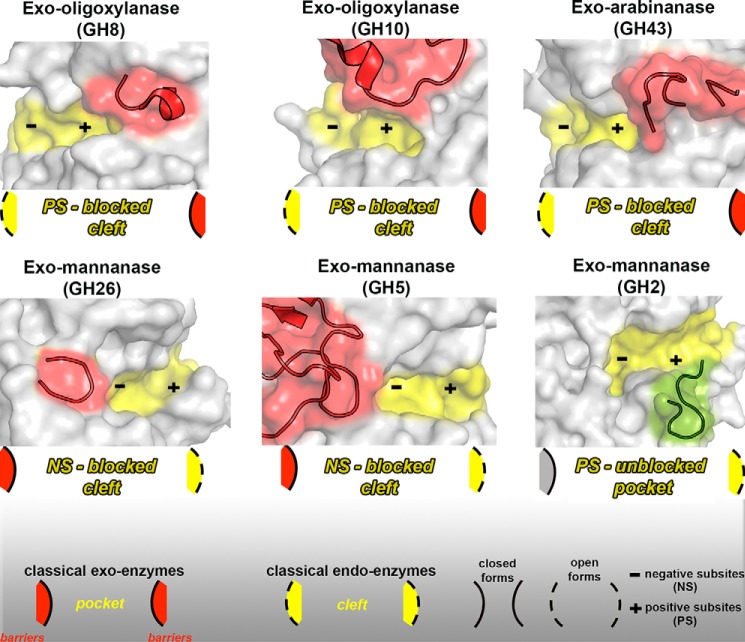
Interestingly, most exo-activities on polymeric substrates and oligosaccharides reported so far for CAZymes seem to have a similar evolutionary strategy involving the transition from an endo-acting enzyme to one with exo-activity, such as in the GH8 (34), GH10 (21), and GH43 (35) families. In all three of these examples, the insertion of a loop blocked the positive subsites (Fig. 11), leading to an exo-cleavage pattern. The active site topology of these proteins could be classified as blocked clefts, because the negative-subsite region remains open, whereas the positive region is clogged (Fig. 11). The evolution of exo-β-mannanase activity in GH26 and GH5 families also seems to follow a similar strategy but, in these cases, via the insertion of a blocking loop at the negative-subsite region (32, 20). In all of these cases, the exo-activity probably originates from the emergence of steric barriers in one of the sides of a cleft-like active site. XacMan2A is the first member of a typical family of enzymes with catalytic preference for side-chain substitutions and short oligosaccharides that assumes an exo-activity against polymeric substrates via the removal of steric impediments in a pocket-like active site (Fig. 11).
Figure 11.
Molecular mechanisms adopted by exo-enzymes derived from classical endo- or exo-GH members. The GH8 exo-oligoxylanase (PDB code 1WU4 (34)) presents an emergent loop (red cartoon) formed by the residues Ser317–Leu323 that blocks the positive-subsite region, resulting in a blocked cleft (represented in the scheme). A similar mechanism is adopted by the GH10 exo-oligoxylanase (PDB code 4PMV (21)) (residues Val267–Pro303) and by the GH43 exo-arabinanase (PDB code 4KCB (35)) (residues Arg203–Asn231). In the GH26 (PDB code 2VX5 (32)) and GH5 (PDB code 1UUQ (20)) exo-mannanases, a steric barrier imposed by the emergent loops (Arg201–Asn231 and Trp378–Phe412, respectively) at the negative-subsite regions confers the exo mode of action and also results in a blocked cleft. In the XacMan2A (PDB code 6BYC), a nonblocking loop (green cartoon) (residues Gly534–Thr544) at the positive-subsite region disrupts the classical pocket found in GH2 enzymes and gives rise to extra positive subsites. All emergent blocking loops are represented in red cartoons. Active-site cavities are highlighted in yellow. Negative (NS) and positive subsites (PS) are represented by a minus and plus sign, respectively.
