Abstract
The tumor microenvironment is increasingly recognized as an active participant in tumor progression. A recent pan-cancer genomic profile analysis has revealed that gene signatures representing components of the tumor microenvironment are robust predictors of survival. A stromal gene signature representing fibroblasts and extracellular matrix components has been associated with good survival in diffuse large B-cell lymphoma (DLBCL). Paradoxically, a closely related gene signature has been shown to correlate with poor survival in carcinomas, including breast, ovarian, pancreatic, and colorectal cancer. To date, there has been no explanation for this paradoxical inverse correlation with survival outcomes in DLBCL and carcinomas. Using public gene data sets, we confirm that the DLBCL stromal gene signature is associated with good survival in DLBCL and several other B-cell lymphomas while it is associated with poor survival in ovarian cancer and several other solid tumors. We show that the DLBCL stromal gene signature is enriched in lymphoid fibroblasts in normal lymph nodes and in cancer-associated fibroblasts (CAFs) in ovarian cancer. Based on these findings, we propose several possible mechanisms by which CAFs may contribute to opposite survival outcomes in B-cell lymphomas and carcinomas.
Keywords: B cells, B-cell lymphoma, CAFs, cancer-associated fibroblasts, DLBCL, gene signature, ovarian cancer, tumor microenvironment
Introduction
During the past decade, gene expression profile analyses of frozen tumor pieces have been widely used to quantify various biological characteristics of malignant tumor cells and the microenvironment in which they reside. Individual biological characteristics and dominant molecular pathways in tumors are frequently associated with expression of a defined set of genes, known as a gene expression signature. Since phenotypic features represented by gene expression signatures are sometimes associated with clinical features, such as the length of survival of cancer patients or their response to therapy, gene expression signatures can be used as quantitative predictors of clinical outcomes. A recent pan-cancer PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) analysis revealed that genes in the tumor microenvironment are better predictors of survival than genes expressed in malignant tumor cells (Gentles et al., 2015). The two most prominent components in the microenvironment of solid tumors are fibroblasts and immune cells (Aran et al., 2017). Generally, in carcinomas, genes expressed in fibroblasts are associated with poor survival while genes expressed in immune cells, particularly leukocytes, are associated with good survival (Gentles et al., 2015). Tumor infiltrating lymphocytes (TILs) and tertiary lymphoid structures (TLS) are generally associated with improved clinical outcomes as evidenced by the improved overall survival and disease-free survival in various types of tumors (Fridman et al., 2012; Dieu-Nosjean et al., 2014; Barnes and Amir, 2017). However, depending on the type of tumor, tumor stage, and location of TILs within the tumor (tumor bed, invasive margin and stroma), different types of TILs have been associated with both positive and negative prognosis. For example, cytotoxic CD8+ T cells, memory T cells, and CD4+ T helper cells are generally associated with a better prognosis, whereas T regulatory cells, tumor associated macrophages, and myeloid-derived suppressor cells are associated with poor prognosis and can promote tumor progression (Fridman et al., 2012; Kitamura et al., 2015; Barnes and Amir, 2017). Furthermore, fibroblasts in the tumor microenvironment are phenotypically heterogeneous and may exhibit both a pro- and anti-tumorigenic phenotype (Augsten, 2014). Thus, the tumor microenvironment is a complex network of interaction between tumor cells and components of the stroma, including the extracellular matrix (ECM), and it is currently unclear which factors in the tumor microenvironment control the quantity and distribution of different immune cell subtypes. Specifically, it is unknown if fibroblasts and immune cells affect prognosis independently or through an interdependent interaction.
The functional interaction between fibroblasts and immune cells has been most thoroughly studied in normal lymph nodes and the spleen, where specialized fibroblasts produce ECM to form a network that allows for lymphocyte movement along the matrix in response to chemokine signaling. The presence of lymphoid fibroblasts is necessary for functional attraction, retention, compartmentalization, and survival of immune cells (Koning and Mebius, 2012). Lymphoid fibroblasts are crucial for lymphocyte homeostasis as well as controlling and expanding the lymphocyte pool (Mueller and Germain, 2009). Lymphoid fibroblasts are also key players in mediating functional immune cell interactions in the lymph nodes through direct contact or via secreted molecules (Chang and Turley, 2015). Follicular dendritic cells (FDC) attract B cells to the germinal center (GC) by secreting C-X-C motif chemokine ligand 13 (CXCL13), while marginal reticular cells (MRC) use a network of follicular conduits to deliver antigens to cognate B cells (Chang and Turley, 2015). By secreting C-C motif chemokine ligands 19 and 21 (CCL19 and CCL21), fibroblastic reticular cells (FRC) recruit mature dendritic cells (DC) and naïve B and T cells to promote cell-cell interactions within the T cell zone (Mueller and Germain, 2009; Brown and Turley, 2015; Fletcher et al., 2015). Recent studies have shown that FRC are important for B-cell homeostasis (Cremasco et al., 2014). This function has been previously ascribed to FDC, however, cell-specific depletion experiments demonstrated that only FRC are crucial for B-cell survival. The mechanism by which FRC support B-cell survival is not entirely clear, but it is thought to involve crosstalk with B cells to control the boundaries of primary B-cell follicles (Cyster, 2010; Mionnet et al., 2013; Cremasco et al., 2014).
Similar to lymphoid fibroblasts in normal lymph nodes, cancer-associated fibroblasts (CAFs) are stromal cells that produce ECM, provide scaffolding, and exert regulatory functions through growth factors, cytokines, and chemokines that can promote tumor growth, angiogenesis, invasion, and metastasis (Kalluri and Zeisberg, 2006; Levental et al., 2009; Lu et al., 2012; Spano and Zollo, 2012; Harper and Sainson, 2014). Recent studies provide evidence that CAFs can also directly or indirectly contribute to immune cell fate and survival (Harper and Sainson, 2014; Costa et al., 2018; Mariathasan et al., 2018; Tauriello et al., 2018). It has recently been shown that a gene signature representing activated CAFs is present in most epithelial tumors (Jia et al., 2016) despite the diversity of resident fibroblasts in different organs and the presence of multiple fibroblast populations within a single tumor type (Costa et al., 2018). Activated CAFs in breast cancer, and possibly in other carcinomas, are associated with immunosuppressive populations of T lymphocytes (Costa et al., 2018). It is unclear if activated CAFs in carcinomas are also associated with immunosuppressive populations of B cells due to poorly defined markers for such cells (Sarvaria et al., 2017). Moreover, studies investigating the associations of B cell subsets with tumor progression using defined B-cell markers have produced conflicting results even within the same tumor type (Guy et al., 2016). An insufficient understanding of the roles of B cells in carcinomas has hindered the development of rational clinical trials targeting B-cells in carcinomas. The remarkable success of B-cell depletion with the cluster of differentiation 20 (CD20) monoclonal antibody, rituximab, in lymphomas and rheumatoid arthritis has sparked interest in rituximab and other B-cell targeted antibodies as possible therapies in carcinomas (Gunderson and Coussens, 2013). Although many carcinomas have significant B cell infiltration (Germain et al., 2014), clinical trials have shown limited benefits of B-cell depletion in carcinomas (Barbera-Guillem et al., 2000; Aklilu et al., 2004), possibly because B cells can have pro-tumorigenic or anti-tumorigenic properties depending on their maturation stage and other conditions that have not yet been defined (Sarvaria et al., 2017).
