Abstract
In grasses, two pathways that generate diverse and numerous 21-nt (premeiotic) and 24-nt (meiotic) phased siRNAs are highly enriched in anthers, the male reproductive organs. These “phasiRNAs” are analogous to mammalian piRNAs, yet their functions and evolutionary origins remain largely unknown. The 24-nt meiotic phasiRNAs have only been described in grasses, wherein their biogenesis is dependent on a specialized Dicer (DCL5). To assess how evolution gave rise to this pathway, we examined reproductive phasiRNA pathways in nongrass monocots: garden asparagus, daylily, and lily. The common ancestors of these species diverged approximately 115–117 million years ago (MYA). We found that premeiotic 21-nt and meiotic 24-nt phasiRNAs were abundant in all three species and displayed spatial localization and temporal dynamics similar to grasses. The miR2275-triggered pathway was also present, yielding 24-nt reproductive phasiRNAs, and thus originated more than 117 MYA. In asparagus, unlike in grasses, these siRNAs are largely derived from inverted repeats (IRs); analyses in lily identified thousands of precursor loci, and many were also predicted to form foldback substrates for Dicer processing. Additionally, reproductive phasiRNAs were present in female reproductive organs and thus may function in both male and female germinal development. These data describe several distinct mechanisms of production for 24-nt meiotic phasiRNAs and provide new insights into the evolution of reproductive phasiRNA pathways in monocots.
In flowering plants, small RNA (sRNA) pathways are active in reproductive tissues (Borges and Martienssen 2015). Two pathways associated with the male germline have been described in grasses and generate diverse and abundant phased, secondary siRNAs (phasiRNAs) (Johnson et al. 2009; Zhai et al. 2015; Fei et al. 2016). These phasiRNAs are produced from 5′-capped and polyadenylated, noncoding precursors (“PHAS” transcripts) transcribed by RNA polymerase II (Pol II) from nonrepetitive loci. Their production is triggered by two 22-nt miRNAs—miR2118 for 21-nt phasiRNAs and miR2275 for 24-nt phasiRNAs—that direct cleavage of PHAS transcripts, setting a consistent 5′ terminus for each PHAS precursor. The 3′ mRNA fragments are converted to double-stranded RNA by RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), and are processed by DCL4 and DCL5 to yield phased 21- and 24-nt siRNAs, respectively (Song et al. 2012a).
The “premeiotic” miR2118-triggered 21-nt phasiRNAs are abundant during the specification of cell fate in anthers, originating from the epidermal layer (Zhai et al. 2015). A genetic lesion in a 21-nt PHAS locus underlies photoperiod-sensitive male sterility in rice (Fan et al. 2016). miR2275-triggered 24-nt phasiRNAs accumulate later, during meiosis, in the tapetum and germinal cells, and they persist into the differentiation and maturation of pollen; their production is dependent on normal tapetal cells (Nonomura et al. 2007; Zhai et al. 2015). There are parallels with mammalian PIWI-associated RNAs (piRNAs): They originate from nonrepetitive loci, are generated in two different size classes with distinct developmental timing, and generate abundant and diverse siRNAs (Nonomura et al. 2007; Johnson et al. 2009; Zhai et al. 2015). Recently, piRNAs were also shown to be phased (Han et al. 2015; Mohn et al. 2015).
The functions of plant reproductive phasiRNAs are as yet unknown, but their role in male reproductive success is evident. For example, a mutant in MEIOSIS ARRESTED AT LEPTOTENE (MEL1), encoding an Argonaute protein in rice, arrests in early meiosis and has an abnormal tapetum and anomalous pollen mother cells (PMC) (Nonomura et al. 2007; Komiya et al. 2014). MEL1 selectively binds 21-nt phasiRNAs (Komiya et al. 2014), indicating that premeiotic phasiRNA functions are required for male fertility. Variants of two loci generating 21-nt phasiRNAs are the basis for photoperiod and temperature sensitive male sterility in rice (Fan et al. 2016). The 24-nt meiotic phasiRNAs are also apparently required for male fertility (Ono et al. 2018).
The evolutionary origins of plant reproductive phasiRNAs are also poorly characterized, with little known outside of their presence in the grasses (Poaceae, Poales). In this context, 24-nt meiotic phasiRNAs are particularly interesting as their biogenesis depends on a Dicer (DCL3) homolog, DCL5, originally known as DCL3b (Margis et al. 2006), apparently specialized for their production, yet not well described outside of grass genomes. Also, miR2275, the specialized trigger for these siRNAs, is poorly characterized outside of the grasses. The 21-nt premeiotic phasiRNAs are triggered by members of the miR2118/482 superfamily, which in many other plant genomes triggers phasiRNAs from NB-LRR pathogen-defense genes (Zhai et al. 2011; Zhang et al. 2016). The evolutionary route by which miR2118 shifted to targeting of noncoding PHAS precursors in a highly restricted spatiotemporal manner is not known. Thus, significant questions remain about when and how in plant evolution reproductive phasiRNAs emerged and took on the characteristics that are exhibited in grasses. We used garden asparagus (Asparagus officinalis) and other monocots (daylily, Hemerocallis lilioasphodelus, and Lilium maculatum) to investigate reproductive phasiRNAs as these lineages diverged from grasses over 110 million years ago (Hedges et al. 2015; Magallón et al. 2015).
Results
Asparagus flowers yield abundant reproductive phasiRNAs
To characterize phasiRNAs and their miRNA triggers in garden asparagus, we generated sRNA libraries from vegetative tissues and sequential stages of reproductive tissues, including pre- to post-meiotic anthers (Supplemental Table S1). In total, we generated 23 sRNA libraries combined with 15 published but not extensively characterized libraries (Harkess et al. 2017). We recently annotated 166 miRNA precursors (105 unique miRNAs from 78 families) in asparagus, including known/conserved and novel miRNAs (Harkess et al. 2017). Our expanded data and investigation yielded another 15 conserved and 46 lineage-specific 21- and 22-nt miRNAs (Supplemental Table S2; Supplemental Fig. S1). Among conserved miRNAs, we observed a high degree of accumulation patterns across developmental stages including fertile anthers and aborted pistils (Supplemental Fig. S2A). We observed significant disparity between premeiotic flower buds and meiotic anthers, primarily due to seven miRNA families that were enriched (fold change ≥4, P ≤ 0.05) including miR160, miR166, miR171, miR319, and miR390, all known to play roles in flowering (Mallory et al. 2004; Nagasaki et al. 2007; Rubio-Somoza and Weigel 2011; Schommer et al. 2012; Curaba et al. 2013). Two triggers of reproductive phasiRNAs—miR2118 and miR2275—were preferentially expressed and highly enriched in all the reproductive libraries (Supplemental Fig. S1). In the asparagus genome, five miR2118 members are generated from three loci (Supplemental Fig. S3); two members (miR2118d/e) accumulate in both vegetative and reproductive stages (Supplemental Figs. S1, S2B), whereas the other three clustered members (miR2118a/b/c) (Supplemental Fig. S3, Clust2) display premeiotic enrichment in anthers (Supplemental Figs. S1, S2B).
