Abstract
Background
Copy number alterations form prognostic molecular subtypes of glioblastoma with clear differences in median overall survival. In this study, we leverage molecular data from several glioblastoma cohorts to define the distribution of copy number subtypes across random cohorts as well as cohorts with selection biases for patients with inherently better outcome.
Methods
Copy number subtype frequency was established for 4 glioblastoma patient cohorts. Two randomly selected cohorts include The Cancer Genome Atlas (TCGA) and the German Glioma Network (GGN). Two more selective cohorts include the phase II trial ARTE in elderly patients with newly diagnosed glioblastoma and a multi-institutional cohort focused on paired resected initial/recurrent glioblastoma. The paired initial/recurrent cohort also had exome data available, which allowed for evaluation of multidimensional scaling analysis.
Results
Smaller selective glioblastoma cohorts are enriched for copy number subtypes that are associated with better survival, reflecting the selection of patients who do well enough to enter a clinical trial or who are deemed well enough to undergo resection at recurrence. Adding exome data to copy number data provides additional data reflective of outcome.
Conclusions
The overall outcome for diffuse glioma patients is predicted by DNA structure at initial tumor resection. Molecular signature shifts across glioblastoma populations reflect the inherent bias of patient selection toward longer survival in clinical trials. Therefore it may be important to include molecular profiling, including copy number, when enrolling patients for clinical trials in order to balance arms and extrapolate relevance to the general glioblastoma population.
Keywords: biomarkers, clinical trials, glioblastoma, molecular profiling
Importance of the study
Glioblastoma is the most common primary malignant neoplasm of the central nervous system. Patient outcome is generally poor, but can be predicted by copy number alterations. DNA copy number profiling across cohorts reflects the inherent bias of clinical trial selection toward patients with better outcome. Understanding the distribution of molecular signatures across cohorts suggests a role for molecular profiling to be incorporated up front in clinical trials and studies focused upon tumor recurrence.
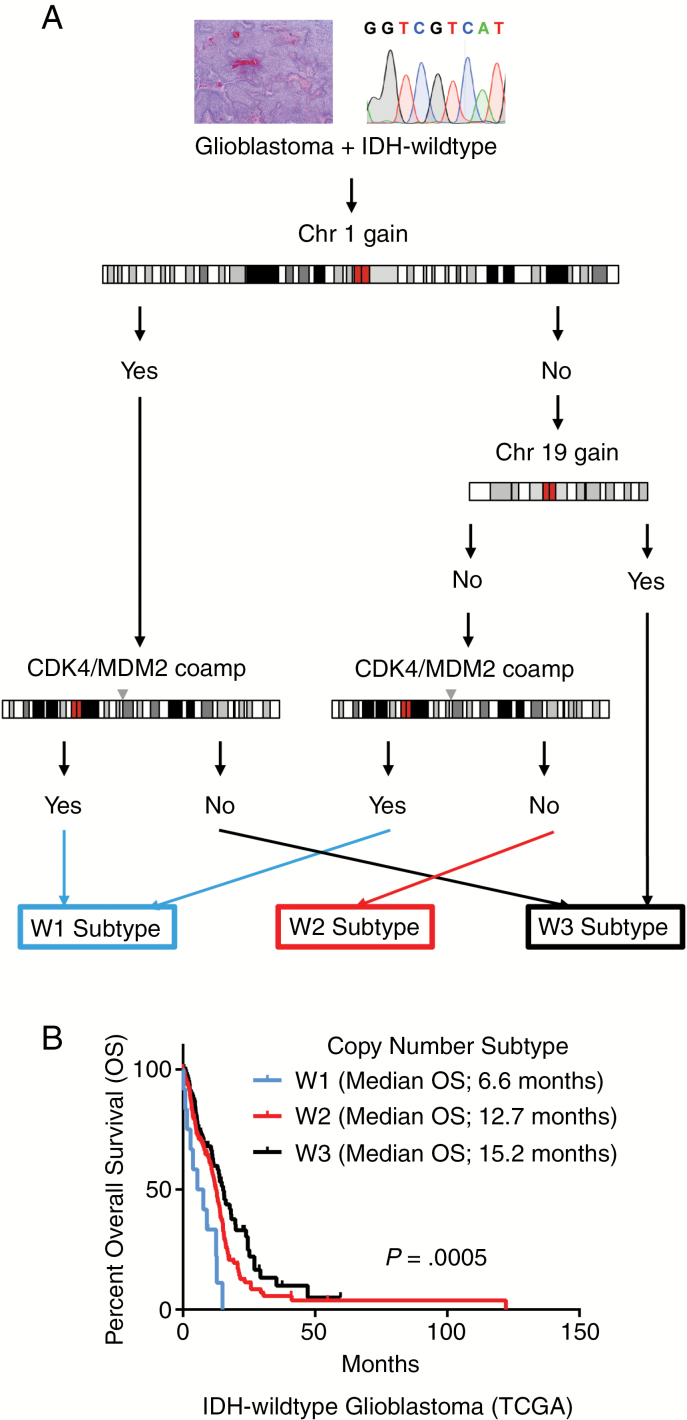
