Abstract
Objective:
Emerging evidence indicates that long non-coding RNAs (lncRNAs) are critical in carcinogenesis and progression of ovarian cancer. This study aimed to explore the functions and molecular mechanisms of plasmacytoma variant translocation I (PVT1) in ovarian cancer
Methods:
PVT1 and miR-214 were detected by qRT-PCR assays in ovarian cancer tissues and cells. The cell proliferation, migration, and invasion abilities were detected by cell functional experiments, respectively. Western blot assay was performed to detect epithelial-mesenchymal transition (EMT) markers. MiR-214 expression regulated by PVT1 was studied by RNA immunoprecipitation (RIP) and chromatin immunoprecipitation (ChIP) assays.
Results:
The expression of PVT1 was up-regulated in ovarian cancer tissues and cell lines. Elevated PVT1 expression was associated with advanced stage and indicated poor prognosis for ovarian cancer patients. The knockdown of PVT1 impaired SKOV3 cell proliferation, migration, and invasion in vitro. The promotion of ovarian cancer progression by PVT1 involved in regulation of the epithelial-mesenchymal transition process and PVT1 interaction with EZH2 represses miR-214 expression in ovarian cancer cells.
Conclusions:
PVT1 plays an important role in ovarian cancer tumorigenesis, which might be as a novel diagnostic marker and therapeutic target for ovarian cancer.
Keywords: Long non-coding RNA, PVT1, ovarian cancer, miR-214
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecologic malignancy. There were about 22,440 newly confirmed cases of ovarian cancer and estimated 14,080 EOC-related deaths in the United States in 20171. More than 70% cases of patients with ovarian cancer are diagnosed in an advanced stage (FIGO stage III or IV) owing to nonspecific symptoms in the early stage and because of the lack of effective screening methods2. The mortality rate among EOC patients remains high and the 5-year overall survival (OS) is only 20%–40% for advanced disease, although some progress has been achieved in EOC treatment3.
MicroRNA (miRNA) and long non-coding RNA (lncRNA) are two important types of non-coding RNA (ncRNA), which play multiple roles in the initiation and progression of various cancers4,5. Some miRNAs and lncRNAs may be useful biomarkers for the early detection and response to treatment of some cancers6. MiRNA is a small non-coding RNA comprising 20–22 nucleotides (nt). It can suppress gene expression by binding with the 3′-untranslated regions (3′-UTRs) of mRNAs and causing mRNA degradation or repression of translation7. LncRNAs are longer than 200 nt in length. They display limited protein-coding potential and have been shown to regulate a variety of cellular processes, such as cell proliferation, apoptosis, cell migration, and tumor metastasis8. The underlying mechanisms of lncRNAs are varied and include acting as competitive endogenous RNAs (ceRNAs) similar to miRNA sponges, chromatin remodeling, and histone-protein modification9.
Hirata et al.10 reported that lncRNA MALAT1 and miR-205 could interact and repress each other, finally acting as crucial regulators for the function of renal cancer. Wan et al.11 also reported that lncRNA PVT1 promoted non-small cell lung cancer cell proliferation through binding to the EZH2 protein and epigenetically regulating LATS2 expression.
The gene encoding plasmacytoma variant translocation I (PVT1) is 1,716 nt in length and it is located in the chr8q24.21 region12. PVT1 has been identified as an oncogene owing to its contribution to the phenotype of multiple cancers13-15. However, the role of lncRNA PVT1 in ovarian cancer is unclear.
In this study, we evaluated the expression of PVT1 and miR-214 in ovarian cancer and normal ovarian tissues. The relationship of PVT1 and miR-214 expression with the clinicopathologic characteristics and prognosis of EOC patients were analyzed. The analyses were intended to clarify the effects of PVT1 on the malignant behaviors of ovarian cancer cells in vitro. The mechanism experiments indicated that PVT1 may recruit EZH2 to the miR-214 promoter and inhibit miR-214 expression. This study will help clarify the role of PVT1 in ovarian cancer progression and its potential as a therapeutic target.
Material and methods
Patient collection
This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Informed consent was obtained from all patients prior to their enrollment in the present study.
A total of 231 patients with EOC, who underwent cytoreductive abdominal surgery between January 2012 and December 2014 were enrolled. Clinical information of all patients was acquired from medical records. The inclusion criteria were: tumor histology originating from epithelial tissue; staging according to the International Federation of Gynecology and Obstetrics (FIGO), including stage I, II, III, and IV; and defined surgical strategies. All patients underwent total hysterectomy with bilateral salpingo-oophorectomy, omentectomy, and appendectomy that were supplemented with partial resection of infiltrated intestine or bowel, peritonectomy, or splenectomy for the purposes of optical cytoreduction. Systemic or sampling lymphadenectomy was performed only in cases when optimal cytoreduction was achieved or in the presence of bulky nodes. Adjuvant therapeutic strategies for all patients included chemotherapy with a platinum-paclitaxel regimen, six standard courses of carboplatin (area under the curve, 5–7.5) and paclitaxel 135–175 mg/m2. The adjuvant treatment was modified according to patients' general status and was used in all cases. Clinical characterizations of patients with ovarian cancer are presented in Table 1. Additionally, the control group consisted of 58 normal ovarian tissue samples that were retrieved from peri-menopausal women during hysterectomy with bilateral salpingo-oophorectomy due to benign uterine disease (uterine leiomyoma).
