Abstract
The cell surface of the oral pathogen Tannerella forsythia is heavily glycosylated with a unique, complex decasaccharide that is O-glycosidically linked to the bacterium’s abundant surface (S-) layer, as well as other proteins. The S-layer glycoproteins are virulence factors of T. forsythia and there is evidence that protein O-glycosylation underpins the bacterium’s pathogenicity. To elucidate the protein O-glycosylation pathway, genes suspected of encoding pathway components were first identified in the genome sequence of the ATCC 43037 type strain, revealing a 27-kb gene cluster that was shown to be polycistronic. Using a gene deletion approach targeted at predicted glycosyltransferases (Gtfs) and methyltransferases encoded in this gene cluster, in combination with mass spectrometry of the protein-released O-glycans, we show that the gene cluster encodes the species-specific part of the T. forsythia ATCC 43037 decasaccharide and that this is assembled step-wise on a pentasaccharide core. The core was previously proposed to be conserved within the Bacteroidetes phylum, to which T. forsythia is affiliated, and its biosynthesis is encoded elsewhere on the bacterial genome. Next, to assess the prevalence of protein O-glycosylation among Tannerella sp., the publicly available genome sequences of six T. forsythia strains were compared, revealing gene clusters of similar size and organization as found in the ATCC 43037 type strain. The corresponding region in the genome of a periodontal health-associated Tannerella isolate showed a different gene composition lacking most of the genes commonly found in the pathogenic strains. Finally, we investigated whether differential cell surface glycosylation impacts T. forsythia’s overall immunogenicity. Release of proinflammatory cytokines by dendritic cells (DCs) upon stimulation with defined Gtf-deficient mutants of the type strain was measured and their T cell-priming potential post-stimulation was explored. This revealed that the O-glycan is pivotal to modulating DC effector functions, with the T. forsythia-specific glycan portion suppressing and the pentasaccharide core activating a Th17 response. We conclude that complex protein O-glycosylation is a hallmark of pathogenic T. forsythia strains and propose it as a valuable target for the design of novel antimicrobials against periodontitis.
Keywords: carbohydrate-active enzymes, glycosyltransferase, immunogenicity, locus for glycosylation, methyltransferase, periodontitis, S-layer
Introduction
Protein glycosylation in bacteria is a frequent modification of secreted and cell-surface proteins, such as flagella, pili, autotransporters, and surface (S-) layer proteins (Upreti et al., 2003; Schäffer and Messner, 2017). The biological roles of these glycoproteins strongly depend on the bacteria’s environmental context and cannot be predicted a priori (Varki et al., 2017). In several cases, general protein glycosylation systems are employed, yielding a suite of proteins with diverse locations and functionalities that carry one or more copies of an identical glycan (Schäffer and Messner, 2017). In bacterial genomes, the genetic information governing protein glycosylation is frequently organized in protein glycosylation gene clusters (Nothaft and Szymanski, 2010), which encode nucleotide sugar pathways genes, genes for Gtfs, glycan processing and modifying enzymes, ligases, and transporters. Based on our current knowledge, in bacteria, O-linked protein glycosylation (where the glycan is linked to Ser, Thr, or Tyr residues of the protein) seems to be more prevalent than N-linked protein glycosylation (where the glycan is bound to Asn) (Schäffer and Messner, 2017). Most protein O-glycosylation systems investigated so far secrete virulence factors or translocate glycoproteins to the bacterial cell surface, exemplified with Campylobacter spp. (Szymanski et al., 1999, 2003), Neisseria spp. (Ku et al., 2009; Vik et al., 2009; Hartley et al., 2011), Bacteroides spp. (Fletcher et al., 2009), Actinomycetes (Espitia et al., 2010), Francisella tularensis (Egge-Jacobsen et al., 2011), Acinetobacter spp. (Iwashkiw et al., 2012; Lees-Miller et al., 2013; Harding et al., 2015), Burkholderia cepacia (Lithgow et al., 2014), Ralstonia solanacearum (Elhenawy et al., 2016), and Tannerella forsythia (Posch et al., 2011).
Tannerella forsythia is a Gram-negative pathogen affiliated to the Bacteroidetes phylum of bacteria which utilizes a general protein O-glycosylation system to decorate several of its proteins with a so far unique, complex decasaccharide. The most abundant cellular proteins targeted by this system are the S-layer proteins TfsA and TfsB, which self-assemble on the bacterium’s cell surface into a two-dimensional crystalline monolayer (Sekot et al., 2012). The T. forsythia O-glycan (Figure 1A) is bound to distinct Ser and Thr residues within the three-amino acid motif D(S/T)(A/I/L/M/T/V) (Posch et al., 2011). The glycan is strain-specifically decorated with a modified terminal nonulosonic acid, which can be either a Pse as shown for the ATCC 43037 type strain, or a Leg exemplified by strain UB4 (Friedrich et al., 2017). Besides these sialic acid mimics, other unique S-layer glycan sugars present in the T. forsythia O-glycan are α-L-fucose (Fuc), Dig, Xyl, N-acetyl mannosaminuronic acid (ManNAcA), and N-acetyl mannosaminuronamide (ManNAcCONH2) (Posch et al., 2011).
FIGURE 1.
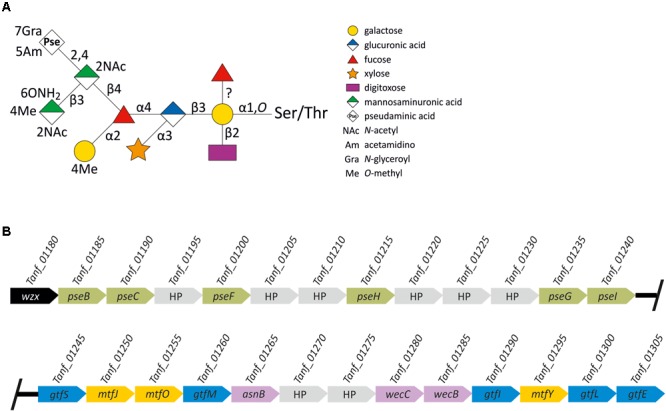
(A) Scheme of the T. forsythia ATCC 43037 S-layer O-glycan structure. Monosaccharide symbols are shown according to the Symbol Nomenclature for Glycans (SNFG) (Varki et al., 2015). Please note that the position of the branching Fuc remained unclear (Posch et al., 2011) until it was determined in the course of this study to be on the reducing-end Gal. (B) Scheme of the 27-kb protein O-glycosylation gene cluster of T. forsythia ATCC 43037. Wzx (black), flippase; pseBCFHGI (green), CMP-Pse biosynthesis genes; gtfSMILE (blue), Gtf genes; mtfJOY (yellow), Mtf genes; asnB (putative asparagine synthetase B), wecC (UDP-N-acetyl-D-mannosamine dehydrogenase) and wecB (UDP-N-acetylglucosamine 2-epimerase) (purple); hypothetical proteins, HP (gray). Genes are not drawn to scale.
To investigate the biosynthesis of the T. forsythia O-glycan, in a previous study, parts of an S-layer protein O-glycosylation gene locus were identified in the T. forsythia ATCC 43037 genome, and clustering of highly homologous genes was observed in different Bacteroidetes species (Posch et al., 2011). Based on that and a successful protein cross-glycosylation experiment between T. forsythia and Bacteroides fragilis (Posch et al., 2013), which both belong to the Bacteroidetes phylum, the presence of a phylum-wide protein O-glycosylation system was proposed (Coyne et al., 2013). An antiserum raised to a defined, truncated glycan of B. fragilis reacted with all Bacteroidetes species tested, including T. forsythia, but not with the full glycan, suggesting a discrimination between a core glycan and a species-specific glycan portion (Coyne et al., 2013).
In its native environment, T. forsythia thrives in a polymicrobial biofilm community that constitutes what is clinically described as oral plaque (Socransky et al., 1998; Holt and Ebersole, 2005). Tannerella forsythia is recognized as a key periodontal pathogen following the polymicrobial synergy and dysbiosis model of periodontal disease etiology (Hajishengallis and Lamont, 2012; Lamont and Hajishengallis, 2015). In this model, low abundance keystone pathogens are crucial as they initially subvert host immune responses, further leading to homeostasis breakdown in the oral cavity and destructive inflammation. Recent studies provided evidence that T. forsythia employs its unique cell surface to colonize its niche within the polymicrobial biofilm and to orchestrate the immune response of resident host tissue and the immune system (Bloch et al., 2017, 2018). In fact, the two S-layer glycoproteins (Sabet et al., 2003; Sharma, 2010; Sekot et al., 2011), as well as a leucine-rich repeat outer membrane glycoprotein BspA (Sharma et al., 2005), are among the bacterium’s virulence factors. The S-layer is strongly antigenic and mediates hemagglutination, as well as adherence to and invasion of human gingival epithelial cells (Sabet et al., 2003; Lee et al., 2006; Sakakibara et al., 2007). Studies with human macrophages and gingival fibroblasts demonstrated that the S-layer attenuates the host immune response by evading recognition by the innate immune system, at least at the early stage of infection (Sekot et al., 2011). There are indications that specifically the S-layer O-glycan is crucial for the modulation of host immunity through Th17 suppression (Settem et al., 2013). A very recent study suggests a role specifically of the modified Pse residue (Pse5Am7Gra) present at terminal position on the T. forsythia ATCC 43037 O-glycan in facilitating immune evasion by dampening the response of epithelial tissues to initial infection (Bloch et al., 2018). In addition to its immunological relevance, it was shown that the glycosylated T. forsythia S-layer plays a role in the bacterium’s biofilm life-style (Honma et al., 2007; Bloch et al., 2017; Friedrich et al., 2017).
These data indicate that the T. forsythia O-glycan and, hence, its biosynthesis pathway could be valuable targets in efforts to interfere with the establishment of periodontitis, which continues to be the most frequent inflammatory disease of bacterial origin world-wide (Hajishengallis, 2015). Thus, this present study was designed to obtain insight into the T. forsythia O-glycoprotein biosynthesis pathway and its involvement in underpinning the bacterium’s pathogenicity. Specifically, emphasis was focused on (i) sequence comparison of the general protein O-glycosylation gene cluster of the T. forsythia ATCC 43037 type strain with what is found in other T. forsythia strains for which genome sequences were available in public databases; (ii) transcriptional analysis of the protein O-glycosylation gene cluster of T forsythia ATCC 43037; (iii) construction of defined T. forsythia mutants deficient in predicted Gtfs and Mtfs encoded in the gene cluster and subsequent analysis of the O-glycans by MS to delineate the roles of the individual enzymes in glycan biosynthesis; and (iv) dissecting an O-glycan structure–function relationship in the immune response of DCs upon stimulation with the T. forsythia wild-type versus defined glycosylation-deficient mutants.
