Abstract
In the present report, we carried out clinical-scale editing in adult mobilized CD34+ hematopoietic stem and progenitor cells (HSPCs) using zinc-finger nuclease-mediated disruption of BCL11a to upregulate the expression of γ-globin (fetal hemoglobin). In these cells, disruption of the erythroid-specific enhancer of the BCL11A gene increased endogenous γ-globin expression to levels that reached or exceeded those observed following knockout of the BCL11A coding region without negatively affecting survival or in vivo long-term proliferation of edited HSPCs and other lineages. In addition, BCL11A enhancer modification in mobilized CD34+ cells from patients with β-thalassemia major resulted in a readily detectable γ-globin increase with a preferential increase in G-gamma, leading to an improved phenotype and, likely, a survival advantage for maturing erythroid cells after editing. Furthermore, we documented that both normal and β-thalassemia HSPCs not only can be efficiently expanded ex vivo after editing but can also be successfully edited post-expansion, resulting in enhanced early in vivo engraftment compared with unexpanded cells. Overall, this work highlights a novel and effective treatment strategy for correcting the β-thalassemia phenotype by genome editing.
Keywords: hemoglobinopathies, BCL11a, genome editing, thalassemia, HbF reactivation, zinc finger nucleases
Introduction
Patients with severe β-globin thalassemias require life-long red blood cell transfusions along with iron chelation treatment. The only curative option is allogeneic hematopoietic cell transplantation, available only to a subset of patients having a suitable donor.1 For patients without an allogeneic hematopoietic cell transplantation option, recent molecular approaches employing gene-modified autologous cells are emerging as therapeutic alternatives. Gene addition protocols through viral vectors, using autologous stem cells, have been studied in vitro for the last 20 years, and since 2006, several small clinical trials have been either completed or initiated.2, 3 These trials offer clear promise with significant clinical improvement in a proportion of patients.
Although innovative protocols for capturing mainly highly purified genetically engineered cells transduced with lentiviral vectors have been proposed to improve efficiency4, significant challenges remain, and the long-term safety risks associated with the use of viral vectors are being currently evaluated. Apart from β- or γ-globin gene addition in hematopoietic stem cells by traditional lentiviral vectors, several other protocols aiming to increase or reactivate fetal hemoglobin (HbF) have been explored because this outcome would be applicable for amelioration of all β-globin disorders, irrespective of the genetic mutation. Tested strategies for HbF reactivation involve either forced chromatin looping, mediated by a lentiviral vector expressing the looping factor LDB1 linked to a zinc-finger protein binding the γ-globin promoter,5 the re-creation of γ-globin promoter mutations leading to hereditary persistence of HbF (HPFH) conditions,6, 7 or downregulation or inhibition of trans-factors involved in γ-globin silencing8 or disruption of their binding regions.9, 10, 11 Attempts to faithfully recreate HPFH γ-globin promoter mutations through gene editing have been associated with low efficiency in normal adult CD34+ cells ex vivo because less efficient homology- or microhomology-mediated repair is required.6, 7 By contrast, attempts to induce nuclease-mediated inactivation of HbF “silencers” (i.e BCL11A,12, 13, 14 LRF,8 or KLF115) can be more efficient because they require repair of DSBs with non-homologous end joining (NHEJ). One of several approaches targeting HbF reawakening is suppression of an important developmental silencer of γ-globin, BCL11A. This approach has shown efficiency in all previous studies, and it is favored for several reasons. BCL11A inactivation through NHEJ repair is more efficient than the homology-mediated repair needed to recreate known HPFH sites in the γ-globin promoter, and inactivation of the erythroid-specific BCL11a enhancer13, 14, 16 alleviates concerns about the effect of BCL11a inactivation in non-erythroid cells because BCL11A deficiency in murine models has been associated with impaired B lymphocyte and hematopoietic stem cell (HSC) development17, 18 or about influencing aspects of erythroid differentiation/maturation raised by targeting other transcription factors involved in silencing (i.e KLF1 and ZBTB7A). Finally, it is known that patients with BCL11A haploinsufficiency have increased levels of HbF at levels likely to be therapeutic for patients with β-globin disorders.19, 20 Lentivirus-mediated erythroid-specific expression of a BCL11A shRNA reflects an additional efficient strategy for HbF upregulation,21, 22 although this approach may be hampered by cytotoxicity triggered via endogenous microRNA dysregulation23, 24 and is coupled with theoretical long-term concerns of lentiviral vector genotoxicity and silencing because of genomic position effects.25
In the present study, we explored the functional consequences of zinc-finger nuclease (ZFN)-induced inactivation of the BCL11A-erythroid enhancer in both normal and thalassemic mobilized CD34+ cells in vitro and in vivo, before and after implementation of our expansion protocols,26 to enhance engraftment of edited HSCs. A similar approach using an updated ZFN architecture is currently being evaluated in a phase 1/2 clinical trial.
Results
Efficient Editing of the BCL11a Enhancer and BCL11a Exon2 in Adult Mobilized CD34+ Cells
In the first set of our experiments, we sought to provide detailed comparisons of the effect of disrupting BCL11A with early-stage ZFN pairs targeting either BCL11A exon 2 or the BCL11A erythroid enhancer using mobilized peripheral blood CD34+ cells under clinical-scale conditions because these cells represent the optimal cell source for gene modification therapies for β-thalassemia.27, 28
