Abstract
The epidermal growth factor (EGF) receptor is a classical receptor tyrosine kinase with an extracellular ligand-binding domain and an intracellular kinase domain. Mutations in the EGF receptor have been shown to drive uncontrolled cell growth and are associated with a number of different tumors. Two different types of ATP-competitive EGF receptor tyrosine kinase inhibitors have been identified that bind to either the active (type I) or inactive (type II) conformation of the kinase domain. Despite the fact that both types of inhibitors block tyrosine kinase activity, they exhibit differential efficacies in different tumor types. Here, we show that in addition to inhibiting kinase activity, these inhibitors allosterically modulate ligand binding. Our data suggest that the conformations of the EGF receptor extracellular domain and intracellular kinase domain are coupled and that these conformations exist in equilibrium. Allosteric regulators, such as the small-molecule tyrosine kinase inhibitors, as well as mutations in the EGF receptor itself, shift the conformational equilibrium among the active and inactive species, leading to changes in EGF receptor-binding affinity. Our studies also reveal unexpected positive cooperativity between EGF receptor subunits in dimers formed in the presence of type II inhibitors. These findings indicate that there is strong functional coupling between the intracellular and extracellular domains of this receptor. Such coupling may impact the therapeutic synergy between small-molecule tyrosine kinase inhibitors and monoclonal antibodies in vivo.
Keywords: epidermal growth factor (EGF), epidermal growth factor receptor (EGFR), tyrosine-protein kinase (tyrosine kinase), growth factor, inhibitor, erlotinib, lapatinib, Ligand binding
Introduction
The epidermal growth factor (EGF)2 receptor is a transmembrane receptor tyrosine kinase that is the founding member of the ErbB receptor family (1, 2). It contains an extracellular ligand-binding domain, a single-pass transmembrane domain, an intracellular tyrosine kinase domain, and an intrinsically disordered C-terminal tail (3). The signal of ligand binding is transduced through the structure of the receptor and results in the activation of the intracellular tyrosine kinase domain. Phosphorylation of tyrosines on the C-terminal tail of the EGF receptor (4–6) generates binding sites for the Src homology 2 (SH2) and PTB domain-containing proteins that mediate the downstream effects of growth factor binding (7).
In the absence of ligand, the EGF receptor is thought to exist as a monomer in which the extracellular domain adopts a closed, tethered conformation (8). This closed conformation is mediated by interactions between the dimerization arm in subdomain II and the tethering arm in subdomain IV of the extracellular domain. Upon ligand binding, this tether is released and the receptor opens into an extended conformation. This exposes the dimerization arm in subdomain II, allowing two EGF receptor monomers to form a back-to-back dimer via their now-exposed dimerization arms (9, 10). The EGF receptor also appears to form ligand-independent dimers but consensus is lacking regarding the structure of these dimers (11, 12).
EGF-induced dimerization of the extracellular domain induces the formation of an asymmetric dimer of the intracellular kinase domains (13). In this dimer, the C-terminal lobe of the activator kinase interacts with the N-terminal lobe of the receiver kinase resulting in the activation of the receiver kinase. Mutations that ablate this interface also abolish receptor tyrosine kinase activity. The kinase domains can also form a symmetric “head-to-head” dimer but this species is catalytically inactive (14).
ATP-competitive, small-molecule inhibitors of the EGF receptor kinase have been developed and many of them are in widespread clinical use (15). Two types of inhibitors have been identified (16). Type I inhibitors, such as erlotinib, bind to the active conformation of the kinase domain in which the important α-C helix is swung in to the active site (17). Type II inhibitors, such as lapatinib, bind to the inactive conformation of the kinase domain in which the α-C helix is swung out (18). Despite the fact that both inhibitors block EGF receptor tyrosine kinase activity, they show differential efficacy in different tumor types (19–21). This suggests that they possess other properties, besides kinase inhibition, that distinguish their effects on tumorigenic forms of the EGF receptor.
In this study, we show that in addition to inhibiting EGF receptor kinase activity, these inhibitors modulate the function of the extracellular ligand-binding domain with erlotinib increasing and lapatinib decreasing receptor-binding affinity. Our data suggest that the conformations of the EGF receptor extracellular domain and intracellular kinase domain are coupled and that these high affinity and low affinity conformations exist in an equilibrium that can be shifted by small-molecule tyrosine kinase inhibitors. Our studies also reveal unexpected positive cooperativity between EGF receptor subunits in dimers formed in the presence of type II inhibitors and some EGF receptor mutants. These findings indicate that there is strong structural and functional coupling between the intracellular and extracellular domains of this receptor. Knowledge of this coupling could be exploited to enhance the efficacy of combinations of extracellular-directed and kinase-directed EGF receptor therapeutics.
Results
Erlotinib and lapatinib alter the ligand-binding properties of the EGF receptor
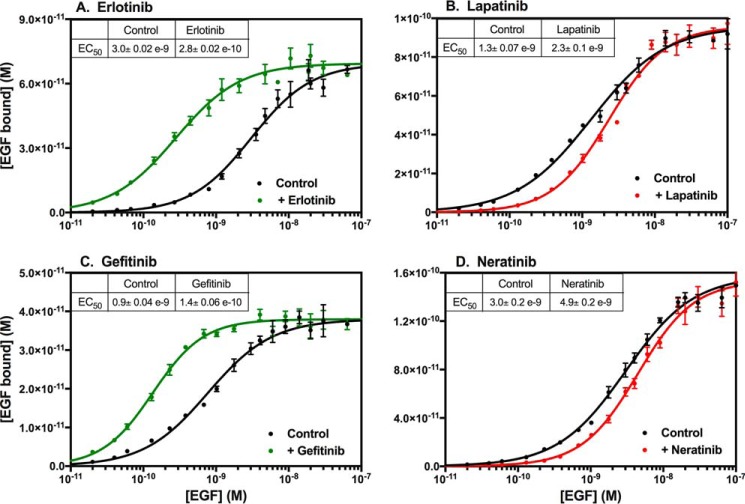
Treatment of CHO cells expressing the EGF receptor with the tyrosine kinase inhibitor, erlotinib, was found to significantly increase the affinity of the receptor for EGF (Fig. 1A) from an EC50 of ∼3 nm in untreated cells to an EC50 of ∼0.3 nm after treatment with erlotinib. To determine whether this effect was due to the inhibition of receptor kinase activity, we also assessed the effect on EGF binding of lapatinib, another small-molecule inhibitor of the EGF receptor kinase. Treatment of cells with lapatinib led to a modest decrease in the affinity of EGF for its receptor (Fig. 1B).
Figure 1.
Effect of EGF receptor tyrosine kinase inhibitors on EGF affinity. CHO cells expressing the WT EGF receptor were treated for 30 min at 37 °C without or with the indicated inhibitor prior to binding of 125I-EGF overnight at 4 °C. The indicated inhibitor was also included in the binding incubation medium. Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of three times.
Erlotinib is a type I inhibitor that binds to the active conformation of the EGF receptor kinase (17), whereas lapatinib is a type II inhibitor that binds to the inactive conformation of the kinase (18). To determine whether the different effects of erlotinib and lapatinib on EGF-binding affinity were due to the different conformations of the kinase domain to which they bound, we assessed the effects of gefitinib and neratinib on 125I-EGF binding.
The data in Fig. 1C demonstrate that, like erlotinib, the type I inhibitor, gefitinib (22), also increased the affinity of the receptor for EGF. An increase in EGF binding due to treatment with gefitinib has been observed previously (23). Neratinib, a type II inhibitor (24), recapitulated the effects of lapatinib, modestly reducing the affinity of the receptor for EGF (Fig. 1D). These data are consistent with the conclusion that the effects of these four inhibitors on EGF binding are related to their ability to shift the EGF receptor kinase domain into either the active (erlotinib and gefitinib) or inactive (lapatinib and neratinib) conformations. This implies that the conformation of the intracellular kinase domain modulates the function of the extracellular ligand-binding domain.
