Abstract
Protein secondary structure prediction can provide important information for protein 3D structure prediction and protein functions. Deep learning offers a new opportunity to significantly improve prediction accuracy. In this paper, a new deep neural network architecture, named the Deep inception-inside-inception (Deep3I) network, is proposed for protein secondary structure prediction and implemented as a software tool MUFOLD-SS. The input to MUFOLD-SS is a carefully designed feature matrix corresponding to the primary amino acid sequence of a protein, which consists of a rich set of information derived from individual amino acid, as well as the context of the protein sequence. Specifically, the feature matrix is a composition of physio-chemical properties of amino acids, PSI-BLAST profile, and HHBlits profile. MUFOLD-SS is composed of a sequence of nested inception modules and maps the input matrix to either eight states or three states of secondary structures. The architecture of MUFOLD-SS enables effective processing of local and global interactions between amino acids in making accurate prediction. In extensive experiments on multiple datasets, MUFOLD-SS outperformed the best existing methods and other deep neural networks significantly. MUFold-SS can be downloaded from http://dslsrv8.cs.missouri.edu/~cf797/MUFoldSS/download.html.
Keywords: Protein Structure Prediction, Protein Secondary Structure, Deep Neural Networks, Deep Learning
1. INTRODUCTION
Protein tertiary (3D) structure prediction from amino acid sequence is a very challenging problem in computational biology1. Protein secondary structure prediction is an important step in reaching the goal. If secondary structure can be predicted accurately, the information is very useful for various tertiary structure related predictions, such as solvent accessibility prediction2, protein disorder prediction3, and protein tertiary structure prediction4. Protein secondary structures can also help identify protein function domains and may guide the rational design of site-specific mutation experiments5.
Protein secondary structure prediction is often evaluated by the Q3 accuracy - the accuracy of a three-class classification, i.e., helix (H), strand (E) and coil (C), which is simply the percentage of correctly predicted secondary structure positions in the protein sequences. In the 1980s, the Q3 accuracy was below 60%. In the 1990s, the Q3 accuracy reached 70% because of the use of additional protein evolutionary information, such as the position-specific score matrices (PSSM). Since then, the Q3 accuracy has gradually improved to around 80% on benchmark datasets. Another commonly used performance measure is the Q8 accuracy -the accuracy of an eight-class classification: 310-helix (G), α-helix (H), π-helix (I), β-strand (E), β-bridge (B), β-turn (T), bend (S), and loop or irregular (L)6.
The previous protein secondary structure methods can be divided into two main categories: template-based methods7 using known protein structures as templates or template-free methods8. Although template-based methods often perform better than template-free methods, in the Critical Assessment of Protein Structure Prediction (CASP), a community-wide experiment for protein structure prediction, typically no homologous templates could be found for hard protein targets and template-based methods did not work well. For template-free methods, several machine-learning methods, such as neural networks7, 8, hidden Markov chain9, support vector machine10 were used. Secondary structure prediction often uses features such as physicochemical propensities, solvent accessibilities and evolutionary information obtained from the protein sequence profiles as the input. Overall, the Q3 accuracy has gradually improved to 76–80% and the Q8 accuracy has gradually improved to over 70%7–10.
The earlier machine-learning methods for protein secondary structure prediction cannot effectively account for non-local interactions between residues that are close in the 3D space but far from each other in their sequence positions10. Many existing techniques rely on sliding windows of size 10–20 amino acid to capture some “short to intermediate” non-local interactions11–12. Recent deep-learning approach offers great potential over previous methods in handling long range non-local interactions. As shown in “Supplemental Materials“, our deep-learning method significantly outperformed methods using sliding windows, which only reflect local interactions, especially in the beta-protein category, whose secondary structures depends more on the long-range interactions.
The contributions of this work include: (1) A new very deep learning network architecture, Deep3I, was proposed to effectively process both local and global interactions between amino acids in making accurate secondary structure prediction. Extensive experimental results show that the new method outperforms the best existing methods and other deep neural networks significantly. (2) A software tool MUFOLD-SS was implemented based on the new method. This tool can predict the protein secondary structure fast and accurately.
Protein secondary structure prediction has been a very active research area in the past 30 years. A small improvement of its accuracy can have a significant impact on many related research problems and software tools.
Various machine learning methods, including different neural networks, have been used by previous protein secondary structure predictors. Over the years, the reported Q3 accuracy on benchmark datasets went from 69.7% by the PHD server13 in 1993, to 76.5% by PSIPRED14, to 80% by structural property prediction with integrated neural networks (SPINE)11, to 79–80% by SSPro using bidirectional recurrent neural networks15, and to 82% by structural property prediction with integrated deep neural network (SPIDER2) 3.
