Abstract
About half of hypertrophic and dilated cardiomyopathies cases have been recognized as genetic diseases with mutations in sarcomeric proteins. The sarcomeric proteins are involved in cardiomyocyte contractility and its regulation, and play a structural role. Mutations in non-sarcomeric proteins may induce changes in cell signaling pathways that modify contractile response of heart muscle. These facts strongly suggest that contractile dysfunction plays a central role in initiation and progression of cardiomyopathies. In fact, abnormalities in contractile mechanics of myofibrils have been discovered. However, it has not been revealed how these mutations increase risk for cardiomyopathy and cause the disease. Much research has been done and still much is being done to understand how the mechanism works. Here, we review the facts of cardiac myofilament contractility in patients with cardiomyopathy and heart failure.
Keywords: human heart muscle contractility, dilated cardiomyopathy, hypertrophic cardiomyopathy, troponin phosphorylation, cardiomyopathy mutations, Ca2+-sensitivity, length dependent activation
1. Introduction
The main function of the heart is pumping the blood around the body according to the organism’s physiological requirements [1]. Heart contraction is initiated by the action potential propagated from sinoatrial node pacemaker cells. Membrane depolarization opens voltage-gated L-type Ca2+ channels in the cytoplasmic membrane. The initial influx of Ca2+ further stimulates efflux of Ca2+ from sarcoplasmic reticulum via ryanodine receptor type-2 channels [2,3]. Ca2+ binds to troponin C of the troponin complex, which is composed of three subunits: troponin C, I, and T. Troponin is attached to tropomyosin, which lies along the actin filament, and regulates its position depending on the Ca2+ concentration [4,5]. Tropomyosin blocks myosin binding sites on actin at low Ca2+ concentration. When [Ca2+] increases, tropomyosin releases the sites and promotes interaction of myosin with actin. Uptake of Ca2+ from cytoplasm by sarcoplasmic/endoplasmic reticulum Ca2+-ATPase Na+/Ca2+ exchanger stimulates relaxation [2,3]. It is widely accepted that myofilament Ca2+ sensitivity is regulated via phosphorylation of troponin I (TnI) at Ser 23 and 24 [6,7,8]. The phosphorylation of other proteins, such as myosin binding protein C [9] and regulatory light chain of myosin, also can modulate Ca2+ sensitivity [10,11,12] but to a lesser degree.
Cardiomyopathy is a disease of the heart muscle leading to cardiac dysfunction with characteristic pathological remodeling and eventually to heart failure. Besides cardiomyopathies, heart failure can be caused by other conditions, particularly by coronary artery disease and hypertension which are the leading causes of cardiac death [13]. There are several different types of cardiomyopathies: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive (RC), and arrhythmogenic right ventricular (ARVC) [14,15,16,17,18]. Hypertrophic cardiomyopathy, DCM, and ARVC have been recognized as genetic diseases [19,20].
Dilated cardiomyopathy and HCM are the most common types of cardiomyopathies and they have been intensively studied. Hypertrophic cardiomyopathy affects one in 500 of the population and DCM affects one in 250–500 [17]. They both have been implicated with a high risk of sudden cardiac death, where DCM accounts for 30–40% of heart failure [13,19,21]. Dilated cardiomyopathy and HCM decrease stroke volume and cardiac output but in different ways [22]. Hypertrophic cardiomyopathy is characterized by a pathologically hypertrophied (wall thickness > 15 mm) left ventricle and septum and without left ventricular cavity dilatation. This leads to a decrease of the intraventricular dimension and blood flow obstruction resulting in diastolic dysfunction. In about 10% of patients, HCM may progress further to a dilated phenotype [23]. The morphologic macroscopic changes do not always become apparent. Myocyte disarray exceeding 5–10% is the recognized trademark for HCM diagnosis [24]. On the contrary, the left ventricle of a DCM heart is dilated and the walls become thinner [25]. The heart cannot contract properly and left ventricular ejection fraction is reduced (<45%; systolic dysfunction). Hypertrophic cardiomyopathy and DCM can show morphological variations [23,25,26,27] and often can be misdiagnosed.
Dilated cardiomyopathy can be initiated by different conditions: autoimmune diseases (diabetes, thyroid disease), infections, and toxins (alcohol, chemotherapy), high blood pressure, heart attack or coronary artery disease [28,29]. It should be noted that the case of dilated cardiomyopathy which develops after a myocardial infarction or ischemia is often classified as a separate disease entity: ischemic cardiomyopathy (ICM) [25,28,29,30,31]. The environmental cases can also have genetic predisposition [32,33]. If known cause is not identified, DCM is called idiopathic DCM. Idiopathic DCM accounts for 50% of all cases [25,34,35] and its prevalence in children is higher (66%) [36]. Family screening shows that 30–50% of all idiopathic DCM cases are familial [19]. Whole genome sequencing analysis revealed that mutations were the main underlying cause of the disease in about 50% of patients diagnosed with idiopathic DCM. Dilated cardiomyopathy is a polygenetic disease with mutations found in sarcomeric, structural, and other protein genes; mutations in 57 genes have been linked to DCM [20]. Where mutations in the titin gene (TTN) are the most frequent (11.6–21.4%) [37,38,39] and account for ~25–27.6% of familial [37,40] and 11.6–18% of sporadic [37,38] genetic cases. The genes of sarcomeric proteins more frequently identified as genetic cause are MYH7 (β-cardiac myosin heavy chain), TPM1 (tropomyosin alpha-1 chain), and the genes of cardiac troponins TNNT2 (troponin T), TNNI3 (troponin I), and TNNC1 (troponin C).
In the case of HCM, pathogenic mutations, which were found in 37.9–63.2% cases [41,42,43,44,45,46,47], are located mostly in the genes of nine sarcomeric proteins: MYH7, MYBPC3 (cardiac myosin binding protein C), TNNT2, TNNI3, TNNC1, TPM1, ACTC (cardiac actin), MYL2 (ventricular myosin light chain 2, LC2), and MYL3 (myosin light chain 3) [42,43,48,49,50]. Where 74–85.1% of all mutations are in the genes of MYBPC3 (36.2–54.8%) and MYH7 (25–48.9%) [42,43,44,45,46,47].
In our review we summarize the changes of contractile property of human cardiac muscle associated with DCM and HCM.
2. Approaches and Parameters to Estimate Contractility
It is recognized that myofibril contractile dysfunction plays a central role in initiation and progression of cardiac disease. However, how pathogenic mutations increase risk of cardiomyopathies or cause the diseases is unclear. The common explanation is that mutations in the contractile and regulatory proteins of sarcomere disturb muscle contraction. Mutations in titin change viscoelasticity properties, and mutations in other non-contractile proteins may induce defects in cell signaling pathways that modify cardiac response. But it seems that the mechanism is complex and there is no model that explains the mechanism of disease.
Numerous studies have been done using patient heart samples as well as animal models of cardiomyopathies and chimeric protein constructs with recombinant proteins from different sources to understand the diseases. However, in this review we would like to summarize the experimental data obtained only from human heart muscle samples. Skinned muscle strips, isolated cardiomyocytes, and myofibrils obtained from frozen patient hearts [51] were used to study heart contractility [52,53,54]. As DCM and HCM affect the left ventricle more, most of the research has been done on left ventricular and septum samples, and not much is known about whether there are significant abnormalities in the contractility of the atriums and right ventricle.
The fundamental parameters used to describe muscle contractility are: the force generating capacity (Fmax) and Ca2+-sensitivity (EC50; and nH is a parameter characterizing the steepness of the sigmoidal curve). Furthermore, the fast kinetics of myofilament contractility can be measured only in a single myofibril, due to the relatively slow diffusion in samples more than a few microns thick, such as cardiomyocytes. This technique is far more advantageous in respect to the understanding of the molecular mechanisms of cardiomyopathies as it also provides us with data on myosin cross-bridge kinetics and the mechanisms of activation and relaxation in a single muscle myofibril [53,54]. Once [Ca2+] reaches a certain threshold, force rapidly develops until it reaches plateau (Fmax). The time course of force development can be fitted with a single exponential function characterized by the rate constant kACT. The activation rate is highly dependent on the concentration of Ca2+ in activating solution.
Relaxation has two phases: slow and fast. The first phase is nearly isotonic, with a slow linear decline in tension, and it lasts about 100 ms. The slow phase can be fit with a linear function and characterized by the rate constant kLIN, which is calculated from the slope value of the linear fit by normalizing it to the maximum tension (), and the duration time tLIN. After the latent period, the slow phase translates into an exponential force decay phase. The rate constants of the exponential force development (kACT) and the fast phase of relaxation (kREL) are fitted with a single exponential function (, where the parameter k, (kACT, kTR or kREL) characterize the rate of the course [55]. In order to determine the rate of myofibril activation in thick specimens, such as cardiomyocytes, a fast length release following quick restretch is applied to cardiac cells or muscle strips and the rate of force redevelopment (kTR) that is comparable to kACT is measured [56].
3. Contractile Properties of HCM and DCM Hearts
3.1. Decreased Force Capacity
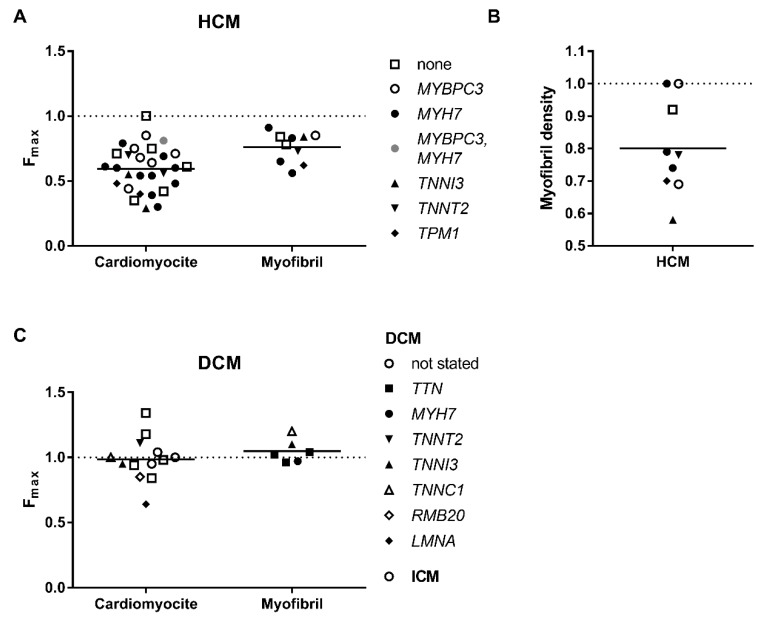
Hypertrophic cardiomyopathy samples produce significantly lower strength per muscle cross-sectional areas (Figure 1A). The decrease in force in muscle tissue and isolated cardiomyocytes can be partially explained by myofibril disarray [14] and decrease of myofibril density (Figure 1B), but also due to the contractile defects. In fact, the maximal force developed by cardiomyocytes from HCM hearts was lower almost in all studied cases (Figure 1A, Table 1). The mean Fmax was 41% lower than that of healthy donor hearts. At the same time, myofibril density was 20% lower in HCM cardiomyocytes [14,57,58,59]. Furthermore, the force produced by single myofibrils from HCM hearts with an identified genetic disease-causing mutation, irrespective of gene (MYBPC3, MYH7, TPM1, TNNT2), was also lower by 24% than that of control heart myofibrils (Figure 1A). This can suggest that there is some abnormality in cross-bridge cycling kinetics. However, the effect does not have to be purely due to mutation. Other factors as haploinsufficiency and secondary changes such as an altered protein phosphorylation [60], protein oxidation [61,62,63], and changes in protein isoform expression (see review [64]) cannot be excluded. Indeed, a number of samples with no mutation found in the genes of sarcomeric proteins also showed significantly lower (by 22%) myofibrillar force [57].
