Summary
Mitochondrial inheritance, genome maintenance, and metabolic adaptation depend on organelle fission by Dynamin-Related Protein 1 (DRP1) and its mitochondrial receptors. DRP1 receptors include the paralogs Mitochondrial Dynamics 49 and 51 (MID49/MID51) and Mitochondrial Fission Factor (MFF), but the mechanisms by which these proteins recruit and regulate DRP1 are unknown. Here we present a cryoEM structure of human, full-length DRP1 coassembled with MID49 and an analysis of structure- and disease-based mutations. We report that GTP induces a remarkable elongation and rotation of the G-domain, Bundle-Signaling Element (BSE) and connecting hinge loops of DRP1. In this conformation, a network of multivalent interactions promotes polymerization of a linear DRP1 filament with MID49/MID51. Following coassembly, GTP hydrolysis and exchange lead to MID receptor dissociation, filament shortening and curling of DRP1 oligomers into constricted and closed rings. Together, these views of full-length, receptor- and nucleotide-bound conformations reveal how DRP1 performs mechanical work through nucleotide-driven allostery.
Introduction
Fragmentation of the mitochondrial reticulum disperses units of the organelle during cell division1,2, coordinates morphological adaptation with metabolic demand3,4, and quarantines damaged units for turnover5,6. Recent work also led to the discovery of the role mitochondrial fission plays in regulated cell death pathways7–9, brain development and synaptic function10,11, and how certain pathogens disrupt these processes and hijack mitochondrial resources12,13. Finally, there is a growing understanding of how inter-organelle contacts between the ER and mitochondria initiate mitochondrial fission14,15, and how this process impacts mitochondrial genome duplication and integrity16,17. The master regulator that unites these processes across eukaryotic evolution is the membrane-remodeling GTPase DRP12,18,19.
DRP1 is necessary but not sufficient for mitochondrial fission because receptors must recruit the enzyme to the Outer Mitochondrial Membrane (OMM). In mammals, these receptors include the paralogs MItochondrial Dynamics proteins MID49 and MID51 and the Mitochondrial Fission Factor, MFF20–24. Following receptor-dependent recruitment, DRP1 assembles into polymers that encircle mitochondria and, via poorly understood mechanisms, channels energy from GTP binding, hydrolysis, and nucleotide exchange into a mechanochemical constriction8,23,25–28. In addition to DRP1 and its OMM receptors, a recent study revealed that a second member of the dynamin-family of GTPases, dynamin-2/DNM2, may enact the final fission event downstream of DRP1-driven constriction of a mitochondrial tubule29. Thus, mitochondrial division is a stepwise reaction regulated by DRP1 receptor binding, oligomerization and guanine nucleotide-dependent conformational dynamics.
We and others have reported that the OMM receptors MFF or MID49/51 are independently sufficient to recruit DRP1 to divide mitochondria20,21,23,30. Previous work has suggested that the mitochondrial receptor in yeast, Mdv1p, can coassemble with the DRP1 homolog Dnm1p31. We observed that MID49/51 coassembled with DRP1 to form a copolymer with altered properties, compared with DRP1-only oligomers23. While these results suggest that organelle receptors could nucleate and alter the architecture of a dynamin polymer, the organization and functions of such a coassembly in organelle constriction remain unclear.
Here, we report structural snapshots of DRP1-driven constriction through a MID49/51-dependent reaction. We used cryoEM to observe how nucleotide binding to the G-domain induces conformational changes that allosterically propagate through the BSE to open and elongate DRP1 and expose multiple surfaces required for receptor-binding and polymerization. 3D reconstruction revealed how MID49/51 binding stabilized an arrangement of GTP-bound DRP1 tetramers and nucleated polymerization of a linear co-filament. Then, we reconstituted a path-dependent reaction to observe how GTP hydrolysis and nucleotide exchange lead to conformational constriction by the DRP1 polymer. Specifically, when DRP1 subunits within the MID49/51 cofilament were allowed to exchange and hydrolyze GTP, they released the OMM receptors and the polymers shortened while curling into closed rings. Finally, we designed phosphomimetic, structure-based and disease-causing mutations to validate our structural models and to dissect the allosteric rearrangements that govern curling of linear strings into closed and constricted rings following receptor dissociation.
Results and Discussion
To date, many structural studies of dynamin-family proteins have relied upon mutated or truncated constructs to facilitate crystallization. We purified wild-type, full-length human DRP1 including the N-terminal GTPase domain (G-domain), Bundle Signaling Element (BSE), and four-helix bundle known as the stalk (Fig. 1a). This construct also contained the lipid binding ~100 amino acid region referred to as the variable domain (VD) that resides between the third and fourth α-helices of the stalk, analogous to the Pleckstrin Homology (PH) domain found in endocytic dynamin proteins. A crystal structure of a nucleotide-free and truncated DRP1 mutant revealed the organization of these domains and an overall similarity with the structure of nucleotide-free endocytic dynamin25,32,33. We also purified soluble truncations of MID49 and MID51 engineered to lack their N-terminal transmembrane anchors but include the cytoplasmic nucleotidyltransferase-like domain and the “Dynamin Recruitment Region” (DRR) required for DRP1 binding (Fig 1a)20,34–37.
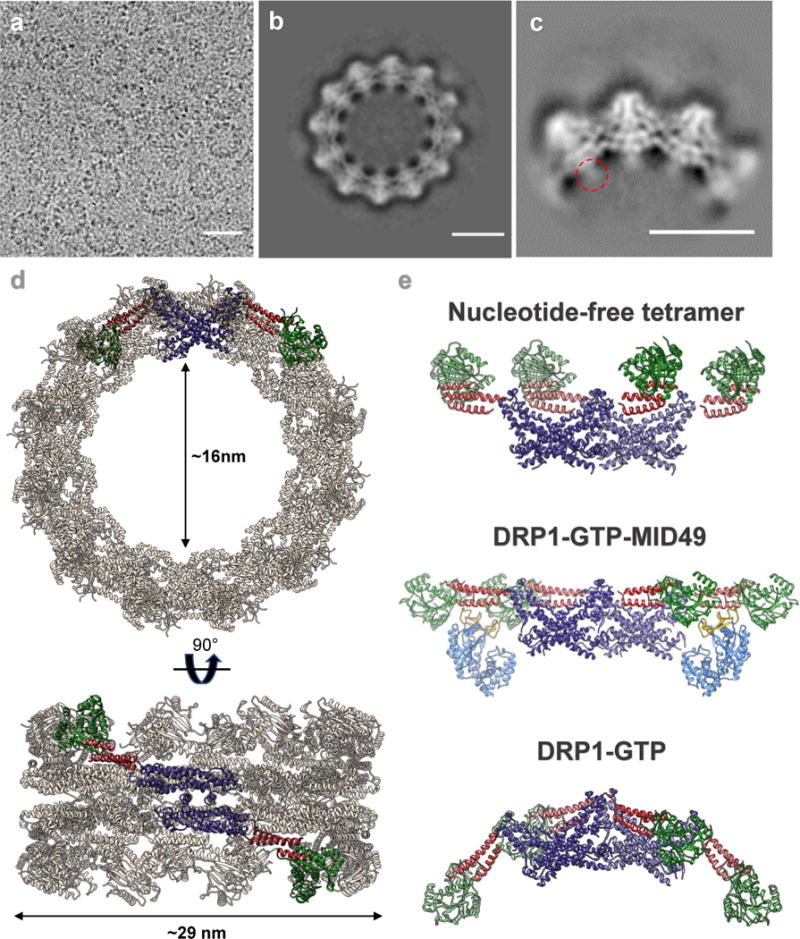
Figure 1. Architecture of the DRP1-MID49 linear filament.
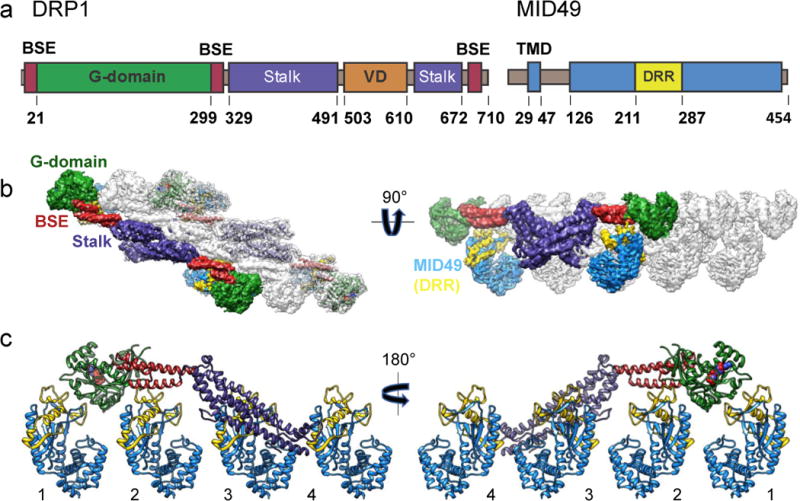
(a) DRP1 and MID49 domain arrangements. (b) Density map and atomic models for DRP1 and MID49126-454. Green: G-domain, Red: Bundle Signaling Element (BSE), Purple: Stalk, Blue: MID49, Yellow: Dynamin Recruitment Region (DRR) of MID49. (c) Each DRP1 chain contacts four different MID49 molecules through receptor interfaces 1-4, as numbered.
