Abstract
Genetic variation within hepatitis B surface antigen (HBsAg), in particular within the major hydrophobic region (MHR), is related to immune/vaccine and test failures and can have a significant impact on the vaccination and diagnosis of acute infection. This study shows, for the first time, variation among acute cases and compares the amino acid variation within the HBsAg between acute and chronic infections. We analyzed the virus isolated from 1231 acute and 585 chronic cases reported to an anonymized public health surveillance database between 2004 and 2014 in The Netherlands. HBsAg analysis revealed the circulation of 6 genotypes (Gt); GtA was the dominant genotype followed by GtD among both acute (68.2% and 17.4%, respectively) and chronic (34.9% and 34.2%, respectively) cases. Variation was the highest among chronic strains compared to that among acute strains. Both acute and chronic GtD showed the highest variation compared to that of other genotypes (P < .01). Substitutions within the MHR were found in 8.5% of the acute strains and 18.6% of the chronic strains. Specific MHR substitutions described to have an impact on vaccine/immune escape and/or HBsAg test failure were found among 4.1% of the acute strains and 7.0% of the chronic strains. In conclusion, we show a high variation of HBsAg among acute and chronic hepatitis B virus–infected cases in The Netherlands, in particular among those infected with GtD, and compare, for the first time, variation in frequencies between acute and chronic cases. Additional studies on the impact of these variations on vaccination and test failure need to be conducted, as well as whether HBsAg false–negative variants have been missed.
Keywords: amino acid variation, hepatitis B surface antigen, hepatitis B virus, major hydrophobic region, test failure, vaccination
1. INTRODUCTION
An estimated 250 to 260 million people in the world are reported to be hepatitis B surface antigen (HBsAg) positive.1 Since the introduction of hepatitis B virus (HBV) vaccination in the 1980s, the incidence of acute HBV infection dropped tremendously worldwide.2, 3 In The Netherlands, HBV infections are statutorily but anonymously notified to the Dutch registry for notifiable infectious diseases (OSIRIS) by the municipal health services. The incidence of acute infections was low, with 0.6 notifications per 100 000 population in 2016.4 The estimated prevalence of chronic HBV infection in The Netherlands is 0.2% to 0.5%.5, 6
HBV is classified in 10 HBV genotypes, designated A to J, based on phylogenetic analysis of the S gene and full‐length genome.7 The different types have a distinct geographical distribution, with genotype A commonly found in North America and Europe and genotype D in North Africa and the Middle East. The dominant types in The Netherlands have been reported to be genotype A, frequently detected among men who have sex with men (MSM), and genotype D, commonly isolated from immigrants from the Mediterranean region.8, 9 HBV is a double‐stranded DNA virus with a genome length of ~3.2 kb. The genome encodes for an overlapping open reading frame and translates into 3 antigens (core [HBcAg], surface [HBsAg], and the e antigen [HBeAg]), the DNA polymerase, and gene x, which acts as a transactivator for replication.
The HBV genome is one of the most variable among DNA viruses. In particular, variation in the major hydrophobic region (MHR; amino acids 100 to 160) of the HBsAg has been related to reduced or absent immune/vaccine responses (immune/vaccine escape), may impact the detection of HBsAg, or lead to reactivation in persons with previously resolved infection or immunosuppressed persons (often receiving immunosuppressive treatment) with stable chronic infection.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 This study shows, for the first time, variation among acute HBV infections as well as a comparison of the HBsAg amino acid variation between acute and chronic infections.
2. METHODS
2.1. Study population
In The Netherlands, all acute HBV infections are anonymously notified to OSIRIS by the municipal health services. OSIRIS is a public health database used for surveillance. Clinical, patient, and test data on HBV markers are not reported, which did not allow further analysis of the relation of levels of different HBV markers and substitutions.
Acute cases are reported to OSIRIS based on a positive HBsAg test result and/or an anti‐HBc IgM result (when available). Chronic cases are notifiable when a positive HBsAg or HBV‐DNA positivity is diagnosed in The Netherlands for the first time. Interviews are conducted to ascertain risk exposures and to conduct source‐tracing investigations and/or partner notification.
Since 2004, blood samples from all acute cases reported to OSIRIS were requested for typing. Since 2010, samples from chronic cases with risk behavior (ie, tested as part of the HBV vaccination program for behavioral risk groups, or with the reported transmission route of MSM sexual contact) were also requested for typing. All samples that were successfully typed in the HBsAg between 2004 and 2014 were studied. Samples that were successfully typed in the HBsAg from chronic cases with risk behavior notified before 2010 were obtained from several epidemiological studies5, 8, 9, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 conducted at the National Institute for Public Health and the Environment (RIVM) and municipal health services.
2.2. Isolation, amplification, and sequencing of HBsAg
DNA was isolated from 200 µL of serum by manual extraction using the QIAamp DNA Mini and Blood Mini kit (Qiagen, Venlo, The Netherlands) (samples from 2004 to 2006) or by automated extraction using the LC Nucleic Acid isolation kit (Roche, Almere, The Netherlands) (samples from 2007 to 2014). DNA was eluted in 50 µL of elution buffer and amplified in a nested PCR using a thermocycler (GeneAmp 9700, Perkin Elmer, Waltham, MA), as previously described.31, 35
A 656‐nucleotide fragment was amplified, which encompassed the first 203 of the 226 amino acids of the HBsAg and amino acid 6 to 209 of the reverse transcriptase (RT) of the polymerase. The sequence reaction was performed in house or by BaseClear (Leiden, The Netherlands) with Big Dye Terminator (Life Technologies, Carlsbad, CA).
