Wang et al. show that Dicer-like 2 (DCL2) is the major Dicer in tomato defense against tobacco mosaic virus (TMV) and potato virus X (PVX) and that it is involved in the biogenesis of endogenous 22-nt sRNA.
Keywords: RNA silencing, tomato, Dicer, miRNA, virus
Abstract
Tomato Dicer-like2 (slDCL2) is a key component of resistance pathways against potato virus X (PVX) and tobacco mosaic virus (TMV). It is also required for production of endogenous small RNAs, including miR6026 and other noncanonical microRNAs (miRNAs). The slDCL2 mRNAs are targets of these slDCL2-dependent RNAs in a feedback loop that was disrupted by target mimic RNAs of miR6026. In lines expressing these RNAs, there was correspondingly enhanced resistance against PVX and TMV. These findings illustrate a novel miRNA pathway in plants and a crop protection strategy in which miRNA target mimicry elevates expression of defense-related mRNAs.
RNA silencing plays important roles in plant development, genome stability, and antiviral resistance (Baulcombe 2004). There are multiple pathways of RNA silencing in plants in which small RNAs (sRNAs), ranging from 20 to 24 nucleotides (nt), bind to Argonaute (AGO) effector proteins. The AGO ribonucleoprotein then anneals to a target RNA through Watson–Crick base pairing with an outcome that depends on the nature of the target RNA. If the target RNA is cytoplasmic, there is post-transcriptional gene silencing (PTGS) through cleavage of target RNA and/or translation inhibition (Rogers and Chen 2013), whereas, with nuclear RNA, there may be epigenetic effects in which AGOs recruit DNA/chromatin-modifying factors (Law and Jacobsen 2011).
In plants, the cytoplasmic pathways of RNA silencing involve two different types of sRNA: siRNAs and microRNAs (miRNA). The siRNAs are produced from various types of dsRNA, whereas miRNAs have a hairpin-like RNA precursor in which there are mismatches in the regions of base pairing. In both instances, the processing enzyme is referred to as Dicer-like (DCL).
The siRNA and miRNA pathways interconnect through a complex mechanism involving secondary siRNA production (Allen et al. 2005; Li et al. 2011; Zhai et al. 2011). In the “two-hit” model, a transcript with dual miRNA target sites is the secondary siRNA precursor (Allen et al. 2005), whereas, in the “one-hit” model, the miRNA is typically 22 nt rather than the canonical 21 nt (Chen et al. 2010; Cuperus et al. 2010). In both models, the miRNAs may mediate cleavage of their target RNA, as in the normal miRNA pathway, but there are several additional steps. First, the 3′ cleavage product of the target RNA is converted into a double-stranded form by an RNA-dependent RNA polymerase 6 (RDR6) RNA. The dsRNA is then processed by DCL4 to secondary siRNAs (Allen et al. 2005; Gasciolli et al. 2005).
A distinct feature of these secondary RNAs is that they align predominantly to their template RNA in a phased register in which the first position is opposite position 10 of the initiator miRNA. In some instances, the phasing register has a 21-nt spacing (Allen et al. 2005; Zhai et al. 2011; Creasey et al. 2014), but, in male reproductive organs of monocots, there are also phased secondary sRNAs with a 24-nt register (Johnson et al. 2009).
There are two mechanisms for 22-nt sRNA production. One mechanism, for miRNAs, is based on DCL1 that typically produces 21-nt sRNAs but generates 22-nt products if the precursor RNA has an asymmetric bulge in the base-paired region. Presumably, the bulge allows 22-nt RNA to be accommodated between the DCL active sites on either side of the mature miRNA. The second mechanism of 22-nt sRNA production involves DCL2 and is independent of bulges in the precursor RNA. DCL2 in Arabidopsis can process endogenous and viral dsRNA into 22-nt sRNAs when other DCLs, especially DCL4, are absent (Xie et al. 2004; Gasciolli et al. 2005; Bouché et al. 2006). More importantly, DCL2 plays a primary role in transgene silencing, especially in sense transgene-induced silencing and transitivity of hairpin-induced transgene silencing (Mlotshwa et al. 2008; Parent et al. 2015). There is also a role of DCL2 in systematic spreading of transitive silencing between cells and through the vascular system (Taochy et al. 2017; Wu et al. 2017).
In addition to involvement in transgene and viral RNA silencing, there could be secondary sRNA cascades affecting endogenous gene expression and antivirus defense that are also dependent on DCL2 and endogenous 22-nt sRNAs. To investigate this possibility, we analyzed DCL2 isoforms in tomato and found that slDCL2a and slDCL2b are the most abundantly expressed genes in the four-member slDCL2 family. From sldcl2ab mutants, we conclude that DCL2 is the major Dicer in tomato defense against tobacco mosaic virus (TMV) and potato virus X (PVX) and that it is involved in the biogenesis of endogenous 22-nt sRNA. The 24-nt sRNA pathways are also influenced by DCL2, both positively and negatively. Among the endogenous 22-nt sRNAs are miRNAs, including miR6026 that targets slDCL2 mRNA and triggers secondary sRNA production. We disrupted this regulatory feedback loop with target mimic RNAs of miR6026 so that slDCL2 was up-regulated and antivirus resistance was enhanced. We propose that disruption of this and other defense-related miRNAs could be used as part of an integrated pest management strategy to protect crops against viruses and other pathogens.