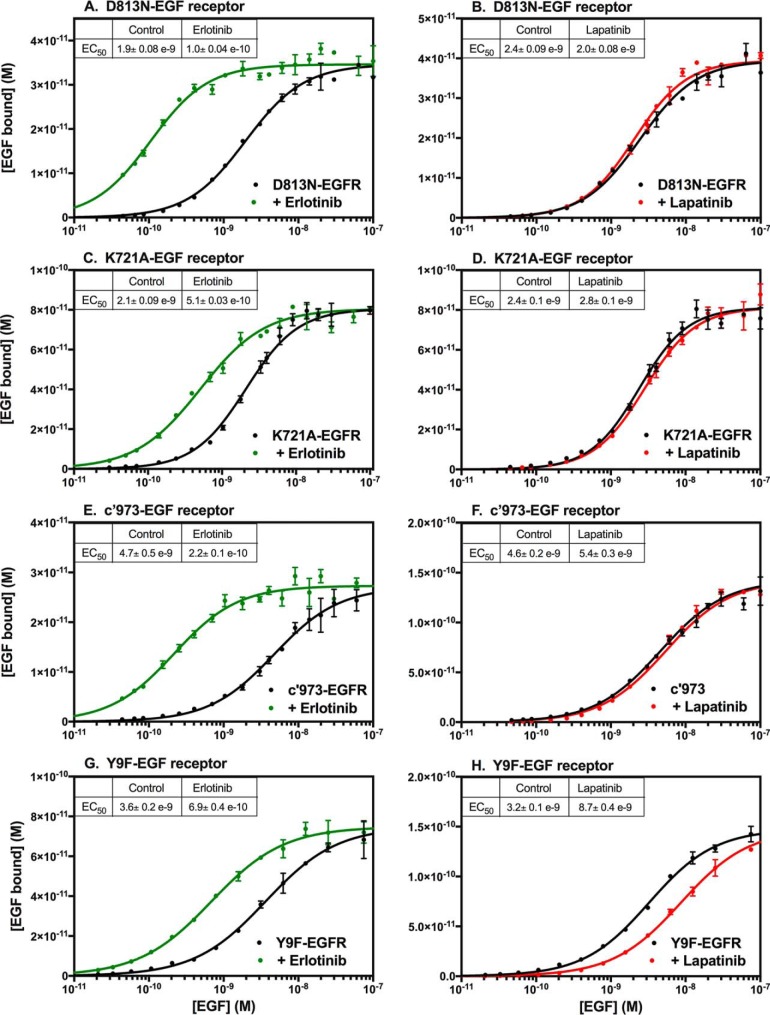
To determine whether either of these effects on receptor binding were related to inhibition of kinase activity, we assessed the effects of erlotinib and lapatinib on two kinase-dead versions of the EGF receptor, the D813N-EGF receptor (Fig. 2, A and B) and the K721A-EGF receptor (Fig. 2, C and D). As can be seen from Fig. 2, A and C, an erlotinib-induced increase in EGF affinity was still seen in both kinase-dead receptors. Thus, the observed increase in affinity upon treatment with erlotinib appears to be independent of its ability to inhibit the kinase activity of the EGF receptor. By contrast, the effect of lapatinib was largely abolished in the kinase-dead receptors (Fig. 2, B and D).
Figure 2.
Effect of erlotinib and lapatinib on EGF affinity in EGF receptor mutants. CHO cells expressing the D813N-EGF receptor (A and B), the K721A-EGF receptor (C and D), the c′973-EGF receptor (E and F), or the Y9F-EGF receptor (G and H) were treated for 30 min at 37 °C without (black lines) or with erlotinib (green lines) or lapatinib (red lines) prior to binding of 125I-EGF overnight at 4 °C. The indicated inhibitor was also included in the binding incubation medium. Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of three times.
Although these data suggest that the effects of lapatinib may be due to its ability to inhibit the kinase activity of the EGF receptor, the results with other mutants suggest that the situation is more complicated than that. The data in Fig. 2, E and F, show the response of the c′973-EGF receptor to treatment with erlotinib or lapatinib. The c′973-EGF receptor is truncated at residue 973, just before the first phosphorylatable tyrosine in the C-terminal tail. This truncation removes most of the C-terminal tail, precluding any autophosphorylation of the receptor. However, the kinase remains active and is, in fact, transforming (25). As was observed with the two kinase-dead receptors, the c′973-EGF receptor showed a large increase in affinity following treatment with erlotinib but no change in affinity upon treatment with lapatinib. Thus, the lack of effect of lapatinib on EGF affinity in the two kinase-dead receptors cannot be attributed to their lack of kinase activity because lapatinib also failed to reduce ligand affinity in the kinase-active c′973-EGF receptor.
The failure of lapatinib to induce a decrease in the affinity of the c′973-EGF receptor may be related to the absence of the C-terminal tail in this mutant. The Y9F-EGF receptor is a mutant in which all nine tyrosines in the C-terminal tail have been replaced with phenylalanines. As a result, the tail cannot undergo autophosphorylation (26). Nevertheless, this receptor showed both an increase in affinity in the presence of erlotinib (Fig. 2G) and a decrease in affinity in the presence of lapatinib (Fig. 2H). Thus, the presence of the C-terminal tail, but not its phosphorylation, is permissive for the lapatinib-induced decrease in EGF affinity.
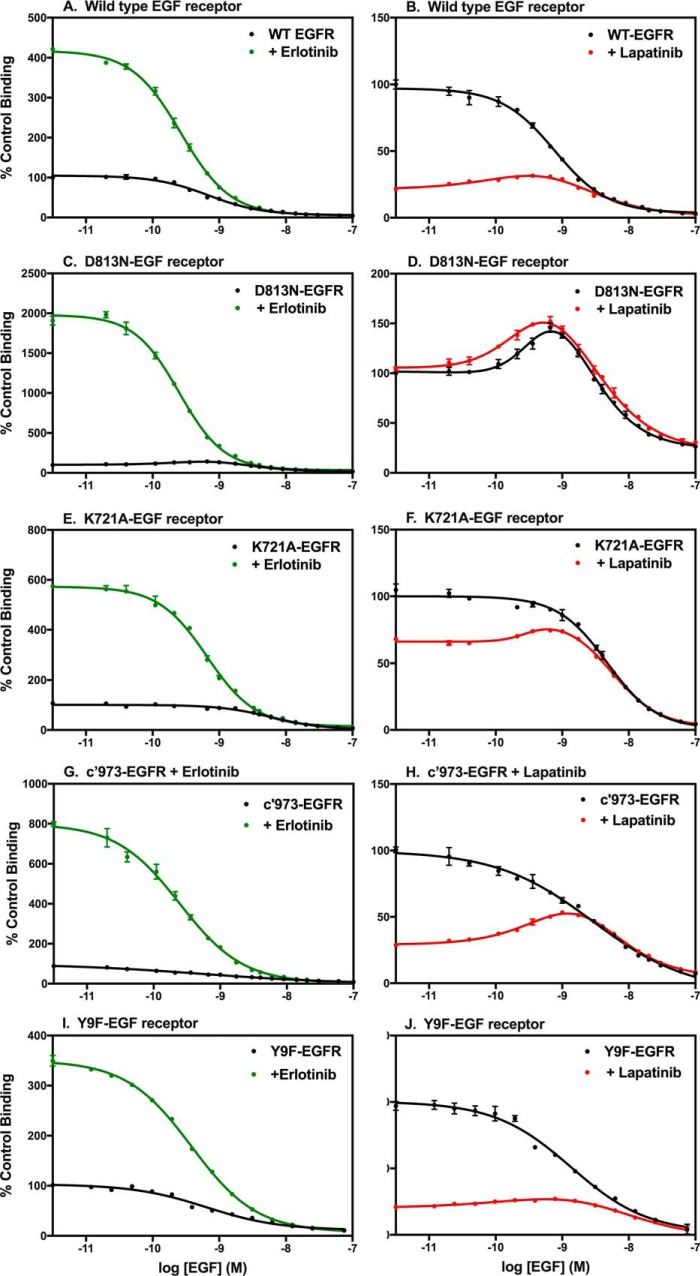
Although not obvious from the saturation-binding curves shown in Figs. 1 and 2, lapatinib was also found to induce an allosteric change in ligand binding by the EGF receptor. This is apparent when the data are plotted as 125I-EGF competition-binding curves, in which a fixed concentration of 125I-EGF is competed off the receptor by the addition of increasing concentrations of unlabeled EGF. In contrast to the saturation-binding curves, this format highlights differences seen at low concentrations of competing ligand. This is because the number of counts of 125I-EGF bound is greatest when little or no cold competitor has been added.
Typically, there is a monotonic decline in the binding of 125I-EGF as the concentration of unlabeled EGF is increased. This is observed in the WT EGF receptor both in the absence or presence of erlotinib (Fig. 3A). The increase in 125I-EGF bound at low doses of unlabeled EGF in the presence of erlotinib is due to the erlotinib-induced increase in the overall affinity of the receptor for EGF as seen in the saturation plots (Fig. 1A). This same pattern of a monotonic decrease in 125I-EGF binding as the dose of unlabeled competitor increases was observed for the D813N-EGF receptor (Fig. 3C), the K721A-EGF receptor (Fig. 3E), the c′973-EGF receptor (Fig. 3G), and the Y9F-EGF receptor (Fig. 3I) when assayed in the presence of erlotinib.