In recent years, significant improvement has also been achieved on Q8 accuracy by deep neural networks. In 16, deep convolutional neural networks integrated with a conditional random field was proposed and achieved 68.3% Q8 accuracy and 82.3% Q3 accuracy on the benchmark CB513 dataset. In 17, a deep neural network with multi-scale convolutional layers followed by three stacked bidirectional recurrent layers was proposed and reached 69.7% Q8 accuracy on the same test dataset. In 18, deep convolutional neural networks with next-step conditioning technique were proposed and obtained 71.4% Q8 accuracy. In 12, long short term memory bidirectional recurrent neural networks (LSTM-bRNN) were applied to this problem and a tool named SPIDER3 was developed, achieving 84% Q3 accuracy and a reduction of 5% and 10% in the mean absolute error for Psi-Phi angle prediction on the dataset used in their previous studies 3. These successes encouraged us to explore more advanced deep-learning architectures. The current work improves upon the previous methods through the development and application of new neural network architectures, including Residual networks19, inception networks20, and Batch Normalization21. Other than the prediction accuracy, secondary structure prediction provides an ideal testbed for exploring and testing these state-of-the-art deep learning methods.
2. MATERIALS AND METHODS
2.1. Problem formulation
Protein secondary structure prediction is that given the amino acid sequence, also called primary sequence, of a protein, predict the secondary structure type of each amino acid. In three-class classification, the secondary structure type is one out of three, helix (H), strand (E) and coil (C). In eight-class classification, the secondary structure type is one out of eight: (G, H, I, E, B, T, S, L).
To make accurate prediction, it is important to provide useful input features to machine learning methods. In our method, we carefully design a feature matrix corresponding to the primary amino acid sequence of a protein, which consists of a rich set of information derived from individual amino acid, as well as the context of the protein sequence.
Specifically, the feature matrix is a composition of physio-chemical properties of amino acids, PSI-BLAST profile, and HHBlits profile. Each amino acid in the protein sequence is represented as a vector of 8 real numbers ranging from −1 to 1. The vector consists of the seven physio-chemical properties as in 22 plus a number of value 0 or 1 representing the existence of an amino acid at this position as an input (called NoSeq label). The reason of adding the NoSeq label is because the proposed deep neural networks are designed to take a fixed size input, such as a sequence of length 700 in our experiment. Then, to run a protein sequence shorter than 700 through the network, the protein sequence will be padded at the end with 0 values and the NoSeq label is set to 1. For example, if the length of a protein is 500, then the first 500 rows have NoSeq label set to 0, and the last 200 rows have 0 values in the first 7 columns and 1 in the last column.
The second set of useful features comes from the protein profiles generated using PSI-BLAST 23. In our experiments, the PSI-BLAST software parameters were set to (evalue: 0.001; num_iterations: 3; db: UniRef50) to generate PSSM, which was then transformed by the sigmoid function into the range (0, 1). Each amino acid in the protein sequence is represented as a vector of 21 real numbers ranging from 0 to 1, representing the 20 amino acids PSSM value plus a NoSeq label in the last column. The feature vectors of the first 5 amino acids are shown. The third through nineteenth columns are omitted.
The third set of useful features comes from the protein profiles generated using HHBlits24. To have a fair comparison with tools published previously, in our experiments, the HHBlits software used an older version of the sequence database, uniprot20_2013_03, which can be downloaded from http://wwwuser.gwdg.de/~compbiol/data/hhsuite/databases/hhsuite_dbs/. Again, the profile values were transformed by the sigmoid function into the range (0, 1). Each amino acid in the protein sequence is represented as a vector of 31 real numbers, of which 30 from amino acids HHM Profile values and 1 NoSeq label in the last column.
The three sets of features can be combined into one feature vector of length 58 for each amino acid as input to our proposed deep neural networks. For the deep neural networks that take fixed size input, e.g., sequence of length 700, the input feature matrix would be of size 700 by 58. If a protein is shorter than 700, the input matrix would be padded with NoSeq rows at the end to make it full size. If a protein is longer than 700, it would be split into segments less than 700.
For our new deep neural networks, the predicted secondary structure output of a protein sequence is represented as a fixed size matrix, such as a 700 by 9 matrix for 8-state labels plus 1 NoSeq label) matrix or a 700 by 4 matrix for 3-state labels plus 1 NoSeq label. One of the first 8 columns have a value 1 (one hot encoding of the target class), while the others are 0.
