Abstract
Hepatitis A virus (HAV) and hepatitis E virus (HEV) are significant human pathogens and are responsible for a substantial proportion of cases of severe acute hepatitis worldwide. Genetically, both viruses are heterogeneous and are classified into several genotypes that differ in their geographical distribution and risk group association. There is, however, little evidence that variants of HAV or HEV differ antigenically or in their propensity to cause severe disease. Genetically more divergent but primarily hepatotropic variants of both HAV and HEV have been found in several mammalian species, those of HAV being classified into eight species within the genus Hepatovirus in the virus family Picornaviridae. HEV is classified as a member of the species Orthohepevirus A in the virus family Hepeviridae, a species that additionally contains viruses infecting pigs, rabbits, and a variety of other mammalian species. Other species (Orthohepevirus B–D) infect a wide range of other mammalian species including rodents and bats.
Hepatitis A virus (HAV) and hepatitis E virus (HEV) show moderate genetic diversity, both being classified into several genotypes infecting humans, and several additional species infecting a wide range of other mammalian hosts. The genetic diversity of both viruses has been extensively used as a tool to investigate their molecular epidemiology and transmission. Further studies have sought to determine the existence of differences between genotypes in their clinical presentations and pathogenicity and will be reviewed.
CLASSIFICATION
HAV and HEV are positive-stranded RNA viruses that share a propensity to cause liver disease in humans. However, the two viruses are quite distinct in terms of their replication strategy, virion structure, and taxonomy (see Kenney and Meng 2018; McKnight and Lemon 2018). HAV is classified as a member of the Hepatovirus genus in the large family Picornaviridae (Zell et al. 2017). Although HAV is spread by the fecal–oral route, it is not known whether HAV replicates within tissues of the gastrointestinal tract, and its disease manifestations arise from the spread of virus infection to the liver.
HEV is classified as a member of the Orthohepevirus genus in the family Hepeviridae (Emerson and Purcell 2003; Purdy et al. 2015). Virions of hepeviruses are structurally distinct from picornaviruses (see Kenney and Meng 2018) and their structural proteins are translated from a separate subgenomic RNA expressed during virus replication rather than as part of a single polyprotein as in the picornaviruses. Although there is detectable homology between the RNA-dependent RNA polymerase and helicase genes of picornaviruses and hepeviruses, they have been assigned to different RNA virus supergroups (I and III, respectively) (Koonin 1991), indicating an extremely distant evolutionary relationship between them.
DIVERSITY AND TAXONOMY OF HAV AND RELATED VIRUSES
Hepatovirus Species
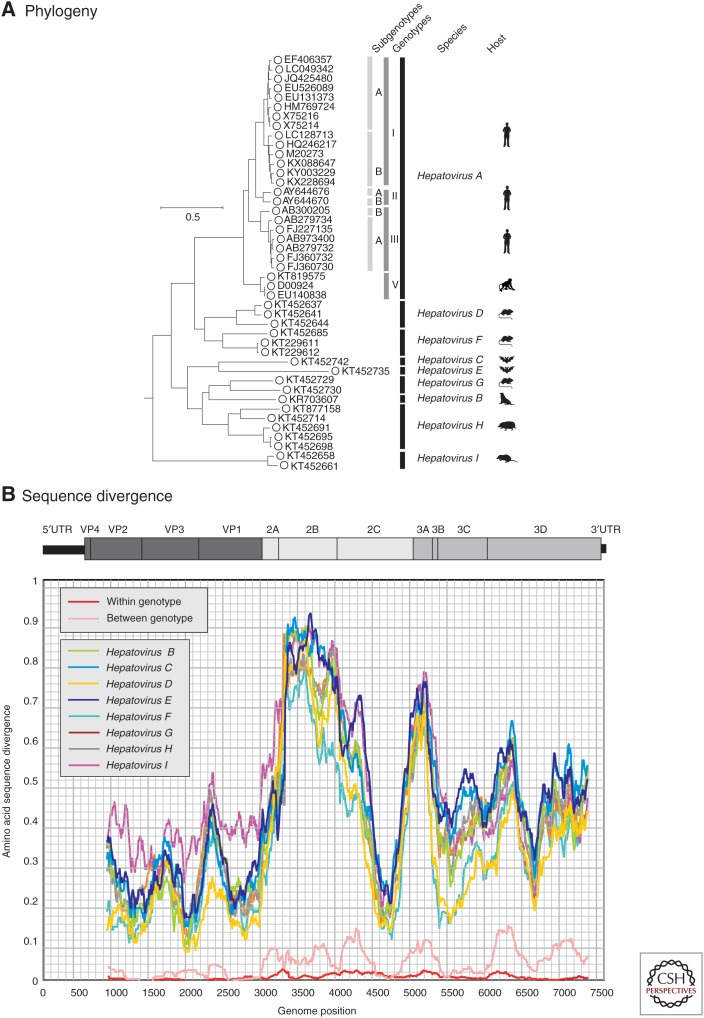
Until very recently, the genus Hepatovirus included a single species, HAV, whose members comprise a group of viruses known to infect humans and various nonhuman primate species. The species was renamed Hepatovirus A in 2014 for consistency with nomenclature conventions adopted elsewhere in the picornavirus family. Since 2015, however, a wide range of further, genetically divergent hepatoviruses have been described in seals (Anthony et al. 2015), shrews, and hedgehogs (order Eulipotyphla), several different rodent (Rodentia) and bat (Chiroptera) host species (Drexler et al. 2015), and, most recently, woodchucks (Yu et al. 2016; and see Sander et al. 2018). Collectively, these viruses are substantially more divergent from each other and from HAV than are existing human and nonhuman primate HAV strains, consistent with them being assigned to several additional species. Using translated sequences from the structural gene region, VP2, variants showing >7% divergence from all other variants might be assigned to as many as 13 new species (Drexler et al. 2015). In 2017, the ICTV accepted the proposal to create new species to classify those hepatovirus variants for which (near-) complete genome sequences were available and that clustered separately on phylogenetic analysis. Accordingly, the genus Hepatovirus now includes nine species (Hepatovirus A–I) based on phylogenetic groupings (Fig. 1). Although distance thresholds have not been specified in the current species definition, nucleotide sequence divergence ranges from 3.8% among members of Hepatovirus A to 29% among members of Hepatovirus G (mean intraspecies divergence of 16%), compared with 30%–42% among members of different species (mean, 38%). The respective amino acid sequence divergence ranges are 0.8%–26% (mean, 11.6%) compared with 31%–48% (mean, 41%).
Figure 1.
Phylogeny and sequence divergence of hepatitis A viruses (HAVs). (A) Phylogenetic analysis of the complete coding sequences of representative variants of HAV (species A) and of all available sequences of other nonhuman hepatoviruses infecting other mammalian species, labeled as in the key. (The coding region spans positions 723–7407 in the HAV-MBB sequences, M20273 [Paul et al. 1987].) The tree was constructed by maximum likelihood using an optimal substitution model (general time reversible and γ distribution). Robustness of branches was indicated by bootstrap resampling supported by ≥70 from 100 replicate samples. The tree was rooted using sequences from the most closely similar genus of picornaviruses (Tremovirus: KT880668, AY275539, KF979338, AJ225173, and AY517471). The host origins of the different species are indicated diagrammatically and correspond to the following species: Hepatovirus B, seals (Phoca vitulina vitulina); Hepatovirus C, bats (Miniopterus cf. manavi); Hepatovirus D, rodents (Microtus arvalis, Myodes glareolus); Hepatovirus E, bats (Lophuromys sikapusi); Hepatovirus F, rodents (Marmota himalayana, Sigmodon mascotensis); Hepatovirus G, bats (Rhinolophus landeri, Coleura afra); Hepatovirus H, hedgehogs, tupias, and bats (Erinaceus europaeus, Tupaia belangeri chinensis, Eidolon helvum); and Hepatovirus I, shrews (Sorex araneus). (B) Scan of amino acid sequence divergence between HAV and hepatovirus species B–I, and comparison with between and among genotype divergence of HAV (red and pink lines). The distances represent mean values for available sequences of each species and were calculated for sequential fragments spanning >90 codons, incrementing by nine codons between data points.