Figure 3.
125I-EGF competition-binding curves of WT and mutant EGF receptors treated without or with erlotinib or lapatinib. CHO cells expressing the indicated EGF receptors were treated without (black lines) or with erlotinib (green lines) or lapatinib (red lines) prior to and during incubation with labeled EGF. Increasing concentrations of unlabeled EGF were added to different wells. The data are normalized to control binding. Control binding represents the number of counts of 125I-EGF bound by the untreated cells in the absence of any additional unlabeled EGF. Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of three times.
The EGF receptor shows heterogeneity in its binding affinity for EGF, exhibiting both high affinity (∼200 pm) and low affinity (∼3 nm) binding sites (27–30). These affinity states are interconvertible and are due to the formation of predimers as well as to the presence of negative cooperativity in the system (28). At low concentrations of EGF, binding takes place almost exclusively at the high affinity site. Thus, the increase in counts bound by erlotinib-treated cells at low doses of EGF suggests that erlotinib increases the fraction of receptors in the high affinity state rather than increasing the affinity of the low affinity form of the receptor.
Unexpectedly, treatment of cells expressing the WT EGF receptor with lapatinib resulted in a bell-shaped competition-binding curve (Fig. 3B), an indication of the presence of positive cooperativity. Positive cooperativity is seen when the binding of a ligand to the first site on a dimer enhances the affinity of the second site on the dimer for that ligand. The large decrease in counts bound (∼75%) at low concentrations of unlabeled, competing ligand in the lapatinib-treated condition is a reflection of the lapatinib-induced decrease in affinity observed in the saturation-binding curves. As with erlotinib, the difference between the control and lapatinib-treated curves was largest at low doses of competitor but minimal at higher concentrations of unlabeled competitor. Thus, like erlotinib, lapatinib appears to exert its effect by reducing the fraction of high affinity sites so the overall population average affinity is decreased.
This bell-shaped binding behavior was relatively subtle in the WT receptor but was very obvious in the kinase-dead D813N-EGF receptor (Fig. 3D). This mutant exhibited bell-shaped competition-binding curves both in the absence and presence of lapatinib. For this kinase-dead mutant, the addition of lapatinib did not substantially reduce 125I-EGF binding at low concentrations of unlabeled ligand, consistent with the absence of a lapatinib-induced decrease in binding affinity (Fig. 2B). Interestingly, the kinase-dead K721A-EGF receptor exhibited a bell-shaped curve only in the presence of lapatinib (Fig. 3F) and showed only a modest decrease (∼30%) in the amount of 125I-EGF bound initially as compared with the WT receptor. Thus, the two kinase-dead versions of the EGF receptor exhibited distinct properties. This may be due to differences in the structural effects of the two mutations. D813N mutates a residue important for catalysis (31), whereas K721A removes a residue that stabilizes the active configuration of the kinase domain (13).
Both the c′973-EGF receptor and Y9F-EGF receptor exhibited behavior very similar to that of the WT EGF receptor, showing a bell-shaped competition-binding curve only in the presence of lapatinib and an ∼70% decrease in 125I-EGF binding upon addition of lapatinib to the cells (Fig. 3H). Thus, the ability of lapatinib to induce positive cooperativity in EGF binding is independent of the status of the C-terminal tail.
Effects of kinase inhibitors require receptor dimerization
Erlotinib has been shown to induce kinase-mediated dimerization of the EGF receptor (32–34). The presence of positive cooperativity in EGF binding in lapatinib-treated cells implies that lapatinib allows the formation of some type of EGF receptor dimer. Therefore, we next examined the influence of mutations in the subunit–subunit interfaces in the EGF receptor on the ability of these inhibitors to modulate ligand binding.
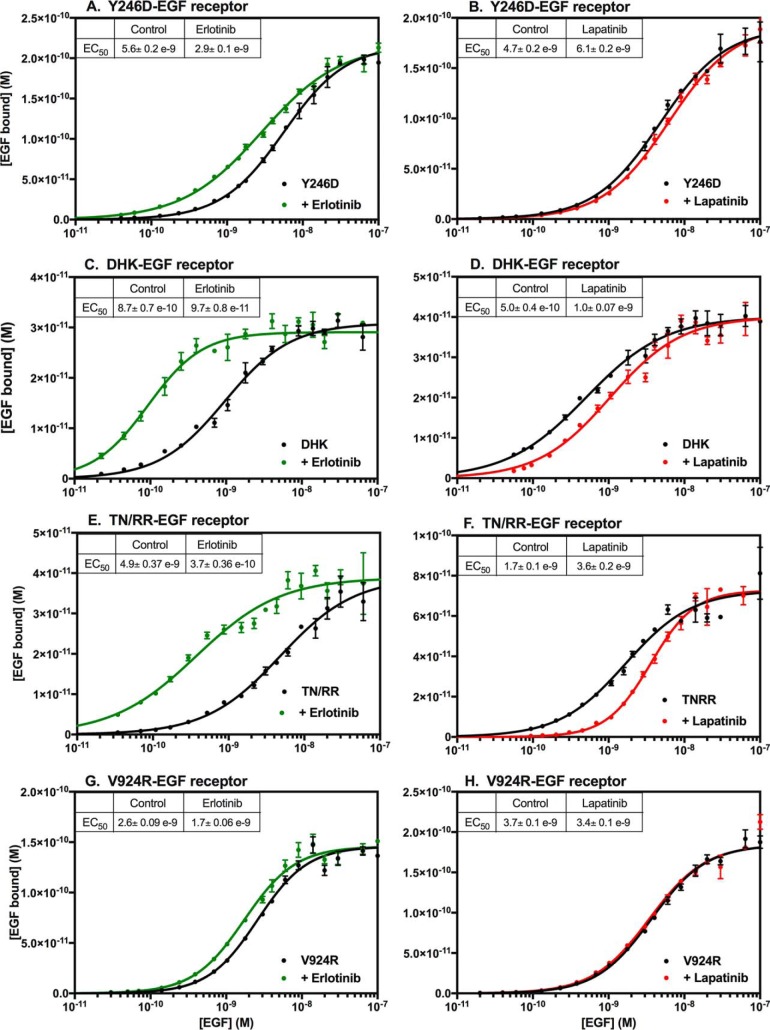
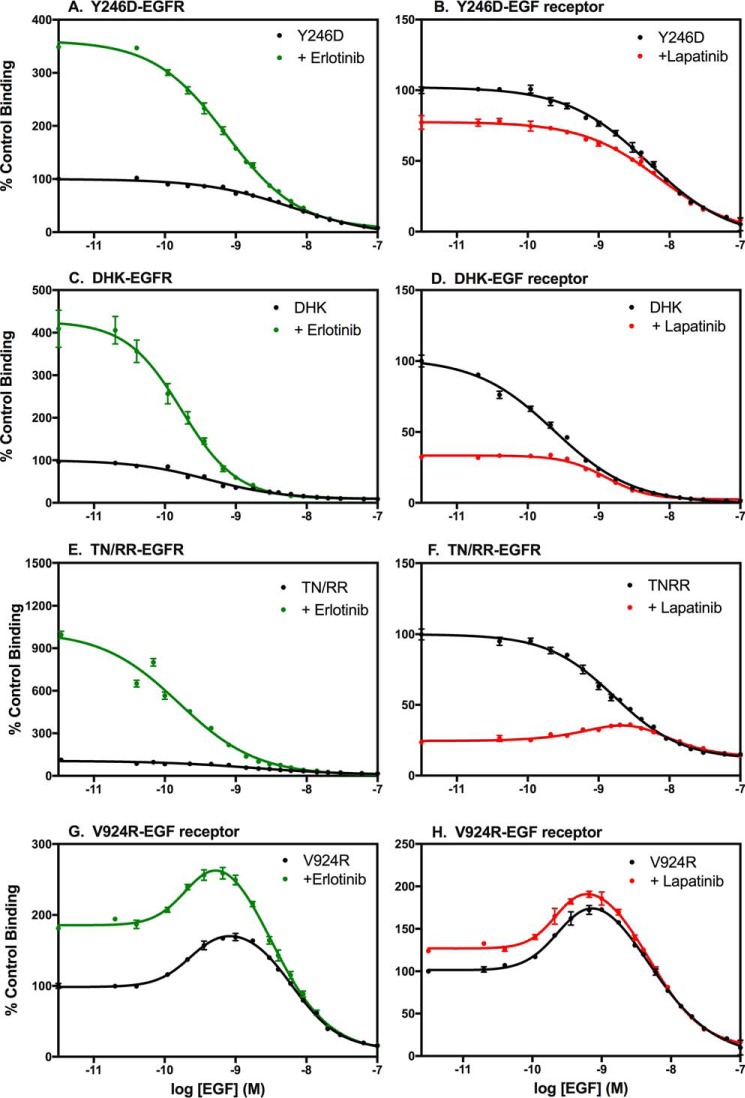
The Y246D-EGF receptor has a mutation in the dimerization arm of extracellular subdomain II that largely abolishes the ability of the receptor to form back-to-back dimers upon binding ligand (9, 10). As shown in Fig. 4A, the Y246D mutation markedly diminished, but did not eliminate, the ability of erlotinib to enhance the affinity of EGF. The effect of lapatinib on EGF binding also appeared to be blunted by the Y246D mutation (Fig. 4B). These data suggest that dimerization of the extracellular domain contributes to the effects of these inhibitors on EGF binding.