2.2. New deep inception networks for protein secondary structure prediction
In this section, a new deep inception network architecture is proposed for protein secondary structure. The architecture consists of a sequence of inception modules followed by a number of convolution and fully connected dense layers.
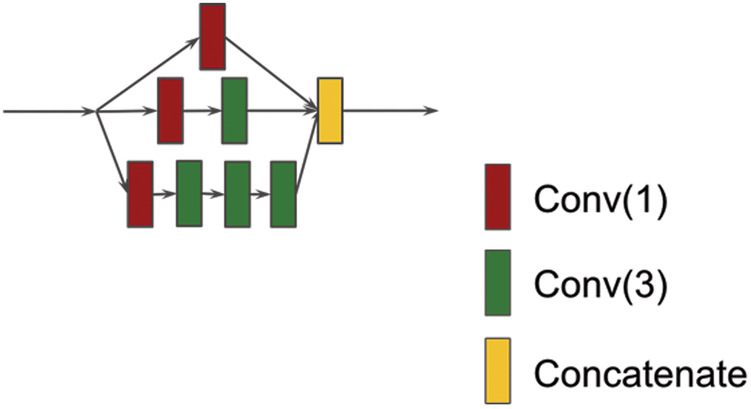
Figure 1 shows the basic inception module 20. The inception model was used for image recognition and achieved the state-of-the-art performance. It consists of multiple convolution operations (usually with different convolutional window sizes) in parallel and concatenates the resulting feature maps before going into next layer. Usually, different convolutional window sizes can be 1×1, 3×3, 5×5, 7×7 along with a 3×3 max pooling (see Supplemental Materials). The 1×1 convolution is used to reduce the feature map dimensionality and the max pooling is used for extracting image region features. In our protein secondary structure prediction experiment, the max pooling is not suitable because the sequence length will change and hence it was not applied in our study. The inception module consists of several parallel convolutional networks that can extract diverse features from input. In this work, an inception network is applied to extract both local and non-local interactions between amino acids and a hierarchical layer of convolutions with small spatial filters is used to be computationally efficient. By stacking convolution operations on top of one another, the network has more ability to extract non-local interaction of residues.
Figure 1.
An Inception module. The red square “Conv(1)” stands for convolution operation using window size 1 and the number of filters is 100. The green square “Conv(3)” stands for convolution operation using window size 3 and the number of filters is 100. The yellow square “Concatenate” stands for feature concatenation.
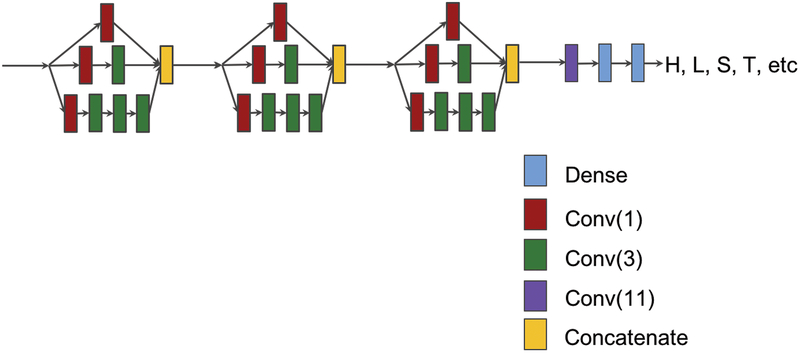
A deep inception network consists of a sequence of inception modules. Figure 2 shows a network with three inception modules followed by a convolutional layer and two dense layers. A dense layer is a fully connected neural network layer. The input of the network is the feature matrix for a protein sequence, such as a matrix of size 700 by 58, and the output is the target label matrix. Different numbers of inception modules were tried in our experiments to find appropriate values for our prediction tasks. The deep neural networks were implemented, trained, and experimented using TensorFlow25 and Keras26 (https://github.com/fchollet/keras).
Figure 2.
A deep inception network consisting of three inception modules, followed by one convolution and two fully-connected dense layers.
2.3. New deep inception-inside-Inception networks for protein secondary structure prediction (Deep3I)
In this section, a new deep inception-inside-inception network architecture (Deep3I) is presented. The architecture extends deep inception networks through nested inception modules.