Results and Discussion
CRISPR mutants of slDCL2
The tomato genome encodes four isoforms of slDCL2 (Bai et al. 2012), of which two (slDCL2a and slDCL2b) are highly expressed in young leaves and two (slDCL2c and slDCL2d) are barely detectable by RT–PCR (Supplemental Fig. S1). To investigate the functions of slDCL2, we designed a pair of CRISPR small guide RNAs (sgRNAs) to target the RNase III domain closest to the C terminus of slDCL2a and slDCL2b (Supplemental Fig. S2A) and transformed them into tomato as part of an integrated construct with Cas9. Of the several T0 plants with edited slDCL2a or slDCL2b, we selected two for further analysis. Both plants carried the same deletion of two codons and an isoleucine-to-valine substitution in the slDCL2a RNase domain but had different premature stop codons in slDCL2b (Supplemental Fig. S2B,C). These alleles (sldcl2a-1, sldcl2b-1, and sldcl2b-2) have mutations in a highly conserved region of DCL2 (Supplemental Fig. S2D), and it is likely that they would encode nonfunctional proteins.
The selfed progeny of these T0 plants in which the CRISPR/Cas9 transgene was lost by segregation are sldcl2a-1 sldcl2b-1, sldcl2a-1 sldcl2b-2, sldcl2a-1, sldcl2b-1, or sldcl2b-2, depending on whether they carried one or both of the mutations in the homozygous condition (Supplemental Table S1). Transcript levels of slDCL2a and slDCL2b were similar or slightly higher in sldcl2a-1 sldcl2b-1/sldcl2a-1 sldcl2b-2 (referred to here as dcl2ab) than in M82 wild type (Supplemental Fig. S3), indicating that the mutations do not cause transcript degradation through mRNA decay. There was similarly no effect on other DCL mRNAs (Supplemental Fig. S3), and it is therefore unlikely that the loss of DCL2 results in compensating changes in other DCLs.
slDCL2a/slDCL2b affects endogenous 22- and 24-nt sRNA accumulation
Although DCL2 plays crucial roles in sense transgene-induced silencing and transitivity of hairpin-induced transgene silencing (Mlotshwa et al. 2008), it may also back up DCL4 in the production of sRNAs from viral and endogenous RNAs (Gasciolli et al. 2005; Bouché et al. 2006; Wu et al. 2017). Until now, however, there has been no analysis of sRNA from endogenous loci dependent on DCL2 in the presence of DCL4 that could be informative about a DCL4-independent role.
To investigate this possibility, we sequenced sRNA in wild-type and dcl2ab tomato leaves and found the predominant size class of sRNA was 24 nt with small amounts of 21-nt species (Fig. 1A) in both genotypes. The lack of a severe development phenotype (Supplemental Fig. S4) in dcl2ab indicates that the mutations did not lead to loss of essential sRNA species. Of the 642,444 sRNAs, there were 2432 (0.4%) with differential accumulation between wild type and dcl2ab, of which 1779 were less abundant in dcl2ab (slDCL2-dependent sRNA-D2 sRNA) and 653 were more abundant (slDCL2-inhibited sRNA-D2i sRNA) (Fig. 1B; Supplemental Tables S2, S3). The D2 sRNAs were predominantly 22 nt (Fig. 1C), consistent with the role of DCL2 in 22-nt sRNA biogenesis, as is likely in Arabidopsis (Gasciolli et al. 2005; Bouché et al. 2006). In addition, there was a minor 24-nt peak (Fig. 1C) in both D2 and D2i sRNA that implies positive and negative roles of DCL2 in 24-nt sRNA pathways.
Figure 1.
