In this study, Zinoviev et al. investigated how translational GTPases (GTPBPs) function in mRNA surveillance and ribosome-associated quality control. They demonstrate that GTPBP1 possesses eEF1A-like elongation activity, delivering cognate aa-tRNA to the ribosomal A site in a GTP-dependent manner, and that GTPBP2's binding to GTP was stimulated by Phe-tRNAPhe, lacked elongation activity, and did not stimulate exosomal degradation. Their results indicate that GTPBP1 and GTPBP2 have different functions.
Keywords: GTPBP1, GTPBP2, trGTPase, translation elongation, 80S ribosome, RNA exosome
Abstract
GTP-binding protein 1 (GTPBP1) and GTPBP2 comprise a divergent group of translational GTPases with obscure functions, which are most closely related to eEF1A, eRF3, and Hbs1. Although recent reports implicated GTPBPs in mRNA surveillance and ribosome-associated quality control, how they perform these functions remains unknown. Here, we demonstrate that GTPBP1 possesses eEF1A-like elongation activity, delivering cognate aminoacyl-transfer RNA (aa-tRNA) to the ribosomal A site in a GTP-dependent manner. It also stimulates exosomal degradation of mRNAs in elongation complexes. The kinetics of GTPBP1-mediated elongation argues against its functioning in elongation per se but supports involvement in mRNA surveillance. Thus, GTP hydrolysis by GTPBP1 is not followed by rapid peptide bond formation, suggesting that after hydrolysis, GTPBP1 retains aa-tRNA, delaying its accommodation in the A site. In physiological settings, this would cause ribosome stalling, enabling GTPBP1 to elicit quality control programs; e.g., by recruiting the exosome. GTPBP1 can also deliver deacylated tRNA to the A site, indicating that it might function via interaction with deacylated tRNA, which accumulates during stresses. Although GTPBP2's binding to GTP was stimulated by Phe-tRNAPhe, suggesting that its function might also involve interaction with aa-tRNA, GTPBP2 lacked elongation activity and did not stimulate exosomal degradation, indicating that GTPBP1 and GTPBP2 have different functions.
Translational GTPases (trGTPases) are a superfamily of proteins that participate in all stages of translation. They belong to the P-loop superclass of GTPases containing a highly conserved GTPase (G) domain followed by two β-barrel domains and can be divided into four families: EF1, SelB, EF2, and IF2 (Leipe et al. 2002). The EF1 family is represented by bacterial and eukaryotic elongation factors EF-Tu and eEF1A (which deliver cognate aminoacyl-transfer RNAs [aa-tRNAs] to the A site of the ribosome during elongation), the eukaryotic release factor eRF3 (which promotes binding of eRF1 to the A site during termination), and the mRNA surveillance factor Hbs1 (which delivers to the A site a paralog of eRF1 called Pelota and is involved in “no-go” and “nonstop” mRNA decay) (Dever and Green 2012; Jackson et al. 2012; Maracci and Rodnina 2016). The SelB family contains bacterial and eukaryotic selenocysteine tRNA-specific elongation factors SelB and eEFSec and the γ subunit of eukaryotic initiation factor eIF2, which promotes binding of initiator tRNA to the P site of the ribosome (Jackson et al. 2010; Maracci and Rodnina 2016). The EF2 family is exemplified by the bacterial and eukaryotic elongation factors EF-G and eEF2 and the bacterial release factor RF3 (Dever and Green 2012; Maracci and Rodnina 2016). The last family, IF2, includes bacterial and eukaryotic initiation factors IF2 and eIF5B (Jackson et al. 2010; Maracci and Rodnina 2016).
GTP-binding proteins (GTPBPs) form a relatively divergent group within the SelB family, which is also closely related to eEF1A, eRF3, and Hbs1 (Atkinson 2015). GTPBPs exist in all major groups of eukaryotes and are widespread in archaea. Many eukaryotes encode two copies of GTPBPs: GTPBP1 and GTPBP2 (Senju and Nishimura 1997; Kudo et al. 2000; Senju et al. 2000; Watanabe et al. 2000; Girardot et al. 2004). Human GTPBP1 and GTPBP2 share 68% sequence similarity and have 39%–41% similarity with eEF1A, eRF3, and Hbs1, with the strongest conservation in the G1–G5 boxes and the switch I and II regions in the G domain (Supplemental Fig. S1). The greatest sequence variability occurs within the GTPBPs’ specific N-terminal and C-terminal extensions.
The trGTPases that are most closely related to GTPBPs (eEF1A, eEFSec, eRF3, and Hbs1) have a common function of delivering their binding partners to the A site in a GTP-dependent manner. These GTPases and their partners bind to the A site in the form of a ternary complex with GTP. Binding to the ribosome triggers GTP hydrolysis, after which the GTPase dissociates, and the partner accommodates in the A site. Thus, the ability to bind the partner is determined by the nucleotide-bound state of the GTPase, and, in turn, association with the partner affects the nucleotide-binding affinity of the GTPase. The ribosome and the binding partner together act as a composite GTPase-activating complex. Subsequent GDP-to-GTP exchange either occurs spontaneously (in the case of SelB, eRF3, and Hbs1) or requires a specific guanine nucleotide exchange factor (GEF; in the case of eEF1A) (Pisareva et al. 2006; Gromadski et al. 2007; Graille et al. 2008).
Despite their widespread distribution, the function of GTPBPs remains obscure. Initial studies focused on their tissue specificity and the effect of their knockout in mice. The mRNA levels for GTPBP1 and GTPBP2 vary in mouse tissues. GTPBP1 mRNA is most abundant in the brain (Senju and Nishimura 1997), whereas the highest levels of GTPBP2 mRNA occur in the testes and thymus (Watanabe et al. 2000). The levels of both mRNAs are elevated after treatment of macrophages with IFN-γ, suggesting a role for GTPBPs in innate immunity (Senju and Nishimura 1997; Kudo et al. 2000). However, knockout mice lacking GTPBP1 or GTPBP2 do not show any phenotypic alterations (Senju et al. 2000; Ishimura et al. 2014). Two recent studies implicated these proteins in mRNA surveillance and ribosome-associated quality control mechanisms. In one of them, rat GTPBP1 was found in an RNA-destabilizing protein complex bound to the 3′ untranslated region (UTR) of the mRNA encoding Aanat, an enzyme involved in melatonin synthesis in the pineal gland (Woo et al. 2011). GTPBP1 did not interact with the mRNA directly but mainly via the RNA exosome, a multisubunit protein complex that has endoribonuclease and (3′ → 5′) exoribonuclease activity and participates in various RNA processing and degradation pathways (Zinder and Lima 2017). GTPBP1 knockout mice had higher levels of Aanat mRNA. This prompted a suggestion that GTPBP1 might direct the exosome toward mRNA targets, although no mechanism for this activity was proposed. In the second study, GTPBP2 was reported to interact with Pelota and was suggested to be involved in disassembly of ribosomes that are stalled due to the presence of nonfunctional tRNA (Ishimura et al. 2014). Thus, disruption of GTPBP2 in combination with a defect in expression of the main tRNAArg-UCU isodecoder in the mouse brain resulted in neurodegeneration, which was linked to excessive ribosome pausing at AGA codons. Ribosome stalling in the brains of these mice also led to activation of the eIF2α-specific GCN2 kinase prior to the onset of neurodegeneration (Ishimura et al. 2016). GTPBP2 mutations have been linked to neurodegeneration in humans as well (Jaberi et al. 2016). However, in Xenopus embryos, GTPBP2 was reported to bind to the transcription modulator SMAD1 and take part in BMP/SMAD1 signaling (Kirmizitas et al. 2014) as well as interact with Axin and act as a positive regulator of Wnt signaling (Gillis et al. 2016).
Until now, no biochemical data have been reported that could shed light on the mechanisms of action of GTPBP1 and GTPBP2. Their affinity to guanine nucleotides has not been determined, and it is not known whether they are active GTPases and, if so, what the mechanism of their activation is. It is also unknown whether they have binding partners that fulfill a role similar to that played by aa-tRNA, eRF1, and Pelota for eEF1A, eRF3, and Hbs1, respectively. Here, we applied integrated biochemical, kinetic, and in vitro reconstitution approaches to elucidate the functions of mammalian GTPBPs.
Results
Preparation of GTPBP1 and GTPBP2
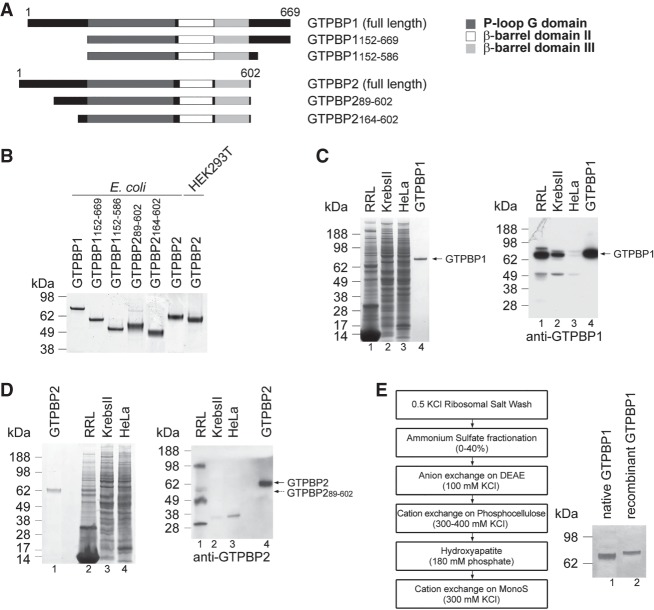
His-tagged full-length human GTPBP1 and GTPBP2; truncated GTPBP1152–669, GTPBP1152–586, and GTPBP2164–602 lacking the N-terminal and C-terminal extensions; and GTPBP289–602 corresponding to the translation product that initiates at a downstream AUG codon (suggested to be the active form of the protein) (Fig. 1A; Kirmizitas et al. 2014) were expressed in Escherichia coli (Fig. 1B). Flag-tagged full-length GTPBP2 was expressed in HEK293T cells (Fig. 1B). To obtain native GTPBPs, their presence in rabbit reticulocyte lysate (RRL), Krebs II, and HeLa cell extracts was assayed by Western blotting. Full-length GTPBP1 was relatively abundant in RRL (Fig. 1C), but full-length GTPBP2 was not detected in any lysate, and the amount of the protein in RRL that may correspond to its shorter form was very low (Fig. 1D). Therefore, we could purify only native GTPBP1 (Fig. 1E).
