Abstract
ATP-binding cassette (ABC) transporters hydrolyze ATP to transport a wide range of substrates. Fusarium graminearum is a major causal agent of Fusarium head blight, which is a severe disease in wheat worldwide. FgABCC9 (FG05_07325) encodes an ABC-C (ABC transporter family C) transporter in F. graminearum, which was highly expressed during the infection in wheat and was up-regulated by the plant defense hormone salicylic acid (SA) and the fungicide tebuconazole. The predicted tertiary structure of the FgABCC9 protein was consistent with the schematic of the ABC exporter. Deletion of FgABCC9 resulted in decreased mycelial growth, increased sensitivity to SA and tebuconazole, reduced accumulation of deoxynivalenol (DON), and less pathogenicity towards wheat. Re-introduction of a functional FgABCC9 gene into ΔFgABCC9 recovered the phenotypes of the wild type strain. Transgenic expression of FgABCC9 in Arabidopsis thaliana increased the accumulation of SA in its leaves without activating SA signaling, which suggests that FgABCC9 functions as an SA exporter. Taken together, FgABCC9 encodes an ABC exporter, which is critical for fungal exportation of SA, response to tebuconazole, mycelial growth, and pathogenicity towards wheat.
Keywords: ABC transporter, SA, Fusarium head blight, mycotoxin
1. Introduction
Fusarium head blight (FHB) is a severe disease of wheat in the world, which is mainly caused by the ascomycete fungus Fusarium graminearum [1,2,3]. FHB causes yield loss and contamination of seeds by the accumulation of trichothecene mycotoxins (mainly deoxynivalenol [DON]), which threatens food and feed security [4,5]. FHB has not been well-controlled due to the complexity of wheat resistance to F. graminearum. Therefore, it is necessary to understand the molecular mechanism of interaction between F. graminearum and wheat, which would be valuable for designing a proper strategy for FHB disease management.
ATP-binding cassette (ABC) transporters hydrolyze ATP to transport a wide range of substrates and can be broadly categorized as importers or exporters in the cell membrane depending on the direction of transportation relative to the cytoplasm [6,7]. Members of this superfamily have conserved domain architecture and have been divided into seven families (A–G) in fungi, plants, and animals [8,9,10]. Recent studies demonstrated that the ABC transporter family C (ABC-C) as a cell membrane protein is important for fungal pathogens in response to fungicide [11,12,13].
In the genome of F. graminearum, there are 16 genes belonging to ABC-C [9]. Many of them are expressed at much higher or lower levels during the infection process when compared with their expression in an axenic culture [14,15,16]. FgABCC15 (FG05_10995) played important and diverse roles in both fungicide resistance and pathogenesis of F. graminearum [9,15]. Considering the importance of ABC-C transporters and the hazards of FHB, it is still necessary to characterize other ABC-C transporters in F. graminearum.
Salicylic acid (SA) is a critical plant defense hormone, which contributes to plant defense against a wide range of pathogens with biotrophic and hemibiotrophic lifestyles [17,18,19]. SA triggers systemic acquired resistance (SAR) in plants and upregulates the expression of a set of genes encoding pathogenesis-related (PR) proteins in tobacco and Arabidopsis thaliana [20,21].
The contribution of SA to the wheat/F. graminearum interaction remains unclear. Infection of wheat spikes with F. graminearum resulted in a significant increase in the accumulation of SA [22], which suggests that SA is important in wheat defense against F. graminearum. Over-expression of Nonexpresser of PR Genes 1 (NPR1) of A. thaliana enhanced the FHB resistance level in transgenic wheat [23]. SA has a significant and direct impact on F. graminearum by reducing the efficiency of germination and growth [24]. Nevertheless, exogenous application of SA in wheat spikes cannot increase its resistance against FHB as expected, which possibly resulted from the capacity of F. graminearum to metabolize and export SA [24]. However, the molecular mechanism on the exportation of SA in F. graminearum is still not well understood.
In this paper, we characterized an ABC-C transporter gene (FG05_07325) in F. graminearum. It was named FgABCC9 following Kovalchuk and Driessen [9]. FgABCC9 was highly expressed during the infection of wheat and was upregulated by SA and by the fungicide tebuconazole [12,22,25]. The objective of this research was to understand the mechanism for the exportation of SA in F. graminearum by analyzing the function of FgABCC9 and elevating its effect on fungal mycelial growth and pathogenicity towards wheat. This research would be helpful for understanding the contribution of SA to the wheat/F. graminearum interaction.
2. Results
2.1. Sequence Analysis
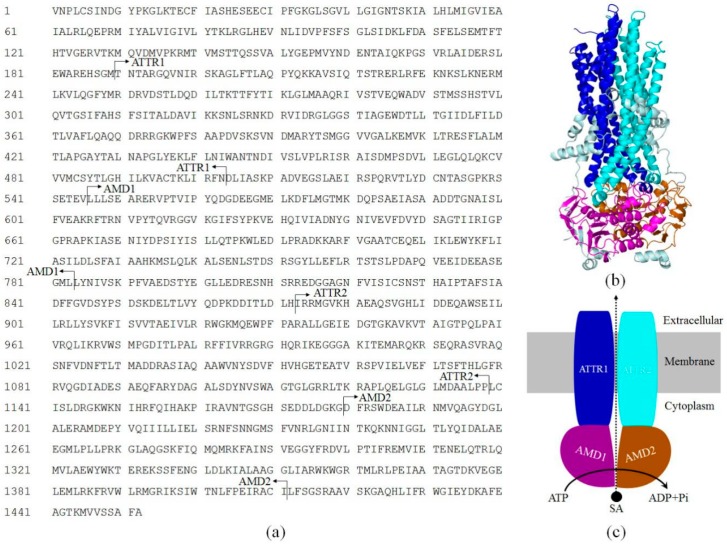
FgABCC9 gene is 4533 bp in length with 3 exons and 2 introns and its open reading frame is 4359 bp. Its deduced amino acid sequence (Appendix Figure A1a) includes the ABC transporter transmembrane region 1 (ATTR1, pfam00116, 190-503 aa), the ABC transporter C family MRP domain 1 (AMD1, cd03250, 546-783 aa), the ABC transporter transmembrane region 2 (ATTR2, pfam00664, 873-1138 aa), and the ABC transporter C family MRP domain 2 (AMD2, cd03250, 1180-1411 aa). The predicted tertiary structure of FgABCC9 was shown in Appendix Figure A1b, which is consistent with the model structure (Appendix Figure A1c) for ABC exporters [26] indicating that FgABCC9 encodes an ABC exporter.
