Abstract
Prostate cancer often metastasizes to the bone, leading to morbidity and mortality. While metastasis‐associated protein 1 (MTA1) is highly overexpressed in metastatic tumors and bone metastatic lesions, its exact role in the development of metastasis is unknown. Here, we report the role of MTA1 in prostate cancer progression and bone metastasis in vitro and in vivo. We found that MTA1 silencing diminished formation of bone metastases and impaired tumor growth in intracardiac and subcutaneous prostate cancer xenografts, respectively. This was attributed to reduced colony formation, invasion, and migration capabilities of MTA1 knockdown cells. Mechanistic studies revealed that MTA1 silencing led to a significant decrease in the expression of cathepsin B (CTSB), a cysteine protease critical for bone metastasis, with an expected increase in the levels of E‐cadherin in both cells and xenograft tumors. Moreover, meta‐analysis of clinical samples indicated a positive correlation between MTA1 and CTSB. Together, these results demonstrate the critical role of MTA1 as an upstream regulator of CTSB‐mediated events associated with cell invasiveness and raise the possibility that targeting MTA1/CTSB signaling in the tumor may prevent the development of bone metastasis in prostate cancer.
Keywords: bone metastasis, cathepsin B, intracardiac xenografts, luciferase imaging, MTA1, prostate cancer
Abbreviations
- ChIP‐Seq
chromatin immunoprecipitation sequencing
- CTSB
cathepsin B
- E‐cad
E‐cadherin
- EMT
epithelial‐to‐mesenchymal transition
- IHC
immunohistochemistry
- MMP
matrix metalloproteinase (enzyme)
- MTA1
metastasis‐associated protein 1
- MUC1
mucin 1
- NS
nonsilencing
- NuRD
nucleosome remodeling and deacetylation (complex)
- PI3K
phosphoinositide 3‐kinase
- PTEN
phosphatase and tensin homolog deleted in chromosome 10
- R26
ROSA26 locus on mouse genome
- RPMI 1640
Roswell Park Memorial Institute (medium)
- s.c.
subcutaneous
- SAHA
suberoylanilide hydroxamic acid
1. Introduction
Bone metastases occur in about 80% of patients with advanced castrate‐resistant prostate cancer. As a major cause of morbidity and mortality in patients with prostate cancer, bone metastases are of great clinical concern. The mechanisms of trafficking and homing of aggressive prostate cancer cells into the bone have been previously investigated (Futakuchi et al., 2016; Sakamoto and Ichikawa, 2014; Ziaee et al., 2015). However, the exact molecular mechanisms involved remain elusive, which restricts the development of new and targeted therapeutics. Identification of bone metastasis driver molecules in the primary prostate tumors might help to target them at early stages of the disease, which can provide a shift of the disease progression toward indolent rather than the aggressive disease.
In our previous studies, we have identified metastasis‐associated protein 1 (MTA1) as a novel factor involved in ‘vicious cycle’ of prostate cancer cells interacting with bone cells, and hypothesized that MTA1 may play a specific role in prostate cancer bone metastasis (Kai et al., 2011). Our following studies validated high MTA1 expression in advanced metastatic prostate cancer and bone lesions of patients with prostate cancer (Dias et al., 2013a; Kai et al., 2011).
MTA1 belongs to the MTA family of proteins that are critical participants of the nucleosome remodeling and deacetylation (NuRD) multiprotein complex, which is involved in chromatin remodeling and gene silencing (Toh et al., 1994; Xue et al., 1998). Overexpression of MTA1 has been reported in various cancers (Levenson et al., 2014). We and others have demonstrated the correlation of MTA1 expression with clinicopathological measures of prostate cancer progression: high Gleason score, development of castrate‐resistant disease, recurrence, and bone metastasis (Dias et al., 2013a; Hofer et al., 2006; Kai et al., 2011). We have extensively reported on the critical role of MTA1 and its signaling at all stages of prostate cancer development and progression (Levenson et al., 2014). In addition to our initial demonstration of MTA1's functional role in vitro and in xenografts (Dhar et al., 2015a; Kai et al., 2010, 2011; Li et al., 2013), we recently demonstrated that MTA1 overexpression promotes molecular changes associated with inflammation‐mediated cancer initiation, epithelial‐to‐mesenchymal transition (EMT), and cancer progression linked to cell metastatic potential and angiogenesis in a multistage transgenic model of prostate cancer that closely mimics human disease (Butt et al., 2017; Dhar et al., 2016, 2017). Altogether, MTA1 is increasingly being recognized as an important upstream multifunctional regulator that affects various cellular responses, including cell adhesion, migration, and invasion in prostate cancer. However, the exact role of MTA1 and its downstream signaling mechanisms that mediate MTA1‐activated prostate tumor cell invasion and colonization into the bone are not well understood.
In this study, we systematically analyzed the in vitro and in vivo role of MTA1 in prostate tumor cell metastatic potential, tumor growth, and formation of metastasis using subcutaneous (s.c.) and experimental bone metastasis xenograft mouse models. We showed that MTA1 ablation profoundly suppressed the growth of prostate tumors and the ability of tumor cells to colonize the bone by inhibiting their migratory and invasive properties through repression of cathepsin B (CTSB), a cysteine protease that plays a critical role in bone metastasis (Podgorski et al., 2005; Withana et al., 2012).
2. Materials and methods
2.1. Cell culture
Prostate cancer cells, PC3M and LNCaP, were maintained in RPMI 1640 media containing 10% FBS as described previously (Butt et al., 2017; Dhar et al., 2016; Dias et al., 2013a,b; Li et al., 2013). PC3M cells expressing luciferase (PC3M‐Luc) were established, expanded, and tested for their luciferase activity using an IVIS Spectrum (PerkinElmer, Waltham, MA, USA). Cells were maintained in an incubator at 37 °C with 5% CO2. Cells were authenticated using short tandem repeat profiling at Research Technology Support Facility, Michigan State University.