Figure 1.
Purification of GTPBP1 and GTPBP2. (A) Domain organization of full-length and truncated GTPBP1 and GTPBP2. (B) Corresponding purified recombinant proteins, analyzed by SDS-PAGE. (C,D) The presence of GTPBP1 (C) and GTPBP2 (D) in RRL, Krebs II, and HeLa cell extracts, analyzed by SDS-PAGE followed by SimplyBlue staining (left panels) or Western blotting (right panels). (E, left panel) The scheme of GTPBP1 purification from RRL. (Right panel) Purified native and His-tagged recombinant GTPBP1 resolved by SDS-PAGE. Recombinant GTPBP1 migrates more slowly than the native factor due to the presence of the N-terminal His6 tag.
Interaction of GTPBP1 with guanine nucleotides
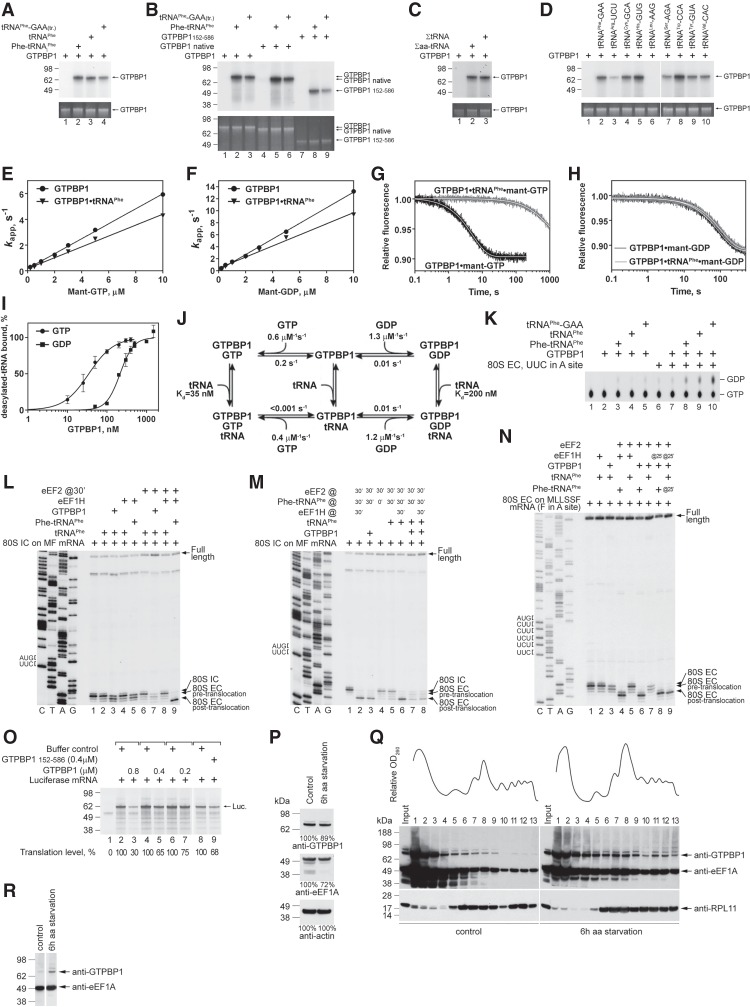
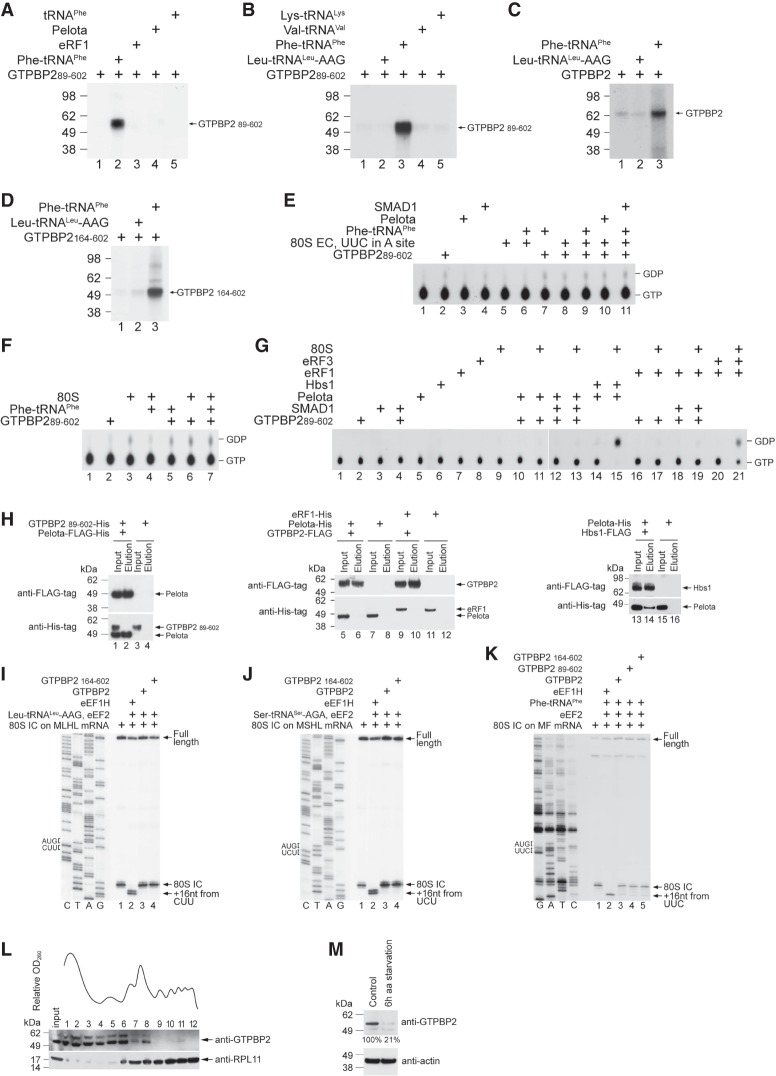
To identify the functional partner of GTPBP1, we tested how aa-tRNA, eRF1, or Pelota affects its GTP-binding activity using a UV cross-linking assay. The efficiency of GTP cross-linking with recombinant GTPBP1 alone was low but was strongly increased by aminoacylated initiator and more so by elongator tRNAs (e.g., in vitro transcribed Leu-tRNALeu-AAG and native yeast Phe-tRNAPhe) (Fig. 2A,B, lane 2). Strong stimulation of GTP cross-linking by aa-tRNA was also observed for native GTPBP1 and truncation mutants (Fig. 2C,D). GTP cross-linking was not enhanced by eRF1 or Pelota (Fig. 2B, lanes 3,4), and no direct interaction between GTPBP1 and eRF1 or Pelota was observed in pull-down assays (data not shown).
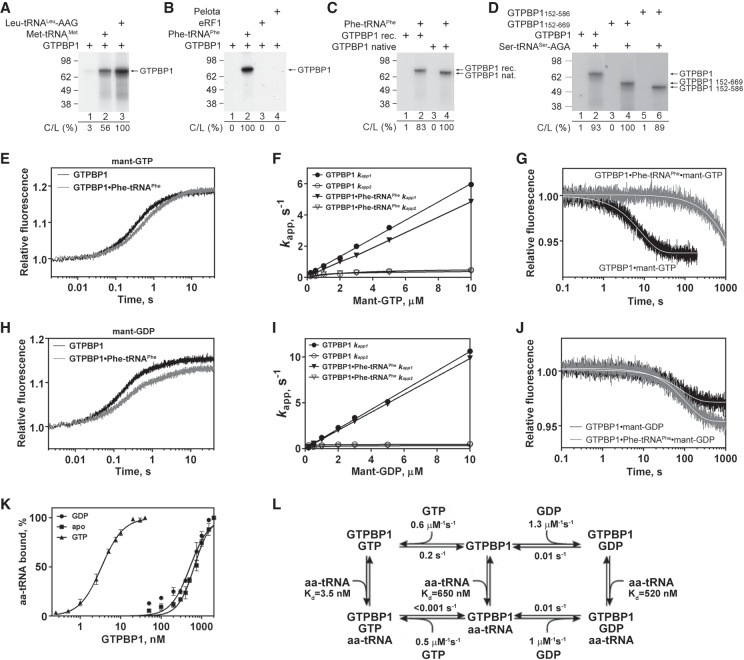
Figure 2.
Interaction of GTPBP1 with guanine nucleotides. (A–D) UV cross-linking of different forms of GTPBP1 to [α-32P]GTP depending on the presence of aa-tRNAs, Pelota, and eRF1, as indicated. Cross-linked proteins were analyzed by SDS-PAGE followed by autoradiography. The efficiency of cross-linking (C/L) was quantified by phosphorimager and normalized to the condition with the highest cross-linking. (E–J) Binding kinetics of methylanthraniloyl GTP (mant-GTP) and mant-GDP to GTPBP1, monitored by stopped-flow. (E,H) Time courses of the association of 5 µM mant-GTP (E) and 5 µM mant-GDP (H) with 50 nM GTPBP1 in the absence or presence of 150 nM Phe-tRNAPhe. (F,I) Concentration dependence of apparent rates kapp1 and kapp2 of mant-GTP (F) and mant-GDP (I) binding to GTPBP1 in the absence or presence of Phe-tRNAPhe. (G,J) Time courses of the dissociation of mant-GTP (G) or mant-GDP (J) from GTPBP1 alone or in the presence of Phe-tRNAPhe upon chase with 500 µM GTP or 500 µM GDP, respectively. (K) Association of GTPBP1 with [35S]Cys-tRNACys depending on the presence of guanine nucleotides, assayed by filter binding. (L) Kinetic scheme of the interaction of GTPBP1 with guanine nucleotides and aa-tRNA.
