Abstract
Alcohol use disorder (AUD) affects millions of people and costs nearly 250 billion dollars annually. Few effective FDA-approved treatments exist, and more are needed. AUDs have a strong heritability, but only a few genes have been identified with a large effect size on disease phenotype. Genome wide association studies (GWASs) have identified common variants with low effect sizes, most of which are in non-coding regions of the genome. Animal models frequently fail to recapitulate key molecular features of neuropsychiatric disease due to the polygenic nature of the disease, partial conservation of coding regions, and significant disparity in non-coding regions. By contrast, human induced pluripotent stem cells (hiPSC) derived from patients provide a powerful platform for evaluating genes identified by GWAS and modeling complex interactions in the human genome. hiPSCs can be differentiated into a wide variety of human cells, including neurons, glia and hepatic cells, which are compatible with numerous functional assays and genome editing techniques. In this review, we focus on current applications and future directions of patient hiPSC-derived CNS cells for modeling AUDs in addition to highlighting successful applications of hiPSCs in polygenic neuropsychiatric diseases.
Keywords: AUD, hiPSCs, GWAS, addiction, neuropsychiatric disease
INTRODUCTION
In 2013, alcohol use disorder (AUD) affected >16.3 million people over the age of 18 (Murray and Lopez, 2013) and cost society nearly $250 billion in 2010 (Sacks et al., 2015). For the past 20+ years, evidence has been accumulating for a strong genetic component to AUD. Family and twin-based studies estimated a 45%-65% heritability (Heath et al., 1997; Kendler et al., 1994; Pickens et al., 1991), and a recent meta-analysis also estimated the heritability to be ~49% (Verhulst et al., 2015). In animal studies, selectively bred (Bell et al., 2012) and genetically engineered rodent models (Crabbe et al., 2006) of alcohol consumption also support a genetic component to AUD. In addition to genetics, environmental exposure also contributes to disease development and severity. For example, early life stress has been shown to play a significant role in AUD (Clarke et al., 2011). Heritability of neuropsychiatric disorders like AUD is notoriously complex and has been the subject of intense investigation (for reviews, see Geschwind and Flint, 2015; McConnell et al., 2017). Studying AUD from a genetic perspective is particularly challenging due to variation in severity of disease phenotype. For example, an individual could meet either two to three of eleven diagnostic criteria in the DSM-V for a “mild” diagnosis, 4-5 for a “moderate” diagnosis, or six or more for a “severe” diagnosis (American Psychiatric Association, 2013). Such clinical heterogeneity is likely reflective of the genetic complexity underlying AUD. Identification of genes involved in AUD is crucial for understanding disease mechanisms and designing targeted treatments.
Genome-wide association studies (GWASs) seek to identify common variants, usually single nucleotide polymorphisms (SNPs), that are associated with common phenotypes (for review of GWAS, see: Bush and Moore, 2012). Most GWAS data have been obtained through chip-based microarray technologies such as Illumina® or Affymetrix®, while targeted sequencing approaches have been used to follow up regions of interest identified in chip microarrays (DiStefano and Taverna, 2011). GWAS studies are correlational and any effects of most SNPs on disease phenotype are small, so large sample sizes are necessary to produce enough power to meet a stringent P value (e.g., 5 × 10−8) and to estimate SNP effect size. Currently, meta-analysis, combining multiple datasets, is used to generate the largest possible sample.
SNPs may occur within protein-coding sequences, introns, regulatory regions, or between genes. Missense variants or other alterations of coding sequences can have profound effects on protein function. For example, linkage analysis of cystic fibrosis inheritance in families has identified multiple mutations in CFTR (cystic fibrosis transmembrane conductance regulator), with the most common variant resulting in the deletion of a phenylalanine (Kerem et al., 1989). Some SNPs reside in the coding region but do not change the amino acid sequence (i.e., synonymous) whereas others are found in non-coding regions, as is the case for many common variants associated with AUD (Clarke et al., 2011, 2017; Zuo et al., 2014). Here, the functional consequence of a disease-associated SNP is more difficult to assess and could result in a change in the expression level of a disease-relevant protein or a change in splicing/regulation of a transcript. Such SNPs may constitute expression quantitative trait loci (eQTLs), which have profound effects on transcript abundance. However, association of a SNP with a phenotype does not necessitate that the SNP plays a causal role; this may be an indication that the SNP is in linkage disequilibrium (LD) with an unidentified causal variant. Because many variants are not found in conserved protein-coding sequences or may be in LD with unknown sequences, functional studies in human cells are needed to examine the significance of GWAS findings and unravel functionality (see Figure 1 for GWAS to hiPSC-based study pipeline).
Figure 1. Flow-chart of sample collection and processing pipeline.
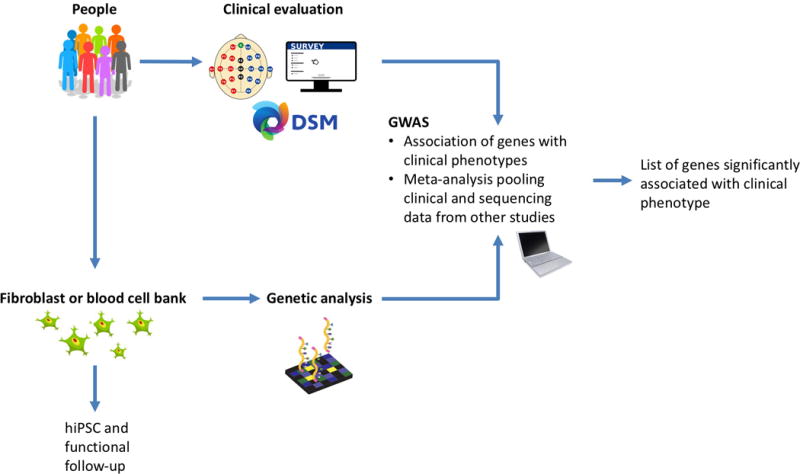
A diverse patient cohort is clinically evaluated, at which time behavioral or EEG endophenotypes may be measured. Skin or blood samples are collected from patients, part of which is subject to genetic sequencing or fingerprinting for GWAS analysis and part of which may be reprogrammed into hiPSCs. GWAS analysis yields a list of significantly associated loci labelled with the nearest gene, which are then investigated through functional studies.
Elucidating the functional consequences of genetic variants and how they contribute to disease risk is critical for understanding neuropsychiatric diseases such as AUD. Animal models, particularly in rodents, have been widely implemented to investigate genetic findings from human studies, but they have several key limitations. While rodent models of psychiatric disease provide important behavioral insight, they typically focus on manipulating a single gene, and often fail to recapitulate the full scope of molecular and cellular phenotypes. This is perhaps best illustrated by the plethora of Alzheimer’s disease rodent models, none of which demonstrates combination of plaques, tangles, and neurodegeneration found in human post-mortem tissue (Elder et al., 2010), unless mutant genes are expressed in combination, a condition not found in humans (Oddo et al., 2003). The lack of adequate animal models hampers clinical studies, with many potential therapies failing in early phase clinical trials (Becker and Greig, 2010). In the context of AUD, animal models typically focus on consumption phenotypes (Lynch et al., 2010). However, recent genetic data suggest that the shared heritability for consumption (Bierut et al., 2012; Clarke et al., 2017) and alcohol dependence (Clarke et al., 2011; Mbarek et al., 2015; Zuo et al., 2014) is low. Furthermore, animal models typically focus on modeling variants of protein-coding genes, but a large number of AUD-linked SNPs are not in coding sequences (Zuo et al., 2014) and non-coding sequences tend to be species specific (Batzoglou et al., 2000).
Collecting sufficient quantity or quality of human brain cells for biochemical and genetic analyses has been difficult. The development of human induced pluripotent stem cells (hiPSCs) provides a strategy to overcome this and is a compelling complement to animal models. hiPSCs are derived from donor somatic cells by reprogramming into a pluripotent state (Takahashi et al., 2007). These stem cells can then be differentiated into a desired cell type, e.g. an excitatory neuron. Once differentiated, cells derived from hiPSCs have the inherent advantage of containing all genes expressed endogenously under the correct tissue-specific promoters as well as non-coding regulatory sequences. Although variability within the whole genome, among multiple donors, poses a challenge to understanding single gene effects, hiPSCs are amenable to a variety of gene editing methods for probing individual or combinatorial gene contributions to a disease. Isogenic lines, which are genetically manipulated cells that have a single genetic background, can be generated to isolate the effects of one or more variants among simple variants or single SNPs. Gene knockouts in isogenic lines can be created to investigate the effects of reduced gene expression.
It is generally believed that specific cell types and neuronal circuits within the brain mediate the acute response to alcohol as well as the long-term changes that occur in AUD. Advances in the stem cell field has enabled creation of different types of central nervous system (CNS) cells, including excitatory forebrain neurons (Bardy et al., 2015; Chanda et al., 2013; Ho et al., 2016), dopaminergic neurons (Caiazzo et al., 2011; Swistowski et al., 2010), serotonergic neurons (Vadodaria et al., 2017), mixed populations of inhibitory and excitatory neurons (Nadadhur et al., 2017), medial ganglionic eminence cells (Ahn et al., 2016), astrocytes (TCW et al. 2017), oligodendrocytes (Ehrlich et al., 2017), microglia (Abud et al., 2017), brain microvascular endothelial cells and pericytes (Yamamizu et al., 2017), as well as three dimensional (3D) cell culture (including neurospheres and brain organoids) (Choi et al., 2014; Paşca et al., 2015). These cells have already been successfully applied to model complex polygenic diseases such as Alzheimer’s disease (Liao et al., 2016; Takamatsu et al., 2014; Wray et al., 2013), schizophrenia (Brennand et al., 2011; K. Brennand et al., 2015; Toyoshima et al., 2016), and bipolar disorder (Mertens et al., 2015b), demonstrating distinct cellular and molecular phenotypes. Furthermore, co-culture systems have great potential for elucidating the contribution of a cell type to a disease phenotype as well as cell-autonomous and non-autonomous effects. Thus, hiPSC studies show great promise for helping to elucidate the genetic underpinnings of human diseases such as AUD.
In this critical review we discuss GWAS studies of AUDs and the potential of hiPSC derived cells for investigating key genetic findings. We also provide some examples of successful applications of hiPSCs for modeling other genetically complex neuropsychiatric diseases. We conclude with an overview of the advantages, caveats, and future directions of using hiPSCs for studying AUD.