Structure of the DRP1-MID49 cofilament
Incubating equimolar ratios of DRP1 with soluble MID49126-454, MID51132-463, or both proteins together, in the presence of Mg2+, GTP or GTP analogs GMPPCP/GTPγS, but not other nucleotides, resulted in cofilament assembly (Extended Data Fig. 1). We focused on the filaments formed with MID49126-454 in the presence of GMPPCP and determined their structure from cryoEM images to an average resolution of 4.2Å (from ~3.5Å to ~8Å, Extended Data Figs. 2–6 and Supplementary Table 1). 3D reconstruction revealed a polymer comprised of three equivalent faces that meet through defined vertices to form a triangular assembly (Extended Data Figs 2–3). A combination of helical reconstruction, segmentation, and single-particle alignment and averaging resolved the elongated DRP1 subunits bound stoichiometrically to MID49126-454, but without assignable density for the majority of the variable domain (Fig. 1b, Extended Data Fig. 2–6). Surprisingly, each chain of DRP1 bound MID49 via four different surfaces, and each MID49 in turn bound four DRP1 molecules to yield a vast interaction network (Figs. 1b-c, Extended Data Figs. 2, 3a-c). MID49 binding to four DRP1 molecules stabilized a linear arrangement of inter-DRP1 interfaces reminiscent of those observed for other dynamin-family proteins (Fig. 1b, Extended Data Figs. 2c, 3d)25,32,33,38–40. We refer to the four distinct surfaces of DRP1 that contribute to MID49/51 binding as receptor interfaces 1 through 4 (Fig. 1c).
Figure 3. Nucleotide-driven allosteric elongation of DRP1 exposes MID49/51 receptor binding sites.
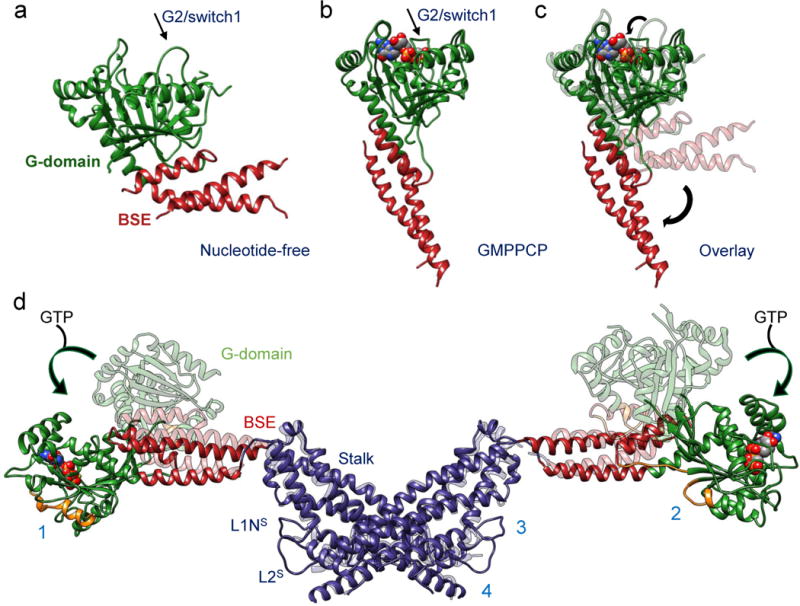
(a) nucleotide-free state of DRP1 G-domain and BSE as seen in a crystal structure (PDB ID: 4BEJ). Arrow points to the G2/switch1 loop. (b) GMPPCP-bound G-domain-BSE conformation determined by cryoEM. (c) Overlay of A and B. Curved arrows highlight the closing of the G2/switch 1 “lid” and the opening of the BSE “wrist”. For comparison, the G2/Switch1 loop from 4BEJ was chosen from the only chain (B) in which it was completely resolved. (d) Global conformational change induced by nucleotide binding. Rotation and translation of the G-domain and BSE elongates the dimer and exposes receptor interfaces 1 and 2 (annotated on separate monomers for clarity). The surfaces of the G-domains that engage the receptors are rendered orange in the nucleotide-bound and elongated conformation.
Structure-based mutants disrupt DRP1-MID49 assembly
MID49’s DRR motif occupied the space between two neighboring G-domains and contacted both via receptor interfaces 1 and 2 (buried surface areas of ~530Å2 and ~200 Å2, respectively, Figs. 1b-c, 2a). The precise spacing required for this bivalent G-domain interaction explains why prior mutagenesis efforts suggested that the size and topology of the β4–α4 loop, rather than its exact sequence (which differs between MID49 and MID51) are critical determinants of binding34,35,37. Nevertheless, the structure indicated that R235 of MID49 within the DRR (analogous with R243 in MID51, Extended data Fig. 7) makes key contacts between two neighboring G-domains (Fig. 2a). Accordingly, the MID49R235E point mutant could not coassemble with DRP1 (Fig. 2d-f). In addition, we mutated conserved DRP1 residues involved in receptor interface 1—the largest interface. Both the D190A mutation, which should neutralize a salt bridge with the receptor, and the D221A mutation, which should alter the conformation of a key loop within receptor interface 1, prevented coassembly with MID49 (Fig. 2a). These substitutions also altered DRP1’s self-assembly properties, suggesting pleiotropic effects on nucleotide handling and receptor binding (Extended Data Fig. 8)41. Specifically, D190 and D221 are also involved in nucleotide-dependent G-domain dimerization (PDB:3W6O), leading to the intriguing possibility that MID49 and MID51 may modulate GTP-dependent conformational dynamics.
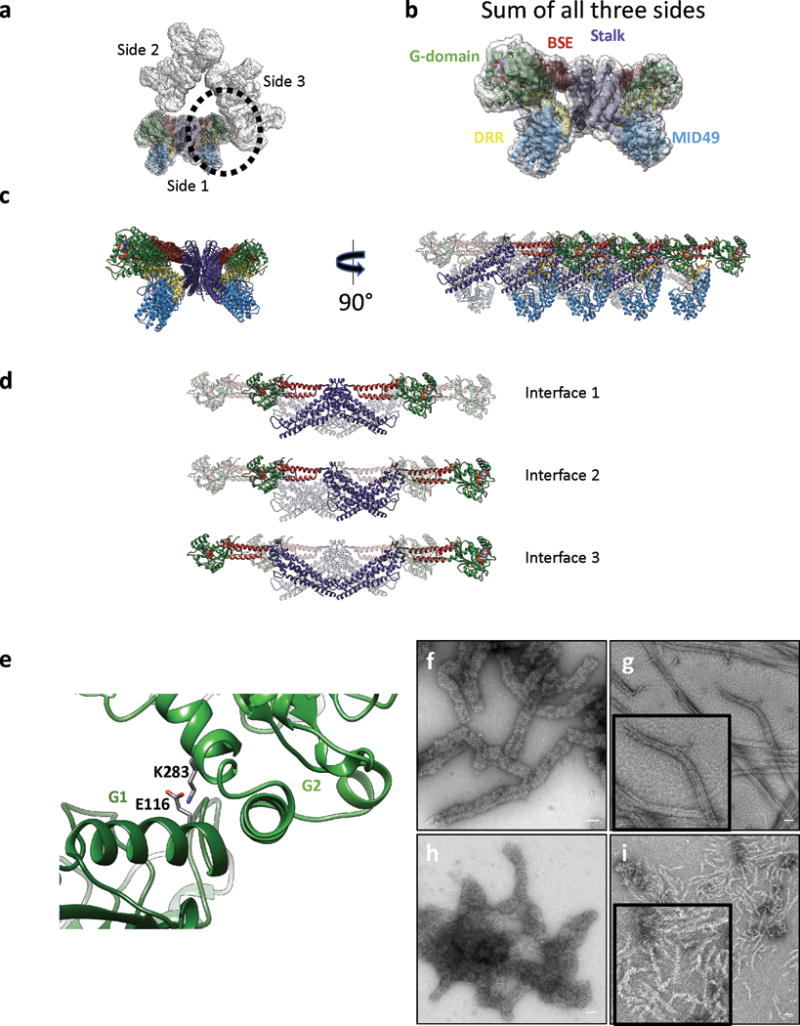
Figure 2. Key DRP1-MID49/51 receptor interfaces and regulatory phosphorylation site.
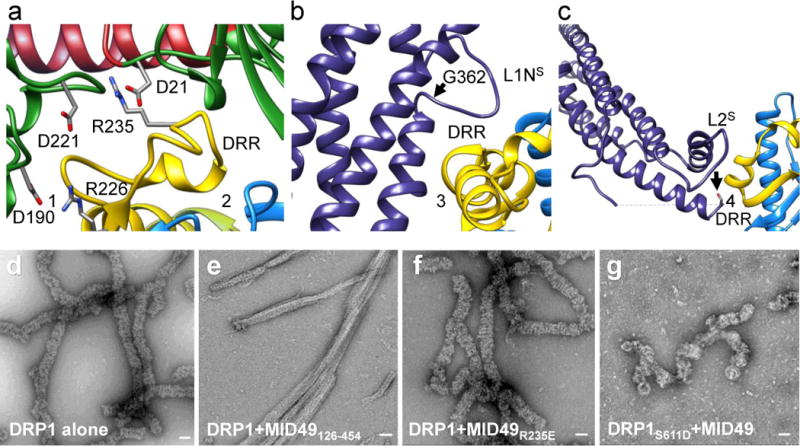
(a) Receptor interfaces 1 and 2. Green ribbons on either side of the DRR come from two separate G-domains. The residues involved in key interactions for the interfaces are shown as sticks. (b) Rotated view of receptor interface 3. Disease-associated DRP1 residue G362 (arrow) supports the conformation of the L1Ns loop essential for linear copolymerization with MID49. (c) Receptor interface 4 with MID49. Arrow points to the DRP1 phosphorylation site Ser611, at the variable domain-stalk junction. A dashed line indicates the unresolved amino acids for the variable domain. (d) DRP1-only assemblies. (e) DRP1 assemblies with wild type MID49126-454. (f) DRP1 assemblies with the MID126-454 mutant R235E that resemble those of DRP1 alone as in (d). (g) DRP1 assemblies of the DRP1 mutant S611D with MID49126-454 that resemble assemblies of DRP1 in isolation. Bars = 50 nm.