We next examined loci generating phasiRNAs in garden asparagus, coupling the sRNA analysis with the identification of the potential miRNA triggers (Methods). We identified 29 loci generating 21-nt phasiRNAs and 42 loci generating 24-nt phasiRNAs. Among the “21-PHAS” loci, 23 were vegetatively expressed and 15 overlapped annotated protein-coding genes (Supplemental Table S3). Only three reproductive-enriched 21-PHAS loci were derived from candidate long, noncoding RNAs (lncRNAs) (Supplemental Table S3), substantially lower than the hundreds or thousands seen in maize or rice (Zhai et al. 2015; Fei et al. 2016). In contrast, all 42 24-PHAS loci were highly enriched in reproductive tissues, peaked at meiosis, and correlated with miR2275 abundances (Fig. 1A). This spatiotemporal pattern is similar to those of the grasses (Komiya et al. 2014; Zhai et al. 2015). We also noted that some 24-nt phasiRNAs were abundant in aborted pistils (female organs); prior work had associated these phasiRNAs only with male reproductive organs (Zhai et al. 2015). From six sequential stages of fertile pistils from female flowers (Supplemental Table S1), we identified 22 24-PHAS loci (Supplemental Table S4), a subset of those identified from anthers and aborted male pistils (Supplemental Fig. S2C,D). Unlike anthers, the 24-nt phasiRNAs in fertile pistils lacked a clear temporal abundance peak, most likely due to variation in the sampled stages. Overall, and unlike the lncRNA precursors of grasses (Johnson et al. 2009; Zhai et al. 2015), we found 30% (n = 18) of 24-PHAS loci in asparagus overlap on either strand annotated protein-coding genes (Supplemental Table S4).
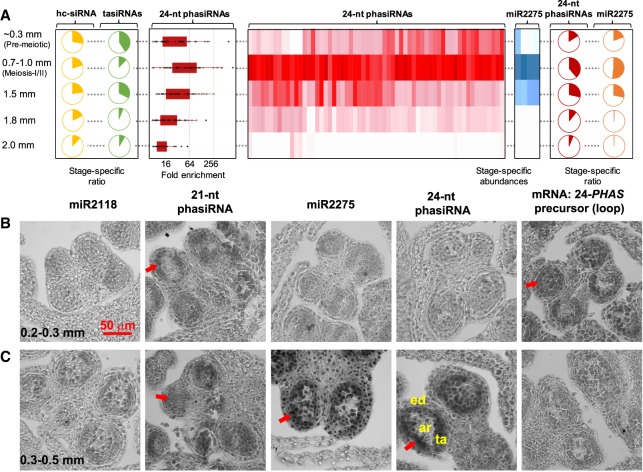
Figure 1.
Reproductive phasiRNAs and their triggers in Asparagus. (A) Heat maps depicting abundance of 24-nt phasiRNAs (red) and their triggers, miR2275 (blue), in developing anthers. Both heat maps are clustered on their similarity of expression. Pie charts on the left or right represent the proportion of all small RNA abundances comprised by the 24-nt phasiRNAs (red), miR2275 (orange), hc-siRNAs (yellow), and TAS3 tasiRNAs (green) across anther developmental stages. The 24-nt phasiRNA accumulation pattern is distinct from both hc-siRNAs and TAS3 tasiRNAs. Box-whisker plots indicate enrichment (log2) of Asparagus 24-nt phasiRNA abundance from all PHAS loci in the meiotic anther compared to the vegetative sample (leaf). (B,C) Small RNA in situ hybridization with probes for the following, from left to right: (1) miR2118; (2) a premeiotic phasiRNA from locus 21-PHAS-9; (3) miR2275; and (4) a meiotic IR-related phasiRNA from locus 24-PHAS-31. The right-most images show mRNA in situ hybridizations with probes for the 24-PHAS-31 precursor. In each panel, signal is dark gray, and cell boundaries appear in light gray. Red arrows indicate small RNA signal. The locations of the epidermis (ed), tapetum (ta), and archesporial cells (ar) are indicated. The scale bar indicates 50 µm, for all images. (B) Images from premeiotic anthers, sized 0.2–0.3 mm. (C) Images from meiotic-stage anthers, sized 0.3–0.5 mm.
To characterize the PHAS precursors, we sequenced from meiotic asparagus anthers a set of 275,565 putative full-length, polyadenylated transcripts using single molecule real time (SMRT) sequencing. These data included 5671 annotated genes, at least 23,797 novel isoforms, and 1771 unannotated loci, including transcripts overlapping the majority of both 21- and 24-PHAS loci (Supplemental Table S4). We built a classifier that categorizes transcripts as coding, noncoding, or of unknown coding potential (TUCP), scored using Coding Potential Calculator (Kong et al. 2007) and Coding-Potential Assessment Tool (Wang et al. 2013; Methods). More than 62% of 21-PHAS loci and 42% of 24-PHAS loci overlapped predicted protein-coding transcripts on either strand (Supplemental Tables S3, S4), and the latter is an unexpected result given that the 24-PHAS precursors in grasses are thought to be lncRNAs (Johnson et al. 2009; Zhai et al. 2015). In contrast, the production of 21-nt phasiRNAs from protein-coding genes is well described for vegetative tissues in both eudicots and gymnosperms (Zhai et al. 2011; Shivaprasad et al. 2012; Arikit et al. 2014; Xia et al. 2015).
In maize, reproductive phasiRNAs display distinct spatiotemporal patterns of accumulation (Zhai et al. 2015); therefore, we decided to assess whether this is also the case in asparagus. We performed small RNA in situ hybridizations using probes for the following: (1) miR2118a, the most abundant member in asparagus; (2) a premeiotic 21-nt phasiRNA; (3) miR2275; and (4) a meiotic IR-derived 24-nt phasiRNA. All of these garden asparagus sRNAs displayed distinct spatiotemporal patterns (Fig. 1B,C). miR2118, the trigger of limited reproductive-enriched 21-PHAS loci, colocalized with phasiRNAs in the middle layer, tapetum, and archesporial (ar) cells (Fig. 1B). Matching the maize meiotic 24-nt phasiRNAs that require normal tapetal differentiation and localize in the tapetum and germinal cells, asparagus 24-nt phasiRNAs were enriched in meiotic stages and predominantly localized in the tapetum and meiocytes, precisely where miR2275 was present (Fig. 1C).
MicroRNA triggers and biogenesis components of 21- and 24-nt phasiRNAs in garden asparagus
We next investigated the miRNA triggers of phasiRNAs, focusing on (1) reproductive 21-PHAS and (2) 24-PHAS loci. We first generated predicted and PARE-supported target sites using sPARTA (Kakrana et al. 2014), followed by an exhaustive search for miRNA triggers (Methods). Of 29 21-PHAS loci, we found triggers for 16, of which 14 were miR2118 family members. Target loci included the three reproductive-specific 21-PHAS loci mentioned previously, three NB-LRR genes, and four other protein-coding loci. Isoforms (22-nt) of miR167 and miR390 triggered 21-nt phasiRNAs from an AUXIN-RESPONSE FACTOR (ARF) gene and an intergenic locus, respectively, the latter likely a TAS3 locus (Axtell et al. 2006). The numbers of phased NB-LRRs and TAS3 loci identified in the asparagus genome are low (n = 3), relative to other plant genomes in which tens to hundreds of NB-LRRs generating phasiRNAs were reported (Zhai et al. 2011; Shivaprasad et al. 2012; Arikit et al. 2014; Xia et al. 2015).
For the reproductive 24-PHAS loci, our analyses failed to identify an miRNA trigger, including miR2275—surprising, given that all reported 24-nt phasiRNAs in maize and rice are triggered by miR2775 family members (Johnson et al. 2009; Song et al. 2012a; Zhai et al. 2015). Target analysis in asparagus for miR2275 did identify 15 intergenic and 10 protein-coding targets, supported by PARE data; although none of these targets corresponded to 24-PHAS loci, weakly abundant, anther-enriched 22-nt phasiRNAs were found at three intergenic targets (Supplemental Table S3). No such miR2275-triggered 22-PHAS loci were found in maize and rice, using published data (Zhai et al. 2015; Fei et al. 2016). Next, we tested the remote possibility of these 22-nt phasiRNAs triggering 24-nt phasiRNAs in a tertiary cycle, perhaps analogous to piRNA production in insects (Brennecke et al. 2007). For 101 22-nt phasiRNAs, 29,594 potential target sites (score <4) were identified, among which 1430 had matched PARE reads at any abundance level, but only one target site passed stringent filters. This target site and others found using relaxed filters showed no evidence of 21-, 22-, or 24-nt phasiRNAs. These observations left us with no clear functional explanation for the 22-PHAS loci; perhaps more importantly, there were evidently no miRNA triggers of the 42 reproductive 24-PHAS loci identified in asparagus.
