Abstract
Exposure to organophosphorus toxicants (OP) can have chronic adverse effects that are not explained by inhibition of acetylcholinesterase, the cause of acute OP toxicity. We therefore hypothesized that OP-induced chronic illness is initiated by the formation of organophosphorus adducts on lysine residues in proteins, followed by protein cross-linking and aggregation. Here, Western blots revealed that exposure to the OP chlorpyrifos oxon converted porcine tubulin from its original 55-kDa mass to high-molecular-weight aggregates. Liquid chromatography–tandem MS analysis of trypsin-digested samples identified several diethoxyphospho-lysine residues in the OP-treated tubulin. Using a search approach based on the Batch Tag program, we identified cross-linked peptides and found that these chemically activated lysines reacted with acidic amino acid residues creating γ-glutamyl-ϵ-lysine or aspartyl-ϵ-lysine isopeptide bonds between β- and α-tubulin. Of note, these cross-linked tubulin molecules accounted for the high-molecular-weight aggregates. To the best of our knowledge, this is the first report indicating that chlorpyrifos oxon–exposed tubulin protein forms intermolecular cross-links with other tubulin molecules, resulting in high-molecular-weight protein aggregates. It is tempting to speculate that chronic illness from OP exposure may be explained by a mechanism that starts with OP adduct formation on protein lysines followed by protein cross-linking. We further speculate that OP-modified or cross-linked tubulin can impair axonal transport, reduce neuron connections, and result in neurotoxicity.
Keywords: tubulin, neurotoxin, protein misfolding, mass spectrometry (MS), Western blot, crosslink, isopeptide, neurotoxicity, organophosphate, protein aggregation
Introduction
The accepted mechanism for acute toxicity from exposure to organophosphorus toxicants is inhibition of acetylcholinesterase (AChE)3 activity (1). AChE activity returns to normal levels within 1 month, but chronic illness can persist for a lifetime. Persistent conditions that develop after acute intoxication can be divided into three categories. The first is organophosphate-induced delayed neuropathy, which is initiated by inhibition and aging of neuropathy target esterase (2). Symptoms include muscular incoordination, numbness, tingling, fatigue, severe muscular weakness, and paralysis of the lower limbs (3). These are associated with degeneration of axons, along with demyelination. Only a subset of organophosphorus toxicants (OP), including leptophos, diisopropylfluorophosphate, mipafox, and tri-o-cresyl phosphate cause this condition (4). The second category has been described as a post-traumatic stress disorder and is characterized by decreases in intellectual functioning, academic skills, and flexibility of thinking. In addition there is memory loss and loss of simple motor skills. Subjects also develop problems with depression, irritability, confusion, and social withdrawal (4). The physiological basis for these persistent conditions remains unknown.
A third source of persistent symptomology comes from repeated exposure to low levels of organophosphorus pesticides, levels that are too low to observe acute symptoms of inhibition of AChE activity. This can lead to chronic illness in adults manifested as deficits in memory, attention, reaction time, and fine motor skills, plus emotional difficulties (5–8). Children exposed in utero to the OP chlorpyrifos can exhibit developmental abnormalities (9–11). Studies in cultured cells and laboratory rodents exposed to low levels of OP have provided evidence that multiple pathways are potentially affected by low doses of chlorpyrifos (CPS) and its active metabolite, chlorpyrifos oxon (CPO). These include altered receptor levels, adenylyl cyclase activity, and signal transduction activity, increased phosphorylation of Ca2+/cAMP-response element-binding protein, and impaired axonal transport (12–14). These studies did not identify molecular targets for CPS or CPO and did not propose a mechanism for neurotoxicity.
Covalent modification of noncholinesterase proteins may provide a rational basis for these symptomatic observations. Before 2007, it was generally accepted that OP reacted primarily with the active site serine of esterases and proteases. A limited number of reports had appeared describing reactions of OP with tyrosine in proteins such as human serum albumin (15), bromelain (16), papain (17), and hen egg white lysozyme (18). Beginning in 2007 we confirmed and amplified these observations using MS. We found OP adducts on both tyrosine and lysine in tubulin, albumin, casein, aprotinin, and keratin (19–23). The OP in these studies included soman, sarin, chlorpyrifos oxon, dichlorvos, diisopropylfluorophosphate, and a biotinylated OP probe, FP-biotin. Other laboratories have reported tyrosine adducts on albumin and other proteins (24–30).
Schmidt et al. (31) demonstrated that phosphonylation of lysine residues in ubiquitin by VX (O-ethyl methyl S-2-(diisopropylaminoethyl)phosphonothioate)) induced intramolecular cyclization through the formation of isopeptide bonds with nearby glutamate residues in the same protein subunit. The Schmidt et al. (31) report led us to the idea that CPO could induce cross-linking between subunits of tubulin. We chose to work with tubulin because tubulin is the major constituent of microtubules in the brain, microtubules being critical for cell morphology and cell division, and because of our previous results that showed modification of microtubule structure in mice treated with CPS (32). We searched for intermolecular cross-links between subunits because such cross-links would be expected to cause protein aggregation. We reasoned that aggregation of tubulin would disrupt microtubule structure, and would be a likely explanation for the altered microtubule structure we observed in mouse brain.
In the present report we treated porcine tubulin with CPO and examined the products for modifications by SDS-PAGE, Western blotting, and MS. SDS-PAGE and Western blotting revealed that treatment with CPO led to extensive tubulin aggregation. Mass spectrometry identified diethoxyphospho-lysine on both α- and β-tubulin. In addition, it showed that lysine residues susceptible to CPO-labeling were involved in isopeptide cross-links with glutamate and aspartate. Cross-links were found between residues on different peptides within the same form of tubulin and between residues on different forms of tubulin.
Results
Chlorpyrifos oxon induced aggregation of tubulin
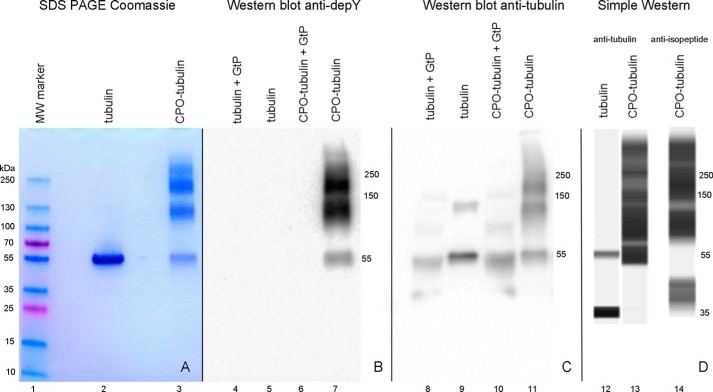
Fig. 1 shows that treatment of tubulin with 1.5 mm CPO leads to protein aggregation. Untreated control tubulin has a band at 55 kDa (lanes 2 and 9). Extensive aggregation can be seen for CPO-treated tubulin on the Coomassie-stained SDS-PAGE (lane 3), on the anti-diethoxyphosphotyrosine Western blotting (lane 7), on the anti-tubulin Western blotting (lane 11), and on the Simple Western using either the anti-tubulin antibody (lane 13) or the anti-isopeptide antibody (lane 14). A weak band at 130 kDa in lane 9 for untreated tubulin on the anti-tubulin Western blotting had no isopeptide cross-links. Treatment of tubulin or CPO-tubulin with γ-glutamyltranspeptidase reduced the size of the aggregate bands (lane 10), suggesting that isopeptide bonds are involved in aggregate formation. Taken together, these results argue that CPO promotes isopeptide bond cross-linking of tubulin monomers to make multimers.
Figure 1.
SDS-PAGE stained with Coomassie Blue (lanes 1–3), Western blotting on PVDF membrane probed with anti-diethoxyphosphotyrosine antibody depY (lanes 4–7), Western blotting probed with anti-tubulin antibody (lanes 8–11), and a capillary electrophoresis Simple Western image (lanes 12–14) probed with anti-tubulin antibody (lanes 12 and 13) or anti-isopeptide antibody (lane 14). All samples were reduced with DTT prior to electrophoresis. The band at 35 kDa for untreated tubulin (lane 12) was an artifact of the Simple Western procedure.
The anti-tubulin Western blotting reveals that after γ-glutamyltranspeptidase treatment the tubulin monomer (lane 8) is slightly smaller than untreated tubulin (lane 9). This suggests that some proteolysis may have taken place. We found that the commercial γ-glutamyltranspeptidase preparation was highly contaminated with other proteins.