1.
The characteristics of EOC patients
| Characteristics | Case (n, %) |
| *Others: others include the endometrioid, clear cell, and undifferentiated ovarian cancers.
**FIGO: Federation International of Gynecology and Obstetrics. | |
| Age at surgery (years) | Media: 51; range: 28–76 |
| ≤51 | 136 (58.9) |
| >51 | 95 (41.1) |
| Body mass index (kg/m2) | 24.18±3.78 |
| Menopausal status | |
| Yes | 154 (66.7) |
| No | 77 (33.3) |
| Pathologic type | |
| Serous | 116 (50.2) |
| Mucous and others* | 115 (49.8) |
| Histologic grade | |
| G1–2 | 141 (61.0) |
| G3 | 90 (39.0) |
| FIGO stage** | |
| I–II | 81 (35.1) |
| III–IV | 150 (64.9) |
| Lymph nodes metastasis | |
| No | 186 (80.5) |
| Yes | 45 (19.5) |
| Residual tumor (cm) | |
| <1 | 187 (81.0) |
| ≥ 1 | 44 (19.0) |
| Ascites volume (mL) | Media: 1000; range: 200–7000 |
| ≤1000 | 157 (68.0) |
| >1000 | 74 (32.0) |
| Serum CA125 (U/mL) | Media: 675; range: 23–5400 |
| ≤675 | 140 (60.6) |
| >675 | 91 (39.4) |
RNA extraction and real-time quantitative PCR analysis (RT-qPCR)
Total RNA was extracted from cultured cells or tissues using TRIzol Reagent (Takara Bio, Dalian, China). The reaction mixture (20 μL) containing 1 μg total RNA was reversely transcribed to cDNA using PrimeScript RT-polymerase (Takara Bio). RNA expression levels were measured by RT-qPCR using SYBR Green PCR Master Mix (Takara Bio). U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as internal controls. The reactions were performed using a model 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Comparative quantification was determined using the 2–ΔΔCt method. The primers used were PVT1-forward: 5′-TGAGAACTGTCCTTACGTGACC-3′.
PVT1-reverse: 5′-AGAGCACCAAGACTGGCTCT-3′.
MiR-214-forward: 5′-ACA GCA GGC ACA GAC AGG CAG U-3′.
Preparation of cells
Ovarian cancer cell lines (HO8910, SKOV3, ES2, SW626, and A2780) were purchased from the Chinese Academy of Sciences (Shanghai, China). A2780, HO8910, and ES2 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS). SW626 cells were cultured in Leibovitz’s L-15 medium (ATCC, Manassas, VA, USA) with 10% FBS. SKOV3 cells were cultured in McCoy’s 5A (Gibco, USA) supplemented with 10% FBS (Invitrogen). Human primary cultured ovarian cells (PriCells, Wuhan, China) were gifted by Prof. Xiubao Ren (Tianjin Medical University Cancer Institute and Hospital). The cells were cultured in DMEM (ATCC) supplemented with 10% FBS, penicillin 100 U/mL, and streptomycin 100 mg/mL in humidified air at 37°C in an atmosphere of 5% CO2.
Cell transfection
Cells (1×105) were seeded into each well of 6-well plates and incubated for 24 h. Cells were then transfected with PVT1 small interfering RNA (siRNA) oligos, siRNA negative control (si-NC), miR-214 inhibitor, or miR-NC oligos using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions.
Cell proliferation assay
SKOV3 cells (1×104 per well) transfected with si-NC or si-PVT1 were cultured in 96-well plates. Proliferation was assessed using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) at 1, 2, 3, 4, and 5 days according to the manufacturer’s instructions. Absorbance was determined at 450 nm using an Elx800 reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Wound healing assay
Cells were cultured in McCoy’s 5A media containing 5% FBS in 35 mm Petri dishes. Once cells reach 90% confluence media was aspirated and cells were washed twice with PBS and seeded in starvation media of McCoy’s 5A and 1% FBS. After overnight culture, media were discarded and cells were washed twice with PBS. Using a 200 μL pipette tip three separate wounds were created across the monolayer cultures perpendicular to the bottom line. The distances of wound were determined at different time intervals (0, 24, and 48 h).
Cell invasion assay
Cell invasion was assessed by Transwell chambers precoated with Matrigel (BD, Franklin Lakes, NJ, USA). After incubation for 48 hours, cells on the upper surface of the Transwell inserts were removed and cells on the lower surface were fixed and then stained with 0.5% crystal violet solution. Cells were counted in five random fields in each well.
Western blot analysis
Proteins from cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then immunodetection was performed with standard techniques. Antibodies to E-cadherin, vimentin, β-catenin, Snail, Slug, and β-actin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Signals were visualized with SuperSignal® West Pico Chemoluminescent Substrate (Pierce, Rockford, IL, USA) by exposure to films.
RNA immunoprecipitation (RIP)
The RIP assay was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) following manufacturer’s instructions. Total cell lysate was used for immunoprecipitation with EZH2 antibody. Rabbit IgG antibody was used as the control.
