Abstract
Chromatin remodelers use bromodomains (BDs) to recognize histones. Polybromo 1 (PBRM1 or BAF180) is hypothesized to function as the nucleosome-recognition subunit of the PBAF chromatin-remodeling complex and is frequently mutated in clear cell renal cell carcinoma (ccRCC). Previous studies have applied in vitro methods to explore the binding specificities of the six individual PBRM1 BDs. However, BD targeting to histones and the influence of neighboring BD on nucleosome recognition have not been well characterized. Here, using histone microarrays and intact nucleosomes to investigate the histone-binding characteristics of the six PBRM1 BDs individually and combined, we demonstrate that BD2 and BD4 of PBRM1 mediate binding to acetylated histone peptides and to modified recombinant and cellular nucleosomes. Moreover, we show that neighboring BDs variably modulate these chromatin interactions, with BD1 and BD5 enhancing nucleosome interactions of BD2 and BD4, respectively, whereas BD3 attenuated these interactions. We also found that binding pocket missense mutations in BD4 observed in ccRCC disrupt PBRM1–chromatin interactions and that these mutations in BD4, but not similar mutations in BD2, in the context of full-length PBRM1, accelerate ccRCC cell proliferation. Taken together, our biochemical and mutational analyses have identified BD4 as being critically important for maintaining proper PBRM1 function and demonstrate that BD4 mutations increase ccRCC cell growth. Because of the link between PBRM1 status and sensitivity to immune checkpoint inhibitor treatment, these data also suggest the relevance of BD4 as a potential clinical target.
Keywords: cancer, chromatin, chromatin regulation, chromatin structure, histones, histone acetylation, histone methylation, histone modification, bromodomain, PBRM1, BAF180
Introduction
Chromatin compaction regulates DNA accessibility. In response to stimuli, cells alter chromatin through covalent modification of histones and the activity of chromatin remodelers. ϵ-N-acetylation of lysine residues is one of the most abundant histone tail modifications and is often associated with transcriptional activation (1). The interaction of proteins with acetylated histones is typically mediated by evolutionarily conserved bromodomains (BDs)2 (2, 3). Sixty-one BDs are found across a diverse array of human proteins, including many chromatin modifiers. BDs are made up of four α-helices linked by two loops to form a hydrophobic cavity. Although BDs share a conserved structure, they display large sequence variation and recognize a range of acetylated histones (4). A conserved asparagine at the C terminus of helix αB mediates interaction with the acetyl group (2, 5, 6).
The original histone code hypothesis proposed that protein interactions with chromatin are mediated through recognition of specific patterns of histone posttranslational modifications (PTMs) (7). The functional redundancy and promiscuous binding of BDs to acetylated lysines suggest a multivalent model in which linked domains mediate cooperative interactions with combinations of histone PTMs (8). Significantly, BDs often exist in tandem with other chromatin recognition domains, most commonly a plant homeodomain (PHD) or second BD (4, 8), suggesting that multivalency may be common in proteins that contain these domains. Most studies of BDs have focused on individual domains, without consideration for the effect of neighboring reader domains. However, several studies support that cooperative interactions can mediate multivalent interactions (8–11). For example, BPTF, a member of the NURF chromatin-remodeling complex, contains a BD and PHD that mediate interaction with nucleosomes harboring H3K4me3 and H4K16ac (12). Tandem domains can also demonstrate autoregulatory function (13). Because of relationships between these protein domains, consideration of BD context is critical when assessing the targeting specificity of these proteins.
PBRM1 (polybromo-1, PB1, BAF180), a member of the PBAF SWI/SNF chromatin-remodeling complex, is unique among chromatin reader proteins as it contains six tandem BDs. No other protein contains more than two BDs, and other components of the SWI/SNF complex contain at most a single BD. The presence of multiple BDs suggests that PBRM1 may act as the nucleosome recognition subunit of the PBAF complex (14). PBRM1 is mutated in ∼40% of clear cell renal cell carcinomas (ccRCCs) (15, 16). However, the role of PBRM1 loss in ccRCC development remains unclear. How the multiple BDs of PBRM1 mediate chromatin interaction would provide insights into the function of PBRM1 and could suggest a role for PBRM1 mutation in the development of ccRCC.
Previous studies have explored the binding of individual PBRM1 BDs to histones' tail sequences using synthetic peptides (4, 6, 17, 18). However, because biologically relevant recognition of the histone tails is in context of a multimeric nucleosome, synthetic peptides may offer a restricted representation of native binding interactions. Also, because these studies were performed with individual BDs, the relationship between neighboring BDs on histone recognition has remained unexplored. The reliance on synthetic peptides, experimental variation, and a low affinity for individual BDs to acetyl lysine residues (18) have limited the robustness and consistency of prior studies.
In this study, we explored the binding characteristics of the six PBRM1 BDs, both individually and in tandem, to histone peptides and to intact nucleosomes. We assessed the impact of kidney cancer–associated mutations in the context of individual and tandem BDs, and full-length PBRM1 to highlight regions within the BDs that are critical for PBRM1 function. We observed selective binding of individual BDs as well as the influence of neighboring BD on peptides and nucleosome interactions, together suggesting a unique role for BD4.
Results
Amino acid sequence classifies PBRM1 bromodomains
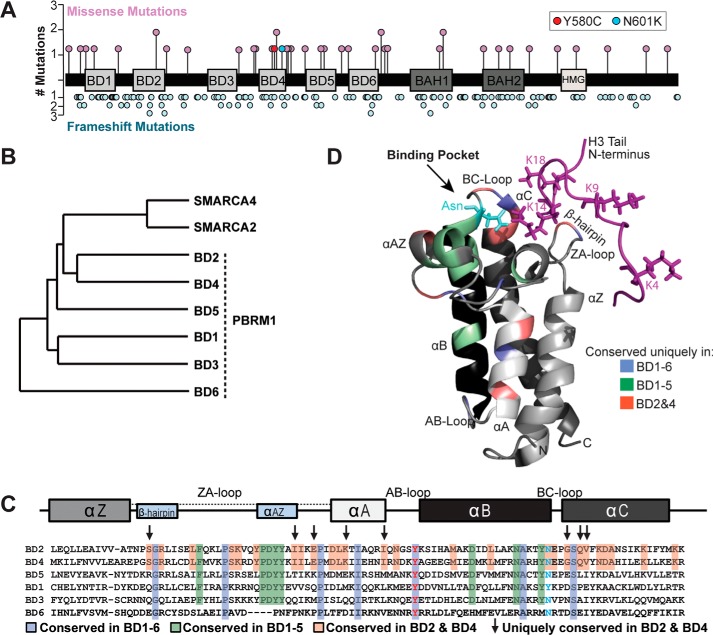
We compiled PBRM1 mutations observed in kidney cancers from several studies, including The Cancer Genome Atlas KIRC project (19–22). We focused on the nonsynonymous missense mutations (17% of total) based on the hypothesis that these mutations would be located in cancer-relevant functional domains in PBRM1. In contrast to frameshift mutations that were distributed throughout the gene, missense mutations tended to cluster within the BDs, specifically in and proximal to BD4 (p < 0.05 by permutation) (Fig. 1A). BD4 harbored five missense mutations, including two at highly conserved amino acids (Y580C and N601K). No missense mutations were identified in BD3. One mutation was identified in BD2, I252R, a nonconserved residue.
