Abstract
Browning of adipose tissue has been prescribed as a potential way to treat obesity, marked by the upregulation of uncoupling protein 1 (Ucp1). Several reports have suggested that histone deacetylase (HDAC) might regulate Ucp1 by remodelling chromatin structure, although the mechanism remains unclear. Herein, we investigate the effect of β-adrenergic receptor (β-AR) activation on the chromatin state of beige adipocyte. β-AR-stimulated Ucp1 expression via cold (in vivo) and isoproterenol (in vitro) resulted in acetylation of histone activation mark H3K27. H3K27 acetylation was also seen within Ucp1 promoter upon isoproterenol addition, favouring open chromatin for Ucp1 transcriptional activation. This result was found to be associated with the downregulation of class I HDAC mRNA, particularly Hdac3 and Hdac8. Further investigation showed that although HDAC8 activity decreased, Ucp1 expression was not altered when HDAC8 was activated or inhibited. In contrast, HDAC3 mRNA and protein levels were simultaneously downregulated upon isoproterenol addition, resulting in reduced recruitment of HDAC3 to the Ucp1 enhancer region, causing an increased H3K27 acetylation for Ucp1 upregulation. The importance of HDAC3 inhibition was confirmed through the enhanced Ucp1 expression when the cells were treated with HDAC3 inhibitor. This study highlights the novel mechanism of HDAC3-regulated Ucp1 expression during β-AR stimulation.
Keywords: beige adipocyte, β-adrenergic receptor, histone deacetylase, histone acetylation, uncoupling protein 1
1. Introduction
Recently, browning of white adipose tissue (WAT) has garnered attention as a potent target for obesity. The term “browning” originated from the distinct characteristics of WAT and brown adipose tissue (BAT). WAT shows low oxidative capacity to support the storage of excess energy as triglyceride, while by contrast, BAT shows high oxidative capacity due to high mitochondrial content. Beige adipocyte develop within WAT in response to β-adrenergic receptor (β-AR) stimulation, such as cold conditions to activate a thermogenesis program to produce heat by increasing energy expenditure [1,2]. This phenomenon would be beneficial for obesity, as WAT has a relatively large mass, and therefore any change in cell physiology in this tissue may affect whole body metabolism. Once activated, beige adipocyte exhibit similar functional thermogenic characteristics as BAT, and thus browning is marked by upregulation of browning-fat specific genes, such as uncoupling protein 1 (Ucp1) [1,3,4]. The increased Ucp1 expression in beige adipose tissue is especially evident in the inguinal region where basal levels of Ucp1 are very low [5,6]. Many efforts have been made to pharmacologically stimulate the thermogenesis program through β-AR agonists, especially β3-AR. However, β3-AR agonists lack efficacy in human translational studies [7,8,9]. Hence, finding a new molecular target to induce browning is needed.
Epigenetic modification has emerged as a new approach to treat diseases including cancer and obesity, by the remodelling of chromatin structure through mechanisms such as acetylation of lysine on histones. Histone is a protein that acts as a spool that packages DNA into nucleosomes and chromatin. Acetylated histone provides binding sites for transcription factors by neutralizing histone positive charges and loosening the interaction between histone and DNA, thus promoting gene expression [10]. Acetylation of histone 3 lysine 27 (H3K27ac) is known as a potent activation mark for browning-related genes, such as Ucp1 [11,12,13,14]. H3K27ac distinguish active enhancers by allowing higher DNA access (open chromatin), which favours transcriptional activation [14,15,16]. Conversely, histone deacetylation condenses chromatin structure, suppressing gene expression [17,18]. Deacetylation of histone is mainly mediated by enzymes called histone deacetylase (HDAC) [18]. So far, there are 18 mammalian classical Zn2+-dependent HDAC that have been characterized and divided into four classes (class I–IV) based on similarity in structure to yeast HDAC [18]. Although structurally similar, the function of each HDAC homologue might differ depending on the complex HDAC formed.
Interestingly, there is accumulating evidence that HDAC inhibitor (HDI) can stimulate browning of adipose tissue in BAT and WAT [11,12,19,20,21,22,23,24] which suggest the inhibition of HDAC might be closely related to browning. However, the detailed mechanism is still unclear and how β-AR stimulation might regulate HDAC activity is unknown. Indeed, in this study we show for the first time that β-AR stimulation also trigger similar HDAC inhibitory activity that benefited Ucp1 expression in beige adipocyte.
2. Results
2.1. β-AR Stimulation-Induced Acetylation of Histone 3 Lysine 27 Favouring Open Chromatin Structure in the Ucp1 Promoter Region
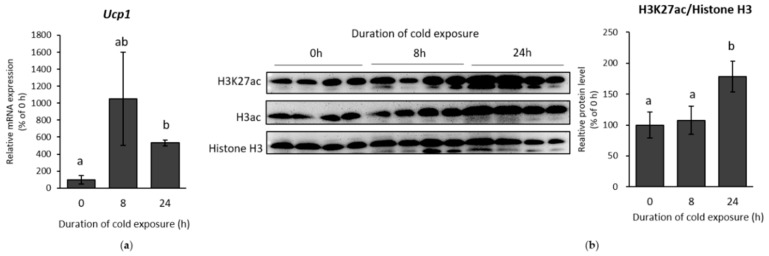
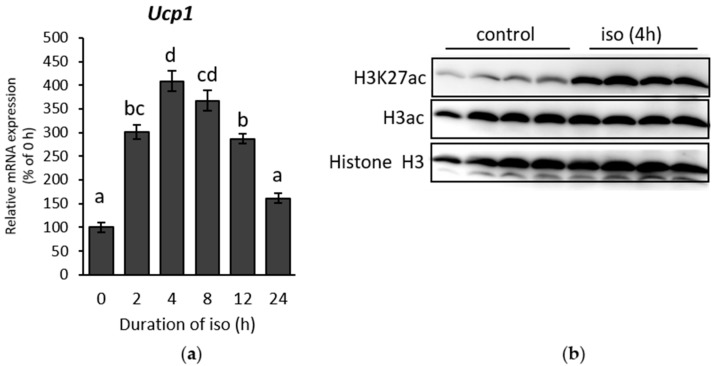
β-AR were first stimulated by exposing mice to cold (10 °C) for 8 or 24 h. Inguinal white adipose tissue (IWAT) were then harvested. Browning in IWAT was successfully stimulated, indicated by the upregulation of Ucp1 expression (Figure 1A). H3K27ac, a signature of histone activation, was also induced after 24 h cold exposure (Figure 1B). Furthermore, total histone H3 acetylation (H3ac), which includes several lysine sites, was also increased, suggesting a major transcriptional activation of genes during cold exposure. Similar results from the initial stage of cold exposure (0–24 h) in IWAT were also obtained in immortalized primary inguinal white adipose tissue cell (IWAT cell) treated with the β-AR agonist isoproterenol. The treatment showed an increment of Ucp1 in time-dependent manner until 4 h (Figure 2A). On the other side, H3K27ac was also significantly increased when Ucp1 was maximally expressed after 4 h isoproterenol addition (Figure 2B).
