Abstract
Repetitive DNA, formerly referred to by the misnomer “junk DNA”, comprises a majority of the human genome. One class of this DNA, alpha satellite, comprises up to 10% of the genome. Alpha satellite is enriched at all human centromere regions and is competent for de novo centromere assembly. Because of its highly repetitive nature, alpha satellite has been difficult to achieve genome assemblies at centromeres using traditional next generation sequencing approaches, and thus, centromeres represent gaps in the current human genome assembly. Moreover, alpha satellite DNA is transcribed into repetitive non-coding RNA and contributes to a large portion of the transcriptome. Recent efforts to characterize these transcripts and their function have uncovered pivotal roles for satellite RNA in genome stability, including silencing “selfish” DNA elements and recruiting centromere and kinetochore proteins. This review will describe the genomic and epigenetic features of alpha satellite DNA, discuss recent findings of noncoding transcripts produced from distinct alpha satellite arrays, and address current progress in the functional understanding of this oft-neglected repetitive sequence. We will discuss unique challenges of studying human satellite DNAs and RNAs and point toward new technologies that will continue to advance our understanding of this largely untapped portion of the genome.
Keywords: satellite, centromere, kinetochore, variation, transcription, non-coding RNA, repetitive DNA, epiallele
Introduction
Repetitive DNA can be classified into two groups based on structure: tandem or interspersed. Tandem repeats are clusters of individual sequence units that are adjacent to one another and organized as either direct repeats (head-to-tail) or inverted repeats (head-to-head and tail-to-tail). Interspersed repeats lacking iterated, hierarchical structure are scattered throughout the genome and are nonadjacent. Repetitive DNA can also be classified by the level of repetition: highly repetitive or middle repetitive. These two fractions were initially distinguished by their differential reassociation rates (C0t values) after high temperature melting, with highly repetitive sequences, such as telomeres and satellite DNA, reannealing more quickly than moderately repetitive DNA, such as retrotransposons and rDNA genes (Britten and Kohne, 1968). Satellite DNA was first identified in the 1970s as a distinct low-density buoyancy band that separated from bulk genomic DNA in cesium chloride density gradients (Yasmineh and Yunis, 1974). This class of DNA encompasses many types of highly repetitive tandem repeats. Satellite DNA is generally classified by three major characteristics: 1) repeat unit size, 2) sequence composition, and 3) total block or array length. Here, we will focus on the major form of satellite DNA in the human genome, alpha satellite. This sequence is predominantly enriched in and around primary constrictions and contributes to essential chromosomal functions such as centromere and kinetochore assembly and heterochromatin formation.
Alpha satellite DNA is composed of fundamental 171bp monomeric repeat units. It is present as either higher-order repeat units (HORs) that are composed of organized, tandemly repeated 171bp monomers or stretches of divergent monomers that lack any overarching organizational pattern (Willard, 1985, Waye and Willard, 1987, Alexandrov et al., 1993b, Rudd et al., 2003b) (Fig. 1a). These two types of alpha satellite DNA are typically located near one another, with unordered monomeric alpha satellite often sandwiched between a large block of HOR alpha satellite and chromosome arms (Schueler et al., 2001, Rudd et al., 2003b, Ross et al., 2005) (Fig. 1a).
Figure 1. Array and chromosome-specific organization of alpha satellite DNA.
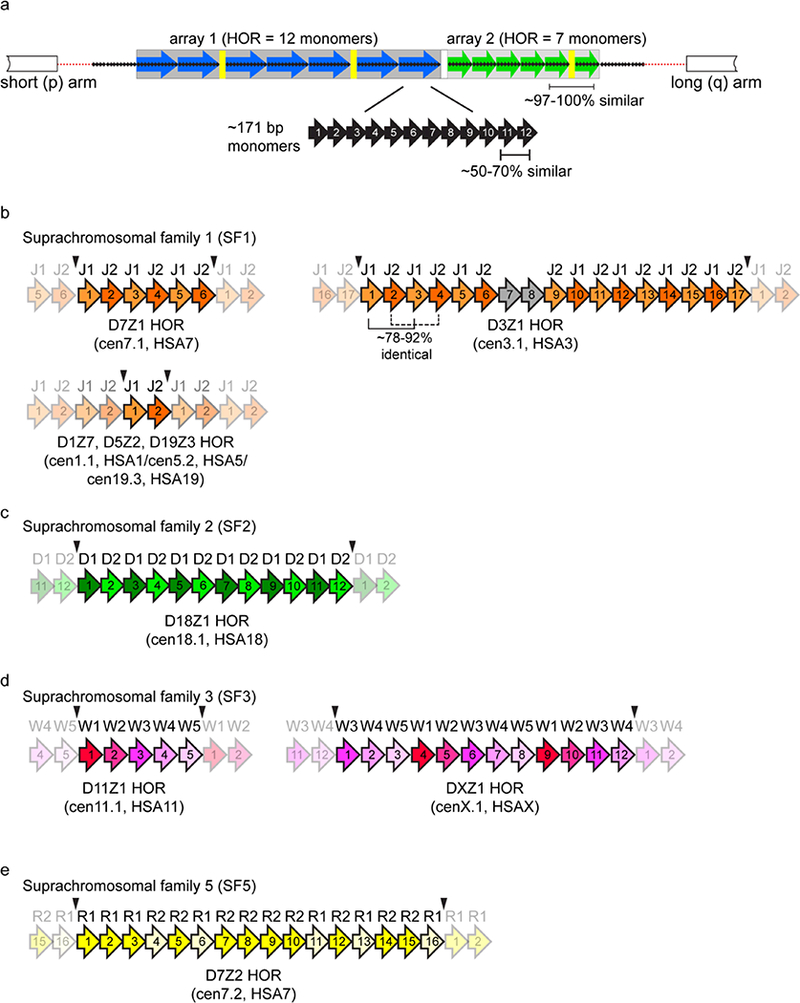
a) Schematic of the general organization of alpha satellite DNA arrays at human centromere regions. Human chromosomes can have either one or more distinct higher order repeat (HOR) arrays. HORs are array- and chromosome-specific. A defined number of individual monomers (black arrows) that are 50–70% identical in sequence are arranged tandemly to form a HOR unit; shown here as either a 12 monomer HOR (blue array) or 7 monomer HOR (green array). Monomers are numbered by their position within the HOR and not based on their homology between two distinct HORs. The HORs are repeated hundreds to thousands of times to create homogenous arrays in which HOR within a given array are 97–100% identical. The HOR array is flanked by degenerate alpha satellite DNA monomers (small black arrays) that lack hierarchical structure and separate the HOR array from the chromosome arrays. HOR arrays are interrupted by other repetitive elements, such as transposable elements (TEs, yellow) but the extent of TE distribution across arrays is unclear due the lack of linear, contiguous assemblies of endogenous alpha satellite arrays.
b) Alpha satellite HOR arrays have been classified into suprachromosomal families (SF) that are related based on monomer type and organization. SF1 arrays are organized as alternating dimers of J1 and J2 monomers (D7Z1, cen7.1), although variation in the regular organization of monomers occurs on some chromosomes, like the D3Z1 (cen3.1) array of Homo sapiens chromosome 3 (HSA3). Additionally, the HORs can be shared among chromosomes, such as the D1Z7 (cen1.1) array that is also present as D5Z2 (cen5.2) on human chromosome 5 (HSA5) and D19Z3 (cen19.3) on HSA19. Each array-specific HOR unit is operationally defined by restriction enzyme sites (black arrowheads) that demarcate the last monomer of one HOR unit and the first monomers of the next HOR unit. Opaque shading illustrates the linear, reiterated nature of HOR units.
c) SF2 is composed of a different dimeric structure based on D1 and D2 monomers. D18Z1 (cen18.1) on HSA18 has SF2 organization.
d) SF3 is based on a pentameric organization of monomers W1-W5. D11Z1 (cen11.1) is an example of a perfect pentameric HOR unit, while DXZ1 has an irregular organization of W1-W5 monomers.
e) SF5 arrays are defined by R1 and R2 monomers, although they largely lack the dimeric organization observed for SF1 and SF2 arrays. Some arrays have HOR unit structure, such as the D7Z2 (cen7.2) array of HSA7.
D_chromosome_Z_number is the original Human Genome Project locus definition of alpha satellite arrays. The newer UCSC Genome Browser annotations of distinct HOR arrays (cen_chromosome number.array number) are also included.
HOR alpha satellite arrays are comprised of a defined number of divergent 171bp monomers arranged head-to-tail (Willard, 1985) (Fig. 1a) The individual monomers within a HOR unit have 50–70% identity and can be distinguished such that HOR unit length is determined by where the next monomer shows nearly total sequence identity to the first monomer in the HOR (Fig 1a). Outside of the higher order arrays, monomers are randomly arranged and span the region between the homogeneous array and the chromosome arm (Fig. 1a). Monomeric alpha satellite is often interspersed with repetitive elements, such as transposable elements and other types of satellite DNA, such as satellite I and gamma satellite DNA (Trowell et al., 1993, Schueler et al., 2001, Kim et al., 2009) (Fig. 1a). Although HOR alpha satellite arrays are largely homogenous, they are often punctuated by transposable elements, either between HOR units or within the units themselves (Schueler et al., 2005, Miga, 2015, Jain et al., 2018).
HOR units of alpha satellite DNA have been operationally defined by restriction enzyme sites that cut usually once within the HOR and demarcate the last monomer of one HOR and the first monomer of the next HOR [reviewed by (Willard and Waye, 1987b)] (Fig. 1b-e). On each chromosome, HOR units are repeated, largely uninterrupted, hundreds to thousands of times, resulting in a large, linear and homogeneous array of highly identical copies of tandem HOR units (Aldrup-MacDonald et al., 2016). The large alpha satellite array at the centromere is a genetic locus and has been designated and referenced using the following nomenclature: DNA segment (D), chromosomal assignment (#), complexity of DNA (Z for repetitive), and sequential number (1, 2, 3...) to confer uniqueness of DNA segment (Willard et al., 1985).
Each human chromosome is associated with a unique alpha satellite HOR
Alpha satellite is often thought to be identical across all centromeres of the human karyotype, but in fact, it exhibits several types of variation or polymorphism that illustrate its complexity, distinctive organization within the human genome, and most importantly, its chromosome-specificity. The sequence of a HOR, the number, type, and order of monomers that define the HOR unit, and the overall copy number of the HOR (i.e the number of times the HOR is repeated) confer chromosome-specificity of alpha satellite. HORs within a chromosome-specific array differ in sequence by only a few percent, however, HORs between non-homologous chromosomes are only 50–70% identical (Manuelidis, 1978, Willard, 1985). For instance, 12 monomers comprise the HOR array DXZ1 that defines the centromere of the Homo sapiens X chromosome (HSAX) (Waye and Willard, 1985, Schueler et al., 2001, Miga et al., 2014). Among all copies of HSAX in the population, the 2.0 kb DXZ1 HOR is repeated between 750 and 2100 times, yielding total array size lengths that range from 1.5Mb to 4.2 Mb (Fig. 2). Total array size polymorphisms exist between homologs even within the same individual, and the DXZ1 arrays on the two HSAXs in a female will often differ in overall array size (Wevrick and Willard, 1989). When DXZ1 arrays from unrelated males were compared, no two HSAX chromosomes showed identical sizes or haplotypes (Mahtani and Willard, 1990). Similar inter-homolog and inter-individual array size polymorphisms exist for alpha satellite arrays on autosomes, such that array lengths represent a continuum of sizes that can vary 10- to 20-fold (Fig. 2; Table 1). Despite inter-homolog/inter-individual variation, alpha satellite array size polymorphisms are heritable and largely stable in meiosis, such that the segregation of specific homologs can be tracked through families based solely on alpha satellite array sizes (Wevrick and Willard, 1989, Marcais et al., 1991, Mahtani and Willard, 1998) (Fig. 3a, b). Likewise, the identity of specific human chromosomes that have been moved to somatic cell hybrid backgrounds can be verified by alpha satellite array size polymorphisms (Aldrup-MacDonald et al., 2016). These centromeric polymorphisms are useful markers for monitoring inheritance of individual chromosomes (Fig. 3b).
