Abstract
Heterodimeric KIF3AC and KIF3AB, two members of the mammalian kinesin-2 family, generate force for microtubule plus end–directed cargo transport. However, the advantage of heterodimeric kinesins over homodimeric ones is not well-understood. We showed previously that microtubule association for entry into a processive run was similar in rate for KIF3AC and KIF3AB at ∼7.0 μm−1 s−1. Yet, for engineered homodimers of KIF3AA and KIF3BB, this constant is significantly faster at 11 μm−1 s−1 and much slower for KIF3CC at 2.1 μm−1 s−1. These results led us to hypothesize that heterodimerization of KIF3A with KIF3C and KIF3A with KIF3B altered the intrinsic catalytic properties of each motor domain. Here, we tested this hypothesis by using presteady-state stopped-flow kinetics and mathematical modeling. Surprisingly, the modeling revealed that the catalytic properties of KIF3A and KIF3B/C were altered upon microtubule binding, yet each motor domain retained its relative intrinsic kinetics for ADP release and subsequent ATP binding and the nucleotide-promoted transitions thereafter. These results are consistent with the interpretation that for KIF3AB, each motor head is catalytically similar and therefore each step is approximately equivalent. In contrast, for KIF3AC the results predict that the processive steps will alternate between a fast step for KIF3A followed by a slow step for KIF3C resulting in asymmetric stepping during a processive run. This study reveals the impact of heterodimerization of the motor polypeptides for microtubule association to start the processive run and the fundamental differences in the motile properties of KIF3C compared with KIF3A and KIF3B.
Keywords: computational biology, intracellular trafficking, microtubule, neuron, mathematical modeling, ATPase, pre-steady-state kinetics, processivity, ciliogenesis, intraflagellar transport
Introduction
Kinesin-2 is a unique family of processive kinesins because it contains both homodimeric and heterodimeric motors involved in microtubule plus end–directed cargo transport (reviewed in Refs. 1–4). The mammalian heterodimeric kinesins result from three gene products: KIF3A, KIF3B, and KIF3C to form heterodimeric KIF3AB and KIF3AC (5–9). KIF3AB and its orthologs form a heterotrimeric complex by association with KAP, a distinctive adaptor protein for cargo linkage because it is largely composed of armadillo repeats (10–15). It is the armadillo repeats that provide specificity of the interaction between KIF3AB and KAP and between KIF3AB-KAP and its cargo. KIF3AB-KAP transports multimeric protein complexes (designated intraflagellar transport (IFT) particles)6 into the cilium and has also been linked to cilia-dependent signal transduction pathways including the Hedgehog signaling pathway (16, 17). Moreover, KIF3A, KIF3B, and KAP are all essential genes (18–23), and the transport role of KIF3AB-KAP in ciliogenesis is considered the reason that KIF3AB-KAP is essential for development and the basis of similarity with other heterotrimeric kinesin-2 motor proteins.
In contrast, there is not strong evidence that mammalian KIF3AC binds KAP, and KIF3C lacks the sequence similarity to KIF3AB at the putative C-terminal KAP-binding region (2, 11, 14, 24). Despite similarities in sequence and structure, KIF3AC appears to function specifically in neurons, whereas heterotrimeric KIF3AB-KAP is more ubiquitously expressed and transports ciliary IFT particles, melanosomes, and organelles as well as cell signaling and cell adhesion molecules (21, 23, 25–32). There are significant gaps in our understanding of KIF3AC cargo transport in neurons. A common KIF3AC adaptor for cargo linkage has not yet been identified, and the identity of the KIF3AC-specific dendritic organelles that KIF3AC transports are also unknown. In addition, KIF3C exhibits a signature motif conserved in mammals, a 25-residue insert in loop L11 of the catalytic motor domain, which is enriched in glycines and serines, and this L11 insert is not present in other kinesins regardless of species (7, 33–35). The distinctive L11 domain of KIF3C has been implicated in regulating processivity for KIF3AC (35) and microtubule dynamics for homodimeric KIF3CC (36).
Like other processive kinesins, KIF3AB and KIF3AC use an asymmetric hand-over-hand stepping mechanism to move along a microtubule with each 8-nm step coupled to one ATP turnover (37–43). Yet unlike other processive kinesins, KIF3AB's motility is highly sensitive to hindering loads resulting in detachment at low stall forces (43, 44). This property is encoded in the catalytic motor domains rather than the neck linker or coiled-coil, and has physiological implications in that in vivo, the detachment of KIF3AB-KAP under load may serve as a mechanism to navigate roadblocks, side-step to another microtubule track, and/or regulate IFT particle transport for ciliary tip length control and structure (45–48).
To explore the contributions of each motor domain to the motile properties of heterodimeric KIF3AC compared with KIF3AB, single-molecule motility studies were pursued that compared engineered homodimers of KIF3AA, KIF3BB, and KIF3CC to the heterodimers (35). KIF3AA and KIF3BB step rapidly with rates of 293 and 328 nm/s, respectively and more similar to KIF3AB, which has a velocity of 246 nm/s (Table 1). In contrast, the velocity of KIF3CC was exceedingly slow at 7.5 nm/s with KIF3AC at 186 nm/s, whose faster rate was thought to be due to the properties of KIF3A (35). Similarly, previous presteady-state kinetics results revealed that microtubule association for KIF3AA and KIF3BB at ∼11 μm−1 s−1 coupled with ADP release at ∼80 s−1 were both fast, yet microtubule association for KIF3AB was 7 μm−1 s−1 with ADP release at 40 s−1 (Table 1) (49). The surprise was that microtubule association for KIF3AC was 6.6 μm−1 s−1 with ADP release at 51 s−1 (50). Yet even though microtubule association by KIF3AC was similar to the constant for KIF3AB, microtubule association for KIF3CC was 2.1 μm−1 s−1 with ADP release at 7.6 s−1, both of which are extremely slow (50). Additional evidence that heterodimerization may alter the intrinsic catalytic properties of each motor domain as defined by the engineered homodimers KIF3AA, KIF3BB, and KIF3CC was supported by experimental results that showed that no mixtures of KIF3AA plus KIF3CC or KIF3AA plus KIF3BB could recapitulate the microtubule association and ADP release kinetics of KIF3AC or KIF3AB (49, 50).
Table 1.
| KIF3AB | KIF3AC | KIF3AA | KIF3BB | KIF3CC | |||
|---|---|---|---|---|---|---|---|
| MT association | k+1 | μm−1s−1 | 7.0 ± 0.4 | 6.6 ± 0.2 | 11.4 ± 0.6 | 11.9 ± 0.1 | 2.1 ± 0.1 |
| k−1 | s−1 | 0.8 ± 0.4 | − | 2.4 ± 0.9 | − | 0.6 ± 0.1 | |
| ADP release | k+2 | s−1 | 39.6 ± 0.9a | 42.5 ± 0.9a | 77.7 ± 1.4 | 80.2 ± 2.5 | 7.6 ± 0.1 |
| K1/2,MT | μm | 3.8 ± 0.3a | 7.5 ± 0.4a | 4.4 ± 0.2 | 4.0 ± 0.4 | 1.7 ± 0.1 | |
| MantATP binding | k+3 | μm−1s−1 | 7.5 ± 0.5 | 11.0 ± 0.6 | 16.0 ± 0.5 | 7.5 ± 0.5 | 0.68 ± 0.04 |
| k−3 | s−1 | 46.1 ± 5.5 | 21.4 ± 7.2 | 10.5 ± 5.1 | 46.1 ± 5.5 | 7.7 ± 0.4 | |
| ATP-isomerization | k+4 | s−1 | 84.0 ± 1.9 | 81.0 ± 1.0 | ND | ND | ND |
| ATP hydrolysis | k+5 | s−1 | 33.0 ± 2.5b | 69.1 ± 1.2 | ND | ND | ND |
| Amax | 3.0 ± 0.2 per site | 0.77 ± 0.02 per site | ND | ND | ND | ||
| Steady-state | kcat | s−1 | 31.7 ± 1.2 | 21.5 ± 0.3 | 34.7 ± 0.5 | 32.1 ± 0.4 | 1.1 ± 0.02 |
| Km,ATP | μm | 122.9 ± 15.0 | 138.1 ± 12.8 | 47.7 ± 0.1 | 71.4 ± 4.0 | 4.8 ± 0.5 | |
| K1/2,MT | μm | 0.14 ± 0.01 | 0.23 ± 0.03 | 0.19 ± 0.002 | 0.14 ± 0.005 | 0.04 ± 0.007 | |
| Single molecule velocity | − | nm/s | 246.2 ± 11.1 | 186.5 ± 5.6 | 293.2 ± 4.2 | 327.6 ± 7.2 | 7.5 ± 0.4 |
The general hypothesis has been that heterodimerization provides diversity for adaptor and cargo binding, yet the results to date for KIF3AC and KIF3AB clearly show that despite high sequence similarity in their motor domains, KIF3A and KIF3B are both catalytically fast, yet KIF3C is extremely slow (35, 49–51). These puzzling results motivated this study to ask the following questions. 1) How do the properties of KIF3C affect the ATPase properties of KIF3AC? 2) How does KIF3AC stepping during a processive run differ from processive stepping by KIF3AB? 3) Can we understand the mechanism by which KIF3A accelerates KIF3C and KIF3C slows KIF3A within the heterodimer?
