Abstract
Longan is an important fruit tree in the subtropical region of Southeast Asia and Australia. However, its blooming and its yield are susceptible to stresses such as droughts, high salinity, and high and low temperature. To date, the molecular mechanisms of abiotic stress tolerance and flower induction in longan have not been elucidated. WRKY transcription factors (TFs), which have been studied in various plant species, play important regulatory roles in plant growth, development, and responses to stresses. However, there is no report about WRKYs in longan. In this study, we identified 55 WRKY genes with the conserved WRKY domain and zinc finger motif in the longan genome. Based on the structural features of WRKY proteins and topology of the phylogenetic tree, the longan WRKY (DlWRKY) family was classified into three major groups (I–III) and five subgroups (IIa–IIe) in group II. Tissue expression analysis showed that 25 DlWRKYs were highly expressed in almost all organs, suggesting that these genes may be important for plant growth and organ development in longan. Comparative RNA-seq and qRT-PCR-based gene expression analysis revealed that 18 DlWRKY genes showed a specific expression during three stages of flower induction in “Sijimi” (“SJ”), which exhibited the “perpetual flowering” (PF) habit, indicating that these 18 DlWRKY genes may be involved in the flower induction and the genetic control of the perpetual flowering trait in longan. Furthermore, the RT-qPCR analysis illustrated the significant variation of 27, 18, 15, 17, 27, and 23 DlWRKY genes under SA (Salicylic acid), MeJA (Methyl Jasmonate), heat, cold, drought, or high salinity treatment, respectively, implicating that they might be stress- or hormone-responsive genes. In summary, we systematically and comprehensively analyzed the structure, evolution, and expression pattern of the DlWRKY genes. The results presented here increase our understanding of the WRKY family in fruit trees and provide a basis for the further elucidation of the biological function of DlWRKY genes in longan.
Keywords: longan, WRKY, expression analysis, flower induction, abiotic stress
1. Introduction
Longan (Dimocarpuslongan Lour.) is an important subtropical fruit tree in the family Sapindaceae, which is grown in many subtropical and tropical countries with most of the production in Southeast Asia and Australia [1]. Biennial bearing is the most serious problem that affects longan fruit products. Among the factors that affect D. longan fruit yield, the difficulty and unstableness to blossom is one of the most challenging problems [2]. Floral bud induction of D. longan requires favorable conditions such as a period of low temperature (vernalization), suitable salinity, and dry conditions. To obtain a stable high yield, off-season flowering in longan is achieved by chemical treatment with potassium chlorate (KClO3) application [3,4]. Nevertheless, the induction effect varies in different regions and varieties. Therefore, the study of the molecular regulatory mechanisms of flower induction and abiotic stress tolerance in longan is particularly important for understanding and solving the problems associated with fruit yield. However, due to the long generation time and lack of genome information, knowledge of the molecular regulatory mechanisms of flower induction and abiotic stress tolerance in longan is scarce.
As an important developmental process in the plant life cycle, flowering is directly linked to production whenever seeds or fruits are harvested [5]. The molecular and genetic bases of flowering have been well studied in Arabidopsis thaliana [6,7,8]. There are at least five major flowering pathways in Arabidopsis, including the photoperiod, autonomous, vernalization, gibberellin (GA), and aging pathways [9]. These pathways activate or inhibit floral transformation through a series of flower integron genes, such as the flowering locus T (FT), flowering locus C (FLC), and constans (CO) [10]. In addition, several transcription factors (TFs), such as MADS-domain TFs [11], NACs [12], MYBs [13], and DREBs [14], participate in the signaling of flowering regulation. As the seventh largest TF family in flowering plants, many WRKY genes are also involved in the determination of flowering time [15]. For example, in A. thaliana, the lines over-express GsWRKY20, MlWRKY12, and WRKY71 in the flowers earlier than in the wild-type [16,17,18]. A recent research study found that two WRKY proteins (AtWRKY12 and AtWRKY13) played opposite functions in controlling the flowering time under short-day conditions in A. thaliana partly through mediating the effect of GA3. The wrky12 mutant exhibits late flowering and the wrky13 mutant shows earlier flowering than that of the wild-type [19].
Abiotic stresses such as drought, heat, salt, and cold are the major causes of declined crop productivity worldwide. At the molecular level, several TFs, such as AP2/EREBP, NAC, WRKY, bZIP, MYB, and bHLH play a vital role in regulating downstream genes to protect plants from these stresses [20]. As one of the largest TF families in plants, the WRKY TFs also play pivotal roles in regulating many abiotic stress reactions [15]. In Arabidopsis, some of the AtWRKYs respond strongly to various abiotic stresses, such as salinity, drought, and cold [21,22,23,24]. In rice, 11 OsWRKY genes showed variable responses to salt, polyethylene glycol (PEG), and cold or heat stresses [25]. Overexpression of OsWRKY47 increased both the drought tolerance and yield compared with wild-type plants [26]. In mulberry, Morus013217 and Morus002784 show high accumulation in response to cold and salt stresses. Morus005757 shows significant up-regulation in response to dehydration stress, salinity stress, and SA and ABA (Abscisic acid) treatments [27]. Similar results were also found in wheat, common bean [28], grape [29], pineapple [30], soybean [31], moso bamboo [32], Caragana intermedia [33], peanut [34], and broomcorn millet [35]. These observations suggest that studying the WRKY gene families may provide valuable insights into the mechanism underlying abiotic stress tolerance in plants. As perennials growing in the subtropical and tropical area, some abiotic stresses, such as drought, heat, salt, and cold often have an adverse effect on the growth and yield of longan. However, given the lack of genome information, the identified and functions of WRKY genes in longan are still unknown.
In the present study, we performed a genome-wide identification of WRKY TFs in longan and analyzed their gene structures, conserved motifs, and expression patterns in nine different tissues. This work also determined the expression profiles of longan WRKY (DlWRKY) in three flowering stages of two longan cultivars and measured their transcript abundance in response to different phytohormone treatments and various abiotic stresses. This study provides a basis for future studies on DlWRKY gene family evolution and function.
2. Results
2.1. Identification of WRKY Gene Family in Longan
To extensively identify the WRKY genes in longan, whole-genome scanning was used to identify the genes which contain the particular domain by both the hidden Markov model (HMM) and Blastn search methods. In total, 59 candidate WRKY genes were identified (Table S1). After the WRKY domain scanning and sequence alignment, three genes (Dlo_007676.1, Dlo_032703.1, and Dlo_028398.1) without a complete predicted WRKY domain and one redundant gene (Dlo_037584.1) were removed. Finally, 55 DlWRKY genes were determined in the longan genome (Table 1). According to their chromosome locations, the 55 DlWRKY genes were designated DlWRKY1–DlWRKY55. In addition, the basic properties of DlWRKY genes, including the length of the full-length sequence, open reading frame (ORF), protein sequence, molecular weight (MW), and PI, were systematically evaluated (Table 1). The average length of these DlWRKY genes was 2417 bp and the length mainly centered on the range of 892 bp (DlWRKY12) to 5385 bp (DlWRKY36). Meanwhile, the length of the ORF was mainly distributed from 480 bp (DlWRKY12 and DlWRKY34) to 3813 bp (DlWRKY36), with an average of 1237 bp. The length of the protein sequences ranged from 160 AA (DlWRKY12 and DlWRKY34) to 1271 AA (DlWRKY36), with an average of 411 AA. The protein MW ranged from 18.10 kDa (DlWRKY34) to 143.77 kDa (DlWRKY36), with an average of 44.73 kDa. The predicted isoelectric point of the DlWRKY proteins varied from 4.62 (DlWRKY22) to 9.77 (DlWRKY13), with an average of 7.11.
Table 1.
The information of the DlWRKY gene family.