Figure 2. Chromosome-specific alpha satellite arrays in the human population are polymorphic in size.

The number of monomers that comprise a HOR differs between chromosomes, conferring chromosome specificity. For example, DXZ1 (HSAX) is defined by a 12 monomer (12-mer) HOR (2kb), while D7Z1 (HSA7) is defined by a 6-mer (1kb) HOR. Within the population and even between homologs of the same individual, the total array size (i.e. the number of times a HOR is repeated) is different. The reported sizes of DXZ1 on single HSAX chromosomes range from 1.3Mb (650 copies of DXZ1 HOR) to 4.2Mb (2100 copies). Likewise, total array size of D7Z1 ranges from 1.5 – 3.8Mb, such that in a given individual, D7Z1 on one HSA7 homolog may be 1.8Mb and 3.5Mb on the other homolog.
Table 1.
Classification of higher order repeat (HOR) alpha satellite arrays on human chromosomes.
| Chrom | Array/Locus Name |
Ucsc designation |
SF | HOR size (kb) |
HOR Monomer # |
Total array size (Mb)* |
Centromere Function | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HAC assay |
CENP ChlPa |
CENP IFa |
Epiallele | |||||||
| 1 | D1Z7b | cen1.1_cen5.2_cen19.3 | 1 | 0.34 | 2 | 0.4–1.1 | n.d. | yes | yes | yes |
| 1 | D1Z5 | cen1.2 | 3 | 1.9 | 11 | 0.4–1.5 | n.d. | yes | yes | yes |
| 2 | D2Z1 | cen2.1 | 2 | 1.4 | 8 | 1.0–2.9 | n.d. | yes | yes | n.a. |
| 3 | D3Z1 | cen3.1 | 1 | 2.9 | 17 | 1.0–3.1 | n.d. | yes | yes | n.a. |
| 4 | D4Z1 | cen4.1 | 2 | 3.2 | 19 | 2.3–4.4 | n.d. | yes | yes | n.a. |
| 5 | D5Z1c | cen5.1 | 5 | 2.2 | 13 | n.d. | no | no | no | n.d. |
| 5 | D5Z2b | cen5.2_cen1.1_cen19.3 | 1 | 0.34 | 2 | n.d. | n.d. | yes | yes | n.d. |
| 6 | D6Z1 | cen6.1 | 1 | 3.0 | 18 | 2.3–37 | n.d. | yes | yes | n.a. |
| 7 | D7Z1 | cen7.1 | 1 | 1.0 | 6 | 1.5–3.8 | n.d. | yes | yes | yes |
| 7 | D7Z2 | cen7.2 | 5 | 2.7 | 16 | 0.1 −0.6 | yes | yes | yes | yes |
| 8 | D8Z2 | cen8.1 | 2 | 1.9 | 11 | 1.5–2.2 | n.d. | yes | yes | n.d. |
| 9 | D9Z4 | cen9.1 | 2 | 1.2 | 7 | 1.8–2.2 | n.d. | yes | yes | n.a. |
| 10 | D10Z1 | cen10.1 | 1 | 1.4 | 8 | 1.4–2.5 | n.d. | yes | yes | n.a. |
| 10 | D10Z? | cen10.2 | 1 | 3.0 | 18 | n.d. | n.d. | n.d. | n.d. | n.d. |
| 11 | D11Z1 | cen11.1 | 3 | 0.85 | 5 | 1.8–4.8 | yes | yes | yes | n.d. |
| 11 | D11Z? | cen11.2 | 3 | 2.0 | 12 | n.d. | n.d. | n.d. | n.d. | n.d. |
| 12 | D12Z3 | cen12.1 | 1 | 1.4 | 8 | 2.2–4.3 | n.d. | yes | yes | n.a. |
| 13 | D13Z1d | cen13.1_cen21.1 | 2 | 1.9 | 11 | 1.1 −3.0 | n.d. | yes | yes | n.a. |
| 14 | D14Z9e | cen14.2_cen22.1 | 2 | 1.4 | 8 | 2.0 (avg) | n.d. | yes | yes | n.a. |
| 15 | D15Z3 | cen15.1 | 5 | 2.4 | 14 | 0.1 −0.8 | n.d | no | no | n.d. |
| 15 | D15Z4 | cen15.2 | 2 | 4.4 | 26 | 1.1 −2.7 | n.d. | yes | yes | n.d. |
| 16 | D16Z2 | cen16.1 | 1 | 1.7 | 10 | 0.4–2.0 | n.d. | yes | yes | n.d |
| 17 | D17Z1 | cen17.1 | 3 | 2.7 | 16 | 1.1 −4.2 | yes | yes | yes | yes |
| 17 | D17Z1-B | cen17.2 | 3 | 2.4 | 14 | 0.5–1.5 | yes | yes | yes | yes |
| 17 | D17Z1-C | cen17.3 | 3 | 2.4 | 14 | 0.5–1.5 | n.d. | no | yes | n.d. |
| 18 | D18Z1 | cen18.1 | 2 | 2.0 | 12 | 1.3–2.5 | n.d. | yes | yes | n.d. |
| 18 | D18Z2 | cen18.2 | 2 | 1.7 | 10 | 0.4–2.8 | n.d. | no | no | n.d. |
| 19 | D19Z1c | cen19.1_cen5.1 | 5 | 2.9 | 17 | n.d. | no | no | no | n.d. |
| 19 | D19Z3b | cen19.3_cen1.1_cen5.2 | 1 | 0.34 | 2 | n.d. | n.d. | yes | yes | n.d. |
| 20 | D20Z1 | cen20.1 | 2 | 2.7 | 16 | n.d. | n.d. | n.d. | n.d. | n.d. |
| 20 | D20Z2 | cen20.2 | 2 | 1.0 | 6 | 0.5–1 | n.d. | yes | yes | n.d. |
| 21 | D21Z1d | cen21.1_cen13.1 | 2 | 1.9 | 11 | o CO I o |
yes | yes | yes | n.a. |
| 22 | D22Z1e | cen22.1_cen14.2 | 2 | 1.4 | 8 | n.d. | n.d. | yes | yes | n.d. |
| 22 | D22Z2 | cen22.2 | 2 | 2.0/2.7 | 12/16 | 2.0 (avg) | n.d. | yes | yes | n.d. |
| X | DXZ1 | cenX.1 | 3 | 2.0/2.2 | 12/13 | 1.3–4.2 | yes | yes | yes | n.a. |
| Y | DYZ3 | cenY.1 | 4 | 5.8 | 34 | 0.2–1.2 | no | yes | yes | n.a. |
sizes compiled from the literature using pulsed-field gel electrophoresis-Southern blotting, quantitative FISH, and/or sequence reads;
on endogenous chromosomes
share the same HOR sequence
share the same HOR sequence
share same HOR sequence
share same HOR sequence
n.a., not applicable
n.d., not determined
Figure 3. Stability of total alpha satellite array sizes and their use as chromosomal markers.
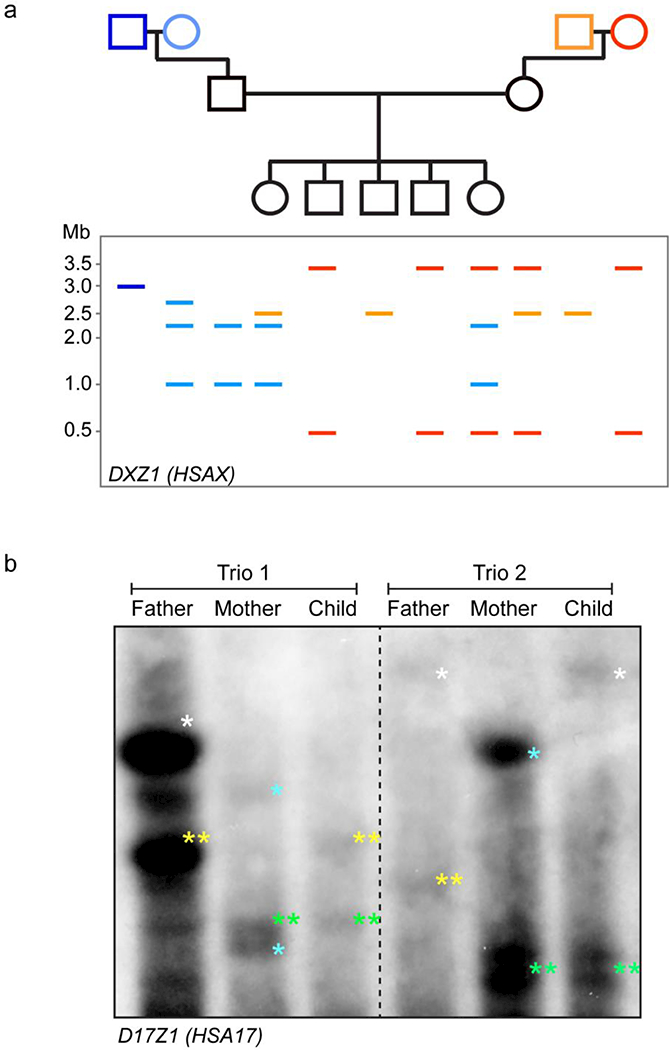
a) Cartoon representation of a multigenerational pedigree and pulsed field gel electrophoresis (PFGE)-Southem blotting analysis of DXZ1 total array sizes. High molecular weight (HMW) DNA can be cut with enzymes that release the multi-megabase array as one or a few high molecular weight fragments that are resolved over many days using PFGE. Southern blotting with a probe specific to DXZ1 will reveal the unique sizes of DXZ1 arrays in each individual. Males (squares) typically show a single band or two bands (dark blue or light orange) that can be added together to yield total array size. Females will exhibit additional bands (red or light blue) since they have two HSAX chromosomes. Each HSAX can be tracked through the family based on DXZ1 array sizes. These type of analyses have revealed the extreme stability of alpha satellite array sizes as well as their usefulness as genetic markers in familial studies (Wevrick and Willard, 1989).
b) PFGE-Southern blot of D17Z1 arrays sizes and segregation of specific HSA17 chromosomes through two different families (trios). The fathers’ and mothers’ D17Z1 alleles in each family are marked by different colored asterisks and the homolog that each child inherited can be tracked by the size of the D17Z1 bands.
Organization of alpha satellite into suprachromosomal subfamilies based on sequence variation and monomer organization
Alpha satellite monomers differ in sequence by 10–40%, depending on their sequence identity to the first described human alpha satellite sequences (Wu and Manuelidis, 1980). Although any two adjacent monomers may differ significantly in sequence, similarities in monomer sequence and order, but not the total number of monomers in a HOR unit, are shared among different chromosomes. From sequence analysis of hundreds of individual monomers, twelve consensus alpha satellite monomers have been designated: J1, J2, D1, D2, W1, W2, W3, W4, W5, M1, R1, and R2 (Alexandrov et al., 1988, Alexandrov et al., 1991, Alexandrov et al., 1993b, Rosandic et al., 2006, Shepelev et al., 2015). These monomers fall into five suprachromosomal groups or families, that are defined by sequence homology and linear order of the monomers that create a HOR that is similar and can even be shared between chromosomes (Table 1). The three main suprachromosomal families (SF1–3) represent the majority of “functional” alpha satellite HORs found at the centromere core (i.e. kinetochore forming region). SF1–3 represent two dimeric and one pentameric HOR configurations. SF4 and SF5 are monomer families that usually flank the functional HOR arrays and separate them from the chromosome arms (Alexandrov et al., 1993b, Shepelev et al., 2009). SF4 is purely monomeric in structure (i.e. does not form HOR units), however, SF5 monomers can be organized into HOR units, but can also exhibit an irregular organization lacking HOR structure (see below) (Rosandic et al., 2006).