In 2016, the World Health Organization (WHO) introduced a classification of CNS tumors that incorporate molecular signatures with traditional histopathology to arrive at “integrated” diagnostic diffuse glioma entities.1 The molecular components of this classification scheme largely involve knowing the mutational status of isocitrate dehydrogenase (IDH) and codeletion of whole chromosome arms 1p and 19q. Integrating these limited molecular alterations into the traditional classification system of diffuse glioma yielded a better predictor of clinical outcome than histopathology alone. However, there is still an issue with the current classification system in that WHO grading of diffuse gliomas is still determined solely by histopathological features (including mitotic activity, necrosis, and microvascular proliferation) without taking molecular features into account. Recent attempts have been made to define molecular alterations (beyond IDH and 1p/19q) which may provide prognostic information beyond histological grading. Our group2 and others3,4 have shown that diffuse glioma survival is associated with whole chromosome and gene level copy number alterations. Copy number alterations predict patient survival in diffuse gliomas, and subtypes with clear differences in median survival can be derived from copy number profiling.2 We have previously shown that data from The Cancer Genome Atlas (TCGA) and the German Glioma Network (GGN) on IDH-wildtype glioblastoma, WHO grade IV, can be used to further stratify 3 distinct prognostic DNA copy number subtypes: W1 (worst survival), W2 (intermediate survival), and W3 (best survival) (Fig. 1).2 These copy number subtypes are determined by relatively few foci: gain of whole chromosome 1, gain of whole chromosome 19, and coamplification of cyclin-dependent kinase 4/murine double minute 2 (CDK4/MDM2) (Fig. 1). These prognostic copy number subtypes do not overlap with other described molecular transcriptional or methylation subtypes.5–7 Given the implications for prognosis, we sought to determine copy number subtype distributions across glioblastoma populations while using the GGN and TCGA datasets as reflective of the general population. We identified 2 additional prospective cohorts that had an inherent bias toward including patients with better functional status and survival, which is not reflective of the general population. The first cohort is from the randomized phase II ARTE trial, which explored the efficacy of bevacizumab as an adjunct to hypofractionated radiotherapy in patients 65 years or older with newly diagnosed O6-methylguanine-DNA methyltransferase unmethylated glioblastoma.8 The second cohort is that of paired initial and recurrent glioblastoma, where inclusion required that the patient survived long enough, and was deemed appropriate, to have a second surgery.9 The paired initial/recurrent glioblastoma cohort had exome data available in addition to copy number analysis, which allows for further insights into risk stratification by molecular profiling.
Fig. 1.
