Abstract
Hibernation is an exceptional physiological response to a hostile environment, characterized by a seasonal period of torpor cycles involving dramatic reductions of body temperature and metabolism, and arousal back to normothermia. As the mechanisms regulating hibernation are still poorly understood, here we analysed the expression of genes involved in energy homeostasis, torpor regulation, and daily or seasonal timing using digital droplet PCR in various central and peripheral tissues sampled at different stages of torpor/arousal cycles in the European hamster. During torpor, the hypothalamus exhibited strongly down-regulated gene expression, suggesting that hypothalamic functions were reduced during this period of low metabolic activity. During both torpor and arousal, many structures (notably the brown adipose tissue) exhibited altered expression of deiodinases, potentially leading to reduced tissular triiodothyronine availability. During the arousal phase, all analysed tissues showed increased expression of the core clock genes Per1 and Per2. Overall, our data indicated that the hypothalamus and brown adipose tissue were the tissues most affected during the torpor/arousal cycle, and that clock genes may play critical roles in resetting the body’s clocks at the beginning of the active period.
Introduction
Animals living in the wild must cope with seasonal variations in ambient temperature (Ta) and day length (photoperiod), which impact food availability. These seasonal variations necessitate major physiological adaptations, particularly for endotherms that have to maintain a relatively constant body temperature (Tb). As winter approaches, animals adapt to the decreasing Ta, photoperiod, and food resources by limiting their energy expenditure in various ways, including decreasing their metabolism, inhibiting reproduction, and insulating their body. The most extreme and efficient strategy is hibernation, an adaptive event observed in mammalian groups, including monotremes, bats, primates, and rodents1,2. Hibernation reportedly gives these animals a better chance of survival2.
Hibernation is an exceptional physiological phenomenon comprising multiple phases within a two-switch model1,3,4. The summer-winter switch enables phases of controlled heterothermy that are required to enter a state of torpor. The second switch occurs within the winter period, and controls a series of hypothermia periods—torpor bouts, lasting from a few hours to several days depending on the species—that are interrupted by short spontaneous interbouts of normothermia. Torpor bouts are characterized by low Tb and drastically reduced metabolic activity, with diminution of heartbeat and respiration, altered fuel utilization, and reduction of costly cell processes. Arousal phases involve rapid rewarming and increased metabolic activity. During the normothermic interbouts, Tb and metabolic activity return to a basal level, possibly to clear metabolic waste5. Hibernators are divided into two categories based on their energy saving strategy: fat-storing species (e.g. the marmot, Marmota marmota) exhibit an extensive fattening period prior to hibernation, while food-storing species (e.g. the European hamster, Cricetus cricetus) hoard food in a burrow and feed between torpor bouts. Although these strategies require different metabolic adaptations, they both involve fasting states that last from a few hours to several days or months6–8.
In many species, the time of hibernation is controlled by seasonal changes in photoperiod and temperature9. Photoperiodic cycles influence the levels of melatonin, a neurohormone that is secreted by the pineal gland only during the night, with correspondingly greater production during long winter nights10. Photoperiod-related changes in circulating melatonin are fundamental for seasonal functions, including hibernation11. Intracerebroventricular infusion of melatonin prolongs hibernation bouts in golden-mantled ground squirrels (Citellus lateralis)12 and treatment with the melatonin antagonist S22153 reduces total hibernation duration in Syrian hamster (Cricetus auratus)13. Studies in mammals have characterized two melatonin receptors with high-affinity binding, MT1 and MT2, and one with low-affinity binding, MT3 (identified as quinone reductase 2; QR2)14. Additionally, GPR50—an orphan receptor derived from Mel1c in amphibians and birds15,16—is reportedly involved in torpor in mice17.
MT1 is present at the highest density in the pars tuberalis, a region of the pituitary stalk where melatonin regulates the expression of thyroid-stimulating hormone (TSH) by specific thyrotrope cells18. TSH secreted during a long photoperiod reaches TSH receptors expressed by the tanycytes, specialized glial cells lining the wall of the basal part of the third ventricle and sending processes in the mediobasal hypothalamus and median eminence. There, TSH increases deiodinase 2 (Dio2) and inhibits Dio3 expression within the tanycytes, resulting in increased local levels of active triiodothyronine (T3)19–25. It is believed that melatonin effects on seasonal functions are enacted through seasonal changes in hypothalamic T311,26. This likely includes hibernation, since central administration of T3 reduces daily torpor in the Siberian hamster (Phodopus sungorus)27. Rhythmic melatonin expression and other biological rhythms depend on a circadian system comprising a master clock located in the suprachiasmatic nucleus (SCN), and secondary clocks within other central and peripheral structures28. Circadian rhythms are generated by transcriptional/translational feedback loops involving core clock genes, such as Bmal1, Clock, Cry, Per, and Rev-Erbα29. While the roles of circadian clocks during hibernation are unknown, during the torpor bout in hibernating European hamsters, clock genes (including Per1, Per2, and Bmal1) stop ticking and melatonin remains at a constant low level30.
During prolonged fasting in hibernating animals, energy is supplied almost exclusively by lipid oxidation, and this switch in fuel utilization from carbohydrates to lipids must be highly regulated31. During hibernation, the primary source of endogenous glucose source becomes hepatic gluconeogenesis using lactate, pyruvate, glycerol, and amino acids32. In white adipose tissue, triglycerides are catabolized into glycerol and fatty acids, which are released into circulation33. Glycerol is converted into glucose, while fatty acids are metabolized into β-hydroxybutyrate and acetoacetate. These ketone bodies and glucose are the only fuel substrates for the brain34. This process also involves upregulation of silent information regulator 1 (SIRT1), which reduces adipogenesis by inhibiting the important adipogenic transcription factor peroxisome proliferator-activated receptor gamma (PPAR-γ), and stimulates hepatic gluconeogenesis by promoting deacetylation of PPARγ coactivator-1 alpha (PGC-1α)35,36. Peroxisome proliferator-activated receptor alpha (PPARα) also influences energy store management by stimulating hepatic fatty acid oxidation37. PPARα regulates the liver-derived fibroblast growth factor 21 (FGF21), which plays a major role in adaptive starvation responses, stimulating lipolysis in white adipose tissue and ketogenesis in liver tissue, and promoting torpor38,39. Finally, previous studies indicate that thioredoxin-interacting protein (TXNIP)—a negative regulator of thioredoxin that is involved in hypothalamic homeostasis and hepatic gluconeogenesis40,41—is up-regulated in the hypothalamus during torpor in GPR50 KO mice42, Siberian hamsters42, and ground squirrels43.
Another essential part of hibernation is the arousal from torpor bouts, in which brown adipose tissue (BAT) plays a major role by increasing Tb through non-shivering thermogenesis. Thermogenesis in BAT occurs due to the mitochondrial uncoupling protein 1 (UCP1), which uncouples the mitochondrial respiratory chain by catalysing the regulated re-entry of protons into the matrix, leading to reduced ATP synthesis and generation of heat44. Indeed, in the thirteen-lined ground squirrel (Ictidomys tridecemlineatus), UCP1 is upregulated to a similar degree during both torpor and arousal compared to in the summer active state45,46.
Although studies suggest general reductions of cellular metabolism in torpid states, the cellular and molecular pathways that drive the successive drops and rises in metabolic activity remain poorly understood. One approach to elucidating these mechanisms is to search for genes that display differential expressing patterns according to hibernation state. This approach implies that hibernators have no specific set of genes dedicated to hibernation, but rather rely on differential expression of genes that exist in most mammals47. Such gene profiling could be performed using an unbiased wide transcriptomic analysis, such RNASeq43,45,48, or a candidate genes approach49. In our present study, we aimed to establish a molecular signature of hibernation by analysing the expression of genes with potential involvement in energy homeostasis, torpor regulation, and daily or seasonal timing, in eight central and peripheral organs sampled during three different hibernation phases in the European hamster.
Results
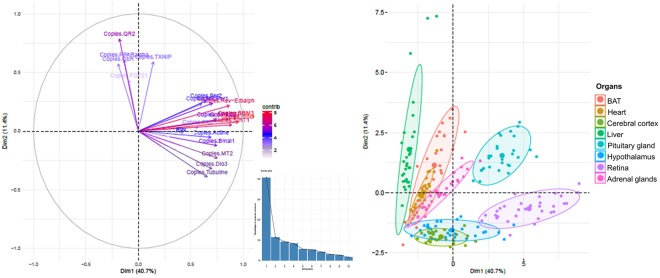
Eight central and peripheral organs (cerebral cortex, hypothalamus, pituitary gland, retina, liver, heart, brown adipose tissue, and adrenal glands) were sampled at three different hibernation phases (normothermia, torpor, and arousal) in male European hamsters. The ddPCR results provided the absolute count of RNA copies encoding melatonin receptors, thyroid metabolism enzymes, clock proteins, and selected proteins involved in energy homeostasis (a summary of the median, quartile and interquartile of absolute RNA copies of each gene in each organ at each hibernation state is given in Table S1). The principal component analysis (PCA) of these ddPCR data revealed that the major sources of variance were associated with the organ effect which groups were separated in the space drawn by the two first components (Dim1&2), explaining respectively 40.7% and 11.4% of variance; as shown in the Fig. 1. Then, Kruskal-Wallis analysis was performed on genes expressed for each organ during the different hibernation phases, revealing 61 significant adjusted P value (Benjamini-Hochberg) corresponding to 18 genes according to the hibernation state (Table 1). Thus, the median ratio between all hibernation phases (torpor versus normothermia, arousal versus normothermia and arousal versus torpor) was calculated for each genes and organs in order to more easily visualize gene expression variation (Table 2). All categories of genes and all organs displayed genomic modifications associated with the hibernation states although with some differences between groups.
Figure 1.
Major sources of variance among the organs, the genes and the hibernation stages of European hamster. The major sources of variance among all the paramaeters were determined by principal component analysis (PCA) of the ddPCR data, are associated with the organ effect. All groups were separated in the space drawn by the two first components (Dim1&2, explaining respectively 40.7% and 11.4% of variance). The variable contribution to the variance is represented with a colour gradient (contrib).
Table 1.
List of the 61-significant adjusted P value (Benjamini-Hochberg) corresponding to 18 genes according to the hibernation state (Kruskal-Wallis, adjusted P-values ≤ 0,05) in a tissue-by-tissue analysis.
| Organs | Genes | Adjusted P values | Organs | Genes | Adjusted P values |
|---|---|---|---|---|---|
| Liver | MT2 | 0.006 | Heart | Tubulin | 0.019 |
| Liver | ObR | 0.006 | Pituitary gland | Dio2 | 0.025 |
| Liver | Tubulin | 0.006 | Pituitary gland | Per2 | 0.025 |
| BAT | Dio2 | 0.008 | Pituitary gland | QR2 | 0.025 |
| BAT | Per2 | 0.008 | Retina | Bmal1 | 0.028 |
| Retina | FGF21 | 0.008 | Retina | MT2 | 0.028 |
| Retina | Per2 | 0.009 | Retina | Per1 | 0.028 |
| Retina | PPARα | 0.009 | Retina | QR2 | 0.028 |
| Hypothalamus | PGC1α | 0.01 | Retina | Tubulin | 0.028 |
| Hypothalamus | PPARα | 0.01 | Hypothalamus | Per1 | 0.031 |
| Hypothalamus | Tubulin | 0.01 | Pituitary gland | Bmal1 | 0.032 |
| Hypothalamus | Actin | 0.011 | Pituitary gland | Tubulin | 0.032 |
| Hypothalamus | G6PD | 0.011 | Adrenal glands | Per1 | 0.036 |
| Hypothalamus | Per2 | 0.011 | Cortex | Per1 | 0.037 |
| Hypothalamus | Bmal1 | 0.011 | BAT | Per1 | 0.038 |
| Hypothalamus | RBM3 | 0.011 | Hypothalamus | TXNIP | 0.038 |
| Hypothalamus | Rev-Erbα | 0.011 | Heart | Rev-Erbα | 0.039 |
| Hypothalamus | MT2 | 0.012 | Pituitary gland | Rev-Erbα | 0.041 |
| BAT | PPARα | 0.013 | Pituitary gland | Clock | 0.041 |
| Adrenal glands | FGF21 | 0.013 | Pituitary gland | FGF21 | 0.041 |
| Adrenal glands | Per2 | 0.013 | Pituitary gland | TXNIP | 0.041 |
| Cortex | Per2 | 0.014 | Liver | Per2 | 0.041 |
| BAT | Dio3 | 0.014 | Heart | Actin | 0.042 |
| Hypothalamus | Clock | 0.015 | Heart | Dio2 | 0.042 |
| Hypothalamus | QR2 | 0.015 | Heart | TXNIP | 0.042 |
| Hypothalamus | SIRT1 | 0.015 | Hypothalamus | FGF21 | 0.042 |
| BAT | MT2 | 0.016 | BAT | SIRT1 | 0.045 |
| Liver | Per1 | 0.018 | Liver | FGF21 | 0.048 |
| Heart | Per1 | 0.019 | Cortex | Dio2 | 0.049 |
| Heart | Per2 | 0.020 |
Organs/genes combinations are ranked by adjusted P -values.