Figure 1.
Histone acetylation state under cold stimulation in inguinal white adipose tissue (IWAT). (a) Uncoupling protein 1 (Ucp1) expression and (b) histone 3 lysine 27 acetylation (H3K27ac) level from the mice exposed to cold (10 °C) at different time points. Protein band were quantified by ImageJ. Data are presented as mean ± S.E.M. (error bars). n = 3–6 in each group. Different letters indicate significant difference (p < 0.05) according to one-way ANOVA followed by the Tukey-Kramer multiple comparison test. Same letters indicate non-significant difference.
Figure 2.
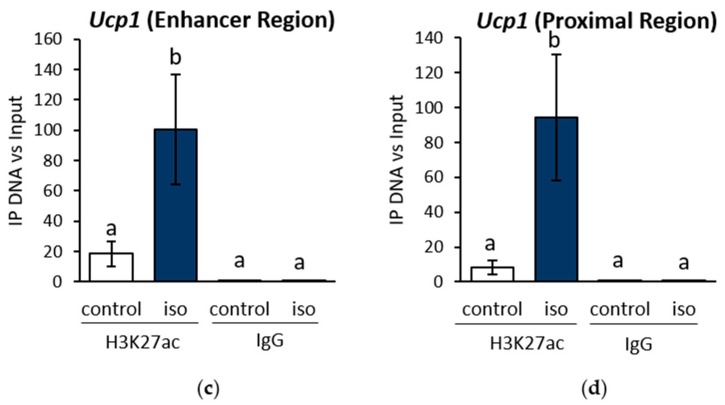
Histone acetylation state under β-adrenergic receptor (β-AR) stimulation in IWAT cell. (a) Ucp1 expression after induction by 10 µM β-AR agonist isoproterenol (iso) over time, and H3K27ac level in (b) whole cells, (c) Ucp1 enhancer region, and (d) Ucp1 proximal region after induction by 10 µM β-AR agonist isoproterenol (iso) for 4 h. IgG act as a mock control. Data are presented as mean ± S.E.M. (error bars). n = 4–6 in each group. Different letters indicate significant difference (p < 0.05) according to one-way ANOVA followed by the Tukey-Kramer multiple comparison test. Same letters indicate non-significant difference.
To further confirm the association between the increased histone acetylation in H3K27 and Ucp1 upregulation during β-AR stimulation, a chromatin immunoprecipitation (ChIP) assay was performed to investigate if H3K27ac also increases within the Ucp1 promoter region in IWAT cell. Two notable sites at the Ucp1 promoter region (the enhancer and proximal regions) were selected to investigate histone acetylation in H3K27 and thus predict Ucp1 chromatin state. The results showed that H3K27ac was considerably increased both in the Ucp1 enhancer (Figure 2C) and proximal (Figure 2D) regions. In addition, peroxisome proliferator-activated receptor gamma coactivator 1 alpha (Pgc1α) mRNA upregulation upon β-AR stimulation, was also accompanied by an increase of H3K27ac in the Pgc1α of the cAMP response element (CRE) region, although the difference was not significant (Figure S1). This result may indicate that H3K27ac regulates not only Ucp1 but other browning genes. Nevertheless, these results concluded an open chromatin structure for Ucp1 transcriptional activation as marked by a significant increase of H3K27ac during β-AR stimulation in beige adipocyte.
2.2. β-AR-Stimulated Ucp1 Transcriptional Activation Is Associated with Inhibition of Class I But not Class II HDAC in IWAT Cell
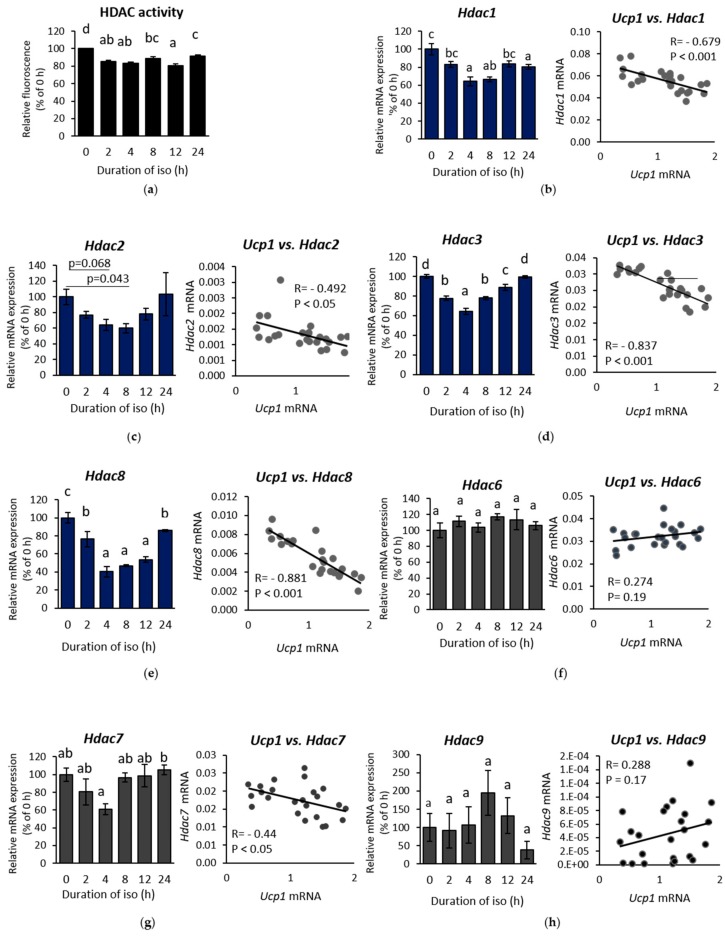
After confirming favourable chromatin state for Ucp1 expression during β-AR stimulation, we investigated the role of HDAC, one of the main regulators of histone acetylation. HDAC activity was suppressed under isoproterenol induction (Figure 3A), displaying the opposite pattern to the level of Ucp1 measured in the time-course experiment (Figure 2A). These data established a negative correlation between Ucp1 and HDAC activity. To investigate the regulation behind HDAC inhibitory activity, we first examined the mRNA levels of class I and II HDACs, as HDIs, which previously reported to induce browning, mainly inhibit these two classes [17,25,26]. Almost all class I HDAC mRNAs, except Hdac2, were suppressed significantly upon isoproterenol treatment (Figure 3B–E, left side). In addition, Pearson’s correlation showed a strong negative correlation between class I HDAC mRNAs and Ucp1 expression (Figure 3B–E, right side), which was consistent with the result from the measurement of HDAC activity (Figure 3A). On the contrary, class II HDAC mRNAs were barely altered and showed weaker correlation with Ucp1 (Figure 3F–H). These results accentuate the importance of class I HDAC specificity, as confirmed by the remarkable increase of Ucp1 expression after treatment with the class I HDAC inhibitor MS275 (Figure S2A), while also significantly decreasing HDAC activity (Figure S2B) in IWAT cell. Yet, among class I HDAC, Hdac3 and Hdac8 showed the highest correlation with Ucp1 (Figure 3D,E right side).
