Abstract
Interleukin (IL)-2, IL-4, IL-7, IL-9, IL-15, and IL-21 form a family of cytokines based on their sharing the common cytokine receptor γ chain (γc), which was originally discovered as the third receptor component of the IL-2 receptor, IL-2Rγ. The IL2RG gene is located on the X chromosome and is mutated in humans with X-linked severe combined immunodeficiency (XSCID). The breadth of the defects in XSCID could not be explained solely by defects in IL-2 signaling, and it is now clear that γc is a shared receptor component of the six cytokines noted above, making XSCID a disease of defective cytokine signaling. Janus kinase (JAK)3 associates with γc, and JAK3-deficient SCID phenocopies XSCID, findings that served to stimulate the development of JAK3 inhibitors as immunosuppressants. γc family cytokines collectively control broad aspects of lymphocyte development, growth, differentiation, and survival, and these cytokines are clinically important, related to allergic and autoimmune diseases and cancer as well as immunodeficiency. In this review, we discuss the actions of these cytokines, their critical biological roles and signaling pathways, focusing mainly on JAK/STAT (signal transducers and activators of transcription) signaling, and how this information is now being used in clinical therapeutic efforts.
Interleukin (IL)-2 is the prototype member of the γ chain (γc) family of cytokines. Initially identified as an activity present in the conditioned medium from normal human lymphocytes cultured with phytohemagglutinin (PHA) that could support the long-term in vitro culture of normal human T cells, IL-2 was initially known as T-cell growth factor (TCGF) (Morgan et al. 1976), but then subsequently renamed as IL-2 (Mizel and Farrar 1979). The cloning of complementary DNAs (cDNAs) encoding human (Taniguchi et al. 1983) and mouse (Kashima et al. 1985) IL-2 and the production of recombinant IL-2 allowed investigators to discover additional actions of IL-2 on T, B, and natural killer (NK) cells. Subsequent cloning of the human IL-2 receptor α chain (Leonard et al. 1984; Nikaido et al. 1984; Cosman et al. 1984) revealed that it had a short cytoplasmic tail with only 13 amino acids, making it unlikely to transduce IL-2 signals. This led to the search for additional IL-2 receptor components and the identification of IL-2Rβ (Sharon et al. 1986; Tsudo et al. 1986; Dukovich et al. 1987; Teshigawara et al. 1987) and subsequently IL-2Rγ (Takeshita et al. 1990; Saito et al. 1991) with eventual cloning of cDNAs encoding IL-2Rβ (Hatakeyama et al. 1989) and IL-2Rγ (Takeshita et al. 1992).
IL2RG was localized to the X chromosome at Xq13, the disease locus for X-linked severe combined immunodeficiency ([XSCID], also known as SCIDX1), which then led to the discovery that IL2RG mutations indeed cause XSCID (Noguchi et al. 1993b), a disease characterized by the absence of T and NK cells with nonfunctional B cells (Fischer et al. 2005). This finding was immediately important as it allowed earlier and more precise diagnosis of XSCID and also paved the way for successful gene therapy (Leonard 2001; Hacein-Bey-Abina et al. 2002). However, there were other major scientific implications of the XSCID discovery as well. Given that T- and NK-cell development was normal in IL2-deficient patients (Pahwa et al. 1989; Weinberg and Parkman 1990) and Il2 knockout (KO) mice (Schorle et al. 1991), it was hypothesized that IL-2Rγ was a shared receptor component for other cytokines as well (Noguchi et al. 1993b), leading to the eventual demonstration that IL-2Rγ is indeed a shared receptor component for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 (Rochman et al. 2009). Thus, it was renamed as the common cytokine receptor γc (Noguchi et al. 1993a; Russell et al. 1993), and cytokines using γc are now known as γc family cytokines. The inactivation of signaling by six cytokines in XSCID underscores that it is indeed a disease of defective cytokine signaling (Leonard 1996).
γc family cytokines all share similar three-dimensional structural features and are four α-helix-bundle type I cytokines (Bazan 1990). Although all of these cytokines were initially discovered based on specific actions for either the development or function of T, B, and NK cells (except for IL-21, which was identified based on its binding to an orphan receptor, as will be discussed below), we now know that each cytokine is pleiotropic with broad roles in the development of immune cells or related to immune responses, including some actions beyond the immune systems.
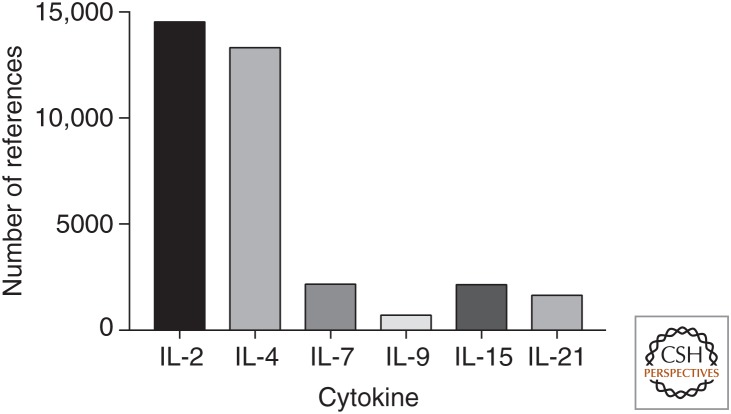
In this review, we discuss the molecular and cellular biology of this family of cytokines, their signaling pathways, actions, and the interplay among them during the development of immune cells and immune responses. We will also discuss the emerging promising approaches for rationally modulating the actions of these cytokines for treating patients with immunodeficiency, autoimmune disorders, infectious diseases, allergic conditions, and malignancies. Needless to say, the number of studies performed and wealth of information on γc family cytokines is enormous, with a huge number of publications in the field (see Fig. 1 for the number of publications just in the period from 2010 to 2017). We have necessarily been selective in our discussion, trying to highlight important early studies as well as some of the exciting progress in this field, and apologize in advance for being unable to cite large numbers of superb studies on these cytokines. However, many other articles in this collection also cover aspects of γc family cytokines, and the reader is directed to those as well.
Figure 1.
PubMed search results of γ chain (γc) family cytokines between 2010 and 2017. The search was performed using EndNote X7.7.1 with the key words IL-2 or interleukin 2, IL-4 or interleukin 4, IL-7 or interleukin 7, IL-9 or interleukin 9, IL-15 or interleukin 15, and IL-21 or interleukin 21, respectively, under Abstract, 2010–2017 under Year, and English under Language.
γc FAMILY CYTOKINES—AN OVERVIEW
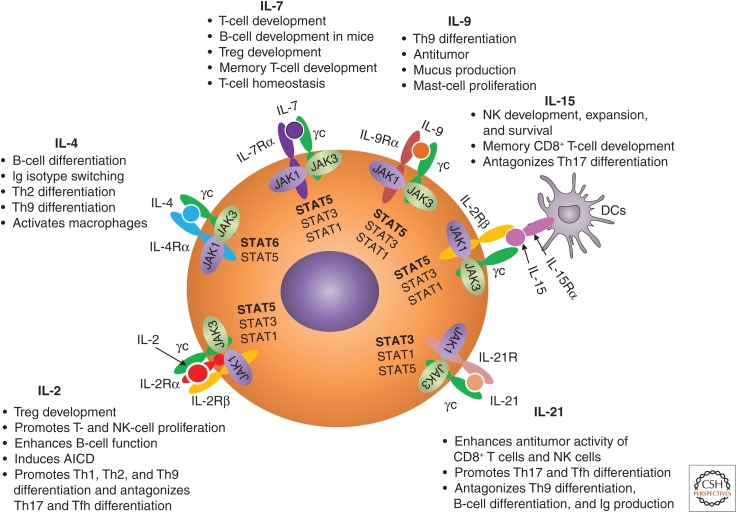
γc family cytokines collectively mediate biological actions on a range of immune cells (Fig. 2). CD4+ T cells are the main producers of IL-2 in response to T-cell receptor (TCR) stimulation, whereas CD8+ T cells, NK cells, and NK T (NKT) cells can also produce IL-2 but at much lower levels (Liao et al. 2013). Although IL-2 was initially discovered as a T-cell growth factor (Morgan et al. 1976), it can also promote the growth and differentiation of B cells that are stimulated by anti-immunoglobulin (Ig)M or CD40 ligand (Armitage et al. 1995) and promote NK-cell proliferation and enhance NK-cell cytotoxicity (Siegel et al. 1987). In addition to its potent proliferative activity for T cells in vitro, IL-2 can induce activation-induced cell death (AICD) (Lenardo 1991) of CD4+ T cells, which is important for the maintenance of peripheral self-tolerance. In addition to its actions as a T-cell growth factor, an essential role of IL-2 in vivo is to promote the development and maintenance of regulatory T (Treg) cells whose suppressive activity is vital to control pathologic inflammatory responses (Malek et al. 2002). In addition, IL-2 promotes the differentiation of T helper (Th)1 (Liao et al. 2011), Th2 (Zhu et al. 2003; Cote-Sierra et al. 2004; Liao et al. 2008), and Th9 (Liao et al. 2014) cells, whereas it suppresses the differentiation of Th17 (Laurence et al. 2007; Liao et al. 2011) and T follicular helper (Tfh) (Ballesteros-Tato et al. 2012; Johnston et al. 2012; Oestreich et al. 2012) cells. As is discussed below, IL-2 has been extensively used as an anticancer agent and for modulating the immune responses.
Figure 2.
Schematic of γ chain (γc) family cytokines and their receptors. Shown are how Janus kinase (JAK)1 and JAK3 associate with each receptor, the signal transducers and activators of transcription (STAT) proteins activated by each cytokine, and the major actions of these cytokines on the development and function immune cells. The STAT proteins predominantly activated by each cytokine are in bold. IL, Interleukin; Th, T helper; Tfh, T follicular helper; Treg, regulatory T; NK, natural killer; Ig, immunoglobulin; DCs, dendritic cells; AICD, activation-induced cell death.
IL-4 was first identified as a B-cell differentiation factor(s) produced by T cells, which induces Ig isotype switch (Howard et al. 1982; Isakson et al. 1982). IL-4 is a signature cytokine for Th2 responses that are essential for the control of extracellular parasites infection and contribute to allergic reactions (Paul 2015). IL-4 also induces the generation of M2 (M-IL-4) macrophages, which are essential for macrophage-mediated control of infection with the protozoan Trypanosoma cruzi (Wirth et al. 1989). In addition, IL-4 promotes the generation of IL-9+ IL-10+ T cells in the presence of transforming growth factor (TGF)-β, which are subsequently designated as Th9 cells and suppress TGF-β-induced Foxp3+ Treg-cell generation (Dardalhon et al. 2008). With its key roles in mediating allergic responses, modulating IL-4 activity is now of considerable therapeutic interest (see below).
In contrast to the production of IL-2 and IL-4 by T cells, IL-7 was discovered as a stromal-cell-derived factor that supported the growth of pre-B cells (Namen et al. 1988; Goodwin et al. 1989). IL-7 signaling also plays an essential, nonredundant role in the development of T cells in humans (Puel et al. 1998; Giliani et al. 2005) and of both B and T cells in mice (Peschon et al. 1994; von Freeden-Jeffry et al. 1995). Unlike other γc family cytokines, the expression level of IL-7 is relatively stable. IL-7 by itself does not promote proliferation of naïve T cells but is essential to ensure sustained expression of antiapoptotic proteins BCL2 and MCL1, and thereby long-term in vivo survival of T cells (Rathmell et al. 2001; Opferman et al. 2003; Sprent and Surh 2011). To maintain long-term memory-T-cell survival, IL-7-induced expression of the glycerol channel aquaporin 9 in antigen-specific memory CD8+ T cells is essential for IL-7-directed glycerol uptake, and this is required for triglyceride synthesis and lipid storage to maintain the longevity of memory CD8+ T cells after viral clearance (Cui et al. 2015). Although IL-7 is not required for the development of conventional NK cells (He and Malek 1996; Puel et al. 1998), it plays a critical role in the homeostasis of thymic NK cells (Vosshenrich et al. 2006). Interestingly, either osteoblast ablation or deletion of Il7 in osteoblasts of adult mice results in significantly lower numbers of common lymphoid progenitors (CLPs) without affecting hematopoietic stem-cell numbers, and administration of IL-7 can restore normal numbers of CLPs (Terashima et al. 2016).
IL-9 was discovered as a T-cell-derived growth factor for certain Th-cell clones in the absence of either antigen or antigen-presenting cells (APCs) (Uyttenhove et al. 1988; Schmitt et al. 1989) and then shown to also be a growth factor for bone marrow mast cells (Hultner et al. 1990). Although IL-9 was initially considered to be a Th2 cytokine (Gessner et al. 1993), it is now recognized as the signature cytokine for Th9 cells, whose differentiation is induced when naïve CD4+ T cells are cultured with IL-2, IL-4, and TGF-β (Schmitt et al. 1994) or when Th2 cells are cultured with TGF-β (Dardalhon et al. 2008; Veldhoen et al. 2008). IL-9 can enhance IL-4-induced IgE and IgG production (Dugas et al. 1993; Petit-Frere et al. 1993), supports innate lymphoid-cell (ILC) survival, and induces cytokine production by these cells (Wilhelm et al. 2011; Turner et al. 2013). IL-9 can also be produced by Th17 cells and can promote the expansion of these cells (Elyaman et al. 2009; Nowak et al. 2009). Expression of IL-9 in NKT cells can be enhanced by IL-2 but not IL-15 (Lauwerys et al. 2000). As discussed below, IL-9 also has anticancer activity.
IL-15 was codiscovered as a T-cell growth factor activity present in the supernatants from a simian kidney epithelial line CV-1/EBNA (Grabstein et al. 1994) and HTLV-1-transformed HUT-102 leukemia cells (Bamford et al. 1994; Burton et al. 1994). Although many different cell types can express IL-15 messenger RNA (mRNA), IL-15 protein is mainly produced by dendritic cells (DCs) and monocytes in response to Toll-like receptor (TLR) activation and binds to receptors on these cells (Waldmann 2006). IL-15 plays critical roles in the development and/or maintenance of memory CD8+ T cells and preferentially can expand central memory phenotype T cells in vivo, with Il15−/− or Il15ra−/− mice having profound loss of memory phenotype CD8+ T cells, intestinal intraepithelial lymphocytes, NKT cells, and NK cells (Lodolce et al. 1998; Kennedy et al. 2000). Although IL-2 and IL-15 both share IL-2Rβ and γc and activate the same Janus kinase (JAK)1/JAK3/signal transducers and activators of transcription (STAT)5 pathway, IL-15 can inhibit IL-2-induced AICD, and the addition of blocking antibodies to IL-15 restores IL-2-induced AICD of these CD4+ T cells (Marks-Konczalik et al. 2000). As discussed below, IL-15 is under active evaluation as an anticancer agent.
IL-21 was identified by expression cloning (Parrish-Novak et al. 2000) as the ligand for a novel “orphan” type I cytokine receptor, originally also denoted as “novel interleukin receptor” (NILR) in addition to the IL-21R (Ozaki et al. 2000; Parrish-Novak et al. 2000). Interestingly, the genes encoding IL-2 and IL-21 are adjacent on human chromosome 4q27 and mouse chromosome 3. IL-21 has significant homology with IL-2, IL-4, and IL-15 (Parrish-Novak et al. 2000), is expressed by Tfh, NKT, Th1, Th2, and Th17 cells, is the key γc family cytokine that contributes to Tfh-cell differentiation in vivo, and it can also promote Th17 differentiation (reviewed in Spolski and Leonard 2014). Although IL-21 alone does not show proliferative activity for either B or T cells, it can potentiate the proliferation of human B cells stimulated by anti-CD40 (Parrish-Novak et al. 2000). IL-21 plays complex roles in the differentiation and function of B cells. For example, IL-21 can promote B-cell proliferation in the presence of anti-CD40 or anti-IgM, whereas it can induce death of resting B cells or B cells stimulated with lipopolysaccharide (LPS) or CpG (Mehta et al. 2003; Jin et al. 2004; Ozaki et al. 2004). IL-21 transgenic mice or wild-type (WT) mice overexpressing IL-21 by hydrodynamic-based gene delivery of IL-21 plasmid DNA show increased numbers of immature B cells, memory B cells, and plasma cells, with elevated serum IgG and IgM levels (Ozaki et al. 2004). IL-21, together with IL-4, plays a vital role in B-cell differentiation and Ig production (Ozaki et al. 2002). In addition to its direct role in the inhibition of Treg differentiation by suppressing Foxp3 expression (Nurieva et al. 2007), IL-21 can diminish Treg homeostasis through inhibiting IL-2 production by T cells (Attridge et al. 2012). IL-21 can also increase the numbers of CD56+ CD16high human NK cells generated from CD34+ human hematopoietic progenitor cells cultured with IL-15 and Flt3 ligand (FLt3L) (Parrish-Novak et al. 2000). IL-21 by itself does not significantly promote the growth of naïve or memory CD8+ T cells, but it can greatly synergize with IL-15 and to a lesser extent with IL-7, but not with IL-2, to enhance the proliferation of these cells (Zeng et al. 2005). These cells develop normally in mice lacking IL-21R, indicating that this cytokine is not required for their development but rather contributes to their expansion. As discussed below, IL-21 is a potent inducer of IL-10 and can drive the production of regulatory B cells and, moreover, IL-21 promotes autoimmune disease and has anticancer activity.
RECEPTORS USED BY γc FAMILY CYTOKINES
Most of the receptors for γc family cytokines (IL-2Rβ, IL-4Rα, IL-7Rα, IL-9Rα, and IL-21R, and γc) are type I cytokine receptors and share an approximately 200-amino-acid-long cytokine-binding homology region (CHR) consisting of two fibronectin type III (FNIII) domains connected by a linker (Bazan 1990). These proteins have four conserved cysteine residues at the N-terminal domain, which can form interstrand disulfide bonds, and a WSXWS (tryptophan-serine-any amino acid-tryptophan-serine) motif near the C terminus (Bazan 1990; Wang et al. 2009). IL-2Rα and IL-15Rα do not contain the CHR module and instead have “Sushi” domains that mediate ligand binding but with very different affinities (Rickert et al. 2005; Lorenzen et al. 2006). Interestingly, in addition to sharing γc, IL-2 and IL-15 receptors additionally share IL-2Rβ (Bamford et al. 1994; Giri et al. 1994).
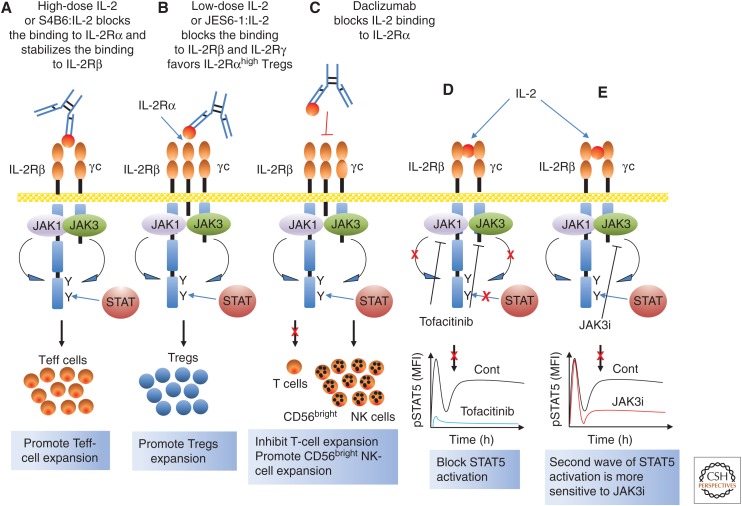
IL-2 receptors are expressed mainly by lymphoid cells. In the absence of stimulation, IL-2Rα is mainly expressed on Treg cells and not expressed on naïve T cells, but it is potently induced on T-cell activation (Leonard et al. 1985; Malek and Castro 2010; Liao et al. 2013). IL-2Rβ and γc are constitutively expressed on some resting T cells, especially NK cells, CD8+ T cells, B cells, macrophages, monocytes, and DCs. Like IL-2Rα, IL-2Rβ can be further induced on stimulation of these cells, either by antigen or by IL-2. Three IL-2 receptor chains, IL-2Rα, IL-2Rβ, and IL-2Rγ, form three different types of IL-2 receptors, binding IL-2 with low affinity (IL-2Rα only, Kd ≈ 10 nm), intermediate affinity (IL-2Rβ + IL-2Rγ, Kd ≈ 1 nm), and high affinity (IL-2Rα + IL-2Rβ + IL-2Rγ, Kd ≈ 10 pm) (Malek and Castro 2010; Liao et al. 2013). Although IL-2Rα and IL-2Rβ together can form pseudo-high-affinity IL-2 receptors (Kd ≈ 100 pm), they cannot transduce IL-2 signals because of the lack of γc (Arima et al. 1992). The intermediate-affinity and high-affinity IL-2 receptors are the functional receptors (Malek and Castro 2010; Liao et al. 2013). Although IL-2 mainly signals via IL-2 receptor chains coexpressed on the same cell (cis signaling), antigen-specific CD25+ mature DCs that lack IL-2Rβ can trans-present IL-2 to CD25− T cells, which can be blocked by declizumab, a humanized antihuman CD25 antibody (Wuest et al. 2011).