Materials and Methods
Bacterial Strains and Cultivation Conditions
Tannerella forsythia ATCC 43037 (American Type Culture Collection – ATCC, Manassas, VA, United States) (Tanner et al., 1986; Friedrich et al., 2015) and its mutants (Table 1) were grown anaerobically in brain heart infusion (BHI) media (Oxoid, Basingstoke, United Kingdom), supplemented with yeast extract (Sigma-Aldrich, Vienna, Austria), L-cysteine (Sigma-Aldrich), hemin (Sigma-Aldrich), menadione (Sigma-Aldrich), N-acetylmuramic acid (Carbosynth, Compton, United Kingdom), and horse serum (Thermo Fisher Scientific, Vienna, Austria) as described previously (Tomek et al., 2017). Media were supplemented with 50 μg/ml gentamicin, 5 μg/ml Erm or 10 μg/ml Cat, when appropriate.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype and use or description | Source or reference |
|---|---|---|
| Escherichia coli strain | ||
| DH5α | F- Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ- thi-1 gyrA96 relA1; cloning strain | Invitrogen, Austria |
| Tannerella forsythia strains | ||
| ATCC 43037 | Type strain, wild-type | ATCC; Friedrich et al., 2015 |
| ATCC 43037 ΔTanf_01245 | ΔTanf_01245::ermF; knock-out strain of Tanf_01245 | Tomek et al., 2017 |
| ATCC 43037 ΔTanf_01245+ | ΔTanf_01245::Tanf_01245 cat; reconstituted knock-out strain | Tomek et al., 2017 |
| ATCC 43037 ΔTanf_01250 | ΔTanf_01250::ermF; knock-out strain of Tanf_01250 | This work |
| ATCC 43037 ΔTanf_01255 | ΔTanf_01255::ermF; knock-out strain of Tanf_01255 | This work |
| ATCC 43037 ΔTanf_01260 | ΔTanf_01260::ermF; knock-out strain of Tanf_01260 | This work |
| ATCC 43037 ΔTanf_01260+ | ΔTanf_01260::Tanf_01260 cat; reconstituted knock-out strain | This work |
| ATCC 43037 ΔTanf_01290 | ΔTanf_01290::ermF; knock-out strain of Tanf_01290 | This work |
| ATCC 43037 ΔTanf_01290+ | ΔTanf_01290::Tanf_01290 cat; reconstituted knock-out strain | This work |
| ATCC 43037 ΔTanf_01295 | ΔTanf_01295::ermF; knock-out strain of Tanf_01295 | This work |
| ATCC 43037 ΔTanf_01300 | ΔTanf_01300::ermF; knock-out strain of Tanf_01300 | This work |
| ATCC 43037 ΔTanf_01300+ | ΔTanf_01300::Tanf_01300 cat; reconstituted knock-out strain | This work |
| ATCC 43037 ΔTanf_01305 | ΔTanf_01305::(Perm)-ermF; knock-out strain of Tanf_01305 | This work |
| ATCC 43037 ΔTanf_01305+ | ΔTanf_01305::Tanf_01305 cat; reconstituted knock-out strain | This work |
| Plasmids | ||
| pJET1.2/blunt | Cloning vector; ampR | Thermo Fisher Scientific |
| pJET/ΔTF0955ko | Vector for amplification of the erythromycin resistance gene | Tomek et al., 2014 |
| pEXALV | Vector for amplification of the chloramphenicol resistance gene | Zarschler et al., 2009 |
| pJET1.2/ΔTanf_01250 | Tanf_01250 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01250+ | Cassette for reconstitution of ΔTanf_01250; ampR catR | This work |
| pJET1.2/ΔTanf_01255 | Tanf_01255 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01260 | Tanf_01260 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01260+ | Cassette for reconstitution of ΔTanf_01260; ampR catR | This work |
| pJET1.2/ΔTanf_01290 | Tanf_01290 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01290+ | Cassette for reconstitution of ΔTanf_01290; ampR catR | This work |
| pJET1.2/ΔTanf_01295 | Tanf_01295 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01300 | Tanf_01300 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01300+ | Cassette for reconstitution of ΔTanf_01300; ampR catR | This work |
| pJET1.2/ΔTanf_01305 | Tanf_01305 knock-out cassette; ampR ermFR | This work |
| pJET1.2/ΔTanf_01305+ | Cassette for reconstitution of ΔTanf_01305; ampR catR | This work |
Escherichia coli strains (Table 1) were grown under standard conditions in LB medium supplemented with 100 μg/ml ampicillin, when appropriate.
Identification of a Protein O-Glycosylation Gene Cluster in T. forsythia
Up- and downstream regions of the previously identified protein O-glycosylation gene locus (Posch et al., 2011) were bioinformatically inspected using the genome sequence of the T. forsythia ATCC 43037 type strain as a basis (Friedrich et al., 2015). Information on putative gene functions were obtained through homology searches using the NCBI BLAST suite1 (Altschul et al., 1997) and the PFAM database2 (Finn et al., 2016).
An alignment of protein O-glycosylation gene clusters from different T. forsythia strains for which genome sequences were publically available was generated using the software MultiGeneBlast 1.1.13 (Camacho et al., 2009; Medema et al., 2013) in “homology search” mode. Genomes and annotations included in the analysis are listed in Supplementary Table S1. The genome assembly from T. forsythia FDC 92A2 was used as query, restricted to the genomic region from coordinates 1,135,922 to 1,166,078. The database was built from the assemblies of strains ATCC 43037, KS16, 3313, UB4, UB20, UB22 and Tannerella sp. HOT-286 (phylotype BU063). The assemblies were downloaded as GenBank flat files (gbff) from the NCBI ftp server3. RefSeq assemblies were downloaded for genomes FDC 92A2, ATCC 43037, KS16, 3313 and BU063. For strains UB4, UB20, and UB22, GenBank assemblies containing annotations generated in the context of the initial characterization of these genomes were used (Stafford et al., 2016). The RefSeq gbff files had to be adapted in order to provide unique protein identifiers in regions of interest as input for MultiGeneBlast. Minimal sequence identity (-minpercid) and minimal sequence coverage (-minseqcov) for protein alignments were both set to 50%, and default parameters -syntenyweight 0.5, -distancekb 20, -hitspergene 250 were used. For generating the BU063 track, an additional run with lower thresholds, i.e., 10% sequence identity and 10% sequence coverage, was performed. MultiGeneBlast runs were carried out under Linux Ubuntu 16.04; Linux shell commands were used for editing input files. The graphic files generated by MultiGeneBlast were combined and edited.
RNA Purification and Reverse Transcription-PCR
Total RNA was extracted from T. forsythia ATCC 43037 using the PureLink RNA Mini Kit (Thermo Fisher Scientific) and subsequently treated with RNase-free DNase (PureLink DNase Set, Thermo Fisher Scientific) to remove DNA contamination. cDNA was generated using the MultiScribe Reverse Transcriptase from the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) using 500 ng of total RNA. One tenth of each cDNA reaction mixture was used as template for PCR using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific). The synthesized cDNA was amplified by PCR with primer pairs spanning stepwise neighboring genes, starting at the 5′ end of the gene cluster with Tanf_01180-Tanf_01185 and ending with Tanf_01300-Tanf_01305 (Supplementary Figure S1). Primer pairs and the size of the corresponding transcripts are indicated for each transcript in Supplementary Figure S1 and primer sequences are listed in Supplementary Table S2. As a positive control, genomic DNA was used, and DNase treated RNA without the cDNA-generating step served as a control for contamination of total RNA with chromosomal DNA.
Construction of Glycosyltransferase- and Methyltransferase-Deficient Mutants
Four putative Gtf genes (Tanf_01260; Tanf_01290; Tanf_01300; Tanf_01305) and three putative Mtf genes (Tanf_01250; Tanf_01255; Tanf_01295) encoded in the T. forsythia ATCC 43037 genome were individually knocked-out by homologous recombination, using a knock-out cassette deleting the selected gene. Clones viable on Erm-containing BHI-agar plates (0.8% w/v) were selected and further tested by PCR for correct integration of the knock-out cassette (Supplementary Figures S2–S8). A detailed description of the construction of the knock-out cassettes is given in the Supplementary Material. In short, approximately 1-kbp up- and downstream homology regions were joined to the Erm-resistance gene (PermF, 1093-bp; ermF, 805-bp) by overlap-extension (OE)-PCR and subsequently blunt-end cloned into the plasmid pJET1.2 (Thermo Fisher Scientific). Knock-out cassettes were transformed into electrocompetent T. forsythia cells, which were regenerated overnight and plated on Erm-containing BHI selection plates. Single colonies were picked and used for inoculation of BHI medium. Once bacterial growth was visible, genomic DNA was isolated (Cheng and Jiang, 2006) and the loss of the selected gene (log) was tested by PCR (Supplementary Figures S2–S8). Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific) or Herculase II Fusion DNA Polymerase (Agilent Technologies, Vienna, Austria) were used for PCR amplification according to the manufacturer’s instructions. Oligonucleotides (Thermo Fisher Scientific) used are listed in Supplementary Tables S3, S4.
To exclude possible polar effects leading to an altered transcription of downstream genes, Gtf-deficient mutants were complemented with the native gene, in combination with a Cat resistance gene (chloramphenicol acetyl transferase, cat; 650 bp) for selection. For this purpose, the approximately 1-kbp homologous upstream region plus the associated native Gtf gene were joined to the cat resistance gene using OE-PCR and subsequently blunt-end cloned into the plasmid pJET1.2. Using the artificially introduced restriction sites SphI and KpnI, the approximately 1-kbp downstream homologous region was added, completing the reconstitution cassettes for Tanf_01260 and Tanf_01290 (Supplementary Figures S2, S3). The downstream region of the Tanf_01300 reconstitution cassette was cloned via KpnI and NdeI restriction sites, while KpnI and BamHI were used for the Tanf_01305 reconstitution cassette (Supplementary Figures S4, S5).
SDS-PAGE and Western-Blotting
SDS-PAGE of crude cell extracts was carried out according to Laemmli (1970). 7.5% SDS-PA gels were prepared according to a standard protocol. Proteins were visualized with colloidal Coomassie Brilliant Blue (CBB) R-250 (Serva, Heidelberg, Germany) or transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Vienna, Austria) for Western-blot analysis. Polyclonal rabbit antisera raised against the recombinant S-layer proteins TfsA (α-TfsA) and TfsB (α-TfsB) (Sekot et al., 2012) were used as primary antibodies in combination with a monoclonal goat α-rabbit secondary antibody labeled with IRDye 800CW (LI-COR Biosciences, Lincoln, NE, United States). S-Layer protein bands were visualized at 800 nm using an Odyssey Infrared Imaging System (LI-COR Biosciences).
S-Layer O-Glycan Preparation and Liquid Chromatography ESI-MS
O-Glycans were released from the glycosylated S-layer proteins TfsA and TfsB by in-gel reductive β-elimination and purified by preparative PGC-HPLC as described previously (Posch et al., 2011; Tomek et al., 2014). The glycan mixture was analyzed using a Dionex Ultimate 3000 system directly linked to an ion trap instrument (amaZon speed ETD, Bruker, Germany) equipped with the standard ESI source in positive ion, Data Dependent Acquisition (DAA) mode (performing MS/MS on signals based on their intensity and LC elution). MS-scans were recorded over an m/z range 450–1650; ICC target was set to 100,000 and maximum accumulation time to 200 ms. The top 10 highest peaks were selected for fragmentation with an absolute intensity threshold above 50,000. Instrument calibration was performed using ESI Tuning Mix (Agilent Technologies) as by the manufacturer’s recommendations. LC separation of released O-glycans was performed on a Thermo Hypercarb separation column (5 μm particle size; 100 mm × 0.360 mm). A gradient from 1 to 21% solvent B in solvent A (solvent A, 65 mM ammonium formate buffer, pH 3.0; solvent B, 100% acetonitrile) over 20 min was applied, followed by a 10-min gradient from 21% B to 50% B, at a flow rate of 6 μl/min. Data were evaluated manually using the DataAnalysis 4.0 software (Bruker) and Glycoworkbench 2.1 build 146 (Ceroni et al., 2008).