Members of these new hepatovirus species share a number of genome characteristics with HAV, such as low G+C content (37.0%–37.8% within HAV genotypes) compared with 33.7%–39.3% in members of the new species. They also share the massive suppression of CpG dinucleotide frequencies (10%–34% of frequencies expected from their G+C content). Although there is some variability in genome length and positions of cleavage sites for the different structural and nonstructural proteins, members of all hepatovirus species generally show comparable genome organization and features. The latter includes their universal possession of internal ribosomal entry sites (IRES) in the 5′-untranslated region (5′UTR), of which most resemble the type III IRES of HAV in sequence and predicted structure (Anthony et al. 2015; Drexler et al. 2015; Yu et al. 2016). However, viruses belonging to Hepatovirus C and Hepatovirus E (infecting a bat and rodent species, respectively) possessed a type IV IRES (Drexler et al. 2015), structurally resembling those of certain other picornavirus genera (e.g., Tremovirus, Duck hepatitis virus, and Sapelovirus). Hepatovirus species are hepatotropic (Drexler et al. 2015), infect a wide range of hosts, but show limited evidence for host specificity. Although members of the species Hepatovirus A are restricted to humans and primates (macaques and African green monkey), related hosts such as hedgehogs and shrews (order Eulipotyphla) are infected with members of other hepatovirus species (Hepatovirus H and Hepatovirus I), as are rodents and bats. Conversely, members of a single species (Hepatovirus H) can infect three quite different hosts—hedgehogs (Erinaceus europaeus), tupias (Tupaia belangeri chinensis), and bats (Eidolon helvum). Although the distribution of hepatovirus species is consistent with frequent jumps among hosts, it has recently been shown that host range may be restricted by incompatibilities in the interaction of the virus and the host innate immune system (Hirai-Yuki et al. 2016). HAV is unable to replicate in mice because it is unable to evade the cytosolic pattern recognition receptor, MAVS (see Hirai-Yuki et al. 2018), suggesting that major jumps in host species may require substantial adaptive changes in the virus. It is very likely that a much wider range of HAV-like viruses will be found in other mammalian species in the future and animal-associated hepatoviruses may conceivably represent zoonotic sources of infections in humans.
HAV Diversity and Genotypes
Most information on diversity within hepatovirus species is available for Hepatovirus A. HAV shows a modest degree of genetic diversity, with an average of ∼10% nucleotide sequence divergence (2.2% amino acid sequence divergence) among coding region sequences of available (nearly) complete genome sequences. Somewhat higher estimates are obtained once epidemiologically linked HAV variants are excluded (12% and 3.7%, respectively). Using sample dates to estimate substitution rates of HAV over time, it was concluded from Bayesian analysis of complete genome sequences that the common ancestor of all variants belonging to Hepatovirus A might have existed 2000–3000 years ago, while the subset of HAV variants infecting humans were somewhat less diverse and may have a more recent common ancestor (∼1500 years ago) (Kulkarni et al. 2009). Underpinning these historically relatively remote dates was a substitution rate of 1.7–2.0 × 10−4 substitutions per site per year (SSY) for whole-genome sequences (Kulkarni et al. 2009) and 9.8 × 10−4 SSY for VP1 (Moratorio et al. 2007). These rates are lower than estimates for VP1 genes of other picornaviruses, for example, 4–14 × 10−3 SSY for foot and mouth disease virus (genus Aphthovirus) (Cottam et al. 2006) and 13.5 × 10−3 for EV-A71 (genus Enterovirus) (Brown et al. 1999) and indeed most other RNA viruses. The reasons for such a low substitution rate in HAV are not known. Putting these estimates into historical context, whether the dating of the divergence of currently circulating strains of HAV to some time after the collapse of the Roman Empire and before the rise of Islam has any epidemiological or historical correlates remains undetermined.
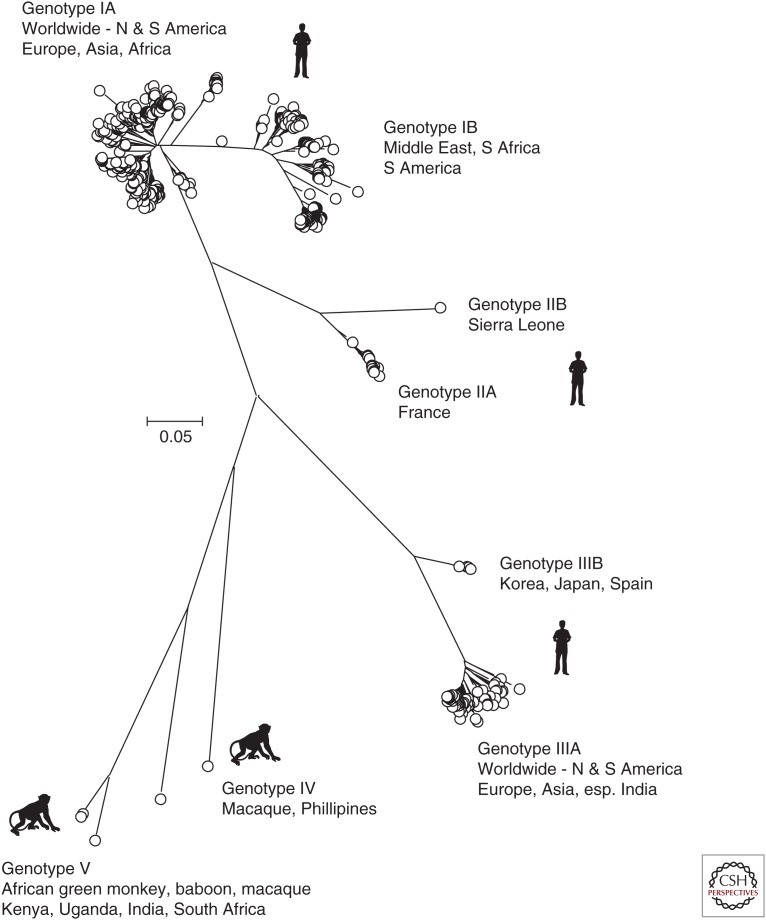
Phylogenetic analysis of all available HAV VP1 sequences on GenBank (June 14, 2017) shows clustering of variants into the previously assigned genotypes (gt)I–III infecting humans and IV and V infecting nonhuman primates (Fig. 2) (Robertson et al. 1992; Costa-Mattioli et al. 2002). The original assignments were based on a 168-nucleotide sequence at the VP1/2A boundary, but the subsequent use of whole VP1 sequences produces a more robust phylogenetic tree and has led to the incorporation of what was originally described as gtVII from Sierra Leone as a member of gtII (Lu et al. 2004). Variants of HAV infecting humans are all assigned to gtI–III and are phylogenetically distinct from nonhuman primate HAV strains, assigned as gtIV, V, and VI (Fig. 2; no VP1 sequence is available for the macaque-derived gtVI). Although sampling of primates is highly limited, current evidence indicates that a wide range of Old World monkey (OWM) species may carry simian strains of HAV. gtIV was isolated from a cynomolgus macaque in the Philippines (Nainan et al. 1991), whereas variants classified as gtV have been found in three different OWM species and locations (African green monkey, Kenya; olive baboon, Uganda; rhesus macaque, India) (Nainan et al. 1991; Arankalle and Ramakrishnan 2009; Bennett et al. 2016). gtVI originated from a cynomolgus macaque originally from Indonesia (Costa-Mattioli et al. 2002).