Figure 1.
Maximum force of contraction by different types of preparations: from hearts of patients with HCM (A) and DCM (C). As can be seen, the maximal force generated by cardiomyocytes as well as myofibrils is lower in the HCM samples. The force is not diminished in DCM samples. (B) Density of myofibrils in cells were measured in some HCM samples. Each data point represents a different experimental group where the symbols indicate genes where mutations were found. See also Table 1 and Table 2. All values are normalised to those of donor heart muscle (dashed line).
Table 1.
Contractile characteristics of hypertrophic cardiomyopathy heart muscle.
| Mutation, Gene Expression and Mutated (mut) Allele Expression | Phosphorylation Level (Cardiomyopathy/Healthy) | Titin Isoforms and Passive Stiffness (Cardiomyopathy/Healthy) | Maximal Force and Myofibril Density (Cardiomyopathy/Healthy) | Contractile Kinetics Parameters (Cardiomyopathy/Healthy) | Ca2-Sensitivity of Force (Cardiomyopathy/Healthy) | Length Dependent Activation Changes upon Stretch (Cardiomyopathy/Healthy) | Effect PKA on EC50 (in µM) | Patient Information (Sex, Age and Number of Patients) | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expression | Myocyte | Myofibril | Activation | Relaxation | +PKA | +PKA | ||||||||||||||||||||||||
| Diagnosis | Gene with Mutation | Mutation and Type: Truncated (T) or Not (NT) | Gene | Mut Allele | TnI | MyBP-C | LC2 | N2AB/N2B | Passive Stiffness | F max | Myofibril Density | F max | k ACT | k TR | t LIN | k LIN | k REL | EC 50 | n H | EC 50 | ΔEC50 | ΔF | ΔEC50 | ΔF | ΔEC50 | Sex M/F | Age | N | Refs | |
| HCM | MYBPC3 | g.T2604A+C del at 2605 | T | 0.64 * | 0.34 * | 0.64 | 1 | 1.14 | 0.42 | M | 42 | 1 | [65,66] | |||||||||||||||||
| HCM | MYBPC3 | T | 0.18 | 0.75 | 0.85 * | 0.79 * | 0.96 | 0.57 * | 1.16 | 1.01 | 0.94 | 0.88 * | M/F | 22–69 | 17 | [67] | ||||||||||||||
| HCM | MYBPC3 | T | 0.67 | 0.69 * | 0.82 | 1.46 | 0.71 * | 0.83 * | 0.84 * | 1 | 0.61 * | 1.04 | 1.13 | M/F | 22–69 | 17 | [68] | |||||||||||||
| HCM | MYBPC3 | T/NT | 0.68 * | 0.69 * | 0.85 * | 1 | 1 | MF | 32–69 | 28 | [57] | |||||||||||||||||||
| HCM | MYBPC3 | p.R502W | NT | 0.8 * | 0.75 | 1.10 | 0.35 | M | 23 | 1 | [65] | |||||||||||||||||||
| HCM | MYBPC3 | NT | 0.09 | 0.64 | 0.44 * | 0.56 * | 0.93 | 0.29 * | 0.28 * | 0.18 * | 0.25 * | 1.45 * | M/F | 31–51 | 4 | [67] | ||||||||||||||
| HCM | MYH7 | p.R719Q | NT | 0.79 | 1.16 | 1.01 | F | 27 | 1 | [65] | ||||||||||||||||||||
| HCM | MYH7 | NT | 0.22 | 0.58 | 0.54 * | 0.56 * | 0.88 * | 0.26 * | 0.44 * | 0.42 * | 0.38 * | 0.97 * | M/F | 25–61 | 4–6 | [67] | ||||||||||||||
| fHCM | MYH7 | p.R403Q | NT | 0.35 * | 0.93 | 0.3 | 0.56 * | 1.92 * | 2.02 * | 0.75 * | 4 * | 1.76 * | M | 24 | 1 | [69,70,71] | ||||||||||||||
| fHCM | MYH7 | p.R403Q | NT | 0.5 | 0.6 | 0.83 | 1.84 | 2.9 | 1.77 | M | 35 | 1 | [69,70,71] | |||||||||||||||||
| fHCM | MYH7 | p.R403Q | NT | 0.45 | 0.6 | 0.91 | 1.52 | 2.7 | 1.99 | F | 39 | 1 | [69,70,71] | |||||||||||||||||
| fHCM | MYH7 | p.R723G | NT | 0.7 | 0.54 * | 1.41 * | M | 38, 55 | 2 | [72] | ||||||||||||||||||||
| fHCM | MYH7 | p.R723G | NT | 0.68 | 0.11 * | 0.42 * | 0.42 * | 0.72 * | 0.69 * | 0.74 * | 0.93 | 1 | 1.28 * | 1.44 * | M/F | 53, 55 | 2 | [73,74] | ||||||||||||
| fHCM | MYH7 | p.A200V | NT | 0.53 | 0.48 * | 1.12 * | F | 19 | 1 | [72,75] | ||||||||||||||||||||
| fHCM | MYH7 | NT | 0.39 * | 0.79 * | 0.65 * | 1 | M/F | 19–61 | 11 | [57] | ||||||||||||||||||||
| fHCM | TPM1 | p.I284V | NT | 0.40 * | 0.70 * | 0.62 * | M | 65 | 1 | [57] | ||||||||||||||||||||
| HCM | TPM1 | p.M281T | NT | 0.07 | 0.55 | 0.48 * | 0.67 * | 0.95 | 0.39 * | 0.72 | −0.2 * | 0.06 * | 1.08 * | M | 65 | 1 | [67] | |||||||||||||
| HCM | TNNT2 | p.K280N | NT | 0.96 | 1.07 | 0.70 | 0.80 * | 0.76 * | 0.45 * | 0.34 | 0.30 * | 0.15 * | 0.12 | M | 26 | 1 | [67] | |||||||||||||
| HCM | TNNT2 | p.K280N | NT | 0.56 | 0.78 * | 0.73 * | M | 26 | 2 | [57] | ||||||||||||||||||||
| HCM | TNNI3 | p.R145W | NT | 0.55 * | 0.58 * | 0.84 | M | 46, 66 | 2 | [57] | ||||||||||||||||||||
| HCM | TNNI3 | p.R145W | NT | 0.09 | 0.49 | 0.29 | 0.88 * | 1.18 * | 0.38 * | 0.03 * | 0.34 * | 0.08 * | 0.99 * | M | 46, 66 | 2 | [67] | |||||||||||||
| HCM | MYH7(2) MYBPC3(1) | NT | 0.25 * | 0.14 * | 0.91 | 0.70 | 0.85 | 0.61 * | 1 | 1.1 * | 0.49 * | 1.26 | M/F | 23–61 | 6 | [65,70] | ||||||||||||||
| HCM | none | 0.18 | 0.4 | 0.81 * | 0.86 * | 0.93 | 0.49 * | 1.2 | 1.06 * | 1.2 | 0.73 | M/F | 46–75 | 3–7 | [67] | |||||||||||||||
| HCM | none | 1 | 0.71 * | 0.92 * | 0.78 * | 1 | M/F | 35–75 | 3–14 | [57] | ||||||||||||||||||||
| HCM | none | 0.60 * | 0.54 * | 1 | 0.75 * | 0.8 * | 0.81 * | 1 | 0.49 * | 0.94 | 0.97 | M/F | 46–75 | 11 | [68] | |||||||||||||||
| HCM | none | 1.0 | 0.84 | 1.01 | 2.1 | 1.59 | M/F | 35–72 | 9 | [71] | ||||||||||||||||||||
| HCM | none | 0.61 | 1.09 | 0.54 | M | 59 | 1 | [65] | ||||||||||||||||||||||
| HCM | none | 0.35 | 1.05 | 0.45 | F | 61 | 1 | [65] | ||||||||||||||||||||||
| HCM | none | 0.42 | 1.12 | 0.49 | M | 58 | 1 | [65] | ||||||||||||||||||||||
The value changes were normalized to healthy control hearts (Cardiomyopathy/Healthy) if not otherwise specified. The parameters of length dependent activation are shown as a difference in EC50 and Fmax between long and short sarcomere lengths. Passive stiffness was evaluated as a Young’s modulus or as a passive tension generated by muscle at stretch. In some cases, approximate values are given. * p < 0.05. HCM—hypertrophic cardiomyopathy, fHCM—familial HCM, PKA—protein kinase A. The table cells are highlighted according to the value changes compared to control: yellow—no changes, green—the value decreased, blue—the value increased.
With respect to DCM samples, neither cardiomyocytes nor myofibrils produce force significantly different from that of donor hearts, except in one study with mutation in LMNA (lamin A/C; p.(R331Q)) [78,79] (Figure 1C, Table 2), where Fmax was lower by 37% and that was explained by the decrease in myofibril density (Table 2). This means that systolic dysfunction observed in DCM hearts is not caused by the decrease in force production, but is rather due to ventricular dilation. The end-diastolic volume increase causes a decrease in left ventricular ejection fraction [91]. A decrease in myofibril density was also observed in the case of end-stage pediatric cardiomyopathy [58] and congenital dilated cardiomyopathy with mutation in the structural protein filamin-C [92].