Figure 1.
PBRM1 mutation rate and sequence conservation varies across bromodomains. A, schematic diagram of PBRM1 marked with missense and frameshift mutations found in kidney cancer. B, dendrogram demonstrating amino acid sequence conservation of PBRM1 and other SWI/SNF complex–associated bromodomains. C, sequence alignment of PBRM1 bromodomains. Shading and arrows reflect the degree of conservation between bromodomains. Residues commonly mutated in ccRCC are shown with red and blue text. Bromodomain structural elements marked above the alignments. D, conserved residues overlaid on an NMR-derived secondary structure of PBRM1 BD2 in association with histone H3 tail (purple).
We then evaluated the amino acid sequence conservation between the six PBRM1 BDs. Hierarchical clustering analysis was performed (23). The comparison included the BDs from SMARCA4 (BRG1) and SMARCA2 (BRM), the other BD-containing components of the SWI/SNF complex. The BDs segregated into three clusters (Fig. 1B). BD2 and BD4 shared the greatest sequence similarity (50.7%) and grouped together with the SMARCA2 and SMARCA4 BDs. BD1 clustered with BD3 (42.3%); BD5 clustered with BD2 (39%) and BD4 (42%). BD6 was most distinct, exhibiting less than 26% sequence similarity to the other BDs. We next identified those amino acids that influenced clustering (Fig. 1C). Alignment of the six BDs revealed that seven amino acids were conserved across all BDs. Among these was a tyrosine residue within the AB-loop that is the site of a missense mutation in ccRCC (Y580C in BD4). Because BD6 shares the least sequence similarity to the other BDs, we focused on residues conserved between BDs 1–5. Residues conserved between these BDs are enriched in the ZA-loop and termini of helix αB, regions that help form the hydrophobic pocket. Amino acids that distinguish BD2 and BD4 were clustered within the ZA-loop and αA and αC helices.
To determine how amino acids may direct the function of each BD, we mapped conserved amino acids onto a published NMR solution structure of the BD2/H3K14ac complex (Fig. 1D) (18). Conserved residues cluster in and around the BD binding pocket. Three of the residues shared uniquely by BD2 and BD4 reside within the region of αC helix that forms the hydrophobic pocket. These residues are proximal to the conserved asparagine that likely mediates interaction with acetylated H3K14 and have been shown to help establish histone interactions (18). Two amino acids shared by BD2 and BD4 are in the ZA-loop predicted to be near the N terminus of the histone tail. In contrast, BD1, BD3, and BD5 share fewer conserved residues at regions that interface with the histone tail (Fig. S1), suggesting that these BDs may have weaker binding interactions with histone tails.
PBRM1 bromodomain-binding specificities reflect variation in primary sequence
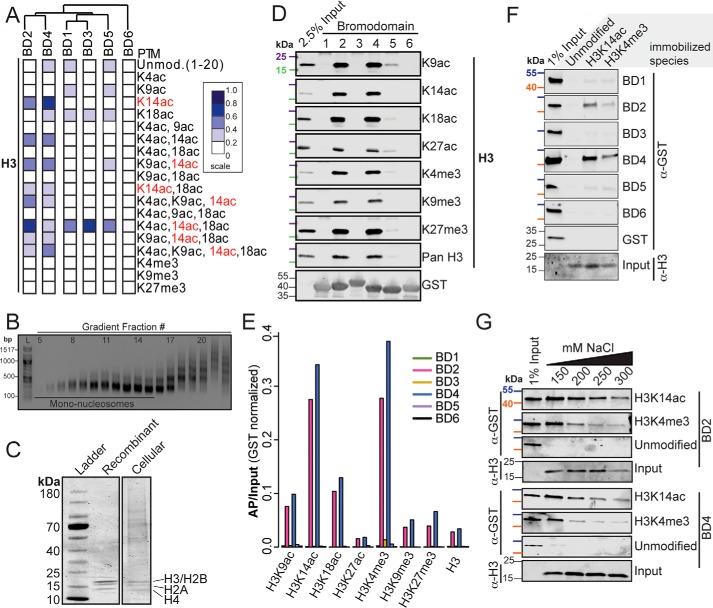
To test for an association between sequence composition and histone recognition, we examined the binding of each BD to histone PTMs using a microarray platform. The histone microarrays consist of >200 synthetic peptides reflecting the core histone tails (H2A, H2B, H3, and H4) modified with one or more known PTMs (24). GST-tagged BDs were incubated with the arrays, and interactions were quantified with a fluorescent anti-GST antibody (Fig. 2A). Binding was rank-ordered relative to the highest signal in the array. BD2 and BD4 recognized a similar pattern of H3 PTMs, with a marked preference for peptides containing H3K14ac alone or in combination with other acetylated amino acids. Acetylation at other sites was not associated with BD2 or BD4 binding. BD1, BD3, and BD5 weakly interacted with H3K14ac modified peptides when in combination with H3K4ac and H3K18ac. Minimal binding of BD2 and BD4 to the unmodified H3 tail was detected, demonstrating that binding is to the histone tail modification, not the tail itself. BDs 1 and 5 weakly bound to various monoacetylated peptides, including H3K18ac. However, this binding intensity was similar to that of the unmodified tail. BD6 demonstrated minimal binding to all peptides. The overall pattern of binding to modified H3 histone tail peptides reflected their sequence conservation (Fig. 1B), suggesting that the BD sequence determines the function of the individual domains.
Figure 2.
PBRM1 BD2 and BD4 recognize H3K14ac and H3K4me3. A, quantification of peptide microarray interactions between individual PBRM1 BDs and H3 peptides. Each column represents one peptide array. Normalized mean intensities are rank-ordered within each array. Dendrogram indicates clustering using Pearson correlation. B, agarose gel electrophoresis of sucrose gradient fractions. 100-bp ladder (L) is shown. Fractions pooled as mononucleosomes are also shown. C, Coomassie Blue–stained SDS-PAGE gel of recombinant nucleosomes and cellular extracted nucleosomes. Presence of all four core histones is indicated. D, mononucleosomes selected by GST-tagged PBRM1 BDs were separated by SDS-PAGE and blotted with antibodies that recognize H3 tail PTMs. 2.5% of the total mononucleosome pool was included for reference. 25 kDa (purple) and 15 kDa (green) molecular markers are indicated. E, quantification of signal for BD-associated nucleosomes relative to total nucleosome input (AP/input). F, unmodified or H3K14ac- or H3K4me3-modified recombinant nucleosomes were used to select single BDs. Proteins were separated by SDS-PAGE and blotted with anti-GST antibody and anti-H3 antibody (input). GST-BD input (1%) is shown. 55 kDa (blue) and 40 kDa (orange) molecular markers are indicated. G, unmodified or H3K14ac- or H3K4me3-modified recombinant nucleosomes were incubated with BD2 and BD4 at several concentrations of NaCl. Associated proteins were separated by SDS-PAGE and blotted with anti-GST antibody. 55 kDa (blue) and 40 kDa (orange) molecular markers are indicated.