Discussion
XacMan2A is an unusual GH2 member because of its ability to efficiently hydrolyze the polysaccharide β-mannan. The enzyme showed greater efficiency on β-mannan compared with other mannan-based polymeric substrates, indicating that glucose in the main-chain or galactose substitutions impairs its catalytic activity. Indeed, crystallographic analysis did not show any evidence that galactosyl substituents could be accommodate in the active site. Among the GH2 β-mannosidases characterized so far, the only one that showed some capacity to cleave β-mannan was BtMan2A (22). However, its kcat (0.49 s−1) on β-mannan is very low compared with that observed on pNP-β-Man (128.15 s−1). In contrast, XacMan2A hydrolyzes pNP-β-Man and β-mannan with catalytic rates in the same order of magnitude (vo/[ET] of 24.5 s−1 to pNP-β-Man and 8.8 s−1 to β-mannan; Table 1), characterizing a genuine exo-β-mannanase. Moreover, increasing hydrolysis rates were observed from M2 to M6 for XacMan2A, which was not observed for BtMan2A (22). Quantitative MS kinetics analysis also showed a clear preference of XacMan2A for longer oligosaccharides. In addition, all MOS and β-mannan were hydrolyzed to mannose by XacMan2A, and the cleavage pattern is characteristic of an exo-acting enzyme. Together, these results demonstrate that XacMan2A acts as an exo-β-mannanase.
The unique composition and conformation of the loop Gly534–Thr544 in the positive-subsite region of XacMan2A resulted in a catalytic interface more open and accessible that is probably associated with the capacity to cleave longer substrates. Compared with structurally characterized GH2 β-mannosidases, this loop in XacMan2A is shortened and does not impose a physical barrier, allowing the emergence of a continuous active-site cleft with additional positive subsites. Structural analysis, along with biochemical data, indicates that XacMan2A has only one negative subsite (−1) and, potentially, at least five positive subsites, which fully meets the stereo-chemical conditions for its new mode of action. In the other GH2 enzymes, such as BtMan2A and ThMan2A, “blocking” loops are found in the same region of the active site, and they can be associated with the selection of smaller substrates. The mannose-releasing exo-β-mannanase activity had been observed in a GH5 family member from C. mixtus (CmMan5A) (20). However, the structural determinants for this mode of action in CmMan5A were attributed to the emergence of a loop that blocked the negative-subsite region, whereas in XacMan2A, it is associated with the removal of a steric barrier at the positive-subsite region.
This rare ability of XacMan2A to saccharify β-mannans without relying on endo-β-mannanases represents a simplified carbon-uptake strategy that would benefit the bacterial growth and development during plant infection. This activity is also of great potential in industrial applications involving β-mannan depolymerization, such as biofuel production and food and beverage processing. In addition, this novel molecular mechanism for exo-activity based on the redesign of a pocket-like active site expands our current understanding of the molecular strategies related to functional differentiation in CAZymes.
Experimental procedures
Molecular cloning, mutagenesis, and protein production
A fragment of 2,691 bp from the XAC3075 gene (NCBI accession code AAM37920), encoding for XacMan2A without the predicted signal peptide (28 N-terminal residues), was amplified from the genomic DNA of X. axonopodis pv. citri 306 using standard cloning methods. The amplified DNA was cloned into pGEM-T vector and then subcloned into the restriction enzyme NdeI/XhoI sites of the pET28a vector (Novagen, Madison, WI) with a hexahistidine tag at the N terminus. Site-directed mutagenesis of XacMan2A construct (E575A, E477A, G439C, and G556Y) were performed according to the QuikChange kit (Stratagene, La Jolla, CA). The recombinant proteins were expressed in E. coli BL21 (DE3) cells and purified by metal-affinity and size-exclusion chromatography. Cells were grown at 37 °C in lysogeny broth medium containing kanamycin (50 mg·ml−1) to A600 nm = 0.6, followed by induction with 0.5 mmol·liter−1 isopropyl-thio-β-d-galactopyranoside for 16 h at 20 °C. After centrifugation, cells were resuspended in lysis buffer (20 mmol·liter−1 sodium phosphate (pH 7.4), 300 mmol·liter−1 NaCl, 5 mmol·liter−1 imidazole, 1 mmol·liter−1 phenylmethylsulfonyl fluoride, and 5 mmol·liter−1benzamidine) and incubated on ice with lysozyme (1 mg·ml−1) for 30 min. Bacterial cells were disrupted by sonication, and the soluble fraction was purified by immobilized metal ion affinity chromatography using a 5-ml HiTrap Chelating HP column (GE Healthcare, Little Chalfont, UK), previously charged with Ni2+, coupled to an ÄKTA purifier (GE Healthcare). The proteins were eluted using a linear gradient (0–500 mmol·liter−1) of imidazole at a flow rate of 1 ml·min−1. The eluted fractions were analyzed by SDS-PAGE, and those containing pure proteins were pooled, concentrated by filtration, and submitted to size-exclusion chromatography. Size-exclusion chromatography experiments were carried out at a flow rate of 1 ml·min−1 using a HiLoad 16/60 Superdex 200 pg column (GE Healthcare), previously equilibrated with 20 mmol·liter−1 sodium phosphate buffer, pH 7.4, and 150 mmol·liter−1 NaCl, coupled to an ÄKTA purifier. The purity of samples was analyzed by SDS-PAGE, and protein concentration was determined by absorbance at 280 nm using the molar extinction coefficient calculated from the amino acid composition (http://web.expasy.org/protparam/). Dynamic light scattering was performed in a Dynapro molecular sizing instrument (Wyatt Technology) to evaluate the homogeneity of the purified samples.