GWAS OF ALCOHOL USE DISORDERS
Alcohol (i.e. ethanol) produces differential effects in multiple brain areas, including reward pathways involving the ventral tegmental area, nucleus accumbens, and prefrontal cortex (for review see Harrison et al., 2017). GWAS has the potential to reveal previously unknown genetic variants affected by ethanol or genes modulating those proteins. Multiple GWAS studies have been used to probe alcoholism risk, defining affected cohorts by a variety of criteria, including DSM criteria for alcohol dependence (Bierut et al. 2010; Dick et al. 2008; Edenberg et al. 2010; Kendler et al. 2011), online survey AUD identification test (AUDIT) (Sanchez-Roige et al., 2017), and reward-related theta oscillations (a highly heritable neuroelectric endophenotype) (Clarke et al., 2011, 2017; Kang et al., 2012). However, some SNPs fail to meet significance cutoffs after multiple test correction, because the heterogeneity and complexity of factors contributing to alcohol use disorder risk requires large sample sizes to parse out significant associations (e.g. Treutlein et al., 2009). Several consortia across the world are gathering expanded datasets from alcohol use disorder families, including Australian Twin Family Study of AUDs (OZALC) and Collaboration of Genetics of Alcoholism (COGA) in the United States. For more information about resource access visit https://niaaagenetics.org/. For a comprehensive summary of alcohol dependence genetics see (Edenberg and Foroud, 2014; Tawa et al., 2016).
Among nonsynonymous SNPs consistently meeting significance thresholds for association with alcohol dependence are those in the genes encoding alcohol dehydrogenase (ADH) and alcohol aldehyde dehydrogenase (ALDH) (Bierut et al., 2012; Edenberg, 2007; Sanchez-Roige et al., 2017), which have been previously identified through linkage analysis (Edenberg et al., 2006) and biochemistry (Smith et al., 1971; von Wartburg et al., 1965). These proteins play a key role in alcohol metabolism, and their variants have a profound effect on alcohol consumption in rodents and humans (Edenberg, 2007). Genetically dictated differences in metabolism, either due to ADH or ALDH function, indicate that liver function may contribute AUD manifestation (Smith et al., 1971; von Wartburg et al., 1965). Linkage analysis by COGA has also enabled fine mapping of targeted chromosomal regions (Wang et a. 2004). Eight SNPs associated with alcohol dependence (p<0.01) were identified, with four located near the ACN9 gene, which codes for a mitochondrial intermembrane protein of gluconeogenesis (Dick et al., 2008). Investigation of mitochondrial abnormalities in patient derived neurons containing these SNPs is a potential avenue of research. Additionally, patient-derived liver cells can be generated from hiPSCs (Pashos et al., 2017; Warren et al., 2017) and these may be valuable for studying AUD-related variants linked with alcohol metabolism.
Comparison with a UK BioBank GWAS study of alcohol consumption suggests that genes influencing consumption and dependence may differ (Clarke et al., 2017, 2011). The study used self-reported consumption measures and identified eight independent significantly associated loci, including replicating AHD1B, ADH1C, and ADH5 loci and two loci in KLB (Clarke et al., 2017). KLB is involved in consumption regulation and has been identified in a recent meta-analysis GWAS of European population alcohol consumption (Schumann et al., 2016). Clarke et al. additionally identified four novel loci linked to consumption but not found in dependence studies, including GCKR, CADM2, TNFRSF11A, and PXDN. The divergence of consumption and dependence suggests a fundamental involvement of the brain in driving alcohol related behavior.
Focusing on neurocognitive aspects of AUDs, other groups have investigated the neuroelectric endophenotype of frontal theta reward event related oscillations (EROs) in addition to clinical diagnoses or self-reported measures (Klimesch et al., 2001), (Kamarajan et al., 2006), (Kang et al., 2012). Theta EROs are implicated in inhibitory control, conscious awareness, episodic and recognition memory, and evaluating loss and gain in gambling paradigms (Klimesch et al., 2001). Decrease in reward response theta EROs is a highly heritable endophenotype observed in AUD patients and their adolescent offspring at risk for the disorder (Kamarajan et al., 2006). A family-based study of theta EROs in AUD patients, their offspring, and healthy controls identified several polymorphisms in the KCNJ6 gene, with the most significant SNP (rs702859, imputed) at p = 4.7×10−10 constituting a synonymous mutation in exon 4 and other genotyped and imputed SNPs located in intronic regions (Kang et al., 2012). KCNJ6 encodes the G protein-gated inwardly rectifying potassium channel GIRK2, which plays an important role in regulating neuronal excitability by stabilizing the membrane potential (Lüscher and Slesinger, 2010). KCNJ6 is expressed in cholinergic, GABAergic, glutamatergic, and, most prominently, dopaminergic neurons (del Burgo et al., 2008). Additionally, GIRK2 has been shown to be directly activated by alcohol (Aryal et al., 2009; Bodhinathan and Slesinger, 2014; Glaaser and Slesinger, 2017). Eleven significant SNPs in KCNJ6 were also identified in an association study between adult drinking, adolescent drinking, and early life stress interactions (Clarke et al., 2011). The variant SNPs identified in KCNJ6 do not result in amino acid changes, suggesting potential differences in gene expression or distal regulatory effects on other genes. The theta ERO differences in AUD patients suggest potential differences in neuronal activity, which can be investigated in vitro, providing a phenotype to overlay the complex genetic interactions. Identification of common cellular phenotypes either at baseline or upon treatment with ethanol could identify targets for pharmacotherapy to normalize divergences in electrophysiological activity.
hiPSC MODELS OF ALCOHOL USE AND ALCOHOL-ASSOCIATED GENE VARIANTS
Lieberman and colleagues used hiPSCs to investigate the biological effects of alcohol on human brain cells (Lieberman et al., 2012). hiPSCs derived from four alcohol dependent individuals and three social drinkers were differentiated into a mixed population of neurons and astrocytes, and then exposed to acute or chronic ethanol. Electrophysiological recordings revealed that acute alcohol exposure attenuated NMDA (excitatory) responses in samples from both healthy and alcohol dependent individuals. Chronic exposure, however, led to an upregulation of NMDA subunit transcripts in dependent but not healthy subjects, indicating a compensatory mechanism for NMDA-R attenuation in the transition from acute to chronic alcohol exposure (Lieberman et al., 2012). This pilot study demonstrated the feasibility of studying the effects of alcohol in hiPSC-derived neurons from healthy and AUD-affected individuals.
Focusing on a genetically defined cohort, a subsequent hiPSC study explored a GABRA2 variant (rs279858*C) (Lieberman et al., 2015) which has been repeatedly associated with AUD (Covault et al., 2004, 2008; Edenberg et al., 2004; Ittiwut et al., 2012; Li et al., 2014). GABRA2 encodes the alpha subunit of the pentameric GABAA receptor, and is located in chromosome 4p12, tandem to three other GABAA subunits. rs279858*C is a synonymous mutation in exon 5, with no effect on protein structure. Other GABAA subunits are located on chromosomes 5q34, 15q11, and Xq28 (Steiger and Russek, 2004). mRNA encoding GABAA subunits on 4p12 as well as subunits located on other chromosomes were measured in 36 neural lines from alcoholic and non-alcoholic donors (Lieberman et al., 2015). Carriers of rs279858*C had significantly lower levels of all GABAA subunits located on 4p12, but not other chromosomal regions (Lieberman et al., 2015). However, protein levels in postmortem cortex did not vary by rs279858 genotype, suggesting the polymorphism’s effects may be most profound during neural development (Lieberman et al., 2015). In a more recent study, 24-hour and 7-day exposure of healthy hiPSCs and NPCs to ethanol activated an inflammasome pathway (NLRP3), priming an innate immune-like response and impairing distribution of lysosomes and mitochondria (De Filippis et al., 2016). Additionally, ethanol exposure did not have any effect on proliferative capacity of those cells, but did negatively impact the number of mature neurons that could be generated from exposed NPCs (De Filippis et al., 2016). These studies underscore the capacity of hiPSC-derived cells to delineate developmental components of AUDs.
A recent investigation of GABAA function in a genetically heterogenous cohort of alcohol dependent (AD) and alcohol-exposed control (CTL) subjects revealed no alteration in GABA-evoked current in directly differentiated, excitatory-enriched neurons following chronic alcohol treatment (Lieberman et al., 2018). The lack of GABA adaptation could be due to the absence of GABAergic signaling in the excitatory-enriched neural cultures, since three-week exposure to chronic ethanol significantly increased transcription levels of GABRA1 and GABRG2 in both AD and CTL groups. GABRD only showed a modest increase (24%) for the AD but not the CTL group (Lieberman et al., 2018), which is consistent with GABRD suggested as an AUD-candidate gene (Rodd et al., 2007). The comparison of differential effects of ethanol on human cells derived from healthy and alcohol dependent subjects illustrates how to use hiPSC-derived cells to study AUDs.
EXISTING hiPSC MODELS OF NEUROPSYCHIATRIC DISEASE
Applications of hiPSCs for investigating AUDs are still in their nascent stage, but promising results have been obtained from several hiPSC models of neuropsychiatric disease, including schizophrenia (Toyoshima et al. 2016; Brennand et al. 2015; Lin et al. 2016), bipolar disorder (Mertens et al., 2015b; Stern et al., 2017), and Alzheimer’s disease (Liao et al., 2016; Mahairaki et al., 2014; Yang et al., 2016). Although hiPSC models have been applied to a large number of different types of neurological disorders, we focus here on a polygenic subset.
Nicotine Dependence
Several GWAS studies of human smokers have identified SNPs associated with risk of nicotine dependence (Bierut et al., 2008; Saccone et al., 2009) or cigarette consumption (Caporaso et al., 2009; Consortium et al., 2010). Bierut et al (2008) found a significant SNP (rs16969968, p=0.007) in the cholinergic nicotinic receptor α5 subunit, changing Aspartate (Asp) (major allele) to Asparagine (Asn) (minor allele) at position 398 (Bierut et al., 2008). Genotyping in African American (AA) and European-American (EA) participants revealed differential associations between populations, but the SNP was most significantly associated with nicotine dependence in the combined samples (Bierut et al., 2008). Recently, Oni and colleagues examined hiPSC-derived neurons from patients with and without the CHRNA5 Asn-398 allele (Oni et al., 2016). Patient somatic cells from the Collaborative Genetic Study of Nicotine Dependence (COGEND) were reprogrammed into hiPSC and differentiated into primarily dopaminergic neurons, with about 20% glutamatergic neurons. Neurons containing the Asn-398 allele exhibited an increase in the amplitude and frequency of spontaneous excitatory post synaptic currents, compared to Asp-398 neurons (Oni et al., 2016). Glutamatergic neurons, but not DA neurons, displayed similar baseline electrophysiological characteristics across genotypes, but had an increased excitatory response to nicotine stimulation and faster receptor desensitization in the Asn-398 variant (Oni et al., 2016). Analysis of the dopaminergic neuron transcriptome by RNAseq revealed significant functional enrichment for pathways specific for calcium signaling, axon guidance, and ligand-receptor interaction (Oni et al., 2016). Due to the young age of these hiPSC-derived neurons, these results suggest the CHRNA5 risk variant may underlie predisposition to nicotine dependence (Oni et al., 2016).