BD2 and BD4 mediate primary interactions with H3 tails
Because the interaction of BDs with histone tail peptides may only partially reflect how histone tails are recognized in the context of an intact nucleosome (4, 6, 17, 18), we then assessed BD association with gradient-purified mononucleosomes from micrococcal nuclease (MNase)-treated nuclei (12) (Fig. 2B). We used a range of enzyme concentrations to ensure that the pool of nucleosomes would broadly reflect chromatin states (Fig. S2A). Purified nucleosomes contained all four core histones (Fig. 2C). Bead-bound GST-tagged BDs were incubated with the cellular nucleosomes, and enriched histone PTMs were detected by Western blot analysis (Fig. 2D). We found that BD2 and BD4 strongly interacted with nucleosomes, whereas BD5 only weakly bound nucleosomes. No binding was detected for BD1, BD3, and BD6.
We then identified the PTMs that were enriched on BD-bound nucleosomes (Fig. 2E). Because individual nucleosomes likely harbor multiple PTMs, observed PTMs may directly or indirectly mediate BD binding. Nucleosomes bound by BD2 and BD4 were preferentially marked by H3K14ac, consistent with the peptide results. Bound nucleosomes were also enriched for H3K4me3. Because binding to H3K4me3 was not observed on the peptide array, we hypothesized that BD-bound nucleosomes harbor both H3K14ac and H3K4me3, with binding primarily mediated by acetylation. The use of mononucleosomes excluded the possibility that binding was mediated by neighboring nucleosomes. To directly test the effect of each PTM on nucleosome binding, we then used unmodified recombinant nucleosomes, those exclusively modified by either H3K14ac or H3K4me3 (Fig. 2F, Fig. S2B). No BD bound the unmodified nucleosomes. Only BD2 and BD4 bound nucleosomes harboring H3K14ac. Limited binding to H3K4me3-marked nucleosomes was also detected. Because binding to H3K4me3 was unexpected, we assessed the relative binding affinity to H3K14ac and H3K4me3 by varying the concentration of NaCl in the binding reaction (Fig. 2G). In contrast to unmodified nucleosomes for which interaction was completely disrupted by 150 mm NaCl, BD2 and BD4 interaction with H3K14-acetylated and H3K4-trimethylated nucleosomes persisted to 300 mm NaCl, although increased binding to H3K14ac-modified was observed. Taken together, these data demonstrate that, in vitro, BD2 and BD4 are able to interact with nucleosomes marked with a wide range of PTMs but are mediated through a direct interaction with H3K14ac and/or H3K4me3.
Neighboring bromodomains influence nucleosome binding
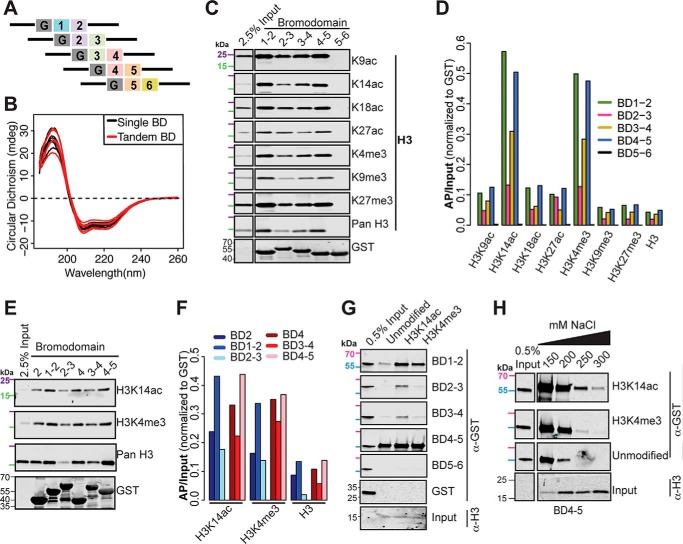
BDs often exist in tandem with other chromatin reader domains to facilitate binding specificity (4, 8). Prior biochemical studies characterizing PBRM1 BD binding specificity have examined individual BDs (4, 6, 17, 18). We hypothesized that neighboring PBRM1 BDs would influence BD2 and BD4 binding. To explore this idea, we generated and FPLC purified five overlapping tandem BDs (Fig. 3A). Circular dichroism (CD) demonstrated similar structures for all single and tandem BDs (Fig. 3B). Consistent with the results from single BDs, only tandem domains that included either BD2 or BD4 bound nucleosomes (Fig. 3C). BD5–6 failed to interact with nucleosomes. Similar to that observed with the individual BDs, the tandem BDs preferentially recognized nucleosomes harboring H3K14ac and H3K4me3 (Fig. 3D). Intriguingly, the combination of BD1 with BD2 and BD5 with BD4 enhanced binding to nucleosomes, whereas the combination of BD3 with either BD2 or BD4 decreased binding.
Figure 3.
Neighboring PBRM1 BDs influence binding of BD2 and BD4. A, schematic of tandem GST-tagged PBRM1 BD constructs, where G stands for GST. B, CD absorbance spectra comparing single (black) and tandem (red) PBRM1 BDs from 190–260 nm. C, GST-tagged tandem BDs were used to select sucrose-gradient purified mononucleosomes. Associated PTMs were detected by Western blotting. 25 kDa (purple) and 15 kDa (green) molecular markers are indicated. D, normalized (AP/input) signal is shown for H3-associated PTMs. E, cellular nucleosome interactions for single and tandem BDs were assessed by Western blotting. 25 kDa (purple) and 15 kDa (green) molecular markers are indicated. F, normalized (AP/input) signal is shown. G, unmodified or H3K14ac- or H3K4me3-marked recombinant nucleosomes were incubated with tandem BDs. Associated proteins were separated by SDS-PAGE and blotted with anti-GST antibody and anti-H3 antibody (input). BD input (0.5%) is shown. 70 kDa (pink) and 55 kDa (light blue) molecular markers are indicated. H, unmodified or H3K14ac- or H3K4me3-marked recombinant nucleosomes were incubated with tandem BDs in the presence of various concentrations of NaCl. Bound proteins were separated by SDS-PAGE and blotted with anti-GST and anti-H3 (input) antibodies. 70 kDa (pink) and 55 kDa (light blue) molecular markers are indicated.
We then directly compared nucleosome binding of individual BDs with tandem BDs (Fig. 3E). We found that BD1–2 showed enhanced binding over BD2 alone, and BD4–5 showed enhanced binding over BD4 alone (Fig. 3F). As BD1 and BD5 do not bind nucleosomes individually (Fig. 2D), the enhanced binding of BD1–2 and BD4–5 suggests that the BDs must cooperate to facilitate histone interactions. Strikingly, the addition of BD3 to either BD2 or BD4 decreased binding below that observed with the individual BD.