Table 2.
The median ratio between all groups (Torpor versus Normothermia, Arousal versus Normothermia and Arousal versus Torpor) was calculated for each genes and organs (N = Normothermia, T = Torpor, A = Arousal).
| Per1 | Bmal1 | Rev-Erbα | Clock | Per2 | MT1 | MT2 | GPR50 | QR2 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | ||
| Cerebral cortex | T vs N | 1,54 | 0,81 | 0,81 | 0,75 | 1,24 | 1,23 | 0,83 | 2,09 | 1,04 |
| A vs N | 2,22 | 0,78 | 0,91 | 0,94 | 2,88 | 1,16 | 1,56 | 3,04 | 0,80 | |
| A vs T | 1,44 | 0,96 | 1,11 | 1,25 | 2,31 | 0,95 | 1,87 | 1,45 | 0,76 | |
| Hypothalamus | T vs N | 1,52 | 0,45 | 0,44 | 0,40 | 0,72 | 0,65 | 0,79 | 0,62 | 0,58 |
| A vs N | 2,54 | 0,51 | 0,48 | 0,63 | 1,94 | 0,72 | 1,60 | 0,89 | 0,42 | |
| A vs T | 1,67 | 1,12 | 1,10 | 1,55 | 2,69 | 1,11 | 2,03 | 1,43 | 0,72 | |
| Pituitary gland | T vs N | 3,48 | 1,22 | 1,38 | 0,86 | 2,42 | 0,68 | 0,91 | 0,61 | 0,86 |
| A vs N | 2,80 | 3,16 | 0,83 | 0,98 | 3,16 | 0,74 | 1,42 | 1,09 | 0,76 | |
| A vs T | 0,80 | 2,59 | 0,60 | 1,14 | 1,31 | 1,08 | 1,56 | 1,79 | 0,88 | |
| Retina | T vs N | 1,49 | 0,93 | 0,87 | 0,79 | 1,14 | 0,84 | 0,96 | 1,30 | 0,92 |
| A vs N | 1,87 | 1,49 | 0,96 | 1,01 | 2,69 | 0,80 | 2,30 | 1,74 | 0,61 | |
| A vs T | 1,25 | 1,60 | 1,11 | 1,28 | 2,36 | 0,95 | 2,39 | 1,34 | 0,66 | |
| Liver | T vs N | 2,74 | 1,51 | 1,02 | 0,92 | 3,13 | 0,50 | 0,28 | 0,46 | 1,04 |
| A vs N | 2,25 | 1,14 | 0,58 | 1,01 | 3,81 | 0,55 | 0,37 | 0,53 | 0,71 | |
| A vs T | 0,82 | 0,75 | 0,57 | 1,10 | 1,22 | 1,10 | 1,31 | 1,13 | 0,68 | |
| BAT | T vs N | 3,11 | 1,28 | 1,49 | 0,76 | 1,27 | 0,57 | 0,43 | 0,49 | 1,12 |
| A vs N | 2,67 | 1,06 | 1,12 | 1,07 | 3,23 | 0,39 | 0,37 | 0,68 | 0,83 | |
| A vs T | 0,86 | 0,83 | 0,75 | 1,41 | 2,55 | 0,69 | 0,86 | 1,39 | 0,74 | |
| Heart | T vs N | 3,72 | 1,08 | 1,08 | 0,88 | 2,04 | 1,25 | 0,63 | 1,05 | 1,19 |
| A vs N | 3,69 | 1,14 | 0,59 | 1,13 | 7,59 | 1,00 | 0,67 | 0,95 | 1,07 | |
| A vs T | 0,99 | 1,06 | 0,54 | 1,29 | 3,72 | 0,79 | 1,06 | 0,90 | 0,90 | |
| Adrenal glands | T vs N | 2,77 | 2,10 | 1,01 | 0,82 | 1,80 | 0,43 | 0,74 | 0,76 | 1,14 |
| A vs N | 3,16 | 2,02 | 1,03 | 1,18 | 3,97 | 0,56 | 0,83 | 1,08 | 0,99 | |
| A vs T | 1,14 | 0,96 | 1,03 | 1,44 | 2,20 | 1,31 | 1,11 | 1,41 | 0,87 | |
| SIRT1 | TXNIP | PPARα | PGC1α | FGF21 | ObR | Dio2 | Dio3 | UCP1 | ||
| Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | Median ratio | ||
| Cerebral cortex | T vs N | 0,83 | 1,21 | 0,61 | 0,50 | 0,95 | 1,11 | 0,67 | 0,96 | |
| A vs N | 1,09 | 0,79 | 1,09 | 0,77 | 1,33 | 1,11 | 0,55 | 0,87 | ||
| A vs T | 1,30 | 0,65 | 1,79 | 1,55 | 1,40 | 1,00 | 0,82 | 0,90 | ||
| Hypothalamus | T vs N | 0,49 | 0,68 | 0,27 | 0,29 | 0,64 | 1,04 | 0,50 | 0,89 | |
| A vs N | 0,63 | 0,52 | 0,72 | 0,56 | 1,19 | 0,63 | 0,61 | 1,00 | ||
| A vs T | 1,30 | 0,76 | 2,73 | 1,91 | 1,84 | 0,61 | 1,23 | 1,13 | ||
| Pituitary gland | T vs N | 1,05 | 1,46 | 0,93 | 1,02 | 2,18 | 0,70 | 0,40 | 0,70 | |
| A vs N | 1,14 | 0,81 | 1,22 | 1,04 | 3,02 | 0,71 | 0,55 | 1,03 | ||
| A vs T | 1,09 | 0,56 | 1,32 | 1,02 | 1,39 | 1,01 | 1,39 | 1,47 | ||
| Retina | T vs N | 0,84 | 1,42 | 0,37 | 0,62 | 2,18 | 0,73 | 0,53 | 0,67 | |
| A vs N | 1,24 | 1,11 | 0,60 | 0,91 | 3,95 | 0,72 | 0,64 | 0,86 | ||
| A vs T | 1,47 | 0,78 | 1,62 | 1,46 | 1,81 | 0,99 | 1,21 | 1,29 | ||
| Liver | T vs N | 1,51 | 1,68 | 0,88 | 1,77 | 0,19 | 2,71 | 0,37 | 0,90 | |
| A vs N | 1,31 | 1,44 | 0,67 | 2,41 | 2,16 | 2,21 | 0,54 | 1,00 | ||
| A vs T | 0,87 | 0,86 | 0,76 | 1,36 | 11,35 | 0,82 | 1,47 | 1,11 | ||
| BAT | T vs N | 0,87 | 0,93 | 0,71 | 0,81 | 0,60 | 0,93 | 0,30 | 0,82 | 1,10 |
| A vs N | 1,46 | 0,87 | 0,48 | 1,50 | 0,78 | 0,60 | 0,23 | 2,34 | 0,80 | |
| A vs T | 1,69 | 0,93 | 0,68 | 1,85 | 1,29 | 0,65 | 0,76 | 2,84 | 0,72 | |
| Heart | T vs N | 1,04 | 0,77 | 0,72 | 0,81 | 1,12 | 0,98 | 0,79 | 1,87 | |
| A vs N | 1,17 | 0,40 | 0,80 | 0,73 | 0,90 | 0,69 | 0,37 | 2,11 | ||
| A vs T | 1,12 | 0,53 | 1,12 | 0,91 | 0,80 | 0,70 | 0,47 | 1,13 | ||
| Adrenal glands | T vs N | 0,94 | 0,71 | 0,80 | 0,58 | 0,46 | 0,63 | 0,16 | 0,99 | |
| A vs N | 1,33 | 0,98 | 1,22 | 1,08 | 2,21 | 0,51 | 0,16 | 1,98 | ||
| A vs T | 1,41 | 1,38 | 1,52 | 1,84 | 4,75 | 0,81 | 1,00 | 2,00 |
Organ-dependent gene profiling during hibernation cycles
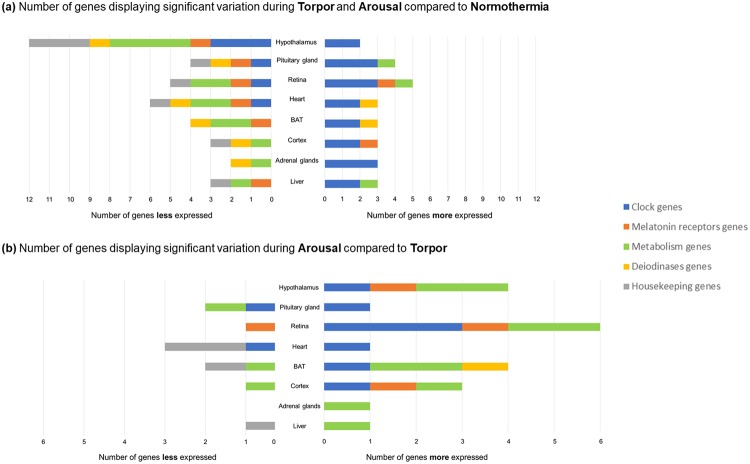
In each central or peripheral organ, numerous genes were differentially expressed according to the hibernation phases (Fig. 2). Hypothalamus samples clearly contained the highest number of genes with significant differences in expression. Large numbers of differential genes were also found in other nervous/neuroendocrine organs, including the pituitary gland and the retina, but not the cortex. On the other hand, genes expressed in the liver and adrenal glands appeared to be the least affected.
Figure 2.
Changes in the expression of functional families of genes in various tissues of the European hamster according to hibernation stage. Data represent the number of genes per functional family that were down-regulated (left side) or up-regulated (right side) in eight central or peripheral tissues. (a) Gene expression during torpor and arousal compared to gene expression during normothermia. (b) Gene expression during arousal compared to gene expression during torpor.
Further examination of whether gene expression was increased or decreased in a given hibernation state revealed that more genes were down-regulated (39 genes) than up-regulated (26 genes) in torpor and arousal compared to in normothermia (Fig. 2a). Moreover, hypothalamus samples exhibited the highest number of down-regulated genes (12 genes) in torpor and arousal compared to normothermia, while only 2 genes were up-regulated. Most of the other organs showed similar numbers of up-regulated and down-regulated genes. A surprisingly low number of genes were differentially expressed when hamsters arose from torpor (Fig. 2b). Overall these results highlighted the strong alteration of hypothalamic genes during hibernation, with down-regulation of a strikingly large number of genes in torpor or arousal as compared to normothermia. During arousal, there is a moderate change in gene expression with the hypothalamus, the BAT and the retina showing the highest number of up-regulated genes.