CD analysis
CD spectroscopy experiments were conducted on a Jasco J-810 CD spectrophotometer (JASCO, Oklahoma City, OK) equipped with a Peltier temperature control using 1-mm path quartz cuvettes. Spectra were acquired with 3.5 μmol·liter−1 XacMan2A in 20 mmol·liter−1 sodium phosphate (pH 7.4) with 150 mmol·liter−1 NaCl at 20 °C. The thermal denaturation of XacMan2A was followed by measurement of the ellipticity changes at 212 nm. For this experiment, a temperature ramp from 20 to 100 °C with a heating rate of 1 °C·min−1 was used. The reversibility of the temperature effect was checked by cooling the denatured sample to 20 °C using the same parameters described above.
SAXS data collection and analysis
SAXS measurements were performed using a monochromatic X-ray beam (λ = 1.488 Å) from the D01A-SAXS2 beamline at the Brazilian Synchrotron Light Laboratory (LNLS, Campinas, Brazil). SAXS measurements for XacMan2A were performed at two different concentrations (2 and 4 mg·ml−1) in 20 mmol·liter−1 sodium phosphate buffer, pH 7.4, with 150 mmol·liter−1 NaCl. The samples were centrifuged for 20 min at 20,000 × g and 4 °C to remove potential aggregates before each measurement. The integration of SAXS patterns was performed using Fit2D (36). The package GNOM (37) was used to evaluate the pair-distance distribution functions p(r). Molecular envelopes were calculated from the experimental SAXS data using the program DAMMIN (38). Averaged models were generated from several runs using DAMAVER suite programs (39). The theoretical scattering curve was calculated from the crystallographic model and compared with the experimental SAXS curves using the program CRYSOL (40). The crystallographic structures were fitted into the corresponding SAXS molecular envelopes using the program SUPCOMB (41).
Protein crystallization
Crystals of native XacMan2A and mutants were grown by the hanging-drop vapor-diffusion method at 18 °C in drops containing 0.5–1.0 μl of protein solution at 7.2 mg·ml−1 and 0.5 μl of the crystallization condition equilibrated against 200 μl of crystallization condition in 48-well plates. The WT enzyme was crystallized in the following conditions: 0.1 mol·liter−1 ammonium acetate, 0.1 mol·liter−1 BisTris, pH 5.5, and 17% (w/v) PEG 10,000; and 0.1 mol·liter−1 ammonium acetate, 0.1 mol·liter−1 BisTris, pH 5.5, and 18% (w/v) PEG 10,000. A complex of XacMan2A with d-mannose (Megazyme, County Wicklow, Ireland) was prepared with the crystal grown in the second crystallization condition, by soaking the crystal into the mother solution containing 100 mmol·liter−1 d-mannose for 4 h. Before data collection, the crystal of native enzyme was soaked in cryoprotectant containing the mother solution with 20% (v/v) PEG 400 and then directly flash-cooled in a nitrogen gas stream at 100 K. Mutant E575A crystal was obtained in 0.1 mol·liter−1 sodium acetate, pH 5.0, 0.2 mol·liter−1 magnesium chloride, and 20% (w/v) PEG 600. Mutant E477A crystal was obtained in 0.1 mol·liter−1 ammonium acetate, 0.1 mol·liter−1 BisTris, pH 5.5, and 18% (w/v) PEG 10,000. Mutant complexes were prepared as described for the WT protein.