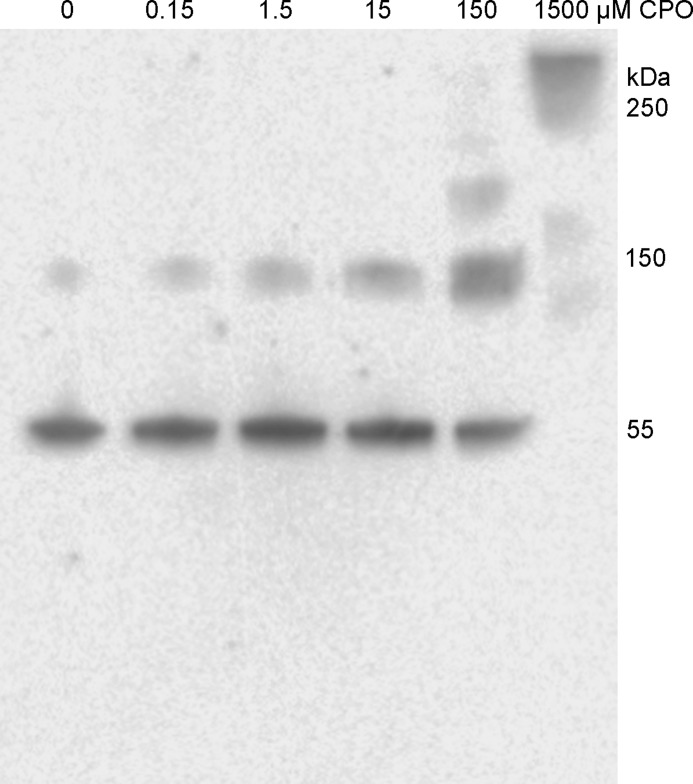
To clarify whether the high molecular species detected in the gels arose from multiple or single cross-linking events that are destabilized enough to form large aggregates we treated 0.25 mg/ml of tubulin with different concentrations of CPO (0, 0.15, 1.5, 15, 150, and 1500 μm). Fig. 2 shows that concentrations of 0.15, 1.5, and 15 μm CPO resulted in increasing amounts of trimer at 130 kDa. Treatment with 150 μm CPO caused higher molecular weight (200 to 300 kDa) aggregates to appear. At 1500 μm all of the tubulin was present as high-molecular-weight aggregates with no monomer and very little trimer (130 kDa). The change in distribution suggests that cross-linking initially forms trimers, but at sufficiently high concentrations of CPO multiple cross-links develop.
Figure 2.
Western blotting on PVDF membrane probed with anti-tubulin antibody. Tubulin (0.25 mg/ml) was treated with 0 to 1500 μm CPO. Each lane of the SDS gel was loaded with 1 μg of tubulin after the sample was reduced with DTT and denatured in the presence of SDS in a boiling water bath.
Organophosphorylated tubulin peptides
Trypsin-digested control tubulin and CPO-treated tubulin samples were separated by ultra HPLC prior to electrospray ionization MS on a Triple TOF 6600 mass spectrometer. The mass spectral data were searched against the uniprot_sprotJan2015.fasta database using the Paragon algorithm from Protein Pilot to identify the organophosphorylated peptides. Candidates were manually sequenced to confirm the Protein Pilot assignment and to identify the labeled amino acids.
We found no diethylphosphate-labeled peptides in control tubulin. In contrast CPO-treated tubulin contained the diethylphosphate-labeled peptides listed in Table 1.
Table 1.
Diethoxyphosphorylated lysine and tyrosine in CPO-treated tubulin
Only peptides with Protein Pilot Confidence values of 95% or greater are listed.
| Peptidea | Tubulin UniProt | Labeled AA | Residue | MS/MS | Ratiob label/total |
|---|---|---|---|---|---|
| QLFHPEQLITGK*EDAANNYAR | P02550 α1A | Lysine | 96 | Yes | 0.099 |
| GHYTIGK*EIIDLVLDR | P02550 | Lysine | 112 | Yes | 0.34 |
| LSVDYGK*K | P02550 | Lysine | 163 | Yes | 0.28 |
| LSVDYGK*K*SK | P02550 | Lysine | 163, 164 | Yes | – |
| HGK*YMACCLLYR | P02550 | Lysine | 311 | Yes | 0.64 |
| DVNAAIATIK*TK | P02550 | Lysine | 336 | Yes | 0.052 |
| DVNAAIATIK*TK*R | P02550 | Lysine | 336, 338 | Yes | – |
| VGINYEPPTVVPGGDLAK*VQR | P02550 | Lysine | 370 | No | 0.35 |
| LDHK*FDLMYAK | P02550 | Lysine | 394 | Yes | 0.33 |
| FDLMYAK*R | P02550 | Lysine | 401 | Yes | 0.31 |
| IHFPLATYAPVISAEK*AYH | Q2XVP4 α1B | Lysine | 280 | Yes | 0.083 |
| K*VGINYQPPTVVPGGDLAK | Q2XVP4 | Lysine | 352 | Yes | 0.043 |
| ISVYYNEATGGK*YVPR | Q767L7 β | Lysine | 58 | No | 0.011 |
| K*GHYTEGAELVDSVLDVVR | Q767L7 | Lysine | 103 | No | 0.19 |
| QLTHSLGGGTGSGMGTLLISK*IR | Q767L7 | Lysine | 154 | Yes | 0.29 |
| IMNTFSVVPSPK* | Q767L7 | Lysine | 174 | Yes | 0.012 |
| TLK*LTTPTYGDLNHLVSATM | Q767L7 | Lysine | 216 | Yes | 0.50 |
| K*LAVNMVPFPR | Q767L7 | Lysine | 252 | Yes | 0.18 |
| MSMK*EVDEQMLNVQNK | Q767L7 | Lysine | 324 | No | 0.18 |
| EVDEQMLNVQNK*NSSYFVEWIPNNVK | Q767L7 | Lysine | 336 | No | 0.011 |
| GLK*MAVTFIGNSTAIQELFK | Q767L7 | Lysine | 362 | Yes | 0.50 |
| STAIQELFK*R | Q767L7 | Lysine | 379 | Yes | 0.27 |
| K*AFLH | Q767L7 | Lysine | 392 | Yes | 1.00 |
| ALTVPELTQQMFDAK*NMMAACDPR | P02554 β | Lysine | 297 | No | 0.0059 |
| EDAANNY*AR | P02550 α1A | Tyrosine | 103 | Yes | 0.35 |
| LSVDY*GK | P02550 | Tyrosine | 161 | Yes | 0.41 |
| LEFSIY*PAPQVSTAVVEPYNSILTTH | P02550 | Tyrosine | 172 | No | 0.14 |
| LDIERPTY*TNLNR | P02550 | Tyrosine | 224 | Yes | 0.50 |
| EFQTNLVPY*PR | P02550 | Tyrosine | 262 | Yes | 0.12 |
| Y*APVISAEK | P02550 | Tyrosine | 272 | Yes | 0.33 |
| Y*MACCLLYR | P02550 | Tyrosine | 312 | Yes | 0.34 |
| VGINY*EPPTVVPGGDLAK | P02550 | Tyrosine | 357 | No | 0.29 |
| AFVHWY*VGEGM | P02550 | Tyrosine | 408 | Yes | 0.18 |
| ISVY*YNEATGGK | Q767L7 β | Tyrosine | 50 | Yes | 0.079 |
| ISVYY*NEATGGK | Q767L7 | Tyrosine | 51 | No | 0.10 |
| ISVYYNEATGGKY*VPR | Q767L7 | Tyrosine | 59 | No | 0.50 |
| GHY*TEGAELVDSVLDVVR | Q767L7 | Tyrosine | 106 | Yes | 0.17 |
| Y*LTVAAVFR | Q767L7 | Tyrosine | 310 | Yes | 0.34 |
| NSSY*FVEWIPNNVK | Q767L7 | Tyrosine | 340 | Yes | 0.28 |
| AFLHWY*TGEGMDEM | Q767L7 | Tyrosine | 398 | Yes | 0.57 |
a Peptides were from a tryptic digestion. Diethoxyphosphorylated residues are followed by an asterisk (*).
b The ratio is the number of times a peptide containing a diethoxyphosphorylated residue appears in the mass spectral data divided by the total number of times a peptide containing that residue appears. Peptides containing 2 labeled lysines are indicated by a dash in the last column.
Tubulin α1A (P02550) and α1B (Q2XVP4) each contain 19 lysines in homologous positions. Database searching with Protein Pilot found 14 of these lysines to be labeled by CPO. Twelve of the labeled peptides were identified with confidence values of 95% or greater. These are shown in Table 1. Peptide IHFPLATYAPVISAEKAYH is unique to α1B by virtue of the Ala to Ile substitution at position 1. The ratio of labeled to unlabeled peptides in this group ranged from 0.043 to 0.64. The sequences and labeled residues for 11 peptides were confirmed by manual sequencing of the mass spectral fragmentation spectra (Table 1). α1A- and α1B-tubulins also contain 19 tyrosines. Thirteen were found to be labeled by CPO, 9 with confidence values of 95% or greater (at ratios of 0.12 to 0.50), of which 7 were confirmed by manual sequencing.