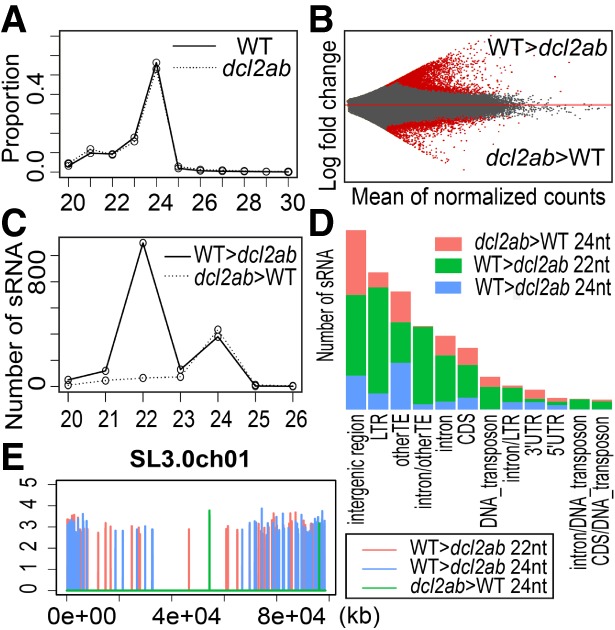
slDCL2a and slDCL2b are responsible for endogenous sRNA biogenesis. High-throughput sequencing data of sRNAs in 24-d-old leaves of wild type and dcl2ab mapped to tomato genome SL3.0. (A) sRNA size profile in wild type and dcl2ab, respectively. (B) MA plot showing differential analysis of sRNAs between wild type and dcl2ab. sRNAs that are differentially expressed (DE) (adjusted P-value < 0.01, calculated by DESeq2) are in red. (C) Length distribution of DE sRNA. (D) Genomic feature analysis of 22-nt D2 sRNA and 24-nt DE sRNA. The Y-axis shows the number of DE sRNA in each category of genomic feature. (E) Chromosomal distributions of 22-nt D2 loci and 24-nt DE loci. The X-axis shows the chromosomal coordinates, and the Y-axis shows the log10 values of sRNA reads number in wild type and dcl2ab for D2 and D2i loci, respectively.
The 22-nt D2 sRNAs were predominantly from transposable elements (TEs) (Fig. 1D), indicating that slDCL2a/ slDCL2b plays a role in genome stability. In contrast, the D2 and D2i 24-nt sRNAs were mostly from intergenic regions (Fig. 1D). There was also a difference in the chromosomal distribution of these differentially expressed (DE) sRNA.
The genomic loci producing 22-nt D2 sRNA revealed by SegmentSeq (Hardcastle et al. 2012) were distributed evenly across chromosomes, whereas those with 24-nt DE sRNAs were in the distal regions (Fig. 1E; Supplemental Fig. S5) that, in the tomato genome, correspond to euchromatin (Tomato Genome Consortium 2012).
Our interpretation of these findings is that slDCL2a/slDCL2b produces 22-nt D2 sRNA directly by cleavage of precursor RNAs. The 24-nt D2 sRNAs are likely to be secondary sRNAs that may be dependent on a 22-nt D2 sRNA, as in monocots (Johnson et al. 2009). According to this idea, the D2 loci with predominantly 22-nt sRNAs (Supplemental Fig. S6) would be largely made up of primary sRNAs generated by DCL2. The D2 loci with predominantly 24-nt sRNA (Supplemental Fig. S6) would be largely secondary sRNA loci. Consistent with the role of DCL2 in the release of 22-nt D2 sRNAs, the precursors of four D2 sRNA loci accumulated at higher levels in dcl2ab than in wild type (Supplemental Fig. S7).
The D2i sRNAs of all size classes are likely to be derived from precursor RNAs that are normally processed by slDCL2 but, in the dcl2ab plants, are available for other DCL proteins (Nagano et al. 2014). Consistent with this proposal, the 24-nt D2i sRNA regions produced sRNA that was predominantly 22 nt in wild-type plants (Supplemental Fig. S8). The precursors of two D2i loci were similarly abundant in wild type and dcl2ab (Supplemental Fig. S7), consistent with processing by DCL2 in wild type into 22-nt sRNA and a substitute DCL producing 24-nt sRNA in dcl2ab.
slDCL2a/slDCL2b-dependent 22-nt miRNAs
To find out whether the D2 22-nt sRNAs included miRNAs, we compared the abundance of tomato miRNA reads in the sRNA sequencing (sRNA-seq) data sets of wild type and dcl2ab. All of the 21-nt miRNAs and most of 22-nt miRNAs did not show statistically significant differences (P < 0.01) (Fig. 2A). The main exception, however, was the 22-nt miRNA: miR6026 (Fig. 2A,B). Consistent with the sRNA-seq data, the level of miR6026 detected by Northern blotting of RNA from 24-d-old leaves was lower than wild type in dcl2b and much lower in dcl2ab (Fig. 2C). By comparison, the 22-nt miR482e was at the same level in wild-type and mutant genotypes (Fig. 2C). In contrast, most miRNAs, including miR482e, were down-regulated in the slDCL1 knockdown line (Kravchik et al. 2014) compared with control, whereas miR6026 was not affected by slDCL1 knockdown (Supplemental Fig. S9). Therefore, we conclude that miR6026 is unusual among miRNAs in that it is slDCL2-dependent.
Figure 2.
