Abstract
DNA methylation patterns in the genome both reflect and help to mediate transcriptional regulatory processes. The digital nature of DNA methylation, present or absent on each allele, makes this assay capable of quantifying events in subpopulations of cells, whereas genome-wide chromatin studies lack the same quantitative capacity. Testing DNA methylation throughout the genome is possible using whole-genome bisulfite sequencing (WGBS), but the high costs associated with the assay have made it impractical for studies involving more than limited numbers of samples. We have optimized a new transposase-based library preparation assay for the Illumina HiSeq X platform suitable for limited amounts of DNA and providing a major cost reduction for WGBS. By incorporating methylated cytosines during fragment end repair, we reveal an end-repair artifact affecting 1%–2% of reads that we can remove analytically. We show that the use of a high (G + C) content spike-in performs better than PhiX in terms of bisulfite sequencing quality. As expected, the loci with transposase-accessible chromatin are DNA hypomethylated and enriched in flanking regions by post-translational modifications of histones usually associated with positive effects on gene expression. Using these transposase-accessible loci to represent the cis-regulatory loci in the genome, we compared the representation of these loci between WGBS and other genome-wide DNA methylation assays, showing WGBS to outperform substantially all of the alternatives. We conclude that it is now technologically and financially feasible to perform WGBS in larger numbers of samples with greater accuracy than previously possible.
The study of cytosine modifications is central to the understanding of how transcriptional regulation is maintained in a cell type, as DNA methylation (5-methylcytosine, 5mC) can be propagated through mitosis to daughter chromatids (Jeltsch 2006) and can therefore mediate the “persistent homeostasis” described by Nanney (1958) as the defining characteristic of cellular epigenetic memory. In human disease susceptibility, evidence for persistent homeostasis is sought at the molecular level, testing whether cells exposed to a past perturbation have retained a memory of that event by undergoing cellular reprogramming (Lappalainen and Greally 2017). The need for such events to be heritable through mitosis in cells undergoing division coupled with the relative technical ease of studying DNA methylation has prompted a large and increasing number of studies of DNA methylation to test whether it could be associated with or even contributing to these phenotypes (Michels et al. 2013). Biochemically, DNA methylation is a term used to describe 5mC, which almost always occurs at cytosines followed by a guanine, the CG (CpG) dinucleotide. This modification occurs genome-wide, generally sparing cis-regulatory sites, especially those where CG dinucleotides cluster densely (CpG islands). Oxidase activity converts 5mC to 5-hydroxymethylcytosine (5hmC), 5-carboxylcytosine (5caC), and 5-formylcytosine (5fC), which occur at very low levels relative to 5mC. There are numerous assays that measure DNA methylation (Ulahannan and Greally 2015), the current gold standard approach using sodium bisulfite conversion to quantify DNA methylation at nucleotide resolution (Clark et al. 1994). Sodium bisulfite converts unmodified cytosine, 5caC, and 5fC to uracil, but both 5mC and 5hmC are resistant to bisulfite conversion, allowing the ratio of converted to unconverted cytosines at a locus to be calculated, representing the proportion of alleles in the cell population with 5mC, as well as the minor fraction of 5hmC.
Epigenetic association studies require testing large numbers of individuals in order to find differences to which statistical significance can be attributed. The choice of assays to use for these studies represents a trade-off between cost and information content. Random fragmentation of the genome followed by bisulfite sequencing allows the whole-genome bisulfite sequencing (WGBS) assay to be performed. However, to get enough coverage to represent cytosines at CG dinucleotides and elsewhere in the genome, the amount of sequencing required has been too costly for most research budgets. The decision has therefore been made to sacrifice comprehensiveness and to survey the genome instead, attempting to focus on the loci believed to be the most informative in the genome. For microarrays, this has involved getting expert input and using genomic annotations to curate the loci represented (Bibikova et al. 2011), while sequencing-based approaches focus on using restriction enzymes that cut at motifs containing CG dinucleotides (Meissner et al. 2005; Smith et al. 2009; Suzuki et al. 2010) or capture enrichment for loci believed to be maximally informative (Li et al. 2015), tending to overrepresent gene promoters and CpG islands (Ulahannan and Greally 2015).
The problem with a single survey design is that it is unlikely to be equally informative across different cell and tissue types. It is also increasingly apparent that changes in DNA methylation are more informative at distal cis-regulatory elements rather than canonical promoters (Schmidl et al. 2009; Aran et al. 2013; Blair et al. 2013; Ko et al. 2013; Hu et al. 2014; Rönnerblad et al. 2014; Taberlay et al. 2014; Zhang et al. 2014). These distal cis-regulatory elements are also notable for being more cell-type–specific in terms of their genomic locations than promoters (Won et al. 2013), increasing the challenge when trying to find a survey assay that is informative for multiple different cell types. We have shown that several of the most common survey assays for DNA methylation grossly underrepresent these distal regulatory loci (Ulahannan and Greally 2015). While we have developed targeted bisulfite sequencing as an interim solution (Li et al. 2015), the capture component of the assay has significant costs, and more sequencing is needed as greater proportions of the genome are captured, with additional costs involved when redesigning for a new cell or tissue type. WGBS is therefore emerging as the optimal strategy for comprehensive DNA methylation studies across different cell types.