2.2. Deletion and Complementation of FgABCC9 in F. graminearum
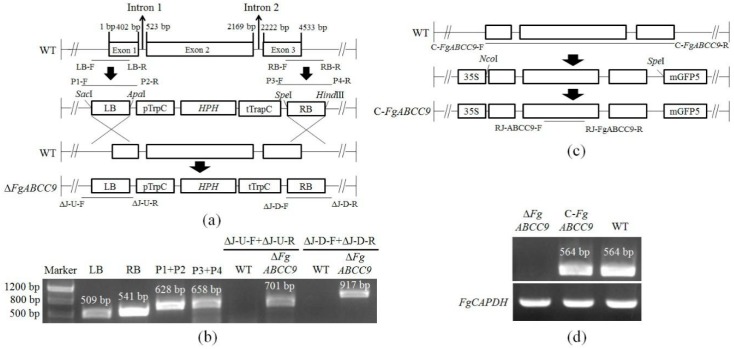
To disrupt the function of FgABCC9 in F. graminearum, the two flanking regions of FgABCC9 were amplified from the genomic DNA of F. graminearum by using P1-F + P2-R and P3-F + P4-R, respectively (Figure 1a), which were then inserted into the pRF-HU2 vector (Figure 1a). Deletion mutants (ΔFgABCC9) were created by replacing the entire FgABCC9 gene with HPH, which is known as a fungal selectable marker gene, using a targeted gene replacement strategy through homologous recombination.
Figure 1.
Construction of deletion and complementation mutants for FgABCC9. (a) The left border (LB) and the right border (RB) were amplified from the wild type (WT) strain to construct the recombinant plasmid. ΔFgABCC9 mutants were then created by homologous recombination between the plasmid and FgABCC9. (b) PCR verification of ΔFgABCC9. (c) Gene sequence of FgABCC9 was amplified from the genomic DNA of WT and ligated into the complementation vector. The T-DNA region of the complementation vector was inserted into the genome of ΔFgABCC9 to create C-FgABCC9. SacI, ApaI, SpeI, HindIII, NcoI, and SpeI show the restriction enzymes used. (d) RT (reverse transcription)-PCR of FgABCC9 in C-FgABCC9 using RJ-ABCC9-F + RJ-ABCC9-R. FgGAPDH was used as reference. The black lines under the elements in (a) and (c) represent the targeted location of primers listed in Table 1. LB-F + LB-R, RB-F + RB-R, C-FgABCC9-F + C-FgABCC9-R were located in the genomic sequence of FgABCC9 of the WT strain. P1-F + P2-R and P3-F + P4-R were mapped in the T-DNA fragment of the pRF-HU2 vector. ΔJ-U-F, ΔJ-U-R, ΔJ-D-F, and ΔJ-D-R were, respectively, positioned into the upstream and downstream of inserted T-DNA sequence of ΔFgABCC9. All the PCR products were verified by sequencing in the commercial company (Qingke, Chengdu, China).
To ensure that the construct was integrated at the intended homologous site (Figure 1b), the primer pairs ΔJ-U-F + ΔJ-U-R and ΔJ-D-F + ΔJ-D-F were used to test ΔFgABCC9, which is shown in Figure 1a. The two primer pairs provided expected PCR bands in ΔFgABCC9 (Figure 1b), which were confirmed by sequencing. These results show that FgABCC9 was successfully removed from the genome of ΔFgABCC9.
To create complementation mutants (C-FgABCC9), a fragment containing the promoter and open reading frame of FgABCC9 was introduced into ΔFgABCC9 (Figure 1c). Reverse transcription PCR (RT-PCR) showed that the FgABCC9 gene was normally expressed in C-FgABCC9 as in WT and no expression was observed in ΔFgABCC9 (Figure 1d).
2.3. Importance of FgABCC9 for Mycelial Growth and Fungal Response to Stress Conditions
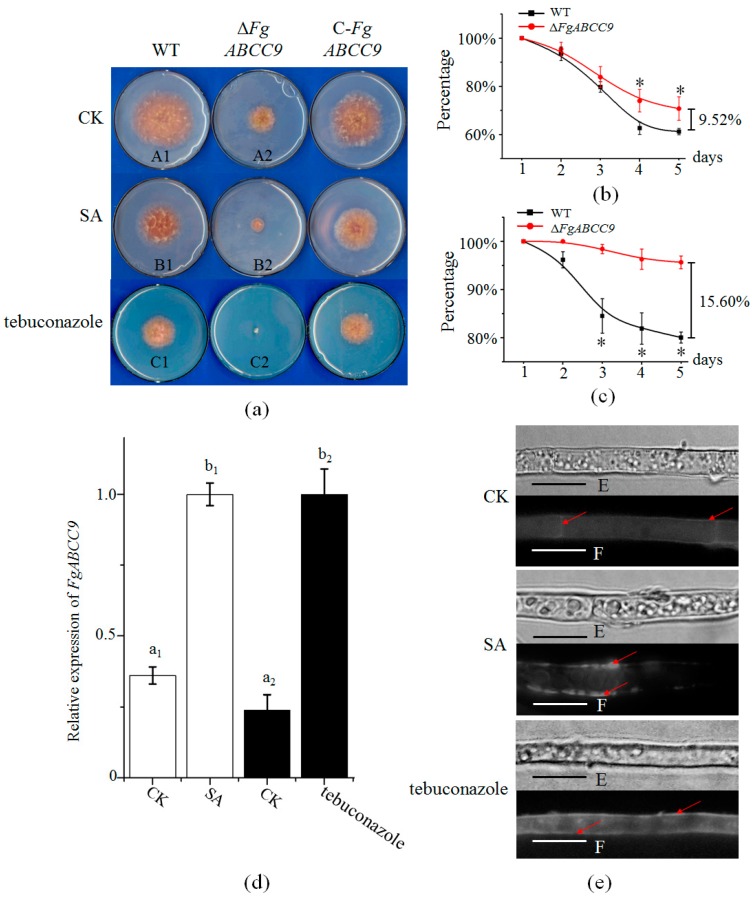
Mycelial growth of WT (wild type), ΔFgABCC9, and C-FgABCC9 strains was compared on mSNA (modified Synthetischer Nährstoffarmer Agar) plates (Figure 2a). ΔFgABCC9 grew much slower than WT and C-FgABCC9. The expression of FgABCC9 could be induced by the plant defense hormone SA and the fungicide tebuconazole (Figure 2d), which was previously reported [12,24]. To clarify whether FgABCC9 was involved in mechanisms alleviating SA and tebuconazole, mycelial growth of WT, ΔFgABCC9 and C-FgABCC9 were elevated on mSNA plates supplemented with 0.9 mM SA and 0.5 mM tebuconazole, respectively (Figure 2a). When compared to WT, ΔFgABCC9 was more sensitive to SA and tebuconazole, which reveals the important role of FgABCC9 in a fungal response to SA and tebuconazole (Figure 2b,c).