Generation of Human Monocyte- and Murine Bone Marrow-Derived Dendritic Cells
Human monocytes (E59; ∼40 × 106 cells) from healthy volunteers (Ethical approval EK 1880/2012 in accordance with the Declaration of Helsinki, Medical University of Vienna, Vienna, Austria; written informed consent was obtained from all volunteers of this study) were thawed at 37°C and placed into AIM V medium (Gibco, Life Technologies, Paisley, United Kingdom) supplemented with 2% human plasma (Octaplas, OP; Octapharma, Zurich, Switzerland). Monocytes were counted and adjusted to a concentration of 0.5 × 106 cells/ml. Dead cells were removed after incubation at 37°C for 1.5 h, while living cells were differentiated with 1000 U/ml GM-CSF and 400 U/ml IL-4. Medium including cytokines was replaced on day 3 and iDCs were harvested on day 5 (Lanzinger et al., 2012).
Murine BMDCs were isolated from C57BL/6 mice (Gallucci et al., 1999). Briefly, femurs and tibias were rinsed with DMEM, supplemented with 10% heat-inactivated fetal calf serum (FCS; PAA, Pasching Austria), 1% penicillin/streptomycin (Gibco), 1% non-essential amino acids (NEAA) (Gibco) and 0.0002% β-mercaptoethanol (Sigma-Aldrich). Erythrocytes were lysed in erolysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA; pH 7.2). The bone marrow was washed, and cells were re-suspended in supplemented DMEM. Cells were cultured in 24-well plates (Iwaki, Japan) at a concentration of 1.0 × 106 cells per well in DMEM supplemented with 10% FCS and in the presence of 5 ng/ml recombinant murine IL-4 (eBioscience, San Diego, CA, United States) and 3 ng/ml recombinant murine GM-CSF (BD Pharmingen, San Diego, CA, United States). Half of the medium (including all supplements) was replaced every second day. Immature DCs (iDs) were harvested on day 6, counted and adjusted to a concentration of 1.0 × 106 cells/ml. All mouse experiments were performed in accordance with the institutional guidelines and approved by the Animal Care and Use Committee of the Medical University of Vienna, Austria (GZ: 856861/2013/16).
Stimulation of Dendritic Cells With Inactivated T. forsythia Cells
Tannerella forsythia wild-type and Gtf-deficient mutants were irradiated three times with UV light using maximum UV energy settings for 1 min, each, (Ultraviolet Crosslinker CL-1000; UVP, Upland, CA, United States) for inactivation. Cell integrity after treatment was verified by scanning electron microscopy using an Inspect S50 scanning electron microscope (FEI, Eindhoven, Netherlands) (Tomek et al., 2014).
Murine and human iDCs were matured in a 48-well plate with inactivated T. forsythia wild-type, ΔTanf_01245 (ΔgtfS), ΔTanf_01290 (ΔgtfI), or ΔTanf_01305 (ΔgtfE) cells, at concentrations of 106, 107 and 108 cfu/ml. iDCs stimulated with 100 ng/ml LPS (E. coli strain O111:B4, Calbiochem, EMD Chemicals, San Diego, CA, United States) served as a positive control. Stimulated DCs were incubated at 37°C for 24 h. Supernatants were frozen for later use, DCs were harvested. DC maturation was monitored by visual and flow cytometric evaluation of typical DC morphology (LSR II flow cytometer, BD Biosciences), and expression of cell-surface markers (α-CD80, α-CD86, α-MHC-II; eBioscience) after 6 h and 24 h, respectively.
Determination of Cytokine Levels by ProcartaPlex Multiplex Immunoassay and ELISA
Multiplex cytokine analysis of human monocyte-derived DC supernatants was performed using ProcartaPlex Multiplex immunoassay (eBioscience) for measuring cytokine secretion of IFN-γ, IL-1α, IL-1β, IL-4, IL-6, IL-10, IL-12p70, IL-23, and TNF-α.
The level of secreted cytokines in murine BMDC supernatants, TNF-α and IL-6, were analyzed using commercial ready-set-go ELISA kits (eBioscience) in 96-well microtiter plates according to the manufacturer’s protocol and measured at 450 nm by a micro plate reader (PerkinElmer, EnSpire Multimode Reader, Waltham, MA, United States).
T Cell-Priming Upon Stimulation of Antigen-Presenting Cells With T. forsythia
Human PBMCs were separated from whole blood by density centrifugation (Lanzinger et al., 2012). PBMCs were cultured in RPMI-1640 medium supplemented with GlutaMAX–I (Gibco) and 2% human plasma (OP, Octapharma). PBMCs were counted with BD Trucount tubes (BD Biosciences), adjusted to a concentration of 1.0 × 107 cells/ml and labeled with CFSE (Invitrogen, Austria). T cell priming upon antigen-presenting cell (APC) stimulation with T. forsythia wild-type and Gtf-deficient mutants was assessed in the mixed PBMC culture. Whole PBMCs stimulated with 50 ng/ml LPS (E. coli O111:B4 LPS, Merck, Darmstadt, Germany) and 103 U/ml human recombinant IFN-γ (PeproTech, Rocky Hill, NJ, United States) served as positive controls. T cell proliferation was calculated as percentage CFSE negative cells of CD3+ cells after 8 days of co-culture (Lyons and Parish, 1994). T cell activation was measured via expression of CD25 (α-CD25; BD Pharmingen) by flow cytometry. CD4+ T cell differentiation (α-CD4; eBiosciences) was assessed by expression of signature transcription factors for Treg (FoxP3; BD Pharmingen) and Th17 (RORγT; eBiosciences) cells as measured by flow cytometry. Th1 differentiation was assessed by expression of the signature transcription factor T-bet (α-Tbet; eBiosciences) as measured by flow cytometry.
Data analysis was made using the Student’s t-test, as appropriate. A p-value below 0.05 was considered to indicate statistical significance.
Results
Genomic Organization of the General Protein O-Glycosylation Gene Cluster of the T. forsythia Type Strain
Previously, a partial protein O-glycosylation gene locus (Posch et al., 2011; Coyne et al., 2013) as well as genes for the biosynthesis of CMP-Pse, which is the activated form of this sugar acid required for its incorporation into the S-layer glycan, (Friedrich et al., 2017), were identified in the genome of T. forsythia ATCC 43037. That genomic region (Friedrich et al., 2015) was now investigated in silico for the presence of further genes with functional annotations as carbohydrate-active enzymes. This revealed a 27-kb protein O-glycosylation gene cluster (Figure 1B). From the 5′ end, the gene cluster starts with a wzx flippase-like gene (Tanf_01180) which is followed by the six genes required for the biosynthesis of CMP-Pse (Friedrich et al., 2017), present in the order pseB (Tanf_01185), pseC (Tanf_01190), pseF (Tanf_01200), pseH (Tanf_01225), pseG (Tanf_01235), pseI (Tanf_01240). ORFs encoding hypothetical proteins of unknown function (HP) intercept this region. Downstream, five genes encoding proteins with Gtf domains (Tanf_01245, Tanf_01260, Tanf_01290, Tanf_01300, Tanf_01305, named gtfS, gtfM, gtfI, gtfL, and gtfE) are present. The corresponding glycosyltransferases GtfS (445 amino acids), GtfM (382 amino acids), GtfI (402 amino acids; GT1 family of Gtfs) and GftL (420 amino acids; GT1 family of Gtfs) exhibit homology to the GT-B type superfamily of Gtfs, while GtfE (255 amino acids; GT2 family of glycosyltransferases) exhibits homology to the GT-A type superfamily.
Further downstream, WecC (Tanf_01280, UDP-N-acetylmannosaminuronic acid dehydrogenase) and WecB (Tanf_01285, UDP-N-acetylglucosamine 2-epimerase) are encoded, predicted to be involved in the biosynthesis of UDP-N-acetylmannosaminuronic acid. The conserved AnsB domain of Tanf_01265 (605 amino acids; predicted asparagine synthetase B) indicates amidotransferase activity of this ORF, putatively involved in the formation of the acetamidino (Am) modification on the C-5 at the Pse residue. A candidate gene for the glyceroyl (Gra) modification at the C-7 of Pse remains elusive. Additionally, within the protein O-glycosylation gene cluster, three genes were identified sharing similarities with SAM-dependent Mtfs; these are Tanf_01250, Tanf_01255 and Tanf_01295, encoding proteins named MtfJ (90 amino acids), MtfO (167 amino acids), and MtfY (213 amino acids), respectively. SAM-dependent domains are present in all of these Mtfs; the typical glycine-rich sequence (motif I; E/DXGXGXG) (Martin and McMillan, 2002; Schubert et al., 2003) is only found within the amino acid sequence of MtfJ.
Inspection of eight further ORFs distributed over the entire protein O-glycosylation gene cluster, i.e., Tanf_01195, Tanf_01205, Tanf_01210, Tanf_01220, Tanf_01225, Tanf_01230, Tanf_01270, and Tanf_01275, did not provide evidence for functions related to carbohydrate metabolism. Thus, these genes were regarded as irrelevant for the present study.
Transcription Analysis of the Protein O-Glycosylation Gene Cluster
To analyze whether the genes encoded by the protein O-glycosylation gene cluster are transcriptionally linked, total RNA from T. forsythia ATCC 43037 cells was extracted and co-transcription of the relevant genes was analyzed using RT-PCR as outlined in Supplementary Figure S1. PCR products of the expected sizes were obtained with primer pairs (Supplementary Table S2) designed to bridge the ends between the ORFs of adjacent genes, yielding amplification products only when co-transcription was occurring. The results revealed that the cluster is transcribed as a polycistronic unit spanning at least the Tanf_01180 to Tanf_01305 genes (Supplementary Figure S1). This implicates that a previously described three-gene “exopolysaccharide synthesis operon” spanning Tanf_01280 to Tanf_01290 (Honma et al., 2007) is part of the contiguous transcription unit of the T. forsythia ATCC 43037 protein O-glycosylation gene cluster.
The General Protein O-Glycosylation Gene Cluster Is a Common Feature of Pathogenic T. forsythia Strains
The species-wide conservation of the general protein O-glycosylation gene cluster was assessed by sequence comparison of the ATCC 43037 type strain with six other publicly available T. forsythia genomes (T. forsythia UB20, FDC 92A2, UB4, KS16, UB22, and 3313; genome accessions are listed in Supplementary Table S1). Prior to that, it was confirmed by sequence homology searches, that all of these genomes possessed orthologs of the ATCC 43037 S-layer genes tfsA (Tanf_03370) and tfsB (Tanf_03375), which are the most abundant glycosylation targets in T. forsythia (Supplementary Table S5).
