Abstract
Background
Pheochromocytomas (PCCs) show the highest degree of heritability in human neoplasms. However, despite the wide number of alterations until now reported in PCCs, it is likely that other susceptibility genes remain still unknown, especially for those PCCs not clearly syndromic.
Methods
Whole exome sequencing of tumor DNA was performed on a set of twelve PCCs clinically defined as sporadic.
Results
About 50% of PCCs examined had somatic mutations on the known susceptibility VHL, NF1, and RET genes. In addition to these driver events, mutations on SYNE1, ABCC10, and RAD54B genes were also detected. Moreover, extremely rare germline variants were present in half of the sporadic PCC samples analyzed, in particular variants of MAX and SAMD9L were detected in the germline of cases wild-type for mutations in the known susceptibility genes.
Conclusions
Additional somatic passenger mutations can be associated with known susceptibility VHL, NF1, and RET genes in PCCs, and a wide number of germline variants with still unknown clinical significance can be detected in these patients. Therefore, many efforts should be aimed to better define the pathogenetic role of all these germline variants for discovering novel potential therapeutic targets for this disease still orphan of effective treatments.
1. Introduction
Pheochromocytomas (PCCs) are rare tumors of the autonomic nervous system that arise from the chromaffin tissue of the adrenal medulla [1]. Most of PCCs are benign; however, approximately 10% of cases are malignant and can develop metastases either at the time of diagnosis or even later after several years, with a highly variable clinical course and a 5-year overall survival rate of 50% [2]. PCCs show the highest degree of heritability in human neoplasms and almost 40% of cases occur within heritable syndromes, including multiple endocrine neoplasia type 2 (MEN2), neurofibromatosis type 1 (NF1), von Hippel Lindau (VHL) disease, and hereditary paraganglioma and familial pheochromocytoma [3]. Germline mutations have been identified in more than 15 well-characterized genes, such as VHL, SDHB, SDHD, NF1, and RET [4].
Despite this well-known inherited basis of PCCs and paragangliomas (PGLs), during the past decades somatic mutations with variable frequency in many genes, including EPAS1 (HIF2α), RET, VHL, RAS, NF1, ATRX, and CSDE1 recurrent somatic copy number alterations and several fusion genes, involving MAML3, BRAF, NGFR, and NF1, have been progressively identified [5–16]. Recently a multiplatform integrated analysis classified PCCs/PGLs into four clinically relevant molecular subtypes: a kinase signaling subtype, a pseudohypoxia subtype, a Wnt-altered subtype driven by MAML3 and CSDE1, and a cortical admixture subtype [16].
Therefore, given this molecular complexity of PCCs, it is likely that other alterations remain still unknown, especially for those PCCs not clearly syndromic. For this purpose, in the present study, we performed whole exome sequencing on a set of 12 clinically sporadic PCCs, with a family history negative for PCCs/PGLs.
2. Materials and Methods
2.1. Patients and Tumor Samples
Fresh tissue specimens of PCC from 12 patients with a family history negative for PCCs/PGLs were collected during the surgical operation, snap-frozen in liquid nitrogen, and stored at −80°C until analysis. Patient characteristics are listed in Table 1. Whole exome sequencing was performed on biological tumor samples on matched peripheral blood samples obtained from all patients. This study was approved by the local institutional ethical committee of S. Orsola-Malpighi hospital (approval number 95/2013/U/Tess). All patients provided written informed consent.
Table 1.
Patient characteristics.