Table 2.
Contractile characteristics of dilated and other cardiomyopathy heart muscles.
| Mutation, Gene Expression and Mutated (mut) Allele Expression | Phosphorylation Level (Cardiomyopathy/Healthy) | Titin Isoforms and Passive Stiffness (Cardiomyopathy/Healthy | Maximal Force and Myofibril Density (Cardiomyopathy/Healthy) | Contractile Kinetics Parameters (Cardiomyopathy/Healthy) | Ca2-Sensitivity of Force (Cardiomyopathy/Healthy) | Length Dependent Activation Changes upon Stretch (Cardiomyopathy/Healthy) | Effect PKA on EC50 (in µM) | Patient Information (Sex, Age and Number of Patients) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expression | Myocyte | Myofibril | Activation | Relaxation | +PKA | +PKA | |||||||||||||||||||||||
| Diagnosis | Gene with Mutation | Mutation and Type: Truncated (T) or Not (NT) | Gene | Mut Protein | TnI | MyBP-C | LC2 | N2AB/ N2B |
Passive Stiffness | F max | Myofibril Density | F max | k ACT | k TR | t LIN | k LIN | k REL | EC 50 | n H | EC 50 | ΔEC50 | ΔF | ΔEC50 | ΔEC50 | Sex M/F | Age | N | Refs | |
| IDCM | TTN | p.(R23464Tfs *41) | T | 1.0 | 0 | 1.05 | 0.63 * | 1.02 | 1.05 | 1.06 | 0.97 | 1.06 | 0.91 | 1.00 | M | 22 | 1 | [76] | |||||||||||
| fDCM | TTN | p.(R23464Tfs*41) | T | 1.0 | 0 | 1.11 | 0.66 * | 0.96 | 1.12 | 0.95 | 0.98 | 0.18 | 1.13 | 1.13 | M | 37 | 1 | [76] | |||||||||||
| fDCM | TTN | p.(Y18923 *) | T | 0.96 | 0 | 1.25 | 0.62 * | 1.04 | 1.15 | 1.16 * | 0.98 | 1.06 | 1.17 | 1.03 | F | 22 | 1 | [76] | |||||||||||
| PPCM | TTN | p.(K15664Vfs*13) | T | 1.85 | 0.4 | F | 1 | [77] | |||||||||||||||||||||
| DCM | LMNA | p.(R331Q) | NT | 0.5 | 1.98 * | 1 | 0.64 * | 0.63 | 0.86 * | 1 | 0.78 | M/F | 45.3 | 3 | [78,79] | ||||||||||||||
| fDCM | RMB20 | p.E913K | NT | 0.13 | 13.89 | 0.8 | 0.85 | 0.67 * | 0.58 | 1.13 | 0.36 | 0.81 | 1.77 | M | 19 | 1 | [80] | ||||||||||||
| fDCM | TNNC1 | p.G159D | NT | 1.13 | 0.49 | 0.13 | 0.89 | 0.59 * | 1 | 1.20 | 0.97 | 0.97 | 0.71 * | 0.35 | 1.17 | 0.60 * | 0.59 * | 1.22 | M | 3 | 1 | [76,81] | |||||||
| fDCM | TNNI3 | p.K36Q | NT | 1.0 | 0.58 * | 1.10 | 1.17 | 1.05 | 0.70 * | 1.39 | 1.30 * | 1.34 | M | 15 | 1 | [76] | |||||||||||||
| IDCM | TNNI3 | p.(R98 *) | T | 0.39 | 0 | 1.08 | 1.56 | 1 | 0.95 | 0.94 * | 0.88 * | 0.66 | M | 46 | 1 | [78,82] | |||||||||||||
| IDCM | TNNT2 | p.(K217del) | NT | 0.51 | 1.08 | 1.64 | 1.3 * | 1.11 | 1.07 | 0.97 | M | 19 | 1 | [78] | |||||||||||||||
| fDCM | MYH7 | p.E1426K | NT | 1.08 | 0.65 * | 0.97 | 1.13 | 1.19 * | 0.70 * | 0.76 | 1.35 * | 0.96 | M | 43 | 1 | [76] | |||||||||||||
| IDCM | not stated | 0.54 | 1.98 | 1 | 0.98 | 0.94 | 1 | 0.73 | M/F | 54.6 | 5 | [78] | |||||||||||||||||
| DCM | not stated | 1.24 * | 0.27 * | 51–56 | 3–4 | [83] | |||||||||||||||||||||||
| DCM | not stated | 0.5 | 0.51 | 1 | 0.96 | 3.37 | M | 45,59 | 2 | [84] | |||||||||||||||||||
| IDCM | not stated | 0.35 * | 0.58 * | 1.62 * | 1 | 0.94 | 0.69 * | 1.12 | 0.76 | 1.09 | 8 | [85] | |||||||||||||||||
| IDCM | not stated | 0.4 * | 0.4 * | 1.1 | 1.18 | 0.88 | 0.67 | 1.33 | 0.8 | 1.26 | M/F | 52–62 | 7 | [86] | |||||||||||||||
| IDCM | not stated | 1.78 | 1.2 | 1.34 | 0.68 * | 0.81 | 0.87 | 0.59 | 0.86 | 1.33 | 3 | [80] | |||||||||||||||||
| IDCM | not stated | 1.14 | F | 41–57 | 6 | [87] | |||||||||||||||||||||||
| PPCM | not stated | 0.24 * | 0.61 * | 1.58 | 1.39 * | 0.84 | 0.65 * | 1.01 | 0.48 * | 0.96 | F | 6 | [85] | ||||||||||||||||
| ICM | not stated | 0.95 | 0.93 | 0.71 * | 0.83 * | 3 | [88] | ||||||||||||||||||||||
| ICM | not stated | 0.5 | 0.72 * | 0.98 | 2.13 | M/F | 41–65 | 7 | [84] | ||||||||||||||||||||
| ICM | not stated | 0.5 | 0.5 * | 0.83 | 1 | 1.04 | 0.70 * | 1.11 | 0.96 | 1.19 | 4 | [85] | |||||||||||||||||
| ICM (7) DCM (2) | not stated | 0.5 | 0.57 * | 1.06 | 1.0 | 0.66 * | 0.99 | 0.98 | 2.55 * | M/F | 41–65 | 10 | [84] | ||||||||||||||||
| HF | not stated | 0.33 * | 0.65 * | 1 | 0.53 * | 0.53 * | 0.96 | 0.63 * | 0.87 | 0.98 | M/F | 42–57 | 4 | [89] | |||||||||||||||
| IRCM | not stated | 0.84 | 1.20 * | 0.05 * | 0.52 * | F | 66 | 1 | [90] | ||||||||||||||||||||
| IRCM | not stated | 1.16 | 1.92 * | 0.07 * | 0.33 * | M | 21 | 1 | [90] | ||||||||||||||||||||
| IDCM (9) NCC (2) | not stated | 0.27 * | 0.4–2.8 | 0.7 * | 0.64 * | 0.67 * | 1 | <18 | 11 | [58] | |||||||||||||||||||
The value changes were normalized to healthy control hearts (Cardiomyopathy/Healthy) if not otherwise specified. The parameters of length dependent activation are shown as a difference in EC50 and Fmax between long and short sarcomere length. Passive stiffness was evaluated as a Young’s modulus or as a passive tension generated by muscle at stretch. In some cases, approximate values are given. * p < 0.05. PPCM—peripartum cardiomyopathy, ICM—ischemic cardiomyopathy, RCM—restrictive cardiomyopathy, HF—heart failure, IDCM—idiopathic DCM, fDCM-familial DCM, IRCM—idiopathic restrictive cardiomyopathy. The table cells are highlighted according to the value changes compared to control: yellow—no changes, green—the value decreased, blue—the value increased.
3.2. Activation and Relaxation Kinetics
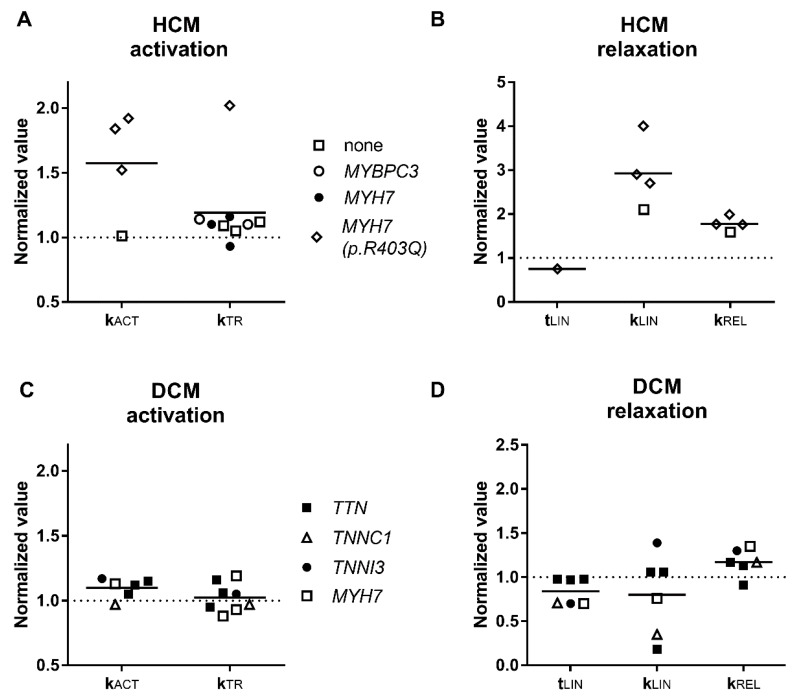
We combined data concerning force development from different papers in Figure 2 and found that the rate of force growth in HCM and DCM samples did not significantly vary from control. Only the samples with mutation in MYH7 R403Q encoding cardiac β-myosin heavy chain, had significantly higher cross bridge turnover rate characterized by the activation and relaxation rate constants [69]. The authors explained it by faster cross-bridge detachment rates. The relaxation kinetics in single R403Q myofibrils was 1.5 times higher than in sarcomere mutation-negative HCM myofibrils and 1.5–2 times higher than in myofibrils from control hearts [71]. Furthermore, it was found 1.6 times higher in the muscle strips of MYH7 R403Q patients compared to control mutation-negative HCM which showed a positive linear correlation with the slope of slow relaxation phase [71].
Figure 2.
Contractile kinetics parameters. Parameters of activation (A,C) and relaxation (B,D) kinetics of HCM and DCM muscle samples. Each data point represents a different experimental group where the symbols indicate genes where mutations were found. All values are normalised to those of donor heart muscle. See also Table 1 and Table 2.
