Yamazaki et al. show that alternative splicing creates two TCF3 isoforms (E12 and E47) and identified two related splicing factors, hnRNPs H1 and F (hnRNP H/F), that regulate TCF3 splicing. Expression of known TCF3 target E-cadherin, critical for maintaining ESC pluripotency, is repressed by E47 but not by E12.
Keywords: alternative splicing, E2A, heterogeneous nuclear ribonucleoprotein, T-cell factor 3, human embryonic stem cell
Abstract
Alternative splicing (AS) plays important roles in embryonic stem cell (ESC) differentiation. In this study, we first identified transcripts that display specific AS patterns in pluripotent human ESCs (hESCs) relative to differentiated cells. One of these encodes T-cell factor 3 (TCF3), a transcription factor that plays important roles in ESC differentiation. AS creates two TCF3 isoforms, E12 and E47, and we identified two related splicing factors, heterogeneous nuclear ribonucleoproteins (hnRNPs) H1 and F (hnRNP H/F), that regulate TCF3 splicing. We found that hnRNP H/F levels are high in hESCs, leading to high E12 expression, but decrease during differentiation, switching splicing to produce elevated E47 levels. Importantly, hnRNP H/F knockdown not only recapitulated the switch in TCF3 AS but also destabilized hESC colonies and induced differentiation. Providing an explanation for this, we show that expression of known TCF3 target E-cadherin, critical for maintaining ESC pluripotency, is repressed by E47 but not by E12.
Alternative splicing (AS) of mRNA precursors plays an important role in regulating embryonic stem cell (ESC) pluripotency and differentiation. Previous studies have reported that multiple AS events influence the activity of transcription and signaling factors implicated in the control of pluripotency genes (Mayshar et al. 2008; Rao et al. 2010; Salomonis et al. 2010). For example, different isoforms of Foxp1 produced from ESC-specific AS have differential effects on the induction of key pluripotency genes such as OCT4 and NANOG (Gabut et al. 2011). Similarly, alternative splice forms of DNMT3B are specific to stem cells, implying that layered and integrated regulation of gene expression occurs at the levels of transcription and splicing (Gopalakrishna-Pillai and Iverson 2011). Several AS regulators have been implicated in stem cell maintenance and differentiation. For example, MBNL proteins, RBFOX2, and SON have been reported to be important regulators of AS in ESCs (Han et al. 2013; Lu et al. 2013; Venables et al. 2013). However, since AS changes during ESC differentiation are profound and are regulated by orchestrating these and/or other, unidentified splicing regulators, further studies are necessary to understand the stem cell states regulated by AS and the precise regulatory mechanisms and pathways involved.
T-cell factor 3 (TCF3; also known as E2A) is a member of the E protein (class I) family of helix–loop–helix (HLH) transcription factors (Murre et al. 1989). More recent studies have revealed that TCF3 plays important roles in both stem cell maintenance and differentiation. In mouse ESCs (mESCs), ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis revealed that TCF3 co-occupies the promoters of many of the genes regulated by Oct4, Sox2, and Nanog (Cole et al. 2008; Yi et al. 2008), countering the action of these factors and stimulating ESC differentiation (Pereira et al. 2006; Wray et al. 2011; Yi et al. 2011). In fact, deletion of TCF3 maintains high expression of pluripotency genes and delays mESC differentiation into three germ cell lines during embryoid body (EB) formation (Yi et al. 2008). Conversely, TCF3 is also able to repress differentiation-associated genes (Tam et al. 2008). In adult skin, TCF3 is expressed in epidermal stem cells located in the hair follicle bulge, activates a progenitor-associated expression program, and inhibits differentiation (Merrill et al. 2001; Nguyen et al. 2006). Thus, TCF3 is thought to be an important regulator of stem cell identity, capable of promoting either stem cell self-renewal or differentiation, depending on the cellular context.
TCF3 exists as two major isoforms that result from mutually exclusive AS. These isoforms, E12 and E47, are related transcription factors that differ only in their basic HLH (bHLH) DNA-binding region and have different dimerization preferences and hence different DNA-binding properties (Sun and Baltimore 1991). This can lead to different functional consequences. For example, one study using knockout mice deficient for E12 or E47 revealed that E47 is essential for developmental progression at the pre-pro-B-cell stage, whereas E12 is dispensable for early B-cell development, commitment, and maintenance (Beck et al. 2009). In cortical neurogenesis, E47 is required for proper neuronal differentiation and layer-specific localization, whereas E12 is dispensable for early corticogenesis (Pfurr et al. 2017). These reports suggest that TCF3 AS plays an important role in a variety of developmental processes. However, possible functional differences between E12 and E47 have not been investigated in the context of stem cell maintenance.
In this study, we used RNA sequencing (RNA-seq) to identify transcripts that display AS patterns specific to pluripotent human ESCs (hESCs) as opposed to differentiated cell states, with the aim of identifying AS events that contribute to the maintenance of pluripotency. TCF3 was among the genes identified, and we found that TCF3 AS is tightly regulated to control the E12 and E47 expression ratio during hESC differentiation. We then used reporter minigenes to identify cis-regulatory elements and showed that two related heterogeneous nuclear ribonucleoproteins (hnRNPs), H1 and F, bound to newly identified exonic splicing silencers (ESSs) in exon 18b (E47), inhibiting its inclusion and thereby promoting E12 expression and maintaining high levels of E12 and low levels of E47 in hESCs. Interestingly, hnRNP H/F levels were found to decrease during differentiation, and knockdown of these proteins both switched TCF3 AS to give rise to elevated levels of E47 and destabilized the compact morphology of cultured hESCs. Providing a possible mechanism, we show that transcription of known TCF3 target gene CDH1 (Pérez-Moreno et al. 2001; Tiwari et al. 2015), which encodes E-cadherin, a cell adhesion molecule critical for colonization and maintenance of ESC pluripotency (Ohgushi et al. 2010; Redmer et al. 2011), is specifically repressed by E47 but not E12. Our results thus establish that hnRNP H/F-mediated regulation of TCF3 AS is an important switch for controlling stem cell pluripotency and differentiation.
Results
RNA-seq identifies AS changes during hESC differentiation
We first wished to identify AS events that may play important regulatory roles in the maintenance of pluripotency and/or differentiation of hESCs. To this end, we performed RNA-seq with RNA isolated from undifferentiated hESCs (H9 cells) and hESCs differentiated into either neural progenitor cells (NPCs) or trophoblast-like cells (TBs) (Fig. 1A). This analysis found 73 splicing changes common to both differentiation pathways (Fig. 1B, top left; Supplemental Table S1). We also compared these data with a published RNA-seq data set (Xie et al. 2013) in which hESCs (H1 cells) were also differentiated into NPCs and TBs, albeit using a different cell line and culture conditions. Since the published data set reflected much greater sequencing depth (Supplemental Fig. S1), 1996 AS events were detected (Fig. 1B, top right; Supplemental Table S2). Comparing the events that changed when hESCs differentiated into both NPCs and TBs from the two different conditions, we found 35 AS events in common (Fig. 1B, bottom; Supplemental Table S3). Of these, eight splicing changes from seven genes (all of which were confirmed by RT–PCR) (Supplemental Fig. S2) were of special interest because these AS events changed in the same way under all differentiation conditions tested, and the average of changes in inclusion levels was >20% (Fig. 1C). Significantly, the top-ranked event was DNMT3B AS, which is specific to stem cells and has been used as biomarker of human pluripotent stem cells (Gopalakrishna-Pillai and Iverson 2011), indicating that our analysis successfully captured stem cell-specific AS events. In this study, we focused on the AS changes affecting TCF3 because, as described above, TCF3 is a well-studied transcription factor that is known to be a key regulator of embryonic development.