| Gene Name | Gene Locus ID | Location | ORF (bp) | Size (aa) | PI | MW (KDa) | Intron | Full Length |
|---|---|---|---|---|---|---|---|---|
| DlWRKY1 | Dlo_000299.1 | scaffold1:3145979:3147233 | 1071 | 356 | 9.63 | 38.76 | 2 | 1255 |
| DlWRKY2 | Dlo_026119.1 | scaffold6:875263:878308 | 894 | 297 | 6.26 | 32.31 | 2 | 3046 |
| DlWRKY3 | Dlo_026149.1 | scaffold6:1127159:1130416 | 1596 | 532 | 7.26 | 57.64 | 3 | 3258 |
| DlWRKY4 | Dlo_026267.1 | scaffold6:2195842:2200859 | 1815 | 605 | 6.66 | 66.08 | 4 | 5018 |
| DlWRKY5 | Dlo_030713.1 | scaffold8:184175:186167 | 1059 | 353 | 5.63 | 39.46 | 2 | 1993 |
| DlWRKY6 | Dlo_002181.1 | scaffold11:1861336:1864296 | 1767 | 589 | 7.23 | 64.37 | 4 | 2961 |
| DlWRKY7 | Dlo_012455.1 | scaffold23:1107291:1111499 | 1914 | 638 | 6.75 | 69.01 | 5 | 4209 |
| DlWRKY8 | Dlo_013053.2 | scaffold24:1070557:1072933 | 1668 | 556 | 6.52 | 61.46 | 4 | 2377 |
| DlWRKY9 | Dlo_015501.2 | scaffold29:1782026:1783019 | 762 | 254 | 8.99 | 28.30 | 4 | 994 |
| DlWRKY10 | dlo_037126.1 | scaffold29:1793158:1794294 | 684 | 228 | 9.02 | 25.58 | 2 | 1146 |
| DlWRKY11 | Dlo_016404.1 | scaffold31:1522675:1524675 | 1026 | 342 | 5.60 | 38.86 | 2 | 2001 |
| DlWRKY12 | Dlo_019125.1 | scaffold38:1882835:1883726 | 480 | 160 | 5.16 | 18.38 | 2 | 892 |
| DlWRKY13 | Dlo_023965.1 | scaffold53:1206068:1207919 | 1035 | 345 | 9.77 | 38.51 | 2 | 1852 |
| DlWRKY14 | Dlo_028963.1 | scaffold71:878665:881164 | 1613 | 471 | 8.87 | 51.80 | 3 | 2500 |
| DlWRKY15 | Dlo_031097.1 | scaffold81:147303:148636 | 972 | 324 | 6.33 | 35.30 | 2 | 1334 |
| DlWRKY16 | Dlo_033905.1 | scaffold98:272537:275137 | 1419 | 473 | 5.82 | 51.20 | 2 | 2601 |
| DlWRKY17 | Dlo_001368.1 | scaffold105:274029:278833 | 1425 | 475 | 6.10 | 52.14 | 4 | 4805 |
| DlWRKY18 | Dlo_003898.1 | scaffold124:605265:607367 | 1053 | 351 | 9.04 | 39.34 | 1 | 2103 |
| DlWRKY19 | Dlo_003928.1 | scaffold124:1058067:1061659 | 633 | 211 | 6.37 | 23.26 | 2 | 3593 |
| DlWRKY20 | Dlo_004435.1 | scaffold129:429868:432959 | 1644 | 548 | 7.41 | 59.78 | 5 | 3092 |
| DlWRKY21 | Dlo_008095.1 | scaffold167:682922:683969 | 714 | 238 | 5.14 | 26.59 | 2 | 1048 |
| DlWRKY22 | Dlo_008126.1 | scaffold168:307774:310141 | 1245 | 415 | 4.62 | 44.95 | 1 | 2368 |
| DlWRKY23 | Dlo_008610.1 | scaffold176:75022:76492 | 1023 | 341 | 8.62 | 38.08 | 4 | 1471 |
| DlWRKY24 | Dlo_009865.1 | scaffold192:233555:234849 | 1071 | 357 | 5.50 | 39.25 | 2 | 1295 |
| DlWRKY25 | Dlo_011410.1 | scaffold213:248908:250725 | 1038 | 346 | 5.93 | 38.65 | 2 | 1818 |
| DlWRKY26 | Dlo_011411.1 | scaffold213:253855:257080 | 1122 | 374 | 6.00 | 40.22 | 2 | 3226 |
| DlWRKY27 | Dlo_012276.1 | scaffold229:13116:15182 | 1005 | 335 | 7.16 | 37.09 | 2 | 2067 |
| DlWRKY28 | Dlo_012878.1 | scaffold238:352167:354143 | 1527 | 509 | 5.89 | 55.49 | 3 | 1977 |
| DlWRKY29 | Dlo_013340.1 | scaffold245:258019:261130 | 696 | 232 | 8.95 | 26.57 | 2 | 3112 |
| DlWRKY30 | Dlo_013413.1 | scaffold247:267246:270528 | 2238 | 746 | 5.59 | 80.60 | 4 | 3283 |
| DlWRKY31 | Dlo_014324.1 | scaffold266:341214:343700 | 663 | 221 | 7.71 | 25.38 | 3 | 2487 |
| DlWRKY32 | Dlo_015139.1 | scaffold286:162902:164294 | 1059 | 353 | 6.32 | 38.46 | 2 | 1393 |
| DlWRKY33 | Dlo_015144.1 | scaffold286:195837:198196 | 615 | 205 | 9.03 | 23.13 | 1 | 2360 |
| DlWRKY34 | Dlo_015224.1 | scaffold287:217068:218497 | 480 | 160 | 9.54 | 18.10 | 1 | 1430 |
| DlWRKY35 | Dlo_016828.1 | scaffold322:63655:67562 | 1326 | 442 | 9.62 | 48.27 | 4 | 3908 |
| DlWRKY36 | Dlo_022548.1 | scaffold487:170363:175747 | 3813 | 1271 | 5.15 | 143.77 | 5 | 5385 |
| DlWRKY37 | Dlo_023098.1 | scaffold502:191885:193351 | 1056 | 352 | 9.46 | 38.46 | 2 | 1467 |
| DlWRKY38 | Dlo_023764.1 | scaffold524:170088:173717 | 1533 | 511 | 8.66 | 55.75 | 3 | 3630 |
| DlWRKY39 | Dlo_025188.1 | scaffold568:191129:193577 | 1530 | 510 | 8.26 | 55.75 | 5 | 2449 |
| DlWRKY40 | Dlo_025974.1 | scaffold597:89062:90386 | 1110 | 370 | 5.07 | 40.99 | 2 | 1325 |
| DlWRKY41 | Dlo_026484.1 | scaffold607:21585:23785 | 1218 | 406 | 6.06 | 45.38 | 4 | 2201 |
| DlWRKY42 | Dlo_027244.2 | scaffold640:85638:89083 | 2298 | 766 | 5.15 | 83.59 | 4 | 3446 |
| DlWRKY43 | Dlo_027361.1 | scaffold648:191661:193182 | 969 | 323 | 9.14 | 36.57 | 2 | 1522 |
| DlWRKY44 | Dlo_027614.1 | scaffold657:107511:111179 | 1521 | 507 | 5.55 | 54.88 | 4 | 3669 |
| DlWRKY45 | Dlo_029034.1 | scaffold711:179562:181398 | 1539 | 513 | 8.27 | 55.15 | 2 | 1837 |
| DlWRKY46 | Dlo_029939.1 | scaffold757:33093:37889 | 1710 | 570 | 6.38 | 61.42 | 5 | 4797 |
| DlWRKY47 | Dlo_031466.1 | scaffold829:42224:45497 | 1023 | 341 | 7.20 | 7.71 | 4 | 3274 |
| DlWRKY48 | Dlo_031469.1 | scaffold829:58277:59797 | 990 | 330 | 9.06 | 36.21 | 3 | 1521 |
| DlWRKY49 | Dlo_031936.1 | scaffold858:266912:269300 | 588 | 196 | 9.46 | 22.05 | 1 | 2389 |
| DlWRKY50 | Dlo_032595.1 | scaffold896:87649:89238 | 1185 | 395 | 6.67 | 43.04 | 2 | 1590 |
| DlWRKY51 | Dlo_033966.1 | scaffold980:88739:90132 | 933 | 311 | 5.14 | 34.86 | 2 | 1394 |
| DlWRKY52 | Dlo_001658.1 | scaffold1077:66972:68290 | 918 | 306 | 6.26 | 33.96 | 4 | 1319 |
| DlWRKY53 | Dlo_002663.1 | scaffold1135:95286:97669 | 1929 | 643 | 5.73 | 70.07 | 4 | 2384 |
| DlWRKY54 | Dlo_004749.1 | scaffold1314:73982:75144 | 795 | 265 | 5.24 | 30.27 | 2 | 1163 |
| DlWRKY55 | Dlo_010873.1 | scaffold2042:2013:3910 | 1023 | 341 | 9.42 | 37.97 | 3 | 1898 |
2.2. Phylogenetic Analysis of DlWRKY
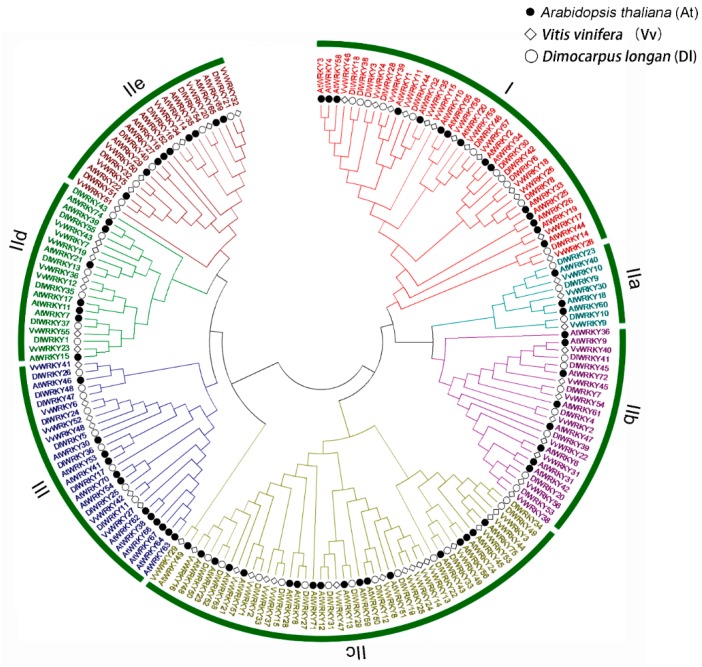
A phylogenetic tree was constructed using the maximum likelihood (ML) method and based on multiple alignments of longan, grape, and Arabidopsis WRKY domain aa sequences. As shown in Figure 1, the phylogenetic results revealed that all the DlWRKY proteins could be categorized into three groups (I, II, and III). Eleven DlWRKY proteins were considered to be group I, which included two WRKY domains and a C2H2 (C–X4–C–X22–23–HXH) zinc finger motif. A total of 35 DlWRKY proteins contained one WRKY domain and a C2H2 (C–X4–5–C–X23–HXH) zinc-binding motif, which were classified as group II. The nine remaining genes were assigned to Group III, which consisted of a single WRKY domain and a C2CH (C–X7–C–X23–HXC) zinc-binding motif. According to the WRKY subgroup classification of Arabidopsis, the DlWRKYs in Group II were further subdivided into five subgroups, including groups IIa (3), IIb (7) IIc (13), IId (6), and IIe (6).