During inspection of the 24-PHAS loci, we observed that many showed a substantial strand bias, inconsistent with the RDR6-dependent biogenesis of PHAS precursors in grasses (Song et al. 2012b). Moreover, we also noticed that an overlap of these 24-PHAS loci with inverted repeats (“IRs”); in other plants like Arabidopsis, IRs are processed by DCL4 or even DCL1 into 21-nt phasiRNAs or miRNAs. Genome-wide, 90% of asparagus 24-PHAS loci corresponded to IRs (for examples, see Fig. 2A; Supplemental Fig. S4). We computed an enrichment or depletion of 24-PHAS overlap with exons, introns, TE-related regions, and IRs, versus random chance, for asparagus, maize, and rice. Both grasses had few 24-PHAS loci overlapping IRs (Fig. 2B; Supplemental Table S5), versus a ∼55-fold enrichment in asparagus. In all three species, there were few 24-PHAS loci in TE-related regions (Fig. 2B; Supplemental Table S5), and they displayed similar distributions of sizes (Fig. 2C), suggesting similar efficiencies of Dicer processing.
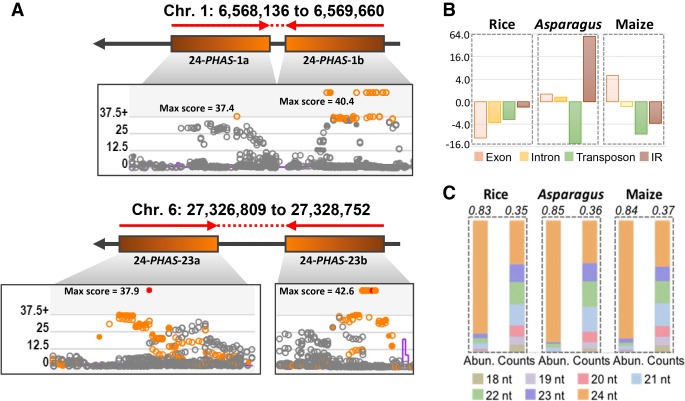
Figure 2.
Many Asparagus 24-PHAS loci are derived from inverted repeats. (A) Genomic organization of two representative IR-type PHAS loci, overlapping with the 5′ and 3′ arm of inverted repeats. Phasing scores from sliding window analysis are presented as shown in our custom web viewer. Loci with score >25 were considered as phased loci. (B) Fold change representing enrichment or depletion of overlap of 24-PHAS loci from rice, Asparagus, and maize, with exons, introns, transposons, and inverted repeats, versus random chance. The complete data are in Supplemental Table S5. (C) Comparison of sRNA abundances and unique sRNA counts of sRNAs produced from 24-PHAS loci from rice, Asparagus, and maize. The values on top represent 2.5% trimmed mean of ratio of abundances or counts of 24-nt phasiRNAs from all 24-PHAS loci of corresponding species.
IR-type 24-PHAS loci were not identified in our earlier work in maize (Zhai et al. 2015), so we sought to better characterize them in maize. Two IRs were found (Supplemental Fig. S5); both had a single, 5′ miR2275 target site. In both cases, the phasiRNAs were precisely spaced at the predicted base of the putative foldback, distal to the loop. In this regard, clust-125 was more unusual, as it also has a tandem repeat of the IR—i.e., two more 24-PHAS loci flanking the foldback, with sequence similarity (>94%) (Supplemental Fig. S5). We concluded that 24-nt phasiRNAs from IR precursors are present in grasses, but they are far more prevalent in asparagus.
Inverted repeat precursors of 21- and 24-nt phasiRNAs in asparagus
To validate the foldback structure of the 24-PHAS precursors in asparagus, we generated full-length, assembled transcripts. We sequenced mRNA from premeiotic and meiotic anthers and from cladode samples (Supplemental Table S1). We assembled these data using two different approaches: (1) a genome-based assembly, and (2) a de novo hybrid assembly using RNA-seq and SMRT libraries. Using the genome-based assembly, we calculated the foldback potential (FP) for transcripts mapped to PHAS loci, and then divided precursors into four categories:
Phased transcripts with full-length or near full-length precursors (>85% coverage of a PHAS locus), that formed at least 240 nt of foldback (FP ≥ 500);
Processed precursors with ≤85% PHAS coverage and high foldback potential (FP ≥ 500);
Near full-length precursors (>85% PHAS coverage) with low foldback potential (FP < 300); and
All other PHAS-matched transcripts, i.e., processed or incomplete precursors.
This analysis identified 74 precursors from 29 unique 24-PHAS loci, 11 represented by at least one category I or II precursor. All 11 precursors were noncoding (one had weak coding potential). Category I/II precursors comprised 38% of 24-PHAS precursors; category IV was 48%, suggesting a high degree of processed transcripts in our data. Only 13.7% of precursors were unstructured or of unknown secondary structure (category III). These 24-PHAS precursors all lacked miR2275 trigger sites.
Unlike 24-PHAS precursors, most 21-PHAS precursors showed an absence of secondary structure (83% category III), with only one IR (category I) 21-PHAS locus (miR2118-triggered) (Supplemental Fig. S6). The presence of an miRNA target site in an IR 21-PHAS precursor is unexpected—foldback RNAs are substrates for Dicer even without RDR6 activity (Dunoyer et al. 2005; Henderson et al. 2006). How the miRNA trigger initiates phasing in an IR, is something not explained by previous models of phasiRNA biogenesis (Arribas-Hernández et al. 2016). Coupled with the preceding observation that asparagus 24-PHAS loci lack miR2275 trigger sites, including IR precursors, we found remarkable diversity in the reproductive phasiRNA precursors of asparagus.
We examined RNA secondary structures in the meiotic anther transcriptome to confirm precursor processing as foldback dsRNAs. For this, we performed sequencing-based structure-mapping (Zheng et al. 2010) to identify paired (dsRNA) and unpaired (single-stranded RNA [ssRNA]) transcripts in meiotic anthers (Supplemental Table S1). We found 22 of 42 24-PHAS loci with a significant (P ≤ 0.05) structure score, consistent with base pairing, while 13 of these 22 showed significant strand specificity (Supplemental Table S6; Supplemental Methods). Comparing these loci with miRNAs, hc-siRNAs, tasiRNAs, or the IR-type 21-PHAS locus (above) revealed varying types of RNA structure. miRNAs exhibited strong secondary structure and strand bias indicative of intramolecular interactions (Fig. 3A); 21-PHAS loci exhibited strong secondary structure with low strand specificity. Furthermore, the 24-PHAS loci showed a strong bias toward intramolecular structure, whereas hc-siRNA-generating RNAs displayed a tendency toward intermolecular structure (Fig. 3A) consistent with earlier reports (Li et al. 2012). For highly structured 24-PHAS loci, we observed two “peaks” separated by a “valley,” supporting formation of the foldback structure from 24-PHAS loci (Fig. 3B).
Figure 3.