The DLBCL Stromal-1 Gene Signature is Inversely Correlated with Survival Outcomes in B-Cell Lymphomas and Other Solid Tumors
Using expression profile analysis of DLBCL biopsy samples from treatment-naïve newly diagnosed patients, Lenz et al. identified two stromal gene signatures, stromal-1 and stromal-2, of which the stromal-1 gene signature was found to be associated with good survival in DLBCL patients (Lenz et al., 2008). However, gene signatures similar to the DLBCL stromal-1 gene signatures have been associated with poor survival in carcinomas, including ovarian cancer (Cheon et al., 2014), breast cancer (Farmer et al., 2009), colorectal cancer (Calon et al., 2015; Isella et al., 2015), and pancreatic cancer (Moffitt et al., 2015). To systematically explore the association of the DLBCL stromal-1 gene signature with survival in cancer patients, we used PRECOG, a pan-cancer database of expression signatures in which each tumor type is represented by multiple independent expression profile data sets and associated survival data. This extensive database is ideal for multi-data set validation of prognostic signatures that have been identified in individual data sets. Using the DLBCL stromal-1 gene signature represented by 50 genes (Lenz et al., 2008), we confirmed that the signature is associated with poor survival in carcinomas and brain tumors and good survival in DLBCL and several other B-cell lymphomas (Table 1). This pattern of inverse association with survival between B-cell lymphomas and carcinomas/brain tumors was specific to the DLBCL stromal-1 gene signature, and was not associated with the DLBCL stromal-2 gene signature represented by 34 genes (Lenz et al., 2008) (data not shown).
Table 1.
DLBCL “stromal-1” signature genes are inversely correlated with survival outcomes in B-cell lymphomas and other malignancies.
| B-cell lymphoma |
Solid tumor |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | BL | CLL | DLBCL | FL | MCL | MM | Bladder | Astro cytoma | Glioma | Colon | Head and neck | Ovarian |
| ACTN1 | -0.928 | -3.216 | -6.211 | -1.901 | -0.94 | 0.658 | 3.312 | 3.22 | 4.557 | 2.36 | 1.988 | 1.552 |
| ADAM12 | 0.746 | -0.084 | -7.809 | -1.749 | -0.866 | -0.395 | 0.537 | 1.653 | 4.405 | 1.675 | 2.051 | 2.99 |
| BGN | 0.842 | 1.309 | -4.115 | -1.775 | 0 | -2.627 | 1.438 | 2.341 | 3.643 | 2.33 | 3.559 | 3.09 |
| CEBPA | -1.516 | -3.127 | -5.644 | -1.639 | 0 | -0.977 | 1.001 | -0.041 | 2.652 | -2.664 | -1.578 | -1.442 |
| COL13A1 | -0.313 | -1.513 | -2.402 | 0.332 | 0 | -0.001 | 2.23 | 2.006 | 1.613 | 2.164 | 1.74 | 0.893 |
| COL16A1 | -0.481 | 0.252 | -3.89 | -0.6 | 0.333 | -0.477 | 2.214 | 2.49 | 5.005 | -0.546 | 1.263 | 4.542 |
| COL1A1 | 0.349 | -1.476 | -4.621 | -1.581 | 0 | -1.951 | 3.592 | 3.326 | 3.77 | 1.544 | 3.354 | 3.929 |
| COL1A2 | -0.097 | -0.879 | -6.264 | -1.605 | 0 | -0.573 | 2.745 | 4.432 | 4.391 | 2.42 | 2.634 | 3.771 |
| COL5A1 | 0.715 | -0.675 | -3.366 | 0.127 | 0 | -0.467 | 1.957 | 3.528 | 4.438 | 2.328 | 3.686 | 3.65 |
| COL5A2 | 0.969 | 1.124 | -3.962 | -1.597 | 0 | -0.777 | 3.47 | 3.588 | 7.322 | 2.437 | 3.26 | 5.256 |
| COL6A2 | 0.677 | -1.368 | -3.719 | -0.749 | -1.415 | 0.14 | 2.369 | 4.591 | 5.693 | 1.301 | 3.12 | 2.11 |
| COL6A3 | 1.194 | -0.129 | -4.502 | -1.442 | 1.37 | 2.684 | 1.282 | 3.005 | 3.071 | 2.403 | 3.141 | 3.178 |
| COL8A2 | -0.212 | -0.894 | -3.046 | 0.069 | 0 | -0.905 | -0.085 | 2.942 | 3.077 | -0.007 | 1.779 | 2.908 |
| CSF2RA | -1.84 | 0 | -2.861 | 0 | 0 | -2.39 | -0.046 | 0.193 | 0 | 0 | 0 | -1.959 |
| CTGF | -0.5 | 0.796 | -5.525 | -0.73 | -1.387 | -0.775 | 1.651 | 1.676 | -1.132 | 2.024 | 2.381 | 2.974 |
| CYR61 | 1.159 | 0.092 | -1.865 | 0.074 | 1.837 | -0.123 | 3.342 | 1.159 | 3.807 | 1.678 | 1.757 | 3.607 |
| DCN | 0.819 | 0.185 | -3.731 | -0.026 | 0 | -0.794 | 0.472 | 1.113 | 2.414 | 1.303 | 0.917 | 4.604 |