Figure 1.
The phylogenetic analysis of the longan WRKY proteins with orthologous members from grape and Arabidopsis. The maximum likelihood phylogenetic tree was constructed by MEGA 6.0. Different groups of DlWRKY proteins are indicated by a circle and the different colors.
2.3. Multiple Sequence Alignment and Structure Analysis
The WRKYGQK sequence is a considerably conservative motif of WRKY proteins and several variants of this conserved WRKY motif have been reported in plants [36], including WRKYGEK, WRKYGKK, WSKYEQK, and WRKYSEK. In the present study, this motif was observed in all longan WRKY proteins and three variants of this motif were also found. The majority of DlWRKY proteins contained the WRKYGQK motif, and WRKYGKK and WKKYRQK were observed in DlWRKY19 and DlWRKY47, respectively. The other remarkably conservative motif was a zinc finger structure which contained two types of zinc finger motifs: C–X4-5–C–X22–23–HXH and C–X7–C–X23–HXC. A total of 46 DlWRKY proteins contained C–X4-5–C–X22–23–HXH, and nine DlWRKY proteins contained C–X7–C–X23–HXC, which all belonged to Group III (Table S1).
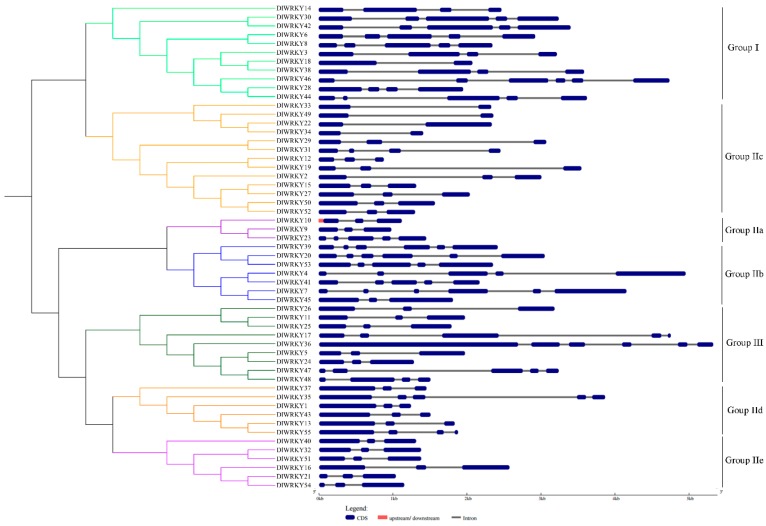
According to the Gene Structure Display Server (GSDS) website, the number of introns was in the range of 1–5 in all the longan WRKY gene families, with most of DlWRKY genes containing 2–4 introns (n = 81.0%). The average number of introns was 2.82. In addition, the phylogenetic analysis of the DlWRKY gene family showed that the genes within the same group generally exhibited a similar exon/intron structure. For example, subgroup IIe contained two introns (Figure 2).
Figure 2.
The unrooted phylogenetic tree (left) and gene structure (right) of 55 DlWRKY proteins. The phylogenetic tree was constructed by MEGA 6.0. The red color indicates the untranslated 5′- and 3′-regions; the blue color indicates exons; and the gray color indicates introns.
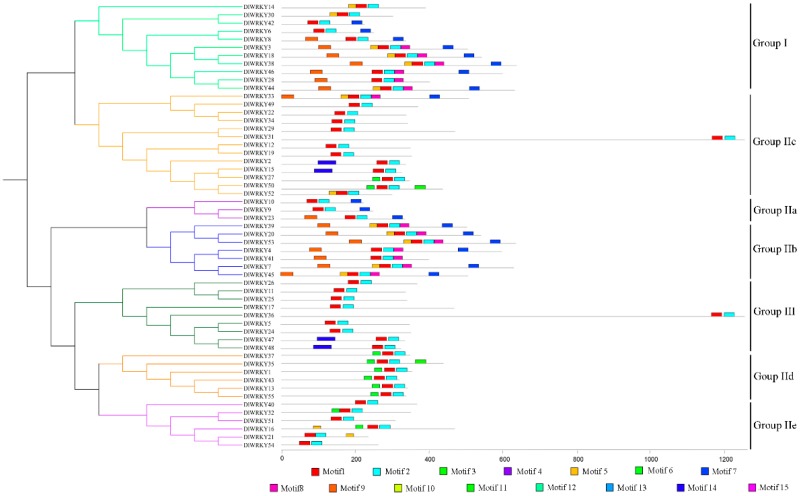
To further understand the similarity and diversity of motif composition among different DlWRKY proteins, a phylogenetic tree based on the full-length DlWRKY proteins was constructed (Figure 3). The motifs in the DlWRKY protein sequences were also predicted using MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi) (Figure 3 and Table S2). A total of 15 motifs were identified to illustrate the WRKY protein structure in longan. The results showed that the number of motifs in DlWRKYs ranged from 2 to 6, and the length of motifs ranged from 21 to 50 amino acids. Among the 15 identified motifs, motifs 1 and 2, characterized as WRKY domains, were broadly distributed across the DlWRKYs.
Figure 3.
The unrooted phylogenetic tree (left) and conserved motifs (right) of 55 DlWRKY proteins. The phylogenetic tree was constructed using the same method used in Figure 2. Different colors represent various groups. MEME was used to predict motifs, and these motifs are represented by boxes.
2.4. Tissue-Specific Expression Patterns of DlWRKY
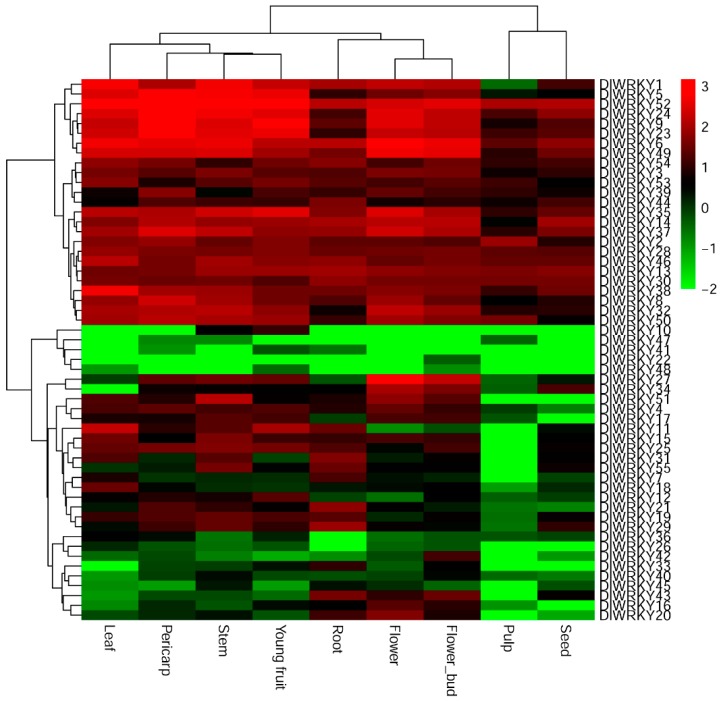
To generate expression profiles of DlWRKY genes under normal conditions, the expression levels of the 55 DlWRKY genes in the root, stem, leaf, seed, young fruit, pulp, pericarp, flower, and flower bud were investigated by the RNA-seq analysis. The log10 (FPKM + 0.01) values of the transcripts were clustered hierarchically and displayed in a heat map (Figure 4 and Table S3). The results showed that 96.36% (53 of 55) of DlWRKYs were expressed in young fruits and 94.55% were expressed in the pericarp, stems, and flower bud. A total of 90.91%, 89.09%, and 81.82% of DlWRKYs were expressed in the flower, leaf, root, and seed, respectively. Only a few DlWRKY genes were detected in pulps (67.27%). Approximately 60% (33 of 55) of the DlWRKY genes were expressed in each tested tissue, in which 25 DlWRKY genes (DlWRKY1, 2, 3, 5, 6, 8, 9, 13, 14, 23, 24, 28, 30, 32, 35, 37, 38, 39, 44, 49, 50, 52, 53, and 54) were highly expressed in at least six longan tissues. In contrast, 12 DlWRKY genes (DlWRKY10, 12, 18, 22, 26, 36, 40, 41, 42, 45, 47, and 48) were expressed at low levels in all tested tissues. Furthermore, DlWRKY22 only displayed a significantly low expression in the flower bud. DlWRKY10, 22, 41, 47, and 48 were preferential accumulation in two or three tissues.
Figure 4.
The heat map of the DlWRKY gene expression profiles in different tissues. The color scale represents the log10 expression values; the red and green colors indicate the higher or lower transcript abundances compared to the relevant control, respectively.