To investigate the kinetics of guanine nucleotides’ binding to GTPBP1, we used the rapid-mixing fluorescence stopped-flow technique using GTP/GDP derivatives carrying the fluorescent methylanthraniloyl group (mant-group). Fluorescence changes were monitored via fluorescence resonance energy transfer (FRET) between tryptophan residues of GTPBP1 and the mant-group (Supplemental Fig. S2). The kinetics of nucleotide binding was measured under pseudo-first-order conditions by mixing 50 nM GTPBP1 with increasing concentrations of mant-GTP or mant-GDP (0.2–10 µM) in the presence or absence of 0.3 µM Phe-tRNAPhe. Time courses were described best by a two-exponential fitting (Fig. 2E,H), yielding the apparent rate constants kapp1 and kapp2. In all cases, kapp1 values increased linearly with mant-GTP and mant-GDP concentrations, indicative of a bimolecular binding reaction (Fig. 2F,I). The association rate constants (k1) (Table 1; Fig. 2L) were calculated as the slope of the linear fit of kapp1 dependence on nucleotide concentration (Fig. 2F,I). The kapp2 values were low and saturated with increasing nucleotide concentrations.
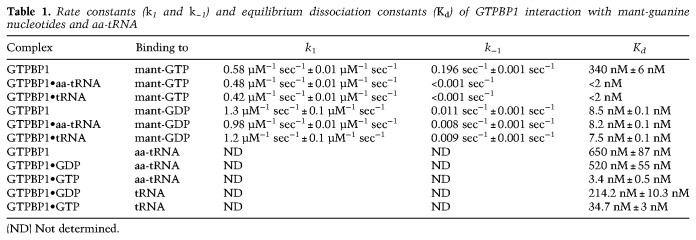
Table 1.
Rate constants (k1 and k−1) and equilibrium dissociation constants (Kd) of GTPBP1 interaction with mant-guanine nucleotides and aa-tRNA
Dissociation rate constants (k−1) were determined from displacement experiments in which GTPBP1•mant-nucleotide complexes prepared in the presence or absence of aa-tRNA were rapidly mixed with a 100-fold molar excess of unlabeled nucleotide over mant-nucleotides. The time courses of nucleotide dissociation were evaluated by single-exponential fitting (Fig. 2G,J), yielding the k−1 value (Table 1; Fig. 2L). Because the second phase appearing during binding was not observed in the displacement experiments, we concluded that it is not part of the main reaction pathway and did not consider it further. The slow downward phase observed upon chasing mant-GTP from GTPBP1 in the presence of aa-tRNA was also observed in the absence of excess unlabeled nucleotide and was therefore due to photobleaching of the mant-group. Hence, we were unable to calculate the precise dissociation rate constant of mant-GTP from the complex and could only estimate the upper limit for the value <0.001 sec−1. The equilibrium dissociation constant (Kd) in each case was calculated from the k−1 to k1 ratio (Table 1; Fig. 2L).
The affinity of GTPBP1 alone for mant-GTP (Kd ∼ 350 nM) was 40-fold lower than for mant-GDP (Kd ∼ 8 nM), which is mainly due to the higher dissociation rate (k−1 ∼ 0.2 sec−1 for mant-GTP vs. k−1 ∼ 0.01 sec−1 for mant-GDP). Addition of aa-tRNA stabilized the binding of GTPBP1 to mant-GTP by over two orders of magnitude (to Kd < 2 nM) by lowering of the dissociation rate constant (k−1 < 0.001 sec−1) but did not influence its affinity for mant-GDP.
The interaction of GTPBP1 with aa-tRNA was studied by filter binding using [35S]Cys-tRNACys (Fig. 2K). The affinity of GTPBP1•GTP for aa-tRNA was high (Kd ∼ 3 nM), similar to that of yeast eEF1A (Gromadski et al. 2007), whereas the affinity of GTPBP1 in the presence of GDP or in the absence of nucleotides was more than two orders of magnitude lower (Kd ∼ 500–650 nM) (Table 1; Fig. 2L).
The GTPase activity of GTPBP1
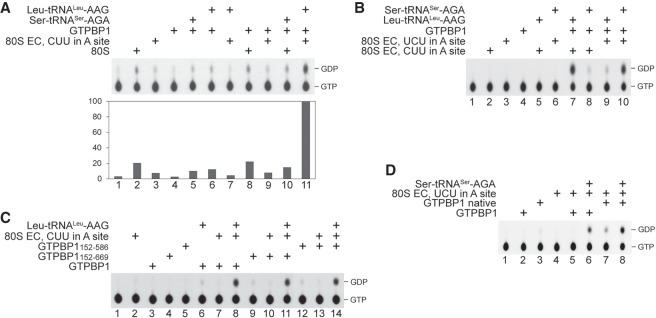
The extremely low intrinsic GTPase activity of GTPBP1 was slightly stimulated by aa-tRNA (Fig. 3A, lanes 4–6). 80S ribosomes alone did not stimulate GTP hydrolysis by GTPBP1 and did not enhance the effect of aa-tRNA (Fig. 3A, cf. lanes 2, 8, and 10). GTP hydrolysis was also not stimulated by elongation complexes (ECs) alone (Fig. 3A, lane 7) or when they were added with noncognate aa-tRNA (Fig. 3B, lanes 8,9). Substantial GTPase activity of GTPBP1 was induced only by the simultaneous presence of ECs and cognate aa-tRNA (Fig. 3A [lane 11], B [lanes 7 and 10]). Recombinant and native GTPBP1 and the truncation mutants required the same conditions for activation (Fig. 3C,D).
Figure 3.
The GTPase activity of GTPBP1. (A–D) GTP hydrolysis by different forms of GTPBP1 in the presence or absence of various combinations of 80S ribosomes, ECs, and aa-tRNAs, assayed by thin-layer chromatography (TLC) and autoradiography. (A) The efficiency of GTP hydrolysis was quantified by phosphorimager and normalized to the condition with the highest GTP hydrolysis.
The elongation activity of GTPBP1
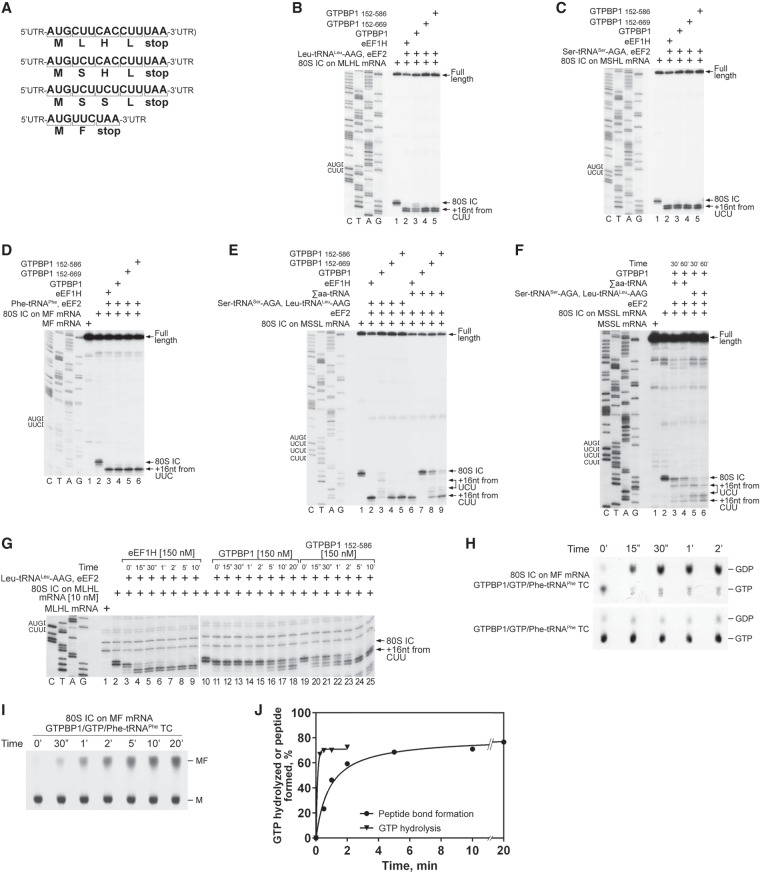
The ability of GTPBP1 to form GTPBP1•GTP•aa-tRNA ternary complexes and the stimulation of its GTPase activity by the presence of ECs and cognate aa-tRNAs suggested that GTPBP1 might have an eEF1A-like elongation activity. We tested this possibility using the in vitro reconstitution approach. Ribosome complexes were assembled from individual translational components on derivatives of β-globin mRNA containing a short ORF followed by a UAA stop codon (Fig. 4A), and the positions of ribosomes on mRNA were mapped by toeprinting.
Figure 4.