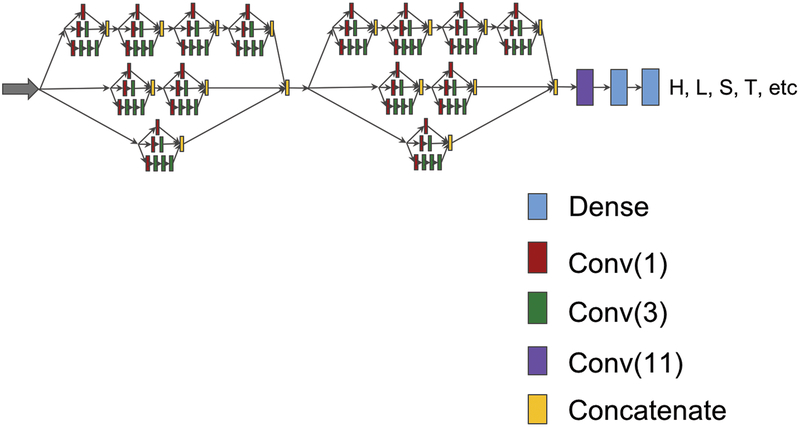
Figure 3 shows a deep inception-inside-inception network (Deep3I) consisting of two Deep3I blocks, followed by a convolutional layer and two dense layers. A Deep3I block is constituted by a recursive construction of an inception model inside another Inception module. Stacked inception modules could extract non-local interactions of residues over a diverse range in a more effective way. Adding more Deep3I blocks is possible but requires more memory.
Figure 3.
A deep inception-inside-inception (Deep3I) network that consists of two Deep3I blocks. Each Deep3I block is a nested inception network self. This network was used to generate the experimental results reported in this paper.
Each convolution layer, such as ‘Conv (3)’ in Figure 3, consists of four operations in sequential order: 1) A one- dimensional convolution operation using the kernel size of three; 2) The Batch normalization technique21 for speeding up the training process and acting as a regularizer; 3) The activation operation, ReLU 27; and 4) The Dropout operation 28 to prevent the neural network from overfitting by randomly dropping neurons during the deep network training process so that the network can avoid co-adapting.
Deep3I networks were implemented, trained, and experimented using TensorFlow and Keras. In our experiments, a large number of network parameters and training parameters were tried. For the results reported, the dropout rate was set at 0.4. During training, the learning rate scheduler from Keras was used to control the learning rate. The initial learning rate was 0.0005, and after every 40 epochs, the learning rate dropped as shown by the following formulas: lrate = 0.0005 * pow (0.8, floor(epoch + 1)/40). An early stopping mechanism from Keras was used to stop the network training when the monitored validation quantity (such as validation loss and/or validation accuracy) stopped improving. The “patience” (number of epochs with no improvement after which training was stopped) was set as a number from 5 to 8. TensorBoard from Keras was used to visualize dynamic graphs of the training and validation metrics.
The new Deep3I networks are different from existing deep neural networks. The networks in 17–18 consist of residual blocks and a multi-scale layer containing convolutional layers with convolution window sizes of 3, 7, and 9 to discover the protein local and global context. Although Deep3I uses convolution window size of 1 or 3, through stacked deep convolution blocks, the network can represent both local and global context well, while maintains efficient computation.
3. RESULTS AND DISCUSSION
In this section, extensive experimental results of the proposed deep neural networks on multiple commonly used datasets and performance comparison with existing methods are presented. Additional benchmark comparisons can be found in Supplemental Materials.
The following five publically available datasets were used in our experiments:
-
1)
CullPDB dataset. CulPDB29 was downloaded on 15 June 2017 with 30% sequence identity cutoff, resolution cutoff 3.0Å and R-factor cutoff 0.25. The raw CullPDB dataset contains 10,423 protein sequences. All sequences with length shorter than 50 or longer than 700 were filtered out, which left the remaining 9972 protein sequences. CD-HIT30 was then used to remove similar sequences between CullPDB and CASP10, CASP11 and CSAP12 datasets. After that, there were 9589 proteins left. Among them, 8 protein sequences were ignored because they are too short to generate PSI-BLAST profile. For the final 9581 proteins, 9000 were randomly selected as the training set and the rest 581 as the validation set.
-
2)
JPRED dataset5. All proteins from the JPRED dataset belong to different super-families, which gives the experimental result a more objective evaluation.
-
3)
CASP datasets. CASP10, CASP11 and CASP12 datasets were downloaded from the official CASP website http://predictioncenter.org. The Critical Assessment of protein Structure Prediction, or CASP, is a bi-annual worldwide experiment for protein structure prediction since 1994. The CASP datasets have been widely used in the bioinformatics community. To get the secondary structure labels of the proteins, the DSSP program6 was used. Some of the PDB files (including T0675, T0677 and T0754) could not generate the DSSP result; so, they were discarded. Some protein sequences (including T0709, T0711, T0816 and T0820) are too short and PSI-BLAST could not generate profiles; so, they were also removed. Finally, 98 out of 103 in CASP10, 83 out of 85 in CASP11 and 40 out of 40 in CASP12 were used as our CASP dataset.