X-ray data collection, structure determination, and refinement
The X-ray data were collected on the MX2 beamline at the LNLS (Campinas, São Paulo, Brazil) using a 1.459 Å wavelength X-ray beam and a PILATUS2M detector (Dectris, Baden-Dattwil, Switzerland). Data were scaled and reduced using XDS (42). The structures were solved by molecular replacement methods using the crystalline structure of the GH2 β-mannosidase from B. thetaiotaomicron (PDB code 2JE8 (22); sequence identity of 39%) as template and MOLREP (43) in the CCP4 suite of programs. The models were further built with the AutoBuild wizard (44) from the PHENIX package, yielding nearly refined structures without internal gaps in the chain. The structures were refined alternating cycles of TLS and restrained refinement using REFMAC (45) or PHENIX Refine (46) and manually inspected using the program COOT (47). TLS groups were generated by the TLSMD server (48). The refined structures were evaluated using the program MolProbity (49). Data collection, processing, and refinement statistics are summarized in Table 2. Graphic representations of structures were generated in PyMOL (50). Structure superposition RMSDs were calculated with PDBeFOLD (51).
The structure factors and atomic coordinates of XacMan2A in native and mannose complex forms were deposited in the Protein Data Bank under the accession codes 6BYC and 6BYE, respectively. The crystallographic data of nucleophile (E575A) and acid-base (E477A) mutants were deposited under the accession codes 6BYG and 6BYI, respectively.
Molecular docking and in silico binding energy evaluation
The complexes between the target protein and MOS (M2, M4, and M6) were built using local small molecule docking onto the active site with AutoDock Vina (52). Based on the top hit, the binary complexes were prepared for molecular dynamics simulations using explicit solvent in YASARA (53) under standard temperature and pressure parameters. The binding energies were calculated throughout the trajectories according to the YAMBER3 force field (54).
Biochemical assays
Aryl glycosidase activity was determined at 40 °C in 40 mmol·liter−1 citrate-phosphate buffer, pH 5.5, containing p-nitrophenyl-β-d-glucopyranoside (pNP-Glu), p-nitrophenyl-β-d-galactopyranoside (pNP-Gal), p-nitrophenyl-β-d-xylopyranoside (pNP-Xyl), p-nitrophenyl-α-l-arabinofuranoside (pNP-AraF), p-nitrophenyl-α-d-mannopyranoside (pNP-α-Man), or pNP-β-Man (Sigma-Aldrich) as substrate in a final volume of 0.3 ml. The reactions were initiated by the addition of the enzyme and interrupted after convenient time intervals by adding 0.3 ml of saturated sodium tetraborate solution. The released p-nitrophenol was measured at 400 nm, using an Infinite® 200 PRO microplate reader (TECAN Group Ltd., Männedorf, Switzerland). The hydrolysis of mannose oligosaccharides, ivory nut mannan, carob galactomannan (low viscosity), konjac glucomannan (low viscosity), sugar beet arabinan, beechwood xylan, carboxymethyl cellulose 4 m (CMC), and barley β-glucan (low viscosity) (all from Megazyme) were evaluated under the same conditions by estimating the reducing sugar released, according to the 3,5-dinitrosalicylic acid method (55). The optimum temperature for enzymatic activity was determined in the range from 10 to 85 °C in 40 mmol·liter−1citrate-phosphate buffer, pH 5.5. The optimum pH was evaluated in McIlvaine buffer (56), ranging from 3.0 to 8.0 at 40 °C. Thermal stability was evaluated by incubating the enzyme in McIlvaine buffer, pH 5.5, for up to 32 h at 40 °C. The residual activity was estimated at 40 °C in 40 mmol·liter−1 citrate-phosphate buffer, pH 5.5, as described above. Reduced MOS samples were previously treated with borohydride and purified with a PD MiniTrapTM G-10 column (GE Healthcare) before enzymatic assays. One unit of enzyme was defined as the amount of enzyme that releases 1 μmol of product per min. Specific activity was defined as units·mg−1 protein (units·mg−1). Kinetic parameters were calculated using nonlinear regression with Origin version 8.1 software (OriginLab, Northampton, MA).