Tubulin β (P02554 and Q767L7) contains 15 lysines. Thirteen lysines were found to be labeled by CPO, 12 with confidence values of 95% or greater (at ratios of 0.0059 to 1.00), of which 7 were confirmed by manual sequencing (Table 1). Peptide ALTVPELTQQMFDAKNMMAACDPR is unique to β P02554 by virtue of the Val to Met substitution at position 11. β-Tubulins also contain 16 tyrosines. Ten tyrosines were found to be labeled by CPO, 7 with confidence values of 95% or greater (at ratios of 0.079 to 0.57), of which 5 were confirmed by manual sequencing. These observations clearly demonstrate that both lysine and tyrosine are labeled by CPO and that the efficiency of labeling varies considerably from residue to residue.
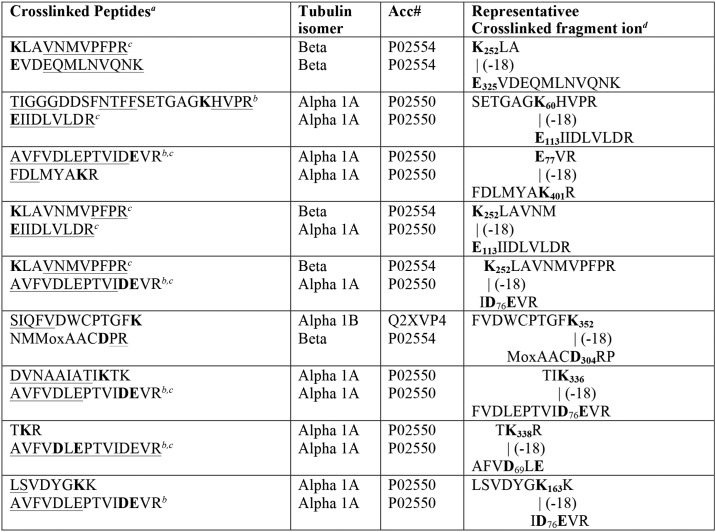
Cross-linked tubulin peptides
We found no isopeptide cross-linked peptides in control tubulin. By contrast, CPO-treated tubulin contained the cross-linked peptides listed in Table 2. Mass spectra of the digests were obtained by electrospray ionization using a Triple TOF 6600 mass spectrometer. Cross-links were identified using the Batch Tag and Search Compare algorithms from ProteinProspector. Batch Tag searches were constrained to find isopeptide cross-links: (a) between lysine and glutamate, aspartate, glutamine, or asparagine; or (b) between lysine and any amino acid. Cross-linked peptide candidates reported by Search Compare were confirmed by manual sequencing. The same isopeptide bonds were found when Batch Tag was configured to identify cross-links between lysine and any amino acid, or between lysine and glutamate, aspartate, glutamine, or asparagine. Cross-links between α- and β-tubulin were observed. When both peptides in a cross-link were from β-tubulin or from α-tubulin, we could not determine whether the peptides came from the same or distinct molecules.
Table 2.
Summary of cross-linked tubulin peptides
a Sequences for the parts of each peptide that were observed in the MS/MS spectrum are underlined (singly-charged sequence and doubly-charged sequence). Residues involved in the crosslink are shown in bold. When there was a choice for labeled residue that could not be resolved, both candidate residues are shown in bold. Cross-linked lysines were diethoxyphosphorylated prior to forming the isopeptide bond.
b An acidic or basic residue near the cross-link promotes isopeptide bond formation.
c This peptide or a portion of this peptide is used in more than one crosslinked pair.
d A crosslinked fragment ion from the MS/MS spectrum is shown. Numbering indicates the position of the residue in the primary sequence. The numbering includes the leader sequence. The mass of the cross-linked peptides is the sum of the two peptides minus 18 Da for the isopeptide linkage.
The MS/MS fragmentation spectrum for each cross-linked peptide contained portions of sequence from each peptide in the pair (underlined in Table 2) plus masses consistent with cross-linked portions from both peptides (a representative fragment ion from the MS/MS spectrum for each cross-linked peptide is shown in Table 2). In most cases the cross-linked fragments appeared in a sequence of doubly-charged masses.
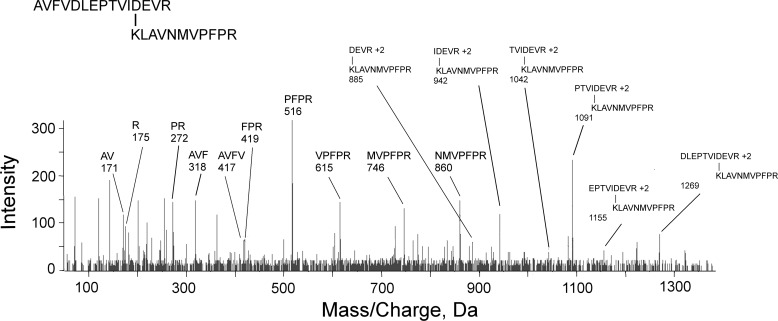
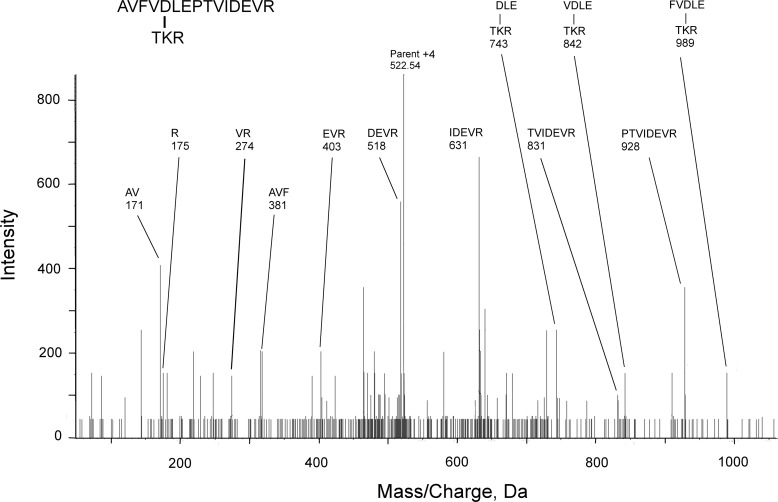
A representative MS/MS fragmentation spectrum for a cross-linked peptide is in Fig. 3. This spectrum is for the cross-linked peptides KLAVNMVPFPR (β, P02554) and AVFVDLEPTVIDEVR (α1A, P02550). It consists of three components: 1) the N-terminal b-series AVFV from AVFVDLEPTVIDEVR; 2) the C-terminal y-series NMVPFPR from KLAVNMVPFPR; and 3) a doubly-charged series for masses, which are too large to correspond to either peptide by itself. The doubly-charged series matches the sequence from AVFVDLEPTVIDEVR. Those masses decrease in the order DLEPTVI, which indicates that this is a y-series. Each observed doubly-charged mass is a combination of the entire mass of KLAVNMVPFPR and a fragment from AVFVDLEPTVIDEVR. Calculating the observed doubly-charged mass from sequence information is illustrated for the 942.86 mass. This mass is equal to the neutral mass for KLAVNMVPFPR, [M0] = 1270.72 Da; plus the neutral mass for the IDEVR peptide, [M0] = 613.33 Da; minus 18 Da for the isopeptide linkage; plus 2 protons; divided by 2. Similar calculations can be made for doubly-charged masses 885, 1042, 1091, 1155, and 1269. Identifying the residues involved in the cross-link is based on the following logic. The only lysine residue in the pair of peptides is Lys-1 from KLAVNMVPFPR, which makes this the lysine component of the isopeptide bond and defines AVFVDLEPTVIDEVR as the peptide containing the acidic component. AVFVDLEPTVIDEVR contains 4 acidic residues that could be the acidic component of the isopeptide bond. Residues Asp-5 and Glu-7 are missing from cross-linked fragments 885 to 1091 m/z and therefore cannot be part of the isopeptide bond. Fig. 3 shows Asp-12 cross-linked to Lys, but this assignment does not rule out the possibility that the cross-link is to the adjacent Glu.
Figure 3.
A mass spectral fragmentation spectrum for the cross-linked peptides AVFVDLEPTVIDEVR from tubulin α1A (P02550) and KLAVNMVPFPR from tubulin β (P02554). Parent ion 739.4 [M + 4H]+4 was not detected in the fragmentation spectrum. Unlabeled masses correspond to internal fragments, immonium ions, and b- or y-ions minus water and/or amine. The mass of cross-linked peptides is decreased by 18 Da due to loss of a water molecule upon formation of the isopeptide bond.