2.3. HBV genotype and variant identification
S‐region sequences were used to determine the genotype of the HBV by comparing them to reference strains using Bionumerics version 7.1 (Sint‐Martens‐Latem, Belgium).36 Phylogenetic trees were constructed using the maximum parsimony method. To assess genetic variation within the HBsAg and RT, we analyzed the appearance and frequency of amino acid sequence variation within the S and RT genes by type by aligning all sequences using Bionumerics version 7.1 to the sequences of the reference HBV types (A to J) assigned by Pourkarim et al.7 Subgenotype‐specific polymorphisms were excluded in the analysis. All sequences were deposited in GenBank (accession numbers: KX659172 to KX659981 and KP243601 to KP244031). For clarity, strains isolated from acute infected cases are referred to as acute strains and strains isolated from chronic cases, as chronic strains.
2.4. Statistical analysis
Statistical analyses were performed using the univariate Χ 2 test. A 2‐sided P < .01 was considered statistically significant.
3. RESULTS
3.1. Epidemiology of HBV genotypes for 2004 to 2014
Between 2004 and 2014, HBV strains isolated from 1231 acutely infected cases and 585 chronically infected cases from behavioral risk groups were successfully sequenced and typed (Figure 1 and Table 1). Genotype (Gt) A was the dominant genotype detected between 2004 and 2014, accounting for 68.2% (n = 839) of the acute cases and 34.9% (n = 161) of the chronic cases with risk behavior (Table 1). A majority of the GtA acute cases (86.2%) were born in The Netherlands, followed by only 1% to 2% born in Surinam, Turkey, and The Netherland Antilles. Similarly, chronic GtA cases were for the most part born in The Netherlands (35.3%), followed by Surinam (9.3%). Over the whole study period, only 2 epidemiological clusters were found, as previously described.37, 38
Figure 1.
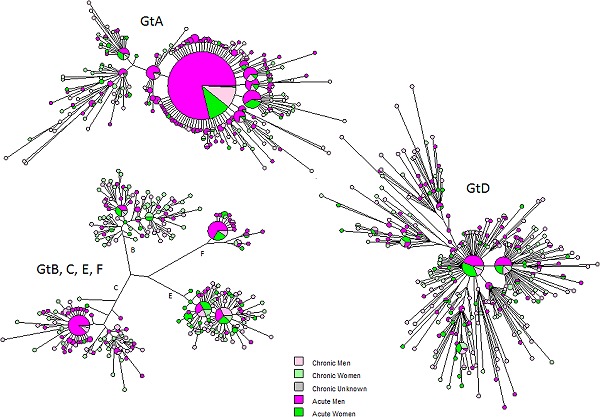
Maximum parsimony tree of the HBsAg of HBV strains isolated from 1232 Dutch acute HBV–infected cases characterized between 2004 and 2014. The cases are color‐coded by infection status and sex. Gt, genotype; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus
Table 1.
Distribution of HBV acute and chronic infected cases (strains) and HBsAg/major hydrophobic region (MHR) variation by genotype between 2004 and 2014
| Genotype | Cases, n (% of all cases) | Strains harboring variation in the HBsAg, n (% of Gt) | Strains harboring variation in MHR, n (% of Gt) | Number of substitutions within the MHR |
|---|---|---|---|---|
| Acute | ||||
| GtA | 839 (68.2) | 100 (11.9) | 23 (2.7) | 28 |
| GtB | 31 (2.5) | 13 (41.9) | 5 (16.1) | 5 |
| GtC | 61 (5.0) | 13 (21.3) | 5 (8.2) | 6 |
| GtD | 214 (17.4) | 88 (41.1) | 64 (29.9) | 73 |
| GtE | 51 (4.1) | 20 (39.2) | 7 (13.7) | 7 |
| GtF | 35 (2.8) | 9 (22.9) | 1 (2.9) | 1 |
| Total | 1231 | 243 (19.7) | 105 (8.5) | 120 |
| Chronic | ||||
| GtA | 204 (34.9) | 86 (42.2) | 24 (11.8) | 30 |
| GtB | 68 (11.6) | 48 (70.6) | 19 (27.9) | 23 |
| GtC | 48 (8.2) | 27 (56.3) | 9 (18.8) | 14 |
| GtD | 200 (34.2) | 110 (55.0) | 46 (23.0) | 70 |
| GtE | 59 (10.1) | 37 (62.7) | 11 (18.6) | 14 |
| GtF | 6 (1) | 1 (16.7) | 0 (‐) | 0 |
| Total | 585 | 309 (52.8) | 109 (18.6) | 151 |
GtD was the second most dominant type and was detected in 17.4% (n = 214) of the acute cases and 34.2% (n = 200) of the chronic cases (Table 1). A majority of the acute gtD cases were born in The Netherlands (66.8%), followed by Turkey (8.4%), whereas this was reversed among chronic cases, where a majority of chronic GtD cases were born in Turkey (33.5%), followed by The Netherlands (12.5%).
For acute GtB, C, D, and E, The Netherlands was the predominant country of birth. In contrast, for chronic GtB and GtC, most cases were born in China (25.4% and 50.0%, respectively), and for GtE, almost all cases (98%) were born in African countries.
A majority of the acute cases were not vaccinated (91%, n = 1117). For 5% (n = 66) vaccination was complete/incomplete, and for 4% (n = 48) vaccination status was unavailable. In contrast to the vaccination status of acute strains, the vaccination status in a majority of the chronic cases notified in OSIRIS (>2010) was unavailable (74%, n = 434). Twenty‐five percent (n = 144) were not vaccinated, and in 1% (n = 7), vaccination was either complete or incomplete. No vaccination data of chronic cases before 2010 could be obtained.