Copy number subtypes for IDH-wildtype glioblastoma. (A) Algorithm for copy number subtype derivation. Copy number subtypes defined by 4 genetic loci: gain of whole chromosome 1 (gChr1), gain of whole chromosome 19 (gChr19), and coamplification of CDK4/MDM2 (caCDK4/MDM2): W1 = [No gChr1 + No gChr19 + caCDK4/MDM2] or [gChr1 + caCDK4/MDM2]; W2 = [No gChr1 + No gChr19 + No caCDK4/MDM2]; W3 = [No gChr1 + gChr19] or [gChr1 + No caCDK4/MDM2]. (B) Overall survival of glioblastoma, copy number subtypes in TCGA dataset. TCGA copy number subtype patient numbers: W1 (n = 12), W2 (n = 157), and W3 (n = 88). P-value determined using Cox proportional hazards regression.
Materials and Methods
Copy Number Data from Glioblastoma Cohorts
Four separate cohorts of IDH-wildtype glioblastoma were analyzed for gain of whole chromosome 1, gain of whole chromosome 19, and CDK4/MDM2 coamplification. Copy number data via GISTIC2.0 scores10 for TCGA glioblastomas (n = 256) were downloaded from the University of California Santa Cruz cancer browser (https://genome-cancer.ucsc.edu/). Clinical data for the glioblastoma dataset of TCGA were obtained from the Genomic Data Commons Data Portal from the National Institutes of Health (NIH).11 Glioblastoma copy number data from the GGN (n = 243) (www.gliomnetzwerk.de) and the ARTE trial8 (n = 59) were derived from 450k methylation array data as previously described using the R package ‘conumee’ (http://bioconductor.org/packages/conumee) applying an adapted algorithm for baseline correction.2,6 Whole chromosomal gains and CDK4/MDM2 coamplifications were determined by log2-scale thresholds of 0.1 and 0.6, respectively. Copy number data from a multi-institutional (The MD Anderson Cancer Center, University of California San Francisco, Istituto Neurologico Carlo Besta, Kyoto University, and Samsung Medical Center) paired initial/recurrent glioblastoma cohort9 (n = 62) were determined from whole exome sequencing using the EXCAVATOR bioinformatics pipeline.12 For all cohorts, molecular data were ascertained in accordance with the World Medical Association Declaration of Helsinki.
Combined Copy Number and Single Nucleotide Data Visualization
Classic multidimensional scaling (MDS) of TCGA glioma data (single nucleotide point mutation and copy number) produced 2-dimensional scatterplots and was performed using R software (version 3.5.0, R Project for Statistical Computing, http://www.r-project.org/) as previously described.2,13 Copy number data (evaluated by GISTIC2.010 thresholds) and single nucleotide variant data (determined by SAVI214) for the paired initial/recurrent glioblastoma cohort9 were mapped onto the MDS plot using TCGA data as a reference set.
Differential Methylation and Gene Expression Analysis
TCGA gene expression and methylation data for glioblastomas were downloaded from the University of California Santa Cruz cancer browser (https://genome-cancer.ucsc.edu/) and analyzed using R software (version 3.5.0, R Project for Statistical Computing, http://www.r-project.org/). Raw counts for chosen RNA-seq samples were obtained from Recount215 and were analyzed for differential expression using R/Bioconductor package DESeq2.16 Significant genes were determined using a cutoff of fold change >2 and adjusted P-value <0.05. Gene methylation was analyzed by applying the Bioconductor packages ‘IlluminaHumanMethylation450kanno.ilmn12.hg19’ (https://bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylation450kanno.ilmn12.hg19.html) and DMRcate (https://bioconductor.org/packages/release/bioc/html/DMRcate.html). Differentially expressed and methylated genes were plotted with principal component analysis. Pathway analysis of differentially methylated genes was performed using TargetMine (http://targetmine.mizuguchilab.org/).
Statistics
Statistical analyses were performed using GraphPad Prism software (version 7.02, https://www.graphpad.com/scientific-software/prism). Kaplan–Meier analysis for overall survival was performed with P-values determined by Cox proportional hazards regression. Other data comparisons were performed using the chi-square test, Mann–Whitney U test, and Fisher’s exact test as indicated.