Figure 4.
Effect of erlotinib and lapatinib on EGF receptors with mutations in subunit–subunit interfaces. CHO cells expressing the Y246D-EGF receptor (A and B), the D563A,H566A,K585A-EGF receptor (C and D), the T548R,N554R-EGF receptor (E and F), or the V924R-EGF receptor (G and H) were treated for 30 min at 37 °C without (black lines) or with erlotinib (green lines) or lapatinib (red lines) prior to binding of 125I-EGF overnight at 4 °C. The indicated inhibitor was also included in the binding incubation medium. Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of two times.
The D563A,H566A,K585A mutation (referred to hereinafter as DHK-EGFR) is in the tethering arm in subdomain IV of the EGF receptor. This triple mutation removes the major interactions in the subdomain II–subdomain IV tether that holds the EGF receptor in the closed conformation (8). As can be seen in Fig. 4C, treatment with erlotinib increased the affinity of the DHK-EGF receptor for EGF ∼9-fold, similar to what is seen in the WT EGF receptor. Treatment of cells expressing the DHK-EGF receptor with lapatinib resulted in a typical ∼2-fold decrease in EGF affinity (Fig. 4D). Thus, based on saturation-binding curves this triple mutation had little effect on ligand binding other than a slight increase in affinity relative to the WT EGF receptor. This effect has been noted previously (8).
The EGF receptor has frequently been reported to form higher order oligomers (35–38). A recent study from the Kuriyan laboratory (38) implicated a portion of extracellular subdomain IV in the formation of EGF receptor multimers. In particular, a mutation in which Thr-548 and Asn-554 were each substituted with Arg (TN/RR-EGF receptor) significantly reduced the formation of multimers. To determine whether the effects of erlotinib or lapatinib were dependent on the ability of the EGF receptor to multimerize, we examined the effect of these inhibitors on the binding of EGF in cells expressing the TN/RR-EGF receptor. The data in Fig. 4, E and F, demonstrate that the behavior of this mutant was very similar to that of the WT EGF receptor, suggesting that the effect of these inhibitors on receptor affinity is not due to an effect on receptor multimerization.
We next determined whether dimerization of the kinase domain was required for the effects of erlotinib and lapatinib on ligand affinity. The V924R mutation of the EGF receptor alters the activator interface of the kinase domain, impairing its ability to form an activated asymmetric dimer (13). The ability of erlotinib to elicit an increase in EGF affinity was almost completely eliminated in the V924R-EGF receptor (Fig. 4G). Likewise, the ability of lapatinib to induce a decrease in receptor affinity was suppressed by this mutation (Fig. 4H). Thus, formation of the asymmetric kinase dimer appears to play a major role in the modulation of ligand-binding affinity by EGF receptor kinase inhibitors.
Fig. 5 shows the corresponding competition-binding curves for these mutants. As expected, the competition-binding curves for the Y246D-EGF receptor were monotonic in the absence of inhibitor and in the presence of erlotinib (Fig. 5A). The fact that erlotinib induced a significant increase in the binding of 125I-EGF at the lowest concentrations of unlabeled EGF competitor is consistent with the modest overall increase in affinity for EGF seen in the saturation-binding curves (Fig. 4A). This suggests that the effect of erlotinib does not depend entirely on the formation of a back-to-back dimer. Lapatinib treatment resulted in only a small decrease in 125I-EGF binding at low doses of competitor, again consistent with the small decrease in overall affinity seen in the saturation-binding curves (Fig. 4B). Notably, lapatinib did not induce a bell-shaped competition-binding curve, implicating the formation of back-to-back dimers in the positive cooperativity observed in the presence of this type II inhibitor.
Figure 5.
125I-EGF competition-binding curves of EGF receptors with mutations in subunit–subunit interfaces treated without or with erlotinib or lapatinib. CHO cells expressing the indicated EGF receptors were treated without (black lines) or with erlotinib (green lines) or lapatinib (red lines) prior to and during incubation with labeled EGF. Increasing concentrations of unlabeled EGF were added to different wells. The data are normalized to control binding. Control binding represents the number of counts of 125I-EGF bound by the untreated cells in the absence of any additional unlabeled EGF. Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of two times.
The data in Fig. 5, C and D, demonstrate that treatment of cells expressing the DHK-EGF receptor with erlotinib increased the amount of 125I-EGF bound at low doses of competitor, whereas treatment with lapatinib reduced the amount of 125I-EGF bound at low doses of competitor. This necessarily follows from the increase and decrease in affinity seen after treatment of these cells with erlotinib or lapatinib, respectively. Interestingly, treatment with lapatinib did not result in a bell-shaped competition-binding curve, suggesting that the positive cooperativity observed in other receptors may be related to release of the intramolecular tether in the extracellular domain of the receptor.
Mutation of the oligomerization interface in the TN/RR-EGF receptor did not affect the response of this receptor to either erlotinib (Fig. 5E) or lapatinib (Fig. 5F). Erlotinib induced a large increase in the amount of 125I-EGF bound at low doses of competitor. Lapatinib treatment resulted in a decrease in binding and produced a bell-shaped competition-binding curve, demonstrating the presence of positive cooperativity in this receptor. Thus, the observed cooperativity does not appear to be due to formation of higher order EGF receptor oligomers.
The competition-binding curves of the V924R-EGF receptor were highly unusual. Like the D813N-EGF receptor, the competition-binding curve for the V924R-EGFR was bell-shaped even in the absence of an inhibitor (Fig. 5G). But surprisingly, whereas there was a small increase in affinity upon addition of erlotinib, as evidenced by the increase in EGF binding, the competition-binding curve remained bell-shaped after treatment with this type I inhibitor. The V924R-EGF receptor was the only mutant that could not be converted to a monotonically decreasing competition-binding curve by the addition of erlotinib. Lapatinib did not significantly affect the competition-binding curve of the V924R-EGF receptor (Fig. 5H). These data suggest that the ability to form the asymmetric dimer is central to the effects of both erlotinib and lapatinib on EGF binding.