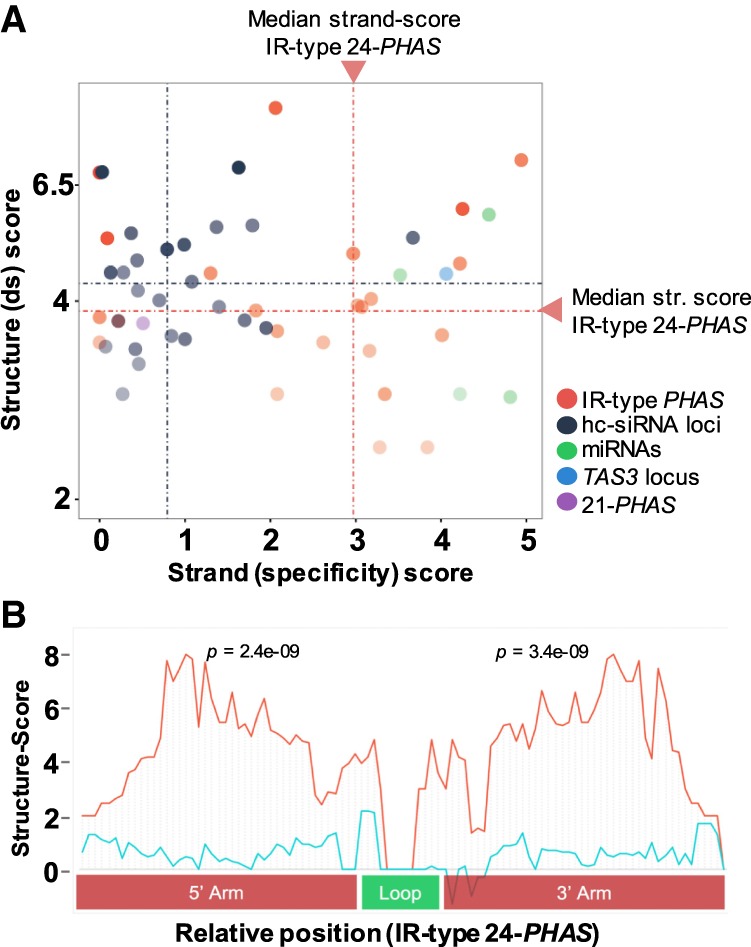
Intramolecular secondary structure at inverted repeat, 24-nt PHAS loci in Asparagus. (A) Scatterplots showing secondary structure scores as function of strand-specificity scores for IR-related loci along with randomly selected hc-siRNA, miRNA, tasiRNA, and IR-type 21-nt loci that passed the coverage cutoff. The dotted line represents score medians: (red) 24-nt PHAS loci; (blue) hc-siRNAs. (B) Consensus of dsRNA structure scores (red) from five IR-type PHAS loci show two statistically significant peaks of paired nucleotides and a “valley” (loop, green) of unpaired nucleotides validating formation of stem–loop structure from these IR-type PHAS transcripts. The five loci for this figure were selected based on high coverage and similar lengths and loop sizes. The control (blue line) represents the mean score from shuffled controls. Methods used to compute secondary structure scores are described in the Supplemental Methods.
The 24-nt phasiRNA pathway exists more broadly in monocots
We extended our study to two more species—daylily (H. lilioasphodelus) and Lilium (L. maculatum, lily). These monocots were selected based on their availability, ease of dissection of anthers, and most importantly, due to their evolutionary distance from Asparagus. Daylily, another member of the Asparagales, and Lilium diverged ∼66 and ∼117 MYA from the most recent common ancestor (MRCA) of Asparagus, respectively (Chase and Reveal 2009; Hedges et al. 2015). We performed large-scale transcriptome sequencing and transcript assembly to compensate for the absence of a genome for both species. mRNA and small RNA (for phasiRNA analysis) data were generated from similarly staged anthers, leaf samples, and pistils in daylily (Methods; Supplemental Fig. S7; Supplemental Table S7). Sequencing to construct transcript assemblies included deep Illumina RNA-seq data for both species plus SMRT-sequencing for Lilium (Supplemental Table S7).
In both daylily and lily, we found miR2118 members highly enriched in reproductive tissues (Fig. 4A; Supplemental Table S8). At least 18 miR2275 members in lily and three miR2275 members in daylily peaked in abundance at premeiotic and meiotic stages (Fig. 4A; Supplemental Fig. S9). Both miR2118 and miR2275 were measurable in daylily pistils (female organs). Neither family was characterized in earlier studies of the basal angiosperm Amborella (Amborella Genome Project 2013) and the early diverging monocot order Alismatales represented by Zostera (Olsen et al. 2016), so we reanalyzed the genomes and published sRNA data for these two species. By comparing mature miRNA sequences from maize and rice to find isomiRs of both families in Zostera and Amborella and investigating these genomes for precursors, we identified at least one candidate locus for both miRNA families in Amborella (Supplemental Table S9). Based on the presence of isomiRs or the candidate loci, we concluded that miR2275 is likely found throughout the monocots and perhaps in other angiosperms; miR2118 originated before the emergence of flowering plants (Zhang et al. 2016).
Figure 4.
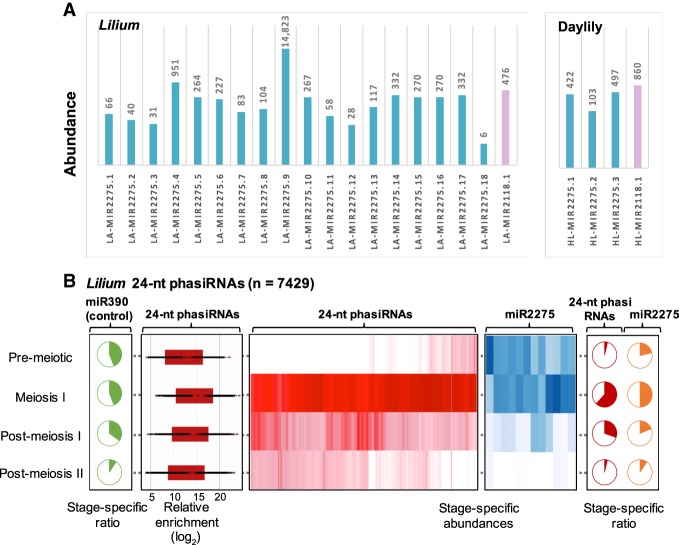
Reproductive PHAS triggers and 24-nt phasiRNAs in Lilium. (A) miR2118 (violet) and miR2275 (blue) family members identified in Lilium and daylily by comparing mature sRNA sequences to members in miRBase (version 21); matches to variants scoring ≤4 were considered as valid candidates. Variants include mismatches between mature sRNAs and miRNAs from miRBASE, plus 5′ or 3′ overhang in the alignment. Values above the bars represent their total abundance (TP30M) in anthers. (B) Heat maps depicting abundance of Lilium 24-nt phasiRNAs (red) and miR2275 triggers (blue) in developing anthers. Both heat maps are clustered on similarity of expression. Pie charts represent the proportion of stage-specific abundances for 24-nt phasiRNAs (red), miR2275 (orange), and miR390 (green), the trigger of tasiRNAs across different anther developmental stages that are included in this study. Box-whisker plot shows enrichment (log2) of Lilium 24-nt phasiRNA abundance from all PHAS loci in the meiotic anther compared to the vegetative sample (leaf).
To characterize reproductive phasiRNAs in lily and daylily, we identified 6277 and 392 24-PHAS transcripts and 158 and six 21-PHAS transcripts, respectively, for the two species. The extraordinarily high number of 24-PHAS transcripts in lily matches the large 18-member miR2275 family. The low numbers of 21-PHAS transcripts might reflect a sampling bias against premeiotic stages. As in the grasses, PHAS precursors were mainly noncoding (Supplemental Table S10) and enriched in premeiotic and meiotic anther stages, respectively (Fig. 4B; Supplemental Fig. S9). We also identified 25 24-PHAS transcripts in developing daylily pistils that largely overlapped the anther 24-PHAS repertoire and were mostly low abundance (Supplemental Table S12), but with the detection of miR2275 (above), confirmed the presence in female organs of 24-nt reproductive phasiRNAs.