| ID | Sex | Age | Tumor size (cm) | HIC characteristics | PASS score |
|---|---|---|---|---|---|
| N47 | F | 63 | 5.0 | Ki-67 2.4% | n.a. |
| N49 | M | 65 | 4.0 | Positive staining for synaptophysin. Ki-67 3.2% | 3 |
| N50 | M | 36 | 4.0 | Positive staining for synaptophysin. S100 protein-positive sustentacular cells. Ki-67 2.2% | 3 |
| N51 | M | 46 | 1.5 | Positive staining for chromogranin A and synaptophysin. S100 protein-positive sustentacular cells. Negative staining for c-kit, EGFr, p53. Ki-67 0.5% | 2 |
| N52 | M | 47 | 3.5 | Positive staining for chromogranin A and synaptophysin. S100 protein-positive sustentacular cells. Negative staining for CD10. Ki-67 0.1% | 4 |
| N53 | F | 58 | 5.4 | Ki-67 1.8% | 5 |
| N54 | F | 46 | 6.0 | Positive staining for chromogranin A and synaptophysin. S100 protein-positive sustentacular cells. Focal positive staining for CD10. Ki-67 0.4% | 6 |
| N55 | F | 30 | 6.0 | Positive staining for chromogranin A and synaptophysin. S100 protein-positive sustentacular cells. Ki-67 0.1% | 4 |
| N56 | M | 41 | 1.7 | Positive staining for synaptophysin. S100 protein-positive sustentacular cells. Negative staining for calretinin. Ki-67 2.2% | 2 |
| N57 | M | 35 | 8.0 | Positive staining for chromogranin A and synaptophysin. Few S100 protein-positive sustentacular cells. Negative staining for calretinin and α-inhibin. Ki-67 5.7% | 4 |
| N62 | M | 59 | 3.0 | Ki-67 1.4%. | 5 |
| N63 | M | 26 | 7.0 | Positive staining for synaptophysin. S100 protein-positive sustentacular cells. Negative staining for calretinin, MEL-A, and α-inhibin. Ki-67 4% | 14 |
2.2. Whole Exome Sequencing
DNA was extracted from peripheral blood and fresh frozen tissue with DNA mini kit (Qiagen, Milan, Italy) following manufacturer's instructions. Whole exome sequencing of tumor DNA was performed on HiScanSQ platform in accordance with Nextera Rapid Exome Enrichment protocol (Illumina, San Diego, California, USA). Briefly, 100 ng of genomic DNA was tagged and fragmented by the Nextera transposome. The Nextera transposome simultaneously fragments the genomic DNA and adds adapter sequences to the ends. The products were then amplified and exome regions were enriched. The enriched libraries were amplified by PCR and quantified using PicoGreen assay (Life Technologies, Milan, Italy).
Paired-end libraries were sequenced at 2 × 100 bp read length using Illumina Sequencing by synthesis (SBS) technology.
2.3. Bioinformatic Analysis
After demultiplexing and FASTQ generation performed with bcltofastq function developed by Illumina, the paired-end reads were trimmed using AdapterRemoval (https://github.com/MikkelSchubert/adapterremoval) with the aim of removing stretches of low-quality bases (<Q10) and Truseq/Nextera rapid capture adapters present in the sequences. The paired-end reads were then aligned on human reference genome hg38 (http://www.genome.ucsc.edu). Data from WES were mapped with Burrows-Wheeler Aligner with the default setting; the PCR and optical duplicates were removed, and Genome Analysis Toolkit (https://software.broadinstitute.org/gatk) was used to locally realign, recalibrate, and call the Ins/del variants, while point mutations were identified with the algorithm MuTect (https://www.broadinstitute.org/cancer/cga/mutect). Single nucleotide variants (SNV) and ins/del were annotated with gene and protein alteration using Annovar (http://annovar.openbioinformatics.org); nonsynonymous and nonsense SNV, frameshift/nonframeshift Indels, and splice site mutations were selected with a threshold read depth ≥ 10x and a variant allele frequency ≥ 0.2. All the variants were filtered to select novel or rare events basing on database of human variability dbSNP (http://www.ncbi.nlm.nih.gov/SNP), 1000Genomes (http://www.1000genomes.org), ExAC (http://exac.broadinstitute.org), and EVS (http://evs.gs.washington.edu/EVS). In-depth evaluation of high confidence somatic variants was performed by verifying the presence of alternate allele on the normal counterpart and manually visualizing each variation with the tview function of SAMmtools. Both somatic mutations and germline variants were searched in COSMIC (Catalog of Somatic Mutations in Cancer; http://cancer.sanger.ac.uk/cosmic), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar), and HGMD (http://www.hgmd.org), and their effect on protein structure and function was predicted with SNPeff, a software that uses three different prediction algorithms (SIFT, Polyphen2, and LRT).
Moreover, based on WES data, the analysis of amplifications and large deletions were performed making a consensus between Control FREEC (http://boevalab.com/FREEC) and ADTEX (http://adtex.sourceforge.net) with paired tumour/matched normal samples. A filtering procedure was applied taking into account the uncertainty value given by Control FREEC (<80%) and the polymorphic copy number variants from the Database of human Genomic Variants (http://dgv.tcag.ca/dgv/app/home).
For germline variants prioritization, all rare (MAF < 0.01) alterations occurring on the known susceptibility genes of PCC and PGL were considered. Moreover, variants with an evident effect on the protein (nonsense and splicing mutations or frameshift ins/del) were prioritized and manually annotated using HGMD and ClinVar database and with literature.