The MS/MS spectra for eight of the nine cross-linked peptides in Table 2 contained doubly-charged fragments from which the cross-linked structure was determined. One spectrum contained only singly-charged fragments. Fig. 4 shows the MS/MS spectrum for the cross-linked peptides AVFVDLEPTVIDEVR (α1A, P02550) and TKR (α1A, P02550). As with the spectra containing doubly-charged species, this spectrum consists of three components: 1) the N-terminal b-series AVF from AVFVDLEPTVIDEVR; 2) the C-terminal y-series PTVIDEVR from AVFVDLEPTVIDEVR; and 3) a singly-charged series that describes the fragmentation of the cross-linked peptide TKR-FVDLE, where the cross-link is between Lys and Asp. Three cross-linked masses are visible. All three contain both an Asp and a Glu residue making either of these residues potential cross-linking partners for the Lys.
Figure 4.
A mass spectral fragmentation spectrum for the cross-linked peptides AVFVDLEPTVIDEVR from tubulin α1A (P02550) and TKR from tubulin α1A (P02550). Most unlabeled masses correspond to internal fragments, immonium ions, and b- or y-ions minus water and/or amine.
All of the lysines involved in these cross-links are also targets for labeling by CPO. All of the lysine partners are either glutamate or aspartate. There is substantial redundancy in the peptides participating in these cross-links. Glutamate 77 or the adjacent aspartate 76, from peptide AVFVDLEPTVIDE77VR, appears in five of the cross-linked pairs, and glutamate 113 from peptide 113EIIDLVLDR is in 2 pairs. On the other side of the cross-link, lysine 252, from peptide 252KLAVNMVPFPR, is part of 3 pairs. Three of the cross-links involve peptides from two different types of tubulin. These cross-links would be expected to promote aggregation. Whether the α-α and β-β cross-links occur between peptides in the same tubulin subunit or between peptides in different subunits is unclear.
Cross-linked peptides in X-ray structure
The cross-linked peptides were located in the X-ray structure of porcine microtubule described by Alushin et al. (33) (Protein Data Bank 3J6G). This structure consists of three rows of tubulin α-β dimers with each row containing three α-β dimers for a total of 18 tubulin subunits total. All possible combinations of α-β, α-α, and β-β interactions are represented. Fig. 5 shows a single α-β pair with two intermolecular cross-links, one between Asp-76 from peptide AVFVDLEPTVID76EVR and Lys-252 from peptide 252KLAVNMVPFPR and another between Glu-113 from peptide 113EIIDLVLDR and Lys-252. Both of these cross-links are located at the interface between the α and β subunits.
Figure 5.
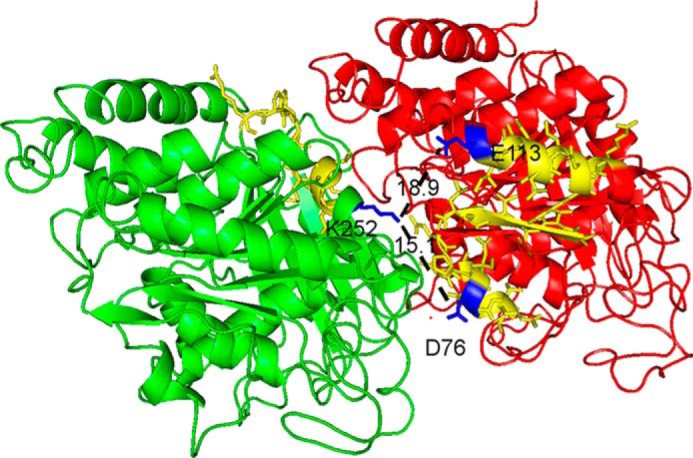
Crystal structure showing the cross-link between Lys-252 on tubulin β and Asp-76 on tubulin α1A, and the cross-link between Lys-252 on tubulin β and Glu-113 on tubulin α1A. β-Tubulin is shown in green, α1A in red. The cross-linked peptides are shown in yellow. Lysine 252, aspartate 76, and glutamate 113 are shown in blue. The distance between Lys-252 and Asp-76 is 15.1 Å and between Lys-252 and Glu-113 is 18.9 Å, indicated by dashed lines.
The distance between Asp-76 and Lys-252 is 15.1 Å. The distance between Glu-113 and Lys-252 is 18.9 Å. Although these distances are long for covalent bond formation, it is worth noting that both KLAVNMVPFPR and AVFVDLEPTVIDEVR are in short α-helices connected via long random coils. EIIDLVLDR is in an α-helix that is connected by two stretches of random coil. All three peptides would be expected to be flexible in solution making it reasonable to suggest that interaction is more likely than the crystal structure would indicate.
Interaction distances for the other cross-linked peptides in Table 2 range from 24 to 54 Å. This observation suggests the microtubule structure in solution is loose enough to allow subunits to move about and interact in ways not apparent from the X-ray structure.
Discussion
Mechanism of cross-link formation
Exposure of tubulin to chlorpyrifos oxon results in the modification of the ϵ-amine of lysine or the aromatic hydroxyl of tyrosine. The chemically-activated lysine residues promoted the formation of isopeptide cross-links. This statement is based on the observation that all of the isopeptide cross-linked peptides we have detected were connected through lysines that were also labeled by CPO. Isopeptide bonds were found between lysine and glutamate or aspartate. Cross-links were formed between peptides within a tubulin subunit or between peptides from different tubulin subunits. See Table 2 for details. The observation of CPO-promoted formation of intermolecular cross-links between peptides is new as of this writing. However, Schmidt et al. (31) reported that VX promoted intramolecular cyclization within ubiquitin subunits by formation of isopeptide bonds between lysine and glutamic acid.
Intermolecular isopeptide bonds have long been known to stabilize protein structure. Isopeptide bond formation is generally the result of enzyme-mediated reactions between the ϵ-amine from lysine on one protein and either glutamate or glutamine on another. Enzymes involved include transglutaminase (34, 35), sortase (36), the sumoylation complex (37), and the ubiquitin complex (38). By contrast, OP-mediated isopeptide bond formation is not catalyzed by an enzyme. Whether produced by an enzyme or by a chemically-activated lysine, isopeptide bonds are insensitive to reduction (39), resistant to hydrolysis by common proteases (40, 41), and resistant to low voltage collision-induced dissociation in MS (42).
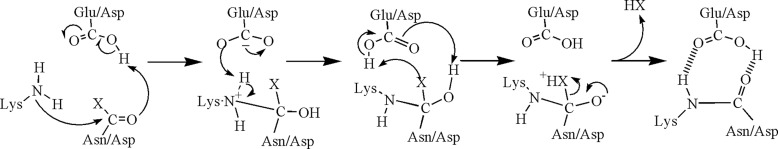
Intramolecular isopeptide bonds are a more recent discovery (43). They were first reported in 2007 (44) for bacterial cell-surface proteins. They differ from the intermolecular isopeptide bonds in that they form autocatalytically. The proposed mechanism involves a nucleophilic attack by an unprotonated lysine amine on the carbonyl carbon from a nearby asparagine or aspartate. Glutamate or aspartate serves as a proton shuttle (Fig. 6). The reaction occurs in a hydrophobic environment that can alter the pKa values of the ionizable groups (43).
Figure 6.
Autocatalytic isopeptide bond formation mechanism, where X is an amine or a hydroxyl (43).
The finding by Schmidt et al. (31) that the organophosphonate VX promotes the formation of intramolecular isopeptide bonds in ubiquitin is another example of an autocatalytic mechanism. Our finding that the organophosphorus toxicant chlorpyrifos oxon promotes intermolecular isopeptide bond formation supports the finding of Schmidt et al. (31). In neither study did the untreated protein exhibit isopeptide bonds. This latter observation lends credence to the proposal that OP-labeling is an essential preliminary step in isopeptide bond formation.
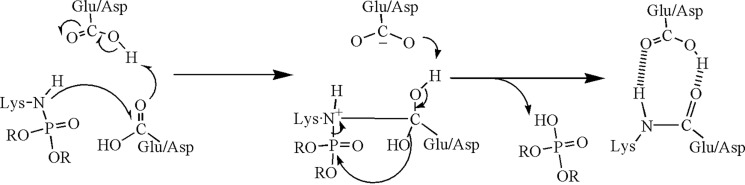
The mechanism of Kang and Baker (43) (Fig. 6) emphasizes the importance of a vicinal acidic group that can shuttle protons and stabilize charged states, thereby promoting the reaction. This concept can be adapted to rationalize OP-promoted isopeptide bond formation. Eight of the 9 cross-linked peptides in Table 2 contain a glutamate or an aspartate close to the cross-linked amino acids. This acidic residue could serve as a proton shuttle similar to the vicinal acidic residue in the Kang mechanism. Because isopeptide bond formation does not occur in the absence of OP it clearly implicates OP in the mechanism of isopeptide bond formation. It is tempting to suggest that OP promotes isopeptide bond formation because the phosphonyl moiety is a better leaving group than hydroxyl or amine in the Kang mechanism. A tentative mechanism is shown in Fig. 7.