To address these questions and gain insight into the motility capability of KIF3AC as a cargo transporter, we repeated the presteady-state kinetics experiments for microtubule association, ADP release, and ATP binding and pursued computational modeling coupled with parameter estimation. The results reveal that the catalytic properties of KIF3A and KIF3C were altered upon microtubule association to be catalytically similar, yet once in the processive run KIF3A remained fast with KIF3C continuing to be slow. The computational modeling also indicated that KIF3A accelerates KIF3C to maintain the processive stepping velocity and steady-state ATP turnover. The results also highlight distinctive differences in KIF3AC compared with KIF3AB despite sequence and structure similarities.
Results
Previously, we determined the presteady-state kinetics for microtubule association and native ADP release for KIF3AC, KIF3AA, and KIF3CC (50). For these experiments, the KIF3 motor was rapidly mixed in the stopped-flow instrument with varying concentrations of microtubules plus 50 μm mantATP, a fluorescent analog of ATP. Upon mixing, the dimeric motor associates with the microtubule, releases its ADP, and subsequently binds mantATP (Fig. 1A, E0–E2). Because mantATP binding is rapid at these conditions (Table 1), the fluorescence signal from mantATP binding acts as a readout of native ADP release after microtubule association (49–52). The results showed that microtubule association for KIF3AA was fast at 11.4 μm−1 s−1 yet slow for KIF3CC at 2.1 μm−1 s−1 with microtubule association for KIF3AC at an intermediate rate of 6.6 μm−1 s−1 (50). Similarly, these experiments revealed that native ADP release for KIF3AA was fast at 77.7 s−1, and slow at 7.6 s−1 for KIF3CC and 51 s−1 for KIF3AC (50). The question to be addressed through computational modeling was whether the observed rate of these constants for KIF3AC was simply the average of the intrinsic catalytic properties of KIF3A and KIF3C as defined by KIF3AA and KIF3CC or alternatively, whether the heterodimerization of the KIF3A and KIF3C polypeptides results in an emergent mechanochemistry not predicted from the kinetics of KIF3AA and KIF3CC.
Figure 1.
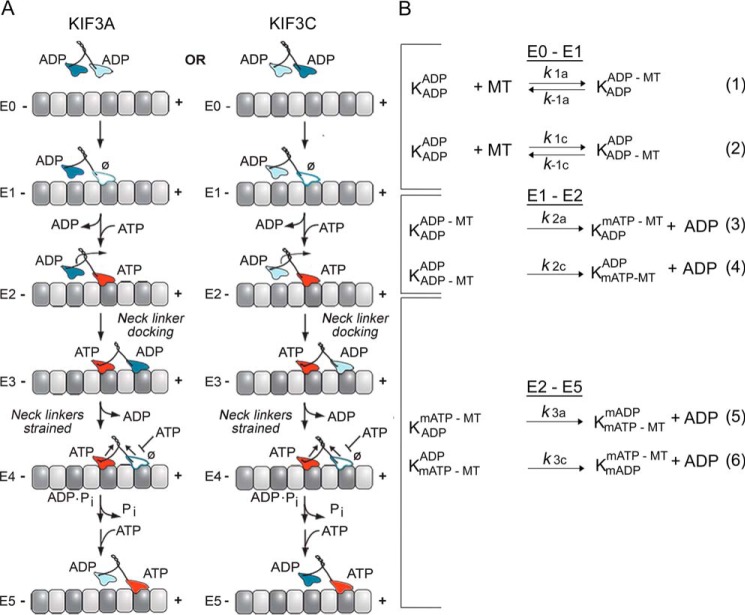
KIF3AC and KIF3AB stepping scheme and model. A, the cycle begins when one head of the kinesin-2 heterodimer collides with the microtubule (E0–E1). Two parallel pathways are shown that represent microtubule association by the KIF3A head (left) or the KIF3C head (right). After microtubule association, ADP is released, and ATP binds rapidly to the nucleotide-binding site (E1–E2). ATP binding at the leading head induces a series of structural transitions by which the rear tethered head moves forward, collides with the microtubule, and releases its ADP (E2–E4). ATP hydrolysis occurs followed by phosphate release coupled with rear head dissociation from the microtubule (E4–E5). The second step is initiated by ATP binding at the leading, partner motor domain (E5). This model makes no assumptions of whether KIF3A or KIF3C initiates the processive run, and two parallel paths are shown based on the motor head that binds the microtubule. B, the computational scheme consists of three steps. The first reaction shows microtubule association (k1, lines 1 and 2) either by the KIF3A or KIF3C. The second step represents ADP release plus mantATP binding (k2, lines 3 and 4), and the third represents the combined transitions following mantATP binding through motor detachment from the microtubule and terminates at mantATP binding by the new leading, partner head (k3, lines 5 and 6). K represents the KIF3 heterodimer, and the superscript and subscript represent the nucleotide and microtubule binding states of each head. mATP, mantATP; mADP, mantADP.
To provide an extensive data set for computational modeling, additional experiments were performed at three concentrations of KIF3AC with microtubule concentrations varying from 1 to 25 μm (Fig. 2). A double exponential function was fit to each fluorescence transient to obtain the observed rates of the fast component and the slow component of fluorescence enhancement as well as the fluorescence amplitudes of each. In addition, the observed rates of the fast component were plotted as a function of microtubule concentration, and the hyperbolic fit to these data provided the rate constant of native ADP release at 42.5 s−1 (Fig. 2G). Therefore, this comprehensive data set to be used for the modeling is consistent with the previously published results (50).
Figure 2.
KIF3AC fluorescence transients and simulations. KIF3AC was rapidly mixed in the stopped-flow instrument with varying concentrations of microtubules plus 50 μm mantATP (25 μm mantATP final concentration). Because the concentration of mantATP results in faster mantATP binding than ADP release, the fluorescence transients represent a readout of native ADP release after motor association with the microtubule (Fig. 1, E0–E2). A, low microtubule concentration family (1–2 μm) at 0.5 μm KIF3AC dimer. B, simulations of the low microtubule concentration family. C, mid-microtubule concentration family (2–4.5 μm) at 0.75 μm KIF3AC dimer. D, simulations of the mid-microtubule concentration family. E, high microtubule concentration family (4.5–18 μm) at 1.25 μm KIF3AC dimer. F, simulations of the high microtubule concentration family. G, a double exponential function was also fit to each transient, and the observed rates of the fast component were plotted as a function of microtubule concentration. The hyperbolic fit to the data provided the maximum rate constant for ADP release at 42.5 ± 0.9 s−1 with a K1/2,MT of 7.5 ± 0.4 μm (mean ± S.E.). Note too that the exponential fit to the transients provided the relative fluorescence amplitude for the fast component at 0.136–0.163 and 0.128–0.173 for the slower component. In simulation panels B, D, and F, the simulations are indicated by solid black lines, and only 100 of 1000 data points for each experimental transient are presented for clarity. The green transient in A, C, and E is the buffer control in the absence of microtubules at each KIF3AC concentration.