Enzymatic activity monitored by MS
Hydrolysis reactions were carried at 40 °C with 5 μl of MOS (M6 and M2), 3 μl of McIlvaine buffer at pH 5.5, 1 μl of water, and 1 μl of a solution of XacMan2A enzyme (final concentration 15 μg/ml). The reactions were initiated by the addition of the enzyme and interrupted after 5 min by adding 40 μl of methanol. Kinetic analysis were performed based on Ge et al. (57) on a Synapt high-definition mass spectrometer (Waters Corp., Milford, CT) at V mode and ESI(+) with a spray voltage maintained at 3.0 kV and heated to 130 °C in the source. The quenched samples were injected in scan mode (m/z 150–1,100) with direct infusion at a flow rate of 20 μl/min. A calibration curve was made to determine the concentrations of the products of the enzymatic reaction. Kinetic parameters were calculated using nonlinear regression with Origin version 8.1 software (OriginLab).
Capillary zone electrophoresis
The products of enzymatic hydrolysis of mannose oligosaccharides with varying degrees of polymerization from 2 to 6 and β-mannan polysaccharides were derivatized with 8-aminopyrene-1,3,6-trisulfonic acid (Sigma-Aldrich) by reductive amination as described previously (58, 59). Capillary zone electrophoresis was performed on a P/ACE MQD capillary electrophoresis system with a laser-induced fluorescence detection system (Beckman Coulter, Brea, CA). An uncoated fused-silica capillary of internal diameter 75 μm and length 20 cm was used for separation of labeled oligosaccharides. The capillary was conditioned with 40 mmol·liter−1 potassium phosphate (pH 2.5), and samples were injected by application of 0.5 p.s.i. for 5 s. Electrophoretic conditions were 20 kV/70–100 mA with reverse polarity at a controlled temperature of 25 °C. 8-Aminopyrene-1,3,6-trisulfonic acid–labeled saccharides were excited at 488 nm, and emission was collected through a 520-nm band pass filter. The electrophoretic behavior of degradation products was compared with mannose oligosaccharide standards (purchased from Sigma-Aldrich and Megazyme).
Author contributions
M. N. D. conducted most of the experiments, analyzed the results, and wrote the manuscript. F. H. M. S. conducted functional experiments, analyzed the data, and participated in manuscript writing. P. S. V. conducted experiments of mutagenesis, analyzed the data, and conducted part of the crystallographic studies. M. A. B. M. performed structure refinement, analyzed results, prepared figures, and wrote the manuscript. L. M. Z. conceived biophysical experiments and analyzed the results. C. R. S. participated in experimental design and results analyses. R. A. S. P. and F. C. G. conducted and analyzed mass spectrometry experiments. R. V. H. and P. S. L. O. performed molecular dynamics simulations and calculated free binding energy parameters. M. T. M. conceived the original idea for the project, analyzed the results, and wrote the paper.
Acknowledgments
We thank the Brazilian Synchrotron Light Laboratory (LNLS) and the Brazilian Biosciences National Laboratory (LNBio) for the provision of time on the MX2 beamline, ROBOLAB, and LEC.
This work was supported by São Paulo Research Foundation (FAPESP) Grant 2015/26982-0 (to M. T. M.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 6BYC, 6BYE, 6BYG, and 6BYI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- GH
- glycoside hydrolase
- pNP-β-Man
- p-nitrophenyl-β-d-mannopyranoside
- pNP-Glu
- p-nitrophenyl-β-d-glucopyranoside
- pNP-Gal
- p-nitrophenyl-β-d-galactopyranoside
- pNP-Xyl
- p-nitrophenyl-β-d-xylopyranoside
- pNP-AraF
- p-nitrophenyl-α-l-arabinofuranoside
- pNP-α-Man
- p-nitrophenyl-α-d-mannopyranoside
- RMSD
- root mean square deviation
- SAXS
- small-angle X-ray scattering
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TLS
- translation/libration/screw
- MOS
- manno-oligosaccharides
- CAZyme
- carbohydrate-active enzyme
- PDB
- Protein Data Bank
- M2
- mannobiose
- M3
- mannotriose
- M4
- mannotetraose
- M5
- mannopentaose
- M6
- mannohexaose.