SF1 has a dimeric organization and is comprised of monomers designated J1 and J2 (Alexandrov et al., 1988, Alexandrov et al., 1991) (Fig. 1b). J1 and J2 monomers share 70% identity; however, all J1 monomers show greater than 80% sequence identity to each other (Alexandrov et al., 1993a). SF1 alpha satellite is present on nine human chromosomes (HSA1, 3, 5, 6, 7, 10, 12, 16, and 19) (Looijenga et al., 1992, Alexandrov et al., 1993b). The typical organization of SF1 is alternating J1 and J2 monomers, with a different total number of J1/J2 monomers creating chromosome-specificity. For example, the HOR unit size of D1Z7 (or cen1.1, the current alpha satellite classification in the human genome assembly hg38) is 340bp (a perfect J1-J2 dimer), but the HOR size for D7Z1 (cen7.1) is 1020bp (6-mer; J1-J2-J1-J2-J1-J2) (Fig. 1b). While HORs within the same SF can differ in unit size (i.e. number of monomers) creating chromosome-specificity, some HORs are shared among more than one chromosome. For example, the same dimeric HOR that defines D1Z7 (cen1.1) is also present on HSA5 as D5Z2 (cen5.2) and HSA19 as D19Z3 (cen19.3) (Fig. 1b). Even within a suprachromosomal group where monomer homology is high, variation exists. For example, the J1-J2 periodicity of the 2.9kb 17-mer HOR of D3Z1 (HSA3) is not perfect and is instead interrupted by two monomers (X1, X2) that lack homology to existing any of the monomer families (Alexandrov et al., 1993a) (Fig. 1b). This departure from the canonical suprachromosomal organization is an excellent example of chromosome-specific structural variation that can occur among alpha satellite arrays.
SF2 is a second dimeric subfamily that is composed of D1 and D2 monomers and is present on eleven chromosomes (HSA2, 4, 8, 9, 13, 14, 15, 18, 20, 21, and 22) (Fig. 1c). D1 and D2 monomers are distinct from J1 and J2 monomers. Within their respective groups, D1 or D2 monomers are on average 88% similar in sequence, while when compared to each other, D1 versus D2 monomers are less similar (Alexandrov et al., 1991). Like SF1, some alpha satellite HORs, like D18Z2 on HSA18 and D8Z2 on HSA8, depart from the alternating pattern of D1/D2 monomers (Rosandic et al., 2006, Shepelev et al., 2015).
SF3 is a “pentameric” subfamily comprised of monomers W1, W2, W3, W4, and W5 (Fig. 1d). These five monomers were initially described as A-E monomers in the first description of the alpha satellite arrays from HSA17 and HSAX (Willard and Waye, 1987a). SF3 is found on four chromosomes (HSA1, 11, 17, and X). However, only the HOR of D11Z1 is organized as a perfect W1-W5 5-mer (Waye et al., 1987a), the 12-mer DXZ1 HOR, 11-mer D1Z7 HOR, and 16-mer D17Z1 exhibit a combination of the pentamer structure with single or double monomer duplications or triplications (Waye and Willard, 1985, 1986b, Alexandrov et al., 2001) (Fig. 1d).
SF4 is an unordered subfamily composed of monomers that were originally defined by a consensus monomer M1 (Alexandrov et al., 1993b). M1 monomers exhibit more sequence identity to D2 and W4 monomers than to the other types of monomers in SF1–3. However, the M1 monomers are classified as a distinct group because they are more homologous to each other (average 81% sequence identity) than they are to similar monomers in other suprachromosomal families. SF4 monomers also do not exhibit higher order periodicity, further emphasizing that they belong to a unique subfamily. Alpha satellite arrays composed of M1 monomers are present on HSA13, 14, 15, 21, and 22 and Y (Alexandrov et al., 1993b). They are positioned adjacent, or peripheral, to larger, higher order arrays that form the centromere core (Vissel and Choo, 1991, 1992). SF4 arrays have been described as “dead” or “inactive” arrays, and yet DYZ3 is in this suprachromosomal family, is organized as a 34-mer HOR, and assembles a functional centromere.
Finally, SF5 is a subfamily characterized by R1 and R2 monomers (Alexandrov et al., 2001). It displays irregular monomer order, rather than an alternating dimeric R1/R2 arrangement (Fig. 1e). SF5 arrays are present on multiple chromosomes, including HSA5, HSA7, HSA15, and HSA19 (Table 1) and are typically smaller in size than SF1–3 alpha satellite arrays. Like SF4, SF5 arrays usually lack HOR unit structure. However, HOR structure is present on a few distinct chromosomes (i.e. 13-mer D5Z1, 16-mer D7Z2), and there is recent evidence that SF5 arrays such as D7Z2 on HSA7 can support centromere function in vitro and in vivo (Hayden et al., 2013, McNulty et al., 2017).
Genomic variation within HORs of specific alpha satellite arrays
The suprachromosomal family classifications illustrate that variation within the alpha satellite DNA is common and complex, due to monomeric differences and chromosome-specific differences in HOR unit size and monomer order and organization. However, on a given chromosome, the primary HOR unit can also exhibit size polymorphisms, such that variant HORs and canonical HORs can both be present within the same array (Durfy and Willard, 1987, Waye et al., 1987c, Choo et al., 1990, Ge et al., 1992, Alexandrov et al., 1993a). HOR size variants are most likely the result of deletions caused by unequal exchange (Waye and Willard, 1986a, b, Warburton et al., 1993).
HSA17 is a premier example of HOR polymorphisms within the D17Z1 array. The predominant HOR unit on D17Z1 is a 16-monomer (16-mer) (Waye and Willard, 1986b, Willard et al., 1986). However, less prevalent 15-mer and 14-mer HORs are present on many D17Z1 arrays, as well as 13-mers, 12-mers, and rare 11-mers (Warburton and Willard, 1995). The 13-mer HOR unit is the most abundant after the 16-mer. The HOR size polymorphisms create D17Z1 haplotypes, with the 16-/15-/14-mer comprising a haplotype (Haplotype I) found on 65% of HSA17s within the population. Arrays that contain 16-/15-/14-mers plus additional 13-mers are present on 35% of HSA17s (Haplotype II) (Waye and Willard, 1986a, Warburton and Willard, 1995). Single nucleotide changes in specific monomers have also been mapped to distinct HOR units. For example, a SNP that creates a HindIII site in monomer 13 of D17Z1 is present in a small subset of 16-mer HORs and in a large number of 13-mer HORs (Warburton and Willard, 1992, 1995).
Alpha satellite arrays on other chromosomes also show HOR size and sequence variation (Waye and Willard, 1986a, Durfy and Willard, 1987, Waye et al., 1987c, Choo et al., 1990, Marcais et al., 1991, Charlieu et al., 1992, Ge et al., 1992, Alexandrov et al., 1993a, Greig et al., 1993, Marcais et al., 1993). For example, within DXZ1, a subset of HORs have acquired a HindIII site and those HORs have been amplified to create a polymorphic domain within the predominantly homogeneous, canonical DXZ1 array (Durfy and Willard, 1987). On HSA8, D8Z2 is present primarily as a 1.9kb HOR, but variant 2.5kb and 3.9kb HORs are also detected along with the 1.9kb HOR within some D8Z2 arrays in the population (Ge et al., 1992). These size and sequence variants, and their spatial relationships to one other within a given HOR array, raise questions regarding the effect of genomic variation on alpha satellite function. On HSA17, HOR variants (SNP and size variants) within D17Z1 are associated with defective kinetochore architecture and the reduced ability to recruit or maintain centromere proteins (Maloney et al., 2012, Aldrup-MacDonald et al., 2016) (see “Centromeric epialleles” section below). Why genomic variation would affect the ability of alpha satellite to form or maintain kinetochore is not clear. Long-range organization or transcription of the HOR units (wild-type versus variant) across the entire alpha satellite array could influence the competence of an alpha satellite array for centromere assembly and kinetochore formation (Sullivan et al., 2017). It is well-established that variation within regulatory and genic regions influences gene expression. Studies that identify and characterize structural and sequence polymorphisms within alpha satellite DNA and their fundamental effects on basic chromosome function will undoubtedly expand our understanding of genomic variation and the function of the non-coding regions of the human genome.
Alpha satellite function: relationship with centromere and kinetochore proteins
Maintenance of human centromere assembly and kinetochore formation is accomplished through the recruitment of ~100 proteins to alpha satellite DNA regions (Musacchio and Desai, 2017). The centromere can be defined as where unique chromatin is assembled that serves as the foundational platform for recruitment of architectural proteins that provide structure to the kinetochore, a multi-subunit protein network that makes attachments to microtubules and moves chromosomes along spindle microtubules during cell division.
CENP-A, the centromere-specific histone variant, and epigenetic marker of centromere identity
The presence of CENP-A at alpha satellite DNA regions distinguishes the centromere from the remainder of the genome. CENP-A was discovered from sera isolated from CREST (Calcinosis, Raynauds phenomenon, Esophogeal dysmotility, Sclerodactyly, Telangiectasia) syndrome patients. Three antigens were biochemically identified and shown to be centromere components by immunostaining of mitotic cells (Earnshaw and Rothfield, 1985). The 17 kDa species was designated CENP-A, while the other two bands were called CENP-B (80 kDa) and CENP-C (140 kDa). Subsequent studies showed that CENP-A co-purified with nucleosome core particles and histones, implicating it as a centromere-specific histone involved in a fundamental chromatin nucleoprotein complex (Palmer et al., 1987, Palmer et al., 1991). CENP-A is present at all endogenous human centromeres and distinguishes the active centromeres of dicentric chromosomes, solidifying its role as a key centromere protein (Vafa and Sullivan, 1997, Warburton et al., 1997, Ando et al., 2002). In humans, the unique association of CENP-A with alpha satellite DNA extends to its deposition into chromatin, not at S phase when most new histones are incorporated into chromatin, but in late M and G1 phases (Shelby et al., 1997, Shelby et al., 2000, Jansen et al., 2007). The uncoupling of CENP-A synthesis in G2 in humans and its deposition in G1 is important for its loading by the CENP-A specific chaperone protein Holliday Junction Recognition Protein (HJURP) (Dunleavy et al., 2009, Bodor et al., 2013). Maintenance of CENP-A within the centromere is thought to be coordinately regulated by the interaction of CENP-A with other centromere proteins, post-translational modification of centromeric histones, and transcription of alpha satellite DNA (Molina et al., 2016, Ohzeki et al., 2016, McNulty et al., 2017). Proper loading and maintenance of CENP-A is bolstered by its interactions with CENP-B and CENP-C and its spatial location within alpha satellite DNA arrays (Fachinetti et al., 2013, Fachinetti et al., 2015, Ross et al., 2016).
CENP-B, an alpha satellite DNA binding protein
CENP-A exists in a constitutive pre-kinetochore complex with CENP-B and CENP-C (Ando et al., 2002). In mammals, CENP-B is an 80 kDa kinetochore protein that binds to the CENP-B box, a 17-bp sequence motif (5’-T/CTCGTTGGAAA/GCGGGA-3’) (Masumoto et al., 1989). The CENP-B box is present in only a subset of alpha satellite monomers (Muro et al., 1992, Ikeno et al., 1994) on all human chromosomes except HSAY (Muro et al., 1992, Haaf and Ward, 1994). The location of CENP-B boxes varies depending on the chromosome-specific HOR and is directly linked to the HOR structure (Fig. 4). Alpha satellite monomers within each suprachromosomal family have specific sequence and higher order characteristics, but all monomers can be broadly classified into two groups based on their identity to the alpha satellite consensus: A-type and B-type monomers (Rosandic et al., 2006). A-type monomers include J1, D2, W4, W5, M1, and R2 monomers, while B-type consist of J2, D1, W1-W3, and R1 monomers. A and B monomers differ in sequence at positions 35–51, a region that correlates with protein binding. B-type monomers contain CENP-B boxes, while A-type monomers contain a binding site for pJα (Rosandic et al., 2006), a protein that has not been well characterized and whose function is unclear. Interestingly, DYZ3 of HSAY completely lacks monomers that contain CENP-B boxes but does contain monomers that have the pJα motif. Since DYZ3 binds CENP-A and other centromere and kinetochore proteins, it is possible that pJα contributes to kinetochore assembly in a way that remains to be fully defined.