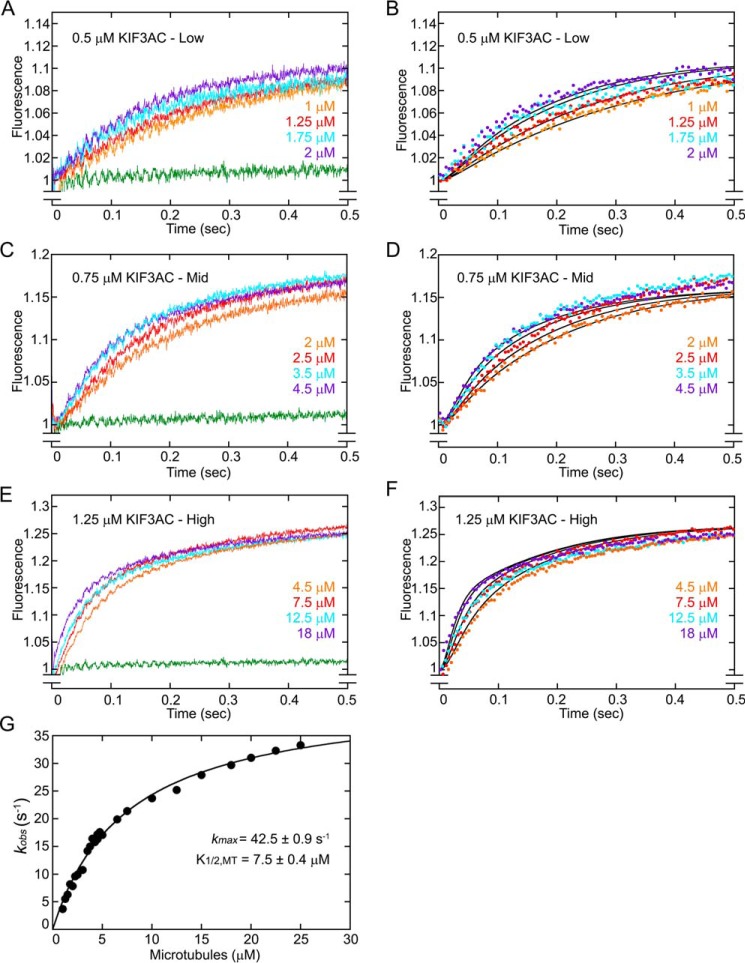
The results presented in Fig. 2 for KIF3AC and Fig. 3 for KIF3AB show that the transients as a function of time plateau indicating that the system has reached a steady-state, which is expected because the mantATP concentration at 25 μm is >10-fold excess of the KIF3 nucleotide-binding sites. These results also imply that the experiment captures entry into the processive ATPase cycle (Fig. 1A, E0–E1) followed by processive stepping once mantATP binds at E2 (Fig. 1A). As the stepping model shows, the first mantATP turnover (Fig. 1A, E2–E5) includes formation of mantADP·Pi and mantADP–bound intermediates. At E5, the lagging tethered head from the first mantATP turnover is occupied by mantADP and the leading head by mantATP for the next mantATP turnover coupled with the second 8-nm step in the processive run. Therefore, the total relative fluorescence observed in the transients in Figs. 2 and 3 is the result of the sum of the fluorescence contribution by KIF3 intermediates bound with mantATP, mantADP·Pi, or mantADP at the active sites during the first mantATP turnover and intermediates with two mant-nucleotides bound beginning at E5 for the second and subsequent mantATP turnovers.
Figure 3.
KIF3AB fluorescence transients and simulations. KIF3AB was rapidly mixed in the stopped-flow instrument with varying concentrations of microtubules plus 50 μm mantATP (25 μm mantATP final) to determine the time course of microtubule association and ADP release. A, low microtubule concentration family (1.75–4 μm) at 0.5 μm KIF3AB dimer. B, simulations of the low microtubule concentration family. C, high microtubule concentration family (5–20 μm) at 1 μm KIF3AB dimer. D, simulations of the high microtubule concentration family. E, a double exponential function was also fit to each transient, and the observed rates of the fast component were plotted as a function of microtubule concentration. The hyperbolic fit to the data provided the maximum rate constant of ADP release at 39.6 ± 0.9 s−1 with a K1/2,MT at 3.8 ± 0.3 μm (mean ± S.E.). In simulation panels B and D, simulations are indicated by solid black lines, and only 100 of 1000 data points for each experimental transient are presented for clarity. The green transient in A and B is the buffer control in the absence of microtubules at each KIF3AB concentration.
In converting the concentrations of mant-nucleotide–bound kinesin intermediates to fluorescence, one potential concern is whether mantADP–bound KIF3 sites produce the same relative fluorescence as mantATP–bound KIF3 sites. If the assumption is true, then the fluorescence amplitude for kinesins saturated with mantATP and mantADP should be double the amplitude for kinesins bound with a single mant-nucleotide. To test the hypothesis that KIF3·mantATP exhibits the same fluorescence output as the intermediates KIF3·mantADP·Pi and KIF3·mantADP, the preformed microtubule·KIF3AC complex was treated with apyrase to remove nucleotide from the active sites and then mixed rapidly in the stopped-flow instrument with either mantATP or mantADP (Fig. 4A). The results show that at three different concentrations of mantATP or mantADP, the resulting transients (mantATP and mantADP) superimpose on each other indicating that the fluorescence emission for KIF3AC during mantATP turnover does not change. Similar results are shown for KIF3AB in Fig. 4B. Therefore, the relative change in fluorescence as a function of time captured by the stopped-flow transients can be correlated with the time-dependent change in concentration of the mant-nucleotide–bound species.
Figure 4.
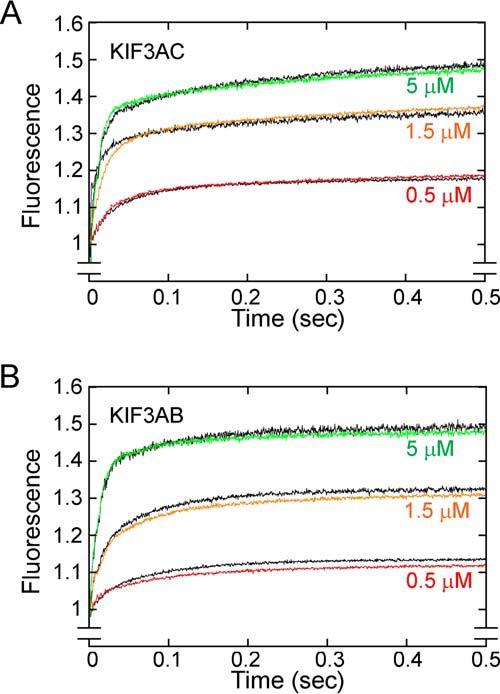
KIF3 mantATP and mantADP titration experiments. A MT·KIF3 complex was rapidly mixed with varying concentrations of mantATP or mantADP (final concentrations of 10 μm MT, 2 μm KIF3 heterodimer, and mantATP/mantADP at 0.5, 1.5, and 5 μm). The fluorescence emission of mantATP–bound KIF3 motors was compared with that of mantADP– KIF3 motors. A, the KIF3AC family of fluorescence transients. B, the KIF3AB family of fluorescence transients. Transients corresponding to titrations of mantATP are colored, and transients corresponding to titrations of mantADP are indicated in black.
Model assumptions for KIF3AC
Prior to model development, it was necessary to make several assumptions. First, for every KIF3AC dimer, only a single head collides with the microtubule for entry into the processive run (Fig. 1A, E0–E1). Second, because there is equal probability of either head binding the microtubule first, two parallel paths must be established: one for microtubule association by KIF3A and the other for microtubule association by KIF3C. Third, because the microtubule association rate constant at one head is largely independent of the microtubule association constant for its partner head, the KIF3AC microtubule association rate constant can be represented by the sum of the parameters for each head. Hence, the sum of the microtubule association parameters for KIF3A and KIF3C should equal the experimental microtubule association rate constant for KIF3AC at ∼6.6 μm−1 s−1 (50) (Table 1, Table S1).