Profiles of functional groups of genes during hibernation cycles
To determine whether some functional groups of genes might be particularly relevant to the hibernation states, separate analyses were performed to establish the hibernation profiles of four categories of genes: clock system, melatonin receptors, thyroid hormone metabolism, and general metabolism.
Clock genes
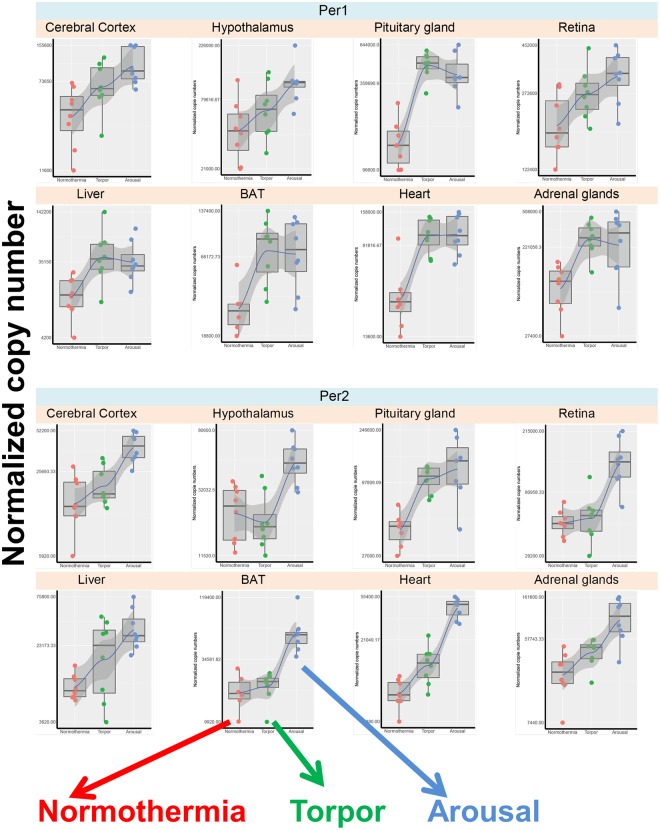
The Per1 and Per2 genes showed large differential expression among the hibernation phases in all organs (Fig. 3a) and these variations appeared not linked to sampling time for most organs (see material and method section). Per2 was strongly up-regulated during arousal compared to in the normothermia state in all investigated organs, and was up-regulated to a less extent during arousal compared to torpor in five organs, including the hypothalamus and BAT. Per1 expression was also markedly up-regulated during arousal in all analysed organs and, to a lesser extent, during torpor compared to during normothermia. In order to better visualize these variations in individual organs across the three hibernation stages, examples of boxplot are given in the Fig. 4. The other measured clock genes showed smaller variations. The Bmal1 mRNA level was higher in arousal compared to normothermia in the pituitary gland and retina, and was higher in arousal compared to torpor in the retina. Clock and Rev-Erbα were down-regulated during arousal and torpor compared to normothermia, particularly in nervous tissues for the Clock gene, and in the hypothalamus and heart for the Rev-Erbα gene. Notably, in the hypothalamus, all clock genes were either down-regulated or unchanged during torpor compared to normothermia, thus confirming the results of whole gene analysis (Fig. 2).
Figure 3.
Changes in the expression of individual genes in various tissues of the European hamster according to hibernation stage. Genes are grouped according to their functional family: clock genes (a), melatonin receptor genes (b), deiodinase genes (c), and metabolism genes (d). In each table, the colour indicates significant up-regulation (green), down-regulation (red), or no change (grey) of the mRNA levels in the eight investigated tissues, after adjustment of their P values. Colour intensity is a function of the level of significance.
Figure 4.
Boxplot presentation of mRNA values of the individual genes Per1 and Per2 across the three hibernation stages in all tissues. Individual samples values for each gene were represented, we also plotted a spline curve on those graphs (without a modulation purpose) to highlight the sense of gene variation across conditions. Normothermia in red, Torpor in green, Arousal in blue.
Melatonin receptors encoding genes
Genes encoding melatonin receptors displayed variations during the hibernation cycle that were strongly dependent on the receptor subtype (Fig. 3b). Notably, the MT2 receptor showed the largest variations, with decreased expression in torpor and arousal compared to in normothermia in peripheral organs (liver, heart, and BAT), but increased expression in arousal compared to torpor in central tissues. The gene encoding QR2 was down-regulated during arousal compared to normothermia in nervous tissue (hypothalamus, retina and pituitary). The orphan receptor GPR50 showed altered expression during arousal compared to normothermia, only in the cortex. MT1 expression did not significantly vary during hibernation in any investigated organ.
Deiodinase genes
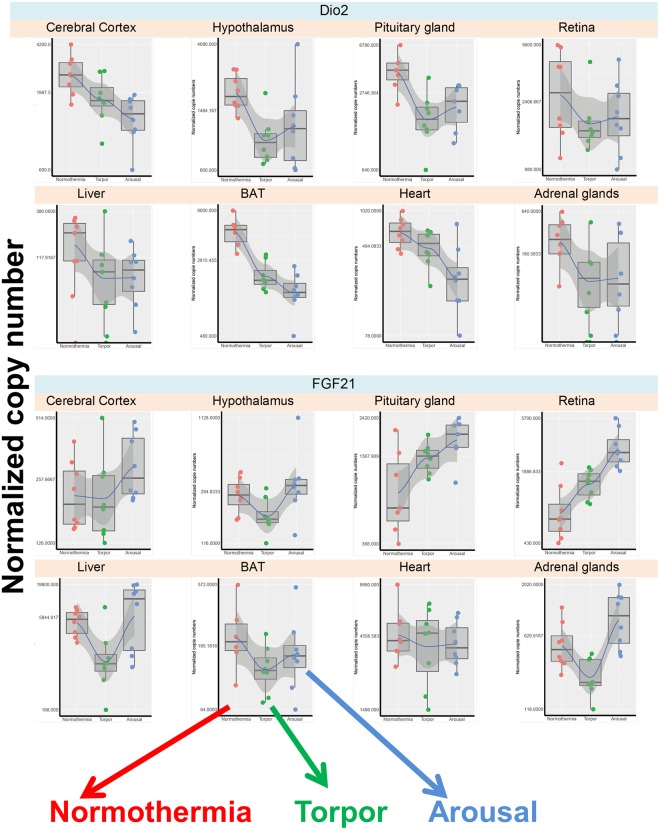
The genes encoding two enzymes involved in the thyroid hormone metabolism, Dio2 and Dio3, displayed different hibernation profiles (Fig. 3c). Dio2 expression was markedly decreased in torpor and arousal compared to in normothermia, particularly in the hypothalamus, pituitary gland, BAT, and adrenal glands (Fig. 5). In contrast, Dio3 expression was increased during arousal compared to during normothermia and torpor in the BAT, and in during arousal compared to normothermia in the heart. Notably in BAT during arousal, Dio2 mRNA was down-regulated in association with up-regulation of Dio3 mRNA, suggesting reduced thyroid metabolism.
Figure 5.
Boxplot presentation of mRNA values of the individual genes Dio2 and FGF21 across the three hibernation stages in all tissues. Individual samples values for each gene were represented, we also plotted a spline curve on those graphs (without a modulation purpose) to highlight the sense of gene variation across conditions. Normothermia in red, Torpor in green, Arousal in blue.
Genes involved in general metabolism
The hibernation profiles of genes involved in general metabolism revealed large differences among genes and organs, making it difficult to illustrate a general scheme (Fig. 3d). A number of genes were down-regulated in torpor compared to normothermia, including FGF21 in the liver, PPARα in central tissues, PGC1α in the hypothalamus and retina, and SIRT1 in the hypothalamus. Overall in the hypothalamus PPARα, PGC1α, and SIRT1 were down-regulated. In contrast, the peripheral organs unexpectedly showed no change in metabolic gene expression between torpor and normothermia, with the exception of FGF21 down-regulation and Ob-R up-regulation in the liver. A number of genes were up-regulated in the arousal state compared to the torpor state, particularly FGF21 in hypothalamus, retina, liver, and adrenal glands; PPARα in central tissues; and PGC1α and SIRT1 in the BAT. Only TXNIP in the hypothalamus and heart, and Ob-R in the BAT were reduced in arousal compared to torpor. Unexpectedly, UCP1 mRNA in the BAT did no vary according to the hibernation stage (not shown). In order to better visualize FGF21 variations in individual organs across the three hibernation stages, examples of boxplot are given in the (Fig. 5).
Housekeeping genes
Although this is not required in the ddPCR approach, we examined classical housekeeping genes to determine whether they were generally altered due to the torpor conditions. Our data (not shown) indicated no organ-dependent or stage-dependant common variations among the three investigated genes: tubulin, actin, and G6PD. These observations indicated that there was no general alteration of gene expression according to hibernation stage in any investigated organ.
Circulating hormones during the hibernation cycles
Some of the hormones related to the functional groups of genes—such as melatonin, T3, T4, glucose, insulin, and leptin—were assayed at the different hibernation states. Although the circulating levels of leptin tended to decrease during torpor, as reported for the Syrian hamster50, overall the circulating concentrations of these hormones did not significantly differ among the three hibernation states possibly due to high inter-individual variations (Supplementary Table S2). Thus, we were unable to examine correlations between changes in circulating hormone levels and changes in gene expression.
Discussion
With the objective to better understand the mechanisms which may control hibernation, we examined the molecular signatures of a set of genes related to daily and seasonal timing, torpor regulation, and energy homeostasis, from eight different organs, in association with three hibernation phases during a winter-like heterotherm period in the well-established hibernating species, the European hamster.
Analysing the overall changes in functional families of genes revealed ubiquitous up-regulation of the core clock genes Per1 and Per2, and to a lesser extent BMAL1 and Clock, upon arousal from torpor. This up-regulation of clock genes is most likely associated with arousal from torpor rather than due to a shift in circadian fluctuations, since we previously determined that circadian clockwork stops oscillating in the European hamster SCN during torpor30 and in most genes we found no apparent, or weak, correlation between the sampling time and the levels of clock gene RNA (a strong correlation between time sampling and Period genes has been observed in the pituitary gland for the expression of Per1 and in both retina and adrenal glands for Per2 expression. In these organs, it is complicated to conclude about the effect of hibernation state on Period genes expression). For the other organs, the absence of correlation strongly suggests that the sampling time did not modify the expression of these genes within the time frame of our experimental setup. The Per gene up-regulation could be explained, among others, by the reported increase of glucocorticoids during arousal51, and correlation between Per gene expressions and glucocorticoid concentration should be measured to test this hypothesis. Our present results further showed an overall down-regulation of Bmal1, Clock, and Rev-Erbα in the hypothalamus of torpid hamsters, further indicating arrest of the circadian clock. The role of the master circadian clock during torpor remains a matter of discussion. Thirteen-lined ground squirrels exhibit markedly increased SCN activity during torpor and arousal, as indicated by c-fos expression52,53. Moreover, in Djungarian hamsters54 and ground squirrels55, SCN lesions do not prevent torpor states but only alter their timing. Altogether, these data indicate that the SCN clock is not essential for torpor occurrence but may play a crucial role in its temporal organization. Moreover, our findings indicate that Per genes may be essential for post-torpor re-entrainment of the clock.