In all of the analyzed T. forsythia genomes, general protein O-glycosylation gene clusters of comparable size, content, and organization were identified (Figure 2). The major difference found between the analyzed gene clusters was in the alternate presence of six genes encoding the biosynthesis pathway for either CMP-Pse (pseBCHGIF in strains ATCC 43037 and UB20) or CMP-Leg (legBCHGIF in strains FDC 92A2, UB4, KS16, and UB22), respectively. In all cases, the corresponding genes are located immediately downstream of a conserved wzx-flippase like gene (Tanf_01180 in strain ATCC 43037). Strain 3313 presents a unique situation, since most components of the CMP-Leg pathway were found, however, legF could not be identified and for legH only a match at 29% sequence identity (covering 95% of the sequence) could be detected. These data leave a prediction of the type of nonulosonic acid in this strain open, if present at all. Furthermore, 10 genes of unknown function that do not have counterparts in the other T. forsythia protein O-glycosylation gene clusters are present in that region of the 3313 genome. Apart from this difference, the region further downstream shares a high degree of similarity with the other protein O-glycosylation gene clusters. This region encodes five predicted glycosyltransferases (GtfSMILE), two UDP-N-acetylmannosaminuronic acid biosynthesis enzymes (WecB, WecC), and the putative carbohydrate modifying enzyme AsnB (Figure 2), as described above for T. forsythia ATCC 43037. With regard to the three Mtfs (MtfJOY) predicted for the type strain, no mtfJ gene was annotated in strain UB22. In strain 3313, a single gene was annotated at the location of mtfJ and mtfO, showing sequence homology to both mtfJ (93% of its length) and mtfO (entire length), suggesting either misannotation or a fused mtfJ–mtfO gene. All CMP-Leg synthesizing strains contained an additional predicted Mtf gene (named mtfX) which does not share sequence homology to the Mtfs MtfJOY. In strain FDC 92A2, there is a transposable element present between the gtfI and mtfY genes.
FIGURE 2.
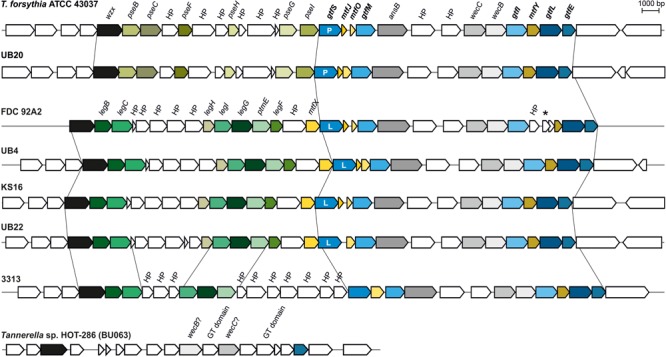
Alignment of protein O-glycosylation gene clusters from different T. forsythia strains showing comparable sizes and gene organizations (drawn to scale). Genes showing sequence identity > 50% and sequence coverage > 50% between strains appear in the same color. The major difference in the analyzed strains are for genes synthesizing either CMP-Pse (pseBCFHGI; light green colors; strains ATCC 43037 and UB20) or CMP-Leg (legBCHIGF, ptmE; dark green colors; strains FDC 92A2, UB4, KS16, UB22). Genes encoding Gtfs (gtfSMILE; blue color), Mtfs (mtfJOY; yellow color) and carbohydrate modifying enzymes (asnB, wecC, wecB; gray color) show high sequence homology between analyzed strains. Genomes of all strains synthesizing CMP-Leg encode an additional putative Mtf gene (mtfX), which does not share sequence homology to other Mtfs located within the cluster. In strain UB22, mtfJ is not predicted and for strain 3313 only five out of seven genes needed for the synthesis of CMP-Leg are predicted confidently. Due to low homology, isolate Tannerella sp. HOT-286 (phylotype BU063) could not be aligned with the other T. forsythia strains; for that isolate, the genomic area between a wzx-like gene and the gtfE gene is shown for comparison. P, Pse transferase; L, Leg transferase; HP, hypothetical protein; the star symbol (∗) indicates a transposable element; genes written in bold letters were investigated in detail in course of this study.
In contrast, the genome sequence of Tannerella sp. HOT-286 (phylotype BU063; genome accession NZ_CP017038.1), which is described as a periodontal health-associated isolate (Beall et al., 2014), showed sequence homology to only the wzx flippase-like gene (46% sequence identity) and the Gtf gene gtfE (69% sequence identity). Apart from weak matches to the wecB and wecC genes, there was no further homology to genes found in the protein O-glycosylation gene cluster of pathogenic T. forsythia strains. Instead, two genes with predicted Gtf domains (locus tags: BCB71_RS10435 and BCB71_RS10420), which are different from the gtfSMILE genes mentioned before, were found in that region of the HOT-286 (phylotype BU063) genome (Figure 2). Further research on this difficult-to-grow Tannerella sp. (Vartoukian, 2016) is needed to assess its glycosylation potential and potential role in periodontal health and disease.
Glycosyltransferases of the Protein O-Glycosylation Gene Cluster Assemble the Species-Specific Glycan
To investigate the principal involvement of the predicted Gtfs GtfSMILE in the biosynthesis of the T. forsythia ATCC 43037 S-layer O-glycan, single gene knock-out mutants of the corresponding genes, i.e., Tanf_01260 (gtfM), Tanf_01290 (gtfI), Tanf_01300 (gtfL) and Tanf_01305 (gtfE), were created (Supplementary Figures S2–S5) - ΔTanf_01245 (ΔgtfS) was available from a previous study (Tomek et al., 2017) - and analyzed by SDS-PAGE and Western-blotting. In comparison to the T. forsythia ATCC 43037 wild-type, in which the glycosylated S-layer proteins TfsA and TfsB migrate on the SDS-PAGE gel at ∼230 kDa (calculated MW of the protein, 135 kDa) and ∼270 kDa (calculated MW of the protein, 152 kDa), respectively, each Gtf-deficient mutant experienced a downshift of these prominent T. forsythia glycoproteins (Figure 3A). Downshifted S-layer glycoproteins suggest a stepwise truncation of the O-glycan from wild-type via ΔgtfS, ΔgtfM, ΔgtfI, ΔgtfL to ΔgtfE. Western-blots probed with α-TfsA and α-TfsB antiserum confirmed the identity of the S-layer glycoproteins in T. forsythia wild-type and mutant strains, with the S-layer glycoproteins of the reconstituted mutants regaining their native SDS-PAGE migration profile (Figure 3B). MS analysis of β-eliminated TfsB O-glycans from the Gtf mutants upon complementation with the native gene (T. forsythia ΔgtfM+, T. forsythia ΔgtfI+, T. forsythia ΔgtfL+, and T. forsythia ΔgtfE+) confirmed the synthesis of the native decasaccharide (Supplementary Figure S9). Data for T. forsythia ΔgtfS+ were published previously (Tomek et al., 2017).
FIGURE 3.
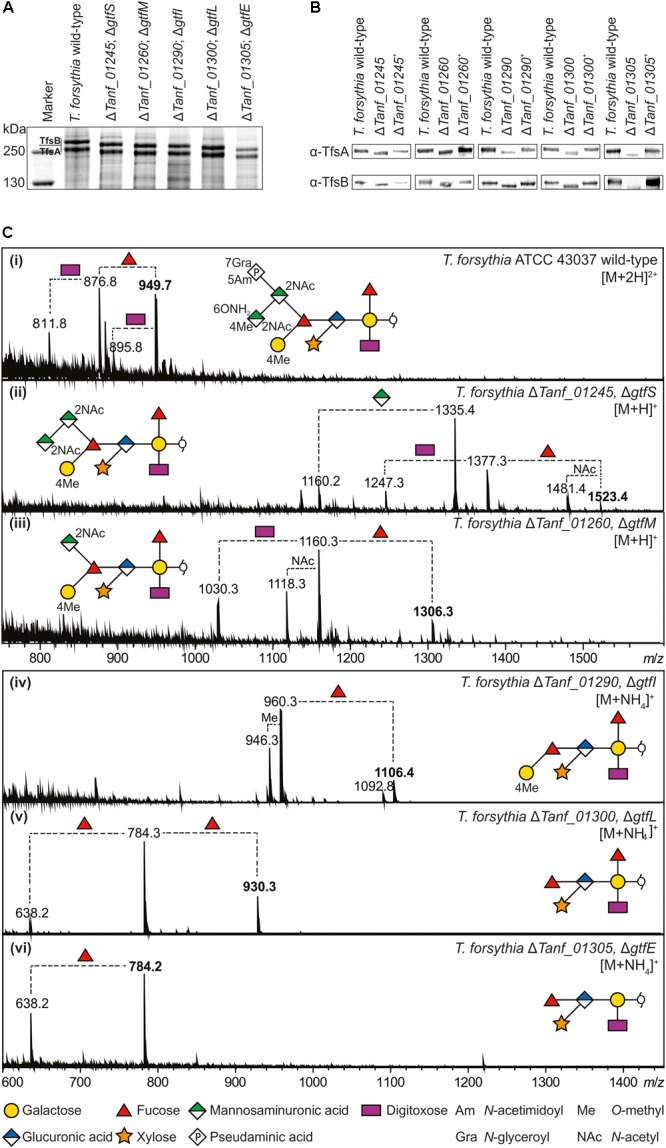
(A) Coomassie Brilliant Blue staining of crude cell extracts from T. forsythia ATCC 43037 wild-type and glycosyltransferase-deficient mutants after separation on a 7.5% SDS-PA gel. The S-layer glycoproteins (labeled TfsA and TfsB) are indicated and the downshifts resulting from glycan truncation can be seen in the mutants. S-layer glycoprotein bands were further processed for MS analyses. PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific) was used as a protein molecular weight marker. (B) Western-blots probed with α-TfsA and α-TfsB antiserum for confirmation of the identity of S-layer glycoproteins. Glycoproteins from all glycosyltransferase-deficient mutants (ΔgtfSMILE) experienced a downshift resulting from glycan truncation, whereas the reconstituted strains (denoted with +) regained wild-type migration, indicating the presence of the complete mature glycan, proving successful recombination. (C, i–vi) ESI-MS sum spectra of β-eliminated TfsB O-glycans from T. forsythia wild-type and mutants. The glycan structures of the signals corresponding to the largest mass (bold m/z values) are shown in SNFG representations (Varki et al., 2015). O-Glycan signals detected for the respective mutants were assigned based on the m/z mass differences corresponding to the loss of individual sugar units and/or modifications.
To obtain insight into the specificity of the Gtfs GtfSMILE in the biosynthesis of the T. forsythia O-glycan, ESI-MS analysis of β-eliminated O-glycans from TfsB (Figure 3C) and TfsA (data not shown) as present in the ΔgtfS, ΔgtfM, ΔgtfI, ΔgtfL, and ΔgtfE mutants was performed. The doubly charged wild-type decasaccharide exhibited an m/z = 949.9 [M+2H]2+, which corresponds to m/z = 1898.8 when calculating the singly charged form thereof, confirming the composition of the previously elucidated O-glycan structure (Posch et al., 2011) (Figure 3C, i). Structure-wise, the position of the branching Fuc residue that has remained ambiguous from our initial investigations could now be determined, using a ΔgtfL mutant (ΔTanf_01300) (see below).