Results
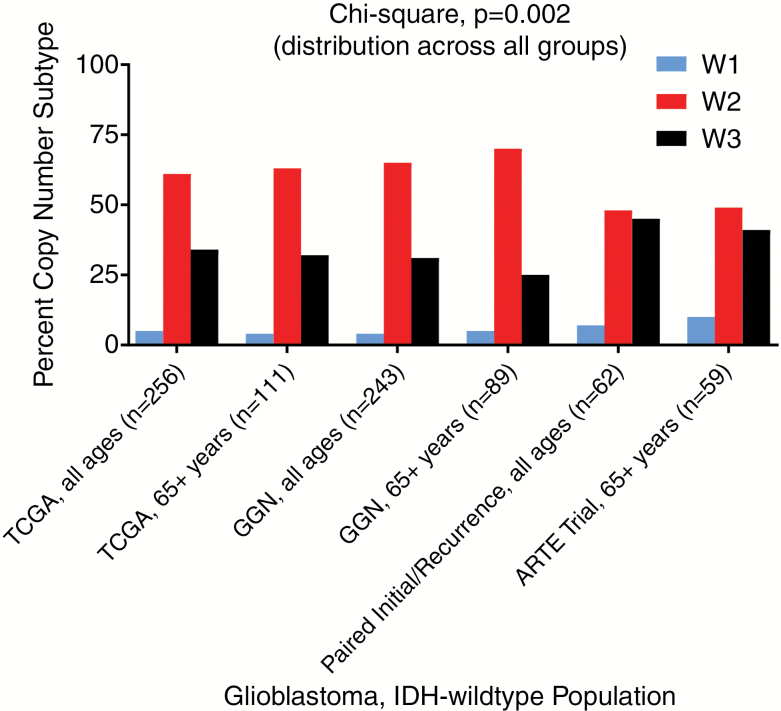
The distribution of copy number subtypes is similar across 2 large datasets of initial glioblastoma across North America (TCGA) and Europe (GGN), suggesting that this is the natural distribution in the population (Fig. 2). Selection bias exists toward a longer-lived glioblastoma subgroup when enrolling patients healthy enough to enter clinical trials or when deciding that patients are well enough for a resection at recurrence. Given the better overall survival of these types of preselected patients, we wondered whether there was evidence of a skew in the population with respect to distribution of the copy number subtypes defined above. In fact, there were 2 such cohorts for analysis. The first cohort is the phase II ARTE trial of hypofractionated radiotherapy with or without bevacizumab in elderly patients with newly diagnosed glioblastoma.8 When compared with the elderly TCGA and GGN general glioblastoma populations, there was a distribution skew toward the better prognostic W3 copy number subtype (Fig. 2). The second prospective cohort is a multi-institutional (The MD Anderson Cancer Center, University of California San Francisco, Istituto Neurologico Carlo Besta, Kyoto University, and Samsung Medical Center) paired initial/recurrent glioma group,9 which investigated glioblastoma patients of all ages who were healthy enough to have a resection at first recurrence. This paired initial/recurrent glioma cohort also showed a skew toward the better prognostic W3 copy number subtype (Fig. 2).
Fig. 2.
Distribution shift of copy number subtypes across glioblastoma cohorts. Distribution plots show increasing percentage of better-performing copy number subtypes in paired initial/recurrent glioblastoma and elderly clinical trial cohorts compared with TCGA and GGN datasets.
