Abstract
Single-cell RNA-seq's (scRNA-seq) unprecedented cellular resolution at a genome-wide scale enables us to address questions about cellular heterogeneity that are inaccessible using methods that average over bulk tissue extracts. However, scRNA-seq data sets also present additional challenges such as high transcript dropout rates, stochastic transcription events, and complex population substructures. Here, we present a single-cell RNA-seq analysis and klustering evaluation (SAKE), a robust method for scRNA-seq analysis that provides quantitative statistical metrics at each step of the analysis pipeline. Comparing SAKE to multiple single-cell analysis methods shows that most methods perform similarly across a wide range of cellular contexts, with SAKE outperforming these methods in the case of large complex populations. We next applied the SAKE algorithms to identify drug-resistant cellular populations as human melanoma cells respond to targeted BRAF inhibitors (BRAFi). Single-cell RNA-seq data from both the Fluidigm C1 and 10x Genomics platforms were analyzed with SAKE to dissect this problem at multiple scales. Data from both platforms indicate that BRAF inhibitor-resistant cells can emerge from rare populations already present before drug application, with SAKE identifying both novel and known markers of resistance. These experimentally validated markers of BRAFi resistance share overlap with previous analyses in different melanoma cell lines, demonstrating the generality of these findings and highlighting the utility of single-cell analysis to elucidate mechanisms of BRAFi resistance.
Compared to bulk RNA-seq, in which expression profiles are the result of averaging over millions of cells that may vary widely, single-cell RNA-seq (scRNA-seq) can be used to investigate the subtle but crucial differences in transcriptomic landscape that differentiate cellular state. Populations of cells that possess very similar gross cellular phenotypes might have remarkably different transcriptome profiles at the single-cell level due to stochastic transcription events, unsynchronized cell-cycle stages, or inherent biological heterogeneity (Grün and van Oudenaarden 2015). Therefore, the standard methods for bulk expression profiling need to be adapted for scRNA-seq analysis. One major concern is the increased levels of noise in the measured transcript abundances. Excessive transcript dropout rates and stochastic bursting events in scRNA-seq data create abundant nondetections, high variability, and complex expression distributions in the data. Therefore, it is important to distinguish low-quality, high-noise samples that are poorly amplified or degraded during library preparation.
Following normalization and quality control procedures, the next step in scRNA-seq analysis involves clustering of the samples to identify a set of gene markers that can segregate cells into distinct groups. Most published scRNA-seq studies have used gene filtering methods developed for bulk RNA-seq, calculating the most variable genes (Klein et al. 2015; Macosko et al. 2015), the most significantly differentially expressed genes (Shalek et al. 2013), or genes that have high contribution to the first few principal components (Satija et al. 2015; Li et al. 2016). This candidate set of markers is then used to identify subpopulations of cells via standard clustering methods. Visualization of these data sets using PCA or t-distributed stochastic neighbor embedding (t-SNE) (van der Maaten and Hinton 2008) can provide qualitative information about the number of clusters present and relative levels of cluster heterogeneity, but does not give a quantitative estimate of how many clusters are present nor whether a given sample belongs with one cluster or another (for additional details about t-SNE and PCA, see Supplemental Material). Finally, proper choice of a clustering algorithm might depend upon the biological context of the samples. For example, most published single-cell clustering tools are optimized either for mixed populations of distinct cells (Kharchenko et al. 2014; Grün et al. 2015; Haghverdi et al. 2015; Satija et al. 2015; Xu and Su 2015; Zeisel et al. 2015) or for time-series data sets that assume a smooth distribution from one cell type to another (Bendall et al. 2014; Marco et al. 2014; Trapnell et al. 2014; Setty et al. 2016). In practice, data sets often include a mixture of cells from distinct cell types as well as related subclusters with significant overlap.
Here, we present an integrated analysis tool that aims to facilitate the analysis of scRNA-seq data, addressing the challenges outlined above. Our single-cell RNA-seq analysis and klustering evaluation (SAKE) method provides several modules that include data preprocessing for quality control, sample clustering, t-SNE visualization of clusters, differential expression between clusters, and functional enrichment analysis. We evaluate the performance of SAKE using several published scRNA-seq data sets that span a range of experimental designs, showing that SAKE successfully identifies known and validated novel cell types from these studies.
Finally, we applied SAKE to characterize the response of individual melanoma cells responding to targeted inhibitors of the BRAF oncogene. Resistance to targeted BRAF inhibitors in melanoma is widespread and presents a barrier to its efficacy as a therapeutic, because a large fraction of melanoma tumors initially respond to BRAF inhibition, but nearly all patients rapidly develop resistance (Müller et al. 2014; Shi et al. 2014; Sun et al. 2014; Perna et al. 2015). We used two single-cell RNA-seq platforms, Fluidigm C1 and 10x Genomics, to follow thousands of individual melanoma cells that have developed resistance to targeted BRAF inhibitors. We show that SAKE recapitulates several known markers of BRAF inhibitor resistance as well as identifying novel markers of resistance that appear in rare populations of cells prior to drug application.
Results
Workflow for analyzing single-cell RNA-seq data with SAKE
The SAKE workflow is designed to robustly categorize gene expression profiles while avoiding unwanted noise. Following the generation of a table of estimated gene abundance counts across samples, the next step in data analysis involves quality control steps to identify poorly amplified and problematic libraries. The SAKE workflow begins with this step (Fig. 1A). Samples with relatively low total transcript counts and gene coverage rates often represent degraded or poorly amplified libraries. These can be identified visually and removed from the sample set before proceeding with downstream analyses. The next step involves trimming the list of input genes to remove low abundance transcripts that suffer most from stochastic dropout events and technical noise issues. Median absolute deviation (MAD) is used as the preferred metric, but custom-filtering criteria can be implemented. SAKE provides a module to generate sample correlation heatmaps for use in evaluating the effects of filtering the gene list.
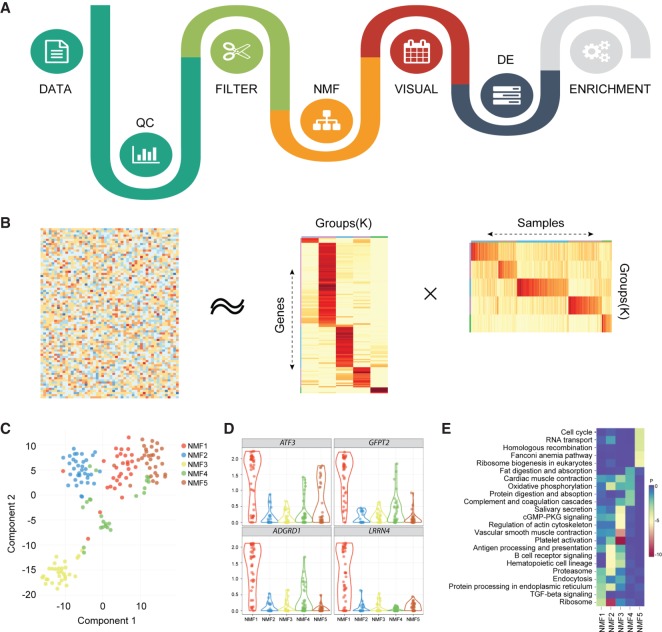
Figure 1.