It has therefore become a major priority to try to harness the advances in technologies that are permitting whole-genome sequencing (WGS) to be performed increasingly cost-efficiently, so that similar cost savings can be applied to WGBS. The release of the Illumina HiSeq X reduced the cost of WGS substantially and has prompted exploration of this platform for WGBS (Raine et al. 2018). In this report, we describe how we developed a new protocol to utilize fully the capabilities of the HiSeq X and of other high-output instruments, such as the HiSeq 4000, and reduce the cost of WGBS, based on the development of a new transposase-based library preparation protocol and optimization of sequencing.
Results
The cost efficiency of the HiSeq X system and of similar systems, such as the HiSeq 4000, in part depends upon the libraries having large inserts that allow 150-bp paired-end sequencing to work effectively without a high fraction of reads overlapping within the insert, a practical problem when using DNA treated with sodium bisulfite, which has a degradative effect. The TruSeq DNA Methylation Kit for WGBS marketed by Illumina uses post-bisulfite adaptor tagging (PBAT) (Miura et al. 2012), for which 75-bp paired-end sequencing is recommended, suitable for the shorter fragments from PBAT libraries. Apart from the insert size issue, the use of patterned flow cells and different base calling software on the HiSeq X system makes a transition from the use of earlier assays and technologies potentially problematic, requiring the optimization of both library preparation and sequencing, as we describe below.
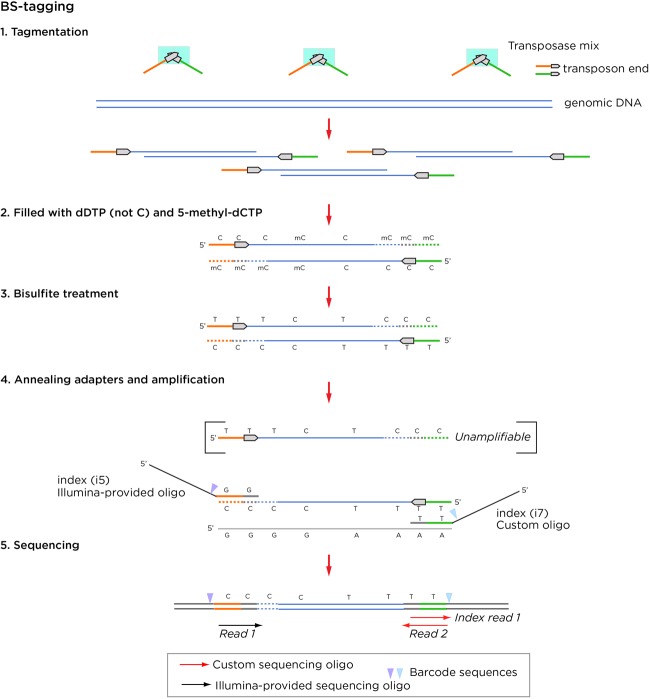
We developed a new transposase-based approach that we call BS (bisulfite)-tagging, illustrated in Figure 1. In its use of transposases, the assay resembles prior tagmentation-based bisulfite library preparation assays (Adey and Shendure 2012; Wang et al. 2013), while the incorporation of 5mC for end repair is comparable with the T-WGBS approach used for very low input DNA amounts (Lu et al. 2015), and generates an initial normal complexity sequence before reading into the (G + C)-depleted bisulfite-converted insert. Unlike T-WGBS, the extra expenses of a modified transposase and premethylated oligonucleotides are not required for BS-tagging.
Figure 1.
Overview of the BS-tagging assay. Standard transposases are used but with a 5mC fill-in. The bisulfite treatment changes the original transposon sequence and prevents it from being amplified, but the fill-in complement with 5mC remains amplifiable. One custom oligonucleotide is needed for this assay.
There are three types of duplicate sequences that can be generated using the X system. The X system-specific problem, because of the use of patterned flow cells, is when a library fragment occupying one well migrates or jumps into an adjacent well, referred to as a proximal duplicate. The second is the PCR duplicate, in which the same library fragment is amplified and is sequenced in different wells, while the third is the separate amplification of each of two complementary strands of DNA (complementary strand duplicate). Duplicates are usually identifiable by having the same start and end positions in the genome, allowing their removal by data filtering. As BS-tagging can only amplify one of the two complementary strands, these should never enter the library to begin with, reducing this source of nonproductive sequencing. We show the duplicate rates for X-WGBS as part of Supplemental Table S1. Our stringent removal of smaller insert size library components in order to minimize overlapping read pairs (and hence, effective coverage) reduces the amount of starting material, probably contributing to the increased PCR duplicate rates compared with WGS, but without the additional penalty of complementary strand duplicates. The insert size of WGBS libraries using PBAT approaches tends to be small, <210 bp (Raine et al. 2018). To optimize insert sizes for the 150-bp paired-end sequencing of the Illumina HiSeq X system, we adjusted the transposase treatment and the bead clean up conditions, as described in the supplied BS-tagging protocol.