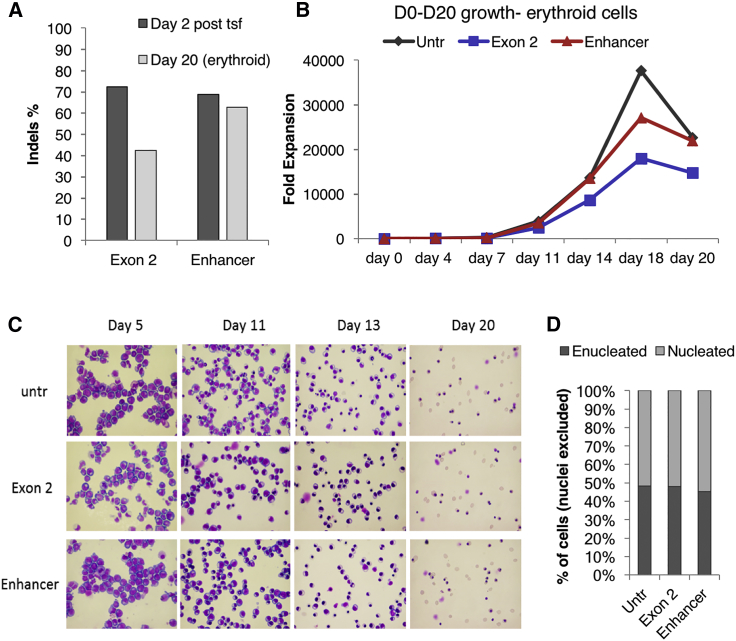
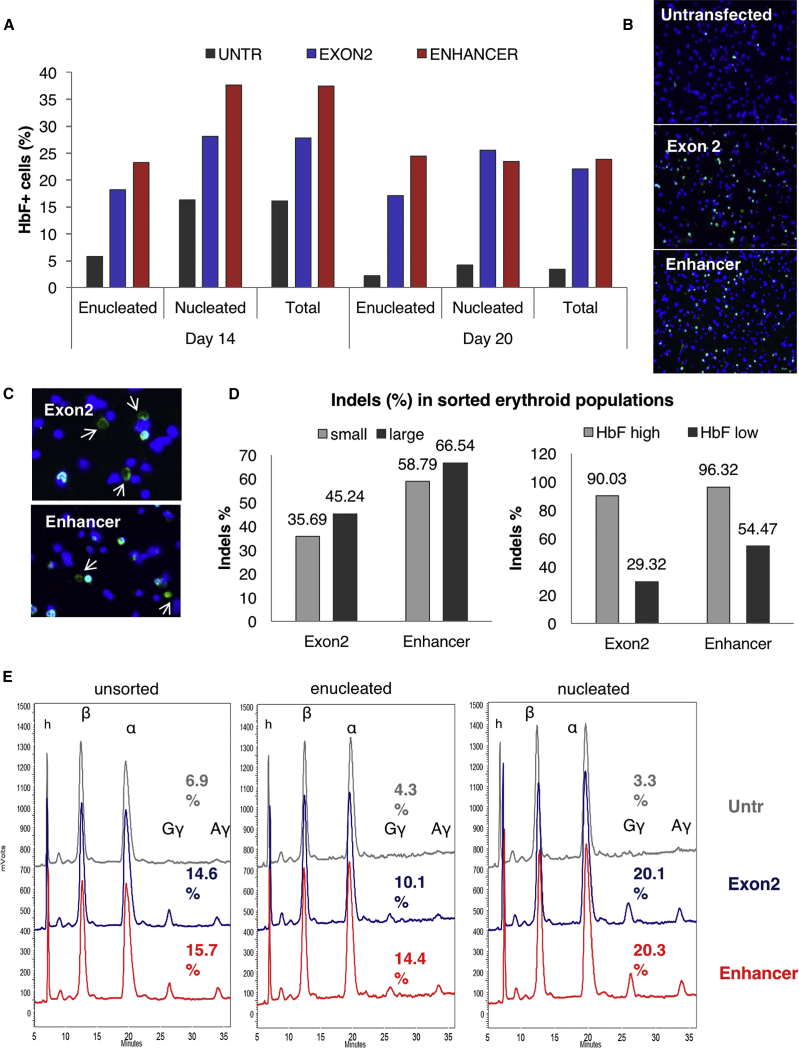
Peripheral blood CD34+ cells from a plerixafor and granulocyte colony-stimulating factor (G-CSF)-mobilized healthy donor were electroporated at clinical scale with ZFN mRNA targeting either BCL11A exon 2 or the BCL11A erythroid enhancer (Figure S1A). Gene modification levels, as assessed by high-throughput DNA sequencing 2 days after electroporation, were 72% for the BCL11A exon 2-modified cells and 68% for the BCL11A enhancer-modified cells (Figure 1A). To assess the effects of genome editing on erythropoiesis, we used a well-established protocol of in vitro erythroid differentiation.29 First we evaluated possible effects of mRNA-ZFN electroporation and genome editing on growth kinetics and in vitro terminal erythroid maturation. Disruption of the BCL11A erythroid enhancer did not measurably affect erythroid differentiation and/or maturation because cell growth, morphology, and enucleation rate were not obviously different from the untransfected sample at all time points tested (Figures 1B–1D, respectively). Expression of the erythroid-specific surface markers glycophorin A and E-cadherin was almost identical between the edited and unedited samples throughout the maturation process (Figure S2), suggesting concurrent differentiation among the three different samples. However, the BCL11A coding region knockout (KO) had a negative effect on cell growth and survival during maturation, as shown by the decreased total cell yield (Figure 1B). In line with this outcome, the total insertion or deletion (indel) levels tended to decrease in BCL11A exon 2 KO erythroid progenitors (42% versus 72% prior to differentiation) throughout the culture. In contrast to BCL11a KO, in enhancer KO, the frequency of edited cells was retained throughout differentiation (63% versus 68% prior to differentiation) (Figure 1A). The effect on HbF reactivation was evaluated by both fluorescence-activated cell sorting (FACS) and immunocytochemistry, and the γ-globin protein levels were assessed by HPLC. The frequency of HbF+ cells detected by FACS and immunofluorescence was significantly increased in both exon 2- and enhancer-edited samples compared with the control (∼25% versus <5% HbF+ cells in enucleated red blood cells, respectively) (Figures 2A and 2B). HbF+ mature enucleated red blood cells were present in both exonic and enhancer BCL11A edited samples, as shown by immunocytochemistry (Figure 2C). The differentiated progeny of the transfected cells was sorted based on γ-globin levels and cell size or maturation level, and the sorted fractions were sequenced for evaluation of indel levels (Figure S2). No evident difference was observed in the indel percentage between the smaller, more mature and larger, less differentiated cells of either group (Figure 2D). More than 95% of the enhancer-targeted alleles were edited in the HbF-high cells, suggesting that BCL11A modification was biallelic in almost every cell with very high γ-globin levels (Figure 2D). As expected, almost no high γ-globin-expressing cells were found in the untransfected sample (data not shown). Reverse-phase HPLC, separating the globin polypeptides, was performed in enucleated and nucleated erythroid cells. Both exon 2- and enhancer-edited nucleated cells presented a 6-fold increase in γ-globin expression (Gγ+Αγ); however, in the enucleated cells, the enhancer ZFN effect was greater than that of the exon ZFN (3.3-fold increase versus 2.3-fold increase, respectively) (Figure 2E; Figure S4). A similar increase in HbF was recently reported for ZFN-edited bone marrow cells.16 Furthermore, qRT-PCR validated this robust effect of BCL11A erythroid enhancer disruption on γ-globin mRNA levels (Figure S5).
Figure 1.
Erythroid Differentiation of Edited and Unedited Wild-Type Cells
(A) Indel percentage 2 days after transfection and on day 20 of erythroid differentiation. (B) Growth of untransfected (untr), exon 2-ZFN-, and enhancer-ZFN-transfected erythroid cells during erythroid differentiation. (C) Cytospin preparations stained with H&E from different time points of erythroid differentiation. (D) Percentage of nucleated over enucleated cells at the endpoint of the culture.
Figure 2.
HbF Reactivation Is Similar between Exon 2 and Enhancer KO
(A) Flow cytometry for HbF+ cells on days 14 and 20 of erythroid differentiation (note the higher levels in immature erythroid cells). (B) Immunocytochemistry in cytospins showing HbF+ cells in both coding and enhancer KO (day 20). (C) Enucleated HbF+ cells present in both edited populations. Blue, DAPI; green, anti-HbF (day 20). (D) Indel percentage of cell subpopulations sorted based on their size (small or large), reflecting maturation levels, and/or based on HbF expression (high or low) (day 20). (E) HPLC traces of unsorted, enucleated, and nucleated cells, showing increases in both γ-globin chains by exonic and enhancer KO (day 20).
Bcl11a Erythroid Enhancer KO Is Sustained Long-Term In Vivo in Both Primary and Secondary Recipients
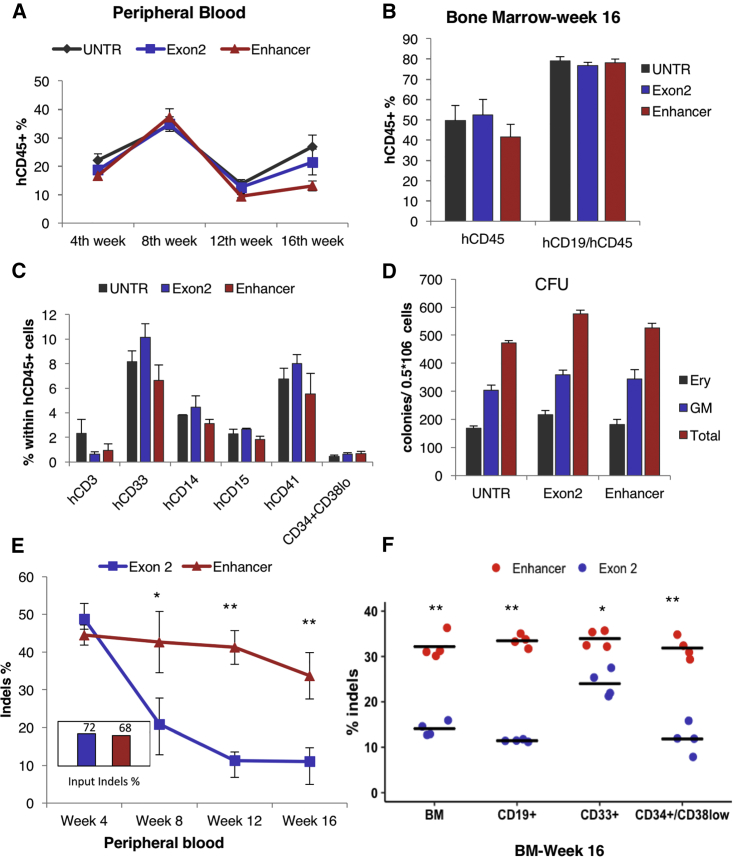
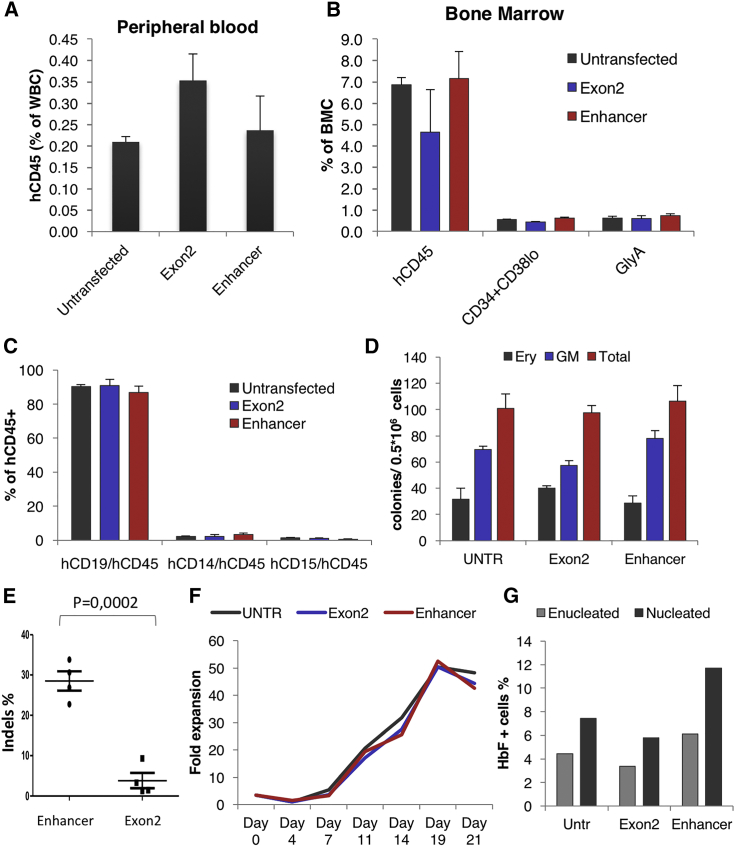