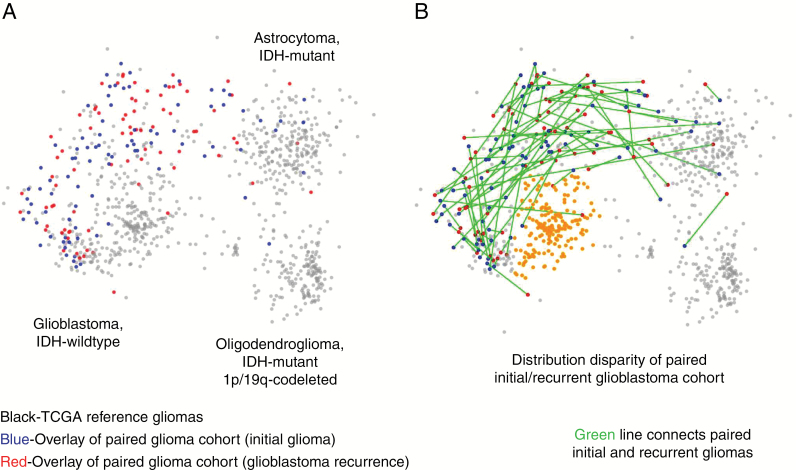
A more granular method to characterize DNA alterations is through MDS analysis by combining copy number with whole exome sequencing.2,13 MDS defines distinct glioma groups with differences in survival as well. Of the 2 selective prospective cohorts, the paired initial/recurrent glioblastoma cohort has DNA exome sequencing available in addition to copy number status and allows for MDS analysis of these patients. To investigate a potential distribution skew with respect to MDS molecular signatures, we overlaid the paired initial/recurrent glioblastoma data cohort9 onto the reference MDS map of TCGA (Fig. 3). As shown in Fig. 3B, 51% of glioblastomas from TCGA exhibit a similar DNA structure that does not correspond to that of those patients deemed appropriate for surgical resection at recurrence. This MDS group of IDH-wildtype glioblastoma largely includes, but is not limited to, glioblastomas that have the poorest survival among MDS regions and is characterized by relatively few regions of chromosomal alterations, with the exception of whole chromosome 7 gain, whole chromosome 10 loss, and loss of chromosome 9p.2 Essentially all patients who do well enough to have a second surgical resection at recurrence arise from the other half of glioblastomas, which tend to have one or more of the following alterations: whole chromosome 1 gain, whole chromosome 19 gain, and/or mutations in TP53.
Fig. 3.
Exome sequencing added to copy number highlights molecular signature of additional poor prognostic group. (A) Multidimensional scaling (MDS) analysis based on whole exome sequencing and copy number alterations shows that mapping of the paired initial/recurrent glioblastoma cohort has uneven spatial distribution when overlaid on the reference dataset of TCGA (TCGA cohort = gray; paired initial glioma = blue; paired recurrent glioma = red). (B) Green edges show the connections of individual paired initial and recurrent gliomas. Orange-colored region highlights tightly clustered area of which 51% of TCGA glioblastomas exist, but only one initial glioma from the paired initial/recurrent glioblastoma dataset.
The data suggest that analysis of recurrent glioblastoma samples is missing the tumors with the most common DNA structure found in the natural population. Further, the data suggest that this global DNA pattern is associated with clinical characteristics that lead surgeons across North America and Asia to not operate on these first recurrent glioblastoma patients. As a point of diagnostic concern, this MDS “poor survival” region cannot be approximated by a differentially expressed gene signature (Supplementary Fig. S1). Differentially methylated gene patterns, outside of the context of copy numbers, are also not sufficient to predict this “poor survival” region (Supplementary Fig. S2). Indeed, determination of the “poor survival” region requires exome sequencing in addition to platform-independent copy number status in order to determine the overall DNA structure. Although differentially methylated genes are not of a diagnostic utility and do not discriminate between MDS regions in our context, biological pathway enrichment analysis of the most significantly differentially methylated genes demonstrates that the “poor survival” group has differentially methylated genes related mostly to cell cycle, and to a lesser extent, immune cell interaction (Supplementary Table S1), consistent with dysregulation of these pathways being associated with more aggressive forms of glioma.2,5,17–24 Clinical characteristics corresponding to this DNA pattern are unclear, but there appears to be no association with median age (P = 0.15; Mann–Whitney U test), male versus female (P = 0.052; Fisher’s exact test), or KPS ≥80 versus <80 (P = 0.42; Fisher’s exact test). It also seems likely that patients with this global DNA pattern are underrepresented in clinical trials, particularly in those focused on first recurrence of glioblastoma.