In comparison to the wild-type, the signal for the largest glycan produced by a ΔgtfS (ΔTanf_01245) mutant was detected at m/z = 1523.4 [M+H]+, which agrees with the m/z value of the T. forsythia O-glycan lacking the terminal Pse5Am7Gra residue (361.2 Da) and one methyl group (14.0 Da). A detailed analysis of the GtfS protein was performed recently in our group and revealed that it has α-2,4 Pse5Am7Gra transferase activity (Tomek et al., 2017) (Figure 3C, ii). The ΔgtfM mutant (ΔTanf_01260) lacks the 4-O-methyl-N-acetylmannosaminuronic acid residue (231.1 Da) in addition to Pse5Am7Gra, making the signal at m/z = 1306.4 [M+H]+ the largest detectable glycan for this mutant (Figure 3C, iii). The MS data together with the T. forsythia ATCC 43037 O-glycan structure (compare with Figure 1A) are, therefore, indicative of GtfM having a β-1,3-N-acetylmannosaminuronic acid transferase activity. The glycan is further truncated upon knocking-out of gtfI (ΔTanf_01290). In this mutant glycan, the three-sugar branch composed of the Pse5Am7Gra, an N-acetyl mannosaminuronic acid and the 4-O-methyl-N-acetyl mannosaminuronic acid residue (in total 809.1 Da), is absent and results in a signal corresponding to the ammonium adduct ion at m/z = 1106.4 [M+NH4]+ (Figure 3C, iv). Thus, the proposed enzymatic function of GtfI is that of a β-1,4-N-acetylmannosaminuronic acid transferase.
The m/z = 930.3 [M+NH4]+ signal of a ΔgtfL mutant (ΔTanf_01300) could initially not be assigned to the published O-glycan structure (Posch et al., 2011). In our previous study, a fragment ion of m/z = 341.0 was tentatively interpreted as the single cleavage C-ion Fuc-MeGal. However, the m/z = 930.3 glycan of the ΔgtfL mutant, which contains two fucoses and lacks the distal Gal, necessitated another interpretation of this fragment (Figure 4), leading to a reconsideration of the location of the second fucose residue. Inspection of the fragment spectrum of m/z = 930.31+ revealed an m/z = 329.0 fragment not present in the monofucosylated m/z = 784.61+ peak (Figure 4). This Y1βY1γ double cleavage ion strongly indicated that a Fuc residue is attached to the reducing-end Gal. This interpretation was supported by the fragment ion m/z = 651.1, which was observed in the product ion spectra of m/z = 930.31+. Thus, these findings prove that the glycan of this mutant is composed of the six innermost sugar residues, and that GtfL acts as an α-1,2 galactosyltransferase (Figures 1A, 3C, v). Simultaneously, β-1,4 N-acetylmannosaminuronic acid transfer activity of GtfI seems to be affected by the deletion of GtfL, since the Pse5Am7Gra-containing trisaccharide branch is also missing in this structure, indicating the requirement of either a distinct composition of the acceptor for recognition by the enzyme or the necessity of association of GtfI with GtfL for enzymatic activity. Finally, fucosyltransferase activity of GtfE is supported by the analysis of a ΔgtfE knock-out mutant (ΔTanf_01305) which reveals an ammonium adduct mass of m/z = 784.2 [M+NH4]+ that represents an O-glycan composed of the five innermost sugar residues (pentasaccharide core) (Figure 3C, vi).
FIGURE 4.
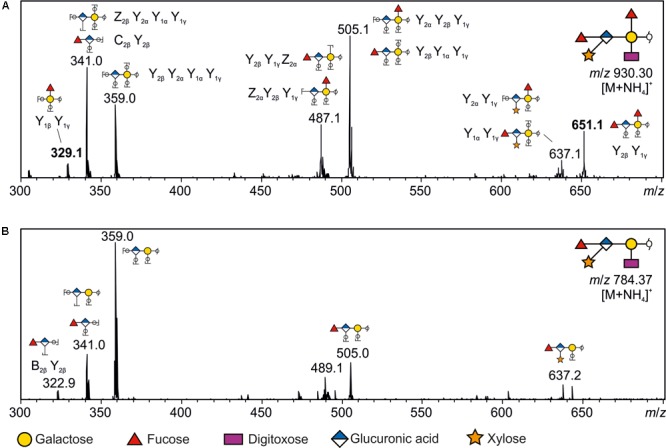
Positive mode CID tandem spectra of reduced O-glycans from glycoproteins in T. forsythiaΔTanf_01300. (A) The m/z = 930.31+ precursor ion corresponds to the glycan composition Gal + GlcA + Xyl + Dig + Fuc2 containing an ammonium adduct [M+NH4]+. Its product ion spectra contained several typical Y/Z and a few B/C ion cleavages that were highly informative to sequence the proposed glycan structure. For example, the C2βY2β (m/z = 341) and B2βY2β (m/z = 322.9; B) ions clearly show that the GlcA is linked to a Fuc at the non-reducing end. Similarly, the reducing end Y1βY1γ double cleavage ion of m/z = 329 is indicative of a Fuc residue linked to the reducing-end Gal. Furthermore, this product ion spectra allows to clearly distinguish the presence of the additional fucose residue, which is lacking in the product ion spectra of m/z 784.371+ (B). Unique fragment ions (m/z = 329 and 651; bold) additionally supporting the location of the additional Fuc residue are only observed in the product ion spectra of m/z = 930.31+. (B) Product ion spectra from [M+NH4]+ ions of m/z = 784.37 corresponding to the composition Gal + GlcA + Xyl + Dig + Fuc. Monosaccharide symbols are according to the Symbol Nomenclature for Glycans (SNFG) (Varki et al., 2015).
These data are complemented by previous results from our group showing that the O-glycan from a wecC knock-out mutant (ΔTanf_01280) lacks the same three-sugar branch as the ΔgtfI mutant (Posch et al., 2011). We concluded that WecC fulfills its predicted role as a UDP-N-acetylmannosaminuronic acid dehydrogenase which works in concert with the predicted UDP-N-acetylglucosamine 2-epimerase WecB (Tanf_01285) to synthesize UDP-N-acetylmannosaminuronic acid prior to its transfer to the branching Fuc residue of the T. forsythia O-glycan structure (compare with Figure 1A), catalyzed by GtfI (Figure 3C, iv).
Summarizing, the T. forsythia protein O-glycosylation gene cluster encodes all necessary information for the assembly of the species-specific portion of the decasaccharide.
Three Methyltransferases Modify the O-Glycan With Two Methyl Groups
While two methylated sugars are present in the T. forsythia ATCC 43037 O-glycan structure, three putative Mtfs are encoded within the protein O-glycosylation gene cluster (MtfJOY). To test the methylation activity and specificity of these predicted enzymes, single gene knock-outs were constructed (Supplementary Figures S7–S9). O-Glycans of mutants, deficient in either the Mtf gene mtfJ (ΔTanf_01250) or mtfO (ΔTanf_01255), experienced a loss of the 4-O-methyl group at the terminal N-acetyl mannosaminuronamide, as MS analyses revealed (Figure 5; red circles indicate the absence of the methyl groups). The expected loss of 14 Da resulted in a doubly charged ion at m/z = 942.9 [M+2H]2+ as detected for the analyzed TfsB (Figure 5) and TfsA (data not shown) O-glycans of either Mtf mutant. While MtfJ and MtfO obviously catalyze methyl transfer to the same sugar residue, these proteins differ in length and do not share significant sequence similarity. In contrast, the O-glycan of the ΔmtfY mutant (ΔTanf_01295) lacked the 4-O-methyl group on the distal Gal residue (Figure 5). The discrimination which of the two methyl groups was affected in the different Mtf mutants was inferred from the occurrence of unique glycan fragments obtained during MS and MS/MS analyses (data not shown).
FIGURE 5.
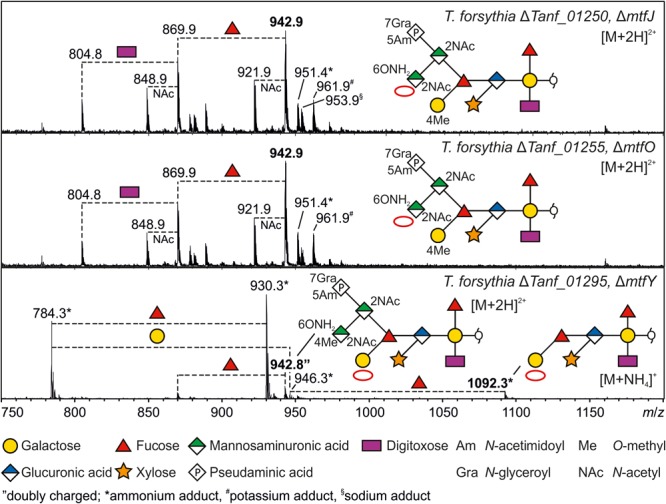
ESI-MS sum spectra of β-eliminated TfsB O-glycans from T. forsythia ATCC 43037 methyltransferase knock-out mutants. The glycan structures of the signals corresponding to the largest mass (bold m/z values) are shown in SNFG representation (Varki et al., 2015). Other O-glycan signals detected for the respective mutants were assigned based on the m/z mass differences corresponding to the loss of individual sugar units and/or modifications. The lack of methyl modifications is indicated by a red circle in the symbolic O-glycan structure representation.
Protein Glycosylation Affects Immunogenicity of T. forsythia
We proceeded to investigate whether differential cell surface protein O-glycosylation as present in the Gtf-deficient mutants of T. forsythia ATCC 43037 impacts the bacterium’s overall immunogenicity. For this endeavor, we selected two mutants with distinct O-glycan compositions for which we expected differential responses of infected immune cells in comparison to the wild-type. This included ΔgtfS, lacking the sialic acid mimic Pse5Am7Gra (Tomek et al., 2017) and ΔgtfE (this study) producing the pentasaccharide core only and, thus, exposing a terminal Fuc residue. Further, ΔgftI which produces the same truncated glycan as ΔwecC (ΔTanf_01280) (Posch et al., 2011) was included as a control, since for that mutant reference immunology data is available in the literature (Settem et al., 2013).
Dendritic cells as central communicators linking innate and adaptive immune responses possess a wide variety of PRRs that allow them to recognize and quickly respond to the presence of opportunistic pathogens (Reis e Sousa, 2006; Merad et al., 2013). The high selectivity of these PRRs enables DCs to fine-tune the outcome of the immune response, depending on the molecular characteristics of the stimulus (Lutz and Schuler, 2002). Therefore, BMDCs and human monocyte-derived DCs were used to explore how the loss of certain sugar residues of the T. forsythia O-glycan affects activation and release of inflammatory cytokines by these cells as well as the subsequent polarization of an adaptive T cell response.