Figure 3.
β-AR stimulation regulates histone deacetylase (HDAC) expression in IWAT cell. (a) HDAC activity, class I HDAC mRNA: (b) Hdac1, (c) Hdac2, (d) Hdac3, and (e) Hdac8; and class II HDAC mRNA: (f) Hdac6 (g) Hdac7, and (h) Hdac9 expression after induction by 10 µM iso in a time-course experiment. HDAC mRNA expression (left side) and its association with Ucp1 (right side) based on Pearson’s correlation. Data are presented as mean ± S.E.M. (error bars). n = 4–8 in each group. Different letters indicate significant differences (p < 0.05) according to one-way ANOVA followed by the Tukey-Kramer multiple comparison test. Same letters indicate non-significant difference.
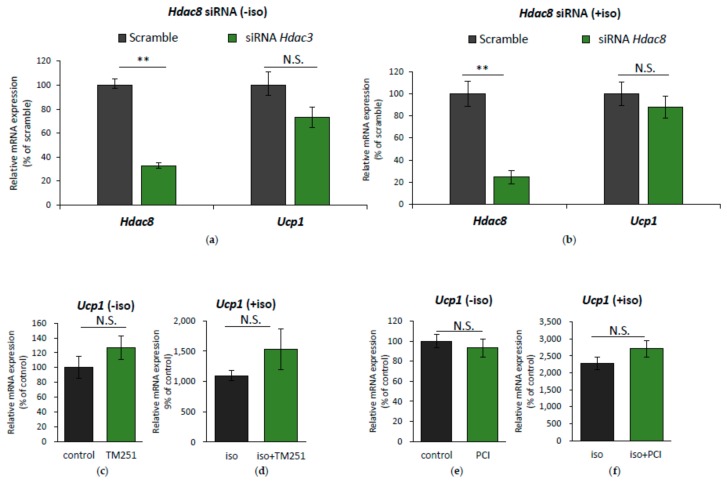
2.3. HDAC8 Might not Be Involved in Ucp1 Regulation in IWAT Cell
Of the two candidates of genes for Ucp1 regulation, Hdac3 and Hdac8, we first investigated the specific role of Hdac8. Initially, IWAT cell was transfected with Hdac8 siRNA to mimic Hdac8 downregulation, as shown by isoproterenol treatment, and Ucp1 expression was measured. However, Hdac8 siRNA interference failed to improve Ucp1 expression, both on a basal level (Figure 4A) or under isoproterenol stimulation (Figure 4B). During isoproterenol induction, although Hdac8 mRNA was downregulated (Figure 3E left side), HDAC8 protein level was barely changed (Figure S3), which might explain why Ucp1 expression was unaffected by Hdac8 siRNA.
Figure 4.
HDAC8 modification has no effect on Ucp1 regulation in IWAT cell. Hdac8 and Ucp1 expression after transfection with Hdac8 siRNA for 24 h without (a) or with (b) 10 µM iso induction for 2 h. Ucp1 expression after treatment with 20 μM HDAC8 activator TM251 without (c) or with (d) 10 µM iso induction for 4 h. Ucp1 expression after treatment with 5 μM HDAC8 inhibitor PCI34051 (PCI) for 24 h without (e) or with (f) 10 µM iso induction at the last 4 h. Data are presented as mean ± S.E.M. (error bars). n = 4–6 in each group. ** indicates significant difference (p < 0.01) according to unpaired-t test. N.S., not significant.
Previous reports have shown that HDAC8 activity is highly regulated through post-translational modification by phosphorylation, mediated by protein kinase A (PKA) [27,28], which is activated in β-AR stimulation [2]. Phosphorylation of HDAC8 resulted in a decrease of HDAC8 activity [27], which was shown upon isoproterenol treatment of IWAT cell (Figure S3), and this was further altered by PKA inhibition by H89. However, as HDAC8 activity was reduced, rescuing its activity by adding the HDAC8 coactivator TM251 barely altered Ucp1 expression (Figure 4C,D). Additionally, IWAT cell was also treated with the selective HDAC8 inhibitor, PCI34051, under isoproterenol induction. Yet, we observed no significant change in Ucp1 expression (Figure 4E,F). These data suggest that HDAC8 might not be involved in promoting Ucp1 expression during β-AR stimulation.
2.4. HDAC3 Inhibition Plays a Major Role in Ucp1 Transcriptional Activation During β-AR Stimulation in IWAT Cell
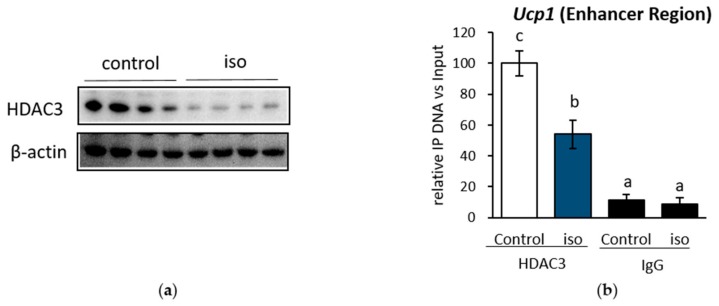
When IWAT cell was treated with isoproterenol, Hdac3 mRNA was significantly downregulated (Figure 3D, left side). Consistently, the level of HDAC3 protein was also reduced (Figure 5A). We investigated whether this decrease in HDAC3 protein level affected HDAC3 recruitment to the Ucp1 enhancer region at the same site of induced histone activation mark H3K27ac, as shown in Figure 2C. The results showed that the HDAC3 recruitment level in the Ucp1 enhancer region (Figure 5B) was inhibited significantly after isoproterenol addition.
Figure 5.