Figure 2.
Effect of FgABCC9 on mycelial growth. (a) Mycelial growth on mSNA (modified Synthetischer Nährstoffarmer Agar) plates with SA and tebuconazole. CK, control treatment. Plates were photographed on the fourth day post inoculation. (b) Percentage of mycelial growth inhibited by SA ([A1-B1]/A1 for WT, [A2-B2]/A2 for ΔFgABCC9). (c) Percentage of mycelial growth inhibited by tebuconazole ([A1-C1]/A1 for WT, [A2-C2]/A2 for ΔFgABCC9). Asterisks represent significance at p < 0.05. A1, B1, and C1 indicate mycelial areas of WT strain under the CK, SA, and tebuconazole treatments, respectively. A2, B2, and C2 indicate mycelial areas of ΔFgABCC9 under the CK, SA, and tebuconazole treatments, respectively. The experiments were repeated three times with 10 plates for each treatment. (d) Expression changes of FgABCC9 under SA and tebuconazole treatments in the WT strain. Different letters above each column indicate significance at p < 0.05. (e) Subcellular localization of the FgABCC9 protein. The fluorescent signal is marked with red arrows. E, Optical microscope. F, fluorescence microscope. Scale bar, 10 µm.
The FgABCC9 protein was usually distributed in the septa zone and the cell membrane (Figure 2e). Consistent with its expression at the RNA level, the accumulation of the FgABCC9 protein in the fungal cell membrane was enhanced when treated with SA and tebuconazole (Figure 2e), which further demonstrates the key role of FgABCC9 in the transportation of SA and tebuconazole in F. graminearum.
2.4. FgABCC9 Affects Pathogenicity and DON Production
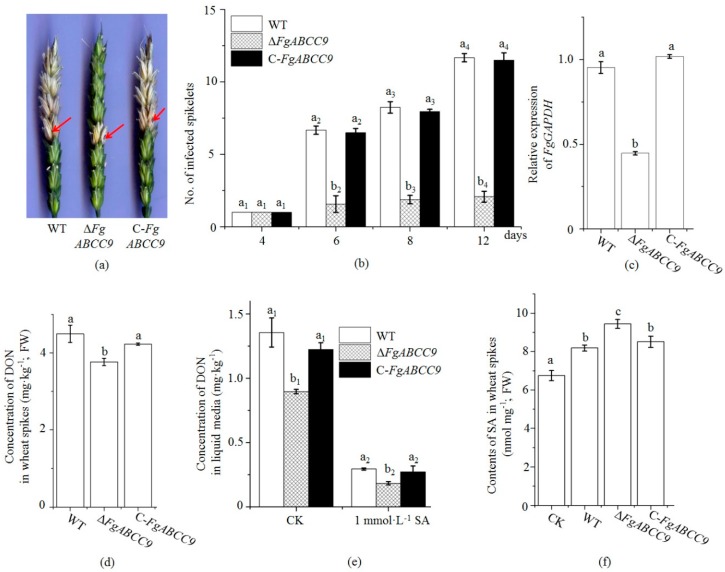
To clarify whether FgABCC9 affected pathogenicity of the F. graminearum, two fully developed florets of a central spikelet were point-inoculated with conidial suspensions of WT, ΔFgABCC9, and C-FgABCC9, respectively. In agreement with its slower mycelial growth on plates and increased sensitivity to SA (Figure 2a), spikes inoculated with ΔFgABCC9 show much less visual disease symptom and a lower fungal biomass when compared with those inoculated with WT and C-FgABCC9 (Figure 3a–c). The concentrations of DON in the liquid culture and in wheat spikes were compared as well. ΔFgABCC9 had a much lower DON production than WT and C-FgABCC9 (Figure 3d,e).
Figure 3.
FgABCC9 affected pathogenesis of F. graminearium in spikes. (a) FHB disease in wheat heads photographed on the eighth day after inoculation. Arrows indicate the inoculated spikelets. (b) the numbers of infected and bleached spikelets on the fourth, sixth, eighth, and twelfth day after inoculation. (c) Relative expression level of FgGAPDH (glyceraldehyde 3-phosphate dehydrogenase gene of F. graminearium, FG05_06257) in wheat spikes inoculated with WT, ΔFgABCC9, and C-FgABCC9, respectively. (d) DON contents in wheat spikes. (e) Concentration of DON in liquid medium. (f) Comparison of the accumulation of SA in wheat spikes after inoculation. Values are the mean ± standard deviation. Different letters above each column indicate a significant difference (p < 0.05). FW, fresh weight.
Considering that FgABCC9 encodes an SA exporter, we compared the contents of SA in spikes inoculated with WT and ΔFgABCC9. Spikes infected with ΔFgABCC9 accumulated a higher concentration of SA (Figure 3f), which suggests that the export of SA by FgABCC9 is critical for the balance between the degradation of wheat endogenous SA by F. graminearum and the toxicity of SA to F. graminearum (reduction in mycelial growth and conidial germination [24]) during the infection in wheat.