Flowchart of SAKE package and example analysis results. (A) Analysis workflow for analyzing single-cell RNA-seq data. (B) A schematic representation of the non-negative matrix factorization (NMF) method shows heatmaps of sample assignment and feature extraction from NMF runs, with dark red indicating high confidence in cluster assignments. (C) A t-SNE plot to compare NMF-assigned groups with t-SNE projections. (D) A table of NMF-identified features (genes defining each cluster) and a box plot of gene expression distributions across NMF-assigned groups. (E) Summary table for GO term enrichment analysis for each NMF-assigned group.
The core of the SAKE clustering algorithm is a module to identify clusters via non-negative matrix factorization (NMF) (Gao and Church 2005; Kim and Park 2007). NMF has successfully been used to identify molecular subtypes in bulk RNA-seq expression profiles in many contexts (Hoadley et al. 2014; The Cancer Genome Atlas Research Network 2014; Moffitt et al. 2015; The Cancer Genome Atlas Network 2015; Yang and Michailidis 2016). Attributes that make NMF particularly appropriate for clustering of single-cell expression data sets include the ability to quantitatively estimate the number of clusters present in each data set, de novo, and the ability to quantitatively estimate the likelihood that each sample belongs to a given cluster.
Briefly, NMF attempts to factor a given gene expression matrix of N samples and M genes into two separate matrices: (1) a matrix of N samples belonging to k clusters (N×k), and (2) a matrix containing the relative importance of each of the M genes in determining whether a sample belongs to each of the k clusters (k×M). This factorization can be attempted for a range of different values of k, with each iteration providing a quantitative measure of the robustness of cluster assignments upon randomization of the starting network. To find the optimal value of clusters, k, we minimize the residuals between the original full gene expression matrix (N×M) and the two factorized matrices (N×k)(k×M) while simultaneously maximizing the cophenetic correlations between actual pairwise sample expression distances and the clustered dendrogram expression distances. SAKE provides a visual representation of matrix residuals and cophenetic correlation coefficients to enable users to select the optimal setting for the number of clusters, k.
Once an optimal number of clusters, k, has been determined, SAKE next performs a larger number of iterations of the NMF algorithm with fixed k, in order to robustly estimate the likelihood that each sample belongs to a given cluster and the relative importance of each marker gene in determining cluster membership. A heatmap of the cluster membership likelihood calculations for an example gene expression matrix is shown in Figure 1B, applied to the data set of Treutlein et al. (2016). The full gene expression matrix is shown on the left of Figure 1B, and the two factorized matrices are at the right. In general, NMF-identified clusters largely agree with t-SNE similarity maps, as shown in Figure 1C, in which each cell on the t-SNE map is colored by SAKE-identified clusters. Users can also check for the enrichment of any gene of interest in each NMF cluster (Fig. 1D). Differential expression analysis between clusters can be evaluated in SAKE using the DESeq2 algorithm (Love et al. 2014). Gene Ontology (GO) Term enrichments and gene set enrichment analysis (GSEA) allow for the identification of functional categories enriched in each NMF cluster (Fig. 1E).
Evaluation of SAKE on published data sets
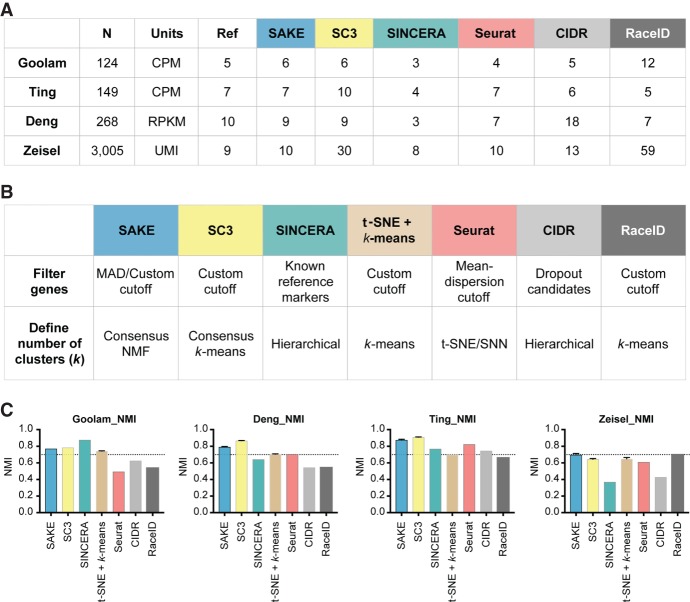
We measured the success of the SAKE pipeline by its ability to reproduce the major findings from recently published scRNA-seq studies that varied in experimental design, number of cells sequenced, and validation methods used (Deng et al. 2014; Ting et al. 2014; Zeisel et al. 2015; Goolam et al. 2016). The sample clustering results from the published scRNA-seq studies served as the reference, and normalized mutual information (NMI, shown in Fig. 2C) or adjusted Rand Index (ARI, shown in Supplemental Fig. S1) was used to evaluate the performance of SAKE compared with a number of recently published algorithms with demonstrated performance for scRNA-seq analysis: SINCERA (Guo et al. 2015), Seurat (Satija et al. 2015), SC3 (Kiselev et al. 2017), CIDR (Lin et al. 2017), and RaceID (Grün et al. 2015). Figure 2A presents a table summarizing the results for each of the three published algorithms on each of the four published data sets, together with the results from SAKE. Figure 2B presents a table that explains the major differences between each of these three algorithms in terms of the methods used for gene feature selection and clustering. For comparison, we also include the results for a “simple” analysis that includes identifying the number of clusters, k, via a t-SNE projection plot, and then running a k-means clustering algorithm to assign samples to each of those k clusters (Fig. 2B,C).
Figure 2.
Data and performance summaries for scRNA-seq software. (A) The number of samples and reported clusters from five published clustering methods (SC3, SINCERA, Seurat, CIDR, and RaceID) compared with SAKE (blue). (B) Key features and techniques used by each method to perform gene filtering and to define the number of clusters. (C) Normalized mutual information (NMI) was used to compare the performance of each method on four published data sets in terms of the ability to recapitulate cluster assignments as given by the initial publication. Error bars were measured by subsampling 90% of the cells from each data set and iterating 1000 times to ensure robust results.