Effect of kinase inhibitors on mutationally activated EGF receptors
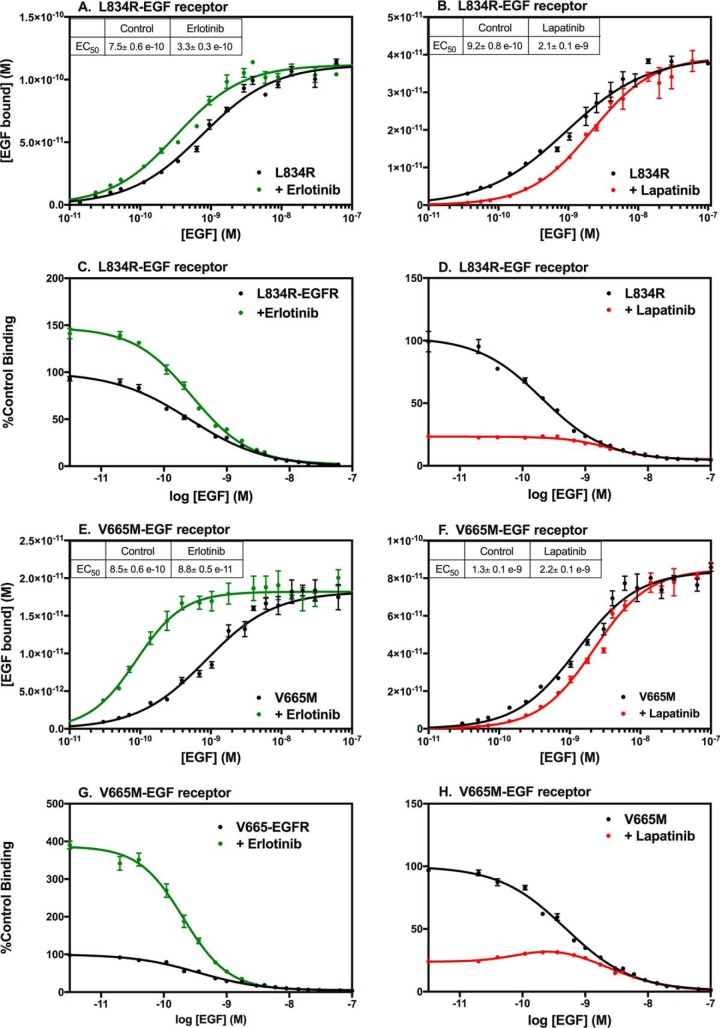
The foregoing data suggest that the effect of erlotinib on EGF binding is due to its ability to promote the formation of the active conformation of the kinase and the activating asymmetric dimer. L834R is a mutation in the kinase domain of the EGF receptor that is commonly seen in lung cancer (39). Shan et al. (40) reported that the L834R mutation activated the EGF receptor kinase by suppressing disorder in the α-C helix, thereby promoting the formation of the active asymmetric kinase dimer. If the ability of erlotinib to increase EGF-binding affinity (41) is due to its ability to order the α-C helix and promote the formation of the asymmetric dimer, we postulated that the effect of erlotinib would be blunted in the L834R-EGF receptor, which already favors the formation of asymmetric dimers under basal conditions.
Fig. 6A shows the effect of erlotinib on the binding of EGF to cells expressing the L834R-EGF receptor. In this cell line, the affinity for EGF in the untreated cells was noticeably higher (∼0.9 nm) than that observed in cells expressing the WT EGF receptor (∼3 nm). Consistent with our hypothesis, treatment with erlotinib resulted in only a 3-fold increase in EGF affinity for the L834R-EGF receptor, substantially less than the ∼10-fold seen in WT receptors. This reduction in the erlotinib effect is seen in the competition-binding curves (Fig. 6C), which are monotonic in the absence or presence of erlotinib. But erlotinib induced a relatively small increase in binding (∼50%) at low doses of cold competitor compared with what is normally seen in the WT EGF receptor (∼400%).
Figure 6.
Saturation-binding curves and 125I-EGF competition-binding curves of mutationally activated EGF receptors. CHO cells expressing the L834R-EGF receptor (A–D) or the V665M-EGF receptor (E–H) were treated without (black lines) or with erlotinib (green lines) or lapatinib (red lines) prior to and during incubation with increasing concentrations of 125I-EGF. Data are shown as saturation-binding curves (A, B, E, and F) or competition-binding curves (C, D, G, and H). Points represent the mean ± S.D. of triplicate determinations in a single experiment, which was repeated a minimum of three times.
Treatment of cells expressing the L834R-EGFR with lapatinib produced an ∼2-fold decrease in the EC50 for EGF (Fig. 6B), similar to what was seen for WT EGF receptors treated with lapatinib. This should be considered the minimum effect of lapatinib in this line as the L834R has a reduced affinity for lapatinib as compared with the WT EGF receptor (41). Interestingly, lapatinib failed to induce a bell-shaped competition-binding curve, suggesting that the mutation overcomes the effect of lapatinib on ligand binding.
Not all transforming mutations blunted the effect of erlotinib. The V665M-EGFR carries a point mutation in the juxtamembrane latch of the receptor that enhances the stability of the asymmetric kinase dimer, but does not affect the conformation of the α-C helix (42). Like the L834R-EGFR, the V665M-EGFR exhibited a slightly increased affinity for EGF in untreated cells (Fig. 6E). But in contrast to the L834R-EGFR, addition of erlotinib to cells expressing the V665M-EGFR resulted in a further ∼10-fold increase in affinity that is typical for the WT EGF receptor (Fig. 6G).
Lapatinib induced an ∼2-fold decrease in affinity in the V665M-EGFR (Fig. 6F) as reflected in the ∼75% decrease of 125I-EGF bound initially in the competition-binding curve (Fig. 6H). In addition, as in the WT EGF receptor, the V665M-EGFR exhibited a bell-shaped competition-binding curve in the presence of lapatinib. Thus, although both the L834R and V665M mutations activate the EGF receptor kinase, only the L834R mutation that affects the conformation of the α-C helix alters the effect of erlotinib on ligand binding.
Discussion
The binding of EGF promotes the dimerization and activation of the EGF receptor kinase domain. The experiments reported here demonstrate that, reciprocally, the binding of small-molecule inhibitors to the kinase domain allosterically regulate the function of the ligand-binding domain of the receptor. Previous studies that assessed the effect of small-molecule inhibitors on the extracellular domain of the EGF receptor looked only for structural changes and found none (43, 44). Our analysis of the effects of these inhibitors on ligand binding clearly demonstrates that there is significant functional linkage between the extracellular domain and the intracellular domain of the EGF receptor.
The binding of EGF to its receptor is known to promote the formation of a back-to-back dimer of the extracellular domains of the receptor. Our finding that the binding affinity for EGF is different in the presence of erlotinib or lapatinib suggests that the dimers formed in the presence of these two inhibitors are different. Indeed, Mi et al. (44) showed that in the presence of erlotinib, EGF induced the formation of a back-to-back dimer of the extracellular domain coupled to a rod-like, asymmetric kinase dimer of the intracellular domain. By contrast, in the presence of lapatinib, the extracellular domain adopted the back-to-back dimer configuration but the kinase domain formed distinct (inactive) globular dimers or unassociated monomers. Our data indicate that even though the extracellular domains appear to form structurally similar dimers when erlotinib or lapatinib are bound to the kinase domain, the distinct conformations of the kinase domains induced by these inhibitors differentially affect the ligand-binding properties of the extracellular domain dimers.
The difference in the conformation of the kinase domains in the erlotinib- versus lapatinib-bound dimers is likely responsible for the difference in allosteric interactions between the two subunits in the dimer. Pretreatment with lapatinib, but not with erlotinib, resulted in bell-shaped competition-binding curves, indicative of positive cooperativity. Positive cooperativity implies the existence of a dimer that binds the first ligand with low affinity but the second ligand with higher affinity. This is exactly the opposite of what is typically seen for the untreated EGF receptor, which exhibits negative cooperativity within receptor dimers (28, 45). This again underscores the functional coupling between the intracellular and extracellular domains of the EGF receptor.
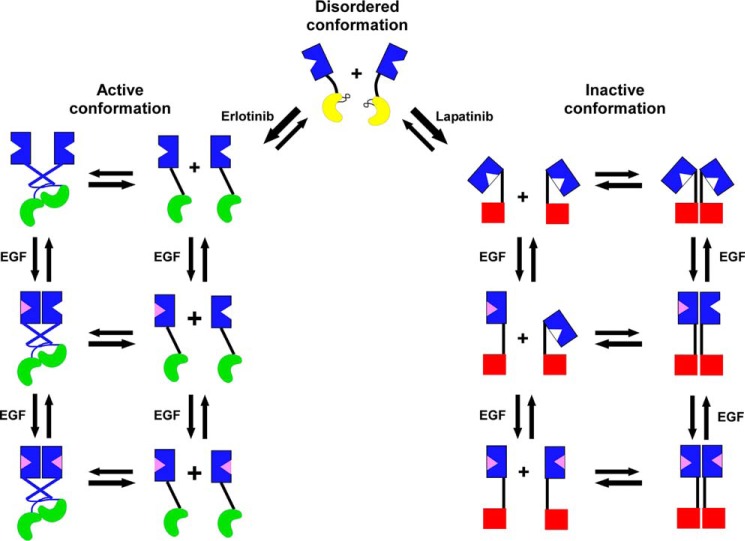
Fig. 7 shows a model of how we envision these kinase inhibitors modulating EGF binding. Erlotinib and gefitinib are type I inhibitors that preferentially bind to the active conformation of the kinase domain (17) and induce the formation of an asymmetric kinase dimer (13, 46) (shown in green in Fig. 7). By contrast, lapatinib and neratinib are type II inhibitors that bind to the inactive conformation of the kinase domain (18) (shown as red squares in Fig. 7).
Figure 7.