Another investigation of this polymorphism in iPSC-derived dopaminergic neurons demonstrated that Asn-398 variant has a decreased sensitivity to the receptor agonists (Deflorio et al., 2017). However, increases in current in response to agonists were observed (Deflorio et al., 2017), consistent with findings in glutamatergic but not dopaminergic neurons by Oni et al., 2016. The difference in findings of the variants’ effects on receptor function by neuron type between these studies could be attributed to differences in iPSC differentiation protocols. While these studies elucidate functional consequences of a GWAS variant, they also highlight the need for replication and standardization of differentiation protocols.
Schizophrenia
Schizophrenia (SCZ) is a debilitating neuropsychiatric disorder with a strong developmental component, numerous perturbations in synaptic and network functions, and a polygenic risk profile (Adriano et al., 2012; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). One of the earliest studies of SCZ used hiPSCs derived neurons from a genetically heterogenous cohort of four SCZ patients and performed gene expression profiling, extensive characterization of neurite outgrowth and synaptic connectivity, as well as pharmacological treatment with antipsychotics (Brennand et al., 2011). SCZ patient cells differentially expressed 596 unique genes and showed deficits in synaptic connectivity, compared to controls, which were partially ameliorated by the antipsychotic drug Loxapine (Brennand et al., 2011). Furthermore, several groups investigated 22q11.2 hemizygous microdeletion, one of the few single loci associated with a large increase in risk for SCZ (see Bassett and Chow, 2008), in hiPSC derived cells. Neurospheres derived from the 22q11.2 deletion patients were smaller in size, had reduced expression levels of markers for cellular proliferation and survival, and an increased astrocyte to neuron ratio, consistent with observations in postmortem brains (Toyoshima et al., 2016). Integrative network analysis of hiPSC-derived neurons from eight cases with 22q11.2 microdeletion and ten healthy controls identified two sub-networks which are disrupted by 22q11.2 deletions: a cell cycle network mediated by CD45, disrupted in embryonic development, and a PRODH-mediated network, potentially contributing to disrupted brain function in adolescence when SCZ symptoms most often begin to emerge (M. Lin et al., 2016). Thus, human SCZ variants can reveal cellular developmental mechanisms linked with the disorder.
Bipolar Disorder
Bipolar disorder (BD) is characterized by recurring episodes of alternating mania and depression, has a complex pathophysiology and a polygenic risk, in addition to reduction in neural volumes and alterations in neurotransmission (Consortium 2009; Boies et al., 2017). Chen et al. (2014) examined changes in gene expression throughout the course of directed differentiation from hiPSCs to mixed-population neurons derived from three patients and three age-matched controls. Overall no significant expression differences were observed in the pluripotent stage, but BD neurons expressed significantly more membrane receptors and ion channels, especially those involved in calcium signaling (Chen et al., 2014). Total RNAseq analyses of three-week-old hippocampal dentate gyrus granule cell-like neurons revealed enhancement in multiple mitochondrial genes in BD neurons. Flow cytometry and mitochondrial membrane potential measurements suggested an enhanced mitochondrial function. Patch-clamp recordings and calcium imaging at baseline and following exposure to lithium, a commonly used pharmacotherapy for BD, revealed lower threshold for action potentials and higher firing frequencies for BD neurons (Mertens et al., 2015b). Interestingly, BD neurons from lithium responders displayed a reduction in hyperexcitability following lithium treatment, in contrast to non-responder neurons (Mertens et al., 2015b). RNAseq analyses showed a partial normalization of mitochondrial gene levels in the lithium responder group (Mertens et al., 2015b). A subsequent hiPSC study of dentate gyrus-like neurons from BD patients confirmed hyperexcitability of BD neurons and differential reductions in hyperexcitability following lithium treatment (Stern et al., 2017). Furthermore, differences in spiking shape were analyzed revealing that lithium responders had a larger sodium current than controls, whereas non responders had smaller sodium currents (Stern et al., 2017). These studies illustrate how selected hiPSC-derived neurons can respond to appropriate medications and predict patient response, which is notoriously variable in concordance with disorder heterogeneity (Drozda et al., 2014).
Alzheimer’s Disease
Humans with Alzheimer’s disease (AD) exhibit progressive cognitive impairment, neurodegeneration, formation of amyloid beta (Aβ) plaques, and neurofibrillary tangles - features that have not been fully recapitulated in animal models (Elder et al., 2010). Linkage studies have uncovered mutations in certain genes causing autosomal dominant forms of AD (ADAD), including presenilin (PSEN1, PSEN2), amyloid precursor protein (APP), and a moderately penetrant polymorphism, APOE-e4 (for AD genetics overview, see (Bagyinszky et al., 2014)). APOE isoforms have differential effects on disease progression: whereas APOE2 is protective and APOE3 is neutral, APOE4 appears to constitute a major risk factor for AD (Strittmatter et al., 1993). Investigation of APOE isoforms in iPSC-derived human neurons has provided novel insights into molecular pathways of neural APOE, such as the differential efficacy of APOE isoforms in activating an unusual MAPK cascade regulating APP gene regulation (Huang et al., 2017). Another study ascribes a neurotoxic function for APOE4 that can be ameliorated with a small-molecular structure corrector, predicting a novel and potentially protective AD therapy (Wang et al., 2018). An innovative single-cell analytical platform for detecting secreted Aβ and soluble amyloid precursor protein-alpha (sAPPα) revealed heterogeneity in secretion profiles from cells harboring APP mutations (Liao et al., 2016). These include subpopulations of cells that secret high levels of Aβ, but not sAPPα, cells that secret both Aβ and sAPPα (enriched for GABAergic markers), and cells that secret Aβ levels (enriched for astrocyte markers) (Liao et al., 2016). Finally, 3D organoid models of AD have been able to recapitulate both amyloid deposition, and hyperphosphorylated tau accumulation in somato-dendritic compartments, both from patients with familial mutations (Raja et al., 2016) and from patients with late onset disease (Lee et al., 2016).
Together, these studies highlight the utility of hiPSC-derived CNS cells for exploring genetically complex neuropsychiatric disorders. Table 1 lists recent publications utilizing hiPSCs for investigating polygenic neuropsychiatric diseases (for reviews, see (Ardhanareeswaran et al., 2017; Prytkova and Brennand, 2017; Shi et al., 2017; Soliman et al., 2017). Though some caveats and limitations exist (discussed below), the rapidly advancing field of hiPSC differentiation and associated analytical platforms is rife with potential.
Table 1.
Abbreviations: DG- dentate gyrus; ELISA- enzyme-linked immunosorbent assay; HPLC- high performance liquid chromatography; MGE- medial ganglionic eminence; VNTR-variable number tandem repeat polymorphisms; VTA – ventral tegmental area
| Publication | Disease | Gene/SNP | Cell Type | Assays | Treatment | Main findings |
|---|---|---|---|---|---|---|
| Lieberman et al., 2012 | Alcohol Use Disorder | – | Mixed population forebrain lineage neurons and astrocytes |
|
Acute and 7-day alcohol exposure |
|
| Lieberman et al., 2015 | Alcohol Use Disorder |
GABRA2 (rs279858*C) |
Mixed population forebrain lineage neurons and astrocytes |
|
– |
|
| De Filippis et al., 2016 | Alcohol Use Disorder | – | NPCs |
|
Chronic alcohol exposure |
|
| Lieberman et al., 2018 | Alcohol Use Disorder | – | Forebrain-type excitatory glutamate neurons |
|
21 day alcohol exposure |
|
| Oni et al 2015 | Nicotine Dependence |
CHRNA5 (rs16969968) |
Dopaminergic and glutamatergic neurons |
|
Acute nicotine |
|
| Deflorio et al., 2017 | Nicotine Dependence |
CHRNA5 (rs16969968) |
Dopaminergic neurons |
|
Acute nicotine & Acetylcholine |
|
| Mertens et al., 2015b | Bipolar Disorder | – | Hippocampal DF-like neurons |
|
Lithium |
|
| Stern et al., 2017 | Bipolar Disorder | – | Hippocampal DG-like neurons |
|
Lithium |
|
| Brennand et al., 2011 | Schizophrenia | – | Mixed population of forebrain lineage neurons |
|
Loxapine |
|
| Toyoshima et al., 2016 | Schizophrenia | 22q11.2; DCGR8 | Forebrain-like neurospheres |
|
– |
|
| Lin et al., 2016 | Schizophrenia | 22q11.2 | Mixed population of glutamatergic and GABAergic neurons |
|
– |
|
| Liao et al., 2016 | Alzheimer’s Disease | APP | Mixed population forebrain lineage neurons |
|
– |
|
| Raja et al., 2016 | Alzheimer’s Disease | APP; PSEN1 | Cortical organoids |
|
β and γ secretase inhibitors |
|
| Huang et al., 2017 | Alzheimer’s Disease | APOE | Mixed population forebrain lineage neurons |
|
suppression of gene expression with CRISPR |
|
| Wang et al. 2018 | Alzheimer’s Disease | APOE |
|
|
|
|
ADVANTAGES OF hiPSC MODELS
Human imaging, postmortem analyses, and animal studies of substance use disorders have demonstrated region-specific changes in neurons, astrocytes, oligodendrocytes, and microglia (for reviews, see Niciu et al., 2014; Zahr and Pfefferbaum, 2017). A significant advantage of hiPSC models is the expanding availability of various CNS cell types that can be derived from patients. This offers a valuable toolbox for exploring human cellular phenotypes within a specific genotype. A variety of reprogramming methods exist for generating specific CNS cells from hiPSCs, including direct induction of hiPSCs (Ahn et al., 2016; Close et al., 2017; Livesey et al., 2016; Lu et al., 2016; Muffat et al., 2016; Swistowski et al., 2010; Tcw et al. 2017b; Yu et al., 2014), neural progenitor cells (NPCs) (Ehrlich et al., 2017; Ho et al., 2016) or fibroblasts (Ladewig et al., 2012; Pang et al., 2011; Vadodaria et al., 2017; Yang et al., 2011). Alternatively, directed differentiation of hiPSCs involves long-term cultures with differentiation-specific growth factors and is generally thought to more closely resemble in vivo conditions but also to produce heterogeneous cell types (Maroof et al., 2013). This approach is suitable for exploring developmental components of AUDs in a mixed excitatory and inhibitory neuronal population. By contrast, induction of cell type specific genes significantly reduces the timeline and increases homogeneity of cell populations (Ho et al., 2016). We summarize some of these protocols in Table 2. Thus one advantage of hiPSCs is the availability of different reprogramming methods which allows the examination of the same genotype in different contexts.