References
- 1. Sandin R. L. (1987) Studies on cell adhesion and concanavalin A-induced agglutination of Candida albicans after mannan extraction. J. Med. Microbiol. 24, 145–150 10.1099/00222615-24-2-145 [DOI] [PubMed] [Google Scholar]
- 2. Yamabhai M., Sak-Ubol S., Srila W., and Haltrich D. (2016) Mannan biotechnology: from biofuels to health. Crit. Rev. Biotechnol. 36, 32–42 10.3109/07388551.2014.923372 [DOI] [PubMed] [Google Scholar]
- 3. Capek P., Kubacková M., Alföldi J., Bilisics L., Lisková D., and Kákoniová D. (2000) Galactoglucomannan from the secondary cell wall of Picea abies L. Karst. Carbohydr. Res. 329, 635–645 10.1016/S0008-6215(00)00210-X [DOI] [PubMed] [Google Scholar]
- 4. Scheller H. V., and Ulvskov P. (2010) Hemicelluloses. Annu. Rev. Plant. Biol. 61, 263–289 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- 5. Moreira L. R., and Filho E. X. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178 10.1007/s00253-008-1423-4 [DOI] [PubMed] [Google Scholar]
- 6. Lavoie J. M., Beauchet R., Berberi V., and Chornet M. (2011) Biorefining lignocellulosic biomass via the feedstock impregnation rapid and sequential steam treatment. Biofuel's Eng. Process Technol. 29, 307–480 [Google Scholar]
- 7. Brett C. T., and Waldren K. (1996) Physiology of plant cell walls. In Topics in Plant Functional Biology (Black M., Charlewood B., eds) Chapman and Hall, London [Google Scholar]
- 8. Chauhan P. S., Sharma P., Puri N., and Gupta N. (2014) A process for reduction in viscosity of coffee extract by enzymatic hydrolysis of mannan. Bioprocess Biosyst. Eng. 37, 1459–1467 10.1007/s00449-013-1118-9 [DOI] [PubMed] [Google Scholar]
- 9. Srivastava P. K., and Kapoor M. (2014) Cost-effective endo-mannanase from Bacillus sp. CFR1601 and its application in generation of oligosaccharides from guar gum and as detergent additive. Prep. Biochem. Biotechnol. 44, 392–417 10.1080/10826068.2013.833108 [DOI] [PubMed] [Google Scholar]
- 10. Dhawan S., and Kaur J. (2007) Microbial mannanases: an overview of production and applications. Crit. Rev. Biotechnol. 27, 197–216 10.1080/07388550701775919 [DOI] [PubMed] [Google Scholar]
- 11. Bettiol J. L. P., Boutique J. P., Gualco L. M. P., and Johnston J. P. (December 28, 2000) European Patent EP1059351
- 12. Mussini F. J., Coto C. A., Goodgame S. D., Lu C., Karimi A. J., Lee J. H., and Waldroup P. W. (2011) Effect of β-mannanase on broiler performance and dry matter output using corn-soyabean meal based diets. Int. J. Pou. Sci. 10, 778–781 10.3923/ijps.2011.778.781 [DOI] [Google Scholar]
- 13. Nunes F. M., Reis A., Domingues M. R., and Coimbra M. A. (2006) Characterization of galactomannan derivatives in roasted coffee beverages. J. Agric. Food Chem. 54, 3428–3439 10.1021/jf052971+ [DOI] [PubMed] [Google Scholar]
- 14. Pee V., Ignatius K. L., Speybroeck V., Michel M. P., and Jozef V. P. (August 28, 2002) European Patent EP0871596B1
- 15. Van Zyl W. H., Rose S. H., Trollope K., and Görgens J. F. (2010) Fungal β-mannanases: mannan hydrolysis, heterologous production and biotechnological applications. Process Biochem. 45, 1203–1213 10.1016/j.procbio.2010.05.011 [DOI] [Google Scholar]
- 16. Parisi G. C., Zilli M., Miani M. P., Carrara M., Bottona E., Verdianelli G., Battaglia G., Desideri S., Faedo A., Marzolino C., Tonon A., Ermani M., and Leandro G. (2002) High-fiber diet supplementation in patients with irritable bowel syndrome (IBS): a multicenter, randomized, open trial comparison between wheat bran diet and partially hydrolyzed guar gum (PHGG). Dig. Dis. Sci. 47, 1697–1704 10.1023/A:1016419906546 [DOI] [PubMed] [Google Scholar]