Chromatin immunoprecipitation (ChIP)
Supernatants were subjected to immunoprecipitation overnight with antibodies against EZH2 (Cell Signaling Technology, Danvers, MA, USA) or with isotype rabbit IgG (Cell Signaling Technology) at 4°C overnight. Chromatin-antibody complexes were isolated by Protein A/G PLUS agarose (Santa Cruz Biotechnology). The crosslinks for the enriched and the input DNA were reversed and the DNA was cleaned by RNase A and proteinase K before phenol/chloroform-purification. For quantification of the enrichment of miR-214 at promoter regions, qRT-PCR was performed using the ABI 7500 Real-Time PCR System (Applied Biosystems). The results are representative of at least three independent experiments.
Statistical analysis
Continuous variables are expressed as the mean ± standard deviation (SD). These variables were analyzed using t-test or one-way ANOVA. Chi-squared and Fisher’s exact tests were applied in the analysis of categorical variables. The Spearman rank correlation was used to assess the degree of correlation between variables. Survival was determined by the Kaplan-Meier method, and the log-rank test was used to determine significance. Factors that were deemed of potential importance by univariate analysis were included in the multivariate analysis. A P <0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS software for Windows, version 20.0 (SPSS Inc., Chicago, IL, USA).
Results
PVT1 and miR-214 expression in ovarian cancer cell lines and tissues
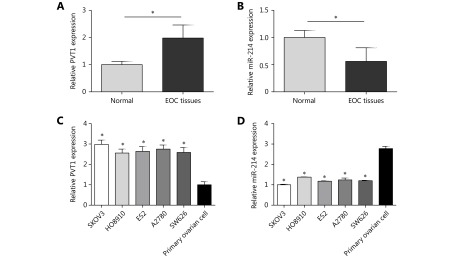
PVT1 and miR-214 were detected by qRT-PCR assay in tissues obtained from 231 EOC patients and 58 normal ovarian tissues. The results showed that the expression of PVT1 was significantly higher in tumor tissues than in normal ovarian tissues (Figure 1A, P<0.05). However, the expression of miR-214 was significantly lower in tumor tissues than in normal ovarian tissues (Figure 1B, P<0.05). The expression of PVT1 was up-regulated in ovarian cancer cell lines compared to that in human primary ovarian cells (P<0.05) and PVT1 expression was the highest in SKOVs3 cells (Figure 1C). On the other hand, the expression of miR-214 was significantly down-regulated in the ovarian cancer cell lines compared to that in human primary ovarian cells (P<0.05) and miR-214 expression was the lowest in SKOV3 cells (Figure 1D). Thus, SKOV3 cells were chosen to perform the subsequent in vitro functional assays.
1.
PVT1 and miR-214 expression in ovarian cancer cell lines and tissues. (A) The expression of PVT1 was significantly higher in tumor tissues compared to normal ovarian tissues. (B) The expression of miR-214 was obviously lower in tumor tissues compared to normal ovarian tissues. (C) The expression of PVT1 was up-regulated in ovarian cancer cell lines (SKOV3, HO8910, ES2, A2780, and SW626) compared to human primary ovarian cells and PVT1 expression was the highest in SKOV3 cells. (D) The expression of miR-214 was down-regulated in ovarian cancer cell lines (SKOV3, HO8910, ES2, A2780, and SW626) compared to human primary ovarian cells and miR-214 expression was the lowest in SKOV3 cells (*P<0.05).
Relationship of PVT1 and miR-214 expression with clinicopathologic features of EOC patients
EOC patients were assigned into two groups (high PVT1 expression vs low PVT1, and high miR-214 expression vs low miR-214 expression) according to the median expression of PVT1 and miR-214 (Table 2). The results indicated that the higher PVT1 expression was associated with advanced FIGO stage (P<0.05). Lower miR-214 expression was related to EOC with a lower differentiated and advanced stage (bothP<0.001). In general, higher expression of PVT1 and lower expression of miR-214 indicated more malignant clinical behavior of EOC.
2.