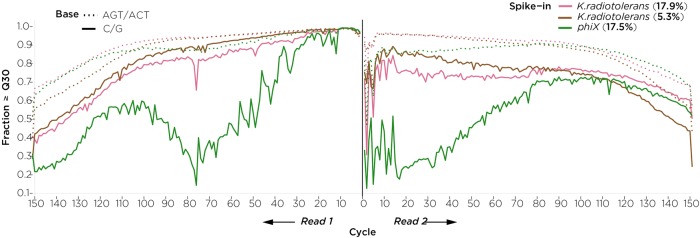
The sequencing was also customized. Early versions of the Illumina software (Toh et al. 2017) (e.g., HiSeq Control Software [HCS] v3.3.39, Real Time Analysis [RTA] v2.7.1) were not designed to handle unbalanced libraries and required a substantial spike-in of PhiX to generate data of reasonable quality. In that setting, we tested whether we could use an alternative source of spike-in DNA with a higher (G + C) content as a more effective balance for the (A + T)-rich bisulfite-converted DNA. We compared Illumina's 44% (G + C) PhiX spike-in (Kircher et al. 2009) with a spike-in library prepared from Kineococcus radiotolerans, which has 74% (G + C) (Bagwell et al. 2008), finding that the K. radiotolerans spike-in performed markedly better than PhiX when both were added at ∼17% proportions, and that even 5.3% K. radiotolerans was enough to restore base quality (Fig. 2). These results suggest the broader possibility that a K. radiotolerans spike-in may be worth exploring as an alternative to PhiX when sequencing not just bisulfite-converted DNA but also extremely (A + T)-rich genomes. The more recent versions of the HiSeq X software (HCS 3.4.0.38, RTA 2.7.7) included a revised Q-table to facilitate sequencing of unbalanced libraries (including WGBS), allowing a 5% PhiX spike-in to generate high-quality sequencing data. However, since PhiX libraries are often slightly different in size than WGBS libraries, and the ExAmp process on the HiSeq X preferentially clusters smaller fragments, it is often difficult to achieve precise proportional representation of the PhiX spike-in in pooled libraries. A (G + C)-rich spike-in such as from K. radiotolerans in the setting of the newer HiSeq X software appears to be more robust to such variation, although we note that the insert sizes of the PhiX and K. radiotolerans libraries differed (Supplemental Fig. S1) and may contribute to some extent to the differences in quality found. We developed a custom indexing primer that allows multiplexing of samples on the X system, including the use of the custom K. radiotolerans spike-in sample. As a control for bisulfite conversion, we used unmethylated lambda DNA as a second spike-in sample.
Figure 2.
Improved quality of whole-genome bisulfite sequencing (WGBS) with Kineococcus radiotolerans used as a spike-in. The plot shows the proportion of reads at or above a quality score of 30 for read 1 (left) and read 2 (right). The pink lines show the performance of K. radiotolerans (G + C = 0.74) added at a proportion of 17.9% (light pink) or 5.3% (dark pink), and PhiX (G + C = 0.44, green). The results are plotted to show the quality for C/G (solid lines) separately from other nucleotides (dotted lines), as the performance of WGBS is especially problematic for C/G nucleotides, a fact missed by plots that do not separate out these nucleotides. The high (G + C) K. radiotolerans spike-in, even at 5.3% proportion, restores C/G quality to levels comparable with the other nucleotides.
We show our analytical pipeline in Supplemental Figure S2 and typical results in Supplemental Table S1. We compared the results of these X-WGBS data with DNA methylation data generated by other approaches in the same cell types. In Supplemental Figure S3, we show the root-mean-square error (RMSE) values for X-WGBS using bwa-meth (Pedersen et al. 2014) and Bismark (Krueger and Andrews 2011), WGBS using older sequencing technology (Lister et al. 2009), reduced representation bisulfite sequencing (RRBS), the SeqCap Epi capture approach that we developed (Li et al. 2015), and microarray-based assays (Infinium HumanMethylation450, Infinium MethylationEPIC, Illumina), showing the DNA methylation patterns of each cell type to cluster separately, concordantly across all assays.
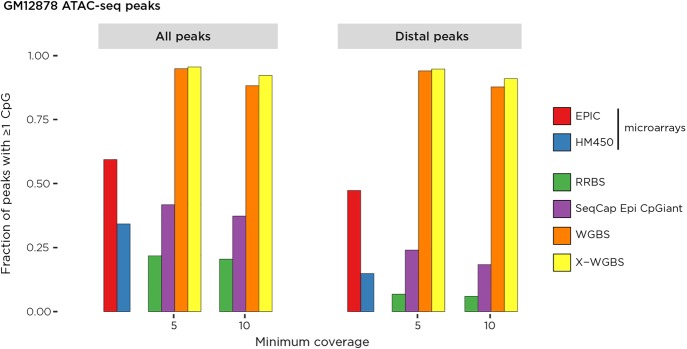
A major rationale for the use of WGBS as opposed to survey assays is the ability to measure DNA methylation at distal cis-regulatory sites in diverse tissue types (Ulahannan and Greally 2015). We tested relative performance by performing the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) (Buenrostro et al. 2013) as well as using data from the ENCODE Project (The ENCODE Project Consortium 2007) to define the cis-regulatory landscape in the GM12878 cell line. The loci of transposase-accessible chromatin coincide with loci of decreased DNA methylation and are flanked by enrichment for histone H3 lysine 4 tri- and monomethylation (H3K4me3, H3K4me1) and histone H3 lysine 27 acetylation (H3K27ac) (Supplemental Fig. S4). The X-WGBS assay reports the large majority of loci of transposase-accessible chromatin in GM12878 cells, whereas RRBS is especially poorly representative of distal (nonpromoter) regulatory loci (<10%) (Fig. 3). The SeqCap Epi CpGiant design has approximately equivalent coverage of cis-regulatory sites as the Infinium HumanMethylation450 microarray, which reflects how both are designed to target the same loci. The Infinium MethylationEPIC microarray, designed to cover more sites than the HumanMethylation450 design, interrogates over half the ATAC-seq peaks overall and just under half of the distal cis-regulatory loci in GM12878 cells.