Studies in various hibernating species report no night-time increase in melatonin during the torpor states, due to SCN clock arrest, with a subsequent rapid restoration of the nocturnal rhythm upon arousal30,56–58. Studies investigating changes in 2-125I-iodomelatonin binding on melatonin receptor density during hibernation in the brains of hedgehogs59 and ground squirrels60 have reported decreased numbers of binding sites in the pars tuberalis during torpor compared to normothermia. Our present data showed that the gene encoding MT1 was not differentially expressed according to hibernation phase in any of the investigated tissues. In contrast, MT2 gene expression increased during arousal in central organs (cortex, hypothalamus, and retina), and decreased during torpor and arousal in peripheral organs (heart, liver and BAT). A recent study investigated how the melatonin receptor antagonist luzindole may influence the brain of the hibernating ground squirrel, and demonstrated that melatonin receptor signalling promotes neuroprotection and optimizes mitochondrial function during arousal from torpor61. Therefore, the increased central MT2 expression observed upon arousal indicates that this receptor may be involved in the neuroprotective function of melatonin during hibernation. This function may be particularly relevant in the retina, which expresses high levels of MT2 and displays profound synaptic remodelling during hibernation62–64. Melatonin also reportedly activates arousal thermogenesis through peripheral actions65–68; however, our data did not reveal significant changes in MT1 expressions in any peripheral organs and a decrease of MT2 in the BAT. Mice bearing a mutation in the melatonin-related receptor GPR50 display enhanced propensity to fasting-induced torpor17,69, but we found that GPR50 gene expression did not vary among the hibernation stages of the European hamster. Similarly, we found no variations in the expression of QR2 (the MT3 binding site14), suggesting either that the enzyme plays a minor role in this context, or that it is not regulated through variation of gene expression. Altogether, our data indicated that the gene encoding MT2 was most substantially altered according to the hibernation cycles, with opposite patterns in central and peripheral organs.
The adaptation to winter physiology, especially torpor, requires reduced availability of thyroid hormones, particularly T326,27,70,71. We show here that the Dio2 enzyme (that converts T4 to T3) was decreased in central structures: hypothalamus and pituitary gland; and peripheral organs: BAT and adrenal glands, during torpor and arousal. The Dio3 enzyme (that degrades T3) was increased during torpor in the heart and BAT. These alterations lead to reduced T3 concentrations during torpor and arousal. Two recent studies in the Djungarian hamster demonstrated that increasing peripheral72 or intra-hypothalamic27 T3 resulted in reduction of torpor induction and torpor bout duration and depth, confirming the pivotal role of T3 in torpor regulation and strengthening our observation of reduced T3 metabolism during torpor.
Torpor induction is associated with a switch in energy utilization from carbohydrates to lipids31. In the present study, we followed the changes in expression of FGF21 and its upstream regulator PPARα, since they reportedly play major roles in adaptive starvation responses and torpor promotion38,39. Indeed, during torpor, FGF21 expression was decreased in the liver, and PPARα expression was decreased in the cortex, hypothalamus, and retina. Upon arousal, FGF21 expression was increased in the hypothalamus, retina, liver, and adrenal glands; and PPARα expression was increased in the cortex, hypothalamus, and retina. SIRT1 and PGC1α are involved in gluconeogenesis and thermogenesis35,73, and both were down-regulated in the hypothalamus during torpor, and up-regulated in the BAT during arousal, likely in response to increasing energy demand. TXNIP is involved in hypothalamic homeostasis and hepatic gluconeogenesis40,41, and is reportedly up-regulated in the hypothalamus during various types of torpor42,43. In our present analysis of the European hamster hypothalamus, TXNIP gene expression was not higher during torpor compared to normothermia, but was significantly reduced during arousal, possibly to adjust the central metabolic demand when the animals awoke from torpor. Altogether, our findings indicated that selected genes involved in metabolic pathways were generally down-regulated during torpor and up-regulated during arousal, particularly in the hypothalamus.
Comparison among the various investigated central and peripheral organs revealed that each organ displayed a specific hibernation gene profile, potentially indicating different functions of these organs during the hibernation cycle. In particular, the hypothalamus and BAT warrant closer analysis. These tissues are thought to play critical roles in hibernation, and previous studies reported thorough transcriptomic analysis at different hibernation stages.
Little is known about the genes responsible for the highly efficient fat burning activity that occurs during the hibernation arousal process. BAT generates heat, notably in newborns and hibernating mammals. BAT thermogenesis involves norepinephrine-dependent β-oxidation of free fatty acids under the control of the hypothalamus via the sympathetic nervous system. It appears that thyroid hormones are also involved in this process since T3 influences BAT thermogenesis either directly or indirectly through central sites. A recent analysis of BAT in the thirteen-lined ground squirrel demonstrates differential expression of 14% of the examined genes across four collection points throughout the year and hibernation stages45. Comparing the transcriptomes of torpid and aroused squirrels revealed that a few genes encoding transcription factors were significantly down-regulated during torpor, notably BHLHE40 which is involved in circadian rhythms74. Our analysis of European hamster BAT confirmed that PGC1α and SIRT1 were up-regulated during arousal compared to torpor. We further observed that the Dio2/Dio3 mRNA ratio decreased during arousal, indicating reduced local availability of T3 in BAT. In agreement with the increase in BHLHE40 in squirrel BAT, we observed higher expression of Per genes during arousal in European hamsters, supporting a putative role of BAT molecular clock machinery during arousal from torpor.
The hypothalamus coordinates a number of biological functions, including thermal and metabolic processes, circadian organization, sleep, reproduction, and the control of both hibernation and daily torpor53,75. During torpor, minimal functional brain activity persists to prevent nervous tissue damage and to allow the animals to rapidly and completely recover from hypothermia. Recent studies involving RNASeq analyses of the hypothalamus and cerebral cortex from thirteen-lined ground squirrels (true hibernator)43, and of the hypothalamus from Djungarian hamsters (daily torpidators)48, at various stages of torpor or interbouts have reported different strategies implemented in the hypothalamus and cerebral cortex during hibernation. In the hypothalamus, the differentially expressed genes are involved in protection against DNA damage, protein turnover through ubiquitination, feeding and satiety signalling, seasonal timing mechanisms, and fuel utilization. In the cerebral cortex, the candidate genes are involved in synapse remodelling and plasticity. Among Djungarian hamsters entering torpor, about 1% of the 27,830 identified genes in the hypothalamus are differentially expressed, most of which are involved in metabolic and cellular functions. The majority of the top 20 most down-regulated genes encoded transcription factors, which may be responsible for the general suppression of protein synthesis during torpor48. In our study, the European hamster’s hypothalamus showed the greatest number of candidate genes that were differentially expressed during the hibernation stages. Most were down-regulated during torpor/arousal compared to normothermia, particularly genes involved in metabolic processes (PPARα, PGC1, SIRT, TXNIP, and Dio2) and genes involved in the core circadian clock (Bmal1, Clock, and Rev-erbα). During arousal from torpor, we observed up-regulation of some metabolic (PPARα and FGF21) and circadian (Per2) genes, as well as MT2. Altogether, our data support the metabolic silencing of the hypothalamus during torpor, and suggest the involvement of new timing components in the hibernation processes.
Studies using various transcriptomic approaches have indicated that a rather limited number of genes are differentially expressed during hibernation, depending on species, organs, and hibernation stages. Thus, it appears likely that posttranscriptional and posttranslational mechanisms are also involved in the dramatic changes in body temperature and in other physiological variables that accompany hibernation. A recent large-scale proteomic analysis also revealed a relative stability of the proteome throughout the extreme physiological changes of hibernation3. Most of the observed protein changes were linked to seasonal changes, i.e. between the homeothermic and heterothermic periods. If even some of the limited differences in protein abundance according to torpor/arousal cycles are attributed to protein modifications, this indicates that the total protein pool is extremely stable across the hibernation season, despite prolonged fasting and wide variations in metabolic levels. This stability is not completely unexpected, since the profound temperature-mediated suppression of both transcription and translation machinery during torpor drastically limits the ability to synthesize new gene products. Across the available proteomic screens of hibernator tissues, cytoskeleton regulation was the most consistent signal, possibly associated with the temperature-induced neural retraction and cytoskeletal depolymerisation. Altogether these results suggest that the most dramatic protein changes associated with the torpor/arousal cycle may be due to post-translational modifications. Notably, protein acetylation appears to be a key component of the torpor/arousal switch3,76.
Hibernation is a key process enabling animals to survive harsh natural conditions. Identifying the complex molecular mechanisms orchestrating the extreme changes in hibernation could provide important contributions to the development of therapeutic strategies to improve medical outcomes in a number of conditions, including hypothermic injury, organ transplantation, stroke recovery, cardiac arrest, muscle disease, and other ischemia/reperfusion insults. Numerous studies have investigated how torpid animals tolerate cerebral ischemia following dramatic reductions of blood flow and oxygen concentration, display reversible rapid and pronounced synaptic flexibility in which synapses retract during torpor and rapidly re-emerge upon arousal, and retain skeletal muscle mass despite prolonged immobilization and lack of nutrition77–79. There remains much to be learned about the cellular, molecular, and systems-wide mechanisms that protect heterothermic mammals during torpor/arousal cycles, which may guide discovery of new therapeutics.
Strikingly, the previously performed transcriptomic, proteomic, and metabolic studies indicate large inter-species and inter-tissue differences in the involved genes and proteins, highlighting the importance of analysing different hibernating species. Our present study is the first to report gene profiling during torpor/arousal cycles in several organs of the European hamster. We report a broad reduction in our candidate gene expression, especially those involved in metabolic activity, within the hypothalamus of torpid hamsters, and a decrease in the genes of thyroid hormone synthesis, notably in the BAT, during the torpor/arousal stages. Furthermore, we observe a general up-regulation of the core circadian clock genes Per1 and Per2 upon arousal, suggesting a possible resetting of body clocks at the start of the active period.
Methods
Animal experimentation was conducted in accordance with the French National Law implementing the European Communities Council Directive 2010/63/EU and the French Directive 2013-118. Animal procedures were reviewed by the local ethical committee (Comité Régional d’Ethique en Matière d’Expérimentation Animale de Strasbourg, CEEA 35) and the official authorization was given on July 2015 by la direction générale de la recherche et de l’innovation under the number #01546.02.
Animals
European hamsters (Cricetus cricetus) are a well-established hibernating species80. The adult male hamsters used in this study were bred in-house (Chronobiotron, UMS 3415). Animals were individually maintained in cages, and food and water were provided ad libitum throughout the study. At the start of the experiment, each hamster was administered isoflurane anaesthesia, and intraperitoneally implanted with a Thermochron iButton (DS1922L; Maxim, Dallas, TX) to monitor its body temperature.