To evaluate the robustness of these results, we randomly selected 90% of the samples for each of the published studies and performed 100 iterations of cluster identification for all of the compared clustering methods. In each iteration, we calculated the normalized mutual information (NMI) and adjusted rand index (ARI) using the cluster assignments in each of the published studies as the reference for accuracy. Results from these 100 runs are presented as bar charts in Figure 2C that display mean NMI for each analysis method with error bars representing standard error. Supplemental Figure S1 shows the results for ARI, which shows largely similar results but can inflate small differences between differently sized clusters. Together these results demonstrate that SAKE is both accurate and robust across a wide range of sample sizes and experimental designs. Overall, each of these algorithmically independent clustering methods gives roughly similar results, with SAKE performance coming out best for experimental designs involving large complex substructures and large numbers of sampled cells. Although the ultimate assignments of each sample to a given cluster is largely similar for each of these algorithms, it is worth noting that SAKE is unique in providing quantitative metrics for the estimated number of clusters present, for the assignment of each sample to a given cluster, and for the relative ability of each gene to act as a marker for cluster membership.
Application of SAKE to human melanoma cell lines
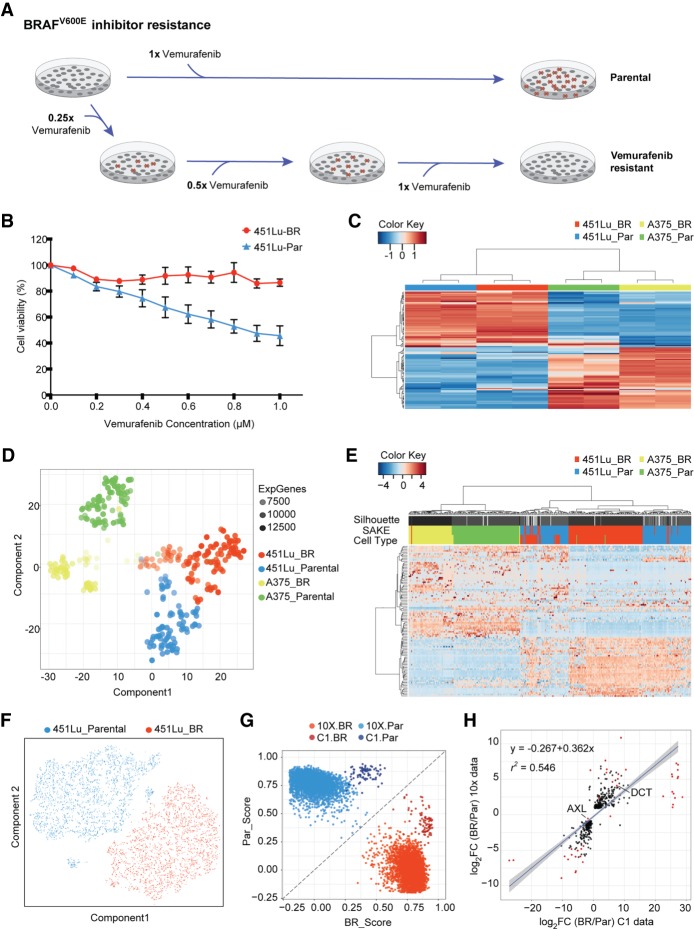
Having demonstrated the success of the SAKE algorithm on published data sets, we next applied the SAKE method to a question that can best be answered by single-cell analysis experiments: How do cancer cells individually respond to targeted therapeutic agents? We first started with cells from a human melanoma cell line, 451Lu, that carries an activating mutation in the BRAF oncogene. These cells were previously demonstrated to be initially responsive to targeted BRAF inhibitor (BRAFi) treatments, but to rapidly acquire resistance to these small molecule BRAF inhibitors (Villanueva et al. 2010). We defined cells in the naïve state before BRAFi treatment as the “parental” population, 451Lu-Par. By gradually increasing the dosage of BRAFi from 0.05 to 1 µm on 451Lu-Par and selecting for cells that survived after each round of treatment, Villanueva et al. (2010) derived a distinct population of BRAFi-resistant cells, 451Lu-BR, that grew stably in a 1 µm concentration of BRAFi (Fig. 3A). Differential response to BRAFi between the 451Lu-Par and 451Lu-BR cells was demonstrated through an MTT assay that measures metabolically active cells 72 h following BRAFi treatment (Fig. 3B).
Figure 3.
Bulk and single-cell RNA-seq were used to study differential drug responses to BRAF inhibitor treatment. (A) Naïve melanoma cells were treated with an increasing dose of the BRAF inhibitor, vemurafenib. Cells that survived after each drug treatment were selected to gradually derive stably BRAFi-resistant cells. (B) Drug sensitivity was measured through the use of MTT assays to assess metabolically active cells 72 h following BRAFi application. (C) Melanoma signature gene sets (Hoek et al. 2008) were used to cluster bulk RNA-seq data from melanoma cell lines. (D) A t-SNE map was used to display the expression profiles from approximately 400 parental and BRAFi-resistant melanoma cells isolated using the Fluidigm C1 platform. The first t-SNE component separates the two cell lines, and the second component distinguishes between parental and BRAFi-resistant cells. (E) Highly expressed and variable genes were used to classify Fluidigm C1 scRNA-seq data. Higher levels of heterogeneity can be observed among 451Lu cells as compared to A375 cells. (F) To determine whether 451Lu cells have more intrinsic heterogeneity, 6545 scRNA-seq transcriptomes were obtained using the 10x Chromium platform. A t-SNE map of this 451Lu 10x data highlights two major groups of cells: parental (blue) and BRAFi resistant (red). (G) In order to compare the 10x and C1 data on the same scale, a scoring system was implemented to determine the Spearman's rank correlation distance of each cell from the centroids of the parental (PAR) and BRAFi-resistant (BR) populations. (H) Differential expression analysis identified genes significantly altered in 451Lu-BR versus 451Lu-Par cells, with log2 fold change values plotted for the C1 data set (horizontal axis) and 10x data set (vertical axis). Genes that have adjusted P < 1 × 10−50 were highlighted in red, with all statistically significant genes (P < 0.01) shown in black, showing highly similar results.