2.5. Evaluation of the Function of FgABCC9 in A. thaliana
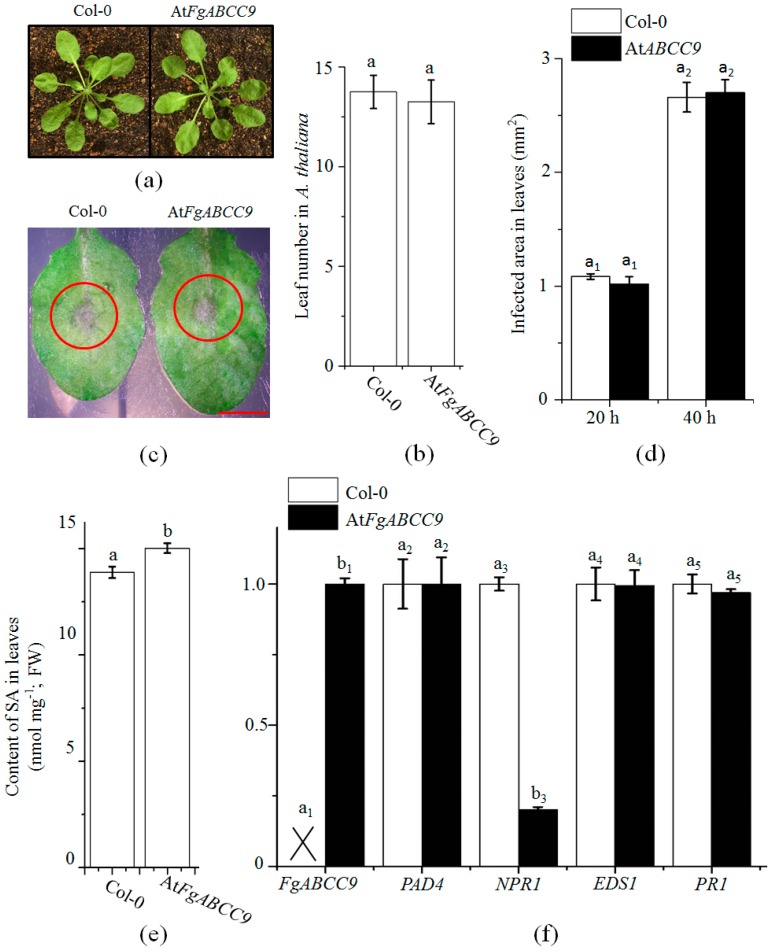
To confirm its function, FgABCC9 was expressed in A. thaliana. The leaves of AtFgABCC9 accumulated more SA than those of Col-0 (Figure 4e) even though there was no visual difference between Col-0 and AtFgABCC9 plants (Figure 4a,b). To make sure the increased accumulation of SA affected expression of SA-related genes, we compared the expression of NPR1, EDS1, PAD4, and PR1 in the leaves of transgenic and WT plants (Figure 4f). NPR1 is a master regulator of SA-mediated transcriptional reprogramming and immunity and it functions as a transcriptional coactivator [27]. EDS1 and PAD4 as part of a central regulator complex contribute to the positive feedback loop of SA accumulation [28]. PR1 is one of the genes that is induced by SA and often used as the marker for SA signaling [29]. Unexpectedly, only the expression of NPR1 was affected, which indicated that transgenic expression of FgABCC9 did not activate SA signaling.
Figure 4.
Evaluation of the function of FgABCC9 in A. thaliana. (a) The WT and AtFgABCC9 plants on the 30th day under SD condition. (b) Comparison of leaf numbers of WT and AtFgABCC9 plants as used in (a). (c) Comparison of visual disease symptoms in leaves of WT and AtFgABCC9 plants infected by F. graminearium at the 48th hour after inoculation. Scale bar, 1 cm (d) Comparison of infected areas in leaves at the 20th and 40th hour after inoculation. (e) Comparison of the contents of SA in leaves (without inoculation) of WT and AtFgABCC9 plants on the 30th day post germination under the SD (short day) condition. (f) Comparison of gene expression in leaves (without inoculation) of WT and AtFgABCC9 plants by qPCR. The cross indicates the absence of FgABCC9 from Col-0. Different letters above each column indicate significance at p < 0.05.
To clarify whether FgABCC9 affected A. thaliana resistance against F. graminearum, the leaves of AtFgABCC9 and Col-0 were point-inoculated with conidial suspension of the WT strain. Consistent with the qPCR data in Figure 4f, there was no significant difference between the resistance levels of AtFgABCC9 and Col-0 (Figure 4c,d).
3. Discussion
The superfamily of the ABC transporter is one of the largest gene families in fungi. It is involved in the transport of a broad range of substrates across biological membranes. ABC transporters, particularly those with a multidrug resistance (MDR) domain or a pleiotropic drug resistance (PDR) domain, are important in the resistance of fungal pathogens to xenobiotics [11]. F. graminearum, which is an ascomycete fungus, is a major causal pathogen for FHB disease in wheat. F. graminearum has 62 ABC transporter genes, which were divided into seven families (A–G) according to domain architecture and phylogenetic data [9]. FgABCG6 (FG05_04580) with a PDR domain plays a role in protecting the fungus from antifungal compounds and helps combat unidentified wheat defense compounds during disease development [30]. FgABCC15 (FG05_10995) with an MRP domain plays important and diverse roles in both fungicide resistance and pathogenesis of F. graminearum [15]. In this study, we characterized a new ABC-C transporter gene with an MRP domain (FgABCC9; FG05_07325). Our data demonstrated that FgABCC9 was an ABC exporter, which was important for the export of SA by F. graminearum under SA stress and for fungal response to tebuconazole and pathogenesis in wheat. Our results together with the previous reports suggested that manipulation of the expression level or activity of ABC transporters would be a hopeful way to manage FHB disease.
SA is one of the key plant defense hormones, which is associated with resistance against F. graminearum in wheat [22,23,24]. Nevertheless, the contribution of SA to wheat/F. graminearum interaction remains unclear. SA has a significant and direct impact on F. graminearum since it reduces the efficiency of germination and mycelial growth at higher concentrations [24] by down regulating the chitin synthase gene FgCHS8 and the cis-12 linoleic acid isomerase gene FgLAI12 [31,32]. Wheat spikes accumulate more SA when infected with F. graminearum [22], which suggests an important role of SA in wheat FHB resistance. Unexpectedly, exogenous application of SA cannot improve the wheat resistance level against FHB possibly due to the fact that F. graminearum can reduce the toxicity of SA by its capacity to metabolize and export SA [24]. This research showed the importance of FgABCC9 as an SA exporter for F. graminearum under SA stress and during infection in wheat. Deletion of the FgABCC9 gene resulted in enhanced fungal sensitivity to SA (Figure 2a), increased accumulation of SA (Figure 3f), lower fungal biomass (Figure 3c), and less disease symptom (Figure 3a,b) in wheat spikes after inoculation. The presence of FgABCC9 was critical for fungal degradation of SA under SA stress (Figure 2a). Therefore, we speculated that ΔFgABCC9 could not efficiently degrade SA in wheat spikes as the WT strain, which results in enhanced accumulation of SA in spikes inoculated with ΔFgABCC9 (Figure 3f). It also suggested that F. graminearum reduced the accumulation of SA in spikes when FgABCC9 functioned normally and made sure that the concentration of endogenous SA was below the toxicity threshold. ΔFgABCC9 might not completely lose the ability to export SA from the cytoplasm to the extracellular space since it is hard to identify all the SA exporters in one experiment. However, even the partial reduction of this ability led to lower fungal biomass and less disease symptom, which indicated the contribution of the SA exporter to deal with SA during the infection of F. graminearum. These results demonstrated that SA did contribute to wheat resistance against F. graminearum.
Transgenic expression of the FgABCC9 gene did not activate the expression of the PR1 that is the marker gene for SA signaling (Figure 4f), even though it increased the concentration of SA in the leaves of A. thaliana (Figure 4e). Considering that FgABCC9 functions as an SA exporter, we speculated that the content of SA within cytoplasm would not increase.