Unexpectedly, MID49 also made contact with the stalk loops of a third and fourth DRP1 molecule through receptor interfaces 3 and 4 (buried surface areas of ~450Å2 and ~230Å2, respectively, Fig. 1c, 2b-c). The DRP1 loops involved in these receptor interfaces, the L1NS and L2S loops, are key determinants of assembly for other dynamin-family oligomers25,32,33,38,40. Receptor interface 3, in particular, involves the conserved loop L1NS and is the site of multiple disease alleles that correlate with elongated mitochondrial morphologies42–44, including G362D and G363D (Fig. 2b, Extended Data Figs. 6b, 6e, 7a). Prior work established that this loop comprises part of the intra-molecular PH domain binding site for the soluble state of endocytic dynamin tetramers (Extended Data Fig. 6f-h)40, and is a determinant of conformational heterogeneity for these and other dynamin-family proteins38,40. The presence of disease alleles near this interface suggests that these mutations may compromise receptor binding and that defects in the recruitment of DRP1 to mitochondria may contribute to pathogenesis. Accordingly, we found that the disease-associated G362D mutant of DRP1 (Fig. 2b) failed to coassemble with MID49 and displayed altered assembly and conformational properties (Extended Data Figs. 6e, 8a, 8i-l).
Receptor interface 4 includes Ser611 (equivalent to Ser637 of isoform 1, Extended data Fig. 7c), an intensively studied phosphorylation site for protein kinase A (PKA) or Ca2+/calmodulin-dependent protein kinase Iα (CaMKIα)45–47. Phosphorylation at this site, as well as phosphomimetic alleles of DRP1, have been reported to inhibit mitochondrial fission, although the mechanism of inhibition has been unclear45,48. Our structure suggests that phosphorylation should inhibit MID49 binding, and accordingly we found that the phosphomimetic mutant, S611D, failed to coassemble with MID49126-454 under the same solution conditions that bound the wild type protein (Fig. 2g-i).
Nucleotide-induced conformational changes enable receptor binding
Understanding the allosteric coupling between nucleotide binding, hydrolysis and exchange and the conformational repertoire of dynamin-family GTPases remains an unmet challenge. We observed that the GMPPCP-bound G-domains and the BSE of DRP1 adopt strikingly different conformations in the cryoEM density compared to the nucleotide-free crystal structure25. In addition to other nucleotide-induced conformational changes within the G-domain, the most salient are the closing of the G2/switch-1 loop to form a closed “lid” over the nucleotide (Figs. 3a-c). Analogous to the conformational change reported for Dynamin-149, the closure of the switch-1 lid propagates through the adjacent beta sheet to push the α-helices of the BSE into an orthogonal position (Supplementary Video 1). When evaluated in the context of an inter-DRP1 stalk interface, the interface-2 “X-shaped” DRP1 dimer, this conformational change is a 90° rotation of the G-domain and a 40Å translation toward the stalk (Fig. 3d, Supplementary Video 2). Two of the four DRP1 surfaces that engage the DRR of MID49/51 (receptor interfaces 1 and 2, Fig. 2a) are inaccessible in the nucleotide-free state but become available for binding upon nucleotide-driven elongation (Fig. 3d, Supplementary Video 2).
GTP-hydrolysis induces filaments to curl into rings
We next evaluated the dynamics of the DRP1+MID49 filaments in the presence of hydrolyzable GTP, rather than the non-hydrolyzable analog used for 3D reconstruction. Following copolymerization in the presence of the non-hydrolyzable analog, we exchanged GMPPCP for GTP through dialysis and followed the reaction using negative stain TEM at sequential time points until the GTP was exhausted. We observed that the linear, three-sided DRP1-MID49 cofilaments disassembled into shorter, single-sided filaments before disassembling entirely upon complete hydrolysis to GDP (Fig. 4). The thinner single-sided filaments seen at intermediate time points resembled the single-sided filaments we observed after mutagenizing a salt bridge that appears to hold the triangular structure together (Extended data fig. 3e-i). The single-sided filaments, moreover, exhibited intriguing dynamics at intermediate time points (Fig. 4b-c). Specifically, upon reaching a reproducibly narrow range of lengths, the nearly linear single-sided filaments spontaneously curled into closed rings of remarkably uniform diameter (Fig. 4c).
Figure 4. Dynamic instability of the DRP1-MID49 linear assembly and curling into closed DRP1 rings.
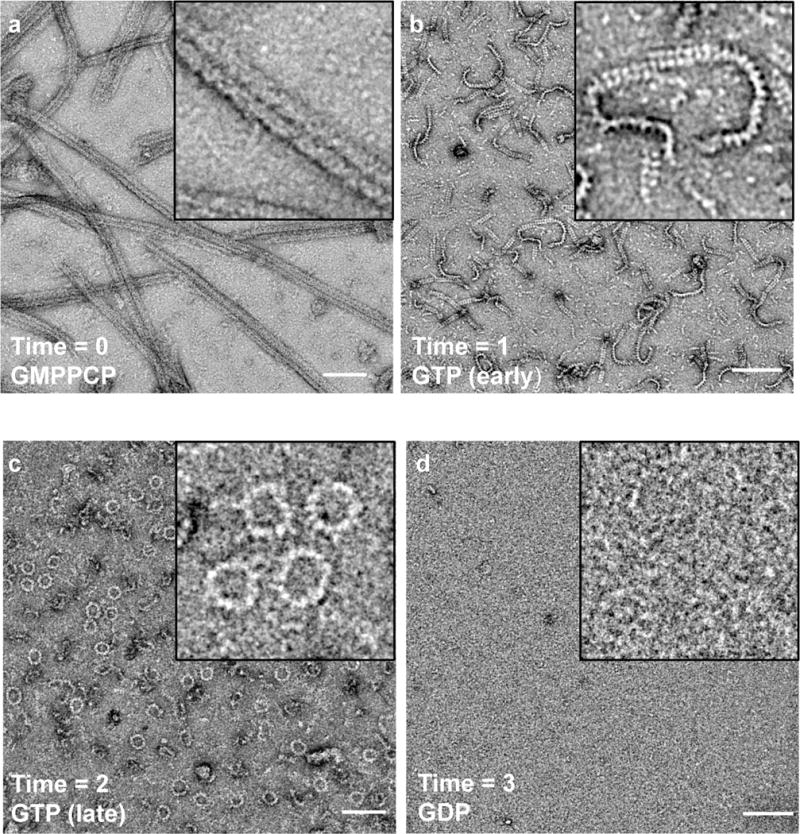
(a) Three-sided DRP1-MID49126-454 linear filaments copolymerized with GMPPCP, as in Extended Data Figures 2–3. (b) Subsequent exchange into GTP leads to disassembly of the three-sided filaments, partial disassembly of the single-sided filaments, and (c) curling of single-sided filaments into closed rings. (d) GTP exhaustion leads to complete oligomer disassembly. Bars = 100nm.
A model for closed DRP1 rings
In a separate but related experiment, we also evaluated the assembly properties of the DRP1 mutant G362D—which disrupts receptor interface 3—with and without MID49126-454. As described above, this disease-associated residue sits at the base of the L1NS loop and this loop is a key inter-stalk interaction between adjacent DRP1 molecules in the linear filament (Figs. 1c, 2b, Extended Data Fig. 6b, e). We found that DRP1G362D purified as a nearly monodisperse and stable dimer, rather than a mix of tetramers and higher order species observed for the wild-type, full-length protein (Extended Data Fig. 8a). In addition, DRP1G362D exclusively formed rings, not filaments, with or without MID49126-454 and in the presence of GTP or GMPPCP (Fig. 5a, Extended Data Figs. 8i-l). These rings resembled those observed with wild-type DRP1 in all respects except that the wild-type protein only formed closed rings from the linear MID49 copolymer through the path-dependent reaction described above (Fig. 4c versus Fig. 5a). We also observed that these apparently closed DRP1G362D rings could constrict liposomes into membrane tubules and circumscribe lipid nanotubes (Extended Data Fig. 9).
Figure 5. Drp1G362D cannot bind MID49 and forms rings exclusively with GMPPCP or GTP.
(a) CryoEM micrograph of Drp1G362D rings. (b) 2D class average of the predominant closed ring that comprises 12 DRP1 dimers. (c) 2D class average of a quarter of the ring revealing the secondary structure elements of the “X”-shaped DRP1 dimer. A red dashed circle indicates density that may be attributable to the variable domain. (d) 3D model of the closed ring. (e) Comparison between DRP1 tetramers observed in the nucleotide-free state (top, PDB ID: 4BEJ), the GMPPCP and MID49126-454-bound linear state (middle), and the bent conformation modeled for the rings. Bar for (a) = 30nm, for (b, c) =100Å.
Switching to the non-hydrolyzable GTP analog GMPPCP with the DRP1G362D rings improved structural homogeneity, presumably because the rings remain dynamic and eventually disassembled upon complete hydrolysis to GDP (Fig. 4). We imaged the GMPPCP-bound Drp1G362D rings using cryoEM and used 2D class averages of the predominant 12-dimer closed ring to model the differences between the linear filaments and the closed rings (Fig. 5, Extended Data Fig. 10). To account for the projected ring density, the G-domain and the BSE of DRP1 must move even further down toward the stalks. Stalk interface-2 remains constant, as revealed by the “X-shaped” dimer seen in projection (Fig. 5d, e). The curvature of the ring, however, dictates that stalk interfaces 1 and 3, and the conformations of the L1NS and L2S loops, must be extensively remodeled to allow a ~30 degree bending per dimer in comparison with the linear DRP1-MID49 copolymer (Fig. 5d-e, Extended Data Fig. 10e-f, Supplementary Videos 2–3). We did not observe any density for MID49 in the wild-type rings that form by curling of the MID49-DRP1 cofilament in the presence of GTP, nor in our higher-resolution analysis of the Drp1G362D rings that form with or without MID49 present (Fig. 5b-c, Extended Data Fig. 8i-l, 10). This suggests that MID49 binding is incompatible with the curled state of the ring-shaped oligomer, and that constriction therefore requires receptor dissociation (Supplementary Video 3).