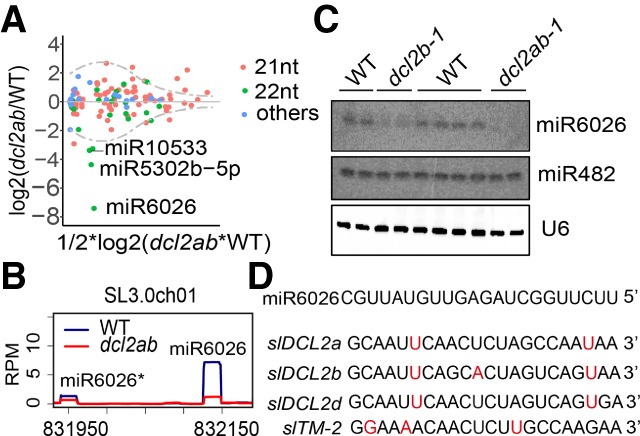
slDCL2a and slDCL2b are required for biogenesis of miR6026. (A) MA plot showing miRNA abundance in wild type and dcl2ab. Different sizes of miRNAs are shown in different colors. miR6026, miR5302b-5p, and miR10533 are marked. (B) sRNA RPM plots showing reduced accumulations of miR6026 in dcl2ab as compared with wild type. Mature miR6026 and miR6026* are marked. (C) Northern blots of miR6026 and miR482e with total RNA extracted from 24-d-old leaves of wild type and dcl2b and dcl2ab mutants, with U6 as a loading control. (D) Sequences of miR6026 and potential targets. Mismatches are shown in red.
The most straightforward interpretation of these data is that the MIR6026 precursor is cleaved by DCL2 to release miR6026. Other possibilities are that other DCL proteins carry out the cleavage and that DCL2 merely stabilizes the processed miR6026 or its precursor. The former possibility is unlikely because DCL2 has no role in stabilizing other 22-nt miRNAs, and the latter can be ruled out because there are 21-nt sRNAs from the stem–loop in the MIR6026 precursor RNA that are unaffected by dcl2ab (Supplemental Fig. S10). An effect on the precursor is also ruled out because there are similar levels of pre-miR6026 in wild type and dcl2ab (Supplemental Fig. S7).
The 22-nt miRNAs in Arabidopsis trigger secondary sRNA production using their RNA targets as a template (Chen et al. 2010; Cuperus et al. 2010), and, correspondingly, in tomato, there were sRNAs corresponding to the miR6026 potential target mRNAs (Fig. 2D; Supplemental Table S4) that include the mRNAs for slDCL2a, slDCL2b, slDCL2d, and a disease resistance gene: slTM2 (Fig. 3; Supplemental Fig. S11; Li et al. 2011; Kravchik et al. 2014; Wu et al. 2016). These sRNAs were predominantly 21 nt in length and aligned with the 3′ side of the miRNA target site (Fig. 3; Supplemental Fig. S11). The sRNAs from slDCL2a/b/d, like many secondary sRNAs, were predominantly in a phased register corresponding to the miRNA-directed cleavage site. This phasing pattern was most pronounced in the regions adjacent to the miRNA target (Fig. 3; Supplemental Fig. S11). The phasing of the slDCL2 sRNAs is less pronounced than with the trans-acting siRNA (tasiRNA) loci in Arabidopsis. To explain this difference, we propose that the 22-nt secondary sRNAs (Supplemental Table S5) from the slDCL2 mRNAs could act in cis and trigger additional rounds of secondary sRNA in various phasing registers.
Figure 3.
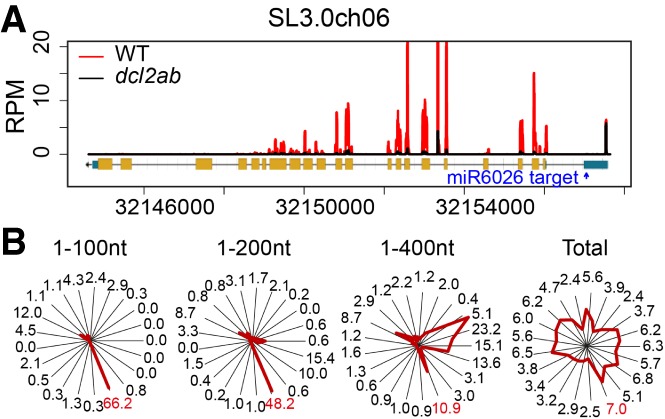
Accumulation of slDCL2a sRNAs is reduced in dcl2ab. (A) sRNA RPM plots showing the levels of slDCL2b sRNAs in wild type and dcl2ab. (Rectangles) Exons; (lines) genes; (cyan) untranslated regions (UTRs); (yellow) ORFs. An arrow marks the direction of transcription. The target site of miR6026 is marked with a blue arrow. (B) The phasing of slDCL2a sRNAs in wild type. Radar plots show the percentages of 21-nt reads corresponding to each of the 21 registers from the whole transcript of slDCL2a or 1–100 nt, 1–200 nt, or 1–400 nt of the slDCL2a transcript (the distance from the miR6026 cleavage position) in wild-type sRNA-seq data. The percentage of register of the 10th position and the miR6026-guided cleavage site between the 10th and 11th nucleotide of miR6026 are marked in red.