Table 2.
Abbreviations: CGE-caudal ganglionic eminence; DG- dentate gyrus; DRN- dorsal raphe nuclei; EB- embryonic body; FACS- fluorescence activated cell sorting; HC- hippocampus; hESC- human embryonic stem cell; HPLC-high performance liquid chromatography; ICC- immunocytochemistry; MGE-medial ganglionic eminence; NPCs-neural progenitor cells; OPC- oligodendrocyte progenitor cell; UPLC-ESI-MS/MS- ultra-performance liquid chromatography–electrospray ionization–tandem mass spectrometry
| Derived cell type | Starting cell type | Reprogramming method | Assays | Publication |
|---|---|---|---|---|
| MGE-like cells | hESCs |
|
|
Close et al. 2017 |
| Excitatory forebrain neurons | hiPSC-derived NPCs |
|
|
Ho et al., 2016b |
| Neurons of different neurotransmitter phenotypes | Human fibroblasts |
|
|
Ladewig et al. 2012 |
| MGE- and CGE-like cells | hiPSCs |
|
|
Anh et al. 2016 |
| Serotonin neurons (rhombomeric segments 2-3 of DRN) | hiPSCs |
|
|
Lu et al. 2016 |
| Serotonin neurons | Human fibroblasts |
|
|
Vadodaria et al. 2016 |
| Dopamine neurons | hiPSCs |
|
|
Swistowski et al. 2010 |
| Dopamine neurons | Human fibroblasts |
|
|
Caiazzo et al. 2011 |
| Astrocytes | hiPSCs-derived NPCs |
|
|
Tcw et al., 2017 |
| Hippocampal DG neurons | hiPSC-derived EBs and NPCs |
|
|
Yu et al. 2014 |
| Microglia-like cells | hiPSCs, with EB/NPC intermediate |
|
|
Muffat et al. 2016 |
| Oligodendrocytes | hiPSCs, with EB/NPC intermediate |
|
|
Livesey et al. 2016 |
| Oligodendrocytes | hiPSC-derived NPCs |
|
|
Ehrlich et al. 2017 |
Though most hiPSC-derived neurons are grown as monolayers, researchers have developed 3D cultures and brain organoids to produce neuronal ensembles that more closely resemble human brain, e.g., layers of cortex (for review, see Quadrato et al., 2016). These cortico-spheroids typically exhibit a mix of neurons and astrocytes (Paşca et al., 2015), and particularly exciting is the recent description of chimeric organoids, which contain integrated excitatory and inhibitory cortical neurons mimicking interneuron migration observed in vivo (Birey et al., 2017; Mariani et al., 2015; Xiang et al., 2017). Finally, a promising development for drug screening applications lies in the recent creation of a human blood brain barrier (BBB) model from hiPSCs that recapitulates drug BBB permeability in humans (Yamamizu et al., 2017). The inherently different function of rodent and human BBB (Aday et al., 2016) accounts for some of the failed CNS therapeutic trials (Alavijeh et al., 2005) as well as being implicated in neurodegenerative and psychiatric diseases (Desai et al., 2007; Saito and Ihara, 2014). These more complex hiPSC-derived cultures provide an important approach for modeling disease specific circuitry and drug delivery.
Lastly, the development of human isogenic lines provides a method for testing specific variants and their effects on a phenotypic trait under conditions of reduced variability. Comparing multiple hiPSCs derived neurons from genetically unrelated humans, or even siblings, can result in large variability in measurements that are unrelated to the disease phenotype (Faulconbridge et al., 2017; Lin et al., 2015). With the discovery of efficient genome-editing techniques (Ran et al., 2013), it is now possible to genetically manipulate hiPSCs with precision and efficiency (see Figure 2) (Bassett, 2017; Bertero et al., 2016). Indeed, several studies have successfully utilized clustered regularly interspaces short palindromic repeats (CRISPR)/Cas9 nuclease genome editing (Wang et al., 2017), CRISPR activation/repression (Ho et al., 2017), and shRNA for manipulating gene expression levels (Bertero et al., 2016; Sancho-Martinez et al., 2016). With these techniques, investigators can manipulate expression of single or several genes, as well as generate control cells with identical genetic (isogenic) backgrounds (for detailed review of genome editing techniques see: Bassett, 2017).
Figure 2. Flow-chart of functional follow-up of GWAS findings using hiPSCs.
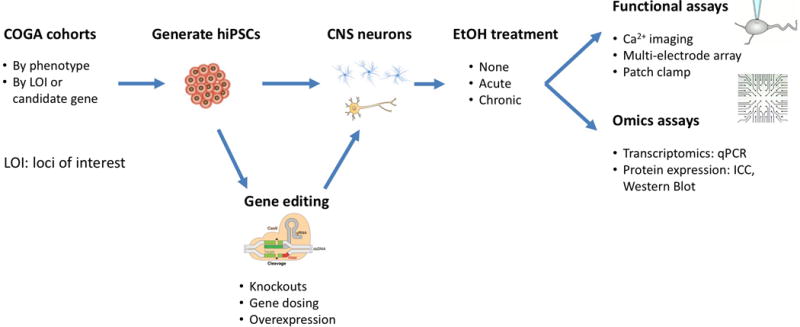
After clinical and genetic evaluation patient cohorts can be divided by phenotype or by significantly associated genes. Patient hiPSCs may be genetically manipulated prior to differentiation into cell types of interest. These cells are then characterized through omics (e.g., RNA, protein, epigenetic for directly induced cell types) and functional assays. Additionally, they may be used as a platform for drug screening, either for investigating effects of therapeutics or drugs of abuse.
ANALYSIS OF hiPSC-DERIVED CELLS
hiPSC-derived neurons are amenable to a variety of functional analyses. Given that perturbations in neuronal excitability are thought to underlie many psychiatric disorders (Anticevic and Murray, 2017), the ability to investigate excitability and network activity of human cells is a crucial step for understanding the disease. Whole-cell patch-clamp electrophysiology provides single-cell measurements of neuronal activity, with details on excitatory and inhibitory synaptic inputs. This is crucial for determining dysfunctions in particular channels implicated in alcoholism (for example, GABAA (Lobo and Harris, 2008). Several techniques are available for examining neuronal network activity, such as with microelectrode array (MEA) and Ca2+ imaging. MEAs can be used to study population activity at different time points throughout development. hiPSC-derived neurons are cultured directly on top of microelectrodes lining the bottom of the plate, and can be subjected to field stimulation or photostimulation techniques (Hales et al., 2010; Obien et al., 2015). Calcium imaging, using cell permeable dye (e.g. Fluo4-AM) or a genetically encoded indicator (e.g. GCaMP6) (Grienberger and Konnerth, 2012), offers single cell resolution as well as information on network activity. Compared to electrophysiology and MEA, Ca2+ imaging has a slower temporal resolution, in which action potential spikes are integrated over time. However, this temporal resolution has been sufficient to monitor activity in neuronal networks (Grienberger and Konnerth, 2012). The development of optical sensors for changes in voltage may overcome this limitation in the future (Marshall and Schnitzer, 2013).
In addition to functional activity studies, hiPSC-derived neurons can be processed for RNAseq to study up and down regulation of transcripts, immunohistochemistry to observe the localization of proteins, and Western analyses to quantitate protein expression levels. Since many alcohol-related variants are believed to be eQTLs (Mamdani et al., 2015; Zhou et al., 2017), genome-wide transcript analysis is likely to reveal regulatory targets and these processes are expected to be cell type specific. With this wide selection of techniques, researchers can elucidate differences in activity in healthy, disease affected, genetically manipulated, and pharmacologically treated human cells. Thus, hiPSC-derived neurons can provide unique insight into developmental events of AUD on a transcriptomic and functional level. Furthermore patient-derived cells are an effective experimental platform for manipulating disease phenotypes and testing pharmacotherapy response.
CAVEATS AND LIMITATIONS OF hiPSC MODELS
While hiPSC models offer a promising approach to studying AUD, there are several limitations worth noting. First, one would ideally study neurons at a mature stage comparable to neurons in adults. However, hiPSC-derived neurons are immature and lack some of the complex connectivity of adult neurons. Recently, transcriptional profiling of neurons and astrocytes derived from hiPSCs indicated these cells most closely resemble fetal cells (Brennand et al. 2015; TCW et al. 2017). Optimization of components in the differentiation culture medium has helped promote the presence of more synaptically mature neurons (Bardy et al., 2015). Second, while it is advantageous to focus on one cell type, the interactions with other cell types play an important role in development and functionality.
This limitation has been recently addressed by the development of co-culture systems and the production of organoids (Quadrato et al., 2016). Co-culture of neurons on rat astrocytes resulted in synchronized network activity within three months of plating, as measured with MEAs (Odawara et al., 2014). Furthermore, co-cultures of human primary astrocytes and a mixed excitatory-inhibitory population of hiPSC-derived cortical neurons can lead to network synchronization and connectivity in three to four weeks (Kuijlaars et al., 2016). Considering distinct electrical activity in EEG and fMRI are associated with alcoholism (Huang et al., 2018; Klimesch et al., 2001; Pandey et al., 2012), this is an important development for modeling predisposition.
Another limitation of hiPSC studies is that connections between neurons are inherently random, unlike neuronal networks that form in vivo. Micropatterning and microfluidic devices offer a potential solution for organizing circuits and exercising granular control over the cells’ microenvironment (for review, see Brunello et al., 2013). Microfluidic local perfusion chambers have been successfully implemented with dissociated primary neurons from rodents. For example, a microfluidic local perfusion chamber has been developed to manipulate distinct synaptic regions, directing synapse formation in distinct parallel rows through microgrooves connecting two distinct neuronal populations (Taylor et al., 2010). hiPSC-derived neurons are compatible with micropatterning for long term growth and circuit organization. Neurons adhere to regularly spaced adherent surfaces with neurites crossing cell-repellant surfaces to form connections (Burbulla et al., 2016). This system facilitates long-term studies of mitochondrial dynamics, axonal transport, and dynamic network formation (Burbulla et al., 2016). Compartmentalized microfluidic devices have been utilized to create interconnections between excitatory, inhibitory, and dopaminergic hiPSC-derived neurons, mimicking reward circuits (Fantuzzo et al., 2017). This is a key development for studying AUDs, considering accumulating evidence pointing towards differential reward processing in alcohol dependent individuals (Kamarajan et al., 2006, 2015; Müller-Oehring et al., 2013).