From July 6th to September 14th 2015, 24 hamsters were subjected to a short photoperiod (SP) comprising 10 hours of light and 14 hours of dark, with the lights on at 8:00 (Zeitgeber ZT0). This induced a winter phenotype characterized by low body weight (mean, 375.6 ± 12 g) and complete gonadal regression confirmed by scrotal palpation. Starting on September 15th, the animals were transferred to a climatic room maintained at 8 °C to induce deep torpor bouts characteristic of European hamster hibernation (Fig. 6). Individual hibernating behavior was followed daily for 3 weeks, after which the animals were euthanized by CO2 inhalation. The animals were euthanized during three different phases of hibernation bouts (n = 8/group): normothermia (stable high Tb of ~36 °C for at least 3 days), torpor (stable low Tb of ~9 °C for at least 3 days), or arousal (increasing Tb between 21–36 °C, 2 hours after the beginning of arousal from torpor). The experiment ended on October 28th. In order to reduce bias related sampling times, we have limited as much as possible the time-window of euthanasia. Hamsters from the normothermia and torpor groups were all sampled between ZT2 and ZT6, but sampling of aroused hamsters was more delicate to control as the objective was to euthanize hamsters exactly 2 hours after the starting of the arousal. We tried to overcome this issue by promoting a gentle hand-warming arousal of the animals at ZT0/ZT1 and looking at their awakening behaviors, however animals started their arousal with different delays and therefore the sampling of this group occurred between ZT4 and ZT9.5. Thus, in order to investigate the potential effect of sampling times on genes most susceptible to variations (Per1 and Per2), we calculated the non-parametric Spearman correlation coefficient for the arousal phase (a non-parametrical method was chosen to be in accordance with the non-normal distribution of ddPCR data.) between time of sampling versus Per1 and time sampling versus Per2 mRNA quantity for each organ. We observed that in most organs Per1 and Per2 expression is not, or weakly correlated, to the time sampling (Spearman correlation coefficient for Per1: cerebral cortex = −0.24; hypothalamus = −0.02; Pituitary gland = −0.91; retina = −0.17; liver = 0; BAT = −0.48; heart = −0.69; adrenal glands = −0.43. Per2: cerebral cortex = 0.38; hypothalamus = 0.29; pituitary gland = 0.07; retina = 0.81; liver = −0.21; BAT = −0.6; heart = −0.62; adrenal glands = −0.76). We observed strong correlation between time sampling and Per1 expression in the pituitary gland and between time sampling and Per2 expression in both retina and adrenal glands. Due to these correlations, it is difficult to conclude on the effect of hibernation state on Period genes expression for these organs. For the other organs, the absence of correlation strongly suggests that the sampling time did not modify the expression of these genes within the time frame of our experimental setup.
Figure 6.
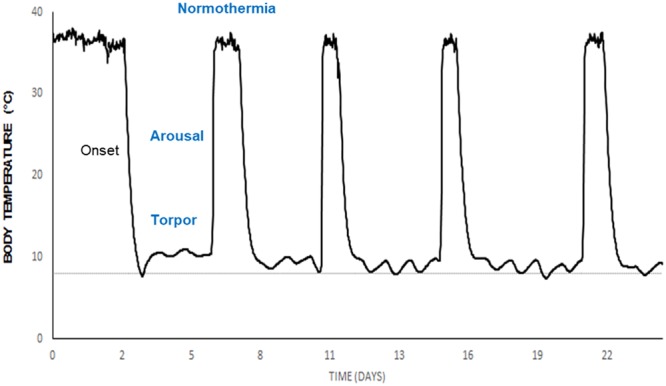
Typical hibernation pattern of a male European hamster. The graph shows body temperature variations, measured by intra-peritoneal iButton, over 25 consecutive days in a hamster maintained with a short photoperiod (10 hours of light and 14 hours of dark) at an ambient temperature of 8 °C (dashed line). Tissues were sampled at three different phases of a characteristic hibernation cycle, as determined by body temperature: torpor (8–10 °C), arousal (21–31 °C), and normothermia (33–36 °C).
Blood was collected and centrifuged at 1,500 g for 15 min, and then the plasma was stored at −20 °C until hormonal assay. Eight central and peripheral organs—namely, the cerebral cortex, hypothalamus, pituitary gland, retina, liver, heart, brown adipose tissue, and adrenal glands—were rapidly removed, rinsed in cold Ringer’s solution, frozen in liquid nitrogen, and then stored at −80 °C until gene analysis.
Prior to this study, a preliminary experiment was performed with fewer animals per group, allowing adjustment of the procedure and selection of tissues and genes of interest. Although the results were generally similar, only the data from the second (2015) experiment are presented in this manuscript, due to their greater statistical strength.
RNA extraction
Total RNA was extracted using the RNeasy® Lipid Tissue Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. Briefly, a sample of each organ (100 mg when possible) was added to 1 mL QIAzol Lysis Reagent, along with one 5-mm stainless steel bead (Qiagen, Valencia, CA, USA). The tissue samples were disrupted, and homogenized for 2 min at 20 Hz using the tissue lyser. Then the homogenates were incubated at room temperature (RT) for 5 min, followed by addition of 200 µL chloroform (Sigma, St. Louis, USA). Next, the homogenates were centrifuged at 12,000 g for 15 min at 4 °C. The upper aqueous phase containing RNA was transferred to a new tube and the final step was performed using the QIAcube protocol: RNeasy lipid Animal tissue (Qiagen, Valencia, CA, USA). RNA quality was measured using Agilent’s 2100 Bioanalyzer system according to manufacturer’s protocol (Agilent, Santa Clara, USA). RIN values are all presented in the Table 3. Total RNA was stored at −80 °C until use.
Table 3.
The RIN of each sample was determined using Agilent’s 2100 Bioanalyzer system.
| Hibernation phases | Animals | Cerebral cortex | Hypothalamus | Pituitary gland | Retina | Liver | BAT | Heart | Adrenal glands | Mean | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Torpor | HE15 | 6,9 | 7,8 | 9,1 | 9,7 | 8,7 | 8,3 | 8,7 | 9,8 | 8,8 | 0,8 |
| HE16 | 8,5 | 8,7 | 9,3 | 9,3 | 8,9 | 8,5 | 8,6 | 9,8 | |||
| HE17 | 7,5 | 8,8 | 9,4 | 9,8 | 8,8 | 9 | 8,6 | 9,8 | |||
| HE24 | 9,2 | 8,6 | 9,7 | 9,6 | 8,7 | 8,8 | 9,1 | 10 | |||
| HE26 | 8,7 | 9,3 | 9,5 | 10 | 6,7 | 8,5 | 8,2 | 10 | |||
| HE10 | 7 | 8,2 | 9,2 | 9,4 | 8,2 | 8,4 | 8,6 | 8,7 | |||
| HE31 | 9,1 | 8,5 | 9,8 | 9,7 | 7,8 | 8,8 | 9,1 | 9,9 | |||
| HE34 | 8,9 | 7,8 | 8,5 | 9,4 | 8 | 7,6 | 8 | 9,7 | |||
| Normothermia | HE9 | 8,4 | 8,7 | 9,6 | 9,8 | 8,3 | 8,8 | 9,8 | 8,7 | 0,7 | |
| HE11 | 8,3 | 7,9 | 8,1 | 9,9 | 8,2 | 8,7 | 8,3 | 9,4 | |||
| HE12 | 7,9 | 7,7 | 9 | 9,7 | 7,4 | 7,8 | 7,9 | 9,7 | |||
| HE19 | 8 | 7,9 | 8,9 | 9,3 | 7,4 | 8,2 | 8 | 9,6 | |||
| HE20 | 8,4 | 9 | 9,2 | 10 | 7,4 | 8,5 | 8,6 | 9,7 | |||
| HE23 | 8,4 | 8,6 | 9,4 | 9,5 | 8,3 | 8,8 | 8,3 | 9,9 | |||
| HE30 | 8,4 | 8,5 | 9,3 | 8,6 | 8,5 | 8,7 | 8 | ||||
| HE33 | 8,5 | 8,9 | 9,1 | 9,8 | 8,2 | 8,7 | 10 | ||||
| Arousal | HE8 | 8,9 | 7 | 9,6 | 9 | 8,6 | 8,7 | 9,6 | 8,8 | 0,8 | |
| HE14 | 8,6 | 8,3 | 9,5 | 9,8 | 6,9 | 8,1 | 8,4 | 9,7 | |||
| HE18 | 8,7 | 7,8 | 7,7 | 8,6 | 8,6 | 8,6 | 8,9 | 9,8 | |||
| HE21 | 8,4 | 8,9 | 8,9 | 9,9 | 8,3 | 7,9 | 8,1 | 8,9 | |||
| HE25 | 8,1 | 8,2 | 8,8 | 9,9 | 7,6 | 8,4 | 8,1 | 9,7 | |||
| HE27 | 9,2 | 8,9 | 9,6 | 10 | 7,8 | 8,3 | 8,3 | 10 | |||
| HE28 | 9 | 9,2 | 9,5 | 9,9 | 8,6 | 8,3 | 9,1 | 9,8 | |||
| HE29 | 8,4 | 9 | 8,3 | 9,6 | 7,5 | 8 | 8,4 | 9,9 | |||
| Mean | 8,39 | 8,43 | 9,10 | 9,65 | 8,08 | 8,39 | 8,51 | 9,63 | |||
| SD | 0,60 | 0,57 | 0,54 | 0,32 | 0,63 | 0,35 | 0,36 | 0,47 |
Cloning genes of interest
Since few Cricetus cricetus gene sequences are published (Database Resources of the National Center for Biotechnology Information), the genes of interest were partially sequenced (about 500 bp) to enable the design of specific ddPCR assays.
European hamster hypothalamus total RNA (5 µg) was converted into cDNA using oligo dT primers and the PrimeScript™ High Fidelity RT-PCR Kit (Takara Bio USA, Mountain View, USA) following the manufacturer’s instructions. The polymerase chain reaction (PCR) was performed using Q5 High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA) following the manufacturer’s protocol. The 50-µL reaction mix comprised 1 µL template cDNA, 5 × Q5 reaction buffer, 10 mM dNTPs, 10 µM forward primer, 10 µM reverse primer, Q5 high-fidelity DNA polymerase 0.02 U/µL, and 5 × Q5 high GC enhancer. Sequences extracted from the National Center for Biotechnology Information (NCBI) were used to design 21 gene-specific primer pairs (Table 4). Amplicons were separated in 1% agarose gels stained with ethidium bromide, and the gel bands were visualized using U Genius (Syngene, Frederick, USA). If multiple bands were observed, the PCR products were purified using the high pure purification kit (Roche Mannheim, Germany). The eluted DNA was inserted into a blunt pJET vector using the CloneJET PCR cloning kit (Thermo Fisher Scientific, Waltham, USA), and transformed into DH10β chemically competent Escherichia Coli cells (NEB, Ipswich, MA, USA).
Table 4.
Sequences of the forward and reverse primers used to clone European hamster genes of interest.