In addition to in vitro experiments, we evaluated the engraftment potential of edited CD34+ cells from normal donors and the subsequent effect on HbF expression in erythroid cells derived from xenografted progeny. Sublethally irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were transplanted with 106 CD34+ cells/recipient (frozen ∼48 hr after electroporation and thawed on the day of injection); 3 samples were used: non-edited CD34+ cells, CD34+ cells edited with a BCL11A-exon2 ZFN pair (72% modification 2 days post-electroporation) and CD34+ cells edited with BCL11A enhancer-targeted ZFNs (68% modification 2 days post-electroporation). The mice were evaluated 4, 8, 12, and 16 weeks after transplantation for the presence of human CD45+ cells in their peripheral blood and in week 16 for bone marrow engraftment of human cells of different lineages. Human chimerism was comparable among the different groups throughout the experiment, both in the peripheral blood (Figure 3A) and bone marrow (Figure 3B), and multilineage reconstitution was observed in all engrafted mice (Figures 3B and 3C). Colony-forming human progenitor cells were also detected in all recipients at the same frequency (Figure 3D). However, evaluation of the indel percentage within the different human cell subpopulations revealed a higher retention of modification at the enhancer versus the coding region both in peripheral blood (Figure 3E) and in sorted bone marrow-derived cells (Figure 3F), a difference that was more prominent in the lymphoid lineage and the HSC compartment (CD34+/CD38−) and less so in the myeloid lineage, suggesting that KO of the coding region severely impairs B cell and HSC development, in contrast to the influence observed following KO of the BCL11a erythroid enhancer (Figure 3F). Similar results in terms of engraftment and indel levels were obtained 20 weeks after transplantation (Figure S6). To assess HbF reactivation and indel frequencies in the engrafted erythroid cells, we performed ex vivo erythroid differentiation using recipient bone marrow cells because the NSG mouse model used for these experiments does not support human erythropoiesis. After culturing bone marrow cells from the engrafted mice, both BCL11A exon and BCL11A enhancer disruption yielded significantly higher HbF+ cell numbers (Figure 4A) and γ-globin protein levels (Figure 4B) compared with the untransfected control in red blood cells derived from human erythroid progenitors from the bone marrow of the engrafted mice. The in vitro erythroid differentiation from recipient bone marrow-derived human cells was similar under all conditions in terms of cell growth and enucleation (Figures 4C and 4D). Evaluation of the editing levels again revealed that the indel percentage in the BCL11a exon-targeted samples was significantly lower than in the enhancer-targeted samples (Figure 4E) and, furthermore, that the indel percentage in the latter was similar to the total bone marrow levels described above (>30%; Figure 4E), arguing strongly against an erythroid lineage-selective impairment by the enhancer KO. After sacrifice, the human cells were enriched from the primary recipients’ bone marrow and subsequently transplanted into secondary recipients. Similar levels of total human cell engraftment were observed again under all conditions in peripheral blood (Figure 5A) and bone marrow (Figure 5B). The majority of engrafted cells were lymphocytes, as expected (Figure 5C); however, a high frequency of hematopoietic progenitors was found in the recipients’ bone marrow, as shown by FACS for the CD34+/CD38− population (Figure 5B) and by colony assays (Figure 5D). Interestingly, among engrafted cells, the enhancer-edited ones again showed significantly higher indel retention than the coding region-edited cells (Figure 5E). After in vitro erythroid differentiation of the human engrafted cells in secondary recipients (Figure 5F), an increased percentage of HbF+ cells was observed in the enhancer-edited sample (Figure 5G). Cumulative evidence so far suggests that enhancer-edited cells engrafted comparably to untransfected cells and that the enhancer disruption is comparable in HbF reactivation with the coding KO but without impairing the viability, multilineage differentiation and reconstitution, and long-term engraftment potential of the genetically modified cells.
Figure 3.
In Vivo Performance of the Edited Cells
(A) Human cell chimerism in the peripheral blood of NSG mice. (B) Human cell chimerism in the bone marrow of primary recipients 16 weeks after transplantation. (C) Multilineage in vivo differentiation of human unedited and edited cells in the mouse bone marrow. (D) Human erythroid (BFU-E) and non-erythroid (CFU-GM) colonies from engrafted cells 16 weeks after transplantation. (E) Indel percentage in the peripheral blood of the primary recipients at different time points 4–16 weeks after transplantation (n = 8 mice per group) and indel percentage of the input cells. (F) Indel percentage in different hematopoietic lineages 16 weeks after transplantation. The error bars represent the SEM of 8 mice. *p < 0.05 and **p < 0.001.
Figure 4.
HbF Reactivation in Ex Vivo Erythroid Cultures from Week 16 In Vivo Engrafted Human CD45+ Cells
(A) HbF+ cell frequency in enucleated and nucleated cells as evaluated by flow cytometry. (B) γ-globin/β-like globin and γ-globin/α-globin percentages as detected by HPLC. (C) Cell growth of erythroid cells. (D) Percentage of enucleation on day 20 of the erythroid culture. (E) Indel percentage of exon 2- and enhancer ZFN-treated cells on day 14 of the erythroid culture. *p < 0.05 versus untransfected (UNTR). The error bars represent the SEM of 4 mice.
Figure 5.
Engraftment of the Edited Cells in Secondary Recipients
(A) Human cell chimerism in the peripheral blood of secondary recipient NSG mice 10 weeks after transplantation. (B) Human CD45, CD34+CD38lo, and GlyA+ cell chimerism in the bone marrow of secondary recipients 10 weeks after transplantation. (C) Multilineage in vivo differentiation of human unedited and edited cells. (D) Human erythroid (BFU-E) and non-erythroid (CFU-GM) colonies from engrafted cells 10 weeks after transplantation. (E) Indel percentage in bone marrow-derived cells 10 weeks after transplantation. (F) Cell growth of the erythroid cells derived from the secondary recipients’ bone marrow. Proliferative expansion here refers to all donor cells irrespective of editing levels. (G) HbF+ cell frequency in the erythroid cultures. For the erythroid differentiation assays, bone marrow cells of all recipients per group were pooled and plated under erythroid conditions. The error bars represent the SEM of 4 mice.