As before, we then used recombinant nucleosomes to assess binding to nucleosomes exclusively modified by H3K14ac or H3K4me3 (Fig. 3G). At low salt concentrations, BD1–2 and 4–5 bound H3K14ac, H3K4me3, and unmodified nucleosomes. BD2–3 and 3–4 weakly bound H3K14ac and H3K4me3. Similar to our previous findings, binding of either BD2 or BD4 to H3K14ac-marked nucleosomes was attenuated by the presence of BD3. Neither BD3 containing tandem BD bound unmodified nucleosomes. BD5–6 failed to bind any nucleosomes. As before, we varied the salt concentration in the binding reaction (Fig. 3H). Increased NaCl fully disrupted BD4–5 binding to the unmodified nucleosomes. Binding to H3K14ac- and H3K4me3-modified nucleosomes persisted at high salt concentration, although more binding to H3K14ac was evident. These data suggest that, in vitro, BD4–5 can bind either H3K14ac or H3K4me3, although binding to H3K14ac is favored.
PBRM1 bromodomains interact with the H4 N terminus
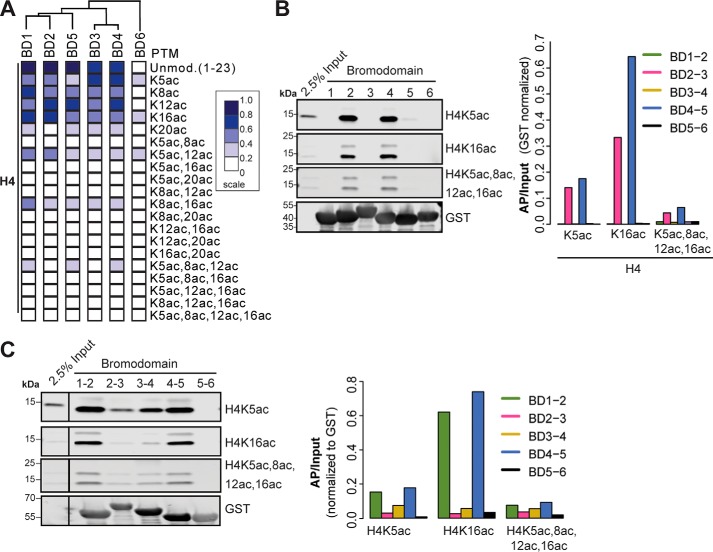
We unexpectedly observed that several of the PBRM1 BDs interact with H4 tail peptides, with the degree of interaction inversely related to acetylation state (Fig. 4A). Assayed on the peptide microarray, BDs 1–5 interacted with the unmodified and monoacetylated N terminus of H4 to various degrees. Limited binding by BDs 1–5 to diacetylated H4K5+K12 and H4K8+K16 was detected. Binding was not detected to triacetylated and tetraacetylated H4 tails. Unlike binding to the H3 tail, the BD binding patterns for H4 did not correspond to sequence conservation, suggesting these interactions may occur outside of the conserved binding pocket. To determine whether nucleosomes bound by the BDs demonstrated selective enrichment for specific H4 tail modifications, we examined three histone H4 modifications: H4K5ac, H4K16ac, and tetraacetylated H4 (H4K5ac, K8ac, K12ac, K16ac) (Fig. 4B). Nucleosomes bound by both BD2 and BD4 were enriched for H4K16ac to a greater extent than H4K5ac or H4 tetraacetyl. Differences between the peptide array and the nucleosome pull-downs may be attributed to the presentation of the PTM in the context of an oligopeptide rather than an intact nucleosome. We then examined enrichment of H4 modifications on cellular nucleosomes bound by the tandem BDs (Fig. 4C). BD1–2 and BD4–5 tandem BDs demonstrated greater binding compared with tandem BDs containing BD3. The tandem BDs also bound nucleosomes enriched for H4K16ac.
Figure 4.
PBRM1 bromodomains interact with H4 tail. A, quantification of peptide microarray interactions between individual PBRM1 BDs and H4 peptides. Each column represents one peptide array. Normalized mean intensities are rank-ordered within each array. Dendrogram indicates clustering using Pearson correlation. B and C, GST-tagged single BDs (B) and tandem BDs (C) were used to select purified mononucleosomes. Associated H4 PTMs were detected by Western blotting. Normalized (AP/input) signal is shown. Input mononucleosomes (2.5%) are shown. The blots demonstrating input signal for H4K5ac and H4K16ac are duplicated in B and C because these figures originated from the same Western blot.
Taken together, our data suggest that BD2 and BD4 are largely responsible for PBRM1 chromatin interactions through a direct interaction with H3K14ac and possibly a weak interaction with H3K4me3. Additionally, we demonstrate that BD1 and BD5 cooperate to enhance these binding interactions, potentially through interaction with an unmodified tail, whereas BD3 may modulate these interactions.
ccRCC BD4 mutants disrupt histone recognition
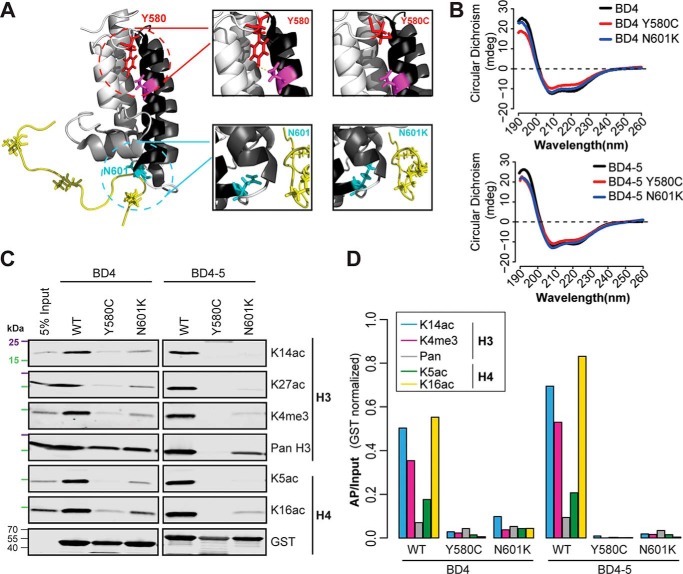
Because BD4 exhibits the highest frequency of missense mutations in ccRCC (Fig. 1A) and plays an important role in mediating nucleosome interaction, we explored the effect of BD4 mutations. Two mutations in ccRCC occur at highly conserved residues. Tyr-580 is in the AB-loop region that interacts with a conserved aspartate in helix αB and is thought to stabilize the loop-helix fold (4). Asn-601 is in the hydrophobic cavity that anchors the BD to the histone tail (Fig. 5A) (2, 5). These mutations were generated in the context of an individual BD and the BD4–5 tandem. CD of the four mutants demonstrated the expected pattern for a highly α-helical protein, similar to the WT protein (Fig. 5B). We next assessed the effect of these mutations on nucleosome recognition (Fig. 5C). Both mutations significantly diminished interaction of the BDs with nucleosomes, including nucleosomes harboring H3K14ac, H3K4me3, and H4K16ac (Fig. 5D). Interestingly, BD4 mutations in the context of tandem BD4–5 more severely disrupted nucleosome interactions than the mutation in the individual BDs, an effect most noticeable for Y580C.