2.2. MTA1 knockdown in prostate cancer cells
PC3M‐Luc cells were transduced with the three GIPZ MTA1 lentiviral shRNA, GIPZ GAPDH lentiviral shRNA, and GIPZ nonsilencing lentiviral shRNA. The GIPZ lentiviral shRNA system (GE Healthcare Dharmacon, Lafayette, CO, USA) contains the puromycin‐resistant gene for selection of transduced cells and TurboGFP for monitoring the selection under the fluorescence microscope. For the preparation of the viral particles, we used the pCMV‐ΔR8.91 packaging plasmid and the pMD.G envelope plasmid (Addgene, Cambridge, MA, USA). Cells were transduced using RPMI 1640 medium, which contained 4 μg·mL−1 polybrene (Sigma‐Aldrich, St. Louis, MO, USA) with lentiviral particles at a multiplicity of infection = 8. On day 2 post‐transduction, selection was initiated with 200 μg·mL−1 puromycin (Sigma‐Aldrich) and GFP‐positive clones were selected and propagated.
2.3. Western blot
Western blot analysis was carried out as described previously (Butt et al., 2017; Dhar et al., 2015a,b, 2016, 2017; Dias et al., 2013a,b; Kai et al., 2010, 2011; Li et al., 2013). The antibodies used in this study are listed in Table S1.
2.4. qRT‐PCR
The total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). qRT‐PCR was performed as described previously (Butt et al., 2017; Dhar et al., 2015a,b, 2016, 2017) on the LightCycler 480 II real‐time PCR instrument (Roche Diagnostics, Basel, Switzerland). All primers for qRT‐PCR are listed in Table S2.
2.5. ChIP‐Seq experiments and analysis
Two hundred milligrams of prostate tissues isolated from prostate‐specific Pten f/+; Pb‐Cre + mouse, which had markedly increased MTA1 levels, was used for ChIP‐Seq assay. ChIP‐Seq was performed using MTA1 antibody (Bethyl Laboratories, Montgomery, TX, USA) at Active Motif (San Diego, CA, USA). Reads were aligned to the mouse genome, and peaks were identified (Dhar et al., 2016, 2017) using SICER algorithm. The ChIP‐Seq tracks presented here were generated using the UCSC Genome Browser.
2.6. Cell proliferation assay
PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells (2 × 103) were seeded in a 35‐mm cell culture dish. The proliferation of these cells was determined by counting the cells every other day over a period of 10 days.
2.7. Colony formation assay
PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells (2 × 103) were seeded in a 35‐mm cell culture dish for the 2‐week observation time. When colonies were freely visible (> 50 cells/colony), cells were fixed with formaldehyde and stained with 0.01% crystal violet solution. Colonies were analyzed by scanning and obtaining the image of each dish in the Bio‐Rad ChemiDoc Imager (Bio‐Rad Laboratories, Hercules, CA, USA). imagetool software (UTHSCSA, San Antonio, TX, USA) was used for counting the number of colonies in each dish.
2.8. Invasion assay
Invasion assay was performed using transwells having permeable support coated with Cultrex basement membrane extract (Corning Life Sciences, Corning, NY, USA) as described previously (Dhar et al., 2017). Briefly, 5 × 103 serum‐starved PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells were seeded in serum‐free media in transwells and incubated for 24 h. After the incubation, cells in the upper membrane surface were scraped off with a cotton tip, whereas cells sticking to lower membrane surface were collected in cell dissociation solution containing calcein AM (Trevigen, Gaithersburg, MD, USA). This solution was then transferred to a black 96‐well plate, and fluorescence was read at 520‐/485‐nm excitation/emission. The relative fluorescence units were converted into cell number using the standard curve plotted with a serial dilution of cell numbers. Relative invasion was quantitated assuming 100% invasion for PC3M‐Luc‐NS control cells.
2.9. Wound healing assay
Motility changes in PC3M‐Luc MTA1 knockdown cells were determined by the wound healing assay as described previously (Dhar et al., 2017). Briefly, 90% confluent cells were starved in low serum media (0.1% serum), after which three separate wounds were scratched through the cells. The wound was imaged at 0, 12, 24, and 48 h using the Olympus CKX41 microscope with the Infinity Analyze software (Lumera Corporation, Bothell, WA, USA). Wound widths were calculated using the imagej software (NIH, Bethesda, MD). Migration index was quantitated assuming 100 for PC3M‐Luc‐NS control cells at 0 h.
2.10. Transgenic mouse model of prostate‐specific MTA1 overexpression
MTA1 transgenic founder mice were generated using CAG‐LoxP‐Stop‐LoxP(LSL)‐2HA‐MTA1‐T2A‐GFP‐pA construct to knock‐in the MTA1 transgene into the mouse ROSA26 (R26) locus (Applied StemCell, Inc., Milpitas, CA, USA). We crossbred these mice with Pb‐Cre4+ mice to obtain prostate‐specific overexpression of MTA1 (R26MTA1/+). Protein lysates from isolated prostate tissues were subjected to immunoblotting to confirm MTA1 overexpression.
2.11. Prostate cancer xenografts: subcutaneous and intracardiac
Foxn1 nu/nu male mice were purchased from Envigo at 5–6 weeks of age. For s.c. xenografts, mice were randomly divided into two groups (n = 5 per group), and PC3M‐Luc‐NS and PC3M‐Luc shMTA1 cells (0.5 × 106) in 100 μL of 50% Matrigel (Corning Life Sciences) were inoculated on the dorsal right flank of mice. Bioluminescent and caliper measurements were taken weekly as described previously (Dhar et al., 2015a,b; Dias et al., 2013a, b; Kai et al., 2011; Li et al., 2013). Calculation of tumor volume by digital vernier caliper measurements was made using formula as before (Dhar et al., 2015b; Kai et al., 2011). The mice were sacrificed at day 35. Tumors were excised from s.c. xenografts, total RNA and protein were isolated, and the remaining tissue was placed in 10% formalin for histological and immunohistochemical (IHC) analysis.