Unlike IL-2 receptor chains, IL-4Rα is expressed on both lymphohematopoietic and non-lymphohematopoietic cells, with ∼300 IL-4 binding sites on resting lymphocytes and 10-fold more upon activation (Paul 2015). IL-4 receptors are also present on macrophages and mast cells (Ohara and Paul 1987). There are two types of IL-4Rs: type I IL-4 receptors on lympho-hematopoietic cells are composed of IL-4Rα and γc (Kondo et al. 1993; Russell et al. 1993), whereas in nonhematopoietic cells, type II IL-4 receptors comprise IL-4Rα plus IL-13Rα1, which is also the functional receptor for IL-13 (Aman et al. 1996). In reconstitution experiments using COS-7 cells, IL-4 binds to IL-4Rα with high-affinity (Kd ≈ 266 pm) and in the presence of γc, the binding affinity is further increased (Kd ≈ 79 pm) (Russell et al. 1993). The direct interaction of the IL-4Rα chain with γc is very weak (Kd in the μm range) (LaPorte et al. 2008).
The IL-7 receptor consists of IL-7Ra and γc (Noguchi et al. 1993a; Kondo et al. 1994), and IL-7Ra) is also a component of the receptor for thymic stromal lymphopoietin (TSLP), in that context cooperating with the direct TSLP-binding protein, TSLPR (Pandey et al. 2000; Park et al. 2000). IL-7Rα is expressed on both hematopoietic cells and cells of nonlymphoid origin (Jiang et al. 2005). The expression of IL-7Rα is dynamically regulated during lymphocyte development and in response to TCR or cytokine stimulation, and IL-7Rα regulation is the major mechanism for regulating IL-7 responses (Mazzucchelli and Durum 2007). IL-7Rα can be induced by FLT3 ligand, glucocorticoids, type I interferons (IFNs), and tumor necrosis factor (TNF), whereas it is suppressed by two γc family cytokines, IL-2 and IL-7, as well as by IL-6 (Xue et al. 2002; Mazzucchelli and Durum 2007). Both high-affinity (Kd ≈ 65 pm) and low-affinity (Kd ≈ 100 nm) IL-7 receptors can be detected on peripheral blood lymphocytes, and a reconstitution experiment in COS-7 cells showed that IL-7 binds to IL-7Rα with an intermediate affinity (Kd ≈ 250 pm) that is enhanced to high affinity when γc is present (Noguchi et al. 1993a).
IL-9 receptors contain IL-9Rα chain and γc (Russell et al. 1994; Kimura et al. 1995). A single class of IL-9 receptor with high affinity (Kd ≈ 100 pm) can be detected on T cells, mast cells, and macrophages (Druez et al. 1990). Interestingly, IL-9Rα is also detected in nonhematopoietic cells, including airway and intestinal epithelial cells and smooth muscle cells (Goswami and Kaplan 2011; Kaplan et al. 2015). The human IL9R gene is located on the X chromosome, and at least four IL9R pseudogenes are localized at the pseudoautosomal region of X and Y chromosomes (Kermouni et al. 1995; Vermeesch et al. 1997). A study of the single-nucleotide polymorphisms (SNPs) shows that a specific haplotype of IL-9Rα gene is protective against wheezing in boys but not in girls and provides weak protection against sensitization to inhalant and/or food allergens (Melen et al. 2004).
IL-15 receptors are composed of IL-15Rα, IL-2Rβ, and γc (Bamford et al. 1994; Giri et al. 1994; Grabstein et al. 1994). IL-15Rα is expressed on a wide range of cells, including immune cells (T cells, B cells, macrophages, and stromal-cell lines) and nonimmune cells (keratinocytes and skeletal muscle cells) (Grabstein et al. 1994). IL-15 binds to IL-15Rα with high affinity (Kd ≈ 25 pm), much higher than the affinity of IL-2 for IL-2Rα, whereas IL-15 binds IL-2Rβ and γc with a Kd ≈ 1 nm, similar to the affinity of IL-2 to IL-2Rβ and γc. Like IL-2Rα, IL-15Rα does not transduce IL-15 signals, but IL-15Rα-expressing APCs, such as DCs and monocytes, can bind IL-15 and trans-present the cytokine to lymphocytes that express IL-2Rβ and γc; this trans signaling is the dominant mode for IL-15 action (Dubois et al. 2002).
IL-21 receptors consist of IL-21Rα chain and γc (Asao et al. 2001). IL-21Rα mRNA is expressed on lymphohematopoietic cells and is potently induced on stimulation of human peripheral blood mononuclear cells with PHA (Ozaki et al. 2000) and TCR (Wu et al. 2005). At the amino acid level, IL-21Rα is most similar to IL-2Rβ (29% identity, 46% similarity), and its cytoplasmic domain is more similar to that of IL-9Rα, whereas its overall domain organization (a single cytokine-binding domain followed by a transmembrane domain and a relatively long cytoplasmic domain) is most similar to IL-4Rα (Parrish-Novak et al. 2002). IL-4Rα and IL-21R are encoded by the adjacent genes (Ozaki et al. 2000).
SIGNALING PATHWAYS USED BY γc FAMILY CYTOKINES
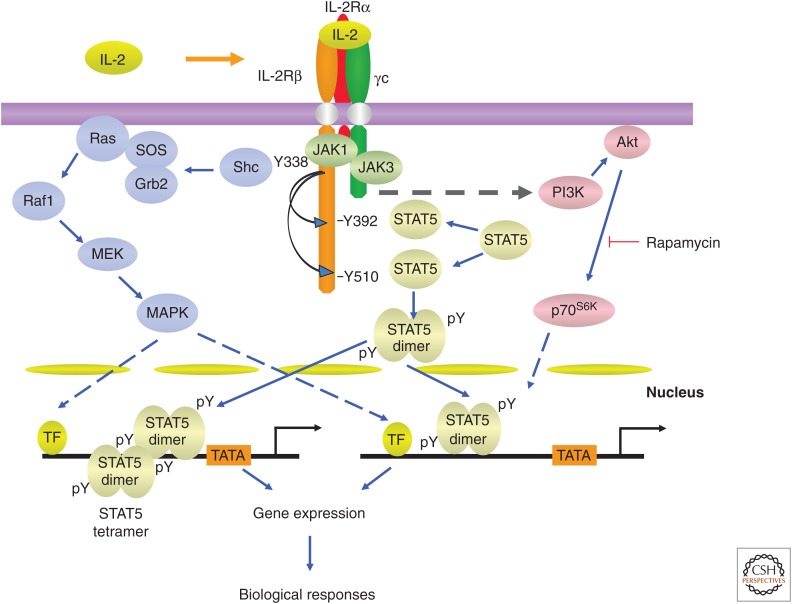
Unlike most growth factor receptors, type I cytokine receptors do not have intrinsic protein kinase activity in their cytoplasmic domains. Thus, the association of nonreceptor JAK1 with the cytokine-specific receptor chains (IL-2Rβ, IL-4Rα, IL-7Rα, IL-9Rα, and IL-21R but not IL-2Rα and IL-15Rα) and JAK3 with γc is essential for transducing the signals induced by γc family cytokines (Fig. 2) (Leonard and O’Shea 1998). Because JAK3 interacts with and is “downstream” from γc (Boussiotis et al. 1994; Russell et al. 1994; Kawahara et al. 1995), it was hypothesized (Russell et al. 1994) and then shown that mutations in JAK3 cause a form of T−B+NK− SCID that phenocopies XSCID (Macchi et al. 1995; Russell et al. 1995). Each γc cytokine induces the juxtaposition of the cytoplasmic domain of its cytokine-specific receptor chain with γc to trigger the activation of JAK1 and JAK3 (Nakamura et al. 1994; Nelson et al. 1994), which then phosphorylates the key tyrosine residues in the cytoplasmic domain of each unique receptor chain (IL-2Rβ, IL-4Rα, IL-7Rα, IL-9Rα, and IL-21Rα), providing the phosphotyrosine docking site(s) for STAT proteins via their SH2 domains; the STAT proteins can then be phosphorylated by JAK kinases (Leonard and O’Shea 1998). Tyrosine-phosphorylated STAT proteins dimerize via bivalent interactions between the C-terminal phosphotyrosine on each monomeric STAT protein and the SH2 domain on the other monomeric STAT protein (Fig. 3). STAT dimers then translocate to the nucleus to bind to IFN-γ-activated site (GAS) motifs, activating the transcription of their target genes (Darnell et al. 1994; Leonard and O’Shea 1998). The docking site(s) for STAT proteins on the cytoplasmic domains of each cytokine receptor determine which STAT protein will be activated by a given cytokine (Lin et al. 1995; Demoulin et al. 1996). For example, IL-2, IL-7, IL-9, and IL-15 predominantly activate STAT5A and STAT5B and to a lesser extent STAT3 and STAT1 (Lin et al. 1995; Demoulin et al. 1999), IL-4 mainly activates STAT6 and to a lesser extent STAT5 (Hou et al. 1994; Quelle et al. 1995; Rolling et al. 1996), and IL-21 activates STAT3 and to a lesser extent STAT1 and STAT5 (Fig. 2) (Zeng et al. 2007; Wan et al. 2013; Wan et al. 2015).
Figure 3.
The interleukin (IL)-2 signaling pathway. Activation of Janus kinase (JAK)/signal transducers and activators of transcription (STAT), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K) pathways by γ chain (γc) family cytokines. As indicated in the text, IL-7 was reported to not activate the MAPK pathway. TF, Transcription factor.
An early study of the IFN-γ enhancer revealed cooperative binding of STAT proteins and the recognition of STAT tetramers. STAT tetramers form based on interactions via their highly conserved N-terminal regions (“N-domains”) in addition to the SH2-phosphotyrosine interactions required for dimerization, thus allowing the dimerization of dimers. Tetramers can bind to less-well-conserved STAT-binding motifs (Vinkemeier et al. 1996; Xu et al. 1996; Soldaini et al. 2000), and the cooperative binding of STAT proteins in this context is caused by N-domain-mediated tetramerization (Vinkemeier et al. 1996; Xu et al. 1996). The crystal structure of the STAT4 N-domain identified the key residues involved in the N-domain-mediated interactions (Vinkemeier et al. 1998; Chen et al. 2003). The nonredundant in vivo function of STAT dimers and tetramers was shown in STAT5 tetramer-deficient mice (Lin et al. 2012). Whereas a complete deletion of both Stat5a and Stat5b results in fetal lethality, at least in part caused by defective erythropoietin-based STAT5 activation and red-cell formation (Socolovsky et al. 1999; Yao et al. 2006), STAT5 tetramer-deficient mice are viable, indicating STAT5 tetramers are dispensable for survival of mice (Lin et al. 2012). Moreover, STAT5 tetramers are not required for the development of B cells, CD4+, CD8+, and CD4+Foxp3+ T cells, but are required for normal numbers of peripheral CD8+ T-cell and NK-cell numbers, in vitro proliferation of CD8+ T cells stimulated by IL-2 or IL-15, and homeostasis of CD4+ and especially CD8+ T cells in lymphopenic hosts (Lin et al. 2012). In addition, STAT5 tetramer-deficient virus antigen-specific CD8+ T cells show decreased expansion in response to lymphocytic choriomeningitis virus (LCMV) or adenovirus 5. Consistent with the potent induction of IL-2Rα mRNA by IL-2, ∼13 major STAT5-binding sites were identified in the Il2ra gene by ChIP-Seq analysis, but only a few of them bind to STAT5 tetramers. Interestingly, IL-2-induced Il2ra mRNA is markedly decreased in STAT5-tetramer-deficient T cells, indicating that STAT5 tetramers are essential for normal Il2ra transcription (Lin et al. 2012). Despite the normal numbers of CD4+Foxp3+ T cells in STAT5 tetramer-deficient mice, IL-2Rα expression in these cells is greatly diminished and STAT5-tetramer-deficient Treg cells show decreased suppressive activity in an adoptive transfer colitis model (Lin et al. 2012). Interestingly, STAT1-tetramer-deficient mice have also been generated and show normal type I IFN responses and antiviral activity, but abolished IFN-γ signaling and antibacterial immunity (Begitt et al. 2014). Thus, there are key roles for STAT1 and STAT5 dimers versus tetramers in mediating cytokine signaling, indicating that targeting the formation of STAT tetramers could be a means of modulating cytokine actions.
JAK/STAT signaling is critical for a broad range of cellular functions, including proliferation, survival, and differentiation, but γc family cytokines, except IL-7, have been shown to activate mitogen-activated protein kinase (MAPK) to promote cell growth, and all γc family cytokines activate phosphatidylinositol 3-kinase (PI3K) to support cell survival (see Fig. 3 for IL-2 activation of these signaling pathways). IL-4 (Wang et al. 1993) and IL-9 (Yin et al. 1995) additionally activate insulin receptor substrates (IRSs) in a manner that is dependent on JAK1 (Yin et al. 1995; Nelms et al. 1999).
To further characterize IL-2 signaling, mass-spectrometry-based quantitative phosphoproteomics were used to identify IL-2 signaling networks in preactivated mouse CD8+ T cells that were cultured in IL-12 to keep the cells viable (Ross et al. 2016). An IL-2-JAK-dependent network appeared to account for most the phosphoproteins identified, including those crucial for the cellular fitness and functions, such as transcription factors, regulators of chromatin structure, mRNA translation machinery, GTPases, vesicle trafficking, and the actin and microtubule cytoskeleton. About 10% of the phosphoproteins identified in these cells were JAK-independent (tofacitinib resistant), comprising those involved in the generation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and the AKT pathway.
Although tofacitinib was developed as a JAK3 inhibitor (Changelian et al. 2003), it also inhibits JAK1 and JAK2 as well as JAK3. A recent study using a more specific JAK3 inhibitor, JAK3i (3000-fold more selective for JAK3 than for JAK1, JAK2, and TYK2), unexpectedly revealed biphasic catalytic activity of JAK3 in STAT5 activation in CD4+ T cells stimulated by IL-2 (Smith et al. 2016). Interestingly, the second wave is required for the expression of cyclins and cell-cycle progression to the S phase, and more sensitive than the first wave of STAT5 activation to JAK3i (Smith et al. 2016). Additional work is needed to clarify the role of the first wave in IL-2 action and whether the second wave is directly or indirectly activated by IL-2. Recently, an additional JAK3-specific inhibitor has been reported (Telliez et al. 2016).
GENE EXPRESSION MEDIATED BY γc FAMILY CYTOKINES
STAT-mediated activation by γc family cytokines is vital for the development and function of immune cells. As discussed below, STAT6 activated by IL-4 and STAT3 activated by IL-21 play essential roles in B-cell differentiation and Ig switching (Linehan et al. 1998; Diehl et al. 2012); STAT5 plays nonredundant roles in T- and B-cell development (IL-7) (Yao et al. 2006), in NK-cell development, expansion, and survival (IL-15) (Yao et al. 2006), and in Treg development (IL-2) (Burchill et al. 2007). STAT5 is also critically important for T-cell homeostasis (IL-7) in peripheral and for T-cell expansion in response to antigen stimulation (IL-2 and IL-15). Furthermore, STAT activation by this family of cytokines is also critical for CD4+ Th-cell differentiation. For example, STAT3 activated by IL-21 is vital for Tfh differentiation (Nurieva et al. 2008), STAT5 activated by IL-2 is required for normal Th1, Th2, and Th9 differentiation (Zhu et al. 2003; Liao et al. 2008, 2011, 2014), and STAT6 activated by IL-4 is essential for Th2 and Th9 differentiation (Kuperman et al. 1998; Goswami et al. 2012).
In addition to promoting CD4+ T-cell differentiation, IL-21-activated STAT3 and IL-2-activated STAT5 show opposing actions on Th9, Th17, and Tfh differentiation (Laurence et al. 2007; Yang et al. 2011; Johnston et al. 2012; Liao et al. 2014). These opposing actions of IL-2 and IL-21 during Th17 differentiation can be achieved either by IL-2-mediated inhibition of Il6ra and Il6st expression or enhanced T-bet (Liao et al. 2011), which should augment binding of RUNX1, thereby diminishing association of RUNX1 with RORγt (Lazarevic et al. 2011), or by competition between STAT3 versus STAT5 for binding to GAS (IFN-γ-activated sequence, TTCN3GAA) motifs on Il17a and Il17f (Yang et al. 2011). Because the GAS motifs recognized by STAT proteins are fairly conserved, except for STAT6 using the motif TTCN4GAA (with N4 instead of N3) (Leonard and O’Shea 1998), binding of STAT3 versus STAT5 is likely influenced by the local concentration of each STAT protein. For Th9 differentiation, BCL6 expression suppressed by IL-2 and enhanced by IL-21 are at least, in part, attributed to their opposing actions (Liao et al. 2014). For Tfh differentiation, induction of BLIMP1 via IL-2-activated STAT5 and maintaining BCL6 levels via IL-21-activated STAT3 are responsible for the opposing actions of these cytokines (Johnston et al. 2012; Oestreich et al. 2012). Thus, STAT proteins activated by γc family cytokines are essential to restrain the inflammation triggered by immune responses and to protect the host from harm by controlling the balance between Treg and Th cells.
In addition to the activation of different STAT proteins by different γc family cytokines, different STAT proteins activated by the same cytokine can also result in distinct biological actions. For example, IL-21 activates both STAT3 and STAT1 in mouse CD4+ T cells, with STAT1 promoting expression of the Ifng and Tbx21 genes and STAT3 inhibiting their expression. Correspondingly, expression of IFNG and TBX21 genes are higher in patients with loss-of-function mutation in STAT3 or with gain-of-function mutation in STAT1 (Wan et al. 2015). Moreover, STAT proteins are involved in the assembly of complexes of transcription factors at enhancers, as first shown for IL-21-induced expression of the Prdm1 gene, which requires STAT3 and IRF4 to act in concert and cooperatively binding during Tfh differentiation (Kwon et al. 2009). Such assembly is essential to initiate and maintain the transcription program during Th differentiation (Ciofani et al. 2012; Vahedi et al. 2012). Moreover, during Th17 differentiation, IL-21 signals via AP1-IRF composite elements (AICEs) (Li et al. 2012) and binding of large complexes of factors (e.g., STAT3, Jun family proteins, basic leucine zipper ATF-like transcription factor (BATF), RORγt, c-Maf, p300, and CTCF) to the regulatory regions of key Th17 genes is required for the initiation and maintenance of the Th17 transcription program (Ciofani et al. 2012).
γc FAMILY CYTOKINES AND T-CELL DEVELOPMENT
The essential roles of γc family cytokines in the development, homeostasis, and survival of T cells are shown by the findings in humans with gene mutations in either the human IL2RG gene encoding γc or the JAK3 gene, as well as in corresponding KO mice. IL-7 is the most critical cytokine for the development of T cells in both humans and mice, with normal T-cell development in Il2 (Schorle et al. 1991), Il4 (Kuhn et al. 1991), Il9 (Townsend et al. 2000), Il15 (Kennedy et al. 2000), and Il21 (Nurieva et al. 2007) KO mice.