2.5. Comparative Expression Profiles of Two Longan Species during the Flowering Process
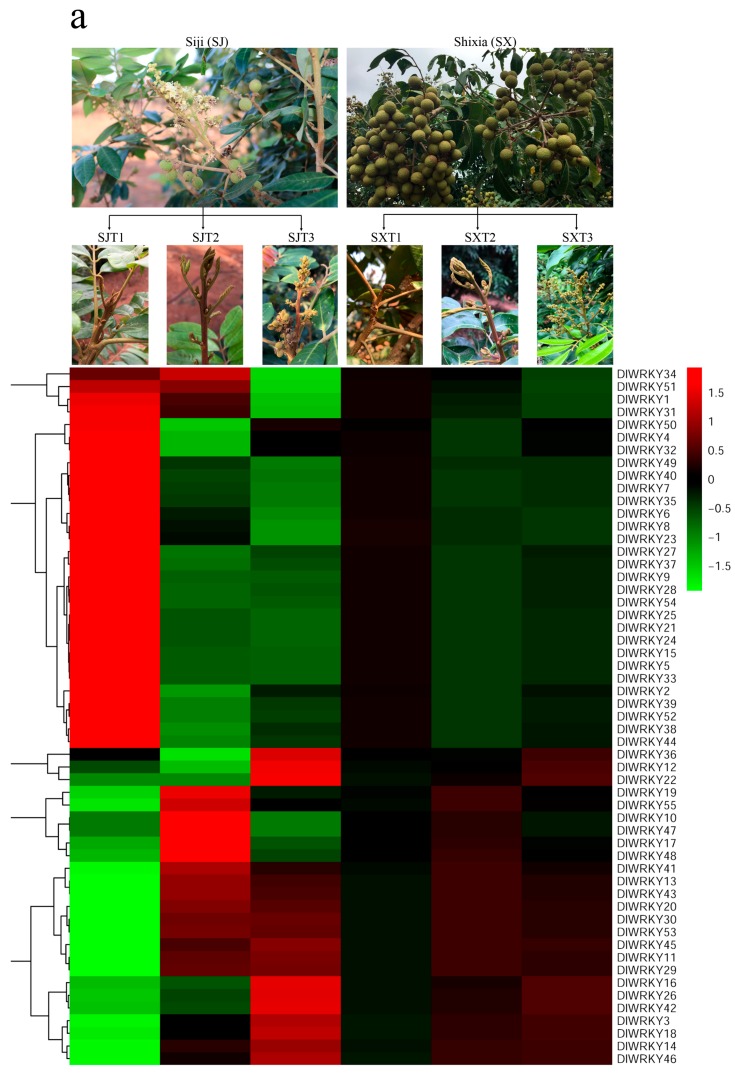
Although the involvement of many WRKY genes has been examined in the control of flowering time [15], the expression of DlWRKY genes during flower induction has not been studied extensively. In the present study, we also analyzed the expression patterns of 55 DlWRKY genes in two longan species during the three flowering stages by RNA-seq analysis (Table S4). Heat maps were constructed based on the log10 (FPKM + 0.01) values for the 55 DlWRKY genes (Figure 5a). Based on the criteria for p-values <0.05 and fold changes ≥2, the DlWRKY genes that were differentially expressed during the three flowering stages of the two longan species were identified. Interestingly, the results showed that all 55 DlWRKY genes were constructively expressed in the three test flowering stages of the “SX” longan, while 18 DlWRKY genes showed a specific expression in the “SJ” longan. Among the 18 DlWRKY genes, 12 (DlWRKY5, 7, 8, 9, 15, 21, 23, 24, 25, 39, 52, and 54) showed a continuously down-regulated expression through the three flowering stages, and four genes (DlWRKY16, 17, 41, and 42) showed an up-regulated expression. Moreover, two genes (DlWRKY10 and 48) showed a transient up-regulation at the second stage and a down-regulation at the third stage.
Figure 5.
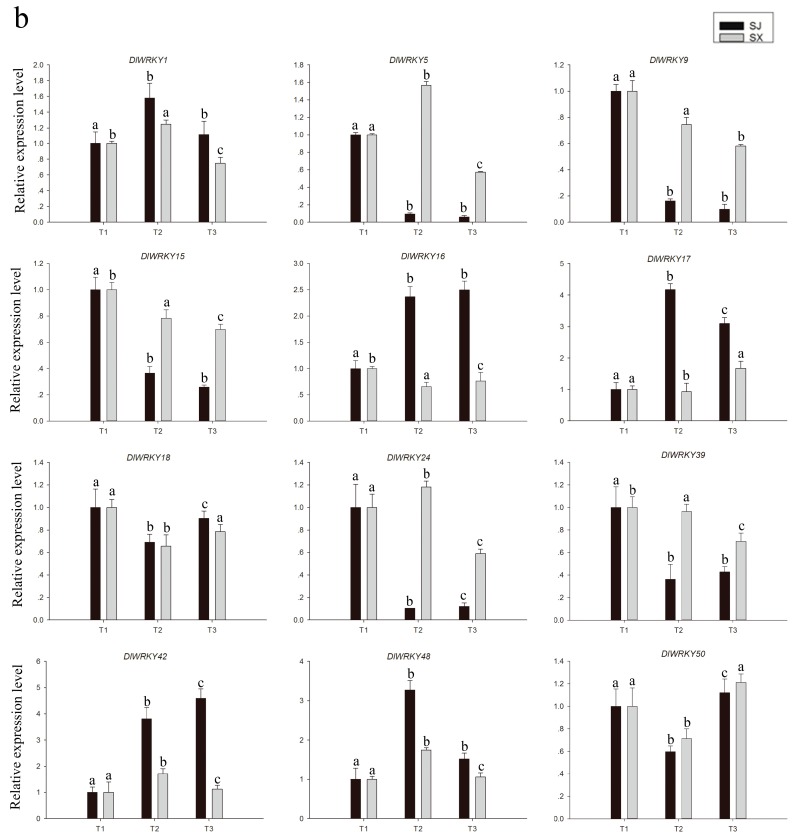
The expression profiles of DlWRKY in two longan species during the floral induction process. (a) A heat map showing the comparative expression level of the WRKY genes in the three flowering stages of“SJ” and “SX”. The color scale represents the log10 expression values. Genes with comparatively low expression values are shown using shades of green, and high expression values are represented using shades of red. The three flowering stages of SJ are indicated by SJT1, SJT2, and SJT3. The three flowering stages of SX are indicated by SXT1, SXT2, and SXT3. (b) Relative expression levels of the twelve DlWRKYs during the three flowering stages of the two longan species by qRT-PCR. For each gene, the relative expression level in T1 (dormant apical bud) was set as one, and the longan actin gene was used as the internal expression control. The data represent the mean ± SD of the three replicates. Values with the same letter were not significantly different when assessed using Duncan’s multiple range test (p < 0.05, n = 3).
To validate the expression levels obtained from the RNA-seq data, twelve DlWRKY genes (DlWRKY1, 5, 9, 15, 16, 17,18,24,39,42, 48, and 50) were selected from the six different longan WRKY groups for the quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis. Consistent with the result of the RNA-seq analysis, the transcript levels of all twelve DlWRKY genes did not exhibit any significant differences in the “SX” longan between the three flowering stages (Figure 5b). In addition, the relative expression level of DlWRKY1, DlWRKY18, and DlWRKY50 did not exhibit any significant differences in ‘"“SJ” during the three flowering stages. The expression levels of DlWRKY16, 17, 42, and 48were up-regulated in the second and third stage. The transcript level of DlWRKY5, 9, 15, 24, and DlWRKY39 was down-regulated in the second and third stages (Figure 5b). In general, the expression levels obtained by qRT-PCR for these genes are similar to the results obtained from the RNA-seq data.