Activation kinetics in all studied DCM myofibrils regardless of mutation was not significantly altered. The situation with relaxation was different. Relaxation of DCM samples with truncating mutations in the TTN gene was not different from control, while relaxation was faster in all other studied myofibrils which had mutations in the genes of contractile proteins: myosin (MYH7 E1426K) and troponins (TNNI3 K36Q and TNNC1 G159D).
Interestingly, that in the case with RCM disease, myofibril relaxation was slower (Table 2), contributing to diastolic ventricular dysfunction [90].
3.3. Elevated Ca2+-Sensitivity
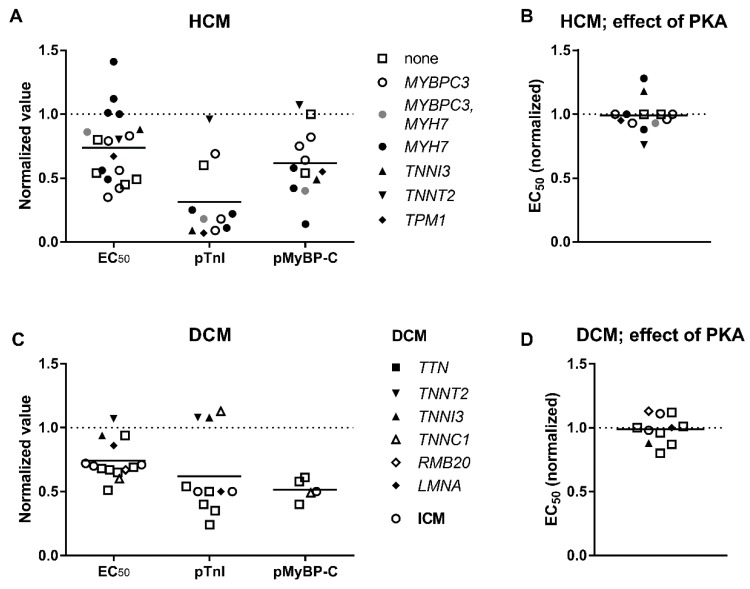
The Ca2+ sensitivity of DCM and HCM patient heart muscle is usually higher (see Figure 3A,C; EC50 (HCM)/EC50 (Donor) = EC50 (DCM)/EC50 (Donor) = 0.74) than in muscle of a healthy donor. There is a perception that the difference in Ca2+-sensitivity is mainly due to TnI dephosphorylation in hearts with cardiomyopathy (Figure 3A,C; 69% and 31% lower in HCM and DCM, respectively) and not because of mutations. In normal donor hearts TnI is highly phosphorylated (0.8–2 mol Pi/mol TnI) and exists mostly in double phosphorylated and single phosphorylated forms. Phosphorylation level is reduced in cardiomyopathies (0.2–1.3 mol Pi/mol TnI) [68,78,84,85,93,94]. But what is true in general may be different for some specific cases. For example, phosphorylation of TnI was found rather high in a number of DCM samples where a mutation was found as the cause of the disease: TNNI3 p.98trunc and TNNT2 p.K217del (~2 mol Pi/mol TnI) [78], ACTC E99K (HCM mutation, 1.61 mol Pi/mol TnI) [95], TNNC1 G159D (1.53 mol Pi/mol TnI) [81], and TNNT2 K280N (1.4–1.6 mol Pi/mol TnI). It may be the case that idiopathic DCM hearts with found mutations may have normal phosphorylation of TnI, whereas hearts without mutation rather have a reduced phosphorylation level. Alongside with TnI, MyBP-C and myosin light chain 2 (or regulatory) phosphorylation also decreased by 1.5–2.5 folds in hearts with cardiomyopathies [67,73,96,97,98]. Phosphorylation of MyBP-C may be responsible for about half of the change in EC50 at low sarcomere length, but not at high sarcomere length, and is an important factor in length-dependent activation [12].
Figure 3.
The concentration of Ca2+ required for half-maximal contraction (EC50) and total phosphorylation level of troponin I (pTnI) and MyBP-C (pMyBP-C) of heart tissue samples. The data are from Table 1 and Table 2. The values for HCM (A,B), DCM (C,D) are normalized to control donor hearts (dashed line). The effect of PKA treatment on Ca2+-sensitivity of heart muscles from HCM (B) and DCM (D) patients. The EC50 values are normalized to the EC50 of normal donor heart samples. PKA eliminates or at least lessens the difference between cardiomyopathic and donor hearts. Each data point represents a different experimental group where the symbols indicate genes where mutations were found.
To discriminate the effect of mutation from the secondary effects caused by dephosphorylation of TnI and MyBP-C, in some experiments skinned muscle samples were treated with protein kinase A (PKA). After PKA treatment, TnI and MyBP-C became phosphorylated to the levels of donor hearts and the difference in EC50 between a donor heart and heart with cardiomyopathy was ether fully eliminated or substantially reduced (Figure 3B,D). Little difference in the phosphorylation level could be responsible for the variance seen between some patient samples and healthy subject samples.
3.4. Uncoupling of TnI Phosphorylation from the Changes in Ca2+-Sensitivity
It is well known that phosphorylation of TnI in a normal heart decreases myofilament Ca2+-sensitivity. A case has been made that loss of this functional modulation of Ca2+-sensitivity through phosphorylation of TnI may play a pivotal role in HCM and DCM development and progression. The experiments using reconstituted thin filamnets in the in vitro motility assay provide a strong support that blunted response is one of the common abnormalities seen for both DCM [76,99,100] and HCM mutations [101,102]. It is interesting, that the effect was observed even if pathogenic mutations were not in thin filaments but in other sarcomeric proteins (myosin and MyBP-C) which were not present in the in vitro test system [76,101]. Unfortunately, the nature of this blunted response to TnI phosphorylation has not yet been identified.
On the level of myofibrils there is no robust support that such uncoupling exists. The uncoupling was found in myofibrils prepared with the use of animal models of DCM [103,104] where mutations E361G in actin and G159D in troponin C uncoupled myofibrillar Ca2+-sensitivity changes from TnI phosphorylation.
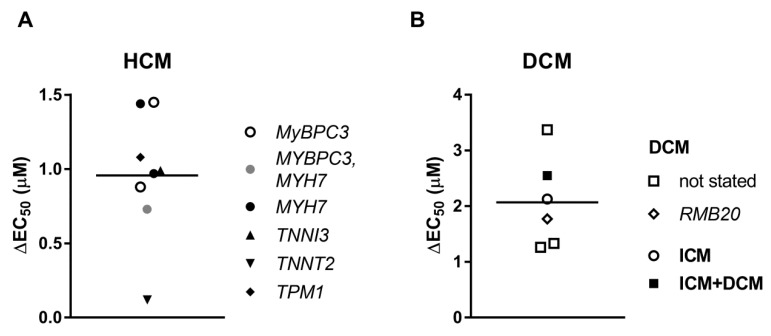
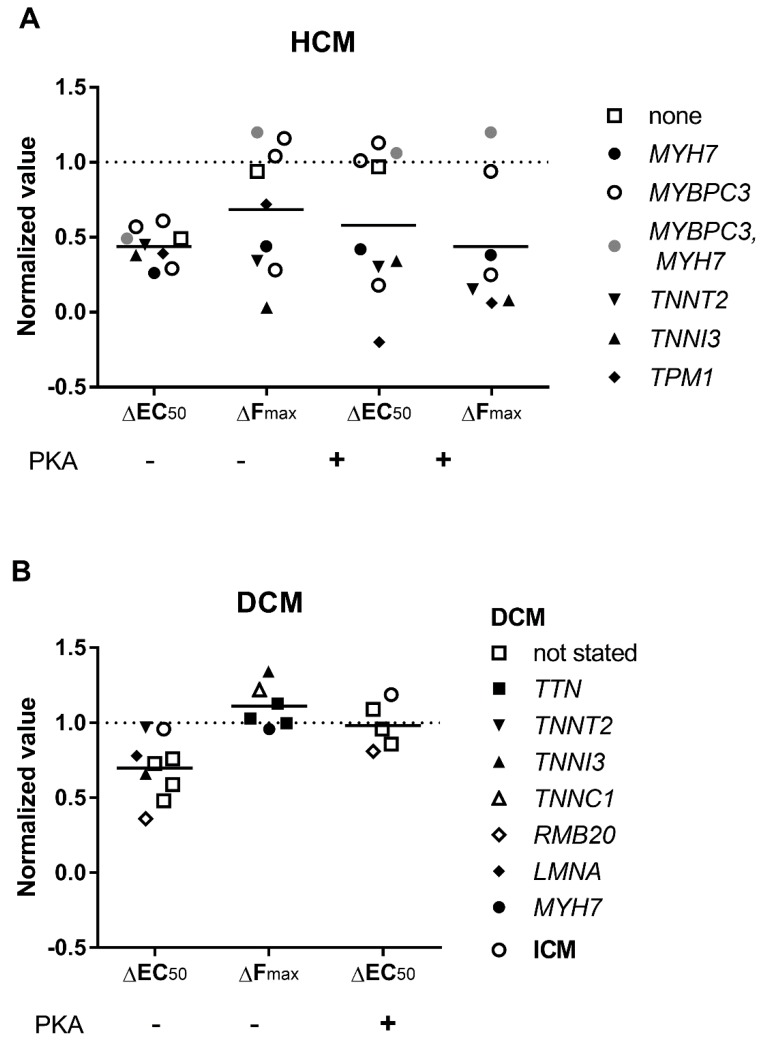
The evidence obtained from the in vitro experiments with isolated proteins and the studies with the use of animal models [105] is not sufficient. Several studies indicate that uncoupling maybe not an intrinsic property of human HCM [67,73] (Figure 4A). Treatment with PKA decreased Ca2+-sensitivity of human cardiomyocytes with HCM mutations in MYBPC3 and MYH7, as well as with mutations in thin filament proteins: TNNI R145W and TNNT2 K280N. The last mutation was identified earlier as an uncoupling mutation [105]. The results plotted on Figure 4 suggest that treatment with PKA has significantly greater effect on Ca2+-sensitivity in DCM samples (DCM ΔEC50 = 1.93 µM; HCM ΔEC50 = 0.96 µM).
Figure 4.
An increase in EC50 after phosphorylation with protein kinase A (PKA) for HCM (A) and DCM (B) muscle samples. Each data point represents a different experimental group where the symbols indicate genes where mutations were found. See also Table 1 and Table 2.
