Abstract
Hippocampal GABAergic interneurons are crucial for cortical network function and have been implicated in psychiatric disorders. We show here that Neuregulin 3 (Nrg3), a relatively little investigated low‐affinity ligand, is a functionally dominant interaction partner of ErbB4 in parvalbumin‐positive (PV) interneurons. Nrg3 and ErbB4 are located pre‐ and postsynaptically, respectively, in excitatory synapses on PV interneurons in vivo. Additionally, we show that ablation of Nrg3 results in a similar phenotype as the one described for ErbB4 ablation, including reduced excitatory synapse numbers on PV interneurons, altered short‐term plasticity, and disinhibition of the hippocampal network. In culture, presynaptic Nrg3 increases excitatory synapse numbers on ErbB4+ interneurons and affects short‐term plasticity. Nrg3 mutant neurons are poor donors of presynaptic terminals in the presence of competing neurons that produce recombinant Nrg3, and this bias requires postsynaptic ErbB4 but not ErbB4 kinase activity. Furthermore, when presented by non‐neuronal cells, Nrg3 induces postsynaptic membrane specialization. Our data indicate that Nrg3 provides adhesive cues that facilitate excitatory neurons to synapse onto ErbB4+ interneurons.
Keywords: ErbB4, GABAergic interneurons, hippocampus, synaptogenesis
Subject Categories: Neuroscience
Introduction
Genetic and environmental factors contribute to the manifestation of neuropsychiatric disorders like schizophrenia. Taking into account the enormous complexity of the human brain, it is difficult to define causative mechanisms. One route toward a better understanding is provided by human genetics, which correlates the occurrence of neuropsychiatric disease with mutations or sequence variants in particular genes (Escudero & Johnstone, 2014; Fromer et al, 2014; Hall et al, 2015). The functional analysis of such genes in model organisms provides an entry point to study the mechanisms and pathophysiology of neuropsychiatric diseases such as schizophrenia.
Neuregulins are a family of growth factors containing an EGF‐like domain that bind and activate tyrosine kinase receptors of the ErbB family. Four different Neuregulin genes (Nrg1, Nrg2, Nrg3, and Nrg4) exist. Neuregulins bind to the ErbB4 receptor with variable affinities and activate its tyrosine kinase with different efficacies (Yarden, 2001). NRG1, NRG3, and ERBB4 mutations and gene variants have been implicated in several neuropsychiatric diseases in humans, but are most frequently associated with schizophrenia in several ethnic groups (Stefansson et al, 2002; Chen et al, 2009; Kao et al, 2010; Greenwood et al, 2011; Morar et al, 2011; Hatzimanolis et al, 2013). ErbB4 is expressed in various neuronal types in the brain where it controls physiology and behavior (Li et al, 2007; Fazzari et al, 2010; Gu et al, 2016; Sun et al, 2016; Geng et al, 2017). In the neocortex and hippocampus, ErbB4 expression is restricted to GABAergic interneurons. ErbB4 expression is particularly high in hippocampal PV inhibitory interneurons, where it is enriched at postsynaptic sites (Vullhorst et al, 2009; Fazzari et al, 2010; Neddens et al, 2011; Mitchell et al, 2013). The mechanisms responsible for this postsynaptic clustering of ErbB4 remain open. Prior work has shown that mutating ErbB4 reduced the number of excitatory synapses on PV inhibitory neurons in the hippocampus, altered synapse function, and resulted in hyperactivity of the cortical circuit (Chen et al, 2010; Fazzari et al, 2010; Del Pino et al, 2013; Yang et al, 2013). In addition, ErbB4 has been assigned both a kinase‐dependent and kinase‐independent role in inhibitory synapse function (Krivosheya et al, 2008; Mitchell et al, 2013). Various responses of ErbB4+ interneurons to recombinant or transgenic overexpression of Nrg1 have been reported both in vitro and in vivo (Abe et al, 2011; Yin et al, 2013; Agarwal et al, 2014; Sun et al, 2016), and cortical ablation of Nrg1 can result in behavioral changes and unbalanced excitatory–inhibitory neurotransmission (Agarwal et al, 2014).
In comparison with Nrg1, little is known about another member of the Neuregulin family, Nrg3, probably because of its low affinity for ErbB4 and its poor signaling activity (Jones et al, 1999; Hobbs et al, 2002). This changed with the discovery that Nrg3 sequence polymorphisms are associated with schizophrenia and other psychiatric disorders (Wang et al, 2008; Chen et al, 2009; Xu et al, 2009; Kao et al, 2010; Morar et al, 2011; Meier et al, 2013). Ablation of Nrg3 and its overexpression in the prefrontal cortex of mice were associated with decreased and increased impulsivity, respectively, while transient overexposure of Nrg3 had lifelong consequences on behavior (Loos et al, 2014; Paterson & Law, 2014). It is interesting to note that Nrg3 is a transmembrane molecule and that transfected Nrg3 was recently observed to be located presynaptically in boutons on cultured inhibitory neurons where ErbB4 is present postsynaptically (Vullhorst et al, 2017). However, the cellular and molecular functions of Nrg3 at the synapse are unknown.
Here, we demonstrate that Nrg3 is a functionally important interaction partner of ErbB4 using biochemical, cell biological, electrophysiological, and genetic analyses. We focused our analyses on excitatory synapses on PV inhibitory neurons in the hippocampus because we observed pronounced Nrg3 and ErbB4 co‐clustering at such synapses in vivo. Our in vivo analysis demonstrates that the loss of Nrg3 weakens the excitatory input to PV neurons, changes the paired pulse ratio at this synapse type, and alters hippocampal network activity. The extensive similarities between the Nrg3 phenotype and the phenotype previously reported for mice that lack ErbB4 in the entire brain or specifically in interneurons indicate that a dominant function of Nrg3 is exerted in excitatory synapses on ErbB4+ interneurons. Our mechanistic analyses performed in cultured neurons show that presynaptic Nrg3 stimulates the formation and/or stabilization of excitatory synapses onto ErbB4+ neurons. Furthermore, Nrg3 enhances the recruitment of synaptophysin, indicating that it participates in synapse maturation, and increases the efficacy of excitatory transmission. The effect of Nrg3 on the number and maturation of excitatory synapses in cultured inhibitory neurons depends on ErbB4, but not on ErbB4 tyrosine kinase activity. In conclusion, Nrg3 enhances synaptogenesis onto inhibitory neurons, and we suggest that it provides adhesive cues that facilitate the selection of ErbB4+ interneurons as synaptic partners of cortical excitatory neurons.
Results
Nrg3 is enriched in excitatory synapses on inhibitory neurons in vivo
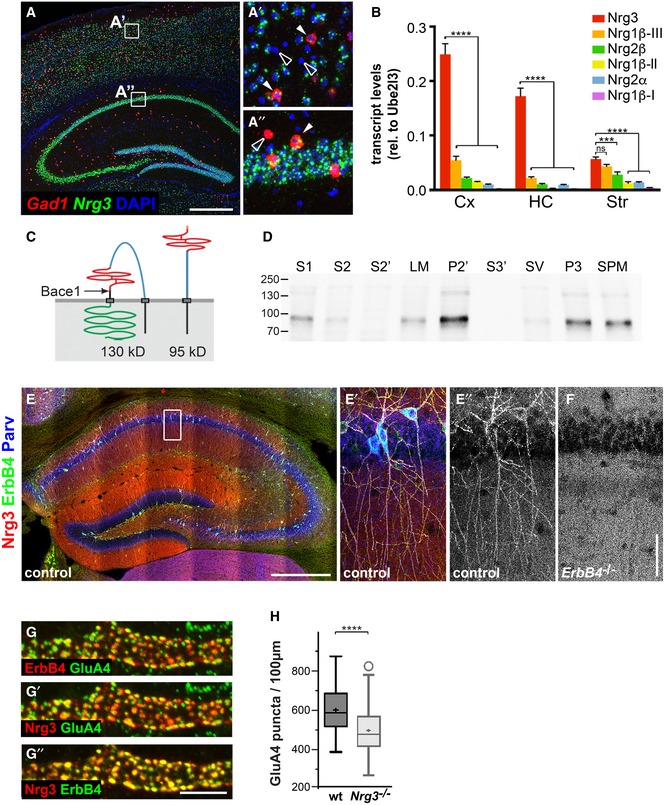
The Nrg3 gene is expressed broadly and abundantly throughout the nervous system (Zhang et al, 1997). In the hippocampus and the cortex, Nrg3 transcripts can be detected in both excitatory and inhibitory neurons (Fig 1A–A″). Nrg3 mRNA levels in the cortex and hippocampus are higher than those of either Nrg1 or Nrg2 (Fig 1B). Nrg3 encodes a transmembrane protein whose domain structure was previously determined (Vullhorst et al, 2017; see Fig 1C for a scheme of the Nrg3 structure). Antibodies directed against Nrg3 detected a protein with a molecular weight of around 95kD in brain extracts of control mice. Subcellular fractionation demonstrated that Nrg3 was enriched in fractions containing synaptic membranes, including synaptosomes (P2′), synaptosomal membranes (P3), and synaptic plasma membranes (SPM) but not in synaptic vesicle or soluble fractions (S2′, S3′) (Fig 1D).
Figure 1. Expression and localization of Nrg3 in vivo .
-
ADouble fluorescence in situ hybridization using Nrg3 (green)‐ and Gad1 (red)‐specific probes; DAPI was used as a counterstain. Nrg3 is present in Gad1‐positive (filled arrowheads) and Gad1‐negative neurons. (A′) and (A″) show higher magnifications of boxed areas indicated in (A). Nrg3‐negative neurons are indicated by open arrowheads; note that many but not all neurons express Nrg3.
-
BComparison of mRNA levels of Nrg3, Nrg1β type III (Nrg1β−III), Nrg2β, Nrg1β type II (Nrg1β−II), Nrg2α, and Nrg1β type I (Nrg1β−I) in the cortex (Cx), hippocampus (HC), and striatum (Str) of juvenile mice (P30–60) by quantitative RT–PCR. Data represent means ± SD of four biological replicates. Two‐way ANOVA with Bonferroni's multiple comparisons test was performed to assess statistical significance (****P < 0.0001, ***P < 0.001, ns = not significant).
-
CSchematic display of the structure of Nrg3 before (left) and after cleavage by Bace1 (right). The following domains of Nrg3 are indicated: black, N‐terminal intracellular domain and transmembrane domain; blue, extracellular domain; red EGF domain; black, stalk region with Bace1 cleavage site indicated by an arrow and C‐terminal transmembrane domain; and green, intracellular domain.
-
DDetection of Nrg3 by Western blot analysis in fractions from brain lysates: S1, crude lysate; S2, cytosol and light membranes; S2′, cytosol; LM, light membranes; P2′, crude synaptosomes; S3′, synaptosomal cytoplasm; SV, synaptic vesicles; P3, synaptosomal membranes; and SPM, synaptic plasma membranes.
-
E, FImmunohistochemical analysis in the hippocampus of wildtype (E, E′, E″) and ErbB4 mutant mice (F) at 2 months of age using Nrg3‐, ErbB4‐, and parvalbumin (Parv)‐specific antibodies. The white box in (E) is displayed magnified in (E′, E″). In control mice, Nrg3 is enriched on dendrites of ErbB4+ PV interneurons, compared to the surrounding neuropil (E′, E″). In ErbB4 mutants, the Nrg3 enrichment on PV interneurons is not apparent (F).
-
GNrg3, ErbB4, and GluA4 were identified by immunohistology and co‐localized in synaptic puncta on dendrites of interneurons in the stratum radiatum of the hippocampal CA1. Note that (G) displays Nrg3/ErbB4, (G′) Nrg3/GluA4, and (G″) ErbB4/GluA4 signals of the same triple‐stained image; false colors were assigned for better signal visualization.
-
HQuantification of GluA4 puncta present on ErbB4+ dendrites in the CA1 stratum radiatum of adult wildtype (wt) and Nrg3 −/− mice (age P90–120). Data are presented as box plots with Tukey's whiskers and outliers; means are indicated by a plus symbol. n = 65 (wt) and n = 62 (Nrg3 −/−) from five animals each. Unpaired t‐test (two‐tailed) with Welch's correction was performed to assess statistical significance (****P < 0.0001).
We used immunohistochemistry to analyze the distribution of Nrg3 in the brains of 2‐month‐old mice. An antibody against Nrg3 detected the protein in the neuropil where it was particularly abundant in the molecular layer of the dentate gyrus and the mossy fiber tract. In addition, puncta with higher levels of Nrg3 were detected. Nrg3 puncta co‐localized with ErbB4 in the dendrites of ErbB4+ PV neurons in the hippocampus (Figs 1E–E″ and EV1; see Fig EV1 for antibody specificity). In the cortex, Nrg3 clusters were observed on PV ErbB4+ interneurons and on a subset of ErbB4+ neurons that did not co‐express PV (Fig EV1). In ErbB4 mutant mice, Nrg3 association with the dendrites of PV interneurons was no longer observed (Fig 1F). This indicates that Nrg3 binds to the dendritic surface of PV interneurons in an ErbB4‐dependent manner.