- 17. Pangsri P., Piwpankaew Y., Ingkakul A., Nitisinprasert S., and Keawsompong S. (2015) Characterization of mannanase from Bacillus circulans NT 6.7 and its application in mannooligosaccharides preparation as prebiotic. Springerplus 4, 771 10.1186/s40064-015-1565-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malgas S., van Dyk J. S., and Pletschke B. I. (2015) A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J. Microbiol. Biotechnol. 31, 1167–1175 10.1007/s11274-015-1878-2 [DOI] [PubMed] [Google Scholar]
- 20. Dias F. M., Vincent F., Pell G., Prates J. A., Centeno M. S., Tailford L. E., Ferreira L. M., Fontes C. M., Davies G. J., and Gilbert H. J. (2004) Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J. Biol. Chem. 279, 25517–25526 10.1074/jbc.M401647200 [DOI] [PubMed] [Google Scholar]
- 21. Santos C. R., Hoffmam Z. B., de Matos Martins V. P., Zanphorlin L. M., de Paula Assis L. H., Honorato R. V., Lopes de Oliveira P. S., Ruller R., and Murakami M. T. (2014) Molecular mechanisms associated with xylan degradation by Xanthomonas plant pathogens. J. Biol. Chem. 289, 32186–32200 10.1074/jbc.M114.605105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tailford L. E., Money V. A., Smith N. L., Dumon C., Davies G. J., and Gilbert H. J. (2007) Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the β-mannosidase, BtMan2A. J. Biol. Chem. 282, 11291–11299 10.1074/jbc.M610964200 [DOI] [PubMed] [Google Scholar]
- 23. Guillotin L., Richet N., and Lafite P., and Daniellou R. (2017) Is the acid/base catalytic residue mutation in β-d-mannosidase DtMan from Dictyoglomus thermophilum sufficient enough to provide thioglycoligase activity? Biochimie 137, 190–196 10.1016/j.biochi.2017.03.020 [DOI] [PubMed] [Google Scholar]
- 24. Nascimento A. S., Muniz J. R., Aparício R., Golubev A. M., and Polikarpov I. (2014) Insights into the structure and function of fungal β-mannosidases from glycoside hydrolase family 2 based on multiple crystal structures of the Trichoderma harzianum enzyme. FEBS J. 281, 4165–4178 10.1111/febs.12894 [DOI] [PubMed] [Google Scholar]
- 25. Jacobson R. H., Zhang X. J., DuBose R. F., and Matthews B. W. (1994) Three-dimensional structure of β-galactosidase from E. coli. Nature 369, 761–766 10.1038/369761a0 [DOI] [PubMed] [Google Scholar]
- 26. Skálová T., Dohnálek J., Spiwok V., Lipovová P., Vondrácková E., Petroková H., Dusková J., Strnad H., Králová B., and Hasek J. (2005) Cold-active β-galactosidase from Arthrobacter sp. C2-2 forms compact 660 kDa hexamers: crystal structure at 1.9 Å resolution. J. Mol. Biol. 353, 282–294 10.1016/j.jmb.2005.08.028 [DOI] [PubMed] [Google Scholar]
- 27. Hashimoto H. (2006) Recent studies of carbohydrate-binding modules. Cell Mol. Life Sci. 63, 2954–2967 10.1007/s00018-006-6195-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koshland D. E. (1953) Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. 28, 416–436 10.1111/j.1469-185X.1953.tb01386.x [DOI] [Google Scholar]
- 29. Davies G., and Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 10.1016/S0969-2126(01)00220-9 [DOI] [PubMed] [Google Scholar]
- 30. Ardèvol R., and Rovira C. (2015) Reaction mechanisms in carbohydrate-active enzymes: glycoside hydrolases and glycosyltransferases: insights from ab initio quantum mechanics/molecular mechanics dynamic simulation. J. Am. Chem. Soc. 137, 7528–7547 10.1021/jacs.5b01156 [DOI] [PubMed] [Google Scholar]
- 31. Jin Y., Petricevic M., John A., Raich L., Jenkins H., Portela De Souza L., Cuskin F., Gilbert H. J., Rovira C., Goddard-Borger E. D., Williams S. J., and Davies G. J. (2016) A β-mannanase with a lysozyme-like fold and a novel molecular catalytic mechanism. ACS Cent. Sci. 2, 896–903 10.1021/acscentsci.6b00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cartmell A., Topakas E., Ducros V. M., Suits M. D., Davies G. J., and Gilbert H. J. (2008) The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J. Biol. Chem. 283, 34403–34413 10.1074/jbc.M804053200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tailford L. E., Offen W. A., Smith N. L., Dumon C., Morland C., Gratien J., Heck M. P., Stick R. V., Blériot Y., Vasella A., Gilbert H. J., and Davies G. J. (2008) Structural and biochemical evidence for a boat-like transition state in β-mannosidases. Nat. Chem. Biol. 4, 306–312 10.1038/nchembio.81 [DOI] [PubMed] [Google Scholar]