| EFEMP2 | 1.823 | 1.113 | -2.797 | 0.307 | 0 | -5.014 | 2.112 | 4.044 | 7.62 | 1.684 | 3.53 | 2.576 |
| EMP2 | -0.057 | 0.044 | -4.122 | 0.147 | 0 | -0.579 | -1.125 | 4.55 | 2.985 | -0.368 | 0.452 | -1.446 |
| FAP | -1.551 | 0.374 | -7.496 | -0.76 | -1.266 | -0.536 | 3.522 | 2.321 | 3.736 | 2.366 | 2.874 | 4.814 |
| FBN1 | 1.125 | 1.079 | -4.907 | -1.854 | 0 | -0.044 | 2.151 | 1.518 | 2.239 | 2.311 | 1.906 | 4.676 |
| FN1 | -1.025 | -0.496 | -5.638 | -1.852 | -1.352 | 2.973 | 3.251 | 2.852 | 5.499 | 2.628 | 2.46 | 4.439 |
| GPNMB | -1.638 | -0.153 | -6.899 | 0.513 | 0 | 1.112 | 1.281 | 3.946 | 5.214 | 1.74 | -2.745 | 1.476 |
| HSPG2 | -0.267 | 2.244 | -2.792 | -1.63 | 0 | 0.845 | -0.02 | 4.261 | 2.989 | 1.313 | 2.108 | 2.396 |
| IL1R1 | -1.566 | -2.791 | -4.858 | -0.432 | 0.804 | -1.789 | -0.186 | 1.194 | 1.217 | 1.275 | 0.897 | -0.137 |
| ITGAV | 0.897 | -2.698 | -6.933 | 0.614 | -2.033 | -0.212 | 0.402 | 0.945 | 0.226 | 2.253 | 1.503 | 1.792 |
| ITGB2 | -1.522 | -2.053 | -5.68 | 0.558 | 0.343 | -1.803 | 0.886 | 0.4 | 4.299 | -0.086 | -2.064 | -2.339 |
| KITLG | 0.896 | -0.172 | -1.923 | 1.04 | -1.197 | 0.454 | 1.113 | -0.331 | 1.091 | 1.164 | -0.721 | -0.504 |
| LAMA4 | 0.445 | 2.207 | -3.683 | 0.453 | 0 | -3.155 | 2.474 | 0.028 | 3.397 | 2.415 | 2.021 | 2.168 |
| LAMB2 | -0.635 | 0.504 | -1.974 | -1.052 | 0 | -0.728 | 0.926 | 1.686 | 5.906 | 0.913 | 1.836 | 2.326 |
| LAMB3 | 1.291 | -1.315 | -2.703 | 0.256 | 0 | 0.265 | -0.927 | 1.977 | 3.542 | 1.516 | 2.039 | -1.966 |
| LOXL1 | -1.453 | -1.007 | -4.202 | -1.287 | 0 | -1.92 | 0.711 | 3.9 | 6.299 | 1.697 | 0.751 | 3.664 |
| LTBP2 | 0.219 | -1.562 | -7.565 | -0.187 | 0 | -1.848 | 2.849 | 1.197 | 3.314 | 0.542 | 2.718 | 1.541 |
| LUM | -0.357 | -1.043 | -5.663 | -0.089 | 0 | -1.859 | 1.442 | 3.796 | 3.723 | 1.447 | 1.428 | 4.841 |
| MFAP2 | 0.862 | 0.01 | -2.835 | 0.608 | 0 | -0.68 | 3.151 | 3.543 | 3.011 | 0.874 | 1.666 | 5.462 |
| MMP14 | -1.105 | 2.746 | -3.319 | 0.69 | 0.681 | -1.647 | 2.046 | 1.787 | 4.691 | 1.786 | 1.168 | 2.297 |
| MMP2 | -1.227 | -0.269 | -5.709 | -1.128 | 0.014 | -0.545 | 0.66 | 1.792 | 3.631 | 1.567 | 3.12 | 3.084 |
| MMP9 | -0.819 | -1.238 | -7.734 | -0.401 | -0.12 | -0.892 | 1.8 | 2.739 | 5.06 | -0.723 | 0.039 | -3.208 |
| PDGFC | 0.62 | -3.08 | -4.268 | 0.632 | 0 | -0.486 | 2.788 | -3.419 | 3.639 | 1.987 | 2.096 | -0.167 |
| PLAU | -1.723 | -1.701 | -7.712 | 0.205 | 0.528 | -0.749 | 2.515 | 2.302 | 4.592 | 0.627 | 1.521 | 2.334 |
| POSTN | 1.565 | 0.675 | -5.031 | -1.266 | -0.77 | -1.157 | 3.246 | 2.76 | 5.46 | 2.632 | 2.092 | 4.696 |
| SDC2 | -0.209 | -1.963 | -3.763 | -0.47 | -0.383 | -0.664 | -1.091 | 1.405 | 5.736 | 2.239 | 1.659 | 1.424 |
| SERPINH1 | -1.173 | 2.067 | -2.912 | -1.224 | 0 | 1.565 | 1.422 | 3.846 | 5.397 | 3.044 | 2.065 | 2.07 |
| SPARC | 0.487 | -3.125 | -7.236 | -1.599 | 1.012 | -2.767 | 2.24 | -1.998 | -0.074 | 2.412 | 2.933 | 4.188 |
| TGFB1I1 | -0.842 | -1.479 | -2.367 | 0.662 | 0 | -1.787 | 1.518 | 2.783 | 4.58 | 1.523 | 3.557 | 4.265 |
| THBS1 | 1.462 | -3.212 | -2.038 | -1.38 | 0.238 | -1.674 | 1.673 | 2.947 | 3.122 | 0.799 | 2.328 | 3.565 |
| TIMP2 | -0.677 | -2.448 | -1.399 | 1.006 | 0.343 | 0.83 | 2.608 | 1.584 | 1.251 | 2.73 | 2.271 | 2.495 |
| VCAN | 1.459 | -3.803 | -3.177 | -0.588 | 0 | -2.078 | 3.133 | -3.546 | -3.171 | 2.264 | 2.238 | 4.277 |
Analysis of the DLBCL “stromal-1” geneset in the PREdiction of Clinical Outcomes from Genomic Profiles (PRECOG) public dataset (https://precog.stanford.edu). Each gene is assigned z scores associated with survival in different cancer types. Scores less than or equal to zero (red) are associated with good survival while positive scores (blue) are associated with poor survival. BL, Burkitt lymphoma; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; MM, multiple myeloma.