Figure 3.
The relative representation of cis-regulatory elements genome-wide by different DNA methylation assays. We use the ATAC-seq peaks as a representation of the cis-regulatory elements in GM12878 cells. These were further subdivided into those distal (>10 kb upstream or >2 kb downstream) from RefSeq genes. The X-WGBS results interrogate at least one CG in the majority of these loci at 5 or 10× coverage, whereas RRBS represents only a small proportion of these loci, with the Infinium HumanMethylation450 microarray and the SeqCap Epi CpGiant system representing similar proportions (as expected for designs targeting the same genomic loci), with the expanded Infinium MethylationEPIC microarray representing a higher proportion of ATAC-seq peaks.
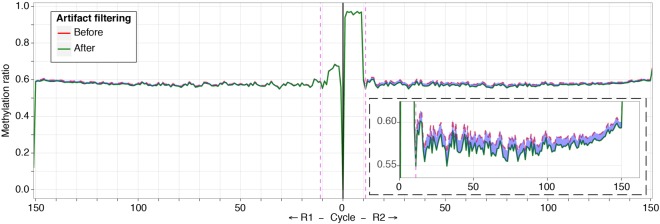
We noted the puzzling finding that lambda DNA appeared to have some unconverted cytosines indicating DNA methylation, which should not be occurring in this organism. When we explored this finding, we found the lack of conversion to occur at all cytosines in both CG and CH contexts throughout a minority of sequence reads. This indicates that our fill-in reaction using 5mC following transposase activity was extending beyond the typical several nucleotides in this subset of molecules (Supplemental Fig. S5). When we then explored the human DNA being simultaneously sequenced, we found that 1%–2% of the reads showed a similar pattern of a complete lack of conversion of all cytosines in all dinucleotide contexts (Fig. 4). This artifact is likely to have occurred in previous bisulfite sequencing studies that involved fragment end repair, but remained unrecognized because of the use of unmodified cytosine in the fill-in reaction. We therefore developed an algorithm (filterFillIn2) to identify four consecutive, unconverted CHH sites in a sequence read, allowing us to filter those for which we have a high suspicion of this technical artifact.
Figure 4.
A subset of reads shows apparent cytosine methylation throughout their lengths, in CG and CH dinucleotide contexts. Our use of 5mC for end repair reveals the expected ∼10-bp increase in the C/T ratio (y-axis) in read 1 (right). However, prompted by our finding of unconverted cytosines in lambda DNA, we tested whether any of the reads had a pattern of unconverted cytosines in all dinucleotide contexts (the expected CG and the uncommon CH contexts). We found a subset of reads in which no cytosines were converted. To categorize these, we developed the filterFillIn2 software to identify four consecutive unconverted cytosines in a CHH context. The effect on DNA methylation values is shown in the C/T plots, the original values in red and following removal in green. The inset shows that ∼1%–2% of the DNA methylation value is accounted for by this artifact.
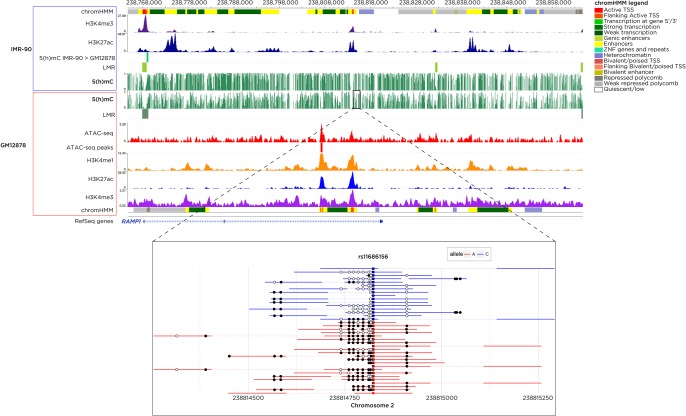
We show that the X-WGBS data reveal the many types of events typically sought in a study of mammalian DNA methylation. In Figure 5, we show examples of low-methylated regions (LMRs) (Feldmann et al. 2013) and differentially methylated regions (DMRs) (Akalin et al. 2012). We also show that the X-WGBS reads can be used to identify allele-specific DNA methylation (ASM) (Kaplow et al. 2015).
Figure 5.
Typical X-WGBS results in IMR-90 and GM12878 cell lines. The green wiggle track represents the proportion of methylated DNA at each CG dinucleotide in a ∼100-kb region on Chromosome 2, with annotations of low-methylated regions (LMRs), histone post-translational modifications, and transposase-accessible chromatin also shown. We also show allele-specific methylation (ASM) at the rs11686156 single nucleotide polymorphism which is heterozygous in GM12878 and flanked by unmethylated DNA for the C allele (blue) and methylated DNA for the A allele (red). A differentially methylated region (DMR), where DNA methylation is higher in IMR-90 than GM12878 cells, is apparent at the site of the LMRs on the left of the region shown.
When we compare the reagent costs for the equivalent amount of WGBS on the X and HiSeq 2500 platforms, the relative cost reduction associated with X-WGBS is approximately fourfold compared to the HiSeq 2500 (2 × 125 bp) and approximately threefold compared with the HiSeq 4000 (2 × 150 bp). This assumes 100 GB of raw data on the 2500 and 4000 at list prices (currently $31.70 and $20.50 per GB, respectively), and 120 GB on the HiSeq X to account for the additional spike-in needed, as well as the elevated platform-specific duplicate rates. Library preparation costs for X-WGBS are similar to standard commercial kits.