Figure 5.
ccRCC-associated mutations decrease recognition of histone PTMs. A, missense mutations (Y580C and N601K) were overlaid on the NMR-derived structure. B, CD absorbance spectra for BD4 and BD4–5 as well as BD4 and BD4–5 Y580C and N601K mutants. C, PBRM1 BDs and mutants were used to pull down cellular mononucleosomes. Bound material was analyzed by immunoblot using antibodies against H3 and H4 PTMs. 25 kDa (purple) and 15 kDa (green) molecular markers are indicated. D, normalized (AP/input) signal is shown.
PBRM1 BD2 and BD4 are required for chromatin interactions
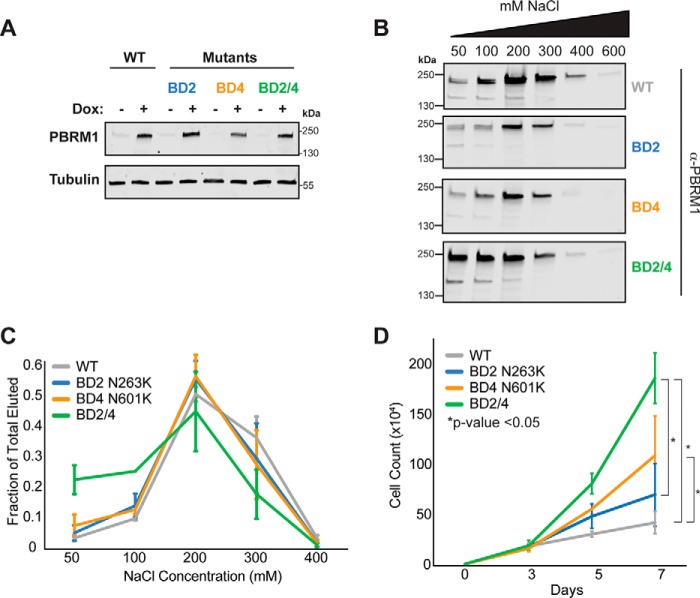
We next assessed how mutations in BD2 and BD4 affect chromatin interactions in the context of full-length PBRM1. Doxycycline-inducible mutants were expressed in a ccRCC cell line (RCC4), which lacks endogenous PBRM1 (Fig. 6A). Following PBRM1 induction, proteins were extracted from nuclei with increasing NaCl concentrations to assess the effect of mutations on chromatin affinity (Fig. 6, B and C). At a low NaCl concentration, PBRM1 and single mutants remained mostly bound to chromatin. Mutation of both BD2 and BD4 (BD2/4), however, significantly decreased binding of PBRM1 to chromatin (Fig. 6C). By 200 mm NaCl virtually all of the BD2/4 mutant was extracted from chromatin, whereas a significant fraction of the BD2 and BD4 single mutants persisted on chromatin (Fig. S3). These data suggest that BD2 and BD4 cooperate to mediate PBRM1 interaction with chromatin.
Figure 6.
Mutation of BD2 and BD4 decreases chromatin association leading to increased cell proliferation. A, induction of full-length PBRM1 mutants in RCC4 cells by doxycycline. Proteins were separated by SDS-PAGE, and PBRM1 expression was detected by immunoblot analysis. B, RCC4 nuclei were collected and extracted with increasing concentrations of NaCl. Eluted proteins were separated by SDS-PAGE, and PBRM1 levels were examined by Western blot analysis. C, quantification of PBRM1 eluted as a fraction of total PBRM1. D, RCC4 cells expressing WT or mutant PBRM1 were counted. p value established by Student's t test.
Mutation of BD2/BD4 increases cell proliferation
We then assessed the biological consequence of mutations in BD2 and BD4 by expressing full-length PBRM1 or mutants in RCC4 cells and examining cell proliferation (Fig. 6D). Because of its diminished chromatin-binding characteristics, we hypothesized that the BD2/4 mutant would have the greatest effect on proliferation. Expression of PBRM1 with a BD4 mutation demonstrated a significantly increased proliferation rate compared with WT PBRM1. In contrast, expressing the BD2 mutant did not affect proliferation. PBRM1 harboring mutations in both BD2 and 4 resulted in the greatest change in proliferation. Together these data suggest that both BD2 and BD4 participate in chromatin interaction. However, phenotypic consequences of PBRM1 mutation were only observed in the context of a BD4 mutation.
Discussion
To understand the role of PBRM1 as the potential targeting subunit of the PBAF complex, we explored the binding specificity of the individual and tandem pairs of BDs and the consequence of cancer-associated mutations within these domains. We demonstrate the importance of BD2 and BD4 in mediating PBRM1 chromatin interactions. BD2 and BD4 share sequence similarities that are reflected in their selective ability to bind modified histone tails. Using peptide microarrays and nucleosome pull-downs we found that both BD2 and BD4 preferentially bound nucleosomes marked by H3K14ac, H3K4me3, and/or H4K16ac. Additionally, we demonstrated that neighboring BDs influence BD2 and BD4 binding. Mutations in BD4 observed in ccRCC disrupted PBRM1 nucleosome interactions and enhanced cell proliferation.