Overall, acute and chronic HBV infections were more frequently reported in men, 79% and 63%, respectively (Figure 1), and the median age of infection among acute cases was 41 years. The median age of infection among chronic cases cannot be assessed as cases are reported as chronically infected and not when (acute) infection has occurred.
3.2. HBsAg variation among acute cases
The HBsAg amino acid sequences of the 6 genotypes were analyzed and compared to their respective reference type.7 Of the 1231 acute strains, 19.7% (n = 242) harbored variation in the HBsAg ranging from a single substitution (14.3%, n = 176) to a combination of 2 to 9 substitutions (5.4%, n = 66). The HBsAg variation among acute GtA strains was the lowest and was significantly lower than that found for GtB, D, and E strains (Table 1 and Figure 2). In total, 100 GtA strains (11.9% of 839 GtA strains; Table 1) harbored HBsAg variation, ranging from a single substitution (9.9%, n = 83) to a combination of 2 to 6 substitutions (2%, n = 17) per strain (Figure 2). In comparison, GtB, C, D, E, and F strains harbored HBsAg variation in 41.9%, 21.3%, 41.1%, 39.2%, and 22.9%, respectively (Table 1). Combinations of substitutions were the most frequently found for GtD acute strains, ranging from 2 to 9 substitutions (14.5%, n = 31 ; Figure 2). Combinations of substitutions were the lowest for GtB, C, E, and F strains and ranged from 2 substitutions per strain (GtF) to 2 to 3 substitutions per strain (GtB and C) and 2 to 5 substitutions per strain (GtE; Figure 2).
Figure 2.
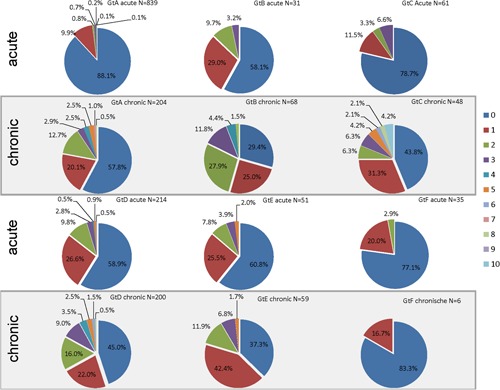
Variation frequency within the HBsAg ranging from a no substitution to 1 to 10 substitutions among acute and chronic strains. The data set for acute cases is a representative set of acute HBV in The Netherlands. The data set for chronic cases only represents those with risk behavior from various risk groups (<2010) and in most cases from MSM (>2010). The HBsAg amino acid sequence variation of the 6 genotypes was compared to the respective reference type 7. HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; MSM, men who have sex with men
In total, we found 105 acute strains (8.5%) with 120 different substitutions divided over 29 positions within the MHR (Table 1 and Figure 3). The MHR substitutions predominantly appeared in a combination of 2 or more substitutions per strain. MHR substitutions were more frequently found among GtD acute strains (n = 64) with 73 different substitutions and accounted for 29.9% of the 214 GtD strains. For GtA, 28 different substitutions were identified among 2.7% of the GtA strains. The number of strains harboring variation in the MHR and the number of MHR substitutions among GtB, C, and E were significantly lower (P < .01) compared to GtD (Table 1). One GtF strain showed only one MHR substitution, which was described as an immune escape substitution having an impact on HBsAg detection: G145R. The patient was fully vaccinated at the time of diagnosis, yet was found to be HBsAg positive. Other substitutions described as immune escape and/or to have been associated with HBsAg test failure were found among GtA, B, D, and E, totaling up to 50 strains harboring these substitutions as a single substitution or in combination (4.1% of all acute strains; Table 2). Apart from the GtF case, 3 other cases (GtD) were found to be vaccinated (1 complete/2 incomplete), yet were HBsAg positive, and were infected with a variant harboring a single MHR substitution (M103I or T118A or P127I), of which only M103I was associated with HBsAg test failure (Table 2). Data on whether the cases were nonresponders could not be assessed as data were anonymized.
Figure 3.
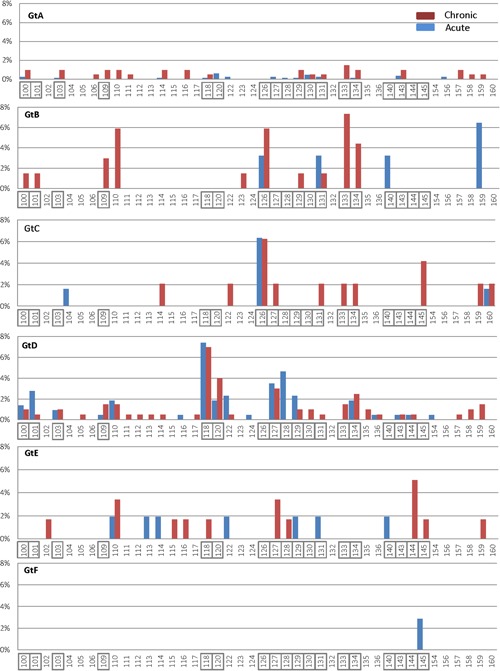
Frequency and distribution of amino acid substitutions within the HBsAg (amino acid residues 1 to 203) of genotypes A to F. Acute strains are shown in blue bars and chronic strains are in red bars. Positions associated with vaccine/immune escape and/or test result are boxed. Gt, genotype; HBsAg, hepatitis B surface antigen
Table 2.