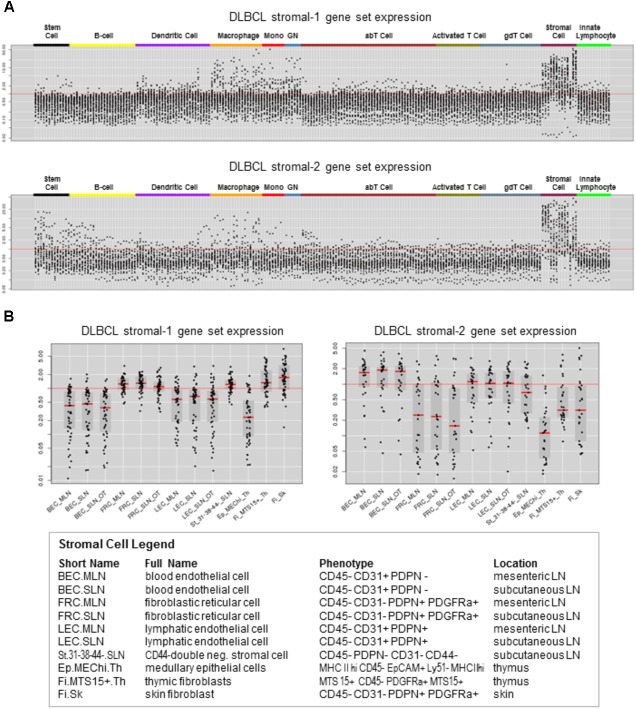
In Normal Lymph Nodes, DLBCL Stromal-1 and Stromal-2 Gene Signatures are Enriched in Stromal Fibroblasts and Endothelial Cells, Respectively
To identify immune cell types that express the DLBCL stromal-1 and stromal-2 signature genes, we looked for enrichment of these genes in the transcriptomes of 249 normal immunological cell types that had been isolated from mice and characterized by the Immunological Genome Project (ImmGen) (Heng and Painter, 2008; Shay and Kang, 2013). This analysis identified stromal cells as the most likely source of both gene signatures, although some of the genes were also expressed in macrophages, monocytes, granulocytes, and stem cells (Figure 1A). Closer examination of the stromal cell subtypes revealed that the DLBCL stromal-1 and stromal-2 signature genes were preferentially expressed in different types of stromal cells. DLBCL stromal-1 signature genes were particularly enriched in cells characterized by expression of podoplanin (PDPN) and platelet-derived growth factor receptor α (PDGFRα), including FRC from mesenteric and subcutaneous lymph nodes and the so-called double-negative stromal cells, while stromal-2 signature genes were enriched in blood and lymphatic endothelial cells (Figure 1B).
FIGURE 1.
DLBCL stromal-1 and stromal-2 signature genes are enriched in different stromal cell types. Expression of the DLBCL stromal-1 and stromal-2 signature genes in the Immunological Genome Project (ImmGen) data set. (A) Gene expression values normalized across 249 mouse immunological cell types. (B) Detailed view of gene expression values normalized to the stromal cell types shown in the legend. The graphs were generated using data from ImmGen (http://www.immgen.org).
The DLBCL Stromal-1 Gene Signature is Enriched in Ovarian CAFs
To identify cells that express the DLBCL stromal-1 and stromal-2 signature genes in an epithelial tumor, we selected ovarian cancer because of the existing microarray data set (GSE40595) in which a large number of ovarian cancers have been laser capture microdissected into epithelial and stromal components (Yeung et al., 2013). For comparison with normal tissue, a small number of samples in this data set were microdissected from the normal ovary epithelium and stroma (Yeung et al., 2013). Our gene signature enrichment analysis revealed strong enrichment of the DLBCL stromal-1 gene signature in CAFs in comparison to cancer cells, normal ovary fibroblasts, and normal ovary epithelial cells (Figure 2). The DLBCL stromal-2 gene signature was enriched in CAFs but also in the normal ovary stroma (Figure 2).
FIGURE 2.
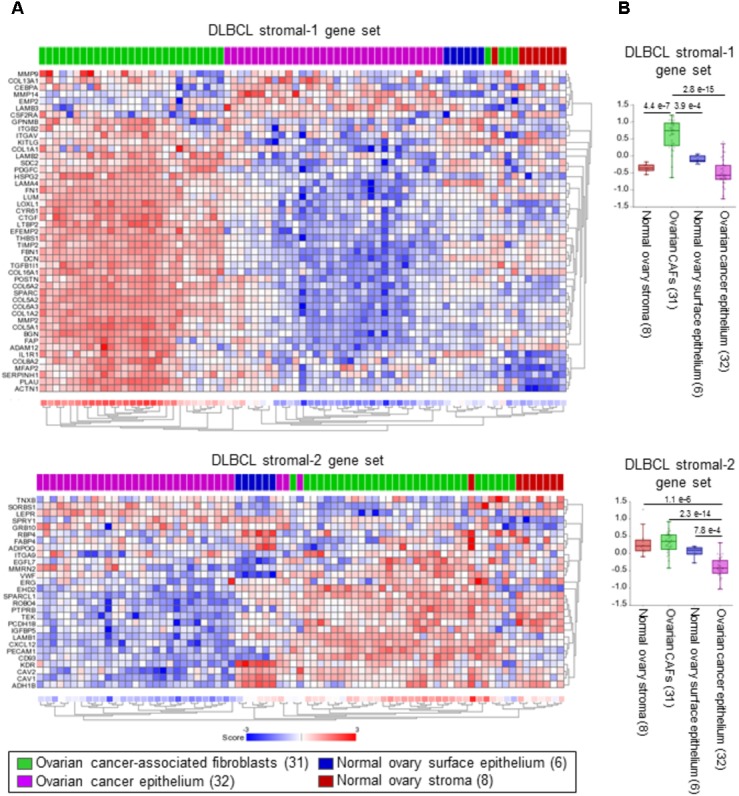
DLBCL stromal-1 signature genes are enriched in cancer-associated fibroblasts (CAFs). (A) Non-centered gene set clustering analysis of the stromal and epithelial cell types in ovarian cancer and the normal ovary in the GSE40595 dataset using the DLBCL stromal-1 and stromal-2 gene sets. The number of samples in each group is indicated in parentheses. The gene set clustering analysis and image acquisition was performed using the R2 Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl). (B) The same data are shown as box dot plots with P-values for differential expression of the DLBCL stromal-1 and stromal-2 gene signatures in different cell types.