BCL11A Editing Efficiently Reactivates HbF in Erythroid Cells of Patients with β0- or β+-Thalassemia
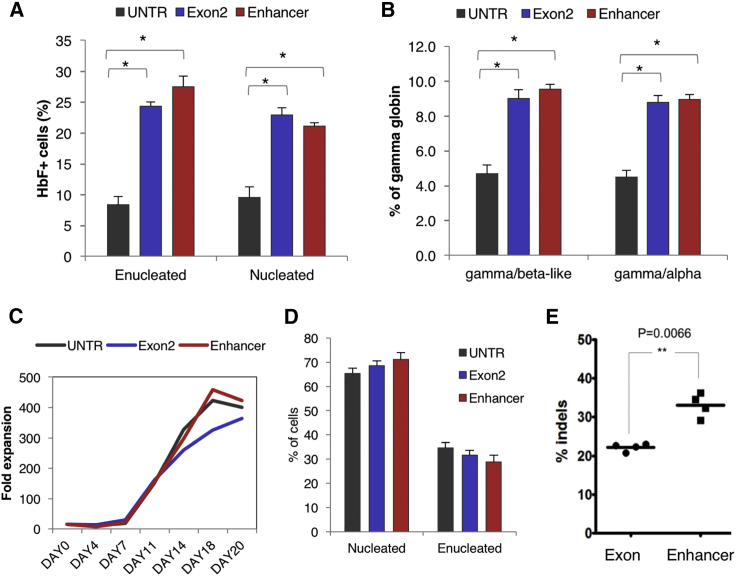
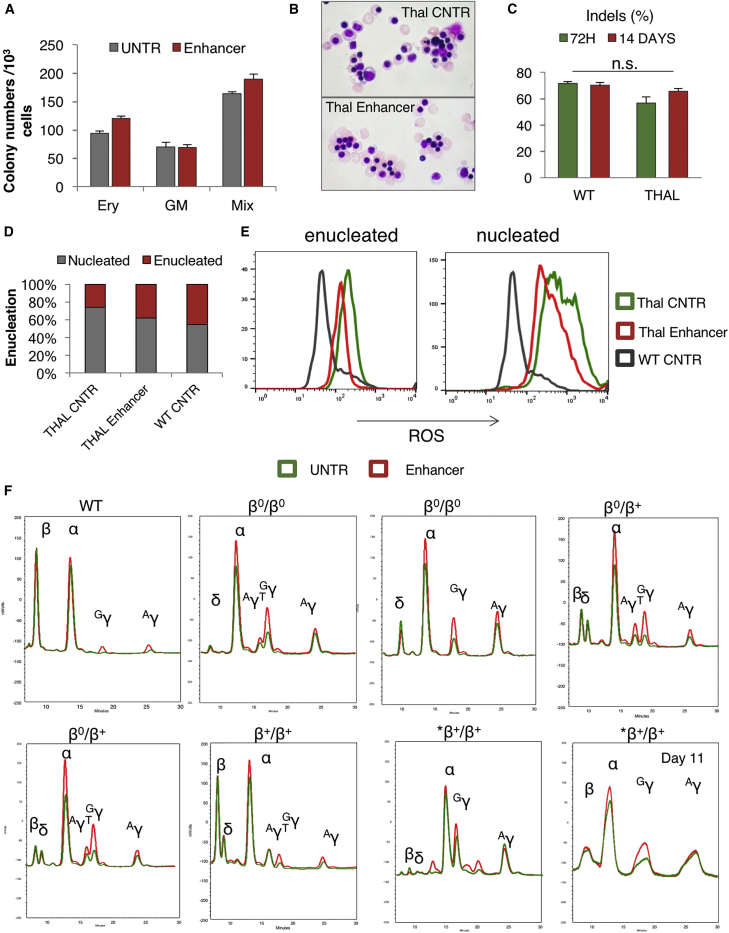
Bcl11A enhancer modification was then tested in mobilized CD34+ cells from thalassemic patients. Specifically, plerixafor- or plerixafor- and G-CSF-mobilized peripheral blood CD34+ cells from two β+/β+-, two β0/β+-, and two β0/β0-thalassemic patients were electroporated with the BCL11A ZFN mRNA. The genotypes, phenotypes, and mobilization methods of the thalassemic patients are listed in Table 1. The ZFN transfection did not affect the clonogenic capacity of the cells (Figure 6A). The edited cells were cultured 48 hr after transfection, under the same 20-day erythroid culture protocol as the wild-type cells described above. The thalassemic erythroid cells grew and, at least partially, matured in vitro without editing (Figure 6B). Of interest, although deep sequencing initially showed indel levels similar to those observed in wild-type cells, there was an increase in the proportion of thalassemic edited cells during the erythroid differentiation (56.7% ± 4.83% on day 3 versus 65.7% ± 2.1% on day 14). This was interpreted as a potential survival advantage of the edited cells; however, the difference was not statistically significant (Figure 6C). Furthermore, BCL11A enhancer editing increased the percentage of enucleated cells at the end of differentiation (Figure 6D), and it reduced the oxidative stress of both enucleated and nucleated cells by reducing the production of reactive oxygen species (ROS) (Figure 6E). At the end of erythroid differentiation, the frequency of HbF+ cells was increased in the edited thalassemic samples (from 82% prior to transfection to more than 95% after editing) (Figure S7). BCL11A enhancer editing resulted in evident γ-globin increase, comparable with the reactivation observed in wild-type cells, as shown by HPLC (Figure 6F). In a sample from a transfusion-dependent β+/β+-thalassemic donor with an IVSII-745/IVSII-745 genotype, β-globin was all but extinguished at the end of differentiation (day 20) (Figure 6F, last panel). Unexpectedly, in all of our thalassemic samples, BCL11A enhancer disruption was found to have a greater effect on Gγ chain expression, whereas the Aγ and AγT30, 31 chains were less affected (Figures S8A and S8B). A significant preferential Gγ chain increase was also observed in wild-type cells from three healthy donors (Figure S8C). Further, in addition to the expected increase in γ-globin protein, comparison of the globin chain peaks shows an increase in the α-globin peak in the enhancer-edited thalassemic cell samples (Figures S8D–S8F). This may indicate that the increase in γ-globin levels is accompanied by stabilization of previously unpaired, and therefore unstable, α-globin protein. On practical grounds, whatever the reasons for the increase (total increase in alpha or decrease in delta), a concomitant increase in stable α-globin protein upon elevation of γ-globin protein would lead to an underestimation of the γ-globin increase when the γ-globin/α-globin ratio is plotted.
Table 1.
Phenotype, Genotype, and Mobilization Scheme of the Thalassemic Patients
| Phenotype | Genotype | Mobilization |
|---|---|---|
| β0/β0 | IVSI-1/IVSI-1 | plerixafor |
| β0/β0 | CD39/CD39 | plerixafor |
| β0/β+ | Cd39/IVSI-110 | G-CSF+plerixafor |
| β0/β+ | CD39/IVSI-110 | G-CSF+plerixafor |
| β+/β+ | IVSI-110/IVSI-110 | plerixafor |
| β+/β+ | IVSII-745/IVSII-745 | plerixafor |
Figure 6.
BCL11A-Erythroid Enhancer KO in Adult Thalassemic Cells from Thalassemia Patients
(A) Erythroid (BFU-E) and non-erythroid (CFU-GM) colonies from untransfected (UNTR) and enhancer-edited thalassemic cells. The error bars represent the SEM of triplicate samples. (B) Morphology of erythroid cells at the endpoint of the erythroid culture (day 18). (C) Indel percentage before and after erythroid differentiation of wild-type (WT) and thalassemic cells. The error bars represent the SEM of 6 thalassemic samples. (D) Percentage of enucleation on day 18 of erythroid differentiation. (E) ROS in normal and thalassemic enucleated and nucleated cells (day 20 of erythroid differentiation). (F) HPLC traces of unedited (green) and enhancer-edited (red) erythroid wild-type and thalassemic cells on day 20 of erythroid differentiation (except for the last bottom graph on day 11, showing a higher presence of β-globin compared with day 20 in the same sample). Genotype, top row (left to right): WT, IVSI-1/IVSI-1, CD39/CD39, CD39/ IVSI-110; bottom row (left to right): CD39/ IVSI-110, IVSI-110/ IVSI-110, IVSII-745/IVSII-745. The data in (A), (D), and (E) are from the *β+/β+ sample. The data in (C) and (F) represent all analyzed samples.
Edited HSPCs Expand Well In Vitro under Specific Expansion Protocols
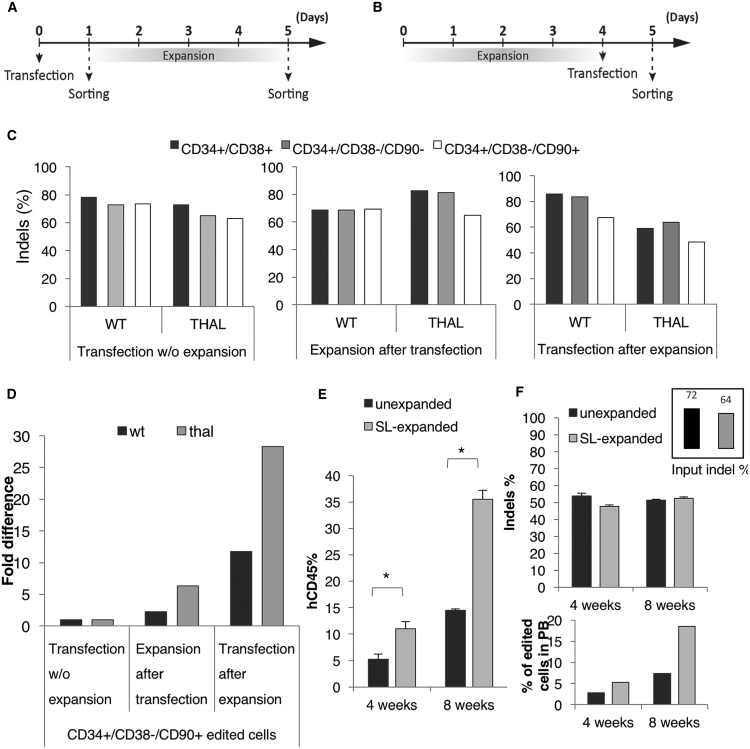
In all gene therapy or genome editing clinical settings, there is a magnified need for large numbers of modified HSCs with the capacity to robustly engraft long-term and to achieve a sustainable therapeutic effect in vivo. We have previously demonstrated that a 5-day culture with a StemRegenin1 and Ly2228820 (SL) small molecule combination can successfully expand highly engraftable CD34+/CD38−/CD90+ cells, even in comparison with their uncultured counterparts.26 Thus, we decided to test the ability of the edited wild-type and thalassemic cells to expand in vitro under these culture conditions.
After the same expansion protocol, several different HSPC subpopulations (CD34+/CD38+, CD34+/CD38−/CD90−, CD34+/CD38−/CD90+) from the BCL11A enhancer-edited wild-type and thalassemic samples were sorted and sequenced (Figure 7A). Furthermore, in additional experiments, we studied the editing efficiency in expanded cells, an approach that could be applied especially when lower cell numbers are available (Figure 7B). Edited cells, especially CD34+/CD38−/CD90+, were efficiently expanded during the 5-day period without any loss of editing efficiency (Figure 7C). Of interest, day 5 expanded cells of the same phenotype could be also successfully edited (Figure 7C). Overall, either expansion of edited cells or transfection of expanded cells yielded significantly higher numbers of edited CD34+/CD38−/CD90+ cells (2.3- and 11.7-fold increase in wild-type cells and 6.3 and 28.3-fold increase in thalassemic cells, respectively) (Figure 7D).