Number of strains with substitutions associated with vaccine escape (VEM) and/or HBsAg test failure located within the major hydrophobic regions (MHRs)
| Acute strains, n | Chronic strains, n | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substitution | A | B | C | D | E | F | A | B | C | D | E | F | Association |
| Y100C | 2 | VEM and HBsAg test failure | |||||||||||
| Q101R | 4 | HBsAg test failure | |||||||||||
| M103I | 1 | 2 | 1 | 2 | HBsAg test failure | ||||||||
| L109I | 1 | VEM and HBsAg test failure | |||||||||||
| T118R | 1 | 1 | VEM and HBsAg test failure | ||||||||||
| P120T/S | 5 | 4 | 8 | VEM and HBsAg test failure | |||||||||
| R122K | 5 | 1 | 1 | HBsAg test failure | |||||||||
| T126I/S/A | 1 | 4 | VEM and HBsAg test failure | ||||||||||
| P127T/L | 2 | 4 | 2 | VEM and HBsAg test failure | |||||||||
| A128V | 1 | 10 | 1 | HBsAg test failure | |||||||||
| Q129R/H | 1 | 2 | 1 | 1 | 1 | 1 | VEM and HBsAg test failure | ||||||
| T131S/I | 1 | 1 | 1 | VEM and HBsAg test failure | |||||||||
| M133I | 3 | 1 | 1 | VEM and HBsAg test failure | |||||||||
| F/Y134N/L | 2 | 2 | 1 | 3 | VEM and HBsAg test failure | ||||||||
| T140I | 1 | VEM and HBsAg test failure | |||||||||||
| S143L | 1 | 1 | VEM and HBsAg test failure | ||||||||||
| D144A/E | 1 | 1 | 3 | VEM and HBsAg test failure | |||||||||
| G145R | 1 | VEM and HBsAg test failure | |||||||||||
| G145A | 1 | VEM and HBsAg test failure | |||||||||||
| E164G | 1 | 1 | HBsAg test failure | ||||||||||
| A168V | 1 | VEM and HBsAg test failure | |||||||||||
| Total | 10 a | 3 | 0 | 39 b | 3 | 1 | 6 | 8 | 4 | 22 c | 4 | 0 | |
M103I/A128V/Q129R was found as a triple substitution in 1 GtA acute strain totaling to 8 acute GtA cases with MHR substitutions.
R122K/Q129R was found as a double substitution in 2 GtD acute strains and Y100C/L109I/T118R was found as a triple substitution in 1 GtD acute strain totaling to 35 acute GtD cases with MHR substitutions.
M103I/P120T/R122K was found as a triple substitution in 1 GtD chronic strain and M103I/P120S was found as a double substitution in 1 GtD chronic strain totaling to 19 acute GtD cases with MHR substitutions.
Gt, genotype; HBsAg, hepatitis B surface antigen; MHR, major hydrophobic region.
Almost all cases were HBsAg positive as this was a reporting criterion to OSIRIS. However, 3 cases (GtD) were HBsAg‐negative harboring an MHR substitution (T118A or A128V or a combination of R122K and Q129R), of which only R122K and A128V were associated with immune/vaccine escape (Table 2). A positive IgM HBcAg test result led to their notification.
3.3. HBsAg variation among chronic infected cases
The HBsAg amino acid sequences of the chronic strains were compared to the respective reference type.7 Of the 585 chronic strains, more than half of the strains (52.8%, n = 309) harbored variation within the HBsAg, ranging from a single substitution (24.4%, n = 143) or a combination of 2 to 10 substitutions per strain (28.4%, n = 166). The variation was high for GtA (42.2%), GtB (70.6%), GtC (56.3%), GtD (55.0%), and GtE (62.7%; Table 1). The genetic HBsAg variation was the lowest for GtF, with only 1 (16.7%) strain (1 substitution outside the MHR; Figure 2).
The combination of substitutions per strain was the highest for GtC and ranged from a combination of 2 to 10 substitutions (25%, n = 12) per strain (Figure 2). This was followed by GtB, GtD, and GtA, with a combination of 2 to 8 substitutions for GtA (22.1%, n = 45) and GtB (45.6%, n = 31) and 2 to 9 for gtD (33.0%, n = 66; Figure 2).
Among the chronic strains, 109 had substitutions (accounting for 151 different substitutions divided over 37 positions) located within the MHR (Table 1 and Figure 3). Similar to acute strains, the MHR substitutions predominantly appeared in a combination of 2 or more substitutions per strain. The number of chronic GtD strains with substitutions located within the MHR (46 strains with 70 substitutions) was significantly (P < .01) higher than that found among GtA (24 strains with 30 substitutions) and other genotypes (Table 1).
Substitutions described as an immune escape substitution and/or to have an impact on HBsAg detection (Table 2) were found among 41 chronic strains (7.0%) that harbored these substitutions as a single substitution or in combination. Only 1 of 7 cases that were vaccinated was infected with a variant (GtA) harboring the substitution F134A that was not associated with vaccine/immune escape. Whether the case was vaccinated before primary infection could not be assessed as data were anonymized.
3.4. RT substitutions
Owing to the overlapping nature of the S gene over the polymerase, sequence analysis of the RT region of the polymerase was additionally conducted to identify antiviral resistance markers.
The acute HBsAg substitutions resulted in 444 different substitutions in the RT region of the polymerase among 335 strains (Table 3). Among the chronic strains, 604 different substitutions were found among 321 chronic cases. Five different substitutions were described to confer antiviral resistance against Lamuvidine, Entecavir, and Telibividune. Substitutions that were found to confer reduced susceptibilty against Adefovir and Tenofovir39, 40 were found in 3 acute strains (GtA (n = 2), and GtD (n = 1) and 5 GtA chronic strains (Table 3). Substitutions L180M and M204V were always found in combination, appearing as double substitutions in 1 acute and 5 chronic GtA strains, and with V173L as a triple substitution in 1 acute GtA strain.