For comparison, BRAFi-resistant cells for a second melanoma cell line also carrying the BRAF V600E mutation, A375, were derived using the same strategy (Supplemental Fig. S2B). Bulk RNA transcriptomes from the two cell lines (Supplemental Table S1) were used to determine gene expression markers that correlated with BRAFi resistance in both populations. However, the samples clustered first by their cell line of origin instead of their BRAFi treatment conditions (Fig. 3C,D). Differential gene expression analysis also showed that there was minimal overlap between the two cell lines in terms of shared genes that marked BRAFi-resistant cells (Supplemental Fig. S2C), suggesting that these two cell lines that shared the same BRAF driver mutation might be using different pathways to enable growth in the presence of BRAF inhibitor molecules.
One possible explanation for the differences between the two melanoma cell lines is that these cell lines possess distinct expression profiles that contribute more to the overall transcriptome landscape than do the genes that drive resistance to BRAFi. Recent results from The Cancer Genome Atlas Consortium described multiple transcriptome subtypes in melanoma patient tumors (The Cancer Genome Atlas Network 2015), similar to the “subtypes” that were previously described for breast cancer and other cancers (Hoadley et al. 2014). We used the SAKE algorithms, which work both for bulk and single-cell RNA-seq data, to determine whether we can reproduce these transcriptome subtype results on the TCGA data sets. These results are shown in Supplemental Figure S3, which displays the SAKE clustering results in Supplemental Figure S3A and the correlations with patient outcome in Supplemental Figure S3B. Comparison to the bulk transcriptomes from our 451Lu and A375 cell lines, and to the full set of melanoma transcriptomes in the Cancer Cell Line Encyclopedia (CCLE) showed that, indeed, these two lines fell into distinct expression clusters, dubbed by the TCGA group as “proliferative” and “invasive” subtypes, respectively (Supplemental Fig. S3C). Moreover, BRAFi-resistant cells from each of the 451Lu and A375 subtypes also remained within the same subtype as their respective parental cell lines, suggesting that subtype switching was not a dominant feature of BRAFi-resistant cells (Supplemental Fig. S2D).
Having confirmed that these melanoma transcriptome subtypes are present in both TCGA patient samples as well as the cultured cell lines described above, we next sought to determine whether these expression profiles would be faithfully represented by each individual cell in the population. Conversely, “subtype” might represent an averaging over many distinct cellular states that was not wholly present in any one cell. To answer this question, we would need to characterize the expression profiles of hundreds to thousands of melanoma cells from each subtype.
SAKE identifies four major groups in Fluidigm/Smart-Seq data sets
Single-cell RNA-seq data were prepared from both the A375 and 451Lu Parental and BRAFi-resistant cell lines using the Fluidigm C1 system to isolate cells and generate single-cell transcriptomes using Smart-Seq-based methods (for details, see Supplemental Methods). We isolated about 100 cells from each of the four conditions (A375-Par, A375-BR, 451Lu-Par, and 451Lu-BR). On average, more than 10,000 genes were detected per library (Supplemental Table S2). These C1 scRNA-seq data sets were first mixed with the bulk data to evaluate the quality of the sequencing results in terms of transcript coverage. Signature gene sets that mark proliferative and invasive subtypes were used for clustering (Hoek et al. 2008). Most single cells recapitulated a similar expression pattern as that seen in the bulk RNA-seq data and did not display a single-cell platform-specific profile (Supplemental Fig. S4A). Moreover, the similarity of the single-cell and bulk expression patterns suggest that the melanoma subtype profiles seen in bulk data sets are also representative of the dominant expression patterns of individual single cells. Additionally, the BRAFi-resistant cells from both the A375 and 451Lu populations appear to remain within their molecular subtypes regardless of sensitivity to BRAFi treatment, as was seen for the bulk transcriptome profiles. Thus, response to BRAFi treatment may be constrained by molecular subtype, but the subtype markers cannot be used to explore BRAFi-driven expression changes.
To explore the gene expression alterations associated with resistance to BRAFi, we next applied the SAKE algorithm to all ∼400 cells from each of the A375 and 451Lu parental and resistance populations. A t-SNE projection of these results can be seen in Figure 3D. The first component of the t-SNE projection separated the two cell lines by their parental cell type (A375 versus 451Lu), whereas the second component separated the cells by their sensitivity to the BRAFi treatment. SAKE identified four distinct clusters among the ∼400 cells in the scRNA-seq data Figure 3D, which largely overlapped with the populations from which these cells derived (Fig. 3E). We did not observe a direct correlation between these four SAKE-identified groups and cell-cycle stages (Supplemental Fig. S4C). However, there were some deviations from these general trends. First, a small fraction of the cells in the parental cell lines appeared to exhibit markers of the resistant populations; conversely, a small fraction of the cells isolated from the resistant populations appeared to still express parental markers (Fig. 3D,E). In addition, the t-SNE plot (Fig. 3D) shows some subclustering within the A375-BR population, but these differences were not robust enough to represent statistically distinct subclusters.
We next asked whether any of the cell populations showed more or less heterogeneity in expression markers either before or after selection for resistance to BRAF inhibitors. Qualitatively, the heatmap of Figure 3E shows that the parental and resistant A375 cells separate cleanly by features identified using the SAKE algorithm. However, the parental and resistant 451Lu cells appear to show more mixing in the cluster dendrogram and more overlap in the expression of SAKE-identified markers that distinguish parental and resistant cells. Sampling a larger number of cells from these populations would enable the distinction between these possibilities.
High-throughput sparse 10x libraries for rare cell identification
We selected the 451Lu parental and resistant populations for higher throughput profiling on the Chromium Single Cell 3′ Solution from 10x Genomics, which provided information for many more cells with shallower coverage (Supplemental Table S2). On average, we obtained about 90,000 reads and more than 5000 expressed genes in approximately 6500 cells (Supplemental Table S2). We first used t-SNE projection maps to get a general overview of the sequencing results, as displayed in Figure 3F. In the 451Lu parental cells, the majority of the cells fell into a single extended cluster, and two outlier groups formed unique subpopulations that were isolated and distinct from the rest of the parental population. These small isolated groups represented cells with extremely low or high transcript coverage rates, according to their UMIs, and were not considered in further analysis. Cells from the 451Lu-BR-resistant population formed a single group and did not have a clear separation into isolated subpopulations or distinct mixing with the parental population (Fig. 3F).
The sequencing depth and the number of expressed transcripts detected from the 10x data showed more variance as well as lower overall transcript coverage than the Fluidigm/Smart-Seq data (Supplemental Table S2), as expected from the lower total read counts sequenced per cell. To assess whether we could combine and compare these two types of data, we used the Nearest Shrunken Centroids method (Tibshirani et al. 2002) to calculate centroids from each population separately for parental and resistant cells and score their similarity to the overall centroid of the population (Supplemental Methods) in terms of how “parental-like” or “BR-like” the cells appeared. Results from both the C1 and 10x data could then be displayed together (Fig. 3G) and indicated that most cells from each platform fell cleanly into one category or another. In addition, we compared the differentially expressed genes between the BRAFi-resistant and parental cells for both C1 and 10x data and identified a largely similar set of commonly altered genes (Fig. 3H; Supplemental Fig. S5A–E), with little platform-dependent effects beyond sequencing depth and the ability to identify rare cells in larger populations.