Figure 7.
Organophosphate-promoted isopeptide bond formation between lysine and glutamate or aspartate. A nearby acidic residue serves as a proton shuttle.
The hydroxyl group on tyrosine can also be covalently modified by CPO. Using LC-MS/MS and the Batch Tag search method we have identified a few cross-linked peptides involving diethoxyphosphotyrosine. These will be described in a separate report.4
Protein aggregation induced by OP
The formation of cross-links between tubulin subunits caused tubulin to aggregate. The extent of aggregation depended on the concentration of CPO used to treat the tubulin. Aggregation was most extensive at 1.5 mm CPO. However, even at 1.5 μm CPO cross-linked trimers were apparent (Fig. 2). The presence of tubulin cross-links explains the CPO-induced high-molecular-weight aggregates observed on SDS-PAGE in Fig. 1. Although the present study focused on tubulin, we expect that cross-linking in vivo is likely to occur between nonidentical proteins, for example, between tubulin and microtubule-associated proteins.
The 1.5 mm CPO concentration used for most of our experiments is high. Low level exposure in humans is expected to correspond to about 25 nm CPO, a dose that would inhibit 50% of the plasma butyrylcholinesterase, but would not be accompanied by toxic symptoms or AChE inhibition. Humans poisoned with chlorpyrifos had up to 12 μm CPO in blood at the time of hospital admission 5 h post-ingestion (45). The cross-linking observed at 1.5 mm CPO in our experiments is far more extensive than could be expected from low dose exposures. The primary purpose of our experiments was to demonstrate that CPO-labeled lysine could promote cross-linking. We labeled with a high level of CPO to ensure that CPO-labeled lysine was formed. Once diethoxyphospholysine has been formed the cross-linking reactions are independent of the CPO concentration. Our experiments show that CPO adducts on lysine are capable of cross-linking proteins. This finding is a proof of that principle. We also showed that 1.5 μm CPO was capable of forming cross-linked trimers. We hypothesize that exposure to lower doses will form only a few cross-links, and that the cross-linked proteins could accumulate as misfolded proteins and lead to neurotoxicity.
Medical relevance of tubulin modification, implications for neurotoxicity
Both simple modification of tubulin by OP and tubulin aggregation would be expected to disrupt the structure and function of tubulin in neurons. This in turn could lead to the neurodevelopmental problems observed in children and the chronic neurotoxicity observed in adults who have been exposed to OP. Tubulin is only an example of proteins that could become aggregated when exposed to OP. OP-mediated cross-linking and aggregation of other proteins is likely to occur.
OP-induced protein aggregation fits well into the Brady hypothesis (46, 47) for neurodegenerative diseases. Brady and co-workers (46, 47) have developed a unifying hypothesis to explain neurodegenerative diseases including Alzheimer's, Parkinson's, Huntington's, and prion diseases as follows. 1) Aggregated proteins stimulate the activity of kinases. 2) The activated kinases phosphorylate motor proteins in excess. 3) The excessively phosphorylated motor proteins drop their cargo. 4) Loss of cargo deprives neurons of constituents that maintain neuron function. 5) Neurons slowly lose their connections to other neurons, which results in 6) clinical symptoms. The Brady hypothesis proposes that specific kinase networks and specific neurons are involved in each disease, but that the overall sequence of events is shared. Aggregated proteins induce excessive phosphorylation of proteins involved in axonal transport, which in turn disrupts fast axonal transport and causes neurotoxicity. We hypothesize that chronic neurotoxicity from low level CPO exposure fits the Brady model, with the difference being that “step 0” is OP-adduct formation.
Despite the proposed connection between the Brady hypothesis and our tubulin results, the correlation is highly speculative. Neurological sequelae due to exposure to CPO might be the consequence of CPO modification/aggregation of other proteins. It is also important to note that all protein aggregates are not the same (48). There are three major types of protein aggregates: amyloid fibrils, amorphous deposits, and native-like deposits. Each is associated with different types of disease. Amorphous deposits and amyloid fibrils are assemblages of small peptides. Native-like deposits derive from fully-folded proteins (48). The tubulin aggregates described in the current work would be most closely related to the native-like deposits. None of the protein aggregates associated with neurodegenerative disorders are believed to require covalent cross-linking for initiation (48).
The published literature on the effects of OP treatment fits the Brady hypothesis by supporting a link between OP exposure, hyperphosphorylation of proteins involved in axonal transport, impaired axonal transport, and chronic illness. Hyperphosphorylation following OP treatment has been reported for neurofilaments (49), Tau protein (50), calcium/cAMP-response element-binding protein (13), tubulin and microtubule-associated protein 2 (51). Studies in cell-free systems and in animals provide evidence for impaired axonal transport and decreased neuron synaptic spine density as consequences of treatment with OP (14, 52, 53). Our results from the current work suggest that chronic neurotoxicity from OP exposure could be initiated by OP-adduct formation followed by isopeptide bond formation and protein aggregation.
Materials and methods
CPO labeling and trypsin digestion of tubulin
One milligram of porcine tubulin (lyophilized powder >99% pure; Cytoskeleton, Denver, CO, catalog number T240) was dissolved in 3.6 ml of 20 mm Tris/Cl buffer, pH 8.5, containing 0.01% sodium azide to give 0.275 mg of tubulin/ml. This was mixed with 2.5 μl of 0.3 m chlorpyrifos oxon (Chem Service, West Chester, PA, catalog number MET-11459B) in ethanol to make 1.5 mm CPO. The reaction was incubated for 8 days at 24 °C. Tubulin remained in solution. Excess CPO was separated from 50 μl of protein by size exclusion chromatography on a Prob Quant G50 micro-column (GE Healthcare catalog number 28-9034-08) while simultaneously changing the buffer to 20 mm ammonium bicarbonate, pH 8. CPO-tubulin in 20 mm ammonium bicarbonate was digested with 0.5 μl of porcine trypsin (0.4 μg/μl, sequencing grade modified; Promega, Madison, WI, catalog number V511C) at 37 °C overnight. The digest was dried and resuspended in 20 μl of 0.1% formic acid in preparation for MS. The digests were stored at −20 °C. Unlabeled, control porcine tubulin was treated similarly and analyzed by LC-MS/MS for diethylphosphate labeling and cross-linked peptides.
Isopeptide hydrolysis
Isopeptide bonds can be selectively hydrolyzed by γ-glutamyltranspeptidase (horse kidney; Sigma, catalog number G9270) (54). Hydrolysis was conducted as described (39). One microgram of CPO-tubulin or untreated porcine tubulin was incubated with 1 unit of γ-glutamyltranspeptidase and 10 mm Gly–Gly peptide (Sigma, catalog number G1002) in 65 μl of 50 mm Tris/Cl, pH 8.0, containing 150 mm sodium chloride at 37 °C for 22 h.
SDS-PAGE
A 4–20% gradient Mini Protean TGX Stain-free 10-well, 0.75-mm thick polyacrylamide precast gel was used (Bio-Rad, catalog number 456-8094) in a Mini Protean Tetra PAGE cell (Bio-Rad, catalog number 1658004). Tank buffer was 60 mm Tris/Cl, pH 8.1. The top buffer was 25 mm Tris glycine, pH 8.4, containing 0.1% SDS. Gels used for Western blotting were loaded with 1 μg of tubulin/well. Gels used for Coomassie staining were loaded with 5 μg of tubulin/well. Before loading, tubulin was mixed in a 1 to 3 ratio with 93.8 mm Tris/Cl, pH 6.8, containing 15% glycerol, 3% SDS, 0.3 m DTT, and 0.0018% bromphenol blue, and heated in a boiling water bath for 3 min. Electrophoresis at room temperature and 150 volts was stopped when the bromphenol blue began to come off the bottom of the gel (1.5 h). Coomassie staining employed 0.025% Coomassie Blue in 50% methanol, 10% acetic acid for about 2 h, followed by destaining in water.