3.5. Length Dependent Activation
A length dependent activation can be specified as a contribution of a several components. At first, the maximal force is a function of sarcomere length. Secondly, an increase in sarcomere length considerably shifts the force-[Ca2+] curve to the left to lower concentrations of Ca2+. The increase in Ca2+-sensitivity, in turn, will enhance the rate of contractility. Additionally, the viscoelastic properties also significantly contribute to the total force production. Neither rate of force development (kACT, kTR) [106] nor relaxation kinetics constants (tLIN, kLIN, kREL) are sarcomere length dependent [76].
It has been shown that length dependent activation was decreased in HCM samples (Figure 5A). The decrease in EC50 for Ca2+ following sarcomere length increase was reduced in HCM samples by about 56%. Furthermore, the length dependence of force was decreased by 32%. Admittedly, it is rather unusual that length dependent increase of force was fully diminished in patients with mutations in the genes of troponins TNNI3 and TNNT2. In DCM muscles, length dependent activation was less affected: the change in ΔEC50 decreased by 30% and the force was unchanged (Figure 5B).
Figure 5.
Length dependent activation in cardiac muscle. An increase in sarcomere length decreases EC50 and increases Fmax. Each data point represents a different experimental group where the symbols indicate genes where mutations were found. The values for HCM (A) and DCM (B) are normalized to control donor hearts (dashed line). The data is from Table 1 and Table 2.
There are a number of facts suggesting that the length dependent shift in Ca2+-sensitivity was blunted in HCM samples due to the low phosphorylation status of the proteins potentially capable to modulate Ca2+-sensitivity: TnI and MyBP-C. Protein kinase A treatment fully restored length dependent activation to a normal level in half of the studies (Figure 5A). A decrease in levels of TnI phosphorylation correlates with a decrease of length dependent activation in DCM samples [48,64] (Figure 5B). MyBP-C phosphorylation (at Ser 275, 284, and 304) was also implied as an important factor capable to modify Ca2+-sensitivity and length dependence activation [77]. In this connection we would like to note the fact that HCM samples with truncations and missense mutations in the MYBPC3 gene show haploinsufficiency of MyBP-C. MyBP-C protein level in these samples was lower by about 30% [66,107,108]. The putative truncated MyBP-C variants were not detected in cardiac tissue samples by Western blot [109]. It suggests protein expression is very low or such mutant proteins undergo degradation in the ubiquitin-proteasome proteolytic pathway [110]. Such overloading of the ubiquitin-proteasome is one of the putative factors to initiate the disease.
3.6. Passive Stiffness
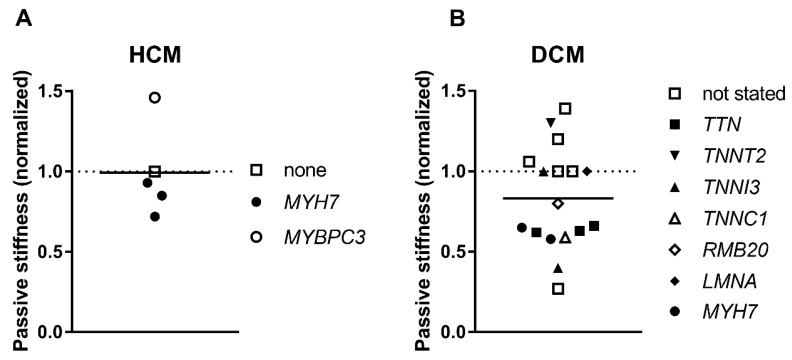
Titin is the major contributor to myofibril elasticity, and it is the main force component during diastole accumulating the potential energy of stretch and discharging it during systole. It was shown that passive myofibrillar stiffness was not substantially changed in HCM (Figure 6A, Table 1). Dilated cardiomyopathy-linked mutations in TTN and other sarcomeric proteins as well as with no found mutations decrease titin elasticity characterized by a decrease in myofibrillar stiffness by about 26% (Figure 6B, Table 2) [76,111,112]. The puzzling thing here is that putative truncated variants were not found in myofibril sarcomeres [76,113]. In some cases, the decrease in passive stiffness was, to some extent, explained by altered titin isoform expression [111,112]. However, in many cases there was no correlation between passive stiffness and differential titin isoform expression (Table 2). The slack or resting sarcomere length of DCM myocytes or myofibrils was matching to control [76,80,85], with a difference only found with RMB20 E913K mutation, where expression of the highly compliant N2BA titin isoform was several times higher [80]. The resting sarcomere length in HCM myocytes was similar to control [69] or slightly shorter in some cases [65].
Figure 6.
Passive stiffness. Passive stiffness was evaluated as a Young’s modulus or as a passive tension generated by muscle at stretch. Each data point represents a different experimental group where the symbols indicate genes where mutations were found. The values for HCM (A) and DCM (B) are normalized to control donor hearts (dashed line). The data is from Table 1 and Table 2.
4. Conclusions
In this review we tried to analyze data from different studies to show some main correlations between the mutations in the sarcomere proteins or their modification associated with different types of cardiomyopathies and the contractile properties of heart muscle from HCM and DCM patients.
Data from Table 1 shows that the inability of HCM cardiac myofibrils to generate the same force as controls is one of the essential differences between HCM and DCM muscles (Figure 1A,C). As defined, maximal force depends on the number of force-generating myosin heads and the rates of myosin cross-bridge attachments and detachments [114]. It is assumed that the increase of cross-bridge detachment rate leads to the detrimental increase in the energetic cost of contraction that may be the underlying mechanism of HCM. This hypothesis requires further research by testing contractility and the efficiency of ATP consumption in human cardiac muscles with different HCM mutations. The decrease of force was not observed for DCM myofibrils, but the rate of relaxation was also higher in samples with mutations in the contractile proteins (Figure 2D) [76].
The diastolic intracellular concentration of Ca2+ is about 0.2 μM and it increases to about 1.5 μM in systole [115,116,117]. An impaired contractility in HCM may be compensated by increased Ca2+-sensitivity regulated via dephosphorylation of TnI. Although an increase in Ca2+-sensitivity is the main feature of all cardiomyopathies and heart failure, dephosphorylation of TnI was the major factor responsible for this increase (Figure 3). Therefore, it is essential here to differentiate the direct effect of mutations in proteins on myofibril contractility from other secondary effects. It was proposed that high myofilament Ca2+-sensitivity slows myofibril relaxation in diastole [8,118,119]. Furthermore, it is interesting to investigate if the coupling between changes in TnI phosphorylation and the Ca2+-sensitivity of force in HCM and DCM myofibrils is reduced, particularly if a mutation was found in thin filaments. Additionally, titin mechanical properties are believed to be involved to the work output characteristic of myocardium. The decrease of titin originated stiffnes can cause systolic dysfunction in DCM heart muscles [76,112].
Funding
This work was funded the British Heart Foundation (PG/17/5/32705) and supported by The Magdi Yacoub Institute (Charity number: 1082750)
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gordan R., Gwathmey J.K., Xie L.H. Autonomic and endocrine control of cardiovascular function. World J. Cardiol. 2015;7:204–214. doi: 10.4330/wjc.v7.i4.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLeod K.T. Recent advances in understanding cardiac contractility in health and disease. F1000Research. 2016;5 doi: 10.12688/f1000research.8661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisner D.A., Caldwell J.L., Kistamas K., Trafford A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017;121:181–195. doi: 10.1161/CIRCRESAHA.117.310230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore J.R., Campbell S.G., Lehman W. Structural determinants of muscle thin filament cooperativity. Arch. Biochem. Biophys. 2016;594:8–17. doi: 10.1016/j.abb.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risi C., Eisner J., Belknap B., Heeley D.H., White H.D., Schroder G.F., Galkin V.E. Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA. 2017;114:6782–6787. doi: 10.1073/pnas.1700868114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Layland J., Solaro R.J., Shah A.M. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Wijnker P.J., Murphy A.M., Stienen G.J., van der Velden J. Troponin I phosphorylation in human myocardium in health and disease. Neth. Heart J. 2014;22:463–469. doi: 10.1007/s12471-014-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marston S.B. Why Is there a Limit to the Changes in Myofilament Ca(2+)-Sensitivity Associated with Myopathy Causing Mutations? Front. Physiol. 2016;7:415. doi: 10.3389/fphys.2016.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig R., Lee K.H., Mun J.Y., Torre I., Luther P.K. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466:425–431. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav S., Szczesna-Cordary D. Pseudophosphorylation of cardiac myosin regulatory light chain: A promising new tool for treatment of cardiomyopathy. Biophys. Rev. 2017;9:57–64. doi: 10.1007/s12551-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampourakis T., Yan Z., Gautel M., Sun Y.B., Irving M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl. Acad. Sci. USA. 2014;111:18763–18768. doi: 10.1073/pnas.1413922112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar M., Govindan S., Zhang M., Khairallah R.J., Martin J.L., Sadayappan S., de Tombe P.P. Cardiac Myosin-binding Protein C and Troponin-I Phosphorylation Independently Modulate Myofilament Length-dependent Activation. J. Biol. Chem. 2015;290:29241–29249. doi: 10.1074/jbc.M115.686790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papadakis M., Sharma S., Cox S., Sheppard M.N., Panoulas V.F., Behr E.R. The magnitude of sudden cardiac death in the young: A death certificate-based review in England and Wales. Europace. 2009;11:1353–1358. doi: 10.1093/europace/eup229. [DOI] [PubMed] [Google Scholar]