-
4)
CB513 benchmark4. This benchmark has been widely used for testing and comparing the performance of secondary structure predictors.
-
5)
Most recently PDB (Protein Data Bank). Because training data used in different tools are not same, it is important to use PDB files that have not been seen by any previous predictors (including ours) to objectively evaluate the performance. For this purpose, the most recently published protein PDB files dated from 2017-7-1 to 2017-8-15 were downloaded. This set contains 614 proteins, each of which share less than 30% sequence identity. Then, 385 proteins with a length between 50 and 700 were kept. To perform a more objective test, each of the 385 proteins was searched against CullPDB using BLAST and was classified into two categories: 1) easy cases where e-value is less than or equal to 0.5; and 2) hard cases which can have no hit or e-value higher than 0.5. After the classification, there are 270 easy cases and 115 hard cases.
In our experiments, the Deep3I configuration as shown in Fig. 3 was used to generate the results reported in this paper. In most cases, the CullPDB dataset was used to train various deep neural networks, while the other datasets were only used in testing and performance comparison with other methods and existing results.
3.1. PSI-BLAST vs. DELTA-BLAST
When generating the sequence profiles, most researchers 4,14–16 chose PSI-BLAST23, which can reliably generate a good protein sequence profiles. Besides PSI-BLAST, other profile search tools are available, such as CS-BLAST31, DELTA_BLAST32, PHI-BLAST33, etc. In this work, the performance between two popular tools were compared: PSI-BLAST vs. DELTA-BLAST on the JPRED dataset to decide which tool to use to generate the input feature vectors for the new Deep3I network. For the PSI-BLAST experiment: both training and test data profiles were generated by setting the e-value at 0.001, num_iterations at 3 and db is UniRef50. The JPRED dataset itself contains a training set of 1348 protein sequences and a test set of 149 protein sequences. The JPRED training dataset was used to train the Deep3I network and the JPRED test dataset was used to report the prediction Q3. The Deep3I network were trained five times from random initial weights, and the average Q3 accuracy and standard deviation are reported in Table 1. The same procedure was used for DELTA-BLAST with its default search database (2017/4/1, with a size of 8.7GB). The comparison of their results is shown in Table 1. Even though the average profile generation time of DELTA-BLAST is faster than PSI-BLAST, the Q3 accuracy of MUFOLD-SS using DELTA-BLAST is not as high as using PSI-BLAST. Hence, PSI-BLAST was chosen to generate profiles in the rest of our experiments.
Table 1.
Comparison of the profile generation execution time and MUFOLD-SS Q3 accuracy using PSI-BLAST and DELTA-BLAST
| Tool | average profile generation time (minutes) | Q3 accuracy % |
|---|---|---|
| PSI-BLAST* | 16.8(+/−10.14) | 84.21(+/−0.49) |
| DELTA-BLAST | 7.56(+/−3.96) | 83.63(+/−0.31) |
Database for PSI-BLAST search was UniRef50 download on 2017/4/12, 8.7GB.
3.2. MUFOLD-SS vs. the Best Existing Methods
Many previous researchers14–16 benchmarked their tools or methods using the dataset CB5134. For a fair comparison, the same CullPDB dataset was used to train and test MUFOLD-SS in the same way as for existing methods reported in previous publications.
Table 2 shows the performance comparison between MUFOLD-SS and four of the best existing methods (SSPro, DeepCNF-SS, DCRNN, and DCNN) in terms of the Q8 prediction accuracy on the CB513 dataset. All these methods were compared under the condition of the same input and features. In this case, only the protein sequence and PSI-BLAST profiles were provided. No additional technique like the next-step condition proposed in 18 was involved. SSPro was run without using template information. The result shows MUFOLD-SS obtained the highest accuracy.
Table 2.
Q8 accuracy (%) comparison between MUFOLD-SS and existing state-of-the-art methods using the same sequence and profile of benchmark CB513.
| Tool | CB513 |
|---|---|
| SSPro (Cheng et al., 2005) | 63.5* |
| DeepCNF-SS (Wang et al., 2016) | 68.3* |
| DCRNN (Li and Yu, 2016) | 69.7* |
| DCNN (Busia et al., 2017) | 70.0** |
| MUFOLD-SS | 70.63 |
results reported by (Wang et al., 2016)
results reported by (Busia et al., 2017)
Tables 3 and 4 show the performance comparison between MUFOLD-SS and the best existing methods (SSPro, PSIPRED, DeepCNF-SS) in terms of the Q3 prediction accuracy on the CASP datasets. PSIPRED could not predict 8-class classification, and thus do not have result on Q8 accuracy.