2.4. Sanger Sequencing
Sequencing of the DNA extracted from tumors and matched peripheral blood samples was performed to validate candidate mutations. Specific PCR assay for the amplification and sequencing of selected genes was designed with Primer Express 3.0 Software (Applied Biosystems, Monza, Italy). PCR products were purified with the Qiaquick PCR purification kit (Qiagen) and sequenced on both strands using the Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems). Sanger Sequencing was performed on ABI 3730 Genetic Analyzer (Applied Biosystems).
2.5. Real-Time PCR
Total RNA was extracted from fresh frozen tissues using the RNeasy spin-column method (Qiagen). RNA was reverse transcribed to cDNA using the Transcriptor First-Strand cDNA Synthesis Kit (Life Technologies) with oligo dT primers. qPCR amplification of genes of interest was performed with real-time LightCycler 480 instrument (Roche). Fold-change was estimated by DDCt method, using ATPS, HPRT, and HMBS genes as housekeeping controls. Primers used were: MAX_FW 5′- GCGATAACGATGACATCGAGGT-3′ and MAX_RV 5′-CCCGCAAACTGTGAAAGCTGT-3′, SAMD9L_FW 5′-AAAGTGAGTGAGTGAGCCCAG-3′ and SAMD9L_RV 5′-CATGCTCTTTGGTCCAGTCT-3′, ATPS_FW 5′-GTCTTCACAGGTCATATGGGGA-3′ and ATPS_RV 5′-ATGGGTCCCACCATATAGAAGG-3′, HMBS_FW 5′-GGCAATGCGGCTGCAA-3′ and HMBS_RV 5′-GGGTACCCACGGAATCAC-3′, HPRT_FW 5′-TGACACTGGCAAAACAATGCA-3′ and HPRT_RV 5′-GGTCCTTTTCACCAGCAAGCT-3′. For detection of cortical admixture profile, expression levels of STAR, CYP2W1, CYP11B2, CYP21A2, and CLND2, genes were evaluated using the following primers: STAR_Fw 5′-TGGGCATCCTTAGCAACCAA-3′ and STAR_Rev 5′-GCCCACATCTGGGACCACTT-3′; CYP2W1_Fw 5′-GTCATGGTCCTCTTGGGGTC-3′ and CYP2W1_Rev 5′-CTCCAGGAGGGTCCTCAGAA-3′; CYP11B2_Fw 5′-TGCATCCCTGCAGGATGAT-3′ and CYP11B2_Rev 5′-GCGACAGCACATCTGGGT-3′; CYP21A2_Fw 5′-AGCCCGACCTCCCCAT-3′ and CYP21A2_Rev 5′-CACCACCACATCTTGCAGCC-3′; CLND2_Fw 5′-CCCCTTGTACTTCGCTCCCC-3′ and CLND2_Rev 5′-AAGCAGCCTCAAGAAGGCATC-3′.
3. Results
Exome sequencing generated a minimum of 45.5 million reads/sample with a mean coverage of RefSeq regions of 44x. Few somatic mutations were identified (an average of 7 mutations per sample) while several copy number alterations were detected, with losses of chr1, chr3, and chr17 being the most recurrent.
3.1. Analysis of Somatic Mutations
The biological effect of somatic mutations was predicted with three bioinformatic tools (Suppl. Table 1). Mutations on susceptibility genes were detected in 6 out of 12 cases and were annotated in COSMIC and HGMD databases. VHL missense mutations were identified in two cases: a p.S65A identified in N51, a mutation already reported in PCC (COSM144970), and a p.Y98H in N56, a mutation reported in ClinVar and HMGD as pathogenic in association with Von Hippel-Lindau syndrome. Two novel somatic alterations were detected in NF1: N62 carried a splicing mutation c.480-1G>C in exon 5 and N55 had a frameshift deletion (p.W784fs) in exon 20. Interestingly, the abovementioned mutations on NF1 and VHL were in regions affected by loss of the wild-type allele (Figure 1). Heterozygous loss of the chromosomal region covering NF1 was detected also in N54; however, no additional mutational event was detected on this gene. RET was found mutated in heterozygosis in two other tumors: a novel exon 11 nonframeshift INDEL (p.L633delinsLCR) was detected in N57 and the hotspot mutation p.M918T (COSM965) in N53.
Figure 1.