Tebuconazole is a sterol demethylation inhibitor and is effective in controlling FHB in agricultural production [33,34]. Recent studies show that the 14-α-demethylase encoded by the Cyp51 gene as a target enzyme affected fungal cell wall sensitivity to tebuconazole. ABC transporters used as efflux pumps were involved in the transportation of tebuconazole in fungi [35,36]. In this study, the expression of the FgABCC9 gene was upregulated by tebuconazole (Figure 2d,e) and deletion of the FgABCC9 gene led to increased sensitivity to tebuconazole on mSNA plates (Figure 2a,c). These results suggest that the FgABCC9 gene is involved in mechanisms alleviating to tebuconazole and that FgABCC9 is an exporter for the fungicide tebuconazole in F. graminearum as well.
FHB threatens food and feed security by contaminating seeds with trichothecene mycotoxins. We previously showed that SA significantly inhibited DON production [24] and SA also significantly upregulated the expression level of FgABCC9 (Figure 2d). In the present study, disruption of the FgABCC9 gene in F. graminearum resulted in reduced accumulation of DON production in wheat spikes and in liquid media (Figure 3d,e). However, the reduced DON contents in spikes inoculated with ΔFgABCC9 was possibly due to the less fungal biomass (Figure 3c). In the liquid media, the same amount of mycelia (0.2 mg) was used, which demonstrates that the expression of the FgABCC9 gene was positively associated with DON production in F. graminearum. It seems likely that the FgABCC9 may not be a regulatory target for SA to downregulate DON production in F. graminearum.
Members in the ABC-C subfamily have been recognized as export pumps for amphiphilic anions especially for conjugates of lipophilic compounds with glutathione or several other non-toxic anionic residues in Saccharomyces cerevisiae [10,37,38,39]. ΔFgABCC9 showed slower mycelial growth on mSNA plates than WT and C-FgABCC9 (Figure 2a), which suggested that FgABCC9 is also an exporter for other materials that are essential for normal growth of F. graminearum.
4. Materials and Methods
4.1. Materials
A virulent isolate of F. graminearum (DAOM180378) was used for the fungal experiment throughout the paper. Triticum aestivum cv. “Roblin” was used for wheat inoculation, which is susceptible to FHB disease. Wheat plants were grown in green houses under 16/8 h day/night cycles at 23/18 °C. Plants were watered as needed and fertilized before sowing with 15-15-15 (N-P-K). Unless specifically noted, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
A. thaliana ecotype Col-0 was used. Seeds were sown on Murashige and Skoog (MS) medium with 0.7% (weight/volume) agar and were kept at 4 °C for 3 d before being moved to a growth chamber. The WT and transgenic plants were grown side-by-side in the same tray to minimize possible variation of growth conditions. Plants were first grown at 22 °C under a short day (SD) condition for 30 d with a 10 h light/14 h dark photoperiod for an extended vegetative growth phase and were then moved to a long day (LD) (16 h light/8 h dark) condition for reproductive growth. The pCAMBIA1302 vector carrying FgABCC9 was frozen and then thawed into the Agrobacterium tumefaciens strain AGL-1 (Tiangen, Beijing, China). It was transformed into the Col-0 through the floral dip method [40]. Six resistant T1 transgenic lines were selected in 50 μg·mL−1 hygromycin (HPH) and verified by using PCR. Phenotypic analyses were performed in the T2 generation and confirmed in the T3 generation. Homozygous plants were used in all experiments.
4.2. Sequence Analysis
Nucleotide sequence of FgABCC9 (FG05_07325) was downloaded from the Ensembl fungi database (Available at: http://fungi.ensembl.org/index.html). Primer Premier 5.0 (Premier Biosoft, Palo Alto, CA, USA) software was used to design PCR primers (Table 1). The analysis of sequence similarity and identification of conserved domains were performed using BLASTp on NCBI (Available at: http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Figure 1a). The 3D-protein structure was predicted by Phyre2 [41] and made by PyMol 1.8 (Figure 1b).
Table 1.
Primers used in this study.
| Primer | Sequence (5′–3′) | Source |
|---|---|---|
| LB-F | GCGGGCCCAGGCTACTATGGCTGTT | This study |
| LB-R | GCGAGCTCAGATGCGAAAGGGTC | This study |
| RB-F | GGAAGCTTCAATCCCGTTGTTCTGGT | This study |
| RB -R | GGACTAGTAGCCTGCTTCGTGTTCC | This study |
| P1-F | CTTTTCTCTTAGGTTTACCCG | [31] |
| P2-R | TAATGCAGGAGTCGCATAAG | [31] |
| P3-F | CCCAAAAAGTGCTCCTTCAA | [31] |
| P4-R | TGTGCTGCAAGGCGATTAA | [31] |
| ΔJ-U-F | CCTCGGGCGGTCTGTTT | This study |
| ΔJ-U-R | TTCGGCGTGGGTATGG | This study |
| ΔJ-D-F | TCCTCGTTCCTGTCTG | This study |
| ΔJ-D-R | CTCCGATGATGAGAAGT | This study |
| C-FgABCC9-F | TGCCATGGTATTACCGTACTTCCCTA | This study |
| C-FgABCC9-R | GGACTAGTCGACTGTCTACTCGACTC | This study |
| CJ-FgABCC9-F | AAGAAGGCAAAGAAAGCAAA | This study |
| CJ-FgABCC9-R | GAACAAGGCCAGTGATGAGA | This study |
| Fg-GAPDH-F | TGACTTGACTGTTCGCCTCGAGAA | [24] |
| Fg-GAPDH-R | ATGGAGGAGTTGGTGTTGCCGTTA | [24] |
| Fg-β-tubulin-F | GTTGATCTCCAAGATCCGTG | [24] |
| Fg-β-tubulin-R | CATGCAAATGTCGTAGAGGG | [24] |
| Fg-elongation Factor1-F | CCTCCAGGATGTCTACAAGA | [24] |
| Fg-elongation Factor1-R | CTCAACGGACTTGACTTCAG | [24] |
| Aox-F | GACTTGTCATGGTAGATGCCTG | [24] |
| Aox-R | CAGGACGAGCATAACCATTCTC | [24] |
| w-GAPDH-F | AACTGTTCATGCCATCACTGCCAC | [24] |
| w-GAPDH-R | AGGACATACCAGTGAGCTTGCCAT | [24] |
| hn-RNP-Q-F | TCACCTTCGCCAAGCTCAGAACTA | [24] |
| hn-RNP-Q-R | AGTTGAACTTGCCCGAAACATGCC | [24] |
| Rj-FgABCC9-F | GCGGGACCACAAGGGATT | This study |
| Rj-FgABCC9-R | GATGTAGGCACCGTAGACAGACC | This study |
| ACT2-F | CTTGCACCAAGCAGCATGAA | [42] |
| ACT2-R | CCGATCCAGACACTGTACTTCCTT | [42] |
| UBQ10-F | GGCCTTGTATAATCCCTGATGAATAAG | [42] |
| UBQ10-R | AAAGAGATAACAGGAACGGAAACATAGT | [42] |
| EF-1α-F | TGAGCACGCTCTTCTTGCTTTCA | [42] |
| EF-1α-R | GGTGGTGGCATCCATCTTGTTACA | [42] |
| EDS1-F | GGATAGAAGATGAATACAAGCC | [43] |
| EDS1-R | ACCTAAGGTTCAGGTATCTGT | [43] |
| PR1F | TGGCTATTCTCGATTTTTAATCG | [29] |
| PR1R | CCATTGCACGTGTTCGCAG | [29] |
| NPR1-F | CATTCTCTCAAAGGCCGACT | [44] |
| NPR1-R | AAGACGTTGAGCAAGTGCAA | [44] |
| PAD4-F | ATGGACGATTGTCGATTCGAG | [43] |
| PAD4-R | CTAAGTCTCCATTGCGTCACT | [43] |
The underlined are cutting sites for restriction enzymes.