Table 3.
Q3 accuracy (%) comparison between MUFOLD-SS and other state-of-the-art methods on CASP datasets.
| CASP10 | CASP11 | CASP12 | |
|---|---|---|---|
| SSpro (w/o template) * | 78.5 | 77.6 | 76.0** |
| SSpro(w/ template) * | 84.2 | 78.4 | 76.6** |
| PSIPRED* | 81.2 | 80.7 | 80.4** |
| DeepCNF-SS** | 83.22 | 82.22 | 81.30 |
| SPIDER3** | 82.6 | 81.5 | 79.87 |
| MUFOLD-SS ** | 86.49 | 85.20 | 83.36 |
results reported by (Wang et al., 2016)
experiment results we conducted
Table 4.
Q8 accuracy (%) comparison between MUFOLD-SS and other state-of-the-art methods on CASP datasets.
| CASP10 | CASP11 | CASP12 | |
|---|---|---|---|
| SSpro(w/o template)* | 64.9 | 65.6 | 63.1** |
| SSpro (w/ template)* | 75.9 | 66.7 | 64.1** |
| DeepCNF-SS** | 72.81 | 71.64 | 69.76 |
| MUFOLD-SS** | 76.47 | 74.51 | 72.1 |
results reported by (Wang et al., 2016)
experiment results we conducted
For all methods, their prediction accuracies on the CASP10 and CASP11 datasets are generally higher than their accuracies on the CASP12 dataset. This is because CASP12 contains more hard cases and the profiles generated are not as good as for those in CASP10 and CSAP11 datasets. Furthermore, MUFOLD-SS outperformed SSPro with template in in all cases, even though SSPro used the template information from similar proteins, whereas MUFOLD-SS did not.
3.3. Effects of Hyper-Parameters of MUFOLD-SS
Tables 5 and 6 show the Q3 and Q8 accuracy comparison between MUFOLD-SS with 1 or 2 blocks and the proposed Deep Inception Networks with 1 to 4 inception modules. The results of Deep Inception Networks with different number of modules are similar, whereas MUFOLD-SS with two blocks is slightly better than that with one block. Both Deep3I networks performed better than Deep inception networks. The reason may be that the ability of inception network in capturing non-local interaction between residues is not as good as MUFOLD-SS. MUFOLD-SS consists of integrated hierarchical inception modules, which gives more ability to extract high level features, i.e. non-local interactions of residues.
Table 5.
Q3 accuracy (%) comparison between MUFOLD-SS with different number of blocks and Deep Inception Networks with different number of modules.
| # of modules | CASP10 | CASP11 | CASP12 | |
|---|---|---|---|---|
| Deep Inception | 1 | 86.05 | 84.13 | 82.48 |
| 2 | 86.12 | 84.31 | 82.80 | |
| 3 | 86.03 | 84.29 | 82.69 | |
| 4 | 86.02 | 84.44 | 82.67 | |
| MUFOLD-SS | 1 | 86.51 | 84.56 | 82.94 |
| 2 | 86.49 | 85.20 | 83.36 |
Table 6.
Q8 accuracy (%) comparison between MUFOLD-SS with different number of blocks and Deep Inception Networks with different number of modules.
| # of modules | CASP10 | CASP11 | CASP12 | |
|---|---|---|---|---|
| Deep Inception | 1 | 75.41 | 73.12 | 71.22 |
| 2 | 75.70 | 73.26 | 71.55 | |
| 3 | 75.14 | 73.33 | 71.31 | |
| 4 | 75.23 | 73.37 | 71.42 | |
| MUFOLD-SS | 1 | 76.03 | 73.74 | 71.54 |
| 2 | 76.47 | 74.51 | 72.1 |
3.4. Results Using Most Recently Released PDB Files
The purpose of this test was to conduct a real-world testing, just like CASP, by using the most recently published PDB protein files dated from 2017-7-1 to 2017-8-15. These PDB files had not been viewed by any previous predictors. As mentioned before, the most recently released PDB proteins were classifier into two categories: the 270 easy cases and 115 hard cases.
MUFOLD-SS is compared with the best available tools in this field, including PSIPRED, SSPro, and the newly developed SPIDER3 on this set of data. Some protein files in this dataset has special amino acid like ‘B’, which causes SPIDER3 prediction failure. To make a fair comparison, proteins containing those special cases were excluded (20 hard cases and 44 easy cases in total) and the remaining proteins were used as test cases to compare the four tools. Again, MUFOLD-SS was trained using the CullPDB dataset and tested on the new dataset.