Deletion of the wild-type allele occurred in PCC samples that carry NF1 or VHL somatic mutations. Alignments of sequencing reads located on NF1 or VHL mutated bases are shown. Green bars represent deleted regions of chr3 for VHL (a, b) and chr17 for NF1 (c, d) detected by copy number analysis. A black arrow indicates the chromosome position of NF1 or VHL and the mutated base on the sequencing reads.
In addition to these driver mutations on known susceptibility genes, other somatic passenger mutations were detected in these 6 tumors. In particular, SYNE1 and ABCC10 were mutated, respectively, in the two RET-mutated cases (N53 and N57), while a missense p.G460S mutation of RAD54B, a gene involved in DNA repair process, affected a NF1 mutated case (N55). On the other side, few somatic mutations were identified in the 6 remaining PCC cases and none was recurrent between samples. N63 carried a p.L114X nonsense mutation in CDC14B, a protein phosphatase involved in DNA damage response. N52 carried two heterozygous missense mutations (SMARCC2 p.P1092R and PRKG1 p.F387) both predicted as pathogenic by bioinformatic predictors.
3.2. Analysis of Germline Rare Variants
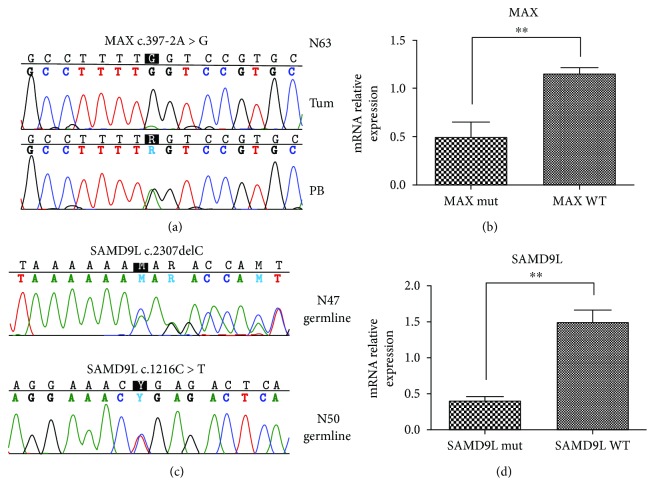
In addition to the abovementioned somatic alterations, rare (ExAc < 0.01) germline variants were identified in five sporadic PCC samples (Suppl. Table 2), among which those occurring in MAX and SAMD9L seem to play an important role in PCC pathogenesis, completing the picture of relevant alterations identified in our cohort (Table 2). In N63, a novel germline variant of MAX (c.397-2A>G), affecting splice site and pathogenic for the protein function, was detected. Noticeably, this patient is a young adult (age 26) and showed the loss of the wild-type allele in the tumor, thus following the Knudson two-hit model (Figure 2(a)), and mRNA level of MAX was found downregulated with respect to the other PCC samples (Figure 2(b)). According to the findings of Fishbein et al. [16], we evaluated the expression level of STAR, CYP11B2, CYP2W1, CYP21A2, and CLND2 with the aim to assess whether a cortical admixture profile was present in this MAX mutated sample. However, a general low expression level of these genes was found in our cohort and no difference between the MAX mutated case and the other PCC samples was detected (data not shown). Conversely, 2 germline variants of SAMD9L were found in other 2 PCC, both wild-type for mutations in known susceptibility genes: a novel p.N769fs frameshift deletion in N47 and a rare (ExAc = 0.2%) nonsense p.R406X in N50 (Figure 2(c)). Evaluation of mRNA level of SAMD9L showed a significant downregulation of the transcript in this two PCC samples (Figure 2(d)). Interestingly, the same gene was found somatically mutated in N54 (p.L1016S), increasing to 3 the number of PCC wild-type cases of our cohort that carried alterations on SAMD9L (Table 2).
Table 2.