4.3. Construction of Deletion and Complementation Mutants
Fresh mycelia cultured on mSNA (1 g KH2PO4, 1 g KNO3, 0.5 g MgSO4, 0.5 g KCl, 1 g glucose, 1 g sucrose and 20 g agar per liter) plates [24] were used to extract the genomic DNA of F. graminearum by using a CTAB method [45]. Disruption of the FgABCC9 gene in F. graminearum was performed, which is shown in Figure 1a. The pRF-HU2 vector [46] was used to create ΔFgABCC9 mutants through homologous recombination. Transformation of F. graminearum was carried out similarly to our previous study [31].
To restore its function, the genomic sequence of FgABCC9 was amplified by primer pair C-FgABCC9-F + C-FgABCC9-R, ligated into pCAMBIA1302 vector, and transformed into ΔFgABCC9 to create C-FgABCC9 mutants. All the C-FgABCC9 mutants were verified by RT (reverse transcription)-PCR by using the primer pair RJ-FgABCC9-F + RJ-FgABCC9-CJ-R (Table 1).
4.4. Conidial and Mycelial Growth Conditions
Conidia were produced in CMC (carboxymethyl cellulose) medium at 28 °C by shaking (180 rpm) for 5 d [47]. The concentration of conidia was determined with a hemocytometer by microscopy.
The effects of SA and tebuconazole on mycelial growth were tested on mSNA plates. SA (0.9 mM SA) and tebuconazole (0.5 mM) were supplemented after autoclaving. A total of 1000 conidia were inoculated on each mSNA plate and the plates were maintained at 28 °C in darkness. Ten replicates were done for each treatment. The growing mycelia on mSNA plates were scanned by using the EPSON Perfection V700 Photo (Seiko Epson, Bekasi, Indonesia) on day 5 after inoculation. Thereafter, the area of mycelia was measured by Computer Aided Design (CAD) software (version 2007). The B-spline curve for mycelia growth was calculated as Fang et al. [48].
4.5. Virulence Assay
To determine the effect of FgABCC9 on the pathogenicity of F. graminearum in wheat heads, two flowering florets of a central spikelet of one head were each inoculated with 1 × 103 conidia. The inoculated heads were sprayed with water and enclosed with a plastic wrap for 48 h at 25 °C. Wheat plants were placed in a controlled-environment room at 25 °C. FHB symptoms were assessed 4 to 12 d after inoculation. Ten plants were used per treatment.
To examine whether FgABCC9 affected pathogenicity of F. graminearum in leaves of A. thaliana, 1 × 103 conidia was point inoculated in the middle of leaves wounded by brush. The inoculated leaves were maintained on MS medium at 25 °C for 48 h. Subsequently, the inoculated leaves were scanned by using the EPSON Perfection V700 Photo and ground to powder in liquid nitrogen.
To test whether FgABCC9 was related to the production of mycotoxin DON in liquid media, a two-stage protocol from References [24,49] was used. The concentration of DON was measured by using the DON ELISA kit (Beacon, Saco, ME, USA) and the Multiskan Spectrum (Thermo Scientific, Vantaa, Finland).
To test whether FgABCC9 affected the production of DON in wheat spikes, two florets of fully developed spikelets at mid-anthesis were each inoculated with 1 × 103 conidia. After inoculation, the wheat plants were placed in a room as described above. The inoculated spikelets were harvested at the 48th hour (for the following gene expression analysis) and on day 8 after inoculation (for DON analysis). The spikelets were ground into a fine powder in liquid nitrogen. DON was extracted from 100 mg spikelet powder with one mL sterile water at 4 °C for 12 h. Three biological replicates were conducted with at least ten heads per treatment.
4.6. Gene Expression Analysis
Total RNA was extracted from fresh powders of mycelia, wheat spikelets, and A. thaliana leaves by using the E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek, Norcross, GA, USA) in accordance with the manufacturer’s instructions. RNA was reverse transcribed using the PrimeScript™ RT Reagent Kit with genomic DNA Eraser (Takara, Dalian, China) following the manufacturer’s protocol.
The primer pair Rj-FgABCC9-F + Rj-FgABCC9-R was used to measure the expression level of FgABCC9 in F. graminearum and A. thaliana. Gene expression of PR1, EDS1, PAD4, and NPR1 was detected in the leaves of A. thaliana by using PR1-F + PR1-R, EDS1-F + EDS1-R, PAD4-F + PAD4-R, and NPR1-F + NPR1-R, respectively (Table 1). The FgGAPDH (FG05_06257), β-tubulin (FG05_09530) and elongation factor 1 (FG05_08811) genes were used as reference genes when performing qPCR for the samples of F. graminearum [24]. The relative amount of F. graminearum was estimated by measuring the expression level of FgGAPDH in wheat spike tissue by using qPCR with normalization to three wheat reference genes (w-GAPDH (glyceraldehyde-3-phosphate dehydrogenase gene in wheat), NCBI UniGene Ta.66461; Aox (aldehyde oxidase gene), Ta.6172; hn-RNP-Q (heterogeneous nuclear ribonucleoprotein Q gene), Ta.10105) [24]. The ACT2, UBQ10, and EF-1α genes [42] were used as reference genes when doing qPCR in the samples of A. thaliana. qPCR was performed by a MyiQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). All of the primers mentioned above were listed in Table 1.