The modeling also requires that the relative change in fluorescence emission as a function of time correlate with the time-dependent change in concentration of the mant-nucleotide–bound species during mantATP turnover. The results in Fig. 4 show conclusively that this assumption is valid. Therefore, the fluorescence output for the E5 intermediate where one head is occupied by mantADP and the other by mantATP was modeled as two times the fluorescence output of the E2 intermediate with only one mant-nucleotide–bound species.
Note that after ATP binding to form the E2 intermediate, some steps involve the partner head also (Fig. 1A, E2–E5). For the KIF3A pathway, the ATP-promoted structural transitions on KIF3A include movement of the KIF3C tethered head forward to its next microtubule-binding site followed by ADP release (E2–E4). For E4–E5, KIF3A hydrolyzes ATP followed by phosphate release and dissociation of the now tethered KIF3A head, and the now leading KIF3C head binds ATP. Note that the KIF3C pathway is similar but with the roles for the two heads interchanged.
The computational model
To test the hypothesis that heterodimerization alters the catalytic properties of KIF3A and KIF3C from their intrinsic properties within KIF3AC, we developed and implemented a nonlinear ODE model (Fig. S1C) to simulate the kinetics captured by the fluorescence transients in Fig. 2. The model was framed in the context of a kinesin stepping cycle to analyze microtubule association followed by native ADP release with subsequent ATP binding (E0–E2, Fig. 1A). Fig. 1B shows the model scheme that was used but in the context of kinesin intermediates with additional details in the supporting information. In Fig. 1B, reactions 1 and 2 represent the two parallel pathways of microtubule association for KIF3A (k1a and k−1a) and KIF3C (k1c and k−1c), respectively. ADP release and subsequent mantATP binding (Fig. 1A, E1–E2) is represented by reaction 3 for KIF3A (k2a) and reaction 4 for KIF3C (k2c). The steps for the ATP-promoted isomerization and subsequent transitions including neck linker docking, ATP hydrolysis, phosphate release, and tethered head detachment (Fig. 1A, E2–E5) are collapsed into a single first-order transition and designated by reactions 5 (k3a) and 6 (k3c) in Fig. 1B.
Fig. 1A, E2–E4 shows that after mantATP binding to the leading head, a series of structural transitions occurs to move the tethered, lagging ADP head forward to its next microtubule-binding site, where it binds the microtubule and releases ADP to form the E4 intermediate. The mantATP-promoted isomerization includes neck linker docking and rearrangement of the residues at the active site to form the E4 intermediate poised for ATP hydrolysis (49–51). MantATP hydrolysis occurs followed by phosphate release resulting in detachment of the rearward head with mantADP at the active site. The new leading head is now able to rapidly bind mantATP, resulting in the leading head occupied by mantATP and the rearward tethered head occupied by mantADP (Fig. 1A, E5). Experimentally it was determined that ATP binding occurs rapidly with the ATP-promoted isomerization at 81 s−1 and ATP hydrolysis at 69 s−1 (50) (Table 1). Because the ATP-promoted isomerization is coupled with ATP hydrolysis, one occurs rapidly and the other is limiting, but only the slower step can be quantified. For the computational model, the steps after E2 through E5 were collapsed into a single first-order rate constant, k3a for the KIF3A pathway and k3c for the KIF3C pathway.
The model terminates in a kinesin state where all sites are saturated with either mantATP or mantADP. Note that the E4 intermediate shows a state in which the leading head is nucleotide-free, therefore theoretically we should visualize the accumulation of this transient intermediate by a decline in the fluorescence emission. However, there was no observable decline in fluorescence in either the KIF3AC (Fig. 2, A, C, and E) or KIF3AB (Fig. 3, A and C) transients. We therefore conclude that the lifetime of the E4 intermediate is too short to accumulate sufficiently to detect a decline in fluorescence. For these reasons, we terminated the model at a state where all sites are saturated with mantATP or mantADP. If all sites were not saturated, then the estimates of the fluorescence parameters λ and 2·λ will be able to compensate for any deviations from this assumption. Additional evidence in support of this conclusion resulted from the fit of each KIF3AC transient to a double exponential function (Fig. 2, A, C, and E) to examine the relative amplitudes of each component. The relative amplitude for the fast component varied from 0.136 to 0.163 and was approximately equal to the amplitude of the slower component that varied from 0.128 to 0.173.
Once the reaction scheme was defined, we proceeded to develop the model shown in Fig. S1C. The model was comprised of 8 ODEs, derived from component balances, where each ODE represents the change in concentration of a kinesin state or nucleotide as a function of time. The time derivative is represented by a dot above the symbol representing the individual concentrations. The equation for (F) in Fig. S1C describes the fluorescence output.
Entry into the processive run for KIF3A and KIF3C is an emergent property of heterodimerization
For microtubule association, a rate constant of 6.6 μm−1 s−1 for KIF3AC could result from either KIF3A and KIF3C binding the microtubule similarly at rates of 3.3 μm−1 s−1 or from KIF3A colliding at a faster rate than KIF3C. Following parameter estimation, the results are consistent with a model in which there is an equal probability of entering the processive run by either KIF3A or KIF3C, and the kinetics of microtubule association are similar for both heads at ∼3.3 μm−1 s−1 (Table 2). This was surprising based on the extreme difference in the intrinsic catalytic properties of homodimeric KIF3AA and KIF3CC and reflects that within the heterodimer, the microtubule association kinetics of KIF3A were slowed, whereas the kinetics of KIF3C were accelerated.
Table 2.
KIF3AC and KIF3AB model parameter estimation
| Parameter | Estimate | Lower limit | Upper limit |
|---|---|---|---|
| KIF3AC | |||
| k1a (μm−1s−1) | 3.31 | 1.00a,b | 4.20 |
| k1c (μm−1s−1) | 3.29 | 2.40 | 5.70a,c |
| k2a (s−1) | 77.7 | ||
| k2c (s−1) | 7.6 | ||
| k3a (s−1) | 90.9 | 87.3 | 93.9 |
| k3c (s−1) | 13.0 | 10.1 | 16.6 |
| λ | 0.1069 | 0.1059 | 0.1083 |
| Training MSE | 1.45 × 10−4 | ||
| Validation MSE | 2.11 × 10−4 | ||
| KIF3AB | |||
| k1a (μm−1s−1) | 4.20 | 4.02 | 4.40 |
| k1b (μm−1s−1) | 4.20 | 4.02 | 4.40 |
| k2a (s−1) | 89.9 | ||
| k2b (s−1) | 80.2 | ||
| k3a (s−1) | 38.3 | 34.2 | 43.1 |
| k3b (s−1) | 38.3 | 34.2 | 43.1 |
| λ | 0.1104 | 0.1093 | 0.1115 |
| Training MSE | 1.09 × 10−4 | ||
| Validation MSE | 1.39 × 10−4 | ||
a Parameter was practically non-identifiable, and either a lower bound or upper bound could not be determined.
b KIF3A lower limit was set as the intrinsic KIF3C MT association rate.
c KIF3C upper limit was set as the intrinsic KIF3A MT association rate.
KIF3A and KIF3C maintain their intrinsic catalytic properties for ADP release
When we conducted the computational modeling, the simulations best captured the fluorescence transients when the ADP release rates for KIF3A and KIF3C were fast and slow at 77.7 and 7.6 s−1, respectively (see supporting information for details on the computational modeling; Tables S1–S3 and Figs. S1 and S2). In contrast, when the same simulations were conducted by fixing KIF3A and KIF3C to the same ADP release rate of 42.5 s−1 for KIF3AC, parameter estimation resulted in the KIF3A microtubule association parameter being significantly slower than that of KIF3C. However, KIF3A can either have faster catalytic properties than KIF3C or both heads can have similar properties (Table S2). Therefore, the modeling suggests that KIF3A and KIF3C maintain their intrinsic catalytic properties for ADP release (Fig. 2).