Somatic and germline alterations identified in our cohort that could support PCC pathogenesis. Annotations concerning Exac frequency, COSMIC, ClinVar, and biological effect predictions are shown for each variant.
| Sample | Gene | Somatic/germline | Position | Exon | cDna | Protein | Type | dbSNP | ExAC freq | COSMIC | ClinVar | Effect prediction§ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N47 | SAMD9L | Germline | chr7:93133665 | 5 | c.2307delC | p.N769fs | Frameshift | Novel | — | Yes | — | Pathogenic |
| N50 | SAMD9L | Germline | chr7:93134756 | 6 | c.1216C>T | p.R406X | Stop gain | rs150070697 | 0.002 | Yes∗ | — | Pathogenic |
| N54 | SAMD9L | Somatic | chr7:93132925 | 4 | c.3047T>C | p.L1016S | Missense | Novel | na | — | — | Pathogenic |
| N63 | MAX | Germline SNV + somatic loss | chr14:65076665 | 7 | c.397-2A>G | na | Splicing | Novel | — | Yes | — | Pathogenic |
| N51 | VHL | Somatic + loss | chr3:10142040 | 1 | c.193T>G | p.S65A | Missense | Novel | — | Yes ∗ | — | Pathogenic |
| N56 | VHL | Somatic + loss | chr3:10142139 | 1 | c.292T>C | p.Y98H | Missense | rs5030809 | — | Yes | Pathogenic | Pathogenic |
| N55 | NF1 | Somatic + loss | chr17:31227548 | 20 | c.2351delG | p.W784fs | Frameshift | Novel | — | Yes | — | Pathogenic |
| N62 | NF1 | Somatic + loss | chr17:31169890 | 5 | c.480-1G>C | na | Splicing | Novel | — | Yes | — | Pathogenic |
| N53 | RET | Somatic | chr10:43121968 | 16 | c.2753T>C | p.M918T | Missense | rs74799832 | — | Yes ∗ | Pathogenic | Pathogenic |
| N57 | RET | Somatic | chr10:43114499 | 11 | c.1899_1900insTGCCGC | p.L633delinsLCR | Nonframeshift insertion | Novel | — | Yes ∗ | — | Pathogenic |
∗COSMIC record that totally match with the mutation identified; § defined pathogenic if the mutation has been classified as pathogenic or deleterious by at least two out three predictors used (SIFT, Polyphen2, and LRT).
Figure 2.
(a) Loss of heterozygosity of germline MAX mutation detected on N63 tumor. Chromatograms showing homozygous status of c.397-2A>G mutation in the tumor of N63 (upper panel), while in the germline it is heterozygous (lower panel). (b) MAX mRNA relative expression of MAX mutant case (N63) in comparison to the other PCC cases. P value was estimated with unpaired t-test (∗∗ P < 0.01). (c) Validation of SNV detected on SAMD9L gene in 2 PCC samples. Germline c.2307delC and c.1216C>T heterozygous mutations detected in both tumor and peripheral blood of N47 and N50, respectively. (d) SAMD9L mRNA relative expression of the 2 SAMD9L mutant cases (N47 and N50) in comparison to the other PCC cases. P value was estimated with unpaired t-test (∗∗ P < 0.01).
On the contrary, the role of the remaining germline variants (suppl. Table 2) is not clear. In N50, in addition to SAMD9L alteration, a heterozygous stop gain of BRCA2 (p.K3326X) was detected as constitutive. This variant is recorded in ClinVar as “benign”; however, it is found at low frequency in healthy individuals (ExAc allele frequency = 0.7%). BRCA2 mRNA expression level was evaluated, but it was found not altered in this case with respect to the other PCC cases (data not shown). Finally, the rare germline variants of ATRX and KTM2D, identified in N56, and of MDH2, in N53, are missense variants occurring in association with a well-defined somatic mutation on a susceptibility gene (VHL in N56 and RET in N53) and then it is not clear whether they could have a role on the tumor onset.
4. Discussion
In this study, we performed whole exome sequencing on a set of twelve PCCs, clinically defined as sporadic, and we found that 50% of PCCs examined had somatic mutations on the known susceptibility VHL, NF1, and RET genes. In addition to these driver mutations, other somatic passenger mutations were detected. In particular, SYNE1 and ABCC10 were mutated, respectively, in the two RET-mutated cases, while a missense p.G460S mutation of RAD54B affected a NF1 mutated case, suggesting these other events may play a potential role on PCCs pathogenesis and development. Nuclear envelope 1 (SYNE1) gene encodes several different isoforms involved in a variety of cellular processes including cytokinesis, Golgi function, and nuclear organization and structural integrity and positioning of the nucleus [17, 18]. Mutations of SYNE1 have been found in colorectal cancer, glioblastoma, and ovarian cancer, and methylation of the gene was also frequently found in lung adenocarcinoma and colorectal cancer [19–23]. On the other side, RAD54B is a telomere-related gene involved in DNA repair process, and coding-missense changes of this gene have been found in familial breast cancer cases not explained by mutations in the best-known high susceptibility genes BRCA1 and BRCA2 [24]. These findings may be even more relevant in PCCs, given the highest degree of heritability of this disease.