Figure 4. Distribution of CENP-B boxes within different types of alpha satellite HORS.
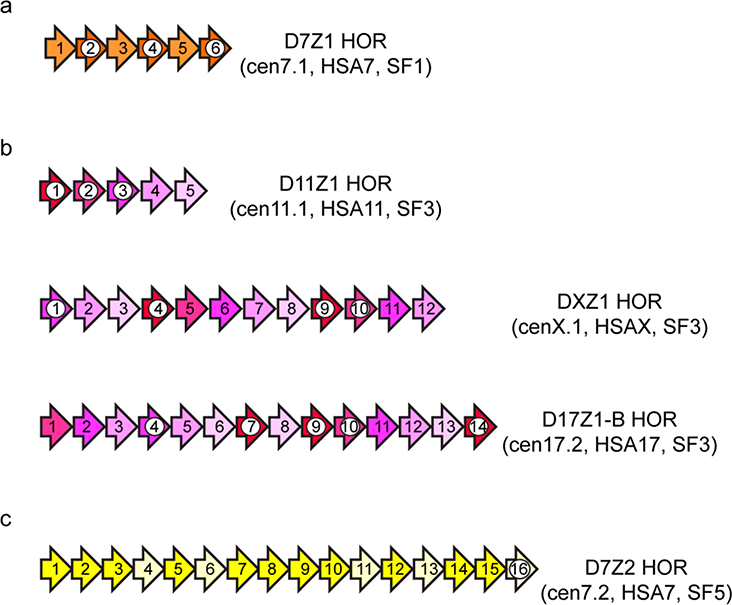
a) Not all alpha satellite monomers contain the 17-bp binding motif of the centromere protein CENP-B. Some HORs, like D7Z1, have a regular, alternating pattern of CENP-B boxes, so that reiteration of the HOR unit yields a large array with dense numbers of CENP-B boxes (monomers with white circles).
b) Pentameric SF3 arrays, like that of D11Z1 (cenll.l), DXZ1 (cenX.l), and the smaller array of D17Z1-B (cenl7.2), have the same number of CENP-B boxes, or more, as the dimeric arrays, but the CENP-B boxes are irregularly spaced. These arrays with irregularly spaced CENP-B boxes are equally competent for centromere assembly.
c) Some HOR arrays, like that of D7Z2 (cen7.2, SF5) have few or no CENP-B boxes and were thought to be “dead arrays” that lack centromere potential. However, D7Z2 (cen7.2) that only has one CENP-B box in monomer 16 can recruit centromere proteins (Thakur and Henikoff, 2018) and assemble a functional centromere (Hayden et al., 2013, McNulty et al., 2017).
Until recently, CENP-B has not been thought to play a functional role in centromeric chromatin, since it is often present at centromeres that have been inactivated (Earnshaw et al., 1989, Sullivan and Schwartz, 1995). It is also present within the additional HOR alpha satellite arrays of multi-array chromosomes, like HSA7 and HSA17. There has been recent, renewed interest in the role of CENP-B in centromere chromatin establishment, structure, or maintenance. New centromere formation depends on CENP-B containing alpha satellite DNA (Ohzeki et al., 2002, Okada et al., 2007). Moreover, CENP-B is thought to position CENP-A nucleosomes and to stabilize CENP-A and CENP-C within centromeric chromatin (Yoda et al., 1998, Okada et al., 2007, Hasson et al., 2013, Fachinetti et al., 2015).
It has been broadly proposed that the presence of CENP-B in alternate monomers in dimeric HOR subfamilies SF1 and SF2 confers enhanced binding and integrity of the constitutive centromere associated-network complex (CCAN). The density of CENP-B boxes within HOR alpha satellite has been correlated with stronger CENP-A enrichment, with the conclusions that dimeric arrays will exhibit the highest CENP-B box density (Thakur and Henikoff, 2018). However, within a given HOR, regardless of suprachromosomal family organization, the CENP-B box is present in only a subset of monomers. It is true that in a HOR array like D7Z1 that has a dimeric configuration (SF1), CENP-B boxes are present in every other monomer (Fig. 4a). However, the alternating arrangement of CENP-B boxes is not the rule. In fact, the distribution of the CENP-B boxes is unique to each chromosome-specific HOR. Within pentameric HORs of SF3, CENP-B boxes are irregularly spaced on HSA11, HSA17, and HSAX (Fig. 4b). The density of CENP-B boxes will also be influenced by total array size, such that a 2Mb array of D7Z1 (dimeric, SF1) will have the same number of CENP-B boxes as a 2Mb array of D11Z1 (pentameric, SF3) that has irregularly spaced CENP-B boxes. Moreover, a lower density of CENP-B boxes within an alpha satellite array does not disqualify it for centromere assembly. Centromeres readily form at the minor array D17Z1-B on epiallele chromosome HSA17 even when the major array D17Z1 has up to three times the number of CENP-B boxes (Aldrup-MacDonald et al., 2016). Likewise, centromere proteins can be enriched at the 16-mer HOR of D7Z2 (SF5) on HSA7 (Thakur and Henikoff, 2018), and centromere assembly at D7Z2 has been shown by HAC assays and on endogenous chromosomes, even though D7Z2 contains a single CENP-B box and neighboring D7Z1 contains many CENP-B boxes (Hayden et al., 2013, McNulty et al., 2017) (Fig. 4c). These findings suggest that arrays with even a few CENP-B boxes are sufficient to confer centromere competence to an array, but also raise the possibility that other aspects of alpha satellite DNA (or RNA) are required for centromere assembly.
CENP-C, a DNA and RNA-binding protein that provides structural integrity and links the inner and outer kinetochore
CENP-C is a member of the constitutive centromere-associated network (CCAN) that links the inner and outer kinetochore and is important for CENP-A recruitment and kinetochore maturation. CENP-C is thought to stabilize CENP-A nucleosomes, through coordinated interactions with CENP-B and CENP-N, a subunit of the CENP-L-N complex (Carroll et al., 2009, Guo et al., 2017, Cao et al., 2018). CENP-C, along with CCAN component CENP-T, provide direct bridges of the inner kinetochore to NDC80/HEC1 in the outer kinetochore (Musacchio and Desai, 2017) CENP-C binds to both alpha satellite DNA and RNA (Politi et al., 2002, Trazzi et al., 2002, Du et al., 2010, Shono et al., 2015, McNulty et al., 2017). CENP-B and CENP-C associate with the same type of alpha satellite DNA (i.e. HOR), but are spatially distinct, suggesting that they interact with distinct HORs or different regions of the same HOR.
Centromeric epialleles: the co-existence of multiple, functionally distinct HOR arrays on single human chromosomes
Each human chromosome contains at least one unique alpha satellite HOR array, with the exceptions of two pairs of chromosomes: HSA13/HSA21 and HSA14/HSA22 (Devilee et al., 1986, Jorgensen et al., 1988, Trowell et al., 1993). HSA13 and HSA21 share the same alpha satellite HOR unit (D13Z1/D21Z1; previously designated αRI, Genbank accession D29750), while the primary alpha satellite array on HSA14 and HSA22 (D14Z1/D22Z1; formerly αXT, Genbank accession M22273) is largely identical. However, centromere regions of these and other chromosomes also contain additional alpha satellite arrays that are distinct from the primary array (Waye et al., 1987b, Choo et al., 1990, Wevrick and Willard, 1991, Vissel and Choo, 1992, Trowell et al., 1993, Pironon et al., 2010). As previously mentioned, distinct alpha satellite arrays on a chromosome can be classified in the same or different suprachromosomal family and are distinguished by monomer sequence and HOR length (i.e. monomer number within the HOR unit). More than half of the chromosomes in the human karyotype (i.e. HSA1, HSA5, HSA7, HSA15, HSA17, HSA18, HSA20) (Choo et al., 1990, Wevrick and Willard, 1991, Slee et al., 2011, Rosenbloom et al., 2015, Shepelev et al., 2015) have more than one HOR array. For instance, HSA17 has three arrays, D17Z1, D17Z1-B, and D17Z1-C; all are SF3 HOR arrays (Rosandic et al., 2006, Shepelev et al., 2009). However, HSA5 contains two arrays D5Z1 (dimeric HOR, SF1) and D5Z2 (monomeric, SF5). Although the presence of multiple arrays on the same chromosome has led to the suggestion that one array is “active/live” and the other is “inactive/dead” (Shepelev et al., 2015), in vitro and in vivo functional studies show that arrays from different suprachromosomal subfamilies can support centromere assembly (Pironon et al., 2010, Hayden et al., 2013, McNulty et al., 2017). Within the population, several chromosomes with multiple alpha satellite arrays often exhibit variation in the alpha satellite site of centromere assembly. These centromeric epialleles have been identified on several multi-array chromosomes including HSA1, HSA7, HSA17, and HSA19 (Pironon et al., 2010, Maloney et al., 2012, Aldrup-MacDonald et al., 2016, McNulty et al., 2017). On HSA17 for example, either D17Z1 or D71Z1-B can be the site of centromere assembly, and in the same individual, one homolog can assemble the centromere at D17Z1 while centromere assembly occurs at D17Z1-B on the other homolog (Maloney et al., 2012). To date, no endogenous chromosomes have been identified in which centromere assembly and kinetochore formation occurs at both arrays simultaneously. Kinetochore formation at the secondary HOR array D17Z1-B is highly correlated with genomic variation (size, sequence) at the larger, primary array D17Z1 (Aldrup-MacDonald et al., 2016). Centromeric epialleles highlight the functional plasticity of alpha satellite and the impact of genomic variation even within highly repetitive DNA arrays.
Alpha satellite DNA and de novo centromere assembly: human artificial chromosomes (HACs)
The identification in the 1980s of alpha satellite arrays at primary constrictions implied that these sequences contributed to centromere function. However, the strongest evidence linking alpha satellite DNA to human centromere assembly came from two distinct chromosome engineering approaches. In the first, successive rounds of telomere-mediated chromosomal truncation were used to modify the X chromosome (HSAX) and Y (HSAY) chromosome, generating a series of derivative chromosomes that, after each round of targeted deletion, contained less HSAX or HSAY chromosome arm material (Brown et al., 1994, Farr et al., 1995, Mills et al., 1999). The smallest HSAX and HSAY minichromosomes to remain mitotically stable contained the alpha satellite DNA arrays DXZ1 and DYZ3, respectively. Since these pioneering studies, additional chromosomes have been truncated to minimal segregation units and used as minichromosomes to study chromosome stability or to house genes to be used in therapeutic applications. These studies strongly connected alpha satellite DNA as the sequence largely responsible for centromere function and chromosome stability.
Complementary experiments performed by two groups took a de novo approach to define sequences required for centromere assembly. Early studies tested the ability of alpha satellite DNA to nucleate functional centromeres by introducing cosmids containing alpha satellite DNA from HSA17 into African green monkey (AGM) cells (Haaf et al., 1992). These experiments resulted in integration of the alpha satellite construct into AGM chromosomes rather than forming an independent chromosome. In subsequent studies, large blocks (100–1000kb) of cloned or synthetic alpha satellite sequences from D17Z1, D21Z1, DYZ3, and DXZ1 were retrofitted onto linear yeast artificial chromosome (YAC) or circular bacterial artificial chromosome (BAC) vectors. Introduction of these artificial chromosome assembly constructs into a human cell line yielded autonomous chromosomes termed human artificial chromosomes (HACs) (Harrington et al., 1997, Ikeno et al., 1998, Masumoto et al., 1998, Schueler et al., 2001, Rudd et al., 2003a) (Fig. 5). HACs containing alpha satellite DNA have been shown to recruit centromere proteins and be continuously stable for over 6 months. Importantly, these studies showed that higher order alpha satellite DNA containing CENP-B boxes, but not higher order arrays lacking the CENP-B binding motif or unordered alpha satellite monomers, could form stable HACs (Fig. 5). Subsequent 2nd and 3rd generation HACs have been created that contain alpha satellite in addition to tetracycline operator (tetO) or lac operon (lacO) sequences (Kononenko et al., 2013, Lee et al., 2013b). The tetO and lacO sequences are bound with high affinity by the tet repressor (tetR) and lac repressor (LacI), respectively, that can be fused to different proteins to track movement and copy number of the HAC (GFP-LacI) or to manipulate the chromatin or protein composition of the HAC (Lee et al., 2013a, Pesenti et al., 2018). With the latter approach, the efficiency of centromere assembly on alpha satellite can be enhanced or inhibited and expression of genes located close to alpha satellite DNA on the HAC can be tested (Kononenko et al., 2013).