Discussion
We show how a combination of modifications to the library preparation, sequencing, and analysis can make WGBS substantially more cost-efficient and accurate. The use of transposases for library preparation coupled with the preservation of large inserts allows a restricted amount of starting DNA to be used while exploiting the long read capability of the Illumina HiSeq X. The inclusion of a custom oligonucleotide gave us the potential to add an additional index that in turn permitted the exploration of the performance of the high (G + C) K. radiotolerans spike-in DNA, associated with better sequencing performance than the standard PhiX spike-in. The selective amplification of one strand had the benefit of reducing redundant sequencing due to the generation of one of the species of potential duplicates at the first step of library preparation. Analytically, our detection of what appeared to be CH methylation throughout individual reads highlights for the first time an artifact that may have gone unrecognized in prior bisulfite sequencing experiments involving end-repair. It can be inferred that a small proportion of the DNA molecules is completely single-stranded at the fill-in stage, resulting in the incorporation of 5mC throughout the molecule and not just the ∼10 bp typically repaired (Fig. 4). Why such a subpopulation of ssDNA molecules should exist at the library preparation stage is not clear; there are no prior reports associating this with the use of transposases, although it should be recognized that this would normally be difficult to recognize to be occurring. In prior protocols using unmodified cytosine for the fill-in, the effect would have been to inflate slightly the proportion of unmethylated cytosines. The subset of affected molecules can be identified when using 5mC for the fill-in by identifying the reads with multiple consecutive methylated cytosines in a CH context, for which we developed the filterFillIn2 software, available in the Supplemental Methods and at GitHub (https://github.com/will-NYGC/bstag).
There is increasing pressure to develop a comprehensive and affordable DNA methylation assay that can be applied in phenotypic association studies. Comprehensiveness is essential so that we are always able to test informative cis-regulatory sites in any given tissue. The substantial variability of cis-regulatory site locations between cell and tissue types (Won et al. 2013) makes a single fixed design surveying the genome less than optimally comprehensive. Furthermore, bisulfite sequencing captures a lot of features about DNA methylation substantially better than is possible using survey approaches. For example, in Figure 5, we demonstrate LMRs (Feldmann et al. 2013), DMRs (Akalin et al. 2012), and ASM (Kaplow et al. 2015). What is apparent at the DMR in the figure is that it reflects the LMR being wider in IMR-90 than in GM12878. This is apparent at multiple loci when browsing these results, and appears to reflect an observation made by Hodges and colleagues several years ago, that changes in DNA methylation between cell types reflects a spreading and contraction of hypomethylated regions around a core that remains constitutively hypomethylated across cell types (Hodges et al. 2011). This is also reminiscent of the model of CpG island shores being more variably methylated than the constitutively hypomethylated CpG islands that they flank (Irizarry et al. 2009). The other major reason for studying DNA methylation as opposed to chromatin components is that DNA methylation can be measured quantitatively across the cells in the population, allowing detection of changes of transcriptional regulation patterns involving minor proportions of cells, whereas chromatin immunoprecipitation followed by sequencing (ChIP-seq) is substantially less quantitative, although capable of identifying allele-specific chromatin states (Ding et al. 2014). Changes involving small proportions of cells is the typical outcome of epigenetic association studies (Lappalainen and Greally 2017), making DNA methylation the better choice than ChIP-seq for sensitive detection of these changes.
When testing human samples, the amount of DNA may be relatively limited. As the bisulfite treatment is performed following the transposase-mediated library preparation, we not only require less input DNA to make the library, it would be possible to split the sample at this stage and use part of the library for whole-genome sequencing and the other for the remainder of the BS-tagging protocol. Concurrent sequencing to genotype the sample is going to be very valuable given the recognition that a substantial proportion of variation of DNA methylation between individuals is attributable to DNA sequence polymorphism (Lappalainen and Greally 2017). The protocol can also be used for 5hmC studies if oxBS-seq (Booth et al. 2012) were to be performed on an aliquot of the same library or can be used for subsequent capture and targeted bisulfite sequencing or 5hmC studies, as we have previously described (Li et al. 2015). The cost savings associated with the use of the Illumina HiSeq X system should translate to the newer Illumina NovaSeq system, which, as is also the case for the HiSeq 4000, is similar to the X technology in terms of the use of patterned flow cells and chemistries, assuming the RTA software is similarly tolerant of unbalanced libraries as recent versions of the HiSeq X software. The published experience with the NovaSeq system indicates that further optimization is required before it can be used reliably for WGBS (Raine et al. 2018). Furthermore, as the BS-tagging library preparation approach should be highly amenable to automation, scaled use of the assay should realize still further savings. This decrease in the cost of the WGBS assay now allows it to be considered as a first-line approach in population studies associating DNA methylation changes with phenotypes. We conclude that, while adapting WGBS to the HiSeq X system created unique challenges, these can be overcome with BS-tagging, allowing more cost-effective mammalian WGBS than previously possible.