HDAC3 protein level was decreased under β-AR stimulation in IWAT cell. (a) HDAC3 protein level, and its recruitment level in (b) the Ucp1 enhancer region after treatment with 10 μM iso for 4 h. Data are presented as mean ± S.E.M. (error bars). n = 4–6 in each group. β-actin act as loading control and IgG as a mock control. Different letters indicate significant differences (p < 0.05) according to one-way ANOVA followed by the Tukey-Kramer multiple comparison test. Same letters indicate non-significant difference.
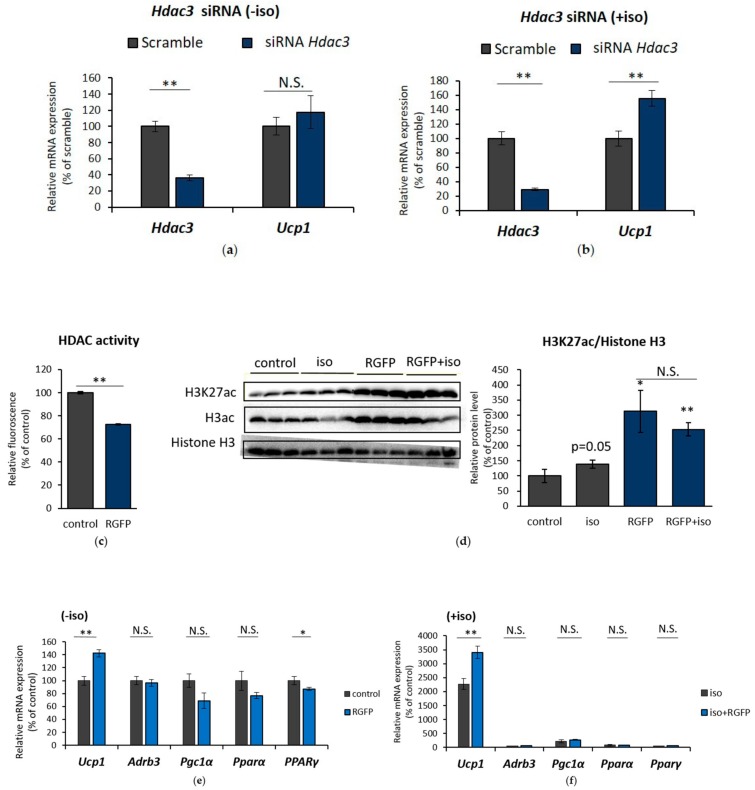
Next, to specifically examine the role of Hdac3 mRNA downregulation on Ucp1 expression, Hdac3 expression was interrupted by transfecting Hdac3 siRNA into IWAT cell. Although not at the basal level (Figure 6A), Hdac3 siRNA successfully increased Ucp1 expression under isoproterenol stimulation (Figure 6B). Next, to further confirm the significance of HDAC3-specific inhibition of histone acetylation and Ucp1 regulation, a selective inhibitor of HDAC3 (RGFP966) was used in IWAT cell. HDAC3 inhibitor treatment showed a significant decrease in HDAC activity (Figure 6C) and escalated H3K27 acetylation (Figure 6D). Unlike Hdac3 siRNA, HDAC3 inhibitor alone was sufficient to induce Ucp1 expression even at the basal level (Figure 6E), while also successfully enhancing Ucp1 under isoproterenol induction (Figure 6F). Interestingly, neither adrenoceptor beta 3 (Adrb3), nor several influential coactivators of Ucp1, such as Pgc1α, PPARα, and PPARγ benefitted upon HDAC3 inhibition (Figure 6E,F), although class I HDAC inhibition by MS275 significantly upregulated Adrb3 (Figure S2A). These results suggest that HDAC3 might be specifically and directly involved in Ucp1 upregulation during β-AR stimulation.
Figure 6.
HDAC3 inhibition resulted in Ucp1 upregulation in IWAT cell. Hdac3 and Ucp1 expression after transfection with Hdac3 siRNA for 24 h without (a) or with (b) 10 µM iso induction for 2 h. (c) HDAC activity after treatment with HDAC3 inhibitor RGFP966 (RGFP) for 24 h. (d) H3K27ac level after induction by 10 µM iso for 4 h, 5 μM RGFP for 24 h, or both compounds. Protein band were quantified by ImageJ. Browning-related gene expression after treatment with 5 μM RGFP for 24 h without (e) or with (f) 10 µM iso induction in the last 4 h. Data are presented as mean ± S.E.M. (error bars). n = 3–6 in each group. *, ** indicate significant differences at p < 0.05 and p < 0.01, respectively, according to unpaired-t test. N.S., not significant.
3. Discussion
The favourable open chromatin structure for Ucp1 transcriptional activation after β-AR stimulation has been previously reported in cultured brown adipocyte, demonstrated by a significant increase of H3K27ac in both the Ucp1 and Pgc1α promoter regions [12]. However, which factors regulate behind this phenomenon is still unclear. The potential of HDAC, as one of the main regulators of histone acetylation, to regulate browning in adipose tissue has been suggested by several studies that tested HDAC inhibitory compounds in both BAT and WAT [11,20,24]. Moreover, many HDIs have shown to be beneficial in alleviating obesity and various diseases [20,21,22,23,29,30,31,32,33]. All of these reports suggest the possibility of β-AR activation to also possess HDAC inhibitory activity which could explain the escalated H3K27ac phenomenon under this stimulation. Indeed, we showed for the first time that HDAC was inhibited during β-AR stimulation in beige adipocyte (IWAT cell) and thus played an active role in Ucp1 regulation, notably through HDAC3. HDAC3 was consistently suppressed at both the mRNA and protein levels, which resulted in the lower HDAC3 recruitment to the Ucp1 enhancer region. The reduced HDAC3 recruitment thus directly cause an increased acetylation of H3K27 in the same site of Ucp1 enhancer region and promote Ucp1 transcriptional activation, as shown in this study. The Ucp1 enhancer region (~2631–2343 bp upstream) is known as the peroxisome proliferator-activated receptor (PPAR) response element coactivated by PGC1α [34,35]. The suppression of HDAC3 in this region might promote PGC1α coactivator complex binding to the Ucp1 enhancer and thus activate Ucp1 transcription.