Discussion
We note that with an inner diameter of ~16nm, the closed ring may be sufficient to sever a double-membrane mitochondrion if both the outer and inner membranes are compressed together. Alternatively, if inner membrane fission is separable and precedes outer membrane fission, a 16nm diameter suggests that a single membrane tubule would be stabilized by these rings. The structures we observe in vitro may therefore correspond with a highly-constricted but pre-fission state observed in vitro50 and in living cells when another dynamin-family protein, dynamin-2, is depleted29. Constriction by DRP1, therefore, may stabilize the high degree of membrane curvature that is suitable for the recruitment and final fission event catalyzed by additional dynamin-family enzymes.
Together, these findings establish four advances. First, our cryoEM structure and mutagenesis studies revealed how receptor proteins like MID49/MID51 recruit and stabilize a specific nucleotide-bound conformation of DRP1 and initiate polymerization of a cofilament. We speculate that the nearly linear properties of this polymer have adapted to encircle low-curvature mitochondrial tubules. Second, analysis of the MID49-DRP1 copolymer revealed how guanine nucleotide binding induces a conformational rearrangement to expose an avid network of receptor binding sites. We now understand these nucleotide-driven allosteric transformations in the context of both full-length and oligomeric DRP1. Third, a path-dependent constriction reaction revealed GTP-dependent conformational dynamics. In this reaction, nucleotide exchange and hydrolysis led to MID49/51 receptor dissociation, disassembly from the ends of the linear filament, and concomitant curling of the shortening filaments into closed rings. The requirement for MID49/51 receptor dissociation before constriction may explain how overexpression of the MID receptors inhibits mitochondrial fission21. Fourth and finally, analysis of a disease mutant in the L1NS loop, Drp1G362D, highlights this loop as a cardinal determinant of receptor binding as well as the inter-stalk interactions that govern oligomer geometry. Together, these observations teach us how DRP1 performs mechanical work by curling from linear filaments into closed rings around mitochondria.
Methods
Construct design
Wild type (WT) DRP1 isoform 2 sequence was purchased from DNASU (sequence ID HsCD00043627, UNIPROT identifier: O00429-3, also known as Dlp1a) and was cloned into pET16b plasmid (Novagen) between the Nde1 and BamH1 sites. The vector was kindly provided by the laboratory of Wesley I. Sundquist with a 10X-His tag followed by a PreScission protease site (Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro). Wild-type MID49126-454 sequence was PCR amplified and cloned into pGEX6p1 vector having an N-terminal GST tag followed by a PreScission protease site. Site directed mutagenesis was performed on pET16b-DRP1 and pGEX6p1-MID49126-454 using the Gibson cloning method to introduce mutations51. All constructs were verified using Sanger sequencing.
Protein purification
Protein purification was performed as described52. Briefly, plasmids containing the WT DRP1 or MID49126-454 sequence were transformed in the BL21-DE3 (RIPL) strain of E. coli. The colonies were inoculated in LB culture medium and grown overnight. Secondary inoculations were done the next morning in ZY medium for auto-induction53,54. The cultures were grown to an OD600 of 0.8 at 37°C in baffled flasks and were shifted to 19°C to grow for another 12 hours. The cultures were spun down and the bacterial pellets were used for protein purification immediately or stored at −80°C.
Full length DRP1 WT and mutant variants were purified as described previously for DRP1 WT with modifications25. Briefly, the bacterial pellets were resuspended in buffer A (50 mM HEPES/NaOH (pH 7.5), 400 mM NaCl, 5 mM MgCl2, 40 mM imidazole, 1 mM DTT, 0.5 mg DNase (Roche) and protease inhibitors (10 mM pepstatin, 50 mM PMSF, 0.5 mM aprotinin and 2 mM leupeptin), followed by cell disruption with a probe sonicator. Lysates were cleared by centrifugation at 40,000×g in Beckman JA 25.50 rotors for 60 min at 4°C. The supernatant was filtered using a 0.45 μm filter and applied to Ni-NTA Agarose beads pre-equilibrated with buffer B (50 mM HEPES/NaOH (pH 7.5), 400 mM NaCl, 5 mM MgCl2, 40 mM imidazole, 1 mM DTT). Upon the application of the supernatant, the beads were washed with 20 column volumes each of buffer B and buffer C (50 mM HEPES/NaOH (pH 7.5), 800 mM NaCl, 5 mM MgCl2, 40 mM imidazole, 1 mM DTT, 1 mM ATP, 10 mM KCl) followed by buffer D (50 mM HEPES/NaOH (pH 7.5), 400 mM NaCl, 5 mM MgCl2, 80 mM imidazole, 1 mM DTT, 0.5% (w/v) CHAPS). A final pre-elution wash was done with 20 column volumes of buffer B. Bound DRP1 was eluted with buffer E (50 mM HEPES/NaOH (pH 7.5), 400 mM NaCl, 5 mM MgCl2, 300 mM imidazole, 1 mM DTT) and dialyzed overnight at 4°C against buffer B without imidazole in the presence of PreScission protease to cleave the N-terminal 10X-His tag. The protein was re-applied to a Ni-NTA column pre-equilibrated with dialysis buffer and was observed to bind the column without the 10X-His tag as well. Subsequently, the protein was eluted with buffer B containing 80 mM imidazole.
Pure protein was concentrated with a 30kDa molecular weight cutoff (MWCO) centrifugal concentration device (Millipore). In a final step, DRP1 was purified by size-exclusion chromatography (SEC) on a Superdex-200 column (GE) in buffer F containing 20 mM HEPES/NaOH (pH 7.5), 300 mM NaCl, 2.5 mM MgCl2 and 1 mM DTT. Fractions containing DRP1 were pooled, concentrated, flash-frozen as single use aliquots in liquid nitrogen and stored at −80°C. Exact masses for purified DRP1 proteins were validated by MALDI-TOF mass spectrometry.
MID49126-454 was purified as described with the following modifications37. pGEX6p1-MID49126-454 plasmid DNA (human, UNIPROT identifier: Isoform 1 Q96C03-1, also known as MIEF2) was transformed in BL21 (DE3) RIPL cells. The colonies were grown overnight in LB medium and secondary cultures were grown in ZY medium. Cells were grown to an OD600 of 0.8-1, collected by centrifugation and processed immediately or stored at −80°C as described above. The bacterial pellets were lysed as described above in MID-buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT and 0.1% (v/v) Triton X-100). The lysates were pre-cleared at 40,000×g and filtered using a 0.45 μm filter before applying to 3 ml glutathione sepharose beads (GE). After overnight binding to beads, the unbound protein was removed and the beads were washed using 20 column volumes each of MID-buffer A and MID-buffer B (50 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT). The protein was eluted with MID-buffer C (50 mM Tris pH 8.0, 500 mM NaCl, 5% glycerol, 1 mM DTT and 20 mM reduced glutathione). The eluate was cleaved overnight with PreScission protease while dialyzing against MID-buffer D (20 mM Tris pH 8.0, 100 mM NaCl, 5% glycerol, 1 mM DTT). Cleaved protein was further purified using ion-exchange chromatography using a Q sepharose (GE) column. The low salt buffer for ion-exchange was the same as MID-buffer D and the high salt buffer was MID-buffer E (20 mM Tris pH 8.0, 1 M NaCl, 5% glycerol, 1 mM DTT). The relevant MID49126-454 fractions were pooled, concentrated and further purified using an SEC column pre-equilibrated with MID-buffer F (20 mM Tris pH 8.0, 200 mM NaCl, 5% glycerol, 1 mM DTT). The fractions containing MID49126-454 were pooled, concentrated, flash-frozen in liquid nitrogen and stored as single use aliquots at −80°C.
Filament assembly, EM sample preparation, data acquisition and processing
To assemble DRP1-MID49126-454 filaments, the proteins were mixed to a final concentration of 2 μM each and kept for an hour at room temperature. The mixture was dialyzed against assembly buffer- 20 mM HEPES pH 7.5, 50 mM KCl, 3 mM MgCl2, 1 mM DTT and 200 μM GMPPCP with or without 0.2% octyl-glucopyranoside (Anatrace). The filaments were observed using negative stain TEM or cryoEM after vitrification. Under these conditions, the mutant DRP1S611D failed to coassemble with MID49, but upon further lowering the ionic strength to 25mM KCl, DRP1S611D displayed detectable but greatly reduced coassembly compared to WT protein. For vitrification, the sample was applied to Quantifoil holey carbon grids (R2/2) using a Vitrobot Mark III with 3.5 μl sample, 5 seconds blotting time and a 0 mm offset at 19°C and 100% humidity. Images were collected on an FEI T30 Polara operating at 300kV at a magnification of 31000X. Images were recorded on a Gatan K2 summit camera in super resolution mode for a final binned pixel size of 1.22 Å/pixel. The movies were dose fractionated, contained 30-40 frames, had a total exposure time of 6-8 seconds with 0.2 seconds per frame and a per-frame dose of 1.1 to 1.4 electrons/Å2. SerialEM was used to automate data collection55. The defocus range was 0.8-3 μm under focus. The data was motion corrected and dose-weighted using UCSF Motioncor256. CTF parameter estimation on the non-dose-weighted but motion-corrected stacks was done using CTFFIND4 and GCTF57,58.