DCT marks cells with intrinsic resistance in 451Lu melanoma cells
In the t-SNE project map of the C1 data set (Fig. 3D), a small number of 451Lu-Par cells can be seen located near the 451Lu-BR population and distinct from the rest of the 451Lu-Par populations (Fig. 3D,E). We wanted to test whether these rare parental cells that are exhibiting similar transcriptomic profiles to BRAFi-resistant cells would also display less sensitivity to the BRAFi drug treatment. This would be consistent with an intrinsically resistant population already present in the parental cells.
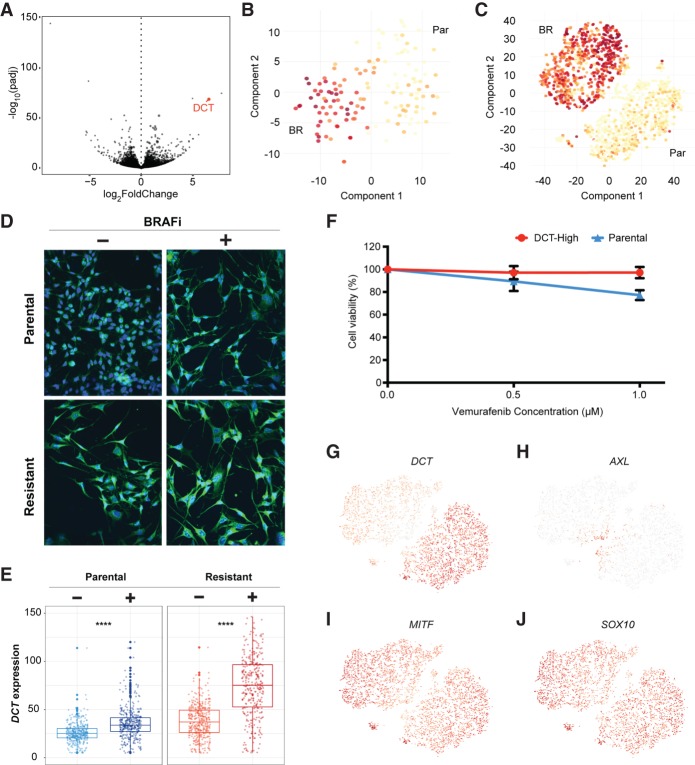
We used a standard differential expression analysis method, DESeq2 (Love et al. 2014), to determine the list of statistically enriched genes in the parental and resistant 451Lu data, requiring twofold mean expression changes and P < 0.05 (Fig. 4A). Although many significantly altered genes showed highly variable levels of gene expression across the two platforms (Supplemental Fig. S6A,B; Supplemental Table S3), DCT was one of the highest confidence candidates that showed a consistent expression pattern between parental and resistant cells in both the Fluidigm C1 and 10x data sets (Fig. 4B,C; Supplemental Fig. S6A,B). Parental cells with high DCT protein expression were isolated by FACS and tested for differential drug response following BRAF inhibitor challenge. These DCT-high parental cells showed a significantly reduced response to BRAF inhibitors, as measured by MTT assays for metabolically active cells 48 h after plating the cells in media containing 1 µM BRAFi compounds (Fig. 4F). This is consistent with DCT protein expression marking parental cells with reduced sensitivity to BRAF inhibitor treatment. To further confirm this, we stained 451Lu-Par cells with DCT antibody and checked for the proportion of cells that survived after BRAF inhibitor treatment. If DCT-high parental cells show better tolerance to BRAF inhibition, there should be a higher percentage of DCT-positive cells remaining following application of BRAF inhibitor treatments to the naïve parental population. Consistent with this, few DCT-positive cells can be seen in the parental population prior to addition of BRAFi compounds to the media (Fig. 4D, left panels). Following treatment, the remaining cells can be seen to largely be DCT positive (Fig. 4D, right panels). As a positive control, most cells from the derived BRAFi-resistant 451Lu population express high levels of DCT (Fig. 4E).
Figure 4.
DCT marks 451Lu melanoma cells intrinsically resistant to BRAFi drug treatments. (A) A Volcano plot displays adjusted −log10(P-value) versus log2FoldChange between BRAFi-resistant and parental cells, which identified DCT as one of the strongest markers of the resistant population. t-SNE maps were used to display the DCT expression levels from data generated through Fluidigm C1 platform (B), and the 10x Genomics sequencing technologies (C). (D,E) Most of the resistant cells had higher levels of DCT compared with the parental cells. Cells were stained with a fluorescent-labeled DCT antibody (green) and counterstained with DAPI (blue). BRAFi-resistant cells show higher expression of DCT at the protein level, quantified in the box plots of E. (F) MTT assays measured metabolically active cells 48 h after application of BRAFi to the media. DCT-positive cells, isolated by FACS from the 451Lu parental population, show significantly reduced response to BRAFi and higher survival rates. (G) DCT shows a binary expression pattern, with high levels in BRAFi-resistant single cells. (H–J) Previously published markers of BRAFi resistance do not show similar binary expression patterns across cell populations.
We additionally checked a list of previously published genes that have been identified as associated with resistance to BRAF inhibitors. Most of these genes did not show a binary pattern similar to DCT, with dominant expression in one of the two populations (Fig. 4G). Instead, most of these published resistance marker genes showed a curious pattern of expression that was most differential at the interface between the two populations (Fig. 4H–J; Supplemental Fig. S7). One possible explanation for this expression pattern would present a model in which expression of some genes is required to initiate the process of developing drug resistance, but this expression is not required for maintenance of the drug-resistant state. The level of support for this model is explored next.
Support for a transient transcriptional state allowing acquired drug resistance
A recently published study used bulk RNA-seq and high-throughput single-cell RNA FISH techniques to identify a panel of signature genes associated with BRAF inhibitor resistance in human melanoma cells (Shaffer et al. 2017). This study proposed that selected genes identified previously in the literature, such as WNT5A, AXL, EGFR, PDGFRB, and JUN, could mark a transitional stage in which cells are better able to develop resistance to BRAFi treatment, a state they dubbed “pre-resistant.” After removal of BRAF inhibitors, these intermediate cells were more likely to revert to their naïve parental state and become drug sensitive again. Intrigued by this finding, we checked the expression distributions for a subset of these transient markers in our data (Supplemental Fig. S7). Although we did not see enrichment for these markers in any isolated outlier groups on the t-SNE maps, these markers were enriched at the tip of the parental population that was proximal to the resistant populations as well as at the tip of the resistant population that was proximal to the parental cluster.