We examined IR PHAS precursors in daylily and Lilium and found at least 131 24-PHAS transcripts predicted to form long foldbacks with high complementarity (hereafter, “foldback-PHAS” transcripts) (Fig. 5A; Supplemental Fig. S10). If the precursors are rapidly processed into small RNAs, full-length mRNAs may be rare in our data (like category III and IV RNAs, above), obfuscating the detection of intramolecular secondary structures. Therefore, we computationally inferred pairs of PHAS transcripts that could both form stem–loops if part of the same mRNA, and that also exhibit a Dicer-generated 2-nt overhang of overlapped phasiRNAs (Methods). In lily and daylily, 2888 and 87 of 24-PHAS transcripts matched these criteria (Fig. 5B). Collapsing the complete set of precursors based on sequence similarity and overlap (Supplemental Table S13), and compared to maize (Supplemental Table S14), lily displays a substantially larger (>25-fold) set of meiotic 24-PHAS precursors. We used sRNA fluorescent in situ hybridization (FISH) to examine their spatial distribution in anther cells. Both types of precursors (IR-type and foldback), and their 24-nt phasiRNA products, localized in tapetal and archesporial cells and enriched at meiosis (Supplemental Fig. S11). Thus, the spatial and temporal characteristics of 24-nt phasiRNAs are conserved for at least some 120 million years since the last common ancestor of the Poales, Asparagales, and Liliales lineages.
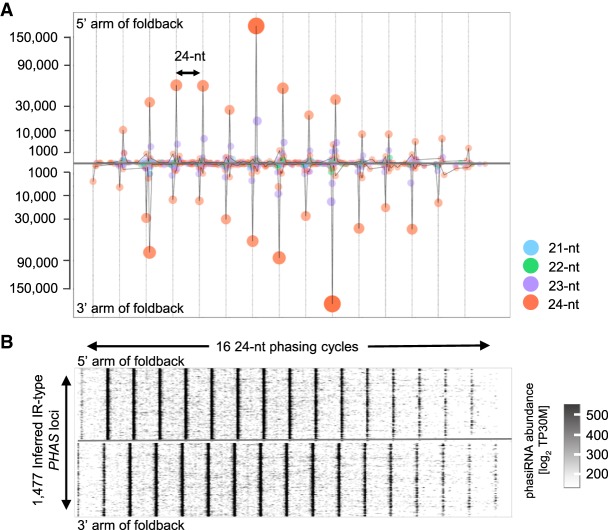
Figure 5.
Foldback and IR PHAS precursor transcripts yield abundant phasiRNAs. (A) Summed sRNA abundances from 5′ and 3′ arms of 50 foldback-PHAS transcripts show a clear 24-nt phasing with a 2-nt overhang. Representative foldback-PHAS with foldback score >500, arm length >384 (eight or more phases) were used to generate this distribution plot. (B) Scatterplot of sRNA abundances from 5′ and 3′ arm of inferred IR-type PHAS transcripts (n = 1477) show a strong 24-nt phasing pattern with a 2-nt overhang between the paired arms.
We analyzed the PHAS precursors from daylily and lily in more detail, predicting miR2118 and miR2275 target sites, and validating miRNA triggers using sPARTA (Kakrana et al. 2014). Triggers were evident for 1098 and 29 of 24-PHAS precursors in lily and daylily, respectively, and for 22 (23.9%) of the 21-PHAS precursors in lily (Supplemental Tables S11–S13). The miRNA triggers for seven of the 22 lily premeiotic 21-PHAS transcripts were, unexpectedly, miR2275 (Supplemental Table S11), and were “in phase” with the first phasiRNA. For these cases, perhaps the unexpected switch of presumed DCL4 versus DCL5 processing (21- versus 24-nt phasiRNAs) depends on distinct spatiotemporal boundaries of precursor expression and not on recruitment by miR2118 or miR2275.
Among the IR 24-PHAS precursors, unlike asparagus, a large number of lily and daylily transcripts had miR2275 target sites (about one-third of precursors in lily, and one-fifth of precursors in daylily) (Supplemental Tables S11, S13). Since 22-nt miRNA targeting leads to RDR6 recruitment, unnecessary for dsRNA precursors formed from IRs, and directs initiation of sRNA phasing, the occurrence of IR/foldback transcripts with miRNA target sites was unexpected. We manually examined a subset of IR precursors (Supplemental Fig. S13), and these lacked sRNAs upstream of the miR2275 target site, consistent with its functionality as a trigger. Precursors with the target site on the stem showed sRNA variability (18- to 23-nt) in the first phasiRNA cycle, especially on the 3′ arm, lacking dominant 24-nt phasiRNAs (Fig. 6A,B, Supplemental Fig. S12; Supplemental Table S15). These siRNAs were largely absent from precursors with miR2275 sites overlapping the dsRNA-ssRNA junction (Fig. 6C), as were siRNAs upstream of the trigger site on the paired 5′ and 3′ arms. These observations suggest that 5′ and 3′ unpaired (ssRNA) ends are removed, either together—perhaps in the absence of miRNA trigger, like pri-miRNA processing—or sequentially, with the 5′ arm first removed via miR2275-directed cleavage, followed by trimming of the 3′ arm by an unknown mechanism, consequently releasing the stem–loop structure for subsequent processing by DCL5. Perhaps trimming could occur as with metazoan miRNA precursors, i.e., recognition of the ssRNA-dsRNA junction (Han et al. 2006; Kim et al. 2009), or as with piRNAs, in which a 3′ to 5′ exonuclease trims excessive nucleotides (Izumi et al. 2016; Tang et al. 2016).
Figure 6.
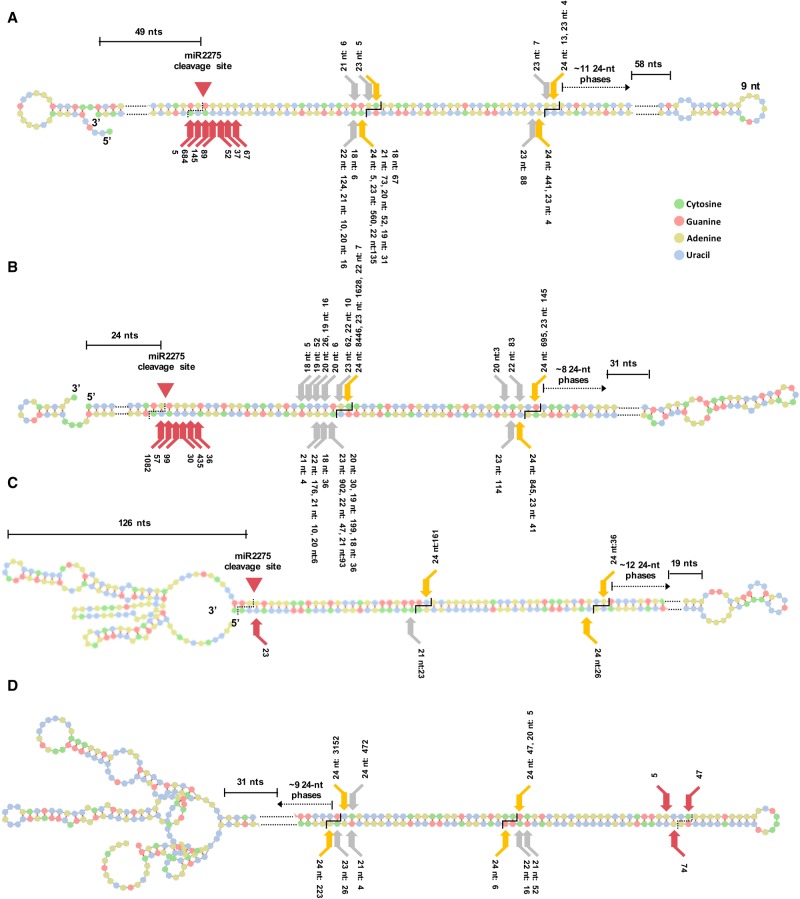
Processing of miR2275-triggered foldback PHAS precursors in Lilium. (A,B) Two representative miR2275-triggered foldback PHAS precursor transcripts in Lilium: 24-PHAS-5 and 24-PHAS-1681. (C) Precursor for PHAS-2398 with no unpaired 3′ arm. (D) Precursor for foldback PHAS-4395 putatively processed from loop-to-base. The cuts leading to release of 24-nt phased siRNAs are shown as orange arrows, whereas those that generate siRNAs of other sizes are indicated as gray arrows. Red arrows indicate 3′ termini of sRNAs of different sizes, at positions on the foldback mRNA distal to the miR2275 target site; the sRNA abundance (in TP30M) is indicated numerically, from the meiotic-stage anther library. Counts beside red arrows represent cut frequencies computed from sRNA data, i.e., abundances. In A and B, the miR2275 cleavage site is 49 and 24 nt inside the dsRNA region (as shown), whereas in C, the cleavage site is 126 nt from the 5′ terminus of the precursor.