Figure 7.
Expansion of Normal and Thalassemic Cells before or after Editing
(A) Flow chart describing the expansion after transfection protocol. Cells sorted on day 1 (without expansion) were used as a control. (B) Flow chart describing the transfection after expansion protocol. (C) Indel levels in different sorted progenitor and stem cell populations from the three editing protocols. (D) Fold increase of the total numbers of CD34+/CD38−/CD90+ edited cells. (E) Human chimerism of unexpanded and SR1+Ly (SL) expanded enhancer-edited normal cells. (F) Indel percentage in the peripheral blood of primary recipients (top) and estimation of the percentage of edited cells in peripheral blood (bottom). Top left: the frequency of edited total input cells. *p < 0.001 versus unexpanded.
Subsequently, we tested the engraftability of wild-type BCL11A enhancer-edited expanded cells. Sublethally irradiated NSG mice received either unexpanded enhancer-edited cells 2 days after transfection or the 5-day small molecule expanded progeny from the same number of unexpanded cells. At several time points after transplantation, human chimerism was tested. We observed that the expanded cells achieved a more than 2-fold higher engraftment level compared with their unexpanded counterparts (Figure 7E). Sequencing of the engrafted cells showed an almost identical indel frequency between the expanded and unexpanded populations. However, because the overall number of edited cells engrafted was much higher after the ex vivo expansion (Figure 7F), these results could potentially translate into a higher clinical benefit.
Discussion
Editing of autologous cells has emerged as a new promising therapeutic strategy for treatment of patients with β-globin disorders. A leading approach applicable to all types of mutations is the reactivation of HbF. ZFN-based genome editing allows precise modification of the genome to reactivate HbF by targeting a specific 5-bp GATAA motif that is a key regulatory element in the BCL11A erythroid enhancer.13, 14 BCL11A is a key regulator of HbF suppression, and by targeting this specific regulatory element, we can selectively knock out BCL11A expression in the erythroid lineage to reactivate HbF expression while retaining the ability of BCL11A to function in support of HSC function and lymphopoiesis. Our current data using mobilized peripheral blood cells build on and expand previous findings comparing BCL11A exonic versus enhancer disruption data using bone marrow CD34+ cell sources.16 In the present study, lower concentrations of ZFNs achieved similar or even higher editing compared with the higher ZFN amounts reported previously, and, in line with their conclusions, we show impaired erythroid cell proliferation only in the BCL11A coding region KO in erythroid cultures. We also present an extensive long-term in vivo comparison between editing of BCL11A exon 2 and the erythroid enhancer. High retention of enhancer modification was observed in primary and secondary transplants in all cell lineages, with HbF increases well maintained in these recipients. In contrast, editing of exon 2 was significantly reduced in vivo, with especially low levels of edited cells detected in secondary recipients.
In addition, by performing clinical-scale editing, we achieved high editing efficiency for both the coding and the enhancer region in contrast to previously reported results.32 Furthermore, the high editing levels observed in our bulk cell population are superior to recently reported editing levels in unsorted bulk in vitro cultures with unclear in vivo performance.10 Current efforts are also being directed toward ZFN designs that increase enhancer disruption activity and cleavage selectivity to yield off-target profiles that are below state-of-the-field detection strategies.33
In addition to clinical-scale editing, for the first time, important information regarding the editing efficiency and phenotypic correction of cells from thalassemic patients is provided. So far, the use of genome editing on thalassemic cells has not been explored, with the exception of gene correction in thalassemic induced pluripotent stem cells (iPSCs).34, 35, 36 Although presently iPSCs represent tools for screening different approaches, their lack of engraftment potential and inefficiency for terminal maturation are major hurdles to overcome. Here we show a high BCL11A-erythroid enhancer disruption rate in thalassemic cells by an early stage ZFN pair that efficiently increases cellular HbF protein levels. This increase balances the α-globin/β-like globin ratio, resulting in an improved terminal erythroid enucleation rate. Terminal differentiation was improved by decreased red cell oxidative damage because ROS were significantly reduced after editing. Oxidative damage by ROS is generated by hemichrome formation because of an excess of aggregated α-globin chains and is believed to be one of the main contributors to cell death, tissue damage, and hypercoagulability in thalassemic patients.37, 38 A decrease in ROS levels in a thalassemic mouse model has also been shown to significantly improve their clinical picture.39 In conclusion, our detailed evaluation of maturation parameters in thalassemic cells after editing uncovered beneficial features of edited cells.
Beyond the above observations, the fact that we have, for the first time, assessed thalassemic samples for reactivation of fetal Hb (HbF) through gene editing provides additional insights. For example, in cells with the β0/β0 genotype, it is very difficult to gauge the effectiveness of the HbF increase when edited cells are compared with control unedited samples because these also produce significant and variable amounts of HbF under in vitro conditions, as documented previously.40 Assessment of protein levels at terminal maturation stages, including enucleated red blood cells (RBCs), is of critical importance for comparative purposes. Furthermore, the observed increase in α-globin protein levels after editing (Figure 6F; Figure S8E) and the absence of an adult β-globin signal make the two usual readouts of HbF activation, increase in the γ-globin over α-globin and γ-globin over beta-like globin protein signals, much less informative than in wild-type cells. Nevertheless, it was encouraging to document clear increases in γ-globin peaks (Figure 6F; Figure S8F) as well as increases in α-globin peaks for the first time in edited thalassemia CD34+ cells.
In a short in vitro culture system, it is hard to assess the selective survival advantage of edited cells with higher HbF content that is expected to occur in vivo after a longer time period. Nevertheless, because the in vitro editing conditions can be further optimized,41 our present data encourage further in vitro studies using edited patient samples to explore whether that may potentially predict in vivo outcomes.
Despite certain ambiguities in assessing the HbF increase in vitro after editing, a detailed assessment of protein levels unveiled additional novel observations using the thalassemic samples. The preferential increase in Gγ compared with Aγ in all homozygous β-thalassemic samples when BCL11A was reduced is intriguing. This observation is reminiscent of earlier findings. Previous studies have found that the Gγ/Aγ ratio in peripheral red blood cells of β-thalassemia homozygotes is generally 3:2.42 Moreover in a subsequent study, it was documented that the Gγ/Gγ+Aγ ratio in the bone marrow of these patients is lower than in peripheral blood, leading to the speculation that cells with mainly Gγ somehow had a preferential survival.43 Because non-edited thalassemic samples in our current cultures had more Aγ compared with the edited ones (Figure S8A), it would appear that the increase in HbF in the BCL11A enhancer-edited samples is accompanied by a coordinate increase in Gγ. In our earlier studies of in vitro cultures of fetal liver, neonatal, and adult samples, we concluded that changes in Gγ/Gγ+Aγ ratios in these samples are regulated independent of the total amount of fetal globin, but the specific regulatory influences were unclear.44 Of further interest is that, in our patients with haploinsufficiency of BCL11A, there was also an increase in the Gγ/Aγ ratio in their peripheral blood,19 pointing to a BCL11A-related finding in non-thalassemic samples as well. A similar tendency was also observed in normal CD34+ cells (Figure 2E). Whether such an increase is only observed with diminished BCL11A but not with other modalities that increase HbF needs to be pursued in subsequent studies. Nevertheless, the differential effect on the two γ-globin promoters could be interpreted to suggest direct binding of the developmental regulator BCL11A on the γ-globin promoters and, possibly, with a differential affinity dependent on additional nucleosome remodeling deacetylase (NuRD) complex members.
For editing autologous samples for transplantation (i.e CD34+ cells), it has been widely recognized that a major goal is the modification of a sufficient number of engraftable long-term HSCs. Because the latter are in very low proportions (0.01% or less) in the mobilized samples available, efficient expansion of these cells ex vivo is a highly desirable means of achieving that goal. Toward this end, we used a previously successful protocol to expand engraftable HSCs.26 Under these conditions, we have now shown, by using both normal and thalassemic samples, that we can not only significantly expand ex vivo cells with an engraftable edited HSC phenotype within 5 days but also efficiently edit HSCs after expansion. Although, by this method, larger-scale transfections and higher amounts of ZFN mRNA are required, the expanded, more primitive CD34+/CD38− cells, if originally insufficient in numbers, could be purified and edited efficiently in isolation without the co-presence of large numbers of the CD34+/CD38+ population.45 As recently proposed,4 the edited HSCs could then be co-transplanted with unedited progenitor or precursor cells. The current studies provide a promising roadmap for efficient autologous editing and expansion of the HSPC pool in β-thalassemia patients who may not efficiently mobilize, providing a therapeutic modality for a broader spectrum of this population. The finding that engraftable HSPC numbers can be augmented could serve other patient populations with hemoglobin disorders, including sickle cell disease.