Figure 1.
Identification of human stem cell-specific splicing changes. (A) Schematic of the identification of hESC-specific splicing events. (B) The top left Venn diagram shows the overlap of altered splicing events between two differentiation pathways in H9 hESCs (this study), and the top right Venn diagram shows the overlap in H1 hESCs (Xie et al. 2013). (Bottom) Venn diagram showing the overlap of splicing changes between two different cultures. (C) List of identified stem cell-specific AS changes. These splicing events switched in the same way in both pathways and both cultures, and the average of changes in inclusion levels in both pathways was >20% (Inc_Level >0.2).
TCF3 AS changes during early differentiation
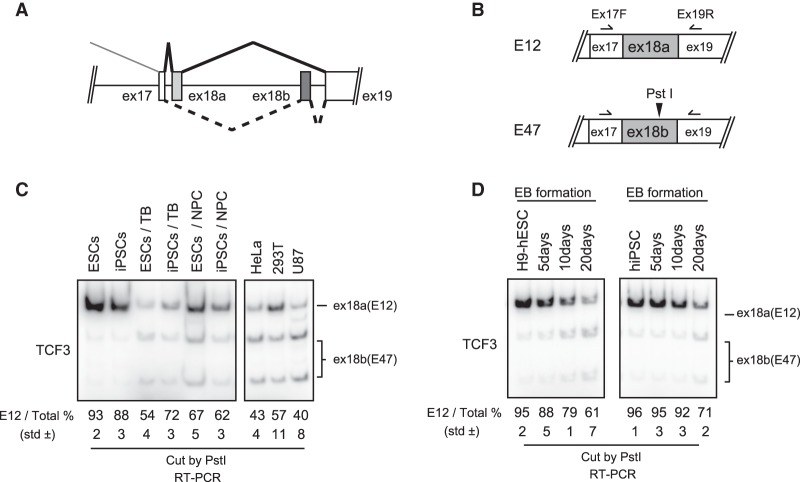
The TCF3 gene encodes the highly related proteins E12 and E47. These result from mutually exclusive splicing of two alternative 18th exons (RefSeq NM_003200), which we refer to as exons 18a and 18b (Fig. 2A). Our RNA-seq analysis indicated that exon 18b inclusion increased in H9-derived TBs and NPCs compared with undifferentiated H9 hESCs (Fig. 1C). Since the two mutually exclusive exons are the same size and display high sequence similarity (74% identity) (Supplemental Fig. S4), we confirmed the splicing switch using RT–PCR followed by exon-specific restriction digestion (Fig. 2B). Consistent with the RNA-seq data, exon 18a is efficiently included in undifferentiated H9 hESCs (Fig. 2C, lane 1), while exon 18b is favored in differentiated cells (Fig. 2C, lanes 3,5). To examine whether the TCF3 AS changes indeed reflect pluripotency, we examined the AS state of TCF3 in undifferentiated human induced pluripotent stem cells (hiPSCs) and hiPSC-derived NPCs and TBs. Notably, similar AS changes were observed in hiPSCs (Fig. 2C, lanes 2,4,6). The AS state of TCF3 mRNA in several transformed human cell lines was also tested, and, as expected, exon 18a inclusion was relatively low compared with the pluripotent stem cells (Fig. 2C, right column). We also investigated AS changes of TCF3 transcripts in early development using an EB formation assay. The AS switch from exon 18a to 18b was observed in EB formation using both H9 hESCs and iPSCs (Fig. 2D). Together, these results suggest that predominant inclusion of TCF3 exon 18a is a characteristic of self-renewing pluripotent hESCs and iPSCs.
Figure 2.
AS switches TCF3 exon 18a (E12) to exon 18b (E47) during differentiation of human pluripotent cells. (A) Schematic representation of TCF3 splicing. (B) Scheme for assaying E12/E47 ratio. (C) E12/E47 ratio in a variety of types of cells was determined by 32P RT–PCR. Total RNA was extracted from hESC H9 cells, human iPSCs, H9-derived trophectoderm, H9-derived NPCs, human iPSC-derived trophectoderm, human iPSC-derived NPCs, and four human immortalized cell lines of diverse origin, as indicated. RT–PCR products were digested by PstI and resolved on a 5% PAGE gel. The E12 percentage is indicated below. The standard deviation (SD) from biological triplicate data is also indicated. (D) E12/E47 ratio during EB formation, analyzed by 32P RT–PCR. H9 cells and human iPSCs aggregated to form EBs, and total RNA was extracted at the indicated time points. RT–PCR products were digested by PstI and resolved on a 5% PAGE gel. The E12 percentage is indicated below.
Identification of AS exonic regulatory elements in TCF3 pre-mRNA
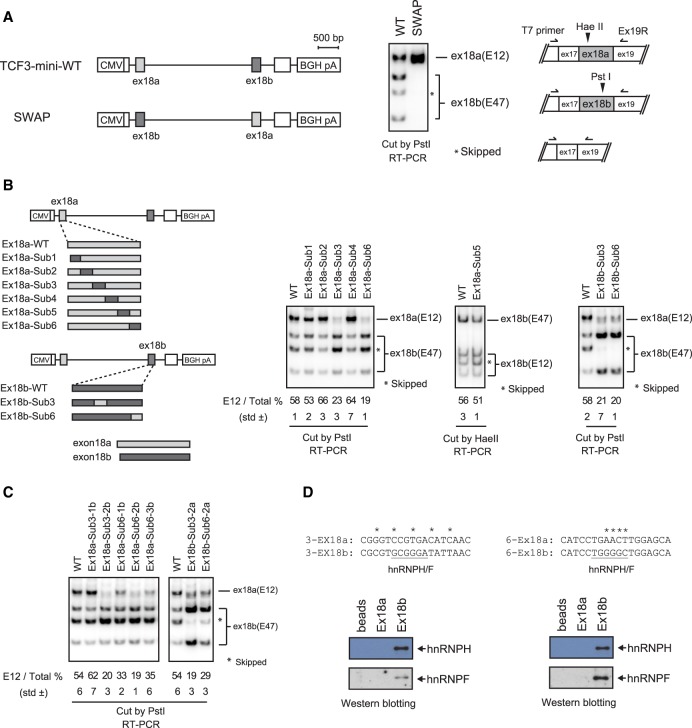
To investigate the TCF3 AS mechanism, we next established a minigene splicing construct containing TCF3 sequences from exons 17 to 19. This construct recapitulates TCF3 mutually exclusive splicing of exons 18a and 18b (Supplemental Fig. S4A), and we therefore used it to investigate whether cis-elements in exon 18a and/or 18b function in TCF3 splicing regulation. First, we precisely swapped exons 18a and 18b to determine whether exon 18 sequences contain position-dependent regulatory elements. Interestingly, exon swapping disrupted TCF3 AS by completely preventing exon 18b inclusion, leading to 100% inclusion of the swapped exon 18a (Fig. 3A). This suggests that exon 18a and/or 18b contain exonic regulatory elements and that the positions of such elements can influence TCF3 AS regulation.