For intracardiac xenografts, mice were randomly divided into two groups (n = 5 per group), and PC3M‐Luc‐NS or PC3M‐Luc‐shMTA1 cells (0.1 × 106) in 100 μL of 1× PBS were injected into the heart's left ventricle of anesthetized mice. Bioluminescent measurements were taken immediately and then weekly as described previously (Dhar et al., 2015a,b; Dias et al., 2013b; Kai et al., 2011; Li et al., 2013). The mice were sacrificed at day 34. The femur, tibia, maxilla, and mandible bones were harvested and placed in 10% formalin for bone histological analysis.
Housing and care of all animals were in accordance with the guidelines established by the University's Institutional Animal Care and Use Committee.
2.12. Histopathology and immunohistochemistry
Four‐micrometer‐thick formalin‐fixed paraffin‐embedded tumor and bone sections were processed and stained with H&E by Reveal Biosciences (San Diego, CA, USA). Immunohistochemical staining with Ki67, MTA1, CTSB, and E‐cad (antibodies are listed in Table S1) was performed as per the protocol described previously (Butt et al., 2017; Dhar et al., 2015a,b, 2016, 2017; Dias et al., 2013a; Kai et al., 2011; Li et al., 2013). The Vectastain ABC Elite Kit and ImmPACT DAB Kit (Vector Laboratories, Burlingame, CA, USA) were used to visualize staining. Images were recorded on an EVOS XL Core microscope (Thermo Fisher Scientific, Waltham, CA, USA). Ki67‐stained nuclei were quantitated using imagej software (NIH, Bethesda, MD).
2.13. Clinical samples
Fresh prostatectomy specimens were obtained immediately after surgery, and core biopsies were taken under sterile conditions using Coaxial Achieve Automatic Biopsy System (14G × 11 cm) as described previously (Dhar et al., 2017). The biopsy specimens were collected in vials and snap‐frozen. Two to three core biopsies were pooled together to extract protein for immunoblotting. All human studies were approved by University's Institutional Review Board. The experiments were undertaken with the understanding and written consent of each subject. The study methodologies confirmed to the standards set by the Declaration of Helsinki.
2.14. Statistical analysis
The differences between the groups were analyzed by the unpaired t‐test or one‐way ANOVA or two‐way ANOVA based on the experiment design. All tests were performed using graphpad prism 7 software (GraphPad Software, La Jolla, CA, USA). P < 0.05 was considered statistically significant. All data are cumulative of at least three independent experiments.
3. Results
3.1. MTA1 depletion impairs formation of bone metastasis, tumor growth, and progression in prostate cancer xenografts
To allow for noninvasive bioluminescence monitoring of bone metastases in vivo, we labeled the PC3M cells with luciferase (PC3M‐Luc) and further utilized these cells to establish cells expressing MTA1 (PC3M‐Luc‐NS) or silenced for MTA1 (PC3M‐Luc‐shMTA1). We chose the shMTA1#3 clone of PC3M cells silenced for MTA1 for our experiments based on its top knockdown efficiency among the tested shRNA clones and its in vitro luciferase activity equal to control PC3M‐Luc‐NS cells (Fig. S1).
PC3M cells were able to develop visceral metastasis in orthotopic prostate cancer xenografts, but no metastasis to the bone was detected (K. Li, S. Dias, S. Dhar, M. Ivanovic & A. S. Levenson, unpublished data). Therefore, to delineate the role of MTA1 in bone metastasis, we utilized the experimental bone metastasis model with injections of cells into the left cardiac ventricle of athymic mice (Chong Seow Khoon, 2015; Holzapfel et al., 2014; Miftakhova et al., 2016; Rosol et al., 2004). Metastases were monitored by weekly bioluminescence imaging until day 34, when mice were sacrificed. We found bioluminescent signal localizing only to the jawbones (maxilla and mandible). Representative image (Fig. 1A) and quantitation of normalized total flux (Fig. 1B) showed statistically significant reduction in jawbone‐localized bioluminescent signal in PC3M‐Luc shMTA1 compared to NS control xenografts, suggesting that MTA1 depletion significantly compromised the colonization of PC3M prostate cancer cells to the bone. H&E staining of jawbones showed that prostate cancer cells infiltrated the bone and formed metastatic tumor foci (Fig. 1C). Notably, the NS control metastatic bone lesions exhibited areas of intense bony remodeling (Fig. 1C, magnified image). Further, we found that tumor foci in NS control bone metastatic lesions occupied a greater percentage of the histological section area compared to shMTA1 lesions (Fig. 1C, D), although results were statistically not significant. These data suggest that cells lacking MTA1 had impaired ability to colonize and form bone metastasis compared to cells expressing high levels of MTA1.
Figure 1.
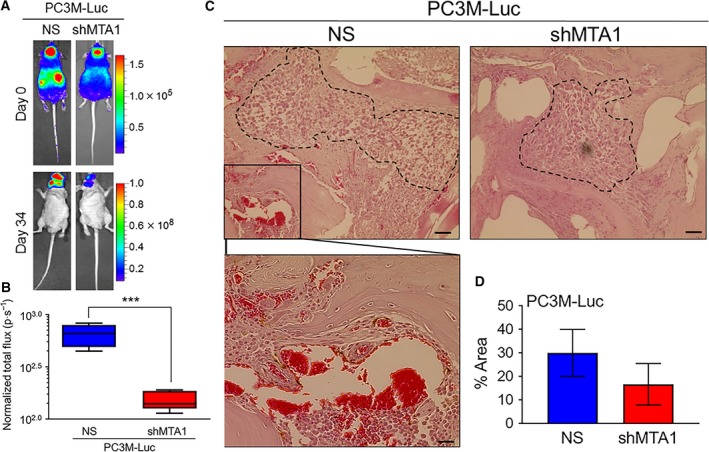
MTA1 knockdown suppresses formation of bone metastasis in experimental bone metastasis xenografts. (A) PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells were injected into the left ventricle of male athymic mice (n = 5) to colonize the bone. Bioluminescent images of representative tumor metastasis at day 0 (dorsal view) and day 34 (ventral view) are shown. (B) Quantitative analysis of bioluminescent signals of metastasis is shown in normalized total flux (photon per sec). ***P < 0.001 (unpaired t‐test). (C) Representative histology for bone metastatic lesions from two groups of mice is shown. Scale bars, 50 μm. Scale bar (magnified image), 10 μm. (D) Quantitation of the metastatic carcinoma area. Data represent the mean ± SEM of metastatic area from three mice in each group.