The later steps in the ATPase cycle (Fig. 1, E2–E5) including ATP hydrolysis followed by phosphate release coupled to motor head detachment from the microtubule, and mantATP binding to the new leading head at E5 can be collapsed into a single first-order rate constant. Although these steps have not yet been determined experimentally for KIF3AA and KIF3CC, the steady-state kcat for KIF3CC is ∼1 s−1, which suggests that there is likely a slow, rate-limiting step downstream of ADP release (Fig. 1, E2–E5). Because the catalytic properties of KIF3AA are fast, the intrinsic rate constant for the KIF3A pathway was assumed to be significantly faster than the KIF3C pathway. Through iterative estimation and simulation, the results reveal that the composite constants were 90.9 and 13.0 s−1 for the combined steps in the KIF3A and KIF3C pathways, respectively (Table 2). Because the composite constant for the KIF3A or KIF3C pathways includes ADP release by the opposite tethered head, the results here suggest that ADP release by tethered head KIF3C within the KIF3A pathway of the stepping cycle (Fig. 1A, E2–E4) must be faster than ADP release by KIF3C during entry into the stepping cycle (Fig. 1A, E2). If ADP release by KIF3C was the same rate during entry into and within the processive run, the rate constant for composite steps E2–E5 for the KIF3A pathway would be expected to be significantly slower. In addition, the stepping velocity and kcat for KIF3CC are ∼1 s−1, whereas the model predicts that the composite steps in the KIF3C pathway occur at a rate of 13 s−1 (Table 2). Therefore, these results are consistent with the hypothesis that KIF3A accelerates KIF3C (Fig. 1A, E2–E5).
KIF3A and KIF3B associate with the microtubule with similar, but slower rates than determined for KIF3AA and KIF3BB
Previous stopped-flow experiments showed that microtubule association by KIF3AB was 7 μm−1 s−1, yet the results for KIF3AA and KIF3BB were much faster at ∼12 μm−1 s−1 (Table 1) (49, 51). Moreover, the kinetics of ADP release for KIF3AB at ∼34 s−1 was approximately half as fast as that for KIF3AA and KIF3BB, each at ∼80 s−1 (Table 1) (49, 51). These results suggest that the catalytic properties of KIF3A and KIF3B within KIF3AB must be different from their intrinsic properties within KIF3AA and KIF3BB. To test this hypothesis and determine the catalytic properties of KIF3A and KIF3B within KIF3AB, additional experiments were performed to measure the time course of microtubule association followed by native ADP release by taking advantage of the fluorescence signal of mantATP binding (Fig. 3). An ODE model of the same structure that was used for KIF3AC (Fig. 1B, Fig. S1) was applied to the fluorescence transients. The transients in Fig. 3 reveal that the fast component of fluorescence enhancement is not well-separated from the slow component, consistent with the interpretation that the rates of KIF3A and KIF3B are similar. Moreover, the fluorescence amplitude saturates as expected based on the concentration of mantATP at 25 μm relative to the concentration of the KIF3AB heterodimer at 0.5 and 1 μm. A double exponential function was fit to each transient, and Fig. 3E shows the observed rates of the fast component of ADP release as a function of microtubule concentration. The hyperbolic fit to the data provided the maximum rate constant of native ADP release for KIF3AB at 39.6 s−1 (49), and it was used to guide the modeling. These results are consistent with previous experiments (49).
The KIF3AB microtubule association rate constant of 7 μm−1 s−1 could only result from KIF3A and KIF3B colliding with the microtubule at slower rates than within their respective homodimers KIF3AA and KIF3BB. If both heads retained their intrinsic properties for microtubule association, then the expected KIF3AB microtubule association rate would be ∼13 μm−1 s−1. In contrast, the computational model results suggest that KIF3A and KIF3B associate with the microtubule at rates of 4.2 μm−1 s−1 for each, which is slower than their intrinsic microtubule association rates within KIF3AA and KIF3BB (Table 2). Therefore, heterodimerization alters the microtubule association properties of KIF3A and KIF3B such that they are slower than their intrinsic properties for entry into the processive run.
The kinetics of ADP release for KIF3A and KIF3B after microtubule association are similar and fast
The KIF3AB experimental results presented in Fig. 3 revealed that the maximum rate constant for ADP release during entry into the processive run (E0–E1, Fig. 1A) is 39.6 s−1, which is approximately half the rate constant for KIF3AA and KIF3BB at 80 s−1 for each (Table 1) (49, 51). Although the model predicts that microtubule association is altered by heterodimerization, the release of ADP from each head of KIF3AB is consistent with the intrinsic kinetics of KIF3A and KIF3B as defined by KIF3AA and KIF3BB (Table 2). Alternative models where both heads are fixed to the same ADP release rate of ∼40 s−1 were ruled out because of a poor goodness of fit for the simulations to the fluorescence transients (Table S3). We conclude that the fluorescence output in the transients cannot be used to accurately estimate the contribution of each individual head, and therefore the experimental rate constant for ADP release was underestimated. Evidence in support of this interpretation is that the double exponential fit to the transients was unable to provide similar amplitudes for the fast and slow components of the double exponential.
The KIF3AB model reveals that processive stepping is controlled by a rate-limiting nucleotide-dependent transition
To explore the series of transitions after mantATP binding for KIF3AB (E3–E5, Fig. 1A), we assumed that the composite rate constant (k3) was similar for the KIF3A and KIF3B pathway and fast at ∼100 s−1 based on the parameter value for the KIF3A pathway of 90.9 s−1 in KIF3AC (Table 2). However, the model best captured the fluorescence transients when the transitions downstream of ADP release/mantATP binding occurred at rates of 38 s−1 (Table 2). These results suggest that the composite steps within E2–E5 in the KIF3A and KIF3B pathways in KIF3AB has a slower composite rate than that modeled for the KIF3A pathway in KIF3AC. Furthermore, the modeling predicts that the identity of the partner head of KIF3A affects the rate at which the KIF3A pathway proceeds through the series of transitions from E2 to E5 in Fig. 1A.
The velocity of processive stepping is controlled differently in KIF3AB and KIF3AC
The parameters presented in Table 2 indicate that the velocity at which KIF3AB steps along the microtubule will be controlled by a rate-limiting transition after ATP binding for entry into the processive run in part because each head within the heterodimer behaves similarly (43). In contrast, the processive stepping of KIF3AC is controlled differently and by the catalytic properties of KIF3C during the processive run. For KIF3AC, the time required to proceed through these transitions is defined as the transit time t and can be calculated by using the equation: t = 1/k. For the KIF3A pathway, the mantATP-promoted isomerization through dissociation results in a total transit time of 0.011 s. Similarly, for the KIF3C pathway, the transit time for the mantATP-promoted isomerization through dissociation is 0.0769 s. Because these transit times correspond to the time required for KIF3AC to take two 8-nm steps, the average velocity can be represented as 16 nm divided by the total transit time for KIF3AC (0.011 + 0.079 s = 0.0879 s or 182 nm/s). This velocity predicted by the modeling is consistent with the single-molecule stepping velocity of 186 nm/s (35). In addition, we also propose that within the KIF3AC heterodimer, KIF3A accelerates the transitions of KIF3C to some degree because the predicted rate of the transitions from E2 to E5 within the KIF3C pathway is 13 s−1 compared with ∼1 s−1 from the single-molecule homodimeric KIF3CC velocity of 7.5 nm/s. In contrast, the computational model for KIF3AB shows that each step of KIF3A and KIF3B will be comparable during the processive run. Table 2 shows that these parameters for KIF3A and KIF3B are each 38.3 s−1, which provides a total transit time of 0.0522 s for two 8-nm steps. The model thus predicts that the velocity of KIF3AB is 306 nm/s, which approximates the single-molecule velocity of 246 nm/s determined experimentally (Table 1) (35).
Goodness of fit of the model to the experimental data
Examination of the simulations for KIF3AC (Fig. 2) and KIF3AB (Fig. 3) reveal that the simulations do not perfectly superimpose on the fluorescence transients. This is due in part to the inherent error in these types of experiments with microtubules varying from 1 to 25 μm and the large number of transients used for the modeling. For example, the KIF3AB model slightly over-predicts the relative fluorescence output for low microtubule concentrations (Fig. 3B), whereas it marginally under-predicts the relative fluorescence values for high microtubule concentrations (Fig. 3D). Although the fit for low or high microtubule concentrations can be improved for individual experimental transients, this approach comes at the expense of worsening the fit for the other case. As such, the results present a good trade-off that ensures a reasonable fit for all of the data sets rather than an excellent fit for some data sets and a poor fit for others.