Correlation of PVT1 and miR-214 expression with clinicopathologic characteristics of EOC patients
| Characteristics | n | PVT1 expression, n | P | MiR-214 expression, n | P | ||
| Low | High | Low | High | ||||
| *Others: others include the endometrioid, clear cell, and undifferentiated ovarian cancers. | |||||||
| Age (years) | 0.679 | 0.885 | |||||
| ≤51 | 136 | 61 | 75 | 76 | 60 | ||
| >51 | 95 | 40 | 55 | 54 | 41 | ||
| Menopause | 0.302 | 0.453 | |||||
| Yes | 154 | 71 | 83 | 84 | 70 | ||
| No | 77 | 30 | 47 | 46 | 31 | ||
| Pathologic type | 0.940 | 0.129 | |||||
| Serous | 116 | 51 | 65 | 71 | 45 | ||
| Mucous and others* | 115 | 50 | 65 | 59 | 56 | ||
| Histologic grade | 0.924 | <0.001 | |||||
| G1–2 | 141 | 62 | 79 | 63 | 78 | ||
| G3 | 90 | 39 | 51 | 67 | 23 | ||
| FIGO stage | <0.001 | <0.001 | |||||
| I–II | 81 | 53 | 28 | 29 | 52 | ||
| III–IV | 150 | 48 | 102 | 101 | 49 | ||
| LN metastasis | 0.436 | 0.266 | |||||
| No | 186 | 79 | 107 | 108 | 78 | ||
| Yes | 45 | 22 | 23 | 22 | 23 | ||
| Residual tumor (cm) | 0.552 | 0.204 | |||||
| <1 | 187 | 80 | 107 | 109 | 78 | ||
| ≥ 1 | 44 | 21 | 23 | 21 | 23 | ||
| Ascites volume (mL) | 0.216 | 0.232 | |||||
| ≤1000 | 157 | 73 | 84 | 83 | 72 | ||
| >1000 | 74 | 28 | 46 | 47 | 29 | ||
| Serum CA125 (U/mL) | 0.548 | 0.831 | |||||
| ≤675 | 140 | 59 | 81 | 78 | 62 | ||
| >675 | 91 | 42 | 49 | 52 | 39 | ||
Correlation between PVT1 and miR-214 expression in EOC
Among 101 EOC tissues with low PVT1 expression, 49 cases (48.5%) displayed low expression of miR-214. The remaining 51.5% displayed high expression of miR-214. Of 130 EOC tissues with high PVT1 expression, 81 (62.3%) displayed low expression of miR-214. Thus, there was a negative relationship between PVT1 and miR-214 expression (correlation coefficient=0.137, P<0.05;Table 3).
3.
The relationship between PVT1 and miR-214 expression in EOC tissues
| PVT1 expression, n (%) | Total cases | ρ | P | ||
| Low | High | ||||
| MiR-214 expression | 0.137 | 0.036 | |||
| Low | 49 (48.5) | 81 (62.3) | 130 | ||
| High | 52 (51.5) | 49 (37.7) | 101 | ||
| Total cases | 101 | 130 | 231 | ||
Survival analysis of PVT1 and miR-214 expression among EOC patients
The results indicated the shorter median progression-free survival (PFS) and OS were associated with lymph node metastasis, advanced stage, high PVT1 expression, and lower miR-214 expression by univariate analysis (P<0.05;Table 3). Moreover, EOC patients with poorly differentiated tumors (G3) had a shorter median PFS (P=0.025; Table 4).
4.
Univariate and multivariate survival analyses of the prognostic factors for overall and disease-free survival in EOC patients
| Variable | n | Progression-free survival | Overall survival | |||||||||||
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||||
| Media of PFS | Pa | HR | 95% CI For HR | Pb | Media of OS | Pa | HR | 95% CI For HR | Pb | |||||
| Pa: P value, log rank test; HR: hazard ratio; CI: confidence interval; Pb: P value, Cox regression; CR: complete response; PR: partial response; SD: table disease; PD: progressive disease; NA: not applicable. *Others: others include the endometrioid, clear cell, and undifferentiated ovarian cancers. | ||||||||||||||
| Age (years) | 0.621 | NA | NA | NA | 0.676 | NA | NA | NA | ||||||
| ≤51 | 136 | 30 | 42 | |||||||||||
| >51 | 95 | 31 | 44 | |||||||||||
| Menopause | 0.373 | NA | NA | NA | 0.488 | NA | NA | NA | ||||||
| Yes | 154 | 30 | 43 | |||||||||||
| No | 77 | 32 | 44 | |||||||||||
| Pathologic type | 0.978 | NA | NA | NA | 0.849 | NA | NA | NA | ||||||
| Serous | 116 | 30 | 41 | |||||||||||
| Mucous and others* | 115 | 31 | 44 | |||||||||||
| Histologic grade | 0.025 | 1.083 | 0.802–
1.462 |
0.604 | 0.114 | NA | NA | NA | ||||||
| G1–2 | 141 | 42 | 44 | |||||||||||