2.6. Differential Expression of DlWRKY Genes in Response to Stress and Hormonal Treatments
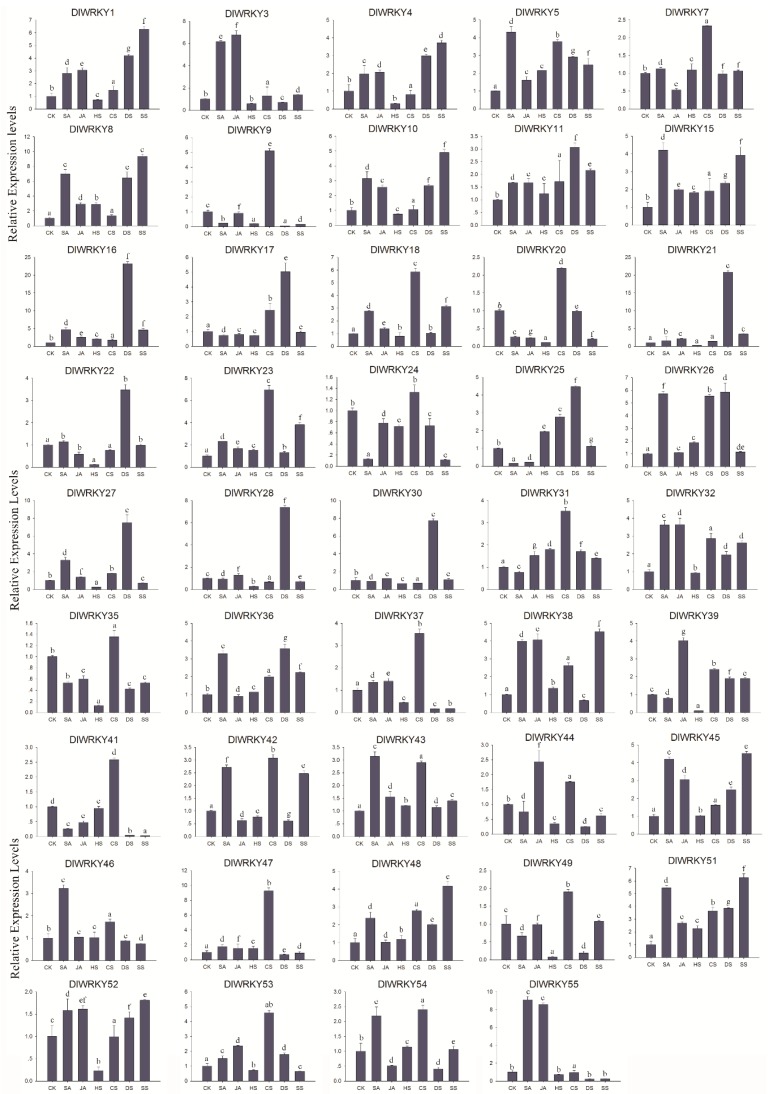
The expression patterns of 55 DlWRKY genes were investigated in response to hormonal and various stresses by using qRT-PCR. As shown in Figure 6 and Figure S1, the majority of the DlWRKY genes (44 of 55) were up-regulated or down-regulated by >2-flod under at least one tested treatment, while eleven genes (DlWRKY2, 6, 12, 13, 14, 19, 29, 33, 34, 40, and 50) showed no significant differential expression in response to the given treatments. The SA treatment induced the expression of the 22 DlWRKY genes (DlWRKY1, 3, 5, 8, 10, 15, 16, 18, 23, 26, 27, 32, 36, 38, 42, 43, 45, 46, 48, 51, 54, and 55) but reduced the expression of five DlWRKY genes (DlWRKY9, 20, 24, 25, and 41). Fifteen DlWRKY genes (DlWRKY1, 3, 4, 8, 10, 16, 21, 32, 38, 39, 44, 45, 51, 53, and 55) were up-regulated, and three (DlWRKY20, 25, and 41) were down-regulated by MeJA treatment. For heat treatment, 11 (DlWRKY4, 9, 20, 27, 28, 35, 37, 39, 44, 49, and 52) and 4 (DlWRKY5, 8, 16, and 51) genes were down-regulated or up-regulated, respectively. A total of 17 DlWRKY (DlWRKY5, 7, 9, 17, 18, 20, 23, 25, 26, 31, 37, 39, 41, 42, 47, 51, and 54) genes showed up-regulated expressions, and no genes were down-regulated by cold treatment. Under the drought treatment, 20 (DlWRKY1, 4, 5, 8, 10, 11, 15, 16, 17, 21, 22, 25, 26, 27, 28, 30, 36, 45, 48, and 51) and 7 DlWRKY genes (DlWRKY9, 35, 37, 41, 44, 49, and 54) were up-regulated or down-regulated, respectively. Eighteen (DlWRKY1, 4, 5, 8, 10, 11, 15, 16, 18, 21, 23, 32, 36, 38, 42, 45, 48, and 51) and five DlWRKY genes (DlWRKY9, 20, 24, 37, and 41) were up-regulated or down-regulated, respectively, under high salinity treatment.
Figure 6.
The expression patterns of the selected DlWRKY genes under various hormonal and abiotic stresses. The x-axis indicates various treatments and the y-axis indicates the relative expression level. Error bars were obtained from three independent biological replicates. Values with the same letter were not significantly different when assessed using Duncan’s multiple range test (p < 0.05, n = 3). SA represents salicylic acid, JA represents jasmonic acid, HS represents heat stress, CS represents cold stress, DS represents drought stress, and SS represents salinity stress.
2.7. Analysis Related Cis-Elements in the Candidate DlWRKY Genes
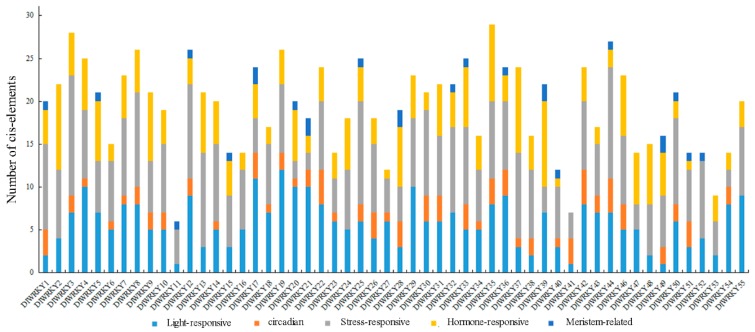
To analyze the potential function of DlWRKY genes in response to various responses, the cis-elements in the promoter region of the DlWRKY genes were further analyzed. Among these 55 genes, 54 genes could perform cis-elements analysis except DlWRKY45, which only contain 270 promoter bases. All the DlWRKY genes shared the light-responsive boxes and stress-responsive boxes in their promoter. Hormone-related cis-elements, such as AuxRR-core, TCA-element, CGTCA-motif, GARE-motif, P-box, and ERE (Ethylene-responsive element), existed in the promoter of all DlWRKY genes except DlWRKY11, DlWRKY41, and DlWRKY52. Additionally, circadian-related cis-elements were found in the promoter of 39 DlWRKY genes and Meristem-related cis-elements were only presented in the promoter of 20 DlWRKY genes (Figure 7, Tables S5 and S6).
Figure 7.
The predicted cis-elements in the promoter of the DlWRKY genes. The 1.5 kb sequences of 55 DlWRKY genes were analyzed with the PlantCARE software.
3. Discussion
The WRKY proteins, an important transcription factor superfamily which is involved in plant development and stress responses, have been widely detected in various organisms from single-celled green algae to monocots and dicots [15]. Recently, the successful genome sequencing of longan makes it possible to analyze WRKY TFs at the whole-genome level [37]. The present study is the first to identify and characterize WRKY proteins from whole-genome sequences of longan.
In this study, we identified 59 candidate WRKY genes in the longan genome (471.88 Mb) using the HMM and Blastn search methods. These genes included 58 DlWRKYs, which were also found by Lin et al. [37], and one gene Dlo_022548.1 (DlWRKY36) found in our study. Finally, after the WRKY domain scanning and sequence alignment, 55 DlWRKY genes were determined in the longan genome (Table 1). The number of WRKY genes in longan was similar to those found in grape (59 VvWRKYs), whose genome size is 487 Mb, which is similar to that of the longan genome [29]. However, the size of the WRKY family in longan is smaller than that in A. thaliana (72), Oryza sativa ssp. Indica (102), and the common bean (88), although their genome sizes are similar (O. sativa ssp. Indica, 466 Mb; common bean, 587 Mb) or even smaller (A. thaliana, 119 Mb) than the longan genome size (Table S7) [28,38,39]. Therefore, the number of WRKY family members is not necessarily correlated with the genome size. Previous studies showed that the only group I WRKYs are present in green algae and all WRKY genes originated from the group I C-terminal WRKY domains, whereas group II members were evolved in the common ancestor of land plants, and Group III members emerged in the common ancestor of seed plants [15]. In addition, as a newly defined and the most dynamic group with many duplication events, the differences in the number of WRKY genes in Group III are the primary cause of the sizes of WRKY gene families [40]. In the present study, the differences in the number of WRKY genes between longan and Arabidopsis mainly existed in groups IIc and III, indicating that the group IIc and III WRKY genes may play important roles in the functional evolution of DlWRKYs.
According to the classification scheme for the WRKY family of Eulgem et al. [41], the DlWRKY proteins were divided into three distinct clusters: groups I, II, and III. Group II proteins were further divided into five distinct groups: a–e (Figure 1 and Table 1). In addition, subgroup IIc contained the largest number of WRKY proteins. These results were consistent with the results observed in other species [28,29,42,43,44]. The WRKY motif was fairly conserved in longan WRKY proteins, and three variants of this motif were observed. All the DlWRKYs, except DlWRKY19 and DlWRKY47, possessed WRKYGQK. DlWRKY19, which belonged to subgroup IIc, possessed WRKYGKK. DlWRKY19, which belonged to subgroup III, possessed WKKYRQK. In the common bean, the variants WRKYGKK, WRKYGEK, WKKYEDK, and WKKYCEDK are mainly observed in subgroup IIc [28]; in mulberry, WRKYGKK is detected in subgroup IIb [27]. Moreover, in rice, nine variants, most of which belong to groups III and IIc, are observed [45]. Previous studies showed that these variations of the WRKYGQK motif might change the DNA binding specificities of downstream target genes, and WRKY genes with the variations of the WRKYGQK motif may recognize binding sequences other than the W-box element ((C/T)TGAC(C/T)) [15]. Hence, the result suggested that DlWRKY19 and DlWRKY47 may possess different binding specificities and functions from those of other DlWRKY proteins.