Discussion
The computational modeling revealed that there is an equal probability that either KIF3A or KIF3C can start the processive run, and there was no evidence of an intrinsic bias within the catalytic motor domains. Moreover, the results also showed that the microtubule association rate constant was approximately the same for KIF3A and KIF3C, which was completely unexpected based on the kinetics for KIF3AA and KIF3CC (50). Previously, we proposed that microtubule association would be determined by the intrinsic properties of KIF3A versus KIF3C, and then once in the processive run, an emergent mechanochemistry would result whereby each step would be the same regardless of whether the microtubule step was by KIF3A or KIF3C. Yet the computational model predicts quite convincingly that after microtubule association and once in the processive run, the motile properties are controlled by the relative intrinsic catalytic properties of either KIF3A or KIF3C (Table 2). Based on these results, we propose that once in the processive run, the steps would alternate between a fast step for KIF3A and a slow step for KIF3C in the absence of hindering load.
The developed model also captured the dynamics of KIF3AB with the overall interpretations similar to those of Andreasson et al. (43) in which the intrinsic catalytic properties of KIF3A and KIF3B are similar and fast, and therefore, each step in the processive run by KIF3AB would be similar. However, the computational modeling indicated that entry into the processive run by either KIF3A or KIF3B was also an emergent property with each at ∼4.2 μm−1 s−1 (Table 2) rather than 11–13 μm−1 s−1 determined for KIF3AA and KIF3BB (Table 1) (49). The computational model also showed that each kinetic step after microtubule association was similar for KIF3A and KIF3B, therefore, predicting that during a processive run, each interaction with the microtubule would occur at the same rate regardless of if it were by KIF3A or KIF3B. These results also support the findings of Andreasson et al. (43) in which they concluded that the motile properties of KIF3AB and its response to hindering load were encoded within the catalytic motor domains rather than the neck linker or coiled-coil domain.
Although this study did address aspects of our mechanistic questions for KIF3AC, there are many questions ahead including why is KIF3C so slow yet the motor domain sequences of KIF3A and KIF3C are so similar? Moreover, we do not yet know whether KIF3AC responds to hindering load as KIF3AB does and detaches from the microtubule more readily, or alternatively, whether KIF3AC is more similar to kinesin-2 KIF17 and kinesin-1, in which stepping can occur against appreciable hindering loads (45). This latter question is especially intriguing because the sequences of helix α6, the neck linker, and helix α7 of KIF3A, KIF3B, KIF3C, and KIF17 are almost identical (4), indicating that the force sensing motif(s) are within the catalytic motor domains.
Although we know that KIF3AC is expressed predominantly in neurons and acts as a cargo transporter (8, 9, 34), we do not yet know the cargos that KIF3AC transports. In contrast, KIF3AB is ubiquitously expressed including neurons (2–4, 34), but our best understanding of KIF3AB transport is for intraflagellar transport in cilia. One testable hypothesis for future investigation is whether KIF3AC and KIF3AB in neurons read the tubulin code differently based on posttranslational modifications of the axonal versus dendritic microtubules to bias selective transport of cargos to enter axons or dendrites (34, 53–55).
In summary, mathematical modeling has provided new insights to understand the motile properties of KIF3AC and KIF3AB. These results revealed that microtubule association for KIF3AC and KIF3AB is an emergent property due to heterodimerization, and once in the processive run, the rates of the nucleotide-dependent transitions result from the relative intrinsic catalytic properties of the motor domains. Moreover, the results predict that the rate of stepping of KIF3A and KIF3B will be approximately equivalent, yet for KIF3AC, the cycle results in an asymmetric walk in which processive steps will alternate between a fast step for KIF3A followed by a slow step for KIF3C in the absence of hindering load.
Experimental procedures
Kinesin-2 KIF3AC and KIF3AB heterodimeric motors
The Mus musculus KIF3A, KIF3B, and KIF3C plasmids for expression of heterodimeric KIF3AB and KIF3AC were described previously in detail (35). When expressed, KIF3A of heterodimeric KIF3AC includes the native N-terminal motor domain, neck-linker, and helix α7 (Met1–Leu374) to initiate coiled-coil formation, followed by the end-binding protein 1 (EB1) dimerization motif (DFYFGKLRNIELICQENEGENDPVLQRIVDILYATDE) to stabilize the dimer, and the tobacco etch virus (TEV) protease site with the cleavable Strep Tag (TTSENLYFQGASNWSHPQFEK). KIF3C when expressed includes the native motor domain, neck-linker, and helix α7 (Met1–Leu396), followed by the EB1 dimerization motif, and C-terminal TEV protease cleavable His8 tag (TTSENLYFQGASHHHHHHHH). For heterodimeric KIF3AB, KIF3A includes the native motor domain, neck-linker, and helix α7 (Met1–Glu376), an acidic heterodimerization motif (LEKEIAALEKEIAALEK), and a C-terminal TEV protease cleavable StrepII tag (51, 56, 57). KIF3B includes the native motor domain, native neck-linker, and helix α7 (Met1–Lys371), a basic heterodimerization motif (LKEKIAALKEKIAALKE), and a C-terminal TEV protease cleavable His8 tag (51). For consistency with prior studies using heterodimeric KIF3AB, a synthetic heterodimerization domain was used rather than the EB1 domain. Note that there is no significant difference in the motility properties for motors containing an EB1 or a synthetic heterodimerization domain (35). For the experiments reported here, the motors were not treated with TEV protease.
KIF3 protein expression and purification
The KIF3 heterodimeric motors were expressed using the Escherichia coli BL21-CodonPlus DE3 (RIL) cell line (Stratagene La Jolla, CA) by co-transformation of two plasmids and selection on LB plates containing 100 μg/ml of ampicillin, 50 μg/ml of kanamycin, and 10 μg/ml of chloramphenicol (35, 51). The KIF3AC or KIF3AB cell lysate was clarified by centrifugation and loaded onto a HisTrap FF nickel-nitrilotriacetic acid column (GE Healthcare) to select for the C-terminal His8 tag of KIF3B or KIF3C. Following elution from the column, fractions were analyzed by SDS-PAGE, and fractions enriched in the heterodimer were selected. Subsequently, the protein was loaded on the StrepTactinTM column (GE Healthcare), which selected for the C-terminal Strep-tagged KIF3A. The fractions were analyzed by SDS-PAGE, and those in which the KIF3A:KIF3B/C ratio was 1:1 were pooled. The KIF3 motor proteins were further dialyzed in ATPase buffer (20 mm HEPES, pH 7.2, with KOH, 5 mm magnesium acetate, 0.1 mm EDTA, 0.1 mm EGTA, 50 mm potassium acetate, 1 mm DTT, and 5% sucrose) at 4 °C, and clarified by ultracentrifugation. The purified KIF3AB and KIF3AC heterodimers were evaluated by analytical gel filtration chromatography (SuperoseTM 10/300, GE Healthcare) and SDS-PAGE to confirm purification of stable heterodimers at a 1:1 stoichiometry of KIF3A to KIF3B or KIF3A to KIF3C (35). Aliquots were frozen in liquid N2 for storage at −80 °C. The predicted molecular weight based on amino acid sequence of KIF3AC is 98,317 and KIF3AB is 92,131. Before each experiment, aliquots of KIF3AB or KIF3AC were thawed and clarified: 10 min at 4 °C (Beckman Coulter TLX Optima Ultracentrifuge, TLA-100 rotor, 313,000 × g). Protein concentration was determined using the Bio-Rad Protein Assay with IgG as the protein standard.
The experiments were performed at 25 °C in ATPase buffer: 20 mm HEPES, pH 7.2, with KOH, 5 mm magnesium acetate, 0.1 mm EDTA, 0.1 mm EGTA, 50 mm potassium acetate, 1 mm DTT, plus 5% sucrose. The morning of each experiment, bovine tubulin was cold-depolymerized, clarified, and polymerized with 1 mm MgGTP at 37 °C, followed by stabilization with 40 μm paclitaxel. The microtubule concentrations reported represent the paclitaxel-stabilized α,β-tubulin polymer (MT). The reported concentrations of ATP, GTP, and the nucleotide analogs 2′-(or 3′)-O-(N-methylanthraniloyl)ATP/ADP (mantATP/mantADP) include an equivalent concentration of magnesium acetate. MantATP and mantADP were purchased from Invitrogen as the isomeric mixture.