Schematic diagram of the proposed effects of erlotinib and lapatinib on the conformation and dimerization of the EGF receptor. The ordered, active conformation (green) and ordered, inactive conformation (red) of the kinase domain are in equilibrium with an intermediate conformation of the kinase domain in which the α-C helix is disordered (yellow). Erlotinib stabilizes the active conformation of the kinase domain, which can form asymmetric dimers and which is linked to the open conformation of the extracellular domain. Lapatinib stabilizes the inactive conformation of the kinase domain, which forms symmetric dimers, which are linked to the tethered conformation of the extracellular domain.
We interpret our data in the context of the report of Shan et al. (40) that EGF receptor kinase monomers exist in an equilibrium of three different conformations: (i) an active conformation of the kinase in which the α-C helix is ordered and swung in, (ii) a Src/CDK2-like inactive conformation in which the α-C helix is ordered but swung out and, (iii) an intermediate conformation in which the α-C helix is in a disordered conformation. We propose that, in the absence of ligand, the two ordered conformations of the intracellular kinase domain are linked to distinct conformations of the extracellular ligand-binding domain that exhibit different affinities for EGF. Specifically, the active conformation of the kinase is linked to an untethered, largely extended conformation of the extracellular domain that has a high affinity for EGF and the inactive conformation of the kinase is linked to a closed, likely tethered, conformation of the extracellular domain that has a low affinity for EGF. The disordered conformation of the kinase domain is associated with an intermediate conformation of the extracellular domain that may be untethered and can adopt either the open or closed conformation.
The population of EGF receptors in any given cells is an equilibrium mixture of all conformations of the receptor and this population exhibits binding characteristics that reflect the distribution of the receptor between high and low affinity forms. Type I inhibitors shift the equilibrium toward the active conformation of the kinase linked to the high affinity form of the extracellular domain. This leads to a population average increase in affinity. Type II inhibitors shift the equilibrium toward the inactive conformation of the kinase that is linked to the low affinity form of the kinase domain. This decreases the population average affinity. Mutations in the EGF receptor, such as V924R or L834R, can also shift the position of the basal equilibrium among these species, leading to changes in the apparent effect of erlotinib or lapatinib on receptor affinity.
A variety of data support the assignment of the extended form of the extracellular domain to the active conformation of the kinase domain. First, the EGF receptor is thought to be held in the closed conformation through the subdomain II–subdomain IV tether (8). It has been estimated that this tethering energy is enough to generate a 5–30-fold increase in affinity and such affinity increases have been documented when the tether is removed or mutated (8, 47, 48). The 10-fold increase in affinity that we see in the presence of erlotinib is in this range, consistent with the hypothesis that erlotinib destabilizes the subdomain II–subdomain IV tethering interaction, allowing the receptor to adopt a more open, extended conformation. In addition, tyrphostin, another type I tyrosine kinase inhibitor, has been shown to induce the formation of an untethered, transitional form of the EGF receptor that is recognized by mAb806 (49). mAb806 binds to an epitope encompassing residues 287–302 that is hidden in both the tethered and dimerized forms of the extracellular domain but available in the extended monomeric receptor (50). Finally, the L834R mutation has been shown to increase the tendency of the kinase domain to adopt its active conformation (40). Furthermore, Valley et al. (51) have reported that the L834R-EGF receptor is dimerized and its extracellular domain is in a more extended form than that of the WT receptor. In our experiments, treatment of the L834R-EGF receptor with erlotinib induced a smaller than usual increase in ligand affinity, suggesting that the L834R mutation and erlotinib treatment have similar and overlapping effects on receptor conformation and affinity. Together, these data suggest that type I inhibitors and mutations that alter the α-C helix and promote asymmetric dimer formation, induce the release of tethering interactions in the extracellular domain, allowing the extracellular domain of the receptor to adopt a more extended conformation.
Few data are available regarding the conformation of the ligand-free extracellular domain of the EGF receptor when the kinase domain is occupied by lapatinib. The inhibitor does not induce dimerization of the receptor (33, 46) so the conformation must be similar to that of the unoccupied EGF receptor monomer. In the absence of EGF, the soluble extracellular domain adopts the closed tethered conformation (8, 52). Bessman et al. (12) showed that Fc-mediated dimerization of the EGF receptor does not enforce the traditional back-to-back extended conformation on the soluble extracellular domain. Rather, it results in the formation of a mixture of conformations that contain the tethered form of the extracellular domain of the receptor. The closed form of the receptor would exhibit a lower affinity for EGF as some of the binding energy would be expended to open the receptor into a conformation that can bind ligand with both subdomains I and III. It therefore seems likely that the lapatinib-bound form of the receptor is linked to a closed form of the extracellular domain.
The results with the c′973-EGF receptor suggest that the C-terminal tail of the receptor is involved in setting the position of the conformational equilibrium of the receptor. In the absence of the tail, the population average affinity was low and not reduced by lapatinib, suggesting that the receptor had shifted toward its inactive conformation. By contrast, the nonphosphorylatable Y9F-EGF receptor behaved similarly to the WT receptor, indicating that the physical presence of the tail, not its phosphorylation, regulates the position of the conformational equilibrium of the receptor. This was unexpected because the C-terminal tail has been reported to exert an inhibitory influence on the EGF receptor kinase (33, 53). However, it is possible that in the absence of the tail, the binding of an inhibitory protein such as Mig6 (54) is facilitated and this shifts the equilibrium toward the inactive conformation.
The presence of positive cooperativity in lapatinib-bound dimers but not in erlotinib-bound dimers clearly suggests that the conformation of the kinase domain, and in particular the position of the α-C helix, significantly alters the subunit–subunit interactions in EGF-induced dimers. Binding of EGF to a lapatinib-occupied EGF receptor monomer occurs with low affinity and must induce formation of a singly occupied dimer in which the kinase domain forms an inactive symmetric dimer (44). The binding of EGF would stabilize the ligand-occupied subunit in its open, extended conformation. This may lead to conformational changes that weaken the tether in its partner receptor or may simply trap the other subunit when it momentarily flickers into an open conformation. This open singly occupied dimer must have a higher affinity for EGF than does the closed monomeric form. This would give rise to the observed positive cooperativity.
The V924R-EGF receptor exhibited positive cooperativity even in the absence of lapatinib. This is likely because this receptor can never form a proper asymmetric kinase dimer and so is found almost exclusively in the inactive conformation. Indeed, Mi et al. (44) showed that the EGF-bound V924R-EGF receptor was coupled to symmetric kinase dimers, just like the lapatinib-bound WT receptor. The D813N mutant also showed striking positive cooperativity in the absence of lapatinib. This suggests that the D813N mutation stabilizes the kinase domain in the Src/CDK2-like inactive state. In the structure of the symmetric kinase dimer (14), Asp-813 is close to Asp-831. If Asp-813 is converted to Asn, an Asn-813–Asp-831 hydrogen bond could form that would stabilize the inactive conformation of the kinase domain, potentially favoring symmetric dimer formation. Unlike the V924R-EGF receptor, the D813N-EGF receptor can form a stable asymmetric dimer and is shifted into this conformation by erlotinib.
The concept that a particular conformation of the intracellular kinase domain is linked to a specific conformation of the extracellular domain could aid in the rational design of combination therapies directed against the EGF receptor. For example, the inhibitory activity of lapatinib is likely to be enhanced by antibodies that stabilize the tethered form of the extracellular domain (55). By contrast, lapatinib treatment would impede the efficacy of antibodies that bind to the open conformation of the EGF receptor (50). The opposite would pertain to erlotinib, which has already been shown to increase the binding of mAb806, an antibody that is directed against an epitope available only in the untethered form of the EGF receptor (49). Thus, attention to structural coupling in the EGF receptor could enhance the synergy of extracellularly directed and intracellularly directed EGF receptor therapeutics.
Experimental procedures
Materials
The CHO-K1 Tet-On cell line, the pBI-Tet vector, and doxycycline were from Clontech (Mountain View, CA). FetalPlex was from Gemini Bioproducts. EGF was from Biomedical Technologies. Na125I was from Perkin Elmer. Erlotinib, gefitinib, and neratinib were from Selleck Chemicals. Lapatinib was from VWR. Cetuximab and pertuzumab were obtained from the Barnes-Jewish Hospital pharmacy.
Cells and tissue culture
All EGF receptors were expressed in CHO-K1 cells from pBI-Tet, using the Tet-on induction system from Clontech. The construction of these lines has been reported previously (28, 56–58). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FetalPlex, 1000 μg/ml of penicillin/streptomycin, 100 μg/ml of G418, and 100 μg/ml of hygromycin. Cells were plated onto lysine-coated 6-well dishes 24 h prior to use. The concentration of doxycycline used to induce receptor expression varied among cell lines but was chosen to yield a level of expression of about 200,000–300,000 receptors/cell.