The elongation activity of GTPBP1. (A) Schematic representation of MLHL-STOP, MSHL-STOP, MSSL-STOP, and MF-STOP mRNAs. (B–D) Toeprinting analysis of the activity of eEF1H and different forms of GTPBP1 in one-cycle elongation on MLHL-STOP, MSHL-STOP, and MF-STOP mRNAs with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native yeast Phe-tRNAPhe, respectively. (E,F) The activity of eEF1H and different forms of GTPBP1 in three-cycle elongation on MSSL-STOP mRNA with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native total tRNA (Σaa-tRNAs), assayed by toeprinting. (G) Time courses of one-cycle elongation by eEF1H, full-length GTPBP1, and GTPBP1152–586 on MLHL-STOP mRNA with cognate in vitro transcribed Leu-tRNALeu-AAG, assayed by toeprinting. (B–G) The positions of 80S initiation complexes (ICs) and ECs are indicated by arrows at the right. Lanes C, T, A, and G depict the corresponding DNA sequences. (H–J) Time courses of GTP hydrolysis and peptide bond formation during GTPBP1-mediated elongation on MF mRNA. Reaction mixtures containing 100 nM purified 80S ICs formed on MF mRNA with [35S]Met-tRNAiMet and 100 nM purified GTPBP1•[α-32P]GTP•Phe-tRNAPhe ternary complexes were incubated at 37°C. Aliquots were removed at different time points, and GTP hydrolysis (H) and formation of the [35S]MF dipeptide (I) were analyzed by TLC. (J) The efficiency of GTP hydrolysis and formation of the peptide bond were quantified by phosphorimager and calculated taking into account the activity of the 80S IC preparations (see Supplemental Fig. S4C,D).
GTPBP1 and its truncation mutants promoted one-cycle elongation on all tested codons in the presence of cognate in vitro transcribed or native aa-tRNAs [shown for CUU(L), UCU(S), and UUC(F) codons in Fig. 4B–D]. The activities of full-length recombinant and native proteins were similar (Supplemental Fig. S3A) but were lower than those of the truncated variants, which was particularly evident on the CUU(L) codon (Fig. 4B, cf. lanes 3 and 4,5). The difference between the full-length and truncated GTPBP1 was even higher during three-cycle elongation even though we used cognate in vitro transcribed aa-tRNAs and excess GTPBP1 so that nucleotide exchange on GTPBP1 should not be a limiting factor (Fig. 4E, cf. lanes 3 and 4,5). When cognate tRNAs were replaced by native total tRNA (Σaa-tRNAs), the activity of the N-terminally and C-terminally truncated GTPBP1152–586 fell substantially below that of eEF1H, and the activity of the N-terminally truncated GTPBP1152–669 was reduced even further (Fig. 4E, lanes 6,8,9). Almost no elongation with Σaa-tRNAs was observed for the full-length protein (Fig. 4E, lane 7), indicating that, in contrast to eEF1H, GTPBP1 was not able to sort efficiently through excess noncognate and near-cognate aa-tRNAs. The efficiency of three-cycle elongation by the full-length GTPBP1 was increased by extending incubation from 10 min to 30 or 60 min (Fig. 4, cf. E and F). We also tested whether GTPBP1's activity can be stimulated by eEF1B, the GEF for eEF1A. eEF1B (Supplemental Fig. S3B) did not bind to GTPBP1 (Supplemental Fig. S3C) and did not influence its GTP-binding or elongation activities (Supplemental Fig. S3D–F).
The rate of elongation was quite different with eEF1H and GTPBP1. One-cycle elongation on the CUU(L) codon was completed within 15 sec in the presence of eEF1H, compared with 2 and 20 min in the presence of GTPBP1152–586 and full-length GTPBP1, respectively (Fig. 4G). Similar relative activities of eEF1H and different forms of GTPBP1 (eEF1H > GTPBP1152–586 > GTPBP1full–length) were observed on the UUC(F) and GUG(V) codons (data not shown).
To study why the elongation by GTPBP1 is so slow, we compared the rates of GTP hydrolysis and peptide bond formation during one-cycle elongation on the UUC(F) codon. Purified 80S initiation complexes (ICs) formed on MF mRNA were incubated with equimolar amounts of GTPBP1•GTP•Phe-tRNAPhe ternary complexes, and GTP hydrolysis and synthesis of the MF dipeptide were analyzed by thin-layer chromatography (TLC). Whereas GTP hydrolysis was nearly complete within 15 sec (Fig. 4H,J), formation of dipeptides required several minutes (Fig. 4I,J), in accordance with the results of the toeprinting experiments (Fig. 4G). Dipeptide synthesis was strictly GTPBP1- and GTP-dependent and did not occur by nonenzymatic loading of Phe-tRNAPhe to the A site (Supplemental Fig. S4A) or when GTP was replaced by GMPPNP (Supplemental Fig. S4B). Thus, the rate-limiting step was downstream from GTP hydrolysis and may involve slow Pi release and/or inefficient dissociation of GTPBP1 from aa-tRNA on the ribosome.
Taken together, our results indicate that, like eEF1A, GTPBP1 can deliver cognate aa-tRNA to the A site. However, the elongation activity of GTPBP1 is much lower than that of eEF1A and is negatively regulated by its N-terminal and C-terminal extensions.
Functional interaction of GTPBP1 with deacylated tRNA
Strikingly, cross-linking of GTP to full-length native or recombinant GTPBP1 and to GTPBP1152–586 was also stimulated by deacylated native yeast or human in vitro transcribed tRNAPhe (Fig. 5A,B). Other tested deacylated in vitro transcribed tRNAs (except for tRNALeu) and native ΣtRNAs showed similar stimulation (Fig. 5C,D). Notably, the elongation activity of GTPBP1 was also lower with the in vitro transcribed Leu-tRNALeu (Fig. 4B). tRNALeu possesses a characteristic long variable loop, which could potentially affect the interaction of tRNALeu with GTPBP1, particularly if the folding of the loop is compromised by the lack of modification in the in vitro transcribed tRNA. Regarding the fact that in vitro transcribed and native tRNALeu may be functionally somewhat different (Pisarev et al. 2010), we cannot exclude that native tRNALeu could also stimulate cross-linking of GTP to GTPBP1. The affinity of guanine nucleotides to GTPBP1 in the presence of yeast tRNAPhe was measured using rapid kinetics as described above (Fig. 5E–H). The presence of tRNAPhe significantly increased the affinity of mant-GTP to GTPBP1 (Kd < 2 nM) by lowering the dissociation rate constant of GTP (k−1 < 0.001 sec−1) (Fig. 5J), similar to the effect observed with Phe-tRNAPhe (Table 1; Fig. 2). On the other hand, the binding and dissociation kinetics and, as a consequence, the affinity of GTPBP1 to mant-GDP remained largely independent of tRNAPhe (Fig. 5J). To confirm that the interaction with GTPBP1 was not exclusive to yeast tRNAPhe, we tested the dissociation of mant-GTP from GTPBP1 in the presence of equimolar amounts of deacylated human ΣtRNAs to ensure that all individual tRNA species in the mixture were bound (Supplemental Fig. S5A). Under these conditions, 80% of GTPBP1 was bound tightly to mant-GTP (k−2 < 0.002 sec−1), confirming that binding to most (if not all) human deacylated tRNA species increases the affinity of GTPBP1 to mant-GTP in a similar manner. The remaining 20% of GTPBP1 displayed a faster dissociation of mant-GTP (k−1 ∼ 0.12 sec−1), suggesting that this portion remained unbound by tRNA. The interaction of GTPBP1 with deacylated tRNA was studied by filter binding using in vitro transcribed [32P]tRNAPhe (Fig. 5I). The affinity of GTPBP1•GTP for deacylated tRNA (Kd ∼ 35 nM) was again much higher than the affinity of GTPBP1 in the presence of GDP (Kd ∼ 214 nM) (Fig. 5J; Table 1).
Figure 5.
The activities of GTPBP1 in the presence of deacylated tRNA. (A–D) UV cross-linking of different forms of GTPBP1 to [α-32P]GTP in the presence or absence of aminoacylated and deacylated tRNAs, as indicated. Cross-linked proteins were analyzed by SDS-PAGE followed by autoradiography. (Bottom panels) To ensure equal loading, GTPBP1 levels were analyzed by fluorescent Sypro staining. (E–H) Kinetics of binding of mant-GTP and mant-GDP to GTPBP1. (E,F) Concentration dependence of kapp1 of mant-GTP (E) and mant-GDP (F) binding to GTPBP1 in the presence or absence of tRNAPhe (data in the absence of tRNAPhe are the same as in Fig. 2F,I). (G,H) Time courses of the dissociation of mant-GTP (G) or mant-GDP (H) from GTPBP1 alone or in the presence of tRNAPhe upon chase with 500 µM GTP or 500 µM GDP, respectively. (I) Association of GTPBP1 with [32P]tRNAPhe depending on the presence of guanine nucleotides, assayed by filter binding. (J) The kinetic scheme of the interaction of GTPBP1 with guanine nucleotides and deacylated tRNA. (K) GTP hydrolysis by full-length GTPBP1 in the presence or absence of combinations of ECs and aminoacylated/deacylated tRNAs, assayed by TLC and autoradiography. (L–N) Toeprinting analysis of the activity of GTPBP1 and eEF1H in one-cycle elongation on MF-STOP (L,M) and MLLSSF-STOP (N) mRNAs with native yeast aminoacylated and deacylated tRNAPhe, as indicated. The positions of 80S ICs and pretranslocated and translocated ECs are indicated at the right. Lanes C, T, A, and G depict the corresponding DNA sequences. (O) The influence of increasing amounts of GTPBP1 on in vitro translation of uncapped luciferase mRNA in RRL. Translation products were quantified by autoradiography relative to those synthesized in the absence of GTPBP1 but in the presence of an equivalent volume of buffer (defined as 100%). Equal loading was confirmed by SimplyBlue staining (not shown). (P) The abundance of GTPBP1 and eEF1A in HEK293 cells under normal growth conditions and following 6 h of amino acid starvation, assayed by Western blotting. (Q) Polysome profiles and ribosomal association of GTPBP1 and eEF1A in HEK293 cells under normal growth conditions and following 6 h of amino acid starvation, assayed by Western blotting. (R) Relative amounts of GTPBP1 and eEF1A associated with ribosomes in HEK293 cells under normal growth conditions and following 6 h of amino acid starvation. Polysomal fractions #12 were normalized with respect to eEF1A levels, assayed by Western blotting.
