Abstract
In C. crescentus, iron metabolism is mainly controlled by the transcription factor Fur (ferric uptake regulator). Iron-bound Fur represses genes related to iron uptake and can directly activate the expression of genes for iron-containing proteins. In this work, we used total RNA sequencing (RNA-seq) of wild type C. crescentus growing in minimal medium under iron limitation and a fur mutant strain to expand the known Fur regulon, and to identify novel iron-regulated genes. The RNA-seq of cultures treated with the iron chelator 2-2-dypiridyl (DP) allowed identifying 256 upregulated genes and 236 downregulated genes, being 176 and 204 newly identified, respectively. Sixteen transcription factors and seven sRNAs were upregulated in iron limitation, suggesting that the response to low iron triggers a complex regulatory network. Notably, lexA along with most of its target genes were upregulated, suggesting that DP treatment caused DNA damage, and the SOS DNA repair response was activated in a RecA-dependent manner, as confirmed by RT-qPCR. Fluorescence microscopy assays using an oxidation-sensitive dye showed that wild type cells in iron limitation and the fur mutant were under endogenous oxidative stress, and a direct measurement of cellular H2O2 showed that cells in iron-limited media present a higher amount of endogenous H2O2. A mutagenesis assay using the rpoB gene as a reporter showed that iron limitation led to an increase in the mutagenesis rate. These results showed that iron deficiency causes C. crescentus cells to suffer oxidative stress and to activate the SOS response, indicating an increase in DNA damage.
Keywords: iron limitation, DNA damage, Caulobacter crescentus, oxidative stress, RNA-seq analysis
Introduction
The metal iron is an essential micronutrient for bacterial growth. Under physiological conditions, it exists mainly in one of two redox states: the ferrous (Fe2+) form and the ferric (Fe3+) form. Iron is a prosthetic component for incorporation into proteins, and participates in important biological processes such as N2 fixation, tricarboxylic acid cycle, respiration, gene regulation, and DNA synthesis (Beinert et al., 1997; Andrews et al., 2003). Iron is not freely available in the environment since ferric iron is extremely insoluble, so bacteria have evolved strategies to ensure their physiological demands. Iron limitation leads to increased iron influx, the reduction in activity of non-essential iron enzymes, the activation of iron-independent pathways and the mobilization of protein-associated stored iron, as from ferritins. On the other hand, excess iron induces genes involved in its efflux and reduction of iron uptake to keep intracellular iron levels under strict regulation (Chandrangsu et al., 2017). This control, along with the activation of oxidative stress response, is an essential factor to allow the aerobic style of life (Touati, 2000).
C. crescentus is an aerobic free-living alphaproteobacterium that grows in oligotrophic aquatic environments, and despite apparently not synthesizing its own siderophores for iron uptake, is able to acquire them through TonB-dependent transporters (Balhesteros et al., 2017). In C. crescentus, four TonB-dependent transporters predicted to be involved in iron acquisition are regulated by Fur, and one of them was characterized as the hemin/hemoglobin transporter HutA (da Silva Neto et al., 2009; Balhesteros et al., 2017).
In most bacteria, the transcriptional regulator Fur tightly controls iron homeostasis (Fillat, 2014). Iron-bound Fur acts as transcriptional repressor under iron sufficient conditions, inhibiting the expression of genes for iron uptake and for non-essential iron enzymes (Andrews et al., 2003; Lee and Helmann, 2007). In several bacteria the repression by iron-Fur is released under iron limitation, allowing the expression of a small RNA (called RyhB in Escherichia coli, FsrA in Bacillus subtilis, PrrF1 and PrrF2 in Pseudomonas aeruginosa, NrrF in Neisseria meningitidis, and Bc_KC_sr1 and Bc_KC_sr2 in Burkholderia cenocepacia KC-01), which facilitates the degradation of the mRNA of non-essential iron enzymes (Masse and Gottesman, 2002; Wilderman et al., 2004; Masse et al., 2005, 2007; Mellin et al., 2007; Gaballa et al., 2008; Ghosh et al., 2017; Jackson et al., 2017).
Previously, a detailed in silico analysis of the promoter regions of the nuoA-N operon (NADH dehydrogenase), acnA (aconitase) and sdhCBAD (succinate dehydrogenase), confirmed by electrophoresis mobility shift assays, showed that Fur directly binds to these regulatory regions and activates the expression of these genes in response to iron (da Silva Neto et al., 2009), revealing that in C. crescentus the mechanism of iron control of metabolism is different than that of E. coli. Global transcriptional analyses using DNA microarrays allowed to define the Fur regulon, identifying 42 genes repressed and 27 genes activated by Fe-Fur (da Silva Neto et al., 2013).
In C. crescentus, Fur also activates the low oxygen signaling network FixK, FixT and FtrB cascade, suggesting that in C. crescentus iron regulation also can be sensitive to O2 levels (da Silva Neto et al., 2013). In E. coli, an anaerobic facultative bacterium, Fur activity is modulated by O2 levels (Beauchene et al., 2015). It has been reported that in anaerobic conditions the labile iron pool is higher than in aerobic conditions (Beauchene et al., 2017). Under anaerobic condition the upregulation of feoABC transport system facilitates the Fe2+ import increasing the labile iron pool, which is required to enhance the Fur regulon in these conditions (Beauchene et al., 2015, 2017).
The previous transcriptional analysis also showed that several genes have an iron-dependent and Fur-independent regulation in C. crescentus, such as those encoding the heat shock sigma factor RpoH and the Fe–S cluster biogenesis operon containing iscR (da Silva Neto et al., 2013). However, the tiling array platform used consisted of primers located mainly around the start codons of annotated protein-coding genes. As previous works using high-throughput RNA sequencing have identified an expressive number of non-coding RNAs that could participate in gene regulation (Landt et al., 2008; Schrader et al., 2014), it is important to identify those that participate in the regulatory network that control the response to iron levels.
In order to further understand the consequences of iron limitation, we carried out an RNA-seq-based transcriptomic analysis seeking to identify new genes responsive to iron limitation and belonging to the Fur regulon, as well as iron-regulated regulatory RNAs. Our analyses showed that under iron limitation several stress response genes are induced, and the cells are in a state of oxidative stress. Moreover, the results indicate that in C. crescentus under iron limitation, the genes involved in the SOS response are induced in a RecA-dependent manner, suggesting that in this condition the cells undergo DNA damage.
Materials and Methods
Bacterial Strains and Growth Conditions
The Caulobacter crescentus strains used were: NA1000 (wild-type) (Evinger and Agabian, 1977), SP0057 (fur mutant) (da Silva Neto et al., 2009), GM10 (recA mutant) (Galhardo et al., 2005), and NA1000 and GM10 harboring the pRKlacZ290 vector containing the imuA promoter (imuA/lacZ fusion) (Galhardo et al., 2005). Cultures were grown aerobically at 30°C in minimal medium (M2), which contains 10 μM FeSO4 (Ely, 1991). Iron-limiting conditions were obtained by adding the iron chelator 2,2-dipyridyl (DP) (Sigma, 100 μM) to the M2 medium (DP-treated M2) or by using a modified M2 medium without iron sulfate (iron-limited M2). Excess iron conditions were achieved by supplementing M2 medium with 100 μM FeSO4 instead of 10 μM FeSO4. The growth curves in each condition are shown in Supplementary Figure S1.
RNA-seq Expression Profiling
The NA1000 and SP0057 strains (pre-cultivated in 3 ml M2 overnight) were diluted in duplicate into 10 ml M2 to an OD600 nm of 0.1 and incubated at 30°C with agitation (250 rpm) until reaching OD600 nm of 0.5. At this time DP was added to a final concentration of 100 μM to one aliquot of the NA1000 culture and these were further incubated for 2 h. Total RNA was isolated from independent cultures (1 ml) treated with RNAprotect Bacteria Reagent (Qiagen) and purified with Qiagen RNeasy Plus Mini Kit (Qiagen). RNA samples were quantified using NanoDrop 1000 (Thermo Scientific) and submitted to the removal of ribosomal RNA using Ribo-Zero Magnetic kit (Illumina). The quality of the isolated RNA and the percentage of rRNA present were checked with RNA 6000 Pico Kit using Agilent 2100 Bioanalyzer (Agilent). Complementary DNA was generated according to instructions of TruSeq RNA Sample Prep Kit (Illumina). The cDNA libraries were normalized to 4 nM using the Kapa Biosystems kit (Kapa Biosystems) for library quantitation prior to cluster generation. The samples were sequenced using the MiSeq (Illumina) platform according to the manufacturer’s instructions. RNA-seq experiments were carried out in biological duplicates.
cDNA reads were quality checked using Bioconductor’s 3.5 (Huber et al., 2015) Rqc 1.10.2 and trimmed using Trimmomatic 0.36 (Bolger et al., 2014) to remove adapters and low quality ends. Pre-processed reads were aligned to C. crescentus NA1000 reference genome using Bowtie 1.2 (Langmead et al., 2009) with the “-m 1” option to keep only uniquely mapped reads. Alignment files were further processed by SAMtools 1.3.1 (Li et al., 2009) and read counting was performed for each gene feature using GenomicAlignments 1.12.1 (Lawrence et al., 2013). The count matrix was input in DESeq2 1.16.1 (Love et al., 2014) and analyzed using group design. We generated two contrasts to find differentially expressed genes: (i) wt DP-treated for 2 h vs. wt and (ii) fur mutant vs. wt. Genes were considered differentially expressed if satisfying |log2fold change|≥ 1 and q-value < 0.05. We used information available in KEGG pathways (Kanehisa et al., 2017) to group differentially expressed genes in functional categories and also performed manual curation.