We have previously reported on MTA1 knockdown inhibiting the tumor growth and progression of the LNCaP and DU145 s.c. prostate cancer xenografts (Kai et al., 2011). To determine the contribution of MTA1 to the tumor growth and progression of the PC3M aggressive prostate cancer cells in vivo, PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells were injected s.c. into the right flank of athymic male nude mice. Both bioluminescent imaging and caliper measurements were taken to monitor tumor growth and progression weekly. Bioluminescence measurement of tumors showed drastically reduced tumor growth in PC3M‐Luc‐shMTA1 compared to PC3M‐Luc‐NS control xenografts (Fig. 2A), as evident by significant differences in photon emission by day 23 (Fig. 2B). As after a certain point of time s.c. tumors give nonconsistent bioluminescent measurements due to limitations of photon emission detection in large tumors with necrotic areas (Dhar et al., 2015a,b; Dias et al., 2013b; Kim et al., 2010; Rehemtulla et al., 2000), caliper‐based measurements, which were feasible after day 14, became more reliable. Figure 2C, D shows highly significant differences in tumor volumes between control and shMTA1 xenografts by day 35, when mice were sacrificed. Immunohistochemical analysis of tumors removed from xenografts revealed that the tumors formed from the PC3M‐Luc‐shMTA1 cells had lower proliferative index compared to PC3M‐Luc‐NS controls, as indicated by decreased number of Ki67‐positive cells in PC3M‐Luc‐shMTA1 tumors (Fig. 2E, F). These data indicate that silencing of MTA1 impairs tumor growth and progression in PC3M prostate cancer xenografts.
Figure 2.
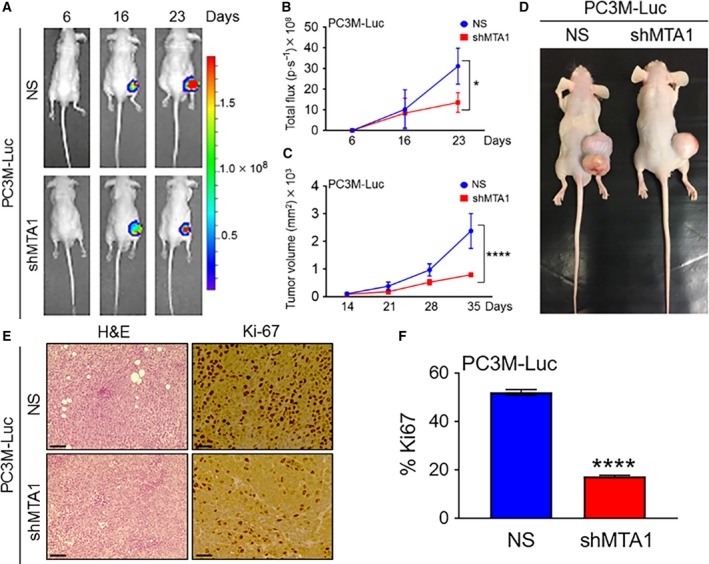
MTA1 knockdown effectively diminishes tumor growth and progression in s.c. tumor xenografts. (A) PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells were injected s.c. into the male athymic mice (n = 5). Dorsal views of bioluminescent (BL) images of representative tumor xenografts at days 6, 16, and 23 are shown. (B) Quantitative analysis of tumor light emission from day 6 after cancer cell injections until day 23 is shown in Total Flux (photon per sec). *P < 0.05 (two‐way ANOVA). (C) Tumor growth was monitored using a digital caliper once a week starting at day 14. ****P < 0.0001 (two‐way ANOVA). (D) Representative mouse tumor xenografts showing MTA1 depletion effect on tumor growth. (E) H&E‐stained sections (scale bars, 100 μm) and IHC analyses (scale bars, 50 μm) for proliferative activity (Ki67) in tumor tissues from PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 xenografts. (F) Quantitation of proliferative index in five randomly selected fields for each tissue. Data represent the mean ± SEM of five fields from five mice. ****P < 0.0001 (unpaired t‐test).
The changes in body weight of mice from both the NS control and shMTA1 groups were not significantly different during the intracardiac and s.c. xenograft experiments (data not shown).
3.2. MTA1 depletion suppresses the metastatic potential of human prostate cancer cells
To determine the cellular mechanisms through which depletion of MTA1 impedes formation of bone metastasis, we performed cell proliferation, colony formation, invasion, and wound healing assay in vitro using PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells. Although the PC3M‐Luc cells silenced for MTA1 (shMTA1) proliferated at a rate slightly slower than the control cells (NS), the differences in proliferation were not statistically significant (Fig. 3A). Notably, however, MTA1 knockdown inhibited the colony‐forming ability of PC3M‐Luc cells, as is evident by a significant decrease in the number of colonies formed by shMTA1 compared to the NS control cells (Fig. 3B, C). Further, MTA1 silencing in PC3M cells significantly altered their invasive and wound healing ability, an in vitro measure of cell metastatic potential. PC3M‐Luc cells silenced for MTA1 (shMTA1) exhibited about 90% reduction in invasiveness (Fig. 3D) and about 20% decrease in wound closure (Fig. 3E, F) as compared to NS controls. These data reveal that MTA1 silencing decreases metastatic potential of PC3M‐Luc cells in vitro, which in turn may be responsible, at least in part, for the reduced formation of bone metastases in intracardiac xenografts generated using cells lacking MTA1.
Figure 3.