Epigenetic contributions, known to be important in AUD, pose another major challenge to utilizing hiPSC models. Incomplete tissue-specific erasure and aberrant de novo methylation during reprogramming somatic cells have effects on conversion efficiency and gene expression (Kim et al., 2010, 2011; Lister et al., 2011; Nazor et al., 2012; Oni and Hart, 2016). Epigenetic reset during conversion of somatic cells to a pluripotent state, even with residual methylation signatures, erases age-related marks (Lapasset et al., 2011) and may have an impact on other epigenetic marks contributing to AUD phenotype. One work-around for this could be to directly induce neurons from fibroblasts, which retain age-related epigenetic marks (Mertens et al., 2015a). In AUD, alcohol metabolism has a profound effect on epigenetic markers (Boschen et al., 2018; Krishnan et al., 2014; Zakhari, 2013), which may vary between CNS and peripheral cells, and careful consideration of reprogramming methods and epigenetic marks is essential for adequate disease modeling.
Verification of patient genotype and understanding of copy number variation is another essential step with hiPSC based studies. For example, characterization of hiPSCs from patients with 9p24.1 duplications/triplications, a copy number variant associated with psychotic disorders (Malhotra et al., 2011), revealed chromosomal instability during reprogramming in the form of extrachromosomal marker element rearrangement. Due to mosaicism in the patient derived fibroblasts, conversion to pluripotency serendipitously generated non-carrier isogenic controls (Tcw et al., 2017). Therefore, investigation in hiPSCs necessitates karyotyping to ensure absence of chromosomal abnormalities.
Lastly, variability in hiPSC derived neurons even amongst control subjects can be a concern for disease modeling (Choi et al., 2015; Kilpinen et al., 2017; Kyttälä et al., 2016). For example, a recent study exhaustively characterized differentiation capacity, cellular morphology, and copy number alterations through genome wide profiling of 711 hiPSC lines from 301 healthy individuals revealing that most variation results from differences between individuals (Kilpinen et al., 2017). Designing a hiPSC study with sufficient statistical power would require large samples sizes, which is impractical both in terms of cost and lack of techniques for high-throughput functional analyses. However, implementation of genetically-engineered hiPSCs can provide a reasonable solution to this limitation (see above).
CONCLUDING REMARKS
The complex genetic architecture of alcohol use and dependence results in profound clinical heterogeneity and difficulty in developing effective treatments. Animal models fail to recapitulate cellular and molecular complexities of human neurons, astrocytes, microglia, and blood brain barrier and accordingly do not capture all disease features, particularly those linked with noncoding genome sequences. Understanding precise molecular events underlying the disease and how an individual’s genetic background contributes to the disease state is essential to move into the age of personalized medicine for neuropsychiatric disorders. Despite their fairly recent development, patient derived hiPSCs have already been utilized to model a variety of psychiatric and neurodegenerative disorders. In addition to the key advantage of being human and patient specific, the ability to differentiate hiPSC into any cell type enables probing differences in cellular function and modeling of cellular interactions. In this review, we have focused on CNS cell types, but hiPSCs have also been reprogrammed into hepatocytes and adipocytes for investigation of liver function (Pashos et al., 2017; Warren et al., 2017), which is key for studying AUD. Furthermore, recent developments in genome editing techniques provide a unique opportunity to manipulate human genes in human cells and to tease apart complex genetic interactions contributing to disease phenotypes.
In conclusion, collaborative studies for understanding genetic risk of alcohol abuse disorders such as COGA are in a prime position to use hiPSCs to study AUD. These genetic studies have collected large numbers of carefully characterized and individually genotyped cells in repositories, suitable for hiPSCs derivation. The combination of thoroughly characterized clinical phenotypes, risk association genotypes, and functional investigations in patient cells, including gene editing and drug screening, will provide an unprecedented level of individual- and population-based resolution of polygenic disease mechanisms. Future functional studies will need to adapt hiPSC-derived neurons to more high-throughput analyses, such as with 384-well plate imaging or electrophysiological instruments, to greatly increase the number of control and AUD subjects studied. Such multi-level disease characterization will help identify novel therapeutic targets and bring the age of personalized medicine for psychiatric disease to fruition.
Acknowledgments
Some of the studies cited in this review were supported by the NIAAA (Collaborative Study on the Genetics of Alcoholism, U10 AA008401; R01 AA018734). Special thanks to Julia TCW for early discussions and Howard Edenberg for comments on this manuscript.
Footnotes
DR. PAUL SLESINGER (Orcid ID : 0000-0002-3868-7528)
References
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW, Blurton-Jones M. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aday S, Cecchelli R, Hallier-Vanuxeem D, Dehouck MP, Ferreira L. Stem Cell-Based Human Blood–Brain Barrier Models for Drug Discovery and Delivery. Trends Biotechnol. 2016;34:382–393. doi: 10.1016/j.tibtech.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Adriano F, Caltagirone C, Spalletta G. Hippocampal Volume Reduction in First-Episode and Chronic Schizophrenia: A Review and Meta-Analysis. The Neuroscientist. 2012;18:180–200. doi: 10.1177/1073858410395147. [DOI] [PubMed] [Google Scholar]
- Ahn S, Kim TG, Kim KS, Chung S. Differentiation of human pluripotent stem cells into Medial Ganglionic Eminence vs. Caudal Ganglionic Eminence cells. Methods San Diego Calif. 2016;101:103–112. doi: 10.1016/j.ymeth.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavijeh MS, Chishty M, Qaiser MZ, Palmer AM. Drug metabolism and pharmacokinetics, the blood-brain barrier, and central nervous system drug discovery. NeuroRX. 2005;2:554–571. doi: 10.1602/neurorx.2.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. American Psychiatric Association; Washington DC: 2013. [Google Scholar]
- Anticevic A, Murray JD. Rebalancing Altered Computations: Considering the Role of Neural Excitation and Inhibition Balance Across the Psychiatric Spectrum. Biol. Psychiatry, Cortical Excitation-Inhibition Balance and Dysfunction in Psychiatric Disorders. 2017;81:816–817. doi: 10.1016/j.biopsych.2017.03.019. [DOI] [PubMed] [Google Scholar]
- Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A, Vaccarino FM. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat Rev Neurol. 2017;13:265. doi: 10.1038/nrneurol.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S, Slesinger PA. A Discrete Alcohol Pocket Involved in GIRK Channel Activation. Nat Neurosci. 2009;12:988–995. doi: 10.1038/nn.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagyinszky E, Youn YC, An SSA, Kim S. The genetics of Alzheimer’s disease. Clin. Interv. Aging. 2014;9:535–551. doi: 10.2147/CIA.S51571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C, van den Hurk M, Eames T, Marchand C, Hernandez RV, Kellogg M, Gorris M, Galet B, Palomares V, Brown J, Bang AG, Mertens J, Böhnke L, Boyer L, Simon S, Gage FH. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc Natl Acad Sci U S A. 2015;112:E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm. Genome. 2017;28:348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EWC. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. Human and Mouse Gene Structure: Comparative Analysis and Application to Exon Prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RE, Greig NH. Lost in Translation: Neuropsychiatric drug developments. Sci Transl Med. 2010;2:61rv6. doi: 10.1126/scitranslmed.3000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Sable HJK, Colombo G, Hyytia P, Rodd ZA, Lumeng L. Animal models for medications development targeting alcohol abuse using selectively bred rat lines: Neurobiological and pharmacological validity. Pharmacol Biochem Behav. 2012;103:119–155. doi: 10.1016/j.pbb.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero A, Pawlowski M, Ortmann D, Snijders K, Yiangou L, Cardoso de Brito M, Brown S, Bernard WG, Cooper JD, Giacomelli E, Gambardella L, Hannan NRF, Iyer D, Sampaziotis F, Serrano F, Zonneveld MCF, Sinha S, Kotter M, Vallier L. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Dev Camb Engl. 2016;143:4405–4418. doi: 10.1242/dev.138081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nöthen MM, Nurnberger JI, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, Horton WJ, Morgan SD, Breslau N, Budde J, Cloninger CR, Dick DM, Foroud T, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Kuperman S, Madden PAF, Mayo K, Nurnberger J, Pomerleau O, Porjesz B, Reyes O, Schuckit M, Swan G, Tischfield JA, Edenberg HJ, Rice JP, Goate AM. Variants in the Nicotinic Receptors Alter the Risk for Nicotine Dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP. Assembly of functionally integrated human forebrain spheroids. Nature advance online publication. 2017 doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed]
- Bodhinathan K, Slesinger PA. Alcohol modulation of G-protein-gated inwardly rectifying potassium channels: from binding to therapeutics. Front Physiol. 2014;5 doi: 10.3389/fphys.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boies S, Mérette C, Paccalet T, Maziade M, Bureau A. Polygenic risk scores distinguish patients from non-affected adult relatives and from normal controls in schizophrenia and bipolar disorder multi-affected kindreds. Am J Med Genet B Neuropsychiatr Genet. 2017 doi: 10.1002/ajmg.b.32614. n/a-n/a. [DOI] [PubMed]