Possible Mechanisms by Which CAFs Contribute to Inverse Survival Outcomes in B-Cell Lymphomas and Carcinomas
It is unusual for a gene signature to be associated with inverse survival outcomes in B-cell lymphomas and carcinomas. This is unlikely to be a technical error related to microarray technology as several individual genes from the DLBCL stromal-1 signature have been validated as predictors of good survival in DLBCL by independent technologies, such as immunohistochemistry and qPCR in formalin-fixed paraffin-embedded tissues (Lossos et al., 2004; Meyer et al., 2011; Tekin et al., 2016). Similarly, various technologies have been used to validate many of the signature genes as predictors of poor survival in carcinomas (Farmer et al., 2009; Cheon et al., 2014; Calon et al., 2015; Isella et al., 2015; Moffitt et al., 2015; Jia et al., 2016). While the mechanism by which the DLBCL stromal-1 signature genes could contribute to good survival in DLBCL is still unclear, multiple mechanisms by which CAFs contribute to poor outcomes in carcinomas have been proposed, including the promotion of tumor growth, angiogenesis, invasion and metastasis, the provision of protective niches for cancer stem cells, and the obstruction of access of chemotherapies and immunotherapies (Jain, 2013; Kalluri, 2016). Here, we will specifically focus on the possible direct or indirect roles of CAFs that could contribute to inverse survival outcomes in DLBCL and carcinomas.
Cancer-associated fibroblasts share structural and molecular features with the reticular fiber networks of secondary lymphoid organs, which are known to guide and compartmentalize specific immune cell types and play key roles in mediating functional immune cell interactions (Acton et al., 2012; Astarita et al., 2012; Cremasco et al., 2014; Chang and Turley, 2015; Fletcher et al., 2015; Turley et al., 2015). However, in addition to being sites in which immune responses are initiated, secondary lymphoid organs are also sites that foster immune privilege that prevents autoimmunity by inducing tolerance and deleting autoreactive T cells, suppressing effector T cell proliferation, and supporting regulatory T cells (Fletcher et al., 2011, 2014, 2015; Brown and Turley, 2015). Currently, lymph node fibroblasts are being explored for their therapeutic potential to circumvent unwanted inflammation in autoimmune diseases, sepsis, and graft rejection after organ transplantation (Fletcher et al., 2011, 2014, 2015). Based on the molecular similarity between CAFs and lymph node fibroblasts, we propose that CAFs primarily play an immunosuppressive role in tumors using similar molecular mechanisms to those used by lymph node fibroblasts in regulating immune cell tolerance and homeostasis. In support of this hypothesis, CAF-derived factors have been shown to contribute to immune editing in vivo to avoid tumor detection and rejection by the host immune system (Stover et al., 2007; Kraman et al., 2010). Specific to B cells, several in vitro models have shown the ability of different types of fibroblasts to modulate B cell differentiation, activation, and function. Adipose tissue-derived fibroblasts have been shown to suppress plasmablast formation and induce formation of regulatory B cells (Franquesa et al., 2015) while rheumatoid synovial fibroblasts have been shown to induce immunoglobulin (Ig) class-switch recombination and IgG/IgA production in IgD+ B cells (Bombardieri et al., 2011). We envision that the immunoregulatory functions of CAFs may lead to improved survival in DLBCL and other B-cell lymphomas where malignant cells themselves are subject to functional alteration. In contrast, immunosuppression by CAFs in carcinomas may lead to an ineffective immune defense against malignant cells, which is associated with poor survival.
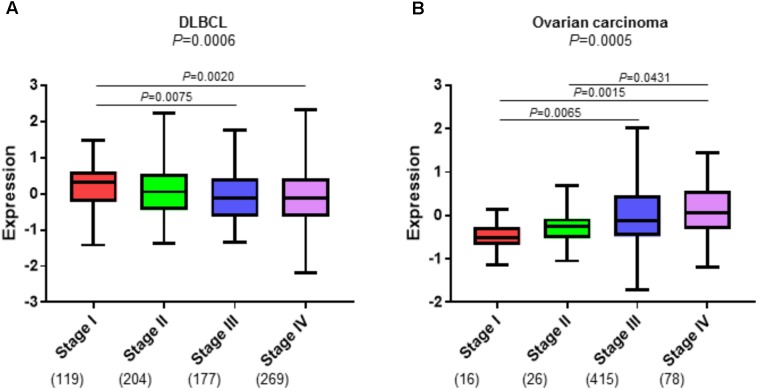
Cancer-associated fibroblasts are also capable of modifying the immune landscape by selective attraction, recruitment, retention, activation, and suppression of different immune cell types (Karin, 2010; Raz and Erez, 2013; Harper and Sainson, 2014). Recent studies provide evidence that CAFs can directly contribute to immune cell fate and survival (Harper and Sainson, 2014). In mouse models, CAFs have been shown to attract macrophages, neutrophils, and subsets of T cells that promote tumor progression (Silzle et al., 2003; Grum-Schwensen et al., 2010; Elkabets et al., 2011). One possible underlying mechanism for the association of the DLBCL stromal-1 gene signature with good survival in patients with DLBCL is that fibroblasts and the associated ECM attract and trap malignant B cells thereby impeding their spread to new anatomical locations. We show a small but consistent inverse association of the DLBCL stromal-1 gene signature expression with DLBCL tumor stage (a measure of lymph node groups and extranodal sites to which malignant cells have metastasized) (Figure 3A). The decrease in stromal gene signature expression in the later stages of DLBCL may indicate that the stroma plays a role in localizing the lymphoma cells to the lymph nodes during the earlier stages of the disease. In contrast, DLBCL stromal-1 gene signature expression is typically increased with increased tumor stage in epithelial carcinomas, such as ovarian cancer (Figure 3B). The increase in CAFs in the later stages of carcinomas may prevent immune cells from reaching the tumor parenchyma by trapping the immune cells in the stroma thereby preventing an anti-tumor response. A recent study of immune cell infiltration in metastatic urothelial carcinomas showed that patients whose tumors were classified as immune-excluded (immune cells localized in the CAF-rich stroma) had increased disease progression and decreased response to immunotherapy (Mariathasan et al., 2018). Therefore, we hypothesize that CAFs aid in retaining DLBCL in the lymph node, which is associated with better prognosis, whereas in carcinomas CAFs trap immune cells, which is associated with decreased anti-tumor immune activity and a worse prognosis.
FIGURE 3.