Figure 5. Alpha satellite sequence requirements for HAC formation.

HACs are commonly generated by transfection of BACs containing alpha satellite sequence into human cell lines. BACs containing alpha satellite HORs (blue arrays) with CENP-B boxes (white monomer arrows) are the only material sufficient to form a HAC. The resulting HAC contains multimerized BAC sequence. A centromere forms on a portion of the HAC and, like endogenous chromosomes, contains both CENP-A (purple) and H3K4me2/H3K36me2 (red) nucleosomes and is flanked by pericentromeric heterochromatin (green). The centromere can form on both alpha satellite sequence and vector sequence. In contrast, BACs containing alpha satellite HORs that lack CENP-B boxes or monomeric alpha satellite are not sufficient to form stable HACs and are often observed to integrate into chromosome arms,
Chromatin signatures of alpha satellite DNA regions
Genomic DNA is packaged into chromatin through the wrapping of DNA around two copies of each core histone (H2A, H2B, H3, and H4) (Kornberg, 1974). Chromatin can be further compacted by the action of chromatin remodeling proteins. Genomic regions that contain genes are typically packaged into euchromatin that is characterized by more loosely packed nucleosomes and DNase and transcription factor accessibility. Gene-poor regions of the genome are conversely packaged into heterochromatin that is largely refractory to transcription or exhibits distinctive association-dissociation kinetics with transcription factors. Post-translational modifications to histone tails act as signals to recruit appropriate chromatin remodeling proteins and transcription factors to distinct genomic locations. Specific histone modifications demarcate euchromatin versus constitutive heterochromatin. For instance, H3K4 and H3K36 di- and tri-methylation (H3K4me2/3, H3K36me2/3), H3 acetylation (K9, K14), and H4 acetylation (K5, K8, K12, K16) are markers of transcriptionally active, open chromatin (Peterson and Laniel, 2004). Conversely, H3K9me2/3 and H3K27me3 are histone modifications associated with repressive facultative or constitutive heterochromatin. Studies using immunocytological approaches combined with chromatin immunoprecipitation (ChIP) surprisingly revealed that alpha satellite DNA is assembled into different types of chromatin, sometimes on the same array (Lam et al., 2006, Mravinac et al., 2009, Ohzeki et al., 2012, Bailey et al., 2016).
Historically, mammalian repetitive DNA has been considered heterochromatic. However, centromere regions exhibit a histone modification pattern that is distinct from both euchromatin and heterochromatin (Fig. 6a). Centromeric chromatin is defined by the presence of interspersed nucleosomes that contain the canonical histone H3 and the centromere-specific H3 variant CENP-A (Blower et al., 2002) This unique arrangement of interspersed H3 and CENP-A nucleosomes has been termed “centrochromatin” (Sullivan and Karpen, 2004). The H3 histones within centrochromatin contain high levels of H3K4me2 and H3K36me2, two histone modifications associated with transcriptionally permissive chromatin (Lam et al., 2006, Bergmann et al., 2011). Acetylated histone modifications typically present in euchromatin are only transiently associated with centrochromatin and are thought to be important for new CENP-A loading and maintain a boundary to prevent encroachment of heterochromatin into centrochromatin (Molina et al., 2016, Ohzeki et al., 2016, Shang et al., 2016).
Figure 6. Alpha satellite transcription and non-coding RNAs play distinct roles at the centromere and pericentromere throughout the cell cycle.
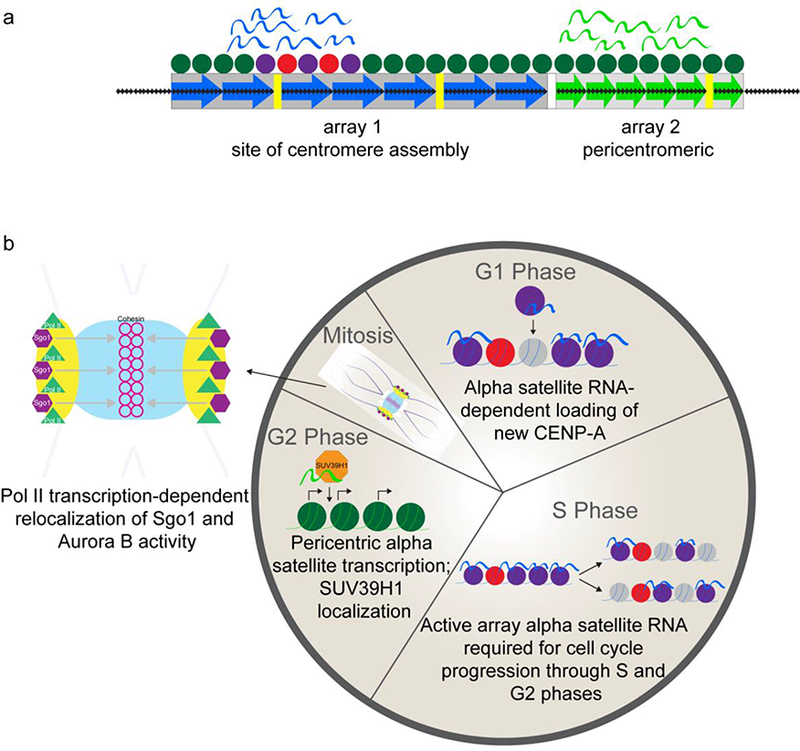
a) Schematic of the dual transcription observed at active and inactive alpha satellite DNA arrays at human centromere regions. The CENP-A domain (red and purple circles) forms on a portion of array 1 (blue arrows) and RNAs produced from this array (blue ribbons) remain associated with the centromere. Adjacent to array 1, array 2 (green arrows) is pericentromeric and associated with heterochromatic nucleosomes (green circles) but, like array 1, produces alpha satellite RNAs (green ribbons) that localize in cis.
b) Summary diagram of the proposed roles of alpha satellite transcription and the resulting non-coding RNAs at each stage of the cell cycle. Alpha satellite RNAs produced from the active array help load new CENP-A at the centromere in early G1. In S phase, CENP-A is distributed semi-conservatively to each daughter strand. Although a precise role for alpha satellite transcription or RNA has not yet been elucidated, the presence of these transcripts is required for normal cell cycle progression through S and G2 phases. Alpha satellite transcription at inactive, pericentric arrays is thought to occur in G2 phase, shortly before the onset of mitosis. These RNAs are required for SUV39H1 (orange octagons) localization to the pericentromere. Sgo1 and Aurora B are both key players in mitosis and have been identified as alpha satellite RNA binding partners. RNAP II-dependent transcription of alpha satellite is involved in relocalizmg Sgo1 (purpl e hexagons) from the kinetochore to cohesin (pink rings) in the inner centromere.
Centrochromatin is adjacent to pericentric heterochromatin enriched for modifications of H3K9me2, H3K9me3, and H3K27me3 (Lam et al., 2006, Ohzeki et al., 2016). In humans, approximately 35% of a given alpha satellite array is assembled into centrochromatin, and heterochromatin forms on the remainder of the array (Lam et al., 2006, Mravinac et al., 2009, Sullivan et al., 2011, Bailey et al., 2016) (Fig. 6a) The boundaries between centrochromatin and heterochromatin within a single alpha satellite array are not clear. Because alpha satellite array sizes are polymorphic, total CENP-A domain sizes vary with alpha satellite size, and the amount of flanking heterochromatin also differs among homologous centromeres. Heterochromatin itself may act as a large chromatin boundary between the core centromere on alpha satellite and the chromosome arms, since depletion or removal of heterochromatin allows centrochromatin to spread and/or chromatin domains to reposition on alpha satellite (Mravinac et al., 2009, Sullivan et al., 2011, Sullivan et al., 2016). Similar to endogenous centromeres, HAC centromeres are assembled into centrochromatin that is flanked by heterochromatin (Lam et al., 2006, Nakano et al., 2008, Ohzeki et al., 2012, Moralli et al., 2013) (Fig. 5). HACs contain nonalpha satellite DNA, including resistance genes and vector sequences. Spreading of centrochromatin onto these neighboring sequences has been observed, suggesting that a continuous domain can assemble even in the absence of a continuous alpha satellite domain. Demarcation of heterochromatin and centrochromatin on HACs and the assembly of new CENP-A appear to be controlled by the interplay of heterochromatin formation by SUV39H1/2 that is antagonized by modification of nearby centrochromatin via the acetyltransferase KAT7/HBO1/MYST2 (Ohzeki et al., 2016). The recruitment of these chromatin modifying enzymes may be controlled by protein-protein interactions and/or by RNAs produced from alpha satellite regions (see below) (Johnson et al., 2017, McNulty et al., 2017).
Transcription of alpha satellite DNA
The enrichment of repetitive regions within heterochromatin has supported the idea that these sequences are transcriptionally silent. However, active transcription appears to be a general feature of many satellite DNAs, including alpha satellite DNA. In fact, satellite RNAs are abundant in mammalian cells and often stably associated with chromatin (Hall et al., 2014). Our understanding from recent studies is that the characteristics and functions of these transcripts are important for specific chromosomal functions, as well as in development and responses to cell stress and cancer.
Non-coding RNAs involved in centromere and kinetochore assembly and function
Studying human centromeric transcription presents a unique challenge due to the structural organization of the centromere. On single alpha satellite HOR array chromosomes, such as HSAX, alpha satellite DNA is incorporated into centrochromatin, where the kinetochore will form, as well as into pericentric heterochromatin. As a result, studies of bulk alpha satellite RNA are unable to determine the chromatin domain from which the RNA originated (centromere or pericentromere), underscoring the need to incorporate protein-association information when analyzing alpha satellite DNA and RNA. Moreover, the existence of multiple distinct arrays on a single chromosome (see “Centromeric epialleles” section above) further complicates the study of the role of alpha satellite DNA in centromere and pericentromere function.
In general, alpha satellite transcripts have been described in many human cell types, although reports of localization, length, binding partners, and function have varied (Wong et al., 2007, Chan et al., 2012, Ideue et al., 2014, Quenet and Dalai, 2014, Liu et al., 2015, McNulty et al., 2017). Initial reports of alpha satellite RNA localization suggested that transcripts were confined to the nucleolus until their relocalization to the centromere at the onset of mitosis via CENP-C (Wong et al., 2007). Alpha satellite has also been reported to localize to centromeres in both interphase and metaphase (Ideue et al., 2014, Quenet and Dalal, 2014, McNulty et al., 2017), co-localizing with key centromere proteins, like CENP-A. Perhaps the most uncertain characteristic of alpha satellite RNA is its binding partners and overall function at the centromere. Two proteins, Aurora B and Sgo1, directly involved in the progression of cell division via dynamic coordination of spindle microtubule attachment and sister chromatid separation, respectively, appear to be regulated by alpha satellite transcription and alpha satellite RNA (Ideue et al., 2014, Liu et al., 2015) (Fig. 6b). The act of RNAPII transcription is required to localize Sgo1 from the outer kinetochore to the inner centromere (Liu et al., 2015). The RNAPII-dependent relocalization of Sgo1 is necessary for full centromeric cohesion. Alpha satellite RNA itself directly associates with Aurora B and alpha satellite RNA depletion leads to abnormal cell shape and errors in cell division (Ideue et al., 2014). Similar results were observed after both minor and major satellite depletion in mouse cells. These studies suggested that alpha satellite transcription and RNA in general are required for proper cell function, but were unable to discriminate between the effects of loss of centromeric RNAs vs pericentromeric RNAs or to demonstrate specific changes in centromere protein recruitment.