DCL1 functions to cut unpaired ends of miRNA foldback precursors in plants (Cuperus et al. 2011; Bologna et al. 2013); we analyzed foldback PHAS precursors for their inferred direction of processing. Based on phasiRNA abundances, we identified 10 representative loci displaying loop-to-base processing (Supplemental Methods; Supplemental Table S13). These precursors lacked a miR2275 target site and showed no major variation or “raggedness” in the processing of the first, loop-side phasiRNA (Fig. 6D) compared to foldback precursors likely processed from a miR2275 site (“miR2275-to-loop”). Thus, ragged processing could result from inconsistent trimming of unpaired ends. This leaves unaddressed the question of which enzyme makes this first, loop-side cut of these reproductive 24-PHAS foldback precursors, perhaps any of the five monocot Dicers (Supplemental Fig. S14B).
To assess the possibility that a Dicer other than DCL5 might make the first cut in an IR transcript, we examined IRs in Arabidopsis. We used public Arabidopsis sRNA data (Lee et al. 2012; Jeong et al. 2013; Li et al. 2015) and focused on two known IR loci: IR71 and IR2039 (Henderson et al. 2006). In wild type, the IR transcripts were mainly processed into 21-, 22-, and 24-nt sRNA species (Supplemental Fig. S13A), products of DCL4, DCL2, and DCL3, respectively. In dcl3 and dcl2/3/4 mutants, 24-nt sRNAs were largely absent, with a slight accumulation of 21-nt sRNAs (Supplemental Fig. S13B), suggesting processing by DCL4 and DCL1 in the absence of DCL3 and DCL2. The 24-nt sRNAs were not impacted in nrpe1, nrpd1, and rdr2 indicating independence from RdDM (Supplemental Fig. S13B). dcl1 showed a strong reduction in levels of all sRNA size classes (Supplemental Fig. S13B), suggesting a primary role of DCL1, perhaps related to its activity in cleaving the pri-miRNA stem–loops to release mature miRNA duplexes. These observations suggest hierarchical processing of IR loci in Arabidopsis, first by DCL1 and subsequently by DCL2, DCL3, and DCL4. We hypothesize that DCL1 might similarly initiate some DCL5-processed IR transcripts that yield 24-nt reproductive phasiRNAs.
Protein partners of the 24-nt phasiRNA pathway: Grass AGO proteins are not entirely representative of monocots
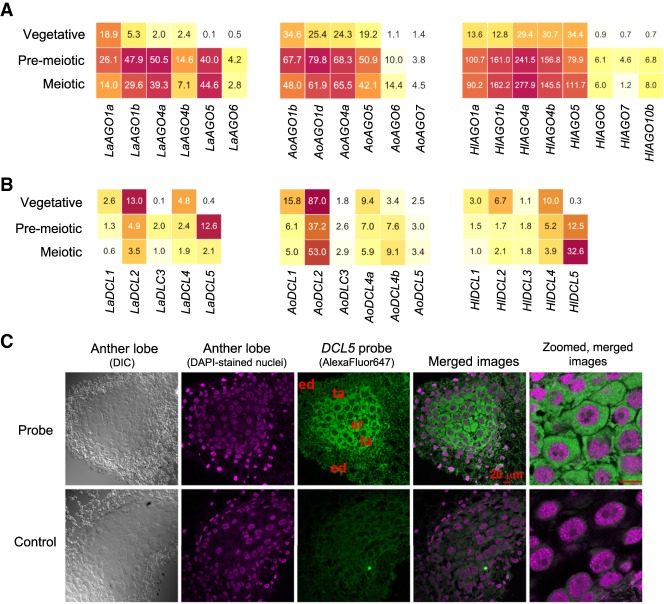
We investigated the Argonaute family members, binding partners of small RNAs, identifying 12, nine, and eight AGO proteins for asparagus, daylily, and lily, respectively (Supplemental Fig. S14A). AGO candidates known for roles in reproductive phasiRNA functions are AGO5 (i.e., rice MEL1) (Komiya et al. 2014), and AGO1d, AGO2b, and AGO18 for 24-nt phasiRNAs, based on transcriptome profiling and spatial localization in maize and rice (Zhai et al. 2015; Fei et al. 2016). In our asparagus, lily, and daylily data, AGO5 members were present and consistently enriched in premeiotic or meiotic anthers. AGO18 was missing from the genome of garden asparagus and transcriptome assemblies of all three species plus those of Z. marina and A. trichopoda (Supplemental Fig. S14A), consistent with its possible emergence in grasses (Zhang et al. 2015). We also found robust expression plus enrichment in premeiotic and meiotic anthers for a homolog of AGO4, known to bind 24-nt heterochromatic siRNAs (Mallory and Vaucheret 2010; Wang et al. 2011). Reproductive-enriched AGO1 family members were also identified, consistent with rice (Fei et al. 2016). Thus, based on transcriptional enrichment (Fig. 7A; Supplemental Table S16) and the Argonaute gene family phylogeny (Supplemental Fig. S14A), we hypothesize that there are at least two candidate AGOs (AGO1 and AGO4) with conserved roles in 24-nt phasiRNA biogenesis or activity.
Figure 7.
Dicer-like (DCL) gene family members and their expression in Asparagus, daylily and Lilium. (A) Heat map representing the expression profile of Asparagus, daylily, and Lilium AGO members. A phylogeny of AGO members is provided in Supplemental Figure S13. (B) Heat map of DCL abundances for three monocots for Dicer-like transcripts that were reliably detected (>1 FPKM) in at least one of three anther stages or the vegetative material. A phylogeny of DCL members is also provided in Supplemental Figure S13. (C) FISH localizing DCL5 transcripts in the cytoplasmic area of the tapetum and archesporial cells in meiotic-stage anthers from Lilium. The green channel (AF647) indicates DCL5 mRNA localization. DAPI (pink) shows the stained nucleus. In the middle image, the locations of the endothecium (ed), tapetum (ta), and archesporial cells (ar) are indicated. (Scale bar) 20 µm for all images.
Finally, we examined Dicer-like (DCL) genes. DCL4 and DCL5 were in all three species (Supplemental Fig. S14B; Supplemental Table S16). DCL4 showed high abundance in anthers. We could not detect asparagus DCL5 using in situ localization experiments, and it was absent from the high-quality full-length transcripts from SMRT-sequencing, presumably due to its low expression levels in asparagus (Fig. 7B; Supplemental Table S16). Given this low abundance, we verified DCL5 expression in asparagus using quantitative RT-PCR (Supplemental Fig. S14C). Its expression was not restricted to anthers, and its transcript abundances were similar in the vegetative (cladode) and reproductive tissues. Thus, DCL5 transcription profiles varied among the three monocots we studied, as daylily DCL5 displayed peak expression at meiosis, whereas Lilium DCL5 abundance peaked at a premeiotic stage, although measured with limited meiotic stages. Analyzed using FISH, lily DCL5 transcripts colocalized with the 24-PHAS precursors and phasiRNAs in tapetal and archesporial cells (Supplemental Fig. S11; Fig. 7C), matching the patterns described in maize (Zhai et al. 2015). Therefore, although there is variation across species for DCL5 expression, at least some nongrass monocots retain reproductive enrichment of the primary biogenesis partner of 24-nt reproductive phasiRNAs, suggesting this may be have been the case in the last common ancestor of the Liliales, Asparagales, and Poales.