Table 7 shows the performance comparison of the 4 tools. Again, MUFOLD-SS outperformed the other state-of-the-art tools in both easy and hard cases. Different from traditional neural networks that use a sliding window of neighbor residues to scan over a protein sequence in making prediction, MUFOLD-SS takes the entire protein sequence (up to 700 amino acids) as the input and processes local and global interactions simultaneously. It is remarkable that MUFOLD-SS can outperform SPIDER3 in both easy and hard cases. Another advantage of MUFOLD-SS is that it can generate prediction for 3-class and 8-class classification at the same time, while some other tools like Psipred and SPIDER3 can predict only 3-class or 8-class classification. Last but not the least, MUFOLD-SS can even outperform SSPro with template in hard cases. (For hard cases, SSPro with template get 81.88% in Q3 and 71.8% in Q8.
Table 7.
Q3 (%) and Q8 (%) compared with MUFOLD-SS and other state-of-the-art tools using recently PDB.
| Easy case | Hard case | |||
|---|---|---|---|---|
| Q3 | Q8 | Q3 | Q8 | |
| PSIPRED | 82.55 | N/A | 80.76 | N/A |
| SSPro | 78.76 | 66.53 | 76.01 | 63.14 |
| SPIDER3 | 86.23 | N/A | 82.64 | N/A |
| MUFOLD-SS | 88.20 | 78.65 | 83.37 | 72.84 |
4. CONCLUSION AND FUTURE WORK
In this work, a new deep neural network architecture Deep3I was proposed for protein secondary structure prediction. Extensive experimental results show that Deep3I’s implementation MUFOLD-SS obtained more accurate predictions than the best state-of-the-art methods and tools. The experiments were designed carefully, and the datasets used in training, e.g., the CullPDB dataset, were processed to remove any significantly similar sequences with the test sets using CD-HIT to avoid any bias. Compared to previous deep-learning methods for protein secondary structure prediction, this work uses a more sophisticated, yet efficient, deep-learning architecture. MUFOLD-SS utilizes hierarchical deep Inception blocks to effectively process local and non-local interactions of residues. It will provide the research community a powerful prediction tool for secondary structures.
In the future work, we will apply MUFOLD-SS to predict other protein structure related properties, such as backbone torsion angles, solvent accessibility, contact number, and protein order/disorder region. These predicted features are also useful for protein tertiary structure prediction and protein model quality assessment.
Supplementary Material
Table 8.
Comparison of mean absolute errors (MAE) of Psi-Phi angle prediction between MUFOLD-SS and SPIDER3.
| Angle | CASP10 | CASP11 | CASP12 | |
|---|---|---|---|---|
| SPIDER3 | Psi | 31.13 | 33.04 | 35.60 |
| Phi | 19.26 | 20.45 | 21.12 | |
| MUFOLD-SS | Psi | 25.74 | 27.18 | 30.16 |
| Phi | 17.62 | 18.88 | 19.64 |
5. Acknowledgements
This work was partially supported by National Institutes of Health Grant R01-GM100701. The high-performance computing infrastructure was partially supported by National Science Foundation under grand number CNS-1429294.
Grant sponsor: NIH Grant number: R01GM100701; NSF Grant number: CNS1429294
6. REFERENCES
- 1.Yaseen A and Li Y, 2014. Context-based features enhance protein secondary structure prediction accuracy. Journal of chemical information and modeling, 54(3), pp.992–1002. [DOI] [PubMed] [Google Scholar]
- 2.Adamczak R, Porollo A and Meller J, 2004. Accurate prediction of solvent accessibility using neural networks–based regression. Proteins: Structure, Function, and Bioinformatics, 56(4), pp.753–767. [DOI] [PubMed] [Google Scholar]
- 3.Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, Sattar A, Yang Y and Zhou Y, 2015. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Scientific reports, 5, p.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J and Troyanskaya O, 2014, January Deep supervised and convolutional generative stochastic network for protein secondary structure prediction. In International Conference on Machine Learning (pp. 745–753). [Google Scholar]
- 5.Drozdetskiy A, Cole C, Procter J and Barton GJ, 2015. JPred4: a protein secondary structure prediction server. Nucleic acids research, 43(W1), pp.W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabsch W and Sander C, 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22(12), pp.2577–2637. [DOI] [PubMed] [Google Scholar]
- 7.Yaseen A and Li Y, 2014. Context-based features enhance protein secondary structure prediction accuracy. Journal of chemical information and modeling, 54(3), pp.992–1002. [DOI] [PubMed] [Google Scholar]