IL-7Rα is mainly expressed on CLPs, pre–T cells, and thymic and peripheral CD4+ and CD8+ single-positive T cells. The key roles for IL-7 in T-cell development are to maintain the expression of the antiapoptotic protein, BCL2, for double-negative (DN) thymocyte survival, and of Rag1 for the rearrangement of T-cell antigen receptors in thymic CD4−CD8− DN T cells (Mazzucchelli and Durum 2007). The absence of γδ T cells in Il7ra−/− mice and the observation that they cannot be restored by expression of a Bcl2 transgene show the indispensable role of IL-7 in RAG-mediated rearrangement of TCR-γ (Maki et al. 1996; Mazzucchelli and Durum 2007). Regulation of the accessibility of the Tcrg locus to recruit STAT5 proteins activated by IL-7 and other cofactors is attributed to the essential role of IL-7 in T-cell development (Ye et al. 2001). In addition, there is a profound decrease in αβ T-cell numbers in Il7−/− or Il7ra−/− mice (Peschon et al. 1994; von Freeden-Jeffry et al. 1995), but normal T-cell numbers could be restored by a Bcl2 transgene or partially restored by the deletion of the gene encoding proapoptotic BAX in Il7ra−/− mice (Akashi et al. 1997; Khaled et al. 2002). The absence of γδ T cells but presence of some αβ T cells results from the lack of initiation of recombination of the Tcrg locus, with normal recombination of Vβ2 but greatly reduced recombination of Vβ14 (Schlissel et al. 2000).
Interestingly, the defect in T-cell development appears somewhat less severe in Il7−/− mice than in Il7ra−/− mice (Peschon et al. 1994; von Freeden-Jeffry et al. 1995), and TSLP also uses IL-7Rα as a receptor component, even though it is not a γc family cytokine. Despite normal development of T cells in Crlf2−/− (TSLPR KO) mice, TSLP contributes to T-cell development to some degree as Il2rg−/yCrlf2−/− mice display a greater defect in T-cell development than observed in Il2rg−/y mice (Al-Shami et al. 2004).
The levels of expression of cytokine receptors is well-correlated with mature T-cell homeostasis. For example, naïve T-cell homeostasis is predominantly dependent on survival signals provided by IL-7, with high expression of IL-7Rα but very low expression of IL-2Rα and IL-2Rβ on these cells. TCR activation of naïve T cells results in decreased expression of IL-7Rα but potently induces IL-2Rα and IL-2Rβ, and IL-2 and IL-15 can also support memory T-cell homeostasis (Surh and Sprent 2008).
γc FAMILY CYTOKINES AND Treg-CELL DEVELOPMENT AND BIOLOGY
Approximately 10% of peripheral mature CD4+ T cells express IL-2Rα (CD25), and these Treg cells show potent suppressive activity as shown by the ability of adoptively transferred CD4+ CD25+ T cells to prevent CD4+CD25− T-cell-induced autoimmunity in athymic nude mice (Sakaguchi et al. 1995). Naturally occurring Treg cells constitutively express the forkhead family transcription factor FOXP3. Treg cells play essential roles in the maintenance of a balanced immunity under various physiological and pathophysiological conditions (Ohkura et al. 2013; Li and Rudensky 2016). For example, they are essential for suppressing autoreactive T cells that have escaped negative selection in the thymus, controlling potentially harmful excessive immune responses, and suppressing the antitumor immunity that helps tumors to escape immune surveillance. The gene encoding FOXP3 is localized on the X chromosome, and FOXP3 is vital for the development and function of Treg cells. Mutation of the FOXP3 gene in humans causes severe autoimmune disease (Bennett et al. 2001), and a truncation mutation of the mouse Foxp3 gene caused by a frameshift attributed to an insertion of two nucleotides in exon VIII (Scurfy mouse) results in fatal lymphoproliferative disorders (Brunkow et al. 2001). FOXP3 is required for the development of these cells in the thymus, as shown by the complete absence of CD4+CD25+ Treg cells in Foxp3-deficient mice (Fontenot et al. 2003). STAT5 proteins are required for optimal expression of FOXP3 in vitro, with binding of STAT5 to three GAS motifs in the Foxp3 gene from CD4+CD25+ but not from CD4+CD25− splenic T cells (Yao et al. 2007).
Finding that Il2, Il2ra, and Il2rb KO mice do not show defective T-cell development but rather have lymphoproliferative autoimmune disorders caused by dysregulated expansion of effector phenotype CD4+ T cells (Sadlack et al. 1993; Suzuki et al. 1995; Willerford et al. 1995) was unexpected, given that IL-2 can potently promote the in vitro proliferation of T cells that express IL-2 receptors. However, this can be explained by markedly decreased Treg-cell numbers in these mice. Importantly, the severe autoimmunity in Il2rb−/− mice can be corrected either by thymic-specific expression of IL-2Rβ or by adoptive transfer of normal Treg cells into Il2rb−/− newborn mice (Malek et al. 2000, 2002; Malek and Castro 2010), demonstrating the important roles of IL-2 in the development and/or homeostasis of Treg cells in vivo. Subsequent studies showed that IL-2 signaling is critical for the expansion and survival of Treg cells (Bayer et al. 2005, 2007; Fontenot et al. 2005; Setoguchi et al. 2005) but is not required for their suppressive function, given that the residual Treg cells from Il2−/− or Il2ra−/− mice show similar suppressive activity in vitro as cells from WT mice (Fontenot et al. 2005). Together with TGF-β, IL-2 also plays important roles in the generation of inducible Treg (iTreg) cells from naïve CD4+ T cells, especially in peripheral mucosal tissues like lungs and gut (Davidson et al. 2007; Zheng et al. 2007).
Foxp3+ Treg cells are essentially absent in thymus and spleen of Il2rg−/−, Jak3−/−, or Stat5−/− mice (Fontenot et al. 2005; Mayack and Berg 2006; Yao et al. 2007), indicating the vital roles of γc family cytokines in the development and homeostasis of Treg cells in vivo. However, although CD4+Foxp3+ Treg cells are markedly decreased in Il2−/−, Il2ra−/−, or Il2rb−/− mice, some Treg cells are still present, suggesting the involvement of other γc family cytokine(s) in maintaining normal Treg numbers. Indeed, there is a moderate decrease of both thymic and splenic CD4+Foxp3+ Treg cells in Il7ra−/− mice (Burchill et al. 2007), and the severity of the Treg defects in Il2rb−/−Il7ra−/− double KO mice is similar to that observed in Il2rg−/Y mice, whereas Il2rb−/−Il4ra−/− double KO mice had more Treg cells than Il2rg−/Y mice (Bayer et al. 2008; Vang et al. 2008), indicating that IL-7R-dependent signaling also contributes to normal Treg numbers. In addition, both IL-7 and IL-15, but not TSLP, can induce the differentiation of Treg cells from CD4+CD25+Foxp3− thymic Treg progenitor cells in vitro, although less potently than IL-2 (Vang et al. 2008). IL-9 was also reported to enhance in vitro Treg function, as a neutralizing antibody to IL-9 blocked such a function (Elyaman et al. 2009). Consistent with this finding, Il9ra−/− mice show decreased in vivo Treg function in a mouse experimental autoimmune encephalomyelitis (EAE) model, and injection of anti-IL-9-blocking antibodies in WT mice also have diminished Treg function (Elyaman et al. 2009).
THE ROLES OF γc FAMILY CYTOKINES IN CD4+ T HELPER–CELL DIFFERENTIATION
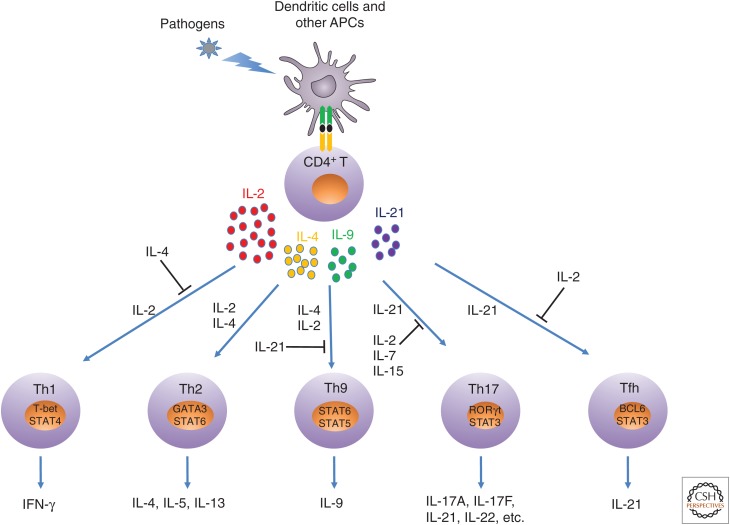
The adaptive immune system plays major roles in eliminating pathogens and protecting the host from harm. Key components of this system include peripheral naïve CD4+ T cells, which in response to antigen stimulation can differentiate into at least five types of Th cells, including Th1, Th2, Th9, Th17, and Tfh cells (Fig. 4). Which cells form is determined based on the type and strength of the antigen encountered, the presence of cytokines produced by innate immune cells, key transcriptional profiles, and the signature cytokines produced by these cells. Although IL-4 and IL-9 are the only γc family cytokines to serve as signature cytokines (for Th2 and Th9, respectively), other γc family cytokines play indispensable roles in these processes as well, either to facilitate or antagonize a given differentiation process (Fig. 4).
Figure 4.
Schematic of roles of γ chain (γc) family cytokines in T helper (Th) differentiation. On antigen stimulation, CD4+ T cells can produce interleukin (IL)-2, IL-4, IL-9, and IL-21. Under different differentiation conditions, each γc family cytokine can either promote or suppress a given differentiation process. APCs, antigen-presenting cells; Tfh, T follicular helper; STAT, signal transducers and activators of transcription; IFN, interferon.
Th1 responses are critical for controlling intracellular pathogens and serve major roles in developing inflammatory disorders. STAT4-dependent IL-12-induced T-bet expression is a key step for the differentiation of naïve CD4+ T cells into Th1 cells, ensuring expression of IFN-γ, and suppressing the expression of IL-4 and IL-17. Among γc family cytokines, IL-2 plays critical roles in promoting Th1 differentiation. IFN-γ production is markedly impaired in Jak3−/− or Stat5fl/flCD2-Cre+ CD4+ T cells, despite intact IFN-γ-STAT1 and IL-12-STAT4 signaling pathways in these cells. Moreover, antibodies to IL-2, IL-2Rα, and IL-2Rβ also greatly diminish IFN-γ expression under Th1 differentiation conditions in WT CD4+ T cells (Shi et al. 2008). IL-7 can partially restore the decreased IFN-γ expression when IL-2 signaling is blocked, but the effect is only partial, correlating with less potent STAT5 activation by IL-7 than by IL-2. IL-2 induces the expression of IL-12Rβ2 as well as IFN-γ and T-bet, at least in part via STAT5 binding to the regulatory regions of these genes (Liao et al. 2011). The importance of IL-2-induced IL-12Rβ2 expression in Th1 differentiation is further shown by the ability of retroviral transduction of IL-12Rβ2 into Il2−/− CD4+ T cells to restore impaired Th1 differentiation in these cells. Interestingly, the anti-inflammatory cytokine IL-27 inhibits Th1 differentiation, limiting the production of IL-2 during Th1 differentiation (Villarino et al. 2006).
Th2 differentiation is critical for eliminating extracellular parasites and involves the production of the signature Th2 cytokines IL-4, IL-5, and IL-13. Together, these cytokines play important roles in Ig isotype switch, IgG1 and IgE production, and mediate allergic inflammatory responses, including asthma and atopic dermatitis. Both the sustained expression of GATA3, at least in part via STAT6 (activated by IL-4) and STAT5 proteins (activated by IL-2 or potentially other γc family cytokines), are essential to initiate and maintain Th2 differentiation (Zhu et al. 2003, 2004; Pai et al. 2004). Early findings that the neutralization of IL-2 inhibited in vivo IL-4 production and that overexpression of constitutively activated STAT5A restored defective IL-4 production in the absence of IL-2 signaling demonstrate the indispensable role of IL-2 for normal Th2 differentiation (Cote-Sierra et al. 2004). These results were extended by the findings that early during the Th2 differentiation, IL-2 potently induces the expression of IL-4Rα chain in both WT and Il4−/− CD4+ T cells activated by TCR stimulation, and that this is critical for the cells to respond to IL-4 for sustained GATA3 expression. The induction of IL-4Rα by IL-2 is attributed to the direct binding of STAT5 proteins to the GAS motifs in the first intron of the Il4ra gene, and, under Th2 differentiation conditions, retroviral transduction of Il4ra cDNA into Il2−/− CD4+ T cells could fully restore the defective IL-4 expression in these cells (Liao et al. 2008). These findings together show essential roles for both IL-4 and IL-2 for normal Th2 differentiation.
Th9 cells express the signature cytokine IL-9, which is involved in inflammatory disorders, including allergic inflammation, autoimmunity, and antitumor immunity. A number of transcription factors are required for Th9 differentiation, including SMAD family proteins, E-26-specific (ETS) family proteins, IRF4, and the BATF (Kaplan et al. 2015). IL-4 and IL-2 are the key γc family cytokines required for Th9 differentiation, as evidenced by the greatly diminished IL-9 expression in Stat6−/− (Goswami et al. 2012) or Il2−/− CD4+ T cells (Liao et al. 2014). Although IL-7, IL-9, and IL-15 can also induce IL-9 expression in Il2−/− CD4+ T cells in the presence of IL-4 and TGF-β, they are much less potent than IL-2 (Liao et al. 2014). Both IL-4-STAT6 and IL-2-STAT5 pathways promote Th9 differentiation through the regulation of different transcription factors. IL-4-STAT6 induces IRF4 to increase IL-9 expression while suppressing the expression of FOXP3 and T-bet to inhibit Treg and Th1 differentiation, respectively (Goswami et al. 2012), whereas IL-2-STAT5 suppresses BCL6 to antagonize the potent inhibitory action of IL-21 on Th9 differentiation, indicating that IL-2-STAT5 and IL-21-STAT3 signaling pathways differentially regulate Th9 differentiation via their opposing effect on BCL6 expression (Liao et al. 2014). In addition, IL-2-STAT5 can promote IL-9 production by directly regulating IRF4 and IL-9 expression (Gomez-Rodriguez et al. 2016). Furthermore, the progressive loss of IL-9 production during in vitro Th9 differentiation is observed in WT but not in Stat3−/− cells, suggesting that the diminished IL-9 results from IL-10-induced activation of STAT3 (Ulrich et al. 2017).
In a mouse melanoma model, IL-9 and Th9 cells show potent antitumor activity (Lu et al. 2012; Purwar et al. 2012). IL-1β can also potentiate Th9 differentiation via activation of STAT1, which in turn induces the expression of IRF1 and thereby IRF1 binding to regulatory regions in both the Il9 and Il21 genes, with augmented expression of both IL-9 and IL-21. The indispensable role of IL-21 in the antitumor activity of Th9 cells is shown by the ability of an anti-IL-21 antibody to abolish the antitumor activity of Th9 cells induced by IL-1β (Vegran et al. 2014).
Both Th2 and Th9 cells are involved in the clearance of parasite infections, but IL-9 is expressed in vivo earlier during Nippostrongylus brasiliensis infection than are IL-4, IL-5, and IL-13 (Licona-Limon et al. 2013). In fact, IL-9 expression is required for IL-5 and IL-13 expression, with impaired expression of these cytokines in Il9−/− mice. Moreover, adoptive transfer of Th9 cells, but not Th2 cells, improves worm expulsion in Rag2−/− mice and the administration of neutralizing IL-9 antibody to these mice abolishes the ability of adoptively transferred Th9 cells to clear worm infection, leading to the conclusion that Th9 cells more efficiently clear worm infection than Th2 cells (Licona-Limon et al. 2013).
Th17 cells express transcription factor RORγt and the signature cytokines IL-17A and IL-17F, as well as other cytokines, to eliminate extracellular pathogens and fungal infections. Early during Th17 differentiation, the cooperative binding of BATF and IRF4 to their target genes plays crucial roles for chromatin accessibility and the subsequent binding of STAT3 activated by IL-6 and IL-21 and other transcription factors to initiate the Th17 transcriptional program (Ciofani et al. 2012; Li et al. 2012). Depending on how Th17 cells are generated, they can be either pathogenic and mediate autoimmune disorders or nonpathogenic and protect the host from harm caused by inflammation. For example, nonpathogenic Th17 cells are generated in the presence of IL-6 and TGF-β (McGeachy et al. 2007), whereas pathogenic Th17 cells are generated when proinflammatory IL-23 is additionally present, thereby inhibiting Th17 cells from producing IL-10 and up-regulating granulocyte macrophage colony-stimulating factor (GM-CSF) (Codarri et al. 2011; El-Behi et al. 2011). In contrast to the positive effects of IL-2 on Th1, Th2, and Th9 differentiation described above, IL-2 suppresses Th17 differentiation as shown by the observation that Il2−/− CD4+ T cells or blocking IL-2 signaling in WT CD4+ T cells results in increased Th17 differentiation (Laurence et al. 2007). Similarly, IL-15 can also inhibit Th17 differentiation as evidenced by the lower IL-17A expression observed when IL-15 is added and increased IL-17A seen when anti-IL-15 antibody is added to neutralize IL-15 produced by APC or as is observed in Il15−/− or Il15rα−/− CD4+ T cells (Pandiyan et al. 2012). The inhibitory effects of IL-2 and IL-15 on Th17 differentiation can potentially be attributed to the competition of the shared binding sites in Il17 by STAT3 activated by IL-6 and IL-21 versus STAT5 activated by IL-2 and IL-15 (Laurence et al. 2007; Yang et al. 2011; Pandiyan et al. 2012) as well as to the suppressive effect of IL-2 on the expression of IL-6Rα and the IL-6 signal-transducing molecule, gp130, which is shown by the finding that retroviral transduction of gp130 can partially reverse the inhibitory effect of IL-2 on Th17 differentiation (Liao et al. 2011). In contrast to its inhibitory actions on Th17 differentiation, IL-2 can expand Th17 cells isolated from peripheral blood of healthy human donors or patients with scleritis and increase IL-17 expression in these cells (Amadi-Obi et al. 2007). It was also reported that IL-2, IL-7, and IL-15 can each enhance the expression of IL-17A, IL-17F, IL-22, and IL-26 in CCR6+, but not in CCR6−, human CD4+CD45RO+ CD25− memory T cells, and this was attributed to the ability of these cytokines to activate PI3K, as inhibition of PI3K signaling selectively abolished the expression of IL-17 signature cytokines induced by these γc family cytokines (Wan et al. 2011). In addition, memory phenotype Th17 cells isolated from human peripheral blood constitutively express low levels of IL-2, and an IL-2-neutralizing antibody can induce apoptosis of these cells, indicating a critical role for IL-2 in Th17-cell survival as well (Yu et al. 2011). Although IL-6 and TGF-β can initiate Th17 differentiation, IL-21 produced by TCR stimulation also contribute to Th17 differentiation, with impaired in vitro differentiation of Th17 cells in Il21−/− CD4+ T cells; IL-21 can also partially compensate for the absence of IL-6 signaling during Th17 differentiation (Korn et al. 2007; Nurieva et al. 2007; Zhou et al. 2007).
Tfh cells are localized in germinal centers and are characterized by coexpression of transcription factor BCL6, inhibitory receptor PD1, IL-21, the chemokine receptor CXCR5, and costimulatory protein ICOS, but these cells do not express BLIMP1 (Crotty 2014; Qi 2016). Tfh cells play crucial roles in T-cell-dependent humoral immunity by directing B-cell differentiation into plasma cells, the selection of affinity-matured antibody-producing B cells, Ig isotype switch, and production of Ig (Crotty 2014; Qi 2016). BCL6 is the key regulator that promotes Tfh differentiation, whereas BLIMP1 antagonizes BCL6 and thus suppresses Tfh differentiation (Johnston et al. 2009). ICOS ligand, IL-6, IL-21, and STAT3 are all required for the development of Tfh cells; indeed, the lack of any of these proteins in mice impairs the development of CD4+CXCR5+ T cells after immunization (Nurieva et al. 2008). Interestingly, IL-21 uniquely up-regulates the expression of both BCL6 and BLIMP1, which result in different outcomes: germinal center B-cell maintenance mediated by BCL6 (Johnston et al. 2009; Yu et al. 2009) versus plasma-cell differentiation mediated by BLIMP1 (Shaffer et al. 2002). IL-21-induced BLIMP1 expression requires the cooperative binding of STAT3 and IRF4 in the regulatory region of Prdm1 (gene encoding for BLIMP1) (Kwon et al. 2009). In contrast, IL-2 potently suppresses Tfh-cell differentiation by activating STAT5 and thereby inducing BLIMP1, which down-regulates BCL6 (Johnston et al. 2009, 2012; Ballesteros-Tato et al. 2012; Oestreich et al. 2012). In addition, quenching IL-2 from activated CD4+ T cells by soluble IL-2Rα produced by Tfh-cell-priming ICOSLhiCD25+ DCs favors Tfh-cell differentiation at the follicle outer T-cell zone (Li et al. 2016). Interestingly, IL-2-activated AKT and mTORc1 kinases play a critical role in determining Th1 versus Tfh differentiation. The lower levels of IL-2 expression in Tfh cells are correlated with lower proliferation, glycolysis, and mitochondrial respiration than is observed in Th1 cells during acute LCMV infection (Ray et al. 2015). Down-regulation of BCL6 in Tfh cells late after immunization correlates with decreased proliferation and increased IL-7 responses, in part via increased IL-7Rα expression in these cells (Kitano et al. 2011); however, unlike IL-2, IL-7 does not increase BLIMP1 expression (McDonald et al. 2016).