| Genes | Reference sequences for primers designa | Forward Primer Sequencesb | Reverse Primer Sequencesc | Cricetus cricetus GenBank Accession Numbersd |
|---|---|---|---|---|
| Per1 | NM_001034125.1: Rattus norvegicus period circadian clock 1 (Per1) | TGTGCACCCCTGGAGCCGCAAGG | TTTCTTGGCCCCCACAGGAACTG | MG598318: [Cricetus cricetus] period circadian clock 1 (Per1) mRNA, partial CDS |
| Bmal1 | AB012600: Rattus norvegicus mRNA for BMAL1b | TAAAACGGATATAACCCCTGGGC CTGCCCTCTGGAGAAGGTGGCCC | TCTGGTTCCCCCTGGAATGCCTG ACCCAGCCCCGCATCTGCTTCCA | MG598320: [Cricetus cricetus] BmalI aryl hydrocarbon receptor nuclear translocator like (Arntl) mRNA, partial CDS |
| Rev-Erbα | XM_003498212.2: Cricetulus griseus nuclear receptor subfamily 1, group D, member 2 (Nr1d2) | GCTCTAACTCTGATGCCAACGG | GCTTTTGAGGTTTTCTTGCTCCAG | MG598307: [Cricetus cricetus] Nuclear receptor subfamily 1 group D member 2 (Nr1d2) mRNA, partial CDS |
| Clock | XM_016980269: Cricetulus griseus clock circadian regulator (Clock) | TCAATTGTTGACAGAGATGACAGTAG | TCTATTGTTCCTCGAAGCATGTGAC | MG598315: [Cricetus cricetus] circadian locomotor output cycles protein kaput (Clock) mRNA, partial CDS |
| Per2 | XM_007622995: Cricetulus griseus period circadian clock 2 (Per2) | ACTGTGATGACAATGGGAAGGAGCT | ATGGAGGCAACTTGGTTAGAGATGT | MG598316: [Cricetus cricetus] Period circadian protein 2 (Per2) mRNA, partial CDS |
| MT 1 | U14110.1: Phodopus sungorus melatonin receptor (Mel-1a) | ATGAAGGGCAATGGTAGCACTCTGCTCAATGCC | CCGTATATAATTGCATTGAGGCAGCTG | MG598322: [Cricetus cricetus] Melatonin receptor 1 A (Mtnr1a) mRNA, complete CDS |
| MT 2 | NM_145712.2: Mus musculus melatonin receptor 1B (Mtnr1b) | TTGTTTGTGGTGAGTCTGGTCTTGG | GCCCATAGACAATGACGTTAAGGCAG | MG598323: [Cricetus cricetus] Melatonin receptor 1B (Mtnr1b) mRNA, partial CDS |
| GPR50 | XM_007631612: Cricetulus griseus G protein-coupled receptor 50 (Gpr50) | CCGAACTGGCTGTATCTTGCAG | TCATACAGCCATCTCATCAGAA | MG598317: [Cricetus cricetus] G protein-coupled receptor 50 (Gpr50) mRNA, partial CDS |
| QR2 | XM_007638944.1: Cricetulus griseus NAD(P)H dehydrogenase, quinone 2 (Nqo2) | TGGCAGGTAAGAAAGTGCTCATC/TCACTGGTTCCCTCTCTAATCCTG | TCTTCAGCCGCTTCGCCCATGATGC/TCTTCAGCCGCTTCGCCCATGATGC | KT992792: [Cricetus cricetus] NAD(P)H dehydrogenase quinone 2 mRNA, partial CDS |
| Tubulin | NM_001243978: Cricetulus griseus tubulin, alpha 1 A (Tuba1a) | ACACCTTCTTCAGTGAGACAGGCG | CCCAAAGATGTCAATGCTGCC | MG598321: [Cricetus cricetus] tubulin alpha 1B (Tuba1b) mRNA, partial CDS |
| Actin | NM_001244575: Cricetulus griseus actin beta (Actb) | CCCATTGAACACGGCATTGTC | CGACATCCGCAAAGACCTCTATG | MG598319: [Cricetus cricetus] actin beta (Actb) mRNA, partial CDS |
| SIRT1 | XM_005070811.1: Mesocricetus auratus sirtuin 1 (Sirt1) | GTCATAGGTTAGGTGGTGAATATGCC | CACAGGAACTAGAGGATAAGATGTCGTC | MG598314: [Cricetus cricetus] sirtuin 1 (Sirt1) mRNA, partial CDS |
| TXNIP | XM_003498621.2: Cricetulus griseus thioredoxin interacting protein (Txnip) | CGACTCAGGAGGCAAAGAAAAAC | CAATCACCAGGGGAAGGTCAAG | MG598310: [Cricetus cricetus] Thioredoxin interacting protein (Txnip) mRNA, partial CDS. |
| PPARα | XM_007621010.1: Cricetulus griseus peroxisome proliferator-activated receptor alpha (Ppara) | GAATAAGTGCCAATACTGCCGC | CATACGCTATCAGCATCCCGTC | MG598312: [Cricetus cricetus] Peroxisome proliferator-activated receptor alpha (Ppara) mRNA, partial CDS |
| PGC1α | XM_007620649.1: Cricetulus griseus peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (Ppargc1a) | TTTGATGTGTCGCCTTCTTGC | GGTGTAACGGTAGGTGATGAAACC | MG598311: [Cricetus cricetus] Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (Ppargc1a) mRNA, partial CDS. |
| FGF21 | XM_007638697.1: Cricetulus griseus fibroblast growth factor 21 (Fgf21) | TGGACTGGATGAAATCTGGAGTTG | AAGGTCCCACCATGCTCAGTGG | MG598309: [Cricetus cricetus] Fibroblast growth factor 21 (Fgf21) mRNA, partial CDS |
| Ob-R | XM_007632623.1: Cricetulus griseus leptin receptor (Lepr) | GCCTGTCTTTCCAGAGAATAACCTTC | CGGCACTCACTTTACTCATTGGC | MG598308: [Cricetus cricetus] Leptin receptor (Lepr) mRNA, partial CDS |
| UCP1 | NM_001281332.1: Mesocricetus auratus uncoupling protein 1 (Ucp1), mRNA | TCTACGATACTGTCCAGGAGTACTTC | CAGTCCACCGTCTGCCTCGACT | MG598313: [Cricetus cricetus] Uncoupling protein 1 (UCP1) mRNA, partial CDS |
To obtain the sequences of the unpublished genes of interest, forwardb and reversec primers were designed using published sequencesa. These sequences have been submitted to GenBankd.
Forward and reverse sequencing reactions were performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) using vector primers for amplification. Sequencing products were purified using the BigDye XTerminator® Purification Kit (Thermo Fisher Scientific, Waltham, USA), and analysed using an ABI 3730 XL Automated Sequencer (Applied Biosystems). Data analysis was performed using Sequencher® version 5.4.6 DNA sequence analysis software (Gene Codes Corporation, Ann Arbor, MI, USA). The Cricetus cricetus sequences of the nineteen non-published genes of interest were partially cloned and sequenced, and the results have been submitted to GenBank (Table 4).
Digital droplet PCR
Primers and probes for the digital droplet PCR assay were designed using the Universal Probe Library (UPL) assay design centre: https://lifescience.roche.com/en_fr/brands/universalprobe-library.html (Roche Mannheim, Germany). Previously cloned Cricetus cricetus sequences were used as references. For technical reasons, custom assays from Biorad (Hercules, CA, USA) were used for the clock and fgf21 genes.
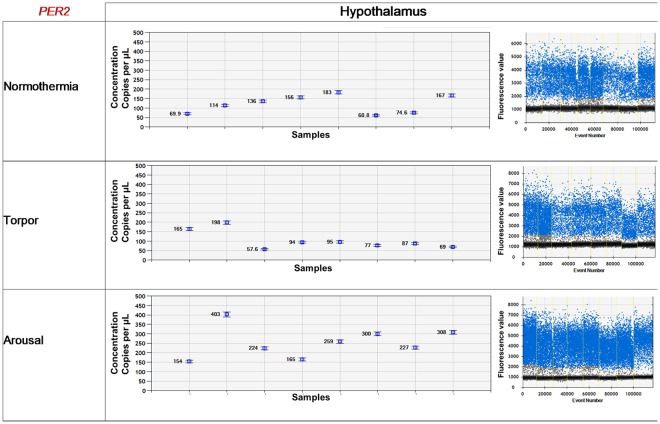
RNA samples were directly partitioned using the One-Step RT-ddPCR Advanced Kit for Probes (Biorad, Hercules, USA). One-step RT-PCR reactions were carried out in a total volume of 22 µL, including 2–3 µL RNA (based on the RNA concentration), 5.5 µL super mix, 2.2 µL reverse transcriptase (20 U/µL), 1.1 µL DTT (300 mM), 0.6 µL forward and reverse primers, 0.55 µL probes, and molecular grade RNAse-free water (Table 5). Droplet generation was performed using an automated droplet generator (Biorad, Hercules, CA, USA) following the manufacturer’s recommendations. The thermal cycling conditions comprised 60 min reverse transcription at 50 °C, and 10 min enzyme activation at 95 °C; followed by 40 cycles of denaturation at 95 °C for 30 sec, and extension at the specific gene annealing temperature for 70 sec (Table 5); and finally 10 min enzyme deactivation at 98 °C using the CFX96 touch real-time PCR detection system (Biorad, Hercules, CA, USA). All steps used a ramp rate of 2 °C/sec. Subsequently, the droplets were analysed using the QX200 droplet reader (Biorad, Hercules, CA, USA). Figure 7 shows an example of ddPCR results for Per2 mRNA data measured in the hypothalamus. Each droplet from the sample is plotted on a 1-D graph of fluorescence intensity versus droplet number.
Table 5.
Primer/probe sequences, RNA quantity, and annealing temperatures for digital droplet PCR (ddPCR).
| Genes | Probesa | Probe Referencesb | Forward Primer Sequencesc | Primer Reverse Sequencesd | RNA Quantity (ng)e | Annealing temperature (°C)f |
|---|---|---|---|---|---|---|
| Per1 | UPL 63 | 4688627001 | CCAGCACAACAAAGCGTAAA | TCAGAGGCTGAGGAAGCAGT | 1 | 57 |
| Bmal1 | UPL 56 | 4688538001 | CCAACCTTCCCACAGCTTAC | CCTGGAATGCCTGGAACA | 5 | 57 |
| Rev-Erbα | UPL 150 | 4694368001 | TGTCTGTCAGTGGGAATGTCA | CTCTGTTTCTCACGCTTAGGAAT | 1 | 57 |
| Clock | AATGAAGTTACACTCTCAGATACAT | NA | CCACAAGATCAGATGGTA | TAGCGATCATGACAGATG | 50 | 51 |
| Per2 | UPL 161 | 4694481001 | CTTCTTGTCTGCAGGGAGGT | TGTCCTTATCAGTTCTTTGTGTGC | 10 | 55 |
| MT 1 | UPL 145 | 4694317001 | CCCTCTGCTACGTGTTCCTG | GAGTTCCGGTTTGCAGATTG | 150 | 59 |
| MT 2 | UPL 131 | 4694155001 | TGTGGTGAGTCTGGTCTTGG | AGGATCAGTGGGTAAGGGTACA | 100 | 57 |
| GPR50 | UPL 86 | 4689119001 | GCTGGCTCTTCCTCTAAGCA | GGCTGGTAGCAGGCTTAATG | 150 | 55 |
| QR2 | UPL 68 | 4688678001 | AAGACAGCTCTGACCAGTGACA | CTAGATCAGCTTCTTGCACCTTC | 5 | 59 |
| Tubulin | UPL 78 | 4689011001 | GAGCGGCTCTCTGTCGATTA | GGGGCTGGGTAGATGGAG | 0.01 | 59 |
| G6PD | UPL 30 | 4687639001 | TGTGGCAAAGCCCTGAAT | TGCCACATCTCGGAACTGTA | 5 | 55 |
| Actin | UPL 9 | 4685075001 | GCTATGAGCTGCCTGATGG | GGCTGGAAAAGAGCCTCA | 0.1 | 57 |
| SIRT1 | UPL 68 | 4688678001 | GAAAGTGCTGGCCCAATAGA | GATTACCATCAAGCCGCTTACTA | 5 | 57 |
| TXNIP | UPL 125 | 4693604001 | CCTTGCTGATCTATGTTAGTGTCC | TCACCAGGGGAAGGTCAA | 1 | 57 |
| PPARα | UPL 56 | 4688538001 | CGGTGTGTATGAAGCCATATTC | ATCAGCATCCCGTCTTTGTT | 1 | 57 |
| PGC1α | UPL 41 | 4688007001 | GTAGGCCCAGGTATGACAGC | CCTTTCAGATTCCCGTTTCTC | 1 | 57 |
| FGF21 | ACACTGAAGTCCACCTGG | NA | ACCTCTACACAGATGACA | GGTTGTTGGCAAAGAAC | 100 | 58 |
| Ob-R | UPL 113 | 4693477001 | CGCAGGAGATCAGACCAATC | ATTGATGGCCAGAACCGTAA | 100 | 57 |
| Dio2 | UPL 22 | 4686969001 | CCACCTTTCACTAGGCAACTG | AGTCGGCCACTGATGAGAAC | 10 | 57 |
| Dio3 | UPL 135 | 4694198001 | GCACCTAACTCGGAGGTCAT | ATAGTCGAGGATGCGCTGTC | 100 | 57 |
| RBM3 | UPL 65 | 4688643001 | TGGAAGCGGAAGATATGACA | TCTCTGGACCGCCCATATC | 1 | 57 |
| UCP1 | UPL 21 | 4686942001 | GGCAACCTACTGAGGTCGTG | ATCGGGGTTTGATCCCATA | 0.1 | 57 |
Each ddPCR reaction requires probesa,b, forwardc and reversed primers, and determination of the optimal RNA quantitye and annealing temperaturef were determined.