Figure 3.
hnRNP H/F proteins bind exonic regulatory elements in exon 18b. (A, left) Diagram of wild-type and SWAP constructs in which the positions of exon 18a and exon 18b are switched. (Right) 32P RT–PCR assays of RNAs extracted from transfected HeLa cells using T7 primer and Ex19R. PCR products were digested with PstI. (B, left) Diagram of point-swapping mutant constructs. The details of a series of mutants are described in Supplemental Figure S4. (Right) 32P RT–PCR assays of RNAs extracted from transfected HeLa cells using T7 primer and Ex19R. PCR products were digested with PstI or HaeII. The E12 (exon18a inclusion) percentage was calculated by dividing exon 18a signal by the sum of exon 18a and 18b signals and is indicated below. (C) 32P RT–PCR assays of additional point-swapping mutants in sections 3 and 6 in HeLa cells. (D, top) Sequence comparison of sections 3-2 and 6-2. The putative hnRNP H/F-binding sites are underlined. The RNA affinity assay used biotinylated 3-2 and 6-2 short RNA oligonucleotides. Isolated proteins were analyzed by Western blot using the indicated antibodies.
To identify the exon 18 regulatory elements, we constructed further mutated minigenes. We first divided both exon 18a and 18b into six sections (Supplemental Fig. S4B) and swapped each of these sections between exon 18a and 18b. Strikingly, replacement of sections 3 and 6 of exon 18a with the corresponding exon 18b sequences strongly reduced inclusion of the modified exon 18a. To confirm this, we constructed reciprocal mutants that replaced regions 3 and 6 of exon 18b with the corresponding exon 18a sequences. As expected, the swapped exon 18b was now efficiently included (Fig. 3B). These results allowed us to construct additional minigene derivatives dividing sections 3 and 6 into smaller regions (underlined in Supplemental Fig. S4). We first divided section 3 into two smaller sections, and Ex18a-Sub3-2b, which replaced only 5 nucleotides (nt) of exon 18a with 18b sequence, almost completely prevented 18a inclusion (Fig. 3C, third lane). For section 6, we produced three smaller sections, and all of the replacements reduced inclusion of modified exon 18a—most effectively Ex18a-Sub6-2b, which replaced 4 nt (Fig. 3C, third through fifth lanes). Consistent with this, reciprocal mutants that replaced regions 3-2 and 6-2 of exon 18b with the corresponding exon 18a sequences showed that the modified exon 18b was efficiently included (Fig. 3C, right). The above splicing assays identified two critical sequences in exon 18b that control differential exon 18a and 18b inclusion, likely by functioning as ESSs (Fig. 3D, top).
We next performed additional splicing assays using a series of TCF3 deletion mutants to examine whether the corresponding sequences in exon 18a contain sequences that contribute to regulation. While, as expected, deletion of exon 18b sequences containing the ESSs identified in the swapping experiments above increased exon 18b inclusion, deletion of the corresponding sequences in exon 18a did not affect exon 18a inclusion (Supplemental Fig. S5). These results confirm that the splicing changes observed with the swap mutants indeed arose from ESSs in exon 18b and that the corresponding sequences in exon 18a do not contain sequences that enhance its splicing.
We next wished to identify RNA-binding proteins (RBPs) that recognize these exonic regulatory elements. To this end, we first evaluated all reported RBP sequence motifs by using the SpliceAid 2 database (Piva et al. 2012). Notably, GC-rich motifs recognized by the related RBPs hnRNP H1 and hnRNP F were found in both exon 18b elements but not in the corresponding exon 18a sequences. We next performed RNA affinity assays using nuclear extracts (NEs) from H9 cells and biotin-labeled 18-nt RNA oligonucleotides corresponding to each of the sequences from both exons to determine whether hnRNP H1 and/or hnRNP F indeed binds the exon 18b sequences. Silver-stained SDS-PAGE gels revealed numerous proteins binding to both RNAs, although bands with the approximate size of hnRNP H/F were detected with the exon 18b but not 18a sequences (Supplemental Fig. S6). Western blot confirmed that hnRNP H1 and hnRNP F indeed bound to the exon 18b but not 18a regulatory sequences (Fig. 3D). These results suggest that hnRNP H/F are potential regulators of TCF3 AS.
hnRNP H/F occupancy on exon 18b and overall levels decrease during differentiation
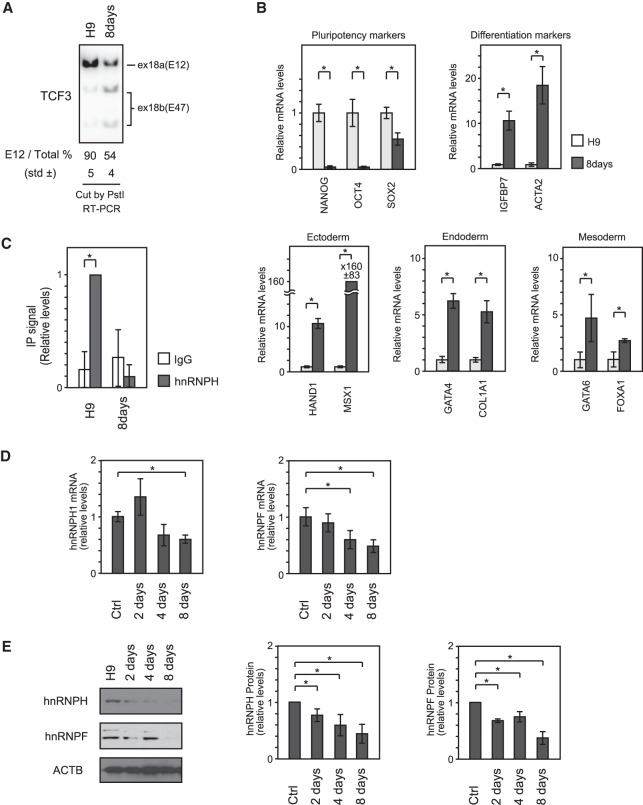
Given the above in vitro evidence that hnRNP H1 and hnRNP F bind regulatory elements in exon 18b in hESC extracts, we next examined whether hnRNP H1 binds these sequences in H9 cells and, if so, whether occupancy changes during differentiation. To this end, we performed CLIP (cross-linking immunoprecipitation) to measure binding to exon 18b. H9 cells were heterogeneously differentiated as a monolayer by culturing on Matrigel in EB-stimulating medium for 8 d. As expected, we observed the TCF3 AS switch from exon 18a to 18b (Fig. 4A) as well as a decrease of stem cell markers and an increase of differentiation markers (Fig. 4B). Following UV cross-linking, hnRNP H1 was immunoprecipitated from cell extracts prepared from undifferentiated and 8-d differentiated cells. After reversal of cross-linking, immunoprecipitated RNA was measured by RT-qPCR using primers spanning the two exonic regulatory elements in exon 18b. As shown in Figure 4C, hnRNP H1 binding to exon 18b was observed in undifferentiated H9 cells, but the signal was greatly reduced in the differentiated cells. Similar experiments to measure hnRNP F levels were unsuccessful, as the antibodies used did not produce signals.