First, the surface expression of DC maturation markers, including MHC-II, CD80 and CD86, upon stimulation by T. forsythia wild-type and the ΔgtfS, ΔgtfI, and ΔgtfE mutants was investigated. Over a period of 24 h, all of these activation markers were up-regulated in response to stimulation and did not reveal any differences between the tested strains (Supplementary Figure S10A). As this suggested no effect on the overall activation of DCs, their functionality was tested next. While secretion of most pro-inflammatory cytokines was not altered between the wild-type and the Gtf-deficient mutants, ΔgftE resulted in a profound increase in IL-1β, IL-12, and IL-23 release by human monocyte-derived DCs over 24 h post-stimulation (Figures 6A and Supplementary Figure S10B), and in TNF-α and IL-6, as observed with mouse-derived DCs (Supplementary Figure S11). Striking differences were observed with regard to IL-1β production; while ΔgftE enhanced secretion of this key inflammatory cytokine in human DCs, ΔgftI and ΔgftS yielded a significant decrease when compared to the wild-type (Figure 6A). A similar effect was observed for IL-10 (Figure 6A), which acts as an important immunosuppressive mediator and is required to dampen ongoing inflammatory responses.
FIGURE 6.
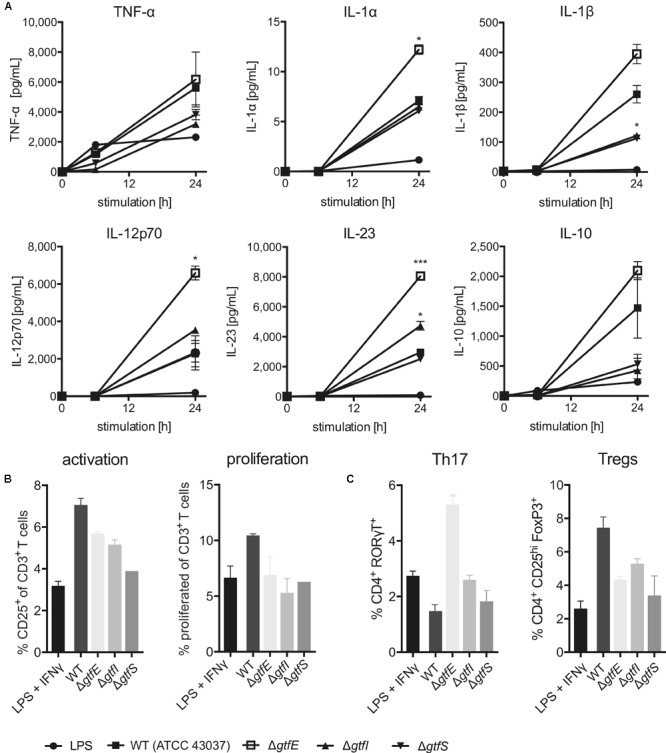
Effects of protein O-glycosylation on T. forsythia immunogenicity. (A) Secretion of inflammatory cytokines by human DCs upon stimulation with T. forsythia wild-type (WT) and glycosyltransferase-deficient mutants (T. forsythia ΔgtfE, ΔgtfI, and ΔgtfS) as measured in culture supernatants by ProcartaPlex Multiplex Immunoassay. (B,C) T cell-priming upon APC stimulation with T. forsythia wild-type and glycosyltransferase-deficient mutants was assessed by culturing PBMCs. (B) T cell activation was measured via expression of CD25 by flow cytometry. Cells were pre-gated for live CD3+ cells, T cell proliferation was assessed after 8 days by CFSE dilution,. (C) CD4+ T cell differentiation was assessed by expression of signature transcription factors for Th17 (RORγT) and Treg (FoxP3) cells as measured by flow cytometry. All data are presented as mean ± SEM of triplicate determinations. One representative out of three (A) and two (B,C), respectively, independent experiments is shown. Statistically significant differences in (A) are indicated as ∗p < 0.05 and ∗∗∗p < 0.001 (Student’s t-test). LPS, E. coli O111:B4 LPS.
These results prompted us to explore the T cell-priming potential of antigen-presenting cells (APCs) upon stimulation by different T. forsythia glycosylation mutants. We therefore cultured PBMCs, containing APCs and T cells, for 8 days in the presence of different T. forsythia strains. With regard to both activation (CD25 up-regulation) and proliferation, a decline in T cell activation upon stimulation with all Gtf-deficient mutants as compared to the wild-type could be observed (Figure 6B). However, dissecting the polarization of the resulting T cell response, we observed a remarkable induction of RORγT-expressing CD4+ T cells by ΔgftE-stimulated DCs, whereas only a minor effect was seen for the other tested strains (Figure 6C). This suggests a trend toward enhanced differentiation of Th17 cells in response to T. forsythia glycan truncation, an effect that has been previously reported to be of relevance for the progression of periodontitis (Settem et al., 2013). Critically, this observation is reinforced by the profound increase in IL-23 and IL-1β we found upon human DC stimulation with ΔgftE (Figure 6A), as these cytokines represent key mediators of Th17 polarization (Iwakura and Ishigame, 2006; Sutton et al., 2009). The differentiation of Tregs was similarly reduced by all truncated glycans in comparison to the wild-type (Figure 6C), indicating an overall involvement of the O-glycan in maintaining a suppressive T cell environment. Moreover, a minor trend toward decreased induction of Th1 (IFN-γ+) cells could be observed (Supplementary Figure S10C), which corresponds to a slightly reduced IFN-γ secretion by human DCs stimulated with T. forsythia Gtf-deficient mutants (Supplementary Figure S10B).
Discussion
Periodontitis is an inflammatory disease that is highly prevalent among the adult population world-wide. It involves the periodontal pathogens T. forsythia, Porphyromonas gingivalis, and Treponema denticola, together constituting the so-called “red complex” of bacteria (Kassebaum et al., 2014; Hajishengallis, 2015). In T. forsythia, the prominent and unique S-layer glycoproteins TfsA and TfsB are virulence factors (Sharma, 2010). While previous studies were designed to elucidate the S-layer O-glycan structure (Posch et al., 2011; Friedrich et al., 2017), the S-layer ultrastructure (Sekot et al., 2012; Oh et al., 2013), and the immunological properties of the S-layer glycoproteins (Yoneda et al., 2003; Sakakibara et al., 2007; Sekot et al., 2011; Chinthamani et al., 2017), only few data is available on the impact of T. forsythia’s cell surface glycosylation on the modulation of host immunity (Settem et al., 2013, 2014). This is surprising given the fact that T. forsythia O-glycans are highly abundant on the bacterial cell surface due to their display via the S-layer matrix (Sekot et al., 2012), making them prone to act at the bacterium-immune interface.
To provide a basis for validating T. forsythia’s S-layer protein O-glycosylation as a possible target against periodontitis, within the frame of this study (i) the prevalence of an underlying protein O-glycosylation gene cluster in the genomes of different T. forsythia strains was assessed, (ii) insights into the O-glycan biosynthesis pathway were obtained, and (iii) a relationship between O-glycan structure and immunogenicity was delineated.
In this study, the general protein O-glycosylation gene cluster of T. forsythia ATCC 43037 was extended to span 27-kb on the bacterium’s genome (Friedrich et al., 2015). For 18 out of the 26 ORFs, carbohydrate metabolism-related functions were predicted. These include, a Wzx-like flippase for translocation of the glycan moiety to the periplasmic space prior to protein transfer, six enzymes needed for the synthesis of CMP-Pse (PseBCFHGI) as well as two enzymes for the synthesis of UDP-linked N-acetyl mannosaminuronic acid (WecB, WecC), five Gtfs (GtfSMILE), three Mtfs (MtfJOY), and one ORF putatively involved in the Am-modification of the Pse residue (Figure 1B).
Genome comparisons of T. forsythia strains revealed that all seven strains included in this study have the genetic potential for protein O-glycosylation (Figure 2). A high degree of conservation of the protein O-glycosylation gene clusters with regard to content and organization of genes became evident, with a possible grouping of strains according to the type of terminal nonulosonic acid synthesized (Pse versus Leg), with only one strain presenting an ambiguous situation. As a common feature, the O-glycosylation gene clusters in all analyzed strains start with orthologs of the Gtf genes gtfE (Tanf_01305) and gtfL (Tanf_01300) and terminate with an ortholog of the flippase gene wzx (Tanf_01180), which is identical to the situation found in 24 other Bacteroides species (Coyne et al., 2013). While orthologs of wzx and gtfE are also predicted in the genome of the periodontal health-associated isolate Tannerella sp. HOT-286 (Beall et al., 2014), none of the intervening genes related to carbohydrate metabolism as present in the pathogenic strains are confidently predicted. Thus, it is conceivable that the potential of complex protein O-glycosylation is a specific trait of periodontitis-associated T. forsythia strains, especially when considering that cell surface glycosylation in bacteria can modulate immune responses during pathogenesis (Szymanski and Wren, 2005; Lebeer et al., 2010; Valguarnera et al., 2016).
Experimentally, we have determined the roles of five Gtfs and three Mtfs encoded in the protein O-glycosylation gene cluster by analysing the protein-released glycans of the respective knock-out mutants. We could establish an assembly line for the T. forsythia-specific portion of the O-glycan by subsequent involvement of the Gtfs GtfELIMS (Figure 7). While the underlying genetic information is encoded in the gene cluster, the information for the assembly of the pentasaccharide core would be encoded elsewhere on the bacterial genome. A putative O-OTase transferring the glycan onto the acceptor proteins could not be identified for T. forsythia, so far. This leaves the question open of whether this O-glycan is synthesized by an OTase-dependent or OTase-independent mechanism (Choi et al., 2010; Grass et al., 2010; Schwarz et al., 2011; Iwashkiw et al., 2013). Coyne et al. (2013) suggested two possible models for protein O-glycosylation in Bacteroidetes, exemplified with B. fragilis. According to the first model, the glycan core and the species-specific glycan would be built separately on an undecaprenyl-phosphate (undP) lipid carrier and flipped to the periplasmic side of the cytoplasmic membrane by individual Wzx flippases. The core glycan would be linked to the protein by an as yet unidentified OTase, followed by linking of the species-specific glycan portion to the core by an O-antigen ligase-like protein. Alternatively, the species-specific glycan might be added to the core in the cytoplasm and then a single flippase would transfer the entire glycan to the periplasm for addition to proteins (Fletcher et al., 2009; Coyne et al., 2013). Considering our experimental set-up we cannot make any conclusion as to which of the two models would be valid for T. forsythia.
FIGURE 7.
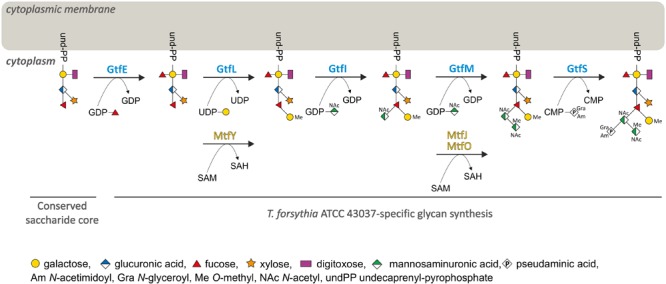
Model for the biosynthesis of the species-specific portion of the T. forsythia ATCC 43037 O-glycan. Upon synthesis of the pentasaccharide core on an undP lipid carrier, the first carbohydrate residue of the species-specific glycan is a Fuc residue conferred by GtfE. The glycan is elongated with a Gal residue which is transferred by GtfL and methylated by MtfY. The assembly of the three sugar branch, consisting of a ManNAcA residue (transferred by GtfI), a ManNAcCONH2 residue (GtfM), which is methylated by either MtfJ or MtfO, and a Pse5Am7Gra residue (transferred via GtfS), completes the synthesis of the decasaccharide.