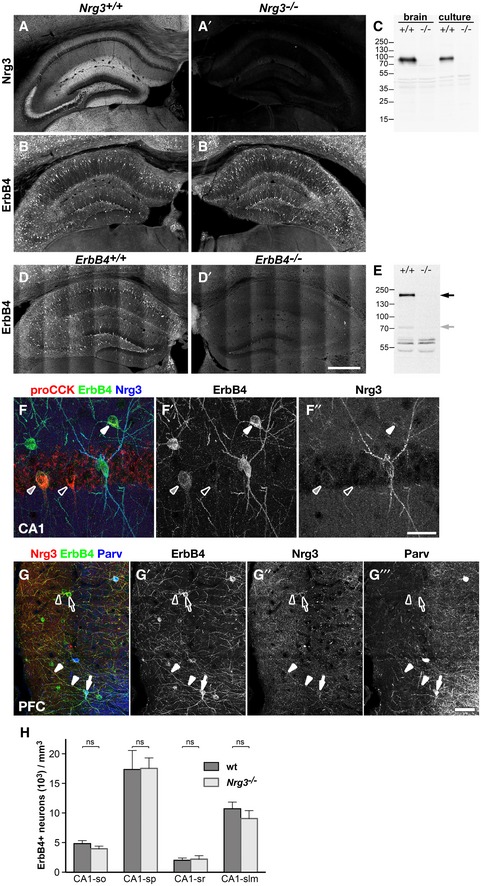
Figure EV1. Specificity of anti‐Nrg3 and anti‐ErbB4 antibodies, Nrg3/ErbB4 co‐clustering on PV‐negative neurons, and ErbB4 interneuron numbers in the hippocampus of Nrg3 mutant mice.
-
A, BHippocampus sections from wildtype (A, B) and Nrg3 −/− mice (A′, B′), age P35, were immunostained for Nrg3 (A, A′) and ErbB4 (B, B′). Nrg3 is widely distributed throughout the neuropil with higher levels being associated with ErbB4+ interneurons (A). Nrg3 immunostaining is abolished in Nrg3 mutant tissue (A′), ErbB4 levels are unchanged (B, B′).
-
CWestern blot analysis of lysates from cortical tissue (brain) and cultured cortical neurons (culture). The anti‐Nrg3 antibody detects a main band of about 95 kD (arrow) in lysates from wildtype (+/+) but not from Nrg3 mutant mice (−/−).
-
DHippocampus sections from wildtype (D) and heart‐rescued ErbB4 mutant mice (D′), 2 months of age, were immunostained for ErbB4. ErbB4 immunostaining is detected in wildtype (D) but not ErbB4 mutant hippocampus (D′).
-
EWestern blot analysis of lysates from cortical tissue. The anti‐ErbB4 antibody detects a main band of 180 kD (black arrow) and a minor band of about 70 kD (gray arrow) in lysates from wildtype (+/+) but not from ErbB4 mutant mice (−/−).
-
FHippocampus section was immunostained against pro‐CCK, ErbB4, and Nrg3; shown is the stratum pyramidale of CA1. CCK+ interneurons express variable amounts of ErbB4 as indicated by white (high expression), gray (middle), and open arrowheads (no detectable ErbB4). Association of Nrg3 with soma or dendrites of CCK+ interneurons is not detectable.
-
GSection of the prefrontal cortex was immunostained against Nrg3, ErbB4, and parvalbumin (Parv). Nrg3 is co‐localized with ErbB4 on soma and dendrites of PV‐positive (filled arrow and arrowheads) and PV‐negative neurons (open arrow and arrowheads).
-
HDensities of ErbB4+ interneurons in different layers of the hippocampal CA1 region are identical in adult wildtype and Nrg3 mutant mice at postnatal days 90–120; so, stratum oriens; sp, stratum pyramidale; sr, stratum radiatum; slm, stratum lacunosum‐moleculare. Data represent mean ± SD of six biological replicates for each genotype. Two‐way ANOVA with Bonferroni's multiple comparisons test was performed to assess statistical significance (ns = not significant). Scale bars: 500 μm (D′), 40 μm (F″), 50 μm (G‴).
Source data are available online for this figure.
Nrg3/ErbB4 co‐localization in puncta was particularly obvious on dendrites of PV interneurons in the stratum radiatum of the CA1 hippocampus, where 95.8 ± 3.4% of Nrg3 puncta contacted clustered ErbB4, and vice versa, 93.1 ± 6.4% of ErbB4 clusters contacted Nrg3 puncta (573 puncta analyzed in 12 dendritic segments from 6 neurons). These Nrg3/ErbB4 puncta also co‐localized with the AMPA receptor subunit GluA4 (Fig 1G–G″), a marker for excitatory synapses on fast‐spiking interneurons (Geiger et al, 1995). We then examined excitatory synapse numbers on interneurons in the hippocampal CA1 region in control and Nrg3 −/− mice. Quantification of GluA4+ puncta on the dendrites of PV interneurons in the stratum radiatum of the hippocampal CA1 region demonstrated a significant reduction in Nrg3 −/− mice (Fig 1H). However, the number and overall distribution of ErbB4+ interneurons in the hippocampus was apparently unchanged (Fig EV1). The role of ErbB4 in the formation and function of excitatory synapses on PV interneurons has been well documented (Fazzari et al, 2010). Since we observed a similar reduction in the number of excitatory synapses on PV interneurons in Nrg3 −/− brains in vivo as the one reported for ErbB4 mutants, we concentrated our further investigations on the cell biological mechanisms of Nrg3 function in this synapse type.
Presynaptic Nrg3 promotes ErbB4 clustering and synapse formation
We used cultures of hippocampal neurons to further investigate the mechanisms of Nrg3 synaptic function. Nrg3 distribution was analyzed in neurons cultured for 21–23 days (Fig 2A and A′). We observed clusters of Nrg3/ErbB4 on the dendrites of both PV‐positive and PV‐negative neurons in culture (Fig EV2). Most synaptic Nrg3 clusters (81.6 ± 1.4%, mean ± SEM) co‐localized with ErbB4 and, similarly, most (80.8 ± 1.6%) ErbB4 puncta co‐localized with Nrg3 (42 dendrites from 30 neurons were quantified in three independent experiments). Next, we analyzed the presence of endogenous Nrg3 in excitatory and inhibitory synapses identified by antibodies against the vesicular glutamate transporters vGlut1/2 and the vesicular GABA transporter vGAT, respectively. Among the vGlut1/2+ and vGAT+ synapses, about 54.1 ± 3.2% and 21.5 ± 1.4% contained Nrg3, respectively (Fig 2B). Nrg3 levels in vGAT+ synapses were lower than the levels observed in vGlut1/2+ synapses (Fig 2C). The comparatively low number of inhibitory synapses and the small ratio of those containing Nrg3 provide an explanation for our difficulties detecting Nrg3 in inhibitory synapses in vivo. To assess whether Nrg3 affects synaptogenesis in cultured neurons, we counted the number of excitatory vGlut1/2+ synapses on the dendrites of PV interneurons (Fig 2D). Similarly to the in vivo situation, we observed a significant reduction in the number of excitatory synapses in Nrg3 −/− compared to control cultures.
Figure 2. Nrg3 locates to synapses on ErbB4+ interneurons in vitro .
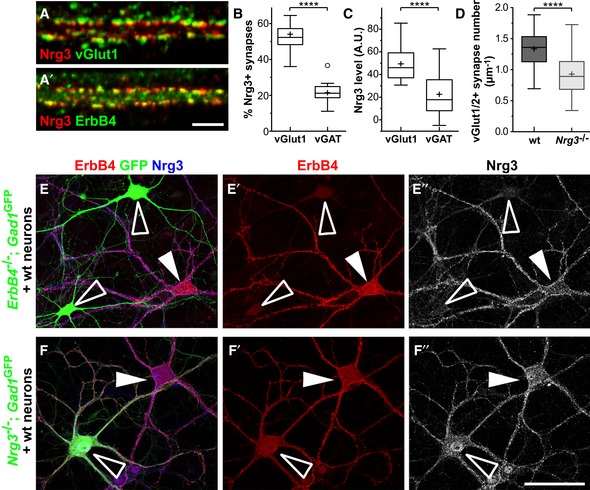
- Hippocampal neurons cultured for 21 days were analyzed by immunocytochemistry using antibodies against Nrg3, ErbB4, and vGlut1, demonstrating co‐clustering of Nrg3 and ErbB4 in excitatory synapses. Note that (A) displays Nrg3/vGlut1 and (A′) Nrg3/ErbB4 signals of the same triple‐stained image.
- Quantification of the proportions of vGlut1/2+ and vGAT+ presynaptic boutons that are positive for Nrg3 (two‐tailed Wilcoxon test, ****P < 0.0001, n = 18 from two independent experiments).
- Quantification of Nrg3 immunofluorescence levels in vGlut1/2+ and vGAT+ presynaptic boutons that are positive for Nrg3 (A.U., arbitrary units, two‐tailed Wilcoxon test, ****P < 0.0001, n = 17 from two independent experiments).
- Quantification of vGlut1/2+ synapse numbers on secondary dendrites of PV interneurons in cultures from wildtype (wt) and Nrg3 −/− mice (21–24 days in culture). Data from n = 27 (wt) and n = 26 (Nrg3 −/−) neurons from three independent experiments were analyzed using two‐tailed Mann–Whitney U‐test, ****P < 0.0001.
- Immunocytochemical analysis of a mixed culture containing neurons of wildtype and ErbB4 −/−;Gad1/Gad67‐GFP animals triple‐stained against ErbB4, GFP, and Nrg3. To improve the visibility, ErbB4 and Nrg3 signals are also shown separately (E′, E″). ErbB4+ interneurons from wildtype animals are indicated by filled arrowheads, and interneurons from ErbB4 −/− mice were identified by GFP and are indicated by open arrowheads.
- Immunocytochemical analysis of a mixed culture containing neurons from wildtype and Nrg3 −/− ;Gad1/Gad67‐GFP animals triple‐stained against ErbB4, GFP, and Nrg3. To improve visibility, ErbB4 and Nrg3 signals are also shown separately (F′, F″). ErbB4+ interneurons from Nrg3 −/− animals are GFP‐positive and are indicated by open arrowheads; ErbB4+ interneurons from wildtype animals are GFP‐negative and are indicated by filled arrowheads.
Figure EV2. Nrg3/ErbB4 co‐clustering in cultured PV‐positive and PV‐negative interneurons.
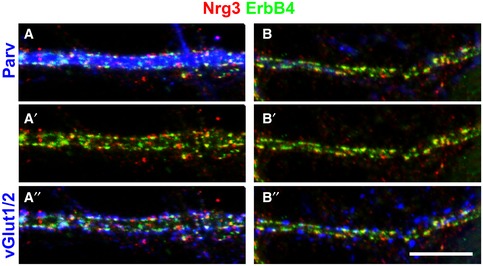
-
A, BNeurons, cultured for 23 days, were co‐immunostained with antibodies against Nrg3, ErbB4, parvalbumin (Parv), and vGlut1/2. Nrg3 and ErbB4 co‐clustered on the dendrites of PV‐positive (A) and PV‐negative neurons (B). Note that (A–A″) and (B–B″) display false colors of the same images. Scale bar: 10 μm (B″).
We next tested whether recruitment of Nrg3 to synaptic sites in culture also depends on ErbB4. For this, we used cultured neurons from ErbB4 −/− mice that carried one Gad1/Gad67‐GFP allele (Tamamaki et al, 2003). The Gad1/Gad67‐GFP allele is expressed by inhibitory neurons, which allowed their identification by GFP. We also mixed neurons from wildtype mice into these cultures. Interneurons from wildtype animals were identified by staining for ErbB4, while interneurons from ErbB4 −/− animals were identified by GFP. In these mixed cultures, Nrg3 puncta were present on ErbB4+ but not on GFP‐positive ErbB4 −/− interneurons (Fig 2E–E″). Thus, in vitro and in vivo, Nrg3 recruitment to synapses on inhibitory neurons depends on ErbB4.
Nrg3 is also expressed by GABAergic interneurons (Fig 1A). This raises the question of whether clustered Nrg3 detected on dendrites is derived from the postsynaptic cell. To address this, we again used mixed neuronal cultures, i.e., cultures containing neurons from Nrg3 −/− mice that carried one Gad1/Gad67‐GFP allele mixed with neurons from wildtype mice. Immunohistochemical analysis demonstrated indistinguishable punctate patterns of Nrg3 on GFP‐positive Nrg3 −/− and GFP‐negative wildtype neurons (Fig 2F–F″), thus demonstrating that Nrg3 is produced presynaptically and provided in trans.
We subsequently used lentivirus‐infected cultures of hippocampal neurons, the lentivirus co‐expressing Nrg3 and synaptophysin fused to cyan fluorescent protein (SypCFP) (Fig EV3). Nrg3 expression was driven by the synapsin (Syn1) promoter. In vGlut1/2+ synapses, the virally produced Nrg3 was present at a 2.2‐fold higher level than the endogenous Nrg3 protein as determined by immunofluorescence (Fig EV3). We used the Nrg3/SypCFP lentivirus at a titer that infected 10–20% of the cultured neurons. In these cultures, infected and non‐infected neurons formed synapses, and these two types of synapses were distinguished by the presence or absence of SypCFP, respectively (see scheme in Fig 3A). Most SypCFP+ puncta were stained with antibodies against vGlut1/2 and thus represented excitatory synapses (mean ± SD: 79.2 ± 11.5% and 60.3 ± 14.3% of the SypCFP+ puncta on ErbB4‐positive and ErbB4‐negative neurons, respectively; n = 27 neurons from four independent experiments; see also Fig 3B–B″). The SypCFP+ puncta apposed clustered glutamate receptors detected by an anti‐GluA antibody (Fig EV3). A low number of SypCFP+ boutons contained the vesicular GABA transporter vGAT and therefore correspond to inhibitory synapses (4.6 ± 5.1% and 8.8 ± 9.4% of SypCFP+ puncta on ErbB4+ and ErbB4− neurons were vGAT+, respectively; n = 28 neurons from four independent experiments). These transduced cultures provided an experimental strategy that allowed us to compare and quantify excitatory synapses generated by neurons that presented/did not present Nrg3 in a single culture.