Filaments were boxed using the program e2helixboxer.py from the EMAN2 suite59. Particle coordinates were used to extract discrete particles using RELION 1.3-1.460 and all further processing was done within the RELION suite. Multiple rounds of 2D classification identified the well-ordered segments. 3D autorefine was run using a customized Relion1.2 version with the IHRSR algorithm implemented61,62. The consensus helical structure was used to classify the particles without refining helical symmetry (using RELION 1.4), resulting in 2 major classes that differed slightly in rise and twist (Extended data figure 2c). Particles from each class were selected and independently refined again with helical RELION 1.2 and IHRSR. Analysis of these reconstructions revealed that each structure was comprised of three linear filaments that bundle together to form a structure that resembled a triangle in cross-section (Extended data figures 2–3). The vertices of the triangle are formed through asymmetric interactions between the G-domains in adjacent filaments. The triangular arrangement of the bundled helices is unlikely to correspond to a biologically meaningful architecture, and this structure cannot form if the MID49 receptor is embedded in the outer mitochondrial membrane.
To further improve signal-to-noise, each of the three filaments in each independent half-map was segmented, extracted, resampled on a common grid and summed using UCSF Chimera63–67. The respective symmetrized but unfiltered half maps from each class were again aligned to a common grid and summed according to the C2 symmetry axis of the DRP1 dimer. In a last step, relion_postprocess was used to add the resulting and fully symmetrized half maps (Extended data figure 2c). These half maps and the final summed map, with differential B-factor sharpening per region (Extended data figures 2c, 4–5), were used for atomic modeling using Rosetta as described below.
For the projection structure of the DRP1G362D rings, 2 μM protein was mixed 1:1 molar ratio with MID49126-454 and was allowed to sit at room temperature for an hour. The mixture was dialyzed against the assembly buffer (without detergent) overnight. The sample was collected after 12 hours and vitrified using ultra-thin 3 nm carbon support films (Ted Pella). For vitrification, a Mark III vitrobot was used with 3.5 μl sample, 0 mm offset, 100% humidity and 3.5 seconds blot time. The images were collected using an FEI TF20 microscope and SerialEM for automated data collection. The data were recorded with a Gatan K2 camera operating in super resolution mode to collect dose fractionated movie stacks with a final binned pixel size of 1.234 Å/pixel. 40 frames were collected per stack (0.2 seconds per frame and 1.42 electrons/Å2). The movie stacks were motion corrected and the parameters of the transfer function were estimated as described above. Approximately 2000 particles were picked manually for initial 2D classification in RELION 1.4 and these averages were used as templates for further particle picking by Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/). Final 2D averages of the entire rings versus quarter segments of the rings were computed using Relion1.4.
Liposome and Nanotube reactions
Liposomes were made as described before52. Briefly, 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) was purchased from Avanti Polar Lipids. DOPS dissolved in chloroform was dried under a steady stream of nitrogen and dried under vacuum for an hour. The dried lipid film was resuspended in n-hexane and dried again under nitrogen. The resulting lipid film was dried under vacuum for 4 hours and was finally resuspended in 20 mM HEPES pH 7.5 and 150 mM KCl. The same protocol was followed for making nanotubes, where the mixture contained 60% D-galactosyl-ß-1,1′ N-nervonoyl-D-erythro-sphingosine (Galactosyl Ceramide), 30% DOPS and 10% Ni2+-NTA DOGS (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] Nickel Salt).
For assembly reactions of DRP1G362D over lipid, 0.5-2μM protein was incubated with liposomes or nanotubes for an hour and dialyzed against the assembly buffer without detergent.
Model building
The general procedure for atomic model interpretation and validation using Rosetta were performed as described68. To obtain an initial model for DRP1, the crystal structure of nucleotide-free DRP1 (PDB ID: 4BEJ)25 was used for the stalk region and DRP1 G domain-BSE structures bound to GMPPCP (PDB ID: 3W6O) were used for the G-domain and BSE regions. Density-guided model completion for DRP1 was carried out with RosettaCM69 using this hybridization of DRP1 crystal structures. A converged solution appeared from the low-energy ensemble of the complete models generated by RosettaCM. However, among the low-energy ensemble, residues 503-610 were found to be extremely flexible without cryoEM density constraints and therefore were omitted for further coordinate refinement. For MID49, the highly homologous mouse MID49 crystal structure (81.3% identity, PDB ID: 4WOY, Extended Data Fig. 5c-d)37 was used to generate a homology model using RosettaCM and used as the starting model.
To enable fragment-based, density-guided model refinement with missing residues (503-610, DRP1), Rosetta iterative local rebuilding tool was customized to disallow backbone rebuilding at breaks within a single chain. Multiple rounds of refinement were done for each component against one half map (training map), and the other half map (validation map) was used to monitor overfitting according to the detailed procedure described in Wang et al.68.
With the refined model of DRP1 and MID49, we further refined the model in the context of a full assembly that included 8 identical copies of each protein, Mg2+ and nucleotide which included all possible inter-domain molecular interactions in the filament (Extended data figure 5a-b). Pseudo-symmetry was used70 to enable and facilitate the energy evaluations of all neighboring interactions around the asymmetric unit (Green model, shown in Extended data figure 5a) for final model refinement of the full assembly. To this end, refinement was done against the training map. Finally, the half maps were used to determine a weight for the density map that did not introduce overfitting. Using the weight and with the symmetry imposed, the whole assembly of DRP1 and MID49 was refined in the full map, followed by B-factors refinement71. Finally, quantification of buried surface area and the number and nature of the bonds involved for each DRP1-MID49 interaction interface modeled by Rosetta were performed with the PISA server (http://www.ebi.ac.uk/pdbe/pisa/).
Visual evaluations of the model-to-map correspondence was carried out in UCSF Chimera using unfiltered and unsharpened maps, maps uniformly sharpened with a range of ad hoc B-factors, and maps processed with a model-based local sharpening and local low-pass filtering procedure to optimize contrast and the visibility of high-resolution features of the map72.
To build a molecular model for the closed 12-dimer DRP1 rings, we used the diameter, thickness, and angles revealed by the 2D cryoEM class averages of the DRP1G362D rings stabilized with GMPPCP. The atomic coordinates determined above using RosettaCM were used to build the ring in sections, first with repeating dimers of the interface-2 “X-shaped” stalk, then the BSE and finally the G-domains and the angles between these sections were iteratively adjusted until calculated projections of the molecular model corresponded with the features of the experimental projection densities. Both the top (Fig 5b-c) and the side view (Extended data Figure 10b) were used as constraints. The complete atomic model of ring was finally refined in Phenix73 to minimize clashes.
Extended Data
Extended Data Figure 1. DRP1 and MID49 assembly states.
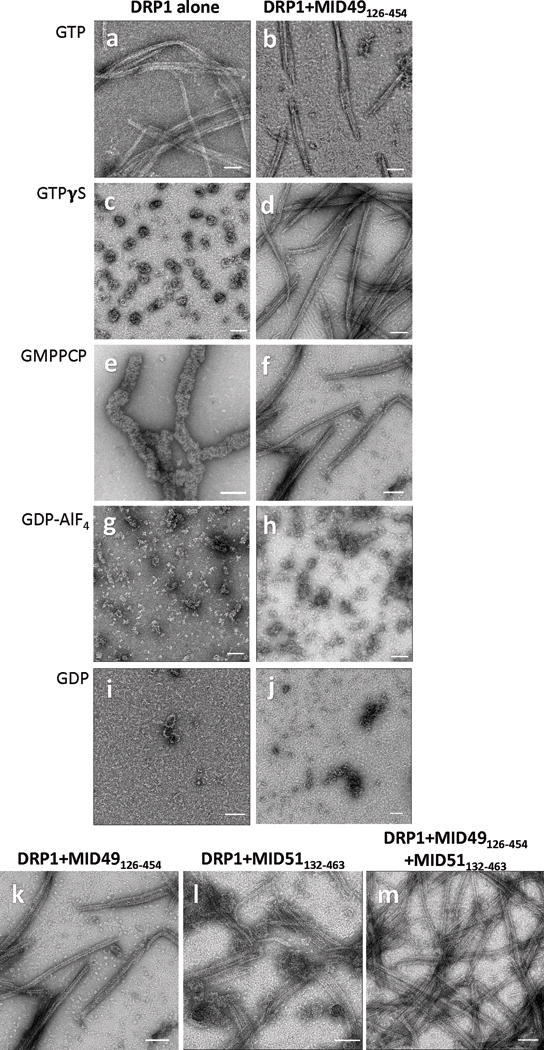
(a-j) DRP1 assembly states visualized with negative stain electron microscopy in the presence of different guanine nucleotides and MID49126-454. Both proteins incubated at 2μM concentration. Bars = 100nm.
(k-m) MID49126-454 and MID51132-463 form indistinguishable assemblies with DRP1. (k) DRP1 plus MID49126-454 and GMPPCP, (l) DRP1 plus MID51132-463 and GMPPCP, (m) DRP1 plus both equimolar MID49 and MID51. Bars = 100nm.
Extended Data Figure 2. CryoEM and 3D Reconstruction.