Discussion
Shifts in the distribution pattern of molecular subtypes across glioblastoma cohorts have overall implications for clinical trial design. In the paired glioblastoma cohort and the ARTE trial cohort, there is a selection bias for longer overall survival compared with population-based studies, as reflected in the molecular marker status shown. As such, comparison of any clinical trial treatment strategy, or institutional outcomes, may be very difficult to interpret in the absence of knowing molecular subtype distributions of their patient populations. Molecular profiling to inform clinical trial structure is of increasing importance,25 and it would be ideal to stratify for DNA copy number subtypes identified at the initial resection upon randomization in a clinical trial. Failure to do so may contribute to occasional discordance between phase II and phase III clinical trial outcomes. The data suggest that historical controls that do not account for molecular signatures are insufficient as comparators for current and future clinical trial enrollment. If molecular subtypes are not addressed up front in clinical trials, it is likely that treatments that may appear effective overall in phase II trials may fail in phase III trials due to unaddressed shifting of molecular distributions, leading to very costly trials. Perhaps molecular profiling including copy numbers could increase the accuracy of phase II trials. For those poorest molecular survival groups, such as W1 copy number subtype, patients could be identified early and put on up-front trials limited to their molecular group. Overall, informing glioblastoma clinical trials by molecular signature status, such as copy number alterations, may lead to more appropriate cohort distributions and therapeutic strategies applicable to the general population.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
Research reported in this publication was supported by the National Cancer Institute of the NIH under award numbers 1U54CA193313 and 1U54CA209997.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Authorship statement
P.J.C. and E.C.H. designed the study. Data collection was performed by P.J.C., R.R., and M.W. Analysis and interpretation of data were done by P.J.C., L.M., H-G.W., S.A., H.B., and E.C.H. All authors participated in drafting, revising, and approval of the manuscript.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.. WHO Classification of Tumours of the Central Nervous System. 4th ed Lyon, France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 2. Cimino PJ, Zager M, McFerrin L, et al. . Multidimensional scaling of diffuse gliomas: application to the 2016 World Health Organization classification system with prognostically relevant molecular subtype discovery. Acta Neuropathol Commun. 2017;5(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee CH, Alpert BO, Sankaranarayanan P, Alter O. GSVD comparison of patient-matched normal and tumor aCGH profiles reveals global copy-number alterations predicting glioblastoma multiforme survival. PLoS One. 2012;7(1):e30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shirahata M, Ono T, Stichel D, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136(1):153–166. [DOI] [PubMed] [Google Scholar]
- 5. Ceccarelli M, Barthel FP, Malta TM, et al. . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 7. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wirsching HG, Tabatabai G, Roelcke U, et al. Bevacizumab plus hypofractionated radiotherapy versus radiotherapy alone in elderly patients with glioblastoma: the randomized, open-label, phase II ARTE trial. Ann Oncol. 2018;29(6):1423–1430. [DOI] [PubMed] [Google Scholar]
- 9. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grossman RL, Heath AP, Ferretti V, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magi A, Tattini L, Cifola I, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14(10):R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolouri H, Zhao LP, Holland EC. Big data visualization identifies the multidimensional molecular landscape of human gliomas. Proc Natl Acad Sci U S A. 2016;113(19):5394–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trifonov V, Pasqualucci L, Tiacci E, Falini B, Rabadan R. SAVI: a statistical algorithm for variant frequency identification. BMC Syst Biol. 2013;7(Suppl 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collado-Torres L, Nellore A, Kammers K, et al. Reproducible RNA-seq analysis using Recount2. Nat Biotechnol. 2017;35(4):319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amankulor NM, Kim Y, Arora S, et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31(8):774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garofalo S, D’Alessandro G, Chece G, et al. Enriched environment reduces glioma growth through immune and non-immune mechanisms in mice. Nat Commun. 2015;6:6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roy DM, Walsh LA, Desrichard A, et al. Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell. 2016;29(5):737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng S, Fu J, Vegesna R, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013;27(13):1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quail DF, Bowman RL, Akkari L, et al. . The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016; 352(6288):aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rutledge WC, Kong J, Gao J, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. 2013;19(18):4951–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanguturi SK, Trippa L, Ramkissoon SH, et al. Leveraging molecular datasets for biomarker-based clinical trial design in glioblastoma. Neuro Oncol. 2017;19(7):908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.