- Boschen KE, Keller SM, Roth TL, Klintsova AY. Epigenetic mechanisms in alcohol- and adversity-induced developmental origins of neurobehavioral functioning. Neurotoxicol Teratol. 2018 doi: 10.1016/j.ntt.2017.12.009. [DOI] [PMC free article] [PubMed]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao S, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modeling schizophrenia using hiPSC neurons. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Marchetto MC, Benvenisty N, Brüstle O, Ebert A, Izpisua Belmonte JC, Kaykas A, Lancaster MA, Livesey FJ, McConnell MJ, McKay RD, Morrow EM, Muotri AR, Panchision DM, Rubin LL, Sawa A, Soldner F, Song H, Studer L, Temple S, Vaccarino FM, Wu J, Vanderhaeghen P, Gage FH, Jaenisch R. Creating Patient-Specific Neural Cells for the In Vitro Study of Brain Disorders. Stem Cell Rep. 2015;5:933–945. doi: 10.1016/j.stemcr.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello CA, Jokinen V, Sakha P, Terazono H, Nomura F, Kaneko T, Lauri SE, Franssila S, Rivera C, Yasuda K, Huttunen HJ. Microtechnologies to fuel neurobiological research with nanometer precision. J Nanobiotechnology. 2013;11:11. doi: 10.1186/1477-3155-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulla LF, Beaumont KG, Mrksich M, Krainc D. Micropatterning Facilitates the Long-Term Growth and Analysis of iPSC-Derived Individual Human Neurons and Neuronal Networks. Adv Healthc Mater. 2016;5:1894–1903. doi: 10.1002/adhm.201500900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Moore JH. Chapter 11: Genome-Wide Association Studies. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, Ziegler RG, Hunter DJ, Chanock S, Hankinson S, Kraft P, Bergen AW. Genome-Wide and Candidate Gene Association Study of Cigarette Smoking Behaviors. PLOS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Marro S, Wernig M, Südhof TC. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc Natl Acad Sci U S A. 2013;110:16622–16627. doi: 10.1073/pnas.1316240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, DeLong CJ, Bame M, Rajapakse I, Herron TJ, McInnis MG, O’Shea KS. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry. 2014;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ, Hochedlinger K. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen J, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, McIntosh AM. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117) Mol Psychiatry. 2017 doi: 10.1038/mp.2017.153. [DOI] [PMC free article] [PubMed]
- Clarke TK, Laucht M, Ridinger M, Wodarz N, Rietschel M, Maier W, Lathrop M, Lourdusamy A, Zimmermann US, Desrivieres S, Schumann G. KCNJ6 is Associated with Adult Alcohol Dependence and Involved in Gene × Early Life Stress Interactions in Adolescent Alcohol Drinking. Neuropsychopharmacology. 2011;36:1142–1148. doi: 10.1038/npp.2010.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close JL, Yao Z, Levi BP, Miller JA, Bakken TE, Menon V, Ting JT, Wall A, Krostag AR, Thomsen ER, Nelson AM, Mich JK, Hodge RD, Shehata SI, Glass IA, Bort S, Shapovalova NV, Ngo NK, Grimley JS, Phillips JW, Thompson CL, Ramanathan S, Lein E. Single-Cell Profiling of an In Vitro Model of Human Interneuron Development Reveals Temporal Dynamics of Cell Type Production and Maturation. Neuron. 2017;93:1035–1048.e5. doi: 10.1016/j.neuron.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.I.S. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T.T. and G. Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D, Bernardinelli L, Mannucci PM, Mauri F, Merlini PA, Absher D, Assimes TL, Fortmann SP, Iribarren C, Knowles JW, Quertermous T, Ferrucci L, Tanaka T, Bis JC, Furberg CD, Haritunians T, McKnight B, Psaty BM, Taylor KD, Thacker EL, Almgren P, Groop L, Ladenvall C, Boehnke M, Jackson AU, Mohlke KL, Stringham HM, Tuomilehto J, Benjamin EJ, Hwang S-J, Levy D, Preis SR, Vasan RS, Duan J, Gejman PV, Levinson DF, Sanders AR, Shi J, Lips EH, McKay JD, Agudo A, Barzan L, Bencko V, Benhamou S, Castellsagué X, Canova C, Conway DI, Fabianova E, Foretova L, Janout V, Healy CM, Holcátová I, Kjaerheim K, Lagiou P, Lissowska J, Lowry R, Macfarlane TV, Mates D, Richiardi L, Rudnai P, Szeszenia-Dabrowska N, Zaridze D, Znaor A, Lathrop M, Brennan P, Bandinelli S, Frayling TM, Guralnik JM, Milaneschi Y, Perry JRB, Altshuler D, Elosua R, Kathiresan S, Lucas G, Melander O, O’Donnell CJ, Salomaa V, Schwartz SM, Voight BF, Penninx BW, Smit JH, Vogelzangs N, Boomsma DI, Geus EJC, de Vink JM, Willemsen G, Chanock SJ, Gu F, Hankinson SE, Hunter DJ, Hofman A, Tiemeier H, Uitterlinden AG, Duijn CM, van Walter S, Chasman DI, Everett BM, Paré G, Ridker PM, Li MD, Maes HH, Audrain-McGovern J, Posthuma D, Thornton LM, Lerman C, Kaprio J, Rose JE, Ioannidis JPA, Kraft P, Lin D-Y, Sullivan PF. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- De Filippis L, Halikere A, McGowan H, Moore JC, Tischfield JA, Hart RP, Pang ZP. Ethanol-mediated activation of the NLRP3 inflammasome in iPS cells and iPS cells-derived neural progenitor cells. Mol Brain. 2016;9 doi: 10.1186/s13041-016-0221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflorio C, Blanchard S, Carisì MC, Bohl D, Maskos U. Human polymorphisms in nicotinic receptors: a functional analysis in iPS-derived dopaminergic neurons. FASEB J Off Publ Fed Am Soc Exp Biol. 2017;31:828–839. doi: 10.1096/fj.201600932R. [DOI] [PubMed] [Google Scholar]
- del Burgo LS, Cortes R, Mengod G, Zarate J, Echevarria E, Salles J. Distribution and neurochemical characterization of neurons expressing GIRK channels in the rat brain. J Comp Neurol. 2008;510:581–606. doi: 10.1002/cne.21810. [DOI] [PubMed] [Google Scholar]
- Desai BS, Monahan AJ, Carvey PM, Hendey B. Blood–Brain Barrier Pathology in Alzheimer’s and Parkinson’s Disease: Implications for Drug Therapy. Cell Transplant. 2007;16:285–299. doi: 10.3727/000000007783464731. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, Budde J, Saccone N, Foroud T, Nurnberger J, Xuei X, Conneally PM, Schuckit M, Almasy L, Crowe R, Kuperman S, Kramer J, Tischfield JA, Hesselbrock V, Edenberg HJ, Porjesz B, Rice JP, Bierut L, Goate A. A Systematic SNP Screen to Fine-Map Alcohol Dependence Genes on Chromosome 7 Identifies Association with a Novel Susceptibility Gene ACN9. Biol Psychiatry. 2008;63 doi: 10.1016/j.biopsych.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStefano JK, Taverna DM. Disease Gene Identification, Methods in Molecular Biology. Humana Press; Totowa, NJ: 2011. Technological Issues and Experimental Design of Gene Association Studies; pp. 3–16. [DOI] [PubMed] [Google Scholar]
- Drozda K, Müller DJ, Bishop JR. Pharmacogenomic Testing for Neuropsychiatric Drugs: Current Status of Drug Labeling, Guidelines for Using Genetic Information, and Test Options. Pharmacotherapy. 2014;34:166–184. doi: 10.1002/phar.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The Genetics of Alcohol Metabolism: Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase Variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. Chapter 32 - Genetics of alcoholism. In: Sullivan EV, Pfefferbaum A, editors. Handbook of Clinical Neurology, Alcohol and the Nervous System. Elsevier; 2014. pp. 561–571. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Rice JP, Schuckit MA, Taylor R, Webb BT, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Mozafari S, Glatza M, Starost L, Velychko S, Hallmann AL, Cui QL, Schambach A, Kim KP, Bachelin C, Marteyn A, Hargus G, Johnson RM, Antel J, Sterneckert J, Zaehres H, Schöler HR, Evercooren ABV, Kuhlmann T. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci. 2017;114:E2243–E2252. doi: 10.1073/pnas.1614412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder GA, Gama Sosa MA, De Gasperi R. Transgenic Mouse Models of Alzheimer’s Disease. Mt Sinai J Med N Y. 2010;77:69–81. doi: 10.1002/msj.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantuzzo JA, De Filippis L, McGowan H, Yang N, Ng YH, Halikere A, Liu JJ, Hart RP, Wernig M, Zahn JD, Pang ZP. μNeurocircuitry: Establishing in vitro models of neurocircuits with human neurons. Technology. 2017;5:87–97. doi: 10.1142/S2339547817500054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulconbridge A, Alderton A, Mann A, White A, Leha A, Goncalves A, Lamond AI, Kolb-Kokocinski A, Kathuria A, Kirton CM, Agu CA, Bensaddek D, Gaffney DJ, Danovi D, McCarthy D, Birney E, Soares F, Watt FM, Casale FP, Kilpinen H, Streeter I, Alasoo K, Clarke L, Vallier L, Patel M, Moens N, Culley OJ, Stegle O, Harrison PW, Danecek P, Beales P, Nelson R, Halai R, Durbin R, Meleckyte R, Harper S, Bala S, McCarthy SA, Patel SR, Ashford S, Afzal V, Ouwehand WH, Memari Y. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaaser IW, Slesinger PA. Dual activation of neuronal G protein-gated inwardly rectifying potassium (GIRK) channels by cholesterol and alcohol. Sci Rep. 2017;7 doi: 10.1038/s41598-017-04681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Konnerth A. Imaging Calcium in Neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Hales CM, Rolston JD, Potter SM. How to Culture, Record and Stimulate Neuronal Networks on Micro-electrode Arrays (MEAs) J Vis Exp JoVE. 2010 doi: 10.3791/2056. [DOI] [PMC free article] [PubMed]
- Harrison NL, Skelly MJ, Grosserode EK, Lowes DC, Zeric T, Phister S, Salling MC. Effects of acute alcohol on excitability in the CNS. Neuropharmacology, Alcoholism. 2017;122:36–45. doi: 10.1016/j.neuropharm.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PaF, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Ho SM, Hartley BJ, Flaherty E, Rajarajan P, Abdelaal R, Obiorah I, Barretto N, Muhammad H, Phatnani HP, Akbarian S, Brennand KJ. Evaluating Synthetic Activation and Repression of Neuropsychiatric-Related Genes in hiPSC-Derived NPCs, Neurons, and Astrocytes. Stem Cell Rep. 2017;9:615–628. doi: 10.1016/j.stemcr.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM, Hartley BJ, Tcw J, Beaumont M, Stafford K, Slesinger PA, Brennand KJ. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods San Diego Calif. 2016a;101:113–124. doi: 10.1016/j.ymeth.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM, Hartley BJ, Tcw J, Beaumont M, Stafford K, Slesinger PA, Brennand KJ. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods San Diego Calif. 2016b;101:113–124. doi: 10.1016/j.ymeth.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mohan A, Ridder D, Sunaert S, Vanneste S. The neural correlates of the unified percept of alcohol-related craving: a fMRI and EEG study. Sci Rep. 2018;8:923. doi: 10.1038/s41598-017-18471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YWA, Zhou B, Wernig M, Südhof TC. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell. 2017;168:427–441.e21. doi: 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiwut C, Yang BZ, Kranzler HR, Anton RF, Hirunsatit R, Weiss RD, Covault J, Farrer LA, Gelernter J. GABRG1 and GABRA2 variation associated with alcohol dependence in African Americans. Alcohol Clin Exp Res. 2012;36:588–593. doi: 10.1111/j.1530-0277.2011.01637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarajan C, Pandey AK, Chorlian DB, Manz N, Stimus AT, Bauer LO, Hesselbrock VM, Schuckit MA, Kuperman S, Kramer J, Porjesz B. Reward processing deficits and impulsivity in high-risk offspring of alcoholics: A study of event-related potentials during a monetary gambling task. Int J Psychophysiol Off J Int Organ Psychophysiol. 2015;98:182–200. doi: 10.1016/j.ijpsycho.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B, Jones K, Chorlian D, Padmanabhapillai A, Rangaswamy M, Stimus A, Begleiter H. Event-related oscillations in offspring of alcoholics: Neuro-cognitive disinhibition as a risk for alcoholism. Biol Psychiatry. 2006;59:625–634. doi: 10.1016/j.biopsych.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B. Family-based Genome-wide Association Study of Frontal Theta Oscillations Identifies Potassium Channel Gene KCNJ6. Genes Brain Behav. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide Association Analysis of Symptoms of Alcohol Dependence in the Molecular Genetics of Schizophrenia (MGS2) Control Sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature advance online publication. 2017 doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee M, Ji H, Ehrlich L, Yabuuchi A, Takeuchi A, Cunniff K, Hongguang H, Mckinney-Freeman S, Naveiras O, Yoon T, Irizarry R, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin S, Weissman I, Feinberg A, Daley G. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lee H, Kim HJ, Bang E, Lee SH, Lee DW, Woo DC, Choi CB, Hong KS, Lee C, Choe BY. In vivo and ex vivo evidence for ketamine-induced hyperglutamatergic activity in the cerebral cortex of the rat: Potential relevance to schizophrenia. NMR Biomed. 2011;24:1235–1242. doi: 10.1002/nbm.1681. [DOI] [PubMed] [Google Scholar]