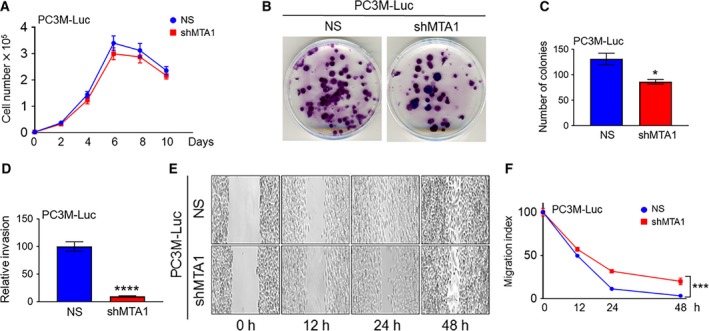
MTA1 knockdown reduces metastatic potential of PC3M‐Luc prostate cancer cells. (A) Proliferation assay of PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells. (B) Representative images of colony formation ability of PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells and (C) quantitation of colonies are shown. Data represent the mean ± SEM of three independent experiments. *P < 0.05 (unpaired t‐test). (D) Invasion assay in PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells. Data represent the mean ± SEM of three independent experiments. ****P < 0.0001 (unpaired t‐test). (E) Representative images of wound healing assay to compare migration ability of PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells and (F) quantitation of wound widths as migration index are shown. Data represent the mean ± SEM of six separate wounds and three independent experiments. ***P < 0.001 (two‐way ANOVA).
3.3. MTA1 knockdown reduces cathepsin B in prostate cancer cells and tumor xenografts
In our previous studies, we have identified key genes involved in the ‘vicious cycle’ interaction between prostate cancer cells and bone cells in vitro (Kai et al., 2011). This ‘bone metastasis signature’ comprised eight genes including vascular endothelial growth factor‐c; matrix metalloproteinases MMP2, MMP10, and MMP11; cathepsins B and K (CTSB and CTSK); mucin 1 (MUC1); and MTA1. In an attempt to dissect molecular mechanisms through which MTA1 promotes bone metastasis, we performed integrative analysis of our bone metastasis signature data along with our MTA1 ChIP‐Seq data and found CTSB as a strong potential candidate responsible for MTA1‐driven invasiveness (Fig. 4A). Furthermore, our MTA1 ChIP‐Seq data demonstrated that pharmacological inhibition of MTA1 (MTA1i) resulted in reduced CTSB promoter occupancy by MTA1, suggesting a possible transcriptional regulation of CTSB by MTA1 (Fig. 4B).
Figure 4.
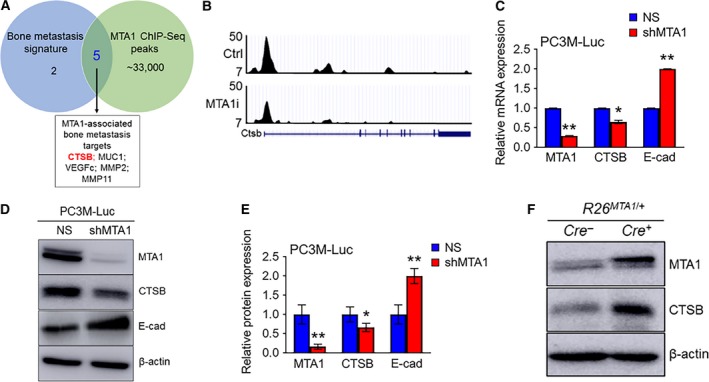
MTA1 knockdown reduces cathepsin B mRNA and protein levels in PC3M‐Luc prostate cancer cells. (A) Venn diagram depicting a number of genes selected as bone metastatic signature (Kai et al., 2011), number of peaks identified by MTA1 ChIP‐Seq (Dhar et al., 2016), and overlapping MTA1‐associated bone metastasis targets of interest. (B) Comparative analysis of ChIP‐Seq data on MTA1 binding in the prostate tissue of Pten +/− mice expressing high levels of MTA1 (Ctrl, upper panel) and reduced levels of MTA1 (MTA1i, lower panel). Representative MTA1 ChIP‐Seq track for CTSB gene loci at 10‐kb resolution is shown. (C) qRT‐PCR of MTA1, CTSB, and E‐Cad mRNA levels in PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells. (D) Immunoblots of MTA1, CTSB, and E‐cad in PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 cells. β‐Actin was used as a loading control. (E) Quantitation of immunoblot signals. Data represent the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01 (two‐way ANOVA). (F) Immunoblots of MTA1 and CTSB in prostate tissues from 13‐week‐old Pb‐Cre +; R26 MTA 1/+ mice and Cre‐negative normal prostate control.
To further characterize the MTA1‐CTSB interrelationship, we examined the expression of cathepsin B in PC3M‐Luc control (NS) and MTA1 knockdown (shMTA1) cells. As shown in Fig. 4C–E, both mRNA and protein levels of CTSB were significantly decreased in PC3M‐Luc cells silenced for MTA1 (shMTA1) compared to control (NS) cells. In agreement with our previous reported finding of MTA1 and E‐cadherin (E‐cad) inverse relationship in prostate cancer (Dhar et al., 2016, 2017), shMTA1 cells showed an expected increase in E‐cad levels compared to NS control cells (Fig. 4C–E). Additionally, LNCaP cells silenced for MTA1 also exhibited reduced levels of CTSB (Fig. S2). Conversely, prostate tissues from our prostate‐specific MTA1‐overexpressing mice (R26MTA1/+; Cre4+), which express significantly reduced levels of E‐cad (Dhar et al., 2017), also exhibited significantly increased CTSB levels (Fig. 4F), thereby further strengthening our observation of a MTA1‐CTSB positive relationship.
Of note, consistent with our in vitro data, qRT‐PCR and immunoblot analysis revealed that the mRNA and protein expressions of CTSB were significantly reduced while E‐cad mRNA and protein levels were considerably increased in the tumor tissues derived from MTA1 knockdown (shMTA1) cells compared to NS controls (Fig. 5A–C). Immunohistochemical analysis of these tumors showed an expected decreased MTA1 nuclear staining in MTA1 knockdown (shMTA1) compared to intense staining in control (NS) tissues. Accordingly, we detected reduced CTSB cytoplasmic protein levels and elevated E‐cad membrane‐bound staining in MTA1 knockdown (shMTA1) compared to MTA1‐expressing (NS) xenografts (Fig. 5D). These data suggest that positive regulation of CTSB expression by MTA1 in prostate tumor cells may be, at least in part, responsible for promotion of prostate tumor growth and bone metastasis formation.