Figure 2.
Unrooted phylogenetic analysis of the 625 available hepatitis A virus (HAV) sequences in the VP1 region. Positions 2220–3234 numbered as in Fig. 1. The tree was constructed by neighbor joining of maximum composite likelihood distances as implemented in the program, MEGA6 (Tamura et al. 2013). Bootstrap resampling was performed as described in Figure 1. Sequences were selected for analysis based on being >90% complete in the VP1 coding region and lacking internal stop codons.
The phylogenetic clustering of sequences into different genotypes is supported by differences in sequence divergence between and within genotype groupings. Analyzing available (near-) complete genome sequences, pairwise nucleotide distances between gtI–III range from 12.2% to 21.9% (mean, 18.2%), whereas diversity within genotypes ranges from 0.3% to 6.5% (mean, 4.3%). Mean distances among subtypes IA/IB, IIA/IIB, and IIIA/IIIB are intermediate (9.3%, 9.6%, and 11.8%, respectively). These ranges are comparable to those determined previously for different subgenomic regions, such as VP1 (Endo et al. 2007).
Genotype Distributions
Geographically and numerically, infections with gtIA predominate worldwide, with a smaller number of gtIB and IIIA sequences represented in the VP1 dataset (Fig. 2). gtIA predominates in almost all countries where surveillance data is available, and includes North and South America, Europe, and large areas of Asia and Africa (Robertson et al. 1992; Costa-Mattioli et al. 2003; Nainan et al. 2006). gtIB circulates extensively in the Middle East (Normann et al. 2008; Nejati et al. 2012) and South Africa (Taylor 1997), and has been detected among environmental surveillance samples in Brazil. gtII was identified among samples from France and Sierra Leone in the 1990s (Robertson et al. 1992), but has only been rarely reported since. Although less frequently detected than gtI, gtIIIA, and gtIIIB, variants of HAV are also distributed globally, being reported throughout the United States, Europe, and Asia (Robertson et al. 1992; Nainan et al. 2006; Endo et al. 2007). The underlying reasons for the unequal frequencies of infections with different HAV genotypes and their somewhat different geographical distributions are uncertain.
Biological Significance of Hepatovirus Diversity
There is very limited information currently available on the existence of possible biological differences among viruses belonging to different hepatovirus species. They are all hepatotropic, judging from the detection of high levels of viral RNA and replication intermediates in the livers of seals, bats, rodents, hedgehogs, and shrews (Anthony et al. 2015; Drexler et al. 2015; Yu et al. 2016). There is, however, some preliminary evidence for a broader tissue distribution in bats (Drexler et al. 2015). There is substantial amino acid sequence divergence among capsid genes of members of different species (Fig. 1B), greater than 15%–20% over most of P1, although with some variability among genotypes. This degree of divergence is consistent with the general lack of serological cross-reactivity described among rodent, shrew, hedgehog, and bat sera and HAV antigens (Drexler et al. 2015). The strong serological cross-reactivity of a small number of bat sera could potentially be attributable to their previous exposure to HAV or a close relative.
Compared with the extensive divergence among sequences of viruses belonging to different hepatovirus species, variants of HAV (Hepatovirus A) show only minimal amino acid sequence divergence within and between genotypes. It is generally considered that human genotypes of HAV are a single serotype (Lemon et al. 1992); even the relatively divergent HAV gtIB and V showed almost identical neutralization profiles with panels of monoclonal antibodies (Brown et al. 1989; Crevat et al. 1990). Vaccines for HAV may thus be equally effective across the range of genotypes documented to date.
There is similarly little evidence for major differences in clinical outcomes of infections with different HAV genotypes, as might be expected from their high degree of genetic conservation. However, severe fulminant hepatitis associated with a particular genotype IA variant been described in a number of Japanese patients, suggesting the potential existence of strain-associated differences in pathogenicity (Fujiwara et al. 2001; Miura et al. 2017). A cohort study of Korean HAV-infected patients reported that gtIII may be associated with higher levels of biochemical markers of liver damage and lower platelet counts than gtIA, even though the overall disease severity (duration of hospital stay, liver failure, mortality) were comparable (Kim et al. 2013).
DIVERSITY AND TAXONOMY OF HEV AND RELATED VIRUSES
When hepatitis E was first described as an enterically transmitted epidemic in New Delhi in 1955, following fecal contamination of drinking water (Viswanathan 1957), the causative agent was presumed to be a picornavirus. Subsequent studies of virus particles revealed it to be more like a calicivirus, but once a complete genome sequence was obtained (Tam et al. 1991; Huang et al. 1992; Tsarev et al. 1992), the virus was reassigned to its own genus Hepatitis E-like viruses (renamed Hepevirus in 2004, and Orthohepevirus A in 2015), and placed in its own family, Hepeviridae, in 2009.
Like HAV, HEV has a nonenveloped virion and a positive-sense RNA genome, but unlike members of the Picornaviridae, it encodes a single capsid protein and its genome comprises multiple, separately transcribed genes. A short 5′-noncoding region is followed by a long open reading frame (ORF)1 that encodes methyltransferase, Y, papain-like cysteine protease, X, helicase, and RNA-dependent RNA polymerase domains (see Kenney and Meng 2018). ORF1 is immediately followed by ORF2, encoding the capsid protein, while overlapping the 5′-end of ORF2 is another, shorter reading frame, ORF3, which encodes a protein with multiple functions, including one possibly acting as an ion channel. At the 3′-end is a short noncoding region of variable length followed by a poly(A) tail.
Orthohepevirus Species
The current taxonomy of viruses within the genus Orthohepevirus is based on the phylogenetic analysis and distribution of sequence distances for subgenomic regions of the virus genome (Purdy et al. 2015; Smith et al. 2015b). For example, phylogenetic analysis of a conserved region of the capsid protein supports the assignment of viruses into four species, named Orthohepevirus A–D, together with additional unassigned variants (Fig. 3). HEV, in the species Orthohepevirus A, includes isolates from humans, pigs, and deer. At present, the only hepeviruses known to infect humans are those belonging to Orthohepevirus A. Human infection is normally asymptomatic or very mild, with few individuals developing an acute, self-limiting hepatitis. Severe or fulminant hepatitis is a rare outcome except in pregnant women in which mortality can be significant. Infection can become chronic when immune function is suppressed. Infection of pigs is asymptomatic, but HEV-infected dolphins had elevated liver transaminase levels and signs of malaise (Montalvo Villalba et al. 2017).
Figure 3.
Phylogeny and sequence divergence of Orthohepevirus species. (A) Phylogenetic relationships of Orthohepevirus species. A conserved region of the capsid protein (nucleotide positions 5485–6497, numbered according to M73218) was aligned for representative isolates and the amino acid sequence used to generate a maximum likelihood tree based on the LG model with a γ distribution of evolutionary rates among sites as implemented in the program MEGA6 (Tamura et al. 2013). Branches supported in >70% of bootstrap replicates are indicated. Genotypes, virus species, and host species are indicated. (B) Scan of amino acid diversity across the Orthohepevirus genome. The sequences used to produce Figure 3A were analyzed as described in Figure 1B.