Figure 7.
Typical example of digital droplet PCR (ddPCR) mRNA quantification. Using ddPCR, the exact number of Per2 mRNA copies was quantified in hypothalamus tissue samples obtained from European hamsters at three different stages of the hibernation cycle (n = 8 from each stage): normothermia, torpor, and arousal. The graph on the left side depicts the calculated concentration of Per2 mRNA copies/µL for each animal in each hibernation stage. The graph on the right side shows the fluorescence intensity of droplets in terms of the droplet number for each sample. QuantaSoft software was used to calculate the numbers of droplets containing Per2 mRNA (blue) or not containing Per2 mRNA (grey) in each sample.
Hormone analysis
Circulating hormone concentrations were determined by radioimmunoassay (RIA) or enzyme-linked immunosorbent assay (ELISA). Plasma melatonin was assayed by RIA after chloroform extraction, as previously validated in European hamsters81. The thyroid hormones T3 and T4 were extracted with a 2:1 mixture of chloroform:methanol, purified by anion exchange chromatography, eluted with 70% acetic acid, and then measured by RIA as previously described82. Plasma leptin was measured using a direct multi-species RIA kit (EMD Millipore, Billerica, MA, USA), insulin using a hamster ELISA kit (Crystal Chem, Downers Grove, USA), and plasma glucose concentration using the colorimetric glucose GOD-PAP method (Biolabo, Maizy, France).
Data and statistical analysis
Raw ddPCR data were analysed using QuantaSoft software v. 1.5.38.1118 (Biorad, Hercules, USA). Raw data were normalized against the initial quantity to obtain equivalent copy numbers for 100 ng total RNA. Statistical analyses were performed using R Program Writer software 3.3.1 (http://www.r-project.org/). Within-dataset variability was explored using two-dimensional principal component analysis (PCA) with the FactoMineR package, to identify the subset of genes showing the greatest differential expression in relation to several factors, including the organ and the hibernation phase. Data analysis was performed for each organ which was the main factors influencing expression of the genes.
Using R software, Kruskal-Wallis analysis was performed on normalized counts to determine the global effect of hibernation phases on individual gene expression in a given tissue type. Then raw P values from each model were adjusted for multiple testing, using Benjamini-Hochberg correction to control the false discovery rate. Genes were considered differentially expressed if the adjusted P values were below 0.05. Dunn’s post-hoc analysis test was used to compare further subgroups of hibernation phases.
The results of hormone measurements are presented as mean ± SD. Data were analysed using the Kruskal-Wallis test, followed by Dunn’s multiple comparison test. Statistical significance was set at P ≤ 0.05. Statistical analyses were performed using PRISM (GraphPad Software Inc., San Diego, CA, USA).
Electronic supplementary material
Acknowledgements
We thank Dr. Pierre Ducrot for his contribution to our data analysis. We would like to thank Dr Véronique Raverot for the melatonin RIA.
Author Contributions
C.G., B.B., D.C., A.G., C.D., G.L., C.B., I.R.F., J.P.S., O.N., performed research; C.G., D.V., J.A.B., V.S. and S.P.G. analysed data; and G.C., J.A.B., S.P.G. and V.S. designed the research and wrote the paper.
Data availability
The datasets generated and analysed during the current study are available from the corresponding author (Jean A Boutin jean.boutin@servier.com) upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sophie-Pénélope Guénin and Valérie Simonneaux contributed equally.
Contributor Information
Jean A. Boutin, Email: jean.boutin@servier.com
Valérie Simonneaux, Email: simonneaux@inci-cnrs.unistra.fr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31506-2.
References
- 1.Heldmaier G, Ortmann S, Elvert R. Natural hypometabolism during hibernation and daily torpor in mammals. Respir. Physiol. Neurobiol. 2004;141:317–329. doi: 10.1016/j.resp.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Geiser F, Turbill C. Hibernation and daily torpor minimize mammalian extinctions. Naturwissenschaften. 2009;96:1235–1240. doi: 10.1007/s00114-009-0583-0. [DOI] [PubMed] [Google Scholar]
- 3.Grabek KR, Martin SL, Hindle AG. Proteomics approaches shed new light on hibernation physiology. J. Comp. Physiol. B. 2015;185:607–627. doi: 10.1007/s00360-015-0905-9. [DOI] [PubMed] [Google Scholar]
- 4.Serkova NJ, Rose JC, Epperson LE, Carey HV, Martin SL. Quantitative analysis of liver metabolites in three stages of the circannual hibernation cycle in 13-lined ground squirrels by NMR. Physiol. Genomics. 2007;31:15–24. doi: 10.1152/physiolgenomics.00028.2007. [DOI] [PubMed] [Google Scholar]
- 5.Geiser, F. Hibernation: Endotherms. In eLS (ed. John Wiley & Sons, Ltd) (John Wiley & Sons, Ltd, 2011).
- 6.Geiser F. Metabolic Rate and Body Temperature Reduction During Hibernation and Daily Torpor. Annu. Rev. Physiol. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- 7.Florant GL, et al. Fat-cell mass, serum leptin and adiponectin changes during weight gain and loss in yellow-bellied marmots (Marmota flaviventris) J. Comp. Physiol. B. 2004;174:633–639. doi: 10.1007/s00360-004-0454-0. [DOI] [PubMed] [Google Scholar]
- 8.Humphries MM, Thomas DW, Kramer DL. The Role of Energy Availability in Mammalian Hibernation: A Cost/Benefit Approach. Physiol. Biochem. Zool. 2003;76:165–179. doi: 10.1086/367950. [DOI] [PubMed] [Google Scholar]
- 9.Canguilhem B, Petrovic A. [Effects of photoperiod and ambient temperature on circannual rhythms of body weight and adrenal cortex activity in European hamster (Cricetus cricetus)] Arch. Sci. Physiol. (Paris) 1974;28:113–126. [PubMed] [Google Scholar]
- 10.Simonneaux V, Ribelayga C. Generation of the melatonin endocrine message in mammals: A review of the complex regulation of melatonin synthesis by norepinephrine, peptides, and other pineal transmitters. Pharmacol. Rev. 2003;55:325–395. doi: 10.1124/pr.55.2.2. [DOI] [PubMed] [Google Scholar]
- 11.Hazlerigg D, Simonneaux V. Chapter 34. Seasonal Regulation of Reproduction in Mammals. In Knobil and Neill’s Physiology of Reproduction: Two-Volume Set. 2015;2:1575–1604. doi: 10.1016/B978-0-12-397175-3.00034-X. [DOI] [Google Scholar]
- 12.Stanton TL, Daley JC, III, Salzman SK. Prolongation of hibernation bout duration by continuous intracerebroventricular infusion of melatonin in hibernating ground squirrels. Brain Res. 1987;413:350–355. doi: 10.1016/0006-8993(87)91027-4. [DOI] [PubMed] [Google Scholar]
- 13.Pitrosky B, Delagrange P, Rettori MC, Pévet P. S22153, a melatonin antagonist, dissociates different aspects of photoperiodic responses in Syrian hamsters. Behav. Brain Res. 2003;138:145–152. doi: 10.1016/S0166-4328(02)00235-8. [DOI] [PubMed] [Google Scholar]
- 14.Nosjean O, et al. Identification of the melatonin-binding site MT3 as the quinone reductase 2. J. Biol. Chem. 2000;275:31311–31317. doi: 10.1074/jbc.M005141200. [DOI] [PubMed] [Google Scholar]
- 15.Dufourny L, et al. GPR50 is the mammalian ortholog of Mel1c: Evidence of rapid evolution in mammals. BMC Evol. Biol. 2008;8:105. doi: 10.1186/1471-2148-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier C, et al. Characterization of the Mel1c melatoninergic receptor in platypus (Ornithorhynchus anatinus) PLOS ONE. 2018;13:e0191904. doi: 10.1371/journal.pone.0191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechtold DA, et al. A role for the melatonin-related receptor GPR5050 in leptin signaling, adaptive thermogenesis, and torpor. Curr. Biol. 2012;22:70–77. doi: 10.1016/j.cub.2011.11.043. [DOI] [PubMed] [Google Scholar]
- 18.Dardente H, Klosen P, Pévet P, Masson-Pévet M. MT1 melatonin receptor mRNA expressing cells in the pars tuberalis of the European hamster: effect of photoperiod. J. Neuroendocrinol. 2003;15:778–786. doi: 10.1046/j.1365-2826.2003.01060.x. [DOI] [PubMed] [Google Scholar]
- 19.Dardente H, Hazlerigg DG, Ebling FJ. Thyroid hormone and seasonal rhythmicity. Thyroid Endocrinol. 2014;5:19. doi: 10.3389/fendo.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis, J. E. & Ebling, F. J. P. Tanycytes As Regulators of Seasonal Cycles in Neuroendocrine Function. Front. Neurol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 21.Hanon EA, et al. Effect of photoperiod on the thyroid-stimulating hormone neuroendocrine system in the european hamster (Cricetus cricetus) J. Neuroendocrinol. 2010;22:51–55. doi: 10.1111/j.1365-2826.2009.01937.x. [DOI] [PubMed] [Google Scholar]
- 22.Nishiwaki-Ohkawa T, Yoshimura T. Molecular basis for regulating seasonal reproduction in vertebrates. J. Endocrinol. 2016;229:R117–R127. doi: 10.1530/JOE-16-0066. [DOI] [PubMed] [Google Scholar]
- 23.Hanon EA, et al. Ancestral TSH Mechanism Signals Summer in a Photoperiodic Mammal. Curr. Biol. 2008;18:1147–1152. doi: 10.1016/j.cub.2008.06.076. [DOI] [PubMed] [Google Scholar]
- 24.Nakao N, et al. Thyrotrophin in the pars tuberalis triggers photoperiodic response. Nature. 2008;452:317–322. doi: 10.1038/nature06738. [DOI] [PubMed] [Google Scholar]
- 25.Ono H, et al. Involvement of thyrotropin in photoperiodic signal transduction in mice. Proc. Natl. Acad. Sci. 2008;105:18238–18242. doi: 10.1073/pnas.0808952105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy M, et al. Effects of manipulating hypothalamic triiodothyronine concentrations on seasonal body weight and torpor cycles in siberian hamsters. Endocrinology. 2012;153:101–112. doi: 10.1210/en.2011-1249. [DOI] [PubMed] [Google Scholar]
- 27.Bank JHH, et al. Gene expression analysis and microdialysis suggest hypothalamic triiodothyronine (T3) gates daily torpor in Djungarian hamsters (Phodopus sungorus) J. Comp. Physiol. B. 2017;187:857–868. doi: 10.1007/s00360-017-1086-5. [DOI] [PubMed] [Google Scholar]
- 28.Dibner C, Schibler U, Albrecht U. The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 29.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 30.Revel FG, et al. The circadian clock stops ticking during deep hibernation in the European hamster. Proc. Natl. Acad. Sci. 2007;104:13816–13820. doi: 10.1073/pnas.0704699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck CL, Barnes BM. Effects of ambient temperature on metabolic rate, respiratory quotient, and torpor in an arctic hibernator. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2000;279:R255–R262. doi: 10.1152/ajpregu.2000.279.1.R255. [DOI] [PubMed] [Google Scholar]