- 8.Pollastri G and Mclysaght A, 2004. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics, 21(8), pp.1719–1720. [DOI] [PubMed] [Google Scholar]
- 9.Asai K, Hayamizu S and Handa KI, 1993. Prediction of protein secondary structure by the hidden Markov model. Bioinformatics, 9(2), pp.141–146. [DOI] [PubMed] [Google Scholar]
- 10.Ward JJ, McGuffin LJ, Buxton BF and Jones DT, 2003. Secondary structure prediction with support vector machines. Bioinformatics, 19(13), pp.1650–1655. [DOI] [PubMed] [Google Scholar]
- 11.Dor O and Zhou Y, 2007. Achieving 80% ten-fold cross-validated accuracy for secondary structure prediction by large-scale training. Proteins: Structure, Function, and Bioinformatics, 66(4), pp.838–845. [DOI] [PubMed] [Google Scholar]
- 12.Heffernan R, Yang Y, Paliwal K and Zhou Y, 2017. Capturing Non-Local Interactions by Long Short Term Memory Bidirectional Recurrent Neural Networks for Improving Prediction of Protein Secondary Structure, Backbone Angles, Contact Numbers, and Solvent Accessibility. Bioinformatics, p.btx218. [DOI] [PubMed] [Google Scholar]
- 13.Rost B and Sander C, 1993. Prediction of protein secondary structure at better than 70% accuracy. Journal of molecular biology, 232(2), pp.584–599. [DOI] [PubMed] [Google Scholar]
- 14.Jones DT, 1999. Protein secondary structure prediction based on position-specific scoring matrices. Journal of molecular biology, 292(2), pp.195–202. [DOI] [PubMed] [Google Scholar]
- 15.Cheng J, Randall AZ, Sweredoski MJ and Baldi P, 2005. SCRATCH: a protein structure and structural feature prediction server. Nucleic acids research, 33(Suppl. 2), pp.W72–W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Peng J, Ma J and Xu J, 2016. Protein secondary structure prediction using deep convolutional neural fields. Scientific reports, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z and Yu Y, 2016. Protein secondary structure prediction using cascaded convolutional and recurrent neural networks. arXiv preprint arXiv:1604.07176. [Google Scholar]
- 18.Busia A and Jaitly N, 2017. Next-Step Conditioned Deep Convolutional Neural Networks Improve Protein Secondary Structure Prediction. arXiv preprint arXiv:1702.03865. [Google Scholar]
- 19.He K, Zhang X, Ren S and Sun J, 2016. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770–778). [Google Scholar]
- 20.Szegedy C, Ioffe S, Vanhoucke V and Alemi AA, 2017. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In AAAI (pp. 4278–4284). [Google Scholar]
- 21.Ioffe S and Szegedy C, 2015, June Batch normalization: Accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning (pp. 448–456). [Google Scholar]
- 22.FAUCHÈRE JL, Charton M, Kier LB, Verloop A, and Pliska V (1988). Amino acid side chain parameters for correlation studies in biology and pharmacology. Chemical Biology & Drug Design, 32(4), 269–278. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W and Lipman DJ, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research, 25(17), pp.3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remmert M, Biegert A, Hauser A and Söding J, 2012. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature methods, 9(2), pp.173–175. [DOI] [PubMed] [Google Scholar]
- 25.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M and Ghemawat S, 2016. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467. [Google Scholar]
- 26.Chollet F, 2015. Keras.
- 27.Nair V and Hinton GE, 2010. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th international conference on machine learning (ICML-10) (pp. 807–814). [Google Scholar]
- 28.Srivastava N, Hinton GE, Krizhevsky A, Sutskever I and Salakhutdinov R, 2014. Dropout: a simple way to prevent neural networks from overfitting. Journal of machine learning research, 15(1), pp.1929–1958. [Google Scholar]
- 29.Wang G and Dunbrack RL Jr, 2003. PISCES: a protein sequence culling server. Bioinformatics, 19(12), pp.1589–1591. [DOI] [PubMed] [Google Scholar]
- 30.Fu L, Niu B, Zhu Z, Wu S and Li W, 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28(23), pp. 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biegert A and Söding J, 2009. Sequence context-specific profiles for homology searching. Proceedings of the National Academy of Sciences, 106(10), pp.3770–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ and Madden TL, 2012. Domain enhanced lookup time accelerated BLAST. Biology direct, 7(1), p.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Miller W, Schäffer AA, Madden TL, Lipman DJ, Koonin EV and Altschul SF, 1998. Protein sequence similarity searches using patterns as seeds. Nucleic acids research, 26(17), pp.3986–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.