WRKY family genes play important roles in diverse plant development and shown a tissue-specific expression in many plant species [15,40]. For example, AtWRKY75 exerts a negative effect on root hair development [46]. SUSIBA2 [47] and MINISEED3 [48] play roles in the regulation of seed development. In grape, nearly half of the 59 VvWRKY genes show no significant organ/tissue-related differences in expression, and some clear spatial differences are noted [29]. In mulberry, 13 WRKY genes exhibit the highest expression in the Morus notabilis root tissue. A maximum of 25 WRKYs show the highest expression in the bark tissue, and 10 WRKY genes display the highest expression in other stages [27]. In the present study, the expression profiles of 55 longan WRKY genes in nine longan tissues were ascertained by RNA-seq analysis (Figure 4). The results demonstrated variation in the expression pattern of DlWRKY genes. In total, 25 DlWRKY genes (DlWRKY1, 2, 3, 5, 6, 8, 9, 13, 14, 23, 24, 28, 30, 32, 35, 37, 38, 39, 44, 49, 50, 52, 53, and 54) were highly expressed in at least six longan tissues. As highly expressed genes usually play important roles in plant development [44], we concluded that the 25 highly expressed DlWRKY genes might be important regulatory factors in longan development. It was found that group I and group IId WRKY genes are ancestral to other WRKY genes in plants or algae and are more likely to be constitutively expressed in different tissues [15,40]. For instance, most of the highly expressed SiWRKY genes belonged to group I and IId [40]. Consistent with these studies, in the present study, most of the members of groups I (9 of 11) and IId (4 of 6) were the highly expressed gene. In contrast, 12 DlWRKY genes were expressed at low levels in all tested tissues and these minimally expressed DlWRKY genes were distributed in almost all the WRKY gene subgroups except for IId. Meanwhile, six DlWRKY genes were preferential accumulation in no more than three tissues, implying that these genes might play crucial roles during the development of specific organs. Additionally, these specifically or minimally expressed DlWRKY genes could be induced under environment stimuli. For example, DlWRKY10, 22, 41, and 47 were not detected in leaves under normal conditions, but they were induced by different abiotic stresses (Figure 6). Similar results were also found in other studies [15,40,49].
Perpetual flowering is a crucial trait for fruit trees as it enlarges the production period [50]. To date, the genetic control of PF has been deciphered in several model plants. For example, In Arabidopsis, the PF trait is controlled by PERPETUAL FLOWERING 1 (PEP1), an orthologue of the FLC floral repressor [51]. In the diploid strawberry and rose, the PF trait is due to a mutation in the orthologue of the TERMINAL FLOWER 1 (TFL1) floral repressor [50,52]. Recent studies showed that the PF trait of some cultivated strawberries is genetically controlled by the major FaPFRU locus, which is non-orthologous to TFL1 [53,54]. However, the multi-year delay in the onset of flowering and the long juvenile phase hampers the research of PF traits in perennials, such as longan. Although WRKY TFs regulate various plant developments, only a few data are available on whether WRKY TFs are involved in the flowering time regulation. Meanwhile, as a kind of TF, WRKY genes regulated plant flowering by being directly active or inhibiting the downstream target gene. For example, promoter sequences of FT, LFY, and AP1 harbor W-boxes (TTTGACT/C); AtWRKY71 affects the flowering time of plants by directly regulating these genes [16]. In our study, all the 55 DlWRKY genes were constructively expressed in the three test flower induction process of the “SX” longan, while 18 DlWRKY genes showed a specific expression in the “SJ” longan (Figure 5a). This result indicated that these 18 DlWRKY genes may specifically be involved in the flower induction of “SJ”. In summary, we proposed that these 18 DlWRKY genes may participate in the forming of the longan PF habit, which further studies are required to verify the function of these genes.
WRKY genes play crucial roles in the response to abiotic and biotic stress-induced defense signaling pathways [15]. Numerous studies have demonstrated that WRKY genes are expressed strongly and rapidly in response to particular abiotic stresses [15,22,29,40,52]. Consistent with these previous studies, our study showed that 44 DlWRKY genes (80%) showed up- or down-regulated expression in at least one tested treatment (Figure 6 and Figure S1), thereby highlighting the extensive involvement of WRKY genes in environmental adaptation. SA, JA, and Eth play important roles in biotic and abiotic stresses [55]. Many WRKYs, such as AtWRKY28, AtWRKY46, AtWRKY70, and AtWRKY54, play an important role in SA- and JA-dependent defense signaling pathways [53,56,57]. In the present study, 27 and 18 DlWRKY genes were up- or down-regulated by SA and MeJA treatment, respectively. For example, DlWRKY25, the orthologue of AtWRKY70 and AtWRKY54, was regulated by the SA and JA treatments. AtWRKY25 and AtWRKY33 regulate plant adaptation to salinity stress through an interaction with their upstream or downstream target genes [58]; their orthologue DlWRKY8 in longan was regulated by SA, JA, heat, drought, and salinity. In grape [29], VvWRKY42 and its orthologue DlWRKY11 in our study were up-regulated by salt treatment. Furthermore, we observed same orthologous genes with different expression patterns under stress treatment. DlWRKY44 was down-regulated under drought, and its orthologous gene VvWRKY35 was up-regulated under this stress treatment. DlWRKY19 showed no significant differential expression in response to salinity, and its orthologous gene VvWRKY25 was up-regulated [29]. We speculate that these orthologous genes may be involved in the different signaling pathways in different species. Additionally, only one gene (DlWRKY52) was significantly highly expressed under all abiotic stresses. These results indicated that the different DlWRKYs played different roles in regulating stress response and that further investigation of the functions of these DlWRKY genes is necessary. Differential responses of several WRKYs are regulated by the presence of cis-elements in their promoter region [27,40,49]. For example, Morus013217, which contains three LTREs in its promoter regions showed a strong response to cold stress [27]. Similar results were also found in our study. For instance, four HSEs were found in the promoter regions of DlWRKY2, which showed a strong response to heat stress. DlWRKY36, DlWRKY46, and DlWRKY48 showed responsiveness to SA treatment and their expressions were all up-regulated, and more than two TCA-elements were found in their promoters. While the DlWRKY11 and DlWRKY52 hormone-related cis-elements existed in their promoter, they showed no response to the SA or MeJA treatments (Figure 6 and Table S6). Thus, these cis-elements could provide more evidence of the DlWRKY genes in response to different stresses or hormonal signaling.
4. Materials
4.1. Identification of Longan WRKY Genes
Longan whole-genome sequences, transcript data, and proteins were downloaded from the NCBI Sequence Read Archive (SRA315202) or ftp://climb.genomics.cn/pub/10.5524/100001_101000/100276/ [37]. The HMM profile of the WRKY DNA binding domain (PF03106) which was extracted from the Pfam database (http://pfam.sanger.ac.uk/) was used to obtain the potential members of the longan WRKY genes [59] and used to search the putative WRKY genes from the longan genome with HMMER 3.0 (http://hmmer.janelia.org/) with the default parameters and 0.01 as the cutoff value. Then, all non-redundant longan WRKY protein sequences were selected and the domain was conserved using Simple Modular Architecture Research Tool (http://smart.emblheidelberg.de/) [60].
4.2. Sequence Alignment, Phylogenetic Analysis, and Cis-Elements in the Promoters
The 72 Arabidopsis and 59 grape WRKY proteins described previously [29,38] were obtained from TAIR (http://www.arabidopsis.org/) and NCBI (http://www.ncbi. nlm.nih.gov/), respectively. By using Clustal X version 1.83, the WRKY protein sequences of Arabidopsis and longan were aligned for phylogenetic analysis. Based on this alignment, a bootstrapped ML (Maximum Likelihood) tree was constructed using MEGA (version 6.0) with the bootstrap test replicated 1000 times [61]. To assess the phylogenetic relationships among the members of the longan WRKY gene family, a phylogenetic tree was prepared according to the alignment of only the longan proteins. All DlWRKY transcription factors were classified into subgroups based on their structural features and evolutionary relationships. The 1500-bp sequences upstream of the start codon of the candidate DlWRKY genes were extracted from the longan genome sequences. The PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used for searching the cis-acting elements [62].
4.3. Protein Feature Analysis
The ExPASy online tools (http://expasy.org/tools/) [63] were used to calculate the MW, the number of amino acids, the ORF, ORF length, and isoelectric point (pI) of DlWRKY proteins. The arrangements and the intron and exon junctions of the DlWRKY genes were analyzed by the GSDS, version 2.0 [64]. MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi) [65] was used to analyze the conserved motifs of the DlWRKY proteins with the following optimized parameters: any number of repetitions; maximum number of motifs: 15; and the optimum width of each motif: between 6 and 50 residues.
4.4. Expression Analysis of Longan WRKY Genes in Various Tissues and Different Flowering Stages
The RNA-seq data for analyzing the expression patterns of WRKY genes in different longan tissues were downloaded from the NCBI Sequence Read Archive (GSE84467). Three pairs of nine-year-old “SJ” and “SX” D. longan trees which displayed opposite flowering phenotype were used for comparative expression analysis of DlWRKY during floral induction. All those trees were grown at an experimental orchard in the South Subtropical Crops Research Institute of the Chinese Academy of Tropical Agricultural Science in Zhanjiang (110°16′ E, 21°10′ N), China. Three different kinds of apical buds, including the dormant stage (T1), the emergence of floral primordia stage (T2), and the floral organ formation stage (T3) of “SJ” and “SX”, were used in this study. The samples obtained for the T1, T2, and T3 in “SJ” and “SX” were collected on 20 November 2016, 24 December 2016, and 1 January 2017, respectively. For each sample, we used three biological replicates from three different trees. Each biological replicate contained the mixed buds which were collected from the four cardinal directions of each tree. All samples were collected from 10:00 am to 12:00 am and were frozen immediately in liquid nitrogen and stored at −80 °C. According to the manufacturer’s instructions, the total RNA was extracted by using the quick RNA Isolation Kit (Hua Yue Yang Bio Co., Ltd., Beijing, China) and the genomic DNA residues were removed during RNA extraction. We used an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) to test the RNA concentration and the quality of each sample. The RNA quality was also confirmed by RNase free agarose gel electrophoresis. The RNA-seq experiment was performed as described by our previous study [66]. The RNA-seq data were uploaded to the NCBI Sequence Read Archive (SRS2241241, SRS2241242, SRS2241243, SRS2241244, SRS2241245, SRS2241246, SRS2241247, SRS2241248, SRS2241249, SRS2241250, SRS2241251, SRS2241252, SRS2241253, SRS2241254, SRS2241255, SRS2241256, SRS2241257, and SRS2241258). The fragments per kilobase of the exon model per million mapped values (FPKM) were log10-transformed, and heat maps with hierarchical clustering were exhibited using the software Mev4.9.0 [67].