Related viruses with a similar genome structure have been isolated from adult chickens with an epidemic disease variously described as big liver and spleen disease or hepatitis-splenomegaly syndrome and associated with weight loss, increased mortality, and decreased egg production (Payne et al. 1999; Haqshenas et al. 2001, 2002; Huang et al. 2002). These avian HEVs are members of the species Orthohepevirus B. More recently, additional viruses have been detected in the rat (Johne et al. 2010a,b), musk shrew, ferret, and mink (all members of the species Orthohepevirus C) and in bats (Orthohepevirus D) (Drexler et al. 2012).
Additional variants with similarities to members of the Orthohepevirus genus, but that do not appear to belong to these four species (Fig. 3), have been described from moose (Lin et al. 2014), fox (Bodewes et al. 2013), kestrel (Reuter et al. 2016a), and little egret (Reuter et al. 2016b), although these have yet to be formally classified into individual species. An incomplete virus genome obtained from a metagenomic study of sewage (hepalivirus) may represent an additional group of viruses (Ng et al. 2012). It seems likely that diversity within the Orthohepevirus genus will be revealed to be even more extensive as studies are extended to a wider range of host species.
A more distantly related virus isolated from salmonids (Hedrick et al. 1991; Batts et al. 2011) has been assigned to the species Piscihepevirus A, the sole member of the genus Piscihepevirus.
HEV Diversity and Genotypes
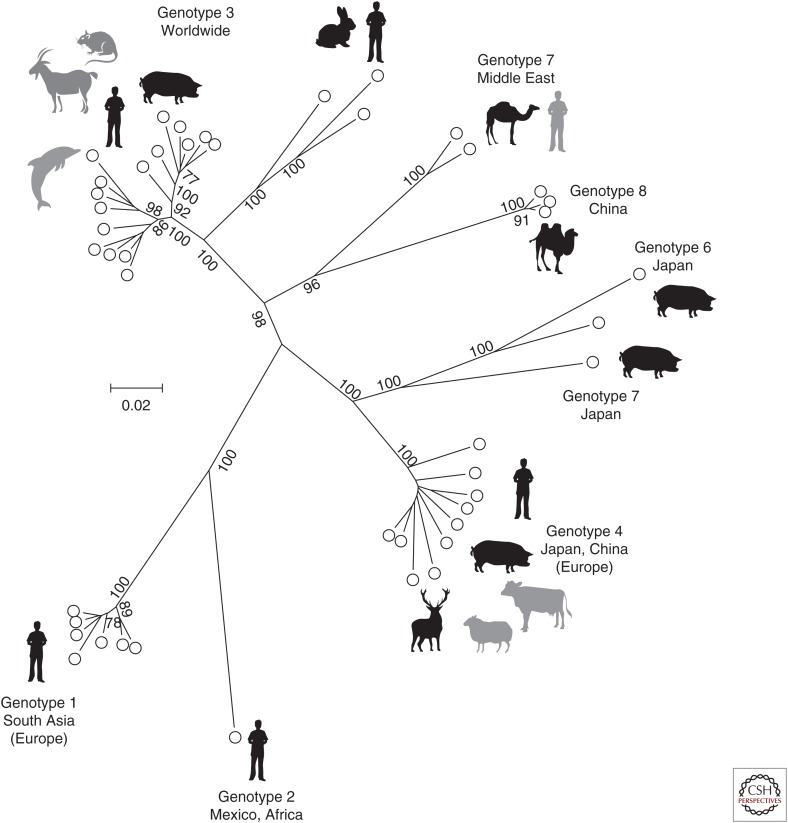
Sequence analyses of Orthohepevirus A isolates from humans around the world and from different host species have revealed the presence of distinct variants that have been classified into several genotypes (gt) and subtypes (Fig. 4). These comprise HEV gt1–4 that have been detected in humans (Lu et al. 2006), gt5 and gt6 that have only been isolated from pigs (Takahashi et al. 2014), gt7 obtained from dromedary camels (Woo et al. 2014) (although there is also one report of human infection) (Lee et al. 2016), and gt8 reported from Bactrian camels (Woo et al. 2016). Numerous subtypes have been described within gt1–4 (Lu et al. 2006), but it is difficult to provide consistent criteria that discriminate viruses that are members of the same subtype from those of different subtypes (Smith et al. 2013a). A particular difficulty applies to variants first isolated from rabbits (Zhao et al. 2009), but now shown to also infect humans (Izopet et al. 2012; Abravanel et al. 2017), and that include characteristic insertions in ORF1. These viruses group phylogenetically with gt3 isolates, but are more divergent from human gt3 variants than these are from each other (Smith et al. 2013a) and are currently considered as belonging to gt3. A similar difficulty applies to gt5 and 6, which could alternatively be considered as subtypes of a single genotype.
Figure 4.
Unrooted phylogenetic analysis of coding sequences of representative members of Orthohepevirus A. Concatenated open reading frame (ORF)1 and ORF2 coding sequences (excluding the hypervariable region) were aligned and used to produce a maximum likelihood tree using a JTT model (with frequencies) and a γ distribution of rate differences among sites, including invariant sites as implemented in the program MEGA6 (Tamura et al. 2013). Branches supported in >70% of bootstrap replicates are indicated. Sequences used were 1a_M73218, 1b_D11092, 1c_X98292, 1d_AY230202, 1e_AY204877, 1f_JF443721, 1_FJ457024, 2a_M74506, 3a_AF082843, 3b_AP003430, 3c_FJ705359, 3e_AB248521, 3_EU360977, 3f_AB369687, 3_EU723513, 3_KJ873911, 3g_AF455784, 3h_JQ013794, 3i_FJ998008, 3j_AY115488, 3_AB290312, 3_AB369689, 3_JQ953664, 3_AB290313, 3ra_FJ906895, 3_JQ013791, 3_KJ013415, 4a_AB197673, 4b_DQ279091, 4c_AB074915, 4d_AJ272108, 4e_AY723745, 4f_AB220974, 4g_AB108537, 4h_GU119961, 4i_DQ450072, 4_AB369688, 5a_AB573435, 6a_AB602441, 6_AB856243, 7a_KJ496143, 7_KJ496144, 8_KX387865, 8_KX387866, and 8_KX387867. Host species are indicated by icons, with less frequent or uncertain species shaded in gray.
A recurring problem in the description of HEV subtypes has been the fact that several were first defined on the basis of subgenomic sequences (Lu et al. 2006). A result of this has been uncertainty about the relation of these variants to viruses sequenced in other genomic regions; there are several examples in the literature in which the same subtype name has been used to identify phylogenetically distinct viruses or where different names have been used for viruses that belong to the same subtype. To address this problem, a list of reference complete genome sequences for each subtype has been proposed by the ICTV Hepeviridae study group and others with an interest in the variation of the Orthohepevirus species (Smith et al. 2016b).
Sequence variability across the HEV genome is relatively constant with few sites of insertion/deletion and a strong bias against nonsynonymous substitution. However, there is a region of unusual nucleotide and amino acid variability within ORF1 between the papain-like cysteine protease and X domains, termed the hypervariable region, which includes an intrinsically disordered region (Purdy et al. 2012) composed of 20%–30% proline and which evolves by both mutation and duplication (Smith et al. 2012). Another notable region is where ORF2 and ORF3 overlap in different reading frames leading to a marked suppression of genetic variability. Finally, attempts to grow HEV in cell culture have led to the identification of HEV strains in which different human coding sequences have been incorporated in frame with ORF1; these can produce viruses that show an enhanced replication ability in vitro compared with that of the virus strains before passage (Nguyen et al. 2012; Shukla et al. 2012).