In the presence of cognate ECs, tRNAPhe also stimulated the GTPase activity of GTPBP1 (Fig. 5K). Stimulation did not occur with tRNALeu (Supplemental Fig. S5B), which did not promote GTP binding (Fig. 5D). To investigate whether GTP hydrolysis is accompanied by accommodation of deacylated tRNA in the A site, we used the toeprinting technique. Incubation of 80S ICs assembled on MF mRNA with Phe-tRNAPhe and eEF1H yielded pretranslocation complexes with the characteristic +1 nucleotide (nt) shift of the most prominent toeprint (Fig. 5L, cf. lanes 1 and 5). A similar shift was observed after incubation of ICs with tRNAPhe and GTPBP1 but not with tRNAPhe and eEF1H (Fig. 5L, lanes 3,4). These results suggest that, in contrast to eEF1H, GTPBP1 can deliver tRNAPhe to the A site. Addition of eEF2 to ribosome complexes with deacylated tRNA in the A site led to the appearance of a faint toeprint that would be consistent with the formation of a small amount of a translocated complex and prominent full-length cDNA (Fig. 5L, cf. lanes 3 and 7), indicating destabilization of ribosomal complexes. Furthermore, preincubation of 80S ICs with GTPBP1 and tRNAPhe inhibited canonical elongation when Phe-tRNAPhe, eEF1H, and eEF2 were added (Fig. 5M, cf. lanes 6 and 8). GTPBP1 and deacylated tRNAPhe behaved similarly on ECs assembled on MSSLLF mRNA that contained MLLSSF-tRNALeu in the P site and a UUC (F) codon in the A site (Fig. 5N). The effect was codon–anti-codon-specific, and preincubation with GTPBP1 and tRNAPhe of 80S ICs formed on MLHL mRNA did not inhibit elongation (Supplemental Fig. S5C). In contrast to GTPBP1, preincubation of 80S ICs assembled on MF mRNA with tRNAPhe and mammalian eEF1H or yeast eEF1A did not affect elongation (Supplemental Fig. S5D). In RRL, GTPBP1 inhibited translation of luciferase mRNA in a dose-dependent manner (Fig. 5O). Although several possible explanations could be envisioned, including severe slowing down of protein synthesis if GTPBP1 becomes able to compete with eEF1A for aa-tRNA or GTPBP1-mediated dissociation of potentially stalled ECs, the most important conclusion from this experiment is that it argues against GTPBP1 functioning as a bona fide elongation factor.
In light of GTPBP1's unique ability to deliver deacylated tRNAs to the A site, we compared polysomal association of GTPBP1 and eEF1A in HEK293 cells under normal and amino acid starvation conditions. The cellular level of eEF1A fell by ∼20%–25%, while the abundance of GTPBP1 decreased marginally after 6 h of amino acid starvation (Fig. 5P). A similar degree of eEF1A depletion occurred during nitrogen starvation in yeast (Kelly and Bedwell 2015). Under normal growth conditions, very little GTPBP1 was bound to polysomes (Fig. 5Q, left panel). Amino acid starvation enhanced polysomal association of both GTPBP1 and eEF1A (Fig. 5Q, right panel), but normalization of polysomal fractions to eEF1A levels revealed that polysomal association of GTPBP1 increased more than that of eEF1A (Fig. 5R).
The activity of GTPBP2
To identify the potential function of GTPBP2, we used the same approach as for GTPBP1. Testing GTP binding to GTPBP1 by UV cross-linking and rapid kinetic approaches yielded qualitatively similar results, validating UV cross-linking as a measure of GTP-binding activity in the experiments described below. GTPBP289–602 alone had a very low GTP-binding activity that was stimulated by native yeast Phe-tRNAPhe (Fig. 6A, lane 2) but not by deacylated tRNAPhe, eRF1, Pelota, or other tested aa-tRNAs, including in vitro transcribed Leu-tRNALeu-AAG as well as native yeast Lys-tRNALys and Val-tRNAVal (Fig. 6A [lanes 3–5], B [lanes 2,4,5]). The full-length GTPBP2 and the N-terminally truncated GTPBP2164–602 showed the same specificity (Fig. 6C,D). However, the GTPase activity of GTPBP2 was not stimulated by Phe-tRNAPhe and cognate ECs (shown for GTPBP289–602 in Fig. 6E). It was also not stimulated by ECs and Pelota (Fig. 6E) or vacant 80S ribosomes with or without Phe-tRNAPhe, Pelota, or eRF1 (shown for GTPBP289–602 in Fig. 6F,G). We also did not observe the reported interaction of GTPBP2 with Pelota (Ishimura et al. 2014) in Flag pull-down assays (Fig. 6H, left and middle panels). In control experiments, Pelota efficiently bound to Hbs1 (Fig. 6H, right panel), and the addition of 80S ribosomes stimulated its GTPase activity (Fig. 6G).
Figure 6.
The activities of GTPBP2. (A–D) UV cross-linking of the full-length and N-terminally truncated GTPBP2 to [α-32P]GTP in the presence or absence of deacylated and aminoacylated tRNAs, Pelota, and eRF1, as indicated. Cross-linked proteins were resolved by SDS-PAGE followed by autoradiography. (E–G) TLC analysis of GTP hydrolysis by GTPBP289–602 in the presence or absence of the indicated combinations of 80S ribosomes, ECs, cognate native yeast Phe-tRNAPhe, Pelota, eRF1, and SMAD1. (G) In control experiments, GTP was hydrolyzed by eRF3 in the presence of 80S ribosomes and eRF1 and by Hbs1 in the presence of 80S ribosomes and Pelota. (H) The interaction between purified recombinant Flag-tagged Pelota and His-tagged GTPBP2 (left), Flag-tagged GTPBP2 and His-tagged Pelota or His-tagged eRF1 (middle), and Flag-tagged Hbs1 and His-tagged Pelota as a positive control (right), assayed by pull-down using anti-Flag agarose beads followed by Western blotting. (I–K) Toeprinting analysis of the activity of eEF1H and different forms of GTPBP2 in one-cycle elongation on MLHL-STOP, MSHL-STOP, and MF-STOP mRNAs with cognate in vitro transcribed Leu-tRNALeu-AAG and Ser-tRNASer-AGA and native yeast Phe-tRNAPhe, respectively. The positions of 80S ICs and ECs are indicated by arrows at the right. Lanes C, T, A, and G depict the corresponding DNA sequences. (L) Polysome profile and ribosomal association of GTPBP2 in HEK293 cells under normal growth conditions, assayed by Western blotting. (M) The abundance of GTPBP2 in HEK293 cells under normal growth conditions and following 6 h of amino acid starvation, assayed by Western blotting.
GTPBP2 did not promote elongation on any tested codon, including UUC(F) (Fig. 6I–K). The transcription factor SMAD1, which was reported to bind to GTPBP2 (Kirmizitas et al. 2014), did not stimulate GTPBP2's GTP-binding (Supplemental Fig. S6A), GTP hydrolysis (Fig. 6E,G), or elongation activities (Supplemental Fig. S6B). The elongation activity of GTPBP2 was also not induced by eEF1B (Supplemental Fig. S6C). A minor amount of GTPBP2 was found in polysomal fractions of HEK293 cells under normal growth conditions, but amino acid starvation severely reduced its abundance (Fig. 6L,M).
In conclusion, the only observed similarity between GTPBP1 and GTPBP2 was the stimulation of their binding to GTP by Phe-tRNAPhe.
GTPBP1 stimulates exosomal degradation of mRNAs engaged in 80S ECs
The suggestion that GTPBP1 directs the exosome toward mRNA targets (Woo et al. 2011) and our observation of its eEF1A-like elongation activity prompted us to investigate the influence of GTPBP1 on exosomal degradation of free and 80S-bound mRNAs.
Eukaryotic RNA exosomes (Zinder and Lima 2017) have a nine-subunit noncatalytic core (Exo9) that consists of a hexameric ring formed by RNase PH-like domain-containing proteins (EXOSC4–EXOSC9) capped by a three-membered ring formed by S1/KH domain-containing proteins (EXOSC1–EXOCS3). The core has a central channel that accommodates ssRNA. The 10th subunit, DIS3, has a processive 3′ → 5′ exoribonuclease activity. Humans encode two DIS3 isoforms: DIS3 and DIS3L. The nuclear RNA exosome also associates with another catalytic subunit, EXOSC10 (RRP6 in yeast), which has a distributive 3′ → 5′ exoribonuclease activity and might also occur in the cytoplasm in humans (Lykke-Andersen et al. 2011). DIS3 binds to the PH-like ring at the opposite side of its cap-binding surface, whereas EXOSC10 resides at the other end of the core. Exosomes also interact with cofactors that target them to specific RNA substrates and pathways. For example, in yeast, nuclear exosomal RNA degradation and rRNA processing are assisted by the TRAMP complex, which contains the DExH-box helicase Mtr4p (SKIV2L2 in mammals), whereas the Ski complex (which contains the Mtr4p/SKIV2L2-like helicase Ski2) assists the cytoplasmic exosome in mRNA turnover and quality control pathways; mammals apparently have orthologous machinery, although precise details vary and characterizations are ongoing (for review, see Kilchert et al. 2016).