CAFs have an inverse association with tumor stage in DLBCL and ovarian carcinoma. Enrichment of the DLBCL stromal-1 gene signature in progression stages I-IV in (A) three DLBCL microarray datasets (GSE10846, GSE87371, and GSE4475) that were combined into one dataset, and (B) The Cancer Genome Atlas (TCGA) ovarian carcinoma dataset (https://cancergenome.nih.gov). The gene signature enrichment analysis was performed using the R2 Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl). The y axis shows relative enrichment of the DLBCL stomal-1 gene signature. The x axis shows tumor stage. The number of samples for each tumor stage is indicated in parentheses.
One of the key modulators of the cancer microenvironment is the multifunctional cytokine, transforming growth factor β (TGFβ). TGFβ induces CAF activation and fibroblast-to-myofibroblast transition with consequent linearization of collagen fibers and stiffening of the ECM. In turn, activated CAFs induce TGFβ signaling to perpetually maintain the activated state (Calon et al., 2014; Beach et al., 2016; Erdogan and Webb, 2017). Consistent with the DLBCL stromal-1 signature representing CAFs, our Ingenuity Pathway Analysis (IPA) of the DLBCL gene signatures implicates TGFβ signaling as the main upstream regulator of the DLBCL stromal-1 gene signature (Table 2). In carcinomas, TGFβ has been shown to promote tumor progression by inhibiting immunosurveillance through multiple mechanisms (Flavell et al., 2010; Sheng et al., 2015), including the recruitment of macrophages (Byrne et al., 2008) and limited efficacy of immunotherapy by excluding CD8+ T cells from the tumor parenchyma (Mariathasan et al., 2018; Tauriello et al., 2018). It is likely that TGFβ also plays an immunosuppressive role in lymphomas. However, TGFβ is also a potent negative regulator of B-cell survival, proliferation, activation, and differentiation (Sanjabi et al., 2017). Stroma-derived TGFβ has been shown to induce senescence and apoptosis in mouse models of B-cell lymphoma (Reimann et al., 2010; Stelling et al., 2018). Thus, the DLBCL stromal-1 gene signature may be primarily associated with tumor-promoting immunosuppression in carcinomas, while the same immunosuppression may lead to the eradication of B cells, which represent the malignant component of B-cell lymphoma.
Table 2.
Upstream regulators of genes in the DLBCL stromal gene signature-1 and stromal gene signature-2.
| Upstream regulator | Molecule type | p-value of overlap |
|---|---|---|
| DLBCL stromal-1 gene signature | ||
| TGFB1 | Growth factor | 4.78E-31 |
| COLQ | Other | 2.70E-20 |
| Bleomycin | Chemical drug | 1.97E-18 |
| SPDEF | Transcription regulator | 2.73E-18 |
| Tgf beta | Group | 3.95E-18 |
| TGFB3 | Growth factor | 8.04E-18 |
| TNF | Cytokine | 1.53E-17 |
| DLBCL stromal-2 gene signature | ||
| KLF2 | Transcription regulator | 1.89E-09 |
| Rosiglitazone | Chemical drug | 5.82E-09 |
| VEGFA | Growth factor | 5.90E-09 |
| PPARG | Ligand-dependent nuclear receptor | 1.36E-08 |
| 10E, 12Z-octadecadienoic acid | Chemical – endogenous Mammalian | 4.98E-08 |
| WNT3A | Cytokine | 6.02E-08 |
| MGEA5 | Enzyme | 1.08E-07 |
The identification of upstream regulators was done using Ingenuity Pathway Analysis (www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/).
Conclusion
Past clinical trials have taught us that successful targeted therapies in one disease do not always yield the desired results in another disease despite the presence of the same target. One example is the poor response of B-cell-infiltrated carcinomas to rituximab, which has shown remarkable success in lymphomas and rheumatoid arthritis. The opposite survival outcomes associated with the presence of stromal cells in B-cell lymphomas and carcinomas should serve as a warning that targeting the tumor microenvironment may produce opposite effects in B-cell lymphomas and carcinomas.
Database Links
GEO Data Sets (https://www.ncbi.nlm.nih.gov/gds) Immunological Genome Project (https://www.immgen.org) PRECOG – PREdiction of Clinical Outcomes from Genomic Profiles (https://precog.stanford.edu) R2: Genomics Analysis and Visualization Platform (http://hgserver1.amc.nl) The Cancer Genome Atlas Project (https://cancergenome.nih.gov).
Author Contributions
SO analyzed the public data sets. SO and MH wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kristy J. Daniels for assistance in manuscript preparation and members of the Women’s Cancer Program for their support.
Abbreviations
- CAF
cancer-associated fibroblast
- CD
cluster of differentiation
- CXCL
C-X-C motif chemokine ligand
- DC
dendritic cells
- DLBCL
diffuse large B-cell lymphoma
- ECM
extracellular matrix
- FDC
follicular dendritic cells
- FRC
fibroblastic reticular cells
- GC
germinal center
- Ig
immunoglobulin
- ImmGen
immunological genome project
- IPA
ingenuity pathway analysis
- MRC
marginal reticular cells
- NK
natural killer
- PDGFRα
platelet-derived growth factor receptor α
- PDPN
podoplanin
- PRECOG
PREdiction of clinical outcomes from genomic profiles
- TCGA
the Cancer genome atlas project
- TGFβ
transforming growth factor β
- TIL
tumor infiltrating lymphocyte
- TLS
tertiary lymphoid structure
Footnotes
Funding. This research was supported by the National Cancer Institute grant R01CA208753 and the Office of the Assistant Secretary of Defense for Health Affairs through the Ovarian Cancer Research Program Awards Nos. W81XWH-17-1-0144 and W81XWH-16-1-0190.