SDS-PAGE Western blotting
A Mini Protean II Western blotting cell (Bio-Rad) was used to transfer the proteins from SDS-PAGE to Immuno-Blot PVDF membrane (Bio-Rad, catalog number 162-0177). The transfer sandwich was assembled according to the manufacturer's instructions. Transfer was performed in 25 mm Tris glycine buffer, pH 8.4, using 70 volts at 6 °C, for 1 h. The membrane was blocked with TBS/Tween (TBST, 20 mm Tris/Cl, pH 7.4, plus 150 mm sodium chloride, 0.05% Tween 20) containing 5% nonfat dry milk (Carnation). The membrane was hybridized with a mouse mAb, 0.27 μg/ml of depY, which targeted diethoxyphosphotyrosine (55) or with a sheep polyclonal that targeted tubulin (Cytoskeleton, catalog number ATN02) in TBST plus 5% nonfat dry milk, for 18 h at 6 °C. After washing with TBST, the membrane was hybridized with a 1 to 5000 dilution of horse anti-mouse IgG conjugated to horseradish peroxidase (Cell Signaling, Danvers, MA, catalog number 7076) or with a 1 to 5000 dilution of donkey anti-sheep IgG conjugated to horseradish peroxidase (The Binding Site Limited, Birmingham, UK, catalog number PP360) in TBS plus 5% nonfat dry milk, for 1.5 h at room temperature.
The labeled proteins were visualized with Super Signal West Dura Extended Duration fluorescent substrate for horseradish peroxidase (Thermo Scientific, Rockford, IL, catalog number 27071). Chemiluminescence was detected with an Azure Biosystems model C600 flat plate scanner (Azure Biosystems, Dublin, CA).
Capillary electrophoresis Western blotting
Simple Western capillary electrophoresis was performed on tubulin and CPO-treated tubulin by RayBiotech, Inc. (Norcross, GA). In one analysis, the primary antibody was a mouse monoclonal IgG1κ anti-isopeptide antibody 81D1C2 (Abcam, Cambridge, MA, catalog number ab422). In another analysis, the primary antibody was sheep polyclonal anti-α/β-tubulin (Cytoskeleton, catalog number ATN02). One microliter of tubulin, at 0.1 mg/ml, was loaded onto the capillary electrophoresis device. After electrophoresis, proteins were immobilized to the capillary wall via a proprietary, photoactivated capture chemistry. Matrix was removed and the primary antibody at a concentration of 0.02 mg/ml was passed through the capillary. Antibody-bound proteins were located using an HRP-conjugated secondary antibody together with a chemiluminescent substrate (ProteinSimple PS-CS01).
Mass spectral data acquisition
Electrospray ionization MS was performed on an AB Sciex 6600 Triple TOF instrument (AB Sciex, Framingham, MA). The peptide preparation (about 1 μg/μl) was acidified with 0.1% formic acid. Five microliters were subjected to HPLC separation using a cHiPLC Nanoflex microchip column (Eksigent, Dublin CA) attached to a splitless Ultra 1D Plus ultra-high pressure chromatography system (Eksigent, Dublin, CA). The Nanoflex microchip system consisted of a replaceable microfluidic trap column (200 μm × 0.5 mm) and separation column (75 μm × 15 cm separation) both packed with ChromXP C18 resin (3 μm, 120 Å particles). Solvents were water/acetonitrile/formic acid (A: 100/0/0.1%; B: 0/100/0.1%). The sample was loaded onto the trap column and washed for 15 min at 2 μl/min with Solvent A to remove salts. The flow rate was reduced to 0.3 μl/min, and the sample was eluted using a 65-min linear gradient ranging from 95% solvent A, 5% solvent B to 70% solvent A, 30% solvent B. Effluent from the HPLC column was sprayed directly into the mass spectrometer. Mass spectra were collected in positive mode, over a mass range of 200 to 3000 Da, using an accumulation time of 250 ms, a collision energy of 10 V, a declustering potential of 70 V, an ion spray potential of 2700 V, and an interface heater temperature of 150 °C. Peptide fragmentation was accomplished by collision-induced dissociation using nitrogen as the collision gas at a pressure of 2 × 10−5 torr. A maximum of 50 parent ions were subjected to fragmentation per cycle. Fragmentation spectra were collected in positive mode, over a mass range of 50–3000 Da, using an accumulation time of 25 ms, a collision energy determined by the software (rolling), and a collision energy spread of ±10 V. Peptides to be fragmented were chosen by an information directed acquisition algorithm using charge states 1 to 4, minimum signal of 100 cps, with no parent ion exclusion.
Protein Pilot analysis of the mass spectral data
Data acquired by the Triple TOF mass spectrometer (*.wiff files) were searched against a Uniprot-Sprot protein database using the Paragon algorithm from Protein Pilot software version 5.0 (AB Sciex). Search parameters included Cys alkylation = none, digestion = trypsin, species = Sus scrofa, ID focus = biological modifications, database = unipro_sprotJan2015.fasta, search effort = thorough ID, and false discovery rate = 0.05. In-house modifications to the Special Factors section included modification of serine, tyrosine, threonine, lysine, histidine, and cysteine by diethoxyphosphate and monoethoxyphosphate. Protein Pilot identified the tubulin peptides including those that were candidates for organophosphate modification, but did not report cross-linked peptide candidates.
Batch Tag (web) analysis of the mass spectral data for cross-linked peptides
The *.wiff mass spectral data included information on cross-linked peptides. The data could be accessed by the Batch Tag (web)/Search Compare algorithms from ProteinProspector (UCSF Mass Spectrometry Facility) as follows. Protein Pilot *.wiff files were converted to a *.mgf file format for use by Batch Tag (web) using Protein Pilot menu item File/Export/MGF Peaklist (56, 57). Batch Tag (web) searches were configured to identify pairs of peptides that were cross-linked via isopeptide bonds. Critical parameters for Batch Tag were chosen based in large part on the recommendations of Trnka et al. (56). Parameters used for all analyses included database = Swissprot.2016.9.6.random.concat, taxonomy = S. scrofa, digest = trypsin, constant modifications = none; protein Mr = 1000 to 70,000, accession number = P02550 (for tubulin α1A), P02554 (for tubulin β), Q767L7 (for tubulin β), and Q2XVP4 (for tubulin α1B), parent Tol = 20 ppm, fragment Tol = 30 ppm, and variable mods = oxidation (M), uncleaved = unchecked, link search type = user defined link. Other parameters were adjusted to match the type of cross-link being searched. For cross-links between lysine and glutamate, aspartate, glutamine, or asparagine: AA table = Asp, Glu, Gln, Asn, and Lys checked, Link AAs = Glu, Asp, Protein C-term → Lys, Protein N-term, and Bridge Elem Comp = H-2O-1. For cross-links between lysine and any amino acid: AA table = all checked, link AAs = Lys, Protein N-term → Ala, Cys, Asp, Glu, Phe, Gly, His, Ile, Lys, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, Tyr, and Bridge Elem Comp = H-2O-1. Search Compare parameters included: accession numbers P02550 (for tubulin α1A), P02554 (for tubulin β), Q767L7 (for tubulin β), and Q2XVP4 (for tubulin α1B), m/z, charge, score difference, coverage, DB peptide, menu = mods in peptide, links, time, cross-linked peptide, report type = cross-linked peptides, and sort type = start residue.
The identity of each cross-linked candidate found by Batch Tag/Search Compare, regardless of the Score or Score Difference value, was checked by manual sequencing using Peak View software (version 1.2, AB Sciex). Peak View reads all of the data in the *.wiff file, including that which is not reported by Protein Pilot. The cross-linked candidate was located based on its observed monoisotopic mass, charge state, and elution time, all of which were obtained from the Search Compare report. Multiple occurrences of a given candidate were summed before the fragmentation spectrum was analyzed.
Identification of a cross-linked peptide is based on the presence of two peptides in the fragmentation spectrum. Each peptide pair that we report exhibited sequence information from both peptides and masses that corresponded to cross-linked portions from both peptides. The cross-linked masses are typically of low abundance but they provide a means for identifying the specific amino acids involved in the cross-link (56, 58). Most of the cross-links were identified by analyzing multiply-charged fragment ions as described by Liu et al. (59).
Author contributions
L. M. S. resources; L. M. S. and O. L. data curation; L. M. S. and O. L. formal analysis; L. M. S. validation; L. M. S. and O. L. investigation; L. M. S. and O. L. methodology; L. M. S. and O. L. writing-original draft; L. M. S. and O. L. writing-review and editing; O. L. conceptualization; O. L. funding acquisition; O. L. project administration.
Acknowledgment
Mass spectrometry data were obtained with the support of the Mass Spectrometry and Proteomics core facility at the University of Nebraska Medical Center.