Table 3.
Distribution of reverse transcriptase substitutions among strains isolated from HBV acute and chronic infected cases by genotype between 2004 and 2014 and substitutions associated with antiviral resistance (AVR)
| Acute | Chronic | |||
|---|---|---|---|---|
| Genotype | n substitutions/strains harboring variation in the RT | Substitution associated with AVR (n strains) a | n substitutions/strains harboring variation in the RT | Substitution associated with AVR (n strains) a |
| GtA | 197/168 | V173L (1) b , L180M (2) c , M204V (2) c | 137/79 | L180M (5) c , M204V (5) c |
| GtB | 19/16 | 74/47 | ||
| GtC | 19/10 | 54/25 | ||
| GtD | 162/108 | V173L (1) | 230/132 | |
| GtE | 35/25 | 105/36 | ||
| GtF | 12/8 | 4/2 | ||
| Total | 444/335 | 604/321 | ||
V173L, L180M, and M204V are associated with lamivudine, telbivudine, and entecavir resistance, respectively; L180M + M204V are associated with lamivudine and telbivudine resistance; V173L is associated with lamivudine resistance.
L180M/M204V/V173L was found as a triple substitution in 1 acute strain.
L180M/M204V was found as a double substitution in 1 acute strain and 5 chronic strains.
AVR, antiviral resistance; Gt, genotype; RT, reverse transcriptase.
3.5. HBsAg stop codons
We identified 7 acute strains (0.6%) and 7 chronic strains (1.2%) that were infected with a variant with a stop codon within the HBsAg gene that could result in truncated expression of the HBsAg: L15STOP (n = 1 acute and n = 2 chronic), residue S61STOP (n = 2 acute), C69STOP (n = 1 acute and n = 1 chronic), W165STOP (n = 1 acute and n = 1 chronic), L175STOP (n = 1 acute), W182STOP (n = 1 acute and n = 1 chronic), and W199 STOP (n = 2 chronic).
Stop codon W182 resulted in the drug‐resistant substitution (V173L) described above. All cases were reported based on a positive HBsAg test result.
4. DISCUSSION
This study shows, for the first time, variation among acute cases as well as a comparison of the genetic variation of the HBsAg and the MHR region between acute and chronic HBV cases diagnosed in The Netherlands. We should point out that the data set for acute cases is a representative set of acute HBV in The Netherlands, whereas the data set for chronic cases represents only those with risk behavior from various risk groups (<2010) and in most cases from MSM (>2010).
We showed that 6 genotypes continue to circulate in The Netherlands,27, 31 of which GtA is the dominant genotype, followed by GtD among both acute and chronic cases. However, GtD showed significantly more variation over the entire HBsAg and within the MHR than GtA or any other genotype among both acute and chronic cases.
Overall, the variation among the chronic strains for all of the genotypes was significantly higher than that for acute strains. Twenty percent of the acute strains harbored variation within the HBsAg, whereas more than half of the chronic strains (53%) harbored variation within the HBsAg. Other studies have shown similar percentages, ranging from 40% to 55% variation, which were mostly among chronic GtD strains.11, 41, 42, 43 The low diversity among GtA acute strains suggests that the transmission of GtA8, 9 is sustained within the MSM community in The Netherlands44, 45 and that there is little transmission from chronic cases leading to new infections. In contrast, the high variation among acute GtB, D, and E most likely represents transmission from chronically infected cases, as shown by Kretzschmar et al.44
Amino acid substitutions within the MHR can influence vaccine effectiveness as well as diagnostic sensitivity by affecting antibody binding either to the virus when infected or to antigens in a specific test.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 HBsAg substitutions have also been related to a decrease in antibody titers affecting test results23, 46 or to reactivation in individuals with previously resolved infection or immunosuppressed individuals with stable chronic infection.24 As such, the circulation of variants, in particular, those affecting immune or vaccination response, is a major public health concern. As the clinical background of the cases, whether the acute infection was resolved or chronic cases were immunosuppressed (by therapy), was unknown, variation as for reactivation could not be taken into account.
In our study, we found 18.6% of chronic strains to harbor a substitution with the MHR, which varied from 0% for GtF to 28% for GtB; this is in line with data from other studies describing a prevalence of these substitutions ranging from 14% to 44% worldwide.11, 47, 48, 49, 50, 51 This was significantly higher compared to acute strains (8.6%), where we show, for the first time, prevalence data on acute strains.
MHR substitution associated with immune escape and HBsAg detection failure were found in 4.1% of all of the acute strains and 7.0% of all chronic strains. Because a majority of the cases with an MHR substitution were not vaccinated and these substitutions have also been described among nonvaccinated individuals, we cannot assess the full impact of these substitutions in this study. In The Netherlands, HBV vaccination of second‐generation migrant children started in 2003 and universal HBV vaccination of all children through the national immunization program started in August 2011. Of concern is that most acutely infected children are asymptomatic, while being at high risk to develop chronic infection. As most chronic cases are detected around 40 years of age (data not shown)31, 52 and as the vaccinated cases are now 4 to 10 years old, it would not be until 30 to 40 years later that the clinical impact of the circulation of these variants would be seen. Furthermore, universal vaccination has been suggested to aid in the selection of vaccine escape variants,50, 53 and infection modeling indicated that the frequency of VEMs will increase in 50 years’ time.54
The immune escape substitution has also been described to be associated with HBsAg test failure.11, 16, 55
In only 3 cases an HBsAg‐negative test result was reported (a positive IgM HBcAg test result led to their notification) that harbored the substitutions T118A, A128V, or a combination of R122K and Q129R.