Interestingly, Hdac3 interference by siRNA did not result in Ucp1 upregulation at the basal level, but improved Ucp1 expression under isoproterenol stimulation, suggesting that Hdac3-regulated Ucp1 transcriptional activation is dependent on β-AR stimulation. HDAC3 inhibitor treatment successfully enhanced Ucp1 expression in both conditions. HDI decrease HDAC activity by disrupting the formation of the HDAC corepressor complex and hence it failed to be recruited to chromatin [36]. The same mechanism similarly happened to HDAC3 recruitment level to Ucp1 promoter when stimulated with isoproterenol, as shown in this report. Thus, the reduction of HDAC3 recruitment level to the chromatin is important for Ucp1 transcriptional activation, as its loss greatly affected histone acetylation state. HDAC3 might also specifically regulate Ucp1 expression, because the treatment of HDAC3 inhibitor barely altered the mRNA expression of other browning markers such as Adrb3, Pgc1α, Pparα, and Pparγ. On the other hand, pan-HDAC inhibitor [20] or class I HDAC inhibitor MS275 (Figure S2) [24] was shown to upregulate not only Ucp1, but also Adrb3 mRNA expression and other browning-related markers.
A previous report has shown that HDAC3-deficient mice underwent significant re-modelling of the WAT metabolic pathway to resemble BAT (browning) without β-AR stimulation [13]. This study thus highlights a negative regulation of HDAC3 to WAT browning. However, the reason why loss of HDAC3 was capable to induce browning remains unclear. Accordingly, our study clarifies the outcome of WAT browning in HDAC3 deficient mice might be because it has the same effect of HDAC3 suppression originated from β-AR stimulation to regulate Ucp1 expression. Interestingly, the other study showed that the ablation of HDAC3 in mice during cold exposure (4 °C, 24 h) failed to activate the thermogenesis program in BAT [37]. In contrast, this study suggests a positive regulation of HDAC3 to browning. The different reports raise a question regarding the positive or negative role of HDAC3 in browning. To address the issue of the opposite role of HDAC3, our study showed that H3K27ac was also differentially regulated during initial and chronic stage of cold exposure. The level of H3K27ac increased at the initial stage of cold challenge (0–24 h, 10 °C) but decreased after long exposure (48–96 h, 10 °C), although Ucp1 expression was stably increased (Figure S4). We thus suggest that the opposite HDAC3 regulation in browning might associate with a different regulation of H3K27ac during initial and chronic stage of cold exposure. Our in vitro study matched the initial stage of H3K27ac of cold stimulation in vivo. In the initial stage of cold exposure, the regulation of histone acetylation could be important for inducing Ucp1 expression, as several studies [11,13], including this report, have suggested. Further research is needed to confirm the regulation of HDAC3 and H3K37ac in relation to Ucp1 expression during chronic cold exposure.
Besides Hdac3, Hdac1, Hdac7, and Hdac8 mRNA also showed a good association with Ucp1 expression. However, HDAC1 and HDAC7 protein level were barely changed under isoproterenol treatment (Figure S5). In addition, HDAC1 specific inhibitor showed a negative effect on Ucp1 expression. (Figure S6). Although HDAC1 deficiency has been reported to be involved in regulating BAT activation [12], it might be regulated differentially in beige adipocyte, as seen in this study. The role of HDAC1 in WAT browning, especially in beige adipocyte, should be confirmed in the future. Interestingly, isoproterenol treatment clearly mediated phosphorylation of HDAC8 through PKA, which has been established to decrease HDAC8 activity and resulted in hyperacetylation of histone H3 and H4 or non-histone protein [27]. It is also known that protein kinase A (PKA) is stimulated during β-AR stimulation, which resulted in the induction of Ucp1 expression [2,3,5,38,39]. However, we found that HDAC8 might not be involved directly in Ucp1 regulation. Additional research is needed to investigate the consequences of HDAC8 downregulation, particularly on non-histone substrates. Apart from 3600 possible acetylation sites (which served as HDAC substrate) that have been identified, 1750 site are non-histone protein [26,40,41,42,43]. Although we have demonstrated the HDAC inhibition in mediating acetylation of histone H3K27, the possibility of HDAC to also induce acetylation of non-histone substrate and their subsequent effect cannot be ruled out completely.
Several pan-HDAC inhibitors have been approved for drug use and more HDAC inhibitors are under clinical trials, intended for cancer treatment [44]. Proportionally, HDAC inhibitor-based therapies have been recognized to be applicable to treat human disease. However, there is a concern over the side effects of pan-HDAC inhibitor [45], shifting the interest to more specific HDAC target. To address this issue, our study showed HDAC3 as a potential, more specific target for WAT browning. Many studies have also concluded HDAC3 as an emerging target for inflammation, insulin-resistance, and type 2 diabetes [31,46,47,48,49,50] which are closely related to obesity. Future study might be directed to the capability of the HDAC3 inhibitor to induce browning of WAT, as our study highly suggested, especially in humans. It has been known that browning of WAT has been targeted not only to treat obesity, but also related metabolic disorders including insulin resistance, inflammation and type-2 diabetes. Accordingly, our study added a solid background of the HDAC3 hidden potential in regulating Ucp1 expression under β-AR stimulation.
4. Materials and Methods
4.1. Materials
All chemicals were obtained from Nacalai Tesque (Nacalai Tesque, Kyoto, Japan), Wako (Wako Pure Chemicals, Osaka, Japan), Corning (Corning, Corning, NY, USA), Qiagen (Qiagen, Hilden, Germany), Invitrogen (Invitrogen, Carlsbad, CA, USA), and Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). TM251 was purchased from Active Motif (Active Motif, Tokyo, Japan). PCI34051, and RGFP966 were acquired from Cayman (Cayman Chemical, Ann Arbor, MI, USA).
4.2. Animal Experiments
Mice were kept in a temperature-controlled room at 23 °C ± 1 °C with a 12 h light/dark cycle and free access to food (standard diet) and water. To stimulate β-AR activation, 14-week-old male C57BL/6N mice (SLC, Shizuoka, Japan) were exposed to cold (10 °C). Mice were sacrificed and inguinal white adipose tissue (IWAT) was harvested for mRNA and protein analysis. The mice were handled in accordance with procedures approved by the Animal Research committee of Kyoto University (Permission number: 29–62; 20 April 2012).
4.3. Cell Culture
Primary pre-adipocyte of IWAT was immortalized by transfecting pBabe-puro largeTcDNA retrovirus containing SV40 largeT antigen. Successfully transfected clone was screened based on puromycin resistance. Immortalized primary inguinal white adipose tissue cell (IWAT cell) was maintained in a humidified 5% CO2 atmosphere at 37 °C using basic medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Maintenance medium consisted of basic medium supplemented with 0.25 μg/mL puromycin. Two days after confluence, cells were differentiated by stimulation with 0.5 mM 1-methyl-3-isobutylxanthine, 0.25 µM dexamethasone, 10 μg/mL insulin, 1 nM triiodo-l-thyronine (T3), 0.5 μM rosiglitazone (Rosi), and 125 µM indomethacin for 48 h. The media was then replaced by growth medium (basic medium containing 5 µg/mL insulin, 1 nM T3, and 0.5 µM Rosi) for another 48 h. After that, the media was changed every 2 days with basic medium supplemented with 5 µg/mL insulin and 1 nM T3. Generally, the differentiation process took 6–7 days. β-AR stimulation was induced by isoproterenol addition in serum free medium.