The sRNAs of slDCL2a, slDCL2b, and slDCL2d are dependent on slRDR6 and partially on slDCL4 (Supplemental Fig. S12), and therefore their biogenesis is like that of the well-known phased siRNA (phasiRNA)/tasiRNA and epigenetically activated siRNA (easiRNA) biogenesis in Arabidopsis (Allen et al. 2005; Zhai et al. 2011; Creasey et al. 2014). A crucial difference, however, is that the miR6026 is DCL2-dependent, and therefore there is a feedback component to the system. The triggers of Arabidopsis phasi/tasiRNA and easiRNAs are 22 nt, similar to miR6026, but are dependent on DCL1 rather than DCL2 (Cuperus et al. 2010).
Our characterization of miR6026 provides the first example of non-DCL1 processed miRNA in plants with a validated target RNA (Supplemental Fig. S9). There are other 22-nt miRNAs, but they are dependent on DCL1 (Cuperus et al. 2010), and their size is determined by an asymmetric bulge in the miRNA/miRNA* (Supplemental Fig. S13). The 22-nt miR6026 has a precursor that is symmetrical in the miRNA/miRNA* region, and its size is likely to be influenced by the DCL2 protein, as for the 22-nt D2 sRNAs, rather than by the structure of the miRNA precursor.
Of the tomato 22-nt miRNAs, there are three others (in addition to miR6026) that have symmetric structures in the miRNA/miRNA* duplex: miR5302b-5p, miR10533, and miR828. The former two are reduced in the dcl2ab samples (Fig. 2A) but not as much as miR6026. We predicted potential target genes of miR5302b-5p and miR10533 (Supplemental Table S6); however, none of them produced sRNA in wild type according to the sRNA-seq data (data not shown). The miR828 is expressed only at low abundance so that differential expression in the dcl2ab mutant cannot be assessed (data not shown), but the phased secondary sRNAs from its TAS4 target locus (Singh et al. 2016) were less abundant in the dcl2ab mutant (Supplemental Fig. S14). It is therefore likely that both of these other 22-nt miRNAs with symmetrical precursors are DCL2-dependent, like miR6026, although it is not clear whether miR5302 or miR10533 triggers secondary siRNA.
The biological function of slDCL2-dependent miR6026
To explore the biological function of miR6026, we transgenically expressed a noncoding RNA with two tandem repeats of a miR6026 target mimic site. Like the endogenous target mimic RNAs of miR399 (Puga et al. 2007), these RNAs were designed to have a 3-nt bulge at positions 10 and 11 of the miRNA-binding site (Fig. 4A). We predicted that this RNA would lock the miR6026 into a nonproductive interaction that would compete with the normal binding to slDCL2a, slDCL2b, slDCL2d, and slTM2 mRNAs.
Figure 4.
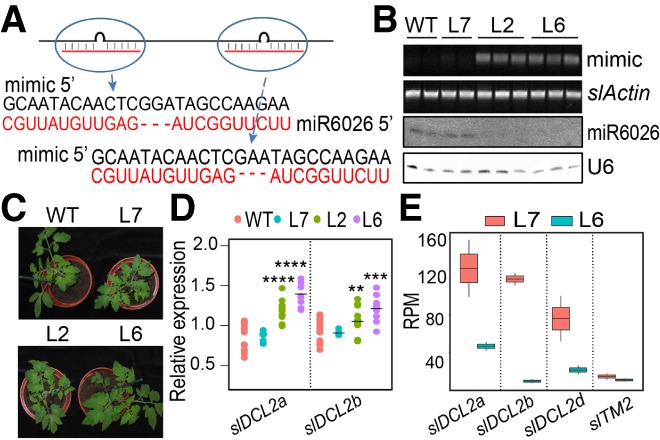
miR6026 targets slDCL2a/b/d transcripts and triggers secondary sRNA production. (A) Diagram of miR6026 target mimic. (B, top to bottom) Semiquantitative RT–PCR of miR6026 mimic RNA in wild-type and different mimicry lines with slActin7 as control. Northern blots of miR6026 with U6 as a loading control. (C) Three-week-old plants of wild type and one weak mimic line (L7) and two strong mimic lines of miR6026 (L2 and L6). (D) Quantitative RT–PCR (qRT–PCR) of slDCL2a and slDCL2b in wild-type and mimicry lines. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (E) RPM plot of total reads mapped to potential target transcripts of miR6026 in a weak mimic line (L7) and a strong mimic line (L6).
Transgenic tomato lines with high (L2 and L6) or low (L7) expression levels of the target mimic RNA grew normally under growth chamber conditions (Fig. 4B,C) and set fruit and seed as well as wild-type plants. There was a negative correlation between miR6026 accumulation and the target mimic RNA in these plants (Fig. 4B), indicating that the target mimic RNA reduces the cognate miRNA abundance, as in other examples (Yan et al. 2012). This target mimic RNA effect was highly specific so that the only affected miRNA in the L6 lines was miR6026 (P-value cutoff, P < 0.05) (Supplemental Fig. S15). Correspondingly, there was up-regulation of slDCL2a and, to a lesser extent, slDCL2b that correlated with the levels of the target mimic RNA (Fig. 4D).