4.7. Quantification of SA
The wheat spikelet and A. thaliana leaf samples for qPCR analysis were utilized for the quantification of SA as well. To prepare samples of A. thaliana, leaves of WT and transgenic plants grown under the SD condition without inoculation were harvested and ground to a fine powder in liquid nitrogen, respectively. Three biological replicates were conducted with at least six plants per treatment. Quantification of SA was performed as described previously [50].
4.8. Microscopic Assay
The C-FgABCC9 strains harboring a green fluorescent protein gene (GFP) tag were used to examine the subcellular localization of the FgABCC9 protein. Hyphae were used to detect GFP fluorescence. The optical microscopic and fluorescence microscopic assays were carried out as shown in Reference [31] by using a Nikon-80i fluorescence microscope (Nikon, Tokyo, Japan).
4.9. Statistical Analysis
Student’s t-test (implemented in DPS version 12.01 software, [51]) was used to test the significance of differences in mycelial growth, SA content, gene expression, DON production, and the level of disease.
Acknowledgments
The authors give special thanks to Thérèse Ouellet of Agriculture and Agri-Food Canada for providing materials.
Appendix A
Figure A1.
Structure of FgABCC9. (a) Distribution of conserved protein domain in the amino acid sequence of FgABCC9. (b) Predicted tertiary structure of FgABCC9 protein. Dark blue, purple, cyan, and brown colors indicate the “ATTR1” (ABC transporter transmembrane region 1; pfam00116; 190-503 aa), “AMD1” (ABC transporter C family MRP (multidrug resistance-associated protein) domain 1; cd03250; 546-783 aa), “ATTR2” (ABC transporter transmembrane region 2; pfam00664; 873-1138 aa), and “AMD2” (ABC transporter C family MRP domain 2; cd03250; 1180-1411 aa), respectively. (c) Schematic of ABC exporters [26].
Author Contributions
P.-F.Q., Y.-Z.Z., C.-H.L., J.Z., Q.C., Z.-R.G., Y.W., B.-J.X., T.Z., Y.-F.J., J.-P.W., C.-Y.Z., X.F., and L.K. performed all the experiments and analyzed the data. P.-F.Q. and Y.-M.W. designed the experiment. X.-J.L., Q.-T.J., and Y.-L.Z. provided key advice. P.-F.Q., Y.-Z.Z., Q.C., Y.-M.W., and Y.-L.Z. wrote the paper. P.-F.Q., Y.-Z.Z., X.-J.L., and Q.-T.J. prepared the figures.
Funding
This research was funded by the National Key Research and Development Program of China (2017YFD0100900) and the National Natural Science Foundation of China (31671677).
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- 1.Goswami R.S., Kistler H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004;5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 2.Walter S., Nicholson P., Doohan F.M. Action and reaction of host and pathogen during Fusarium head blight disease. New Phytol. 2010;185:54–66. doi: 10.1111/j.1469-8137.2009.03041.x. [DOI] [PubMed] [Google Scholar]
- 3.Kazan K., Gardiner D.M., Manners J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012;13:399–413. doi: 10.1111/j.1364-3703.2011.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernando W.G.D., Paulitz T.C., Seaman W.L., Dutilleul P., Miller J.D. Head blight gradients caused by Gibberella zeae from area sources of inoculum in wheat field plots. Phytopathology. 1997;87:414–421. doi: 10.1094/PHYTO.1997.87.4.414. [DOI] [PubMed] [Google Scholar]
- 5.Chandler E.A., SimPson D.R., Thomsett M.A., Nicholson P. Development of PCR assays to Tri7 and Tri13 trichothecene biosynthetic genes, and characterization of chemotypes of Fusarium graminearum, Fusarium culomrum and Fusarium cerealis. Physiol. Mol. Plant Pathol. 2003;62:355–367. doi: 10.1016/S0885-5765(03)00092-4. [DOI] [Google Scholar]
- 6.Higgins C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 7.Dassa E., Bouige P. The ABC of ABCs: A phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 2001;152:211–229. doi: 10.1016/S0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Fernández R., Davies T.G., Coleman J.O., Rea P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 9.Kovalchuk A., Driessen A. Phylogenetic analysis of fungal ABC transporters. BMC Genom. 2010;11:177. doi: 10.1186/1471-2164-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ween M.P., Armstrong M.A., Oehler M.K., Ricciardelli C. The role of ABC transporters in ovarian cancer progression and chemoresistance. Crit. Rev. Oncol./Hematol. 2015;96:220–256. doi: 10.1016/j.critrevonc.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Kretschmer M., Leroch M., Mosbach A., Walker A.S., Fillinger S., Mernke D., Schoonbeek H.J., Pradier J.M., Leroux P., De Waard M.A., et al. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 2009;5:e1000696. doi: 10.1371/journal.ppat.1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becher R., Weihmann F., Deising H.B., Wirsel S.G. Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genom. 2011;12:12–52. doi: 10.1186/1471-2164-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Son H., Lee J., Lee Y.R., Lee Y.W. A putative ABC transporter gene, ZRA1, is required for zearalenone production in Gibberella zeae. Curr. Genet. 2011;57:343–351. doi: 10.1007/s00294-011-0352-4. [DOI] [PubMed] [Google Scholar]
- 14.Stephens A.E., Gardiner D.M., White R.G., Munn A.L., Manners J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant Microbe Interact. 2008;21:1571–1581. doi: 10.1094/MPMI-21-12-1571. [DOI] [PubMed] [Google Scholar]
- 15.Abou Ammar G., Tryono R., Döll K., Karlovsky P., Deising H.B., Wirsel S.G. Identification of ABC transporter genes of Fusarium graminearum with roles in azole tolerance and/or virulence. PLoS ONE. 2013;8:e79042. doi: 10.1371/journal.pone.0079042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosaka A., Ban T., Manickavelu A. Genome-wide transcriptional profiling of wheat infected with Fusarium graminearum. Genom. Data. 2015;5:260–262. doi: 10.1016/j.gdata.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Vos M., Van Oosten V.R., Van Poecke R.M.P., Van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Metraux J.P., Van Loon L.C., Dicke M., et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 18.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 19.Görlach J., Volrath S., Knauf-Beiter G., Hengy G., Beckhove U., Kogel K., Oostendorp M., Staub T., Ward E., Kessmann H., et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahl-Goy P., Metraux J.P., Ryals J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., Chandler D., Slusarenko A., Ward E., Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi P.F., Balcerzak M., Rocheleaub H., Leung W., Wei Y.M., Zheng Y.L., Ouellet T. Jasmonic acid and abscisic acid play important roles in host-pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol. Mol. Plant Pathol. 2016;93:39–48. doi: 10.1016/j.pmpp.2015.12.004. [DOI] [Google Scholar]