4.4. RNA Preparation and Quantification of Gene Expression
RNA was prepared and quantified as previously described [51,52]. Total RNA was extracted from cultured cells in 12-well plates or tissue according to the phenol-chloroform extraction method. RNA expression was quantified by real-time PCR using a LightCycler System (Roche Diagnostics, Mannheim, Germany) with SYBR Green fluorescence signal detection. All mRNA signals were normalized to a 36b4 internal control. The primer sequences are listed in Table 1.
Table 1.
Primers used for RNA quantification.
| Gene | Forward | Reverse |
|---|---|---|
| Ucp1 | 5′-CAAAGTCCGCCTTCAGATCC-3′ | 5′-AGCCGGCTGAGATCTTGTTT-3′ |
| Adrb3 | 5′-GCACCTTAGGTCTCATTATGG-3′ | 5′-GCGAAAGTCCGGGCTGCGGCAGTA-3′ |
| Pgc1α | 5′-CCCTGCCATTGTTAAGACC-3′ | 5′-TGCTGCTGTTCCTGTTTTC-3′ |
| Pparα | 5′-TCGCGTACGGCAATGGCTTTT-3′ | 5′-CTTTCATCCCCAAGCGTAGGAGG-3′ |
| Pparγ | 5′-GGAGATCTCCAGTGATATCGACCA-3′ | 5′-ACGGCTTCTACGGATCGAAAACT-3′ |
| Hdac1 | 5′-CCCATGAAGCCTCACCGAAT-3′ | 5′-CAAACACCGGACAGTCCTCA-3′ |
| Hdac2 | 5′-CTGTCTCGCTGGTGTTTTGC-3′ | 5′-GTCATTTCTTCAGCAGTGGCT-3′ |
| Hdac3 | 5′-ATGTGCCGCTTCCATTCTGA-3′ | 5′-TGGCATGATGTAGACCACCG-3′ |
| Hdac8 | 5′-ACTTGACCGGGGTCATCCTA-3′ | 5′-AACCGCTTGCATCAACACAC-3′ |
| Hdac6 | 5′-CAGCAGGATTTGCCCACCTA-3′ | 5′-TCTCCAGGACCTCCCAGAAG-3′ |
| Hdac7 | 5′-TGGGGGATCCTGAGTACCTG-3′ | 5′-GTCCACCCTCTAAGGCCAAC-3′ |
| Hdac9 | 5′-CCCACCACACATCACTGGAT-3′ | 5′-TCCATCCTTCCGCCTGAGTA-3′ |
| 36B4 | 5′-TCCTTCTTCCAGGCTTTGGG-3′ | 5′-GACACCCTCCAGAAAGCGAG-3′ |
4.5. Immunoblotting
Western blotting was performed as previously described [53]. Briefly, cells or tissue were lysed and protein was collected after centrifugation. Protein concentration was measured using the DC protein assay (Bio-Rad, Hercules, CA, USA). Denatured protein was then separated and transferred to a polyvinylidene difluoride transfer membrane. The membrane was then blocked, washed, and incubated with the corresponding primary antibody, followed by the appropriate secondary antibody. Anti-histone H3 (acetyl K27) (Abcam, Cambridge, UK), anti-acetyl-histone H3 (Millipore, Burlington, MA, USA), anti-histone H3 (Novus biological, Littleton, CO, USA), anti-HDAC3 (Sigma-Aldrich, St. Louis, MO, USA), and anti-β-actin (Cell Signaling Technology, Danvers, MA, USA) were used as primary antibodies. The secondary antibody staining was visualized using a chemiluminescent horseradish peroxidase (HRP) substrate (Millipore, Burlington, MA, USA).
4.6. HDAC Activity Assay
HDAC activity was measured using an HDAC Cell-Based Activity Assay Kit (Cayman Chemical, Ann Arbor, MI, USA) as described in the manufacturer’s instructions. Briefly, 1 × 104 differentiated IWAT cell was plated in a 96-well black plate, clear bottom (Greiner Bio-One, Kremsmünster, Austria) as recommended by the protocol, and let to set for 6 h overnight before stimulation, and then analysed for HDAC activity.
4.7. Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed according to company protocol (Upstate) with some modification. Cells were first fixed in 1% formaldehyde and then quenched by 125 mM glycine. Cells were collected and resuspended in 1% SDS lysis buffer, and then sonicated to shear DNA into 100–1000 bp fragments. The supernatant was collected and subjected to overnight immunoprecipitation with 4 µg H3K27ac antibody (Abcam, Cambridge, UK), 25 µg HDAC3 antibody (Abcam, Cambridge, UK), or rabbit IgG isotype control (Novus Biological, Littleton, CO, USA) as a mock control, together with Magna ChIP™ Protein A + G Magnetic Beads (Millipore, Burlington, MA, USA) at 4 °C in a rotatory shaker, followed by reverse cross-link and protease K digestion. Eluted DNA was then purified using a MinElute PCR Purification Kit (Qiagen, Hilden, Germany) and analysed by real-time PCR. Primer sequences are listed in Table 2.
Table 2.
Primers Used in Chromatin Immunoprecipitation (ChIP) Assay.
| Gene | Forward | Reverse |
|---|---|---|
| Ucp1 enhancer | 5′-CTCCTCTACAGCGTCACAGAGG-3′ | 5-AGTCTGAGGAAAGGGTTGA-3′ |
| Ucp1 proximal | 5′-CCCACTAGCAGCTCTTTGGA-3′ | 5-CTGTGGAGCAGCTCAAAGGT-3′ |
4.8. Small Interfering RNA (siRNA) Transfection
The transfection of siRNA was performed in mature IWAT cell. After differentiation, cells were re-plated for 80% confluence in a 24-well plate. Hdac3 and Hdac8 siRNA (Table 3) transfection was performed according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). Cells were collected 24 h after transfection.
Table 3.
Small Interfering RNA (siRNA).
| Gene | Sequence |
|---|---|
| Hdac3 | CAGCAUGACAUGUGCCGCUUCCAUU |
| Hdac8 | GACGGAAAUUUGACCGUAUUCUCUA |
4.9. Statistical Analysis
All data were analyzed using Student’s t-test or one-way ANOVA followed by Tukey-Kramer test, when variances were heterogeneous. All data are presented as means ± SEM. Differences were considered significant at p < 0.05.
Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (16K07734, 16H02551). A. Yuliana was supported by the Indonesia Endowment Fund for Education (LPDP) by Ministry of Finance, Indonesia. The authors also thank M. Komori and R. Yoshii for kindly providing technical assistance and secretarial support.
Abbreviations
| β-AR | beta-adrenergic receptor |
| BAT | brown adipose tissue |
| ChIP | chromatin immunoprecipitation |
| CRE | cAMP response element |
| H3ac | histone 3 acetylation |
| H3K27 | histone 3 lysine 27 |
| HDAC | histone deacetylase |
| HDI | HDAC inhibitor |
| IWAT | inguinal white adipose tissue |
| PGC1α | peroxisome proliferator-activated receptor gamma coactivator 1 alpha |
| PKA | protein kinase A |
| PPAR | peroxisome proliferator-activated receptor |
| siRNA | small interfering RNA |
| UCP1 | uncoupling protein 1 |
| WAT | white adipose tissue |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/8/2436/s1.
Author Contributions
Conceptualization, A.Y., H.-F.J. and T.G.; Methodology, A.Y. and S.K.; Analysis, A.Y.; Writing-Original Draft Preparation, A.Y.; Writing-Review & Editing, H.-F.J., W.N., H.T., T.A. and T.G.; Supervision, H.F.J., W.N., H.T., T.A., T.K. and T.G.; Funding Acquisition, T.K. and T.G.
Conflicts of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Cannon B., Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Collins S., Yehuda-Shnaidman E., Wang H. Positive and negative control of Ucp1 gene transcription and the role of b-adrenergic signaling networks. Int. J. Obes. 2010;34:S28–S33. doi: 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- 3.Harms M., Seale P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 4.Kalinovich A.V., de Jong J.M.A., Cannon B., Nedergaard J. UCP1 in adipose tissues: Two steps to full browning. Biochimie. 2017;134:127–137. doi: 10.1016/j.biochi.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Xue B., Coulter A., Rim J.S., Koza R.A., Kozak L.P. Transcriptional Synergy and the Regulation of Ucp1 during Brown Adipcytes Induction in White Fat Depots. Mol. Cell. Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mottillo E.P., Balasubramanian P., Lee Y.-H., Weng C., Kershaw E.E., Granneman J.G. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic β3-adrenergic receptor activation. J. Lipid Res. 2014;55:2276–2286. doi: 10.1194/jlr.M050005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey A.L., Formosa M.F., Van Every B., Bertovic D., Eikelis N., Lambert G.W., Kalff V., Duffy S.J., Cherk M.H., Kingwell B.A. Ephedrine activates brown adipose tissue in lean but not obese humans. Diabetologia. 2013;56:147–155. doi: 10.1007/s00125-012-2748-1. [DOI] [PubMed] [Google Scholar]
- 8.Redman L.M., de Jonge L., Fang X., Gamlin B., Recker D., Greenway F.L., Smith S.R., Ravussin E. Lack of an Effect of a Novel β3-Adrenoceptor Agonist, TAK-677, on Energy Metabolism in Obese Individuals: A Double-Blind, Placebo-Controlled Randomized Study. J. Clin. Endocrinol. Metab. 2007;92:527–531. doi: 10.1210/jc.2006-1740. [DOI] [PubMed] [Google Scholar]
- 9.Peng X.R., Gennemark P., O’Mahony G., Bartesaghi S. Unlock the thermogenic potential of adipose tissue: Pharmacological modulation and implications for treatment of diabetes and obesity. Front. Endocrinol. (Lausanne) 2015;6 doi: 10.3389/fendo.2015.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shahbazian M.D., Grunstein M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 11.Galmozzi A., Mitro N., Ferrari A., Gers E., Gilardi F., Godio C., Cermenati G., Gualerzi A., Donetti E., Rotili D., et al. Inhibition of class i histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62:732–742. doi: 10.2337/db12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F., Wu R., Cui X., Zha L., Yu L., Shi H., Xue B. Histone deacetylase 1 (HDAC1) negatively regulates thermogenic program in brown adipocytes via coordinated regulation of histone H3 lysine 27 (H3K27) deacetylation and methylation. J. Biol. Chem. 2016;291:4523–4536. doi: 10.1074/jbc.M115.677930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari A., Longo R., Fiorino E., Silva R., Mitro N., Cermenati G., Gilardi F., Desvergne B., Andolfo A., Magagnotti C., et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 2017;8:93. doi: 10.1038/s41467-017-00182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calo E., Wysocka J. Modification of enhancer chromatin: What, how and why? Mol. Cell. 2013;49:1–24. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche J., Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 2016;121:451–483. doi: 10.1016/j.ejmech.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 18.De Ruijter A.J.M., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B.P. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling C., Groop L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia Y., Hong J., Li H., Hu Y., Jia L., Cai D., Zhao R. Butyrate stimulates adipose lipolysis and mitochondrial OXPHOS through histone hyperacetylation-associated AR3β activation in high-fat diet-induced obese mice. Exp. Physiol. 2016;2:273–281. doi: 10.1113/EP086114. [DOI] [PubMed] [Google Scholar]
- 21.Chriett S., Zerzaihi O., Vidal H., Pirola L. The histone deacetylase inhibitor sodium butyrate improves insulin signalling in palmitate-induced insulin resistance in L6 rat muscle cells through epigenetically-mediated up-regulation of Irs1. Mol. Cell. Endocrinol. 2017;439:224–232. doi: 10.1016/j.mce.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Rumberger J.M., Arch J.R.S., Green A. Butyrate and other short-chain fatty acids increase the rate of lipolysis in 3T3-L1 adipocytes. PeerJ. 2014;2:e611. doi: 10.7717/peerj.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye J. Improving insulin sensitivity with HDAC inhibitor. Diabetes. 2013;62:685–687. doi: 10.2337/db12-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrari A., Fiorino E., Longo R., Barilla S., Mitro N., Cermenati G., Giudici M., Caruso D., Mai A., Guerrini U., et al. Attenuation of diet-induced obesity and induction of white fat browning with a chemical inhibitor of histone deacetylases. Int. J. Obes. (Lond.) 2016:1–32. doi: 10.1038/ijo.2016.191. [DOI] [PubMed] [Google Scholar]
- 25.Chiocca S., Segré C.V. Regulating the regulators: The post-translational code of class i HDAC1 and HDAC2. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihaylova M.M., Shaw R.J. Metabolic reprogramming by class I and II histone deacetylases. Trends Endocrinol. Metab. 2013;24:48–57. doi: 10.1016/j.tem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H., Rezai-Zadeh N., Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol. Cell. Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandl A., Heinzel T., Krämer O.H. Histone deacetylases: Salesmen and customers in the post-translational modification market. Biol. Cell. 2009;101:193–205. doi: 10.1042/BC20080158. [DOI] [PubMed] [Google Scholar]