- Klimesch W, Doppelmayr M, Yonelinas A, Kroll NEA, Lazzara M, Röhm D, Gruber W. Theta synchronization during episodic retrieval: neural correlates of conscious awareness. Cogn Brain Res. 2001;12:33–38. doi: 10.1016/S0926-6410(01)00024-6. [DOI] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TDM, Pandey SC. The Epigenetic Landscape of Alcoholism. Int Rev Neurobiol. 2014;115:75–116. doi: 10.1016/B978-0-12-801311-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijlaars J, Oyelami T, Diels A, Rohrbacher J, Versweyveld S, Meneghello G, Tuefferd M, Verstraelen P, Detrez JR, Verschuuren M, De Vos WH, Meert T, Peeters PJ, Cik M, Nuydens R, Brône B, Verheyen A. Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci Rep. 2016;6:36529. doi: 10.1038/srep36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttälä A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, Van Handel B, Parkkonen O, Sinisalo J, Jalanko A, Hawkins RD, Woods NB, Otonkoski T, Trokovic R. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Rep. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kögler G, Müller FJ, Koch P, Brüstle O. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, Vos JD, Lehmann S, Lemaitre JM. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Sanchez CV, Chen M, Morin PJ, Wells JM, Hanlon EB, Xia W. Three Dimensional Human Neuro-Spheroid Model of Alzheimer’s Disease Based on Differentiated Induced Pluripotent Stem Cells. PLOS ONE. 2016;11:e0163072. doi: 10.1371/journal.pone.0163072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sulovari A, Cheng C, Zhao H, Kranzler HR, Gelernter J. Association of gamma-aminobutyric acid A receptor α2 gene (GABRA2) with alcohol use disorder. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2014;39:907–918. doi: 10.1038/npp.2013.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MC, Muratore CR, Gierahn TM, Sullivan SE, Srikanth P, De Jager PL, Love JC, Young-Pearse TL. Single-Cell Detection of Secreted Aβ and sAPPα from Human IPSC-Derived Neurons and Astrocytes. J Neurosci. 2016;36:1730–1746. doi: 10.1523/JNEUROSCI.2735-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Kranzler HR, Joshi P, Shin DG, Covault J. GABRA2 alcohol dependence risk allele is associated with reduced expression of chromosome 4p12 GABAA subunit genes in human neural cultures. Alcohol Clin Exp Res. 2015;39:1654–1664. doi: 10.1111/acer.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Kranzler HR, Levine ES, Covault J. Examining the effects of alcohol on GABAA receptor mRNA expression and function in neural cultures generated from control and alcohol dependent donor induced pluripotent stem cells. Alcohol. 2018;66:45–53. doi: 10.1016/j.alcohol.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R, Levine ES, Kranzler HR, Abreu C, Covault J. Pilot Study of iPS-derived Neural Cells to Examine Biological Effects of Alcohol on Human Neurons in vitro. Alcohol Clin Exp Res. 2012;36:1678–1687. doi: 10.1111/j.1530-0277.2012.01792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Cai Y, Zhang Q, Zhang W, Nogales-Cadenas RD, Zhang Z. Integrated Post-GWAS Analysis Shed New Light on the Disease Mechanisms of Schizophrenia. Genetics. 2016;204 doi: 10.1534/genetics.116.187195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Swerdel MR, Lazaropoulos MP, Hoffman GS, Toro-Ramos AJ, Wright J, Lederman H, Chen J, Moore JC, Hart RP. Spontaneous ATM Gene Reversion in A-T iPSC to Produce an Isogenic. Cell Line Stem Cell Rep. 2015;5:1097–1108. doi: 10.1016/j.stemcr.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Hrabovsky A, Chen J, Puliafito BR, Gilbert SR, Zheng D, Lachman HM. Integrative transcriptome network analysis of iPSC-derived neurons from schizophrenia and schizoaffective disorder patients with 22q11.2 deletion. BMC Syst Biol. 2016;10 doi: 10.1186/s12918-016-0366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey MR, Magnani D, Cleary EM, Vasistha NA, James OT, Selvaraj BT, Burr K, Story D, Shaw CE, Kind PC, Hardingham GE, Wyllie DJA, Chandran S. Maturation and electrophysiological properties of human pluripotent stem cell-derived oligodendrocytes. Stem Cells Dayt Ohio. 2016;34:1040–1053. doi: 10.1002/stem.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo IA, Harris RA. GABAA receptors and alcohol. Pharmacol Biochem Behav. 2008;90:90–94. doi: 10.1016/j.pbb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhong X, Liu H, Hao L, Huang CTL, Sherafat MA, Jones J, Ayala M, Li L, Zhang SC. Generation of serotonin neurons from human pluripotent stem cells. Nat Biotechnol. 2016;34:89–94. doi: 10.1038/nbt.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, Slesinger PA. Emerging concepts for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11:301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch WJ, Nicholson KL, Dance ME, Morgan RW, Foley PL. Animal Models of Substance Abuse and Addiction: Implications for Science, Animal Welfare, and Society. Comp Med. 2010;60:177–188. [PMC free article] [PubMed] [Google Scholar]
- Mahairaki V, Ryu J, Peters A, Chang Q, Li T, Park TS, Burridge PW, Talbot CC, Asnaghi L, Martin LJ, Zambidis ET, Koliatsos VE. Induced Pluripotent Stem Cells from Familial Alzheimer’s Disease Patients Differentiate into Mature Neurons with Amyloidogenic Properties. Stem Cells Dev. 2014;23:2996–3010. doi: 10.1089/scd.2013.0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, Gill M, Karayiorgou M, Kelsoe JR, Krastoshevsky O, Krause V, Leibenluft E, Levy DL, Makarov V, Bhandari A, Malhotra AK, McMahon FJ, Nöthen MM, Potash JB, Rietschel M, Schulze TG, Sebat J. High Frequencies of De Novo CNVs in Bipolar Disorder and Schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, Hack L, Bigdeli T, van der Vaart AD, Web BT, Bacanu SA, Kalsi G, Consortium, C. Kendler KS, Miles MF, Dick D, Riley BP, Dumur C, Vladimirov VI. Integrating mRNA and miRNA Weighted Gene Co-Expression Networks with eQTLs in the Nucleus Accumbens of Subjects with Alcohol Dependence. PLOS ONE. 2015;10:e0137671. doi: 10.1371/journal.pone.0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko E, Chawarska K, Pelphrey K, Howe J, Vaccarino FM. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, Widmer HR, Eggan K, Goldstein PA, Anderson SA, Studer L. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JD, Schnitzer MJ. Optical Strategies for Sensing Neuronal Voltage Using Quantum Dots and Other Semiconductor Nanocrystals. ACS Nano. 2013;7:4601–4609. doi: 10.1021/nn401410k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, de Moor MHM, Jansen R, Gelernter J, Sherva R, Willemsen G, Boomsma DI, Penninx BW, Vink JM. The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet B Neuropsychiatr Genet. 2015;168:739–748. doi: 10.1002/ajmg.b.32379. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, Ganz J, Jaffe AE, Kwan KY, Kwon M, Lodato MA, Mills RE, Paquola ACM, Rodin RE, Rosenbluh C, Sestan N, Sherman MA, Shin JH, Song S, Straub RE, Thorpe J, Weinberger DR, Urban AE, Zhou B, Gage FH, Lehner T, Senthil G, Walsh CA, Chess A, Courchesne E, Gleeson JG, Kidd JM, Park PJ, Pevsner J, Vaccarino FM. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science. 2017;356 doi: 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, Gonçalves JT, Toda T, Kim Y, Winkler J, Yao J, Hetzer MW, Gage FH. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015a;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, Zheng Y, Diffenderfer KE, Zhang J, Soltani S, Eames T, Schafer ST, Boyer L, Marchetto MC, Nurnberger JI, Calabrese JR, Ødegaard KJ, McCarthy MJ, Zandi PP, Alda M, Alba M, Nievergelt CM, Pharmacogenomics of Bipolar Disorder Study. Mi S, Brennand KJ, Kelsoe JR, Gage FH, Yao J. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015b;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, Jaenisch R. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Oehring EM, Jung YC, Sullivan EV, Hawkes WC, Pfefferbaum A, Schulte T. Midbrain-Driven Emotion and Reward Processing in Alcoholism. Neuropsychopharmacology. 2013;38:1844–1853. doi: 10.1038/npp.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJL, Lopez AD. Measuring the Global Burden of Disease. N Engl J Med. 2013;369:448–457. doi: 10.1056/NEJMra1201534. [DOI] [PubMed] [Google Scholar]