125I-EGF synthesis and binding
125I-EGF was synthesized using the oxidative ICl procedure of Doran and Spar (59). For radioligand binding studies, cells in 6-well dishes were incubated for 30 min at 37 °C in DMEM, 50 mm HEPES, pH 7.4, containing 5 mg/ml of BSA in the absence or presence of 5 μm erlotinib, 10 μm lapatinib, 50 μm gefitinib, or 5 μm neratinib. Cells were then placed on ice, washed twice in ice-cold PBS, and incubated overnight at 4 °C in DMEM, 50 mm HEPES, pH 7.4, 5 mg/ml of BSA, without or with the indicated concentration of inhibitor and containing 20–40 pm 125I-EGF plus increasing concentrations of unlabeled EGF. Pertuzumab (5 μg/ml) was included in all binding assays to preclude heterodimerization of the EGF receptor with the few ErbB2 receptors present in these cells. We have shown that there are no ErbB3 or ErbB4 receptors in these CHO cells (26).
At the end of the incubation, cells were washed three times in ice-cold PBS and the monolayers dissolved in 1 n NaOH. The solution was transferred to tubes, which were counted for 125I in a Beckman Gamma Counter. Assays were done in triplicate.
For competition-binding curves, the data were fit to the log(inhibitor) versus response (variable slope) equation in GraphPad Prism 7,
| (Eq. 1) |
where Y is the cpm of 125I-EGF bound, X is the log of the concentration of cold, competing EGF, top and bottom are the fitted maximum and minimum counts, IC50 is the value of X that achieves half-maximal inhibition, and h is the Hill slope.
For competition-binding curves showing positive cooperativity, the data were fit to the equation for a bell-shaped dose-response curve,
| (Eq. 2) |
where X is the log of the concentration of unlabeled EGF, B0 is the cpm bound in the absence of added competitor, Bmax is the cpm bound at the peak of binding, B∞ is cpm bound at infinite [X], EC50_1 is the EC50 for the first (upward) part of the curve, h1 is the corresponding Hill slope, EC50_2 is the EC50 for the second (downward) part of the curve, and h2 is the corresponding Hill slope.
For saturation-binding curves, nonspecific binding was determined by fitting the raw data to a competition-binding model and using the fitted bottom value as the nonspecific binding. The data corrected for nonspecific binding were converted to direct binding curves using the known specific activity of the 125I-EGF and were fitted to the equation,
| (Eq. 3) |
where Y is ligand binding at concentration X of ligand. Bmax is the amount of ligand bound at saturating concentrations of ligand. EC50 is the concentration of ligand needed to achieve half-maximal binding and h is the Hill slope. Fitting was done using GraphPad Prism 7.
Author contributions
L. J. P. conceptualization; J. L. M.-O. and L. J. P. data curation; J. L. M.-O. and L. J. P. formal analysis; L. J. P. supervision; L. J. P. funding acquisition; J. L. M.-O. and L. J. P. investigation; J. L. M.-O. and L. J. P. methodology; L. J. P. writing-original draft; J. L. M.-O. and L. J. P. writing-review and editing.
This work was supported by National Institutes of Health Grant R01 GM108785 (to L. J. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EGF
- epidermal growth factor
- DMEM
- Dulbecco's modified Eagle's medium.
References
- 1. Ferguson K. M. (2008) Structure-based view of epidermal growth factor receptor regulation. Annu. Rev. Biophys. 37, 353–373 10.1146/annurev.biophys.37.032807.125829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lemmon M. A., Schlessinger J., and Ferguson K. M. (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harbor Perspect. Biol. 6, a020768 10.1101/cshperspect.a020768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J., Downward J., Mayes E. L. V., Whittle N., Waterfield M. D., and Seeburg P. H. (1984) Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature 309, 418–425 10.1038/309418a0 [DOI] [PubMed] [Google Scholar]
- 4. Downward J., Parker P., and Waterfield M. D. (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485 10.1038/311483a0 [DOI] [PubMed] [Google Scholar]
- 5. Hsuan J. J., Totty N., and Waterfield M. D. (1989) Identification of a novel autophosphorylation site (P4) on the epidermal growth factor receptor. Biochem. J. 262, 659–663 10.1042/bj2620659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Margolis B. L., Lax I., Kris R., Dombalagian M., Honegger A. M., Howk R., Givol D., Ullrich A., and Schlessinger J. (1989) All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. J. Biol. Chem. 264, 10667–10671 [PubMed] [Google Scholar]
- 7. Schlessinger J., and Lemmon M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 191, re12 [DOI] [PubMed] [Google Scholar]
- 8. Ferguson K. M., Berger M. B., Mendrola J. M., Cho H.-S., Leahy D. J., and Lemmon M. A. (2003) EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11, 507–517 10.1016/S1097-2765(03)00047-9 [DOI] [PubMed] [Google Scholar]
- 9. Garrett T. P. J., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H.-J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., and Ward C. W. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110, 763–773 10.1016/S0092-8674(02)00940-6 [DOI] [PubMed] [Google Scholar]
- 10. Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J.-H., Saito K., Sakamoto A., Inoue M., Shirouzu M., and Yokoyama S. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 10.1016/S0092-8674(02)00963-7 [DOI] [PubMed] [Google Scholar]
- 11. Arkhipov A., Shan Y., Das R., Endres N. F., Eastwood M. P., Wemmer D. E., Kuriyan J., and Shaw D. E. (2013) Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 10.1016/j.cell.2012.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bessman N. J., Bagchi A., Ferguson K. M., and Lemmon M. A. (2014) Complex relationship between ligand binding and dimerization in the epidermal growth factor receptor. Cell Rep. 9, 1306–1317 10.1016/j.celrep.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang X., Gureasko J., Shen K., Cole P. A., and Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 14. Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., and Kuriyan J. (2009) Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137, 1293–1307 10.1016/j.cell.2009.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu P., Nielsen T. E., and Clausen M. H. (2015) FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 36, 422–439 10.1016/j.tips.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 16. Bose R., and Zhang X. (2009) The ErbB kinase domain: structural perspectives into kinase activation and inhibition. Exp. Cell Res. 315, 649–658 10.1016/j.yexcr.2008.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stamos J., Sliwkowski M. X., and Eigenbrot C. (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 277, 46265–46272 10.1074/jbc.M207135200 [DOI] [PubMed] [Google Scholar]
- 18. Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., Alligood K. J., Rusnak D. W., Gilmer T. M., and Shewchuk L. (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 64, 6652–6659 [DOI] [PubMed] [Google Scholar]
- 19. Vivanco I., Robins H. I., Rohle D., Campos C., Grommes C., Nghiemphu P. L., Kubek S., Oldrini B., Chheda M. G., Yannuzzi N., Tao H., Zhu S., Iwanami A., Kuga D., Dang J., et al. (2012) Differential sensitivity of glioma versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2, 458–471 10.1158/2159-8290.CD-11-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pao W., and Chmielecki J. (2010) Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer 10, 760–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brandes A. A., Franceschi E., Tosoni A., Hegi M. E., and Stupp R. (2008) Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin. Cancer Res. 14, 957–960 10.1158/1078-0432.CCR-07-1810 [DOI] [PubMed] [Google Scholar]
- 22. Yun C.-H., Mengwasser K. E., Toms A. V., Woo M. S., Greulich H., Wong K. K., Meyerson M., and Eck M. J. (2008) The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. U.S.A. 105, 2070–2075 10.1073/pnas.0709662105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lichtner R. B., Menrad A., Sommer A., Klar U., and Schneider M. R. (2001) Signaling-inactive epidermal growth factor receptor/ligand complexes in intact carcinoma cells by quinazoline tyrosine kinase inhibitors. Cancer Res. 61, 5790–5795 [PubMed] [Google Scholar]
- 24. Yun C.-H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., and Eck M. J. (2007) Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11, 217–227 10.1016/j.ccr.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wells A., Welsh J. B., Lazar C. S., Wiley H. S., Gill G. N., and Rosenfeld M. G. (1990) Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science 247, 962–964 10.1126/science.2305263 [DOI] [PubMed] [Google Scholar]