- 14.Hughes S.E., McKenna W.J. New insights into the pathology of inherited cardiomyopathy. Heart. 2005;91:257–264. doi: 10.1136/hrt.2004.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapezzi C., Arbustini E., Caforio A.L., Charron P., Gimeno-Blanes J., Helio T., Linhart A., Mogensen J., Pinto Y., Ristic A., et al. Diagnostic work-up in cardiomyopathies: Bridging the gap between clinical phenotypes and final diagnosis. A position statement from the ESC Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2013;34:1448–1458. doi: 10.1093/eurheartj/ehs397. [DOI] [PubMed] [Google Scholar]
- 16.Towbin J.A., Bowles N.E. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 17.McKenna W.J., Maron B.J., Thiene G. Classification, Epidemiology, and Global Burden of Cardiomyopathies. Circ. Res. 2017;121:722–730. doi: 10.1161/CIRCRESAHA.117.309711. [DOI] [PubMed] [Google Scholar]
- 18.Abelmann W.H., Lorell B.H. The challenge of cardiomyopathy. J. Am. Coll. Cardiol. 1989;13:1219–1239. doi: 10.1016/0735-1097(89)90293-3. [DOI] [PubMed] [Google Scholar]
- 19.Haas J., Frese K.S., Peil B., Kloos W., Keller A., Nietsch R., Feng Z., Muller S., Kayvanpour E., Vogel B., et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur. Heart J. 2015;36:1123–1135. doi: 10.1093/eurheartj/ehu301. [DOI] [PubMed] [Google Scholar]
- 20.Burke M.A., Cook S.A., Seidman J.G., Seidman C.E. Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. J. Am. Coll. Cardiol. 2016;68:2871–2886. doi: 10.1016/j.jacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deo R., Albert C.M. Epidemiology and genetics of sudden cardiac death. Circulation. 2012;125:620–637. doi: 10.1161/CIRCULATIONAHA.111.023838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konstam M.A., Abboud F.M. Ejection Fraction: Misunderstood and Overrated (Changing the Paradigm in Categorizing Heart Failure) Circulation. 2017;135:717–719. doi: 10.1161/CIRCULATIONAHA.116.025795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes S.E. The pathology of hypertrophic cardiomyopathy. Histopathology. 2004;44:412–427. doi: 10.1111/j.1365-2559.2004.01835.x. [DOI] [PubMed] [Google Scholar]
- 24.Basso C., Burke M., Fornes P., Gallagher P.J., de Gouveia R.H., Sheppard M., Thiene G., van der Wal A., Association for European Cardiovascular, Pathology Guidelines for autopsy investigation of sudden cardiac death. Virchows Arch. 2008;452:11–18. doi: 10.1007/s00428-007-0505-5. [DOI] [PubMed] [Google Scholar]
- 25.Mathew T., Williams L., Navaratnam G., Rana B., Wheeler R., Collins K., Harkness A., Jones R., Knight D., O’Gallagher K., et al. Diagnosis and assessment of dilated cardiomyopathy: A guideline protocol from the British Society of Echocardiography. Echo Res. Pract. 2017;4:G1–G13. doi: 10.1530/ERP-16-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantazis A., Vischer A.S., Perez-Tome M.C., Castelletti S. Diagnosis and management of hypertrophic cardiomyopathy. Echo Res. Pract. 2015;2:R45–R53. doi: 10.1530/ERP-15-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankaranarayanan R., Fleming E.J., Garratt C.J. Mimics of Hypertrophic Cardiomyopathy—Diagnostic Clues to Aid Early Identification of Phenocopies. Arrhythm. Electrophysiol. Rev. 2013;2:36–40. doi: 10.15420/aer.2013.2.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozkurt B., Colvin M., Cook J., Cooper L.T., Deswal A., Fonarow G.C., Francis G.S., Lenihan D., Lewis E.F., McNamara D.M., et al. Current Diagnostic and Treatment Strategies for Specific Dilated Cardiomyopathies: A Scientific Statement From the American Heart Association. Circulation. 2016;134:e579–e646. doi: 10.1161/CIR.0000000000000455. [DOI] [PubMed] [Google Scholar]
- 29.Japp A.G., Gulati A., Cook S.A., Cowie M.R., Prasad S.K. The Diagnosis and Evaluation of Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2016;67:2996–3010. doi: 10.1016/j.jacc.2016.03.590. [DOI] [PubMed] [Google Scholar]
- 30.Elliott P., Andersson B., Arbustini E., Bilinska Z., Cecchi F., Charron P., Dubourg O., Kuhl U., Maisch B., McKenna W.J., et al. Classification of the cardiomyopathies: A position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 31.Pinto Y.M., Elliott P.M., Arbustini E., Adler Y., Anastasakis A., Bohm M., Duboc D., Gimeno J., de Groote P., Imazio M., et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: A position statement of the ESC working group on myocardial and pericardial diseases. Eur. Heart J. 2016;37:1850–1858. doi: 10.1093/eurheartj/ehv727. [DOI] [PubMed] [Google Scholar]
- 32.Ware J.S., Amor-Salamanca A., Tayal U., Govind R., Serrano I., Salazar-Mendiguchia J., Garcia-Pinilla J.M., Pascual-Figal D.A., Nunez J., Guzzo-Merello G., et al. Genetic Etiology for Alcohol-Induced Cardiac Toxicity. J. Am. Coll. Cardiol. 2018;71:2293–2302. doi: 10.1016/j.jacc.2018.03.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linschoten M., Teske A.J., Baas A.F., Vink A., Dooijes D., Baars H.F., Asselbergs F.W. Truncating Titin (TTN) Variants in Chemotherapy-Induced Cardiomyopathy. J. Card. Fail. 2017;23:476–479. doi: 10.1016/j.cardfail.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Bakalakos A., Ritsatos K., Anastasakis A. Current perspectives on the diagnosis and management of dilated cardiomyopathy Beyond heart failure: A Cardiomyopathy Clinic Doctor’s point of view. Hellenic J. Cardiol. 2018 doi: 10.1016/j.hjc.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Felker G.M., Thompson R.E., Hare J.M., Hruban R.H., Clemetson D.E., Howard D.L., Baughman K.L., Kasper E.K. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N. Engl. J. Med. 2000;342:1077–1084. doi: 10.1056/NEJM200004133421502. [DOI] [PubMed] [Google Scholar]
- 36.Towbin J.A., Lowe A.M., Colan S.D., Sleeper L.A., Orav E.J., Clunie S., Messere J., Cox G.F., Lurie P.R., Hsu D., et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA. 2006;296:1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 37.Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E., et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer S., de Marvao A., Adami E., Fiedler L.R., Ng B., Khin E., Rackham O.J., van Heesch S., Pua C.J., Kui M., et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 2017;49:46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tayal U., Newsome S., Buchan R., Whiffin N., Halliday B., Lota A., Roberts A., Baksi A.J., Voges I., Midwinter W., et al. Phenotype and Clinical Outcomes of Titin Cardiomyopathy. J. Am. Coll. Cardiol. 2017;70:2264–2274. doi: 10.1016/j.jacc.2017.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Begay R.L., Graw S., Sinagra G., Merlo M., Slavov D., Gowan K., Jones K.L., Barbati G., Spezzacatene A., Brun F., et al. Role of Titin Missense Variants in Dilated Cardiomyopathy. J. Am. Heart Assoc. 2015;4:e002645. doi: 10.1161/JAHA.115.002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Velzen H.G., Schinkel A.F.L., Baart S.J., Oldenburg R.A., Frohn-Mulder I.M.E., van Slegtenhorst M.A., Michels M. Outcomes of Contemporary Family Screening in Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2018;11:e001896. doi: 10.1161/CIRCGEN.117.001896. [DOI] [PubMed] [Google Scholar]
- 42.Mathew J., Zahavich L., Lafreniere-Roula M., Wilson J., George K., Benson L., Bowdin S., Mital S. Utility of genetics for risk stratification in pediatric hypertrophic cardiomyopathy. Clin. Genet. 2018;93:310–319. doi: 10.1111/cge.13157. [DOI] [PubMed] [Google Scholar]
- 43.Lopes L.R., Syrris P., Guttmann O.P., O’Mahony C., Tang H.C., Dalageorgou C., Jenkins S., Hubank M., Monserrat L., McKenna W.J., et al. Novel genotype-phenotype associations demonstrated by high-throughput sequencing in patients with hypertrophic cardiomyopathy. Heart. 2015;101:294–301. doi: 10.1136/heartjnl-2014-306387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Driest S.L., Vasile V.C., Ommen S.R., Will M.L., Tajik A.J., Gersh B.J., Ackerman M.J. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 45.Kaski J.P., Syrris P., Esteban M.T., Jenkins S., Pantazis A., Deanfield J.E., McKenna W.J., Elliott P.M. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 2009;2:436–441. doi: 10.1161/CIRCGENETICS.108.821314. [DOI] [PubMed] [Google Scholar]
- 46.Millat G., Bouvagnet P., Chevalier P., Dauphin C., Jouk P.S., Da Costa A., Prieur F., Bresson J.L., Faivre L., Eicher J.C., et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur. J. Med. Genet. 2010;53:261–267. doi: 10.1016/j.ejmg.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Mazzarotto F., Girolami F., Boschi B., Barlocco F., Tomberli A., Baldini K., Coppini R., Tanini I., Bardi S., Contini E., et al. Defining the diagnostic effectiveness of genes for inclusion in panels: The experience of two decades of genetic testing for hypertrophic cardiomyopathy at a single center. Genet. Med. 2018 doi: 10.1038/s41436-018-0046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Authors/Task Force Members. Elliott P.M., Anastasakis A., Borger M.A., Borggrefe M., Cecchi F., Charron P., Hagege A.A., Lafont A., Limongelli G., et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: The Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur. Heart J. 2014;35:2733–2779. doi: 10.1016/j.rec.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Alfares A.A., Kelly M.A., McDermott G., Funke B.H., Lebo M.S., Baxter S.B., Shen J., McLaughlin H.M., Clark E.H., Babb L.J., et al. Results of clinical genetic testing of 2912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet. Med. 2015;17:880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 50.Mademont-Soler I., Mates J., Yotti R., Espinosa M.A., Perez-Serra A., Fernandez-Avila A.I., Coll M., Mendez I., Iglesias A., Del Olmo B., et al. Additional value of screening for minor genes and copy number variants in hypertrophic cardiomyopathy. PLoS ONE. 2017;12:e0181465. doi: 10.1371/journal.pone.0181465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dos Remedios C.G., Lal S.P., Li A., McNamara J., Keogh A., Macdonald P.S., Cooke R., Ehler E., Knoll R., Marston S.B., et al. The Sydney Heart Bank: Improving translational research while eliminating or reducing the use of animal models of human heart disease. Biophys. Rev. 2017;9:431–441. doi: 10.1007/s12551-017-0305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ait Mou Y., Bollensdorff C., Cazorla O., Magdi Y., de Tombe P.P. Exploring cardiac biophysical properties. Glob. Cardiol. Sci. Pract. 2015;2015:10. doi: 10.5339/gcsp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poggesi C., Tesi C., Stehle R. Sarcomeric determinants of striated muscle relaxation kinetics. Pflugers Arch. 2005;449:505–517. doi: 10.1007/s00424-004-1363-5. [DOI] [PubMed] [Google Scholar]