Concerning 4-O-methylation of the distal Gal and the N-acetyl mannosaminuronamide residue, there is redundancy of enzymes, since the latter residue can be methylated by both MtfJ and MtfO, while MtfY recognizes only the distal Gal residue as a substrate. It remains to be investigated if MtfJ and MtfO might be differentially expressed in T. forsythia as a function of growth conditions or other signals and, generally, if methylation occurs at the nucleotide-sugar level or after sugar transfer to the nascent glycan. Methylation of bacterial glycans is a rarely reported event. Although methylation of the non-reducing-end sugar of a nascent polysaccharide chain has been discussed as a stop-signal for glycan chain elongation (Clarke et al., 2011), a clear determination of its biological function is still missing, especially when occurring at multiple sites of the glycan (Staudacher, 2012). O-Methylated glycans were recently suggested to constitute a conserved target of the fungal and animal innate immune system (Wohlschlager et al., 2014). All three Mtf-deficient T. forsythia mutants from our study might aid in defining the roles of O-methylated sugars in a bacterial context.
In the natural habitat of the oral cavity, T. forsythia finds itself in an area of conflict between the requirement of inflammation to procure nutrients from tissue breakdown and the necessity to evade immune-mediated killing (Settem et al., 2013, 2014). Similar to other inflammatory diseases, disruption of the proper balance between individual subtypes of Th cells contributes to the progression of periodontitis (Baker, 2000). In a previous study using a T. forsythia ΔwecC glycosylation mutant (representing the same glycan structure as the ΔgtfI mutant of the current study), it was found that truncation of the O-glycan translated into a robust Th17 response with the consequence of reduced alveolar bone loss in mice. Additionally, the ΔwecC mutant was increasingly susceptible to neutrophil-mediated clearance. Based on these data a fundamental role of T. forsythia wild-type cell surface glycosylation in restraining the Th17 response and ensuring the persistence of the pathogen in the host was proposed (Settem et al., 2013). The attenuated, Th17-biasing glycosylation mutant devoid of the O-glycan’s trisaccharide branch was effective in blocking P. gingivalis persistence in a periodontitis mouse model (Settem et al., 2014), unraveling the T. forsythia O-glycan as a means for influencing the pathogenesis of periodontitis. However, it is still under debate if Th17 responses have protective or destructive roles in inflammatory diseases, since this might depend on the specificity and phase of the disease (Ohyama et al., 2009).
Using T. forsythia mutants with defined, truncated O-glycans, we attempted to obtain clearer insight into the O-glycan structure–function relationship at the immune interface. Analysis of DC maturation markers did not reveal differential activation by the glycosylation mutants in comparison to the wild-type. In terms of release of proinflammatory cytokines by human DCs, only T. forsythiaΔgftE resulted in a profound increase in TNF-α, IL-6, IL-1β, IL-12, and IL-23 release over 24 h post-stimulation (Figures 6A and Supplementary Figure S11). Especially with the enhanced secretion of IL-1β and IL-23, which are key mediators of Th17 polarization (Iwakura and Ishigame, 2006; Sutton et al., 2009), this mutant showed a striking difference to ΔgftI and ΔgftS where a decrease of these cytokines was found (Figure 6A). In line with these data, also the analysis of the T cell-priming potential of APCs upon stimulation revealed T. forsythiaΔgftE as a unique stimulus. Here, a remarkable induction of RORγT-expressing CD4+ T cells was observed, whereas only a minor effect was seen for the other tested mutants (Figure 6C). This suggests an enhanced differentiation of Th17 cells in response to T. forsythia glycan truncation, which was most pronounced upon stimulation with the pentasaccharide core only. For the ΔgftI mutant, equaling O-glycan structure-wise the ΔwecC mutant for which this effect had been reported before (Settem et al., 2013), a comparably minor effect was seen. We conclude that a more truncated version of the O-glycan is even more favorable for inducing a robust Th17 response. Vice versa, Th17 suppression is maintained regardless of the presence of the residues N-acetyl mannosaminuronic acid, N-acetyl mannosaminuronamide and Pse5Am7Gra. This implicates that the phylum-wide conserved core as present in ΔgftE is responsible for modulating DC effector functions by Th17 activation, while the species-specific glycan portion, either in complete or truncated form, is required for maintaining a Th17 suppressive environment. This might contribute to T. forsythia’s strategy as part of a dysbiotic microbial community to resist immune elimination and create permissive conditions for growth in a nutritionally favorable environment.
Author Contributions
CS and MT conceived the study, designed the experiments, developed the methodology, and wrote the manuscript. MT, DM, MW, VF, BJ, KF, LN, IN, NZ, AE-D, and DK performed the experiments. MT, DM, NZ, JD, AE-D, DK, HH, FA, and CS analyzed the data. All authors read and approved the submitted version of the manuscript. Written informed consent was obtained from all authors of the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge St. Anna Kinderkrebsforschung, Children’s Cancer Research Institute, Medical University of Vienna, Vienna, Austria, for providing access to the immunology facility.
Abbreviations
- Ac
N-acetyl or acetamido
- Am
N-acetimidoyl or acetamidino
- APC
antigen presenting cell
- BHI
brain heart infusion
- BMDC
bone marrow-derived dendritic cell
- Cat
chloramphenicol
- cDNA
copy DNA
- CFSE
carboxyfluorescein succinimidyl ester
- CMP
cytidine-5′-monophosphate
- Dig
digitoxose
- DC
dendritic cell
- DMEM
Dulbecco’s Modified Eagle’s Medium
- Erm
erythromycin
- ESI
electrospray ionization
- Fuc
fucose
- Gal
galactose
- Gc
N-glycolyl
- GDP
guanosine-5′-diphosphate
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- Gra
N-glyceroyl or N-2,3-dihydroxypropionyl or glycerate group
- Gtf
glycosyltransferase
- IL
interleukin
- INF
interferon
- LB
lysogeny broth
- LC
liquid chromatography
- Leg
legionaminic acid (Leg5,7Ac2), 5,7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-galacto-nonulosonic acid
- LFG
locus for glycosylation
- LPS
lipopolysaccharide
- MHC
major histocompatibility complex
- Mtf
methyltransferase
- MS
mass spectrometry
- OP
Octaplas medium
- ORF
open reading frame
- OTase
oligosaccharyltransferase
- PBMC
peripheral blood mononuclear cell
- PGC
porous graphitized carbon
- PRR
pattern recognition receptor
- Pse
pseudaminic acid (Pse5,7Ac2), 5,7-diacetamido-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic acid
- SAM
S-adenosyl-L-methionine
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Th
T helper cell
- TNF
tumor necrosis factor
- Tregs
regulatory T cells
- UDP
uridine-5′-diphosphate
- und-PP
undecaprenyl pyrophosphate
- Xyl
xylose
Funding. This work was supported by the Austrian Science Fund FWF, projects P24317-B22 and I2875-B22 (to CS) and the Doctoral Program “BioToP – Biomolecular Technology of Proteins” W1224. DK is the recipient of an Australian Research Council Future Fellowship (project number FT160100344) funded by the Australian Government.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02008/full#supplementary-material
References
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J. (2000). The role of immune responses in bone loss during periodontal disease. Microbes Infect. 2 1181–1192. 10.1016/S1286-4579(00)01272-7 [DOI] [PubMed] [Google Scholar]
- Beall C. J., Campbell A. G., Dayeh D. M., Griffen A. L., Podar M., Leys E. J. (2014). Single cell genomics of uncultured, health-associated Tannerella BU063 (Oral Taxon 286) and comparison to the closely related pathogen Tannerella forsythia. PLoS One 9:e89398. 10.1371/journal.pone.0089398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch S., Thurnheer T., Murakami Y., Belibasakis G. N., Schäffer C. (2017). Behavior of two Tannerella forsythia strains and their cell surface mutants in multispecies oral biofilms. Mol. Oral Microbiol. 32 404–418. 10.1111/omi.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch S., Zwicker S., Bostanci N., Sjöling A., Bostrom E. A., Belibasakis G. N., et al. (2018). Immune response profiling of primary monocytes and oral keratinocytes to different Tannerella forsythia strains and their cell surface mutants. Mol. Oral Microbiol. 33 155–167. 10.1111/omi.12208 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A., Maass K., Geyer H., Geyer R., Dell A., Haslam S. M. (2008). GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7 1650–1659. 10.1021/pr7008252 [DOI] [PubMed] [Google Scholar]
- Cheng H. R., Jiang N. (2006). Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28 55–59. 10.1007/s10529-005-4688-z [DOI] [PubMed] [Google Scholar]
- Chinthamani S., Settem R. P., Honma K., Kay J. G., Sharma A. (2017). Macrophage inducible C-type lectin (Mincle) recognizes glycosylated surface (S)-layer of the periodontal pathogen Tannerella forsythia. PLoS One 12:e0173394. 10.1371/journal.pone.0173394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-J., Grass S., Paek S., St. Geme J. W., III, Yeo H.-J. (2010). The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS One 5:e15888. 10.1371/journal.pone.0015888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. R., Richards M. R., Greenfield L. K., Hou D., Lowary T. L., Whitfield C. (2011). In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: the chain-length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J. Biol. Chem. 286 41391–41401. 10.1074/jbc.M111.295857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. J., Fletcher C. M., Chatzidaki-Livanis M., Posch G., Schäffer C., Comstock L. E. (2013). Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol. Microbiol. 88 772–783. 10.1111/mmi.12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge-Jacobsen W., Salomonsson E. N., Aas F. E., Forslund A.-L., Winther-Larsen H. C., Maier J., et al. (2011). O-linked glycosylation of the PilA pilin protein of Francisella tularensis: identification of the endogenous protein-targeting oligosaccharyltransferase and characterization of the native oligosaccharide. J. Bacteriol. 193 5487–5497. 10.1128/JB.00383-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W., Scott N. E., Tondo M. L., Orellano E. G., Foster L. J., Feldman M. F. (2016). Protein O-linked glycosylation in the plant pathogen Ralstonia solanacearum. Glycobiology 26 301–311. 10.1093/glycob/cwv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espitia C., Servin-Gonzalez L., Mancilla R. (2010). New insights into protein O-mannosylation in actinomycetes. Mol. Biosyst. 6 775–781. 10.1039/b916394h [DOI] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. (2009). A general O-glycosylation system important to the physiology of a major human intestinal symbiont Bacteroides fragilis. Cell 137 321–331. 10.1016/j.cell.2009.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich V., Janesch B., Windwarder M., Maresch D., Braun M. L., Megson Z. A., et al. (2017). Tannerella forsythia strains display different cell-surface nonulosonic acids: biosynthetic pathway characterization and first insight into biological implications. Glycobiology 27 342–357. 10.1093/glycob/cww129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich V., Pabinger S., Chen T., Messner P., Dewhirst F. E., Schäffer C. (2015). Draft genome sequence of Tannerella forsythia type strain ATCC 43037. Genome Announc. 3:e00660-15. 10.1128/genomeA.00660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci S., Lolkema M., Matzinger P. (1999). Natural adjuvants: endogenous activators of dendritic cells. Nat. Med. 5 1249–1255. 10.1038/15200 [DOI] [PubMed] [Google Scholar]