Diversity within other Orthohepevirus species is much less well characterized. Four closely related genotypes have been described within Orthohepevirus B; two divergent genotypes within Orthohepevirus C are confined to rodents and mustelids, respectively, and it seems likely that multiple genotypes exist within Orthohepevirus D.
Genotype Distributions
The genotype distribution of human isolates of Orthohepevirus A is still incompletely known, with sparse information about virus diversity in many parts of the world, but particularly in Africa where the genotype(s) responsible for epidemic and sporadic infections remains unknown for several countries (Kim et al. 2014). gt1 is associated with epidemics of enterically transmitted hepatitis in the Indian subcontinent and Africa, with cases elsewhere apparently deriving from recent travel to those regions. gt2 has been reported from Mexico and Africa, where it also associated with epidemic spread, but very few isolates are known. To date, gt1 and 2 have only been detected in humans. gt3 is widely distributed in Europe, Africa, North and South America, and in Northeast and Southeast Asia. In these regions, acute (non-travel-associated) infection occurs sporadically with a strong bias toward older men (Lewis et al. 2008), although serological evidence suggests a more general risk of subclinical infection in the whole population, with 25% of blood donors in the Netherlands and Austria having evidence of past infection by the age of 50 (Slot et al. 2013; Fischer et al. 2015). gt4 occurs in Northeast Asia with only occasional reports in Europe (Colson et al. 2016), possibly reflecting the recent introduction of infected pigs from Japan (Nakano et al. 2016). These genotype distributions are mirrored by those in domestic pigs, wild boar, and deer in these regions, the presumption being that human infection results from transmission from these animal reservoirs through consumption of undercooked meat. Such transmission has been shown directly in a few cases (Takahashi et al. 2004; Colson et al. 2010) and suggested by epidemiological studies (Said et al. 2014). This simple picture has been complicated in recent years by the detection of HEV gt3 in goats (Di Martino et al. 2016), rats (Kanai et al. 2012), and, unexpectedly, in bottlenose dolphins (Montalvo Villalba et al. 2017); gt4 has been detected in cows (Hu and Ma 2010; Huang et al. 2016) and sheep (Wu et al. 2015). That patterns of transmission are more complicated than they might seem is suggested by a study of virus diversity among pigs in India. This revealed that all isolates belonged to gt4, whereas human isolates from the same region were all gt1 (Arankalle et al. 2002). In most cases of nonepidemic hepatitis E, the source of human infection is unknown; addressing this should be a priority for future research.
A complication to this description is evidence that the distribution of virus genotypes can change over relatively short time scales. The first isolates of HEV identified in China before the year 2000 were gt1, but this genotype now represents <10% of human infections, with gt4 predominating (Liu et al. 2012). This change may partially reflect improvements in detection methods and interest in HEV.
However, there is also evidence for changes in the distribution of variants within a genotype. For example, in the period 2003–2005, subtype 3f represented 90% of infections in Southwestern France, but by 2012–2014, this had decreased to 65% with the frequency of subtype 3i (previously described as 3c) rising from 5% to 25% (Lhomme et al. 2015). Similarly, a new subtype of gt3 (3c) was introduced to the United Kingdom from about 2008, where within 5 years it became the predominant source of infections (Ijaz et al. 2014). This rapid shift in virus variants may reflect changes in food sourcing and processing, although specific events have yet to be identified. For example, the new variant was not obviously derived from pigs from the United Kingdom; the most closely related, although still distinct, viruses were isolated from a German wild boar and from Italian and French pigs (Grierson et al. 2015).
Biological Significance of HEV Diversity
gt1 and 2 of HEV appear to differ from gt3 and 4 in their ability to be transmitted by the fecal–oral route; only the former genotypes are associated with epidemic spread in the Indian subcontinent and Africa, despite the presence of the other genotypes in these regions. Similarly, although HEV gt3 can be readily detected in human sewage (see references in Smith et al. 2016a), epidemiological studies of HEV infection in Europe do not usually report working with water or contact with untreated sewage as a risk factor. Another difference is that infection with gt1 has been associated with an increased risk of mortality in pregnant women (Jin et al. 2016), but no such association has yet been reported for gt3 or 4, despite the increased awareness of HEV infection as a leading cause of liver disease with a potential impact on pregnancy (Renou et al. 2014; Tabatabai et al. 2014).
There have been occasional reports of HEV genotypes or variants associated with severe outcomes of infection (Takahashi et al. 2009; Jeblaoui et al. 2013). However, a study of virus diversity in hepatitis patients with one or more clinical sign of overt disease, compared with that in blood donors revealed no difference in the distribution of virus subtypes between these two groups for separate studies in France, Germany, the Netherlands, or the United Kingdom (Smith et al. 2015a). An assumption made in this study, that blood donors were asymptomatic, is qualified by the results of a follow-up of HEV-infected U.K. blood donors (Tedder et al. 2016). Although most individuals were healthy at the time of donation, 10% had some signs of illness (fatigue, dark urine, pale stools, or diarrhea), whereas for another 20% these symptoms developed subsequently, with some having elevated transaminase levels. Among these blood donors, there was evidence that those infected with viruses of subtypes 3a, 3b, 3c, 3h, 3i, and 3j (mostly 3c), had a lower virus load and less severe disease than those infected with subtypes 3e, 3f, and 3g. Nevertheless, it does not seem to be the case that virus strains that cause overt hepatitis are distinct from those that result in asymptomatic or relatively mild infections.
A rare outcome of HEV infection is progression to fulminant hepatitis, and there have been suggestions that this might be related to particular genotypes, subtypes or strains of virus (Inoue et al. 2009; Pujhari et al. 2010; Miyashita et al. 2012; Mishra et al. 2013). However, a survey of such reports concluded that fulminant hepatitis could occur after infection with any genotype of HEV, and that once epidemiological linkage of viruses was accounted for, there was no evidence for an association with particular subtypes or with particular mutations or strains (Smith and Simmonds 2015).
There have been few reports of mixed infections with different subtypes of HEV (Moal et al. 2012; Smith et al. 2013a) and it is not yet known whether there is any effect on disease outcome.
CONCLUDING REMARKS
Our understanding of the diversity of HEV has passed through phases of expansion and confusion to what may now be a period of stability in which most virus isolates can be assigned unambiguously to a particular genotype and subtype. In combination with diagnostic methods that are capable of detecting viruses of different genotypes, it should now be possible to address two important questions that relate to HEV variation. First, it should be possible to establish the source of human HEV infections by phylogenetic analysis of virus genomes by relating the subtypes and variants found in human populations with those present in contaminated foods, farmed animals, and environmental sources, with conclusive identification of sources obtained by comparison of hypervariable region sequences. Second, increased awareness of HEV as a major source of acute hepatitis (Kokki et al. 2016) together with more frequent analysis of virus genome sequences may reveal whether particular virus variants are associated with specific disease outcomes. For example, a not-infrequent outcome of HEV infection is the development of neurological symptoms such as Guillain–Barré syndrome, neuralgic amyotrophy, and meningoencephalitis (McLean et al. 2017), but it is unknown whether specific virus variants are responsible for this pathology. Similarly, although the usual treatment for HEV infection is ribavirin, there are currently no reports of the influence of virus genotype on treatment outcomes; this contrasts with hepatitis C virus where the influence of virus genotype on the efficacy of interferon and antiviral treatment has been a fertile topic of research for three decades.