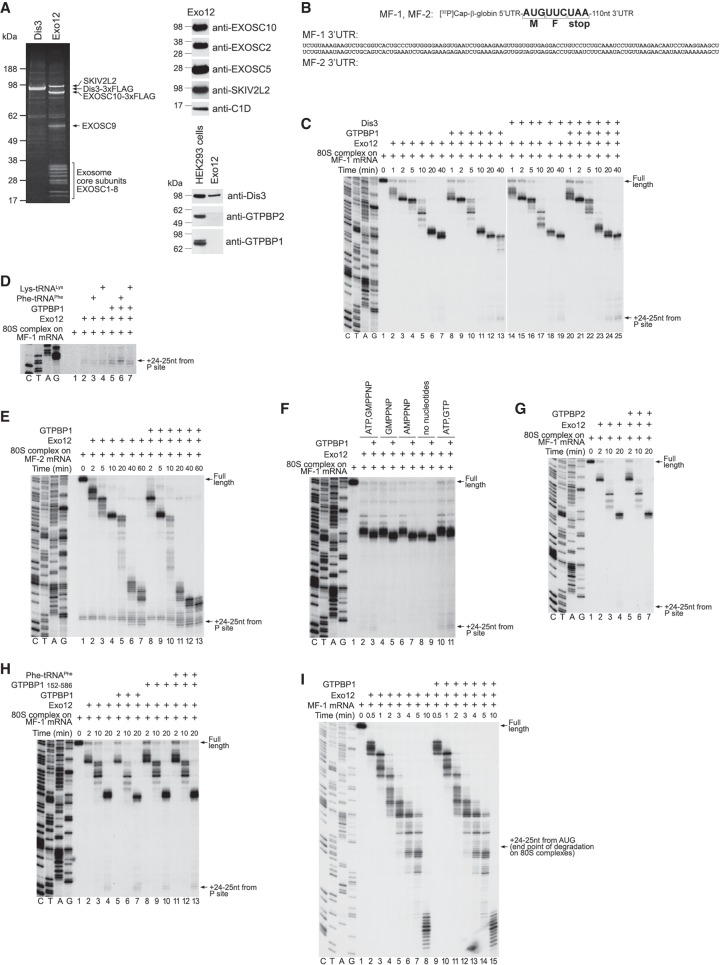
We purified the exosome from HEK293 cells expressing 3xFlag-tagged EXOSC10 (Domanski et al. 2016). The purified exosome (Exo12EXOSC10/SKIV2L2/C1D) comprised the Exo9 core, EXOSC10, SKIV2L2, and C1D that tethers SKIV2L2 to the exosome (Fig. 7A, left and top right panels). The preparation contained only trace amounts of DIS3, consistent with its labile association with the core (Staals et al. 2010; Tomecki et al. 2010; Domanski et al. 2016), and was free of GTPBP1 and GTPBP2 (Fig. 7A, bottom right panel). 3xFlag-tagged DIS3 was purified individually from HEK293 cells (Fig. 7A, left panel), and its exonuclease activity was confirmed (Supplemental Fig. S7A). 80S ICs were assembled on two [32P]cap-labeled mRNAs, MF-1 and MF-2 (Fig. 7B), comprising the β-globin 5′ UTR, Met-Phe-coding region, a UAA stop codon, and distinct 110-nt-long 3′ UTRs (Materials and Methods; Supplemental Fig. S7B).
Figure 7.
The influence of GTPBP1 on exosomal degradation of mRNA. (A) Purified DIS3 and Exo12EXOSC10/SKIV2L2/C1D, analyzed by SDS-PAGE followed by fluorescent Sypro staining (left panel) or Western blotting (right panels). (B) Schematic representation of MF-1 and MF-2 mRNAs showing nucleotide sequences of their 3′ UTRs. (C–E,G,H) RNA degradation intermediates obtained after incubation for the indicated time of 80S ICs assembled on [32P]cap-labeled MF-1 (C,D,G,H) or MF-2 (E) mRNAs with Exo12 and various combinations of GTPBP1, DIS3, GTPBP1152–586, GTPBP2, Phe-tRNAPhe, and Lys-tRNALys in the presence of ATP and GTP. (F) RNA degradation intermediates obtained after incubation of 80S ICs assembled on [32P]cap-labeled MF-1 mRNA with Exo12 for 15 min in the presence or absence of GTPBP1 and the indicated nucleotides. (I) RNA degradation intermediates obtained after incubation of free [32P]cap-labeled MF-1 mRNA with Exo12, ATP, and GTP in the presence or absence of GTPBP1 for the indicated time. (C–H) The position of the final cleavage products (+24–25 nt from the P site) is indicated at the right. (C–I) Lanes C, T, A, and G depict the DNA sequences used for RNA size evaluation.
GTPBP1 stimulated degradation by Exo12 of 80S-engaged MF-1 mRNA (Fig. 7C, cf. lanes 2–7 and 8–13). DIS3 also enhanced Exo12-mediated degradation, albeit to a smaller extent (Fig. 7C, cf. lanes 2–7 and 14–19). Degradation in the presence of DIS3 and GTPBP1 was similar to that observed in the presence of GTPBP1 alone (Fig. 7C, lanes 20–25). The most prominent exosome pauses coincided with the boundaries of mFold-predicted secondary structure elements. In all cases, cleavage continued until +24–25 nt from the P site. The same difference between degradation in the absence and presence of GTPBP1, DIS3, or GTPBP1/DIS3 was observed at shorter incubation times when the exosome had not moved past its first pause (Supplemental Fig. S7C). Stimulation of degradation by GTPBP1 at longer times appeared to be more pronounced. One potential explanation for this fact is that Exo12 might dissociate upon encountering mRNA secondary structure, in which case the presence of GTPBP1 would help reassociation of Exo12 at each pause, thus increasing the difference with the time. In control experiments, GTPBP1 did not degrade mRNA in the absence of the exosome (Supplemental Fig. S7D). Inclusion of cognate Phe-tRNAPhe or tRNAPhe with GTPBP1 did not stimulate degradation further (Supplemental Fig. S7E). tRNAs also did not influence degradation in the absence of GTPBP1 (Supplemental Fig. S7F). Notably, GTPBP1 and cognate Phe-tRNAPhe (but not Lys-tRNALys) altered the relative intensity of +24- and +25-nt cleavage products, enhancing the former (Fig. 7D). This indicates that although Phe-tRNAPhe did not enhance the effect of GTPBP1 on mRNA degradation by Exo12, it was nevertheless delivered to the A site because the change in the relative intensity of +24- and +25-nt cleavage products is consistent with the +1-nt toeprint shift that occurs upon binding of tRNA to the A site (Fig. 5L). GTPBP1 also stimulated degradation by Exo12 in the case of 80S-bound MF-2 mRNA (Fig. 7E). GTPBP1-mediated stimulation of Exo12 occurred in the presence of ATP and GTP, ATP and GMPPNP, only GMPPNP, or only AMPPNP or in the absence of any nucleotide (Fig. 7F). However, degradation reached the end point only in the presence of ATP, likely owing to the activity of SKIV2L2. Stimulation of exosomal degradation was specific for GTPBP1 and was not observed in the presence of GTPBP2 after either long (Fig. 7G) or short (Supplemental Fig. S7G) periods of incubation. There was also no stimulation in the case of the truncated GTPBP1152–586 (Fig. 7H). Importantly, GTPBP1 did not stimulate exosomal degradation of ribosome-unbound mRNA (Fig. 7I).
Discussion
In this study, we focused on two unconventional trGTPases: mammalian GTPBP1 and GTPBP2. Our analysis revealed that they possess distinct biochemical and functional activities.
We found that GTPBP1 shares many functional similarities with other members of the EF1 and SelB families. Like eEF1A, EF-Tu, and SelB, it can form GTPBP1• GTP•aa-tRNA ternary complexes and deliver aa-tRNAs to the A site, thereby promoting elongation. However, GTPBP1 possesses one unique activity: It can interact strongly with GTP and deacylated tRNA and deliver the latter to the A site. In contrast, other trGTPases have a very low affinity to deacylated tRNA in their apo, GDP, and GTP forms (Schulman et al. 1974; Dell et al. 1990; Gromadski et al. 2007; Schümmer et al. 2007).
GTPBP1's nucleotide-binding properties are comparable with those of eEF1A and EF-Tu (Gromadski et al. 2007; Schümmer et al. 2007). The association rate constants of GTP and GDP for these trGTPases are in the range of 0.1–2 µM−1 sec−1, and, at cellular concentrations of GTP (Traut 1994), nucleotide binding is essentially instantaneous. The dissociation rate constants of GTP from GTPBP1 and eEF1A are also similar (0.1–0.2 sec−1) and are somewhat higher than that of EF-Tu (0.03 sec−1). The dissociation rate constants of GDP differ by two orders of magnitude (0.01 sec−1 for GTPBP1, 0.13 sec−1 for eEF1A, and 0.002 sec−1 for EF-Tu). The rather low dissociation rate of GDP from GTPBP1 suggests the requirement for a GEF; however, if such a factor exists, it is likely not eEF1B, as recombinant eEF1B did not bind to GTPBP1 or influence its activity.
Given the 40-fold higher affinity of GTPBP1 to GDP than to GTP and the 10-fold excess of GTP over GDP in the cell (Traut 1994), the factor on its own would exist largely in the GDP-bound form. Association of GTPBP1• GTP with aa-tRNA strongly increases GTPBP1's affinity to GTP. The effect of aa-tRNA on nucleotide binding to GTPBP1 is reminiscent of that of eRF3, whose affinity to GTP is strongly increased by the interaction with eRF1 owing to a lowered dissociation rate constant (Pisareva et al. 2006). Thus, both aa-tRNA and eRF1 act as GTP dissociation inhibitors (TDIs) for their GTPase partners. As in the case of GTPBP1, the affinities of the GTP forms of eEF1A and EF-Tu to aa-tRNA are also in the low nanomolar range (this study; Gromadski et al. 2007; Schümmer et al. 2007; Mittelstaet et al. 2013), and the formation of a stable protein•GTP•aa–tRNA complex appears to be a general feature of all of these trGTPases. GTPBP1•GDP, on the other hand, binds aa-tRNA very weakly (similarly to eEF1A and EF-Tu), and, conversely, the affinity of GTPBP1 to GDP is unaffected by aa-tRNA.
Although GTPBP1 has some elongation activity, taking into account the low cellular levels of GTPBP1 and the inefficiency of GTPBP1-mediated peptide bond formation compared with that mediated by eEF1A, elongation by GTPBP1 is unlikely to be relevant during optimal growth conditions. Consistent with this finding, only a small amount of GTPBP1 was polysome-associated under normal growth conditions. The fact that the elongation activity of GTPBP1 is negatively regulated by its specific N-terminal and C-terminal extensions constitutes further indirect evidence against a role for GTPBP1 in cellular elongation.