Figure 4.
hnRNP H/F expression and recruitment to exonic regulatory elements decrease during differentiation. (A) E12/E47 ratio was examined by 32P RT–PCR. PCR products were digested by PstI. (B) Expression of pluripotency and differentiation markers as determined by quantitative PCR (qPCR) after 8 d of differentiation. The P-value was determined by two-tailed Student's t-test. (*) P < 0.05. (C) CLIP analysis to measure the hnRNP H protein enrichment on exon 18b of TCF3 mRNA by qPCR after 8 d of differentiation. Data were normalized to hnRNP H immunoprecipitation signal in undifferentiated H9 cells. All results represent the means ± SD from at least three independent experiments. (*) P < 0.05. (D) Expression of hnRNP H and hnRNP F during 8 d of differentiation of H9 cells. mRNA levels were determined by qPCR. (E) Protein levels were analyzed by Western blot using the indicated antibodies (left), and the bar graph shows its quantification (right).
A simple explanation for the decreased occupancy of hnRNP H1 on exon 18b following differentiation is that expression levels decrease. We therefore examined both hnRNP H1 and hnRNP F mRNA and protein levels in H9 cells differentiated as described above. We first measured mRNA levels in undifferentiated and 8-d differentiated cells by RT-qPCR. After differentiation, both hnRNP H1 and hnRNP F mRNA levels were significantly reduced—by ∼50% (Fig. 4D). Importantly, Western blot analysis showed even stronger decreases of both proteins in the differentiated cells (Fig. 4E). This might arise from post-transcriptional regulation, as suggested by Grammatikakis et al. (2016).
hnRNP H/F regulate TCF3 AS and help drive stem cell differentiation
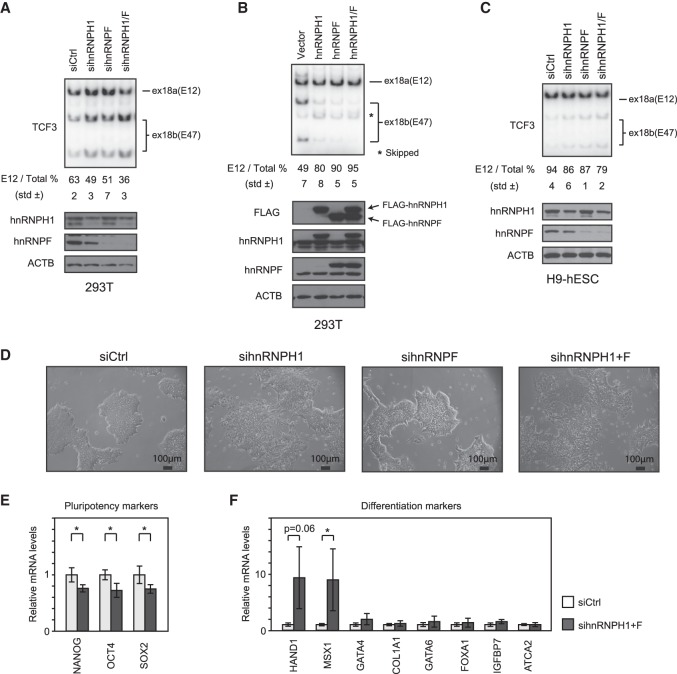
Since our data suggest that hnRNP H/F are involved in TCF3 AS regulation during differentiation, we next tested whether depletion or overexpression of either protein affects TCF3 splicing. We first transfected 293T cells with siRNAs targeting hnRNP H1 and/or F. Knockdown of both proteins resulted in a significant increase in exon 18b inclusion and a corresponding decrease in the E12/E47 ratio—from 63% to 36% (Fig. 5A). Knockdown of hnRNP H/F in HeLa cells behaved similarly (Supplemental Fig. S7). Conversely, overexpression of hnRNP H1 and/or hnRNP F strongly suppressed 18b inclusion in 293T cells cotransfected with the TCF3 minigene described above (Fig. 5B). Previous studies have shown that when hnRNP H1 binds exonic regions, it often functions to repress exon inclusion (Xiao et al. 2009), which is consistent with our results. We next examined whether knockdown of these proteins in H9 hESCs had a similar effect on TCF3 AS. Indeed, knockdown of both proteins resulted in a modest but reproducible and significant increase in exon 18b inclusion and a corresponding decrease in the E12/E47 ratio—from 94% to 79% (Fig. 5C). The relatively small magnitude of the AS switch may reflect the low knockdown efficiency of hnRNP H1 in H9 cells (Fig. 5C, bottom).
Figure 5.
hnRNP H1 and hnRNP F regulate TCF3 splicing and hESC morphology. (A) E12/E47 ratio in hESCs after the indicated siRNA treatment analyzed by 32P RT–PCR. RT–PCR products were digested by PstI and resolved by 5% PAGE. (Top) The E12 percentage is indicated below. (Bottom) Immunoblots showing protein levels after the indicated siRNA treatment. (B) E12/E47 ratio in 293T cells after the indicated siRNA treatment analyzed by 32P RT–PCR. (C) Splicing assays analyzed by 32P RT–PCR using T7 primer and Ex19R. The TCF3 minigene and the indicated expression plasmids were transfected into 293T cells. Total RNA from the transfected cells was used for 32P RT–PCR. (D) Cell morphology of H9 hESCs transfected by the indicated siRNAs. (E) Expression of pluripotency markers in hnRNP H/F knockdown cells, determined by qPCR. (F) Expression of differentiation markers in H9 hESCs after the indicated siRNA treatment, determined by qPCR.
We next examined the H9 hESCs following hnRNP H/F knockdown to determine whether, in addition to the TCF3 AS switch, they underwent morphological changes characteristic of differentiation. It is well known that hESCs must be cultured as cell clumps or colonies because they undergo apoptosis when dissociated (Watanabe et al. 2007 and references therein). Strikingly, following 48 h of siRNA transfection, hnRNP H/F knockdown transformed the cells from a typical, tight epithelium phenotype to a loose, decolonized phenotype (Fig. 5D; Supplemental Fig. S8). This indeed resembles the epithelial–mesenchymal transition (EMT), which is essential for germ layer formation and cell migration in the early vertebrate embryo (Acloque et al. 2009). To characterize this differentiated phenotype in the knockdown cells, we examined expression of several differentiation markers by RT-qPCR. This analysis revealed that although only a limited but significant decrease of stem cell marker expression occurred (Fig. 5E), expression of two markers—HAND1 for extraembryonic ectoderm and MSX1 for embryonic ectoderm—was significantly increased (Fig. 5F). Together, these data suggest that hnRNP H/F knockdown stimulates stem cell differentiation, especially along the pathway to embryonic and extraembryonic ectoderm.
TCF3 splice variants E12 and E47 regulate CDH1 expression differently
We next investigated how hnRNP H/F knockdown might bring about decolonization and differentiation. Colonization of hESCs is known to be maintained by the cell adhesion molecule E-cadherin (Watanabe et al. 2007; Ohgushi et al. 2010), and E-cadherin balances ESC self-renewal and differentiation (Redmer et al. 2011; Huang et al. 2015). Interestingly, CDH1, which encodes E-cadherin, is a known TCF3 transcriptional target (Pérez-Moreno et al. 2001; Tiwari et al. 2015), and we therefore next investigated CDH1 expression levels. Significantly, Western blot and quantitative RT–PCR (qRT–PCR) revealed a decrease in CDH1/E-cadherin protein and mRNA levels following hnRNP H/F knockdown in both H9 hESCs (Fig. 6A,B) and differentiated H9 hESCs (Fig. 6C,D).