4.5. Stress and Hormonal Treatments and Expression Profiling Using qRT-PCR
Twenty-seven one-year-old uniform grafted seedlings of “SJ”, obtained from the South Subtropical Crops Research Institute of the Chinese Academy of Tropical Agricultural Science in Zhanjiang (110°16′E, 21°10′N) were used for stress and hormonal treatments. For hormone treatments, three seedlings were treated with methyl jasmonate (MeJA) or SA solution (100 μM) for 4 h at 28 °C, respectively. Meanwhile, three seedlings sprayed with water were used as a control. For heat and cold stresses, three samples were grown at 42 or 0 °C for 4 h, respectively, and three samples grown at 28 °C were used as a control. All the treatments were performed in a greenhouse. Six leaves were collected from each seedling and all samples were immediately frozen in liquid nitrogen and stored at −80 °C for expression analysis.
According to the manufacturer’s instructions, the total RNA was obtained by using the SuperFast RNA extraction kit (Hua Yue Yang Bio Co.). The first-strand cDNA was synthesized by reverse transcription of the total RNA (500 ng) using PrimeScriptRTase (TaKaRa Biotechnology, Dalian, China). Gene-specific primers were designed according to the DlWRKY gene sequences using Primer Premier 5.0 and checked using Blastn in NCBI (Table S8). In addition, the longan Actin1 gene (Dlo_028674) was used as an internal control for normalization. qRT-PCR was conducted using the LightCycler® 480 Real-Time PCR System (Roche, Germany) and SYBR Green II PCR Master Mix (Takara, Dalian, China). The amplification program was as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Each reaction was performed in three replicates. The relative expression levels of the candidate genes were calculated by the 2–∆∆Ct method. The analysis included cDNA from the three biological samples for each tissue, and all the reactions were run in triplicates. In the comparative expression analysis of the DlWRKY genes, genes that were up- or down-regulated by at least two-fold were considered differentially expressed.
5. Conclusions
It is essential to systematically analyze the function of transcription factors (TFs), since these genes can regulate the expression of many others, resulting in deep physiological modifications. Although WRKY genes have been identified in many other species, the information of longan WRKY is still unknown. In the present study, we conducted a genome-wide identification and analysis of the WRKY genes in longan. A total of 55 DlWRKY genes were identified in the longan genome. Phylogenetic analysis indicated that these 55 DlWRKYs could be divided into seven groups. An RNA-seq-based analysis showed that several of the identified WRKY genes may play various roles in the development of longan tissues. In addition, comparative expression analysis revealed that 18 DlWRKY genes might have participated in the regulation of longan flowering. Our RNA-seq, qRT-PCR, and promoter analyses revealed the gene expression profiles and implied that the response to different stress or hormonal signaling of some DlWRKY may be due to the cis-elements in their promoters. In summary, our results will facilitate further studies into the role of DlWRKY genes in response to abiotic stresses and the development of molecular breeding programs to enhance abiotic stress tolerance and increase yield in longans.
Abbreviation
| HMM | Hidden Markov model |
| NJ | Neighbor-joining |
| GSDS | Gene Structure Display Server |
| MW | The molecular weight |
| ORF | Open reading frame |
| pI | Isoelectric point |
| NCBI | National Center of Biotechnology Information |
| qRT-PCR | Quantitative real-time reverse transcription polymerase chain reaction |
| RNA-seq | RNA sequencing |
| SA | Salicylic acid |
| JA | Jasmonic acid |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/8/2169/s1. The following are available online. Table S1. The candidate WRKY genes and their protein structure found in the longan genome. Table S2. The information for each motif of DlWRKYs. Table S3. FPKM values of DlWRKY genes in nine tissues of longan. Table S4. FPKM values of DlWRKY genes in the three flower induction stages of “SJ” and “SX” longan species. The red color indicates the genes which showed down-regulated expression; the blue color indicates the genes which showed up-regulated expression; and the green color indicates the genes that showed an up-regulated expression in the first two stages and a down-regulated expression in the third stage. Table S5. Details of the cis-elements identified in this study. Table S6. Predicted cis-elements in the promoter of the DlWRKY genes. Table S7. The WRKY gene number and genome size of different species. Table S8. Primers used in quantitative RT-PCR of DlWRKY genes. Figure S1. Expression patterns of selected DlWRKY genes which have no significant difference under various hormonal and abiotic stresses. The x-axis indicates various treatments, and the y-axis indicates the relative expression level. Error bars were obtained from three independent biological replicates.
Author Contributions
D.J., X.S., and S.S. conceived the experiments and D.J. performed the experiments. J.X. Additionally, C.L. analyzed the data, D.J. Additionally, S.S. contributed to the writing of the manuscript, L.L. provided the value comments and revised the grammar of the manuscript. B.S. provided help in the analysis of qRT-PCR. Y.W. prepared samples for RNA sequencing.
Funding
This work was supported by the Natural Science Foundation of China (31572087), the China Litchi and Longan Industry Technology Research System (CARS-32-02), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630062018011) and the Natural Science Foundation of Hainan Province (20163111 and 317243).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Matsumoto T.K. Genes uniquely expressed in vegetative and potassium chlorate induced floral buds of Dimocarpus longan. Plant Sci. 2006;170:500–510. doi: 10.1016/j.plantsci.2005.09.016. [DOI] [Google Scholar]
- 2.You X., Wang L., Liang W., Gai Y., Wang X., Chen W. Floral reversion mechanism in longan (Dimocarpus longan lour.) revealed by proteomic and anatomic analyses. J. Proteom. 2012;75:1099–1118. doi: 10.1016/j.jprot.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Jia T., Wei D., Meng S., Allan A.C., Zeng L. Identification of regulatory genes implicated in continuous flowering of longan (Dimocarpus longan L.) PLoS ONE. 2014;9:e114568. doi: 10.1371/journal.pone.0114568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H.N., Shi S.Y., Li W.C., Shu B., Liu L.Q., Xie J.H., Wei Y.Z. Transcriptome analysis of ‘sijihua’ longan (Dimocarpus longan L.) based on next-generation sequencing technology. J. Hortic. Sci. Biotechnol. 2016;91:180–188. doi: 10.1080/14620316.2015.1133539. [DOI] [Google Scholar]
- 5.Shabala S., Bose J., Hedrich R. Salt bladders: Do they matter? Trends Plant Sci. 2014;19:687–691. doi: 10.1016/j.tplants.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Bluemel M., Dally N., Jung C. Flowering time regulation in crops-what did we learn from arabidopsis? Curr. Opin. Biotechnol. 2015;32:121–129. doi: 10.1016/j.copbio.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 7.Dally N., Xiao K., Holtgräwe D., Jung C. The b2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. USA. 2014;111:10365–10370. doi: 10.1073/pnas.1404829111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 9.Turnbull C. Long-distance regulation of flowering time. J. Exp. Bot. 2011;62:4399–4413. doi: 10.1093/jxb/err191. [DOI] [PubMed] [Google Scholar]
- 10.Srikanth A., Schmid M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smaczniak C., Immink R.G., Muiño J.M., Blanvillain R., Busscher M., Busscher-Lange J., Dinh Q.P., Liu S., Westphal A.H., Boeren S. Characterization of mads-domain transcription factor complexes in arabidopsis flower development. Proc. Natl. Acad. Sci. USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo S.Y., Kim Y., Kim S.Y., Lee J.S., Ahn J.H. Control of flowering time and cold response by a nac-domain protein in arabidopsis. PLoS ONE. 2007;2:e642. doi: 10.1371/journal.pone.0000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin B., Choi G., Yi H., Yang S., Cho I., Kim J., Lee S., Paek N.C., Kim J.H., Song P.S. Atmyb21, a gene encoding a flower-specific transcription factor, is regulated by cop1. Plant J. 2002;30:23–32. doi: 10.1046/j.1365-313X.2002.01264.x. [DOI] [PubMed] [Google Scholar]
- 14.Tong Z., Hong B., Yang Y., Li Q., Ma N., Ma C., Gao J. Overexpression of two chrysanthemum dgdreb1 group genes causing delayed flowering or dwarfism in arabidopsis. Plant Mol. Biol. 2009;71:115–129. doi: 10.1007/s11103-009-9513-y. [DOI] [PubMed] [Google Scholar]
- 15.Chen F., Hu Y., Vannozzi A., Wu K., Cai H., Qin Y., Mullis A., Lin Z., Zhang L. The wrky transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2017;36:311–335. doi: 10.1080/07352689.2018.1441103. [DOI] [Google Scholar]