ADP release upon microtubule collision
To measure the kinetics of entry into the processive run (Fig. 1, E0–E2), KIF3AC or KIF3AB was rapidly mixed in the stopped-flow instrument (KinTek SF300X, KinTek Corp.) with varying concentrations of microtubules plus 50 μm mantATP, final concentration of 25 μm mantATP after mixing (49, 50). At the conditions of the assay, mantATP binding was significantly faster than ADP release after microtubule association (Table 1). Therefore, the enhanced relative fluorescence upon mantATP binding was used as the readout of ADP release (Figs. 2 and 3). Experiments were also performed at different time domains to optimize the experimental design so that the 1000 data points per transient were evenly distributed to capture the fast component and the slower component of fluorescence enhancement. Therefore, the time domain for KIF3AC was 0.5 s and for KIF3AB was 0.2 s.
A double exponential function was fit to each transient to obtain the observed rates and relative amplitudes of each fluorescence component. The observed rates of the fast component were plotted as a function of microtubule concentration. The hyperbolic fit to the data provided the maximum rate constant of native ADP release, and this constant was used to guide the computational modeling (49, 50).
MantATP and MantADP titrations
To determine whether the relative fluorescence amplitude exhibited by microtubule complexes of KIF3AC and KIF3AB changed during mantATP turnover, a series of experiments were pursued to evaluate mantATP binding compared with mantADP binding (Fig. 4). The MT·KIF3AC or MT·KIF3AB complexes were preformed (20 μm MT plus 4 μm KIF3 heterodimer) and treated with 0.02 unit/ml of apyrase (grade VII, Sigma) for 20 min at room temperature. Apyrase converts free ADP to AMP plus Pi. However, the affinity of kinesins for AMP is so weak that apyrase effectively generates a nucleotide-free state (58). Subsequently, the MT·KIF3 complex was rapidly mixed in the stopped-flow instrument with mantATP or mantADP at varying concentrations (1–10 μm) resulting in final concentrations of 2 μm KIF3 heterodimer, 10 μm MT, and mantATP/mantADP at 0.5–5 μm.
Experimental approach for data analysis
The presteady-state experiments in Figs. 2–4 were repeated 5–8 times at varying KIF3 and MT concentrations as well as at different time domains for data collection. Each transient shown is an average of 8–10 individual transients and for each time domain, the stopped-flow instrument collects 1000 data points. To compare the relative fluorescence amplitude of individual transients in each data set, each transient was adjusted by adding or subtracting an offset such that each transient begins at the same point on the y axis, arbitrary set at 1. This normalization approach was guided by the double exponential fit to the transients to identify the point on the y axis at time 0. The transients in Fig. 2 resulted from experiments performed on two separate days with the same stopped-flow settings to obtain an extensive data set, and those in Figs. 3 and 4 resulted from experiments performed on the same day for each data set. The rates for ADP release reported in Figs. 2G and 3E include data from multiple experiments to provide a more extensive range of microtubule concentrations. The hyperbolic fit to the data provides the mean ± S.E.
Author contributions
S. M. Q., D. P. H., J. H., and S. P. G. resources; S. M. Q., J. H., and S. P. G. data curation; S. M. Q., D. P. H., J. H., and S. P. G. formal analysis; S. M. Q., D. P. H., J. H., and S. P. G. validation; S. M. Q., D. P. H., J. H., and S. P. G. investigation; S. M. Q., D. P. H., J. H., and S. P. G. methodology; S. M. Q., D. P. H., and S. P. G. writing-original draft; S. M. Q., D. P. H., J. H., and S. P. G. writing-review and editing; D. P. H. and J. H. software; J. H. and S. P. G. conceptualization; J. H. and S. P. G. supervision; J. H. and S. P. G. funding acquisition; J. H. and S. P. G. project administration.
Supplementary Material
Acknowledgments
We thank Stephanie Guzik-Lendrum and Brandon Bensel for discussions of the experimental work in the context of the modeling.
This work was supported by NIGMS, National Institutes of Health Grants R01-AI110642 (to J. H.) and R37-GM054141 (to S. P. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1 and S2.
- IFT
- intraflagellar transport
- ODE
- ordinary differential equations
- mantATP/mantADP
- 2′-(or 3′)-O-(N-methylanthraniloyl)ATP/ADP
- MT
- microtubule
- EB1
- end-binding protein 1
- TEV
- tobacco etch virus.
References
- 1. Hirokawa N., Niwa S., and Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- 2. Verhey K. J., Kaul N., and Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 10.1146/annurev-biophys-042910-155310 [DOI] [PubMed] [Google Scholar]
- 3. Scholey J. M. (2013) Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 29, 443–469 10.1146/annurev-cellbio-101512-122335 [DOI] [PubMed] [Google Scholar]
- 4. Gilbert S. P., Guzik-Lendrum S., and Rayment I. (2018) Kinesin-2 motors: kinetics and biophysics. J. Biol. Chem. 293, 4510–4518 10.1074/jbc.R117.001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kondo S., Sato-Yoshitake R., Noda Y., Aizawa H., Nakata T., Matsuura Y., and Hirokawa N. (1994) KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125, 1095–1107 10.1083/jcb.125.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1995) KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130, 1387–1399 10.1083/jcb.130.6.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sardella M., Navone F., Rocchi M., Rubartelli A., Viggiano L., Vignali G., Consalez G. G., Sitia R., and Cabibbo A. (1998) KIF3C, a novel member of the kinesin superfamily: sequence, expression, and mapping to human chromosome 2 at 2p23. Genomics 47, 405–408 10.1006/geno.1997.5123 [DOI] [PubMed] [Google Scholar]
- 8. Muresan V., Abramson T., Lyass A., Winter D., Porro E., Hong F., Chamberlin N. L., and Schnapp B. J. (1998) KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell 9, 637–652 10.1091/mbc.9.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Z., and Goldstein L. S. (1998) Characterization of the KIF3C neural kinesin-like motor from mouse. Mol. Biol. Cell 9, 249–261 10.1091/mbc.9.2.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cole D. G., Chinn S. W., Wedaman K. P., Hall K., Vuong T., and Scholey J. M. (1993) Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366, 268–270 10.1038/366268a0 [DOI] [PubMed] [Google Scholar]
- 11. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1996) Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. U.S.A. 93, 8443–8448 10.1073/pnas.93.16.8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wedaman K. P., Meyer D. W., Rashid D. J., Cole D. G., and Scholey J. M. (1996) Sequence and submolecular localization of the 115-kDa accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132, 371–380 10.1083/jcb.132.3.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimizu K., Shirataki H., Honda T., Minami S., and Takai Y. (1998) Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J. Biol. Chem. 273, 6591–6594 10.1074/jbc.273.12.6591 [DOI] [PubMed] [Google Scholar]
- 14. Doodhi H., Ghosal D., Krishnamurthy M., Jana S. C., Shamala D., Bhaduri A., Sowdhamini R., and Ray K. (2009) KAP, the accessory subunit of kinesin-2, binds the predicted coiled-coil stalk of the motor subunits. Biochemistry 48, 2248–2260 10.1021/bi8018338 [DOI] [PubMed] [Google Scholar]
- 15. Gindhart J. G. Jr., and Goldstein L. S. (1996) Armadillo repeats in the SpKAP115 subunit of kinesin-II. Trends Cell Biol. 6, 415–416 10.1016/S0962-8924(96)20037-6 [DOI] [PubMed] [Google Scholar]
- 16. Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., and Anderson K. V. (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- 17. Drummond I. A. (2012) Cilia functions in development. Curr. Opin. Cell Biol. 24, 24–30 10.1016/j.ceb.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., and Hirokawa N. (1998) Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- 19. Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., and Hirokawa N. (1999) Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by KIF3A−/− mice analysis. J. Cell Biol. 145, 825–836 10.1083/jcb.145.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marszalek J. R., Ruiz-Lozano P., Roberts E., Chien K. R., and Goldstein L. S. (1999) Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. U.S.A. 96, 5043–5048 10.1073/pnas.96.9.5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., and Goldstein L. S. (2000) Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 10.1016/S0092-8674(00)00023-4 [DOI] [PubMed] [Google Scholar]