- 34. Fushinobu S., Hidaka M., Honda Y., Wakagi T., Shoun H., and Kitaoka M. (2005) Structural basis for the specificity of the reducing end xylose-releasing exo-oligoxylanase from Bacillus halodurans C-125. J. Biol. Chem. 280, 17180–17186 10.1074/jbc.M413693200 [DOI] [PubMed] [Google Scholar]
- 35. Santos C. R., Polo C. C., Costa M. C. M. F., Nascimento A. F. Z., Meza A. N., Cota J., Hoffmam Z. B., Honorato R. V., Oliveira P. S., Goldman G. H., Gilbert H. J., Prade R. A., Ruller R., Squina F. M., Wong D. W., and Murakami M. T. (2014) Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases. J. Biol. Chem. 289, 7362–7373 10.1074/jbc.M113.537167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hammersley A., Svensson S. O., Hanfland M., Fitch A. N., and Hausermann D. (1996) Two-dimensional detector software: from real detector to idealised image or two-θ scan. High Pressure Res. 14, 235–248 10.1080/08957959608201408 [DOI] [Google Scholar]
- 37. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 10.1107/S0021889892001663 [DOI] [Google Scholar]
- 38. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 10.1016/S0006-3495(99)77443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 10.1107/S0021889803000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Svergun D. I., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 41. Kozin M., and Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 10.1107/S0021889800014126 [DOI] [Google Scholar]
- 42. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 10.1107/S0021889897006766 [DOI] [Google Scholar]
- 44. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 46. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 48. Painte J., and Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 10.1107/S0021889805038987 [DOI] [Google Scholar]
- 49. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 51. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 52. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krieger E., and Vriend G. (2014) YASARA View: molecular graphics for all devices: smartphones to workstations. Bioinformatics 30, 2981–2982 10.1093/bioinformatics/btu426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krieger E., and Vriend G. (2015) New ways to boost molecular dynamics simulations. J. Comput. Chem. 36, 996–1007 10.1002/jcc.23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 10.1021/ac60147a030 [DOI] [Google Scholar]
- 56. McIlvaine T. C. (1921) A buffer solution for colorimetric comparison. J. Biol. Chem. 49, 183–186 [Google Scholar]
- 57. Ge X., Sirich T. L., Beyer M. K., Desaire H., and Leary J. A. (2001) A strategy for the determination of enzyme kinetics using electrospray ionization with an ion trap mass spectopmeter. Anal. Chem. 73, 5078–5082 10.1021/ac0105890 [DOI] [PubMed] [Google Scholar]
- 58. Naran R., Pierce M. L., and Mort A. J. (2007) Detection and identification of rhamnogalacturonan lyase activity in intercellular spaces of expanding cotton cotyledons. Plant J. 50, 95–107 10.1111/j.1365-313X.2007.03033.x [DOI] [PubMed] [Google Scholar]
- 59. Cairo J. P., Oliveira L. C., Uchima C. A., Alvarez T. M., Citadini A. P., Cota J., Leonardo F. C., Costa-Leonardo A. M., Carazzolle M. F., Costa F. F., Pereira G. A., and Squina F. M. (2013) Deciphering the synergism of endogenous glycoside hydrolase families 1 and 9 from Coptotermes gestroi. Insect Biochem. Mol. Biol. 43, 970–981 10.1016/j.ibmb.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 60. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]