| G3 | 90 | 34 | 40 | |||||||||||
| FIGO stage | <0.001 | 2.265 | 1.632–
3.144 |
<0.001 | <0.001 | 1.994 | 1.423–
2.792 |
<0.001 | ||||||
| I–II | 81 | 44 | 58 | |||||||||||
| III –IV | 150 | 24 | 39 | |||||||||||
| LN metastasis | 0.019 | 1.366 | 0.958–
1.947 |
0.085 | 0.035 | 1.274 | 0.887–
1.829 |
0.190 | ||||||
| No | 186 | 31 | 43 | |||||||||||
| Yes | 45 | 24 | 41 | |||||||||||
| Residual tumor (cm) | 0.277 | NA | NA | NA | 0.806 | NA | NA | NA | ||||||
| <1 | 187 | 31 | 43 | |||||||||||
| ≥ 1 | 44 | 30 | 41 | |||||||||||
| Ascites volume (mL) | 0.237 | 0.403 | NA | NA | NA | |||||||||
| ≤1000 | 157 | 40 | NA | NA | NA | 55 | ||||||||
| >1000 | 74 | 36 | 58 | |||||||||||
| Serum CA125 (U/mL) | 0.753 | NA | NA | NA | 0.991 | NA | NA | NA | ||||||
| ≤675 | 140 | 36 | 55 | |||||||||||
| >675 | 91 | 39 | 55 | |||||||||||
| PVT1 expression | 0.016 | 1.111 | 0.819–
1.506 |
0.498 | 0.020 | 1.153 | 0.842–
1.579 |
0.375 | ||||||
| Low | 101 | 34 | 51 | |||||||||||
| High | 130 | 28 | 40 | |||||||||||
| MiR-214 expression | 0.021 | 1.246 | 0.925–
1.680 |
0.148 | 0.034 | 1.193 | 0.880–
1.616 |
0.256 | ||||||
| Low | 130 | 28 | 41 | |||||||||||
| High | 101 | 31 | 44 | |||||||||||
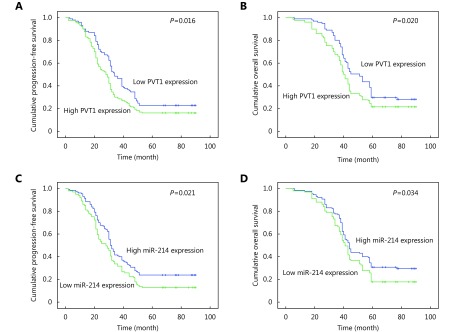
Variables that were significant in the univariate analysis were examined by multivariate analysis. We found the FIGO stage was the independent factor for evaluation of PFS and OS in the Cox proportional hazard model (P<0.001;Table 4). EOC patients with high PVT1 expression and low miR-214 expression displayed shorter PFS and OS than patients with low PVT1 expression and high miR-214 expression, respectively (Figure 2A-D).
2.
Kaplan–Meier analysis for PFS and OS of EOC. (A) High PVT1 expression demonstrated the shorter median of PFS compared to low PVT1 expression (28 vs. 34 months, P=0.016). (B) High PVT1 expression demonstrated the shorter median of OS compared to low PVT1 expression (40 vs. 51 months, P=0.020). (C) Low miR-214 expression demonstrated the shorter median of PFS compared to high miR-214 expression (28 vs. 31 months, P=0.021). (D) Low miR-214 expression demonstrated the shorter median of OS compared to high miR-214 expression (41 vs. 44 months, P=0.034).
Effect of PVT1 knockdown on ovarian cancer cell proliferation, migration, and invasion
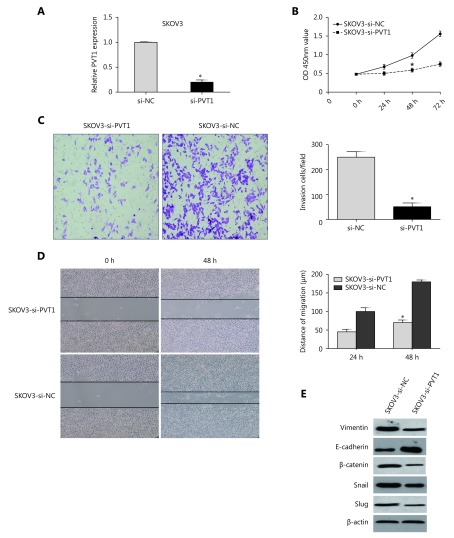
To investigate the functional role of PVT1 in ovarian cancer, si-PVT1 was transfected into SKOV3 cells. Satisfactory repression efficiency was achieved at 48 after transfection (Figure 3A). Then, CCK-8 and wound healing assays showed that knockdown of PVT1 inhibited SKOV3 cell proliferation and migration, respectively (Figure 3B and 3D). Additionally, an invasion assay showed that knockdown of PVT1 dramatically decreased SKOV3 cell invasion (Figure 3C). Furthermore, we detected epithelial-mesenchymal transition (EMT) markers and found that mesenchymal marker proteins (vimentin and β-catenin) and dedicated EMT transcription factors (Snail and Slug) were down-regulated in SKOV3 cells with PVT1-knockdown, while the epithelial marker E-cadherin was up-regulated (Figure 3E).
3.
Knockdown of PVT1 inhibits ovarian cancer cell proliferation, migration, and invasion. (A) Satisfactory repressing efficiency was achieved at 48 h after transfection. (B) CCK-8 assay showed that knockdown of PVT1 inhibited SKOV3 cell proliferation. (C) Invasion assay showed that knockdown of PVT1 dramatically decreased SKOV3 cell invasion. (D) Wound healing assay showed that knockdown of PVT1 inhibited SKOV3 cell migration. (E) Mesenchymal marker proteins (Vimentin and β-catenin) and EMT-mediating transcription factor (Snail and Slug) were down-regulated in SKOV3 cells with PVT1-knockdown, while epithelial marker E-cadherin was up-regulated (*P<0.05).