An sRNA-seq analysis of two independent T1 plants from L6 and L7 confirmed that the level of miR6026 was lower in L6 than L7 (Supplemental Fig. S15) and that there was a parallel effect on secondary sRNA from slDCL2a, slDCL2b, and slDCL2d (Fig. 4E). In contrast, the slTM2 sRNA and mRNA were not affected (Fig. 4E; Supplemental Fig. S16). To further validate miR6026-directed target cleavage, we assayed for miR6026 cleavage sites of slDCL2a, slDCL2b, and slDCL2d mRNAs with 5′-RLM-RACE (RNA ligase-mediated rapid amplification of cDNA ends). In each instance, the predicted cleaved products for miR6026 were amplified in wild-type rather than dcl2ab or target mimic lines (Supplemental Fig. S16).
These findings confirm that miR6026 targets mRNAs of slDCL2a, slDCL2b, and slDCL2d in vivo and that it is responsible for the slDCL2 secondary sRNA production. These findings also explain the higher mRNA levels of slDCL2a and slDCL2b in dcl2ab than in wild type (Supplemental Fig. S3B). The lack of an effect on slTM2 sRNA is likely because other miRNAs target the slTM2 mRNA and could trigger secondary siRNA in the absence of miR6026 (Supplemental Figs. S11F, S17; Li et al. 2011).
The increased abundance of slDCL2a and slDCL2b RNA in L6 (strong mimic line) relative to wild-type lines (Fig. 4D) should influence the levels of endogenous D2 and D2i sRNAs (Fig. 1) and exogenous viral sRNAs (Xie et al. 2004; Bouché et al. 2006). Consistent with these predictions, there was a trend for the endogenous D2 sRNAs to increase more in L6 than in L7 and for the D2i sRNAs to be less abundant (Supplemental Fig. S18). The effect on D2 sRNAs was clearest with the 22-nt species and the 24-nt D2i sRNAs (Supplemental Fig. S18).
The potential involvement of DCL2 in viral sRNA production was supported by the increase in slDCL2b mRNA in TMV- and PVX-infected tomatoes (Fig. 5A) and the enhanced viral symptoms in dcl2ab relative to wild type (Fig. 5B,C). The TMV-infected mutant plants had “wiry” (Lesley 1928) or shoestring-like leaves, whereas infected wild-type plants had chlorosis without leaf distortion (Fig. 5B; Supplemental Fig. S19). PVX-infected dcl2ab plants were dead 2 wk after inoculation, in contrast to the wild-type plants that survived, although with growth stunting and leaf distortion (Fig. 5C). Consistent with the up-regulation of slDCL2b upon virus infection (Fig. 5A), the TMV-infected dcl2b had “wiry” leaves, as with infected dcl2ab (Supplemental Fig. S20). However, at 34 d after infection, the TMV symptoms were milder on dcl2b than on dcl2ab (Supplemental Fig. S20), consistent with a role of slDCL2a in the late stages of antiviral defense.
Figure 5.
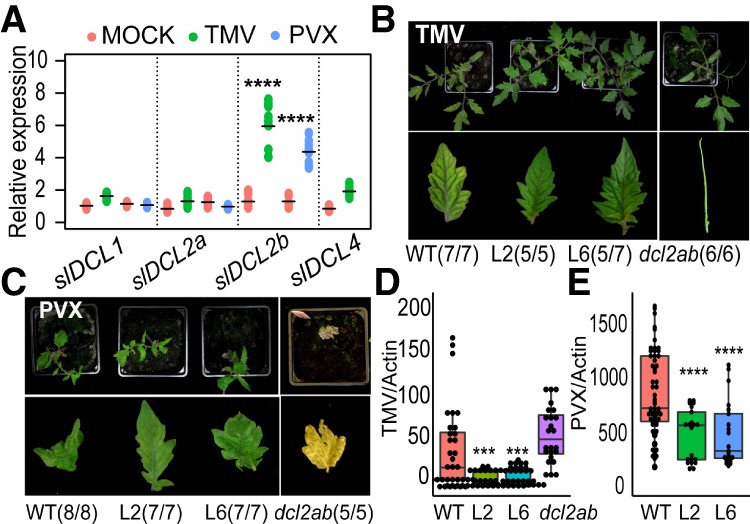
slDCL2a and slDCL2b are required for tomato antivirus defense against TMV and PVX. (A) qRT–PCR results of slDCLs in mock- and virus-infected samples. slActin7 was used as an endogenous control. (B) Two weeks after TMV inoculation, the plants showed chlorosis (wild type [WT]), weak chlorosis (L2 and L6), and “shoestring” (dcl2ab) symptoms of TMV. Numbers in parenthesis show the number of plants with the indicated symptoms and the number of infected plants. (C) Two weeks after PVX inoculation, the plants showed growth stunting and leaf distortion symptoms (wild type [WT] and target mimic lines L2 and L6) or lethality (dcl2ab). Numbers in parenthesis show the number of plants with the indicated symptoms and the number of infected plants. (D,E) Real-time PCR analysis of TMV and PVX in infected plants. slActin7 was used as a control. (***) P < 0.001; (****) P < 0.0001.