Discussion
Reproductive phasiRNAs are the least-characterized small RNA pathway(s) in plants. First described in rice (Johnson et al. 2009), large-scale genetics, imaging, and biochemical studies from rice and maize have defined their pattern of accumulation in developmental time and space (Song et al. 2012a,b; Fei et al. 2013; Komiya et al. 2014; Zhai et al. 2015; Dukowic-Schulze et al. 2016). Their functional importance is clear but not well characterized (Komiya et al. 2014; Fan et al. 2016; Ono et al. 2018). However, their evolutionary origins are as yet uncharacterized. Using precisely staged anthers from diverse monocots, we found both the premeiotic and meiotic phasiRNA pathways. We detected 24-nt phasiRNAs in (female) pistils, consistent with the observation that the rice mel1 mutant is arrested in female gametogenesis (Komiya et al. 2014).
Our work also provided insights into the emergence and functional evolution of phasiRNA triggers, miR2118 and miR2275. Unusual among plant miRNAs, miR2118 has two primary functions in angiosperms, targeting NB-LRRs and triggering premeiotic phasiRNAs; the former is prevalent in gymnosperms and eudicots (Zhai et al. 2011; Shivaprasad et al. 2012; Arikit et al. 2014; Xia et al. 2015). In asparagus, the co-occurrence of temporally distinct reproductive and vegetative miR2118 members targeting both 21-PHAS precursors likely via a pathway conserved in asparagus and grasses (Fig. 8A) and NB-LRRs suggests dual roles for this miRNA. The evolutionary origins of miR2275 are thus far unknown, but we show here that this miRNA functions as the trigger of meiotic reproductive phasiRNAs in diverse monocots, a role thus predating the divergence of Liliales, Asparagales, and Poales lineages. The function of miR2275 in asparagus is somewhat puzzling, as its accumulation pattern mirrors that of grasses, yet asparagus 24-phasiRNAs are produced from IR precursors lacking miR2275 target sites. Therefore, the enrichment of miR2118 and miR2275 in reproductive tissues is largely but not exclusively an indicator of the presence of the grass-like premeiotic and meiotic phasiRNA pathways.
Figure 8.
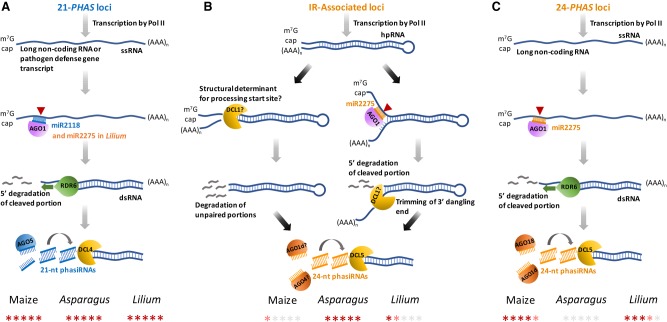
Overview of biogenesis pathways for reproductive phasiRNAs in monocots. (A) The miR2118-triggered 21-nt reproductive phasiRNA pathway, as described in grasses in Song et al. (2012a) and Zhai et al. (2015). (B) The inverted-repeat-type 24-nt phasiRNAs pathway indicated by the data from maize, Asparagus, and Lilium in this study. (C) The miR2275-triggered 24-nt reproductive phasiRNA pathway as described in grasses. At the bottom is an indication for each species of the proportion of 21- or 24-PHAS loci that correspond to each pathway.
Dicers and Argonautes are also key components of phasiRNA biogenesis, yet the diversification and functional evolution of these gene families within the monocots is not well described. AGO18, proposed as an effector molecule for meiotic phasiRNAs (Zhai et al. 2015), is absent from asparagus, daylily, lily, and Zostera. AGO1d and AGO4d were proposed by two independent studies in rice (Fei et al. 2016) and maize (Dukowic-Schulze et al. 2016) to load meiotic phasiRNAs, based on transcriptional enrichment. Paralogs of AGO1 and AGO4 were enriched in meiotic anthers in asparagus, lily, and daylily. A rice ago18 loss-of-function mutant shows no obvious developmental defects (Wu et al. 2015), and AGO4 is well-known to bind 24-nt siRNAs (Wang et al. 2011). Maize AGO4d functions in male and female meiosis, directing heterochromatic CHH and CHG methylation (Singh et al. 2011), and an AGO4 homolog in Arabidopsis, AtAGO9, is a key component of RdDM (Havecker et al. 2010). Also, meiotic phasiRNAs are reported to direct DNA methylation in cis (Dukowic-Schulze et al. 2016). Therefore, an AGO4 paralog is a promising candidate as an effector molecule for meiotic phasiRNAs.
We propose that phased siRNAs detected in maize, rice, asparagus, and lily are analogous. Our conclusion that the loci we have characterized are generated by the same pathways is based on a number of observations that do not rely on sequence identity. First, 24-nt phased siRNAs are quite unique and distinctive (i.e., previously described nowhere other than grass anthers). The current state of our understanding is that plant reproductive phasiRNAs are defined by the following properties: (1) phased production of siRNA/sRNAs from an mRNA precursor; (2) the processing is precise, and thus the phasiRNAs account for the majority of sRNAs at specific locus (i.e., there are few nonphased sRNAs); (3) expression of these phasiRNAs is restricted or at least extraordinarily enriched in premeiotic or meiotic anthers and usually spatially to one or two cell layers within the anther; and (4) there is a clear developmental change in abundance with the phasiRNAs, peaking at premeiotic and/or meiotic stages. The siRNA loci in asparagus, daylily, and Lilium that we describe satisfy all four of these properties; therefore, to us, these indeed are reproductive phasiRNAs. The question might arise as to whether we should expect sequence conservation across phasiRNA clusters in grasses and nongrasses, especially those targeted by miR2275. A detailed examination of these loci found no statistically significant conservation across these species, perhaps because phasiRNAs do not function like miRNAs or tasiRNAs—that is, perhaps the phasiRNA-target interactions are evolutionary dynamic. By way of analogy, piRNAs vary across animals in terms of their biogenesis, functions, overlap with protein-coding genes, specificity to male versus female organs, and the number of encoding loci in the animal genomes that produce them—and there is little sequence conservation across species. Plant reproductive phasiRNAs demonstrate a similar variability.
Our investigation of asparagus small RNAs identified the production of reproductive phasiRNAs from intramolecular duplex mRNAs. Therefore, there are at least three described processes for the production of 24-nt meiotic phasiRNAs in monocots: (1) the canonical process seen in grasses, involving miR2275 and RDR6 (Fig. 8C); (2) IR-derived phasiRNAs from mRNAs lacking a miR2275 target site, noting that these are not secondary siRNAs (Fig. 8B); and (3) production from IRs triggered by miR2275 (Fig. 8B). The grass 24-nt reproductive phasiRNAs and the asparagus phased IR-derived siRNAs appear analogous based on similar spatial localization and temporal dynamics. The precise nature of the triggering mechanism for the IR-derived siRNAs that guides their processing, such as a structural motif, has yet to be determined. In conclusion, our characterization of reproductive phasiRNAs in diverse monocots is consistent with early emergence during monocot or angiosperm evolution, and a broad role of these pathways in germinal development.
Methods
Sample collection, stage-size correlation, Illumina- and SMRT-sequencing, and data preprocessing
Garden asparagus samples were collected from the T.S. Smith and Son's Farm. Flowering lily (Lilium maculatum) and daylily (Hemerocallis lilioasphodelus) were purchased from Home Depot. Anther stages were examined on propidium iodide-stained (Asparagus and Lilium) or cleared tissue (daylily) using confocal microscopy. Small and mRNA libraries were constructed using standard TruSeq protocols (Illumina) (Mathioni et al. 2017). PARE libraries were constructed as published (Zhai et al. 2014). SMRT-seq libraries were prepared according to the manufacturer's protocol for full-length transcript library preparation (Clontech Laboratories, Inc.; Pacific Biosciences [PacBio]). Full-length transcript libraries were fractionated into four pools; 1, 2, 5, and 10 kb. Full-length transcript sequencing was performed on a PacBio RS II (Univ. of Delaware). Raw sequencing data preprocessed used pbtranscript-tofu (v2.3.0, now referred to as IsoSeq3; https://github.com/PacificBiosciences/IsoSeq_SA3nUP) using default settings.