Figure 6.
E47-specific, but not E12-specific, knockdown releases E-cadherin repression. (A,B) E-cadherin protein levels in H9 hESCs analyzed by Western blot (A) and mRNA levels determined by RT-qPCR (B). (C,D) Expression of E-cadherin protein levels during 8 d of differentiation of H9 hESCs analyzed by Western blot (C) and mRNA levels determined by RT-qPCR (D). (E) Expression of CDH1 in H9 hESCs after the indicated siRNA treatment, determined by RT-qPCR and Western blot. (F) Expression of CDH1 in 293T cells after the indicated siRNA treatment, determined by qPCR and Western blot.
We next wished to investigate whether the hnRNP H/F-mediated switch in TCF3 AS might underlie the reduction in CDH1 expression during differentiation. Overexpression of TCF3 (E47) induces the EMT, transforming epithelial-like MDCK cells to a migratory phenotype by repressing CDH1 transcription and stimulating mESC differentiation, although whether this activity is specific to E47 is not known (Pérez-Moreno et al. 2001; Yi et al. 2011). Our finding that hnRNP H/F knockdown favored E47 production and concomitantly induced decolonization of hESCs prompted us to hypothesize that E12 and E47 differently regulate CDH1. To examine this, we transfected H9 hESCs with siRNAs specifically targeting either exon 18a or 18b. Significantly, RT-qPCR revealed that CDH1 mRNA levels increased following E47 knockdown but decreased upon E12 knockdown (Fig. 6E, right). Additionally, Western blots indicated that E-cadherin protein levels increased in the E47 knockdown cells, but no changes in protein levels were observed following E12 knockdown (the difference in relative mRNA and protein levels in the E12 knockdown cells may reflect E-cadherin protein stability and/or a requirement for appropriate cell signaling for protein turnover) (Fig. 6E; Shen et al. 2008). Similar but more striking effects on CDH1 mRNA and protein expression following E47 knockdown were observed in 293T cells (Fig. 6F), perhaps because of higher knockdown efficiency and/or because 293T cells have a higher E47/E12 expression ratio compared with undifferentiated H9 cells.
The above data support our hypothesis that the two TCF3 isoforms differentially regulate CDH1 expression. Importantly, the fact that only E47 acts as a repressor is consistent with the changes in TCF3 AS and E-cadherin expression that occur during hESC differentiation. Together, our data revealed a TCF3 AS switch regulated by hnRNP H/F, which in turn contributes to hESC maintenance and differentiation at least in part by controlling E-cadherin expression.
Discussion
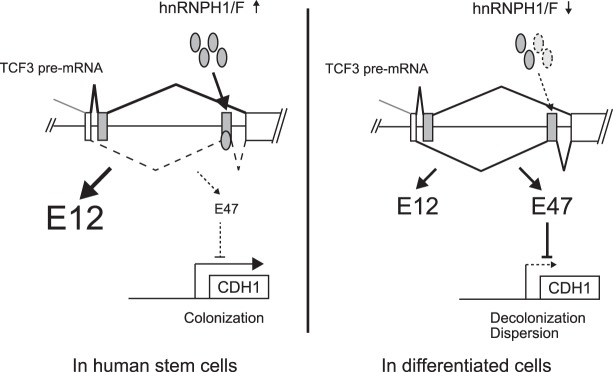
In this study, we identified a number of AS events that have the potential to be important for stem cell identity by comparing multiple RNA-seq data sets and found several interesting stem cell-specific splicing changes in hESCs. One of the most intriguing of these was TCF3 mutually exclusive AS. TCF3 AS was first described almost 30 yr ago (Murre et al. 1989) and since then has been investigated and characterized predominantly in the context of lymphoid development (Miyazaki et al. 2017 and references therein). However, TCF3 was essentially rediscovered as an integral factor in the core regulatory circuitry of pluripotent stem cells, and multiple important roles have been reported, mostly in mESCs (see above; Pereira et al. 2006; Cole et al. 2008; Yi et al. 2008, 2011; Wray et al. 2011). Whereas E12 and E47 are now thought to function by differentially dimerizing with a variety of tissue-specific class II HLH proteins (such as NeuroD1, Ets, Pax, and negative regulatory ID proteins) to control cell type specification and differentiation programs during embryonic development (Wang and Baker 2015 and references therein), possible functional differences between the two isoforms in stem cell maintenance had not been investigated previously. Our study shows not only that these two TCF3 isoforms can differentially regulate a physiologically important target, CDH1, but also that the highly related hnRNPs H1 and F play critical roles in regulating the AS event that generates E12 and E47. Importantly, this contributes to maintenance of the pluripotent state at least in part by controlling E-cadherin expression (Fig. 7). Below, we discuss the newfound roles of hnRNP H/F and TCF3 AS in controlling differentiation of hESCs.
Figure 7.
Model for TCF3 splicing regulation. In stem cells, hnRNP H/F are highly expressed, bind to ESSs in exon 18b, and repress its inclusion. Therefore, E12, which does not repress CDH1 expression, is preferentially expressed, resulting in E-cadherin production and thus E-cadherin-dependent intercellular attachment. In differentiated cells, hnRNP H/F are down-regulated, likely due to reduced expression of the activator c-Myc, resulting in preferential inclusion of exon 18b. E47 expression is thus increased, resulting in CDH1 repression, and the lower E-cadherin levels lead to an altered cell morphology—from stable colonies that have tight intercellular attachments to an invasive and migratory phenotype.
TCF3 is well known to play important roles in controlling differentiation in mESCs. For example, knockdown of TCF3 in mESCs was shown to induce expression of NANOG, OCT4, and other pluripotency genes and result in a permanent undifferentiated state (Pereira et al. 2006; Cole et al. 2008). In light of this, it was unexpected when we found that knockdown of E12 and/or E47 did not result in induction of pluripotent genes in H9 hESCs. We suggest that this discrepancy arises from interspecies differences. There are, in fact, several substantial differences between hESCs and mESCs even though both are pluripotent cells derived from blastocyst embryos (Thomson et al. 1998; Sato et al. 2003). Most relevantly, significant interspecies differences in TCF3 and its target genes exist. For example, a recent study showed that TCF3 targets overlap less with those of the pluripotency regulators OCT4 and NANOG in hESCs than in mESCs and that human TCF3 acts generally on differentiation-specific rather than pluripotency-specific genes (Sierra et al. 2018). In addition, while we found that mutually exclusive AS of TCF3 switches exon 18a (E12) to exon 18b (E47) during hESC differentiation, Salomonis et al. (2010) reported that skipping of exon 4, which creates a short TCF3 isoform, is an mESC-specific AS event not conserved in hESCs even though the mESC-specific TCF3 isoform does regulate pluripotency and expression of early developmental genes. Furthermore, TCF3-responsive elements in the CDH1 promoter are also different between humans and mice. In mice, TCF3 binds a palindromic E-box sequence, which is often recognized by homodimers (Behrens et al. 1991; Pérez-Moreno et al. 2001), whereas multiple single E-box sequences are found in the human gene (Tiwari et al. 2015; Ahn et al. 2016). These observations suggest that regulation of TCF3 AS and TCF3 downstream targets such as CDH1 may explain at least in part the interspecies differences between hESCs and mESCs. Consistent with this, cadherin/catenin-dependent intercellular attachment is crucial for survival of hESCs but not mESCs (Ohgushi et al. 2010).