Of note on this topic, the identification of germline variants in half of the sporadic PCC samples was analyzed, among which those occurring in MAX and SAMD9L genes may be extremely interesting. Indeed, some reports disclose the importance of screening for germline variants also in sporadic cases, especially on susceptibility genes [25, 26], and besides the potential role played in PCCs pathogenesis, the detection of these germline variants in patients clinically defined as sporadic may suggest the existence of unknown multineoplasia hereditary diseases.
In our study, we have identified a variant of MAX (c.397-2A>G), affecting splice site and pathogenic for the protein function, with the loss of the wild-type allele in the tumor as second hit. Of note, this patient is a young adult of 26 years old, and the presence of a germline variant in a known cancer susceptibility gene may suggest that the PCC in this patient could be the first clinical expression of a hereditary disease still undefined. It is already known that around 1% of PCC patients were negative for mutations in the other known susceptibility genes carried a germline mutation affecting MAX [26, 27]. About 20 variants affecting MAX have been already described distributed along the gene, but more frequently involving exons 3 and 4, matching some of the most important residues within the conserved bHLH-Zip domain of MAX. Most mutations lead to truncated proteins, with the expected LOH affecting the remaining wild-type allele of the MAX tumor suppressor gene [26, 27]. Two truncating MAX mutations affecting exon 3 (c.97C>T) and 4 (c.185_186delA) and three missense variants (c.67G>A, c.281T>C, and c.425C>T) located in exons 3, 4, and 5, respectively, have been identified [27]. Furthermore, other mutations affecting the initial methionine (c.2T>A), creating a premature stop codon (c.25del, c.97C>T, c.223C>T, and c.244C>T) or affecting a donor/acceptor splice site (c.171 + 1G>A and c.295 + 1G>T), have been subsequently reported [26]. In addition, 2 deletions were identified: the first caused an inframe loss of 6 highly conserved amino acids within the first helix of the protein (c.140_157del), and the second, spanned the whole gene (c.1-?_483+?del) [26]. The MAX mutant case of our cohort showed a marked downregulation of MAX at mRNA level; however, it did not show a significant modulation of the genes involved in cortical admixture phenotype. Conversely to this finding, two cases of MAX mutant PCC were described to overexpress adrenal cortex markers (including CYP11B2, CYP21A2, and STAR) supporting an association between MAX mutation and the cortical admixture PCC subtype [16]. Further studies on larger cohort will be needed to assess this association.
Moreover, we have found three germline variants in BRCA2 and SAMD9L in other two PCC patients wild-type for mutations in susceptibility genes. Specifically, even if it is a rare variant (ExAc allele frequency = 0.7%) recently described as associated with an increased risk of developing breast and ovarian cancers [28], the heterozygous stop gain of BRCA2 (p.K3326X) is considered as “benign” in ClinVar. In support of this consideration, we did not detect any variation of BRCA2 mRNA expression level. Thus, we cannot draw any conclusion on the role of this variant and PCC onset. On the contrary, the 2 rare germline variants of SAMD9L detected in two sporadic PCC cases were both producing a premature STOP of the protein (a p.N769fs frameshift deletion and a nonsense p.R406X) and were associated with significant downregulation of SAMD9L mRNA, supporting the pathogenicity of these alterations. The function of this gene is not well characterized; however, evidence has accumulated supporting the role of SAMD9L in cell proliferation and tumor suppression. In particular, somatic mutations have been found in hepatitis B-related hepatocellular carcinomas [29] and inactivation of SAMD9L has been recently correlated with myeloid transformation [30]. Interestingly, a somatic p.L1016S mutation of SAMD9L was detected in another PCC sample of our cohort, and additionally a p. P636S (COSM3412478) was reported in one case of TCGA PCC/PGL dataset (http://cancergenome.nih.gov/). Taken together, these data could support a possible role of SAMD9L in PCC biology; however, functional studies will be needed to further assess this hypothesis.
5. Conclusions
Taken together, the discovery of novel germline variants of known cancer-related genes in sporadic PCC patients, wild-type for mutations in susceptibility genes, may suggest that the likely existence of other multineoplasia syndrome. On the contrary, the role of the other rare germline variants ATRX, KTM2D, and MDH2 genes identified in two cases having a well-defined somatic mutation on a susceptibility gene (RET or VHL) remains still unclear.