Footnotes
Editors: Stanley M. Lemon and Christopher Walker
Additional Perspectives on Enteric Hepatitis Viruses available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abravanel F, Lhomme S, El Costa H, Schvartz B, Peron JM, Kamar N, Izopet J. 2017. Rabbit hepatitis E virus infections in humans, France. Emerg Infect Dis 23: 1191–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, St Leger JA, Liang E, Hicks AL, Sanchez-Leon MD, Jain K, Lefkowitch JH, Navarrete-Macias I, Knowles N, Goldstein T, et al. 2015. Discovery of a novel hepatovirus (Phopivirus of Seals) related to human hepatitis A virus. mBio 6: e01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle VA, Ramakrishnan J. 2009. Simian hepatitis A virus derived from a captive rhesus monkey in India is similar to the strain isolated from wild African green monkeys in Kenya. J Viral Hepat 16: 214–218. [DOI] [PubMed] [Google Scholar]
- Arankalle VA, Chobe LP, Joshi MV, Chadha MS, Kundu B, Walimbe AM. 2002. Human and swine hepatitis E viruses from Western India belong to different genotypes. J Hepatol 36: 417–425. [DOI] [PubMed] [Google Scholar]
- Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158: 116–123. [DOI] [PubMed] [Google Scholar]
- Bennett AJ, Sibley SD, Lauck M, Weny G, Hyeroba D, Tumukunde A, Friedrich TC, O’Connor DH, Johnson CA, Rothman JM, et al. 2016. Naturally circulating hepatitis A virus in Olive Baboons, Uganda. Emerg Infect Dis 22: 1308–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R, van der Giessen J, Haagmans BL, Osterhaus AD, Smits SL. 2013. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J Virol 87: 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EA, Jansen RW, Lemon SM. 1989. Characterization of a simian hepatitis A virus (HAV): Antigenic and genetic comparison with human HAV. J Virol 63: 4932–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BA, Oberste MS, Alexander JP, Kennett ML, Pallansch MA. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol 73: 9969–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P, Borentain P, Queyriaux B, Kaba M, Moal V, Gallian P, Heyries L, Raoult D, Gerolami R. 2010. Pig liver sausage as a source of hepatitis E virus transmission to humans. J Infect Dis 202: 825–834. [DOI] [PubMed] [Google Scholar]
- Colson P, Brunet P, Lano G, Moal V. 2016. Hepatitis E virus genotype 4 in Southeastern France: Still around. Liver Int 36: 765–767. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Cristina J, Romero H, Perez-Bercof R, Casane D, Colina R, Garcia L, Vega I, Glikman G, Romanowsky V, et al. 2002. Molecular evolution of hepatitis A virus: A new classification based on the complete VP1 protein. J Virol 76: 9516–9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Di Napoli A, Ferre V, Billaudel S, Perez-Bercoff R, Cristina J. 2003. Genetic variability of hepatitis A virus. J Gen Virol 84: 3191–3201. [DOI] [PubMed] [Google Scholar]
- Cottam EM, Haydon DT, Paton DJ, Gloster J, Wilesmith JW, Ferris NP, Hutchings GH, King DP. 2006. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J Virol 80: 11274–11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevat D, Crance JM, Chevrinais AM, Passagot J, Biziagos E, Somme G, Deloince R. 1990. Monoclonal antibodies against an immunodominant and neutralizing epitope on hepatitis A virus antigen. Arch Virol 113: 95–98. [DOI] [PubMed] [Google Scholar]
- Di Martino B, Di Profio F, Melegari I, Sarchese V, Robetto S, Marsilio F, Martella V. 2016. Detection of hepatitis E virus (HEV) in goats. Virus Res 225: 69–72. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose SM, Adu-Sarkodie Y, Oppong SK, et al. 2012. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86: 9134–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Lukashev AN, van den Brand JM, Gmyl AP, Brunink S, Rasche A, Seggewibeta N, Feng H, Leijten LM, et al. 2015. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci 112: 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU, Purcell RH. 2003. Hepatitis E virus. Rev Med Virol 13: 145–154. [DOI] [PubMed] [Google Scholar]
- Endo K, Inoue J, Takahashi M, Mitsui T, Masuko K, Akahane Y, Okamoto H. 2007. Analysis of the full-length genome of a subgenotype IIIB hepatitis A virus isolate: Primers for broadly reactive PCR and genotypic analysis. J Med Virol 79: 8–17. [DOI] [PubMed] [Google Scholar]
- Fischer C, Hofmann M, Danzer M, Hofer K, Kaar J, Gabriel C. 2015. Seroprevalence and incidence of hepatitis E in blood donors in Upper Austria. PLoS ONE 10: e0119576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Yokosuka O, Fukai K, Imazeki F, Saisho H, Omata M. 2001. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J Hepatol 35: 112–119. [DOI] [PubMed] [Google Scholar]
- Grierson S, Heaney J, Cheney T, Morgan D, Wyllie S, Powell L, Smith D, Ijaz S, Steinbach F, Choudhury B, et al. 2015. Prevalence of hepatitis E virus infection in pigs at the time of slaughter, United Kingdom, 2013. Emerg Infect Dis 21: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqshenas G, Shivaprasad HL, Woolcock PR, Read DH, Meng XJ. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J Gen Virol 82: 2449–2462. [DOI] [PubMed] [Google Scholar]
- Haqshenas G, Huang FF, Fenaux M, Guenette DK, Pierson FW, Larsen CT, Shivaprasad HL, Toth TE, Meng XJ. 2002. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J Gen Virol 83: 2201–2209. [DOI] [PubMed] [Google Scholar]
- Hedrick RPY, Yun S, Wingfield WH. 1991. A small RNA virus isolated from salmonid fishes in California, USA. Can J Fish Aquat Sci 44: 99–104. [Google Scholar]
- Hirai-Yuki A, Hensley L, McGivern DR, Gonzalez-Lopez O, Das A, Feng H, Sun L, Wilson JE, Hu F, Feng Z, et al. 2016. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hirai-Yuki A, Whitmire JK, Joyce M, Tyrrell DL, Lemon SM. 2018. Murine models of hepatitis A virus (HAV) infection. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu GD, Ma X. 2010. Detection and sequences analysis of bovine hepatitis E virus RNA in Xinjiang Autonomous Region. Bing Du Xue Bao 26: 27–32. [PubMed] [Google Scholar]
- Huang CC, Nguyen D, Fernandez J, Yun KY, Fry KE, Bradley DW, Tam AW, Reyes GR. 1992. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 191: 550–558. [DOI] [PubMed] [Google Scholar]
- Huang FF, Haqshenas G, Shivaprasad HL, Guenette DK, Woolcock PR, Larsen CT, Pierson FW, Elvinger F, Toth TE, Meng XJ. 2002. Heterogeneity and seroprevalence of a newly identified avian hepatitis E virus from chickens in the United States. J Clin Microbiol 40: 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Li Y, Yu W, Jing S, Wang J, Long F, He Z, Yang C, Bi Y, Cao W, et al. 2016. Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis. Hepatology 64: 350–359. [DOI] [PubMed] [Google Scholar]
- Ijaz S, Said B, Boxall E, Smit E, Morgan D, Tedder RS. 2014. Indigenous hepatitis E in England and wales from 2003 to 2012: Evidence of an emerging novel phylotype of viruses. J Infect Dis 209: 1212–1218. [DOI] [PubMed] [Google Scholar]