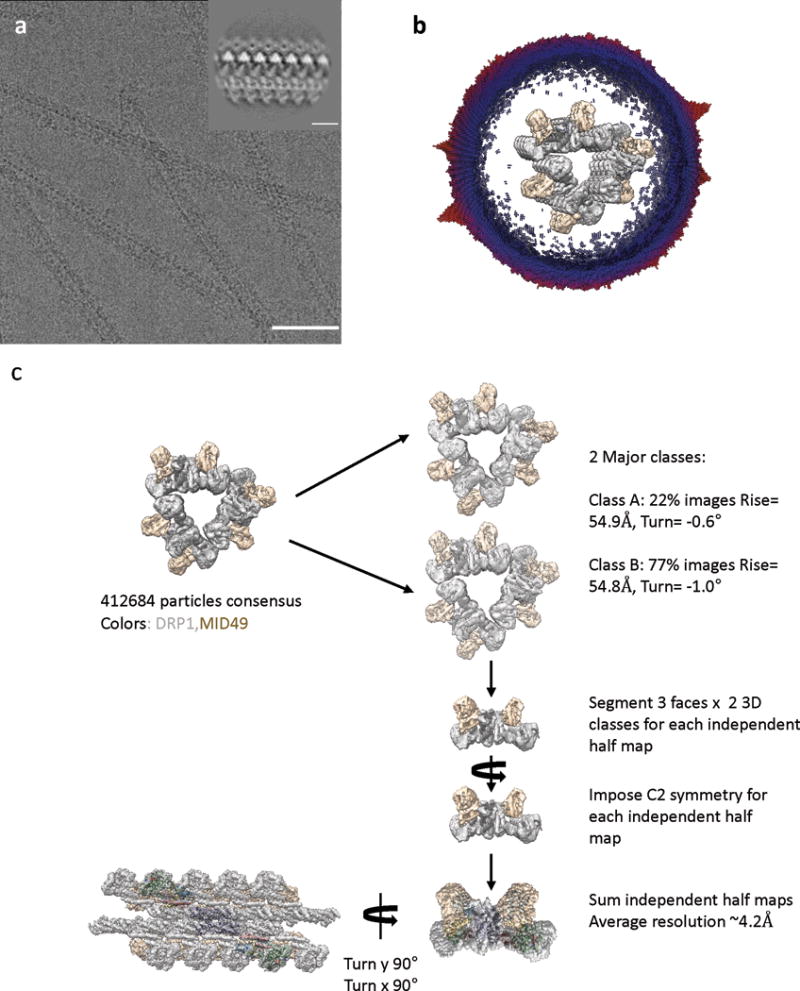
(a) An electron cryo-micrograph of DRP1-MID49126-454 filaments formed with GMPPCP. Bar =100nm. Inset shows a representative 2D class average. Inset bar =10nm.
(b) Cross-section of the 3D reconstruction of the filament and the distribution of views determined during helical reconstruction. The length of the cylinders and the color code correspond with the number of particles for that viewing direction (from few to many, blue to red). The 3D structure has been segmented and colored with DRP1 in grey and MID49 in golden yellow.
(c) Particle numbers and workflow for the reconstruction protocol. DRP1 density is shown in grey and MID49 is in golden yellow.
Extended Data Figure 3. Intra- and Inter-filament Interactions.
(a) The triangular structure seen in cross section. ‘Side 1’ of the triangular structure has the atomic model placed in the density. The G-domain-to-G-domain contact between adjacent sides is circled.
(b) The sum of the 3 sides with the model fit in density.
(c) Ribbon diagram of the same atomic model as in (b). The rotated view shows 8 chains each of DRP1 and MID49. The chains further from the reader are rendered transparent.
(d) An isolated tetramer of DRP1 from the filament, rendered to highlight the stalk interfaces 1, 2 and 3 observed for DRP1 and other dynamin-family of GTPases.
(e) Expanded view of the circular region in (a) illustrating a salt bridge between adjacent G-domains.
(f) DRP1-only wild type polymers with GMPPCP versus (g) DRP1 coassembly with wild type MID49126-454 and GMPPCP and high-magnification inset.
(h) DRP1E116R mutant polymers versus (i) DRP1E116R mutant coassembly with MiD49126-454. Shorter, single-sided filaments predominate. Disordered “triangular assemblies” were also observed, but were much shorter and infrequent compared with the wild type proteins. Bars = 50nm.
Extended Data Figure 4. Resolution Estimates.
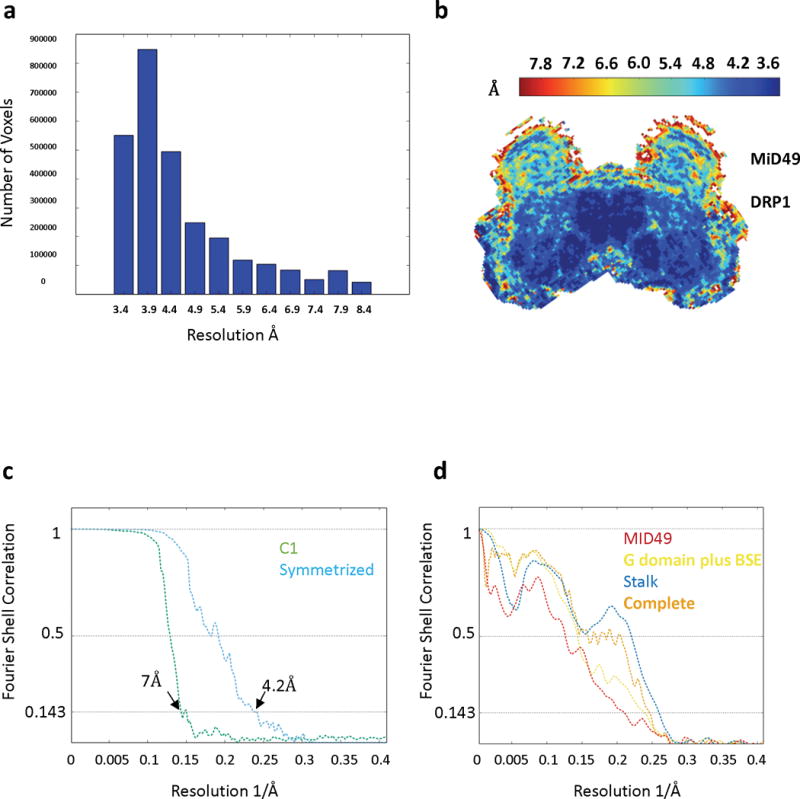
(a-b) Local resolution estimates computed by Resmap74. (a) Histogram of voxel values, (b) Results in A depicted as a heat map of a cross-section through the reconstruction.
(c-d) Fourier Shell Correlation plots for (c) the half-maps with and without symmetry and (d) model-to-map correlations for each sub-region of the structure.
Extended Data Figure 5. Rosetta-based Model Refinement.
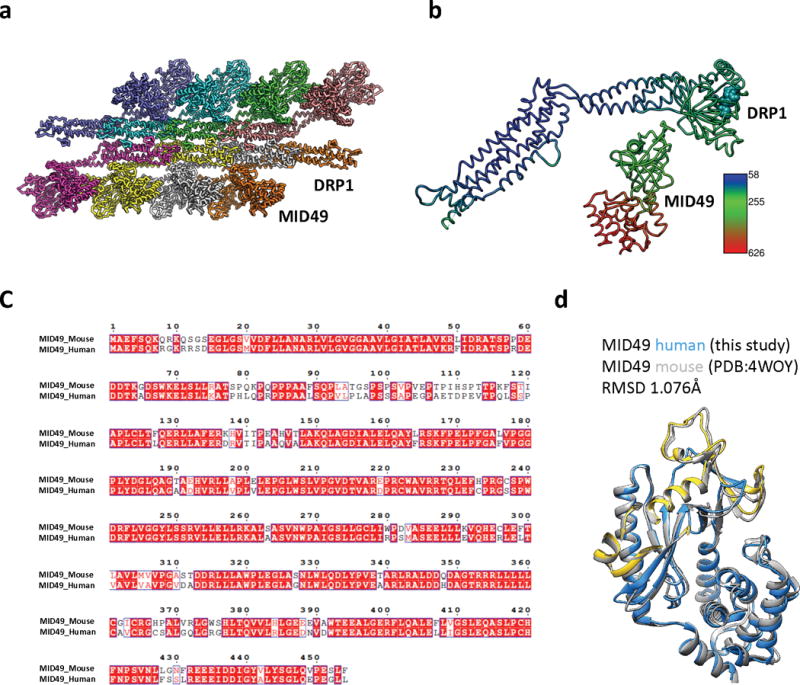
(a): Complete assembly used for Rosetta-based model building with the asymmetric unit shown in green. (b) Atomic B-factors for one asymmetric unit, DRP1 versus MID49 models (ribbon) and bound GMPPCP (space filling).
(c) Sequence alignment between human and mouse MID49 sequences. (d) Overlay of the homology model of human MID49126-454 (blue, with yellow DRR, ribbon) modeled within the cryoEM density overlaid with the mouse MID49 crystal structure (PDB ID: 4WOY, grey ribbon)37 which was used as a constraint. No density attributable to ADP within the nucleotidyltransferase domain was observed.
Extended Data figure 6. Map-to-Model Fits and Role of the L1NS and L2S loops.