TCF3 is also known to play important roles in a variety of differentiation-related processes, but, in these cases, how AS might contribute is unknown. For example, in mice, TCF3 is highly expressed in neuronal stem cells of the subventricular zone niche, and TCF3-null embryos die at embryonic day 9.5 and display defects in neural and mesodermal patterning (Merrill et al. 2004). TCF3 also functions in skin stem cells to maintain an undifferentiated state and, through Wnt signaling, directs these cells along the hair lineage (Nguyen et al. 2006). It also has been reported recently that TCF3 coordinates early heart formation by functioning with other HLH and ID proteins (Cunningham et al. 2017). In addition to developmental functions, TCF3 mutations have been reported to cause disease, such as Burkitt lymphoma (Schmitz et al. 2012). It will be of interest to determine whether any of these TCF3-dependent events are modulated by AS and, if so, whether changes in hnRNP H/F concentrations play a regulatory role.
Previous studies have shown that alterations of hnRNP H1 and/or hnRNP F levels can have physiologically significant effects on AS. For example, hnRNP H/F were found to function in neuronal differentiation by controlling the AS of multiple transcripts (Min et al. 1995; Chou et al. 1999; Han et al. 2005; Wang et al. 2012; Mandler et al. 2014; Xu et al. 2014). hnRNP H1 has been reported to regulate AS in oligodendrocytes (Wang et al. 2012; Mandler et al. 2014) and inclusion of the brain-specific C1 cassette exon of the glutamate NMDA R1 receptor mRNA (Han et al. 2005). Additionally, a recent study showed that a decrease in hnRNP H1 levels stimulates neuronal differentiation in rat PC 12 cells (Grammatikakis et al. 2016). Also supporting a physiologically important role for hnRNP H1 in AS control, we reported recently that the C9ORF72 (C9) GGGGCC expansion, a major cause of amyotrophic lateral sclerosis (ALS)/frontotemporal lobar degeneration (FTD), forms RNA G-quadruplex inclusions that sequester hnRNP H1, which disrupts splicing of hnRNP H1 target transcripts in ALS patient brains (Conlon et al. 2016). Interestingly, consistent with our findings here that hnRNP H1 represses exon 18b inclusion, hence reducing E47 levels, RNA-seq analysis comparing C9ALS and healthy brains (Prudencio et al. 2015) suggested that E47 levels were in fact increased in C9 ALS patient brains. Together, these data suggest that regulation of TCF3 AS by hnRNP H/F may have a profound impact on transitions between cell proliferation and lineage specification—especially during neural development—and may perhaps play a role in neurodegenerative disease.
Our findings also suggest that hnRNP H/F play an important role in stem cell maintenance and differentiation. Coupled with the decrease in hnRNP H/F expression in differentiated H9 cells, our finding that hnRNP H/F knockdown destabilizes the hESC colony phenotype supports a role in maintenance of pluripotency (Eastham et al. 2007; Acloque et al. 2009). Interestingly, a recent study showing that hnRNP F inhibits EMT by regulating CD44 AS in breast cancer (Huang et al. 2017) supports our model, as our data suggest that hnRNP H/F have the potential to regulate the migration step from the hESC pluripotent state to differentiated tissues by releasing cells from E-cadherin-mediated tight intercellular attachments in early development. Additionally, based on expression of characteristic markers, loss of hnRNP H/F leads to ectoderm lineage differentiation. Importantly, here, neuronal cells are generally believed to be differentiated from the ectoderm (Murry and Keller 2008). Together with the fact that hnRNP H/F regulates a variety of AS events in neuronal tissues and hence neural differentiation (see above), hnRNP H/F may function very broadly—from the early ectodermal stage to specific differentiated neural linages.
In conclusion, we showed here that hnRNP H/F are important for maintenance and differentiation of hESCs and that this at least in part reflects a switch in TCF3 AS that leads to repression of CDH1/E-cadherin. Notably, this is consistent with a previous genome-wide RNAi screen using an OCT4 promoter-driven reporter system designed to identify determinants for hESC identity (Chia et al. 2010). Although stem cell transcription factors and cofactors were enriched among the top candidate genes, hnRNP H1 ranked in the top 3% among 21,000 tested genes, consistent with our findings that hnRNP H1 plays a critical role in maintenance and differentiation of hESCs. Furthermore, providing insight into how hnRNP H1 levels are regulated, the Yamanaka factor c-Myc, the levels of which decrease during ESC differentiation (Cartwright et al. 2005), activates hnRNP H1 expression by binding to a noncanonical E-box in the promoter of the hnRNPH1/2 gene in cancer cells (Rauch et al. 2011; Li et al. 2016). Future studies are likely to reveal more functions of hnRNP H/F—and perhaps other RBPs—in controlling AS in hESCs and thereby contributing to the maintenance of pluripotency and/or differentiation of stem cells.
Materials and methods
Cell culture and transient transfection
HeLa and 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). DNA transfection reagent (Biotools) was used for plasmid transient transfection. siRNAs were transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. To achieve satisfactory transfection efficiency, H9 cells were transfected twice with siRNAs. H9 cells were plated at 200,000∼400,000 cells per well on Matrigel-coated 12-well plates in Essential 8 medium (Stem Cell Technologies). After 2 d of culture, H9 cells were transfected with siRNA, transfected again 1 d later, and then harvested 2 d later. Sequences of siRNAs are listed in Supplemental Table S4.
Stem cell maintenance and differentiation
hiPSCs were established as described previously (Wu et al. 2012). A human H9 ESC line (WA09) and hiPSCs were maintained in Essential 8 medium or mTeSR1 (Stem Cell Technologies) on Matrigel-coated (Corning) plates. hESCs (WA-09) were differentiated into either NPCs (Chambers et al. 2009) or TBs (Xu et al. 2002) as described. For EB formation, a modified protocol from Wu et al. (2012) was used. H9 cells and hiPSCs were harvested with ReLeSR (Stem Cell Technologies) and plated on a Nunclon Sphera 35-mm dish (Thermo Scientific) in DMEM/F12 (Gibco) consisting of 15% FBS (BenchMark), 15% knockout serum replacement (Invitrogen), 0.1 mM nonessential amino acids, and 0.5% penicillin and streptomycin. The medium was changed every 2 d. EBs in the supernatant were harvested by centrifugation at 1000 rpm for 5 min (IEC Centra CL2). For CLIP assays, we differentiated H9 cells heterogeneously as a monolayer to cross-link RNA–protein interactions using UV irradiation. H9 cells were plated on Matrigel-coated plates in Essential 8 medium. Three days later, the culture medium was changed to the EB-stimulating medium described above and then cultured for 8 d. Y27632 (Regents Direct) was supplemented on the first 2 d. The medium was changed every 2 d.
RT–PCR, 32P RT–PCR, and RT-qPCR
Total RNA was extracted from tissue culture samples using TRIzol (Invitrogen) according to the manufacturer's instructions. Total RNA (1 µg) was reverse-transcribed using Maxima RT (Thermo Scientific) primed with random hexamers (Invitrogen). RT–PCR was performed using Taq DNA polymerase (Invitrogen). For 32P RT–PCR, 3 µCi of [32P]dCTP was added per 50 µL of PCR reaction. RT-qPCR was performed with Power SYBR using StepOnePlus (Applied Biosystems). qPCR data were analyzed by the ΔΔCT method, normalizing to the ACTB gene for knockdown experiments in HeLa, 293T, and H9 cells. RPL13A was used as a stable control (Vossaert et al. 2013) for 8-d differentiated samples. Primers used in the PCR reactions are listed in Supplemental Table S4.