- Inoue J, Takahashi M, Mizuo H, Suzuki K, Aikawa T, Shimosegawa T, Okamoto H. 2009. Nucleotide substitutions of hepatitis E virus genomes associated with fulminant hepatitis and disease severity. Tohoku J Exp Med 218: 279–284. [DOI] [PubMed] [Google Scholar]
- Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeblaoui A, Haim-Boukobza S, Marchadier E, Mokhtari C, Roque-Afonso AM. 2013. Genotype 4 hepatitis E virus in France: An autochthonous infection with a more severe presentation. Clin Infect Dis 57: e122–e126. [DOI] [PubMed] [Google Scholar]
- Jin H, Zhao Y, Zhang X, Wang B, Liu P. 2016. Case-fatality risk of pregnant women with acute viral hepatitis type E: A systematic review and meta-analysis. Epidemiol Infect 144: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich RG. 2010a. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16: 1452–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Plenge-Bonig A, Hess M, Ulrich RG, Reetz J, Schielke A. 2010b. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol 91: 750–758. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Miyasaka S, Uyama S, Kawami S, Kato-Mori Y, Tsujikawa M, Yunoki M, Nishiyama S, Ikuta K, Hagiwara K. 2012. Hepatitis E virus in Norway rats (Rattus norvegicus) captured around a pig farm. BMC Res Notes 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kenney SP, Meng X-J. 2018. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yeon JE, Baik SK, Kim YS, Kim HS, Park SH, Lee MS, Sohn JH, Lee JW, Choi SK, et al. 2013. Genotypic shift of the hepatitis A virus and its clinical impact on acute hepatitis A in Korea: A nationwide multicenter study. Scand J Infect Dis 45: 811–818. [DOI] [PubMed] [Google Scholar]
- Kim JH, Nelson KE, Panzner U, Kasture Y, Labrique AB, Wierzba TF. 2014. A systematic review of the epidemiology of hepatitis E virus in Africa. BMC Infect Dis 14: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokki I, Smith D, Simmonds P, Ramalingam S, Wellington L, Willocks L, Johannessen I, Harvala H. 2016. Hepatitis E virus is the leading cause of acute viral hepatitis in Lothian, Scotland. New Microbes New Infect 10: 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72: 2197–2206. [DOI] [PubMed] [Google Scholar]
- Kulkarni MA, Walimbe AM, Cherian S, Arankalle VA. 2009. Full length genomes of genotype IIIA hepatitis A virus strains (1995–2008) from India and estimates of the evolutionary rates and ages. Infect Genet Evol 9: 1287–1294. [DOI] [PubMed] [Google Scholar]
- Lee GH, Tan BH, Teo EC, Lim SG, Dan YY, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, et al. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150: 355–357.e353. [DOI] [PubMed] [Google Scholar]
- Lemon SM, Jansen RW, Brown EA. 1992. Genetic, antigenic and biological differences between strains of hepatitis A virus. Vaccine 10: S40–S44. [DOI] [PubMed] [Google Scholar]
- Lewis HC, Boisson S, Ijaz S, Hewitt K, Ngui SL, Boxall E, Teo CG, Morgan D. 2008. Hepatitis E in England and Wales. Emerg Infect Dis 14: 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhomme S, Abravanel F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, Mansuy JM, Rostaing L, Kamar N, Izopet J. 2015. Temporal evolution of the distribution of hepatitis E virus genotypes in Southwestern France. Infect Genet Evol 35: 50–55. [DOI] [PubMed] [Google Scholar]
- Lin J, Norder H, Uhlhorn H, Belak S, Widen F. 2014. Novel hepatitis E like virus found in Swedish moose. J Gen Virol 95: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li L, Wang L, Bu Q, Fu H, Han J, Zhu Y, Lu F, Zhuang H. 2012. Phylogenetic analysis of 626 hepatitis E virus (HEV) isolates from humans and animals in China (1986–2011) showing genotype diversity and zoonotic transmission. Infect Genet Evol 12: 428–434. [DOI] [PubMed] [Google Scholar]
- Lu L, Ching KZ, de Paula VS, Nakano T, Siegl G, Weitz M, Robertson BH. 2004. Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate). J Gen Virol 85: 2943–2952. [DOI] [PubMed] [Google Scholar]
- Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: Genetic diversity, subtypes and zoonosis. Rev Med Virol 16: 5–36. [DOI] [PubMed] [Google Scholar]
- *.McKnight KL, Lemon SM. 2018. Hepatitis A virus genome organization and replication strategy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean BN, Gulliver J, Dalton HR. 2017. Hepatitis E virus and neurological disorders. Pract Neurol 17: 282–288. [DOI] [PubMed] [Google Scholar]
- Mishra N, Walimbe AM, Arankalle VA. 2013. Hepatitis E virus from India exhibits significant amino acid mutations in fulminant hepatic failure patients. Virus Genes 46: 47–53. [DOI] [PubMed] [Google Scholar]
- Miura Y, Kanda T, Yasui S, Takahashi K, Haga Y, Sasaki R, Nakamura M, Wu S, Nakamoto S, Arai M, et al. 2017. Hepatitis A virus genotype IA-infected patient with marked elevation of aspartate aminotransferase levels. Clin J Gastroenterol 10: 52–56. [DOI] [PubMed] [Google Scholar]
- Miyashita K, Kang JH, Saga A, Takahashi K, Shimamura T, Yasumoto A, Fukushima H, Sogabe S, Konishi K, Uchida T, et al. 2012. Three cases of acute or fulminant hepatitis E caused by ingestion of pork meat and entrails in Hokkaido, Japan: Zoonotic food-borne transmission of hepatitis E virus and public health concerns. Hepatol Res 42: 870–878. [DOI] [PubMed] [Google Scholar]
- Moal V, Gerolami R, Colson P. 2012. First human case of co-infection with two different subtypes of hepatitis E virus. Intervirology 55: 484–487. [DOI] [PubMed] [Google Scholar]
- Montalvo Villalba MC, Cruz Martinez D, Ahmad I, Rodriguez Lay LA, Bello Corredor M, Guevara March C, Martinez LS, Martinez-Campo LS, Jameel S. 2017. Hepatitis E virus in bottlenose dolphins Tursiops truncatus. Dis Aquat Organ 123: 13–18. [DOI] [PubMed] [Google Scholar]
- Moratorio G, Costa-Mattioli M, Piovani R, Romero H, Musto H, Cristina J. 2007. Bayesian coalescent inference of hepatitis A virus populations: Evolutionary rates and patterns. J Gen Virol 88: 3039–3042. [DOI] [PubMed] [Google Scholar]
- Nainan OV, Margolis HS, Robertson BH, Balayan M, Brinton MA. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis). J Gen Virol 72: 1685–1689. [DOI] [PubMed] [Google Scholar]
- Nainan OV, Xia G, Vaughan G, Margolis HS. 2006. Diagnosis of hepatitis a virus infection: A molecular approach. Clin Microbiol Rev 19: 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Takahashi K, Takahashi M, Nishigaki Y, Watanabe N, Ishida S, Fujimoto S, Kato H, Okano H, Takei Y, et al. 2016. Investigating the origin and global dispersal history of hepatitis E virus genotype 4 using phylogeographical analysis. Liver Int 36: 31–41. [DOI] [PubMed] [Google Scholar]
- Nejati A, Makvandi M, Samarbafzadeh A, Neisi N, Moradzadegan H. 2012. Molecular epidemiology of hepatitis A virus in patients in the Ahwaz region of Iran. J Med Virol 84: 582–586. [DOI] [PubMed] [Google Scholar]
- Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol 86: 12161–12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Torian U, Faulk K, Mather K, Engle RE, Thompson E, Bonkovsky HL, Emerson SU. 2012. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J Gen Virol 93: 526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normann A, Badur S, Onel D, Kilic A, Sidal M, Larouze B, Massari V, Muller J, Flehmig B. 2008. Acute hepatitis A virus infection in Turkey. J Med Virol 80: 785–790. [DOI] [PubMed] [Google Scholar]