The antiviral activity of DCL2 was confirmed by the sRNA profiles of TMV-infected wild type and dcl2ab. In dcl2ab, the 22-nt viral sRNAs were less abundant than in wild type (Supplemental Fig. S21), whereas 21-nt viral sRNAs were increased (Supplemental Fig. S21), possibly due to compensating activity of DCL4.
Also consistent with an antiviral role of DCL2, the 22-nt sRNAs in TMV-infected L6 were more abundant than in wild-type plants (Supplemental Fig. S21). In addition, there were fainter chlorotic symptoms than in wild type (Fig. 5B; Supplemental Fig. S19), and viral RNA accumulation was reduced (Fig. 5D). There was also less viral RNA accumulation in PVX-infected mimic plants than in wild type (Fig. 5E), although the viral symptoms were similar (Fig. 5C).
The reduction of symptoms or virus infection in the TMV-infected plants is likely due to DCL2/miR6026 because the TMV symptoms in the progeny of L6 × dcl2b or dcl2ab were as severe as in dcl2b and dcl2ab (Supplemental Fig. S22). From these results, we conclude that the DCL2-miR6026 feedback influences the effect of RNA silencing on viral RNA accumulation and viral disease progression in tomato (Supplemental Fig. S23).
This analysis of slDCL2 has both applied and basic science implications. At the applied level, we demonstrated that disease resistance can be enhanced by blocking a miRNA that targets a defense mRNA (Fig. 5). In our L6 lines, this effect was achieved without inhibition of basic features in growth and development (Fig. 4C), although a more detailed phenotypic analysis remains to be carried out. The potential of this approach is also confirmed by findings of enhanced resistance to soybean mosaic virus (SMV) (Bao et al. 2018) and Phytophthora infestans (Jiang et al. 2018) in other lines in which defense mRNAs were elevated by miRNA target site mimics. There are multiple miRNAs, including miR403 (Harvey et al. 2011) and the miR482 family (Shivaprasad et al. 2012), that, like miR6026, target defense proteins. Although the effect of any single defense miRNA mimic may be weak (Fig. 5), it could be that strong resistance can be achieved by combining several of these RNAs in a single line.
The fundamental science advance of this work is through the finding that there are endogenous 22-nt sRNA products of DCL2, including the miRNAs (Figs. 1, 2). The finding of DCL2-dependent siRNAs had been suspected previously from the analysis of DCL2 in Arabidopsis (Xie et al. 2004; Bouché et al. 2006) but not shown directly unless the plants also lacked DCL4. We also show that there are D2 sRNAs and D2i sRNAs that are predominantly 24 nt in length (Fig. 1C–E). The DCL2 effect on 24-nt sRNAs is most likely indirect and could influence the epigenome of tomato through the RNA–direct DNA methylation pathway (Nuthikattu et al. 2013). From these various findings, it is clear that DCL2 is not merely a backup for DCL4 but an important component of the diverse RNA silencing pathways of plants.
Materials and methods
Plant materials
All tomato plants used in this study were Solanum lycopersicum cv M82. CRISPR/Cas9 mutants were obtained by stably transforming tomato plants as described previously (Gouil and Baulcombe 2016). For target mimicry plants, two tandem repeats of miR6026 target mimic were cloned into the gateway construct pGWB402Ω and then transformed into tomato. Primers for cloning and genotyping are listed in Supplemental Table S7.
For virus infection, 3-wk-old plants of Nicotiana benthamiana were rub-inoculated with TMV U1 virion or infiltrated with agrobacteria carrying pGR106 for PVX (Harris et al. 2013). After 1 wk, N. benthamiana leaves were harvested and ground to sap, which was rubbed onto 10-d-old tomato cotyledons for inoculation.
RNA analyses
Total RNA was extracted by the TRIzol method. Northern blotting and sRNA library preparation were performed as described (Shivaprasad et al. 2012). RT–PCR was performed as described (Harris et al. 2013). 5′-RLM-RACE was performed following the instruction of the GeneRacer kit (Life Technologies). Primers are listed in Supplemental Table S7.
Bioinformatics
The sRNA reads were trimmed using Trimgalore and mapped to the Heinz genome SL3.0 or respective genes using Bowtie (Langmead et al. 2009) with specified parameters of -m 1 and -v 0. The bam files were used for differential sRNA analysis by SegmentSeq (Hardcastle et al. 2012) and DESeq2 (Love et al. 2014) after filtering out those sRNAs with less than five sequencing reads in five libraries. The raw sequencing data are available in the NCBI Sequence Read Archive database under accession number SRP127908.