Methods
BS-tagging and analysis
The detailed BS-tagging protocol and the list of custom oligonucleotides and primers are provided in the Supplemental Methods. We used 100 ng of genomic DNA extracted from IMR-90 and GM12878 cells for the BS-tagging library preparation. To monitor bisulfite conversion efficiency, we spiked in 0.5 ng of unmethylated lambda DNA per 100 ng of genomic DNA (Supplemental Fig. S1). The genomic DNA (gDNA) was tagmented at 37°C for 5 min with 25 µL 2× TD buffer, 5 µL transposase, and 0.5 ng of unmethylated lambda DNA in 50-µL reaction volume (Illumina Nextera). The product was purified with Dynabeads MyOne Silaine beads (Life Technologies) with Buffer PB (Qiagen). Following purification, the product was end-filled with 2.5 µL NEB buffer 2 (10×), 1.5 µL Klenow fragment (3′→5′ exo-, NEB), and 2 µL of 5-methyl-dCTP/dGTP/dATP/dTTP mix (Promega) in a 50-µL reaction volume. The product was bisulfite-converted using an EZ DNA Methylation-Gold kit (Zymo Research). Following the bisulfite treatment, the libraries were amplified with the following PCR conditions: 98°C for 30 sec, a total of 10 cycles of 98°C for 10 sec/63°C for 30 sec/3 min at 72°C using 25 µL of KAPA HiFi HotStart Uracil+ Ready Mix (2×), 1.5 µL of Illumina i5-adapter, 1.5 µL of custom i7-adapter, and 5 µL of PCR primer cocktail (Illumina). Subsequently, the libraries were purified using Agencourt AMpure XP beads at 0.6× (of PCR mixture volume) magnetic bead volume to exclude fragments <400 bp. Before running the libraries on massively parallel sequencing, we tested the length distribution of the library using a Bioanalyzer analysis (Agilent) and quantified the product with Qubit Fluorometric Quantitation. Massively parallel sequencing was performed on the Illumina HiSeq X system with HiSeq Control Software (HCS) v3.3.39 and RTA v2.7.1 using PhiX (17.9% or 5.3%) or 5.3% of K. radiotolerans gDNA as a spike-in, or RTA 2.7.7 and HCS v3.4.0.38 with 5% of PhiX as a spike-in.
We show the data analytical pipeline in Supplemental Figure S2. The obtained sequences were adaptor-trimmed using cutadapt v1.9.1 (Martin 2011) (cutadapt -O 1—mask-adaptor -g TATAAGAGACAG -a CTATCTCTTATA -G TATAAGAGATAG -A CTGTCTCTTATA -p <OUTPUT PREFIX> <R1 FASTQ> <R2 FASTQ>). Because the library protocol produces R1 reads with G>A transitions and R2 reads with C>T transitions after bisulfite conversion, short-read alignment to the Homo sapiens (GRCh37/hg19) reference genome was performed with bwa-meth with R2 reads mapped as a typical C>T converted R1 read and R1 reads mapped as a standard G>A converted read (Pedersen et al. 2014) (bwameth.py –prefix <OUTPUT PREFIX> --threads <N> --reference hg19.fa <R2 FASTQ> <R1 FASTQ>). We used hg19 to allow comparisons with microarray data, which are annotated using this older assembly. Given that sequence content is largely unchanged in the current reference sequence (i.e., GRCh38), we do not expect that realigning the reads would significantly affect our conclusions.
The resulting alignments were marked for duplicates using Picard v2.4.1 (default parameters). To calculate strand-specific duplicates, alignments were separated based on their orientation to the reference genome, then MarkDuplicates was run on the two alignment sets separately. Our use of methylated cytosines in end-repair following tagmentation revealed a rare artifact (∼1%–2% of reads) where incorporation of the methylated cytosines extended deeper into the duplex fragment than the typical ∼9–10 bp expected from the transposition footprint. Because this obscures the original methylation state, we designed a custom Perl script, filterFillIn2, which marks reads with four consecutive methylated CHs beyond the first nine bases and excludes those reads from downstream analysis by appending the 0 × 200 bitwise flag, defining them as QC failed (see Supplemental Methods). The remaining sequences were used for downstream analysis.
DNA methylation microarray analysis
Illumina Infinium HumanMethylation450 (HM450) and MethylationEPIC (EPIC) arrays performed at the New York Genome Center (NYGC) were prepared and processed according to manufacturer specifications. Raw IDAT files were processed to Beta values with a custom pipeline utilizing the minfi R package (Aryee et al. 2014) using Illumina background correction (preprocessIllumina function) and subset within array normalization (SWAN) (Maksimovic et al. 2012) (preprocessSWAN function) to correct for probe type bias.
ATAC-seq library preparation
The ATAC-seq libraries were prepared similarly to Buenrostro et al. (2013). We used 50,000 cells from two biological replicates of GM12878, harvested during the exponential growth phase. The cells were spun at 500g for 5 min at 4°C and then washed using 50 µL of cold 1× PBS. Samples were then centrifuged at 500g for 5 min. Cells were lysed in cold lysis buffer (10 mM Tris-HCL, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630) and immediately spun at 500g for 5 min at 4°C. The pellet was then resuspended in the transposase reaction mix (25 µL 2× TD buffer, 2.5 µL transposase, and 22.5 µL nuclease-free water; Illumina Nextera). Following a 30-min incubation at 37°C, the samples were purified using the Zymo DNA Clean and Concentrator purification kit. Following the purification, the libraries were amplified with the following PCR conditions: 72°C for 5 min, 98°C for 30 sec, a total of 10 cycles of 98°C for 10 sec/63°C for 30 sec/1 min for 72°C. Subsequently, the libraries were purified using Agencourt AMpure XP beads; large fragments were filtered by using 0.6× (of PCR mixture volume) magnetic bead volume and taking the supernatant. Primer-dimer and short fragments were removed by collecting bead-associated DNA in a 1:1 (bead solution volume:mixture volume) mix. The two replicates were run on different Illumina HiSeq 2500 flowcells to obtain 100-bp paired-end reads, resulting in a mean of 57 million paired-end reads per sample.