- 29.Henagan T.M., Stefanska B., Fang Z., Navard A.M., Ye J., Lenard N.R., Devarshi P.P. Sodium butyrate epigenetically modulates high-fat diet-induced skeletal muscle mitochondrial adaptation, obesity and insulin resistance through nucleosome positioning. Br. J. Pharmacol. 2015;172:2782–2798. doi: 10.1111/bph.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong J., Jia Y., Pan S., Jia L., Li H., Han Z., Cai D., Zhao R. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget. 2016;7:56071–56082. doi: 10.18632/oncotarget.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leus N.G.J., Van Der Wouden P.E., Van Den Bosch T., Hooghiemstra W.T.R., Ourailidou M.E., Kistemaker L.E.M., Bischoff R., Gosens R., Haisma H.J., Dekker F.J. HDAC 3-selective inhibitor RGFP966 demonstrates anti-inflammatory properties in RAW 264.7 macrophages and mouse precision-cut lung slices by attenuating NF-κB p65 transcriptional activity. Biochem. Pharmacol. 2016;108:58–74. doi: 10.1016/j.bcp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardinale J.P., Sriramula S., Pariaut R., Guggilam A., Mariappan N., Elks C.M., Francis J. HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension. 2010;56:437–444. doi: 10.1161/HYPERTENSIONAHA.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X., Xiao X., Li N., Li Y. Histone deacetylases inhibitors (HDACis) as novel therapeutic application in various clinical diseases. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2017;72:60–72. doi: 10.1016/j.pnpbp.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Villarroya F., Iglesias R., Giralt M. PPARs in the control of uncoupling proteins gene expression. PPAR Res. 2007;2007 doi: 10.1155/2007/74364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Delcuve G.P., Khan D.H., Davie J.R. Roles of histone deacetylases in epigenetic regulation: Emerging paradigms from studies with inhibitors. Clin. Epigenet. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmett M.J., Lim H., Jager J., Richter H.J., Adlanmerini M., Peed L.C., Briggs E.R., Steger D.J., Ma T., Sims C.A., et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546:544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao W., Medvedev A.V., Daniel K.W., Collins S. beta-adrenergic activation of p38 MAP kinase in adipocytes: CAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 map kinase. J. Biol. Chem. 2001;276:27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 39.Chen H.Y., Liu Q., Salter A.M., Lomax M.A. Synergism between cAMP and PPAR γ signalling in the initiation of UCP1 gene expression in HIB1B brown adipocytes. PPAR Res. 2013;2013 doi: 10.1155/2013/476049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulated major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 41.Wolfson N.A., Ann Pitcairn C., Fierke C.A. HDAC8 substrates: Histones and beyond. Biopolymers. 2013;99:112–126. doi: 10.1002/bip.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang W.M., Yao Y.L. Beyond histone and deacetylase: An overview of cytoplasmic histone deacetylases and their nonhistone substrates. J. Biomed. Biotechnol. 2011;2011 doi: 10.1155/2011/146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarti A., Oehme I., Witt O., Oliveira G., Sippl W., Romier C., Pierce R.J., Jung M. HDAC8: A multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 2015;36:481–492. doi: 10.1016/j.tips.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Mottamal M., Zheng S., Huang T.L., Wang G. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules. 2015;20:3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakrabarti A., Melesina J., Kolbinger F.R., Oehme I., Senger J., Witt O., Sippl W., Jung M. Targeting histone deacetylase 8 as a therapeutic approach to cancer and neurodegenerative diseases. Future Med. Chem. 2016;8:1609–1634. doi: 10.4155/fmc-2016-0117. [DOI] [PubMed] [Google Scholar]
- 46.Sharma S., Taliyan R. Histone deacetylase inhibitors: Future therapeutics for insulin resistance and type 2 diabetes. Pharmacol. Res. 2016;113:320–326. doi: 10.1016/j.phrs.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Christensen D.P., Dahllof M., Lundh M., Rasmussen D.N., Nielsen M.D., Billestrup N., Grunnet L.G., Poulsen T.M. Histone Deacetylase (HDAC) Inhibition as a Novel Treatment as Novel Treatment for Ddiabetes Mellitus. Mol. Med. 2011;17:1. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leus N.G.J., Zwinderman M.R.H., Dekker F.J. Histone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammation. Curr. Opin. Chem. Biol. 2016;33:160–168. doi: 10.1016/j.cbpa.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arguelles A.O., Meruvu S., Bowman J.D. Are epigenetic drugs for diabetes and obesity at our door step? Drug Discov. Today. 2016;21:499–509. doi: 10.1016/j.drudis.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Sathishkumar C., Prabu P., Balakumar M., Lenin R., Prabhu D., Anjana R.M., Mohan V., Balasubramanyam M. Augmentation of histone deacetylase 3 (HDAC3) epigenetic signature at the interface of proinflammation and insulin resistance in patients with type 2 diabetes. Clin. Epigenet. 2016;8:125. doi: 10.1186/s13148-016-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirai S., Uemura T., Mizoguchi N., Lee J.Y., Taketani K., Nakano Y., Hoshino S., Tsuge N., Narukami T., Yu R., et al. Diosgenin attenuates inflammatory changes in the interaction between adipocytes and macrophages. Mol. Nutr. Food Res. 2010;54:797–804. doi: 10.1002/mnfr.200900208. [DOI] [PubMed] [Google Scholar]
- 52.Kang M.S., Hirai S., Goto T., Kuroyanagi K., Lee J.Y., Uemura T., Ezaki Y., Takahashi N., Kawada T. Dehydroabietic acid, a phytochemical, acts as ligand for PPARs in macrophages and adipocytes to regulate inflammation. Biochem. Biophys. Res. Commun. 2008;369:333–338. doi: 10.1016/j.bbrc.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Yang H.E., Li Y., Nishimura A., Jheng H.F., Yuliana A., Kitano-Ohue R., Nomura W., Takahashi N., Kim C.S., Yu R., et al. Synthesized enone fatty acids resembling metabolites from gut microbiota suppress macrophage-mediated inflammation in adipocytes. Mol. Nutr. Food Res. 2017;61:1–13. doi: 10.1002/mnfr.201700064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.