Figure EV3. Lentiviral expression of Nrg3 and Nrg3ΔEGF .
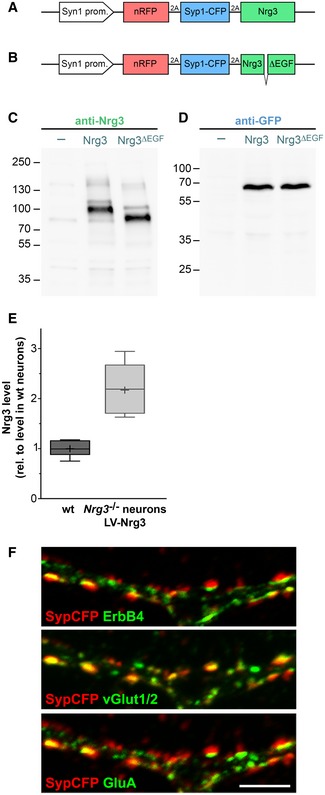
-
A, BSchematic display of the expression cassettes. The synapsin I (Syn1) promoter is used for neuronal expression. Coding sequences for red fluorescence protein with a nuclear localization sequence (nRFP) and a fusion protein of synaptophysin and the cyan fluorescent protein (SypCFP) are followed by coding sequences for Nrg3 (A) or Nrg3ΔEGF (B). nRFP, SypCFP, and Nrg3/Nrg3ΔEGF coding sequences are separated by 2A sequences.
-
C, DComparison of Nrg3 (C) and SypCFP (D) expression levels in Nrg3 −/− neurons that were transduced with the Nrg3/SypCFP (middle lanes) and Nrg3ΔEGF/SypCFP lentiviruses (right lanes) at high titer, respectively, by Western blot analysis using antibodies against Nrg3 (C) and GFP (D). The lanes indicated by (−) show lysates from non‐transduced neurons as negative control. Both lentiviruses express similar protein levels of Nrg3/Nrg3ΔEGF and SypCFP. Note that the main Nrg3 band in lysates from neurons transduced with the Nrg3ΔEGF/SypCFP lentivirus is smaller due to the deletion.
-
EImmunocytochemical comparison of endogenous and lentivirally expressed Nrg3 levels in vGlut1/2+ excitatory synapses on ErbB4+ neurons in cultures of wildtype (wt, left) or Nrg3 −/− neurons transduced with the SypCFP/Nrg3 lentivirus (right). Fluorescence levels were normalized to the mean fluorescence level in wildtype neuron cultures. The Nrg3 lentivirus produced 2.17 ± 0.14‐fold higher Nrg3 levels than wt neurons, and n = 13 and 11 neurons were analyzed in cultures from wildtype and Nrg3 mutants, respectively. Data are presented as box plots with Tukey's whiskers and outliers. Plus symbols denote the mean.
-
FHippocampal Nrg3 −/− neurons after transduction with lentivirus expressing SypCFP/Nrg3 were analyzed by immunocytochemistry using antibodies against CFP, ErbB4, vGlut1/2, and GluA. Panels show the same image and display SypCFP/ErbB4, SypCFP/vGlut1/2, and SypCFP/GluA signals, respectively. The images were assigned false colors for better visualization of the signals. SypCFP, ErbB4, vGlut1/2, and GluA co‐localize in excitatory synapses. Scale bar: 5 μm.
Source data are available online for this figure.
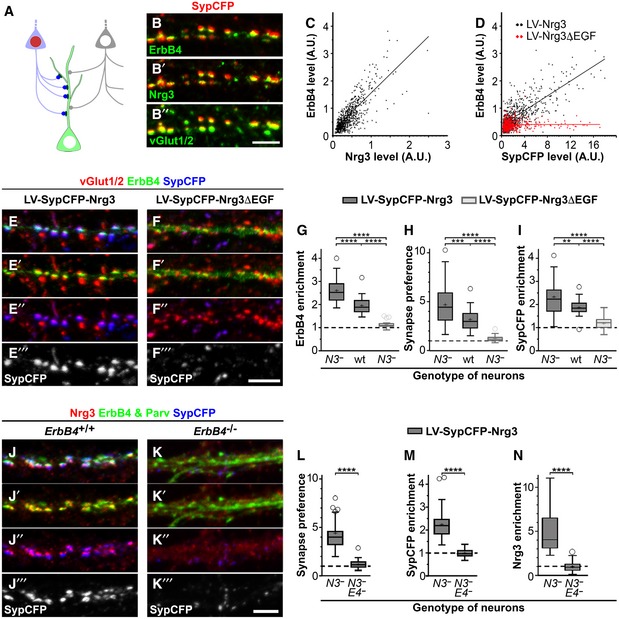
Figure 3. Nrg3 promotes the formation of excitatory synapses onto ErbB4+ interneurons in vitro .
-
ASchematic display of the culture model using hippocampal neurons that were infected by a lentivirus co‐expressing a synaptophysin‐CFP fusion protein (SypCFP) together with either Nrg3 or Nrg3ΔEGF at a low titer so that 10–20% of the hippocampal neurons were transduced with the lentivirus (depicted as a light blue colored neuron with a red nucleus). Presynaptic boutons formed by non‐transduced (depicted as a light gray neuron with a white nucleus) and transduced neurons can be distinguished by the absence or presence of CFP labeling (gray and blue boutons, respectively).
-
BHippocampal Nrg3 −/− neurons after transduction with lentivirus co‐expressing SypCFP/Nrg3 were analyzed by immunocytochemistry. (B) Antibodies against CFP, ErbB4, Nrg3, and vGlut1/2; (B), (B′), and (B″) show the same image and display SypCFP/ErbB4, SypCFP/Nrg3, and SypCFP/vGlut1/2 signals, respectively. The images were assigned false colors for better visualization of the signals. SypCFP, Nrg3, and ErbB4 co‐localize in excitatory vGlut1/2+ synapses.
-
C, DCorrelation analyses of the levels of ErbB4 and Nrg3 (C) or ErbB4 and SypCFP (D) staining in vGlut1/2+ synapses; every dot corresponds to one synapse. Cultured Nrg3 −/− neurons were transduced with either SypCFP/Nrg3 (black in C, D) or SypCFP/Nrg3ΔEGF (red in D) lentiviruses.
-
E, FHippocampal Nrg3 −/− neurons were transduced with (E) SypCFP/Nrg3 or (F) SypCFP/Nrg3ΔEGF viruses and immunostained for vGlut1/2, ErbB4, and SypCFP. Both viruses express similar amounts of protein (Fig EV2). In cultures transduced with the SypCFP/Nrg3 virus, synaptic enrichment of ErbB4 and SypCFP was very pronounced, but not in cultures transduced with the SypCFP/Nrg3ΔEGF virus. Note that (E–E‴) and (F–F‴) display the same images, but color channels were separated for better visualization.
-
GErbB4 enrichment in synapses formed in hippocampal cultures after infection with either the SypCFP/Nrg3 or the SypCFP/Nrg3ΔEGF virus. We compared neuronal cultures of different genotypes (Nrg3 −/− and wildtype cultures indicated by N3 − and wt, respectively). ErbB4 enrichment was calculated as the level of ErbB4 in synapses formed by infected neurons (vGlut1/2+/SypCFP+ boutons) divided by the level of ErbB4 in synapses formed by uninfected neurons (vGlut1/2+/SypCFP‐ boutons). The dashed line indicates a ratio of 1 at which SypCFP‐positive and SypCFP‐negative boutons would behave identical.
-
HQuantification of the preference for neurons expressing SypCFP/Nrg3 or SypCFP/Nrg3ΔEGF to form synapses on ErbB4+ neurons. The synapse preference was calculated as the proportion of vGlut1/2 boutons containing SypCFP that were present on ErbB4‐positive neurons divided by the proportion of such boutons present on ErbB4‐negative neurons.
-
IQuantification of SypCFP enrichment in excitatory synapses on ErbB4+ neurons. Enrichment was determined as the ratio between SypCFP levels in SypCFP+/vGlut1/2 boutons on ErbB4‐positive and ErbB4‐negative neurons.
-
J, KHippocampal neurons from Nrg3 −/− (J) and Nrg3 −/−;ErbB4 −/− (K) mice were transduced with the SypCFP/Nrg3‐expressing virus and immunostained for Nrg3, ErbB4/parvalbumin and SypCFP. Nrg3 and SypCFP were strongly enriched in synapses on dendrites of PV neurons in Nrg3 −/− (J) but not Nrg3 −/−;ErbB4 −/− cultures (K).
-
L–NQuantification of synapse preference (L), enrichment of SypCFP (M) and of Nrg3 (N) in Nrg3 −/− (N3 −) and Nrg3 −/−;ErbB4 −/− (N3 − E4 −) hippocampal cultures transduced with the SypCFP/Nrg3 lentivirus. Synapse preference (L) was calculated as the proportion of vGlut1/2 boutons containing SypCFP present on parvalbumin‐positive neurons divided by the proportion of such boutons present on parvalbumin‐negative neurons. SypCFP enrichment (M) was determined as the ratio of SypCFP levels in SypCFP/vGlut1/2+ boutons present on parvalbumin‐positive and parvalbumin‐negative neurons. Nrg3 enrichment (N) was determined as the ratio of Nrg3 levels in SypCFP+/vGlut1/2+ boutons present on parvalbumin‐positive and parvalbumin‐negative neurons.
We used SypCFP/Nrg3 transduced Nrg3 −/− cultures to measure Nrg3 and ErbB4 levels in vGlut1/2+/SypCFP+ synapses using immunocytochemistry (Fig 3B–B″). The levels of Nrg3 and ErbB4 in synapses correlated (Fig 3C); similarly, levels of SypCFP and ErbB4 correlated (Fig 3D, black dots and black line). Next, we used a lentivirus that co‐expressed Nrg3ΔEGF and SypCFP (Fig EV3). Nrg3ΔEGF lacks the EGF domain and is therefore unable to bind ErbB4 (Jones et al, 1999; Van Zoelen et al, 2000). When the SypCFP/Nrg3ΔEGF virus was transduced in Nrg3 −/− neurons, SypCFP and ErbB4 levels did not correlate in vGlut1/2+ synapses (Fig 3D, red dots and red line). This indicates that the Nrg3 EGF domain is essential for the pronounced ErbB4 recruitment at the synapses. In agreement, previous observations demonstrated that an excess of the soluble Nrg1 EGF domain interferes with synaptic ErbB4 recruitment (Vullhorst et al, 2017). We used a Fiji/ImageJ macro to identify vGlut1/2+ boutons on ErbB4+ inhibitory neurons formed by (i) infected cells (SypCFP‐positive synapses) and (ii) non‐infected cells (SypCFP‐negative synapses). In a second step, we quantified the ratio of ErbB4 protein levels in SypCFP‐positive versus SypCFP‐negative synapses. This demonstrated that ErbB4 recruitment was enhanced when synapses were formed by neurons transduced with the SypCFP/Nrg3 virus, but not when the SypCFP/Nrg3ΔEGF virus was used (Fig 3E and F, quantified in 3G). Enhancement by virally expressed SypCFP/Nrg3 was less pronounced when wildtype cells were transduced with the virus (Fig 3G). From these experiments, we conclude that the amount of postsynaptic ErbB4 correlates with levels of presynaptic Nrg3.
To further analyze the effects of Nrg3 in synaptogenesis, we transduced Nrg3 −/− and wildtype neurons with the SypCFP/Nrg3 lentivirus and quantified numbers of excitatory vGlut1/2+ synapses formed by infected (SypCFP‐positive) and non‐infected (SypCFP‐negative) cells on either ErbB4‐positive or ErbB4‐negative neurons. We calculated the proportions of excitatory synapses formed by infected cells (vGlut1/2+SypCFP+ puncta/all vGlut1/2+ puncta) on ErbB4‐positive and ErbB4‐negative neurons. In Nrg3 −/− and wildtype cultures, this proportion was increased 4.4‐fold and 3‐fold on ErbB4‐positive relative to ErbB4‐negative neurons, respectively (quantified in Fig 3H). Nrg3 −/− neurons expressing Nrg3ΔEGF did not prefer to form synapses on ErbB4‐positive interneurons (Fig 3H). Thus, Nrg3 mutant neurons are poor donors of presynaptic terminals in the presence of competing neurons that produce Nrg3. Moreover, the preference to synapse onto ErbB4+ neurons depends on Nrg3 levels.
Next, we quantified SypCFP levels in vGlut1/2+ presynaptic boutons in neuronal cultures transduced with the SypCFP/Nrg3 lentivirus. When neuron cultures from Nrg3 −/− or wildtype brains were transduced, SypCFP levels were markedly higher in excitatory boutons on ErbB4‐positive than ErbB4‐negative dendrites (Fig 3I). Increased levels of SypCFP in synapses on ErbB4‐positive interneurons depended on the presence of the EGF domain in Nrg3 (Fig 3I). We conclude that Nrg3 increases the recruitment of presynaptic proteins to synapses on ErbB4 neurons.
Therefore, we tested whether Nrg3‐dependent synaptogenesis on interneurons required ErbB4 by comparing Nrg3 −/− and Nrg3 −/−;ErbB4 −/− neuron cultures infected with the SypCFP/Nrg3 lentivirus at a low titer. We used PV antibodies to identify dendrites of inhibitory neurons. This demonstrated that the Nrg3‐dependent preference to synapse onto PV interneurons depended on the presence of ErbB4 (Fig 3J and K quantified in 3L). Furthermore, the increased recruitment of SypCFP and Nrg3 to presynaptic boutons was also ErbB4‐dependent (Fig 3M and N).