- 16.Yu Y., Liu Z., Wang L., Kim S.G., Seo P.J., Qiao M., Wang N., Li S., Cao X., Park C.M. Wrky71 accelerates flowering via the direct activation of flowering locus t and leafy in arabidopsis thaliana. Plant J. 2015;85:96–106. doi: 10.1111/tpj.13092. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y., Chen X., Xie K., Xing Q., Wu Y., Li J., Du C., Sun Z., Guo Z. Dlf1, a wrky transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE. 2014;9:e102529. doi: 10.1371/journal.pone.0102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y., Hu R., Wang H., Cao Y., He G., Fu C., Zhou G. Mlwrky12, a novel miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 2013;212:1–9. doi: 10.1016/j.plantsci.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Li W., Wang H., Yu D. The arabidopsis wrky transcription factors wrky12 and wrky13 oppositely regulate flowering under short-day conditions. Mol. Plant. 2016;9:1492–1503. doi: 10.1016/j.molp.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Kiranmai K., Lokanadha Rao G., Pandurangaiah M., Nareshkumar A., Amaranatha Reddy V., Lokesh U., Venkatesh B., Anthony Johnson A.M., Sudhakar C. A novel wrky transcription factor, muwrky3 (Macrotyloma uniflorum lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 2018;9:346. doi: 10.3389/fpls.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., Fu Q., Chen L., Huang W., Yu D. Arabidopsis thaliana wrky25, wrky26, and wrky33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 22.Han C., Lai Z., Shi J., Yong X., Chen Z., Xu X. Roles of arabidopsis wrky18, wrky40 and wrky60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010;10:281. doi: 10.1186/1471-2229-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilian J., Whitehead D.J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. The atgenexpress global stress expression data set: Protocols, evaluation and model data analysis of uv-b light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 24.Seki M., Narusaka M., Ishida J., Nanjo T., Fujita M., Oono Y., Kamiya A., Nakajima M., Enju A., Sakurai T. Monitoring the expression profiles of 7000 arabidopsis genes under drought, cold and high-salinity stresses using a full-length cdna microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 25.Qiu Y., Jing S., Fu J., Li L., Yu D. Cloning and analysis of expression profile of 13 wrky genes in rice. Sci. Bull. 2004;49:2159–2168. doi: 10.1360/982004-183. [DOI] [Google Scholar]
- 26.Raineri J., Wang S., Peleg Z., Blumwald E., Chan R.L. The rice transcription factor oswrky47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 2015;88:401–413. doi: 10.1007/s11103-015-0329-7. [DOI] [PubMed] [Google Scholar]
- 27.Baranwal V.K., Negi N., Khurana P. Genome-wide identification and structural, functional and evolutionary analysis of wrky components of mulberry. Sci. Rep. 2016;6:30794. doi: 10.1038/srep30794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J., Chen J., Wang L., Wang S. Genome-wide investigation of wrky transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 2017;8:380. doi: 10.3389/fpls.2017.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C., Guo R., Xu X., Gao M., Li X., Song J., Zheng Y., Wang X. Evolution and expression analysis of the grape (Vitis vinifera L.) wrky gene family. J. Exp. Bot. 2014;65:1513–1528. doi: 10.1093/jxb/eru007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie T., Chen C., Li C., Liu J., Liu C., He Y. Genome-wide investigation of wrky gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018;19:490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Zhou Y., Chi Y., Fan B., Chen Z. Characterization of soybean wrky gene family and identification of soybean wrky genes that promote resistance to soybean cyst nematode. Sci. Rep. 2017;7:17804. doi: 10.1038/s41598-017-18235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L., Mu S., Cheng Z., Cheng Y., Zhang Y., Miao Y., Hou C., Li X., Gao J. Characterization and expression analysis of the wrky gene family in moso bamboo. Sci. Rep. 2017;7:6675. doi: 10.1038/s41598-017-06701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y., Mao M., Wan D., Yang Q., Yang F., Mandlaa, Li G., Wang R. Identification of thewrkygene family and functional analysis of two genes in caragana intermedia. BMC Plant Biol. 2018;18:31. doi: 10.1186/s12870-018-1235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song H., Wang P., Lin J.Y., Zhao C., Bi Y., Wang X. Genome-wide identification and characterization ofwrkygene family in peanut. Front. Plant Sci. 2016;7:534. doi: 10.3389/fpls.2016.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue H., Wang M., Liu S., Du X., Song W., Nie X. Transcriptome-wide identification and expression profiles of the wrky transcription factor family in broomcorn millet (Panicum miliaceum L.) BMC Genom. 2016;17:343. doi: 10.1186/s12864-016-2677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohanta T.K., Park Y.H., Bae H. Novel genomic and evolutionary insight of wrky transcription factors in plant lineage. Sci. Rep. 2016;6:37309. doi: 10.1038/srep37309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y., Min J., Lai R., Wu Z., Chen Y., Yu L., Cheng C., Jin Y., Tian Q., Liu Q. Genome-wide sequencing of longan (Dimocarpus longan lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience. 2017;6:1–14. doi: 10.1093/gigascience/gix023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu X., Mao Z., Yu H., Zhang Y., Jiang W., Ling J., Huang S., Xie B. Genome-wide analysis of wrky gene family in cucumis sativus. BMC Genom. 2011;12:471. doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J., Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. Indica) Science. 2002;296:1937–1942. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 40.Li D., Liu P., Yu J., Wang L., Dossa K., Zhang Y., Zhou R., Wei X., Zhang X. Genome-wide analysis of wrky gene family in the sesame genome and identification of the wrky genes involved in responses to abiotic stresses. BMC Plant Biol. 2017;17:152. doi: 10.1186/s12870-017-1099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. The wrky superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 42.Shuai L., Luo C., Zhu L., Sha R., Qu S., Cai B., Wang S. Identification and expression analysis of wrky transcription factor genes in response to fungal pathogen and hormone treatments in apple (Malus domestica) J. Plant Biol. 2017;60:215–230. [Google Scholar]
- 43.Zhi Z., Yang L., Wang D., Huang Q., Mo Y., Xie G. Gene structures, evolution and transcriptional profiling of the wrky gene family in castor bean (Ricinus communis L.) PLoS ONE. 2016;11:e0148243. doi: 10.1371/journal.pone.0148243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Y., Jalalahammed G., Yu J., Yao Z., Ruan M., Ye Q., Li Z., Wang R., Feng K., Zhou G. Putative wrkys associated with regulation of fruit ripening revealed by detailed expression analysis of the wrky gene family in pepper. Sci. Rep. 2016;6:39000. doi: 10.1038/srep39000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Wang L. The wrky transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evolut. Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rishmawi L., Hülskamp M. Non-cell-autonomous regulation of root hair patterning genes by wrky75 in arabidopsis thaliana. Plant Physiol. 2014;165:186. doi: 10.1104/pp.113.233775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun C., Palmqvist S., Olsson H., Borén M., Ahlandsberg S., Jansson C. A novel wrky transcription factor, susiba2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. Miniseed3 (mini3), a wrky family gene, and haiku2 (iku2), a leucine-rich repeat (lrr) kinase gene, are regulators of seed size in arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Li H., Yang Y., Wang Y., Mo Y., Zhang R., Zhang Y., Ma J., Wei C., Zhang X. Identification and expression analyses ofwrkygenes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus) PLoS ONE. 2018;13:e0191308. doi: 10.1371/journal.pone.0191308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwata H., Gaston A., Remay A., Thouroude T., Jeauffre J., Kawamura K., Oyant L.H.S., Araki T., Denoyes B., Foucher F. The tfl1 homologue ksn is a regulator of continuous flowering in rose and strawberry. Plant J. Cell Mol. Biol. 2012;69:116–125. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- 51.Vincent C. Pep1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- 52.Koskela E.A., Hytönen T. Mutation in terminal flower1 reverses the photoperiodic requirement for flowering in the wild strawberry fragaria vesca. Plant Physiol. 2012;159:1043–1054. doi: 10.1104/pp.112.196659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perrotte J., Gaston A., Potier A., Petit A., Rothan C., Denoyes B. Narrowing down the single homoeologous fapfru locus controlling flowering in cultivated octoploid strawberry using a selective mapping strategy. Plant Biotechnol. J. 2016;14:2176–2189. doi: 10.1111/pbi.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaston A., Perrotte J., Lerceteauköhler E., Rousseaugueutin M., Petit A., Hernould M., Rothan C., Denoyes B. Pfru, a single dominant locus regulates the balance between sexual and asexual plant reproduction in cultivated strawberry. J. Exp. Bot. 2013;64:1837–1848. doi: 10.1093/jxb/ert047. [DOI] [PubMed] [Google Scholar]
- 55.Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Besseau S., Li J., Palva E.T. Wrky54 and wrky70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012;63:2667–2679. doi: 10.1093/jxb/err450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Brader G., Palva E.T. The wrky70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang Y., Deyholos M.K. Functional characterization of arabidopsis nacl-inducible wrky25 and wrky33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009;69:91–105. doi: 10.1007/s11103-008-9408-3. [DOI] [PubMed] [Google Scholar]
- 59.Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E., Gavin O.L., Gunasekaran P., Ceric G., Forslund K. The pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. Smart 4.0: Towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evolut. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. Expasy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo A.Y., Zhu Q.H., Chen X., Luo J.C. Gsds: A gene structure display server. Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 65.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jue D., Sang X., Liu L., Shu B., Wang Y., Xie J., Liu C., Shi S. The ubiquitin-conjugating enzyme gene family in longan (Dimocarpus longan lour.): Genome-wide identification and gene expression during flower induction and abiotic stress responses. Molecules. 2018;23:662. doi: 10.3390/molecules23030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saeed A., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M. Tm4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.