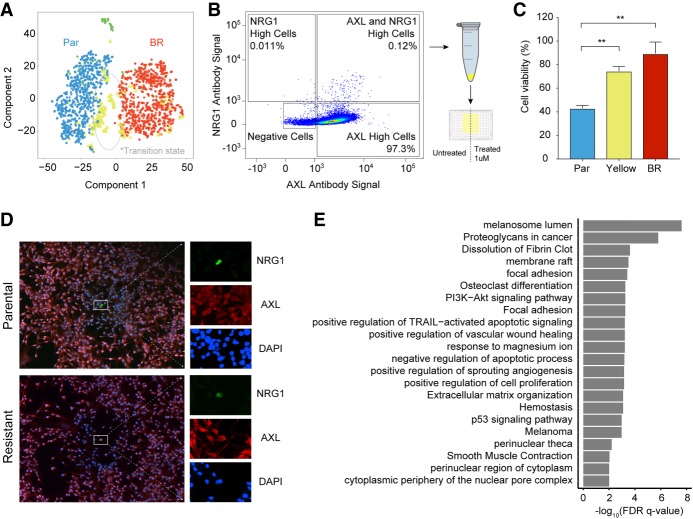
We ran SAKE to determine whether SAKE would also identify the presence of this unique “cluster” of candidate transient cells based solely on their transcriptional profile without any prior knowledge from the marker lists. SAKE reported three major clusters from the entire 10x data set: parental cells, resistant cells, and a new population of cells that sat at the interface between these two major clusters (marked in yellow on Fig. 5A and triangles in Supplemental Fig. S9A–F). The cells in this yellow intermediate population were marked by high levels of AXL, JUN, NGFR, WNT5A, FGFR1, and NRG1 (Supplemental Figs. S8, S9), which is consistent with the expression patterns displayed by the transient population previously identified by Shaffer et al. (2017). This is consistent with the hypothesis that some melanoma cells acquire resistance by transiting through a transient “pre-resistant” state marked by high levels of MAPK pathway genes. SAKE identified several additional genes that mark this candidate “transitional” population, which are given in Supplemental Table S4. Although these cells do not occupy an isolated subcluster on the t-SNE projection map, the SAKE algorithm did identify this population as a robust subcluster, highlighting the success of SAKE at finding small subpopulations in high dimensional data. We ran each of the five other methods listed in Figure 2 on the 10x data, finding wide disagreement on the number of clusters present (Supplemental Fig. S11A,B). Moreover, no other method identified the yellow cells as an isolated population present in both parental and BRAFi-resistant cells, although several methods did identify clusters enriched for the yellow cell population (Supplemental Fig. S11D).
Figure 5.
Transcriptome profiles identified an intermediate state between parental and resistant cells. (A) A t-SNE map displays SAKE-identified groups from the 10x data set of 451Lu cells. One group of cells (yellow) marked a potential transitional state during the acquisition of BRAFi resistance, present in both parental and BRAFi-resistant populations. (B) FACS analysis was used to identify cells doubly positive for two of the markers expressed in the proposed transitional state, NRG1 and AXL. These rare cells represented <1% of the 451Lu Parental population. (C) CellTiter-Glo assays were used to assess the BRAFi sensitivity of AXL/NRG1 doubly positive cells (yellow), which showed significantly increased survival in a 1 µM dose of BRAFi compared with the rest of the 451Lu Parental population (blue). (D) Confocal images of doubly positive AXL/NRG1 cells (green) show these cells are rare in both the 451Lu Parental and 451Lu BR populations. (E) Differentially expressed genes between cells in the transitional state and the rest of the 451Lu Parental cells were used for gene set enrichment analysis.
We next sought to determine whether the yellow cells are truly a distinct population with different phenotypic properties. Indeed, if these cells are truly marking a pre-resistant population, then they should be less sensitive to BRAFi treatment. Therefore, we used two of the markers identified by SAKE, AXL, and NRG1, to select the candidate pre-resistant cells from the 451Lu parental population. Cells that showed high levels of both AXL and NRG1 were rare, representing <1% of the total population, as shown in flow Figure 5B and in confocal images obtained in Figure 5D. These doubly positive cells were sorted from the larger population and tested for their sensitivity to BRAFi treatment using a CellTiter-Glo assay (Supplemental Methods; Supplemental Fig. S10) to measure metabolically active cells 72 h after plating the cells in media containing 1 µM BRAFi compounds. The yellow cells marked by high levels of both AXL and NRG1 showed a significantly reduced response (and higher growth rate) in the presence of BRAFi compounds than the rest of the parental population (Fig. 5C) at levels that were near those of the stably resistant 451Lu populations. Thus, SAKE was able to identify markers of cells that do display reduced sensitivity to the BRAF inhibitors, even when these markers do not persist as a dominant expression pattern in the fully resistant population. This is consistent with the patterns seen in the transient populations of pre-resistant cells identified previously (Supplemental Fig. S12; Shaffer et al. 2017).
Although we have shown that the yellow cell population does have both transcriptional and phenotypic differences from the rest of the 451Lu parental population, we cannot formally rule out the possibility that the yellow cells are in a transient rather than transitional state. Better evidence would come from obtaining both CNV data and gene expression profiles from the same cells in order to determine whether the yellow cells are genetically related to each other. As a proxy for true CNV profiles, we generated inferred genomic copy number data from the RNA-seq profiles following a computational method previously established (Patel et al. 2014; Tirosh et al. 2016). To ensure that the CNV profiles we inferred using this method match what would have been found as directly measured from the DNA, we additionally isolated DNA from bulk cell populations from each of the parental and resistant populations and generated DNA libraries directly (Supplemental Table S4; Supplemental Methods). We found a high degree of concordance between the CNV profiles derived from the scRNA-seq data and the bulk DNA-seq data (Supplemental Fig. S13). The inferred CNV patterns showed two major clones that corresponded to the parental and resistant cell populations. Depletions on Chr9p and amplifications on Chr1p, Chr7p, Chr11q, Chr20q, and Chr22q were signatures for the resistant cells, matching what has been reported in other CNV profiles using bulk melanoma samples (Beroukhim et al. 2010). Beyond the two dominant clones, these cells were further classified into eight subgroups, or “clonal lineages.” However, none of these eight subgroups of cells showed an enrichment for the cells that are identified as having the “transitional” transcriptome patterns (yellow bars in Supplemental Fig. S13A), which could be found in several of the eight subgroups. This suggested that these cells were derived from several unrelated clonal lineages and were likely not derived from a single strictly heritable lineage. This is consistent with the hypothesis that nongenetic effects contribute to the acquisition of resistance to BRAF inhibitors in melanoma cells and is consistent with previous reports (Shaffer et al. 2017; Sharma et al. 2017).