This work was supported by Fred and Pamela Buffett Cancer Center Support Grant P30CA036727 from the National Institutes of Health, NCI. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
L. M. Schopfer and O. Lockridge, unpublished data.
- AChE
- acetylcholinesterase
- CPO
- chlorpyrifos oxon
- CPS
- chlorpyrifos
- OP
- organophosphorus toxicants
- PVDF
- polyvinylidine fluoride
- depY
- anti-diethoxyphosphotyrosine antibody
- VX
- O-ethyl methyl S-2-(diisopropylaminoethyl)phosphonothioate).
References
- 1. Casida J. E., and Quistad G. B. (2004) Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem. Res. Toxicol. 17, 983–998 10.1021/tx0499259 [DOI] [PubMed] [Google Scholar]
- 2. Richardson R. J., Hein N. D., Wijeyesakere S. J., Fink J. K., and Makhaeva G. F. (2013) Neuropathy target esterase (NTE): overview and future. Chem. Biol. Interact. 203, 238–244 10.1016/j.cbi.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 3. Lotti M., and Moretto A. (2005) Organophosphate-induced delayed polyneuropathy. Toxicol. Rev. 24, 37–49 10.2165/00139709-200524010-00003 [DOI] [PubMed] [Google Scholar]
- 4. Brown M. A., and Brix K. A. (1998) Review of health consequences from high-, intermediate- and low-level exposure to organophosphorus nerve agents. J. Appl. Toxicol. 18, 393–408 10.1002/(SICI)1099-1263(199811/12)18:6%3C393::AID-JAT528%3E3.0.CO%3B2-0 [DOI] [PubMed] [Google Scholar]
- 5. Ross S. M., McManus I. C., Harrison V., and Mason O. (2013) Neurobehavioral problems following low-level exposure to organophosphate pesticides: a systematic and meta-analytic review. Crit. Rev. Toxicol. 43, 21–44 10.3109/10408444.2012.738645 [DOI] [PubMed] [Google Scholar]
- 6. Hayden K. M., Norton M. C., Darcey D., Ostbye T., Zandi P. P., Breitner J. C., Welsh-Bohmer K. A., and Cache County Study Investigators (2010) Occupational exposure to pesticides increases the risk of incident AD: the Cache County study. Neurology 74, 1524–1530 10.1212/WNL.0b013e3181dd4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hancock D. B., Martin E. R., Mayhew G. M., Stajich J. M., Jewett R., Stacy M. A., Scott B. L., Vance J. M., and Scott W. K. (2008) Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol. 8, 6 10.1186/1471-2377-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamel F., Tanner C., Umbach D., Hoppin J., Alavanja M., Blair A., Comyns K., Goldman S., Korell M., Langston J., Ross G., and Sandler D. (2007) Pesticide exposure and self-reported Parkinson's disease in the agricultural health study. Am. J. Epidemiol. 165, 364–374 [DOI] [PubMed] [Google Scholar]
- 9. Rauh V. A., Garfinkel R., Perera F. P., Andrews H. F., Hoepner L., Barr D. B., Whitehead R., Tang D., and Whyatt R. W. (2006) Impact of prenatal chlorpyrifos exposure on neurodevelopment in the first 3 years of life among inner-city children. Pediatrics 118, e1845–e1859 10.1542/peds.2006-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rauh V. A., Perera F. P., Horton M. K., Whyatt R. M., Bansal R., Hao X., Liu J., Barr D. B., Slotkin T. A., and Peterson B. S. (2012) Brain anomalies in children exposed prenatally to a common organophosphate pesticide. Proc. Natl. Acad. Sci. U.S.A. 109, 7871–7876 10.1073/pnas.1203396109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rauh V. A., Garcia W. E., Whyatt R. M., Horton M. K., Barr D. B., and Louis E. D. (2015) Prenatal exposure to the organophosphate pesticide chlorpyrifos and childhood tremor. Neurotoxicology 51, 80–86 10.1016/j.neuro.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer A., Seidler F. J., Cousins M. M., and Slotkin T. A. (2003) Developmental neurotoxicity elicited by gestational exposure to chlorpyrifos: when is adenylyl cyclase a target? Environ. Health Perspect. 111, 1871–1876 10.1289/ehp.6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schuh R. A., Lein P. J., Beckles R. A., and Jett D. A. (2002) Noncholinesterase mechanisms of chlorpyrifos neurotoxicity: altered phosphorylation of Ca2+/cAMP response element binding protein in cultured neurons. Toxicol. Appl. Pharmacol. 182, 176–185 10.1006/taap.2002.9445 [DOI] [PubMed] [Google Scholar]
- 14. Hernandez C. M., Beck W. D., Naughton S. X., Poddar I., Adam B. L., Yanasak N., Middleton C., and Terry A. V. Jr. (2015) Repeated exposure to chlorpyrifos leads to prolonged impairments of axonal transport in the living rodent brain. Neurotoxicology 47, 17–26 10.1016/j.neuro.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 15. Sanger F. (1963) Amino-acid sequences in the active centers of certain enzymes. Proc. Chem. Soc. 5, 76–83 [Google Scholar]
- 16. Murachi T., Inagami T., and Yasui M. (1965) Evidence for alkylphosphorylation of tyrosyl residues of stem bromelain by diisopropylphosphorofluoridate. Biochemistry 4, 2815–2825 10.1021/bi00888a036 [DOI] [PubMed] [Google Scholar]
- 17. Chaiken I. M., and Smith E. L. (1969) Reaction of a specific tyrosine residue of papain with diisopropylfluorophosphate. J. Biol. Chem. 244, 4247–4250 [PubMed] [Google Scholar]
- 18. Murachi T., Miyake T., and Yamasaki N. (1970) Alkylphosphorylation of hen egg-white lysozyme by diisopropylphosphorofluoridate. J. Biochem. 68, 239–244 10.1093/oxfordjournals.jbchem.a129351 [DOI] [PubMed] [Google Scholar]
- 19. Grigoryan H., Li B., Xue W., Grigoryan M., Schopfer L. M., and Lockridge O. (2009) Mass spectral characterization of organophosphate-labeled lysine in peptides. Anal. Biochem. 394, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grigoryan H., Schopfer L. M., Thompson C. M., Terry A. V., Masson P., and Lockridge O. (2008) Mass spectrometry identifies covalent binding of soman, sarin, chlorpyrifos oxon, diisopropyl fluorophosphate, and FP-biotin to tyrosines on tubulin: a potential mechanism of long term toxicity by organophosphorus agents. Chem. Biol. Interact. 175, 180–186, PMCID:PMC2577157 10.1016/j.cbi.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lockridge O., and Schopfer L. M. (2010) Review of tyrosine and lysine as new motifs for organophosphate binding to proteins that have no active site serine. Chem. Biol. Interact. 187, 344–348 10.1016/j.cbi.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li B., Schopfer L. M., Hinrichs S. H., Masson P., and Lockridge O. (2007) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay for organophosphorus toxicants bound to human albumin at Tyr411. Anal. Biochem. 361, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onder S., Schopfer L. M., Tacal O., Blake T. A., Johnson R. C., and Lockridge O. (2018) Mass spectral detection of diethoxyphospho-tyrosine adducts on proteins from HEK293 cells using monoclonal antibody depY for enrichment. Chem. Res. Toxicol. 31, 520–530 10.1021/acs.chemrestox.8b00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bui-Nguyen T. M., Dennis W. E., Jackson D. A., Stallings J. D., and Lewis J. A. (2014) Detection of dichlorvos adducts in a hepatocyte cell line. J. Proteome Res. 13, 3583–3595 10.1021/pr5000076 [DOI] [PubMed] [Google Scholar]
- 25. Chen S., Zhang J., Lumley L., and Cashman J. R. (2013) Immunodetection of serum albumin adducts as biomarkers for organophosphorus exposure. J. Pharmacol. Exp. Ther. 344, 531–541 10.1124/jpet.112.201368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. John H., Breyer F., Thumfart J. O., Höchstetter H., and Thiermann H. (2010) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for detection and identification of albumin phosphorylation by organophosphorus pesticides and G- and V-type nerve agents. Anal. Bioanal. Chem. 398, 2677–2691 10.1007/s00216-010-4076-y [DOI] [PubMed] [Google Scholar]
- 27. Noort D., Hulst A. G., van Zuylen A., van Rijssel E., and van der Schans M. J. (2009) Covalent binding of organophosphorothioates to albumin: a new perspective for OP-pesticide biomonitoring? Arch. Toxicol. 83, 1031–1036 10.1007/s00204-009-0456-5 [DOI] [PubMed] [Google Scholar]