References
- Acton S. E., Astarita J. L., Malhotra D., Lukacs-Kornek V., Franz B., Hess P. R., et al. (2012). Podoplanin-rich stromal networks induce dendritic cell motility via activation of the C-type lectin receptor CLEC-2. Immunity 37 276–289. 10.1016/j.immuni.2012.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aklilu M., Stadler W. M., Markiewicz M., Vogelzang N. J., Mahowald M., Johnson M., et al. (2004). Depletion of normal B cells with rituximab as an adjunct to IL-2 therapy for renal cell carcinoma and melanoma. Ann. Oncol. 15 1109–1114. 10.1093/annonc/mdh280 [DOI] [PubMed] [Google Scholar]
- Aran D., Hu Z., Butte A. J. (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18:220. 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarita J. L., Acton S. E., Turley S. J. (2012). Podoplanin: emerging functions in development, the immune system, and cancer. Front. Immunol. 3:283. 10.3389/fimmu.2012.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten M. (2014). Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4:62. 10.3389/fonc.2014.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera-Guillem E., Nelson M. B., Barr B., Nyhus J. K., May K. F., Jr., Feng L., et al. (2000). B lymphocyte pathology in human colorectal cancer. Experimental and clinical therapeutic effects of partial B cell depletion. Cancer Immunol. Immunother. 48 541–549. 10.1007/PL00006672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T. A., Amir E. (2017). HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br. J. Cancer 117 451–460. 10.1038/bjc.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach J. A., Aspuria P. J., Cheon D. J., Lawrenson K., Agadjanian H., Walsh C. S., et al. (2016). Sphingosine kinase 1 is required for TGF-beta mediated fibroblastto- myofibroblast differentiation in ovarian cancer. Oncotarget 7 4167–4182. 10.18632/oncotarget.6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardieri M., Kam N. W., Brentano F., Choi K., Filer A., Kyburz D., et al. (2011). A BAFF/APRIL-dependent TLR3-stimulated pathway enhances the capacity of rheumatoid synovial fibroblasts to induce AID expression and Ig class-switching in B cells. Ann. Rheum. Dis. 70 1857–1865. 10.1136/ard.2011.150219 [DOI] [PubMed] [Google Scholar]
- Brown F. D., Turley S. J. (2015). Fibroblastic reticular cells: organization and regulation of the T lymphocyte life cycle. J. Immunol. 194 1389–1394. 10.4049/jimmunol.1402520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S. N., Knox M. C., Halliday G. M. (2008). TGFbeta is responsible for skin tumour infiltration by macrophages enabling the tumours to escape immune destruction. Immunol. Cell Biol. 86 92–97. 10.1038/sj.icb.7100116 [DOI] [PubMed] [Google Scholar]
- Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., et al. (2015). Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47 320–329. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- Calon A., Tauriello D. V., Batlle E. (2014). TGF-beta in CAF-mediated tumor growth and metastasis. Semin. Cancer Biol. 25 15–22. 10.1016/j.semcancer.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Chang J. E., Turley S. J. (2015). Stromal infrastructure of the lymph node and coordination of immunity. Trends Immunol. 36 30–39. 10.1016/j.it.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Cheon D. J., Tong Y., Sim M. S., Dering J., Berel D., Cui X., et al. (2014). A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin. Cancer Res. 20 711–723. 10.1158/1078-0432.CCR-13-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., et al. (2018). Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 33 463.e10–479.e10. 10.1016/j.ccell.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Cremasco V., Woodruff M. C., Onder L., Cupovic J., Nieves-Bonilla J. M., Schildberg F. A., et al. (2014). B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat. Immunol. 15 973–981. 10.1038/ni.2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J. G. (2010). B cell follicles and antigen encounters of the third kind. Nat. Immunol. 11 989–996. 10.1038/ni.1946 [DOI] [PubMed] [Google Scholar]
- Dieu-Nosjean M. C., Goc J., Giraldo N. A., Sautes-Fridman C., Fridman W. H. (2014). Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 35 571–580. 10.1016/j.it.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Elkabets M., Gifford A. M., Scheel C., Nilsson B., Reinhardt F., Bray M. A., et al. (2011). Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J. Clin. Invest. 121 784–799. 10.1172/JCI43757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan B., Webb D. J. (2017). Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 45 229–236. 10.1042/bst20160387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer P., Bonnefoi H., Anderle P., Cameron D., Wirapati P., Becette V., et al. (2009). A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 15 68–74. 10.1038/nm.1908 [DOI] [PubMed] [Google Scholar]
- Flavell R. A., Sanjabi S., Wrzesinski S. H., Licona-Limon P. (2010). The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 10 554–567. 10.1038/nri2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher A. L., Acton S. E., Knoblich K. (2015). Lymph node fibroblastic reticular cells in health and disease. Nat. Rev. Immunol. 15 350–361. 10.1038/nri3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher A. L., Elman J. S., Astarita J., Murray R., Saeidi N., D’Rozario J., et al. (2014). Lymph node fibroblastic reticular cell transplants show robust therapeutic efficacy in high-mortality murine sepsis. Sci. Transl. Med. 6:249ra109. 10.1126/scitranslmed.3009377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher A. L., Malhotra D., Turley S. J. (2011). Lymph node stroma broaden the peripheral tolerance paradigm. Trends Immunol. 32 12–18. 10.1016/j.it.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquesa M., Mensah F. K., Huizinga R., Strini T., Boon L., Lombardo E., et al. (2015). Human adipose tissue-derived mesenchymal stem cells abrogate plasmablast formation and induce regulatory B cells independently of T helper cells. Stem Cells 33 880–891. 10.1002/stem.1881 [DOI] [PubMed] [Google Scholar]
- Fridman W. H., Pages F., Sautes-Fridman C., Galon J. (2012). The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12 298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- Gentles A. J., Newman A. M., Liu C. L., Bratman S. V., Feng W., Kim D., et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21 938–945. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain C., Gnjatic S., Tamzalit F., Knockaert S., Remark R., Goc J., et al. (2014). Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am. J. Respir. Crit. Care Med. 189 832–844. 10.1164/rccm.201309-1611OC [DOI] [PubMed] [Google Scholar]
- Grum-Schwensen B., Klingelhofer J., Grigorian M., Almholt K., Nielsen B. S., Lukanidin E., et al. (2010). Lung metastasis fails in MMTV-PyMT oncomice lacking S100A4 due to a T-cell deficiency in primary tumors. Cancer Res. 70 936–947. 10.1158/0008-5472.CAN-09-3220 [DOI] [PubMed] [Google Scholar]