Our study highlights the importance of BD4 for PBRM1 function. Although BD2 and BD4 mediate histone interactions, the ccRCC missense mutation observed in BD2 (I252R) did not affect BD2 binding to nucleosomes (data not shown), whereas BD4 mutations (N601K and Y580C) abrogated histone interactions. Further, the pocket mutant, N601K, of BD4 (but not BD2) in full-length PBRM1 affected cell proliferation. We hypothesize that the Y580C mutation of BD4 would have similar effects on proliferation as that seen with the N601K mutation because this mutation is predicted to further disrupt the binding pocket. These data suggest that whereas BD2 and BD4 can mediate interaction with chromatin, BD4–chromatin interaction plays a unique role for PBRM1 function. Remarkably, previous studies using primarily biochemical assays have focused only on BD2 as the primary driver of chromatin interactions via H3K14ac (4, 6, 17, 18). It is possible that the use of intact nucleosomes in our study rather than the oligopeptides used in other studies, together with the low affinity of BDs for histone tails, explains this difference. In the absence of tethering to a histone octamer, tail peptides may not be suitably presented to chromatin-binding proteins. The specific importance of BD4 was also supported by a recent study in which expression of a PBRM1 mutant lacking both BD1 and BD2 functioned similarly to WT PBRM1 to suppress cell proliferation, whereas loss of all six BDs showed increased cell proliferation (25). Together with our data, these results suggest that, in contrast BD2, BD4 may be necessary for PBRM1 function. Studying the six BDs as tandem pairs, we identified relationships between the BDs that influenced their ability to engage nucleosomes. Individual BDs have low binding affinities for acetylated lysine substrates (26), suggesting that multimers of BDs may enhance affinity. Remarkably, we found that three BDs, none of which demonstrated nucleosome association individually, exerted variable effects when in the context of neighboring BDs. Specifically, BD1 and BD5 act cooperatively with BD2 and BD4, respectively, to enhance binding interactions, whereas BD3 decreased binding. It is possible that the enhanced chromatin interaction of the BD1–2 and BD4–5 tandem domains with chromatin is mediated through interactions with the unmodified H4 tail. BDs 1–5 demonstrated weak interaction with unmodified H4 tails, and BD1–2 and 4–5, but not the individual BDs, weakly bound unmodified recombinant nucleosomes. Protein stabilization through interactions with the H4 tail has been shown in the yeast enzyme, Chd1, which requires binding to the H4 tail for efficient nucleosome remodeling. Tetraacetylation of the tail reduces Chd1 activity (27). Although additional experiments are necessary to establish the interaction with H4, BD1 and BD5 could augment nucleosome binding, primarily mediated by BD2 and BD4. In contrast to BD1 and BD5, adding BD3 to BD2 or BD4 diminished binding. This finding is supported by a recent study demonstrating that mutation of the binding pocket of BD3 resulted in a slight, although statistically insignificant, enhancement of PBRM1 chromatin association (6). Collectively, these data suggest that the PBRM1 BDs offer variable roles to enhance or limit chromatin interaction, perhaps resulting in a dynamic interaction across the genome.
We demonstrate that PBRM1 mutants that disrupt chromatin interaction interfere with PBRM1 effects on cell proliferation. This suggests that the BD2- and BD4-mediated chromatin interaction is important for the effects of PBRM1 on cell growth. This conclusion is supported by a recent study (25) demonstrating that expression of a mutant lacking all six BDs increased cell proliferation, whereas expression of a mutant lacking both BD1 and BD2 functioned similarly to WT PBRM1 to suppress cell proliferation. Another recent study suggested that PBRM1 plays a tumor suppressive role by down-regulating expression of genes involved in the cell cycle and increasing the number of cells in G1 (28). Together with these studies, our results link PBRM1–chromatin interaction with regulation of cell proliferation. However, future studies are necessary to better understand how PBRM1 targeting alters the transcriptome.
Because BDs are known to mediate interactions with acetylated histones, we initially considered that the H3K4me3 interaction was indirect. Separating these interactions using cell-derived nucleosomes is challenging because these modifications are both enriched at transcriptionally active transcription start sites (29). However, using recombinant nucleosomes we found that BD2 and BD4 can weakly bind H3K4me3. Based on NMR structure, we speculate that H3K4me3 may weakly interact with the BD outside of the acetyl lysine binding pocket. This observation is a source of ongoing investigation in laboratory. These data suggest that PBRM1 is targeted to transcriptionally active regions of the genome marked with H3K14ac and H3K4me3, whereby BD2 and BD4 mediate the primary interaction with H3K14ac but may also be stabilized through an interaction with H3K4me3. Like BPTF, PBRM1 could engage in multivalent interactions with acetylated and methylated histone tails (12).
Collectively, these data illustrate the distinctiveness of the six PBRM1 BDs to mediate nucleosome interactions and the consequence of ccRCC mutation in the critical domains. We highlight the specific importance of BD4 to facilitate these interactions and that mutations within this domain enhance cell proliferation, which may promote ccRCC development. In addition to limiting cell proliferation, recent studies point to a role of PBRM1 in modulating immune responses. For ccRCC patients and in a mouse melanoma model, the absence of PBRM1 enhances efficacy of immune checkpoint inhibition (30, 31). As BD4 plays an especially significant role in chromatin recognition, it may offer an important therapeutic target.
Experimental procedures
Modeling PBRM1 bromodomains
Mutations in kidney cancer were identified using cBioPortal on March 4, 2018 (Kidney, TCGA, IRC, U Tokyo, MSKCC, Genentech, BGI). The protein sequences of the six PBRM1 BDs were curated from UniProt (Q86U86). Hierarchical clustering was performed by Unweighted Pair Group Method with Arithmetic Mean analysis (23) and the tree diagram was generated by JalView (32). Sequence alignment was performed using Clustal Omega (33). Homology was determined using the percent overlap of the aligned regions.
Preparation and purification of recombinant PBRM1 bromodomains
DNA containing the coding regions for the single and tandem PBRM1 BDs were PCR-amplified from cDNA generated from 293-T mRNA and inserted into a pGEX-6P-1 expression vector. Mutagenesis of BD4 and BD4–5 was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). All constructs were sequenced to confirm accuracy prior to protein purification. Transformed Escherichia coli BL21 cells were grown in Luria-Bertani medium, supplemented with ampicillin (50 μg/ml), and protein expression was induced overnight at 18 °C with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation, resuspended in lysis buffer (1× PBS, 1 mm DTT, 1 mm PMSF), and sonicated. Insoluble proteins were cleared by centrifugation and proteins were purified with GST Agarose (Pierce). Resin was resuspended in elution buffer (50 mm Tris, 10 mm GSH). Proteins were concentrated (EMD Millipore Amicon Ultra-15 concentrators) and dialyzed in storage buffer (20 mm Tris, pH 8, 150 mm NaCl, 1 mm DTT, 10% glycerol). Single BDs correspond to amino acids 31–169 (BD1), 162–348 (BD2), 341–512 (BD3), 497–661(BD4), 654–784 (BD5), and 776–903 (BD6). Tandem BDs correspond to amino acids 31–340 (BD1–2), 165–512 (BD2–3), 341–646 (BD3–4), 498–769 (BD4–5), and 639–890 (BD5–6). Tandem BDs were further purified by FPLC on an AKTAxpress. Samples were examined by SDS-PAGE.
Histone peptide microarrays
Array preparation and protein analysis were performed as described (24, 34). Proteins (1 μm) were incubated with peptide microarrays in PBST with 5% BSA overnight at 4 °C. Arrays were washed with PBS, incubated with a GST antibody (Sigma) for 2 h, then washed with PBS and finally incubated with an Alexa 647 anti-rabbit antibody (Invitrogen). Arrays were scanned (Typhoon Trio+ Imager, GE Healthcare), and the interactions were quantified by fluorescence (ImageQuant array software, GE Healthcare). The signal from each of the six spots for each peptide was averaged and values were normalized to the highest calculated value across all peptides and plotted on a scale from 0 to 1. Heat maps were created using GENE-E and represent the mean of two independent arrays.