Besides their opposing actions on Tfh cells, IL-2 and IL-21 regulate Tfh-cell function through differential actions on follicular regulatory T (Tfr) cells, which suppress Tfh-cell-mediated Ig production by B cells (Sage and Sharpe 2015). In addition to other signature molecules expressed by Tfh cells, Tfr cells additionally express Foxp3+ and high levels of ICOS and PD1 (Chung et al. 2011; Linterman et al. 2011). A transcriptomic analysis of Tfh cells in response to Tfr cells revealed that transcription factors essential for Tfh cells, including Bcl6, Ascl2, and Tcf1, are not affected by Tfr-cell suppression, whereas Prdm1 and especially Il4 and Il21 mRNA levels in Tfh cells are markedly reduced (Sage and Sharpe 2016). In addition, Tfr cells alter Myc signals and mechanistic target of rapamycin (mTOR) pathway in B cells. Importantly, IL-21 or IL-6, but not IL-4, can rescue Tfh- and B-cell functions suppressed by Tfr cells (Sage et al. 2016). In human patients with loss-of-function mutations in IL21RA (Kotlarz et al. 2013; Stepensky et al. 2015), there are increased numbers of IL-2-dependent total Treg and Tfr cells in peripheral blood (Jandl et al. 2017). Correspondingly, in the mouse, IL-21 suppresses the expansion of Tfr cells by down-regulating IL-2Rα expression, which is mediated by BCL6 (Jandl et al. 2017). Thus, IL-2 and IL-21 act either directly on Tfh cells or on Tfr cells to maintain B-cell functions via their opposing actions.
ROLES OF IL-2, IL-7, IL-15, AND IL-21 IN THE DEVELOPMENT AND DIFFERENTIATION OF CD8+ T CELLS
IL-2, IL-7, IL-15, and IL-21 play important roles in the homeostasis, expansion, survival, and function of CD8+ T cells. IL-2 and IL-15 potently promote CD8+ T-cell proliferation and induce effector molecules and inflammatory cytokines (Waldmann 2006; Liao et al. 2013), and IL-7 is essential for the survival and homeostasis of naïve CD8+ T cells (Schluns et al. 2000); IL-15 can also promote naïve CD8+ T-cell homeostasis albeit less potently (Kennedy et al. 2000; Berard et al. 2003). IL-7 also regulates the homeostasis of memory CD8+ T cells after viral infection (Kaech et al. 2003), whereas IL-2 can suppress the homeostasis and function of memory CD8+ T cells by Treg cells (Ku et al. 2000; Murakami et al. 2002; Waldmann 2006). When combined with IL-15 or to a lesser extent with IL-7 but not with IL-2, IL-21 can synergistically expand CD8+ T cells in vitro, and the combination of IL-21 and IL-15 potently increases antigen-specific CD8+ T-cell numbers in vivo and inhibits tumor progression in a B16 melanoma model (Zeng et al. 2005). IL-2 and IL-15 enhance expression of transcription factor eomesodermin (Eomes) by antigen-primed CD8+ T cells and their development into effector CD8+ T cells, whereas IL-21 induces Tcf7 and Lef1 while suppressing Eomes and effector CD8+ T-cell development (Hinrichs et al. 2008). Moreover, antigen-primed CD8+ T cells cultured with IL-21 but not with IL-2 or IL-15 show enhanced antitumor activity when adoptively transferred into B16 melanoma-bearing mice (Hinrichs et al. 2008).
Cytotoxic CD8+ T cells can directly kill virus-infected cells via perforin and granzymes. During LCMV infection, a sustained presence of IL-2 up-regulates Eomes and perforin but down-regulates expression of BCL6 and IL-7Rα, resulting in the expansion of effector cells while suppressing the development of memory cells (Pipkin et al. 2010). Dynamic expression of IL-2Rα in antigen-specific CD8+ T cells appears to correlate with the fate of CD8+ T-cell differentiation in response to LCMV infection. IL-2Rαlow CD8+ T cells expressing high levels of IL-7Rα and CD62L show enhanced survival and are thought to become long-lived functional memory cells, whereas IL-2Rαhigh CD8+ T cells expand more rapidly but are more susceptible to apoptosis (Kalia et al. 2010). In chronic viral infections, including human immunodeficiency virus (HIV) in humans and LCMV in mice, CD8+ T cells become exhausted (Zajac et al. 1998), and this is characterized by their greatly diminished secretion of IL-2 and decreased proliferation but increased expression of the inhibitory receptor PD1 (Barber et al. 2006). Besides PD1, transcription factor BATF is coexpressed, potently suppressing T-cell proliferation and cytokine secretion, whereas BATF small interfering RNA (siRNA) can rescue the defective secretion of IL-2 in T cells from HIV patients (Quigley et al. 2010). Furthermore, in mice chronically infected with LCMV, low-dose IL-2 can enhance CD8+ T-cell responses, decrease PD1 expression, and increase IL-7Rα and CD44 expression on virus-specific CD8+ T cells. Moreover, a combination of IL-2 with anti-PD-L1 greatly enhances virus-specific CD8+ T-cell responses (West et al. 2013).
ROLES OF IL-4, IL-7, AND IL-21 IN THE DEVELOPMENT AND BIOLOGY OF B CELLS
B-cell numbers in Il2rg−/− and Jak3−/− mice are markedly diminished, and the cells that develop are nonfunctional because of the lack of T-cell help, given the defective T-cell development in these animals (Cao et al. 1995; Nosaka et al. 1995). IL-7 is the key γc family cytokine for mouse B-cell development, with a severe block at the early stages of B-cell development in mice deficient in either IL-7 or IL-7Rα, including in CLPs and during the transition from pro-B cells to pre-B cells, with absent mature follicular B cells in both Il7- or Il7r-deficient mice (Peschon et al. 1994; von Freeden-Jeffry et al. 1995). Interestingly, B-cell development is more defective in Il7ra−/− mice than in Il7−/− mice, suggesting that TSLP, which also uses IL-7Rα as a receptor component, might also be involved. However, unlike the observation indicating a role for TSLP in T-cell development, B-cell development is similar in Il7/Tslpr double KO mice and Il7 KO mice, indicating that TSLP may not be involved in B-cell development (Jensen et al. 2008). Unlike its ability to restore αβ T-cell development in Il7r−/− mice, expression of the Bcl2 transgene could not rescue B-cell development defect in the absence of an IL-7 signal (Maraskovsky et al. 1998). However, the Bcl2 transgene could compensate for the markedly decreased expression of antiapoptotic protein MCL1 and restore pro-B-cell development in the absence of STAT5 (Malin et al. 2010). IL-7 produced by bone marrow endothelial cells is crucial in B-cell lymphogenesis, as the conditional deletion of Il7 in mouse mesenchymal progenitor cells results in a decrease of both pro- and pre-B-cell numbers in bone marrow (Cordeiro Gomes et al. 2016). In contrast to its importance for B-cell development in the mouse, IL-7 signaling is not essential for human B-cell development, as patients with mutations in IL7RA have normal B-cell numbers, although these B cells are not functional because of severe defects in T cells (Puel et al. 1998; Giliani et al. 2005). Thus, either IL-7 does not contribute to human B-cell development or there is a redundant pathway.
IL-4 signaling is not involved in B-cell development but rather is crucial for the differentiation of mature B cells, Ig isotype switching, and IgE production, as Il4−/− and Il4ra−/− mice show normal development of T and B cells but diminished IgG1 production, greatly diminished IgE production, and normal levels of IgM and other Ig isotypes after infection with N. brasiliensis (Kuhn et al. 1991; Noben-Trauth et al. 1997). Similarly, the development of T and B cells are normal in Il21r−/− mice but IgG1 levels are greatly diminished, indicating the involvement of IL-21 in Ig switch as well. However, IL-21 is also required for IgG3 production by CD40-activated naïve human B cells (Pene et al. 2004; Avery et al. 2008). Interestingly, IL-4 favors the production of IgG1, whereas IL-21 favors the IgG3 production by CD40-activated naïve human B cells (Avery et al. 2008). However, unlike the mice lacking IL-4 signaling, IgE levels are actually elevated in Il21r−/− mice after immunization (Ozaki et al. 2002), possibly caused by an inhibitory effect of IL-21 on IL-4-induced germ line Cε transcription (Suto et al. 2002). Possible cooperative actions between IL-4 and IL-21 for Ig production are further provided by the findings of a pan-hypogammaglobulinemia and poorly organized germinal centers in mice lacking both IL-4 and IL-21 signaling (Ozaki et al. 2002). These observations indicate that both IL-4 and IL-21 contribute to the differentiation of B cells into plasma cells, Ig isotype switch, and Ig production. In addition to actions of IL-21 on T cells to promote B-cell function, a study using a mouse multiple sclerosis (MS) model showed that IL-21 and CD40 are required for the expansion and maturation of regulatory B cells into IL-10-producing effector “B10” cells to inhibit autoimmune disorders by suppressing effector T (Teff)-cell function (Yoshizaki et al. 2012).
As discussed above, IL-4 and IL-21 are the most important γc family cytokines for promoting Tfh development and function, which are essential for B-cell differentiation and Ig isotype switching and production (Ozaki et al. 2002) and they play nonredundant roles in these processes (Ozaki et al. 2004). However, the kinetics of when each of these two key γc family cytokines is expressed during Tfh differentiation was unclear. By using mice expressing IL-21 and IL-4 reporters, it was shown that Tfh cells in germinal centers go through different transcriptional and functional stages to provide different signals for B-cell differentiation and function in germinal centers. For example, cells secreting only IL-21 are most efficient at promoting somatic hypermutation and affinity maturation in B cells, whereas cells expressing only IL-4 are more efficient at promoting plasma-cell differentiation and IgG1 class switch, and cells expressing both IL-4 and IL-21 can serve both functions (Weinstein et al. 2016). During LCMV infection, IL-21 signaling is dispensable for Tfh differentiation but is required for the generation of long-lived plasma cells and sustained antibody production (Rasheed et al. 2013).
Defective T cells in IL2RG- or JAK3-deficient patients can be reconstituted after hematopoietic cell transplantation, but B cells are still not functional in a significant portion of patients who require Ig replacement therapy. Interestingly, the naïve B cells from these patients respond well to IL-4 plus CD40L, but respond poorly to IL-21 plus CD40L, indicating that IL-21 is the major γc family cytokine to initiate humoral immunity in humans (Recher et al. 2011). Consistent with this, IL7R-deficient SCID patients, where IL-21 signaling is intact, show more efficient reconstitution of normal B-cell function after bone marrow transplantation (Buckley 2011).
ROLES OF γc FAMILY CYTOKINES FOR NK-CELL DEVELOPMENT AND CYTOLYTIC ACTIVITY
Unlike T and B cells, NK cells can rapidly secrete inflammatory cytokines, chemokines, and, importantly, proteinases on encountering virus-infected cells or tumor cells to eliminate these cells (Vivier et al. 2011; Cerwenka and Lanier 2016). The essential roles of γc family cytokines in the maturation, expansion, and survival of NK cells are shown by the profoundly decreased numbers of immature and mature NK cells in Il2rg−/Y mice. However, NK progenitor numbers in mutant mice are similar to those in WT mice, indicating that γc family cytokines are dispensable for NK-cell commitment in bone marrow. IL-15 is the principal γc family cytokine for the generation and maintenance of normal numbers of immature and mature NK cells, as Il15−/−, Il15ra−/−, and Il2rb−/− mice each show profoundly decreased NK-cell numbers (Vosshenrich et al. 2005). IL-7 is dispensable for conventional NK-cell development, as mice deficient in either Il7 or Il7ra show normal bone marrow and splenic NK numbers (He and Malek 1996; Moore et al. 1996), but IL-7 plays an important role in thymic NK-cell homeostasis, as there is a higher frequency but lower number of thymic NK cells in Il7−/− mice (Moore et al. 1996) and nearly a complete absence of IL-7Rα+ NK cells in Rag2−/−Il7−/− mice (Vosshenrich et al. 2006).
Although IL-2, IL-4, and IL-21 are dispensable for the generation, differentiation, expansion, and survival of NK cells in vivo, they can nevertheless either promote NK-cell proliferation and survival or enhance NK-cell function. In mice infected with Leishmania major, IL-2 and IL-12, but not IL-4 produced by antigen-specific CD4+ T cells, are required for early IFN-γ production by NK cells, which depends on the presence of CD40/CD40L (Bihl et al. 2010). In response to virus rechallenge after vaccination, IL-2 secreted by human memory T cells can potently induce a sustained cytokine secretion and degranulation by NK cells, indicating the important roles of IL-2-induced NK-cell function in the initial control of the viral infection after vaccination (Horowitz et al. 2010). Wiskott–Aldrich syndrome protein (WASp) is an actin regulator, and NK cells from patients with loss-of-function mutations in WASp cannot form lytic synapses because of decreased degranulation and expression of IFN-γ (Huang et al. 2005; Orange et al. 2011). IL-2-treated WASp KO NK cells can rescue the defective NK function to eliminate major histocompatibility complex (MHC) class I negative hematopoietic tumor cells (Kritikou et al. 2016). Overexpression of IL-4 in mice results in altered expression of cell-surface markers and function of conventional NK cells via a cooperation with macrophage-produced IL-15, with lower expression of CD11b and IL-18Rα but increased expression of IFN-γ, IL-10, and GM-CSF. These IL-4-stimulated NK cells show higher cytotoxicity than conventional NK cells, and NK cells with a phenotype similar to the IL-4-stimulated NK cells are found in mice following infection with N. brasiliensis (Kiniwa et al. 2016), suggesting the involvement of NK cells in Th2 responses during the clearance of this parasite.
IL-21 can promote differentiation and expansion of bone marrow human NK progenitors in the presence of Flt3L and IL-15, and it can also enhance NK cytotoxicity, albeit less potently than IL-2 or IL-15, and IL-21 has a cooperative effect when combined with IL-2 or IL-15 (Parrish-Novak et al. 2000). NK function in human patients with loss-of-function mutations in IL-21R is variable, ranging from normal to profoundly impaired NK-cell cytotoxicity (Kotlarz et al. 2013); the reason(s) for this variation in NK function remains unclear. Interestingly, IL-21-dependent expansion of the IFN-γ-producing memory-like NK cells during bacille Calmett–Guérin (BCG) vaccination is important for protective immunity against Mycobacterium tuberculosis (Venkatasubramanian et al. 2017).
ROLE OF IL-4 IN MACROPHAGE BIOLOGY
Activation of macrophages by cytokines and TLRs is important for host defense and immunity. IL-4-activated macrophages play a role in type 2 immunity in allergic inflammation, helminth infection, and wound healing, and these cells are referred to as M2 or M(IL-4) macrophages (Van Dyken and Locksley 2013; Wynn and Vannella 2016; Eming et al. 2017). M(IL-4) macrophages also play a critical role in wound repair following helminth infection (Chen et al. 2012) and are involved in the repair of infarcts in the adult mouse heart (Shiraishi et al. 2016). Tumor-associated macrophages (TAMs) share the immune tolerance and immunosuppressive phenotype with M2 macrophages and are associated with tumor progression (Wynn et al. 2013). Local injection of IL-21 into tumors can suppress the expression of genes associated with M2 macrophages in TAMs, including Vegf and Tgfb. In contrast, Ccl2 expression is associated with tumor-inhibiting M1 macrophages, which then act through CD8+ T cells to enhance antitumor therapy (Xu et al. 2015).
ROLES OF IL-15 AND IL-21 IN DENDRITIC CELL FUNCTION
DCs play vital roles in both innate and adaptive immunities and can be divided in two major types based on the cell-surface markers and their functions, including conventional DCs (cDCs) and plasmacytoid DCs (pDCs) (Pulendran 2015; Durai and Murphy 2016). cDCs, which can be subdivided into cDC1 and cDC2 cells and express high levels of MHC class II molecules, are professional APCs, whereas pDCs rapidly secrete high levels of type I IFNs on encountering pathogens mediated by activation of TLRs. γc family cytokines critically regulate the development and function of DCs. For example, IL-15 can promote the differentiation of monocytes into Langerhans cells in the presence of GM-CSF (Mohamadzadeh et al. 2001), a subset of immature DCs, and can support the maturation of monocytes into DCs (Saikh et al. 2001). IL-15 also promotes DC activation and maturation, whereas IL-21 inhibits these processes and blocks the maturation and activation of LPS-induced DCs (Brandt et al. 2003). In addition, IL-21 induces apoptosis of splenic cDCs but not GM-CSF-induced DCs, and this is dependent on its activation of STAT3 and increased expression of proapoptotic protein BIM. GM-CSF via STAT5 activation can antagonize the apoptotic effect of IL-21 on cDCs (Wan et al. 2013). Although IL-21 does not affect expression of type I IFNs, IL-6, or TNF-α nor the maturation of pDCs, it can potently induce granzyme B (GZMB) expression in human pDCs via a STAT3-dependent pathway. Increased GZMB production induced by IL-21 in pDC is, at least in part, responsible for the inhibition of CD4+ T-cell proliferation mediated by TLR-activated pDCs (Karrich et al. 2013).
γc FAMILY CYTOKINES AND AUTOIMMUNE DISEASES: KEY ROLES FOR IL-2 AND IL-21
As discussed above, γc family cytokines are essential for the development of T and NK cells and the function of B cells and other immune cells and are involved in every aspect of the immune response. Balanced actions of these cytokines are crucial for protecting the host from harm, not only by pathogens but also by inflammation caused by an immune response. Accumulating evidence has shown that effector CD4+ T cells play key roles in mediating autoimmune pathology, whereas Tregs play important roles in controlling the immune response (Grant et al. 2015; Suarez-Fueyo et al. 2017). Dysregulated signals by γc family cytokines are significantly associated with the progression and outcome of the human autoimmune disorders (Spolski and Leonard 2014; Tangye 2015; Suarez-Fueyo et al. 2017); thus, modulating γc family signals is an important means to manage autoimmunity.
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease of unknown cause, characterized by massive production of autoantibodies and proinflammatory cytokines and can affect every organ. A study of 1318 SLE patients and 1318 matched controls found that two of three SNPs in the introns of IL21 were significantly associated with SLE (Sawalha et al. 2008). Another study in two large cohorts found that one of the 17 SNPs in the IL21RA gene was significantly associated with SLE (Webb et al. 2009). There is also a significant association of the decreased expression of IL-21Rα in peripheral B cells with nephritis and high-titer anti-double-stranded DNA antibodies in SLE patients. B cells from some SLE patients fail to proliferate in response to IL-21 plus anti-CD40 antibody stimulation (Mitoma et al. 2005). Increased IL-21 mRNA levels were also detected in skin biopsies of SLE patients (Caruso et al. 2009). In addition to this association in human SLE, IL-21 was shown to play a vital role in the development of SLE in mouse models. In the BXSB-Yaa mouse model of SLE, both IL-10 and IL-21 levels are elevated and the increased IL-21 is not produced by Th17 cells but instead is from ICOS+CD4+ T cells, and increased IL-10 production by T cells requires IL-21 (Pot et al. 2009; Spolski et al. 2009). Importantly, Il21r−/− BXSB-Yaa mice have decreased antinuclear antibodies, do not develop histological and immunological characteristics of SLE, and survive for more than 250 days (Bubier et al. 2009), and blocking IL-21 signaling in SLE-prone MRL-Faslpr mice with an IL-21R-Fc fusion protein can reduce disease progression (Herber et al. 2007). IL-21 also plays a pathological role in mouse experimental autoimmune uveitis (EAU), which shares the pathological features with human uveitis and is a Th17-cell-related disease, with attenuated EAU in Il21r−/− mice as compared with WT mice (Wang et al. 2011).