Real-Time PCR
The gene expression profile was assessed by RT-qPCR. Cultures pre-cultivated in 3 ml M2 overnight were diluted into two aliquots of 10 ml M2 to an OD600 nm of 0.05 and incubated at 30°C with agitation (250 rpm) until reaching OD600 nm of 0.3. At this time DP was added to one aliquot to a final concentration of 100 μM and these were further incubated for 2 h. When iron limited M2 was used, the overnight culture was diluted in 10 ml M2, and when the cultures reached an OD600 nm of 0.3 they were centrifuged for 20 min at 5,000 ×g and resuspended in the same volume of iron limited M2 (no iron added). One of the aliquots received FeSO4 to a final concentration of 10 μM, and the cultures were incubated at 30°C until the appropriate time. Total RNA was isolated from 5 ml cultures either in M2, iron limited M2 (with no iron addition) or DP-treated M2 for 2 h as indicated for each experiment. RNA was purified using TRIzol® RNA (Invitrogen), treated with deoxyribonuclease I (Invitrogen) and complementary DNA was synthesized using the Superscript III First-Strand Synthesis System kit for RT-PCR (Invitrogen) with random hexamers. RT-qPCR reactions were performed on the Step One Plus Real Time PCR System (Applied Biosystems), using SYBR Green PCR Master Mix (Applied Biosystems) as detection reagent, in technical triplicates. RT-qPCR reactions were carried out in a final volume of 12 μl Power SYBR Green PCR master mix containing 10 μM of each primer, and 50 ng.μl-1 cDNA. The conditions were 95°C for 10 min, followed by 40 cycles comprising 95°C for 15 s and 60°C for 1 min. In order to confirm the amplification of a single product, dissociation curves were generated at the end of each PCR cycle. The 2-ΔΔCt method was used to calculate the relative change in gene expression of each gene (Pfaffl, 2001), and the analysis was performed using the StepOne Software v.2.2. Specific primers were designed using primer3 software (Untergasser et al., 2012) for each gene for validation of RNA-seq data and gene CCNA_03876, coding the transcription termination factor Rho, used as reference control (Supplementary Table S1). All determinations were performed in biological duplicates and at least in technical triplicates and the mean values ± standard deviation (SD) are reported.
β-Galactosidase Activity Assays
C. crescentus NA1000 or ΔrecA strains harboring the pRKlacZ290 vector containing the imuA promoter (imuA/lacZ fusion) were grown in M2 medium up to midlog phase (OD600 nm of 0.2). Each culture of NA1000 was divided into four equal aliquots, and one was left without additions throughout the experiment. To two other aliquots, either 100 μM FeSO4 (called iron-added M2) or 100 μM DP (called DP-treated M2) were added. The fourth aliquot was centrifuged and the cells were suspended in M2 medium without iron (called iron-limited M2). In the assays with the ΔrecA strain, the culture was divided in two aliquots, one without additions and the second received 100 μM DP. The cultures were further incubated during the appropriate times and the β-galactosidase activity was determined (Miller, 1972).
Microscopy
C. crescentus NA1000 and the fur mutant were grown in M2 medium up to midlog phase (OD600 nm of 0.2–0.3). Each C. crescentus NA1000 culture was divided into equal aliquots, and one was left without additions throughout the experiment. To one other aliquot, DP to 100 μM was added and the cultures were further incubated for 2 h. One aliquot was centrifuged and the cells were suspended in the same volume of M2 medium without iron (iron-limited M2) for 4 h. To one of the aliquots H2O2 was added to 10 mM and incubated for 15 min. In the assays with the fur strain, the culture was divided in two aliquots, one was left without additions and the second received DP as above. Dihydrorhodamine 123 (Sigma D1054) was added to the cultures to a final concentration of 20 μM and the cultures were further incubated for 60 min. After the cells were washed they were resuspended in phosphate-buffered saline solution. The slides were observed using a fluorescein filter in a Nikon Eclipse TiE microscope, using a 100× objective and images were captured in an Andor EM CCD i-Xon camera.
Survival and Mutagenesis Tests
For the mutagenesis tests, cultures of the C. crescentus NA1000 and recA strain were grown for 10 h in 10 ml of M2 medium. The cells from each culture were pelleted by centrifugation for 20 min at 5,000 ×g and resuspended in iron limited M2 (no iron added) to the same previous volume. The cultures were adjusted to OD600nm of 0.2, and divided into two 5 ml aliquots. One of the aliquots received FeSO4 to a final concentration of 10 μM, and the cultures were incubated at 30°C with agitation for 12 h. One ml of each culture was centrifuged at 5,000 ×g and the cells were resuspended in 1 ml M2. Two hundred microliters were diluted in 1 ml M2 and incubated at 30°C with agitation for 24 h. Serial dilutions and plating on PYE were performed to determine the total number of viable cells. To determine the number of Rif-resistant mutants, 1 ml of each culture was plated on PYE medium containing 100 μg/ml rifampicin.
For the UV survival tests, C. crescentus NA1000, SP0057 and GM10 overnight cultures were diluted to an optical density at 600nm (OD600nm) of 0.05 in M2 medium. Each culture was divided into three equal aliquots, and to one (control) no additions were made throughout the experiment. One aliquot of the NA1000 culture was pre-incubated with 100 μM DP for 2 h. One aliquot was centrifuged and the cells were suspended in iron-limited M2 for the appropriate times. After the respective times, each culture was irradiated with 120 J/cm2 UV light (NA1000), 60 J/cm2 (SP0057) and 10 J/cm2 (GM10). Serial dilutions from each control and treated cultures were plated on M2 plates, and these were incubated in the dark for 48 h at 30°C for CFU counting.
Analysis by HPLC-ESI/MS/MS of 8-oxo-dG
DNA extraction and enzymatic hydrolysis was carried out as described in Sanchez et al. (2018) with the following modifications. The bacterial pellet was resuspended in lysis buffer containing 0.1 mM desferroxamine. Treatment with 60 μL of proteinase K (20 g/L) was performed together with 675 μL of a 10% (w/v) solution of SDS for 2 h at 37°C. The DNA pellet was solubilized in a 3 mM solution of desferroxamine mesylate, and concentration was measured by A260 nm. For the hydrolysis of 30 μg of DNA, 5 μl of 3 M sodium acetate buffer pH 5.0 and 4 U of nuclease P1 were added to the sample, which was incubated at 37°C for 30 min at 450 rpm. Following this step, 5 μl of 3 M Tris-Cl buffer pH 7.4 and 5 μl of phosphatase buffer containing 100 mM Tris-Cl, 5 mM MgCl2, 0.2 mM ZnCl2 and 50% (v/v) glycerol were added. After addition of 2 U of alkaline phosphatase, the reaction mixture was incubated at 37°C for 1 h at 450 rpm.
Online HPLC/ESI/MS-MS analyses were carried out as described in Garcia et al. (2010) using an API-4000 QTRAP mass spectrometer (Applied Biosystems, Foster City, CA, United States). 8-oxo-dG in the DNA samples were detected by selected reaction monitoring (SRM) using an analytical column (Luna C18(2), 250 mm × 4.6 mm i.d., 5 μm, Phenomenex, Torrance, CA, United States). 8-oxo-dG was eluted from this column with a gradient of water and acetonitrile containing 0.1% formic acid with the following method: from 0 to 10 min, 5 to 12% acetonitrile, and 0.5 to 0.25 mL/min; from 10 to 15 min, 12% acetonitrile and 0.25 mL/min; from 15 to 40 min, 12 to 40% acetonitrile and 0.25 mL/min; from 40 to 41 min, 40 to 95% acetonitrile and 0.25 to 0.5 mL/min; from 41 to 45 min, 95% acetonitrile and 0.5 mL/min; from 45 to 46 min, 95 to 5% acetonitrile and 0.5 mL/min; from 46 to 50 min, 5% acetonitrile and 0.5 mL/min. An isocratic pump was used to simultaneously load a second column [Luna C18(2), 150 mm × 2 mm i.d., 3 μm, Phenomenex, Torrance, CA, United States]. The DNA hydrolysates containing 3.6 fmol of the [15N5]-8-oxo-dG internal standards were injected into the system described above. The [M + H]+ ions corresponding to the m/z values 284.1/167.9 (8-oxo-dG) and 289.1/172.9 ([15N5]-8-oxo-dG), were monitored with a dwell time of 150 ms.