ATAC-seq peak calling
Sequenced reads were aligned to the Homo sapiens (hg19) reference genome using the Burrows-Wheeler Aligner version 0.7.13 (bwa mem –M –t <N> hg19.idxbase <R2 FASTQ> <R1 FASTQ>) (Li and Durbin 2009). Uniquely mapped reads were retained using SAMtools v0.1.19, followed by removal of mitochondrial reads by picard-tools v1.92 (Li et al. 2009). Finally, duplicate reads were removed using both picard-tools (MarkDuplicates.jar) and SAMtools (samtools rmdup). Read1 reads were shifted using BEDTools 2 v2.26.0 (Quinlan and Hall 2010), as previously performed (Buenrostro et al. 2013), before calling peaks for each replicate using MACS2 v2.1.0 (macs2 callpeak --nomodel --nolambda -g 3e9 --keep-dup “all” --slocal 10000 -t <INPUT FILE> -n <OUTPUT PREFIX>) (Quinlan and Hall 2010; Feng et al. 2012). Irreproducible discovery rates (IDRs) were found for the overlapping peaks using the method previously described (Li et al. 2011). Peaks with an IDR of less than 0.05 were filtered by consensus blacklisted regions and mitochondria homologous regions. Finally, the remaining peaks were retained for analysis.
Access and analysis of public data
Previously published whole-genome bisulfite and SeqCap Epi data were downloaded from Gene Expression Omnibus (GEO) (GSE16256) and DNA Data Bank of Japan (DDBJ) (SRP049215), respectively, and passed through the same analytical pipeline as our BS-tagging data, except without flipping R1 and R2 during alignment or excluding fill-in artifacts. For SeqCap Epi data, duplicates were also retained for downstream analysis. For the public RRBS and HM450 data, preprocessed methylation ratios were obtained from the UCSC Table Browser. Histone modification ChIP-seq data sets (GSE16256) were downloaded from the NIH ENCODE/Roadmap Epigenomics Project data matrix (https://www.encodeproject.org/). Replicates were consolidated into a single high coverage data set. FASTQs were adaptor-trimmed using Trim Galore! (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), then aligned to the hg19 reference genome using BWA-MEM (Li and Durbin 2009), both with default parameters. Alignments were further processed using the Genome Analysis Toolkit pipeline, performing insertion/deletion realignment and base quality score recalibration (DePristo et al. 2011), in accordance with the recommended best practices (https://software.broadinstitute.org/gatk/best-practices/). Duplicates were marked using Picard v2.4.1. Read fragment estimation was performed with phantompeakqualtools (https://github.com/kundajelab/phantompeakqualtools) and used to extend alignments to estimated fragment size, then piled up, ignoring nonunique and duplicate alignments, to determine the ChIP signal. Signals were normalized by library size (number or reads aligned to autosomal chromosomes), then scaled to the equivalent of 1× coverage using deepTools (Ramírez et al. 2014) (bamCoverage –bam <BAM> --minMappingQuality 20 –binSize 1 –smoothLength 0 –ignoreForNormalization chrX chrY chrM –ignoreDuplicates –numberOfProcessors <N> --samFlagExclude 512 –normalizeTo1x 2451960000 –outFileName <bigwig output>).
Integrated data analysis
Comparison of DNA methylation ratios was performed at CpG dinucleotides covered at ≥5× in the sequencing data. Heat maps of RMSE values were generated and clustered by complete-linkage based on Euclidean distance (Supplemental Fig. S3). To assess various assay signals across putative regulatory regions defined by the ATAC-seq data (Supplemental Fig. S4), we focused only on distal accessibility peaks, defined as those ATAC peaks falling outside 10 kb upstream of or 2 kb downstream from a gene (UCSC Known Genes canonical set). The ATAC-seq transposition signal was calculated based on read alignments centered at the putative transposase insertion point position using only R1 of the read pairs to prevent double counting of insertion instances. For signal heat maps (Supplemental Fig. S4), we further restricted the analysis to ATAC peaks with an IDR score ≤0.016 to reduce the set to the top candidates that were of a more manageable size for plotting. Mean assay signal was obtained at 50-bp adjacent bins tiled across a region ±5 kb around summits of these selected ATAC-seq peaks. The WashU EpiGenome Browser (http://epigenomegateway.wustl.edu/browser/) was used to visualize user-provided and public data sets, including the 15-state ChromHMM classifications (Ernst and Kellis 2012). Candidates for allele-specific methylation were selected based on previously implicated transversions (Kaplow et al. 2015) and manually inspected for allele-specific methylation.
Computational requirements
The core analysis steps of preprocessing raw FASTQ files, bisulfite-aware short-read alignment and DNA methylation genotyping were performed on the New York Genome Center (NYGC) high-performance compute cluster requiring roughly ∼300 core hours for a 30× bisulfite genome on Intel Xeon 2.60 GHz CPUs with >48 GB of available memory. Additional analysis time was required to perform various quality control steps, including but not limited to bisulfite conversion rate estimation, coverage uniformity assessment, and mean coverage estimation (both genome-wide and across cis-regulatory loci). Downstream analysis such as differential methylation detection and hypo-/hypermethylated domain prediction required further computational time.