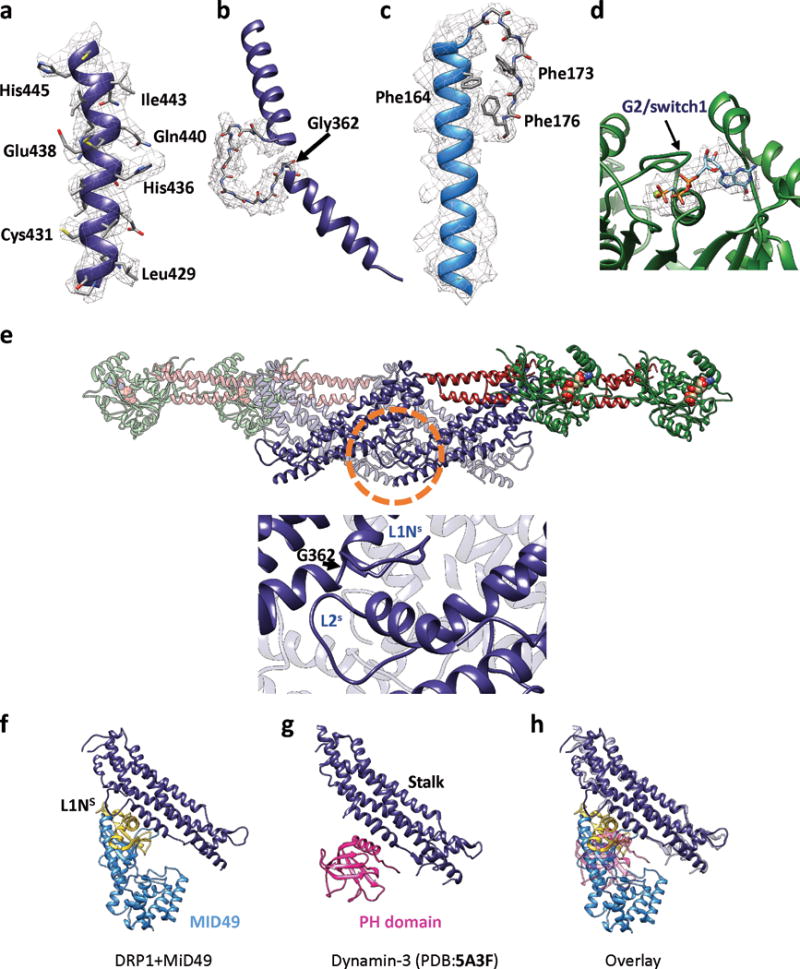
Examples of models fit within B-factor sharpened cryoEM density for (a) a helix from the DRP1 stalk, (b) the backbone of the L1NS loop from the stalk, (c) an elongated helix and turn found in MID49, and (d) GMPPCP and Mg within the G-domain. (e) Roles of L1NS and L2S in linear filament formation. Upper panel: An isolated Drp1 tetramer from the cryoEM model. The circled region is expanded in the lower panel. Lower: interactions that the conserved loop L1Ns makes within the assembly. G362 is highlighted. (f) DRP1 stalk and MID49 at receptor interface-3, (g) Dyn3 stalk plus PH domain from PDB 5A3F40, (h) Overlay of (f) and (g).
Extended Data Figure 7. Sequence Conservation and Key Interaction Sites.
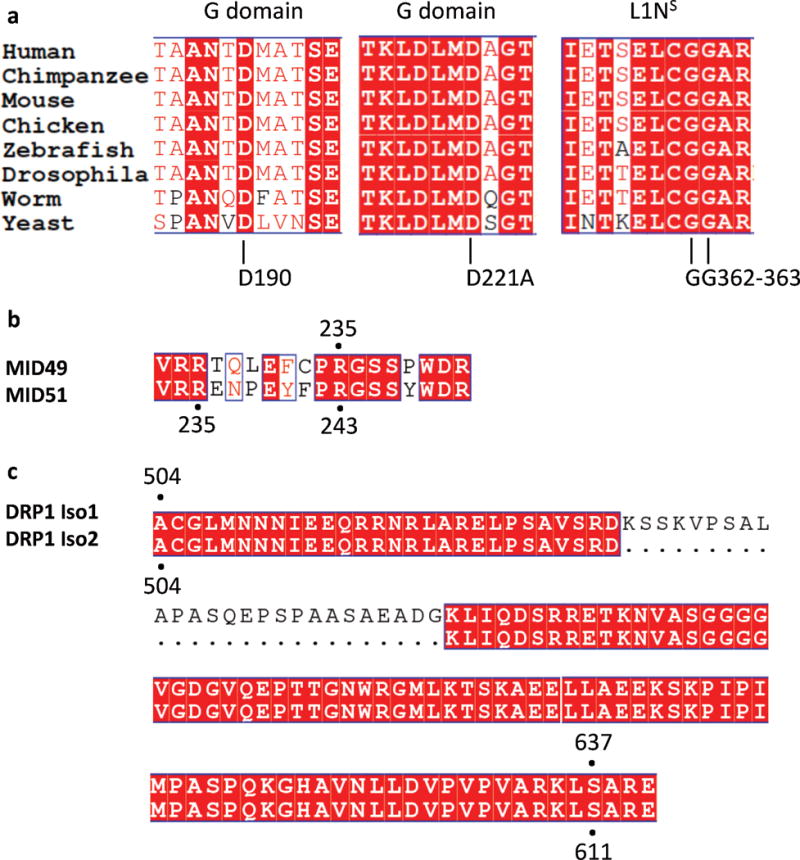
(a) Multiple sequence alignment of the regions near and including the DRP1 residues mutated in this study: D190, D221 and G362, G363. The residue numbers apply to human DRP1, isoform 2 (UNIPROT identifier: O00429-3 which is also known as DLP1a).
(b) Sequence alignment of MID49 and MID51 at the region around residue R235 of MID49. R235 of MID49 corresponds to R243 of MID51.
(c) Sequence alignment of DRP1 Isoform 1 vs Isoform 2 showing the correspondence of S637 (Isoform 1) and S611 (Isoform 2).
Extended Data Figure 8. Biochemical and Structural Characterization of Mutants.
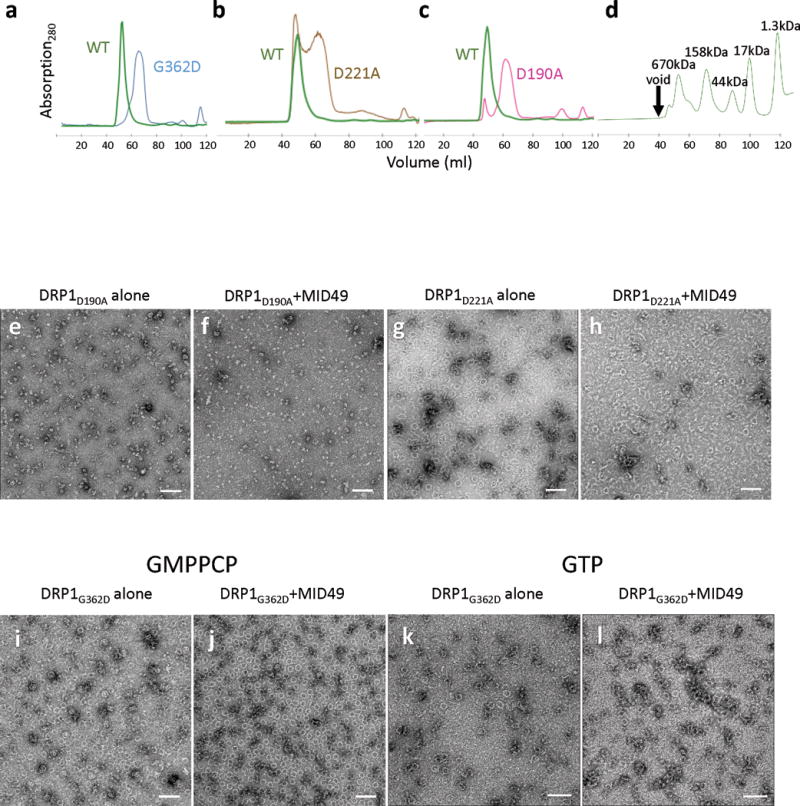
(a-d) Size exclusion chromatography traces for DRP1 wild-type and mutants used in the study. (a) Comparison between wild type (WT) versus G362D, (b) WT versus D221A, and (c) WT versus D190A. (d) Gel filtration standards with annotated molecular weights from the same column and FPLC system.
(e-h) DRP1 assembly and coassembly reactions with GMPPCP for (e) DRP1D190A alone, (f) DRP1D190A+MID49126-454, (g) DRP1D221A alone and (h) DRP1D221A+MID49126-454. Bars = 100nm.
(i-l) DRP1G362D assembly and coassembly reactions with GMPPCP or GTP. DRP1G362D forms rings but not linear filaments: i,k) without MID49, j,l) with MID49, with i, j) GMPPCP or k, l) GTP. Bars = 100 nm.
Extended Data Figure 9. DRP1G362D Rings on Model Membranes.
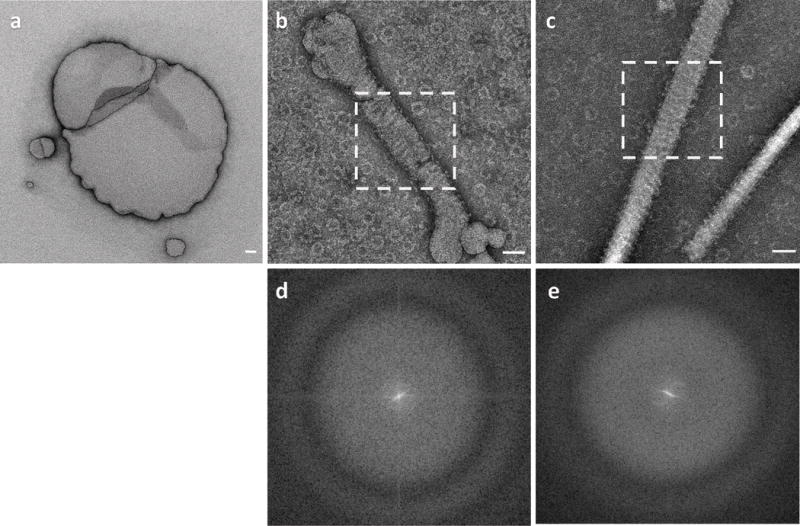
(a) DOPS liposomes used in the study.
(b) DOPS liposomes after incubation with DRP1G362D showing ring-like assemblies on the membrane and in the background.
(c) Lipid nanotubes incubated with DRP1G362D.
(d) Power spectrum of the area shown within the dashed square in (b).
(e) Power spectrum of the area shown within the dashed square in (c). In both (d) and (e), layer lines indicative of helical geometry are not detectable.
Bars = 50nm.
Extended Data Figure 10. DRP1G362D forms 12-dimer closed rings.