Nrg3 functions do not require ErbB4 tyrosine kinase activity
Nrg3 was previously shown to bind exclusively to ErbB4 or ErbB4/ErbB2 heterodimers and to be a poor signaling molecule in cancer cells (Hobbs et al, 2002). We tested the signaling capacity of Nrg3 in hippocampal neurons using soluble His6‐tagged EGF domains of Nrg1 and Nrg3 produced in 293T cells. The proteins were partially purified, and their concentration and purity were estimated by gel electrophoresis (see Materials and Methods). At a 10 nM concentration, the EGF domain of Nrg1 induced a strong tyrosine phosphorylation of ErbB4 and increased phosphorylation of Erk and Akt (Fig 4A and B). The EGF domain of Nrg3 did not stimulate ErbB4 phosphorylation, not even at 2,000 nM when mild increases in pErk and pAkt levels were observed (increase of 33 ± 4% and 9 ± 8%, respectively). Thus, Nrg3 poorly activates ErbB4 tyrosine kinase signaling in hippocampal neurons.
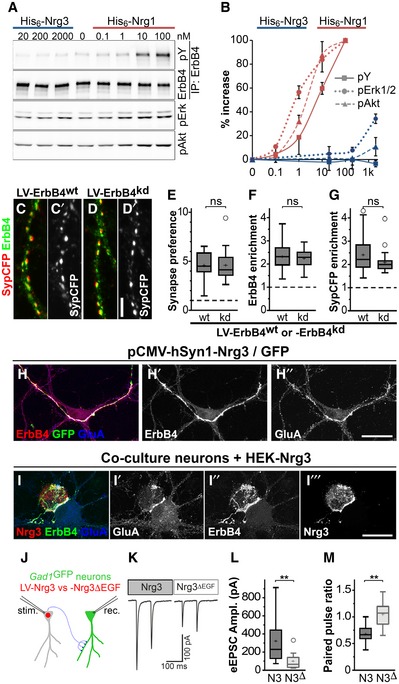
Figure 4. ErbB4 but not ErbB4 kinase activity mediates Nrg3 functions in synaptogenesis.
-
AHis6‐tagged EGF domains of Nrg1β and Nrg3 were added to neuronal cultures at the indicated concentrations. Western blot analyses of ErbB4 immunoprecipitates using antibodies against phospho‐tyrosine (pY) and ErbB4, and of whole lysates using antibodies against pErk1/2 and pAkt.
-
BQuantification of Nrg1 and Nrg3‐dependent changes in phosphorylation of ErbB4, Erk1/2 and Akt normalized to input, where 100% corresponds to the maximal change observed for His6‐Nrg1β. Data represent mean ± SD from three independent experiments.
-
C, DNeurons from Nrg3 −/−;ErbB4 flox/− ;vGAT‐Cre mice were transduced with a lentivirus expressing either (C) ErbB4wt or (D) ErbB4kd (kinase‐dead ErbB4) in a Cre‐dependent manner, and with a second virus at a low titer expressing Nrg3/SypCFP. Immunocytochemical analysis of ErbB4 and SypCFP; (C, C′) and (D, D′) each show the same image, and (C′, D′) show the SypCFP signal.
-
E–GQuantification of Synapse preference toward ErbB4+ neurons (E), ErbB4 enrichment (F), and SypCFP enrichment (G) in synapses on neurons expressing wildtype ErbB4 (wt) or ErbB4kd (kd).
-
HNrg3 −/− neuron cultures were transfected with a plasmid that strongly overexpresses Nrg3 and GFP and immunostained with antibodies against ErbB4, GFP, and GluA; (H) shows the triple‐stained image, while (H′) and (H″) only show ErbB4 and GluA signals, respectively.
-
ICo‐cultures of Nrg3 −/− neurons with HEK293 cells expressing Nrg3. Cultures were stained with antibodies against Nrg3, ErbB4, and GluA. Note that Nrg3, ErbB4, and GluA are enriched at the contact sites between Nrg3‐expressing HEK293 cells and ErbB4+ neurons.
-
JSchematic diagram of the strategy used to obtain paired recordings in cultures obtained from Nrg3 −/−;Gad1/Gad67GFP mice. Inhibitory neurons were identified by GFP expression. The cultures were transduced with lentiviruses expressing either Nrg3 or Nrg3ΔEGF (N3 and N3Δ, respectively) together with nuclear red fluorescence protein (nRFP). Transduced neurons, identified by nuclear RFP fluorescence, were stimulated, and evoked responses were recorded from interneurons that were GFP‐positive and nRFP‐negative at 12–15 days in culture.
-
K–MSample traces of paired recordings (K), quantification of the amplitude of the evoked excitatory postsynaptic currents (eEPSCs) (L), and quantification of paired pulse ratios (M).
Next, we asked whether Nrg3 requires ErbB4 signaling for its role in synapse formation. Tyrosine kinases require an intact ATP‐binding site for their enzymatic activity. Thus, mutation of the ErbB4 ATP‐binding site (K751M) abolishes its kinase activity (Prickett et al, 2009), and is subsequently named kinase‐dead ErbB4. We used neuronal cultures from Nrg3 −/− ;ErbB4flox/−;vGAT‐cre mice. Inhibitory vGAT+ interneurons from these mice did not produce endogenous ErbB4. Wildtype or kinase‐dead ErbB4 was re‐expressed in inhibitory interneurons using cre‐dependent lentiviruses. Additionally, these neurons were transduced with a virus expressing SypCFP/Nrg3 at a low titer (Fig 4C and D). As before, synapses formed by Nrg3‐transduced (SypCFP+) and non‐transduced (SypCFP‐negative) neurons were compared. Regardless of whether wildtype or kinase‐dead ErbB4 was expressed, SypCFP/Nrg3 neurons preferred to synapse on inhibitory ErbB4+ interneurons (Fig 4E). Furthermore, ErbB4 and presynaptic SypCFP were enriched to similar extents in synapses on wildtype or kinase‐dead ErbB4‐expressing interneurons (Fig 4F and G). Thus, ErbB4 is needed for Nrg3‐dependent effects on synapse formation and maturation, but ErbB4 kinase activity is dispensable.
Interestingly, when we strongly overexpressed Nrg3 together with GFP in neurons using the CMV/synapsin‐1 promoter, extended contacts between GFP+ axons and dendrites of ErbB4+ neurons were formed. ErbB4 and AMPA receptors (GluA) accumulated at these unusual contacts (Fig 4H–H″) that were neither formed with ErbB4‐negative neurons nor when Nrg3ΔEGF was overexpressed. This result suggests that Nrg3/ErbB4 interactions provide adhesive cues.
To test this further, we expressed Nrg3 or Nrg3ΔEGF in non‐neuronal cells (HEK293) and asked whether the Nrg3 presented by HEK293 cells is able to induce postsynaptic specializations in co‐cultured neurons. GluA, ErbB4, and Nrg3 were recruited to contacts between Nrg3‐producing HEK cells and ErbB4+ interneurons (Fig 4I–I‴). This was not observed when the contacting neuron was ErbB4‐negative or when the HEK cells expressed Nrg3ΔEGF (Fig EV4). Thus, remarkably, Nrg3 presented by non‐neuronal cells to interneurons induces ErbB4 recruitment and postsynaptic specialization.
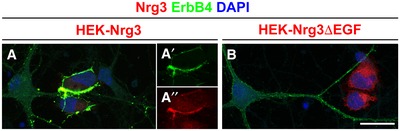
Figure EV4. Interaction of Nrg3‐ and Nrg3ΔEGF‐expressing HEK293 cells with ErbB4+ neurons.
-
A, BNeurons from Nrg3 mutant mice were co‐cultured with (A) Nrg3‐ and (B) Nrg3ΔEGF‐expressing HEK293 cells and immunostained for Nrg3/Nrg3ΔEGF (red) and ErbB4 (green). DAPI was used to counterstain nuclei (blue). Nrg3 and ErbB4 are highly enriched in contact areas between Nrg3‐expressing HEK cells and ErbB4+ interneurons (A); (A′ and A″) show lower exposures of the immunofluorescence signals of ErbB4 and Nrg3. Note that Nrg3ΔEGF and ErbB4 remained evenly distributed when an ErbB4+ interneuron was contacted by an Nrg3ΔEGF‐expressing HEK cell (B). Scale bar: 25 μm
Nrg3 modulates synaptic plasticity and strength in culture
We next tested whether the presence of Nrg3 changed the functional properties of synapses. For this, we used cultured neurons from Nrg3 −/− mice that additionally carried one Gad1/Gad67‐GFP allele (Tamamaki et al, 2003). Cultures were infected with a lentivirus encoding nuclear red fluorescent protein (nRFP) and Nrg3 or Nrg3ΔEGF. We conducted paired recordings, in which nRFP+/GFP− excitatory neurons were stimulated and postsynaptic currents were recorded from connected nRFP−/GFP+ inhibitory neurons (schematically shown in Fig 4J). The amplitude of the evoked excitatory postsynaptic current was markedly higher when Nrg3 was expressed in the presynaptic cell (Fig 4K and L). Furthermore, the paired pulse ratio, i.e., the ratio between the postsynaptic responses to two consecutive presynaptic stimulations, was strongly depressed if presynaptic cells expressed Nrg3, but not if they expressed Nrg3ΔEGF (Fig 4M). Thus, presynaptic Nrg3 influences the electrophysiological properties of synapses and modulates both synapse strength and short‐term plasticity.
Nrg3 changes excitatory input into inhibitory neurons in hippocampal slices
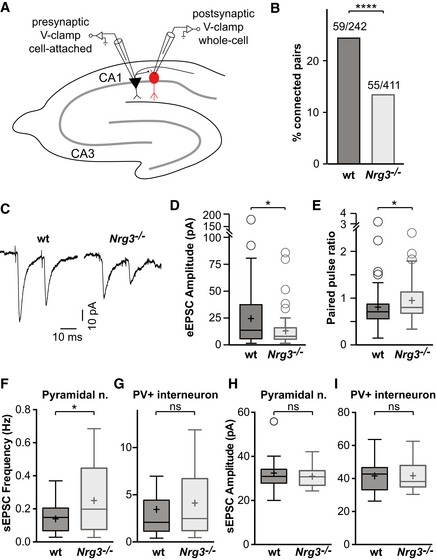
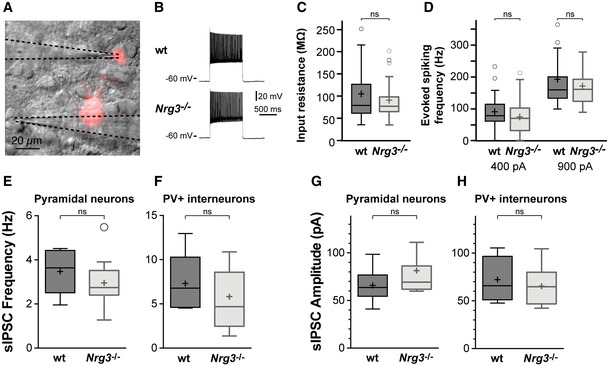
We next directly analyzed the functional connectivity of inhibitory neurons in acute hippocampal slices from Nrg3 −/− and wildtype mice. For this, PV interneurons were genetically labeled with tdTomato (Parv‐cre;Ai14 mice), and connected pyramidal and tdTomato+ neuron pairs were identified in the CA1 region of the hippocampus (schematically shown in Fig 5A). A substantially reduced proportion of pyramidal neurons were connected to PV interneurons in Nrg3 −/− compared to wildtype brain slices (Fig 5B), which is in accordance with the loss of excitatory synapses on PV neurons. Furthermore, in paired recordings the amplitudes of evoked excitatory postsynaptic currents in PV interneurons were markedly reduced in acute Nrg3 −/− slices (Fig 5C and D). Finally, the paired pulse ratio was moderately increased in Nrg3 −/− slices, indicating altered short‐term plasticity at connections between pyramidal neurons and PV interneurons in the hippocampus (Fig 5E). Other parameters of PV interneurons were unchanged, i.e., input resistance (84 ± 4 vs. 116 ± 20 MΩ, P > 0.5) and maximal firing frequency (146 ± 50 Hz vs 162 ± 75 Hz, P > 0.5; Fig EV5). Thus, Nrg3 ablation changes the functional properties of synapses between pyramidal neurons and PV interneurons, while the basic physiology of interneurons remains intact. Notably, reciprocal changes of synaptic properties were observed in slices from Nrg3 mutants and after Nrg3 overexpression in cultured neurons.
Figure 5. Reduced connectivity of ErbB4+ PV interneurons in Nrg3 mutants.
-
ASchematic outline of the connectivity analyses performed in hippocampal slices. PV inhibitory cells (shown in red) were identified using a genetically encoded fluorescence marker (ParvCre;Ai14flox) and recorded from. Neighboring pyramidal neurons were stimulated to probe for connectivity to the recorded interneuron.
-
BQuantification of the proportion of connected vs. unconnected pyramidal neurons in slices from wildtype (wt) and Nrg3 mutants. Data are presented as bars, and the numbers of connected/tested neuron pairs from 20 wildtype and 30 mutant mice are displayed. Chi‐square test was performed to assess statistical significance (****P < 0.0001).
-
CExample traces of evoked excitatory postsynaptic currents (eEPSCs) averaged over 50 sweeps.
-
D, EQuantification of the peak amplitude of the first eEPSC (D) and of the paired pulse ratio (E).
-
F–IsEPSC recordings from CA1 pyramidal (F, H) and PV neurons (G, I) in acute slices. sEPSC frequencies (F, G) and amplitudes (H, I) are shown.