H2O2 Dosage Assays
Colonies from the NA1000 strain were inoculated in 3 ml of M2 medium and grown for 16 h at 30°C with agitation. These two independent cultures (biological replicas) were then diluted to an OD600nm of 0.1 in 15 ml of M2 and incubated at 30°C with agitation until reaching an OD of 0.25. The cultures were divided into four 7 ml aliquots, and cells were pelleted by centrifugation for 20 min at 5,000 rpm. Cells from two aliquots were suspended in M2 and from the other two in iron limited M2 (no iron added) in the same previous volume. One culture in M2 and one culture in iron limited M2 received DP to a final concentration of 100 μM for each replica. The cultures were incubated for 3 h at 30°C with agitation. For the assays 1.5 ml from each culture was centrifuged at 12,000 ×g and the cells resuspended in 100 μl of 20 mM Tris-Cl pH 7.4.
Determination of the amount of H2O2 was carried out as described in Barros et al. (2003). Briefly, 50 μl of each bacterial suspensions were added to 3 ml of a solution containing 50 μM Amplex Red (Thermo Scientific) and 1.0 U/ml horseradish peroxidase (HRP; Sigma-Aldrich), in 20 mM Tris-Cl pH 7.4. The rate of Amplex Red oxidation to the fluorescent product (resorufin) was analyzed at 25°C using a Cary Eclipse (Varian) fluorescence spectrophotometer equipped with continuous stirring.
Statistical Analyses
Error bars shown on graphs indicate standard deviations or standard error, as indicated. Statistical significance was tested by one-way ANOVA using the Tukey post-test for multiple comparisons or by t-test when only two conditions were being compared.
Results
Expanding the Low Iron Stimulon in C. crescentus
Previous works using DNA microarrays and bioinformatics have characterized the iron limitation response in C. crescentus in the rich amino acid-based medium PYE (da Silva Neto et al., 2009, 2013). In that case, PYE was supplemented with 100 μM FeSO4 or 100 μM DP, respectively, to allow identification of differentially expressed genes. In order to further characterize the response to iron limitation of this oligotrophic bacterium, RNA-seq-based global transcriptional analysis was performed from cells grown in M2 minimal medium. The RNA-seq experiments were carried out using RNA samples prepared from two independent biological cultures. To define the C. crescentus iron-regulated genes we compared the transcriptome of wt cells grown in M2 treated or not with the iron chelator 2′,2-dypiridyl for 2 h (DP-treated). The addition of DP significantly altered the expression of 492 genes (256 upregulated genes and 236 downregulated genes) (Figure 1). To identify the genes regulated by Fur, we compared the transcriptome of wt cells with the fur mutant in M2. Overall, 93 genes were differentially expressed in the fur mutant (40 upregulated genes and 53 downregulated genes) (Figure 1).
FIGURE 1.
Overview of the C. crescentus iron-regulated and Fur-regulated genes identified by RNA-seq analyses. The Venn diagrams indicate the number of up-regulated (green) and down-regulated (red) genes from experiments comparing wild type cells exposed to DP-treated M2 versus M2 or comparing the fur mutant strain versus wild type strain both in M2. Differentially expressed genes were considered those with log2 fold change ≥1 or ≤1 and q-value < 0.05.
We found 23 genes upregulated and 38 genes downregulated both in DP-treated condition and in the fur mutant (Supplementary Table S2). Seventy-four percent (17 genes) of the upregulated genes were genes previously identified by DNA microarray analysis in PYE, and six genes are new. Of these, four genes encoded hypothetical proteins, one a serine protease (CCNA_01341) and another the amelogenin/CpxP-related protein (CCNA_03997). Of the 38 downregulated genes, 11 genes were previously known as iron- and Fur-regulated in PYE, and 17 genes belong to the regulon controlled by FixL-FixK-FixT, among them the cytbb3 operon (CCNA_01467-68-69-70-71-72-73), CCNA_01475 encoding the family outer membrane cation channel (ompW) and CCNA_01476-77 that encode the CRP-family transcription regulator (ftrB) and oxygen-independent coproporphyrinogen-III oxidase (hemN). In agreement with the results obtained with DNA microarrays, these data indicate that Fur might activate, probably indirectly, the oxygen signaling network.
In general, the Fur regulon determined in M2 (23 genes) was smaller than that previously determined in PYE (42 genes), while the number of genes induced only in DP-treated condition was larger in M2 (233 genes versus 66 genes upregulated in PYE). Interestingly, some genes that are regulated by Fur and were highly responsive to DP-treatment in PYE (da Silva Neto et al., 2009, 2013) did not show an increase in their transcript levels in DP-treated M2 medium. For example, CCNA_03023 that encodes a TonB-dependent receptor and CCNA_02275 that were upregulated gene in DP-treated PYE (15.3- and 64.8-fold, respectively) were below the cutoff in our experimental conditions. Still, these genes were upregulated in the fur mutant (CCNA_03023, 3.3-fold and CCNA_02275 5-fold), confirming that their expression is under Fur regulation, although the levels of expression are differently modulated according to the culture medium (Supplementary Table S2).
Eight previously predicted sRNAs (Landt et al., 2008; Schrader et al., 2014) were identified as differentially expressed in our experiments. One of these (R0117) was upregulated both in DP-treated condition (16.3-fold) and in the fur mutant (3.6-fold). This gene encodes a 1-kb RNA that maps to a region where previously annotated transcripts were shown to be regulated by Fur (da Silva Neto et al., 2013). One sRNA gene (R0088) was upregulated only in the fur mutant (7.7-fold), while six (R0180, R0119, R0116, R0049, R0080, and R0077) were upregulated only in DP-treated condition, varying from 3.6-fold for R0077 to 45.7-fold for R0180 (Supplementary Table S2). We could not identify a conserved Fur-box upstream of any of the sRNAs, including R0088, suggesting that it is indirectly regulated by Fur. The other six sRNAs could be important for the iron limitation response, in a Fur-independent manner, possibly regulating the expression of subsets of genes.
Several Transcription Factors Are Induced in Iron Limitation
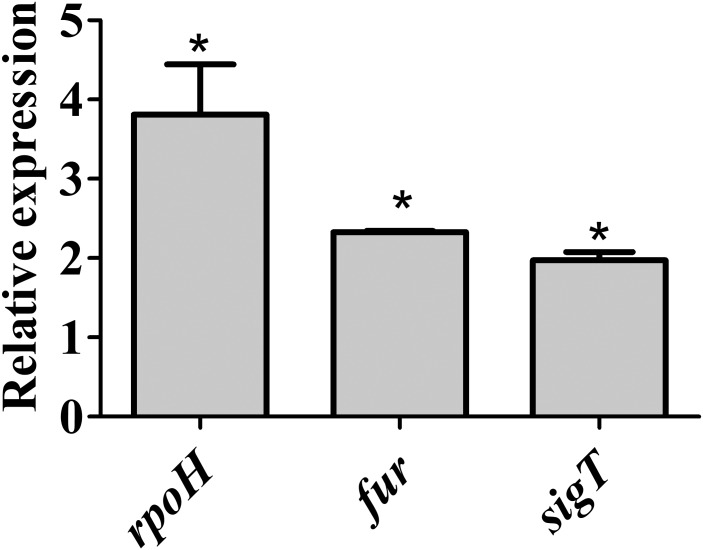
In M2 treated with DP the induction of 16 genes encoding transcription regulators was observed (Supplementary Table S2), of which several are involved in controlling stress response in C. crescentus. These include the RNA polymerase sigma factor rpoH (8.5-fold) and three ECF sigma factors (σE, σT, and σU, 5.9-, 5.5-, and 12.7-fold, respectively) as well as the σT regulatory systems LovR/LovK (lovR, 5.3-fold) and PhyR/PhyK/NepR (3.9-, 4.8-, and 6.7-fold, respectively) (Foreman et al., 2012). While the RpoH and several heat shock genes were previously identified as induced in DP-treated PYE (da Silva Neto et al., 2013), the other systems were identified only in the conditions used in this work. Surprisingly, the lexA gene encoding the SOS system regulator was also induced in DP-treated condition (8.4-fold), along with many genes belonging to the LexA regulon that will be discussed below (Supplementary Table S2). fur was not classified as upregulated in RNA-seq analysis although it presented a twofold induction, because it did not attend our statistical criteria. To further verify the expression of these regulators under iron limitation, we used RT-qPCR to measure the transcript levels of fur, rpoH, and sigT. The results show that all genes were induced in iron limitation generated by resuspending the cells in M2 without added iron (Figure 2).
FIGURE 2.
Expression of fur, sigT, and rpoH in wild type strain under iron limitation. Gene expression was determined by RT-qPCR using RNA from cells grown in iron limited M2 (for 4 h) relative to the expression in M2. Bars with asterisks (∗) are significantly different to wt growing in M2 medium (P < 0.05) by Student’s t-test.