Materials and Methods
Healthy and Thalassemic Donor Collection, Culture, and Editing
Mobilized leukapheresis products were procured from healthy donors following a mobilization regimen of 10 μg G-CSF/kg/day for 5 days and 240 μg plerixafor/kg/day on the evenings of days 4 and 5. On days 5 and 6 of mobilization, peripheral blood mononuclear cells were collected by apheresis with a COBE Spectra apheresis system. During apheresis collection, 150 mL of autologous plasma was collected and then added to the cell collection to sustain cell viability during transit and storage. CD34+ cell purification was performed as described by Yannaki et al.46 Platelet depletion on the leukapheresis product was performed using the Lovo cell processor device (Fresenius Kabi) before enrichment for CD34+ cells using the CliniMACS Plus instrument (Miltenyi Biotec). The collected cells were cultured in CD34 culture medium at 1 × 106 cells/mL in a VueLife 750-C1 gas-permeable bag (Saint Gobain, Gaithersburg, MD). For large-scale transfections, CD34+ HSPCs were washed with MaxCyte electroporation buffer and split into three experimental conditions: 1 × 108 cells were left untransfected, 1.98 × 108 cells were transfected with SB-BCLmR mRNA (targeting exon 2) in 1 × 108 cells/mL at a dose of 120 μg/mL, and 1.75 × 108 cells were transfected with 46801-2a-47923 mRNA (targeting the erythroid enhancer) in 1 × 108 cells/mL at a dose of 120 μg/mL. CD34 cell cultures were then transiently incubated in a 30°C, 5% CO2 incubator for 18–24 hr. Subsequently, cells were diluted to 1 × 106 cells/mL in CD34 culture medium and incubated at 37°C, 5% CO2. The cell viability was maintained above 70% throughout the process. The conditions for the collection of the thalassemic cells were described in two previous clinical trials.46, 47 Small-scale transfections of 2 × 105 cells were performed 2 days after thawing the cells using a BTX electroporator (ECM 830, Holliston, MA; voltage, 250 V; pulse length, 5 ms) in 100 μL of BTX Express electroporation solution with 8 μg of the 46801-2a-47923 mRNA. The thalassemia patient samples had been frozen twice before thawing and transfection, probably resulting in lower cell survival and editing efficiencies. The CD34 culture medium consisted of Stem Span H3000 supplemented with CC110 cytokine cocktail (STEMCELL Technologies). For ex vivo expansion, normal and thalassemic cells were cultured under serum-free conditions in the medium described above, supplemented with 1 μM StemRegenin1 (SR1, Cellagen Technology) and 100 nM Ly2228820 (Ly, Selleckchem). For colony formation assays (colony-forming unit [CFU]-granulocyte macrophage [GM] and burst-forming unit-erythroid [BFU-E]), cells were plated at a density of 750–2,000 cells/mL in semi-solid methylcellulose-based medium containing cytokines, ColonyGEL (ReachBio).
Zinc Finger Nucleases
ZFN pairs were expressed from mRNAs containing 2a self-cleaving peptide linkers. mRNAs were generated by in vitro transcription at Eufets (Idar-Oberstein, Germany).
The Bcl11a enhancer-targeting ZFN mRNA 46801-2a-47923 contained the following ZFNs:
46801: DQSNLRA RPYTLRL SGYNLEN TSGSLTR DQSNLRA AQCCLFH
Binding site: AGCAACtGTTAGCTTGCAC (small letters refer to bases skipped by the DNA binding domain)
47923: RSDHLTQ QSGHLAR QKGTLGE QSSDLSR RRDNLHS
Binding site: CAGGCTCCAGGAAGG.
The BCL11a exon 2-targeting mRNA SB-BCLmR contained the following ZFNs:
44490: RSDHLTQ DRSALAR RSDSLSR DRSVRTK RSDHLSA QRSNLKV
Binding site: CAACGGGCCGTGGTCTGG
43490: DRSNLSR LRQNLIM LQSQLNR RSDHLSR QSGNLAR QRNDRKS
Binding site: CCAGAAGGGGATCATGAC.
MiSeq Deep Sequencing
The efficiency of ZFN-mediated gene modification was assessed by deep DNA sequencing. The region of interest (containing the ZFN binding sites within the BCL11A locus) was PCR-amplified, and the level of modification was determined by paired-end deep sequencing on an Illumina MiSeq. To generate libraries compatible with the Illumina MiSeq sequencing platform, adaptors, barcodes, and a flow cell binder (short DNA sequence) were attached to the target-specific amplicons using two sets of fusion primers in sequential PCRs. The primers used for the MiSeq Adaptor PCR are shown in Table 2.
Table 2.
The Primers Used for the MiSeq Adaptor PCR
| BCL11A Enhancer Primers | |
|---|---|
| miBcl_59105f | ACA CGA CGC TCT TCC GAT CTN NNN CCA GTG CAA AGT CCA TAC AGG |
| miBcl_59304r | GAC GTG TGC TCT TCC GAT CTG TCT GCC AGT CCT CTT CTA CC |
| BCL11A Exon 2 Primers | |
| miBCL11A7700_08f | ACACGACGCTCTTCCGATCTNNNNCAATGGGAAGTTCATCTGGCAC |
| miBCL11A7700_08r | GACGTGTGCTCTTCCGATCTCCGAGCCTCTTGAAGCCATT |
The underlined portions are the ZFN target-specific sequences.
Insertion or Deletion Quantitation
Loci were PCR-amplified from genomic DNA (gDNA), and the levels of modification were determined by paired-end deep sequencing on an Illumina MiSeq. Paired sequences were merged via SeqPrep (https://github.com/jstjohn/SeqPrep). Merged FASTQ reads were filtered on the following criteria: quality score o of 15 or higher at all positions; the 5′- and 3′-terminal 23 bp must match the expected amplicon exactly, the read must not map to a different locus in the target genome as determined by Bowtie2 with default settings,48 and deletions must be less than 70% of the amplicon size or less than 70 bp long. Filtered sequences were then aligned to the expected sequence using the Needleman-Wunsch algorithm, with penalties set to prioritize single insertions or deletions. Contiguous gaps in either the target or query alignment were counted as indels.
Erythroid Differentiation
For erythroid differentiation of both wild-type and thalassemic CD34+ cells, we used a previously described three-step differentiation protocol.29 Briefly, 2 days after transfection, 2–5 × 104 CD34+ cells were cultured for 7 days in Step-1 medium consisting of IMDM (Gibco) supplemented with 1× GlutaMAX, 100 U/mL penicillin/streptomycin, 5% human AB+ plasma, 330 mg/mL human holo-transferrin, 20 mg/mL human insulin, 2 U/mL heparin, 1 mM hydrocortisone (all four from Sigma-Aldrich), 3 U/mL recombinant human erythropoietin (EPO) (Janssen Products), 100 ng/mL stem cell factor (SCF; PeproTech), and 5 ng/mL interleukin-3 (IL-3) (PeproTech). On day 7 (step 2), hydrocortisone and IL-3 were removed from the medium, and the cells were cultured for 4 days. On day 11, SCF was also removed, and the cells were cultured for 7–10 more days until enucleation was observed. For erythroid differentiation of human progenitor cells engrafted in NSG recipients, 2–3 × 105 hCD45+ cells from recipient bone marrow were cultured under the same conditions as described above. For the evaluation of maturation, erythroid cells were collected at different time points for cytospin preparation. Cytoslides were then stained with PROTOCOL Hema 3 Fixative and Solution (Thermo Fisher Scientific), a method comparable with the Wright-Giemsa stain.
TaqMan/qRT-PCR Analysis of Globin mRNA Ratios
The ratios of γ-globin mRNA to α-globin mRNA and γ-globin mRNA to β-globin mRNA were assessed to evaluate the effects of ZFN activity on globin mRNA levels. Whole-cell RNA was isolated from in vitro-generated erythrocytes using the High Pure RNA Isolation Kit (Roche). The levels of mRNA for the individual globin genes (α-globin, β-globin, and γ-globin) were then measured by real-time qRT-PCR on the CFX96 C1000 Touch Real-Time PCR Detection System (Bio-Rad) using the following commercially available probe sets: α-globin (HBA)- Hs00361191_g1, β-globin (HBB)-Hs00758889_s1, and γ-globin (HBG)-Hs00361131_g1 (Applied Biosystems). Each sample was measured twice, thus yielding a technical replicate. The data were analyzed using the Bio-Rad CFX Manager software. The data from this approach can be used to accurately quantitate, for example, the change in the γ-globin mRNA/β-globin mRNA ratio that occurs following a given experimental treatment. By its nature, this method does not yield the absolute ratio of the number of γ-globin mRNA molecules to the number of β-globin mRNA molecules.