- 28. Crow B. S., Pantazides B. G., Quiñones-Gonzalez J., Garton J. W., Carter M. D., Perez J. W., Watson C. M., Tomcik D. J., Crenshaw M. D., Brewer B. N., Riches J. R., Stubbs S. J., Read R. W., Evans R. A., Thomas J. D., Blake T. A., and Johnson R. C. (2014) Simultaneous measurement of tabun, sarin, soman, cyclosarin, VR, VX, and VM adducts to tyrosine in blood products by isotope dilution UHPLC-MS/MS. Anal. Chem. 86, 10397–10405 10.1021/ac502886c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Williams N. H., Harrison J. M., Read R. W., and Black R. M. (2007) Phosphylated tyrosine in albumin as a biomarker of exposure to organophosphorus nerve agents. Arch. Toxicol. 81, 627–639 10.1007/s00204-007-0191-8 [DOI] [PubMed] [Google Scholar]
- 30. John H., van der Schans M. J., Koller M., Spruit H. E. T., Worek F., Thiermann H., and Noort D. (2018) Fatal sarin poisoning in Syria 2013: forensic verification within an international laboratory network. Forensic Toxicol. 36, 61–71 10.1007/s11419-017-0376-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt C., Breyer F., Blum M. M., Thiermann H., Worek F., and John H. (2014) V-type nerve agents phosphonylate ubiquitin at biologically relevant lysine residues and induce intramolecular cyclization by an isopeptide bond. Anal. Bioanal. Chem. 406, 5171–5185 10.1007/s00216-014-7706-y [DOI] [PubMed] [Google Scholar]
- 32. Jiang W., Duysen E. G., Hansen H., Shlyakhtenko L., Schopfer L. M., and Lockridge O. (2010) Mice treated with chlorpyrifos or chlorpyrifos oxon have organophosphorylated tubulin in the brain and disrupted microtubule structures, suggesting a role for tubulin in neurotoxicity associated with exposure to organophosphorus agents. Toxicol. Sci. 115, 183–193 10.1093/toxsci/kfq032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alushin G. M., Lander G. C., Kellogg E. H., Zhang R., Baker D., and Nogales E. (2014) High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157, 1117–1129 10.1016/j.cell.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gundemir S., Colak G., Tucholski J., and Johnson G. V. (2012) Transglutaminase 2: a molecular Swiss army knife. Biochim. Biophys. Acta 1823, 406–419 10.1016/j.bbamcr.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lorand L., and Graham R. M. (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 10.1038/nrm1014 [DOI] [PubMed] [Google Scholar]
- 36. Marraffini L. A., Dedent A. C., and Schneewind O. (2006) Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70, 192–221 10.1128/MMBR.70.1.192-221.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 10.1016/j.molcel.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 38. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- 39. Recktenwald C. V., and Hansson G. C. (2016) The reduction-insensitive bonds of the MUC2 mucin are isopeptide bonds. J. Biol. Chem. 291, 13580–13590 10.1074/jbc.M116.726406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tarcsa E., and Fesus L. (1990) Determination of epsilon (gamma-glutamyl)lysine crosslink in proteins using phenylisothiocyanate derivatization and high-pressure liquid chromatographic separation. Anal. Biochem. 186, 135–140 10.1016/0003-2697(90)90586-X [DOI] [PubMed] [Google Scholar]
- 41. Nemes Z., Petrovski G., and Fésüs L. (2005) Tools for the detection and quantitation of protein transglutamination. Anal. Biochem. 342, 1–10 10.1016/j.ab.2004.10.015 [DOI] [PubMed] [Google Scholar]
- 42. Nemes Z., Devreese B., Steinert P. M., Van Beeumen J., and Fésüs L. (2004) Cross-linking of ubiquitin, HSP27, parkin, and α-synuclein by γ-glutamyl-ϵ-lysine bonds in Alzheimer's neurofibrillary tangles. FASEB J. 18, 1135–1137 10.1096/fj.04-1493fje [DOI] [PubMed] [Google Scholar]
- 43. Kang H. J., and Baker E. N. (2011) Intramolecular isopeptide bonds: protein crosslinks built for stress? Trends Biochem. Sci. 36, 229–237 10.1016/j.tibs.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 44. Kang H. J., Coulibaly F., Clow F., Proft T., and Baker E. N. (2007) Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 318, 1625–1628 10.1126/science.1145806 [DOI] [PubMed] [Google Scholar]
- 45. Eyer F., Roberts D. M., Buckley N. A., Eddleston M., Thiermann H., Worek F., and Eyer P. (2009) Extreme variability in the formation of chlorpyrifos oxon (CPO) in patients poisoned by chlorpyrifos (CPF). Biochem. Pharmacol. 78, 531–537 10.1016/j.bcp.2009.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zamponi E., Buratti F., Cataldi G., Caicedo H. H., Song Y., Jungbauer L. M., LaDu M. J., Bisbal M., Lorenzo A., Ma J., Helguera P. R., Morfini G. A., Brady S. T., and Pigino G. F. (2017) Prion protein inhibits fast axonal transport through a mechanism involving casein kinase 2. PLoS ONE 12, e0188340 10.1371/journal.pone.0188340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brady S. T., and Morfini G. A. (2017) Regulation of motor proteins, axonal transport deficits and adult-onset neurodegenerative diseases. Neurobiol. Dis. 105, 273–282 10.1016/j.nbd.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 49. Gupta R. P., and Abou-Donia M. B. (1995) Neurofilament phosphorylation and [125I]calmodulin binding by Ca2+/calmodulin-dependent protein kinase in the brain subcellular fractions of diisopropyl phosphorofluoridate (DFP)-treated hen. Neurochem. Res. 20, 1095–1105 10.1007/BF00995565 [DOI] [PubMed] [Google Scholar]
- 50. Gupta R. P., and Abou-Donia M. B. (1999) Tau phosphorylation by diisopropyl phosphorofluoridate (DFP)-treated hen brain supernatant inhibits its binding with microtubules: role of Ca2+/calmodulin-dependent protein kinase II in tau phosphorylation. Arch. Biochem. Biophys. 365, 268–278 10.1006/abbi.1999.1165 [DOI] [PubMed] [Google Scholar]
- 51. Choudhary S., Joshi K., and Gill K. D. (2001) Possible role of enhanced microtubule phosphorylation in dichlorvos induced delayed neurotoxicity in rat. Brain Res. 897, 60–70 10.1016/S0006-8993(00)03222-4 [DOI] [PubMed] [Google Scholar]
- 52. Gearhart D. A., Sickles D. W., Buccafusco J. J., Prendergast M. A., and Terry A. V. Jr. (2007) Chlorpyrifos, chlorpyrifos-oxon, and diisopropylfluorophosphate inhibit kinesin-dependent microtubule motility. Toxicol. Appl. Pharmacol. 218, 20–29 10.1016/j.taap.2006.10.008 [DOI] [PubMed] [Google Scholar]
- 53. Speed H. E., Blaiss C. A., Kim A., Haws M. E., Melvin N. R., Jennings M., Eisch A. J., and Powell C. M. (2012) Delayed reduction of hippocampal synaptic transmission and spines following exposure to repeated subclinical doses of organophosphorus pesticide in adult mice. Toxicol. Sci. 125, 196–208 10.1093/toxsci/kfr253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Allison R. D., and Meister A. (1981) Evidence that transpeptidation is a significant function of γ-glutamyl transpeptidase. J. Biol. Chem. 256, 2988–2992 [PubMed] [Google Scholar]
- 55. Onder S., Dafferner A. J., Schopfer L. M., Xiao G., Yerramalla U., Tacal O., Blake T. A., Johnson R. C., and Lockridge O. (2017) Monoclonal antibody that recognizes diethoxyphosphotyrosine-modified proteins and peptides independent of surrounding amino acids. Chem. Res. Toxicol. 30, 2218–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trnka M. J., Baker P. R., Robinson P. J., Burlingame A. L., and Chalkley R. J. (2014) Matching cross-linked peptide spectra: only as good as the worse identification. Mol. Cell. Proteomics 13, 420–434 10.1074/mcp.M113.034009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chalkley R. J., Baker P. R., Hansen K. C., Medzihradszky K. F., Allen N. P., Rexach M., and Burlingame A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer: I. how much of the data is theoretically interpretable by search engines? Mol. Cell. Proteomics 4, 1189–1193 10.1074/mcp.D500001-MCP200 [DOI] [PubMed] [Google Scholar]
- 58. Schilling B., Row R. H., Gibson B. W., Guo X., and Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass. Spectrom. 14, 834–850 10.1016/S1044-0305(03)00327-1 [DOI] [PubMed] [Google Scholar]
- 59. Liu M., Zhang Z., Zang T., Spahr C., Cheetham J., Ren D., and Zhou Z. S. (2013) Discovery of undefined protein cross-linking chemistry: a comprehensive methodology utilizing 18O-labeling and mass spectrometry. Anal. Chem. 85, 5900–5908 10.1021/ac400666p [DOI] [PMC free article] [PubMed] [Google Scholar]