Iron Deficiency Induces the SOS Response
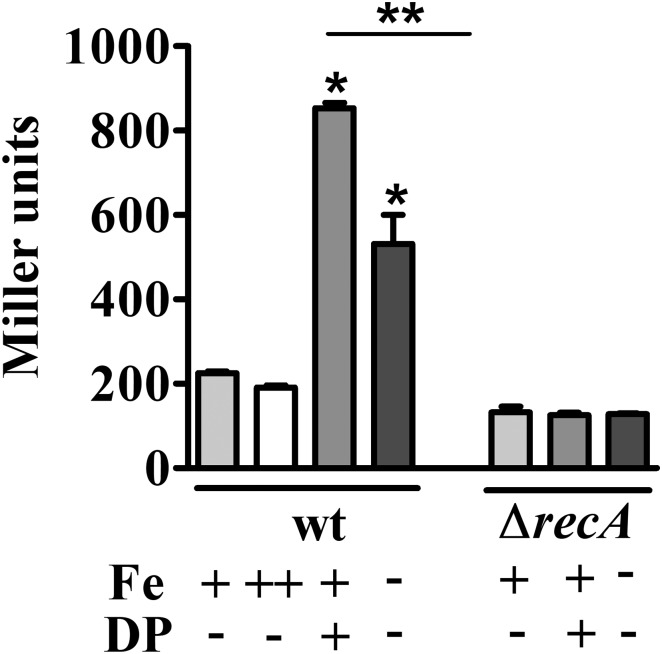
To discriminate whether the upregulation of genes belonging to the SOS response was caused by iron limitation or due to a secondary effect of the iron chelator used, the expression of the imuA-imuB-dnaE2 operon was measured by β-galactosidase activity using a transcriptional fusion to lacZ (PimuA-lacZ) (Figure 3). This operon is induced in response to DNA lesions in a RecA-dependent manner, and the three genes are required for the error-prone processing of DNA lesions (Galhardo et al., 2005). The results showed that imuA is induced under iron limitation conditions in both DP-treated M2 (containing the chelator DP) and in iron-limited M2 (no iron added), compared to M2 or iron-supplemented M2. The levels of β-galactosidase activity were slightly different between DP-treated M2 and iron-limited M2, but both were higher than the controls. These results indicated that the SOS response is being activated by iron limitation, suggesting that DNA damage is occurring in this condition. In order to verify if imuA induction was in fact responding to the SOS regulators, we analyzed imuA expression in the recA mutant (Figure 3). The results showed that the induction of imuA under iron limitation is dependent on RecA, indicating that it was induced by the SOS regulatory system.
FIGURE 3.
Determination of iron-dependent regulation of imuA expression. Expression was determined by β-galactosidase activity assays of each strain harboring imuA/lacZ transcriptional fusions. Cultures were grown in either M2 (Fe+/DP–), iron-supplemented M2 (with 100 μM FeSO4, Fe++/DP–), DP-treated M2 for 4 h (with 100 μM DP, Fe+/DP+) or iron-limited M2 (Fe–/DP–) for 4 h. The ΔrecA mutant was grown in the same conditions, except for iron supplemented M2. Bars with asterisks (∗) are significantly different to wt grown in M2; and (∗∗) indicates that results from DP-treated M2 and iron-limited M2 are significantly different (P < 0.05) by one-way ANOVA test.
Iron Deficiency Generates Oxidative Stress
The genes encoding the catalase-peroxidase and two superoxide dismutases, as well as several genes coding for oxidative stress response (glutaredoxin, glutathione S-transferase, peroxiredoxin, glutathione synthetase, thioredoxin) were upregulated upon DP-treated conditions (Supplementary Table S2). To determine if the reason for the activation of SOS response could be a state of oxidative stress, we tested if iron limitation could cause oxidative stress in C. crescentus.
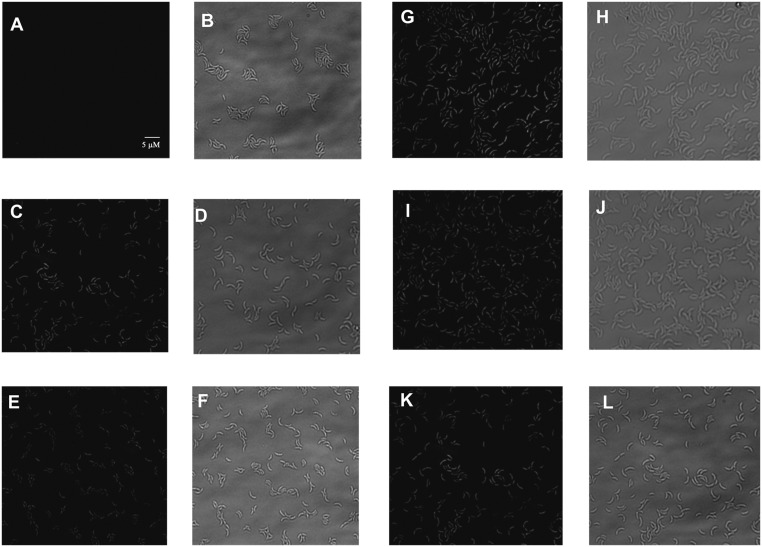
Firstly, this was done by incubating exponential phase cells with dihydrorhodamine 123. This compound is able to penetrate the cells and becomes fluorescent as a result of intracellular oxidation. The cells were analyzed by fluorescence microscopy (Figures 4A,C,E,G,I,K) and light microscopy (Figures 4B,D,F,H,J,L) to verify the amount of cells with visible fluorescence. While the wt cells in M2 showed no fluorescence (Figures 4A,B), when exposed to 100 μM DP (Figures 4C,D) or grown in iron-limited medium (Figures 4E,F) all cells were fluorescent after 2 h, confirming that iron limitation leads to an oxidative state. Interestingly, the fur mutant showed fluorescence both in M2 medium (Figures 4G,H) and in the presence of 100 μM DP (Figures 4I,J), suggesting that Fur could be involved in preventing oxidative stress in C. crescentus. As a positive control, wt cells were incubated with H2O2 for 15 min (Figures 4K,L).
FIGURE 4.
Determination of the oxidative stress state of C. crescentus NA1000 in response to iron levels. Wild type NA1000 strain cultures were grown in M2 (A,B), DP-treated M2 (DP for 2 h) (C,D), or iron-limited M2 for 4 h (E,F); and cultures of the fur mutant were grown in M2 (G,H) or DP-treated M2 for 2 h (I,J). As a control, NA1000 cultures received H2O2 for 5 mM and incubated for 15 min (K,L). Samples were treated with dihydrorhodamine 123 and analyzed by fluorescence microscopy using a fluorescein filter (A,C,E,G,I,K) and under light microscopy (B,D,F,H,J,L).
As a second confirmation of these results, determination of H2O2 in NA1000 cultures was carried out by measuring the rate of oxidation of Amplex Red in the presence of HRP. Cells from NA1000 cultures grown either in M2, or incubated for 3 h in iron limited M2 (no iron added), DP-treated M2 or DP-treated iron limited M2 were used for H2O2 determination (Table 1). The results showed that in the three cultures subject to iron limitation the rate of Amplex Red oxidation was 2- to 2.5-fold higher than in M2 (P < 0.05), indicating that there is more endogenous H2O2, which could be measured in intact cells since it is freely diffusible.
Table 1.
Determination of hydrogen peroxide generated in C. crescentus NA1000 strain in different growth conditions.
| Growth condition | pmol H2O2/DO/minc |
|---|---|
| M2 | 20.81 ± 2.43 |
| Iron limited M2a,d | 51.50 ± 3.17 |
| DP-treated M2b,d | 49.50 ± 4.30 |
| DP-treated iron limited M2a,b,d | 42.91 ± 1.67 |
aThe cells pellets were resuspended in iron limited M2 (no iron added) in the same previous volume. See “Materials and Methods” section for details. bThe culture was incubated with 100 μM DP for 3 h. cThe results are the average of two independent assays ±SD. dSignificantly different from the M2 growth condition (P < 0.05) by one-way ANOVA test.
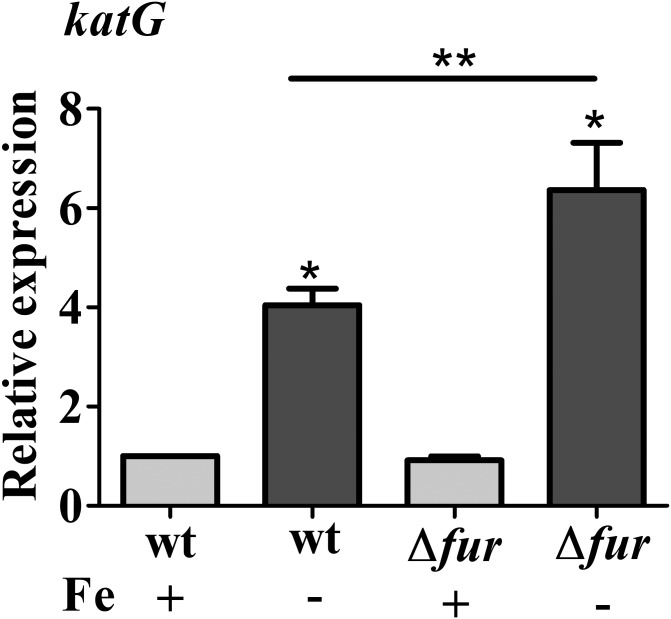
Previously, it was shown that OxyR activates the expression the C. crescentus katG gene in response to hydrogen peroxide, and katG was not regulated by Fur (Italiani et al., 2011). To determine whether the oxidative stress observed in the fur mutant and in the wt cells under iron limitation could lead to the OxyR-mediated response, the expression of katG was measured by RT-qPCR (Figure 5). However, there is still an increase in katG expression in the fur mutant in iron limited M2 as compared to M2, to a similar extent as in wt, suggesting that iron limitation is causing this induction.