Mononucleosome preparation
Nucleosomes were prepared as described (12). Briefly, following a 1 h treatment with 1 μm vorinostat, 293-T cells were washed twice with 1× PBS and resuspended (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 340 mm sucrose, 10% (v/v) glycerol, 0.5 mm PMSF, 5 mm 2-mercaptoethanol, 1× Roche protease inhibitor mixture, 0.5 mg/ml vorinostat). Cells were lysed using 0.1% Triton X-100 detergent. Nuclei were separated through a sucrose gradient followed by an in nucleo MNase digestion and nucleosome recovery. Cells were digested with 0.25, 0.5, 1, 2, and 4 units of MNase (Worthington) per 70 μg of DNA for 8 min. Reactions were stopped using 1.3 mm EGTA. Fractions from each treatment were evaluated by agarose gel electrophoresis to confirm proper digestion. For further purification and isolation of mononucleosomes, samples were applied to a second sucrose cushion and ultracentrifuged for 17 h, 27,000 rpm in SW40 rotor, 4 °C. Fractions were collected and separated by agarose gel electrophoresis. Fractions containing only mononucleosomes were collected and concentrated (Amicon concentrators, EMD Millipore) and brought to 5% glycerol.
Cellular nucleosome pull-downs
GST nucleosome pull-downs were performed by applying 10 μg of nucleosomes on a 10 μl bed volume of 500 pmols pre-immobilized protein on GSH agarose (Pierce). Nucleosomes and BD-bound resin were incubated for 2 h at 4 °C in 1× PBS, 0.1% Nonidet P-40, 0.5% BSA. Resin was washed with binding buffer. Proteins were eluted using 2× SDS-loading buffer and separated on a 4–15% gradient SDS-PAGE (Bio-Rad). Proteins were transferred to a nitrocellulose membrane and immunoblotted with antibodies (GST, MA4-004, Thermo; total H3, 13-0001, Epicypher; H3K9ac, 07-352, EMD Millipore; H3K14ac, ab82501, Abcam; H3K18ac, 39130, Active Motif; H3K27ac, ab4729, Abcam; H3K4me3, 07-473, EMD Millipore; H3K9me3, ab8898, Abcam; H3K27me3, ab6002, Abcam; H4K5ac, ab51997, Abcam; H4K16ac, ab109463, Abcam; panH4ac, 05-858, EMD Millipore) followed by anti-mouse and anti-rabbit IRDye secondary antibodies (LI-COR Biosciences, Lincoln, NE). Signal was quantified (Odyssey IR imager, LI-COR Biosciences), and densitometry analysis was performed (ImageStudio version 2.0). Signal was normalized to GST input and plotted as a fraction of total nucleosome input. At least three replicates were performed for each pull-down.
Recombinant nucleosome pull-downs
Recombinant nucleosome pull-downs were performed by applying 250 pmols of GST-BD on a 10-μl bed volume of 500 pmols pre-immobilized nucleosomes (H3K4me3, Epicypher 16-0316; H3K14ac, Epicypher 16-0343; unmodified, Epicypher 16-0006) on streptavidin magnetic beads (Pierce). Nucleosomes and BD-bound resin were incubated for 2 h at 4 °C in 50 mm Tris-HCl, pH 8.0, 0.1% Nonidet P-40, 150 mm NaCl. Beads were washed with binding buffer. Proteins were eluted using 2× SDS-loading buffer and separated on a 4–15% gradient SDS-PAGE (Bio-Rad). Proteins were transferred to a nitrocellulose membrane and immunoblotted with antibodies (GST, MA4-004, Thermo; total H3, 13-0001, Epicypher) followed by anti-mouse/anti-rabbit IRDye secondary antibodies (LI-COR Biosciences). Pull-downs with different NaCl concentrations are indicated individually. For these reactions, the entire pull-down was performed in the specified NaCl concentration.
CD spectroscopy
Proteins were diluted to a concentration of 0.5 μg/μl in 20 mm sodium phosphate (pH 7.0), 150 mm sodium fluoride, and 0.2 mm tris(2-carboxyethyl)phosphine. CD spectra were generated using a 0.1-cm quartz cuvette (Chirascan Plus, Applied Photophysics Inc.) Assays were conducted at 20 °C from 190 to 260 nm with a step size of 0.5 nm. Background absorbance measured using the buffer alone was subtracted from sample spectra. The CD signal is represented in millidegrees.
Preparation and lentiviral infection of full-length PBRM1 cell lines
Site-directed mutagenesis was performed on pBabepuro-BAF180 (Addgene plasmid no. 41078) and cloned into tetracycline inducible lentiviral vector TetO-FUW (Addgene plasmid no. 20323) and used in combination with pLenti CMV rtTA3 Hygro (Addgene plasmid no. 26730). Plasmid sequences were checked by Sanger sequencing. Lentivirus for plasmids were produced from transfection of 293-T cells with constructs and packaging vectors (pVSVG, pRRE, pRSV). Supernatant was collected after 48 h and concentrated with Lenti-X Concentrator (Clontech). RCC4 cells were infected with CMV lentivirus for 48 h and treated with hygromycin (200 μg/ml). After 1 week of selection, cells were infected with WT and mutant TetO constructs. Cells were treated with puromycin (0.6 μg/ml) 48 h post infection and selected for 1 week. Cells were induced with doxycycline (1 μg/ml) for 72 h and PBRM1 expression was assessed by Western blot analysis (Bethyl A301-591A).
Salt fractionations
RCC4 cells were treated with doxycycline (1 μg/ml) for 72 h. Salt fractionations were performed as described in Porter et al. (6). Ten million RCC4 cells were used for each salt fractionation. Proteins were transferred to a nitrocellulose membrane and immunoblotted with antibodies (GST, MA4-004, Thermo) followed by anti-mouse IRDye secondary antibodies (LI-COR Biosciences). Experiments were performed in triplicate. Signal was plotted as a fraction of total PBRM1 eluted.
Cell proliferation assays
RCC4 cells (2 × 104) were plated in 6-cm tissue culture dishes on day 0 with 1 μg/ml of doxycycline. Cells were trypsinized at days 3, 5, and 7 and counted using a Bio-Rad Tc20 Automated Cell Counter over 7 days. Experiments were performed in triplicate.
Author contributions
M. J. S., A. W. M., and I. J. D. data curation; M. J. S., E.K. S., E. S. H., L. E. S., B. D. S., and I. J. D. formal analysis; M. J. S. validation; M. J. S., E. K. S., A. W. M., E. S. H., and L. E. S. investigation; M. J. S. and I. J. D. visualization; M. J. S. and B. D. S. methodology; M. J. S. and I. J. D. writing-original draft; M. J. S., A. W. M., E. S. H., L. E. S., B. D. S., and I. J. D. writing-review and editing; B. D. S. and I. J. D. supervision; B. D. S. and I. J. D. funding acquisition; I. J. D. conceptualization; I. J. D. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Ashutosh Tripathy (University of North Carolina Macromolecular Interactions Facility) for help collecting the CD data.
This work was supported by National Institutes of Health Grants CA166447 (I. J. D.), CA198482 (I. J. D. and B. D. S.), and GM126900 (B. D. S.); the Corn-Hammond Fund for Pediatric Oncology (I. J. D.) and a University of North Carolina Lineberger Developmental Grants. This work was also supported by Institutional Research and Academic Career Development Award GM000678 (to E. K. S.) granted by the National Institute of General Medical Sciences, division of Training, Workforce Development, and Diversity. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3 and Tables S1 and S2.