Figure EV5. Electrophysiological properties of PV interneurons in the hippocampal CA1 region of wildtype and Nrg3 mutant mice.
-
AOverlay of an infrared DIC image of the stratum pyramidale of the hippocampal CA1 with tdTomato fluorescence in a slice from a Parv Cre/+ ;Ai14 flox/+ mouse. Dotted lines highlight the stimulation and recording electrodes.
-
BExample traces of the firing pattern of wildtype and Nrg3 −/− PV interneurons. A current of 900 pA was injected for both patterns.
-
CInput resistance of PV interneurons in slices from wildtype and Nrg3 −/− mice. Data from n = 59 (wt) and n = 55 (Nrg3 −/−) neurons from 20 wt and 30 mutant mice are displayed. Mann–Whitney U‐test (unpaired, two‐tailed) was performed to assess statistical significance (ns, not significant).
-
DEvoked firing frequency in PV interneurons from wildtype and Nrg3 −/− mice after current injection of 400 pA (left) and 900 pA (right). Data from n = 59 (400 pA, wt), n = 55 (400 pA, Nrg3 −/−), n = 17 (900 pA, wt), and n = 41 (900 pA, Nrg3 −/−) neurons are displayed. Mann–Whitney U‐test (unpaired, two‐tailed) was performed to assess statistical significance (ns, not significant). No differences were detected in input resistance and evoked firing frequency between wildtype and Nrg3 −/− mice.
-
E–HsIPSC recordings of pyramidal (E, G) and PV neurons (F, H) in acute slices. sIPSC frequencies (E, F) and amplitudes (G, H) are shown. Data from n = 6 wildtype and n = 11 Nrg3 −/− pyramidal neurons and from n = 4 wildtype and n = 7 Nrg3 −/− PV interneurons were analyzed using an unpaired two‐tailed t‐test, and no significant (ns) differences were detected.
We hypothesized that a loss of excitatory synapses onto interneurons leads to a disinhibition of the hippocampal network. Indeed, such disinhibition was previously reported in mice that lack ErbB4 in interneurons (Del Pino et al, 2013). To test whether hippocampal neurons were more active in slices of Nrg3 −/− mice, we recorded spontaneous excitatory postsynaptic currents (sEPSCs; Fig 5F–I). The frequency of sEPSCs, but not their amplitude, was increased in pyramidal cells of Nrg3 −/− mice, indicating that these neurons received more excitatory drive than in controls (Fig 5F and H). sEPSC frequency and amplitude were not significantly changed in PV interneurons, although we observed a trend toward increased frequency (Fig 5G and I). Recordings of spontaneous inhibitory postsynaptic currents (sIPSCs) in pyramidal cells and PV interneurons performed in pharmacological isolation were unchanged, suggesting that the observed disinhibition is based largely upon a reduction in glutamatergic recruitment of interneurons (Fig EV5). Together, these experiments indicate that the loss of excitatory synapses onto interneurons results in increased activity of the hippocampal network in acute slice preparations of Nrg3 mutants, which was further supported by network analysis.
Nrg3 and hippocampal network function
To examine the impact of the altered connectivity on hippocampal network activity in vivo, we compared local field potential recordings in the CA1 region of awake Nrg3 −/− and wildtype mice. Recordings from Nrg3 −/− mice displayed abnormal, fast rising and large amplitude (2–15 mV) network spikes that were never observed in wildtype mice (Fig 6A). When spontaneous forepaw movement was monitored in parallel (Zhao et al, 2016), we observed that the abnormal network spikes were present during both periods of forepaw movement and quiet wakefulness in Nrg3 −/− mice (Fig 6A). Thus, neuronal network synchrony is disrupted in Nrg3 −/− mice. We next analyzed network activity during spontaneous forepaw movement. Fast Fourier transformation analysis of the local field potential recordings (Fig 6B–D) revealed that high‐frequency gamma band activity (30–80 Hz) was increased in Nrg3 −/− mice. This frequency band has been previously associated with inhibitory neuron signaling (Buzsaki & Wang, 2012). Finally, we examined sharp wave ripple oscillations, a hallmark of hippocampal activity in resting mice that is thought to be generated by local synaptic interactions in the hippocampus (Maier et al, 2011). Ripple oscillations were present in Nrg3 −/− mice but occurred only rarely (Fig 6E and F) and had a lower characteristic peak frequency (Fig 6G and H). We suggest that the reduced excitatory connectivity onto PV interneurons in Nrg3 mutant mice leads to altered neuronal synchrony and dysfunctions at the hippocampal network level.
Figure 6. Disruption of hippocampal activity in awake Nrg3 −/− mice.
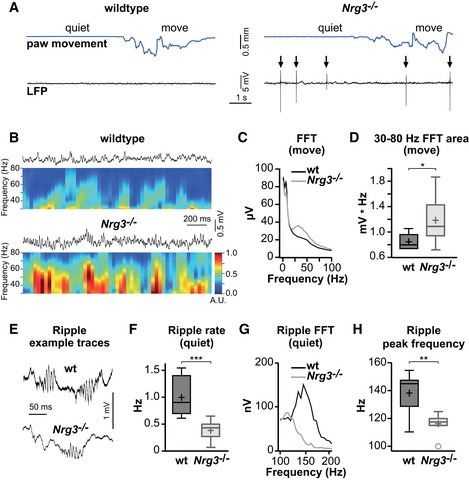
- Examples of local field potential (LFP) recordings (black) from the CA1 region of the hippocampus of a wildtype (wt, left) and a Nrg3 −/− mouse (right). Movement of the forepaw was monitored in parallel and is shown on top (blue). Note the appearance of unusual network spikes in the Nrg3 −/− mouse (arrows).
- Example LFP recordings of wildtype and Nrg3 −/− mice during paw movement and, below, the corresponding normalized spectrogram of the fast Fourier transform (FFT) over time, colored in arbitrary units (A.U.).
- Population mean FFT from 9 wildtype and 8 Nrg3 −/− mice during movement.
- Gamma band activity during paw movement quantified as the area of the FFT from 30 to 80 Hz.
- Example LFP traces showing sharp wave ripple oscillations from resting wildtype and Nrg3 −/− mice.
- Quantification of the occurrence rate of sharp wave ripple oscillations in wildtype and Nrg3 −/− mice.
- Population mean FFT of sharp wave ripples in quiet wildtype and Nrg3 −/− mice.
- Quantification of peak frequencies of ripples in wildtype and Nrg3 −/− mice.
Discussion
Synaptic dysfunction has been implicated in various neuropsychiatric disorders such as schizophrenia and autism (van Spronsen & Hoogenraad, 2010). Genomewide association studies in humans as well as behavioral analyses of mutant mice have implicated Nrg3 in neuropsychiatric disorders (Wang et al, 2008; Chen et al, 2009; Xu et al, 2009; Kao et al, 2010; Morar et al, 2011; Meier et al, 2013). Here, we demonstrate that Nrg3 has a role in excitatory synaptogenesis and synaptic function in hippocampal interneurons and that it is the crucial interaction partner of ErbB4 in PV interneurons. We show that Nrg3 in presynaptic cells recruits ErbB4 to the postsynaptic site, and vice versa that ErbB4 recruits Nrg3 to the presynapse. Nrg3 enhanced synaptogenesis on ErbB4+ hippocampal interneurons in culture. Further, loss of Nrg3 reduced synapse numbers in vivo and in vitro. It should be noted that Nrg3/ErbB4 interaction might enhance formation and/or contribute to the stabilization of excitatory synapses that together determine synapse numbers. In cultured neurons, the role of Nrg3 in synapse formation and recruitment of pre‐ and postsynaptic proteins did depend on ErbB4 but was independent of ErbB4 kinase activity. Furthermore, strong overexpression of Nrg3 induced the formation of long contacts between ErbB4+ dendrites and Nrg3+ axons. These observations indicate that Nrg3/ErbB4 interaction provides adhesive cues. We conclude that Nrg3 is a critical interaction partner of ErbB4 in interneurons and that Nrg3/ErbB4 interactions are necessary for the correct development and function of excitatory synapses onto interneurons.
Synapse specificity
We show here that Nrg3 promotes synaptogenesis as well as pre‐ and postsynaptic specialization of excitatory synapses on hippocampal interneurons. Thus, Nrg3 is a determinant of the inhibitory neuron connectivity. Adhesive interactions between pre‐ and postsynaptic neurons provide a basis for synaptogenesis. Our data indicate that Nrg3/ErbB4 interactions provide such adhesive cues. Neurexins and neuroligins are the best‐studied example of adhesive molecules that are key organizers of synapse formation and function in the brain (Chen et al, 2017; Polepalli et al, 2017). We suggest that the Nrg3/ErbB4 interaction provides an analogous key and lock system. When neurons produced different relative Nrg3 levels in culture (i.e., endogenous Nrg3 levels versus the levels observed in neurons that express endogenous plus lentivirally expressed Nrg3), the neurons that expressed more Nrg3 were better donors of presynaptic terminals and thus had a competitive advantage to form synapses on ErbB4+ interneurons. Thus, different levels of Nrg3 might be presented by different neuron types in vivo and this might modulate their propensity to synapse onto ErbB4+ interneurons.
In addition, we show here that Nrg3 affects the functional properties of excitatory synapses on GABAergic interneurons. Synapses are continuously restructured throughout life, which is important for all aspects of brain function (Fu & Zuo, 2011; Berry & Nedivi, 2017). The fact that Nrg3 changes the paired pulse ratio of excitatory postsynaptic currents at synapses in vivo and in vitro indicates that Nrg3/ErbB4 also plays a role in short‐term plasticity. Pre‐ and postsynaptic recruitment of proteins to the synapse is well known to modulate functional synaptic properties. Our observation that Nrg3 enhances the recruitment of pre‐ and postsynaptic proteins in a dose‐dependent manner indicates that differences in the levels of presented Nrg3 might also regulate the functional properties of synapses.
We also observed clear co‐clustering of Nrg3 and ErbB4 in excitatory synapses on PV interneurons in the hippocampus and cortex, where PV neurons are known to express particularly high ErbB4 levels in vivo. In hippocampal neuron cultures, Nrg3/ErbB4 co‐clustering can be observed in all ErbB4+ neurons, i.e., PV‐positive and PV‐negative interneurons. Nrg3 is also expressed by a subset of inhibitory neurons and we observed low levels of Nrg3 staining in a small proportion of inhibitory synapses in hippocampal neuron cultures. Therefore, an important topic for future investigation will be to investigate whether and how Nrg3 and ErbB4 function in other synapse types, for instance, in inhibitory synapses.
Nrg3, synaptic excitation of inhibitory neurons, and hippocampal network activity
We show here that there is a reduction in excitatory synapses on PV interneurons in Nrg3 mutant mice, which was revealed by histological analyses and paired recordings that directly assessed the monosynaptic connectivity between pyramidal and PV interneurons. Reduced excitatory input onto interneurons was previously shown to cause disinhibition of pyramidal cells (Del Pino et al, 2013). We also observed this in our Nrg3 mutant mice. Additional evidence for a change in the balance of excitation and inhibition comes from our analysis of network activity in live animals that demonstrates remarkable large amplitude network spikes. Furthermore, the power of gamma oscillations in the hippocampus of Nrg3 mutant mice is strongly increased. Gamma oscillations represent a network phenomenon that involves distant inputs to hippocampus, and depend on pyramidal and PV neurons but also on additional local cell types (Veit et al, 2017). Recent studies have linked reduced glutamatergic excitatory drive to interneurons with an increase in gamma activity, for example, in ErbB4 mutant mice (Del Pino et al, 2013) and in mice in which the NR1 NMDA receptor subunit was ablated in PV neurons (Korotkova et al, 2010; Carlen et al, 2012). Thus, reduced excitatory input to PV neurons, but also other changes, might contribute to the increase in gamma activity observed in Nrg3 mutant mice. Mutation of Nrg3 in specific neuronal types as well as manipulations that distinguish cell‐autonomous and paracrine functions of Nrg3 will be required to unambiguously assign physiological changes observed in vivo to particular synapse types. Large amplitude network spikes and increased power of gamma oscillations in the hippocampus have been also reported in ErbB4 mutant mice, which supports the notion that Nrg3 is a functionally critical ErbB4 interaction partner.
Nrg1, Nrg3, and ErbB4 signaling
Nrg3 shares structural similarities with the type III isoform of Nrg1. The precursor forms of type III Nrg1 and Nrg3 pass the membrane twice, are processed by Bace1 in the juxtamembrane region, remain membrane‐associated after Bace1‐proteolysis, and their EGF domains are presented extracellularly (Vullhorst et al, 2017). However, Nrg3 and Nrg1 differ remarkably in their signaling activity. We show here that even at micromolar concentrations, the EGF domain of Nrg3 does not induce efficient tyrosine phosphorylation of ErbB4 in primary neurons, which is in marked contrast to the strong signaling activity of Nrg1. This is in accordance with previous reports that found little Nrg3 signaling activity in cancer cell lines (Hobbs et al, 2002). Further, among the known ligands of ErbB4, Nrg3 was reported to have unusually low affinity (Jones et al, 1999). It is interesting to note that affinities between cell adhesion receptors and their ligands are typically lower than affinities of signaling molecules. The low affinity and poor signaling activity are in accordance with a role of Nrg3 as a presynaptic adhesion molecule.