Plasmids constructs
hnRNP H1 and hnRNP F genes were amplified from HeLa cDNA and cloned in pcDNA3 with 3xFlag tag using either BamHI and XbaI or BamH1 and XhoI for mammalian expression. To construct the human TCF3 minigene reporter, TCF3 exon 17, exon 18a, exon 18b, exon 19, and flanking regions were cloned by PCR from 293T genomic DNA and inserted into pcDNA3 (Invitrogen) using HindIII and EcoRI. For the SWAP construct, exon 18a fragments and TCF3 minigene sequence without the exon 18b region were amplified by PCR and ligated using T4 PNK and T4 ligase (New England Biolabs). These steps were repeated to replace exon 18a with exon 18b. A series of swapping mutations was introduced by PCR-based site-directed mutagenesis. Primers used in the mutagenesis are listed in Supplemental Table S4.
Minigene splicing assays
One-hundred nanograms of wild-type or mutated minigene vectors was transfected into HeLa cells. To examine the effects of exogenous expression of hnRNP H1 and hnRNP F, 500 ng of empty vector and/or expression vectors was transfected with 50 ng of TCF3 minigene. Forty-eight hours after transfection, cells were collected, and the E12/E47 ratio was analyzed using 32P RT–PCR followed by PstI digestion. Digested PCR products were separated on a 5% PAGE gel. ImageQuant TL (GE healthcare) was used for quantification.
Western blotting
Cells were lysed with SDS sample buffer (50 mM Tris at pH 7.4, 100 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 1× proteinase inhibitor cocktail [Biotools]), resolved by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with primary antibodies detecting CDH1 (1:1000; Cell Signaling), hnRNP H1 (1:1000; Bethyl Laboratories), hnRNP F (1:1500; Santa Cruz Biotechnology), GAPDH (1:20,000; Sigma), or ACTB (1:30,000; Sigma) overnight at 4°C. Secondary antibodies HRP-conjugated anti-mouse and anti-rabbit IgG (Sigma) were used at 1:10,000 for 2 h at room temperature. Following washes with PBST (1× PBS, 0.1% Tween-20), ECL-Prime (GE Healthcare) was used to visualize the chemiluminescence signal, and ImageJ software was used for quantification.
CLIP analysis
Self-renewing H9 cells and differentiated H9 cells plated on Matrigel were cross-linked by 400 mJ/cm2 of 254-nm UV using a UV Stratalinker 1800 (Stratagene). Cross-linked H9 cells were collected by scraping, and total cell lysates were prepared in lysis buffer (50 mM Tris at pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1× proteinase inhibitor cocktail [Biotools]). For immunoprecipitation, 5 µg of anti-hnRNP H antibody (Invitrogen) or rabbit IgG (Santa Cruz Biotechnology) was incubated with 40 µL of protein G/A magnetic beads (Pierce) for 1 h at room temperature. Beads were then washed twice with lysis buffer. Total cell lysates were treated with 3 U of DNase RQ1 (Promega) per sample and 0.2 U of RNase I per sample for exactly 30 min at 37°C. Lysates were then centrifuged at 20,000g for 20 min at 4°C, and the supernatants were mixed and incubated with a specific antibody or IgG-coated beads for 4 h at 4°C. After washing twice with high-salt wash buffer (50 mM Tris at pH 7.4, 500 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) and twice with lysis buffer, cross-linked RNAs were incubated with 1 mg/mL Proteinase K (Upstate Biotechnology) in PK buffer (50 mM Tris at pH 7.4, 50 mM NaCl, 10 mM EDTA) for 30 min at 37°C. Samples were then incubated with an equal volume of PK buffer containing 7 M urea for 30 min at 37°C. Eluted RNAs were phenol/chloroform-extracted and EtOH-precipitated, redisolved, treated with DNase RQ1, and purified by phenol/chloroform extraction and EtOH precipitation. Purified RNAs were analyzed by RT-qPCR.
RNA affinity pull-down assay
The 5′ biotinylated 18 RNA oligonucleotides were purchased from IDT. NEs from H9 hESCs were prepared as described previously (Kleiman and Manley 2001). Three nanomoles of biotinylated RNA was incubated with 100 µL of streptavidin-agarose beads (Sigma) overnight at 4°C with rotation. Incubated beads then were washed twice with binding buffer (10 mM Tris at pH 7.4, 0.5 mM EDTA, 500 mM NaCl) and twice with buffer D (20 mM HEPES at pH 7.9, 20% glycerol, 300 mM KCl, 0.2 mM EDTA, 0.5 mM DTT). RNA-coated beads were then mixed and rotated with NEs from H9 hESCs for 4 h at 4°C. After washing beads twice with buffer D and twice with buffer D without glycerol, precipitated proteins were eluted by SDS sample buffer and analyzed by Western blot.
RNA-seq
hESCs (WA-09) were cultured on mouse embryonic fibroblast feeder cells and differentiated into either NPCs (Chambers et al. 2009) or TBs (Xu et al. 2002) as described. RNA from two biological replicates of the initial hESC culture and the resultant NPC and TB cultures were subjected to Illumina high-throughput sequencing, yielding a total of ∼59 million 76-nt reads from hESC and NPC samples and ∼70 million reads from the TB samples. Reads were aligned to the human genome using Bowtie (Langmead et al. 2009). In all samples, >77% of the reads aligned to a unique locus (Supplemental Fig. S1). Changes in AS were detected using MATS software (version 3.0.8), which uses Bowtie to map reads to the exon junction database (Shen et al. 2012). Annotation files were prepared using TopHat (version 2.0.13) and Cufflinks (version 2.2.1) to include unannotated splicing events (Trapnell et al. 2012). AS changes were selected under highly stringent selection conditions (P-value ≤ 0.0005; false discovery rate ≤ 0.05). Published data sets acquired from different culture conditions were downloaded from Sequence Read Archive (SRA) accession number SRP000941 (Xie et al. 2013) and processed in the same way.
Supplementary Material
Acknowledgments
We thank Erin Conlon for discussion regarding hnRNP H1 functions, Kaori Yamazaki for help with RT–PCR, and members of the Manley laboratory for discussions. This work was supported by National Institutes of Health (NIH) R35 GM118136 and, in the early stages, New York State Stem Cell Science (NYSTEM) C024299, SDH-N08G-278 (to J.L.M.), NIH R21 HD081682 (to C.-W.L.), C026399, and core grant P30 CA008748 (to S.M.C. and L.S.).
Author contributions: T.Y. and J.L.M. conceived the study and experiments described, and T.Y. performed/oversaw all of the experiments. L.L. and A.A.-Z. carried out TCF3 minigene splicing assays. D.L. and V.F. performed RNA-seq analysis. L.S. and S.M.C. prepared human stem cell samples for RNA-seq. C.-W.L. and P.L.Y. established hiPSCs and prepared samples. T.Y. wrote the first draft, and J.L.M. contributed to manuscript preparation.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.316984.118.