- 54.Stehle R., Solzin J., Iorga B., Poggesi C. Insights into the kinetics of Ca2+-regulated contraction and relaxation from myofibril studies. Pflugers Arch. 2009;458:337–357. doi: 10.1007/s00424-008-0630-2. [DOI] [PubMed] [Google Scholar]
- 55.Vikhorev P.G., Ferenczi M.A., Marston S.B. Instrumentation to study myofibril mechanics from static to artificial simulations of cardiac cycle. MethodsX. 2016;3:156–170. doi: 10.1016/j.mex.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brenner B. Effect of Ca2+ on cross-bridge turnover kinetics in skinned single rabbit psoas fibers: Implications for regulation of muscle contraction. Proc. Natl. Acad. Sci. USA. 1988;85:3265–3269. doi: 10.1073/pnas.85.9.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Witjas-Paalberends E.R., Piroddi N., Stam K., van Dijk S.J., Oliviera V.S., Ferrara C., Scellini B., Hazebroek M., ten Cate F.J., van Slegtenhorst M., et al. Mutations in MYH7 reduce the force generating capacity of sarcomeres in human familial hypertrophic cardiomyopathy. Cardiovasc. Res. 2013;99:432–441. doi: 10.1093/cvr/cvt119. [DOI] [PubMed] [Google Scholar]
- 58.Bollen I.A.E., van der Meulen M., de Goede K., Kuster D.W.D., Dalinghaus M., van der Velden J. Cardiomyocyte Hypocontractility and Reduced Myofibril Density in End-Stage Pediatric Cardiomyopathy. Front. Physiol. 2017;8:1103. doi: 10.3389/fphys.2017.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmer G., Zimmermann R., Hess O.M., Schneider J., Kubler W., Krayenbuehl H.P., Hagl S., Mall G. Decreased concentration of myofibrils and myofiber hypertrophy are structural determinants of impaired left ventricular function in patients with chronic heart diseases: A multiple logistic regression analysis. J. Am. Coll. Cardiol. 1992;20:1135–1142. doi: 10.1016/0735-1097(92)90369-X. [DOI] [PubMed] [Google Scholar]
- 60.Van Dijk S.J., Holewijn R.A., Tebeest A., Dos Remedios C., Stienen G.J., van der Velden J. A piece of the human heart: Variance of protein phosphorylation in left ventricular samples from end-stage primary cardiomyopathy patients. J. Muscle Res. Cell Motil. 2009;30:299–302. doi: 10.1007/s10974-010-9205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Z., Yan L.J. Protein Oxidative Modifications: Beneficial Roles in Disease and Health. J. Biochem. Pharmacol. Res. 2013;1:15–26. [PMC free article] [PubMed] [Google Scholar]
- 62.Wijnker P.J.M., Sequeira V., Kuster D.W.D., Velden J.V. Hypertrophic Cardiomyopathy: A Vicious Cycle Triggered by Sarcomere Mutations and Secondary Disease Hits. Antioxid. Redox Signal. 2018 doi: 10.1089/ars.2017.7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Predmore J.M., Wang P., Davis F., Bartolone S., Westfall M.V., Dyke D.B., Pagani F., Powell S.R., Day S.M. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin Z., Ren J., Guo W. Sarcomeric protein isoform transitions in cardiac muscle: A journey to heart failure. Biochim. Biophys. Acta. 2015;1852:47–52. doi: 10.1016/j.bbadis.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoskins A.C., Jacques A., Bardswell S.C., McKenna W.J., Tsang V., dos Remedios C.G., Ehler E., Adams K., Jalilzadeh S., Avkiran M., et al. Normal passive viscoelasticity but abnormal myofibrillar force generation in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2010;49:737–745. doi: 10.1016/j.yjmcc.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marston S., Copeland O., Jacques A., Livesey K., Tsang V., McKenna W.J., Jalilzadeh S., Carballo S., Redwood C., Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ. Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 67.Sequeira V., Wijnker P.J., Nijenkamp L.L., Kuster D.W., Najafi A., Witjas-Paalberends E.R., Regan J.A., Boontje N., Ten Cate F.J., Germans T., et al. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ. Res. 2013;112:1491–1505. doi: 10.1161/CIRCRESAHA.111.300436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Dijk S.J., Paalberends E.R., Najafi A., Michels M., Sadayappan S., Carrier L., Boontje N.M., Kuster D.W., van Slegtenhorst M., Dooijes D., et al. Contractile dysfunction irrespective of the mutant protein in human hypertrophic cardiomyopathy with normal systolic function. Circ. Heart Fail. 2012;5:36–46. doi: 10.1161/CIRCHEARTFAILURE.111.963702. [DOI] [PubMed] [Google Scholar]
- 69.Belus A., Piroddi N., Scellini B., Tesi C., D’Amati G., Girolami F., Yacoub M., Cecchi F., Olivotto I., Poggesi C. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J. Physiol. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piroddi N., Belus A., Scellini B., Tesi C., Giunti G., Cerbai E., Mugelli A., Poggesi C. Tension generation and relaxation in single myofibrils from human atrial and ventricular myocardium. Pflugers Arch. 2007;454:63–73. doi: 10.1007/s00424-006-0181-3. [DOI] [PubMed] [Google Scholar]
- 71.Witjas-Paalberends E.R., Ferrara C., Scellini B., Piroddi N., Montag J., Tesi C., Stienen G.J., Michels M., Ho C.Y., Kraft T., et al. Faster cross-bridge detachment and increased tension cost in human hypertrophic cardiomyopathy with the R403Q MYH7 mutation. J. Physiol. 2014;592:3257–3272. doi: 10.1113/jphysiol.2014.274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montag J., Kowalski K., Makul M., Ernstberger P., Radocaj A., Beck J., Becker E., Tripathi S., Keyser B., Muhlfeld C., et al. Burst-Like Transcription of Mutant and Wildtype MYH7-Alleles as Possible Origin of Cell-to-Cell Contractile Imbalance in Hypertrophic Cardiomyopathy. Front. Physiol. 2018;9:359. doi: 10.3389/fphys.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraft T., Witjas-Paalberends E.R., Boontje N.M., Tripathi S., Brandis A., Montag J., Hodgkinson J.L., Francino A., Navarro-Lopez F., Brenner B., et al. Familial hypertrophic cardiomyopathy: Functional effects of myosin mutation R723G in cardiomyocytes. J. Mol. Cell. Cardiol. 2013;57:13–22. doi: 10.1016/j.yjmcc.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 74.Tripathi S., Schultz I., Becker E., Montag J., Borchert B., Francino A., Navarro-Lopez F., Perrot A., Ozcelik C., Osterziel K.J., et al. Unequal allelic expression of wild-type and mutated beta-myosin in familial hypertrophic cardiomyopathy. Basic Res. Cardiol. 2011;106:1041–1055. doi: 10.1007/s00395-011-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montag J., Syring M., Rose J., Weber A.L., Ernstberger P., Mayer A.K., Becker E., Keyser B., Dos Remedios C., Perrot A., et al. Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy. J. Muscle Res. Cell Motil. 2017;38:291–302. doi: 10.1007/s10974-017-9486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vikhorev P.G., Smoktunowicz N., Munster A.B., Copeland O., Kostin S., Montgiraud C., Messer A.E., Toliat M.R., Li A., Dos Remedios C.G., et al. Abnormal contractility in human heart myofibrils from patients with dilated cardiomyopathy due to mutations in TTN and contractile protein genes. Sci. Rep. 2017;7:14829. doi: 10.1038/s41598-017-13675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Spaendonck-Zwarts K.Y., Posafalvi A., van den Berg M.P., Hilfiker-Kleiner D., Bollen I.A., Sliwa K., Alders M., Almomani R., van Langen I.M., van der Meer P., et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur. Heart J. 2014;35:2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 78.Bollen I.A.E., Schuldt M., Harakalova M., Vink A., Asselbergs F.W., Pinto J.R., Kruger M., Kuster D.W.D., van der Velden J. Genotype-specific pathogenic effects in human dilated cardiomyopathy. J. Physiol. 2017;595:4677–4693. doi: 10.1113/JP274145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoorntje E.T., Bollen I.A., Barge-Schaapveld D.Q., van Tienen F.H., Te Meerman G.J., Jansweijer J.A., van Essen A.J., Volders P.G., Constantinescu A.A., van den Akker P.C., et al. Lamin A/C-Related Cardiac Disease: Late Onset With a Variable and Mild Phenotype in a Large Cohort of Patients With the Lamin A/C p.(Arg331Gln) Founder Mutation. Circ. Cardiovasc. Genet. 2017;10:e001631. doi: 10.1161/CIRCGENETICS.116.001631. [DOI] [PubMed] [Google Scholar]
- 80.Beqqali A., Bollen I.A., Rasmussen T.B., van den Hoogenhof M.M., van Deutekom H.W., Schafer S., Haas J., Meder B., Sorensen K.E., van Oort R.J., et al. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc. Res. 2016;112:452–463. doi: 10.1093/cvr/cvw192. [DOI] [PubMed] [Google Scholar]
- 81.Dyer E.C., Jacques A.M., Hoskins A.C., Ward D.G., Gallon C.E., Messer A.E., Kaski J.P., Burch M., Kentish J.C., Marston S.B. Functional analysis of a unique troponin c mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circ. Heart Fail. 2009;2:456–464. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 82.Vallins W.J., Brand N.J., Dabhade N., Butler-Browne G., Yacoub M.H., Barton P.J. Molecular cloning of human cardiac troponin I using polymerase chain reaction. FEBS Lett. 1990;270:57–61. doi: 10.1016/0014-5793(90)81234-F. [DOI] [PubMed] [Google Scholar]
- 83.Chaturvedi R.R., Herron T., Simmons R., Shore D., Kumar P., Sethia B., Chua F., Vassiliadis E., Kentish J.C. Passive stiffness of myocardium from congenital heart disease and implications for diastole. Circulation. 2010;121:979–988. doi: 10.1161/CIRCULATIONAHA.109.850677. [DOI] [PubMed] [Google Scholar]