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disorder characterized by autoantibody-induced joint inflammation and systemic inflammation. A number of proinflammatory cytokines are associated with the development and disease activity of RA, although the exact etiology of RA is still not understood. IL-21 is associated with pathogenesis of RA. For example, expression of IL-21Rα was detected in synovial fibroblasts and synovial macrophages of RA patients (Jungel et al. 2004), and IL-21 expression in synovial fluid and peripheral blood in RA patients correlates with the presence of Th17 cells (Niu et al. 2010). In addition, increased plasma IL-21 level correlates with the disease activity of RA (Rasmussen et al. 2010). Interestingly, treatment with tocilizumab, a blocking antibody to the IL-6 receptor, selectively reduced IL-21 and IgG4 anti-CCP autoantibody levels, with an improved RA disease activity (Carbone et al. 2013). Based on mass cytometry, multidimensional cytometry, transcriptomics, and functional assays, a marked increase was noted in PD1hiCXCR5−CD4+ T cells in the synovium of RA patients. These cells express IL-21, CXCL13, ICOS, and MAF and induce plasma-cell differentiation to promote B-cell responses and Ig production only within pathologically inflamed nonlymphoid tissues (Rao et al. 2017). Autoantibodies can be detected years before the onset of RA, but titers do not well correlate with disease activity. A strong correlation was noted between the expression levels of IL-17 and increased Th17-cell numbers and the systemic disease activity in both the onset and the progression of RA (Leipe et al. 2010). In a mouse RA model, IL-23 can suppress the expression of St6gal1, which encodes for a rate-limiting enzyme to control the IgG glycosylation in antibody-producing cells during plasma-cell development. The suppressive effect of St6gal1 by IL-23 is mediated by IL-21 and IL-22, but not by IL-17A, IL-17F, or GM-CSF (Pfeifle et al. 2017).
Although the etiology for MS is not fully understood, infiltration of the central nervous system (CNS) by autoreactive T cells is important for the initiation of chronic inflammation and neurodegeneration causing damage to myelin and axons. Among the factors associated with MS, Th1- and Th17-mediated inflammation in both peripheral tissues and CNS play important roles in the pathogenesis of MS. Among proinflammatory cytokines and chemokines aberrantly produced during inflammation, IL-2 and IL-21 are the only γc family cytokines. Interestingly, genome-wide association studies (GWAS) reveal that genetic variations in IL2RA, IL7, IL7R, STAT3, STAT4, and TYK2 are risk factors that confer susceptibility in MS patients (International Multiple Sclerosis Genetics et al. 2011, 2013). Furthermore, in mouse EAE, a mouse model of MS, mice with Stat3-deficient CD4+ T cells are resistant to EAE owing to defective Th17 differentiation (Liu et al. 2008), and, unexpectedly, mice with Stat5 deleted in CD4+ T cells also show diminished development of EAE because of their impaired expression of GM-CSF in CD4+ T cells activated by IL-7 (Sheng et al. 2014). To dampen IL-2’s potent proliferative activity on IL-2Rαhigh Teff cells during inflammation mediated by T cells, a humanized neutralizing IgG1 monoclonal antibody to IL-2Rα (daclizumab) was developed (Queen et al. 1989). Daclizumab is well tolerated and results in a significant improvement in treating relapsing-remitting or secondary progressive forms of MS (Bielekova et al. 2004), which account for a majority of MS patients. Daclizumab treatment causes a gradual decrease in circulating T-cell numbers and significantly expands CD56bright NK-cell numbers, which correlates with a positive response to daclizumab therapy (Bielekova et al. 2006; Wynn et al. 2010). In 2016, daclizumab (Zinbryta; previously known as daclizumab high-yield process) was approved by the European Medicines Agency and the U.S. Food and Drug Administration (FDA) for the treatment of relapsing forms of MS in adults (Shirley 2017a).
Inflammatory bowel diseases (IBDs) are characterized by chronic inflammation in the intestine and are mediated by increased proinflammatory cytokine secretion. Environmental, genetic, and commensal microbial factors are involved in the development of IBD. Crohn’s disease (CD) and ulcerative colitis (UC) are two major forms of IBD and a meta-analysis identified 163 IBD susceptibility loci, including signaling molecules for cytokines STAT1, STAT3, STAT4, and JAK2 and γc family cytokines and cytokine receptors IL2, IL21, IL2RA, and IL15RA (Jostins et al. 2012). In addition to genetic and environmental factors, T cells play important roles in the outcome of IBD. As discussed above, mice with defective IL-2 signaling spontaneously develop severe colitis (Sadlack et al. 1993; Suzuki et al. 1995; Willerford et al. 1995). A homozygous mutation of IL21 (c.T147C, p.Leu49Pro) was identified in early-onset IBD patients with decreased circulating B-cell numbers, including naïve and memory B cells and increased transitional B-cell numbers. Although the mutant IL-21 was predicted to have decreased protein stability, in vitro–produced IL-21Leu49Pro cannot activate STAT3, promote normal B-cell proliferation, nor mediate B-cell activation in vitro, suggesting that the mutant IL-21 may not be able to stably bind to the IL-21 receptor. Indeed, the B cells from these patients show normal B-cell activation and Ig class switch in response to either WT IL-21 plus CD40L or IL-4 plus CD40L stimulation, indicating that the defective B-cell function is not caused by intrinsic B-cell defects (Salzer et al. 2014).
Type I diabetes (T1D) is a chronic autoimmune disease characterized by the loss of insulin-producing pancreatic β cells, and both genetic susceptibility and environmental factors contribute to the onset of T1D. Inflammatory processes driven by self-reactive T cells in response to infectious agents and commensal organisms can either be pathogenic or protective (Herold et al. 2013). In human GWAS, among γc family cytokine and cytokine receptor gene loci, IL2RA, IL2, IL21, and IL7RA are risk loci for T1D patients (Todd et al. 2007; Concannon et al. 2009). The essential role of IL-2 in Treg cells was further shown by the findings that the ratios of Treg and Teff cells progressively decrease in inflamed pancreatic islets but not in pancreatic lymph nodes. A low dose of IL-2 promotes survival of Treg cells and protects nonobese diabetic (NOD) mice from T1D onset (Tang et al. 2008; Grinberg-Bleyer et al. 2010). In addition, injection of NOD mice with anti-IL-7Rα antibody alone can reverse established T1D (Lee et al. 2012). Treg cells from either healthy individuals or from T1D patients can respond to IL-2 at a 10-fold lower concentration than that required for memory T cells (Yu et al. 2015). Low-dose IL-2 mainly increases Treg-cell numbers and suppresses IFN-γ production by pancreas-infiltrating T cells, whereas anti-IL-7Rα antibody reduces Th1 and Tc1-cell numbers and increases PD1 expression in Teff cells. In addition, circulating anti-IL-2 autoantibodies are also detected in NOD mice and patients with T1D, and their titer is positively correlated with age and disease onset (Perol et al. 2016). The Il2/Il21 loci are within the diabetes-associated Idd3 locus, and increased levels of IL-21 were detected in NOD mice, suggesting that IL-21 is a pathogenic cytokine in this model of T1D. Indeed, NOD mice crossed with Il21r−/− mice show low lymphocyte infiltration into the pancreas and reduced Th17 cells, indicating critical roles of IL-21 in the development of T1D in NOD mice (Spolski et al. 2008; McGuire et al. 2009). Incubation of bone marrow from NOD mice with CpG can induce a B-cell population that resembles innate pro-B cells (Montandon et al. 2013). Adoptive transfer of these CpG-induced pro-B cells can protect NOD mice from disease onset, which is attributed to the ability of these CpG-pro-B to suppress the proliferation of Teff cells and induce their apoptosis. Furthermore, the expression of IL-21 by Teff cells cocultured with CpG-induced pro-B cells was markedly reduced, but the expression of IL-2 and IL-10 were increased (Montandon et al. 2013).
MODULATION OF γc FAMILY CYTOKINE SIGNALING AND THERAPEUTIC RAMIFICATIONS
Given their indispensable roles in the development and function of T, B, and NK cells and broad actions on the immune response, rationally modulating the actions of γc family cytokines has become an important approach for managing patients with a range of diseases, including immunodeficiency, allergy, infection, autoimmune disorders, transplant rejection, and cancer. For example, giving cytokines can enhance their actions, whereas blocking antibodies against cytokines or cytokine receptors, or small molecules that either inhibit cytokine production or kinase activity like JAK inhibitors, can dampen their actions to protect the host from harm mediated by overreactive immune responses. Indeed, γc family cytokines have been effective in completed and/or ongoing clinical trials for cancer immunotherapy (Rosenberg 2014; Spolski and Leonard 2014; Waldmann 2015; Pulliam et al. 2016; Tran et al. 2017). Moreover, blocking IL-4 signaling is effective for treating adult patients with moderate-to-severe atopic dermatitis (Wenzel et al. 2013; Beck et al. 2014) and adult patients with uncontrolled persistent asthma (Wenzel et al. 2016).
IL-2’s potent ability to expand Teff cells and enhance NK-cell cytotoxicity led to the use of high-dose recombinant IL-2 (proleukin) to treat metastatic melanoma and renal-cell carcinoma. Although 15% of the patients with melanoma or renal-cell carcinoma responded well to the treatment and some of them had a long-term tumor-free period, the use of high-dose IL-2 therapy was limited because of its severe side effects, including vascular leak syndrome (VLS) and increased serum creatinine and bilirubin levels (Rosenberg 2014). However, adoptive cell transfer of autologous tumor-infiltrating lymphocytes (TILs) expanded in vitro by IL-2 also results in significant improvement in treating patients with metastatic melanoma (Rosenberg 2014; Tran et al. 2017).
The discovery of IL-2’s essential role in Treg function led to observations that low-dose IL-2 preferentially expands Treg cells in vivo, thereby suppressing graft-versus-host disease (GVHD) (Koreth et al. 2011; Asano et al. 2017) and autoimmune disorders, like T1D (Hartemann et al. 2013; Yu et al. 2015) and SLE (He et al. 2016). Importantly, when combined with IL-2, two monoclonal antibodies against mouse IL-2 can selectively mimic either high-dose or low-dose IL-2 treatment. The IL-2–JES6-1 complexes, like low-dose IL-2, can expand Treg and the IL-2–S4B6 complexes, similar to high-dose IL-2, can expand Teff cells (Fig. 5) (Boyman et al. 2006). The reason for the differential effects of these two anti-IL-2 antibodies has been elucidated by structural and biochemical studies (Spangler et al. 2015). JES6-1–IL-2 complexes sterically block the interaction of IL-2 with IL-2Rβ and γc and lower the affinity of IL-2–IL-2Rα interaction to favor IL-2Rαhigh cells, including Treg cells. In contrast, S4B6–IL-2 complexes sterically block the interaction of IL-2 with IL-2Rα, resulting in increased affinity and stability of IL-2–IL-2Rβ interaction to favor the expansion of IL-2Rαlow T cells (Spangler et al. 2015).
Figure 5.
Modulation of interleukin (IL)-2 signaling by antibodies or Janus kinase (JAK) inhibitors. (A) High-dose IL-2 or IL-2 complexed with anti-IL-2 antibody S4B6 preferentially promotes the expansion of effector T cells to enhance the immune response, whereas (B) low-dose IL-2 or IL-2 complexed with anti-IL-2 antibody JES6-1 preferentially expands Treg cells to suppress the immune response. (C) The humanized anti-IL-2Rα antibody, daclizumab, can block IL-2 binding to IL-2Rα to favor the expansion of CD56bright natural killer (NK) cells while diminishing T-cell expansion. (D) The JAK inhibitor, tofacitinib, can potently inhibit signal transducers and activators of transcription (STAT) activation by γ chain (γc) family cytokines because of its inhibitory effect on JAK3 and JAK1, whereas (E) JAK3i more efficiently inhibits JAK3 and blocks the second wave of STAT5 activation and proliferation of CD4+ T cells stimulated with IL-2.
To extend these mouse studies into human cancer immunotherapy clinical trials, a monoclonal antihuman IL-2 antibody, NARA1, has been used. NARA1 binds IL-2Rα with high affinity, therefore blocking the interaction of IL-2 with IL-2Rα (Arenas-Ramirez et al. 2016). In vivo administration of IL-2–NARA1 complexes into mice preferentially expands CD8+ T, CD44hiCD8+ T, and NK cells over CD25+ CD4+ T cells, resulting in superior antimelanoma CD8+ T-cell responses than those achieved with high-dose IL-2. In another study, adoptive transfer of tumor-reactive CD8+ T cells treated with IL-2–anti-IL-2 complexes, but not IL-15-soluble IL-15Rα complexes, showed sustained antitumor immunity (Su et al. 2015); this is attributed to a vigorous and sustained expansion of IL-2–anti-IL-2 complex-treated cells in vivo versus a poor and transient expansion of IL-15-soluble IL-15Rα-treated cells. The sustained expression of high-level IL-2Rα on these tumor-reactive CD8+ T cells is caused by IL-2Rα’s ability to sustain IL-2 signaling by recycling IL-2 to the cell surface. Consistent with this finding and the critical role of IL-15 in maintaining memory CD8+ T cells in vivo, membrane-bound chimeric IL-15 on CD19-specific chimeric antigen receptor (CAR)+ T cells can maintain long-lived CD45RO−CCR7+CD95+ T cells in vivo; these cells most resemble T-memory stem cells and show potent antitumor activity for established CD19+ leukemias (Hurton et al. 2016). STAT5 activated by IL-15 plays a role in maintaining these cells in vivo even after tumor clearance.
The detailed analyses of the IL-2–IL-2 receptor complexes also led to the creation of IL-2 molecules that can differentially act on different T-cell populations by changing the binding affinity of IL-2 with one of the IL-2 receptor chains (Fig. 6). For example, “super-2” (also known as H9) is more potent than WT IL-2 based on its augmented affinity for IL-2Rβ, where it no longer requires the presence of IL-2Rα to mediate STAT5 activation (Levin et al. 2012). H9 more potently expands cytotoxic T cells than Treg cells and shows less-severe VLS as assessed by pulmonary edema in mice than WT IL-2. A number of H9 variants were also generated, which retain high-affinity binding for IL-2Rβ but have altered binding affinity for γc, thereby outcompeting endogenous IL-2 and IL-15 and showing distinct activities on different T cells (Fig. 6) (Mitra et al. 2015). As such, these molecules are partial agonists, and indeed represent the first partial agonists for a type I cytokine. Interestingly, H9-RET, which contains three amino acid substitutions at the IL-2–γc interface, can promote the proliferation of preactivated but not freshly isolated CD8+ T cells; whereas H9-RETR, with four amino acid substitutions at the interface, only minimally activates STAT5 and does not promote T-cell proliferation. However, H9-RETR can potently antagonize IL-2 or IL-15 activity—if anything—more potently than blocking antibodies to IL-2Rα or IL-2Rβ and similar to the combination of such agents, so that it inhibits IL-2-induced NK cytolytic activity, prolongs survival in a mouse GVHD model, and inhibits spontaneous proliferation of malignant cells from patients with the chronic/smoldering form adult T-cell leukemia (Mitra et al. 2015).
Figure 6.
An interleukin (IL)-2 superkine and IL-2 partial agonists. (A) An IL-2 superkine (super-IL-2, H9) (green triangle) shows enhanced affinity for IL-2Rβ and more potent activity. (B) By retaining the high-affinity binding for IL-2Rβ of H9 (in green) but by changing additional amino acids (in red) on H9 to reduce the affinity for γc, a number of partial agonists with different potencies for activating IL-2 signaling pathways were created as well as a nonagonist (H9-RETR) that could potently inhibit the actions of both IL-2 and IL-15. (C) Schematic of full, partial, and nonagonists of IL-2. (Based on Fig. 1A in Mitra et al. 2015.)
Engineered IL-4 superkines have also been generated. A type I IL-4 receptor–selected IL-4 superkine shows a much higher binding affinity for IL-2Rγ chain and is 3- to 10-fold more potent than WT IL-4 to activate STAT6 phosphorylation and cytokine production, whereas a variant selected with high affinity to IL-13Rα1 more potently induces differentiation of monocyte-derived DCs (Junttila et al. 2012). Dupilumab (developed by Regeneron Pharmaceuticals and Sanofi) is a fully humanized IgG4 monoclonal antibody to IL-4Rα that inhibits signaling by both IL-4 and IL-13 and has been approved by the FDA for treating adult patients with moderate-to-severe atopic dermatitis (Chang and Nadeau 2017; Shirley 2017b). Collectively, these results show that using monoclonal antibodies, engineered cytokines, and potentially small molecules are ways to fine-tune cytokine signal strength and selectively promote certain functions of immune cells. Such molecules represent next-generation approaches to treat infectious, allergic, and autoimmune disorders, and cancer.
Given the key roles of JAKs in mediating cytokine signaling, including by γc family cytokines, a number of JAK inhibitors have been developed to suppress cytokine-mediated inflammation. Because of their potent immunosuppressive activity, these JAK inhibitors have been tested in clinical trials for autoimmune disorders, including psoriasis, diabetic nephropathy, atopic dermatitis, myelofibrosis, juvenile idiopathic arthritis (JIA), RA, SLE, and IBD (Winthrop 2017). Among them, tofacitinib preferentially inhibits JAK1 and JAK3 and to a lesser extent JAK2 (Changelian et al. 2003; Meyer et al. 2010) and was approved by the FDA for treating RA and is being tested in a phase III trial for psoriasis and UC and in a phase I trial for JIA (Winthrop 2017). In a large cohort phase III trial, tofacitinib showed promising efficacy as an induction and maintenance therapy for patients with moderate-to-severe UC (Sandborn et al. 2017).
Thus, a wide variety of approaches to more efficiently modulate γc family cytokine signals have been and are being developed, which hopefully will further enhance the treatment of patients with various immune disorders and cancers. For example, high-dose IL-2 and super IL-2 (H9) can potently promote Teff-cell expansion and function to treat malignancies; low-dose IL-2 and H9 agonist variants preferentially expand Treg cells to better treat autoimmune disorders like T1D; the humanized anti-IL-2Rα antibody, daclizumab, can block IL-2 binding to IL-2Rα to inhibit T-cell expansion while serving to expand CD56bright NK cells because of enhanced availability of IL-2; and the H9-RETR variant can block IL-2 and IL-15 binding to IL-2Rβ, thus suppressing the growth of chronic/smoldering acute T-cell leukemia ex vivo. In mice, anti-IL-2 antibody S4B6 complexed with IL-2 mimics the action of high-dose IL-2, whereas anti-IL-2 antibody JES6-1 complexed with IL-2 mimics the action of low-dose IL-2. Moreover, in addition to tofacitinib, which can inhibit JAK1 and JAK2 in addition to JAK3, more specific JAK3 inhibitors have been developed but have not yet been evaluated clinically. Moreover, it is possible that specific inhibitors for STAT proteins may also be useful immunosuppressants.
CONCLUSIONS
In this review, we have discussed the roles of γc family cytokines, including their biological actions, mechanisms of signaling and gene regulation, and the therapeutic applications and potential of administering or blocking the actions of these cytokines. The study of this system has had profound impact, from elucidating the basis of human inherited immunodeficiency, spawning new medications (e.g., IL-2 and JAK inhibitors), and providing the basis for the first human gene therapy (for XSCID), as well as providing tremendous insight into the molecular basis of gene regulation and cell differentiation. Future mechanistic studies will be invaluable for greater understanding of the basic science of these cytokines as well as evolving new therapeutic approaches to a range of diseases. Furthermore, the approaches used for modulation of IL-2 signals and the lessons learned may be broadly applicable to other γc family cytokines as well as other cytokines and growth factors, with implications for scientific investigation and potentially for therapeutic advances as well.