FIGURE 5.
Expression of katG in response to iron limitation. Expression was determined by RT-qPCR from NA1000 (wt) and fur mutant (Δfur) cultures grown in M2 or iron limited M2 (no iron added) for 4 h. Bars with asterisks (∗) are significantly different from wt cells in M2 medium (∗∗) indicates that the results of Δfur and Δfur in iron limited M2 are significantly different from each other (P < 0.05 by one-way ANOVA test).
Interestingly, several toxin-antitoxin encoding genes were highly upregulated in DP-treated cells, such as the ParD2/ParE2 (22.8- and 23.3-fold induction, respectively), the antitoxin protein RelB3 (23.6-fold), the HigB toxin (17.2-fold) and the DNA replication inhibitor toxin SocB/antitoxin SocA (17.2- and 14.0-fold induction, respectively). Previous work has shown that ψ-parDE2 and relBE3 are induced in response to oxidative stress (Fiebig et al., 2010). The higB gene is repressed by LexA, and HigB plays a role in mediating the intensity of the response to antibiotics together with the SOS response, in a regulatory interplay between LexA, HigBA, and AcrAB2-NodT (Kirkpatrick et al., 2016). In fact, in the DP-treated cells the acrAB2 genes were also induced threefold to fivefold.
The SOS Response Is Induced Under Iron Deficiency
The activation of SOS response in DP-treated C. crescentus cultures suggests that DNA damage is being generated in this condition. To establish the role of Fur in the SOS signaling network, the transcripts levels of lexA and imuA were measured by RT-qPCR in wt and the fur mutant cells grown in either M2 medium or DP-treated M2 for 2 h (Figure 6A). In DP-treated NA1000 cultures, both transcripts were upregulated twofold (lexA) and fourfold (imuA) in wt cells, whereas in the fur mutant the expression was increased 3- and 16-fold comparing to wt in M2 medium, respectively. This induction is not observed in the recA mutant (Figure 6A). Since the fur mutant in M2 is under oxidative stress, but does not show an upregulation of the SOS response, this suggests that iron deficiency, and not the oxidative stress generated, is triggering the activation of SOS response. In fact, when both were grown in DP-treated medium, the SOS response is highly induced. The results also indicate that Fur may have a protective role against the resulting DNA damage, since the induction of both genes in the fur mutant was much higher than in the wt. To confirm that this effect was due to the iron limitation, the assays were repeated with the NA1000 strain grown in M2 or iron limited M2 (no iron added) (Figure 6B). The results were the same as with the DP-treated cultures, confirming that iron limitation is causing this response.
FIGURE 6.
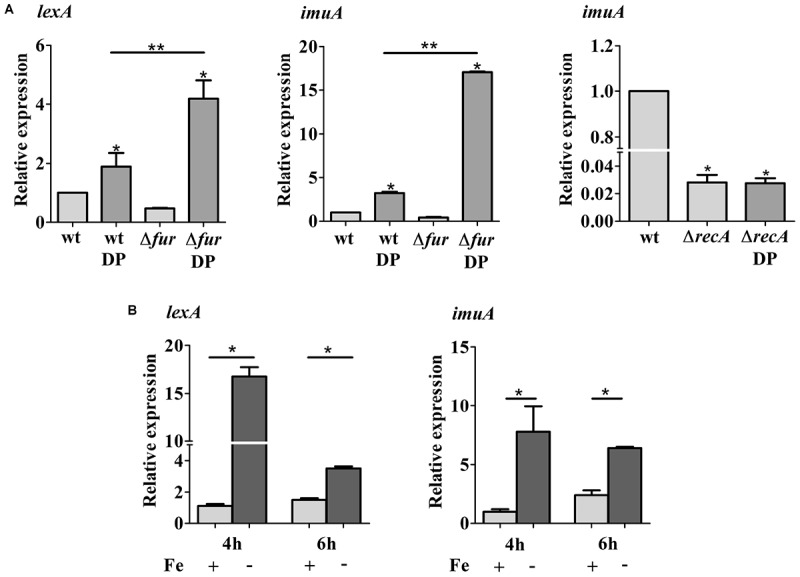
Expression of lexA and imuA. (A) Gene expression was assessed by RT-qPCR from wild type NA1000 (wt), the Fur mutant (Δfur) and recA mutant (ΔrecA) cultures grown in M2 and DP-treated M2 (DP) for 2 h. (B) Gene expression was assessed by RT-qPCR from NA1000 cultures grown in M2 and iron limited M2 (–Fe) for 4 h and 6 h. Bars with asterisks (∗) are significantly different from wt in M2 medium; (∗∗) indicates that the results. Gene expression was assessed by RT-qPCR from wild type NA1000 (wt), the Fur mutant (Δfur) and recA mutant (ΔrecA) cultures grown in M2 and DP-treated M2 (DP) are significantly different from each other (P < 0.05) by one-way ANOVA test.
In order to directly measure the increase in the levels of DNA damage that could be occurring in response to iron deprivation, we measured the amount of 8-oxo-dG in the DNA of cells incubated in M2 or iron limited M2 (no iron added) for 12 h. The results did not show any significant difference between the two treatments (M2, 8.45 ± 5.4 8-oxo-dG/105 dG, and M2 with no iron 8.18 ± 2.71 8-oxo-dG/105 dG), although the levels of 8-oxo-dG detected were at the limit of detection. However, we have to consider that this type of base alteration may not be the most indicative for Caulobacter, and other types of oxidative lesions may be occurring.
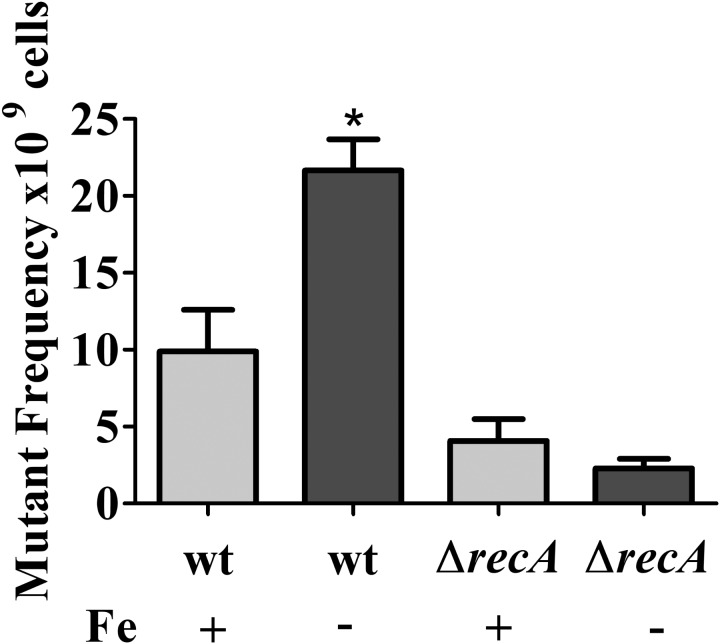
In the mutagenesis tests, cultures of NA1000 and recA strains grown in M2 or iron limited M2 for 12 h were plated on PYE containing 100 μg/ml rifampicin. The results showed that the NA1000 cultures grown in iron limited medium presented an increase in the amount of Rif-resistant mutants (Figure 7), but this is not the case for the recA mutant. These results indicate that iron deprivation leads to an increase in mutagenesis in C. crescentus, mediated by the SOS response. These results were consistent with the upregulation of the SOS genes in the same conditions, suggesting that increase in mutagenesis might be a result of error-prone DNA polymerases activity.
FIGURE 7.
Effect of iron limitation on mutagenesis. Midlog phase cultures of C. crescentus NA1000 were grown in M2 (Fe+) or iron limited M2 (Fe–) (see the section “Materials and Methods”), and the cultures were incubated for 12 h. Cultures were plated on PYE to determine the total CFU counts. To determine the number of Rif-resistant mutants, 1 ml of each culture after treatment was plated on PYE with 100 μg/ml rifampicin. The relative mutant frequency was calculated by dividing the average of the Rif-resistant CFU by the average of the total CFU. The experiments were done in at least six independent biological assays with two technical replicas each. Bars with asterisks (∗) are significantly different to the wt control (P < 0.05) by one-way ANOVA test. Bars indicate standard error.