Test failure also holds true for variants with a stop codon. Even though a stop codon can lead to truncated HBsAg, thereby leading to false‐negative test results,56 we found 14 cases that contained a stop codon within the HBsAg gene both upstream and downstream of the MHR, all with HBsAg‐positive test results. Similar findings were reported by Pourkarim et al,41 who found no relation between a stop codon and a false‐negative test result.
Our study comprises, for the most part, HBsAg‐positive cases, as this is one of the reporting criteria, and we therefore may have or are missing cases that indeed cannot be detected by current assays. It should be noted that current HBV surface antigen assays are able to detect most MHR variants that previously failed detection, and indeed the substitutions found in our study were among the acute strains detected through a positive HBsAg test result. However, the detection level may vary per assay, and data on which assays were used on reported cases are unavailable (various laboratories use different assays), which limits analysis of the impact of substitutions found in our assay and those possibly missed. Further studies are thus needed to assess the efficacy of the different tests used and to study the extent of HBsAg false–negative variants (not reported). Furthermore, the standardized testing algorithm for HBV in most laboratories is an HBsAg and an anti‐HBc test. When either is or both are positive, additional testing is conducted, but what is tested varies between laboratories. In the case of sole anti–HBc‐positive (HBsAg/anti–HB‐negative) additional testing of anti‐HBe, HBeAg and anti‐HBs should be conducted to exclude immune escape or test failure.
As the region of the S gene analyzed overlaps with the RT gene of the polymerase, variation of the RT gene could be analyzed for antiviral resistance with 0.4% of the acute strains and 1.0% of the chronic strains having a substitution associated with antiviral resistance.39, 40 However, data on treatment are not included in the database, and further conclusions could not be drawn from these data.
The anonymized data also limited the analysis of different HBV markers, which are not reported on HBsAg variation frequency; it has been shown that the levels of different HBV markers can influence the variation frequency.57, 58 In addition, various laboratories use different assays, and the data on HBsAg level/DNA level that may vary between assays57 cannot be assessed using retrospective data and a new prospective study with standardized platforms needs to be conducted.
In conclusion, this study is the first to report on HBsAg variation among acute cases and to make a comparison of HBV acute and chronically infected cases in The Netherlands. The study shows significant differences of variation between both acute and chronic cases. These findings indicate the necessity of monitoring and studying amino acid substitutions, which can have an impact on vaccine effectiveness and HBsAg testing. Furthermore, the diagnostic testing algorithms/assays should be evaluated further to exclude underreporting of false HBsAg‐negative cases.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
ACKNOWLEDGMENTS
We would like to thank Susan Hahné and Audrey King for critically reading the manuscript and the laboratory staff of Infectious diseases Diagnosis and Screening of the Center for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, The Netherlands, for typing all the serum samples. This study was supported by the Ministry of Health, Welfare and Sport, The Netherlands. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Cremer J, Hofstraat SHI, van Heiningen F, Veldhuijzen IK, van Benthem BHB, Benschop KSM. Genetic variation of hepatitis B surface antigen among acute and chronic hepatitis B virus infections in The Netherlands. J Med Virol. 2018;90:1576‐1585. 10.1002/jmv.25232
References
REFERENCES
- 1. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386(10003):1546‐1555. [DOI] [PubMed] [Google Scholar]
- 2. Zanetti AR, Van Damme P, Shouval D. The global impact of vaccination against hepatitis B: a historical overview. Vaccine. 2008;26(49):6266‐6273. [DOI] [PubMed] [Google Scholar]
- 3. Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373(9663):582‐592. [DOI] [PubMed] [Google Scholar]
- 4. Visser MvA F, van Oeffelen AAM, van den Broek IVF, et al. Sexually transmitted infections, including HIV, in the Netherlands in 2016 National Institute for Public Health and the Environment; Bilthoven, 2017. The Netherlands: National Institute for Public Health and the Environment; 2017. [Google Scholar]
- 5. Hahné SJM, De Melker HE, Kretzschmar M, et al. Prevalence of hepatitis B virus infection in The Netherlands in 1996 and 2007. Epidemiol Infect. 2012;140(8):1469‐1480. [DOI] [PubMed] [Google Scholar]
- 6. Marschall T, Kretzschmar M, Mangen MJJ, Schalm S. High impact of migration on the prevalence of chronic hepatitis B in the Netherlands. Eur J Gastroenterol Hepatol. 2008;20(12):1214‐1225. [DOI] [PubMed] [Google Scholar]
- 7. Pourkarim MR, Amini‐Bavil‐Olyaee S, Kurbanov F, Van Ranst M, Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol. 2014;20(23):7152‐7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Houdt R, Sonder GJB, Dukers NHTM, et al. Impact of a targeted hepatitis B vaccination program in Amsterdam, The Netherlands. Vaccine. 2007;25(14):2698‐2705. [DOI] [PubMed] [Google Scholar]