PVT1 inhibits miR-214 expression by interacting with EZH2 in ovarian cancer cells
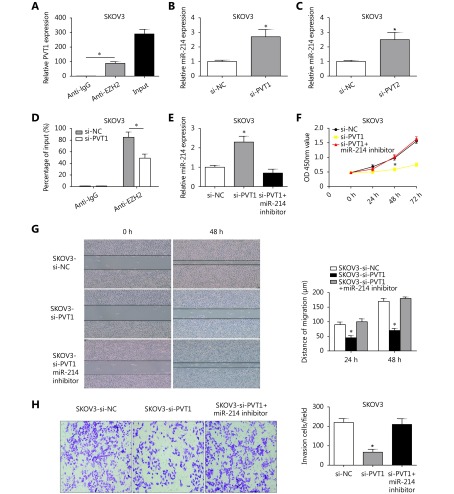
To further detect whether PVT1 affects gene expression by recruiting EZH2 to its target genes, the RIP assay was conducted using an antibody against EZH2 in SKOV3 cells. The results showed that PVT1 was enriched in the anti-EZH2 RIP fraction compared to that in the anti-IgG fraction in SKOV3 cells (Figure 4A). Therefore, these results indicated PVT1 was significantly associated with EZH2. Furthermore, PVT1 knock-down led to the up-regulation of miR-214 in SKOV3 cells (Figure 4B). Additionally, miR-214 was significantly up-regulated after si-EZH2 was transfected into SKOV3 cells (Figure 4C). To evaluate whether PVT1 represses miR-214 mRNA expression by recruiting EZH2 to the promoter of miR-214, the ChIP assay was performed with SKOV3 cells. The results showed that PVT1 silencing decreased the binding ability between EZH2 and miR-214 promoter compared to the IgG group (Figure 4D).
4.
PVT1 inhibits the miR-214 expression by interacting with EZH2 in ovarian cancer cells. (A) PVT1 was enriched in the anti-EZH2 RIP fraction compared to the anti-IgG fraction in SKOV3 cells. (B) After PVT1 was knocked down, miR-214 was up-regulated in SKOV3 cells. (C) MiR-214 was significantly up-regulated after si-EZH2 was transfected into SKOV3 cells. (D) Chromatin immunoprecipitation (ChIP) method proved that PVT1 silencing decreased the binding ability between EZH2 and miR-214 promoter compared to IgG group. (E) The expression of miR-214 was decreased in PVT1 repressing SKOV3 cells using miR-214 inhibitor. (F) CCK-8 assay showed that si-PVT1 inhibited the proliferation of SKOV3 cells and miR-214 inhibitor could counteract the si-PVT1 inhibiting effect of SKOV3 cells. (G) Wound healing assay showed that si-PVT1 inhibited the migration of SKOV3 cells. However, miR-214 inhibitor could counteract the si-PVT1 inhibiting effect in SKOV3 cells. (H) Invasion assay showed that si-PVT1 inhibited the invasion of SKOV3 cells. However, miR-214 inhibitor could counteract the si-PVT1 inhibiting effect in SKOV3 cells (*P<0.05).
To dissect the effect of miR-214 binding to PVT1 on ovarian cancer progression, the expression of miR-214 was decreased in PVT1 repressing SKOV3 cells using miR-214 inhibitor (Figure 4E). By using the CCK-8 assay, we found that si-PVT1 inhibited the proliferation of SKOV3 cells and that the miR-214 inhibitor could counteract the si-PVT1 inhibiting effect in SKOV3 cells (Figure 4F). Wound healing assay and the invasion assay showed that si-PVT1 inhibited the migration and invasion of SKOV3 cells (Figure 4G and 4H). However, miR-214 inhibitor could counteract the si-PVT1 inhibiting effect in SKOV3 cells (Figure 4G and 4H). These results demonstrated that PVT1 promotes the progression of ovarian cancer by silencing miR-214. The potential mechanism was likely that PVT1 prompted the binding of EZH2 to the miR-214 promoter, with the resulting inhibition of miR-214 expression.
Discussion
LncRNAs are transcripts of more than 200 nt in length with limited protein encoding functions. They have been implicated in the molecular mechanisms of carcinogenesis16. As regulators of genetic information flow that interact with the epigenetic, transcriptional, and posttranscriptional pathways, lncRNAs promote tumor formation, progression, and metastasis in many human malignancies17. MiRNAs are another vital class of small ncRNAs, which are involved in some pathological progresses, especially cancer18. The model of miRNA action involves binding to target mRNAs via sequence complementarities, which results in mRNA destabilization and translation inhibition19.
In this study, we found that the expression of PVT1 was significantly higher in tumor tissues than in normal ovarian tissues. On the contrary, the expression of miR-214 was obviously lower in tumor tissues than in normal ovarian tissues. Similar results were also found in ovarian cancer cell lines in vitro. Moreover, higher PVT1 expression was associated with advanced FIGO stage. Lower miR-214 expression was related to EOC featuring lower tumor differentiation and an advanced stage. In general, higher expression of PVT1 and lower expression of miR-214 indicated a clinically more malignant EOC, with a negative relationship between the expression of PVT1 and miR-214 in EOC tissues. Importantly, univariate survival analysis revealed that EOC patients with high PVT1 expression and low miR-214 expression were more likely to have shorter PFS and OS than those with low PVT1 expression and high miR-214 expression. Our results were quite consistent with those of previous studies. Zhang et al.20 reported that PVT1 was up-regulated in cervical cancer tissues. Enhanced expression of PVT1 was associated with larger tumor size, advanced stage, and poor prognosis of patients with cervical cancer. Kong et al.14 indicated the overexpression of PVT1 was suggestive of a poor prognosis of gastric cancer and promoted cell proliferation through epigenetic regulation of p15 and p16. Similarly, Takahashi et al.21 demonstrated that amplification of PVT1 is involved in poor prognosis via apoptosis inhibition in colorectal cancer.