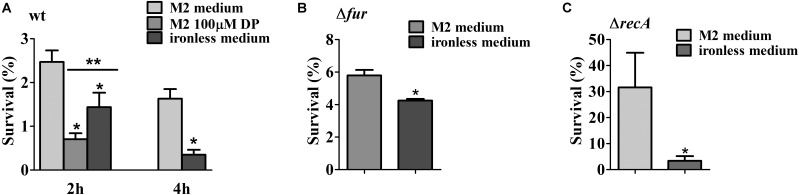
The activation of DNA repair enzymes protects the DNA against exogenous physical agents like UV light. In order to verify if cells under iron limitation were more resistant to UV light-induced damage due to the activation of the SOS response, midlog phase cultures of the NA1000 strain grown in M2 were divided into aliquots. One was left without addition, one received 100 μM DP for 2 h, and in two samples cells were centrifuged and resuspended in iron-limited M2 and further incubated for either 2 h or 4 h (Figure 8A). The cultures were then irradiated with 120 J/cm2 UV light, and the results showed that cells that were in iron limitation were more sensitive to UV than those without treatment. The same result was obtained with the fur mutant, (irradiated with 60 J/cm2 UV light), and the recA mutant (irradiated with 10 J/cm2 UV light) grown in M2 or iron limited M2 (Figures 8B,C). This indicates that the previous activation of the SOS response in iron limitation is not only not sufficient to mount a protection against damage caused by UV light, but the cells being already stressed contributes for a poor response to UV-generated stress.
FIGURE 8.
Survival test of the NA1000 strain exposed to UV light. (A) NA1000 cultures were grown either in M2, DP-treated M2 for 2 h or iron-limited M2 for 2 h or 4 h and then irradiated with 120 J/cm2 UV-light. (B) SP0057 cultures were grown either in M2, or iron-limited M2 for 4 h and then irradiated with 60 J/cm2 UV-light. (C) GM10 cultures were grown either in M2, or iron-limited M2 for 4 h and then irradiated with 10 J/cm2 UV-light. Survival was determined relative to the colony counts at time 0 h, before irradiation (100%). The results shown are the average of three independent experiments. Bars with asterisks (∗) are significantly different to M2 medium; (∗∗) indicates that DP-treated M2 and iron-limited M2 are significantly different (P < 0.05) by one-way ANOVA test (A) or Student’s t-test (B,C).
Discussion
The Fur regulon in C. crescentus grown in rich amino acid-based medium was extensively studied and its targets were determined in previous works (da Silva Neto et al., 2009, 2013). However, in minimal medium cells are in a metabolic biosynthetic mode, more similar to the oligotrophic environments where this bacterium thrives, so the consequences of iron limitation for C. crescentus grown in these conditions were investigated. The previous whole transcriptome study was carried out in PYE medium that received either 100 μM FeSO4 or of 100 μM DP, which allowed the Fur regulon to be almost completely identified. In this work, the M2 medium was compared with DP-treated M2 medium, and allowed a complementary identification of iron limitation-induced genes. As expected, in defined M2 medium the genes for nitrogen and amino acid metabolism that were differentially expressed in DP-treated PYE were not identified, showing that the growth conditions affect the iron limitation response.
Among the iron limitation-induced genes were those encoding seven predicted small RNAs (sRNAs) (Landt et al., 2008; Schrader et al., 2014), being only one differentially expressed in the fur mutant compared to wt. The regulatory networks of several bacteria show that Fur is directly regulating the expression of iron-regulated regulatory RNAs (Masse and Gottesman, 2002; Wilderman et al., 2004; Masse et al., 2005, 2007; Mellin et al., 2007; Gaballa et al., 2008; Ghosh et al., 2017; Jackson et al., 2017). Although in this work we could detect only one Fur-regulated sRNA, we cannot exclude the possibility that there are some sRNAs that are regulated by Fur in other medium conditions. One caveat of the method used is that the RNA-seq was not strand-specific, therefore we may not detect all the differentially expressed sRNAs genes when they overlap with other genes. A more thorough investigation will bring light to this matter.
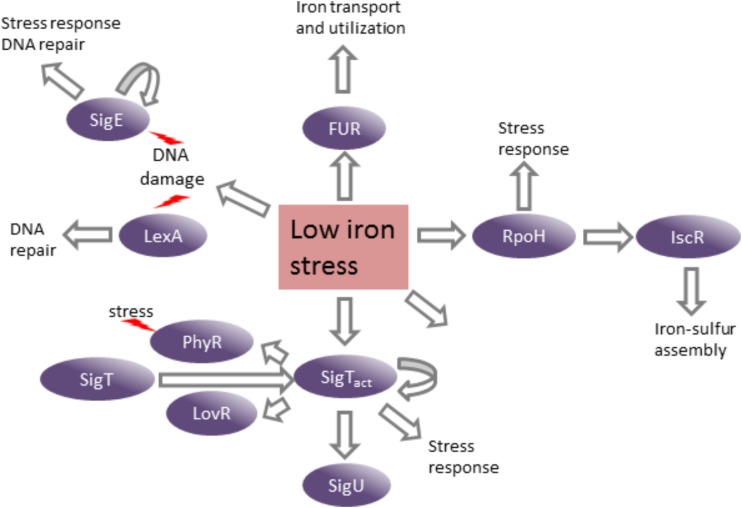
The genes encoding several transcription factors were upregulated in the DP-treated medium in a Fur-independent manner, which could be mediating the regulation of subsets of genes in response to the stress generated by iron limitation (Figure 9). Three extracytoplasmic function (ECF) sigma factors (σT, σU, and σE) regulate genes involved in protecting cells against stress, mainly oxidative damage. It has been shown that the σE is responsive to singlet oxygen, UV-A, cadmium, and organic hydroperoxide (Lourenço and Gomes, 2009). The SigT regulon is important for the response to osmotic and oxidative stresses (Alvarez-Martinez et al., 2007; Britos et al., 2011; Lourenço et al., 2011). Although sigT and most of its known regulon fell below our cutoff criterion in the fur mutant, sigT and sigU were highly activated in the DP-treated medium.
FIGURE 9.
Consequences of low iron stress on C. crescentus gene expression. The induction of several transcription factors in response to stress generated by iron limitation leads to the activation of their regulons and distinct stress responses. Curved arrows indicate autoregulation.
The induction of several genes related to oxidative stress response was in agreement with an endogenous oxidative stress state identified in C. crescentus cells under iron limitation. Since katG, the genes for superoxide dismutases, and several redoxins were upregulated in iron limitation, this indicates that the oxidative stress observed is caused by reactive oxygen species (ROS) of different types. The increase of oxidative stress under iron limitation has also been observed in the cyanobacterium Anabaena sp., but not in E. coli or B. subtilis. Iron starved Anabaena PCC7120 exhibit 100-fold increase of ROS compared to non-starved cells (Latifi et al., 2005, 2009). Interestingly, a striking difference is that Caulobacter and Anabaena are strict aerobes, while E. coli and B. subtilis can grow in anaerobiosis. In Anabaena PCC7120, the FurA regulator has been shown to control genes related to protection against oxidative stress (Gonzalez et al., 2016). Contrasting with this, E. coli does not exhibit a significant increase in the ROS levels under iron limitation (Latifi et al., 2005), and it has been proposed that Fur represses iron uptake as an adaptive response to protect against hydroxyl radicals generated by Fenton chemistry (Faulkner and Helmann, 2011). In B. subtilis the upregulation of genes encoding enzymes for peroxide stress response (ahpCF, mrgA, and katA) was observed in iron limitation (Baichoo et al., 2002); however, there was no increase of ROS levels in cells in these conditions (Latifi et al., 2005). The activation of these genes in B. subtilis can be explained by the fact that fur belongs to the PerR regulon (Baichoo et al., 2002; Faulkner and Helmann, 2011). Under iron deficiency PerR becomes an inactive repressor leading to the activation of fur, ahpCF, mrgA, and katA gene expression (Faulkner and Helmann, 2011).
Interestingly, the gene encoding the ferritin-like DNA-binding protein Dps was induced specifically in DP-treated conditions. In E. coli Dps confers protection against oxidative stress caused by H2O2, and it was proposed that this is due its ability to bind DNA and inhibit the Fenton reaction. In C. crescentus, dps is regulated by SigT (Lourenço et al., 2011) and Dps has an important role in protecting the cell from oxidative stress (de Castro Ferreira et al., 2016). However, while in E. coli dps is induced by starvation and regulated by OxyR under oxidative stress (Almiron et al., 1992; Altuvia et al., 1994), in C. crescentus its regulation is not responsive to hydrogen peroxide, and is independent of Fur and OxyR (de Castro Ferreira et al., 2016). Its regulatory pattern indicates that its main role in C. crescentus is protecting the DNA against the damage generated in low iron condition.
The most remarkable difference in the stimulon determined in this work was the induction of the SOS regulon. The presence of single stranded DNA activates the recombinase RecA that stimulates the autocatalytic cleavage of the transcriptional repressor LexA, inducing the SOS regulon that is comprised of DNA repair genes. The SOS regulon in C. crescentus was previously characterized (da Rocha et al., 2008; Modell et al., 2011). Twenty-two genes involved in DNA repair (18 belonging to SOS regulon) were upregulated in DP-treated medium, including lexA and recA. We showed that the iron limitation induction of imuA, as an SOS indicator gene, was dependent on RecA, so the induction of the SOS regulon was caused by the presence of DNA damage. Moreover, two genes (CCNA_01596 and CCNA_02930) that are induced by genotoxic treatments even though they do not belong to the SOS regulon are also upregulated in response to DP treatment (da Rocha et al., 2008; Modell et al., 2011, 2014), suggesting the response to DNA damage is broader than just the SOS system. Unexpectedly, despite the increase in expression of some genes belonging to the SOS regulon, cells grown in iron deficiency were more sensitive to UV. This could be explained by a distinct spectrum of specificity toward DNA lesions among the DNA repair systems, and different thresholds of induction. The response to UV damage is carried out mainly by the UvrABC system, which is part of the SOS response, but these genes (CCNA_02673, CCNA_02975, and CCNA_03076) were not induced in the DP-treated condition. In fact, these genes are poorly induced even in the absence of the lexA repressor (uvrA, 3.5-fold; uvrC, 1.7-fold, and uvrB not induced) (da Rocha et al., 2008). The DNA lesions caused by oxidative stress are distinct from the typical UV-generated DNA lesions, requiring a specific set of repair enzymes, and the concomitant exposure to oxidative stress (generated by iron limitation) and UV is probably excessive for the cell responses.