We show here that presynaptic Nrg3 efficiently clusters ErbB4 in the postsynaptic membrane independently of ErbB4 kinase activity. Clustered ErbB4 might interact with the high‐affinity ligands like Nrg1 or Nrg2. In such competing situations, low‐affinity Nrg3 that possesses poor signaling activity could be displaced in subsets of ErbB4 molecules by a high‐affinity ligand and activate the clustered ErbB4 tyrosine kinase, thereby modulating synaptic function. Recombinant Nrg1 regulates PSD95 stability, fast‐spiking interneuron activity, and the activity of glutamate receptors and voltage‐gated sodium and potassium channels (Bjarnadottir et al, 2007; Abe et al, 2011; Li et al, 2011; Ting et al, 2011; Janssen et al, 2012; Yao et al, 2013), and all of these functions were suggested to be mediated by ErbB4 signaling. Thus, it is possible that Nrg3 and Nrg1/Nrg2 have distinct synaptic functions, which differentially depend on ErbB4 tyrosine kinase signaling.
We observed in vitro that neurons expressing Nrg3 have a pronounced bias to synapse onto ErbB4‐positive rather than onto ErbB4‐negative neurons. Interestingly, this bias does not rely on the classical tyrosine kinase signaling activity of ErbB4, which supports the notion that adhesive Nrg3/ErbB4 interactions might provide the mechanism for the synaptic bias. Furthermore, we also noted that Nrg3‐dependent synaptophysin recruitment was independent of the ErbB4 tyrosine kinase activity. In accordance, overexpressed kinase‐dead ErbB4 was previously noted to modulate synaptic maturation (Krivosheya et al, 2008). While this paper was under revision, an ErbB4‐independent cell‐autonomous role of Nrg3 in glutamatergic transmission was reported (Wang et al, 2018). Future work will be required to distinguish the relative contribution of ErbB4‐dependent and ErbB4‐independent functions of Nrg3. It should however be noted that ErbB4 (Fazzari et al, 2010; Del Pino et al, 2013) and Nrg3 mutant mice display extensive phenotypic similarities, suggesting that ErbB4 depends on Nrg3 to exert its roles in inhibitory connectivity, synaptic function, and hippocampal network activity. We conclude that despite its low affinity, Nrg3 is an important ErbB4 interaction partner for synaptogenesis and synapse function.
Materials and methods
Animals
All experiments were done in accordance with the guidelines and policies of the European Union and the Max Delbrueck Center for Molecular Medicine and the Charité Berlin, Germany, and approved by the Berlin animal ethic committee.
The following mutant mouse strains were used in this study: Nrg3, Nrg3 <tm1.1Cbm> (Loos et al, 2014); heart‐rescued ErbB4, ErbB4 <tm1Grl>;Tg(Myh6‐ERBB4)HT2Gass (Tidcombe et al, 2003); ErbB4flox, ErbB4 <tm1Fej> (Long et al, 2003); Parv‐Cre, Pvalb<tm1(cre)Arbr> (Hippenmeyer et al, 2005); Gad67‐GFP, Gad1 <tm1.1Tama> (Tamamaki et al, 2003); Ai14, Gt(ROSA)26Sor<tm14(CAG‐tdTomato)>Hze (Madisen et al, 2010); and vGAT‐Cre, Tg(Slc32a1‐cre)2.1Hzo (Chao et al, 2010). All strains were maintained on a C57BL/6 background.
Quantitative RT–PCR analyses of Neuregulin transcript levels in brain
Cortex, hippocampus, and striatum were dissected from juvenile mice (P30–60) and quickly frozen on dry ice. RNA was isolated using TRIzol reagent (ThermoFisher, Waltham, USA), and single‐strand cDNA was transcribed. qPCR was performed and analyzed in CFX96 Real‐Time System (Bio‐Rad, Hercules, USA). PCR products were subcloned into pGEM‐T Easy (Promega, Madison, USA), and their identity was confirmed by sequencing. Dilution series of linearized plasmids were used as standard curves to compare numbers of different transcripts. PCR primers for neuregulins, Ube2l3, and the AmpR gene β‐lactamase (bla) are listed in Table 1.
Table 1.
qPCR primer details
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Nrg1β‐I | AAGGCGACCCGAGCCCAGCATTG | TTACGTAGTTTTGGCAACGATCACCAGT |
| Nrg1β‐II | CCACTCAGCCTTCCCGTCCT | CGTAGTTTTGGCAACGATCACCAGT |
| Nrg1β‐III | CAGGAACTCAGCCACAAACAACAGA | CGTAGTTTTGGCAACGATCACCAGT |
| Nrg2α | ATTCGCATCAAGTATGGCAATGGC | GCTTAGGATCTGGCATGTACAATCGC |
| Nrg2β | CAACCGGAGTCGTGATATTCGCAT | CCGGTGTATCCCACAGGACACTTG |
| Nrg3 | ACAAGGACCTGGCGTATTGTCTCA | AGATGGACATGGCTCTTCATCAAGCTC |
| Ube2l3 | GGTCTGTCTGCCAGTCATTAGTGC | GGGGTCATTCACCAGTGCTATGAG |
| bla (AmpR) | CTCAGCGATCTGTCTATTTCGT | TTACTCTAGCTTCCCGGCAAC |
Production of recombinant His6‐Nrg1β and His6‐Nrg3
Sequences of Nrg1β and Nrg3 encoding their EGF domains up to the Bace1‐cleavage site were fused downstream to the signal sequence and His6‐tag encoding sequences in the expression vector pCEP‐Pu/BM40 (Kohfeldt et al, 1997). HEK293‐6E cells were transiently transfected, and secreted His6‐Nrg1β and His6‐Nrg3 were enriched from the medium on TALON Metal Affinity Resin (Clontech, Mountain View, USA). The concentrations of His6‐Nrg1β and His6‐Nrg3 were estimated from the total protein concentration (Pierce BCA Protein Assay kit, ThermoFisher Scientific, Schwerte, Germany) and purity calculated from optical density scans of Coomassie‐stained electrophoresis gels.
Generation of anti‐ErbB4 antisera
Coding sequences for the C‐terminal tail of mouse ErbB4 (aa 975–1292 of NP_034284.1) were inserted behind His6‐tag coding sequences in the expression vector pET14b (Novagen, Madison, USA). Protein was produced in the bacterial strain BL21(DE3)pLysS and purified from inclusion bodies on TALON Metal Affinity Resin under denaturing conditions in the presence of 6M urea. Three rabbits and three guinea‐pigs were used for immunization (Charles River, Sulzfeld, Germany). Antisera were tested for specificity on material from wildtype and ErbB4 mutant animals (Fig EV1).
Neuron culture and lentiviral transduction
Hippocampal neurons were cultured on poly‐L‐lysine‐coated coverslips (PrimeGlass, Forstinning, Germany) over an astrocyte feeder layer in NBA medium (Neurobasal A supplemented with GlutaMAX and 2% B‐27) as described (Kaech & Banker, 2006). Hippocampal neurons were plated at a density of 70,000 cells/well in 12‐well plates containing the coverslips. Ninety minutes after plating, coverslips were transferred upside down to wells containing an astrocyte feeder layer. 5–10,000 IU/well of lentiviruses encoding nuclear red fluorescence protein (nRFP), synaptophysin‐CFP fusion protein, and Nrg3 or Nrg3ΔEGF separated by 2A sequences resulted in the transduction of 10–20% of neurons. Neuron‐specific lentiviral expression was ensured by the use of the Syn1 promoter. Cre‐dependent lentiviruses encoding ErbB4 and ErbB4kd (Prickett et al, 2009) were used at 50–100,000 IU per well. All lentiviruses were constructed and produced by the Viral Core Facility of Charité‐Universitätsmedizin, Berlin, Germany. Neuron cultures transduced with lentiviruses were fixed after 14–18 days with 4% PFA in PBS for 20 min at room temperature. For the quantification of synapses on wildtype and Nrg3 −/− PV interneurons, cultures were fixed after 21–24 days in culture, which improved parvalbumin expression. Secondary dendrites of PV+ interneurons were imaged and analyzed.
For co‐culture experiments, HEK293 cells were transfected with pcDNA3.1(puro)‐Nrg3 or pcDNA3.1(puro)‐Nrg3ΔEGF. A Bace1 expression vector was co‐transfected to improve Nrg3 processing and surface localization. Cells were selected using puromycin and FACS‐sorted for surface expressed Nrg3. Nrg3‐ and Nrg3ΔEGF‐expressing HEK cells were plated on top of neurons at 7 days in culture. Co‐cultures were fixed 3 days later.
For biochemical experiments, ganglionic eminences from brains of embryonic day 13–14 mice were used to increase the number of ErbB4+ interneurons in culture. The cells were plated on poly‐ornithine‐coated tissue culture dishes at a density of 3 million cells per 10‐cm dish in serum‐free NBA medium; 50% of the medium was astrocyte conditioned. Neurons were stimulated with recombinant His6‐Nrg1 and His6‐Nrg3 at day 7 for 8 min, washed briefly with cold PBS, and lysed in RIPA buffer (150 mM NaCl, 1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, Roche Complete protease inhibitor and Sigma phosphatase inhibitor cocktails 2 and 3).
Fractionation of brain extracts, Western blot analysis, and immunohistology
Brain fractionation was performed as described (Fiuza et al, 2013). In brief, cortices from adult mice were homogenized, and the homogenate was cleared by low‐speed centrifugation (1000 g for 10 min). The supernatant (crude lysate, S1) was separated by centrifugation at 10,000 g for 15 min into supernatant S2 containing cytosol and light membranes, and pellet P2 containing crude synaptosomes. S2 was centrifuged at 100,000 g for 20 min, resulting in a supernatant containing the cytosol fraction (S2′) and a pellet containing the light membranes fraction (LM). P2 was resuspended and re‐pelleted at 10,000 g for 15 min, resulting in washed synaptosomes (P2′). Synaptosomes were hypotonically lysed, and the lysates were centrifuged at 25,000 g for 20 min, producing the lysed synaptosomal membrane fraction in pellet P3 and a supernatant S3, which was centrifuged again at 165,000 g for 3 h to pellet synaptic vesicles (SV), and a supernatant of the soluble synaptosomal fraction (S3′). P3 was resuspended and layered on top of a discontinuous sucrose gradient of 0.8, 1.0, and 1.25 M sucrose. The gradient was centrifuged for 3 h at 150,000 g, and protein was recovered from the 1.0/1.25 M sucrose interphase, diluted, and pelleted for 30 min at 150,000 g, resulting in the synaptic plasma membrane fraction (SPM). For Western blot detection of Nrg3 in these fractions, 25 μg of fractions S1, S2, and S2′ and 15 μg of all other fractions except S3′ were used for electrophoresis. S3′ contained very little protein, and therefore, a volume equivalent to fraction P2′ was used.
Western blot analysis was performed using brain and cell lysates prepared in RIPA buffer. Protein concentrations were determined using the Pierce BCA Protein Assay kit. 750 μg of lysate protein was used for ErbB4 immunoprecipitation with 1 μl of guinea‐pig antiserum and 50 μl of Protein A‐Dynabeads (Life Technologies, Darmstadt, Germany).
For immunohistochemistry, brains were fixed with 4% PFA in 0.1 Na‐phosphate buffer for 4–5 h at 4°C. They were washed several times with cold PBS and cryo‐protected using 30% sucrose in PBS before embedding and freezing them in 20% gelatin/20% sucrose in PBS. 50‐μm sections were cut on a sliding microtome and stained as floating sections. For GluA4 immunostainings, sections were pretreated with pepsin as described (Fukaya & Watanabe, 2000). Primary antibodies are listed in Table 2. Secondary antibodies raised in donkey, highly cross‐adsorbed, and conjugated to DL405, Alexa488, DL488, Cy3, Alexa647, and DL647 fluorophores were purchased from Dianova, Hamburg, Germany.
Table 2.
List of antibodies
| Specificity | Host species | Vendor, Cat. # | Usage |
|---|---|---|---|
| Akt | Rabbit | Cell Signaling, #9272 | WB: 1:1,000 |
| ErbB4 | Mouse | Thermo, MA5‐12888 | ICC: 1:300 |
| ErbB4 | Rabbit & guinea‐pig | Described in Material and Methods |
ICC, IHC, WB: 1:10,000 IP: 1 μl antiserum |
| Erk1/2 | Rabbit | Cell Signaling, #9102 | WB: 1:1,000 |
| GFP | Chick | Thermo, A10262 | ICC: 1:1,000 |
| GluA | Mouse | SySy, 182411 | ICC: 1:300 |
| GluA4 | Guinea‐pig | Fontier Sc., GluA4N‐GP‐Af640‐1 | IHC: 1:300 |
| Nrg3 | Goat | Neuromics, GT15021 | ICC, IHC WB: 1:3,000 |
| Nrg3 | Rabbit | Santa Cruz, SC‐67002 | ICC, IHC WB: 1:3,000 |
| Parvalbumin | Rabbit | Swant, PV‐25 | IHC, ICC: 1:3,000 |
| Phospho‐Erk1/2 | Rabbit | Cell Signaling, #4370 | WB: 1:1,000 |
| Phospho‐Akt | Rabbit | Cell Signaling, #9271 | WB: 1:1,000 |
| Phospho‐tyrosine | Mouse | Santa Cruz, SC‐7020 | WB: 1:1,000 |
| Pro‐CCK | Rabbit | Gift from Andrea Varro | IHC: 1:10,000 |
| RFP | Rabbit | Antibodies‐online, ABIN129578 | ICC: 1:2,000 |
| vGAT | Rabbit | SySy, 131002 | ICC: 1:1,000 |
| vGlut1 | Guinea‐pig | SySy, 135304 | ICC: 1:1,000 |
| vGlut2 | Guinea‐pig | SySy, 135404 | ICC: 1:300 |
Imaging and image analysis
All images of immunostained neurons and tissue slices were acquired using a LSM700 (Zeiss, Jena, Germany) confocal microscope with a motorized stage and 20×, 40×, and 100× objectives. To acquire overview images of the hippocampus, the 20× objective and the tile scan and stitch functions of the ZEN2010 software were used. For display of immunostainings, brightness and contrast were adjusted globally using Adobe Photoshop.