Although the pathogenetic role of all these variants is still not known, due to the large number of susceptibility genes implicated in the diagnosis of inherited PCCs and PGLs, we confirm also what other authors stated that the next-generation sequencing technology is ideally suited for carrying out genetic screening of these individuals [31]. Moreover, the high degree of heritability of PCCs and the wide number of germline variants described suggest the likely need of a more extended genetic counselling and the type and duration of the surveillance program of patients affected by PCCs with variants of unknown significance. Finally, these findings may underlie the possible occurrence of novel hereditary diseases that remain still undefined.
In conclusion, additional somatic passenger mutations can be associated to known susceptibility VHL, NF1, and RET genes in PCCs and a wide number of germline variants with still unknown clinical significance can be detected in these patients. Therefore, many efforts should be aimed to better define the pathogenetic role of all these germline variants for discovering novel potential therapeutic targets for this disease still orphan of effective treatments.
Data Availability
All sequencing data are available upon request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
Milena Urbini and Margherita Nannini contributed equally to this work.
Supplementary Materials
Supplementary Table 1: list of somatic mutations identified in our cohort. Annotations concerning ExAC frequencies, COSMIC, and effect prediction are shown for each variant. In yellow are highlighted mutations affecting PCC susceptibility genes. Supplementary Table 2: list of rare germline variants identified. Annotations concerning Exac frequency, COSMIC, ClinVar, and biological effect predictions (SIFT, Polyphen2, and LRT) are shown for each variant.
References
- 1.Lenders J. W. M., Eisenhofer G., Mannelli M., Pacak K. Phaeochromocytoma. Lancet. 2005;366(9486):665–675. doi: 10.1016/S0140-6736(05)67139-5. [DOI] [PubMed] [Google Scholar]
- 2.Eisenhofer G., Bornstein S. R., Brouwers F. M., et al. Malignant pheochromocytoma: current status and initiatives for future progress. Endocrine-Related Cancer. 2004;11(3):423–436. doi: 10.1677/erc.1.00829. [DOI] [PubMed] [Google Scholar]
- 3.Dahia P. L. M. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nature Reviews Cancer. 2014;14(2):108–119. doi: 10.1038/nrc3648. [DOI] [PubMed] [Google Scholar]
- 4.Favier J., Amar L., Gimenez-Roqueplo A. P. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nature Reviews. Endocrinology. 2015;11(2):101–111. doi: 10.1038/nrendo.2014.188. [DOI] [PubMed] [Google Scholar]
- 5.Burnichon N., Vescovo L., Amar L., et al. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Human Molecular Genetics. 2011;20(20):3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 6.Burnichon N., Buffet A., Parfait B., et al. Somatic NF1 inactivation is a frequent event in sporadic pheochromocytoma. Human Molecular Genetics. 2012;21(26):5397–5405. doi: 10.1093/hmg/dds374. [DOI] [PubMed] [Google Scholar]
- 7.Cho N. H., Lee H. W., Lim S. Y., Kang S., Jung W. Y., Park C. S. Genetic aberrance of sporadic MEN 2A component tumours: analysis of RET. Pathology. 2005;37(1):10–13. doi: 10.1080/00313020400024816. [DOI] [PubMed] [Google Scholar]
- 8.Comino-Méndez I., de Cubas A. A., Bernal C., et al. Tumoral EPAS1 (HIF2A) mutations explain sporadic pheochromocytoma and paraganglioma in the absence of erythrocytosis. Human Molecular Genetics. 2013;22(11):2169–2176. doi: 10.1093/hmg/ddt069. [DOI] [PubMed] [Google Scholar]
- 9.Crona J., Delgado Verdugo A., Maharjan R., et al. Somatic mutations in H-RAS in sporadic pheochromocytoma and paraganglioma identified by exome sequencing. The Journal of Clinical Endocrinology and Metabolism. 2013;98(7):E1266–E1271. doi: 10.1210/jc.2012-4257. [DOI] [PubMed] [Google Scholar]
- 10.Fishbein L., Khare S., Wubbenhorst B., et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nature Communications. 2015;6(1):p. 6140. doi: 10.1038/ncomms7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hrasćan R., Pećina-Slaus N., Martić T. N., et al. Analysis of selected genes in neuroendocrine tumours: insulinomas and phaeochromocytomas. Journal of Neuroendocrinology. 2008;20(8):1015–1022. doi: 10.1111/j.1365-2826.2008.01755.x. [DOI] [PubMed] [Google Scholar]