HPLC
Individual globin chain levels were quantified on a Shimadzu Prominence instrument with an SPD-10AV diode array detector and an LC-10AT binary pump (Shimadzu, Kyoto, Japan). A gradient mixture of 0.1% trifluoroacetic acid in water/acetonitrile was applied at a rate of 1 mL/min using a Vydac C4 reverse-phase column (Grace Discovery Sciences, Albany, OR). The IVSII-745/IVSII-745 *β+/β+ samples (Figure 6F) were analyzed on alternate columns from Hichrom (UK). Adult and fetal globin chains were identified by comparing their mobility with that of reference samples (adult peripheral blood and cord blood).
Flow Cytometry
Erythroid cells were collected at different time points and stained with e-Cadherin (67A4), a marker of early erythropoiesis,49, 50 and GlyA (JC159) (Dako), a marker of late erythropoiesis. For evaluation of enucleation, the cells were washed and stained with NucRed (Thermo Fisher Scientific) according to the manufacturer’s instructions. To evaluate the frequency of HbF+ cells, erythroid cells were fixed with 4% formaldehyde (Sigma-Aldrich), permeabilized with ice-cold acetone (Thermo Fisher Scientific), and stained with the 51-7 anti-g-globin antibody (Santa Cruz Biotechnology). For the ROS assay, 2–5 × 105 cells from the erythroid cultures (day 20) were washed and stained with CM-H2DCFDA (Invitrogen) according to the manufacturer’s instructions. Cell sorting for enucleated and nucleated cells was performed using a BD FACS Aria II (BD Biosciences). All data acquired were analyzed using the FlowJo software. Cells from the expansion cultures were washed and stained with the following fluorochrome-conjugated antibodies: CD34 (8G12) and CD38 (HIT2) (BD Biosciences) and CD90 (5E10) (Miltenyi Biotec) and acquired in BD FACS-Calibur.
Xenotransplantation
Eight-week-old NSG mice were sublethally irradiated (300 rad) and transplanted with 1 × 106 unedited and edited CD34+ cells. Donor chimerism was assessed every 4 weeks as the frequency of human cells in total white blood cells by flow cytometry after red blood cell lysis and staining with human CD45 (BD Biosciences). Upon sacrifice, 16 or 20 weeks after transplantation, the chimeric bone marrow was harvested and assayed for level of engraftment and multilineage reconstitution after staining with the following antibodies: CD45 (2D1), CD19 (HIB19), CD33 (P67.6), CD34 (8G12), and CD38 (HIT2) (BD Biosciences) and CD41 (VIPL3) (Invitrogen). For the secondary transplantations, bone marrow cells were enriched for human CD45+ cells by immunomagnetic separation (hCD45 microbeads, Miltenyi Biotec), and the same number of human cells (5 × 106) was injected into each secondary recipient. Secondary recipients were sacrificed 10 weeks after transplantation. All in vivo experiments were conducted with approval from the institutional animal care and use committee.
Statistical Analysis
Results are expressed as mean – SE, and statistical significance was determined by ANOVA with Bonferroni correction using Minitab v.16 or with Student’s t test for pairwise comparisons; p < 0.05 was considered statistically significant.
Author Contributions
Writing – Original Draft, N.P.; Methodology, N.P. and A.R.; Investigation, N.P., A.R., S.P., Y.Z., and D.D.; Writing- Review & Editing, A.R., E.Y., D.N.L., M.C.H., and T.P.; Resources, E.Y.; Conceptualization, F.D.U. and T.P.; Supervision, T.P.
Conflicts of Interest
A.R., F.D.U., and M.C.H. are or were full-time employees and equity owners of Sangamo Therapeutics when these studies were conducted. D.L. was a full-time employee and equity owner of Bioverativ when these studies were conducted.
Acknowledgments
This study was supported by NIH grant R01-DK101328–01A1 (to T.P.). N.P. was supported by the Cooley's Anemia Foundation.
Footnotes
Supplemental Information includes eight figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.08.003.
Supplemental Information
References
- 1.Higgs D.R., Engel J.D., Stamatoyannopoulos G. Thalassaemia. Lancet. 2012;379:373–383. doi: 10.1016/S0140-6736(11)60283-3. [DOI] [PubMed] [Google Scholar]
- 2.Negre O., Eggimann A.-V., Beuzard Y., Ribeil J.A., Bourget P., Borwornpinyo S., Hongeng S., Hacein-Bey S., Cavazzana M., Leboulch P., Payen E. Gene Therapy of the β-Hemoglobinopathies by Lentiviral Transfer of the β(A(T87Q))-Globin Gene. Hum. Gene Ther. 2016;27:148–165. doi: 10.1089/hum.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters M.C., Rasko J., Hongeng S., Kwiatkowsi J., Schiller G.J., Kletzel M., Ho P.J., Vichinsky E., von Kalle C., Cavazzana M. Update of Results from the Northstar Study (HGB-204): A Phase 1/2 Study of Gene Therapy for Beta-Thalassemia Major Via Transplantation of Autologous Hematopoietic Stem Cells Transduced Ex-Vivo with a Lentiviral Beta AT87Q-Globin Vector (LentiGlobin BB305) Blood. 2015;126:201. [Google Scholar]
- 4.Zonari E., Desantis G., Petrillo C., Boccalatte F.E., Lidonnici M.R., Kajaste-Rudnitski A., Aiuti A., Ferrari G., Naldini L., Gentner B. Efficient Ex Vivo Engineering and Expansion of Highly Purified Human Hematopoietic Stem and Progenitor Cell Populations for Gene Therapy. Stem Cell Reports. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breda L., Motta I., Lourenco S., Gemmo C., Deng W., Rupon J.W., Abdulmalik O.Y., Manwani D., Blobel G.A., Rivella S. Forced chromatin looping raises fetal hemoglobin in adult sickle cells to higher levels than pharmacologic inducers. Blood. 2016;128:1139–1143. doi: 10.1182/blood-2016-01-691089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traxler E.A., Yao Y., Wang Y.-D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C., Weiss M.J. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wienert B., Funnell A.P.W., Norton L.J., Pearson R.C., Wilkinson-White L.E., Lester K., Vadolas J., Porteus M.H., Matthews J.M., Quinlan K.G., Crossley M. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat. Commun. 2015;6:7085. doi: 10.1038/ncomms8085. [DOI] [PubMed] [Google Scholar]
- 8.Masuda T., Wang X., Maeda M., Canver M.C., Sher F., Funnell A.P., Fisher C., Suciu M., Martyn G.E., Norton L.J. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science. 2016;351,:285–289. doi: 10.1126/science.aad3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martyn G.E., Wienert B., Yang L., Shah M., Norton L.J., Burdach J., Kurita R., Nakamura Y., Pearson R.C.M., Funnell A.P.W. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat. Genet. 2018;50:498–503. doi: 10.1038/s41588-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 10.Antoniani C., Meneghini V., Lattanzi A., Felix T., Romano O., Magrin E., Weber L., Pavani G., El Hoss S., Kurita R. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human β-globin locus. Blood. 2018;131:1960–1973. doi: 10.1182/blood-2017-10-811505. [DOI] [PubMed] [Google Scholar]
- 11.Liu N., Hargreaves V.V., Zhu Q., Kurland J.V., Hong J., Kim W., Sher F., Macias-Trevino C., Rogers J.M., Kurita R. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell. 2018;173:430–442.e17. doi: 10.1016/j.cell.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science. 2008;189:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 13.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vierstra J., Reik A., Chang K.-H., Stehling-Sun S., Zhou Y., Hinkley S.J., Paschon D.E., Zhang L., Psatha N., Bendana Y.R. Functional footprinting of regulatory DNA. Nat. Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wienert B., Martyn G.E., Kurita R., Nakamura Y., Quinlan K.G.R., Crossley M. KLF1 drives the expression of fetal hemoglobin in British HPFH. Blood. 2017;130:803–807. doi: 10.1182/blood-2017-02-767400. [DOI] [PubMed] [Google Scholar]