To count Nrg3, ErbB4, and GluA4+ puncta in pepsin‐treated brain slices, z‐stack images of dendrites in the CA1 stratum radiatum of the intermediate hippocampus were taken. Images were processed in ImageJ. GluA4+ puncta were counted blind to genotype.
Lentivirus‐transduced neurons were immunostained for Nrg3, ErbB4, GFP, and vGlut1/2 and imaged using the 40× objective, 1× zoom, and 2,048 × 2,048 pixel resolution. Particular care was taken that no channel became saturated. Images containing ErbB4+ neurons were analyzed automatically using a macro for Fiji/ImageJ: vGlut1/2+ puncta were identified using the AdaptiveThreshold plug‐in (using = Mean, from = 15, then = −10) and the Analyze Particles function (size = 30–250, circularity = 0.80–1.00). This results in a list of identified vGlut1/2+ puncta, called Particles in Fiji/ImageJ, that are rounded and have a diameter range of 0.5–1.4 μm. Typically, between 2,000 and 4,000 puncta were identified in a single sample image. To identify which of these puncta were overlapping or in close proximity to ErbB4+ neurons, the ErbB4 channel was auto‐thresholded, expanded by 5px (corresponding to ≈ 0.4 μm), and the Measure Particles function was applied. vGlut1/2+ puncta with a measured MAX of 255 were considered in contact with ErbB4+ neurons. To identify which of the vGlut1/2+ puncta derived from virus‐transduced SypCFP+ neurons, the GFP channel was thresholded using the AdaptiveThreshold plug‐in. The Measure Particles function was applied to this image. vGlut1/2+ puncta with a measured MAX of 255 were considered to be SypCFP+ and to derive from virus‐transduced neurons. Lastly, fluorescence levels in the individual channels of the original image were measured for all vGlut1/2+ puncta using the Measure Particles function. All measurements from a single sample image were imported into Excel (Microsoft Office). vGlut1/2+ puncta were sorted into four groups: (i) C+ group corresponding to puncta with contact to ErbB4 and positive for SypCFP; (ii) C− group corresponding to puncta with contact to ErbB4 and negative for SypCFP; (iii) N+ group corresponding to puncta not in contact with ErbB4 but positive for SypCFP; and (iv) N‐ group corresponding to puncta not in contact with ErbB4 and negative for SypCFP. For the puncta in each group, the fluorescence readings were averaged. For the comparison of Nrg3 −/− and Nrg3 −/−;ErbB4 −/− neurons, interneurons were stained with antibodies against ErbB4 and parvalbumin.
The following values were calculated for every sample image:
Synapse preference (Sp) is a measure for the increased likelihood of a virus‐transduced neuron to synapse on an ErbB4+ neuron and was calculated using the formula, in which [ ] refers to the number of puncta in each group:
The SypCFP enrichment factor (SypCFP‐Ef) is a measure for the increase in presynaptic SypCFP levels in synapses on ErbB4+ neurons as compared to synapses on ErbB4− neurons and was calculated as:
The ErbB4 enrichment factor (ErbB4‐Ef) is a measure for the increase of postsynaptic ErbB4 levels in synapses with boutons from transduced neurons as compared to synapses with untransduced neurons and was calculated as:
Images from at least three independent experiments were analyzed. The use of ratios for quantification facilitated the pooling of numbers from different experiments because they compensated for variability in staining intensity.
Similarly, for counting of vGlut1/2+ and vGAT+ synapses on ErbB4/PV+ neurons in culture, presynaptic boutons were identified and counted using the AdaptiveThreshold macro and the Analyze Particles function in Fiji/ImageJ.
Paired recordings from cultured neurons
Nrg3 −/−;Gad1/Gad67‐GFP/+ were transduced with lentiviruses expressing either nRFP‐Nrg3 or nRFP‐Nrg3ΔEGF. GFP fluorescence marks interneurons of which all with large soma express ErbB4, and nuclear red fluorescence marks virus‐infected neurons. Paired recordings were performed using the whole‐cell patch‐clamp technique in voltage‐clamp mode after 12–15 days of culture. The extracellular solution consisted of (in mM) 115 NaCl, 3 KCl, 10 HEPES, 5 glucose, 2 CaCl2, and 1 MgCl2, pH 7.3, osmolality 260 mOsmol/kg. The patch pipettes were filled with intracellular solution composed of (mM) 3 NaCl, 90 KCl, 5 EGTA, 5 HEPES, 5 glucose, 0.5 CaCl2, and 4 MgCl2, pH 7.3, osmolality 220 mOsmol/kg. The resistance of patch pipettes was 3–5 MΩ. Both cells were clamped at a holding potential (Vh) of −80 mV. Evoked postsynaptic currents in the GFP+ interneuron were induced by two depolarizing pulses from the Vh (−80 mV) to 0 mV (2 ms, interpulse interval 100 ms) in the presynaptic neuron (nRFP+, Gad67GFP−). Recordings were made using an EPC‐9 (HEKA Electronics, Lambrecht/Pfalz, Germany). Signals were sampled at a rate of 10 kHz and analyzed offline using WinTida 5.24 (HEKA Electronics, Lambrecht/Pfalz, Germany). More than three independent experiments were performed and analyzed blind to the lentivirus used.
Electrophysiology on hippocampal slices
Mice aged between P20 and P37 were anesthetized with isoflurane and decapitated. Immediately, the brain was removed and transferred to ice‐cold sucrose in artificial cerebrospinal fluid (ACSF), oxygenated with carbogen (95% O2, 5% CO2). ACSF consisted of (in mM) 87 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 10 d‐glucose, 75 sucrose, 0.5 CaCl2, and 3 MgCl2, 310 mOsmol. Experiments were performed and analyzed blind to genotype.
We performed paired recordings in the hippocampal CA1 region from slices of ParvCre/+;Ai14flox/+ littermate mice that were wildtype or mutant for Nrg3. Slices were transferred to the recording chamber and continually superfused with the extracellular solution containing (in mM) 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 10 glucose, 2 CaCl2, and 1 MgCl2 (saturated with 95% O2/5% CO2). The temperature in the recording chamber was 34 ± 2°C. PV interneurons were targeted based on their tdTomato expression and confirmed by their firing pattern, while pyramidal neurons were targeted based on their shape under DIC illumination (Fig EV5). For whole‐cell patch‐clamp recordings of PV interneurons, the electrode was filled with an intracellular solution containing (in mM) 10 HEPES, 6 KCl, 135 K‐gluconate, 2 MgCl2, 0.2 EGTA, 2 Na2ATP, 0.5 Na2GTP, and 5 Na2‐phosphocreatine, pH 7.2 (adjusted with a solution of KOH), 283 mOsm. For cell‐attached recordings and stimulation of pyramidal neurons, the presynaptic patch pipette was filled with (in mM) 141 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2 CaCl2, and 1 MgCl2, pH 7.3 (adjusted with a solution of NaOH), 275 mOsm. Patch pipettes had resistances of 2–4 MΩ for whole‐cell recording of PV interneurons and 4–8 MΩ for cell‐attached recordings of pyramidal neurons. Multiclamp 700B amplifiers (Axon Instruments, Foster City, USA) were used for current‐ and voltage‐clamp recordings. Data were recorded and filtered at 10 kHz, digitized at 20 kHz, and monitored online using p‐clamp software. Postsynaptic PV interneurons were held in voltage‐clamp configuration at −70 mV, while putative presynaptic pyramidal neurons were held in the voltage‐clamp configuration in a cell‐attached mode technique, which allowed us to quickly screen for connection with still high selectivity (Barbour & Isope, 2000). In voltage clamp, the presynaptic pyramidal neuron was stimulated with two brief positive pulses applied within the electrode, 300 mV to 1V for 0.3 ms to 0.5 ms, separated by 20‐ms delay. EPSC amplitudes of the evoked responses in the postsynaptic cell were measured at the peak of the response. After averaging 50 trials, a pair was defined as connected when the average of the EPSC peak current (4–5 ms after the stimulation) was crossing the threshold of 1.5 standard deviation of the mean baseline. The amplitude of the first EPSC and the paired pulse ratio (PPR), which was determined as EPSC2/EPSC1, were calculated as average of the first 50 responses.
sEPSCs were recorded from pyramidal neurons and PV interneurons in Nrg3 +/+ and Nrg3 −/− littermate mice that carried additional ParvCre and Ai14flox alleles in artificial cerebrospinal fluid. The patch pipette was filled with 10 HEPES, 6 KCl, 135 K‐gluconate, 2 MgCl2, 0.2 EGTA, 2 Na2ATP, 0.5 Na2GTP, and 5 Na2‐phosphocreatine, pH 7.2 (adjusted with a solution of KOH), at an osmolarity of 283 mOsm. sIPSCs were recorded from pyramidal cells and PV interneurons using artificial cerebrospinal fluid containing CNQX (20 μM) and D‐AP5 (50 μM), with a patch pipette containing (in mM) 140 KCl, 10 HEPES, 2 MgCl2, 0.2 EGTA, 2 Na2ATP, 0.5 Na2GTP, 5 Na2‐phosphocreatine, and 10 QX314 (pH 7.2, 290 mOsm).
In vivo electrophysiology
Recordings were previously described (Maier et al, 2011). Briefly, wildtype and Nrg3−/− littermates were implanted with a lightweight metal head holder under isoflurane anesthesia (1.5–2.0% in O2). A recording chamber was built from dental cement (Paladur, Heraeus Kulzer, Helsingborg, Sweden) on the left hemisphere. After 2 days of recovery followed by habituation to head and arm restrain, a craniotomy was performed (−2.5 mm posterior from bregma, +2 mm lateral). Animals (P40–52 on the day of the experiment) were allowed to recover in their home cage for 5 h. The right forepaw was lightly tethered to the recording platform with digits 2–5 overhanging the platform edge. A force‐feedback movement sensor arm (Dual‐Mode Lever Arm Systems 300‐C, Aurora Scientific, Aurora, USA) was positioned in contact with the glabrous skin of the digits to provide an online monitor of digit movement. Local field potential (LFP) recordings were made with low‐resistance (3.5–5.5 MΩ) glass pipettes filled with Ringer's solution and an Axon Multiclamp 700B amplifier. LFPs were filtered between 0.1 Hz and 10 kHz and digitized at 20 kHz (ITC18; HEKA Electronics) under the control of Igor Pro (Wavemetrics, Portland, USA). To identify CA1, the LFP pipette was positioned at 1383 ± 64 μm where clear ripple activity was detected on a TDS2024C oscilloscope screen (Tektronix, Beaverton, USA). Recordings and analyses were conducted blind to genotype. Network spikes in Nrg3 mutants were multiphasic, rapidly rising events that exceeded a threshold of ≥ 0.5 V with variable amplitude (Fig 6A). We used the paw movement signal to identify 2‐second segments of quiet wakefulness (absence of movement, Quiet) and active movement (Move). The spectral content of the LFP was visualized with the MATLAB (MathWorks, Natick, USA) spectrogram function, and the gamma band (30–80 Hz) power was analyzed by measuring the area under the fast Fourier transform (FFT). The isolation and frequency analysis of sharp wave ripples followed the procedures as described (Maier et al, 2011) and used FFT analysis with 5 Hz frequency resolution. The frequency between 100 and 200 Hz with the highest power was defined as ripple peak frequency.
Statistical analyses and display of data
Data are presented as box plots, showing the first quartile, median, and third quartile, with Tukey's whiskers and outliers; means are indicated by “+”. Statistical analyses were performed using Prism 5 (GraphPad, La Jolla, USA), SigmaStat v11.0 (Systat Software, San Jose, USA), MATLAB (MathWorks, Natick, USA), or Igor Pro (Wavemetrics, Portland, USA). Details of statistical analyses are given in the legends.
Data availability
The data that support the findings of this study, anti‐ErbB4 antibodies, and Nrg3 mutant mice are available from the corresponding author on reasonable request.
Code availability
The ImageJ and Excel macro codes are available from the corresponding author on reasonable request.
Author contributions
TM performed and analyzed most neuron culture, histological, and biochemical experiments, and KP and MES helped with these experiments. RJ, SB, and BCV performed and analyzed electrophysiological experiments on neuron culture, hippocampal slices, and awake mice, respectively. CK performed analyses of the data obtained on hippocampal slices and helped with some of these experiments. FR, JRPG, and JFAP supervised electrophysiology experiments. DG performed behavioral experiments. CB supervised the project, and CB and TM conceived the study and wrote the manuscript together. All authors read and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Acknowledgements
We thank Petra Stallerow for help with the animal husbandry, Mandy Terne and Maria Braunschweig for technical support, and Elijah Lowenstein and Pierre‐Louis Ruffault for reading the manuscript (all MDC). We are very thankful for the scientific advice from Craig Garner and Christian Rosenmund. This work was funded by the German Science Foundation, SFB 665 (to C.B), and Thyssen Foundation (to J.F.A.P. and C.B.).