As for miR-214, accumulating evidence suggests that miR-214 participation in carcinogenesis is common, although the specific role of miR-214 can vary depending on the type of cancer. Yang et al.22 showed that miR-214 was up-regulated and induced cisplatin resistance in ovarian cancer cells. In both human osteosarcoma23 and nasopharyngeal carcinoma24, miR-214 expression was up-regulated and it was elucidated as an oncogene. However, down-regulation of miR-214 was observed in hepatocellular cancer tissues, indicating an anti-oncogene function of miR-21425.
Recently, lncRNAs were reported to be capable of binding with miRNAs and can prevent them from interacting with their target mRNAs. LncRNAs belong to ceRNAs and their roles and functions in cancer have been described26. Recently, Mitra et al.27 reported that MEG3 can influence the expression of STAT3 by altering miR-21 expression in ovarian carcinoma. In addition, there was another association between lncRNAs and miRNAs, in which lncRNAs are a precursor RNAs for miRNAs. For example, H19 is a precursor RNA for miR-67528. MiR-214 functions as a tumor suppressor in breast cancer29 and colon cancer30, and as an oncogene by targeting PTEN in gastric cancer31, targeting β-catenin in esophageal cancer32, and promoting EMT and metastasis in lung cancer by targeting the suppressor-of–fused protein33. Additionally, miR-214 can interact with lncRNA DANCR, which is overexpressed in stem-like hepatocellular carcinoma (HCC) cells, and serves as a prognostic biomarker for HCC patients34. MiR-214 has also been identified as an oncogene in nasopharyngeal carcinoma by reversing the inhibitory effect of LINC0086 on carcinoma cell growth35.
In this study, in order to further investigate the functional role of PVT1 in ovarian cancer, the PVT1 expression was knocked down in SKOV3 cells. The results showed that knockdown of PVT1 inhibited SKOV3 cell proliferation, migration, and invasion. The mechanism experiments demonstrated that PVT1-mediated progression of ovarian cancer involved cell progression through silencing miR-214 in cancer cells. The likely potential mechanism was the PVT1-mediated binding of EZH2 to the miR-214 promoter and the subsequent inhibition of miR-214 expression. EZH2 is a histone methyltransferase that induces histone H3 lysine 27 trimethylation (H3K27me3) in specific gene loci and then inhibits gene expression. Consistent with our results, Gou et al.12 reported that PVT1 inhibited the miR-214 expression via binding to EZH2 and recruit EZH2 to the promoter of miR-214 promoter in HCC cells.
EMT has been proven to be an important early event of tumor cell metastatic dissemination, in which cells are endowed with a more motile and invasive potential36. Mesenchymal marker proteins (vimentin and β-catenin) and EMT-mediating transcription factors (Snail and Slug) were down-regulated in SKOV3 cells with PVT1-knockdown, while the epithelial marker E-cadherin was up-regulated in our study, which indicates that PVT1 promotes ovarian cancer progression and may be also involved in the regulation of the EMT process. Yuan et al.37 reported that lncRNA-ATB acted as a ceRNA and shared miR-200 response elements with ZEB1, which induced EMT in HCC. Shen et al.36 also demonstrated that lncRNA-PE was upregulated in HCC cell lines and tissues, and acted as a key regulator of the miR-200-EMT pathway to promote HCC progression. The transcriptional factors ZEB1 and ZEB2 induce EMT by repressing the expression of E-cadherin and promote cancer cell proliferation, invasion, and metastasis7. The accurate mechanism of PVT1 in the progression of EMT requires further study.