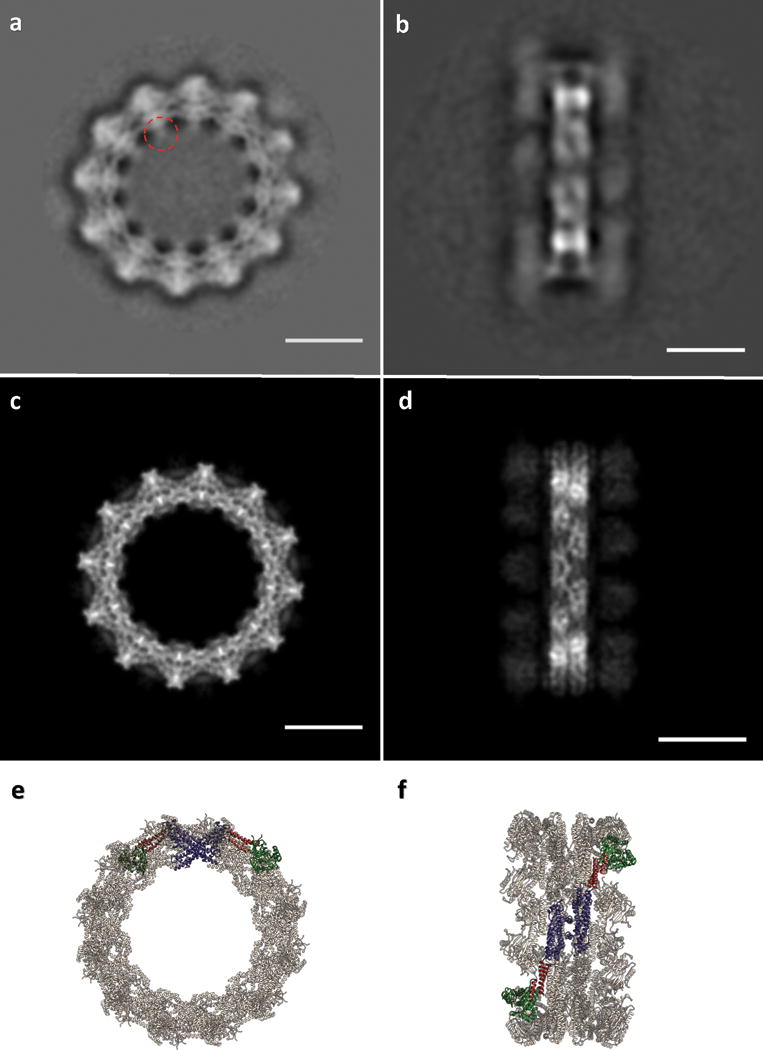
(a) 2D class average of the rings. The red dashed circle indicates density that may be attributable to the variable domain. (b) 2D class average of infrequent, orthogonal or “side” views used as a constraint during model building; (c) “top” and (d) “side” projections of the model; (e) “top” and (f) “side” views of the final model rendered as ribbons. Bars =100Å. Green: G-domain, Red: BSE, Purple: Stalk.
Supplementary Material
Acknowledgments
We thank Michael Braunfeld, Cameron Kennedy, David Bulkley, and Alexander Myasnikov and the UCSF Center for Advanced CryoEM, which is supported in part from NIH grants S10OD020054 and 1S10OD021741 and the Howard Hughes Medical Institute. We also thank the QB3 shared cluster and NIH grant 1S10OD021596-01, Jean-Paul Armache, Nathaniel Talledge for microscopy advice, Charles Greenberg for consulting on structural modeling, Dennis Winge for insightful conversations and Mingyu Gu for facilitating mass spectrometry of proteins. This work was further supported by a Faculty Scholar grant from the Howard Hughes Medical Institute (A.F.), the Searle Scholars Program (A.F.), NIH grant 1DP2GM110772-01 (A.F.), NIH grants GM53466 and GM84970 (J.S.), the Sandler Family Foundation through the UCSF Program for Breakthrough Biomedical Research and the American Asthma Foundation, and the Howard Hughes Medical Institute (R.W., J.S., D.A.). R.W. is a HHMI Fellow of the Life Sciences Research Foundation. A.F. is a Chan Zuckerberg Biohub investigator.
Footnotes
Author Contributions
R.K., J.S. and A.F. conceived of the study. R.K., A.Y. and P.T. performed all experiments. R.K., R.W. and A.F. performed the computational analyses. D.A.A. advised R.W. and R.K. on model building. All authors evaluated the results and edited the manuscript. R.K. and A.F. wrote the manuscript with input from all of the authors. The authors declare no competing interests.
Data Accessibility:
All of the 3D cryoEM density maps associated with this study have be deposited in the EMDB with accession numbers EMD-8874. The atomic coordinates have been deposited in the PDB as 5WP9. Raw data, models and image processing scripts are also available from Adam Frost (adam.frost@ucsf.edu) upon request.
Statistics and Reproducibility
All electron microscopy experiments in this this study were repeated at least three times.
References
- 1.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nature reviews Molecular cell biology. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nature cell biology. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toyama EQ, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science (New York, NY) 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy M, Reddy PH, Iijima M, Sesaki H. Mitochondrial division and fusion in metabolism. Current opinion in cell biology. 2015;33:111–118. doi: 10.1016/j.ceb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 8.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 10.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nature cell biology. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. Journal of Cell Biology. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatel-Chaix L, et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SJ, et al. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science (New York, NY) 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis SC, Uchiyama LF, Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science (New York, NY) 2016;353:aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osman C, Noriega TR, Okreglak V, Fung JC, Walter P. Integrity of the yeast mitochondrial genome, but not its distribution and inheritance, relies on mitochondrial fission and fusion. Proc Natl Acad Sci U S A. 2015;112:E947–956. doi: 10.1073/pnas.1501737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 19.Kraus F, Ryan MT. The constriction and scission machineries involved in mitochondrial fission. Journal of cell science. 2017;130:2953–2960. doi: 10.1242/jcs.199562. [DOI] [PubMed] [Google Scholar]
- 20.Osellame LD, et al. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. Journal of cell science. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer CS, et al. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem. 2013;288:27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer CS, et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO reports. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koirala S, et al. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci U S A. 2013;110:E1342–1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Molecular biology of the cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frohlich C, et al. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. Embo J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. The Journal of cell biology. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daumke O, Praefcke GJ. Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers. 2016;105:580–593. doi: 10.1002/bip.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Molecular biology of the cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science (New York, NY) 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faelber K, et al. Crystal structure of nucleotide-free dynamin. Nature. 2011;477:556–560. doi: 10.1038/nature10369. [DOI] [PubMed] [Google Scholar]
- 33.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter V, et al. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. The Journal of cell biology. 2014;204:477–486. doi: 10.1083/jcb.201311014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loson OC, et al. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 2014;22:367–377. doi: 10.1016/j.str.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Chan DC. Drp1 recruitment by Mff, MiD51 and MiD49. Molecular biology of the cell. 2015;26 doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loson OC, et al. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 2015;24:386–394. doi: 10.1002/pro.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 39.Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J Biol Chem. 2010;285:28419–28424. doi: 10.1074/jbc.R110.145839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reubold TF, et al. Crystal structure of the dynamin tetramer. Nature. 2015;525:404–408. doi: 10.1038/nature14880. [DOI] [PubMed] [Google Scholar]
- 41.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanstone JR, et al. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genet. 2016;24:1084–1088. doi: 10.1038/ejhg.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheffer R, et al. Postnatal microcephaly and pain insensitivity due to a de novo heterozygous DNM1L mutation causing impaired mitochondrial fission and function. Am J Med Genet A. 2016;170:1603–1607. doi: 10.1002/ajmg.a.37624. [DOI] [PubMed] [Google Scholar]
- 44.Chang CR, et al. A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem. 2010;285:32494–32503. doi: 10.1074/jbc.M110.142430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann N Y Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 47.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO reports. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chappie JS, et al. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 2011;147:209–222. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ugarte-Uribe B, Prevost C, Das KK, Bassereau P, Garcia-Saez AJ. Drp1 polymerization stabilizes curved tubular membranes similar to those of constricted mitochondria. Journal of cell science. 2017 doi: 10.1242/jcs.208603. [DOI] [PubMed] [Google Scholar]
- 51.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 52.Kalia R, Talledge N, Frost A. Methods in Cell Biology. Vol. 128. Elsevier Ltd; 2015. Structural and functional studies of membrane remodeling machines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blommel PG, Becker KJ, Duvnjak P, Fox BG. Enhanced Bacterial Protein Expression During Auto- induction Obtained by Alteration of Lac Repressor Dosage and Medium Composition. Biotechnol Prog. 2007;23:585–598. doi: 10.1021/bp070011x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Zheng SQ, Palovcak E, Armache JP, Cheng Y, Agard DA. Anisotropic Correction of Beam-induced Motion for Improved Single- particle Electron Cryo-microscopy. Nat Methods. 2017;14:332–333. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bell JM, Chen M, Baldwin PR, Ludtke SJ. High resolution single particle refinement in EMAN2.1. Methods. 2016;100:25–34. doi: 10.1016/j.ymeth.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres SHW. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egelman EH. Methods in enzymology. Vol. 482. Elsevier Inc.; 2010. Reconstruction of helical filaments and tubes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ge P, et al. Cryo-EM Model of the Bullet-Shaped Vesicular Stomatitis Virus. Science. 2010;327:689–694. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen EF, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Wang RYR, et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife. 2016;5:1–22. doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song Y, Dimaio F, Wang RY, Kim D, Miles C. High resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiMaio F, Leaver-Fay A, Bradley P, Baker D, Andre I. Modeling symmetric Macromolecular structures in Rosetta3. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakobi AJ, Wilmanns M, Sachse C. Model-based local density sharpening of cryo-EM maps. Elife. 2017;6:1–26. doi: 10.7554/eLife.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