Footnotes
Editors: Warren J. Leonard and Robert D. Schreiber
Additional Perspectives on Cytokines available at www.cshperspectives.org
REFERENCES
- Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell 89: 1033–1041. [DOI] [PubMed] [Google Scholar]
- Al-Shami A, Spolski R, Kelly J, Fry T, Schwartzberg PL, Pandey A, Mackall CL, Leonard WJ. 2004. A role for thymic stromal lymphopoietin in CD4+ T cell development. J Exp Med 200: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadi-Obi A, Yu CR, Liu X, Mahdi RM, Clarke GL, Nussenblatt RB, Gery I, Lee YS, Egwuagu CE. 2007. TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Nat Med 13: 711–718. [DOI] [PubMed] [Google Scholar]
- Aman MJ, Tayebi N, Obiri NI, Puri RK, Modi WS, Leonard WJ. 1996. cDNA cloning and characterization of the human interleukin 13 receptor α chain. J Biol Chem 271: 29265–29270. [DOI] [PubMed] [Google Scholar]
- Arenas-Ramirez N, Zou C, Popp S, Zingg D, Brannetti B, Wirth E, Calzascia T, Kovarik J, Sommer L, Zenke G, et al. 2016. Improved cancer immunotherapy by a CD25-mimobody conferring selectivity to human interleukin-2. Sci Transl Med 8: 367ra166. [DOI] [PubMed] [Google Scholar]
- Arima N, Kamio M, Imada K, Hori T, Hattori T, Tsudo M, Okuma M, Uchiyama T. 1992. Pseudo-high affinity interleukin 2 (IL-2) receptor lacks the third component that is essential for functional IL-2 binding and signaling. J Exp Med 176: 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage RJ, Macduff BM, Eisenman J, Paxton R, Grabstein KH. 1995. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J Immunol 154: 483–490. [PubMed] [Google Scholar]
- Asano T, Meguri Y, Yoshioka T, Kishi Y, Iwamoto M, Nakamura M, Sando Y, Yagita H, Koreth J, Kim HT, et al. 2017. PD-1 modulates regulatory T-cell homeostasis during low-dose interleukin-2 therapy. Blood 129: 2186–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asao H, Okuyama C, Kumaki S, Ishii N, Tsuchiya S, Foster D, Sugamura K. 2001. Cutting edge: The common γ-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol 167: 1–5. [DOI] [PubMed] [Google Scholar]
- Attridge K, Wang CJ, Wardzinski L, Kenefeck R, Chamberlain JL, Manzotti C, Kopf M, Walker LS. 2012. IL-21 inhibits T cell IL-2 production and impairs Treg homeostasis. Blood 119: 4656–4664. [DOI] [PubMed] [Google Scholar]
- Avery DT, Bryant VL, Ma CS, de Waal Malefyt R, Tangye SG. 2008. IL-21-induced isotype switching to IgG and IgA by human naïve B cells is differentially regulated by IL-4. J Immunol 181: 1767–1779. [DOI] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. 2012. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford RN, Grant AJ, Burton JD, Peters C, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA. 1994. The interleukin (IL) 2 receptor β chain is shared by IL-2 and a cytokine, provisionally designated IL-T, that stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci 91: 4940–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [DOI] [PubMed] [Google Scholar]
- Bayer AL, Yu A, Adeegbe D, Malek TR. 2005. Essential role for interleukin-2 for CD4+CD25+ T regulatory cell development during the neonatal period. J Exp Med 201: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer AL, Yu A, Malek TR. 2007. Function of the IL-2R for thymic and peripheral CD4+CD25+ Foxp3+ T regulatory cells. J Immunol 178: 4062–4071. [DOI] [PubMed] [Google Scholar]
- Bayer AL, Lee JY, de la Barrera A, Surh CD, Malek TR. 2008. A function for IL-7R for CD4+CD25+Foxp3+ T regulatory cells. J Immunol 181: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan JF. 1990. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci 87: 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck LA, Thaci D, Hamilton JD, Graham NM, Bieber T, Rocklin R, Ming JE, Ren H, Kao R, Simpson E, et al. 2014. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med 371: 130–139. [DOI] [PubMed] [Google Scholar]
- Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, Knobeloch KP, Owen MR, Naumann R, Decker T, et al. 2014. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat Immunol 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27: 20–21. [DOI] [PubMed] [Google Scholar]
- Berard M, Brandt K, Bulfone-Paus S, Tough DF. 2003. IL-15 promotes the survival of naïve and memory phenotype CD8+ T cells. J Immunol 170: 5018–5026. [DOI] [PubMed] [Google Scholar]
- Bielekova B, Richert N, Howard T, Blevins G, Markovic-Plese S, McCartin J, Frank JA, Wurfel J, Ohayon J, Waldmann TA, et al. 2004. Humanized anti-CD25 (daclizumab) inhibits disease activity in multiple sclerosis patients failing to respond to interferon β. Proc Natl Acad Sci 101: 8705–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA, McFarland H, Henkart PA, Martin R. 2006. Regulatory CD56bright natural killer cells mediate immunomodulatory effects of IL-2Rα-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci 103: 5941–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihl F, Pecheur J, Breart B, Poupon G, Cazareth J, Julia V, Glaichenhaus N, Braud VM. 2010. Primed antigen-specific CD4+ T cells are required for NK cell activation in vivo upon Leishmania major infection. J Immunol 185: 2174–2181. [DOI] [PubMed] [Google Scholar]
- Boussiotis VA, Barber DL, Nakarai T, Freeman GJ, Gribben JG, Bernstein GM, D’Andrea AD, Ritz J, Nadler LM. 1994. Prevention of T cell anergy by signaling through the γc chain of the IL-2 receptor. Science 266: 1039–1042. [DOI] [PubMed] [Google Scholar]
- Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. 2006. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311: 1924–1927. [DOI] [PubMed] [Google Scholar]
- Brandt K, Bulfone-Paus S, Foster DC, Ruckert R. 2003. Interleukin-21 inhibits dendritic cell activation and maturation. Blood 102: 4090–4098. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27: 68–73. [DOI] [PubMed] [Google Scholar]
- Bubier JA, Sproule TJ, Foreman O, Spolski R, Shaffer DJ, Morse HC 3rd, Leonard WJ, Roopenian DC. 2009. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc Natl Acad Sci 106: 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley RH. 2011. Transplantation of hematopoietic stem cells in human severe combined immunodeficiency: Longterm outcomes. Immunol Res 49: 25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. 2007. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol 178: 280–290. [DOI] [PubMed] [Google Scholar]
- Burton JD, Bamford RN, Peters C, Grant AJ, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA. 1994. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci 91: 4935–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity 2: 223–238. [DOI] [PubMed] [Google Scholar]
- Carbone G, Wilson A, Diehl SA, Bunn J, Cooper SM, Rincon M. 2013. Interleukin-6 receptor blockade selectively reduces IL-21 production by CD4 T cells and IgG4 autoantibodies in rheumatoid arthritis. Int J Biol Sci 9: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Botti E, Sarra M, Esposito M, Stolfi C, Diluvio L, Giustizieri ML, Pacciani V, Mazzotta A, Campione E, et al. 2009. Involvement of interleukin-21 in the epidermal hyperplasia of psoriasis. Nat Med 15: 1013–1015. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL. 2016. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 16: 112–123. [DOI] [PubMed] [Google Scholar]
- Chang HY, Nadeau KC. 2017. IL-4Rα inhibitor for atopic disease. Cell 170: 222. [DOI] [PubMed] [Google Scholar]
- Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, Rizzuti BJ, Sawyer PS, Perry BD, Brissette WH, et al. 2003. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science 302: 875–878. [DOI] [PubMed] [Google Scholar]
- Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE Jr, Kuriyan J. 2003. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci 12: 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Liu Z, Wu W, Rozo C, Bowdridge S, Millman A, Van Rooijen N, Urban JF Jr, Wynn TA, Gause WC. 2012. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med 18: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med 17: 983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. 2012. A validated regulatory network for Th17 cell specification. Cell 151: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol 12: 560–567. [DOI] [PubMed] [Google Scholar]
- Concannon P, Rich SS, Nepom GT. 2009. Genetics of type 1A diabetes. N Engl J Med 360: 1646–1654. [DOI] [PubMed] [Google Scholar]
- Cordeiro Gomes A, Hara T, Lim VY, Herndler-Brandstetter D, Nevius E, Sugiyama T, Tani-Ichi S, Schlenner S, Richie E, Rodewald HR, et al. 2016. Hematopoietic stem cell niches produce lineage-instructive signals to control multipotent progenitor differentiation. Immunity 45: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosman D, Cerretti DP, Larsen A, Park L, March C, Dower S, Gillis S, Urdal D. 1984. Cloning, sequence and expression of human interleukin-2 receptor. Nature 312: 768–771. [DOI] [PubMed] [Google Scholar]
- Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. 2004. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci 101: 3880–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, Kaech SM. 2015. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161: 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, et al. 2008. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3− effector T cells. Nat Immunol 9: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421. [DOI] [PubMed] [Google Scholar]
- Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. 2007. Cutting Edge: IL-2 is essential for TGF-β-mediated induction of Foxp3+ T regulatory cells. J Immunol 178: 4022–4026. [DOI] [PubMed] [Google Scholar]
- Demoulin JB, Uyttenhove C, Van Roost E, DeLestre B, Donckers D, Van Snick J, Renauld JC. 1996. A single tyrosine of the interleukin-9 (IL-9) receptor is required for STAT activation, antiapoptotic activity, and growth regulation by IL-9. Mol Cell Biol 16: 4710–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoulin JB, Van Roost E, Stevens M, Groner B, Renauld JC. 1999. Distinct roles for STAT1, STAT3, and STAT5 in differentiation gene induction and apoptosis inhibition by interleukin-9. J Biol Chem 274: 25855–25861. [DOI] [PubMed] [Google Scholar]
- Diehl SA, Schmidlin H, Nagasawa M, Blom B, Spits H. 2012. IL-6 triggers IL-21 production by human CD4+ T cells to drive STAT3-dependent plasma cell differentiation in B cells. Immunol Cell Biol 90: 802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druez C, Coulie P, Uyttenhove C, Van Snick J. 1990. Functional and biochemical characterization of mouse P40/IL-9 receptors. J Immunol 145: 2494–2499. [PubMed] [Google Scholar]
- Dubois S, Mariner J, Waldmann TA, Tagaya Y. 2002. IL-15Rα recycles and presents IL-15 in trans to neighboring cells. Immunity 17: 537–547. [DOI] [PubMed] [Google Scholar]
- Dugas B, Renauld JC, Pene J, Bonnefoy JY, Peti-Frere C, Braquet P, Bousquet J, Van Snick J, Mencia-Huerta JM. 1993. Interleukin-9 potentiates the interleukin-4-induced immunoglobulin (IgG, IgM and IgE) production by normal human B lymphocytes. Eur J Immunol 23: 1687–1692. [DOI] [PubMed] [Google Scholar]
- Dukovich M, Wano Y, Le thi Bich T, Katz P, Cullen BR, Kehrl JH, Greene WC. 1987. A second human interleukin-2 binding protein that may be a component of high-affinity interleukin-2 receptors. Nature 327: 518–522. [DOI] [PubMed] [Google Scholar]
- Durai V, Murphy KM. 2016. Functions of murine dendritic cells. Immunity 45: 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. 2011. The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12: 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyaman W, Bradshaw EM, Uyttenhove C, Dardalhon V, Awasthi A, Imitola J, Bettelli E, Oukka M, van Snick J, Renauld JC, et al. 2009. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc Natl Acad Sci 106: 12885–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Wynn TA, Martin P. 2017. Inflammation and metabolism in tissue repair and regeneration. Science 356: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Fischer A, Le Deist F, Hacein-Bey-Abina S, Andre-Schmutz I, Basile Gde S, de Villartay JP, Cavazzana-Calvo M. 2005. Severe combined immunodeficiency. A model disease for molecular immunology and therapy. Immunol Rev 203: 98–109. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4: 330–336. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol 6: 1142–1151. [DOI] [PubMed] [Google Scholar]
- Gessner A, Blum H, Rollinghoff M. 1993. Differential regulation of IL-9-expression after infection with Leishmania major in susceptible and resistant mice. Immunobiology 189: 419–435. [DOI] [PubMed] [Google Scholar]
- Giliani S, Mori L, de Saint Basile G, Le Deist F, Rodriguez-Perez C, Forino C, Mazzolari E, Dupuis S, Elhasid R, Kessel A, et al. 2005. Interleukin-7 receptor α (IL-7Rα) deficiency: Cellular and molecular bases. Analysis of clinical, immunological, and molecular features in 16 novel patients. Immunol Rev 203: 110–126. [DOI] [PubMed] [Google Scholar]
- Giri JG, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park LS, Cosman D, Anderson D. 1994. Utilization of the β and γ chains of the IL-2 receptor by the novel cytokine IL-15. EMBO J 13: 2822–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Meylan F, Handon R, Hayes ET, Anderson SM, Kirby MR, Siegel RM, Schwartzberg PL. 2016. Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4. Nat Commun 7: 10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RG, Lupton S, Schmierer A, Hjerrild KJ, Jerzy R, Clevenger W, Gillis S, Cosman D, Namen AE. 1989. Human interleukin 7: Molecular cloning and growth factor activity on human and murine B-lineage cells. Proc Natl Acad Sci 86: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R, Kaplan MH. 2011. A brief history of IL-9. J Immunol 186: 3283–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, Kaplan MH. 2012. STAT6-dependent regulation of Th9 development. J Immunol 188: 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, et al. 1994. Cloning of a T cell growth factor that interacts with the β chain of the interleukin-2 receptor. Science 264: 965–968. [DOI] [PubMed] [Google Scholar]
- Grant CR, Liberal R, Mieli-Vergani G, Vergani D, Longhi MS. 2015. Regulatory T-cells in autoimmune diseases: Challenges, controversies and—yet—unanswered questions. Autoimmun Rev 14: 105–116. [DOI] [PubMed] [Google Scholar]
- Grinberg-Bleyer Y, Baeyens A, You S, Elhage R, Fourcade G, Gregoire S, Cagnard N, Carpentier W, Tang Q, Bluestone J, et al. 2010. IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J Exp Med 207: 1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, Dupuis-Girod S, et al. 2002. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med 18: 1185–1193. [DOI] [PubMed] [Google Scholar]
- Hartemann A, Bensimon G, Payan CA, Jacqueminet S, Bourron O, Nicolas N, Fonfrede M, Rosenzwajg M, Bernard C, Klatzmann D. 2013. Low-dose interleukin 2 in patients with type 1 diabetes: A phase 1/2 randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol 1: 295–305. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M, Tsudo M, Minamoto S, Kono T, Doi T, Miyata T, Miyasaka M, Taniguchi T. 1989. Interleukin-2 receptor β chain gene: Generation of three receptor forms by cloned human α and β chain cDNA’s. Science 244: 551–556. [DOI] [PubMed] [Google Scholar]
- He YW, Malek TR. 1996. Interleukin-7 receptor α is essential for the development of γδ+ T cells, but not natural killer cells. J Exp Med 184: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang X, Wei Y, Sun X, Chen Y, Deng J, Jin Y, Gan Y, Hu X, Jia R, et al. 2016. Low-dose interleukin-2 treatment selectively modulates CD4+ T cell subsets in patients with systemic lupus erythematosus. Nat Med 22: 991–993. [DOI] [PubMed] [Google Scholar]
- Herber D, Brown TP, Liang S, Young DA, Collins M, Dunussi-Joannopoulos K. 2007. IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J Immunol 178: 3822–3830. [DOI] [PubMed] [Google Scholar]
- Herold KC, Vignali DA, Cooke A, Bluestone JA. 2013. Type 1 diabetes: Translating mechanistic observations into effective clinical outcomes. Nat Rev Immunol 13: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS, Spolski R, Paulos CM, Gattinoni L, Kerstann KW, Palmer DC, Klebanoff CA, Rosenberg SA, Leonard WJ, Restifo NP. 2008. IL-2 and IL-21 confer opposing differentiation programs to CD8+ T cells for adoptive immunotherapy. Blood 111: 5326–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Behrens RH, Okell L, Fooks AR, Riley EM. 2010. NK cells as effectors of acquired immune responses: Effector CD4+ T cell-dependent activation of NK cells following vaccination. J Immunol 185: 2808–2818. [DOI] [PubMed] [Google Scholar]
- Hou J, Schindler U, Henzel WJ, Ho TC, Brasseur M, McKnight SL. 1994. An interleukin-4-induced transcription factor: IL-4 Stat. Science 265: 1701–1706. [DOI] [PubMed] [Google Scholar]
- Howard M, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, Paul WE. 1982. Identification of a T cell-derived B cell growth factor distinct from interleukin 2. J Exp Med 155: 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ochs HD, Dupont B, Vyas YM. 2005. The Wiskott–Aldrich syndrome protein regulates nuclear translocation of NFAT2 and NF-κB (RelA) independently of its role in filamentous actin polymerization and actin cytoskeletal rearrangement. J Immunol 174: 2602–2611. [DOI] [PubMed] [Google Scholar]
- Hultner L, Druez C, Moeller J, Uyttenhove C, Schmitt E, Rude E, Dormer P, Van Snick J. 1990. Mast cell growth-enhancing activity (MEA) is structurally related and functionally identical to the novel mouse T cell growth factor P40/TCGFIII (interleukin 9). Eur J Immunol 20: 1413–1416. [DOI] [PubMed] [Google Scholar]
- Hurton LV, Singh H, Najjar AM, Switzer KC, Mi T, Maiti S, Olivares S, Rabinovich B, Huls H, Forget MA, et al. 2016. Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc Natl Acad Sci 113: E7788–E7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium; Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, et al. 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium; Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, et al. 2013. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakson PC, Pure E, Vitetta ES, Krammer PH. 1982. T cell-derived B cell differentiation factor(s). Effect on the isotype switch of murine B cells. J Exp Med 155: 734–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandl C, Liu SM, Canete PF, Warren J, Hughes WE, Vogelzang A, Webster K, Craig ME, Uzel G, Dent A, et al. 2017. IL-21 restricts T follicular regulatory T cell proliferation through Bcl-6 mediated inhibition of responsiveness to IL-2. Nat Commun 8: 14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen CT, Kharazi S, Boiers C, Cheng M, Lubking A, Sitnicka E, Jacobsen SE. 2008. FLT3 ligand and not TSLP is the key regulator of IL-7-independent B-1 and B-2 B lymphopoiesis. Blood 112: 2297–2304. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Li WQ, Aiello FB, Mazzucchelli R, Asefa B, Khaled AR, Durum SK. 2005. Cell biology of IL-7, a key lymphotrophin. Cytokine Growth Factor Rev 16: 513–533. [DOI] [PubMed] [Google Scholar]
- Jin H, Carrio R, Yu A, Malek TR. 2004. Distinct activation signals determine whether IL-21 induces B cell costimulation, growth arrest, or Bim-dependent apoptosis. J Immunol 173: 657–665. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med 209: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. 2012. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungel A, Distler JH, Kurowska-Stolarska M, Seemayer CA, Seibl R, Forster A, Michel BA, Gay RE, Emmrich F, Gay S, et al. 2004. Expression of interleukin-21 receptor, but not interleukin-21, in synovial fibroblasts and synovial macrophages of patients with rheumatoid arthritis. Arthritis Rheum 50: 1468–1476. [DOI] [PubMed] [Google Scholar]
- Junttila IS, Creusot RJ, Moraga I, Bates DL, Wong MT, Alonso MN, Suhoski MM, Lupardus P, Meier-Schellersheim M, Engleman EG, et al. 2012. Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines. Nat Chem Biol 8: 990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32: 91–103. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Hufford MM, Olson MR. 2015. The development and in vivo function of T helper 9 cells. Nat Rev Immunol 15: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrich JJ, Jachimowski LC, Nagasawa M, Kamp A, Balzarolo M, Wolkers MC, Uittenbogaart CH, Marieke van Ham S, Blom B. 2013. IL-21-stimulated human plasmacytoid dendritic cells secrete granzyme B, which impairs their capacity to induce T-cell proliferation. Blood 121: 3103–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima N, Nishi-Takaoka C, Fujita T, Taki S, Yamada G, Hamuro J, Taniguchi T. 1985. Unique structure of murine interleukin-2 as deduced from cloned cDNAs. Nature 313: 402–404. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Minami Y, Miyazaki T, Ihle JN, Taniguchi T. 1995. Critical role of the interleukin 2 (IL-2) receptor γ-chain-associated Jak3 in the IL-2-induced c-fos and c-myc, but not bcl-2, gene induction. Proc Natl Acad Sci 92: 8724–8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, et al. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med 191: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermouni A, Van Roost E, Arden KC, Vermeesch JR, Weiss S, Godelaine D, Flint J, Lurquin C, Szikora JP, Higgs DR, et al. 1995. The IL-9 receptor gene (IL9R): Genomic structure, chromosomal localization in the pseudoautosomal region of the long arm of the sex chromosomes, and identification of IL9R pseudogenes at 9qter, 10pter, 16pter, and 18pter. Genomics 29: 371–382. [DOI] [PubMed] [Google Scholar]
- Khaled AR, Li WQ, Huang J, Fry TJ, Khaled AS, Mackall CL, Muegge K, Young HA, Durum SK. 2002. Bax deficiency partially corrects interleukin-7 receptor α deficiency. Immunity 17: 561–573. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Takeshita T, Kondo M, Ishii N, Nakamura M, Van Snick J, Sugamura K. 1995. Sharing of the IL-2 receptor γ chain with the functional IL-9 receptor complex. Int Immunol 7: 115–120. [DOI] [PubMed] [Google Scholar]