Discussion
Methods developed for bulk RNA-seq data analysis can be adapted for application to scRNA-seq data, but need to be tailored to address the specific issues inherent in working with noisy, low-coverage, heterogeneous sample sets. We present a processing and analysis pipeline for single-cell RNA-seq data sets, SAKE, that is designed to look for quality control issues specific to scRNA-seq data, identify subclusters present in the cell populations, and evaluate the gene and functional group enrichments in each cluster. Importantly, SAKE provides quantitative estimates of cluster membership alongside qualitative evaluations of cluster relationships via t-SNE projection maps. We show that SAKE can provide very similar single-cell cluster results to those derived from a variety of sample sources and evaluated with very different algorithms. We also show that SAKE provides accurate and robust results for a wide range of experimental designs. Moreover, SAKE operates as a simple, intuitive pipeline in which the parameters are set by quantitative metrics provided by SAKE during run time.
Having verified the success of SAKE on several published scRNA-seq data sets, we next applied this method to uncover the transcriptional alterations that occur as melanoma cells develop resistance to targeted inhibitors of the BRAF oncogene. Single-cell transcriptomes were obtained from both a low-throughput high-depth method (Fluidigm C1, approximately 100 cells per group) as well as a high-throughput shallow sequencing technology (10x genomics, approximately 3500 cells per group). The expression profiles and typical melanoma specific marker levels were very similar between these two data sets. Overall, these two sequencing technologies provided highly concordant results, suggesting that platform-specific effects did not dominate the results.
Identification of rare ‘intrinsically resistant cells’
The data sets from the Fluidigm C1 and 10x platforms were combined to identify statistically differentially expressed genes between the 451Lu parental and resistant populations. Those genes that were highly abundant in the 451Lu resistant cells, with much lower overall expression in the 451Lu parental population, formed a candidate list of resistance marker genes. We next sought to determine whether rare cells in the parental population might be expressing these candidate resistance markers. We validated one novel resistance marker gene, DCT, with high expression in the majority of the BRAFi-resistant cells and low expression in 99% of the parental population. These rare DCT-positive cells were sorted from the parental population and challenged to determine sensitivity to BRAFi treatment. Consistent with DCT marking cells that are more likely to be resistant to BRAFi treatment, the DCT-positive population showed reduced sensitivity to BRAFi challenge. Although one report identified DCT as a potential marker for general resistance to radiotherapy (Pak et al. 2004), DCT has not previously been evaluated for its role in resistance to targeted inhibitors or specifically BRAF inhibitors.
Identification of a ‘transitional state’ of pre-resistant cells
Most of the genes that were identified in previous studies to confer resistance to BRAFi treatment in melanoma did not show a similar binary expression pattern to DCT, with high levels in one population. Instead, a large fraction of these genes showed differential expression just at the boundary between the parental and resistant populations. This expression pattern could be consistent with previously proposed models that suggest a transitional cellular state dubbed “pre-resistance” in which cells transiently express these markers as they develop resistance to BRAF inhibitors (Shaffer et al. 2017; Sharma et al. 2017), but do not continue marker expression once resistance is stably established. We experimentally isolated cells expressing markers of this candidate “pre-resistant” population and verified that they do exhibit reduced sensitivity to BRAFi compounds. In this model, one might expect cells with shared transient transcriptomic patterns, marking the transitional state, to not necessarily share genomic mutational patterns.
Integrating genomic mutational information allows for an explanation of the clonal relationship of the cells that were selected for resistance to BRAF inhibitors. If most cells in the resistant population appear to be more genetically similar than the cells in the parental population, this would imply a bottleneck scenario in which a small number of clones were selected. On the other hand, if resistance can also arise stochastically through nongenetic mechanisms, one would expect many genetically distinct clones with shared transcriptome patterns. Our data seem to support a contribution from both genetic and nongenetic mechanisms. Specifically, the intrinsically resistant DCT-positive cells likely mark a small number of clonally related cells that were innately resistant to BRAFi treatment prior to drug application and which were simply selected from the population. However, our proxy genomic CNV and RNA-seq information showed that the candidate “transitional” cells appear to derive from many distinct clonal lineages. Although these results must be tempered by the caveats imposed by using inferred CNV information from RNA-seq data, these results suggest that the transitional expression pattern was not clonally inherited. Further results that obtain high-throughput single-cell DNA and RNA from the same cells would better settle the question of whether nongenetic mechanisms could offer a path for acquired resistance to targeted inhibitors.
Methods
Cell culture
Parental cell lines A375 and 451Lu were cultured in DMEM media supplemented with 10% FBS and 1% Penicillin-Streptomycin. Resistant cell lines A375-Br and 451Lu-BR3 were cultured in DMEM media supplemented with 10% FBS, 1% Penicillin-Streptomycin, and 1 µM of BRAF inhibitor, PLX-4720 (Selleckchem S1152).
RNA-seq library preparation
Briefly, bulk libraries were created using TruSeq Stranded Total RNA kits. Fluidigm C1 libraries were generated using the Clontech SMART-Seq v4 Ultra Low Input RNA Kit. 10x Genomics platform libraries were generated using the Chromium Single Cell 3′ Chip Kit V2. Extended details about the generation of these libraries are available in the Methods section of the Supplemental Materials.
RNA-seq data processing
The RNA-seq reads were mapped with STAR 2.5.2b (Dobin et al. 2013) to the hg19 reference genome. Although the most recent genome build, GRCh38, was released in 2013, much of the published work used for comparison in this study was mapped to hg19. We thus determined that mapping to hg19 would maintain consistency and was unlikely to cause changes large enough to affect the conclusions of this study. Kallisto 0.42.5 (Bray et al. 2016) was then used for abundance estimation of genes in the UCSC RefSeq gene list. DESeq2 (Love et al. 2014) was used to identify differential expressed genes between conditions. Libraries were normalized to reads per million mapped (RPM). The RPM normalized gene expression matrix was then floored and log-transformed prior to clustering: M′ = log2(M + 1).