Alpha satellite RNAs have also been identified in prenucleosomal complexes (i.e. histone complexes not yet incorporated into DNA to form chromatin) containing CENP-A and HJURP prior to association with centromeric chromatin (Quenet and Dalal, 2014). Active RNAPII was found to be associated with chromatin fibers specifically in early G1, when CENP-A is loaded into chromatin, and to be required for CENP-A and HJURP targeting (Fig. 6b). Prior to assembly into chromatin, CENP-A and HJURP were bound to a 1.3kb putative alpha satellite transcript. Fragments of this sequence colocalized with half of the CENP-A signals visible on chromatin fibers, suggesting that only some centromeres produce the alpha satellite RNA used to recruit new CENP-A. General depletion of alpha satellite RNA using an shRNA to previously published alpha satellite consensus sequences (Waye and Willard, 1987) led to mitotic defects and reduced CENP-A loading, implying an essential role for alpha satellite RNAs in centromere function.
Recently, studies in primary and transformed human cultured cells have shown that alpha satellite arrays produce sequence-specific non-coding transcripts that complex with centromere proteins CENP-A and CENP-C, as well as the alpha satellite DNA binding protein CENP-B (Quenet and Dalal, 2014, McNulty et al., 2017). However, as mentioned previously, human centromere regions often contain multiple, distinct alpha satellite arrays, and even inactive (non-kinetochore forming) alpha satellite arrays produce alpha satellite RNA (Johnson et al., 2017, McNulty et al., 2017) (Fig. 6a). Transcripts from the distinct arrays appear to be parsed into functionally distinct chromatin complexes, since RNA from inactive alpha satellite arrays is not associated with CENP-A or CENP-C. At active, kinetochore-forming arrays, alpha satellite RNA is involved in centromere protein loading (Fig. 6b). The specific regions of the RNAs that interact with CENPs and the binding sites for RNA on these centromere proteins have not yet been identified, although CENP-C is a known RNA-binding protein (Du et al., 2010).
Non-coding alpha satellite RNAs involved in pericentromeric heterochromatin
Early evidence pointing to an RNA component involved in mammalian pericentric heterochromatin maintenance came from the finding that RNase treatment of mammalian nuclei resulted in a loss of heterochromatin and a structural alteration of the pericentromere (Maison et al., 2002). Two histone lysine methyltransferases, SUV39H1 and SUV39H2, are conserved components of heterochromatin that are important regulators of constitutively silent chromatin (Aagaard et al., 1999, Aagaard et al., 2000, Rea et al., 2000, Peters et al., 2001, Peters et al., 2003). SUV39H enzymes catalyze the addition of methyl groups to lysine 9 in histone 3 to form H3K9me2 and H3K9me3. These modified histones serve as binding sites for HP1, that oligomerizes to perpetuate nucleosome chromatin condensation and transcriptional repression (Bannister et al., 2001, Lachner et al., 2001, Canzio et al., 2011). Protein-protein interactions including recruitment of SUV39H by methylated DNA binding proteins establish heterochromatin in non-repetitive regions of the genome (Nan et al., 1997, Fuks et al., 2003). RNA has been implicated as a binding partner of SUV39H, but its role in heterochromatin formation and maintenance have been unclear. Recent evidence suggests that SUV39H1 is bound to single-stranded alpha satellite RNA in human cells (Johnson et al., 2017). Mutations in the nucleic acid-binding region of SUV39H prevent its association with heterochromatin in human cells, suggesting that non-coding satellite RNAs recruit the enzyme to form stable associations with chromatin (Johnson et al., 2017) (Fig. 6b). Therefore, alpha satellite RNAs may provide specificity for SUV39H binding within the pericentromere region and form a scaffold for the formation of constitutive heterochromatin. HP1 may also require RNA to localize to pericentric chromatin, as the hinge region of this protein is known to bind satellite RNA in mouse cells (Muchardt et al., 2002, Maison et al., 2011), although evidence for HP1 localization via direct alpha satellite RNA binding has not yet been reported in human cells.
Alpha satellite RNAs appear to have distinctive roles in normal human cells, particularly within the centromere and pericentromere regions, serving to recruit centromere proteins for kinetochore assembly or to establish and perpetuate heterochromatin. This raises an interesting paradox in that alpha satellite transcripts produced from the same array or from similar adjacent arrays can direct both centromere protein recruitment or heterochromatin maintenance. How do cells distinguish between repetitive RNA destined to have different effects on chromatin assembly and organization? Differences in timing of transcription, unique RNA modifications, phase separation, or transcript length could be factors involved in helping the cell discriminate between these two paths.
Insight into alpha satellite transcription from artificial chromosomes
Studies of HAC chromatin and transcriptional competency suggest that transcription is required for centromere function and the level of transcription is finely tuned. Transcription is thought to occur at relatively low levels on alphoidtetO sequences, however more robust transcription of non-alpha satellite sequences, such as resistance genes used in HAC creation, embedded in centrochromatin has also been observed in HACs (Lam et al., 2006, Nakano et al., 2008). Transcription occurs largely within the centrochromatin domain of HACs rather than the flanking alphoidtetO sequences (Molina et al., 2016).
Tet-repressor-mediated tethering of chromatin modifying enzymes, such as LSD1/2, SUV39H1, and BMI, to tetO-containing HAC centromeres has demonstrated that the introduction of heterochromatic marks to alpha satellite DNA previously assembled in centrochromatin is not compatible with alpha satellite transcription or centromere maintenance (Bergmann et al., 2011, Ohzeki et al., 2012, Molina et al., 2016). Similarly, driving the KRAB repressor domain or its downstream effector KAP1 to HAC centromeres leads to an increase in H3K9me3 levels at the centromere and centromere protein loss and inactivation (Nakano et al., 2008, Cardinale et al., 2009). Together, these studies suggest that HAC centromere function and maintenance relies on transcriptional activity within centrochromatin and that transcription may be involved in preventing heterochromatin spreading into the centromere. Importantly, excessive alpha satellite transcription at HAC centromeres also has detrimental effects on centromere maintenance. Tet-mediated tethering of transcriptional activators VP16 increases HAC transcription 150-fold and leads to a reduction in CENP-A, a transition from centrochromatin to heterochromatin, and eventual kinetochore inactivation and HAC loss (Nakano et al., 2008, Bergmann et al., 2012). More moderate increases (10-fold) in the level of transcription induced by NF-κΒ p65 are compatible with continued centromere function, suggesting that some variation in transcription levels can be tolerated (Bergmann et al., 2012). Interestingly, transcription alone is not sufficient to maintain HAC centromere function. H3K3me2 and H3K9ac marks are specifically required for HAC transcription and heterochromatin antagonization and cannot be substituted with H4K12ac (Molina et al., 2016).
Alpha satellite DNA transcription in stress response, cancer, and genome instability.
Cell stress induces genome-wide changes in cells, including alterations in expression of repetitive DNA and localization of the resulting RNAs to nuclear stress granules (Denegri et al., 2002, Metz et al., 2004, Rizzi et al., 2004, Valgardsdottir et al., 2008). Changes in satellite DNA expression have been reported in response to stress and observed in a variety of human cancers (Denegri et al., 2002, Jolly et al., 2004, Metz et al., 2004, Rizzi et al., 2004, Valgardsdottir et al., 2008, Eymery et al., 2009, Ting et al., 2011, Zhu et al., 2011, Hall et al., 2017). For example, satellite II and satellite III sequences are normally silenced, but are upregulated in response to stress and may have a protective role by regulating mRNA processing and modification. In contrast, loss of satellite II and satellite III silencing does not have a protective effect in cancer cells and severely compromises epigenetic regulation. Alpha satellite overexpression does not appear to be as closely associated with stress conditions or cancer as other types of satellite DNA. Alpha satellite derepression was reported in cells containing a mutant BRCA1 gene, that encodes a protein that normally mediates heterochromatic silencing of satellite DNA (Zhu et al., 2011). In this context, exogenous alpha satellite expression was linked to DNA damage, mitotic errors, and genomic instability. Similar effects on chromosome stability and segregation were reported upon exogenous overexpression of alpha satellite in cultured cells (Chan et al., 2017). However, in a separate study, alpha satellite RNA overexpression was only observed in ~20% of cancer cell lines and BRCA1 mutation status was not correlated with alpha satellite overexpression (Hall et al., 2017). These latter results agree with previous findings showing that alpha satellite transcription is not upregulated in heat-shocked HeLa cells (Eymery et al., 2009). Overall, then, alpha satellite expression seems more tightly controlled, perhaps due to its involvement in kinetochore assembly that may prohibit mis-regulation.
Drivers of alpha satellite transcription and characteristics of transcripts
Mammalian cells contain three RNAPs: RNAP I, II, and III. In general, each polymerase is defined by the type of RNA it transcribes. RNAP I transcribes ribosomal RNA genes other than 5S rRNA, RNAP II transcribes protein-coding genes and microRNAs, and RNAP III transcribes tRNA genes, 5S rRNA genes, and some small nuclear RNAs. There is no consensus for the RNAP responsible for the generation of alpha satellite RNA. Indeed, all three polymerases have been identified as candidates. Given the noted presence of RNAP II at human centromeres and the effects of polymerase inhibition on transcripts, human alpha satellite DNA is thought to be actively transcribed by RNAP II (Chan et al., 2012, Quenet and Dalal, 2014, Liu et al., 2015, McNulty et al., 2017), but the resulting transcripts may depend upon RNAP I for proper localization (Wong et al., 2007). RNAP III-dependent transposable element transcription has also been suggested as a potential promoter of nearby alpha satellite transcription (Klein and O’Neill, 2018). Improved assemblies of repetitive regions could help identify promoter elements and other genetic signatures that can more definitively determine the polymerase involved in repetitive DNA transcription (see “Alpha satellite genomics: moving into a new era” section below).
Little is known about the post-transcriptional processing of alpha satellite RNAs (capping, splicing, polyadenylation). Ideue et al. (2014) have suggested that at least some alpha satellite RNAs lack poly-A tails, but the slow turnover of repetitive RNAs has been documented and suggests that alpha satellite RNA is innately stable (Hall et al., 2014, McNulty et al., 2017). Whether this stability is conferred by the presence of a poly-A tail or another protective mechanism, such as the formation of an RNA-DNA hybrid or post-transcriptional modifications, remains to be elucidated.
Remaining challenges in alpha satellite biology
Alpha satellite genomics: moving into a new era
The extensive reiteration of the HOR structure of alpha satellite has made it a challenge for standard genomic assembly. Short sequence reads corresponding to alpha satellite are abundant in the genome assembly pools, but their exact placement within the linear arrays that stretch for multiple megabases between the chromosome arms is not possible. Thus, in earlier assemblies of the human genome, HOR alpha satellite arrays were not present and the centromeres were presented as assembly gaps. In 2014, graphical, reference models of HOR alpha satellite regions were placed in the centromeric gaps (Miga et al., 2014). The graphical models were built from whole genome sequence (WGS) reads and Markov modeling to construct the most plausible configuration of HOR units. They were not intended to represent the linear organization of specific HOR units on a given chromosome and could not provide long-range organization of an entire alpha satellite array. Excitingly, a newly published study using single molecule nanopore sequencing has reported the successful assembly of contiguous linear alpha satellite DYZ3 sequence spanning the region between the short and long arms of HSAY (Jain et al., 2018). DYZ3 is a small array (0.1–1Mb) and, thus, a logical choice for an initial long sequencing approach. The approach of this tour de force effort was to sequence BACs containing inserts spanning from the entire HSAY centromere, including the intervening alpha satellite and other repetitive sequences. These ground-breaking results, combined with promise of improved nanopore sequence read lengths up to 1Mb, bring into focus the possibility of assembling alpha satellite arrays throughout the human genome, and identifying biologically relevant copy number and sequence variation in alpha satellite regions.