On the other hand, the kinetics of GTP hydrolysis and peptide bond formation mediated by GTPBP1 is supportive of a role of the factor in mRNA surveillance (Woo et al. 2011). Although, similarly to EF-Tu (Rodnina et al. 1995), GTP hydrolysis by GTPBP1 is activated by codon–anti-codon base-pairing in the A site, in contrast to EF-Tu, it is not followed by rapid peptide bond formation. This suggests that after GTP hydrolysis, GTPBP1 likely retains aa-tRNA for a relatively long time, delaying the accommodation of aa-tRNA in the A site. In the physiological setting, this would result in ribosome stalling at the respective codons, enabling GTPBP1 to elicit quality control programs; e.g., by directing the exosome toward mRNA targets.
In the in vitro reconstituted system, we did observe stimulation by GTPBP1 of exosomal degradation of 80S-engaged mRNAs, but it did not depend on GTP and cognate aa-tRNA, suggesting that elongation-uncoupled interaction of GTPBP1 with ribosomal complexes was sufficient to recruit the exosome. However, formation of the GTPBP1•GTP•aa-tRNA ternary complex might be essential in the cellular environment owing to the competition with other translation factors binding to elongating ribosomes. Notably, although the interaction between GTPBP1 and the exosome did not depend on GTP, the GTP-bound state of GTPBP1 was important for stimulation of exosomal degradation in the HeLa cytoplasmic extract (Woo et al. 2011).
Some structural and functional characteristics of GTPBP1 are similar to those of yeast Ski7p, which contains an N-terminal region that bridges the exosome with the Ski complex and a C-terminal eEF1A-like GTPase domain that was suggested to interact with the ribosome (van Hoof et al. 2002; Kowalinski et al. 2015). However, Ski7p does not interact with tRNA, and, although it binds GTP, its GTPase activity has not been observed (Horikawa et al. 2016). The suggested ribosomal interaction of Ski7p has also not been demonstrated. Notably, whereas GTPBP1 was able to stimulate the activity of the EXOSC10-containing exosome, the interaction of Rrp6p (EXOSC10) and Ski7p with the yeast Exo9 core is mutually exclusive (Kowalinski et al. 2016; Liu et al. 2016). Thus, it seems unlikely that GTPBP1 functions as the mammalian counterpart of Ski7p. Moreover, recent studies identified HBS1Lv3 (a splicing isoform of Hbs1 that lacks the GTPase domain) as the mammalian factor that bridges the exosome with the Ski complex and performs at least some functions of Ski7p (Kowalinski et al. 2016; Kalisiak et al. 2017).
In our experiments, exosomal cleavage of 80S-engaged mRNA continued until +24–25 nt from the P site (∼10 nt outside of the mRNA entrance) irrespective of the presence or absence of DIS3. To reach the DIS3 active site for processive degradation, RNA is threaded through the Exo9 central channel, whereas, in the case of RRP6 (EXOSC10), RNA enters the core but is deflected back to reach the active center (e.g., Makino et al. 2015; Zinder et al. 2016). Because the RNA-binding path through the exosome core to the DIS3 active site is ∼30 nt long (Bonneau et al. 2009), even if ribosome-bound mRNA is initially degraded by DIS3, subsequent trimming of mRNA to within 10 nt of the ribosome would have to involve a “handover” to EXOSC10, which is presumably induced by the encounter between the ribosome and the exosome lid (Allmang et al. 1999; Makino et al. 2015). In the case of cytoplasmic exosomes lacking EXOSC10, the end point of exosome-mediated cleavage may occur at a position other than ∼10 nt from the edge of the ribosome. This and many other aspects of the function of the human exosome and GTPBP1 remain to be characterized. Thus, although GTPBP1 interacts with individual recombinant EXOSC2 (Woo et al. 2011), the subunits to which it binds in the context of the intact exosome as well as the elements of GTPBP1 involved in this interaction are currently unknown. The ability of GTPBP1 to function with the cytoplasmic exosome paired with the Ski complex also merits further investigation, which will require the development of approaches for purification of native homogenous mammalian exosomes lacking the EXOSC10 and Ski complexes.
Given the large excess of eEF1A over GTPBP1, GTPBP1-mediated stalling events are unlikely to be frequent during normal growth conditions but may become significant if the activity of eEF1A is impaired; e.g., by its phosphorylation at Ser300, which inhibits association of eEF1A with aa-tRNA (Lin et al. 2010), or phosphorylation of eEF1Bδ at Ser133, which decreases the rate of nucleotide exchange on eEF1A (Sivan et al. 2011). On the other hand, the ability of GTPBP1 to deliver deacylated tRNA to the A site suggests that GTPBP1 might function in mRNA surveillance and quality control mechanisms via the interaction with deacylated tRNA. Consistent with this notion, we observed an increase in polysomal association of GTPBP1 during amino acid starvation. Interestingly, expression of the Drosophila melanogaster GTPBP1 homolog Dgp-1 is up-regulated during oxidative stress, and the disruption of its gene leads to stress resistance (Girardot et al. 2004; Gruenewald et al. 2009). It is well established that stress conditions, including oxidative stress, inhibit translation at the initiation stage by phosphorylation of eIF2α, but recent studies showed that stress also affects elongation (e.g., Gerashchenko et al. 2012; Czech et al. 2013). Notably, deacylated tRNAs can accumulate not only during amino acid starvation but also in response to oxidative stress, when the cleavage of the adenosine residue of the tRNA acceptor end by angiogenin leads to the accumulation of tRNAs that cannot be charged (Czech et al. 2013). It is therefore possible that during oxidative and potentially other stresses, Dgp-1 performs its protective function through interaction with deacylated tRNA, targeting mRNAs for degradation or—in concert with eEF2—promoting the disassembly of stalled ribosomes, as was observed for GTPBP1 in the experiments shown in Figure 5, L–N.
Oxidative stress underlies many neurodegenerative disorders, including Alzheimer's, Parkinson's, and Huntington's diseases. Interestingly, the Dgp-1 gene is up-regulated in Drosophila containing loss-of-function mutations in the parkin gene (Greene et al. 2005), encoding a ubiquitin–protein E3 ligase, mutations in which are a common cause of autosomal recessive juvenile parkinsonism (Abbas et al. 1999). Furthermore, loss-of-function alleles of the Dgp-1 gene reduce the viability of parkin mutants (Greene et al. 2005). The functional link between Dgp-1 and the E3 ligase also points to the potential role of GTPBP1 in quality control mechanisms and in preventing disturbances in protein metabolism.
Although GTPBP1 and GTPBP2 have significant sequence similarity, they appear to have different cellular functions. GTPBP2, for instance, does not interact with the exosome and does not stimulate exosomal mRNA decay (this study; Woo et al. 2011). Even though stimulation of GTP binding to GTPBP2 by Phe-tRNAPhe implies that the mechanism of GTPBP2's action might also involve interaction with aa-tRNA, unlike GTPBP1, GTPBP2 lacks elongation activity. Moreover, in contrast to GTPBP1, only Phe-tRNAPhe was able to stimulate binding of GTP to GTPBP2. The nature of GTPBP2's tRNA specificity and its potential significance constitute an important subject for further investigation.
Materials and methods
Plasmids, purification of factors and ribosomal subunits, and aminoacylation of tRNA are described in the Supplemental Material, which also contains detailed protocols for all experimental procedures.
UV cross-linking
GTPBP1 and GTPBP2 (0.3–0.5 µM) were incubated with 120 nM [α-32P]GTP at 37°C in buffer A (20 mM Tris at pH 7.5, 2.5 mM MgCl2, 100 mM KCl, 0.25 mM spermidine, 2 mM DTT) in the presence of different tRNAs, Pelota, or eRF1. Assembled complexes were irradiated at 254 nm and analyzed by SDS-PAGE followed by autoradiography.
Fluorescence stopped-flow kinetic measurements
Stopped-flow measurements were performed using a SX-20MV apparatus (Applied Photophysics) in buffer A at 25°C. The time courses of mant-GTP/GDP binding to or dissociation from GTPBP1 were monitored via FRET changes between the protein tryptophan residues and the mant-group (excitation: 290 nm; cutoff filter: KV408 [Schott]). Time courses were evaluated with a single or double exponential function using Prism (Graphpad Software).
Filter-binding assay
[35S]Cys-tRNACys or [32P]tRNAPhe (0.3 nM) was incubated with 0.5–2000 nM GTPBP1 in the presence or absence of 0.2 mM GTP or GDP in buffer A at 37°C. The resulting complexes were passed through 0.45-µm nitrocellulose filters. The retained radioactivity was measured by scintillation counting. The curves were fitted to the Hill equation.
GTPase assay
GTPBP1, GTPBP2, eRF3, or Hbs1 (0.25 µM) was incubated with 80 nM [α-32P]GTP in the presence or absence of different combinations of 80S ribosomes, ECs, various tRNAs, eRF1, and Pelota in buffer A at 37°C. Reaction mixtures were analyzed by TLC on polyethyleneimine cellulose and quantified by phosphorimager.
Assembly and analysis of ribosomal complexes by toeprinting
80S ICs were assembled on derivatives of β-globin mRNA using 40S and 60S ribosomal subunits, Met-tRNAiMet, and eIFs essentially as described (Pisarev et al. 2010). To investigate elongation, 80S ICs were incubated with eEF2, various tRNAs, and eEF1H, GTPBP1, or GTPBP2. Resulting ribosomal complexes were analyzed by primer extension.
Comparison of the rates of GTP hydrolysis and peptide bond formation
80S ICs (100 nM) formed on MF mRNA in the presence of [35S]Met-tRNAiMet were incubated with 100 nM purified GTPBP1•[α-32P]GTP•Phe-tRNAPhe complexes in buffer A for different time periods at 37°C. The formation of [α-32P]GDP and [35S]MF was assayed by TLC on polyethyleneimine cellulose and quantified by phosphorimager.