Identification of microRNAs and PHAS loci
Two different computational pipelines for discovery of miRNAs were used: a stringent pipeline for de novo identification and a relaxed pipeline for identification of conserved “known” miRNAs. Stem–loop structure of candidate precursors was visually inspected in both pipelines using CENTROIDFOLD (Sato et al. 2009), and mature miRNAs were matched with miRBASE entries to identify conserved families. Phased siRNA generating (PHAS) loci or precursors and their triggers were identified using PHASIS (Kakrana et al. 2017).
Transcriptome assembly, coding-potential assessment, and identification of encoded proteins
Preprocessed RNA-seq libraries and polished SMRT-seq transcripts were used to generate transcriptome assemblies. For asparagus, ab initio assembly used TopHat and Cufflinks (Trapnell et al. 2012). The de novo hybrid transcriptome assemblies for asparagus and daylily were generated by combining reads from Illumina and SMRT-seq libraries via the “--long-reads” parameter of Trinity (v2.1.1) (Haas et al. 2013). For lily, a de novo transcriptome assembly was generated from Illumina data using Trinity.
Transcripts from all assemblies were annotated independently using Trinotate (Haas et al. 2013). To access the coding potential of transcripts, we built a logical classifier that uses CPC and CPAT scores, along with annotations and classified transcripts into three categories: coding, noncoding, and transcript of unknown potential (TUCP). Species-specific gene models or transcriptome annotations from the Trinotate workflow were manually curated to identify Dicer and Argonaute family members in asparagus, lily, and daylily.
Secondary structure analysis of PHAS loci
The ds- and ss-RNA sequencing libraries were generated and analyzed as previously described (Li et al. 2012). Structure scores (Si) were computed as the generalized log ratio (glog) of normalized dsRNA-seq reads to normalized ssRNA-seq reads. Similarly, strand scores were computed as glog of sense versus antisense ds-RNA sequencing reads.
Fluorescent in situ hybridization (FISH) confocal microscopy
Small RNAs were detected using LNA probes (Exiqon). Samples were vacuum fixed using 4% paraformaldehyde and processed by the A.I. DuPont Hospital for Children histology laboratory for paraffin embedding. Protocols for the prehybridization, hybridization, post-hybridization, and detection steps were as previously described (Javelle and Timmermans 2012). For FISH of DCL5 mRNA, digoxigenin-labeled probes were detected with sheep anti-digoxigenin antibodies (1/500) from Sigma-Aldrich (cat# 11214667001), and then with donkey anti-sheep antibodies conjugated to AlexaFluor647 (1/1000) from Thermo Fisher Scientific (cat# A-21448). Confocal images were taken with a Zeiss LSM880 microscope (Delaware Bioimaging Center) using a C-Apochromat 40X (NA = 1.3) oil immersion objective lens. FISH images were taken under 633 nm excitation and 649–758 nm emission wavelengths.
Data access
Illumina and PacBio sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE112799 for daylily and GSE97978 for lily (libraries detailed in Supplemental Table S7), and GSE112800 for asparagus (libraries detailed in Supplemental Table S1).
Supplementary Material
Acknowledgments
We thank Karol Miaskiewicz from the Delaware Biotechnology Institute for work on the SMRT portal and pbtranscript-tofu, and Mayumi Nakano for assistance with data visualization. We thank Pingchuan Li for assistance with analysis scripts, and John McDonald for discussions on statistical tests. This work was supported by the US National Science Foundation Plant Genome Research Program (NSF PGRP) grants (1649424 and 0922742) to B.C.M. and J.H.L.-M., respectively, a University Competitive Fellow Award (2015–2016) to A.K., and NSF Career Award MCB-1053846 and grant MCB-1243947 to B.D.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.228163.117.
References
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdés-López O, Prince S, Musket TA, Nguyen HT, et al. 2014. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26: 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernández L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P. 2016. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Schapire AL, Zhai J, Chorostecki U, Boisbouvier J, Meyers BC, Palatnik JF. 2013. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res 23: 1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. 2015. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chase MW, Reveal JL. 2009. A phylogenetic classification of the land plants to accompany APG III. Bot J Linn Soc 161: 122–127. [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. 2011. Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Talbot M, Li Z, Helliwell C. 2013. Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley. BMC Plant Biol 13: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukowic-Schulze S, Sundararajan A, Ramaraj T, Kianian S, Pawlowski WP, Mudge J, Chen C. 2016. Novel meiotic miRNAs and indications for a role of phasiRNAs in meiosis. Plant Genet Genomics 7: 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360. [DOI] [PubMed] [Google Scholar]
- Fan Y, Yang J, Mathioni SM, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, et al. 2016. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc Natl Acad Sci 113: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Yang L, Liang W, Zhang D, Meyers BC. 2016. Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J Exp Bot 67: 6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A, et al. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. 2010. The Arabidopsis RNA-directed DNA methylation Argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 38: 721–725. [DOI] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, Tomari Y. 2016. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell 164: 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Timmermans MC. 2012. In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7: 533–541. [DOI] [PubMed] [Google Scholar]
- Jeong DH, Thatcher SR, Brown RS, Zhai J, Park S, Rymarquis LA, Meyers BC, Green PJ. 2013. Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol 162: 1225–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A, Hammond R, Patel P, Nakano M, Meyers BC. 2014. sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res 42: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A, Li P, Patel P, Hammond R, Anand D, Mathioni SM, Meyers BC. 2017. PHASIS: a computational suite for de novo discovery and characterization of phased, siRNA-generating loci and their miRNA triggers. bioRxiv 10.1101/158832. [DOI] [Google Scholar]
- Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura K-I. 2014. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J 78: 385–397. [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Gurazada SG, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. 2012. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. 2012. Global analysis of RNA secondary structure in two metazoans. Cell Rep 1: 69–82. [DOI] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X. 2015. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H. 2010. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. 2004. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM. 2006. The evolution and diversification of Dicers in plants. FEBS Lett 580: 2442–2450. [DOI] [PubMed] [Google Scholar]
- Mathioni SM, Kakrana A, Meyers BC. 2017. Characterization of plant small RNAs by next generation sequencing. Curr Protoc Plant Biol 2: 39–63. [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348: 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. 2007. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci 104: 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2007. A germ cell–specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335. [DOI] [PubMed] [Google Scholar]
- Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, Sasaki T, Nonomura KI. 2018. EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D. 2011. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci 16: 258–264. [DOI] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T. 2009. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res 37: W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Bresso EG, Spinelli SV, Palatnik JF. 2012. Role of microRNA miR319 in plant development. In MicroRNAs in plant development and stress responses (ed. Sunkar R), Vol. 15 of Signaling and communication in plants, pp. 29–47. Springer-Verlag, Berlin Heidelberg, Germany. [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. 2011. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, et al. 2012a. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69: 462–474. [DOI] [PubMed] [Google Scholar]
- Song X, Wang D, Ma L, Chen Z, Li P, Cui X, Liu C, Cao S, Chu C, Tao Y, et al. 2012b. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J 71: 378–389. [DOI] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee HC, Weng Z, Mello CC. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua NH, Wang XJ. 2011. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 67: 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L, et al. 2015. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 4: e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Xu J, Arikit S, Meyers BC. 2015. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol Biol Evol 32: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, Paoli ED, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC. 2014. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67: 84–90. [DOI] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V, Meyers BC. 2015. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci 112: 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xia R, Meyers BC, Walbot V. 2015. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol 27: 84–90. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xia R, Kuang H, Meyers BC. 2016. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol 33: 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD. 2010. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.