MTT and CellTiter-Glo Assays for measuring cell viability in BRAFi
Parental (A375, 451Lu) and Resistant (A375-BR, 451Lu-BR3) cells were plated in 96-well plates at a concentration of 7500–10,000 cells/well and allowed to culture overnight. Once the cells were at 70%–75% confluency, cells were treated in triplicates with different concentrations (0.1–1.0 µM) of PLX-4720 in 200 µL of media for 72 h. After treatment, the media was replaced with 100 µL fresh phenol red free media. MTT assays were then carried using the Vybrant MTT Cell Proliferation Assay Kit from Thermo Fisher Scientific. For CellTiter-Glo assays, the cells were plated at 3000 cells/well in 384-well plates. Otherwise, the same protocol for culture and addition of PLX-4720 was followed prior to viability assays using the manufacturer's instructions (Promega, G7570).
FACS to isolate select populations
Cells were trypsinized and resuspended in FACS buffer (PBS with 0.1% BSA and 0.05% Sodium Azide) and stained with the corresponding antibody. Using a BD FACS Aria, 5–10 million cells were bulk sorted into DMEM media supplemented with 10% FBS and 1% Penicillin-Streptomycin. The sorted cells were then immediately plated on 96-well plates at a concentration of 5000 cells/well. All antibodies were obtained from Santa Cruz Biotechnology: DCT conjugated to Alexa Fluor 488 (SC-74439), AXL conjugated to PE (SC-166269), and NRG1 conjugated to FITC (SC-393006).
Bulk DNA sequencing
DNA was extracted from 3 to 4 million cells using Qiagen DNeasy Blood and Tissue kit. Two micrograms DNA was fragmented using a Covaris sonicator. Libraries for sequencing were prepared by subjecting the resulting DNA fragments to end-repair, 3′ adenylation, and ligation of TruSeq barcoded adapters. A 0.75 ratio of Ampure XP beads to sample was used to select fragments larger than 200 bp. These fragments were further amplified by PCR, cleaned using QIAquick PCR Purification Kit, and then once again size selected using Ampure XP beads with a 0.75 ratio of beads to sample. The final libraries were sequenced using SE76 on an Illumina NextSeq to a depth of 6–7 million reads per sample.
Inferred CNV profiles from RNA-seq data
The inferCNV algorithm (Tirosh et al. 2016) was used to generated inferred copy number profiles from the scRNA-seq data in order to estimate genetic relatedness of the thousands of cells in our 10x data sets, using sliding windows containing 100 consecutive genes. For additional details, see Supplemental Methods.
Imaging
Cells were plated on glass coverslips coated with 0.1% Poly L-Lysine solution and allowed to grow for 24 h. Cells were then treated with a 1.0 µM concentration of PLX-4720 for 72 h. After treatment, cells were washed with PBS and fixed using ice-cold methanol. Fixed cells were blocked with 5% FBS in PBS overnight at 4°C and stained with a 1:50 concentration of Anti-DCT antibody conjugated to Alexa Fluor 488 (SC-74439).
This was followed by NucBlue counterstaining of nuclei according to the manufacturer's manual and mounted on a slide using ProLong Diamond Antifade Mountant. Imaging was performed on a confocal microscope.
Software availability
SAKE is an R-based software package (R Core Team 2017) available for download at https://github.com/naikai/sake and provided as a gzipped tar archive in Supplemental Material. A detailed tutorial for using SAKE is also provided as R markdown documents that allow for instructions, commands, and results to be presented together.
Data access
Illumina sequencing data from this study have been submitted to the NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA427121. These data are also available from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP127319 (bulk RNA-seq), SRP127320 (Fluidigm scRNA-seq), SRP127328 (10x Genomics scRNA-seq), and SRP127299 (DNA WGS).
Supplementary Material
Acknowledgments
Y.J.H. is funded through generous supports from the Ministry of Education in Taiwan and The Florence Gould Foundation. M.G.H. is a scholar of the Rita Allen Foundation. We also acknowledge a grant from Amazon Web Services to D.M., which provided a virtual demo web server for hosting SAKE analyses on the cloud. We thank Meenhard Herlyn and his laboratory members for their many helpful comments, suggestions, and for the gift of the 451Lu-BR cells. We also acknowledge help from the Bioinformatics Shared Resource at Cold Spring Harbor Laboratory, which is supported by a grant from the NIH/NCI.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.234062.117.
References
- Bendall SC, Davis KL, Amir el-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe'er D. 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Church G. 2005. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics 21: 3970–3975. [DOI] [PubMed] [Google Scholar]
- Goolam M, Scialdone A, Graham SJL, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC, Zernicka-Goetz M. 2016. Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165: 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, van Oudenaarden A. 2015. Design and analysis of single-cell sequencing experiments. Cell 163: 799–810. [DOI] [PubMed] [Google Scholar]
- Grün D, Anna L, Lennart K, Kay W, Onur B, Nobuo S, Hans C, van Oudenaarden A. 2015. Single-cell mRNA sequencing reveals rare intestinal cell types. Nature 525: 251–255. [DOI] [PubMed] [Google Scholar]
- Guo M, Wang H, Potter SS, Whitsett JA, Xu Y. 2015. SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput Biol 11: e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Buettner F, Theis FJ. 2015. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31: 2989–2998. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson M, Niu B, McLellan MD, Uzunangelov V, et al. 2014. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Döbbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. 2008. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res 68: 650–656. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. 2014. Bayesian approach to single-cell differential expression analysis. Nat Methods 11: 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park H. 2007. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 23: 1495–1502. [DOI] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, et al. 2017. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods 14: 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161: 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, Berishvili E, Bock C, Kubicek S. 2016. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep 17: 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Troup M, Ho JW. 2017. CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol 18: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC. 2014. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci 111: E5643–E5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, et al. 2015. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 47: 1168–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes Foppen MH, et al. 2014. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 5: 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak BJ, Lee J, Thai BL, Fuchs SY, Shaked Y, Ronai Z, Kerbel RS, Ben-David Y. 2004. Radiation resistance of human melanoma analysed by retroviral insertional mutagenesis reveals a possible role for dopachrome tautomerase. Oncogene 23: 30–38. [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, Iorio F, Alifrangis C, Arends MJ, Bosenberg MW, Bollag G, et al. 2015. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc Natl Acad Sci 112: E536–E545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe'er D. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 34: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, Beqiri M, Sproesser K, Brafford PA, Xiao M, et al. 2017. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. 2017. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168: 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4: 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM, et al. 2014. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508: 118–122. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci 99: 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, et al. 2014. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8: 1905–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. 2016. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539: 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Lee Q, Camp GJ, Mall M, Koh W, Shariati S, Sim S, Neff NF, Skotheim JM, Wernig M, et al. 2016. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J Mach Learn Res 9: 2579–2605. [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. 2010. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Su Z. 2015. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 31: 1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Michailidis G. 2016. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics 32: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.