- 22. Lin F., Hiesberger T., Cordes K., Sinclair A. M., Goldstein L. S., Somlo S., and Igarashi P. (2003) Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. U.S.A. 100, 5286–5291 10.1073/pnas.0836980100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teng J., Rai T., Tanaka Y., Takei Y., Nakata T., Hirasawa M., Kulkarni A. B., and Hirokawa N. (2005) The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat. Cell Biol. 7, 474–482 10.1038/ncb1249 [DOI] [PubMed] [Google Scholar]
- 24. Brunnbauer M., Mueller-Planitz F., Kösem S., Ho T. H., Dombi R., Gebhardt J. C., Rief M., and Okten Z. (2010) Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc. Natl. Acad. Sci. U.S.A. 107, 10460–10465 10.1073/pnas.1005177107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takeda S., Yamazaki H., Seog D. H., Kanai Y., Terada S., and Hirokawa N. (2000) Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J. Cell Biol. 148, 1255–1265 10.1083/jcb.148.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao C., Omori Y., Brodowska K., Kovach P., and Malicki J. (2012) Kinesin-2 family in vertebrate ciliogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 2388–2393 10.1073/pnas.1116035109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaudin R., de Alencar B. C., Jouve M., Bèrre S., Le Bouder E., Schindler M., Varthaman A., Gobert F. X., and Benaroch P. (2012) Critical role for the kinesin KIF3A in the HIV life cycle in primary human macrophages. J. Cell Biol. 199, 467–479 10.1083/jcb.201201144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirokawa N., Tanaka Y., and Okada Y. (2012) Cilia, KIF3 molecular motor and nodal flow. Curr. Opin. Cell Biol. 24, 31–39 10.1016/j.ceb.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 29. Gumy L. F., Chew D. J., Tortosa E., Katrukha E. A., Kapitein L. C., Tolkovsky A. M., Hoogenraad C. C., and Fawcett J. W. (2013) The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 33, 11329–11345 10.1523/JNEUROSCI.5221-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ichinose S., Ogawa T., and Hirokawa N. (2015) Mechanism of activity-dependent cargo loading via the phosphorylation of KIF3A by PKA and CaMKIIa. Neuron 87, 1022–1035 10.1016/j.neuron.2015.08.008 [DOI] [PubMed] [Google Scholar]
- 31. Carpenter B. S., Barry R. L., Verhey K. J., and Allen B. L. (2015) The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci. 128, 1034–1050 10.1242/jcs.162552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruane P. T., Gumy L. F., Bola B., Anderson B., Wozniak M. J., Hoogenraad C. C., and Allan V. J. (2016) Tumour suppressor adenomatous polyposis coli (APC) localisation is regulated by both kinesin-1 and kinesin-2. Sci. Rep. 6, 27456 10.1038/srep27456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakagawa T., Tanaka Y., Matsuoka E., Kondo S., Okada Y., Noda Y., Kanai Y., and Hirokawa N. (1997) Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc. Natl. Acad. Sci. U.S.A. 94, 9654–9659 10.1073/pnas.94.18.9654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang C. F., and Banker G. (2012) The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic 13, 549–564 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guzik-Lendrum S., Rank K. C., Bensel B. M., Taylor K. C., Rayment I., and Gilbert S. P. (2015) Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 109, 1472–1482 10.1016/j.bpj.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guzik-Lendrum S., Rayment I., and Gilbert S. P. (2017) Homodimeric kinesin-2 KIF3CC promotes microtubule dynamics. Biophys. J. 113, 1845–1857 10.1016/j.bpj.2017.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svoboda K., Schmidt C. F., Schnapp B. J., and Block S. M. (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365, 721–727 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- 38. Schnitzer M. J., and Block S. M. (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388, 386–390 10.1038/41111 [DOI] [PubMed] [Google Scholar]
- 39. Hua W., Young E. C., Fleming M. L., and Gelles J. (1997) Coupling of kinesin steps to ATP hydrolysis. Nature 388, 390–393 10.1038/41118 [DOI] [PubMed] [Google Scholar]
- 40. Asbury C. L., Fehr A. N., and Block S. M. (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 10.1126/science.1092985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yildiz A., Tomishige M., Vale R. D., and Selvin P. R. (2004) Kinesin walks hand-over-hand. Science 303, 676–678 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]
- 42. Kaseda K., Higuchi H., and Hirose K. (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 5, 1079–1082 10.1038/ncb1067 [DOI] [PubMed] [Google Scholar]
- 43. Andreasson J. O. L., Shastry S., Hancock W. O., and Block S. M. (2015) The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 25, 1166–1175 10.1016/j.cub.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schroeder H. W. 3rd, Hendricks A. G., Ikeda K., Shuman H., Rodionov V., Ikebe M., Goldman Y. E., and Holzbaur E. L. (2012) Force-dependent detachment of kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys. J. 103, 48–58 10.1016/j.bpj.2012.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Milic B., Andreasson J. O. L., Hogan D. W., and Block S. M. (2017) Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proc. Natl. Acad. Sci. U.S.A. 114, E6830–E6838 10.1073/pnas.1708157114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoeprich G. J., Thompson A. R., McVicker D. P., Hancock W. O., and Berger C. L. (2014) Kinesin's neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 106, 1691–1700 10.1016/j.bpj.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chien A., Shih S. M., Bower R., Tritschler D., Porter M. E., and Yildiz A. (2017) Dynamics of the IFT machinery at the ciliary tip. Elife 6, e28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stepp W. L., Merck G., Mueller-Planitz F., and Ökten Z. (2017) Kinesin-2 motors adapt their stepping behavior for processive transport on axonemes and microtubules. EMBO Rep. 18, 1947–1956 10.15252/embr.201744097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Albracht C. D., Guzik-Lendrum S., Rayment I., and Gilbert S. P. (2016) Heterodimerization of kinesin-2 KIF3AB modulates entry into the processive run. J. Biol. Chem. 291, 23248–23256 10.1074/jbc.M116.752196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang P., Rayment I., and Gilbert S. P. (2016) Fast or slow, either head can start the processive run of kinesin-2 KIF3AC. J. Biol. Chem. 291, 4407–4416 10.1074/jbc.M115.705970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Albracht C. D., Rank K. C., Obrzut S., Rayment I., and Gilbert S. P. (2014) Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289, 27836–27848 10.1074/jbc.M114.583914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen G. Y., Arginteanu D. F., and Hancock W. O. (2015) Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 290, 10274–10294 10.1074/jbc.M114.628032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tas R. P., Chazeau A., Cloin B. M. C., Lambers M. L. A., Hoogenraad C. C., and Kapitein L. C. (2017) Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271 10.1016/j.neuron.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bentley M., and Banker G. (2016) The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci. 17, 611–622 10.1038/nrn.2016.100 [DOI] [PubMed] [Google Scholar]
- 55. Park J. H., and Roll-Mecak A. (2018) The tubulin code in neuronal polarity. Curr. Opin. Neurobiol. 51, 95–102 10.1016/j.conb.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lindhout D. A., Litowski J. R., Mercier P., Hodges R. S., and Sykes B. D. (2004) NMR solution structure of a highly stable de novo heterodimeric coiled-coil. Biopolymers 75, 367–375 10.1002/bip.20150 [DOI] [PubMed] [Google Scholar]
- 57. Rank K. C., Chen C. J., Cope J., Porche K., Hoenger A., Gilbert S. P., and Rayment I. (2012) Kar3Vik1, a member of the kinesin-14 superfamily, shows a novel kinesin microtubule binding pattern. J. Cell Biol. 197, 957–970 10.1083/jcb.201201132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krzysiak T. C., Grabe M., and Gilbert S. P. (2008) Getting in sync with dimeric Eg5: initiation and regulation of the processive run. J. Biol. Chem. 283, 2078–2087 10.1074/jbc.M708354200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.