Data access
The bisulfite sequencing, Illumina HumanMethylation450 BeadChip, and Infinium MethylationEPIC microarray data from the GM12878 and IMR-90 cell lines have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession number GSE103505. filterFillIn2 source code is available in the Supplemental Methods.
Supplementary Material
Acknowledgments
We thank the New York Genome Center (NYGC) personnel, Stefan Pescatore and Harold Swerdlow, and the NYGC production sequencing team for their support, as well as Einstein's Center for Epigenomics. A.D.J. was supported by Einstein's Medical Scientist Training Program National Institutes of Health, National Institute of General Medical Sciences T32 GM007288.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.232587.117.
References
- Adey A, Shendure J. 2012. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res 22: 1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sabato S, Hellman A. 2013. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol 14: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagwell CE, Bhat S, Hawkins GM, Smith BW, Biswas T, Hoover TR, Saunders E, Han CS, Tsodikov OV, Shimkets LJ. 2008. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLoS One 3: e3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295. [DOI] [PubMed] [Google Scholar]
- Blair JD, Yuen RKC, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. 2013. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol Hum Reprod 19: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22: 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Ni Y, Timmer SW, Lee B-K, Battenhouse A, Louzada S, Yang F, Dunham I, Crawford GE, Lieb JD, et al. 2014. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet 10: e1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schübeler D. 2013. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet 9: e1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. 2011. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell 44: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CY, Mohtat D, Yu Y, Ko Y-A, Shenoy N, Bhattacharya S, Izquierdo MC, Park ASD, Giricz O, Vallumsetla N, et al. 2014. Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin Cancer Res 20: 4349–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. 2006. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol 301: 203–225. [DOI] [PubMed] [Google Scholar]
- Kaplow IM, MacIsaac JL, Mah SM, McEwen LM, Kobor MS, Fraser HB. 2015. A pooling-based approach to mapping genetic variants associated with DNA methylation. Genome Res 25: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Stenzel U, Kelso J. 2009. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol 10: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y-A, Mohtat D, Suzuki M, Park ASD, Izquierdo MC, Han SY, Kang HM, Si H, Hostetter T, Pullman JM, et al. 2013. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol 14: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Greally JM. 2017. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 18: 441–451. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ. 2011. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5: 1752–1779. [Google Scholar]
- Li Q, Suzuki M, Wendt J, Patterson N, Eichten SR, Hermanson PJ, Green D, Jeddeloh J, Richmond T, Rosenbaum H, et al. 2015. Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Res 43: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yuan Z, Tan T, Wang J, Zhang J, Luo H-J, Xia Y, Ji W, Gao F. 2015. Improved tagmentation-based whole-genome bisulfite sequencing for input DNA from less than 100 mammalian cells. Epigenomics 7: 47–56. [DOI] [PubMed] [Google Scholar]
- Maksimovic J, Gordon L, Oshlack A. 2012. SWAN: Subset-quantile Within Array Normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol 13: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10. [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. 2005. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 33: 5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Houseman EA, Izzi B, Kelsey KT, Meissner A, et al. 2013. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods 10: 949–955. [DOI] [PubMed] [Google Scholar]
- Miura F, Enomoto Y, Dairiki R, Ito T. 2012. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney DL. 1958. Epigenetic control systems. Proc Natl Acad Sci 44: 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Eyring K, De S, Yang IV, Schwartz DA. 2014. Fast and accurate alignment of long bisulfite-seq reads. arXiv:1401.1129 [q-bio.GN].
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A, Liljedahl U, Nordlund J. 2018. Data quality of whole genome bisulfite sequencing on Illumina platforms. PLoS One 13: e0195972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnerblad M, Andersson R, Olofsson T, Douagi I, Karimi M, Lehmann S, Hoof I, de Hoon M, Itoh M, Nagao-Sato S, et al. 2014. Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood 123: e79–e89. [DOI] [PubMed] [Google Scholar]
- Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, Rehli M. 2009. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res 19: 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. 2009. High-throughput bisulfite sequencing in mammalian genomes. Methods 48: 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM. 2010. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol 11: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. 2014. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res 24: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Shirane K, Miura F, Kubo N, Ichiyanagi K, Hayashi K, Saitou M, Suyama M, Ito T, Sasaki H. 2017. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics 18: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulahannan N, Greally JM. 2015. Genome-wide assays that identify and quantify modified cytosines in human disease studies. Epigenetics Chromatin 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gu L, Adey A, Radlwimmer B, Wang W, Hovestadt V, Bähr M, Wolf S, Shendure J, Eils R, et al. 2013. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc 8: 2022–2032. [DOI] [PubMed] [Google Scholar]
- Won K-J, Zhang X, Wang T, Ding B, Raha D, Snyder M, Ren B, Wang W. 2013. Comparative annotation of functional regions in the human genome using epigenomic data. Nucleic Acids Res 41: 4423–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xing X, Li J, Lowdon RF, Zhou Y, Lin N, Zhang B, Sundaram V, Chiappinelli KB, Hagemann IS, et al. 2014. Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC Genomics 15: 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.