- Gunderson A. J., Coussens L. M. (2013). B cells and their mediators as targets for therapy in solid tumors. Exp. Cell Res. 319 1644–1649. 10.1016/j.yexcr.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy T. V., Terry A. M., Bolton H. A., Hancock D. G., Shklovskaya E., Fazekas de St. Groth B. (2016). Pro- and anti-tumour effects of B cells and antibodies in cancer: a comparison of clinical studies and preclinical models. Cancer Immunol. Immunother. 65 885–896. 10.1007/s00262-016-1848-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J., Sainson R. C. (2014). Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 25 69–77. 10.1016/j.semcancer.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Heng T. S., Painter M. W. (2008). The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 9 1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Isella C., Terrasi A., Bellomo S. E., Petti C., Galatola G., Muratore A., et al. (2015). Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47 312–319. 10.1038/ng.3224 [DOI] [PubMed] [Google Scholar]
- Jain R. K. (2013). Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J. Clin. Oncol. 31 2205–2218. 10.1200/JCO.2012.46.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Liu Z., Deng N., Tan T. Z., Huang R. Y., Taylor-Harding B., et al. (2016). A COL11A1-correlated pan-cancer gene signature of activated fibroblasts for the prioritization of therapeutic targets. Cancer Lett. 382 203–214. 10.1016/j.canlet.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. (2016). The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16 582–598. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- Kalluri R., Zeisberg M. (2006). Fibroblasts in cancer. Nat. Rev. Cancer 6 392–401. 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Karin N. (2010). The multiple faces of CXCL12 (SDF-1alpha) in the regulation of immunity during health and disease. J. Leukoc. Biol. 88 463–473. 10.1189/jlb.0909602 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Qian B. Z., Pollard J. W. (2015). Immune cell promotion of metastasis. Nat. Rev. Immunol. 15 73–86. 10.1038/nri3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning J. J., Mebius R. E. (2012). Interdependence of stromal and immune cells for lymph node function. Trends Immunol. 33 264–270. 10.1016/j.it.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Kraman M., Bambrough P. J., Arnold J. N., Roberts E. W., Magiera L., Jones J. O., et al. (2010). Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330 827–830. 10.1126/science.1195300 [DOI] [PubMed] [Google Scholar]
- Lenz G., Wright G., Dave S. S., Xiao W., Powell J., Zhao H., et al. (2008). Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 359 2313–2323. 10.1056/NEJMoa0802885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., et al. (2009). Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139 891–906. 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossos I. S., Czerwinski D. K., Alizadeh A. A., Wechser M. A., Tibshirani R., Botstein D., et al. (2004). Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 350 1828–1837. 10.1056/NEJMoa032520 [DOI] [PubMed] [Google Scholar]
- Lu P., Weaver V. M., Werb Z. (2012). The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196 395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Turley S. J., Nickles D., Castiglioni A., Yuen K., Wang Y., et al. (2018). TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554 544–548. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P. N., Fu K., Greiner T., Smith L., Delabie J., Gascoyne R., et al. (2011). The stromal cell marker SPARC predicts for survival in patients with diffuse large B-cell lymphoma treated with rituximab. Am. J. Clin. Pathol. 135 54–61. 10.1309/ajcpjx4bjv9nlqhy [DOI] [PubMed] [Google Scholar]
- Mionnet C., Mondor I., Jorquera A., Loosveld M., Maurizio J., Arcangeli M. L., et al. (2013). Identification of a new stromal cell type involved in the regulation of inflamed B cell follicles. PLoS Biol. 11:e1001672. 10.1371/journal.pbio.1001672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt R. A., Marayati R., Flate E. L., Volmar K. E., Loeza S. G., Hoadley K. A., et al. (2015). Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47 1168–1178. 10.1038/ng.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S. N., Germain R. N. (2009). Stromal cell contributions to the homeostasis and functionality of the immune system. Nat. Rev. Immunol. 9 618–629. 10.1038/nri2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz Y., Erez N. (2013). An inflammatory vicious cycle: fibroblasts and immune cell recruitment in cancer. Exp. Cell Res. 319 1596–1603. 10.1016/j.yexcr.2013.03.022 [DOI] [PubMed] [Google Scholar]
- Reimann M., Lee S., Loddenkemper C., Dorr J. R., Tabor V., Aichele P., et al. (2010). Tumor stroma-derived TGF-beta limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17 262–272. 10.1016/j.ccr.2009.12.043 [DOI] [PubMed] [Google Scholar]
- Sanjabi S., Oh S. A., Li M. O. (2017). Regulation of the immune response by TGF-beta: from conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 9:a022236. 10.1101/cshperspect.a022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvaria A., Madrigal J. A., Saudemont A. (2017). B cell regulation in cancer and anti-tumor immunity. Cell Mol. Immunol. 14 662–674. 10.1038/cmi.2017.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay T., Kang J. (2013). Immunological genome project and systems immunology. Trends Immunol. 34 602–609. 10.1016/j.it.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J., Chen W., Zhu H. J. (2015). The immune suppressive function of transforming growth factor-beta (TGF-beta) in human diseases. Growth Factors 33 92–101. 10.3109/08977194.2015.1010645 [DOI] [PubMed] [Google Scholar]
- Silzle T., Kreutz M., Dobler M. A., Brockhoff G., Knuechel R., Kunz-Schughart L. A. (2003). Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur. J. Immunol. 33 1311–1320. 10.1002/eji.200323057 [DOI] [PubMed] [Google Scholar]
- Spano D., Zollo M. (2012). Tumor microenvironment: a main actor in the metastasis process. Clin. Exp. Metastasis 29 381–395. 10.1007/s10585-012-9457-5 [DOI] [PubMed] [Google Scholar]
- Stelling A., Hashwah H., Bertram K., Manz M. G., Tzankov A., Muller A. (2018). The tumor suppressive TGF-beta/SMAD1/S1PR2 signaling axis is recurrently inactivated in diffuse large B-cell lymphoma. Blood 131 2235–2246. 10.1182/blood-2017-10-810630 [DOI] [PubMed] [Google Scholar]
- Stover D. G., Bierie B., Moses H. L. (2007). A delicate balance: TGF-beta and the tumor microenvironment. J. Cell. Biochem. 101 851–861. 10.1002/jcb.21149 [DOI] [PubMed] [Google Scholar]
- Tauriello D. V. F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., et al. (2018). TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554 538–543. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- Tekin N., Omidvar N., Morris T. P., Conget P., Bruna F., Timar B., et al. (2016). Protocol for qRT-PCR analysis from formalin fixed paraffin embedded tissue sections from diffuse large b-cell lymphoma: validation of the six-gene predictor score. Oncotarget 7 83319–83329. 10.18632/oncotarget.13066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley S. J., Cremasco V., Astarita J. L. (2015). Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 15 669–682. 10.1038/nri3902 [DOI] [PubMed] [Google Scholar]
- Yeung T. L., Leung C. S., Wong K. K., Samimi G., Thompson M. S., Liu J., et al. (2013). TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 73 5016–5028. 10.1158/0008-5472.can-13-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]