- 9. Hahné S, van Houdt R, Koedijk F, et al. Selective hepatitis B virus vaccination has reduced hepatitis B virus transmission in the Netherlands. PLoS One. 2013;8(7):e67866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carman WF. The clinical significance of surface antigen variants of hepatitis B virus. J Viral Hepat. 1997;4(Suppl 1):11‐20. [DOI] [PubMed] [Google Scholar]
- 11. Avellón A, Echevarria JM. Frequency of hepatitis B virus ‘a’ determinant variants in unselected Spanish chronic carriers. J Med Virol. 2006;78(1):24‐36. [DOI] [PubMed] [Google Scholar]
- 12. Coleman PF, Jack chen YC, Mushahwar IK. Immunoassay detection of hepatitis B surface antigen mutants. J Med Virol. 1999;59(1):19‐24. [DOI] [PubMed] [Google Scholar]
- 13. Luongo M, Critelli R, Grottola A, et al. Acute hepatitis B caused by a vaccine‐escape HBV strain in vaccinated subject: sequence analysis and therapeutic strategy. J Clin Virol. 2015;62:89‐91. [DOI] [PubMed] [Google Scholar]
- 14. Fischer L, Sterneck M, Müller‐Ruchholtz C, Gish R, Will H. Hepatitis B virus variants associated with clinically severe recurrence after liver transplantation. Transplant Proc. 1999;31(1‐2):492‐493. [DOI] [PubMed] [Google Scholar]
- 15. Zhang YY, Nordenfelt E, Hansson BG. Increasing heterogeneity of the 'a' determinant of HBsAg found in the presumed late phase of chronic hepatitis B virus infection. Scand J Infect Dis. 1996;28(1):9‐15. [DOI] [PubMed] [Google Scholar]
- 16. Ireland JH, O'Donnell B, Basuni AA, et al. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology. 2000;31(5):1176‐1182. [DOI] [PubMed] [Google Scholar]
- 17. Oon CJ, Lim GK, Ye Z, et al. Molecular epidemiology of hepatitis B virus vaccine variants in Singapore. Vaccine. 1995;13(8):699‐702. [DOI] [PubMed] [Google Scholar]
- 18. Wallace LA, Echevarria JE, Echevarria JM, Carman WF. Molecular characterization of envelope antigenic variants of hepatitis B virus from Spain. J Infect Dis. 1994;170(5):1300‐1303. [DOI] [PubMed] [Google Scholar]
- 19. Levicnik‐Stezinar S. Hepatitis B surface antigen escape mutant in a first time blood donor potentially missed by a routine screening assay. Clin Lab. 2004;50(1‐2):49‐51. [PubMed] [Google Scholar]
- 20. Carman WF, Karayiannis P, Waters J, et al. Vaccine‐induced escape mutant of hepatitis B virus. Lancet. 1990;336(8711):325‐329. [DOI] [PubMed] [Google Scholar]
- 21. El Chaar M, Candotti D, Crowther RA, Allain JP. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology. 2010;52(5):1600‐1610. [DOI] [PubMed] [Google Scholar]
- 22. Weber B, Melchior W, Gehrke R, Doerr HW, Berger A, Rabenau H. Hepatitis B virus markers in anti‐HBc only positive individuals. J Med Virol. 2001;64(3):312‐319. [DOI] [PubMed] [Google Scholar]
- 23. Rezaee R, Poorebrahim M, Najafi S, et al. Impacts of the G145R mutation on the structure and immunogenic activity of the hepatitis B surface antigen: A computational analysis. Hepat Mon. 2016;16(7):e39097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salpini R, Colagrossi L, Bellocchi MC, et al. Hepatitis B surface antigen genetic elements critical for immune escape correlate with hepatitis B virus reactivation upon immunosuppression. Hepatology. 2015;61(3):823‐833. [DOI] [PubMed] [Google Scholar]
- 25. Hahné S, Ramsay M, Balogun K, Edmunds WJ, Mortimer P. Incidence and routes of transmission of hepatitis B virus in England and Wales, 1995‐2000: implications for immunisation policy. J Clin Virol. 2004;29(4):211‐220. [DOI] [PubMed] [Google Scholar]
- 26. Hahne S, Wormann T, Kretzschmar M. Migrants and hepatitis B: new strategies for secondary prevention needed. Eur J Public Health. 2009;19(4):439‐439. [DOI] [PubMed] [Google Scholar]
- 27. Hahné SJM, Veldhuijzen IK, Smits LJM, Nagelkerke N, van de Laar MJW. Hepatitis B virus transmission in The Netherlands: a population‐based, hierarchical case‐control study in a very low‐incidence country. Epidemiol Infect. 2008;136(2):184‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Urbanus AT, van Houdt R, van de Laar TJ, Coutinho RA. Viral hepatitis among men who have sex with men, epidemiology and public health consequences. Euro Surveill. 2009;14(47 [DOI] [PubMed] [Google Scholar]
- 29. van Ballegooijen WM, van Houdt R, Bruisten SM, Boot HJ, Coutinho RA, Wallinga J. Molecular sequence data of hepatitis B virus and genetic diversity after vaccination. Am J Epidemiol. 2009;170(12):1455‐1463. [DOI] [PubMed] [Google Scholar]
- 30. van Houdt R, Bruisten SM, Geskus RB, et al. Ongoing transmission of a single hepatitis B virus strain among men having sex with men in Amsterdam. J Viral Hepat. 2010;17(2):108‐114. [DOI] [PubMed] [Google Scholar]
- 31. van Houdt R, Bruisten SM, Koedijk FDH, et al. Molecular epidemiology of acute hepatitis B in the Netherlands in 2004: nationwide survey. J Med Virol. 2007;79(7):895‐901. [DOI] [PubMed] [Google Scholar]
- 32. van Houdt R, Bruisten SM, Speksnijder AGCL, Prins M. Unexpectedly high proportion of drug users and men having sex with men who develop chronic hepatitis B infection. J Hepatol. 2012;57(3):529‐533. [DOI] [PubMed] [Google Scholar]