- Grass S., Lichti C. F., Townsend R. R., Gross J., St Geme J. W., III (2010). The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6:e1000919. 10.1371/journal.ppat.1000919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. (2015). Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15 30–44. 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27 409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. M., Nasr M. A., Kinsella R. L., Scott N. E., Foster L. J., Weber B. S., et al. (2015). Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol. Microbiol. 96 1023–1041. 10.1111/mmi.12986 [DOI] [PubMed] [Google Scholar]
- Hartley M. D., Morrison M. J., Aas F. E., Borud B., Koomey M., Imperiali B. (2011). Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50 4936–4948. 10.1021/bi2003372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. C., Ebersole J. L. (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ”red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 38 72–122. 10.1111/j.1600-0757.2005.00113.x [DOI] [PubMed] [Google Scholar]
- Honma K., Inagaki S., Okuda K., Kuramitsu H. K., Sharma A. (2007). Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42 156–166. 10.1016/j.micpath.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishigame H. (2006). The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 116 1218–1222. 10.1172/JCI28508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw J. A., Fentabil M. A., Faridmoayer A., Mills D. C., Peppler M., Czibener C., et al. (2012). Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb. Cell Fact. 11:13. 10.1186/1475-2859-11-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashkiw J. A., Vozza N. F., Kinsella R. L., Feldman M. F. (2013). Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89 14–28. 10.1111/mmi.12265 [DOI] [PubMed] [Google Scholar]
- Kassebaum N. J., Bernabe E., Dahiya M., Bhandari B., Murray C. J., Marcenes W. (2014). Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J. Dent. Res. 93 1045–1053. 10.1177/0022034514552491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S. C., Schulz B. L., Power P. M., Jennings M. P. (2009). The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem. Biophys. Res. Commun. 378 84–89. 10.1016/j.bbrc.2008.11.025 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lamont R. J., Hajishengallis G. (2015). Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21 172–183. 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzinger M., Jürgens B., Hainz U., Dillinger B., Raberger J., Fuchs D., et al. (2012). Ambivalent effects of dendritic cells displaying prostaglandin E2-induced indoleamine 2,3-dioxygenase. Eur. J. Immunol. 42 1117–1128. 10.1002/eji.201141765 [DOI] [PubMed] [Google Scholar]
- Lebeer S., Vanderleyden J., De Keersmaecker S. C. (2010). Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8 171–184. 10.1038/nrmicro2297 [DOI] [PubMed] [Google Scholar]
- Lee S. W., Sabet M., Um H. S., Yang J., Kim H. C., Zhu W. (2006). Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene 371 102–111. 10.1016/j.gene.2005.11.027 [DOI] [PubMed] [Google Scholar]
- Lees-Miller R. G., Iwashkiw J. A., Scott N. E., Seper A., Vinogradov E., Schild S., et al. (2013). A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89 816–830. 10.1111/mmi.12300 [DOI] [PubMed] [Google Scholar]
- Lithgow K. V., Scott N. E., Iwashkiw J. A., Thomson E. L. S., Foster L. J., Feldman M. F., et al. (2014). A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol. Microbiol. 92 116–137. 10.1111/mmi.12540 [DOI] [PubMed] [Google Scholar]
- Lutz M. B., Schuler G. (2002). Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 23 445–449. 10.1016/S1471-4906(02)02281-0 [DOI] [PubMed] [Google Scholar]
- Lyons A. B., Parish C. R. (1994). Determination of lymphocyte division by flow cytometry. J. Immunol. Methods 171 131–137. 10.1016/0022-1759(94)90236-4 [DOI] [PubMed] [Google Scholar]
- Martin J. L., McMillan F. M. (2002). SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12 783–793. 10.1016/S0959-440X(02)00391-3 [DOI] [PubMed] [Google Scholar]
- Medema M. H., Takano E., Breitling R. (2013). Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30 1218–1223. 10.1093/molbev/mst025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31 563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Szymanski C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8 765–778. 10.1038/nrmicro2383 [DOI] [PubMed] [Google Scholar]
- Oh Y. J., Sekot G., Duman M., Chtcheglova L., Messner P., Peterlik H., et al. (2013). Characterizing the S-layer structure and anti-S-layer antibody recognition on intact Tannerella forsythia cells by scanning probe microscopy and small angle X-ray scattering. J. Mol. Recognit. 26 542–549. 10.1002/jmr.2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama H., Kato-Kogoe N., Kuhara A., Nishimura F., Nakasho K., Yamanegi K., et al. (2009). The involvement of IL-23 and the Th17 pathway in periodontitis. J. Dent. Res. 88 633–638. 10.1177/0022034509339889 [DOI] [PubMed] [Google Scholar]
- Posch G., Pabst M., Brecker L., Altmann F., Messner P., Schäffer C. (2011). Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286 38714–38724. 10.1074/jbc.M111.284893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G., Pabst M., Neumann L., Coyne M. J., Altmann F., Messner P., et al. (2013). ”Cross-glycosylation” of proteins in Bacteroidales species. Glycobiology 23 568–577. 10.1093/glycob/cws172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis e Sousa C. (2006). Dendritic cells in a mature age. Nat. Rev. Immunol. 6 476–483. 10.1038/nri1845 [DOI] [PubMed] [Google Scholar]
- Sabet M., Lee S. W., Nauman R. K., Sims T., Um H.-S. (2003). The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology 149 3617–3627. 10.1099/mic.0.26535-0 [DOI] [PubMed] [Google Scholar]
- Sakakibara J., Nagano K., Murakami Y., Higuchi N., Nakamura H., Shimozato K., et al. (2007). Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153 866–876. 10.1099/mic.0.29275-0 [DOI] [PubMed] [Google Scholar]
- Schäffer C., Messner P. (2017). Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41 49–91. 10.1093/femsre/fuw036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert H. L., Blumenthal R. M., Cheng X. (2003). Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28 329–335. 10.1016/S0968-0004(03)00090-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F., Fan Y. Y., Schubert M., Aebi M. (2011). Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J. Biol. Chem. 286 35267–32274. 10.1074/jbc.M111.277160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G., Posch G., Messner P., Matejka M., Rausch-Fan X., Andrukhov O., et al. (2011). Potential of the Tannerella forsythia S-layer to delay the immune response. J. Dent. Res. 90 109–114. 10.1177/0022034510384622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G., Posch G., Oh Y. J., Zayni S., Mayer H. F., Pum D., et al. (2012). Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch. Microbiol. 194 525–539. 10.1007/s00203-012-0792-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem R. P., Honma K., Nakajima T., Phansopa C., Roy S., Stafford G. P., et al. (2013). A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 6 415–426. 10.1038/mi.2012.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem R. P., Honma K., Sharma A. (2014). Neutrophil mobilization by surface-glycan altered th17-skewing bacteria mitigates periodontal pathogen persistence and associated alveolar bone loss. PLoS One 9:e108030. 10.1371/journal.pone.0108030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. (2010). Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 106–116. 10.1111/j.1600-0757.2009.00332.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Inagaki S., Honma K., Sfintescu C., Baker P. J., Evans R. T. (2005). Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J. Dent. Res. 84 462–467. 10.1177/154405910508400512 [DOI] [PubMed] [Google Scholar]
- Socransky S., Haffajee A., Cugini M., Smith C., Kent R. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25 134–144. 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- Stafford G. P., Chaudhuri R. R., Haraszthy V., Friedrich V., Schäffer C., Ruscitto A., et al. (2016). Draft genome sequences of three clinical isolates of Tannerella forsythia isolated from subgingival plaque from periodontitis patients in the United States. Genome Announc. 4:e01286-16. 10.1128/genomeA.01286-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher E. (2012). Methylation – an uncommon modification of glycans. Biol. Chem. 393 675–685. 10.1515/hsz-2012-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C. E., Lalor S. J., Sweeney C. M., Brereton C. F., Lavelle E. C., Mills K. H. (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31 331–341. 10.1016/j.immuni.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Szymanski C. M., Logan S. M., Linton D., Wren B. W. (2003). Campylobacter – a tale of two protein glycosylation systems. Trends Microbiol. 11 233–238. 10.1016/S0966-842X(03)00079-9 [DOI] [PubMed] [Google Scholar]
- Szymanski C. M., Wren B. W. (2005). Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3 225–237. 10.1038/nrmicro1100 [DOI] [PubMed] [Google Scholar]
- Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999). Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32 1022–1030. 10.1046/j.1365-2958.1999.01415.x [DOI] [PubMed] [Google Scholar]
- Tanner A., Listgarten M., Ebersole J., Strzempko M. (1986). Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int. J. Syst. Evol. Microbiol. 36 213–221. 10.1099/00207713-36-2-213 [DOI] [Google Scholar]
- Tomek M. B., Janesch B., Maresch D., Windwarder M., Altmann F., Messner P., et al. (2017). A pseudaminic acid or a legionaminic acid derivative transferase is strain-specifically implicated in the general protein O-glycosylation system of the periodontal pathogen Tannerella forsythia. Glycobiology 27 555–567. 10.1093/glycob/cwx019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomek M. B., Neumann L., Nimeth I., Koerdt A., Andesner P., Messner P., et al. (2014). The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol. Oral Microbiol. 29 307–320. 10.1111/omi.12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upreti R. K., Kumar M., Shankar V. (2003). Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3 363–379. 10.1002/pmic.200390052 [DOI] [PubMed] [Google Scholar]
- Valguarnera E., Kinsella R. L., Feldman M. F. (2016). Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J. Mol. Biol. 428 3206–3220. 10.1016/j.jmb.2016.04.013 [DOI] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., et al. (2015). Symbol nomenclature for graphical representations of glycans. Glycobiology 25 1323–1324. 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W. (2017). Essentials of Glycobiology. Cold Spring, NY: Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Vartoukian S. R. (2016). Cultivation strategies for growth of uncultivated bacteria. J. Oral Biosci. 58 142–149. 10.1016/j.job.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik Å., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W.et al. (2009). Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 106 4447–4452. 10.1073/pnas.0809504106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlager T., Butschi A., Grassi P., Sutov G., Gauss R., Hauck D., et al. (2014). Methylated glycans as conserved targets of animal and fungal innate defense. Proc. Natl. Acad. Sci. U.S.A. 111 E2787–E2796. 10.1073/pnas.1401176111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M., Hirofuji T., Motooka N., Nozoe K., Shigenaga K., Anan H., et al. (2003). Humoral immune responses to S-layer-like proteins of Bacteroides forsythus. Clin. Diagn. Lab. Immunol. 10 383–387. 10.1128/CDLI.10.3.383-387.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarschler K., Janesch B., Zayni S., Schäffer C., Messner P. (2009). Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl. Environ. Microbiol. 75 3077–3085. 10.1128/AEM.00087-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