References
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. 2009. Epithelial–mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest 119: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn H-J, Lib Jang K, Yeom S, Kwak J, Shin Kim S. 2016. Hepatitis B virus X protein induces epithelial–mesenchymal transition by repressing E-cadherin expression via upregulation of E12/E47. J Gen Virol 97: 134–143. [DOI] [PubMed] [Google Scholar]
- Beck K, Peak MM, Ota T, Nemazee D, Murre C. 2009. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med 206: 2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, Löwrick O, Klein-Hitpass L, Birchmeier W. 1991. The E-cadherin promoter: functional analysis of a G.C-rich region and an epithelial cell-specific palindromic regulatory element. Proc Natl Acad Sci 88: 11495–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, et al. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320. [DOI] [PubMed] [Google Scholar]
- Chou M-Y, Rooke N, Turck CW, Black DL. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol 19: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. 2008. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev 22: 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. 2016. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 5: e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Yu MS, McKeithan WL, Spiering S, Carrette F, Huang C-T, Bushway PJ, Tierney M, Albini S, Giacca M, et al. 2017. Id genes are essential for early heart formation. Genes Dev 31: 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastham AM, Spencer H, Soncin F, Ritson S, Merry CLR, Stern PL, Ward CM. 2007. Epithelial–mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res 67: 11254–11262. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung H-K, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. 2011. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147: 132–146. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna-Pillai S, Iverson LE. 2011. A DNMT3B alternatively spliced exon and encoded peptide are novel biomarkers of human pluripotent stem cells. PLoS ONE 6: e20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I, Zhang P, Panda AC, Kim J, Maudsley S, Abdelmohsen K, Yang X, Martindale JL, Motiño O, Hutchison ER, et al. 2016. Alternative splicing of neuronal differentiation factor TRF2 regulated by HNRNPH1/H2. Cell Rep 15: 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. 2005. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol 3: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung H-K, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. 2013. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-S, Li L, Moalim-Nour L, Jia D, Bai J, Yao Z, Bennett SAL, Figeys D, Wang L. 2015. A regulatory network involving β-Catenin, e-Cadherin, PI3k/Akt, and Slug balances self-renewal and differentiation of human pluripotent stem cells in response to Wnt signaling. Stem Cells 33: 1419–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Harvey SE, Hu X, Cheng C. 2017. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev 31: 2296–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. 2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qian X, Peng L-X, Jiang Y, Hawke DH, Zheng Y, Xia Y, Lee J-H, Cote G, Wang H, et al. 2016. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol 18: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Göke J, Sachs F, Jacques P-É, Liang H, Feng B, Bourque G, Bubulya PA, Ng H-H. 2013. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol 15: 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandler MD, Ku L, Feng Y. 2014. A cytoplasmic quaking I isoform regulates the hnRNP F/H-dependent alternative splicing pathway in myelinating glia. Nucleic Acids Res 42: 7319–7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Rom E, Chumakov I, Kronman A, Yayon A, Benvenisty N. 2008. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells 26: 767–774. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. 2001. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 15: 1688–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, García-García MJ, Anderson KV, Fuchs E. 2004. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development 131: 263–274. [DOI] [PubMed] [Google Scholar]
- Min H, Chan RC, Black DL. 1995. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev 9: 2659–2671. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Miyazaki K, Chen K, Jin Y, Turner J, Moore AJ, Saito R, Yoshida K, Ogawa S, Rodewald H-R, et al. 2017. The E-Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity 46: 818–834.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. 2008. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132: 661–680. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E. 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127: 171–183. [DOI] [PubMed] [Google Scholar]
- Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M, et al. 2010. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 7: 225–239. [DOI] [PubMed] [Google Scholar]
- Pereira L, Yi F, Merrill BJ. 2006. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol 26: 7479–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial–mesenchymal transitions. J Biol Chem 276: 27424–27431. [DOI] [PubMed] [Google Scholar]
- Pfurr S, Chu Y-H, Bohrer C, Greulich F, Beattie R, Mammadzada K, Hils M, Arnold SJ, Taylor V, Schachtrup K, et al. 2017. The E2A splice variant E47 regulates the differentiation of projection neurons via p57(KIP2) during cortical development. Development 144: 3917–3931. [DOI] [PubMed] [Google Scholar]
- Piva F, Giulietti M, Burini AB, Principato G. 2012. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum Mutat 33: 81–85. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P, et al. 2015. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan G-C, Orkin SH. 2010. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol 30: 5364–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch J, Moran-Jones K, Albrecht V, Schwarzl T, Hunter K, Gires O, Kolch W. 2011. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res 71: 4664–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmer T, Diecke S, Grigoryan T, Quiroga-Negreira A, Birchmeier W, Besser D. 2011. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep 12: 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. 2010. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci 107: 10514–10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. 2003. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol 260: 404–413. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. 2012. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hirsch DS, Sasiela CA, Wu WJ. 2008. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J Biol Chem 283: 5127–5137. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. 2012. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-seq data. Nucleic Acids Res 40: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra RA, Hoverter NP, Ramirez RN, Vuong LM, Mortazavi A, Merrill BJ, Waterman ML, Donovan PJ. 2018. TCF7L1 suppresses primitive streak gene expression to support human embryonic stem cell pluripotency. Development 145: dev161075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Baltimore D. 1991. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 64: 459–470. [DOI] [PubMed] [Google Scholar]
- Tam W-L, Lim CY, Han J, Zhang J, Ang Y-S, Ng H-H, Yang H, Lim B. 2008. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26: 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Tiwari I, Yoon M-H, Park B-J, Jang KL. 2015. Hepatitis C virus core protein induces epithelial–mesenchymal transition in human hepatocytes by upregulating E12/E47 levels. Cancer Lett 362: 131–138. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, et al. 2013. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun 4: 2480. [DOI] [PubMed] [Google Scholar]
- Vossaert L, O'Leary T, Van Neste C, Heindryckx B, Vandesompele J, De Sutter P, Deforce D. 2013. Reference loci for RT-qPCR analysis of differentiating human embryonic stem cells. BMC Mol Biol 14: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-H, Baker NE. 2015. E proteins and ID proteins: helix–loop–helix partners in development and disease. Dev Cell 35: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Aslanzadeh V, Papa F, Zhu H, de la Grange P, Cambi F. 2012. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS ONE 7: e51266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, et al. 2007. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25: 681–686. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. 2011. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Seita Y, Zhang X, Lu C-W, Roth MJ. 2012. Antibody-directed lentiviral gene transduction for live-cell monitoring and selection of human iPS and hES cells. PLoS ONE 7: e34778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB. 2009. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R-H, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. 2002. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20: 1261–1264. [DOI] [PubMed] [Google Scholar]
- Xu J, Lu Z, Xu M, Pan L, Deng Y, Xie X, Liu H, Ding S, Hurd YL, Pasternak GW, et al. 2014. A heroin addiction severity-associated intronic single nucleotide polymorphism modulates alternative pre-mRNA splicing of the μ opioid receptor gene OPRM1 via hnRNPH interactions. J Neurosci 34: 11048–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Pereira L, Merrill BJ. 2008. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells 26: 1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Pereira L, Hoffman JA, Shy BR, Yuen CM, Liu DR, Merrill BJ. 2011. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat Cell Biol 13: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.