- 84.Van der Velden J., Papp Z., Zaremba R., Boontje N.M., de Jong J.W., Owen V.J., Burton P.B., Goldmann P., Jaquet K., Stienen G.J. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc. Res. 2003;57:37–47. doi: 10.1016/S0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 85.Bollen I.A.E., Ehler E., Fleischanderl K., Bouwman F., Kempers L., Ricke-Hoch M., Hilfiker-Kleiner D., Dos Remedios C.G., Kruger M., Vink A., et al. Myofilament Remodeling and Function Is More Impaired in Peripartum Cardiomyopathy Compared with Dilated Cardiomyopathy and Ischemic Heart Disease. Am. J. Pathol. 2017;187:2645–2658. doi: 10.1016/j.ajpath.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 86.Kooij V., Saes M., Jaquet K., Zaremba R., Foster D.B., Murphy A.M., Dos Remedios C., van der Velden J., Stienen G.J. Effect of troponin I Ser23/24 phosphorylation on Ca2+-sensitivity in human myocardium depends on the phosphorylation background. J. Mol. Cell. Cardiol. 2010;48:954–963. doi: 10.1016/j.yjmcc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kotter S., Gout L., Von Frieling-Salewsky M., Muller A.E., Helling S., Marcus K., Dos Remedios C., Linke W.A., Kruger M. Differential changes in titin domain phosphorylation increase myofilament stiffness in failing human hearts. Cardiovasc. Res. 2013;99:648–656. doi: 10.1093/cvr/cvt144. [DOI] [PubMed] [Google Scholar]
- 88.Papp Z., van der Velden J., Borbely A., Edes I., Stienen G.J.M. Altered myocardial force generation in end-stage human heart failure. ESC Heart Fail. 2014;1:160–165. doi: 10.1002/ehf2.12020. [DOI] [PubMed] [Google Scholar]
- 89.Mamidi R., Li J., Gresham K.S., Verma S., Doh C.Y., Li A., Lal S., Dos Remedios C.G., Stelzer J.E. Dose-Dependent Effects of the Myosin Activator Omecamtiv Mecarbil on Cross-Bridge Behavior and Force Generation in Failing Human Myocardium. Circ. Heart Fail. 2017;10:e004257. doi: 10.1161/CIRCHEARTFAILURE.117.004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeong M.Y., Lin Y.H., Wennersten S.A., Demos-Davies K.M., Cavasin M.A., Mahaffey J.H., Monzani V., Saripalli C., Mascagni P., Reece T.B., et al. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aao0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshida J., Kawai M., Minai K., Ogawa K., Ogawa T., Yoshimura M. Associations between Left Ventricular Cavity Size and Cardiac Function and Overload Determined by Natriuretic Peptide Levels and a Covariance Structure Analysis. Sci. Rep. 2017;7:2037. doi: 10.1038/s41598-017-02247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reinstein E., Gutierrez-Fernandez A., Tzur S., Bormans C., Marcu S., Tayeb-Fligelman E., Vinkler C., Raas-Rothschild A., Irge D., Landau M., et al. Congenital dilated cardiomyopathy caused by biallelic mutations in Filamin C. Eur. J. Hum. Genet. 2016;24:1792–1796. doi: 10.1038/ejhg.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Messer A.E., Gallon C.E., McKenna W.J., Dos Remedios C.G., Marston S.B. The use of phosphate-affinity SDS-PAGE to measure the cardiac troponin I phosphorylation site distribution in human heart muscle. Proteom. Clin. Appl. 2009;3:1371–1382. doi: 10.1002/prca.200900071. [DOI] [PubMed] [Google Scholar]
- 94.Bodor G.S., Oakeley A.E., Allen P.D., Crimmins D.L., Ladenson J.H., Anderson P.A. Troponin I phosphorylation in the normal and failing adult human heart. Circulation. 1997;96:1495–1500. doi: 10.1161/01.CIR.96.5.1495. [DOI] [PubMed] [Google Scholar]
- 95.Song W., Dyer E., Stuckey D.J., Copeland O., Leung M.C., Bayliss C., Messer A., Wilkinson R., Tremoleda J.L., Schneider M.D., et al. Molecular mechanism of the E99K mutation in cardiac actin (ACTC Gene) that causes apical hypertrophy in man and mouse. J. Biol. Chem. 2011;286:27582–27593. doi: 10.1074/jbc.M111.252320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hamdani N., Kooij V., van Dijk S., Merkus D., Paulus W.J., Remedios C.D., Duncker D.J., Stienen G.J., van der Velden J. Sarcomeric dysfunction in heart failure. Cardiovasc. Res. 2008;77:649–658. doi: 10.1093/cvr/cvm079. [DOI] [PubMed] [Google Scholar]
- 97.Copeland O., Sadayappan S., Messer A.E., Steinen G.J., van der Velden J., Marston S.B. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J. Mol. Cell. Cardiol. 2010;49:1003–1011. doi: 10.1016/j.yjmcc.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 98.Jacques A.M., Briceno N., Messer A.E., Gallon C.E., Jalilzadeh S., Garcia E., Kikonda-Kanda G., Goddard J., Harding S.E., Watkins H., et al. The molecular phenotype of human cardiac myosin associated with hypertrophic obstructive cardiomyopathy. Cardiovasc. Res. 2008;79:481–491. doi: 10.1093/cvr/cvn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Papadaki M., Vikhorev P.G., Marston S.B., Messer A.E. Uncoupling of myofilament Ca2+ sensitivity from troponin I phosphorylation by mutations can be reversed by epigallocatechin-3-gallate. Cardiovasc. Res. 2015;108:99–110. doi: 10.1093/cvr/cvv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Messer A.E., Bayliss C.R., El-Mezgueldi M., Redwood C.S., Ward D.G., Leung M.C., Papadaki M., Dos Remedios C., Marston S.B. Mutations in troponin T associated with Hypertrophic Cardiomyopathy increase Ca(2+)-sensitivity and suppress the modulation of Ca(2+)-sensitivity by troponin I phosphorylation. Arch. Biochem. Biophys. 2016;601:113–120. doi: 10.1016/j.abb.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Messer A.E., Chan J., Daley A., Copeland O., Marston S.B., Connolly D.J. Investigations into the Sarcomeric Protein and Ca(2+)-Regulation Abnormalities Underlying Hypertrophic Cardiomyopathy in Cats (Felix catus) Front. Physiol. 2017;8:348. doi: 10.3389/fphys.2017.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayliss C.R., Jacques A.M., Leung M.C., Ward D.G., Redwood C.S., Gallon C.E., Copeland O., McKenna W.J., Dos Remedios C., Marston S.B., et al. Myofibrillar Ca(2+) sensitivity is uncoupled from troponin I phosphorylation in hypertrophic obstructive cardiomyopathy due to abnormal troponin T. Cardiovasc. Res. 2013;97:500–508. doi: 10.1093/cvr/cvs322. [DOI] [PubMed] [Google Scholar]
- 103.Vikhorev P.G., Song W., Wilkinson R., Copeland O., Messer A.E., Ferenczi M.A., Marston S.B. The dilated cardiomyopathy-causing mutation ACTC E361G in cardiac muscle myofibrils specifically abolishes modulation of Ca(2+) regulation by phosphorylation of troponin I. Biophys. J. 2014;107:2369–2380. doi: 10.1016/j.bpj.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Biesiadecki B.J., Kobayashi T., Walker J.S., Solaro R.J., de Tombe P.P. The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ. Res. 2007;100:1486–1493. doi: 10.1161/01.RES.0000267744.92677.7f. [DOI] [PubMed] [Google Scholar]
- 105.Messer A.E., Marston S.B. Investigating the role of uncoupling of troponin I phosphorylation from changes in myofibrillar Ca(2+)-sensitivity in the pathogenesis of cardiomyopathy. Front. Physiol. 2014;5:315. doi: 10.3389/fphys.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edes I.F., Czuriga D., Csanyi G., Chlopicki S., Recchia F.A., Borbely A., Galajda Z., Edes I., van der Velden J., Stienen G.J., et al. Rate of tension redevelopment is not modulated by sarcomere length in permeabilized human, murine, and porcine cardiomyocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293:R20–R29. doi: 10.1152/ajpregu.00537.2006. [DOI] [PubMed] [Google Scholar]
- 107.McNamara J.W., Li A., Lal S., Bos J.M., Harris S.P., van der Velden J., Ackerman M.J., Cooke R., Dos Remedios C.G. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS ONE. 2017;12:e0180064. doi: 10.1371/journal.pone.0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Dijk S.J., Dooijes D., dos Remedios C., Michels M., Lamers J.M., Winegrad S., Schlossarek S., Carrier L., ten Cate F.J., Stienen G.J., et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: Haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
- 109.Rottbauer W., Gautel M., Zehelein J., Labeit S., Franz W.M., Fischer C., Vollrath B., Mall G., Dietz R., Kubler W., et al. Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J. Clin. Investig. 1997;100:475–482. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gilda J.E., Gomes A.V. Proteasome dysfunction in cardiomyopathies. J. Physiol. 2017;595:4051–4071. doi: 10.1113/JP273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Makarenko I., Opitz C.A., Leake M.C., Neagoe C., Kulke M., Gwathmey J.K., del Monte F., Hajjar R.J., Linke W.A. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 112.Nagueh S.F., Shah G., Wu Y., Torre-Amione G., King N.M., Lahmers S., Witt C.C., Becker K., Labeit S., Granzier H.L. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 113.Roberts A.M., Ware J.S., Herman D.S., Schafer S., Baksi J., Bick A.G., Buchan R.J., Walsh R., John S., Wilkinson S., et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 2015;7:270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rice J.J., Winslow R.L., Hunter W.C. Comparison of putative cooperative mechanisms in cardiac muscle: Length dependence and dynamic responses. Am. J. Physiol. 1999;276:H1734–H1754. doi: 10.1152/ajpheart.1999.276.5.H1734. [DOI] [PubMed] [Google Scholar]
- 115.Beuckelmann D.J., Nabauer M., Erdmann E. Intracellular calcium handling in isolated ventricular myocytes from patients with terminal heart failure. Circulation. 1992;85:1046–1055. doi: 10.1161/01.CIR.85.3.1046. [DOI] [PubMed] [Google Scholar]
- 116.Gwathmey J.K., Slawsky M.T., Hajjar R.J., Briggs G.M., Morgan J.P. Role of intracellular calcium handling in force-interval relationships of human ventricular myocardium. J. Clin. Investig. 1990;85:1599–1613. doi: 10.1172/JCI114611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bayer J.D., Narayan S.M., Lalani G.G., Trayanova N.A. Rate-dependent action potential alternans in human heart failure implicates abnormal intracellular calcium handling. Heart Rhythm. 2010;7:1093–1101. doi: 10.1016/j.hrthm.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dewey S., Xu Q., Gomes A. Static and dynamic properties of the HCM myocardium. J. Mol. Cell. Cardiol. 2010;49:715–718. doi: 10.1016/j.yjmcc.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Poggesi C., Ho C.Y. Muscle dysfunction in hypertrophic cardiomyopathy: What is needed to move to translation? J. Muscle Res. Cell. Motil. 2014;35:37–45. doi: 10.1007/s10974-014-9374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]