Likewise, more thorough sequencing of repetitive RNA is needed to fully define the heterogeneity of these non-coding RNAs and to identify promoters, start sites, and termination sequences. However, assembly and interpretation of RNA sequencing reads relies on complete assemblies of repetitive regions of the genome. Since most repetitive regions have been excluded from current builds of the human genome, efforts to fully characterize repetitive RNA have been stymied. As long-read sequencing technology continues to advance and repetitive regions of the genome are added to the genome assemblies, progress in detailing the repetitive transcriptome is sure to follow and inform efforts to identify the function of these transcripts. Such reads could shed light on the difference between transcripts produced from HOR alpha satellite vs. monomeric alpha satellite and could help discriminate between RNAs produced from centromeric versus pericentric regions. This level of discrimination between HOR and monomeric alpha satellite is not possible with in situ hybridization approaches. With these technological advances, our understanding of repetitive regions of mammalian genomes could soon equal our understanding of coding regions. It is clear, though, that rather than being simply passive regions of the genome or relics of past genomic events, repetitive DNA appears to be an active player in development, homeostasis, and genome stability.
Alpha satellite RNA biology: discriminating between the act of transcription and the role of transcripts themselves
Approaches that deplete specific alpha satellite RNAs, such as shRNA, antisense oligonucleotides (ASO), and dsRNAs, in mammalian cells are effective (Ideue et al., 2014, Quenet and Dalal, 2014, McNulty et al., 2017), but can only address the role of the transcripts. Altering the process of transcription using polymerase inhibitors, through steric hindrance by dCas9-KRAB localization, or tethering chromatin modifiers has confounding effects, including altering chromatin structure and simultaneously reducing the level of RNAs. Presumably, the loss of transcripts themselves could be overcome by expression of alpha satellite RNA from an exogenous locus or plasmid. Artificial expression of alpha satellite RNAs may temporarily increase amounts of exogenous RNA in the nucleus and phenocopy some effects of alpha satellite overexpression (Zhu et al., 2011, Chan et al., 2017), but the movement of these RNAs in trans to chromatin and nuclear bodies also appears important for their function. It has not yet been tested if exogenously expressed alpha satellite RNAs also localize to the same site of endogenous alpha satellite RNA production. There is also the question of how long (i.e. number of copies of the repeat) the exogenously expressed or directly transfected satellite sequence should be. Given the heterogeneity of sequence length described for nearly all satellite RNAs and lack of understanding about non-coding satellite RNA processing, this is not a trivial consideration.
Alpha satellite DNA was first described 40 years ago. Much has been learned about its chromosomal location, organizational structure, and importance in centromere specification and de novo centromere assembly. Its transcription is necessary for formation of unique chromatin domains and interactions with key centromere and chromatin proteins. Despite notable advances in alpha satellite biology over the past three decades, more work lies ahead as the field tackles the challenges of sequencing entire alpha satellite arrays and functionally annotating the types and frequency of size and sequence variants, as well as interspersed functional elements, within populations. As genome assemblies intersect with comparative and functional studies, we will reach a fuller understanding of this complicated and fascinating repetitive sequence and its role in basic biology and medicine.
Acknowledgements:
We thank Megan Aldrup-Macdonald for data contributing to Figure 3 and Karen Miga for helpful discussions and sharing data prior to publication. Our research is supported by National Science Foundation Graduate Research Fellowship DGE-1644868 (S.M.M.) and National Institutes of Health grant R01 GM124041 (B.A.S.).
Abbreviations
- ASO
antisense oligonucleotide
- bp
basepair
- Cas9
CRISPR associated protein 9
- CENP
centromere protein
- ChIP
chromatin immunoprecipitation
- dCas9
nuclease deficient Cas9
- DNA
deoxyribonucleic acid
- dsRNA
double stranded RNA
- GFP
green fluorescent protein
- HAC
human artificial chromosome
- HJURP
Holliday Junction Recognition Protein
- HOR
higher order repeat
- HP1
heterochromatin protein 1
- HSA
Homo sapiens
- kb
kilobase
- kDa
kilodalton
- KRAB
Krüppel associated box
- Mb
megabase
- RNA
ribonucleic acid
- RNAP
RNA polymerase
- shRNA
short hairpin RNA
- tRNA
transfer RNA 4
- VP16
virus protein 16
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Author Contribution Statement
SMM and BAS conceived and jointly wrote the manuscript.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. Embo j 18:1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L, Schmid M, Warburton P, Jenuwein T (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. Journal of cell science 113 ( Pt 5):817–829. [DOI] [PubMed] [Google Scholar]
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA (2016) Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y (2001) Alpha-satellite DNA of primates: old and new families. Chromosoma 110:253–266. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB (1991) Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics 11:15–23. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Mashkova TD, Romanova LY, Yurov YB, Kisselev LL (1993a) Segment substitutions in alpha satellite DNA. Unusual structure of human chromosome 3-specific alpha satellite repeat unit. J Mol Biol 231:516–520. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB (1993b) Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res 21:2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IA, Mitkevich SP, Yurov YB (1988) The phylogeny of human chromosome specific alpha satellites. Chromosoma 96:443–453. [DOI] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002) CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22:2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Shabanowitz J, Lehman SM, Bai DL, Hunt DF, Black BE, Foltz DR (2016) Identification of the Post-translational Modifications Present in Centromeric Chromatin. Mol Cell Proteomics 15:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Martins NM, Larionov V, Masumoto H, Earnshaw WC (2012) HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosome Res 20:505–519. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Developmental cell 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE (2013) Assembly in G1 phase and long term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell 24:923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE (1968) Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science (New York, NY) 161:529–540. [DOI] [PubMed] [Google Scholar]
- Brown KE, Barnett MA, Burgtorf C, Shaw P, Buckle VJ, Brown WR (1994) Dissecting the centromere of the human Y chromosome with cloned telomeric DNA. Human molecular genetics 3:1227–1237. [DOI] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Molecular cell 41:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Zhou K, Zhang Z, Luger K, Straight AF (2018) Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Mol Biol Cell 29:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC (2009) Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell 20:4194–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature cell biology 11:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DYL, Moralli D, Khoja S, Monaco ZL (2017) Noncoding Centromeric RNA Expression Impairs Chromosome Stability in Human and Murine Stem Cells. Disease markers 2017:7506976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH (2012) Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A 109:1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlieu JP, Murgue B, Laurent AM, Marcais B, Bellis M, Roizes G (1992) Discrimination between alpha-satellite DNA sequences from chromosomes 21 and 13 by using polymerase chain reaction. Genomics 14:515–516. [DOI] [PubMed] [Google Scholar]
- Choo KH, Earle E, Vissel B, Filby RG (1990) Identification of two distinct subfamilies of alpha satellite DNA that are highly specific for human chromosome 15. Genomics 7:143–151. [DOI] [PubMed] [Google Scholar]
- Denegri M, Moralli D, Rocchi M, Biggiogera M, Raimondi E, Cobianchi F, De Carli L, Riva S, Biamonti G (2002) Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol Biol Cell 13:2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilee P, Cremer T, Slagboom P, Bakker E, Scholl HP, Hager HD, Stevenson AF, Cornelisse CJ, Pearson PL (1986) Two subsets of human alphoid repetitive DNA show distinct preferential localization in the pericentric regions of chromosomes 13, 18, and 21. Cytogenet Cell Genet 41:193–201. [DOI] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137:485–497. [DOI] [PubMed] [Google Scholar]
- Durfy SJ, Willard HF (1987) Molecular analysis of a polymorphic domain of alpha satellite from the human X chromosome. Am J Hum Genet 41:391–401. [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H 3rd, Stetten G (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98:1–12. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321. [DOI] [PubMed] [Google Scholar]
- Eymery A, Horard B, El Atifi-Borel M, Fourel G, Berger F, Vitte AL, Van den Broeck A, Brambilla E, Fournier A, Callanan M, Gazzeri S, Khochbin S, Rousseaux S, Gilson E, Vourc’h C (2009) A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res 37:6340–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, Cleveland DW (2013) A two-step mechanism for epigenetic specification of centromere identity and function. Nature cell biology 15:1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW (2015) DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Developmental cell 33:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr CJ, Bayne RA, Kipling D, Mills W, Critcher R, Cooke HJ (1995) Generation of a human X-derived minichromosome using telomere-associated chromosome fragmentation. EMBO J 14:5444–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. The Journal of biological chemistry 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- Ge Y, Wagner MJ, Siciliano M, Wells DE (1992) Sequence, higher order repeat structure, and long-range organization of alpha satellite DNA specific to human chromosome 8. Genomics 13:585–593. [DOI] [PubMed] [Google Scholar]
- Greig GM, Warburton PE, Willard HF (1993) Organization and evolution of an alpha satellite DNA subset shared by human chromosomes 13 and 21. J Mol Evol 37:464–475. [DOI] [PubMed] [Google Scholar]
- Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, Cheeseman IM, Black BE (2017) Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat Commun 8:15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Warburton PE, Willard HF (1992) Integration of human alpha-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell 70:681–696. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC (1994) Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Human molecular genetics 3:697–709. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Carone DM, Whitfield TW, Pouliot GP, Fischer A, Jones P, Lawrence JB (2017) Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies. Cell reports 18:2943–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB (2014) Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 156:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15:345–355. [DOI] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE (2013) The octamer is the major form of CENP-A nucleosomes at human centromeres. Nature structural & molecular biology 20:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF (2013) Sequences associated with centromere competency in the human genome. Mol Cell Biol 33:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Cho Y, Nishimura K, Tani T (2014) Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes to cells : devoted to molecular & cellular mechanisms 19:528–538. [DOI] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16:431–439. [DOI] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T (1994) Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Human molecular genetics 3:1245–1257. [DOI] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH (2018) Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol 36:321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O’Neill RJ, Sullivan BA, Straight AF (2017) RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen AL, Kolvraa S, Jones C, Bak AL (1988) A subfamily of alphoid repetitive DNA shared by the NOR-bearing human chromosomes 14 and 22. Genomics 3:100–109. [DOI] [PubMed] [Google Scholar]
- Kim JH, Ebersole T, Kouprina N, Noskov VN, Ohzeki J, Masumoto H, Mravinac B, Sullivan BA, Pavlicek A, Dovat S, Pack SD, Kwon YW, Flanagan PT, Loukinov D, Lobanenkov V, Larionov V (2009) Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome research 19:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SJ, O’Neill RJ (2018) Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res 26:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononenko AV, Lee NC, Earnshaw WC, Kouprina N, Larionov V (2013) Re-engineering an alphoid(tetO)-HAC-based vector to enable high-throughput analyses of gene function. Nucleic Acids Res 41:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD (1974) Chromatin Structure: A Repeating Unit of Histones and DNA. Science (New York, NY) 184:868–871. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120. [DOI] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA (2006) Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A 103:4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee NC, Grimes BR, Samoshkin A, Kononenko AV, Bansal R, Masumoto H, Earnshaw WC, Kouprina N, Larionov V (2013a) A new assay for measuringchromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC cancer 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NC, Kononenko AV, Lee HS, Tolkunova EN, Liskovykh MA, Masumoto H, Earnshaw WC, Tomilin AN, Larionov V, Kouprina N (2013b) Protecting a transgene expression from the HAC-based vector by different chromatin insulators. Cellular and molecular life sciences : CMLS 70:3723–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H (2015) Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Molecular cell 59:426–436. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Oosterhuis JW, Smit VT, Wessels JW, Mollevanger P, Devilee P (1992) Alpha satellite DNAs on chromosomes 10 and 12 are both members of the dimeric suprachromosomal subfamily, but display little identity at the nucleotide sequence level. Genomics 13:1125–1132. [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Willard HF (1990) Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: high-frequency polymorphisms and array size estimate. Genomics 7:607–613. [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Willard HF (1998) Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome research 8:100–110. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30:329–334. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, Almouzni G (2011) SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 43:220–227. [DOI] [PubMed] [Google Scholar]
- Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA (2012) Functional epialleles at an endogenous human centromere. Proc Natl Acad Sci U S A 109:13704–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L (1978) Chromosomal localization of complex and simple repeated human DNAs. Chromosoma 66:23–32. [DOI] [PubMed] [Google Scholar]
- Marcais B, Bellis M, Gerard A, Pages M, Boublik Y, Roizes G (1991) Structural organization and polymorphism of the alpha satellite DNA sequences of chromosomes 13 and 21 as revealed by pulse field gel electrophoresis. Hum Genet 86:311–316. [DOI] [PubMed] [Google Scholar]
- Marcais B, Laurent AM, Charlieu JP, Roizes G (1993) Organization of the variant domains of alpha satellite DNA on human chromosome 21. J Mol Evol 37:171–178. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Ikeno M, Nakano M, Okazaki T, Grimes B, Cooke H, Suzuki N (1998) Assay of centromere function using a human artificial chromosome. Chromosoma 107:406–416. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109:1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SM, Sullivan LL, Sullivan BA (2017) Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Developmental cell 42:226–240.e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz A, Soret J, Vourc’h C, Tazi J, Jolly C (2004) A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. Journal of cell science 117:4551–4558. [DOI] [PubMed] [Google Scholar]
- Miga KH (2015) Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res 23:421–426. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ (2014) Centromere reference models for human chromosomes X and Y satellite arrays. Genome research 24:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills W, Critcher R, Lee C, Farr CJ (1999) Generation of an approximately 2.4 Mb human X centromere-based minichromosome by targeted telomere-associated chromosome fragmentation in DT40. Human molecular genetics 8:751–761. [DOI] [PubMed] [Google Scholar]
- Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC (2016) Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 7:13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moralli D, Jefferson A, Valeria Volpi E, Larin Monaco Z (2013) Comparative study of artificial chromosome centromeres in human and murine cells. Eur J Hum Genet 21:948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sulivan BA (2009) Histone modifications within the human X centromere region. PloS one 4:e6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO reports 3:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T (1992) Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 116:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Desai A (2017) A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H (2008) Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Developmental cell 14:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A (1997) MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471–481. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H (2012) Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J 31:2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H (2016) KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Developmental cell 37:413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131:1287–1300. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A 88:3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL (1987) A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesenti E, Kouprina N, Liskovykh M, Aurich-Costa J, Larionov V, Masumoto H, Earnshaw WC, Molina O (2018) Generation of a Synthetic Human Chromosome with Two Centromeric Domains for Advanced Epigenetic Engineering Studies. ACS synthetic biology 7:1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular cell 12:1577–1589. [DOI] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel M-A (2004) Histones and histone modifications. Current Biology 14:R546–R551. [DOI] [PubMed] [Google Scholar]
- Pironon N, Puechberty J, Roizes G (2010) Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21. BMC Genomics 11:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi V, Perini G, Trazzi S, Pliss A, Raska I, Earnshaw WC, Della Valle G (2002) CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. Journal of cell science 115:2317–2327. [DOI] [PubMed] [Google Scholar]
- Quenet D, Dalal Y (2014) A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife e03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. [DOI] [PubMed] [Google Scholar]
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G (2004) Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 15:543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosandic M, Paar V, Basar I, Gluncic M, Pavin N, Pilas I (2006) CENP-B box and pJalpha sequence distribution in human alpha satellite higher-order repeats (HOR). Chromosome Res 14:735–753. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ (2015) The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43:D670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JE, Woodlief KS, Sullivan BA (2016) Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenetics Chromatin 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RI, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, Dhami P, Ding Y, Dinh H, Dodsworth S, Draper H, Dugan-Rocha S, Dunham A, Dunn M, Durbin KJ, Dutta I, Eades T, Ellwood M, Emery-Cohen A, Errington H, Evans KL, Faulkner L, Francis F, Frankland J, Fraser AE, Galgoczy P, Gilbert J, Gill R, Glockner G, Gregory SG, Gribble S, Griffiths C, Grocock R,Gu Y, Gwilliam R, Hamilton C, Hart EA, Hawes A, Heath PD, Heitmann K, Hennig S,Hernandez J, Hinzmann B, Ho S, Hoffs M, Howden PJ, Huckle EJ, Hume J, Hunt PJ, Hunt AR, Isherwood J, Jacob L, Johnson D, Jones S, de Jong PJ, Joseph SS, Keenan S, Kelly S, Kershaw JK, Khan Z, Kioschis P, Klages S, Knights AJ, Kosiura A, KovarSmith C, Laird GK, Langford C, Lawlor S, Leversha M, Lewis L, Liu W, Lloyd C, Lloyd DM, Loulseged H, Loveland JE, Lovell JD, Lozado R, Lu J, Lyne R, Ma J, Maheshwari M, Matthews LH, McDowall J, McLaren S, McMurray A, Meidl P, Meitinger T, Milne S, Miner G, Mistry SL, Morgan M, Morris S, Muller I, Mullikin JC, Nguyen N, Nordsiek G, Nyakatura G, O’Dell CN, Okwuonu G, Palmer S, Pandian R, Parker D, Parrish J, Pasternak S, Patel D, Pearce AV, Pearson DM, Pelan SE, Perez L, Porter KM, Ramsey Y, Reichwald K, Rhodes S, Ridler KA, Schlessinger D, Schueler MG, Sehra HK, ShawSmith C, Shen H, Sheridan EM, Shownkeen R, Skuce CD, Smith ML, Sotheran EC, Steingruber HE, Steward CA, Storey R, Swann RM, Swarbreck D, Tabor PE, Taudien S, Taylor T, Teague B, Thomas K, Thorpe A, Timms K, Tracey A, Trevanion S, Tromans AC, d’Urso M, Verduzco D, Villasana D, Waldron L, Wall M, Wang Q, Warren J, Warry GL, Wei X, West A, Whitehead SL, Whiteley MN, Wilkinson JE, Willey DL, Williams G, Williams L, Williamson A, Williamson H, Wilming L, Woodmansey RL, Wray PW, Yen J, Zhang J, Zhou J, Zoghbi H, Zorilla S, Buck D, Reinhardt R, Poustka A, Rosenthal A, Lehrach H, Meindl A, Minx PJ, Hillier LW, Willard HF, Wilson RK, Waterston RH, Rice CM, Vaudin M, Coulson A, Nelson DL, Weinstock G, Sulston JE, Durbin R, Hubbard T, Gibbs RA, Beck S, Rogers J, Bentley DR (2005) The DNA sequence of the human X chromosome. Nature 434:325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Mays RW, Schwartz S, Willard HF (2003 a) Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol Cell Biol 23:7689–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF (2003b) Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol 68:141–149. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Dunn JM, Bird CP, Ross MT, Viggiano L, Program NCS, Rocchi M, Willard HF, Green ED (2005) Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci U S A 102:10563–10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF (2001) Genomic and genetic definition of a functional human centromere. Science (New York, NY) 294:109–115. [DOI] [PubMed] [Google Scholar]
- Shang WH, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y, Fujiyama A, Kimura H, Straight AF, Fukagawa T (2016) Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat Commun 7:13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol 151:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF (1997) Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol 136:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA (2009) The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet 5:e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev VA, Uralsky LI, Alexandrov AA, Yurov YB, Rogaev EI, Alexandrov IA (2015) Annotation of suprachromosomal families reveals uncommon types of alpha satellite organization in pericentromeric regions of hg38 human genome assembly. Genom Data 5:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono N, Ohzeki J, Otake K, Martins NM, Nagase T, Kimura H, Larionov V, Earnshaw WC, Masumoto H (2015) CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. Journal of cell science 128:4572–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee RB, Steiner CM, Herbert B-S, Vance GH, Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP, Schindelhauer D, Grimes BR (2011) Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene 31:3244–3253. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature structural & molecular biology 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Human molecular genetics 4:2189–2197. [DOI] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA (2011) Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res 19:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Chew K, Sullivan BA (2017) alpha satellite DNA variation and function of the human centromere. Nucleus (Austin, Tex) 8:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Maloney KA, Towers AJ, Gregory SG, Sullivan BA (2016) Human centromere repositioning within euchromatin after partial chromosome deletion. Chromosome Res 24:451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Henikoff S (2018) Unexpected conformational variations of the human centromeric chromatin complex. Genes Dev 32:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, Haber DA (2011) Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science (New York, NY) 331:593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S, Bernardoni R, Diolaiti D, Politi V, Earnshaw WC, Perini G, Della Valle G (2002) In vivo functional dissection of human inner kinetochore protein CENP-C. J Struct Biol 140:39–48. [DOI] [PubMed] [Google Scholar]
- Trowell HE, Nagy A, Vissel B, Choo KH (1993) Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Human molecular genetics 2:1639–1649. [DOI] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF (1997) Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol 7:897–900. [DOI] [PubMed] [Google Scholar]
- Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G (2008) Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 36:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KH (1991) Four distinct alpha satellite subfamilies shared by human chromosomes 13, 14 and 21. Nucleic Acids Res 19:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KH (1992) Evolutionary relationships of multiple alpha satellite subfamilies in the centromeres of human chromosomes 13, 14, and 21. J Mol Evol 35:137–146. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7:901–904. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Waye JS, Willard HF (1993) Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 13:6520–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Willard HF (1992) PCR amplification of tandemly repeated DNA: analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human alpha satellite DNA. Nucleic Acids Res 20:6033–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Willard HF (1995) Interhomologue sequence variation of alpha satellite DNA from human chromosome 17: evidence for concerted evolution along haplotypic lineages. J Mol Evol 41:1006–1015. [DOI] [PubMed] [Google Scholar]
- Waye JS, Creeper LA, Willard HF (1987a) Organization and evolution of alpha satellite DNA from human chromosome 11. Chromosoma 95:182–188. [DOI] [PubMed] [Google Scholar]
- Waye JS, England SB, Willard HF (1987b) Genomic organization of alpha satellite DNA on human chromosome 7: evidence for two distinct alphoid domains on a single chromosome. Mol Cell Biol 7:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Greig GM, Willard HF (1987c) Detection of novel centromeric polymorphisms associated with alpha satellite DNA from human chromosome 11. Hum Genet 77:151–156. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF (1985) Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res 13:2731–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF (1986a) Molecular analysis of a deletion polymorphism in alpha satellite of human chromosome 17: evidence for homologous unequal crossing-over and subsequent fixation. Nucleic Acids Res 14:6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF (1986b) Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol 6:3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF (1987) Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res 15:7549–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF (1989) Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc Natl Acad Sci U S A 86:9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF (1991) Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res 19:2295–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF (1985) Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet 37:524–532. [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Skolnick MH, Pearson PL, Mandel JL (1985) Report of the Committee on Human Gene Mapping by Recombinant DNA Techniques. Cytogenet Cell Genet 40:360–489. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS (1987a) Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol 25:207–214. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS (1987b) Hierarchical order in chromosome-specific alpha satellite DNA. Trends Genet 3:192–198. [Google Scholar]
- Willard HF, Waye JS, Skolnick MH, Schwartz CE, Powers VE, England SB (1986) Detection of restriction fragment length polymorphisms at the centromeres of human chromosomes by using chromosome-specific alpha satellite DNA probes: implications for development of centromere-based genetic linkage maps. Proc Natl Acad Sci U S A 83:5611–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome research 17:1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Manuelidis L (1980) Sequence definition and organization of a human repeated DNA. J Mol Biol 142:363–386. [DOI] [PubMed] [Google Scholar]
- Yasmineh WG, Yunis JJ (1974) Localization of repeated DNA sequences in CsC1 gradients by hybridization with complementary RNA. Experimental cell research 88:340–344. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T (1998) In vitro assembly of the CENP-B/alpha-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes to cells : devoted to molecular & cellular mechanisms 3:533–548. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]