Figure 5.
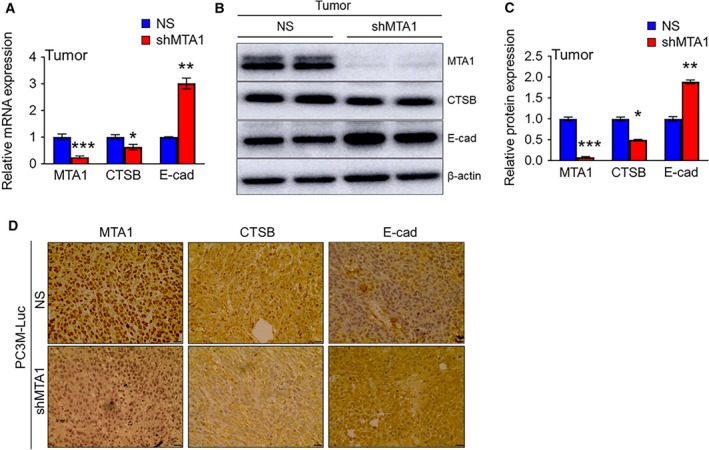
MTA1 knockdown reduces cathepsin B mRNA and protein levels in s.c. tumor tissues. (A) qRT‐PCR of MTA1, CTSB, and E‐cad mRNA levels in the tumor tissues from s.c. xenografts. (B) Immunoblots of MTA1, CTSB, and E‐cad in the tumor tissues from s.c. xenografts. β‐Actin was used as a loading control. (C) Quantitation of immunoblot. Data represent the mean ± SEM of three independent experiments from n = 3 for qRT‐PCR and n = 2 for immunoblot analysis. *P < 0.05; **P < 0.01; ***P < 0.001 (two‐way ANOVA). (D) Comparison of nuclear MTA1 and cytoplasmic CTSB and membrane‐bound E‐cad IHC staining in the tumor tissues from PC3M‐Luc‐NS and PC3M‐Luc‐shMTA1 xenografts. Scale bars, 10 μm.
3.4. MTA1 and CTSB expressions show a strong positive correlation in prostate cancer clinical samples
To explore the clinical significance of the MTA1 and CTSB interrelationship in prostate cancer, we examined the expression of MTA1 and CTSB in a limited cohort of five prostate adenocarcinoma biopsies obtained previously (Dhar et al., 2017). We found a strong positive correlation between MTA1 and CTSB protein expressions: two of three biopsies with high levels of MTA1 had high CTSB expression (Fig. 6A, B). To strengthen our findings from smaller sample numbers, we analyzed the publicly available prostate cancer patient dataset (Wallace et al., 2008) from Oncomine database and found significantly higher levels of both MTA1 and CTSB in tumor samples compared to normal prostate tissues (Fig. 6C). Of note, MTA1 and CTSB exhibited a strong positive correlation from 69 prostate cancer patient samples (Wallace et al., 2008) (Fig. 6D, r = 0.38). In addition, copy number analysis data of the prostate cancer patient samples from independent studies showed an amplification of MTA1 and CTSB (Fig. S3). These data indicate that CTSB is overexpressed in prostate tumors and that this may depend, at least in part, on the MTA1‐associated transcriptional induction of CTSB.
Figure 6.
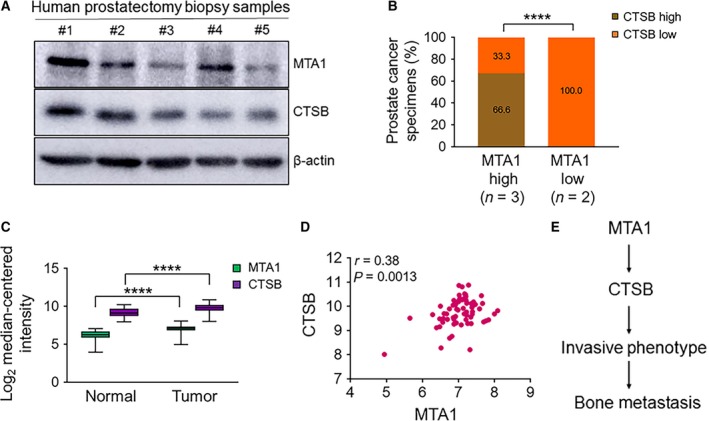
MTA1/CTSB axis in human prostate cancer. (A) Immunoblot analysis for MTA1 and CTSB protein levels in five representative patients. (B) Distribution of MTA1 and CTSB expression in the cohort of patients with prostate cancer analyzed in (A). ****P < 0.0001 (chi square test). (C) Meta‐analysis showing MTA1 and CTSB mRNA overexpression in prostate tumor samples compared to normal tissues from expression array (Wallace et al., 2008) using the Oncomine database. ****P < 0.0001 (two‐way ANOVA). (D) Meta‐analysis of MTA1 and CTSB mRNA expression showing a strong positive correlation in prostate tumor samples from expression array (Wallace et al., 2008) using the Oncomine database. (E) Schematic of the proposed MTA1‐mediated cathepsin B involvement in prostate cancer metastasis to the bone.
4. Discussion
Various mechanisms are associated with metastatic process of primary tumor including interactions of tumor cells with reactive stroma and extracellular matrix and altered cell attachment. Although successful tumor invasion requires degradation of extracellular matrices and EMT by effector molecules such as proteases and cadherins, the key role of upstream master regulators of these effector molecules in the promotion of invasiveness leading to metastasis cannot be undermined.
We have previously demonstrated the key upstream regulatory role of MTA1 in processes involved in development and progression of prostate cancer (Dhar et al., 2016). High levels of MTA1 were detected in bone metastatic lesions of human prostate cancer patients (Dias et al., 2013a; Kai et al., 2011). At the same time, CTSB has been shown to play an important role in invasion and metastasis of prostate cancer (Miyake et al., 2004). Increased CTSB expression and activity were observed in bone tissues colonized by human prostate carcinoma cells (Podgorski et al., 2005). It was suggested that metabolically rich bone microenvironment is responsible for secretion of active CTSB from prostate cancer cells interacting with the bone (Podgorski et al., 2005). Our unpublished data also show induction of CTSB expression in prostate cancer cells, when cocultured with bone cells than when cultured alone. While we fully agree that complex tumor–host interactions can modulate the expression of proteins involved in the ‘vicious cycle’ of bone metastasis, we believe that regulating CTSB expression and activity in the primary tumor might be a way of preventing the development of site metastasis, including the bone.