The fact that iron is a cofactor for several enzymes necessary to help protecting against ROS (e.g., Fe-containing superoxide dismutases and heme-containing catalases/peroxidases) increases the complexity of this matter in what is the cause and what is the effect. The lack of iron could be disrupting the detoxification enzymes activity to a point when the cells start accumulating ROS. On the other hand, this does not seem to be the case in the fur mutant, since in this case iron uptake is constitutively active and these enzymes would be functional. A possible explanation for the increase in oxidative stress in low iron medium and in the fur mutant is an impairment of the respiratory function, mainly due to lower synthesis of the iron–sulfur groups for enzymes of the respiratory chain. In eukaryotic cells dysfunctions that prevent the assembly of mitochondrial iron-sulfur components generate mtDNA instability and ROS, and have been treated with antioxidants (Figueira et al., 2013). It was shown that in C. crescentus Fur directly activates the expression of sdhCBAD (succinate dehydrogenase) and nuoA-N (NADH dehydrogenase) operons (da Silva Neto et al., 2009), and in this work several genes encoding cytochromes were downregulated in DP-treated medium and in the fur mutant, noteworthy the ccoNOQP and cydAB operons, which belong to the FixK regulon. Moreover, in DP-treated medium the operon encoding ATP synthase was also downregulated.
It has been reported in E. coli that the fur mutant showed a significant increase of mutagenesis in aerobic conditions and the addition of ferrozine did not reduce the frequency of mutagenesis, suggesting that this strain is suffering DNA damage (Touati et al., 1995). Furthermore, in the E. coli fur mutant the SOS response was not activated, and the mutagenesis was proposed to be due to recombination defects (Touati, 2000). It was suggested that the fur mutant has a permanent influx of iron, and this intracellular overload of iron could produce DNA damage that the addition of ferrozine was not able to avoid (Touati, 2000). We previously demonstrated that the C. crescentus fur null mutant is highly sensitive to exogenously added H2O2 and tert-butyl hydroperoxide (da Silva Neto et al., 2009). On the contrary to what was seen for E. coli, we observed that under iron limitation wild type C. crescentus generates oxidative stress, and the fur mutant is already in an endogenous oxidative state. Moreover, iron limitation (Figure 5 and Supplementary Figure S2) triggered the induction of SOS system in both strains, suggesting that DNA damage did not occur by excessive iron uptake. Agreeing with this, the SOS response was not induced even when it was exposed to 100 μM of FeSO4 (Supplementary Figure S2). Taken together, these results suggest that in C. crescentus DNA damage is not generated by excess of free iron, although we cannot exclude the possibility that high endogenous iron levels are generating the oxidative stress observed in the fur mutant.
The increase in DNA damage observed in C. crescentus could be a consequence of the oxidative stress generated under iron limitation. Previous studies had shown that the lesions caused by H2O2 in E. coli are dependent on iron concentration, with higher H2O2 concentrations being necessary for the maximal induction of SOS response in low iron condition (Asad and Leitao, 1991; Asad et al., 1997). Furthermore, E. coli lexA and recA mutants are sensitive to H2O2 under iron limitation, suggesting that the induction of SOS response is important when the cells are treated with H2O2 in low iron condition (Asad et al., 1997). We cannot exclude the possibility that the lower levels of intracellular iron could affect not only iron metabolism, but also other ions like copper or zinc, that could lead to DNA damage. It was previously reported that high H2O2 concentration under iron limitation leads to an increase of Cu ions inside the cells causing DNA damage and the activation of SOS response (Almeida et al., 2000). However, we have to take into account that an increase in the oxidative stress was not observed in E. coli under iron limitation. At any case, the induction of DNA repair genes observed did not protect the cells against UV-light exposure, suggesting that either the SOS response is not at its maximum, or there is a synergistic effect of both treatments in generating DNA damage.
The C. crescentus fur mutant in M2 is under oxidative stress, but this does not lead to the induction of SOS response, since the lexA and imuA transcript levels are repressed 0.5-fold in the fur mutant as compared to wild type in iron sufficiency. Moreover, in the DP-treated medium the lexA and imuA transcript levels are twofold and eightfold higher in the fur mutant, respectively, than wt cells in the same condition. These results suggest that Fur is involved in their regulation, probably indirectly. In Anabaena sp 7120 FurA acts as repressor of a single-strand DNA binding protein (SSB) that plays a role in recombination and repair by protecting the single-stranded DNA, indicating a possible role of Fur in DNA repair in cyanobacteria (Kirti et al., 2017).
The results of this work showed here for the first time that iron deprivation is causing oxidative stress and DNA damage in C. crescentus, triggering the activation of several stress response pathways. Further experimentation will be necessary to establish whether these phenotypes are or not related, and whether this may be a common trend to other bacteria.
Data Availability Statement
The genomic datasets analyzed for this study can be found in NCBI Assembly (ASM2200v1; GCF_000022005.1). The RNA-seq datasets generated for this study can be found in the NCBI Sequence Read Archive under accession code SRP136695.
Author Contributions
LL performed the experiments and analyzed the data. LS conducted the RNA-seq experiments. RR, AL, and TK performed the bioinformatics analyses. NMS performed the qRT-PCR experiments. TA performed the H2O2 quantification assays. MS and MHGM performed the 8-oxo-dG quantification assays. MVM designed the experiments and analyzed the data. LL and MVM wrote the manuscript. All authors read, revised, and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Frederico Gueiros Filho and Ana Carolina Cauz for assistance with the microscopy experiments, Luis Eduardo Soares Netto and Fernando Gomes for assistance with the H2O2 determination, José Freire da Silva Neto and Rodrigo Galhardo for revising the manuscript and for helpful comments during the course of this work, and R. Galhardo and members of the Galhardo lab for assistance with the UV and mutagenesis assays.