- 32.Rui L. Energy Metabolism in theLiver. Compr. Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis – A highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahill GF. Fuel metabolism in starvation. Annu. Rev. Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 35.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1|[alpha]| and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 37.Kersten S, et al. Peroxisome proliferator–activated receptor α mediates the adaptive response to fasting. J. Clin. Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badman MK, et al. Hepatic Fibroblast Growth Factor 21 Is Regulated by PPARα and Is a Key Mediator of Hepatic Lipid Metabolism in Ketotic States. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Inagaki T, et al. Endocrine Regulation of the Fasting Response by PPARα-Mediated Induction of Fibroblast Growth Factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Chutkow WA, Patwari P, Yoshioka J, Lee RT. Thioredoxin-interacting Protein (Txnip) Is a Critical Regulator of Hepatic Glucose Production. J. Biol. Chem. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- 41.Blouet C, Liu S-M, Jo Y-H, Chua S, Schwartz GJ. TXNIP in Agrp Neurons Regulates Adiposity, Energy Expenditure, and Central Leptin Sensitivity. J. Neurosci. 2012;32:9870–9877. doi: 10.1523/JNEUROSCI.0353-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hand LE, et al. Induction of the metabolic regulator txnip in fasting-induced and natural torpor. Endocrinology. 2013;154:2081–2091. doi: 10.1210/en.2012-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz C, Hampton M, Andrews MT. Seasonal and Regional Differences in Gene Expression in the Brain of a Hibernating Mammal. PLoS ONE. 2013;8:e58427. doi: 10.1371/journal.pone.0058427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 45.Hampton M, Melvin RG, Andrews MT. Transcriptomic Analysis of Brown Adipose Tissue across the Physiological Extremes of Natural Hibernation. PLoS ONE. 2013;8:e85157. doi: 10.1371/journal.pone.0085157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarpace PJ, Matheny M, Pollock BH, Tumer N. Leptin increases uncoupling protein expression and energy expenditure. Am. J. Physiol. - Endocrinol. Metab. 1997;273:E226–E230. doi: 10.1152/ajpendo.1997.273.1.E226. [DOI] [PubMed] [Google Scholar]
- 47.Srere HK, Wang LC, Martin SL. Central role for differential gene expression in mammalian hibernation. Proc. Natl. Acad. Sci. USA. 1992;89:7119–7123. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cubuk C, Markowsky H, Herwig A. Hypothalamic control systems show differential gene expression during spontaneous daily torpor and fasting-induced torpor in the Djungarian hamster (Phodopus sungorus) PLoS ONE. 2017;12:e0186299. doi: 10.1371/journal.pone.0186299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Hara BF, et al. Gene Expression in the Brain across the Hibernation Cycle. J. Neurosci. 1999;19:3781–3790. doi: 10.1523/JNEUROSCI.19-10-03781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weitten M, Robin J-P, Oudart H, Pévet P, Habold C. Hormonal changes and energy substrate availability during the hibernation cycle of Syrian hamsters. Horm. Behav. 2013;64:611–617. doi: 10.1016/j.yhbeh.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 51.Tahara Y, Shibata S. Entrainment of the mouse circadian clock: Effects of stress, exercise, and nutrition. Free Radic. Biol. Med. 2018;119:129–138. doi: 10.1016/j.freeradbiomed.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 52.Bitting L, et al. C-fos mRNA increases in the ground squirrel suprachiasmatic nucleus during arousal from hibernation. Neurosci. Lett. 1994;165:117–121. doi: 10.1016/0304-3940(94)90723-4. [DOI] [PubMed] [Google Scholar]
- 53.Bratincsák A, et al. Spatial and temporal activation of brain regions in hibernation:c-fos expression during the hibernation bout in thirteen-lined ground squirrel. J. Comp. Neurol. 2007;505:443–458. doi: 10.1002/cne.21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruby NF, Zucker I. Daily torpor in the absence of the suprachiasmatic nucleus in Siberian hamsters. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 1992;263:R353–R362. doi: 10.1152/ajpregu.1992.263.2.R353. [DOI] [PubMed] [Google Scholar]
- 55.Ruby NF, Dark J, Heller HC, Zucker I. Ablation of suprachiasmatic nucleus alters timing of hibernation in ground squirrels. Proc. Natl. Acad. Sci. USA. 1996;93:9864–9868. doi: 10.1073/pnas.93.18.9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Florant GL, Rivera ML, Lawrence AK, Tamarkin L. Plasma melatonin concentrations in hibernating marmots: absence of a plasma melatonin rhythm. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 1984;247:R1062–R1066. doi: 10.1152/ajpregu.1984.247.6.R1062. [DOI] [PubMed] [Google Scholar]
- 57.Darrow JM, Tamarkin L, Duncan MJ, Goldman BD. Pineal melatonin rhythms in female Turkish hamsters: effects of photoperiod and hibernation. Biol. Reprod. 1986;35:74–83. doi: 10.1095/biolreprod35.1.74. [DOI] [PubMed] [Google Scholar]
- 58.Vanĕcek J, Janský L, Illnerová H, Hoffmann K. Arrest of the circadian pacemaker driving the pineal melatonin rhythm in hibernating golden hamsters, Mesocricetus auratus. Comp. Biochem. Physiol. A. 1985;80:21–23. doi: 10.1016/0300-9629(85)90671-1. [DOI] [PubMed] [Google Scholar]
- 59.Gauer F, Masson-Pévet M, Saboureau M, George D, Pévet P. Differential seasonal regulation of melatonin receptor density in the pars tuberalis and the suprachiasmatic nuclei: a study in the hedgehog (Erinaceus europaeus, L.) J. Neuroendocrinol. 1993;5:685–690. doi: 10.1111/j.1365-2826.1993.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 60.Stanton TL, Siuciak JA, Dubocovich ML, Krause DN. The area of 2-[125I]iodomelatonin binding in the pars tuberalis of the ground squirrel is decreased during hibernation. Brain Res. 1991;557:285–288. doi: 10.1016/0006-8993(91)90145-L. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz C, Ballinger MA, Andrews MT. Melatonin receptor signaling contributes to neuroprotection upon arousal from torpor in thirteen-lined ground squirrels. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2015;309:R1292. doi: 10.1152/ajpregu.00292.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luan Y, et al. Integrated transcriptomic and metabolomic analysis reveals adaptive changes of hibernating retinas. J. Cell. Physiol. 2018;233:1434–1445. doi: 10.1002/jcp.26030. [DOI] [PubMed] [Google Scholar]
- 63.Merriman DK, Sajdak BS, Li W, Jones BW. Seasonal and post-trauma remodeling in cone-dominant ground squirrel retina. Exp. Eye Res. 2016;150:90–105. doi: 10.1016/j.exer.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Remé CE, Young RW. The effects of hibernation on cone visual cells in the ground squirrel. Invest. Ophthalmol. Vis. Sci. 1977;16:815–840. [PubMed] [Google Scholar]
- 65.Cipolla‐Neto J, Amaral FG, Afeche SC, Tan DX, Reiter RJ. Melatonin, energy metabolism, and obesity: a review. J. Pineal Res. 2014;56:371–381. doi: 10.1111/jpi.12137. [DOI] [PubMed] [Google Scholar]
- 66.Heldmaier G, Steinlechner S, Rafael J, Vsiansky P. Photoperiodic control and effects of melatonin on nonshivering thermogenesis and brown adipose tissue. Science. 1981;212:917–919. doi: 10.1126/science.7233183. [DOI] [PubMed] [Google Scholar]
- 67.McMillan AC, White MD. Induction of thermogenesis in brown and beige adipose tissues: molecular markers, mild cold exposure and novel therapies. Curr. Opin. Endocrinol. Diabetes Obes. 2015;22:347–352. doi: 10.1097/MED.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 68.Tan D-X, Manchester LC, Fuentes-Broto L, Paredes SD, Reiter RJ. Significance and application of melatonin in the regulation of brown adipose tissue metabolism: relation to human obesity. Obes. Rev. 2011;12:167–188. doi: 10.1111/j.1467-789X.2010.00756.x. [DOI] [PubMed] [Google Scholar]
- 69.Ivanova EA, et al. Altered metabolism in the melatonin-related receptor (GPR50) knockout mouse. Am. J. Physiol. - Endocrinol. Metab. 2008;294:E176–E182. doi: 10.1152/ajpendo.00199.2007. [DOI] [PubMed] [Google Scholar]
- 70.Barrett P, et al. Hypothalamic thyroid hormone catabolism acts as a gatekeeper for the seasonal control of body weight and reproduction. Endocrinology. 2007;148:3608–3617. doi: 10.1210/en.2007-0316. [DOI] [PubMed] [Google Scholar]
- 71.Klosen P, Sébert M-E, Rasri K, Laran-Chich M-P, Simonneaux V. TSH restores a summer phenotype in photoinhibited mammals via the RF-amides RFRP3 and kisspeptin. FASEB J. 2013;27:2677–2686. doi: 10.1096/fj.13-229559. [DOI] [PubMed] [Google Scholar]
- 72.Bank JHH, Kemmling J, Rijntjes E, Wirth EK, Herwig A. Thyroid hormone status affects expression of daily torpor and gene transcription in Djungarian hamsters (Phodopus sungorus) Horm. Behav. 2015;75:120–129. doi: 10.1016/j.yhbeh.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Puigserver P, Spiegelman BM. Peroxisome Proliferator-Activated Receptor-γ Coactivator 1α (PGC-1α): Transcriptional Coactivator and Metabolic Regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 74.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 75.Jastroch, M. et al. Seasonal Control of Mammalian Energy Balance: Recent Advances in the Understanding of Daily Torpor and Hibernation. J. Neuroendocrinol. 28 (2016). [DOI] [PubMed]
- 76.Hindle AG, Grabek KR, Epperson LE, Karimpour-Fard A, Martin SL. Metabolic changes associated with the long winter fast dominate the liver proteome in 13-lined ground squirrels. Physiol. Genomics. 2014;46:348–361. doi: 10.1152/physiolgenomics.00190.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bouma HR, et al. Induction of torpor: mimicking natural metabolic suppression for biomedical applications. J. Cell. Physiol. 2012;227:1285–1290. doi: 10.1002/jcp.22850. [DOI] [PubMed] [Google Scholar]
- 78.Dave KR, Christian SL, Perez-Pinzon MA, Drew KL. Neuroprotection: Lessons from hibernators. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012;162:1–9. doi: 10.1016/j.cbpb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivakine EA, Cohn RD. Maintaining skeletal muscle mass: lessons learned from hibernation. Exp. Physiol. 2014;99:632–637. doi: 10.1113/expphysiol.2013.074344. [DOI] [PubMed] [Google Scholar]
- 80.Canguilhem B, Marx C. Regulation of the body weight of the European Hamster during the annual cycle. Pflugers Arch. 1973;338:169–175. doi: 10.1007/BF00592751. [DOI] [PubMed] [Google Scholar]
- 81.Garidou M-L, Vivien-Roels B, Pévet P, Miguez J, Simonneaux V. Mechanisms regulating the marked seasonal variation in melatonin synthesis in the European hamster pineal gland. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2003;284:R1043–R1052. doi: 10.1152/ajpregu.00457.2002. [DOI] [PubMed] [Google Scholar]
- 82.Reyns GE, Venken K, Morreale de Escobar G, Kühn ER, Darras VM. Dynamics and regulation of intracellular thyroid hormone concentrations in embryonic chicken liver, kidney, brain, and blood. Gen. Comp. Endocrinol. 2003;134:80–87. doi: 10.1016/S0016-6480(03)00220-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author (Jean A Boutin jean.boutin@servier.com) upon reasonable request.