In this study, we have demonstrated that MTA1 promotes prostate tumor growth and formation of bone metastasis, at least in part, by positively regulating CTSB expression in the primary tumor cells. Opposed to effects on CTSB and as expected based on our previous observations (Dhar et al., 2016, 2017), MTA1 negatively regulates E‐cad expression in primary tumor, a hallmark of EMT associated with metastasis (Figiel et al., 2017), indicating the impact of MTA1 in the process of EMT and cancer cell ability to migrate to distant sites.
Marked increase in MTA1 levels, which is an indicator of advanced castrate‐resistant prostate tumors, might be an important factor in endorsing CTSB‐mediated degradation of basement membrane and in the ability of primary prostate tumor cells to undergo E‐cad‐mediated EMT. We believe that a functional interaction between MTA1 and these two downstream pathways is critical for the acquisition of an invasive phenotype by tumor cells, offering new therapeutic approaches for targeting MTA1‐positive high‐risk tumors to prevent metastases. Of note, the fact that our intracardiac shMTA1 xenografts exhibited reduced formation of bone metastasis suggests that it is possible to prevent homing of primary prostate tumor cells to the bone microenvironment by targeting MTA1.
Thus, MTA1 is an attractive target for the treatment of prostate cancer and prevention of bone metastasis. Our group has intensively published on pharmacological inhibition of MTA1 and its associated signaling by natural dietary compounds such as resveratrol and pterostilbene as well as PI3K and histone deacetylase inhibitors resulting in targeted chemoprotective and anticancer effects in various prostate cancer model systems including xenografts and transgenic mice (Butt et al., 2017; Dhar et al., 2015a,b, 2016, 2017; Kai et al., 2010; Kumar et al., 2015; Levenson et al., 2014; Li et al., 2013). Particularly, we have demonstrated that inhibition of MTA1 by pterostilbene restored E‐cad expression in the prostate tissues of the conditional Pten null mice (Dhar et al., 2016). We have also demonstrated the superior MTA1‐targeted anticancer efficacy of pterostilbene/SAHA combination strategy in prostate cancer (Butt et al., 2017).
5. Conclusions
In conclusion, our study provides mechanistic and preclinical insight into the critical role of MTA1 in tumor progression and formation of bone metastasis in prostate cancer, at least in part, through the CTSB‐dependent promotion of metastatic pathways (Fig. 6E). Our results offer the MTA1/CTSB axis as a promising target for novel molecular therapies in prostate cancer bone metastasis.
Author contributions
ASL conceived the project. AK, SD, and ASL designed the research. AK, SD, GC, NAB, and JMS performed experiments. AK, SD, JMS, CRG, and ASL analyzed the data. AK and ASL wrote the manuscript. All authors critically revised and approved the manuscript.
Supporting information
Fig. S1. Establishment of PC3M‐Luc MTA1 knockdown cells.
Fig. S2. MTA1 knockdown reduces cathepsin B protein levels.
Fig. S3. CTSB expression correlates with MTA1 expression in human prostate cancer.
Table S1. Antibodies for Immunoblots and IHC used in this study.
Table S2. Primers for qRT‐PCR used in this study.
Acknowledgements
We are extremely grateful to Dr K Li (UMMC) for establishing PC3M‐Luc cell line and Ms Tanvi Joshi (Pharmacy College, LIU) for immunoblot results of human samples. We thank Dr CR Pound and Dr JM Lage and her pathology team (UMMC) for providing prostate adenocarcinoma biopsy samples. We also appreciate the assistance of Dr KC Vallabhaneni (UMMC) in animal experiments. We are thankful to Drs RL Summers and S Vijayakumar (UMMC) for their support. This study was supported in part by the Department of Defense Prostate Cancer Research Program under Award W81XWH‐13‐1‐0370 to AS Levenson. Views and opinions of, and endorsements by, the author(s) do not reflect those of the US Army of the Department of Defense.
Contributor Information
Avinash Kumar, Email: avinash.kumar@liu.edu.