In vitro translation
Luciferase mRNA (0.4 µg) was translated in 20 µL of RRL (Promega) supplemented with [35S]Met in the presence or absence of GTPBP1. Translation products were resolved by SDS-PAGE and quantified by phosphorimager.
Polysome analysis
HEK293T cells were grown under normal conditions or starved for amino acids for 6 h before being treated with 100 µg/mL cycloheximide and lysed. Supernatants were subjected to 10%–50% sucrose density gradient (SDG) centrifugation followed by Western blotting.
Flag pull-down assays
Different combinations of 200 nM His/Flag-tagged GTPBP2, Hbs1, eRF1, and Pelota were incubated in buffer A for 15 min at 37°C and applied to anti-Flag agarose. Bound proteins were eluted by buffer A containing 0.2 mg/mL 3xFlag peptide and analyzed by Western blotting.
Analysis of mRNA degradation
RNA exosome (Exo12EXOSC10/SKIV2L2/C1D) and DIS3 were purified as described (Domanski et al. 2016). 80S complexes were assembled on AUG codons of [32P]cap-labeled MF mRNAs. 80S complexes or free mRNAs (3 nM) were incubated with combinations of 15 nM RNA exosome, 25 nM DIS3, 50 nM GTPBP1, 50 nM GTPBP2, and 100 nM various tRNAs in the presence or absence of different nucleotides in buffer A at 37°C for the indicated times and resolved in 6% sequencing gels.
Supplementary Material
Acknowledgments
We thank T. Kinzy for the gift of yeast eEF1A, N. Ranjan and O. Geintzer for purified native yeast tRNA, and D. Burakovsky for purified total tRNA from HeLa cells and for optimizing the protocol for phosphorylating residual mant-GDP impurities found in commercial preparations of mant-GTP. This work was supported by National Institutes of Health (NIH) grants GM122602 and GM80623 to T.V.P., NIH grant AI123406 to C.U.T.H., the funding of the Leibniz Prize to M.V.R., and NIH grant HL121779 to A.A.K. and was aided by collaboration with the National Center for Dynamic Interactome Research, supported by NIH grant P41 GM109824.
Author contributions: Biochemical studies of GTPBP1 and GTPBP2 were performed by A.Z. with the supervision of T.V.P, C.U.T.H., and M.V.R. Kinetic analysis of GTPBP1 was performed by A.Z. and A.G. with the supervision of M.V.R. and T.V.P. Polysome analysis was performed by S.J. with the supervision of A.A.K. Experiments on the exosomal degradation of mRNA were performed by A.Z. with the supervision of J.L and T.V.P. The manuscript was written by T.V.P, C.U.T.H., M.V.R., A.Z., A.G., J.L., and A.A.K. All authors approved the final manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.314724.118.
References
- Abbas N, Lücking CB, Ricard S, Dürr A, Bonifati V, De Michele G, Bouley S, Vaughan JR, Gasser T, Marconi R, et al. 1999. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. French Parkinson's Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Hum Mol Genet 8: 567–574. [DOI] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC. 2015. The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life. BMC Genomics 16: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559. [DOI] [PubMed] [Google Scholar]
- Czech A, Wende S, Mörl M, Pan T, Ignatova Z. 2013. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet 9: e1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell VA, Miller DL, Johnson AE. 1990. Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA. A fluorescence study. Biochemistry 29: 1757–1763. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanski M, Upla P, Rice WJ, Molloy KR, Ketaren NE, Stokes DL, Jensen TH, Rout MP, LaCava J. 2016. Purification and analysis of endogenous human RNA exosome complexes. RNA 22: 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, Gladyshev VN. 2012. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci 109: 17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis WQ, Kirmizitas A, Iwasaki Y, Ki DH, Wyrick JM, Thomsen GH. 2016. GTPBP2 is a positive regulator of Wnt signaling and maintains low levels of the Wnt negative regulator Axin. Cell Commun Signal 14: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot F, Monnier V, Tricoire H. 2004. Genome wide analysis of common and specific stress responses in adult Drosophila melanogaster. BMC Genomics 5: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Chaillet M, van Tilbeurgh H. 2008. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. J Biol Chem 283: 7145– 7154. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Andrews LA, Parker TJ, Pallanck LJ. 2005. Genetic and genomic studies of Drosophila parkin mutants implicate oxidative stress and innate immune responses in pathogenesis. Hum Mol Genet 14: 799–811. [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Schümmer T, Strømgaard A, Knudsen CR, Kinzy TG, Rodnina MV. 2007. Kinetics of the interactions between yeast elongation factors 1A and 1Bα, guanine nucleotides, and aminoacyl-tRNA. J Biol Chem 282: 35629–35637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenewald C, Botella JA, Bayersdorfer F, Navarro JA, Schneuwly S. 2009. Hyperoxia-induced neurodegeneration as a tool to identify neuroprotective genes in Drosophila melanogaster. Free Radic Biol Med 46: 1668–1676. [DOI] [PubMed] [Google Scholar]
- Horikawa W, Endo K, Wada M, Ito K. 2016. Mutations in the G-domain of Ski7 cause specific dysfunction in non-stop decay. Sci Rep 6: 29295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. 2014. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Chuang JH, Ackerman SL. 2016. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 5: e14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaberi E, Rohani M, Shahidi GA, Nafissi S, Arefian E, Soleimani M, Rasooli P, Ahmadieh H, Daftarian N, KaramiNejadRanjbar M, et al. 2016. Identification of mutation in GTPBP2 in patients of a family with neurodegeneration accompanied by iron deposition in the brain. Neurobiol Aging 216: e11–e18. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Kalisiak K, Kuliński TM, Tomecki R, Cysewski D, Pietras Z, Chlebowski A, Kowalska K, Dziembowski A. 2017. A short splicing isoform of HBS1L links the cytoplasmic exosome and SKI complexes in humans. Nucleic Acids Res 45: 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SP, Bedwell DM. 2015. Both the autophagy and proteasomal pathways facilitate the Ubp3p-dependent depletion of a subset of translation and RNA turnover factors during nitrogen starvation in Saccharomyces cerevisiae. RNA 21: 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L. 2016. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol 17: 227–239. [DOI] [PubMed] [Google Scholar]
- Kirmizitas A, Gillis WQ, Zhu H, Thomsen GH. 2014. GTPBP2 is required for BMP signaling and mesoderm patterning in Xenopus embryos. Dev Biol 392: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Schuller A, Green R, Conti E. 2015. Saccharomyces cerevisiae Ski7 is a GTP-binding protein adopting the characteristic conformation of active translational GTPases. Structure 23: 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Kögel A, Ebert J, Reichelt P, Stegmann E, Habermann B, Conti E. 2016. Structure of a cytoplasmic 11-subunit RNA exosome complex. Mol Cell 63: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo H, Senju S, Mitsuya H, Nishimura Y. 2000. Mouse and human GTPBP2, newly identified members of the GP-1 family of GTPase. Biochem Biophys Res Commun 272: 456–465. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317: 41–72. [DOI] [PubMed] [Google Scholar]
- Lin KW, Yakymovych I, Jia M, Yakymovych M, Souchelnytskyi S. 2010. Phosphorylation of eEF1A1 at Ser300 by TβR-I results in inhibition of mRNA translation. Curr Biol 20: 1615–1625. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Niu CY, Wu Y, Tan D, Wang Y, Ye MD, Liu Y, Zhao W, Zhou K, Liu QS, et al. 2016. CryoEM structure of yeast cytoplasmic exosome complex. Cell Res 26: 822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. 2011. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol 8: 61–66. [DOI] [PubMed] [Google Scholar]
- Makino DL, Schuch B, Stegmann E, Baumgärtner M, Basquin C, Conti E. 2015. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524: 54–58. [DOI] [PubMed] [Google Scholar]
- Maracci C, Rodnina MV. 2016. Review: translational GTPases. Biopolymers 105: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstaet J, Konevega AL, Rodnina MV. 2013. A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J Am Chem Soc 135: 17031–17038. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. 2006. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J Biol Chem 281: 40224–40235. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Fricke R, Kuhn L, Wintermeyer W. 1995. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J 14: 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman LH, Pelka H, Sundari RM. 1974. Structural requirements for recognition of Escherichia coli initiator and non-initiator transfer ribonucleic acids by bacterial T factor. J Biol Chem 249: 7102–7110. [PubMed] [Google Scholar]
- Schümmer T, Gromadski KB, Rodnina MV. 2007. Mechanism of EF-Ts-catalyzed guanine nucleotide exchange in EF-Tu: contribution of interactions mediated by helix B of EF-Tu. Biochemistry 46: 4977–4984. [DOI] [PubMed] [Google Scholar]
- Senju S, Nishimura Y. 1997. Identification of human and mouse GP-1, a putative member of a novel G-protein family. Biochem Biophys Res Commun 231: 360–364. [DOI] [PubMed] [Google Scholar]
- Senju S, Iyama K, Kudo H, Aizawa S, Nishimura Y. 2000. Immunocytochemical analyses and targeted gene disruption of GTPBP1. Mol Cell Biol 20: 6195–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Aviner R, Elroy-Stein O. 2011. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J Biol Chem 286: 27927–27935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. 2010. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J 29: 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW. 1994. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 140: 1–22. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yoshida K, Hida M, Kato H, Uchida K, Yamaguchi R, Tateyama S, Sugano S. 2000. Cloning, expression analysis, and chromosomal mapping of GTPBP2, a novel member of the G protein family. Gene 256: 51–58. [DOI] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim S, Lee HR, Kang HJ, Chung SJ, Senju S, Nishimura Y, et al. 2011. Modulation of exosome-mediated mRNA turnover by interaction of GTP-binding protein 1 (GTPBP1) with its target mRNAs. FASEB J 25: 2757–2769. [DOI] [PubMed] [Google Scholar]
- Zinder JC, Lima CD. 2017. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Wasmuth EV, Lima CD. 2016. Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol Cell 64: 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.