- 33. van Houdt R, Koedijk FDH, Bruisten SM, et al. Hepatitis B vaccination targeted at behavioural risk groups in the Netherlands: does it work? Vaccine. 2009;27(27):3530‐3535. [DOI] [PubMed] [Google Scholar]
- 34. van Houdt R, van den Berg CHSB, Stolte IG, et al. Two decades of hepatitis B infections among drug users in Amsterdam: are they still a high‐risk group? J Med Virol. 2009;81(7):1163‐1169. [DOI] [PubMed] [Google Scholar]
- 35. Boot HJ, Cremer J, Koedijk FDH, van Ballegooijen WM, Op de coul ELM. Improved tracing of hepatitis B virus transmission chains by phylogenetic analysis based on C region sequences. J Med Virol. 2008;80(2):233‐241. [DOI] [PubMed] [Google Scholar]
- 36. Bartholomeusz A, Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol. 2004;14(1):3‐16. [DOI] [PubMed] [Google Scholar]
- 37. Simoons‐Smit AM, Zaaijer HL, van den Hoek JAR, Cremer J, Boot HJ. Familiar transmission of hepatitis B virus. Tijdschrif t voor Infectieziekten. 2013;8(3):88‐93. [Google Scholar]
- 38. Hoefnagel JGM, Cremer J, Koene RP, Boot HJ. Een niet opgehelderd cluster van acute hepatitis B. Infectieziekten bulletin. 2011;9(22):316‐318. [Google Scholar]
- 39. He X, Wang F, Huang B, Chen P, Zhong L. Detection and analysis of resistance mutations of hepatitis B virus. Int J Clin Exp Med. 2015;8(6):9630‐9639. [PMC free article] [PubMed] [Google Scholar]
- 40. Lim YS. Management of antiviral resistance in chronic hepatitis B. Gut Liver. 2017;11(2):189‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pourkarim MR, Sharifi Z, Soleimani A, et al. Evolutionary analysis of HBV “S” antigen genetic diversity in Iranian blood donors: a nationwide study. J Med Virol. 2014;86(1):144‐155. [DOI] [PubMed] [Google Scholar]
- 42. Song BC, Kim SH, Kim H, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76(2):194‐202. [DOI] [PubMed] [Google Scholar]
- 43. Pourkarim MR, Amini‐Bavil‐Olyaee S, Verbeeck J, et al. Molecular evolutionary analysis and mutational pattern of full‐length genomes of hepatitis B virus isolated from Belgian patients with different clinical manifestations. J Med Virol. 2010;82(3):379‐389. [DOI] [PubMed] [Google Scholar]
- 44. Kretzschmar M, de Wit GA, Smits LJM, van de Laar MJW. Vaccination against hepatitis B in low endemic countries. Epidemiol Infect. 2002;128(2):229‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soetens LC, van Benthem BHB, Urbanus A, et al. Ongoing transmission of hepatitis B virus in rural parts of the Netherlands, 2009‐2013. PLoS One. 2015;10(2):e0117703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamani LN, Yano Y, Utsumi T, et al. Ultradeep sequencing for detection of quasispecies variants in the major hydrophilic region of hepatitis B virus in Indonesian patients. J Clin Microbiol. 2015;53(10):3165‐3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grabarczyk P, Garmiri P, Liszewski G, et al. Molecular and serological characterization of hepatitis B virus genotype A and D infected blood donors in Poland. J Viral Hepat. 2010;17(6):444‐452. [DOI] [PubMed] [Google Scholar]
- 48. Kitab B, El Feydi AE, Afifi R, et al. Hepatitis B genotypes/subgenotypes and MHR variants among Moroccan chronic carriers. J Infect. 2011;63(1):66‐75. [DOI] [PubMed] [Google Scholar]
- 49. Hou J, Wang Z, Cheng J, et al. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen‐negative Chinese carriers. Hepatology. 2001;34(5):1027‐1034. [DOI] [PubMed] [Google Scholar]
- 50. Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30(5):1312‐1317. [DOI] [PubMed] [Google Scholar]
- 51. Mohebbi SR, Amini‐Bavil‐Olyaee S, Zali N, et al. Molecular epidemiology of hepatitis B virus in Iran. Clin Microbiol Infect. 2008;14(9):858‐866. [DOI] [PubMed] [Google Scholar]
- 52. Hahné SJ, Veldhuijzen IK, Wiessing L, Lim TA, Salminen M, Laar M. Infection with hepatitis B and C virus in Europe: a systematic review of prevalence and cost‐effectiveness of screening. BMC Infect Dis. 2013;13:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang MH. Hepatitis B virus mutation in children. Indian J Pediatr. 2006;73(9):803‐807. [DOI] [PubMed] [Google Scholar]
- 54. Wilson JN, Nokes DJ, Carman WF. The predicted pattern of emergence of vaccine‐resistant hepatitis B: a cause for concern? Vaccine. 1999;17(7‐8):973‐978. [DOI] [PubMed] [Google Scholar]
- 55. Zuckerman J, Zuckerman A. Mutations of the surface protein of hepatitis B virus. Antiviral Res. 2003;60(2):75‐78. [DOI] [PubMed] [Google Scholar]
- 56. Banerjee A, Chandra PK, Datta S, et al. Frequency and significance of hepatitis B virus surface gene variant circulating among ‘antiHBc only’ individuals in Eastern India. J Clin Virol. 2007;40(4):312‐317. [DOI] [PubMed] [Google Scholar]
- 57. Thibault V, Servant‐Delmas A, Ly TD, Roque‐Afonso AM, Laperche S. Performance of HBsAg quantification assays for detection of Hepatitis B virus genotypes and diagnostic escape‐variants in clinical samples. J Clin Virol. 2017;89:14‐21. [DOI] [PubMed] [Google Scholar]
- 58. van de Klundert MAA, Cremer J, Kootstra NA, Boot HJ, Zaaijer HL. Comparison of the hepatitis B virus core, surface and polymerase gene substitution rates in chronically infected patients. J Viral Hepat. 2012;19(2):e34‐e40. [DOI] [PubMed] [Google Scholar]