The EMBO Journal (2018) 37: e98858
References
- Abe Y, Namba H, Kato T, Iwakura Y, Nawa H (2011) Neuregulin‐1 signals from the periphery regulate AMPA receptor sensitivity and expression in GABAergic interneurons in developing neocortex. J Neurosci 31: 5699–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Zhang M, Trembak‐Duff I, Unterbarnscheidt T, Radyushkin K, Dibaj P, Martins de Souza D, Boretius S, Brzozka MM, Steffens H, Berning S, Teng Z, Gummert MN, Tantra M, Guest PC, Willig KI, Frahm J, Hell SW, Bahn S, Rossner MJ et al (2014) Dysregulated expression of neuregulin‐1 by cortical pyramidal neurons disrupts synaptic plasticity. Cell Rep 8: 1130–1145 [DOI] [PubMed] [Google Scholar]
- Barbour B, Isope P (2000) Combining loose cell‐attached stimulation and recording. J Neurosci Methods 103: 199–208 [DOI] [PubMed] [Google Scholar]
- Berry KP, Nedivi E (2017) Spine dynamics: are they all the same? Neuron 96: 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnadottir M, Misner DL, Haverfield‐Gross S, Bruun S, Helgason VG, Stefansson H, Sigmundsson A, Firth DR, Nielsen B, Stefansdottir R, Novak TJ, Stefansson K, Gurney ME, Andresson T (2007) Neuregulin1 (NRG1) signaling through Fyn modulates NMDA receptor phosphorylation: differential synaptic function in NRG1 ± knock‐outs compared with wild‐type mice. J Neurosci 27: 4519–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Wang XJ (2012) Mechanisms of gamma oscillations. Annu Rev Neurosci 35: 203–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlen M, Meletis K, Siegle JH, Cardin JA, Futai K, Vierling‐Claassen D, Ruhlmann C, Jones SR, Deisseroth K, Sheng M, Moore CI, Tsai LH (2012) A critical role for NMDA receptors in parvalbumin interneurons for gamma rhythm induction and behavior. Mol Psychiatry 17: 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY (2010) Dysfunction in GABA signalling mediates autism‐like stereotypies and Rett syndrome phenotypes. Nature 468: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Avramopoulos D, Lasseter VK, McGrath JA, Fallin MD, Liang KY, Nestadt G, Feng N, Steel G, Cutting AS, Wolyniec P, Pulver AE, Valle D (2009) Fine mapping on chromosome 10q22‐q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet 84: 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Zhang M, Yin DM, Wen L, Ting A, Wang P, Lu YS, Zhu XH, Li SJ, Wu CY, Wang XM, Lai C, Xiong WC, Mei L, Gao TM (2010) ErbB4 in parvalbumin‐positive interneurons is critical for neuregulin 1 regulation of long‐term potentiation. Proc Natl Acad Sci USA 107: 21818–21823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Jiang M, Zhang B, Gokce O, Sudhof TC (2017) Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron 94: 611–625.e614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pino I, Garcia‐Frigola C, Dehorter N, Brotons‐Mas JR, Alvarez‐Salvado E, Martinez de Lagran M, Ciceri G, Gabaldon MV, Moratal D, Dierssen M, Canals S, Marin O, Rico B (2013) Erbb4 deletion from fast‐spiking interneurons causes schizophrenia‐like phenotypes. Neuron 79: 1152–1168 [DOI] [PubMed] [Google Scholar]
- Escudero I, Johnstone M (2014) Genetics of schizophrenia. Curr Psychiatry Rep 16: 502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari P, Paternain AV, Valiente M, Pla R, Lujan R, Lloyd K, Lerma J, Marin O, Rico B (2010) Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature 464: 1376–1380 [DOI] [PubMed] [Google Scholar]
- Fiuza M, Gonzalez‐Gonzalez I, Perez‐Otano I (2013) GluN3A expression restricts spine maturation via inhibition of GIT1/Rac1 signaling. Proc Natl Acad Sci USA 110: 20807–20812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA et al (2014) De novo mutations in schizophrenia implicate synaptic networks. Nature 506: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Zuo Y (2011) Experience‐dependent structural plasticity in the cortex. Trends Neurosci 34: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Watanabe M (2000) Improved immunohistochemical detection of postsynaptically located PSD‐95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol 426: 572–586 [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H (1995) Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron 15: 193–204 [DOI] [PubMed] [Google Scholar]
- Geng HY, Zhang J, Yang JM, Li Y, Wang N, Ye M, Chen XJ, Lian H, Li XM (2017) Erbb4 deletion from medium spiny neurons of the nucleus accumbens core induces schizophrenia‐like behaviors via elevated GABAA receptor alpha1 subunit expression. J Neurosci 37: 7450–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, Green MF, Gur RE, Gur RC, Hardiman G, Kelsoe JR, Leonard S, Light GA, Nuechterlein KH, Olincy A, Radant AD, Schork NJ, Seidman LJ, Siever LJ, Silverman JM et al (2011) Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 168: 930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Tran T, Murase S, Borrell A, Kirkwood A, Quinlan EM (2016) Neuregulin‐dependent regulation of fast‐spiking interneuron excitability controls the timing of the critical period. J Neurosci 36: 10285–10295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ (2015) Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol Psychiatry 77: 52–58 [DOI] [PubMed] [Google Scholar]
- Hatzimanolis A, McGrath JA, Wang R, Li T, Wong PC, Nestadt G, Wolyniec PS, Valle D, Pulver AE, Avramopoulos D (2013) Multiple variants aggregate in the neuregulin signaling pathway in a subset of schizophrenia patients. Transl Psychiatry 3: e264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S (2005) A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol 3: e159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs SS, Coffing SL, Le AT, Cameron EM, Williams EE, Andrew M, Blommel EN, Hammer RP, Chang H, Riese DJ II (2002) Neuregulin isoforms exhibit distinct patterns of ErbB family receptor activation. Oncogene 21: 8442–8452 [DOI] [PubMed] [Google Scholar]
- Janssen MJ, Leiva‐Salcedo E, Buonanno A (2012) Neuregulin directly decreases voltage‐gated sodium current in hippocampal ErbB4‐expressing interneurons. J Neurosci 32: 13889–13895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JT, Akita RW, Sliwkowski MX (1999) Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett 447: 227–231 [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G (2006) Culturing hippocampal neurons. Nat Protoc 1: 2406–2415 [DOI] [PubMed] [Google Scholar]
- Kao WT, Wang Y, Kleinman JE, Lipska BK, Hyde TM, Weinberger DR, Law AJ (2010) Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci USA 107: 15619–15624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohfeldt E, Maurer P, Vannahme C, Timpl R (1997) Properties of the extracellular calcium binding module of the proteoglycan testican. FEBS Lett 414: 557–561 [DOI] [PubMed] [Google Scholar]
- Korotkova T, Fuchs EC, Ponomarenko A, von Engelhardt J, Monyer H (2010) NMDA receptor ablation on parvalbumin‐positive interneurons impairs hippocampal synchrony, spatial representations, and working memory. Neuron 68: 557–569 [DOI] [PubMed] [Google Scholar]
- Krivosheya D, Tapia L, Levinson JN, Huang K, Kang Y, Hines R, Ting AK, Craig AM, Mei L, Bamji SX, El‐Husseini A (2008) ErbB4‐neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem 283: 32944–32956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Woo RS, Mei L, Malinow R (2007) The neuregulin‐1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron 54: 583–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KX, Lu YM, Xu ZH, Zhang J, Zhu JM, Zhang JM, Cao SX, Chen XJ, Chen Z, Luo JH, Duan S, Li XM (2011) Neuregulin 1 regulates excitability of fast‐spiking neurons through Kv1.1 and acts in epilepsy. Nat Neurosci 15: 267–273 [DOI] [PubMed] [Google Scholar]
- Long W, Wagner KU, Lloyd KC, Binart N, Shillingford JM, Hennighausen L, Jones FE (2003) Impaired differentiation and lactational failure of Erbb4‐deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130: 5257–5268 [DOI] [PubMed] [Google Scholar]
- Loos M, Mueller T, Gouwenberg Y, Wijnands R, van der Loo RJ, Neuro BMPC, Birchmeier C, Smit AB, Spijker S (2014) Neuregulin‐3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry 76: 648–655 [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier N, Tejero‐Cantero A, Dorrn AL, Winterer J, Beed PS, Morris G, Kempter R, Poulet JF, Leibold C, Schmitz D (2011) Coherent phasic excitation during hippocampal ripples. Neuron 72: 137–152 [DOI] [PubMed] [Google Scholar]
- Meier S, Strohmaier J, Breuer R, Mattheisen M, Degenhardt F, Muhleisen TW, Schulze TG, Nothen MM, Cichon S, Rietschel M, Wust S (2013) Neuregulin 3 is associated with attention deficits in schizophrenia and bipolar disorder. Int J Neuropsychopharmacol 16: 549–556 [DOI] [PubMed] [Google Scholar]
- Mitchell RM, Janssen MJ, Karavanova I, Vullhorst D, Furth K, Makusky A, Markey SP, Buonanno A (2013) ErbB4 reduces synaptic GABAA currents independent of its receptor tyrosine kinase activity. Proc Natl Acad Sci USA 110: 19603–19608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morar B, Dragovic M, Waters FA, Chandler D, Kalaydjieva L, Jablensky A (2011) Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol Psychiatry 16: 860–866 [DOI] [PubMed] [Google Scholar]
- Neddens J, Fish KN, Tricoire L, Vullhorst D, Shamir A, Chung W, Lewis DA, McBain CJ, Buonanno A (2011) Conserved interneuron‐specific ErbB4 expression in frontal cortex of rodents, monkeys, and humans: implications for schizophrenia. Biol Psychiatry 70: 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson C, Law AJ (2014) Transient overexposure of neuregulin 3 during early postnatal development impacts selective behaviors in adulthood. PLoS One 9: e104172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polepalli JS, Wu H, Goswami D, Halpern CH, Sudhof TC, Malenka RC (2017) Modulation of excitation on parvalbumin interneurons by neuroligin‐3 regulates the hippocampal network. Nat Neurosci 20: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y (2009) Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet 41: 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, Hoogenraad CC (2010) Synapse pathology in psychiatric and neurologic disease. Curr Neurol Neurosci Rep 10: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ikrar T, Davis MF, Gong N, Zheng X, Luo ZD, Lai C, Mei L, Holmes TC, Gandhi SP, Xu X (2016) Neuregulin‐1/ErbB4 signaling regulates visual cortical plasticity. Neuron 92: 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T (2003) Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67‐GFP knock‐in mouse. J Comp Neurol 467: 60–79 [DOI] [PubMed] [Google Scholar]
- Tidcombe H, Jackson‐Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP (2003) Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci USA 100: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AK, Chen Y, Wen L, Yin DM, Shen C, Tao Y, Liu X, Xiong WC, Mei L (2011) Neuregulin 1 promotes excitatory synapse development and function in GABAergic interneurons. J Neurosci 31: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zoelen EJ, Stortelers C, Lenferink AE, Van de Poll ML (2000) The EGF domain: requirements for binding to receptors of the ErbB family. Vitam Horm 59: 99–131 [DOI] [PubMed] [Google Scholar]
- Veit J, Hakim R, Jadi MP, Sejnowski TJ, Adesnik H (2017) Cortical gamma band synchronization through somatostatin interneurons. Nat Neurosci 20: 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D, Neddens J, Karavanova I, Tricoire L, Petralia RS, McBain CJ, Buonanno A (2009) Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci 29: 12255–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullhorst D, Ahmad T, Karavanova I, Keating C, Buonanno A (2017) Structural Similarities between Neuregulin 1‐3 Isoforms Determine Their Subcellular Distribution and Signaling Mode in Central Neurons. J Neurosci 37: 5232–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Chen JY, Chen ML, Chen CH, Lai IC, Chen TT, Hong CJ, Tsai SJ, Liou YJ (2008) Neuregulin 3 genetic variations and susceptibility to schizophrenia in a Chinese population. Biol Psychiatry 64: 1093–1096 [DOI] [PubMed] [Google Scholar]
- Wang YN, Figueiredo D, Sun XD, Dong ZQ, Chen WB, Cui WP, Liu F, Wang HS, Li HW, Robinson H, Fei EK, Pan BX, Li BM, Xiong WC, Mei L (2018) Controlling of glutamate release by neuregulin3 via inhibiting the assembly of the SNARE complex. Proc Natl Acad Sci USA 115: 2508–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Woodroffe A, Rodriguez‐Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, Gogos JA, Karayiorgou M (2009) Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci USA 106: 16746–16751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JM, Zhang J, Chen XJ, Geng HY, Ye M, Spitzer NC, Luo JH, Duan SM, Li XM (2013) Development of GABA circuitry of fast‐spiking basket interneurons in the medial prefrontal cortex of erbb4‐mutant mice. J Neurosci 33: 19724–19733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JJ, Sun J, Zhao QR, Wang CY, Mei YA (2013) Neuregulin‐1/ErbB4 signaling regulates Kv4.2‐mediated transient outward K+ current through the Akt/mTOR pathway. Am J Physiol Cell Physiol 305: C197–C206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y (2001) The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur J Cancer 37(Suppl 4): S3–S8 [DOI] [PubMed] [Google Scholar]
- Yin DM, Chen YJ, Lu YS, Bean JC, Sathyamurthy A, Shen C, Liu X, Lin TW, Smith CA, Xiong WC, Mei L (2013) Reversal of behavioral deficits and synaptic dysfunction in mice overexpressing neuregulin 1. Neuron 78: 644–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski PJ (1997) Neuregulin‐3 (NRG3): a novel neural tissue‐enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA 94: 9562–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WJ, Kremkow J, Poulet JF (2016) Translaminar cortical membrane potential synchrony in behaving mice. Cell Rep 15: 2387–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Data Availability Statement
The data that support the findings of this study, anti‐ErbB4 antibodies, and Nrg3 mutant mice are available from the corresponding author on reasonable request.