In conclusion, we demonstrate that PVT1 is up-regulated in ovarian cancer. Higher PVT1 expression is associated with advanced FIGO stage and with poor prognosis in ovarian cancer patients. Ovarian cancer cell proliferation, migration, and invasion are inhibited after knockdown of PVT1 expression in vitro. Furthermore, PVT1-mediated promotion of ovarian cancer progression may be involved in the regulation of the EMT process, and PVT1 interacts with EZH2 to repress miR-214 expression in ovarian cancer cells. Our findings facilitate the understanding of PVT1 function in the tumorigenesis of ovarian cancer and indicate that further studies are required to investigate the mechanisms of PVT1/miR-214 in various types of ovarian cancer. Finally, the data point the way towards the development of a novel diagnostic marker and therapeutic target for ovarian cancer.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Zietek A, Bogusiewicz M, Szumilo J, Rechberger T. Opportunistic salpingectomy for prevention of sporadic ovarian cancer - a jump from basic science to clinical practice? Ginekol Pol. 2016;87:467–72. doi: 10.5603/GP.2016.0027. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. doi: 10.1016/j.canlet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Gulia C, Baldassarra S, Signore F, Rigon G, Pizzuti V, Gaffi M, et al. Role of Non-coding RNAs in the etiology of bladder cancer. Genes. 2017;8:339. doi: 10.3390/genes8110339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romano G, Veneziano D, Acunzo M, Croce C M. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–91. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang L. Members of the microRNA-200 family are promising therapeutic targets in cancer. Exp Ther Med. 2017;14:10–7. doi: 10.3892/etm.2017.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Wang R, Zhang K, Chen LB. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med. 2014;18:2425–36. doi: 10.1111/jcmm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZL, Yu C, Zhan L, Pan YZ, Chen L, Sun CY. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–309. [PMC free article] [PubMed] [Google Scholar]
- 10.Hirata H, Hinoda Y, Shahryari V, Deng GR, Nakajima K, Tabatabai ZL, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, et al. Long noncoding RNA PVT1 Promotes Non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression . Mol Cancer Ther. 2016;15:1082–94. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 12.Gou X, Zhao XY, Wang ZR. Long noncoding RNA PVT1 promotes hepatocellular carcinoma progression through regulating miR-214. Cancer Biomark. 2017;20:511–9. doi: 10.3233/CBM-170331. [DOI] [PubMed] [Google Scholar]
- 13.Bao X, Duan JY, Yan YJ, Ma X, Zhang Y, Wang HF, et al. Upregulation of long noncoding RNA PVT1 predicts unfavorable prognosis in patients with clear cell renal cell carcinoma . Cancer Biomark. 2017;21:55–63. doi: 10.3233/CBM-170251. [DOI] [PubMed] [Google Scholar]
- 14.Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16 . Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–90. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 16.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357–66. doi: 10.1016/j.canlet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Zlotorynski E. Small RNAs: new microRNA-like molecules. Nat Rev Mol Cell Biol. 2016;17:396. doi: 10.1038/nrm.2016.84. [DOI] [PubMed] [Google Scholar]
- 19.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76:3666–70. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang SR, Zhang GL, Liu JY. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–58. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers . Br J Cancer. 2014;110:164–71. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 23.Xu ZY, Wang T. miR-214 promotes the proliferation and invasion of osteosarcoma cells through direct suppression of LZTS1. Biochem Biophys Res Commun. 2014;449:190–5. doi: 10.1016/j.bbrc.2014.04.140. [DOI] [PubMed] [Google Scholar]
- 24.Deng M, Ye QR, Qin ZL, Zheng Y, He W, Tang HL, et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumor Biol. 2013;34:1793–800. doi: 10.1007/s13277-013-0718-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhang LL, Guo YJ, Zhao CN, Gao JY. Effects and mechanism of miR-214 on hepatocellular carcinoma. Asian Pac J Trop Med. 2015;8:392–8. doi: 10.1016/S1995-7645(14)60350-3. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y, Lu Y, Lu BJ. MicroRNA and long Non-coding RNA in ovarian carcinoma: translational insights and potential clinical applications. Cancer Invest. 2016;34:465–76. doi: 10.1080/07357907.2016.1227446. [DOI] [PubMed] [Google Scholar]
- 27.Mitra R, Chen X, Greenawalt EJ, Maulik U, Jiang W, Zhao ZM, et al. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat Commun. 2017;8:1604. doi: 10.1038/s41467-017-01781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Dong B, Cao JC, Mao JM, Guan W, Peng Y, et al. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017;8:27582–92. doi: 10.18632/oncotarget.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Tian YF, Li F, Zhao ZR, Jiang X, Zhai CJ, et al. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol Rep. 2016;35:3178–84. doi: 10.3892/or.2016.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long LM, He BF, Huang GQ, Guo YH, Liu YS, Hou JR. microRNA-214 functions as a tumor suppressor in human colon cancer via the suppression of ADP-ribosylation factor-like protein 2. Oncol Lett. 2015;9:645–50. doi: 10.3892/ol.2014.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin RJ, Bai FH, Feng YN, Jiu MN, Liu XG, Bai FY, et al. MicroRNA-214 promotes peritoneal metastasis through regulating PTEN negatively in gastric cancer. Clin Res Hepatol Gastroenterol. 2016;40:748–54. doi: 10.1016/j.clinre.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Xu YH, Lu S. Regulation of β-catenin-mediated esophageal cancer growth and invasion by miR-214. Am J Transl Res. 2015;7:2316–25. [PMC free article] [PubMed] [Google Scholar]
- 33.Long HX, Wang ZY, Chen JY, Xiang T, Li QJ, Diao XW, et al. microRNA-214 promotes epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting the suppressor-of-fused protein (Sufu) Oncotarget. 2015;6:38705–18. doi: 10.18632/oncotarget.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1 . Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 35.Guo J, Ma JQ, Zhao GS, Li GC, Fu YF, Luo YW, et al. Long noncoding RNA LINC0086 functions as a tumor suppressor in nasopharyngeal carcinoma by targeting miR-214. Oncol Res. 2017;25:1189–97. doi: 10.3727/096504017X14865126670075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y, Liu SS, Yuan HY, Ying XM, Fu HJ, Zheng XF. A long non-coding RNA lncRNA-PE promotes invasion and epithelial-mesenchymal transition in hepatocellular carcinoma through the miR-200a/b-ZEB1 pathway. Tumour Biol. 2017;39:1010428317705756. doi: 10.1177/1010428317705756. [DOI] [PubMed] [Google Scholar]
- 37.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]