- Kiniwa T, Enomoto Y, Terazawa N, Omi A, Miyata N, Ishiwata K, Miyajima A. 2016. NK cells activated by Interleukin-4 in cooperation with Interleukin-15 exhibit distinctive characteristics. Proc Natl Acad Sci 113: 10139–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34: 961–972. [DOI] [PubMed] [Google Scholar]
- Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. 1993. Sharing of the interleukin-2 (IL-2) receptor γ chain between receptors for IL-2 and IL-4. Science 262: 1874–1877. [DOI] [PubMed] [Google Scholar]
- Kondo M, Takeshita T, Higuchi M, Nakamura M, Sudo T, Nishikawa S, Sugamura K. 1994. Functional participation of the IL-2 receptor γ chain in IL-7 receptor complexes. Science 263: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Koreth J, Matsuoka K, Kim HT, McDonough SM, Bindra B, Alyea EP 3rd, Armand P, Cutler C, Ho VT, Treister NS, et al. 2011. Interleukin-2 and regulatory T cells in graft-versus-host disease. N Engl J Med 365: 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. 2007. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448: 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlarz D, Zietara N, Uzel G, Weidemann T, Braun CJ, Diestelhorst J, Krawitz PM, Robinson PN, Hecht J, Puchalka J, et al. 2013. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J Exp Med 210: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikou JS, Dahlberg CI, Baptista MA, Wagner AK, Banerjee PP, Gwalani LA, Poli C, Panda SK, Karre K, Kaech SM, et al. 2016. IL-2 in the tumor microenvironment is necessary for Wiskott–Aldrich syndrome protein deficient NK cells to respond to tumors in vivo. Sci Rep 6: 30636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. 2000. Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science 288: 675–678. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Rajewsky K, Muller W. 1991. Generation and analysis of interleukin-4 deficient mice. Science 254: 707–710. [DOI] [PubMed] [Google Scholar]
- Kuperman D, Schofield B, Wills-Karp M, Grusby MJ. 1998. Signal transducer and activator of transcription factor 6 (Stat6)-deficient mice are protected from antigen-induced airway hyperresponsiveness and mucus production. J Exp Med 187: 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. 2008. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26: 371–381. [DOI] [PubMed] [Google Scholar]
- Lauwerys BR, Garot N, Renauld JC, Houssiau FA. 2000. Cytokine production and killer activity of NK/T-NK cells derived with IL-2, IL-15, or the combination of IL-12 and IL-18. J Immunol 165: 1847–1853. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. 2011. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat Immunol 12: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LF, Logronio K, Tu GH, Zhai W, Ni I, Mei L, Dilley J, Yu J, Rajpal A, Brown C, et al. 2012. Anti-IL-7 receptor-α reverses established type 1 diabetes in nonobese diabetic mice by modulating effector T-cell function. Proc Natl Acad Sci 109: 12674–12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe J, Grunke M, Dechant C, Reindl C, Kerzendorf U, Schulze-Koops H, Skapenko A. 2010. Role of Th17 cells in human autoimmune arthritis. Arthritis Rheum 62: 2876–2885. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ. 1991. Interleukin-2 programs mouse αβ T lymphocytes for apoptosis. Nature 353: 858–861. [DOI] [PubMed] [Google Scholar]
- Leonard WJ. 1996. The molecular basis of X-linked severe combined immunodeficiency: Defective cytokine receptor signaling. Annu Rev Med 47: 229–239. [DOI] [PubMed] [Google Scholar]
- Leonard WJ. 2001. Cytokines and immunodeficiency diseases. Nat Rev Immunol 1: 200–208. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, O’Shea JJ. 1998. Jaks and STATs: Biological implications. Annu Rev Immunol 16: 293–322. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Depper JM, Crabtree GR, Rudikoff S, Pumphrey J, Robb RJ, Kronke M, Svetlik PB, Peffer NJ, Waldmann TA, et al. 1984. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature 311: 626–631. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Kronke M, Peffer NJ, Depper JM, Greene WC. 1985. Interleukin 2 receptor gene expression in normal human T lymphocytes. Proc Natl Acad Sci 82: 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AM, Bates DL, Ring AM, Krieg C, Lin JT, Su L, Moraga I, Raeber ME, Bowman GR, Novick P, et al. 2012. Exploiting a natural conformational switch to engineer an interleukin-2 “superkine.” Nature 484: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Rudensky AY. 2016. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol 16: 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. 2012. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature 490: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lu E, Yi T, Cyster JG. 2016. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. 2008. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat Immunol 9: 1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Wang L, Li P, Leonard WJ. 2011. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol 12: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Leonard WJ. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Spolski R, Li P, Du N, West EE, Ren M, Mitra S, Leonard WJ. 2014. Opposing actions of IL-2 and IL-21 on Th9 differentiation correlate with their differential regulation of BCL6 expression. Proc Natl Acad Sci 111: 3508–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licona-Limon P, Henao-Mejia J, Temann AU, Gagliani N, Licona-Limon I, Ishigame H, Hao L, Herbert DR, Flavell RA. 2013. Th9 cells drive host immunity against gastrointestinal worm infection. Immunity 39: 744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S, et al. 1995. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 2: 331–339. [DOI] [PubMed] [Google Scholar]
- Lin JX, Li P, Liu D, Jin HT, He J, Ata Ur Rasheed M, Rochman Y, Wang L, Cui K, Liu C, et al. 2012. Critical role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 36: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton MT. 1998. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J Immunol 161: 302–310. [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med 17: 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lee YS, Yu CR, Egwuagu CE. 2008. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J Immunol 180: 6070–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9: 669–676. [DOI] [PubMed] [Google Scholar]
- Lorenzen I, Dingley AJ, Jacques Y, Grotzinger J. 2006. The structure of the interleukin-15α receptor and its implications for ligand binding. J Biol Chem 281: 6642–6647. [DOI] [PubMed] [Google Scholar]
- Lu Y, Hong S, Li H, Park J, Hong B, Wang L, Zheng Y, Liu Z, Xu J, He J, et al. 2012. Th9 cells promote antitumor immune responses in vivo. J Clin Invest 122: 4160–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O’Shea JJ, et al. 1995. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature 377: 65–68. [DOI] [PubMed] [Google Scholar]
- Maki K, Sunaga S, Komagata Y, Kodaira Y, Mabuchi A, Karasuyama H, Yokomuro K, Miyazaki JI, Ikuta K. 1996. Interleukin 7 receptor-deficient mice lack γδ T cells. Proc Natl Acad Sci 93: 7172–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek TR, Castro I. 2010. Interleukin-2 receptor signaling: At the interface between tolerance and immunity. Immunity 33: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek TR, Porter BO, Codias EK, Scibelli P, Yu A. 2000. Normal lymphoid homeostasis and lack of lethal autoimmunity in mice containing mature T cells with severely impaired IL-2 receptors. J Immunol 164: 2905–2914. [DOI] [PubMed] [Google Scholar]
- Malek TR, Yu A, Vincek V, Scibelli P, Kong L. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rβ-deficient mice. Implications for the nonredundant function of IL-2. Immunity 17: 167–178. [DOI] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. 2010. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 11: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraskovsky E, Peschon JJ, McKenna H, Teepe M, Strasser A. 1998. Overexpression of Bcl-2 does not rescue impaired B lymphopoiesis in IL-7 receptor-deficient mice but can enhance survival of mature B cells. Int Immunol 10: 1367–1375. [DOI] [PubMed] [Google Scholar]
- Marks-Konczalik J, Dubois S, Losi JM, Sabzevari H, Yamada N, Feigenbaum L, Waldmann TA, Tagaya Y. 2000. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc Natl Acad Sci 97: 11445–11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayack SR, Berg LJ. 2006. Cutting edge: An alternative pathway of CD4+ T cell differentiation is induced following activation in the absence of γ-chain-dependent cytokine signals. J Immunol 176: 2059–2063. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli R, Durum SK. 2007. Interleukin-7 receptor expression: Intelligent design. Nat Rev Immunol 7: 144–154. [DOI] [PubMed] [Google Scholar]
- McDonald PW, Read KA, Baker CE, Anderson AE, Powell MD, Ballesteros-Tato A, Oestreich KJ. 2016. IL-7 signalling represses Bcl-6 and the TFH gene program. Nat Commun 7: 10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. 2007. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nat Immunol 8: 1390–1397. [DOI] [PubMed] [Google Scholar]
- McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C. 2009. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci 106: 19438–19443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta DS, Wurster AL, Whitters MJ, Young DA, Collins M, Grusby MJ. 2003. IL-21 induces the apoptosis of resting and activated primary B cells. J Immunol 170: 4111–4118. [DOI] [PubMed] [Google Scholar]
- Melen E, Gullsten H, Zucchelli M, Lindstedt A, Nyberg F, Wickman M, Pershagen G, Kere J. 2004. Sex specific protective effects of interleukin-9 receptor haplotypes on childhood wheezing and sensitisation. J Med Genet 41: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DM, Jesson MI, Li X, Elrick MM, Funckes-Shippy CL, Warner JD, Gross CJ, Dowty ME, Ramaiah SK, Hirsch JL, et al. 2010. Anti-inflammatory activity and neutrophil reductions mediated by the JAK1/JAK3 inhibitor, CP-690,550, in rat adjuvant-induced arthritis. J Inflamm (Lond) 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, Horiuchi T, Kimoto Y, Tsukamoto H, Uchino A, Tamimoto Y, Miyagi Y, Harada M. 2005. Decreased expression of interleukin-21 receptor on peripheral B lymphocytes in systemic lupus erythematosus. Int J Mol Med 16: 609–615. [PubMed] [Google Scholar]
- Mitra S, Ring AM, Amarnath S, Spangler JB, Li P, Ju W, Fischer S, Oh J, Spolski R, Weiskopf K, et al. 2015. Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity 42: 826–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizel SB, Farrar JJ. 1979. Revised nomenclature for antigen-nonspecific T-cell proliferation and helper factors. Cell Immunol 48: 433–436. [DOI] [PubMed] [Google Scholar]
- Mohamadzadeh M, Berard F, Essert G, Chalouni C, Pulendran B, Davoust J, Bridges G, Palucka AK, Banchereau J. 2001. Interleukin 15 skews monocyte differentiation into dendritic cells with features of Langerhans cells. J Exp Med 194: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montandon R, Korniotis S, Layseca-Espinosa E, Gras C, Megret J, Ezine S, Dy M, Zavala F. 2013. Innate pro-B-cell progenitors protect against type 1 diabetes by regulating autoimmune effector T cells. Proc Natl Acad Sci 110: E2199–E2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TA, von Freeden-Jeffry U, Murray R, Zlotnik A. 1996. Inhibition of γδ T cell development and early thymocyte maturation in IL-7−/– mice. J Immunol 157: 2366–2373. [PubMed] [Google Scholar]
- Morgan DA, Ruscetti FW, Gallo R. 1976. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science 193: 1007–1008. [DOI] [PubMed] [Google Scholar]
- Murakami M, Sakamoto A, Bender J, Kappler J, Marrack P. 2002. CD25+CD4+ T cells contribute to the control of memory CD8+ T cells. Proc Natl Acad Sci 99: 8832–8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Russell SM, Mess SA, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard WJ. 1994. Heterodimerization of the IL-2 receptor β- and γ-chain cytoplasmic domains is required for signalling. Nature 369: 330–333. [DOI] [PubMed] [Google Scholar]
- Namen AE, Lupton S, Hjerrild K, Wignall J, Mochizuki DY, Schmierer A, Mosley B, March CJ, Urdal D, Gillis S. 1988. Stimulation of B-cell progenitors by cloned murine interleukin-7. Nature 333: 571–573. [DOI] [PubMed] [Google Scholar]
- Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. 1999. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu Rev Immunol 17: 701–738. [DOI] [PubMed] [Google Scholar]
- Nelson BH, Lord JD, Greenberg PD. 1994. Cytoplasmic domains of the interleukin-2 receptor β and γ chains mediate the signal for T-cell proliferation. Nature 369: 333–336. [DOI] [PubMed] [Google Scholar]
- Nikaido T, Shimizu A, Ishida N, Sabe H, Teshigawara K, Maeda M, Uchiyama T, Yodoi J, Honjo T. 1984. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature 311: 631–635. [DOI] [PubMed] [Google Scholar]
- Niu X, He D, Zhang X, Yue T, Li N, Zhang JZ, Dong C, Chen G. 2010. IL-21 regulates Th17 cells in rheumatoid arthritis. Hum Immunol 71: 334–341. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth N, Shultz LD, Brombacher F, Urban JF Jr, Gu H, Paul WE. 1997. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci 94: 10838–10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, Leonard WJ. 1993a. Interleukin-2 receptor γ chain: A functional component of the interleukin-7 receptor. Science 262: 1877–1880. [DOI] [PubMed] [Google Scholar]
- Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. 1993b. Interleukin-2 receptor γ chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73: 147–157. [DOI] [PubMed] [Google Scholar]
- Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. 1995. Defective lymphoid development in mice lacking Jak3. Science 270: 800–802. [DOI] [PubMed] [Google Scholar]
- Nowak EC, Weaver CT, Turner H, Begum-Haque S, Becher B, Schreiner B, Coyle AJ, Kasper LH, Noelle RJ. 2009. IL-9 as a mediator of Th17-driven inflammatory disease. J Exp Med 206: 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, et al. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448: 480–483. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, et al. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Mohn SE, Weinmann AS. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 13: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara J, Paul WE. 1987. Receptors for B-cell stimulatory factor-1 expressed on cells of haematopoietic lineage. Nature 325: 537–540. [DOI] [PubMed] [Google Scholar]
- Ohkura N, Kitagawa Y, Sakaguchi S. 2013. Development and maintenance of regulatory T cells. Immunity 38: 414–423. [DOI] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426: 671–676. [DOI] [PubMed] [Google Scholar]
- Orange JS, Roy-Ghanta S, Mace EM, Maru S, Rak GD, Sanborn KB, Fasth A, Saltzman R, Paisley A, Monaco-Shawver L, et al. 2011. IL-2 induces a WAVE2-dependent pathway for actin reorganization that enables WASp-independent human NK cell function. J Clin Invest 121: 1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Kikly K, Michalovich D, Young PR, Leonard WJ. 2000. Cloning of a type I cytokine receptor most related to the IL-2 receptor β chain. Proc Natl Acad Sci 97: 11439–11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC III, Liu C, Schwartzberg PL, Leonard WJ. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science 298: 1630–1634. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Ettinger R, Kim HP, Wang G, Qi CF, Hwu P, Shaffer DJ, Akilesh S, Roopenian DC, et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol 173: 5361–5371. [DOI] [PubMed] [Google Scholar]
- Pahwa R, Chatila T, Pahwa S, Paradise C, Day NK, Geha R, Schwartz SA, Slade H, Oyaizu N, Good RA. 1989. Recombinant interleukin 2 therapy in severe combined immunodeficiency disease. Proc Natl Acad Sci 86: 5069–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ho IC. 2004. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci 101: 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Ozaki K, Baumann H, Levin SD, Puel A, Farr AG, Ziegler SF, Leonard WJ, Lodish HF. 2000. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nat Immunol 1: 59–64. [DOI] [PubMed] [Google Scholar]
- Pandiyan P, Yang XP, Saravanamuthu SS, Zheng L, Ishihara S, O’Shea JJ, Lenardo MJ. 2012. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J Immunol 189: 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park LS, Martin U, Garka K, Gliniak B, Di Santo JP, Muller W, Largaespada DA, Copeland NG, Jenkins NA, Farr AG, et al. 2000. Cloning of the murine thymic stromal lymphopoietin (TSLP) receptor: Formation of a functional heteromeric complex requires interleukin 7 receptor. J Exp Med 192: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, West J, et al. 2000. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature 408: 57–63. [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J, Foster DC, Holly RD, Clegg CH. 2002. Interleukin-21 and the IL-21 receptor: Novel effectors of NK and T cell responses. J Leukoc Biol 72: 856–863. [PubMed] [Google Scholar]
- Paul WE. 2015. History of interleukin-4. Cytokine 75: 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene J, Gauchat JF, Lecart S, Drouet E, Guglielmi P, Boulay V, Delwail A, Foster D, Lecron JC, Yssel H. 2004. Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J Immunol 172: 5154–5157. [DOI] [PubMed] [Google Scholar]
- Perol L, Lindner JM, Caudana P, Nunez NG, Baeyens A, Valle A, Sedlik C, Loirat D, Boyer O, Creange A, et al. 2016. Loss of immune tolerance to IL-2 in type 1 diabetes. Nat Commun 7: 13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med 180: 1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit-Frere C, Dugas B, Braquet P, Mencia-Huerta JM. 1993. Interleukin-9 potentiates the interleukin-4-induced IgE and IgG1 release from murine B lymphocytes. Immunology 79: 146–151. [PMC free article] [PubMed] [Google Scholar]
- Pfeifle R, Rothe T, Ipseiz N, Scherer HU, Culemann S, Harre U, Ackermann JA, Seefried M, Kleyer A, Uderhardt S, et al. 2017. Regulation of autoantibody activity by the IL-23-TH17 axis determines the onset of autoimmune disease. Nat Immunol 18: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot C, Jin H, Awasthi A, Liu SM, Lai CY, Madan R, Sharpe AH, Karp CL, Miaw SC, Ho IC, et al. 2009. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol 183: 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Ziegler SF, Buckley RH, Leonard WJ. 1998. Defective IL7R expression in T−B+NK+ severe combined immunodeficiency. Nat Genet 20: 394–397. [DOI] [PubMed] [Google Scholar]
- Pulendran B. 2015. The varieties of immunological experience: Of pathogens, stress, and dendritic cells. Annu Rev Immunol 33: 563–606. [DOI] [PubMed] [Google Scholar]
- Pulliam SR, Uzhachenko RV, Adunyah SE, Shanker A. 2016. Common γ chain cytokines in combinatorial immune strategies against cancer. Immunol Lett 169: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, et al. 2012. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med 18: 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. 2016. T follicular helper cells in space-time. Nat Rev Immunol 16: 612–625. [DOI] [PubMed] [Google Scholar]
- Queen C, Schneider WP, Selick HE, Payne PW, Landolfi NF, Duncan JF, Avdalovic NM, Levitt M, Junghans RP, Waldmann TA. 1989. A humanized antibody that binds to the interleukin 2 receptor. Proc Natl Acad Sci 86: 10029–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle FW, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben SM, Cleveland JL, Pierce JH, Keegan AD, Nelms K, et al. 1995. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol 15: 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, et al. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 16: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, et al. 2017. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed MA, Latner DR, Aubert RD, Gourley T, Spolski R, Davis CW, Langley WA, Ha SJ, Ye L, Sarkar S, et al. 2013. Interleukin-21 is a critical cytokine for the generation of virus-specific long-lived plasma cells. J Virol 87: 7737–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TK, Andersen T, Hvid M, Hetland ML, Horslev-Petersen K, Stengaard-Pedersen K, Holm CK, Deleuran B. 2010. Increased interleukin 21 (IL-21) and IL-23 are associated with increased disease activity and with radiographic status in patients with early rheumatoid arthritis. J Rheumatol 37: 2014–2020. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Farkash EA, Gao W, Thompson CB. 2001. IL-7 enhances the survival and maintains the size of naïve T cells. J Immunol 167: 6869–6876. [DOI] [PubMed] [Google Scholar]
- Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, Laidlaw BJ, Araki K, Ahmed R, Kaech SM, et al. 2015. The interleukin-2-mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity 43: 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recher M, Berglund LJ, Avery DT, Cowan MJ, Gennery AR, Smart J, Peake J, Wong M, Pai SY, Baxi S, et al. 2011. IL-21 is the primary common γ chain-binding cytokine required for human B-cell differentiation in vivo. Blood 118: 6824–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. 2005. The structure of interleukin-2 complexed with its α receptor. Science 308: 1477–1480. [DOI] [PubMed] [Google Scholar]
- Rochman Y, Spolski R, Leonard WJ. 2009. New insights into the regulation of T cells by γc family cytokines. Nat Rev Immunol 9: 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolling C, Treton D, Pellegrini S, Galanaud P, Richard Y. 1996. IL4 and IL13 receptors share the γc chain and activate STAT6, STAT3 and STAT5 proteins in normal human B cells. FEBS Lett 393: 53–56. [DOI] [PubMed] [Google Scholar]