- BD
- bromodomain
- PTM
- posttranslational modification
- ccRCC
- clear cell renal cell carcinoma
- MNase
- micrococcal nuclease
References
- 1. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 2. Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., and Zhou M. M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature. 399, 491–496 10.1038/20974 [DOI] [PubMed] [Google Scholar]
- 3. Zeng L., and Zhou M. M. (2002) Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 513, 124–128 10.1016/S0014-5793(01)03309-9 [DOI] [PubMed] [Google Scholar]
- 4. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J. P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T., Gingras A. C., Arrowsmith C. H., and Knapp S. (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 10.1016/j.cell.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., and Travers A. A. (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19, 6141–6149 10.1093/emboj/19.22.6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Porter E. G., and Dykhuizen E. C. (2017) Individual bromodomains of polybromo-1 contribute to chromatin association and tumor suppression in clear cell renal carcinoma. J. Biol. Chem. 292, 2601–2610 10.1074/jbc.M116.746875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature. 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 8. Ruthenburg A. J., Li H., Patel D. J., and Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 10.1038/nrm2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacobson R. H., Ladurner A. G., King D. S., and Tjian R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 10.1126/science.288.5470.1422 [DOI] [PubMed] [Google Scholar]
- 10. Dey A., Chitsaz F., Abbasi A., Misteli T., and Ozato K. (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 100, 8758–8763 10.1073/pnas.1433065100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y.-J., Umehara T., Inoue M., Saito K., Kigawa T., Jang M.-K., Ozato K., Yokoyama S., Padmanabhan B., and Güntert P. (2008) Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 17, 2174–2179 10.1110/ps.037580.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruthenburg A. J., Li H., Milne T. A., Dewell S., McGinty R. K., Yuen M., Ueberheide B., Dou Y., Muir T. W., Patel D. J., and Allis C. D. (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 145, 692–706 10.1016/j.cell.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. VanDemark A. P., Kasten M. M., Ferris E., Heroux A., Hill C. P., and Cairns B. R. (2007) Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell. 27, 817–828 10.1016/j.molcel.2007.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemon B., Inouye C., King D. S., and Tjian R. (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 414, 924–928 10.1038/414924a [DOI] [PubMed] [Google Scholar]
- 15. Varela I., Tarpey P., Raine K., Huang D., Ong C. K., Stephens P., Davies H., Jones D., Lin M.-L., Teague J., Bignell G., Butler A., Cho J., Dalgliesh G. L., Galappaththige D., et al. (2011) Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 10.1038/nature09639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., Nagata Y., Yoshida K., Kon A., Suzuki Y., Chiba K., et al. (2013) Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 10.1038/ng.2699 [DOI] [PubMed] [Google Scholar]
- 17. Chandrasekaran R., and Thompson M. (2007) Polybromo-1-bromodomains bind histone H3 at specific acetyl-lysine positions. Biochem. Biophys. Res. Commun. 355, 661–666 10.1016/j.bbrc.2007.01.193 [DOI] [PubMed] [Google Scholar]
- 18. Charlop-Powers Z., Zeng L., Zhang Q., and Zhou M.-M. (2010) Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 20, 529–538 10.1038/cr.2010.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hakimi A. A., Ostrovnaya I., Reva B., Schultz N., Chen Y.-B., Gonen M., Liu H., Takeda S., Voss M. H., Tickoo S. K., Reuter V. E., Russo P., Cheng E. H., Sander C., Motzer R. J., and Hsieh J. J. (2013) Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin. Cancer Res. 19, 3259–3267 10.1158/1078-0432.CCR-12-3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Durinck S., Stawiski E. W., Pavía-Jiménez A., Modrusan Z., Kapur P., Jaiswal B. S., Zhang N., Toffessi-Tcheuyap V., Nguyen T. T., Pahuja K. B., Chen Y.-J., Saleem S., Chaudhuri S., Heldens S., Jackson M., et al. (2015) Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat. Genet. 47, 13–21 10.1038/ng.3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerlinger M., Horswell S., Larkin J., Rowan A. J., Salm M. P., Varela I., Fisher R., McGranahan N., Matthews N., Santos C. R., Martinez P., Phillimore B., Begum S., Rabinowitz A., Spencer-Dene B., et al. (2014) Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao J., Aksoy B., Dogrusoz U., and Dresdner G. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokal R., and Michener C. (1958) A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38, 1409–1438 [Google Scholar]
- 24. Rothbart S. B., Krajewski K., Strahl B. D., and Fuchs S. M. (2012) Peptide microarrays to interrogate the “histone code.” Methods Enzymol. 512, 107–135 10.1016/B978-0-12-391940-3.00006-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao W., Li W., Xiao T., Liu X. S., and Kaelin W. G. (2017) Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL−/− clear cell renal carcinoma. Proc. Natl. Acad. Sci. 114, 1027–1032 10.1073/pnas.1619726114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muller S., Filippakopoulos P., and Knapp S. (2011) Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 13, e29 10.1017/S1462399411001992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferreira H., Flaus A., and Owen-Hughes T. (2007) Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 374, 563–579 10.1016/j.jmb.2007.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chowdhury B., Porter E. G., Stewart J. C., Ferreira C. R., Schipma M. J., and Dykhuizen E. C. (2016) PBRM1 regulates the expression of genes involved in metabolism and cell adhesion in renal clear cell carcinoma. PLoS One 11, e0153718 10.1371/journal.pone.0153718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taverna S. D., Ueberheide B. M., Liu Y., Tackett A. J., Diaz R. L., Shabanowitz J., Chait B. T., Hunt D. F., and Allis C. D. (2007) Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. U.S.A. 104, 2086–2091 10.1073/pnas.0610993104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miao D., Margolis C. A., Gao W., Voss M. H., Li W., Martini D. J., Norton C., Bossé D., Wankowicz S. M., Cullen D., Horak C., Wind-Rotolo M., Tracy A., Giannakis M., Hodi F. S., et al. (2018) Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 10.1126/science.aan5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan D., Kobayashi A., Jiang P., de Andrade L. F., Tay R. E., Luoma A. M., Tsoucas D., Qiu X., Lim K., Rao P., Long H. W., Yuan G.-C., Doench J., Brown M., Liu X. S., and Wucherpfennig K. W. (2018) A major chromatin regulator determines resistance of tumor cells to T cell–mediated killing. Science 359, 770–775 10.1126/science.aao1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clamp M., Cuff J., Searle S. M., and Barton G. J. (2004) The Jalview Java alignment editor. Bioinformatics 20, 426–427 10.1093/bioinformatics/btg430 [DOI] [PubMed] [Google Scholar]
- 33. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuchs S. M., Krajewski K., Baker R. W., Miller V. L., and Strahl B. D. (2011) Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr. Biol. 21, 53–58 10.1016/j.cub.2010.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.