- 23.Makandar R., Essig J.S., Schapaugh M.A., Trick H.N., Shah J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 2006;19:123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- 24.Qi P.F., Johnston A., Balcerzak M., Rocheleau H., Harris L.J., Long X.Y., Wei Y.M., Zheng Y.L., Ouellet T. Effect of salicylic acid on Fusarium graminearum, the major causal agent of Fusarium Head Blight in wheat. Fungal Biol. 2012;116:413–426. doi: 10.1016/j.funbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Harris L.J., Balcerzak M., Johnston A., Schneiderman D., Ouellet T. Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016;120:111–123. doi: 10.1016/j.funbio.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Locher K.P. Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pajerowska-Mukhtar K.M., Emerine D.K., Mukhtar M.S. Tell me more: Roles of NPRs in plant immunity. Trends Plant Sci. 2013;18:402–411. doi: 10.1016/j.tplants.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Seyfferth C., Tsuda K. Salicylic acid signal transduction: The initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 2014;5:697. doi: 10.3389/fpls.2014.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giri M.K., Singh N., Banday Z.Z., Singh V., Ram H., Singh D., Chattopadhyay S., Nandi A.K. GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in Arabidopsis thaliana. Plant J. 2017;91:802–815. doi: 10.1111/tpj.13608. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner D.M., Stephens A.E., Munn A.L., Manners J.M. An ABC pleiotropic drug resistance transporter of Fusariumg raminearum with a role in crown and root diseases of wheat. FEMS Microbiol. Lett. 2013;348:36–45. doi: 10.1111/1574-6968.12240. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y.Z., Chen Q., Liu C.H., Liu Y.B., Yi P., Niu K., Wang Y.Q., Wang A.Q., Yu H.Y., Pu Z.E., et al. Chitin synthase gene FgCHS8 affects virulence and fungal cell wall sensitivity to environmental stress in Fusarium graminearum. Fungal Biol. 2016;120:764–774. doi: 10.1016/j.funbio.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y.Z., Wei Z.Z., Liu C.H., Chen Q., Xu B.J., Guo Z.R., Cao Y.L., Wang Y., Han Y.N., Chen C., et al. Linoleic acid isomerase gene FgLAI12 affects sensitivity to salicylic acid, mycelial growth and virulence of Fusarium graminearum. Sci. Rep. 2017;7:46129. doi: 10.1038/srep46129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blandino M., Minelli L., Reyneri A. Strategies for the chemical control of Fusarium head blight: Effect on yield, alveographic parameters and deoxynivalenol contamination in winter wheat grain. Eur. J. Agron. 2006;25:193–201. doi: 10.1016/j.eja.2006.05.001. [DOI] [Google Scholar]
- 34.De Waard M.A., Andrade A.C., Hayashi K., Schoonbeek H.J., Stergiopoulos I., Zwiers L.H. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 2006;62:195–207. doi: 10.1002/ps.1150. [DOI] [PubMed] [Google Scholar]
- 35.Leroux P., Albertini C., Gautier A., Gredt M., Walker A.S. Mutations in the CYP51 gene correlated with changes in sensitivity to sterol 14 alpha-demethylation inhibitors in field isolates of Mycosphaerella graminicola. Pest Manag. Sci. 2007;63:688–698. doi: 10.1002/ps.1390. [DOI] [PubMed] [Google Scholar]
- 36.Luo C.X., Schnabel G. The cytochrome P450 lanosterol 14alpha-demethylase gene is a demethylation inhibitor fungicide resistance determinant in Monilinia fructicola field isolates from Georgia. Appl. Environ. Microbiol. 2008;74:359–366. doi: 10.1128/AEM.02159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzmann D.J., Hallstrom T.C., Voet M., Wysock W., Golin J., Volckaert G., Moye-Rowley W.S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6875–6883. doi: 10.1128/MCB.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Z., Hirata D., Tsuchiya E., Osada H., Miyakawa T. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J. Biol. Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 39.Decottignies A., Grant A.M., Nichols J.W., de Wet H., McIntosh D.B., Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 40.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 43.Tandon G., Jaiswal S., Iquebal M.A., Kumar S., Kaur S., Rai A., Kumar D. Evidence of salicylic acid pathway with EDS1 and PAD4 proteins by molecular dynamics simulation for grape improvement. J. Biomol. Struct. Dyn. 2015;33:2180–2191. doi: 10.1080/07391102.2014.996187. [DOI] [PubMed] [Google Scholar]
- 44.Gatz C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 2013;26:151–159. doi: 10.1094/MPMI-04-12-0078-IA. [DOI] [PubMed] [Google Scholar]
- 45.Lodhi M.A., Ye G.N., Weeden N.F., Reisch B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994;12:6–13. doi: 10.1007/BF02668658. [DOI] [Google Scholar]
- 46.Frandsen R.J., Andersson J.A., Kristensen M.B., Giese H. Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 2008;9:70. doi: 10.1186/1471-2199-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capellini R.A., Peterson J.L. Macroconidium formation in submerged cultures by a non-sporulating strains of Gibberella zeae. Mycologia. 1965;57:962–966. doi: 10.2307/3756895. [DOI] [Google Scholar]
- 48.Fang S., Yan X., Liao H. 3D reconstruction and dynamic modeling of root architecture in situ and its application to crop phosphorus research. Plant J. 2009;60:1096–1108. doi: 10.1111/j.1365-313X.2009.04009.x. [DOI] [PubMed] [Google Scholar]
- 49.Miller D., Blackwell B.A. Biosynthesis of 3-acetyldeoxynivalenol and other metabolites by Fusarium culmorum HLK 1503 in a stirred jar fermentor. Can. J. Bot. 1986;64:1–5. doi: 10.1139/b86-001. [DOI] [Google Scholar]
- 50.Siciliano I., Amaral Carneiro G., Spadaro D., Garibaldi A., Gullino M.L. Jasmonic acid, abscisic acid, and salicylic acid are involved in the phytoalexin responses of rice to Fusarium fujikuroi, a high gibberellin producer pathogen. J. Agric. Food Chem. 2015;63:8134–8142. doi: 10.1021/acs.jafc.5b03018. [DOI] [PubMed] [Google Scholar]
- 51.Tang Q.Y., Zhang C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013;20:254–260. doi: 10.1111/j.1744-7917.2012.01519.x. [DOI] [PubMed] [Google Scholar]