- Paul AV, Tada H, von der Helm K, Wissel T, Kiehn R, Wimmer E, Deinhardt F. 1987. The entire nucleotide sequence of the genome of human hepatitis A virus (isolate MBB). Virus Res 8: 153–171. [DOI] [PubMed] [Google Scholar]
- Payne CJ, Ellis TM, Plant SL, Gregory AR, Wilcox GE. 1999. Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet Microbiol 68: 119–125. [DOI] [PubMed] [Google Scholar]
- Pujhari SK, Kumar S, Ratho RK, Chawla YK, Chakraborti A. 2010. Phylogenetic analysis and subtyping of acute and fulminant strains of hepatitis E virus isolates of North India with reference to disease severity. Arch Virol 155: 1483–1486. [DOI] [PubMed] [Google Scholar]
- Purdy MA, Lara J, Khudyakov YE. 2012. The hepatitis E virus polyproline region is involved in viral adaptation. PLoS ONE 7: e35974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy MA, Harrison TJ, Jameel S, Meng XJ, Okamoto H, Van der Poel WHM, Smith DB, Consortium IR. 2015. ICTV Taxonomy Profile: Hepeviridae. J Gen Virol 96: 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou C, Gobert V, Locher C, Moumen A, Timbely O, Savary J, Roque-Afonso AM. 2014. Prospective study of hepatitis E virus infection among pregnant women in France. Virol J 11: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Boros A, Matics R, Kapusinszky B, Delwart E, Pankovics P. 2016a. Divergent hepatitis E virus in birds of prey, common kestrel (Falco tinnunculus) and red-footed falcon (F. vespertinus), Hungary. Infect Genet Evol 43: 343–346. [DOI] [PubMed] [Google Scholar]
- Reuter G, Boros A, Matics R, Kapusinszky B, Delwart E, Pankovics P. 2016b. A novel avian-like hepatitis E virus in wild aquatic bird, little egret (Egretta garzetta), in Hungary. Infect Genet Evol 46: 74–77. [DOI] [PubMed] [Google Scholar]
- Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, et al. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol 73: 1365–1377. [DOI] [PubMed] [Google Scholar]
- Said B, Ijaz S, Chand MA, Kafatos G, Tedder R, Morgan D. 2014. Hepatitis E virus in England and Wales: Indigenous infection is associated with the consumption of processed pork products. Epidemiol Infect 142: 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sander A-L, Corman VM, Lukashev AN, Drexler JF. 2018. Evolutionary origins of enteric hepatitis viruses. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. 2012. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86: 5697–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot E, Hogema BM, Riezebos-Brilman A, Kok TM, Molier M, Zaaijer HL. 2013. Silent hepatitis E virus infection in Dutch blood donors, 2011 to 2012. Euro Surveill 18: 20550. [DOI] [PubMed] [Google Scholar]
- Smith DB, Simmonds P. 2015. Hepatitis E virus and fulminant hepatitis—A virus or host-specific pathology? Liver Int 35: 1334–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Vanek J, Ramalingam S, Johannessen I, Templeton K, Simmonds P. 2012. Evolution of the hepatitis E virus hypervariable region. J Gen Virol 93: 2408–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Purdy MA, Simmonds P. 2013a. Genetic variability and the classification of hepatitis E virus. J Virol 87: 4161–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Vanek J, Wellington L, Johannessen I, Ramalingam S, Simmonds P. 2013b. Hepatitis E virus mixed infection in immunocompetent patient. Emerg Infect Dis 19: 468–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Ijaz S, Tedder RS, Hogema B, Zaaijer HL, Izopet J, Bradley-Stewart A, Gunson R, Harvala H, Kokki I, et al. 2015a. Variability and pathogenicity of hepatitis E virus genotype 3 variants. J Gen Virol 96: 3255–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2015b. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 96: 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Paddy JO, Simmonds P. 2016a. The use of human sewage screening for community surveillance of hepatitis E virus in the UK. J Med Virol 88: 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, et al. 2016b. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol 97: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabai J, Wenzel JJ, Soboletzki M, Flux C, Navid MH, Schnitzler P. 2014. First case report of an acute hepatitis E subgenotype 3c infection during pregnancy in Germany. J Clin Virol 61: 170–172. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kitajima N, Abe N, Mishiro S. 2004. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330: 501–505. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okamoto H, Abe N, Kawakami M, Matsuda H, Mochida S, Sakugawa H, Suginoshita Y, Watanabe S, Yamamoto K, et al. 2009. Virulent strain of hepatitis E virus genotype 3, Japan. Emerg Infect Dis 15: 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Nishizawa T, Nagashima S, Jirintai S, Kawakami M, Sonoda Y, Suzuki T, Yamamoto S, Shigemoto K, Ashida K, et al. 2014. Molecular characterization of a novel hepatitis E virus (HEV) strain obtained from a wild boar in Japan that is highly divergent from the previously recognized HEV strains. Virus Res 180: 59–69. [DOI] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 185: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MB. 1997. Molecular epidemiology of South African strains of hepatitis A virus: 1982–1996. J Med Virol 51: 273–279. [DOI] [PubMed] [Google Scholar]
- Tedder RS, Tettmar KI, Brailsford SR, Said B, Ushiro-Lumb I, Kitchen A, Morgan D, Lattimore S, Tossell J, Ijaz S, et al. 2016. Virology, serology, and demography of hepatitis E viremic blood donors in South East England. Transfusion 56: 1529–1536. [DOI] [PubMed] [Google Scholar]
- Tsarev SA, Emerson SU, Reyes GR, Tsareva TS, Legters LJ, Malik IA, Iqbal M, Purcell RH. 1992. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci 89: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan R. 1957. Epidemiology. Indian J Med Res 45: 1–29. [PubMed] [Google Scholar]
- Woo PC, Lau SK, Teng JL, Tsang AK, Joseph M, Wong EY, Tang Y, Sivakumar S, Xie J, Bai R, et al. 2014. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis 20: 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Teng JL, Cao KY, Wernery U, Schountz T, Chiu TH, Tsang AK, Wong PC, Wong EY, et al. 2016. New hepatitis E virus genotype in Bactrian camels, Xinjiang, China, 2013. Emerg Infect Dis 22: 2219–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Si F, Jiang C, Li T, Jin M. 2015. Molecular detection of hepatitis E virus in sheep from southern Xinjiang, China. Virus Genes 50: 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JM, Li LL, Zhang CY, Lu S, Ao YY, Gao HC, Xie ZP, Xie GC, Sun XM, Pang LL, et al. 2016. A novel hepatovirus identified in wild woodchuck Marmota himalayana. Sci Rep 6: 22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, et al. 2017. ICTV virus taxonomy profile: Picornaviridae. J Gen Virol 98: 2421–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, et al. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81: 1371–1379. [DOI] [PubMed] [Google Scholar]