Anait S. Levenson, Email: anait.levenson@liu.edu
References
- Butt NA, Kumar A, Dhar S, Rimando AM, Akhtar I, Hancock JC, Lage JM, Pound CR, Lewin JR, Gomez CR et al (2017) Targeting MTA1/HIF‐1alpha signaling by pterostilbene in combination with histone deacetylase inhibitor attenuates prostate cancer progression. Cancer Med 6, 2673–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Seow Khoon M (2015) Experimental models of bone metastasis: opportunities for the study of cancer dormancy. Adv Drug Deliv Rev 94, 141–150. [DOI] [PubMed] [Google Scholar]
- Dhar S, Kumar A, Gomez CR, Akhtar I, Hancock JC, Lage JM, Pound CR and Levenson AS (2017) MTA1‐activated Epi‐microRNA‐22 regulates E‐cadherin and prostate cancer invasiveness. FEBS Lett 591, 924–933. [DOI] [PubMed] [Google Scholar]
- Dhar S, Kumar A, Li K, Tzivion G and Levenson AS (2015a) Resveratrol regulates PTEN/Akt pathway through inhibition of MTA1/HDAC unit of the NuRD complex in prostate cancer. Biochim Biophys Acta 1853, 265–275. [DOI] [PubMed] [Google Scholar]
- Dhar S, Kumar A, Rimando AM, Zhang X and Levenson AS (2015b) Resveratrol and pterostilbene epigenetically restore PTEN expression by targeting oncomiRs of the miR‐17 family in prostate cancer. Oncotarget 6, 27214–27226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Kumar A, Zhang L, Rimando AM, Lage JM, Lewin JR, Atfi A, Zhang X and Levenson AS (2016) Dietary pterostilbene is a novel MTA1‐targeted chemopreventive and therapeutic agent in prostate cancer. Oncotarget 7, 18469–18484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias SJ, Li K, Rimando AM, Dhar S, Mizuno CS, Penman AD and Levenson AS (2013b) Trimethoxy‐resveratrol and piceatannol administered orally suppress and inhibit tumor formation and growth in prostate cancer xenografts. Prostate 73, 1135–1146. [DOI] [PubMed] [Google Scholar]
- Dias SJ, Zhou X, Ivanovic M, Gailey MP, Dhar S, Zhang L, He Z, Penman AD, Vijayakumar S and Levenson AS (2013a) Nuclear MTA1 overexpression is associated with aggressive prostate cancer, recurrence and metastasis in African Americans. Sci Rep 3, 2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel S, Vasseur C, Bruyere F, Rozet F, Maheo K and Fromont G (2017) Clinical significance of epithelial‐mesenchymal transition markers in prostate cancer. Hum Pathol 61, 26–32. [DOI] [PubMed] [Google Scholar]
- Futakuchi M, Fukamachi K and Suzui M (2016) Heterogeneity of tumor cells in the bone microenvironment: mechanisms and therapeutic targets for bone metastasis of prostate or breast cancer. Adv Drug Deliv Rev 99, 206–211. [DOI] [PubMed] [Google Scholar]
- Hofer MD, Tapia C, Browne TJ, Mirlacher M, Sauter G and Rubin MA (2006) Comprehensive analysis of the expression of the metastasis‐associated gene 1 in human neoplastic tissue. Arch Pathol Lab Med 130, 989–996. [DOI] [PubMed] [Google Scholar]
- Holzapfel BM, Wagner F, Loessner D, Holzapfel NP, Thibaudeau L, Crawford R, Ling MT, Clements JA, Russell PJ and Hutmacher DW (2014) Species‐specific homing mechanisms of human prostate cancer metastasis in tissue engineered bone. Biomaterials 35, 4108–4115. [DOI] [PubMed] [Google Scholar]
- Kai L, Samuel SK and Levenson AS (2010) Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer 126, 1538–1548. [DOI] [PubMed] [Google Scholar]
- Kai L, Wang J, Ivanovic M, Chung YT, Laskin WB, Schulze‐Hoepfner F, Mirochnik Y, Satcher RL Jr and Levenson AS (2011) Targeting prostate cancer angiogenesis through metastasis‐associated protein 1 (MTA1). Prostate 71, 268–280. [DOI] [PubMed] [Google Scholar]
- Kim JB, Urban K, Cochran E, Lee S, Ang A, Rice B, Bata A, Campbell K, Coffee R, Gorodinsky A et al (2010) Non‐invasive detection of a small number of bioluminescent cancer cells in vivo. PLoS One 5, e9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dhar S, Rimando AM, Lage JM, Lewin JR, Zhang X and Levenson AS (2015) Epigenetic potential of resveratrol and analogs in preclinical models of prostate cancer. Ann N Y Acad Sci 1348, 1–9. [DOI] [PubMed] [Google Scholar]
- Levenson AS, Kumar A and Zhang X (2014) MTA family of proteins in prostate cancer: biology, significance, and therapeutic opportunities. Cancer Metastasis Rev 33, 929–942. [DOI] [PubMed] [Google Scholar]
- Li K, Dias SJ, Rimando AM, Dhar S, Mizuno CS, Penman AD, Lewin JR and Levenson AS (2013) Pterostilbene acts through metastasis‐associated protein 1 to inhibit tumor growth, progression and metastasis in prostate cancer. PLoS One 8, e57542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miftakhova R, Hedblom A, Semenas J, Robinson B, Simoulis A, Malm J, Rizvanov A, Heery DM, Mongan NP, Maitland NJ et al (2016) Cyclin A1 and P450 aromatase promote metastatic homing and growth of stem‐like prostate cancer cells in the bone marrow. Cancer Res 76, 2453–2464. [DOI] [PubMed] [Google Scholar]
- Miyake H, Hara I and Eto H (2004) Serum level of cathepsin B and its density in men with prostate cancer as novel markers of disease progression. Anticancer Res 24, 2573–2577. [PubMed] [Google Scholar]
- Podgorski I, Linebaugh BE, Sameni M, Jedeszko C, Bhagat S, Cher ML and Sloane BF (2005) Bone microenvironment modulates expression and activity of cathepsin B in prostate cancer. Neoplasia 7, 207–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehemtulla A, Stegman LD, Cardozo SJ, Gupta S, Hall DE, Contag CH and Ross BD (2000) Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia 2, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosol TJ, Tannehill‐Gregg SH, Corn S, Schneider A and McCauley LK (2004) Animal models of bone metastasis. Cancer Treat Res 118, 47–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S and Ichikawa T (2014) Mechanism of prostate cancer invasion and metastasis. Nihon Rinsho 72, 2086–2089. [PubMed] [Google Scholar]
- Toh Y, Pencil SD and Nicolson GL (1994) A novel candidate metastasis‐associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem 269, 22958–22963. [PubMed] [Google Scholar]
- Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA and Ambs S (2008) Tumor immunobiological differences in prostate cancer between African‐American and European‐American men. Cancer Res 68, 927–936. [DOI] [PubMed] [Google Scholar]
- Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K et al (2012) Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res 72, 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J and Wang W (1998) NURD, a novel complex with both ATP‐dependent chromatin‐remodeling and histone deacetylase activities. Mol Cell 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Ziaee S, Chu GC, Huang JM, Sieh S and Chung LW (2015) Prostate cancer metastasis: roles of recruitment and reprogramming, cell signal network and three‐dimensional growth characteristics. Transl Androl Urol 4, 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Establishment of PC3M‐Luc MTA1 knockdown cells.
Fig. S2. MTA1 knockdown reduces cathepsin B protein levels.
Fig. S3. CTSB expression correlates with MTA1 expression in human prostate cancer.
Table S1. Antibodies for Immunoblots and IHC used in this study.
Table S2. Primers for qRT‐PCR used in this study.