Phasing analysis of sRNA was performed by counting 21-nt sRNA mapped to each of 21 registers in each gene. Register 1–21 represents the distance of the sRNA starting site from 5′ of miR6026. Percentages of 21-nt reads corresponding to each of the 21 registers from each gene are shown by radar plots.
Analyses of miRNA were performed based on a tomato miRNA list (Supplemental Table S8) combining 92 miRNAs from miRBase22 and 30 miRNAs from recent publications.
Supplementary Material
Acknowledgments
We are grateful to Dr. John Carr for providing virus material and help. We thank Mel Steer and James Barlow for technical assistance. We thank Dr. Betty Chung, Dr. Sara Lopez-Gomollon, Dr. Claudia dos santos Martinho, Dr. Francisco Navarro, and Dr. Adrian Valli for helpful comments and discussions. We also thank Dr. Ming-Tsung Wu for assistance with RLM-RACE analysis. This work was supported by European Research Council Advanced Investigator grant ERC-2013-AdG 340642 (Transgressive Inheritance in plant Breeding and Evolution [TRIBE]), the Royal Society (RP170001), and the Balzan Foundation. D.C.B. is the Royal Society Edward Penley Abraham Research Professor.
Author contributions: Experiments were designed by Z.W. and D.C.B. and performed by Z.W., A.C.P., W.H.Y., and S.T. Bioinformatics analysis was performed by Z.W. and T.J.H. The manuscript was prepared by Z.W. and D.C.B.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.313601.118.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Bai M, Yang G-S, Chen W-T, Mao Z-C, Kang H-X, Chen G-H, Yang Y-H, Xie B-Y. 2012. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 501: 52–62. [DOI] [PubMed] [Google Scholar]
- Bao D, Ganbaatar O, Cui X, Yu R, Bao W, Falk BW, Wuriyanghan H. 2018. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful soybean mosaic virus infection in soybean. Mol Plant Pathol 19: 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. 2004. RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chen L, Patel K, Li Y, Baulcombe DC, Wu S. 2010. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gouil Q, Baulcombe DC. 2016. DNA methylation signatures of the plant chromomethyltransferases. PLOS Genet 12: e1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA, Baulcombe DC. 2012. Identifying small interfering RNA loci from high-throughput sequencing data. Bioinformatics 28: 457–463. [DOI] [PubMed] [Google Scholar]
- Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC. 2013. Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci 110: 21189–21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJW, Lewsey MG, Patel K, Westwood J, Heimsta S, Baulcombe DC. 2011. An antiviral defense role of AGO2 in plants. PLoS One 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Meng J, Cui J, Sun G, Luan Y. 2018. Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans. Hortic Res 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan G, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchik M, Sunkar R, Damodharan S, Stav R, Zohar M, Isaacson T. 2014. Global and local perturbation of the tomato microRNA pathway by a trans-activated DICER-LIKE 1 mutant. J Exp Bot 65: 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2011. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley MM. 1928. The ‘wiry’ tomato: a recessive mutant form resembling a plant affected with mosaic disease. J Hered 8: 337–344. [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H. 2011. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci 109: 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V. 2008. Dicer-like2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 3: e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T. 2014. Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res 42: 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, Mccue AD, Panda K, Fultz D, Defraia C, Thomas EN, Slotkin RK. 2013. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6. Plant Physiol 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent JS, Bouteiller N, Elmayan T, Vaucheret H. 2015. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81: 223–232. [DOI] [PubMed] [Google Scholar]
- Puga I, Franco-zorrilla M, Todesco M, Mateos I, Paz-ares J, Rubio-somoza I, Leyva A, Weigel D, Garcı JA. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X. 2013. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen H-M, Patel K, Bond DM, Santos BACM, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Saraf S, Dasgupta I, Mukherjee SK. 2016. Identification and validation of a virus-inducible ta-siRNA-generating TAS4 locus in tomato. J Biosci 41: 109–118. [DOI] [PubMed] [Google Scholar]
- Taochy C, Gursanscky NR, Cao J, Fletcher SJ, Dressel U, Mitter N, Tucker MR, Koltunow AM, Bowman JL, Vaucheret H, et al. 2017. A genetic screen for impaired systemic RNAi highlights the crucial role of Dicer-like 2. Plant Physiol 176: 01181.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium G. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Wu Y, Liu C-C, Liu L-W, Ma F-F, Wu X-Y, Wu M, Hang Y-Y, Chen J-Q, Shao Z-Q, et al. 2016. Identification of Arbuscular mycorrhiza (AM)-responsive microRNAs in tomato. Front Plant Sci 7: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YY, Hou BH, Lee WC, Lu SH, Yang CJ, Vaucheret H, Chen HM. 2017. DCL2- and RDR6-dependent transitive silencing of SMXL4 and SMXL5 in Arabidopsis dcl4 mutants causes defective phloem transport and carbohydrate over-accumulation. Plant J 90: 1064–1078. [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G. 2012. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong D, De Paoli E, Park S, Rosen BD, Yan Z, Kitto SL, Grusak MA, Jackson SA, Li Y, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