- Rosenberg SA. 2014. IL-2: The first effective immunotherapy for human cancer. J Immunol 192: 5451–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SH, Rollings C, Anderson KE, Hawkins PT, Stephens LR, Cantrell DA. 2016. Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8+ T cells. Immunity 45: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann MC, Miyajima A, Puri RK, Paul WE, et al. 1993. Interleukin-2 receptor γ chain: A functional component of the interleukin-4 receptor. Science 262: 1880–1883. [DOI] [PubMed] [Google Scholar]
- Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. 1994. Interaction of IL-2R β and γc chains with Jak1 and Jak3: Implications for XSCID and XCID. Science 266: 1042–1045. [DOI] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. 1995. Mutation of Jak3 in a patient with SCID: Essential role of Jak3 in lymphoid development. Science 270: 797–800. [DOI] [PubMed] [Google Scholar]
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. 1993. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75: 253–261. [DOI] [PubMed] [Google Scholar]
- Sage PT, Sharpe AH. 2015. T follicular regulatory cells in the regulation of B cell responses. Trends Immunol 36: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, Sharpe AH. 2016. T follicular regulatory cells. Immunol Rev 271: 246–259. [DOI] [PubMed] [Google Scholar]
- Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, et al. 2016. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat Immunol 17: 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikh KU, Khan AS, Kissner T, Ulrich RG. 2001. IL-15-induced conversion of monocytes to mature dendritic cells. Clin Exp Immunol 126: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Tada H, Sabe H, Honjo T. 1991. Biochemical evidence for a third chain of the interleukin-2 receptor. J Biol Chem 266: 22186–22191. [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. 1995. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor α-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 155: 1151–1164. [PubMed] [Google Scholar]
- Salzer E, Kansu A, Sic H, Majek P, Ikinciogullari A, Dogu FE, Prengemann NK, Santos-Valente E, Pickl WF, Bilic I, et al. 2014. Early-onset inflammatory bowel disease and common variable immunodeficiency-like disease caused by IL-21 deficiency. J Allergy Clin Immunol 133: 1651–1659 e1612. [DOI] [PubMed] [Google Scholar]
- Sandborn WJ, Su C, Sands BE, D’Haens GR, Vermeire S, Schreiber S, Danese S, Feagan BG, Reinisch W, Niezychowski W, et al. 2017. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N Engl J Med 376: 1723–1736. [DOI] [PubMed] [Google Scholar]
- Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, Wakeland EK, Li QZ, Wandstrat AE, Karp DR, et al. 2008. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis 67: 458–461. [DOI] [PubMed] [Google Scholar]
- Schlissel MS, Durum SD, Muegge K. 2000. The interleukin 7 receptor is required for T cell receptor γ locus accessibility to the V(D)J recombinase. J Exp Med 191: 1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns KS, Kieper WC, Jameson SC, Lefrancois L. 2000. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat Immunol 1: 426–432. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Van Brandwijk R, Van Snick J, Siebold B, Rude E. 1989. TCGF III/P40 is produced by naïve murine CD4+ T cells but is not a general T cell growth factor. Eur J Immunol 19: 2167–2170. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, Rude E. 1994. IL-9 production of naïve CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-β and IL-4, and is inhibited by IFN-γ. J Immunol 153: 3989–3996. [PubMed] [Google Scholar]
- Schorle H, Holtschke T, Hunig T, Schimpl A, Horak I. 1991. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature 352: 621–624. [DOI] [PubMed] [Google Scholar]
- Setoguchi R, Hori S, Takahashi T, Sakaguchi S. 2005. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med 201: 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, et al. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17: 51–62. [DOI] [PubMed] [Google Scholar]
- Sharon M, Klausner RD, Cullen BR, Chizzonite R, Leonard WJ. 1986. Novel interleukin-2 receptor subunit detected by cross-linking under high-affinity conditions. Science 234: 859–863. [DOI] [PubMed] [Google Scholar]
- Sheng W, Yang F, Zhou Y, Yang H, Low PY, Kemeny DM, Tan P, Moh A, Kaplan MH, Zhang Y, et al. 2014. STAT5 programs a distinct subset of GM-CSF-producing T helper cells that is essential for autoimmune neuroinflammation. Cell Res 24: 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Lin TH, Appell KC, Berg LJ. 2008. Janus-kinase-3-dependent signals induce chromatin remodeling at the Ifng locus during T helper 1 cell differentiation. Immunity 28: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M, Shintani Y, Shintani Y, Ishida H, Saba R, Yamaguchi A, Adachi H, Yashiro K, Suzuki K. 2016. Alternatively activated macrophages determine repair of the infarcted adult murine heart. J Clin Invest 126: 2151–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley M. 2017a. Daclizumab: A review in relapsing multiple sclerosis. Drugs 77: 447–458. [DOI] [PubMed] [Google Scholar]
- Shirley M. 2017b. Dupilumab: First global approval. Drugs 77: 1115–1121. [DOI] [PubMed] [Google Scholar]
- Siegel JP, Sharon M, Smith PL, Leonard WJ. 1987. The IL-2 receptor β chain (p70): Role in mediating signals for LAK, NK, and proliferative activities. Science 238: 75–78. [DOI] [PubMed] [Google Scholar]
- Smith GA, Uchida K, Weiss A, Taunton J. 2016. Essential biphasic role for JAK3 catalytic activity in IL-2 receptor signaling. Nat Chem Biol 12: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. 1999. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: A direct role for Stat5 in Bcl-XL induction. Cell 98: 181–191. [DOI] [PubMed] [Google Scholar]
- Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ. 2000. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol Cell Biol 20: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler JB, Tomala J, Luca VC, Jude KM, Dong S, Ring AM, Votavova P, Pepper M, Kovar M, Garcia KC. 2015. Antibodies to interleukin-2 elicit selective T cell subset potentiation through distinct conformational mechanisms. Immunity 42: 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Leonard WJ. 2014. Interleukin-21: A double-edged sword with therapeutic potential. Nat Rev Drug Discov 13: 379–395. [DOI] [PubMed] [Google Scholar]
- Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ. 2008. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc Natl Acad Sci 105: 14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Kim HP, Zhu W, Levy DE, Leonard WJ. 2009. IL-21 mediates suppressive effects via its induction of IL-10. J Immunol 182: 2859–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J, Surh CD. 2011. Normal T cell homeostasis: The conversion of naïve cells into memory-phenotype cells. Nat Immunol 12: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepensky P, Keller B, Abuzaitoun O, Shaag A, Yaacov B, Unger S, Seidl M, Rizzi M, Weintraub M, Elpeleg O, et al. 2015. Extending the clinical and immunological phenotype of human interleukin-21 receptor deficiency. Haematologica 100: e72–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su EW, Moore CJ, Suriano S, Johnson CB, Songalia N, Patterson A, Neitzke DJ, Andrijauskaite K, Garrett-Mayer E, Mehrotra S, et al. 2015. IL-2Rα mediates temporal regulation of IL-2 signaling and enhances immunotherapy. Sci Transl Med 7: 311ra170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Fueyo A, Bradley SJ, Klatzmann D, Tsokos GC. 2017. T cells and autoimmune kidney disease. Nat Rev Nephrol 13: 329–343. [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J. 2008. Homeostasis of naïve and memory T cells. Immunity 29: 848–862. [DOI] [PubMed] [Google Scholar]
- Suto A, Nakajima H, Hirose K, Suzuki K, Kagami S, Seto Y, Hoshimoto A, Saito Y, Foster DC, Iwamoto I. 2002. Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line Cɛ transcription of IL-4-stimulated B cells. Blood 100: 4565–4573. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. 1995. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor β. Science 268: 1472–1476. [DOI] [PubMed] [Google Scholar]
- Takeshita T, Asao H, Suzuki J, Sugamura K. 1990. An associated molecule, p64, with high-affinity interleukin 2 receptor. Int Immunol 2: 477–480. [DOI] [PubMed] [Google Scholar]
- Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, Tanaka N, Munakata H, Nakamura M, Sugamura K. 1992. Cloning of the γ chain of the human IL-2 receptor. Science 257: 379–382. [DOI] [PubMed] [Google Scholar]
- Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, Piccirillo CA, Salomon BL, Bluestone JA. 2008. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity 28: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye SG. 2015. Advances in IL-21 biology—Enhancing our understanding of human disease. Curr Opin Immunol 34: 107–115. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Matsui H, Fujita T, Takaoka C, Kashima N, Yoshimoto R, Hamuro J. 1983. Structure and expression of a cloned cDNA for human interleukin-2. Nature 302: 305–310. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Dowty ME, Wang L, Jussif J, Lin T, Li L, Moy E, Balbo P, Li W, Zhao Y, et al. 2016. Discovery of a JAK3-selective inhibitor: Functional differentiation of JAK3-selective inhibition over pan-JAK or JAK1-selective inhibition. ACS Chem Biol 11: 3442–3451. [DOI] [PubMed] [Google Scholar]
- Terashima A, Okamoto K, Nakashima T, Akira S, Ikuta K, Takayanagi H. 2016. Sepsis-induced osteoblast ablation causes immunodeficiency. Immunity 44: 1434–1443. [DOI] [PubMed] [Google Scholar]
- Teshigawara K, Wang HM, Kato K, Smith KA. 1987. Interleukin 2 high-affinity receptor expression requires two distinct binding proteins. J Exp Med 165: 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JM, Fallon GP, Matthews JD, Smith P, Jolin EH, McKenzie NA. 2000. IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity 13: 573–583. [DOI] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Rosenberg SA. 2017. “Final common pathway” of human cancer immunotherapy: Targeting random somatic mutations. Nat Immunol 18: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudo M, Kozak RW, Goldman CK, Waldmann TA. 1986. Demonstration of a non-Tac peptide that binds interleukin 2: A potential participant in a multichain interleukin 2 receptor complex. Proc Natl Acad Sci 83: 9694–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JE, Morrison PJ, Wilhelm C, Wilson M, Ahlfors H, Renauld JC, Panzer U, Helmby H, Stockinger B. 2013. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J Exp Med 210: 2951–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich BJ, Verdan FF, McKenzie AN, Kaplan MH, Olson MR. 2017. STAT3 activation impairs the stability of Th9 cells. J Immunol 198: 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttenhove C, Simpson RJ, Van Snick J. 1988. Functional and structural characterization of P40, a mouse glycoprotein with T-cell growth factor activity. Proc Natl Acad Sci 85: 6934–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. 2012. STATs shape the active enhancer landscape of T cell populations. Cell 151: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. 2013. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: Roles in homeostasis and disease. Annu Rev Immunol 31: 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang KB, Yang J, Mahmud SA, Burchill MA, Vegoe AL, Farrar MA. 2008. IL-2, -7, and -15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J Immunol 181: 3285–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegran F, Berger H, Boidot R, Mignot G, Bruchard M, Dosset M, Chalmin F, Rebe C, Derangere V, Ryffel B, et al. 2014. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat Immunol 15: 758–766. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. 2008. Transforming growth factor-β “reprograms” the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol 9: 1341–1346. [DOI] [PubMed] [Google Scholar]
- Venkatasubramanian S, Cheekatla S, Paidipally P, Tripathi D, Welch E, Tvinnereim AR, Nurieva R, Vankayalapati R. 2017. IL-21-dependent expansion of memory-like NK cells enhances protective immune responses against Mycobacterium tuberculosis. Mucosal Immunol 10: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeesch JR, Petit P, Kermouni A, Renauld JC, Van Den Berghe H, Marynen P. 1997. The IL-9 receptor gene, located in the Xq/Yq pseudoautosomal region, has an autosomal origin, escapes X inactivation and is expressed from the Y. Hum Mol Genet 6: 1–8. [DOI] [PubMed] [Google Scholar]
- Villarino AV, Stumhofer JS, Saris CJ, Kastelein RA, de Sauvage FJ, Hunter CA. 2006. IL-27 limits IL-2 production during Th1 differentiation. J Immunol 176: 237–247. [DOI] [PubMed] [Google Scholar]
- Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE Jr. 1996. DNA binding of in vitro activated Stat1 α, Stat1 β and truncated Stat1: Interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J 15: 5616–5626. [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier U, Moarefi I, Darnell JE Jr, Kuriyan J. 1998. Structure of the amino-terminal protein interaction domain of STAT-4. Science 279: 1048–1052. [DOI] [PubMed] [Google Scholar]
- Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331: 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med 181: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich CA, Ranson T, Samson SI, Corcuff E, Colucci F, Rosmaraki EE, Di Santo JP. 2005. Roles for common cytokine receptor γ-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J Immunol 174: 1213–1221. [DOI] [PubMed] [Google Scholar]
- Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, Corcuff E, Guy-Grand D, Rocha B, Cumano A, et al. 2006. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol 7: 1217–1224. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. 2006. The biology of interleukin-2 and interleukin-15: Implications for cancer therapy and vaccine design. Nat Rev Immunol 6: 595–601. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. 2015. The shared and contrasting roles of IL2 and IL15 in the life and death of normal and neoplastic lymphocytes: Implications for cancer therapy. Cancer Immunol Res 3: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, Sundrud MS, Unutmaz D. 2011. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J Exp Med 208: 1875–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CK, Oh J, Li P, West EE, Wong EA, Andraski AB, Spolski R, Yu ZX, He J, Kelsall BL, et al. 2013. The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity 38: 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CK, Andraski AB, Spolski R, Li P, Kazemian M, Oh J, Samsel L, Swanson PA II, McGavern DB, Sampaio EP, et al. 2015. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc Natl Acad Sci 112: 9394–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LM, Keegan AD, Li W, Lienhard GE, Pacini S, Gutkind JS, Myers MG Jr, Sun XJ, White MF, Aaronson SA, et al. 1993. Common elements in interleukin 4 and insulin signaling pathways in factor-dependent hematopoietic cells. Proc Natl Acad Sci 90: 4032–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. 2009. Structural biology of shared cytokine receptors. Annu Rev Immunol 27: 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu CR, Kim HP, Liao W, Telford WG, Egwuagu CE, Leonard WJ. 2011. Key role for IL-21 in experimental autoimmune uveitis. Proc Natl Acad Sci 108: 9542–9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb R, Merrill JT, Kelly JA, Sestak A, Kaufman KM, Langefeld CD, Ziegler J, Kimberly RP, Edberg JC, Ramsey-Goldman R, et al. 2009. A polymorphism within IL21R confers risk for systemic lupus erythematosus. Arthritis Rheum 60: 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg K, Parkman R. 1990. Severe combined immunodeficiency due to a specific defect in the production of interleukin-2. N Engl J Med 322: 1718–1723. [DOI] [PubMed] [Google Scholar]
- Weinstein JS, Herman EI, Lainez B, Licona-Limon P, Esplugues E, Flavell R, Craft J. 2016. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol 17: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, Wang L, Kirkesseli S, Rocklin R, Bock B, et al. 2013. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med 368: 2455–2466. [DOI] [PubMed] [Google Scholar]
- Wenzel S, Castro M, Corren J, Maspero J, Wang L, Zhang B, Pirozzi G, Sutherland ER, Evans RR, Joish VN, et al. 2016. Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: A randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet 388: 31–44. [DOI] [PubMed] [Google Scholar]
- West EE, Jin HT, Rasheed AU, Penaloza-Macmaster P, Ha SJ, Tan WG, Youngblood B, Freeman GJ, Smith KA, Ahmed R. 2013. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J Clin Invest 123: 2604–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C, Hirota K, Stieglitz B, Van Snick J, Tolaini M, Lahl K, Sparwasser T, Helmby H, Stockinger B. 2011. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat Immunol 12: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. 1995. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity 3: 521–530. [DOI] [PubMed] [Google Scholar]
- Winthrop KL. 2017. The emerging safety profile of JAK inhibitors in rheumatic disease. Nat Rev Rheumatol 13: 234–243. [DOI] [PubMed] [Google Scholar]
- Wirth JJ, Kierszenbaum F, Zlotnik A. 1989. Effects of IL-4 on macrophage functions: Increased uptake and killing of a protozoan parasite (Trypanosoma cruzi). Immunology 66: 296–301. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Kim HP, Xue HH, Liu H, Zhao K, Leonard WJ. 2005. Interleukin-21 receptor gene induction in human T cells is mediated by T-cell receptor-induced Sp1 activity. Mol Cell Biol 25: 9741–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SC, Edwan JH, Martin JF, Han S, Perry JS, Cartagena CM, Matsuura E, Maric D, Waldmann TA, Bielekova B. 2011. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med 17: 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Vannella KM. 2016. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44: 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn D, Kaufman M, Montalban X, Vollmer T, Simon J, Elkins J, O’Neill G, Neyer L, Sheridan J, Wang C, et al. 2010. Daclizumab in active relapsing multiple sclerosis (CHOICE study): A phase 2, randomised, double-blind, placebo-controlled, add-on trial with interferon β. Lancet Neurol 9: 381–390. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Sun YL, Hoey T. 1996. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science 273: 794–797. [DOI] [PubMed] [Google Scholar]
- Xu M, Liu M, Du X, Li S, Li H, Li X, Li Y, Wang Y, Qin Z, Fu YX, et al. 2015. Intratumoral delivery of IL-21 overcomes anti-Her2/Neu resistance through shifting tumor-associated macrophages from M2 to M1 phenotype. J Immunol 194: 4997–5006. [DOI] [PubMed] [Google Scholar]
- Xue HH, Kovanen PE, Pise-Masison CA, Berg M, Radovich MF, Brady JN, Leonard WJ. 2002. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. Proc Natl Acad Sci 99: 13759–13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 12: 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci 103: 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. 2007. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109: 4368–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SK, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, Honjo T, Ikuta K. 2001. The IL-7 receptor controls the accessibility of the TCRγ locus by Stat5 and histone acetylation. Immunity 15: 813–823. [DOI] [PubMed] [Google Scholar]
- Yin T, Keller SR, Quelle FW, Witthuhn BA, Tsang ML, Lienhard GE, Ihle JN, Yang YC. 1995. Interleukin-9 induces tyrosine phosphorylation of insulin receptor substrate-1 via JAK tyrosine kinases. J Biol Chem 270: 20497–20502. [DOI] [PubMed] [Google Scholar]
- Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. 2012. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 491: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31: 457–468. [DOI] [PubMed] [Google Scholar]
- Yu CR, Oh HM, Golestaneh N, Amadi-Obi A, Lee YS, Eseonu A, Mahdi RM, Egwuagu CE. 2011. Persistence of IL-2 expressing Th17 cells in healthy humans and experimental autoimmune uveitis. Eur J Immunol 41: 3495–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Snowhite I, Vendrame F, Rosenzwajg M, Klatzmann D, Pugliese A, Malek TR. 2015. Selective IL-2 responsiveness of regulatory T cells through multiple intrinsic mechanisms supports the use of low-dose IL-2 therapy in type 1 diabetes. Diabetes 64: 2172–2183. [DOI] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188: 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Finkelstein SE, Oh S, Kovanen PE, Hinrichs CS, Pise-Masison CA, Radonovich MF, Brady JN, Restifo NP, et al. 2005. Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J Exp Med 201: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R, Spolski R, Casas E, Zhu W, Levy DE, Leonard WJ. 2007. The molecular basis of IL-21-mediated proliferation. Blood 109: 4135–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. 2007. IL-2 is essential for TGF-β to convert naïve CD4+CD25− cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol 178: 2018–2027. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. 2007. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 8: 967–974. [DOI] [PubMed] [Google Scholar]
- Zhu J, Cote-Sierra J, Guo L, Paul WE. 2003. Stat5 activation plays a critical role in Th2 differentiation. Immunity 19: 739–748. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr, Guo L, Paul WE. 2004. Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat Immunol 5: 1157–1165. [DOI] [PubMed] [Google Scholar]