- Nadadhur AG, Emperador Melero J, Meijer M, Schut D, Jacobs G, Li KW, Hjorth JJJ, Meredith RM, Toonen RF, Van Kesteren RE, Smit AB, Verhage M, Heine VM. Multi-level characterization of balanced inhibitory-excitatory cortical neuron network derived from human pluripotent stem cells. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0178533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Müller FJ, Wang YC, Boscolo FS, Fakunle E, Dumevska B, Lee S, Park HS, Olee T, D’Lima DD, Semechkin R, Parast MM, Galat V, Laslett AL, Schmidt U, Keirstead HS, Loring JF, Laurent LC. Recurrent Variations in DNA Methylation in Human Pluripotent Stem Cells and their Differentiated Derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niciu MJ, Henter ID, Sanacora G, Zarate CA. Glial abnormalities in substance use disorders and depression: Does shared glutamatergic dysfunction contribute to comorbidity? World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2014;15:2–16. doi: 10.3109/15622975.2013.829585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obien MEJ, Deligkaris K, Bullmann T, Bakkum DJ, Frey U. Revealing neuronal function through microelectrode array recordings. Front Neurosci. 2015;8 doi: 10.3389/fnins.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odawara A, Saitoh Y, Alhebshi AH, Gotoh M, Suzuki I. Long-term electrophysiological activity and pharmacological response of a human induced pluripotent stem cell-derived neuron and astrocyte co-culture. Biochem Biophys Res Commun. 2014;443:1176–1181. doi: 10.1016/j.bbrc.2013.12.142. [DOI] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-Transgenic Model of Alzheimer’s Disease with Plaques and Tangles: Intracellular Aβ and Synaptic Dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/S0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- Oni EN, Halikere A, Li G, Toro-Ramos AJ, Swerdel MR, Verpeut JL, Moore JC, Bello NT, Bierut LJ, Goate A, Tischfield JA, Pang ZP, Hart RP. Increased nicotine response in iPSC-derived human neurons carrying the CHRNA5 N398 allele. Sci Rep. 2016;6 doi: 10.1038/srep34341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, Kamarajan C, Rangaswamy M, Porjesz B. Event-Related Oscillations in Alcoholism Research: A Review. J Addict Res Ther Suppl. 2012;7 doi: 10.4172/2155-6105.S7-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Paşca SP. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD, Musunuru K. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 2017;20:558–570.e10. doi: 10.1016/j.stem.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the Inheritance of Alcoholism: A Study of Male and Female Twins. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Prytkova I, Brennand KJ. Prospects for Modeling Abnormal Neuronal Function in Schizophrenia Using Human Induced Pluripotent Stem Cells. Front Cell Neurosci. 2017;11 doi: 10.3389/fncel.2017.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Brown J, Arlotta P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med. 2016;22:1220–1228. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLOS ONE. 2016;11:e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Lumeng L, Jr, N JI, Edenberg HJ, McBride WJ, Niculescu AB. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7:222–256. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF, Grucza RA, Sun L, Duan W, Budde J, Culverhouse RC, Fox L, Hinrichs AL, Steinbach JH, Wu M, Rice JP, Goate AM, Bierut LJ. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med. 2015;49:e73–e79. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- Saito S, Ihara M. New Therapeutic Approaches for Alzheimer’s Disease and Cerebral Amyloid Angiopathy. Front Aging Neurosci. 2014;6 doi: 10.3389/fnagi.2014.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, The 23andMe Research Team. Gray JC, de Wit H, Davis LK, MacKillop J, Palmer AA. Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol. 2017 doi: 10.1111/adb.12574. n/a-n/a. [DOI] [PMC free article] [PubMed]
- Sancho-Martinez I, Nivet E, Xia Y, Hishida T, Aguirre A, Ocampo A, Ma L, Morey R, Krause MN, Zembrzycki A, Ansorge O, Vazquez-Ferrer E, Dubova I, Reddy P, Lam D, Hishida Y, Wu MZ, Esteban CR, O’Leary D, Wahl GM, Verma IM, Laurent LC, Belmonte JCI. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat Commun. 2016;7:10743. doi: 10.1038/ncomms10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hällfors J, Harris SE, Hieber S, Hofer E, Hottenga JJ, Johansson Å, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikäinen L-P, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud A-C, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, de Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen A-L, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin M-R, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimäki T, Liu Yongmei Madden PAF, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, van der Harst P, van Duijn CM, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Yun Rice KM, Desrivières S, Kliewer SA, Mangelsdorf DJ, Müller CP, Levy D, Elliott P. KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A. 2016;113:14372–14377. doi: 10.1073/pnas.1611243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Hopkinson DA, Harris H. Developmental changes and polymorphism in human alcohol dehydrogenase. Ann Hum Genet. 1971;34:251–271. doi: 10.1111/j.1469-1809.1971.tb00238.x. [DOI] [PubMed] [Google Scholar]
- Soliman MA, Aboharb F, Zeltner N, Studer L. Pluripotent stem cells in neuropsychiatric disorders. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.40. [DOI] [PMC free article] [PubMed]
- Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, Harikrishnan K, Elahi S, Freedman D, Brennand KJ, Sarder P, Stachowiak MK. Cerebral organoids reveal early cortical maldevelopment in schizophrenia—computational anatomy and genomics, role of FGFR1. Transl Psychiatry. 2017;7 doi: 10.1038/s41398-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger JL, Russek SJ. GABAA receptors: building the bridge between subunit mRNAs, their promoters, and cognate transcription factors. Pharmacol Ther. 2004;101:259–281. doi: 10.1016/j.pharmthera.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Stern S, Santos R, Marchetto MC, Mendes APD, Rouleau GA, Biesmans S, Wang QW, Yao J, Charnay P, Bang AG, Alda M, Gage FH. Neurons derived from patients with bipolar disorder divide into intrinsically different sub-populations of neurons, predicting the patients’ responsiveness to lithium. Mol Psychiatry. 2017 doi: 10.1038/mp.2016.260. [DOI] [PMC free article] [PubMed]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swistowski A, Peng J, Liu Q, Mali P, Rao MS, Cheng L, Zeng X. Efficient Generation of Functional Dopaminergic Neurons from Human Induced Pluripotent Stem Cells Under Defined Conditions. Stem Cells Dayt Ohio. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takamatsu K, Ikeda T, Haruta M, Matsumura K, Ogi Y, Nakagata N, Uchino M, Ando Y, Nishimura Y, Senju S. Degradation of amyloid beta by human induced pluripotent stem cell-derived macrophages expressing Neprilysin-2. Stem Cell Res. 2014;13:442–453. doi: 10.1016/j.scr.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Tawa EA, Hall SD, Lohoff FW. Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol Oxf Oxfs. 2016;51:507–514. doi: 10.1093/alcalc/agw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66:57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Carvalho CMB, Yuan B, Gu S, Altheimer AN, McCarthy S, Malhotra D, Sebat J, Siegel AJ, Rudolph U, Lupski JR, Levy DL, Brennand KJ. Divergent Levels of Marker Chromosomes in an hiPSC-Based Model of Psychosis. Stem Cell Rep. 2017a;8:519–528. doi: 10.1016/j.stemcr.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, Machlovi SI, Abdelaal R, Karch CM, Phatnani H, Slesinger PA, Zhang B, Goate AM, Brennand KJ. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017b;9:600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Akamatsu W, Okada Y, Ohnishi T, Balan S, Hisano Y, Iwayama Y, Toyota T, Matsumoto T, Itasaka N, Sugiyama S, Tanaka M, Yano M, Dean B, Okano H, Yoshikawa T. Analysis of induced pluripotent stem cells carrying 22q11.2 deletion. Transl Psychiatry. 2016;6:e934. doi: 10.1038/tp.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer W, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nöthen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria KC, Stern S, Marchetto MC, Gage FH. Serotonin in psychiatry: in vitro disease modeling using patient-derived neurons. Cell Tissue Res. 2017:1–10. doi: 10.1007/s00441-017-2670-4. [DOI] [PubMed]
- Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2015;45:1061–1072. doi: 10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wartburg JP, Papenberg J, Aebi H. An atypical human alcohol dehydrogenase. Can J Biochem. 1965;43:889–898. doi: 10.1139/o65-102. [DOI] [PubMed] [Google Scholar]
- Wang C, Najm R, Xu Q, Jeong D, Walker D, Balestra ME, Yoon SY, Yuan H, Li G, Miller ZA, Miller BL, Malloy MJ, Huang Y. Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat Med. 2018;1 doi: 10.1038/s41591-018-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yang L, Grishin D, Rios X, Ye LY, Hu Y, Li K, Zhang D, Church GM, Pu WT. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat Protoc. 2017;12:88–103. doi: 10.1038/nprot.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CR, O’Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, Wakabayashi Y, Morningstar JE, Shi X, Choi J, Xia F, Peters DT, Florido MHC, Tsankov AM, Duberow E, Comisar L, Shay J, Jiang X, Meissner A, Musunuru K, Kathiresan S, Daheron L, Zhu J, Gerszten RE, Deo RC, Vasan RS, O’Donnell CJ, Cowan CA. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell. 2017;20:547–557.e7. doi: 10.1016/j.stem.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:nrg3457. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K, Iwasaki M, Takakubo H, Sakamoto T, Ikuno T, Miyoshi M, Kondo T, Nakao Y, Nakagawa M, Inoue H, Yamashita JK. In Vitro Modeling of Blood-Brain Barrier with Human iPSC-Derived Endothelial Cells, Pericytes, Neurons, and Astrocytes via Notch Signaling. Stem Cell Rep. 2017;8:634–647. doi: 10.1016/j.stemcr.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang J, Zhao H, Ma Y, Shi G, Song J, Tang Y, Li S, Li T, Liu N, Tang F, Gu J, Zhang L, Zhang Z, Zhang X, Jin Y, Le W. Early pathogenic event of Alzheimer’s disease documented in iPSCs from patients with PSEN1 mutations. Oncotarget. 2016;8:7900–7913. doi: 10.18632/oncotarget.13776. https://doi.org/10.18632/oncotarget.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Südhof TC, Wernig M. Induced Neuronal Cells: How to Make and Define a Neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, Silberman P, Silberman G, Jappelli R, Gage FH. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep. 2014;2:295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr NM, Pfefferbaum A. Alcohol’s Effects on the Brain: Neuroimaging Results in Humans and Animal Models. Alcohol Res Curr Rev. 2017;38:183–206. [PMC free article] [PubMed] [Google Scholar]
- Zakhari S. Alcohol Metabolism and Epigenetics Changes. Alcohol Res Curr Rev. 2013;35:6–16. [PMC free article] [PubMed] [Google Scholar]
- Zhou DX, Zhao Y, Baker JA, Gu Q, Hamre KM, Yue J, Jones BC, Cook MN, Lu L. The effect of alcohol on the differential expression of cluster of differentiation 14 gene, associated pathways, and genetic network. PLOS ONE. 2017;12:e0178689. doi: 10.1371/journal.pone.0178689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Lu L, Tan Y, Pan X, Cai Y, Wang X, Hong J, Zhong C, Wang F, Zhang X, Vanderlinden LA, Tabakoff B, Luo X. Genome-wide association discoveries of alcohol dependence. Am J Addict Am Acad Psychiatr Alcohol Addict. 2014;23:526–539. doi: 10.1111/j.1521-0391.2014.12147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