- 26. Gill K., Macdonald-Obermann J. L., and Pike L. J. (2017) Epidermal growth factor receptors containing a single tyrosine in their C-terminal tail bind different effector molecules and are signaling-competent. J. Biol. Chem. 292, 20744–20755 10.1074/jbc.M117.802553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King A. C., and Cuatrecasas P. (1982) Resolution of high and low affinity epidermal growth factor receptors. J. Biol. Chem. 257, 3053–3060 [PubMed] [Google Scholar]
- 28. Macdonald J. L., and Pike L. J. (2008) Heterogeneity in EGF binding affinities arises from negative cooperativity in an aggregating system. Proc. Natl. Acad. Sci. U.S.A. 105, 112–117 10.1073/pnas.0707080105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Magun B. E., Matrisian L. M., and Bowden G. T. (1980) Epidermal growth factor: ability of tumor promoter to alter its degradation, receptor affinity and receptor number. J. Biol. Chem. 255, 6373–6381 [PubMed] [Google Scholar]
- 30. Shoyab M., De Larco J. E., and Todaro G. J. (1979) Biologically active phorbol esters specifically alter affinity of epidermal growth factor membrane receptors. Nature 279, 387–391 10.1038/279387a0 [DOI] [PubMed] [Google Scholar]
- 31. Coker K. J., Staros J. V., and Guyer C. A. (1994) A kinase-negative epidermal growth factor receptor that retains the capacity to stimulate DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 6967–6971 10.1073/pnas.91.15.6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arteaga C. L., Ramsey T. T., Shawver L. K., and Guyer C. A. (1997) Unliganded epidermal growth factor receptor dimerization induced by direct interaction of quinazolines with the ATP binding site. J. Biol. Chem. 272, 23247–23254 10.1074/jbc.272.37.23247 [DOI] [PubMed] [Google Scholar]
- 33. Bublil E. M., Pines G., Patel G., Fruhwirth G., Ng T., and Yarden Y. (2010) Kinase-mediated quasi-dimers of EGFR. FASEB J. 24, 4744–4755 10.1096/fj.10-166199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chantry A. (1995) The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J. Biol. Chem. 270, 3068–3073 [PubMed] [Google Scholar]
- 35. Clayton A. H. A., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., and Burgess A. W. (2005) Ligand-induced dimer-tetramer transition during the activation of cell surface epidermal growth factor receptor: a multidimensional microscopy analysis. J. Biol. Chem. 280, 30392–30399 10.1074/jbc.M504770200 [DOI] [PubMed] [Google Scholar]
- 36. Needham S. R., Roberts S. K., Arkhipov A., Mysore V. P., Tynan C. J., Zanetti-Domingues L. C., Kim E. R., Losasso V., Hirsch M., Rolfe D. J., Clarke D. T., Winn M., Clayton A. H. A., Pike L. J., Parker P. J., et al. (2016) EGFR oligomerization organizes kinase-active dimers into competent signaling platforms. Nature Commun. 7, 13307 10.1038/ncomms13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozer N., Barua D., Henderson C., Nice E. C., Burgess A. W., Hlavacek W. S., and Clayton A. H. A. (2014) Recruitment of the adaptor protein Grb2 to EGFR tetramers. Biochemistry 53, 2594–2604 10.1021/bi500182x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang Y., Bharill S., Karandur D., Peterson S. M., Marita M., Shi X., Kaliszewski M. J., Smith A. W., Isacoff E. Y., and Kuriyan J. (2016) Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife 5, e14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharma S. V., Bell D. W., Settleman J., and Haber D. A. (2007) Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 [DOI] [PubMed] [Google Scholar]
- 40. Shan Y., Eastwood M. P., Zhang X., Kim E. T., Arkhipov A., Dror R. O., Jumper J., Kuriyan J., and Shaw D. E. (2012) Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell 149, 860–870 10.1016/j.cell.2012.02.063 [DOI] [PubMed] [Google Scholar]
- 41. Wang Z., Longo P. A., Tarrant M. K., Kim K., Head S., Leahy D. J., and Cole P. A. (2011) Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat. Struct. Mol. Biol. 18, 1388–1393 10.1038/nsmb.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Red Brewer M., Yun C.-H., Lai D., Lemmon M. A., Eck M. J., and Pao W. (2013) Mechanism for activation of mutated epidermal growth factor receptors in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 110, E3595–E3604 10.1073/pnas.1220050110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu C., Mi L.-Z., Grey M. J., Zhu J., Graef E., Yokoyama S., and Springer T. A. (2010) Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell. Biol. 30, 5432–5443 10.1128/MCB.00742-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mi L.-Z., Lu C., Li Z., Nishida N., Walz T., and Springer T. A. (2011) Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 18, 984–999 10.1038/nsmb.2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarado D., Klein D. E., and Lemmon M. A. (2010) Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell 142, 568–579 10.1016/j.cell.2010.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu C., Mi L.-Z., Schürpf T., Walz T., and Springer T. A. (2012) Mechanisms for kinase-mediated dimerization of the epidermal growth factor receptor. J. Biol. Chem. 287, 38244–38253 10.1074/jbc.M112.414391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dawson J. P., Berger M. B., Lin C.-C., Schlessinger J., Lemmon M. A., and Ferguson K. M. (2005) Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell. Biol. 25, 7734–7742 10.1128/MCB.25.17.7734-7742.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walker F., Orchard S. G., Jorissen R. N., Hall N. E., Zhang H.-H., Hoyne P. A., Adams T. E., Johns T. G., Ward C. W., Garrett T. P., Zhu H. J., Nerrie M., Scott A. M., Nice E. C., and Burgess A. W. (2004) CR1/CR2 interactions modulate the functions of the cell surface epidermal growth factor receptor. J. Biol. Chem. 279, 22387–22398 10.1074/jbc.M401244200 [DOI] [PubMed] [Google Scholar]
- 49. Gan H. K., Walker F., Burgess A. W., Rigopoulos A., Scott A. M., and Johns T. G. (2007) The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. J. Biol. Chem. 282, 2840–2850 10.1074/jbc.M605136200 [DOI] [PubMed] [Google Scholar]
- 50. Johns T. G., Adams T. E., Cochran J. R., Hall N. E., Hoyne P. A., Olsen M. J., Kim Y.-S., Rothacker J., Nice E. C., Walker F., Ritter G., Jungbluth A. A., Old L. J., Ward C. W., Burgess A. W., Wittrup K. D., and Scott A. M. (2004) Identification of the epitope for the epidermal growth factor receptor-specific monoclonal antibody 806 reveals that it preferentially recognizes an untethered form of the receptor. J. Biol. Chem. 279, 30375–30384 10.1074/jbc.M401218200 [DOI] [PubMed] [Google Scholar]
- 51. Valley C. C., Arndt-Jovin D. J., Karedla N., Steinkamp M. P., Chizhik A. I., Hlavacek W. S., Wilson B. S., Lidke K. A., and Lidke D. S. (2015) Enhanced dimerization drives ligand-independent activity of mutant epidermal growth factor receptor in lung cancer. Mol. Biol. Cell 26, 4087–4099 10.1091/mbc.e15-05-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dawson J. P., Bu Z., and Lemmon M. A. (2007) Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure 15, 942–954 10.1016/j.str.2007.06.013 [DOI] [PubMed] [Google Scholar]
- 53. Kovacs E., Das R., Wang Q., Collier T. S., Cantor A., Huang Y., Wong K., Mirza A., Barros T., Grob P., Jura N., Bose R., and Kuriyan J. (2015) Analysis of the role of the C-terminal tail in the regulation of the epidermal growth factor receptor. Mol. Cell. Biol. 35, 3083–3102 10.1128/MCB.00248-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., and Kuriyan J. (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450, 741–744 10.1038/nature05998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmitz K. R., Bagchi A., Roovers R. C., van Bergen en Henegouwen P. M., and Ferguson K. M. (2013) Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure 21, 1214–1224 10.1016/j.str.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adak S., DeAndrade D., and Pike L. J. (2011) The tethering arm of the EGF receptor is required for negative cooperativity and signal transduction. J. Biol. Chem. 286, 1545–1555 10.1074/jbc.M110.182899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Adak S., Yang K. S., Macdonald-Obermann J., and Pike L. J. (2011) The membrane-proximal intracellular domain of the EGF receptor underlies negative cooperativity in ligand binding. J. Biol. Chem. 286, 45146–45155 10.1074/jbc.M111.274175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Macdonald-Obermann J. L., and Pike L. J. (2009) The intracellular juxtamembrane domain of the EGF receptor is responsible for the allosteric regulation of EGF binding. J. Biol. Chem. 284, 13570–13576 10.1074/jbc.M109.001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Doran D. M., and Spar I. L. (1980) Oxidative iodine monochloride iodination technique. J. Immunol. Methods 39, 155–163 10.1016/0022-1759(80)90304-X [DOI] [PubMed] [Google Scholar]