Footnotes
Funding. This work was supported by São Paulo Research Foundation (FAPESP, grant 2014/04046-8) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brazil, grant 306558/2013-0). During the course of this work, RR and NMS were supported by FAPESP masters fellowship grants 2016/06378-3 and 2017/02127-9, respectively, LL by a FAPESP postdoctoral fellowship grant 2015/07386-7, LS by a postdoctoral fellowship grant 2015/1461678 from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), AL by a FAPESP doctorate fellowship grant 2017/03052-2, TK by FAPESP research grant 2015/21038-1, and MVM was partially supported by CNPq-Brazil.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02014/full#supplementary-material
References
- Almeida C. E., Galhardo R. S., Felicio D. L., Cabral-Neto J. B., Leitao A. C. (2000). Copper ions mediate the lethality induced by hydrogen peroxide in low iron conditions in Escherichia coli. Mutat. Res. 460 61–67. 10.1016/S0921-8777(00)00014-8 [DOI] [PubMed] [Google Scholar]
- Almiron M., Link A. J., Furlong D., Kolter R. (1992). A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6 2646–2654. 10.1101/gad.6.12b.2646 [DOI] [PubMed] [Google Scholar]
- Altuvia S., Almiron M., Huisman G., Kolter R., Storz G. (1994). The dps promoter is activated by OxyR during growth and by IHF and sigma(s) in stationary phase. Mol. Microbiol. 13 265–272. 10.1111/j.1365-2958.1994.tb00421.x [DOI] [PubMed] [Google Scholar]
- Alvarez-Martinez C. E., Lourenco R. F., Baldini R. L., Laub M. T., Gomes S. L. (2007). The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol. Microbiol. 66 1240–1255. 10.1111/j.1365-2958.2007.06005.x [DOI] [PubMed] [Google Scholar]
- Andrews S. C., Robinson A. K., Rodriguez-Quinones F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27 215–237. 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- Asad L. M., Asad N. R., Silva A. B., de Almeida C. E., Leitao A. C. (1997). Role of SOS and OxyR systems in the repair of Escherichia coli submitted to hydrogen peroxide under low iron conditions. Biochimie 79 359–364. 10.1016/S0300-9084(97)80030-2 [DOI] [PubMed] [Google Scholar]
- Asad N. R., Leitao A. C. (1991). Effects of metal ion chelators on DNA strand breaks and inactivation produced by hydrogen peroxide in Escherichia coli: detection of iron-independent lesions. J. Bacteriol. 173 2562–2568. 10.1128/jb.173.8.2562-2568.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N., Wang T., Ye R., Helmann J. D. (2002). Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45 1613–1629. 10.1046/j.1365-2958.2002.03113.x [DOI] [PubMed] [Google Scholar]
- Balhesteros H., Shipelskiy Y., Long N. J., Majumdar A., Katz B. B., Santos N. M., et al. (2017). TonB-dependent heme/hemoglobin utilization by Caulobacter crescentus hutA. J. Bacteriol. 199:e00723-16. 10.1128/JB.00723-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros M. H., Netto L. E., Kowaltowski A. J. (2003). H(2)O(2) generation in Saccharomyces cerevisiae respiratory pet mutants: effect of cytochrome c. Free Radic. Biol. Med. 35 179–188. 10.1016/S0891-5849(03)00307-1 [DOI] [PubMed] [Google Scholar]
- Beauchene N. A., Mettert E. L., Moore L. J., Keles S., Willey E. R., Kiley P. J. (2017). O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 114 12261–12266. 10.1073/pnas.1707189114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchene N. A., Myers K. S., Chung D., Park D. M., Weisnicht A. M., Keles S., et al. (2015). Impact of Anaerobiosis on expression of the iron-responsive fur and RyhB regulons. mBio 6:e01947-15. 10.1128/mBio.01947-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H., Holm R. H., Munck E. (1997). Iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277 653–659. 10.1126/science.277.5326.653 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britos L., Abeliuk E., Taverner T., Lipton M., McAdams H., Shapiro L. (2011). Regulatory response to carbon starvation in Caulobacter crescentus. PLoS One 6:e18179. 10.1371/journal.pone.0018179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P., Rensing C., Helmann J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15 338–350. 10.1038/nrmicro.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha R. P., Paquola A. C., Marques Mdo. V., Menck C. F., Galhardo R. S. (2008). Characterization of the SOS regulon of Caulobacter crescentus. J. Bacteriol. 190 1209–1218. 10.1128/JB.01419-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Neto J. F., Braz V. S., Italiani V. C., Marques M. V. (2009). Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 37 4812–4825. 10.1093/nar/gkp509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Neto J. F., Lourenco R. F., Marques M. V. (2013). Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. 10.1186/1471-2164-14-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro Ferreira I. G., Rodrigues M. M., da Silva Neto J. F., Mazzon R. R., do Valle Marques M. (2016). Role and regulation of ferritin-like proteins in iron homeostasis and oxidative stress survival of Caulobacter crescentus. Biometals 29 851–862. 10.1007/s10534-016-9956-y [DOI] [PubMed] [Google Scholar]
- Ely B. (1991). Genetics of Caulobacter crescentus. Methods Enzymol. 204 372–384. 10.1016/0076-6879(91)04019-K [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. (1977). Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner M. J., Helmann J. D. (2011). Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid. Redox Signal. 15 175–189. 10.1089/ars.2010.3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A., Castro Rojas C. M., Siegal-Gaskins D., Crosson S. (2010). Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol. Microbiol. 77 236–251. 10.1111/j.1365-2958.2010.07207.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira T. R., Barros M. H., Camargo A. A., Castilho R. F., Ferreira J. C., Kowaltowski A. J., et al. (2013). Mitochondria as a source of reactive oxygen and nitrogen species: from molecular mechanisms to human health. Antioxid. Redox Signal. 18 2029–2074. 10.1089/ars.2012.4729 [DOI] [PubMed] [Google Scholar]
- Fillat M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546 41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- Foreman R., Fiebig A., Crosson S. (2012). The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J. Bacteriol. 194 3038–3049. 10.1128/JB.00182-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A., Antelmann H., Aguilar C., Khakh S. K., Song K. B., Smaldone G. T., et al. (2008). The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U.S.A. 105 11927–11932. 10.1073/pnas.0711752105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R. S., Rocha R. P., Marques M. V., Menck C. F. (2005). An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 33 2603–2614. 10.1093/nar/gki551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C. C., Freitas F. P., Di Mascio P., Medeiros M. H. (2010). Ultrasensitive simultaneous quantification of 1,N2-etheno-2′-deoxyguanosine and 1,N2-propano-2′-deoxyguanosine in DNA by an online liquid chromatography-electrospray tandem mass spectrometry assay. Chem. Res. Toxicol. 23 1245–1255. 10.1021/tx1001018 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Dureja C., Khatri I., Subramanian S., Raychaudhuri S., Ghosh S. (2017). Identification of novel small RNAs in Burkholderia cenocepacia KC-01 expressed under iron limitation and oxidative stress conditions. Microbiology 10.1099/mic.0.000566 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Bes M. T., Peleato M. L., Fillat M. F. (2016). Expanding the role of FurA as essential global regulator in Cyanobacteria. PLoS One 11:e0151384. 10.1371/journal.pone.0151384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., Carvalho B. S., et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiani V. C., da Silva Neto J. F., Braz V. S., Marques M. V. (2011). Regulation of catalase-peroxidase KatG is OxyR dependent and Fur independent in Caulobacter crescentus. J. Bacteriol. 193 1734–1744. 10.1128/JB.01339-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L. A., Day M., Allen J., Scott E. II, Dyer D. W. (2017). Iron-regulated small RNA expression as Neisseria gonorrhoeae FA 1090 transitions into stationary phase growth. BMC Genomics 18:317. 10.1186/s12864-017-3684-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45 D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C. L., Martins D., Redder P., Frandi A., Mignolet J., Chapalay J. B., et al. (2016). Growth control switch by a DNA-damage-inducible toxin-antitoxin system in Caulobacter crescentus. Nat. Microbiol. 1:16008. 10.1038/nmicrobiol.2016.8 [DOI] [PubMed] [Google Scholar]
- Kirti A., Kumar A., Rajaram H. (2017). Differential regulation of ssb genes in the nitrogen-fixing cyanobacterium, Anabaena sp. strain PCC71201. J. Phycol. 53 322–332. 10.1111/jpy.12500 [DOI] [PubMed] [Google Scholar]
- Landt S. G., Abeliuk E., McGrath P. T., Lesley J. A., McAdams H. H., Shapiro L. (2008). Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 68 600–614. 10.1111/j.1365-2958.2008.06172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Jeanjean R., Lemeille S., Havaux M., Zhang C. C. (2005). Iron starvation leads to oxidative stress in Anabaena sp. strain PCC 7120. J. Bacteriol. 187 6596–6598. 10.1128/JB.187.18.6596-6598.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A., Ruiz M., Zhang C. C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33 258–278. 10.1111/j.1574-6976.2008.00134.x [DOI] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., et al. (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9:e1003118. 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Helmann J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20 485–499. 10.1007/s10534-006-9070-7 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço R. F., Gomes S. L. (2009). The transcriptional response to cadmium, organic hydroperoxide, singlet oxygen and UV-A mediated by the sigmaE-ChrR system in Caulobacter crescentus. Mol. Microbiol. 72 1159–1170. 10.1111/j.1365-2958.2009.06714.x [DOI] [PubMed] [Google Scholar]
- Lourenço R. F., Kohler C., Gomes S. L. (2011). A two-component system, an anti-sigma factor and two paralogous ECF sigma factors are involved in the control of general stress response in Caulobacter crescentus. Mol. Microbiol. 80 1598–1612. 10.1111/j.1365-2958.2011.07668.x [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Salvail H., Desnoyers G., Arguin M. (2007). Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10 140–145. 10.1016/j.mib.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Masse E., Vanderpool C. K., Gottesman S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187 6962–6971. 10.1128/JB.187.20.6962-6971.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin J. R., Goswami S., Grogan S., Tjaden B., Genco C. A. (2007). A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189 3686–3694. 10.1128/JB.01890-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring, NY: Cold Spring Harbor Laboratoy Press. [Google Scholar]
- Modell J. W., Hopkins A. C., Laub M. T. (2011). A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25 1328–1343. 10.1101/gad.2038911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell J. W., Kambara T. K., Perchuk B. S., Laub M. T. (2014). A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12:e1001977. 10.1371/journal.pbio.1001977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. B., Garcia C. C. M., Freitas F. P., Batista G. L., Lopes F. S., Carvalho V. H., et al. (2018). DNA adduct formation in the lungs and brain of rats exposed to low concentrations of [(13)C2]-acetaldehyde. Chem. Res. Toxicol. 31 332–339. 10.1021/acs.chemrestox.8b00016 [DOI] [PubMed] [Google Scholar]
- Schrader J. M., Zhou B., Li G. W., Lasker K., Childers W. S., Williams B., et al. (2014). The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 10:e1004463. 10.1371/journal.pgen.1004463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. (2000). Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373 1–6. 10.1006/abbi.1999.1518 [DOI] [PubMed] [Google Scholar]
- Touati D., Jacques M., Tardat B., Bouchard L., Despied S. (1995). Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177 2305–2314. 10.1128/jb.177.9.2305-2314.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P. J., Sowa N. A., FitzGerald D. J., FitzGerald P. C., Gottesman S., Ochsner U. A., et al. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101 9792–9797. 10.1073/pnas.0403423101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic datasets analyzed for this study can be found in NCBI Assembly (ASM2200v1; GCF_000022005.1). The RNA-seq datasets generated for this study can be found in the NCBI Sequence Read Archive under accession code SRP136695.