- 12.Komminoth P., Kunz E., Hiort O., et al. Detection of RET proto-oncogene point mutations in paraffin-embedded pheochromocytoma specimens by nonradioactive single-strand conformation polymorphism analysis and direct sequencing. The American Journal of Pathology. 1994;145(4):922–929. [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo R. A., Qin Y., Cheng Z. M., et al. Recurrent mutations of chromatin-remodeling genes and kinase receptors in pheochromocytomas and paragangliomas. Clinical Cancer Research. 2016;22(9):2301–2310. doi: 10.1158/1078-0432.CCR-15-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang Z., Yang C., Lorenzo F., et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. The New England Journal of Medicine. 2012;367(10):922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn A., Benn D., Clifton-Bligh R., et al. The genomic landscape of phaeochromocytoma. The Journal of Pathology. 2015;236(1):78–89. doi: 10.1002/path.4503. [DOI] [PubMed] [Google Scholar]
- 16.Fishbein L., Leshchiner I., Walter V., et al. Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31(2):181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan J., Beck K. A. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. Journal of Cell Science. 2004;117(4):619–629. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- 18.Gough L. L., Fan J., Chu S., Winnick S., Beck K. A. Golgi localization of Syne-1. Molecular Biology of the Cell. 2003;14(6):2410–2424. doi: 10.1091/mbc.e02-07-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tessema M., Willink R., Do K., et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25 . Cancer Research. 2008;68(6):1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 20.Schuebel K. E., Chen W., Cope L., et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genetics. 2007;3(9):1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sjoblom T., Jones S., Wood L. D., et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 22.Masica D. L., Karchin R. Correlation of somatic mutation and expression identifies genes important in human glioblastoma progression and survival. Cancer Research. 2011;71(13):4550–4561. doi: 10.1158/0008-5472.CAN-11-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doherty J. A., Rossing M. A., Cushing-Haugen K. L., et al. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium Study. Cancer Epidemiology Biomarkers & Prevention. 2010;19(1):245–250. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanowsky K., Barroso A., Osorio A., Urioste M., Benitez J., Martinez-Delgado B. Mutational analysis of telomere genes in BRCA1/2-negative breast cancer families with very short telomeres. Breast Cancer Research and Treatment. 2012;134(3):1337–1343. doi: 10.1007/s10549-012-2141-2. [DOI] [PubMed] [Google Scholar]
- 25.Welander J., Andreasson A., Juhlin C. C., et al. Rare germline mutations identified by targeted next-generation sequencing of susceptibility genes in pheochromocytoma and paraganglioma. The Journal of Clinical Endocrinology and Metabolism. 2014;99(7):E1352–E1360. doi: 10.1210/jc.2013-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnichon N., Cascon A., Schiavi F., et al. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clinical Cancer Research. 2012;18(10):2828–2837. doi: 10.1158/1078-0432.CCR-12-0160. [DOI] [PubMed] [Google Scholar]
- 27.Comino-Méndez I., Gracia-Aznárez F. J., Schiavi F., et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nature Genetics. 2011;43(7):663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 28.Meeks H. D., Song H., Michailidou K., et al. BRCA2 Polymorphic Stop Codon K3326X and the Risk of Breast, Prostate, and Ovarian Cancers. Journal of the National Cancer Institute. 2015;108(2, article djv315) doi: 10.1093/jnci/djv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Zhai Y. Y., Dai J. H., Li K. Y., Deng Q., Han Z. G. SAMD9L inactivation promotes cell proliferation via facilitating G1-S transition in hepatitis B virus-associated hepatocellular carcinoma. International Journal of Biological Sciences. 2014;10(8):807–816. doi: 10.7150/ijbs.9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagamachi A., Matsui H., Asou H., et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell. 2013;24(3):305–317. doi: 10.1016/j.ccr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 31.NGS in PPGL (NGSnPPGL) Study Group, Toledo R. A., Burnichon N., et al. Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nature Reviews Endocrinology. 2017;13(4):233–247. doi: 10.1038/nrendo.2016.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: list of somatic mutations identified in our cohort. Annotations concerning ExAC frequencies, COSMIC, and effect prediction are shown for each variant. In yellow are highlighted mutations affecting PCC susceptibility genes. Supplementary Table 2: list of rare germline variants identified. Annotations concerning Exac frequency, COSMIC, ClinVar, and biological effect predictions (SIFT, Polyphen2, and LRT) are shown for each variant.
Data Availability Statement
All sequencing data are available upon request to the corresponding author.