- 16.Chang K.-H., Smith S.E., Sullivan T., Chen K., Zhou Q., West J.A., Liu M., Liu Y., Vieira B.F., Sun C. Long-Term Engraftment and Fetal Globin Induction upon BCL11A Gene Editing in Bone-Marrow-Derived CD34+ Hematopoietic Stem and Progenitor Cells. Mol. Ther. Methods Clin. Dev. 2017;4:137–148. doi: 10.1016/j.omtm.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y., Wang J., Khaled W., Burke S., Li P., Chen X., Yang W., Jenkins N.A., Copeland N.G., Zhang S., Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luc S., Huang J., McEldoon J.L., Somuncular E., Li D., Rhodes C., Mamoor S., Hou S., Xu J., Orkin S.H. Bcl11a Deficiency Leads to Hematopoietic Stem Cell Defects with an Aging-like Phenotype. Cell Rep. 2016;16:3181–3194. doi: 10.1016/j.celrep.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funnell A.P.W., Prontera P., Ottaviani V., Piccione M., Giambona A., Maggio A., Ciaffoni F., Stehling-Sun S., Marra M., Masiello F. 2p15-p16.1 microdeletions encompassing and proximal to BCL11A are associated with elevated HbF in addition to neurologic impairment. Blood. 2015;126:89–93. doi: 10.1182/blood-2015-04-638528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias C., Estruch S.B., Graham S.A., McRae J., Sawiak S.J., Hurst J.A., Joss S.K., Holder S.E., Morton J.E., Turner C., DDD Study BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. Am. J. Hum. Genet. 2016;99:253–274. doi: 10.1016/j.ajhg.2016.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brendel C., Guda S., Renella R., Bauer D.E., Canver M.C., Kim Y.J., Heeney M.M., Klatt D., Fogel J., Milsom M.D. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J. Clin. Invest. 2016;126:3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guda S., Brendel C., Renella R., Du P., Bauer D.E., Canver M.C., Grenier J.K., Grimson A.W., Kamran S.C., Thornton J. miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol. Ther. 2015;23:1465–1474. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 24.Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 26.Psatha N., Georgolopoulos G., Phelps S., Papayannopoulou T. Brief Report: A Differential Transcriptomic Profile of Ex Vivo Expanded Adult Human Hematopoietic Stem Cells Empowers Them for Engraftment Better than Their Surface Phenotype. Stem Cells Transl. Med. 2017;6:1852–1858. doi: 10.1002/sctm.17-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yannaki E., Psatha N., Athanasiou E., Karponi G., Constantinou V., Papadopoulou A., Tasouli A., Kaloyannidis P., Batsis I., Arsenakis M. Mobilization of hematopoietic stem cells in a thalassemic mouse model: implications for human gene therapy of thalassemia. Hum. Gene Ther. 2010;21:299–310. doi: 10.1089/hum.2009.077. [DOI] [PubMed] [Google Scholar]
- 28.Karponi G., Psatha N., Lederer C.W., Adair J.E., Zervou F., Zogas N., Kleanthous M., Tsatalas C., Anagnostopoulos A., Sadelain M. Plerixafor+G-CSF-mobilized CD34+ cells represent an optimal graft source for thalassemia gene therapy. Blood. 2015;126:616–619. doi: 10.1182/blood-2015-03-629618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giarratana M.-C., Kobari L., Lapillonne H., Chalmers D., Kiger L., Cynober T., Marden M.C., Wajcman H., Douay L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 30.Saglio G., Ricco G., Mazza U., Camaschella C., Pich P.G., Gianni A.M., Gianazza E., Righetti P.G., Giglioni B., Comi P. Human T gamma globin chain is a variant of A gamma chain (A gamma Sardinia) Proc. Natl. Acad. Sci. USA. 1979;76:3420–3424. doi: 10.1073/pnas.76.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masala B., Manca L. Detection of the common Hb F Sardinia [A gamma (E19)Ile----Thr] variant by isoelectric focusing in normal newborns and in adults affected by elevated fetal hemoglobin syndromes. Clin. Chim. Acta. 1991;198:195–202. doi: 10.1016/0009-8981(91)90353-e. [DOI] [PubMed] [Google Scholar]
- 32.Humbert O., Peterson C.W., Norgaard Z.K., Radtke S., Kiem H.P. A Nonhuman Primate Transplantation Model to Evaluate Hematopoietic Stem Cell Gene Editing Strategies for β-Hemoglobinopathies. Mol. Ther. Methods Clin. Dev. 2017;8:75–86. doi: 10.1016/j.omtm.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., Plati T., Castiello M.C., Sanvito F., Gennery A.R. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan0820. eaan0820. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Yang Y., Kang X., Lin B., Yu Q., Song B., Gao G., Chen Y., Sun X., Li X. One-Step Biallelic and Scarless Correction of a β-Thalassemia Mutation in Patient-Specific iPSCs without Drug Selection. Mol. Ther. Nucleic Acids. 2017;6:57–67. doi: 10.1016/j.omtn.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P., Tong Y., Liu X.Z., Wang T.T., Cheng L., Wang B.Y., Lv X., Huang Y., Liu D.P. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci. Rep. 2015;5:12065. doi: 10.1038/srep12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai L., Bai H., Mahairaki V., Gao Y., He C., Wen Y., Jin Y.C., Wang Y., Pan R.L., Qasba A. A Universal Approach to Correct Various HBB Gene Mutations in Human Stem Cells for Gene Therapy of Beta-Thalassemia and Sickle Cell Disease. Stem Cells Transl. Med. 2018;7:87–97. doi: 10.1002/sctm.17-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rund D., Rachmilewitz E. β-thalassemia. N. Engl. J. Med. 2005;353:1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- 38.Kong Y., Zhou S., Kihm A.J., Katein A.M., Yu X., Gell D.A., Mackay J.P., Adachi K., Foster-Brown L., Louden C.S. Loss of α-hemoglobin-stabilizing protein impairs erythropoiesis and exacerbates β-thalassemia. J. Clin. Invest. 2004;114:1457–1466. doi: 10.1172/JCI21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franco S.S., De Falco L., Ghaffari S., Brugnara C., Sinclair D.A., Matte’ A., Iolascon A., Mohandas N., Bertoldi M., An X. Resveratrol accelerates erythroid maturation by activation of FoxO3 and ameliorates anemia in beta-thalassemic mice. Haematologica. 2014;99:267–275. doi: 10.3324/haematol.2013.090076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breda L., Casu C., Gardenghi S., Bianchi N., Cartegni L., Narla M., Yazdanbakhsh K., Musso M., Manwani D., Little J. Therapeutic hemoglobin levels after gene transfer in β-thalassemia mice and in hematopoietic cells of β-thalassemia and sickle cells disease patients. PLoS ONE. 2012;7:e32345. doi: 10.1371/journal.pone.0032345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes M.C., Reik A., Rebar E.J., Miller J.C., Zhou Y., Zhang L., Li P., Vaidya S. A Potential Therapy for Beta-Thalassemia (ST-400) and Sickle Cell Disease (BIVV003) Blood. 2017;130:2066. [Google Scholar]
- 42.Ricco G., Mazza U., Turi R.M., Pich P.G., Camaschella C., Saglio G., Bernini L.F. Significance of a new type of human fetal hemoglobin carrying a replacement isoleucine replaced by threonine at position 75 )E 19) of the gamma chain. Hum. Genet. 1976;32:305–313. doi: 10.1007/BF00295821. [DOI] [PubMed] [Google Scholar]
- 43.Saglio G., Camaschella C., Guerrasio A., Cambrin G.R., Capaldi A., Pich P.G., Trento M., Mazza U. G gamma and a gamma globin chain synthesis in bone marrow and peripheral blood of beta-thalassaemia homozygotes. Br. J. Haematol. 1982;52:225–231. doi: 10.1111/j.1365-2141.1982.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 44.Papayannopoulou T., Kurachi S., Brice M., Nakamoto B., Stamatoyannopoulos G. Asynchronous synthesis of HbF and HbA during erythroblast maturation. II. Studies of G gamma, A gamma, and beta chain synthesis in individual erythroid clones from neonatal and adult BFU-E cultures. Blood. 1981;57:531–536. [PubMed] [Google Scholar]
- 45.Baldwin K., Urbinati F., Romero Z., Campo-Fernandez B., Kaufman M.L., Cooper A.R., Masiuk K., Hollis R.P., Kohn D.B. Enrichment of human hematopoietic stem/progenitor cells facilitates transduction for stem cell gene therapy. Stem Cells. 2015;33:1532–1542. doi: 10.1002/stem.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yannaki E., Papayannopoulou T., Jonlin E., Zervou F., Karponi G., Xagorari A., Becker P., Psatha N., Batsis I., Kaloyannidis P. Hematopoietic stem cell mobilization for gene therapy of adult patients with severe β-thalassemia: results of clinical trials using G-CSF or plerixafor in splenectomized and nonsplenectomized subjects. Mol. Ther. 2012;20:230–238. doi: 10.1038/mt.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yannaki E., Karponi G., Zervou F., Constantinou V., Bouinta A., Tachynopoulou V., Kotta K., Jonlin E., Papayannopoulou T., Anagnostopoulos A., Stamatoyannopoulos G. Hematopoietic stem cell mobilization for gene therapy: superior mobilization by the combination of granulocyte-colony stimulating factor plus plerixafor in patients with β-thalassemia major. Hum. Gene Ther. 2013;24:852–860. doi: 10.1089/hum.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armeanu S., Bühring H.J., Reuss-Borst M., Müller C.A., Klein G. E-cadherin is functionally involved in the maturation of the erythroid lineage. J. Cell Biol. 1995;131:243–249. doi: 10.1083/jcb.131.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohgami R.S., Chisholm K.M., Ma L., Arber D.A. E-cadherin is a specific marker for erythroid differentiation and has utility, in combination with CD117 and CD34, for enumerating myeloblasts in hematopoietic neoplasms. Am. J. Clin. Pathol. 2014;141:656–664. doi: 10.1309/AJCP8M4QQTAZPGRP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.