Abstract
Emissions from oil fires associated with the Deepwater Horizon explosion and oil discharge that began on April 20, 2010 in the Gulf of Mexico were analyzed chemically to only a limited extent at the time but were shown to induce oxidative damage in vitro and in mice. To extend this work, we burned oil floating on sea water and performed extensive chemical analyses of the emissions [Gullett et al., submitted]. Here we examine the ability of a dichloromethane extract of the particulate material with an aerodynamic size ≤ 2.5 µm (PM2.5) from those emissions to induce oxidative damage in human lung cells in vitro and mutagenicity in 6 strains of Salmonella. The extract had a percentage of extractable organic material (EOM) of 7.0% and increased expression of the heme oxygenase (HMOX1) gene in BEAS-2B cells after exposure for 4 h at 20 µg of EOM/ml. However, the extract did not alter mitochondrial respiration rate as measured by extracellular flux analysis. The extract was most mutagenic in TA100 +S9, indicative of a role for polycyclic aromatic hydrocarbons (PAHs), reflective of the high concentrations of PAHs in the emissions (1 g/kg of oil consumed). The extract had a mutagenicity emission factor of 1.8 ± 0.1 × 105 revertants/megajoulethermal in TA98 +S9, which was greater than that of diesel exhaust and within an order of magnitude of wood burning in residential fireplaces and the open burning of plastic. Thus, organics from PM2.5 of burning oil can induce oxidative responses in human airway epithelial cells and are highly mutagenic.
Keywords: oil-burn emissions, mutagenicity, oxidative damage, Salmonella, mitochondrial respiration, extracellular flux analysis
INTRODUCTION
On April 20, 2010, an explosion and fire on the Deepwater Horizon (DWH) mobile offshore drilling unit resulted in the deaths of 11 workers and discharges of 4.2 million barrels of oil and other substances into the Gulf of Mexico over a period of 87 days [National Commission, 2011]. Along with leakage of oil under water at the wellhead, this event also produced combustion emissions from burning oil at the top of the rig. Limited monitoring by aircraft for 2 days and from nearby ships provided some data on the concentrations of particulate material (PM), volatile organic compounds (VOCs) [Middlebrook et al., 2012], organic aerosols [de Gouw et al., 2011], and black carbon [Perring et al., 2011] in these emissions.
Shortly after the explosion, surface oil was collected by boom-towing trawlers, and the U.S. Coast Guard conducted 411 burns of this collected oil to remove it from the surface of the Gulf [Mabile and Allen, 2010]. These burns reduced the total mass loading of polycyclic aromatic hydrocarbons (PAHs) from the burned oil to the Gulf of Mexico by 89% [Stout and Payne, 2016]. Nonetheless, large amounts of bioavailable PAHs [Allan et al., 2012] were volatilized [Tidwell et al., 2015]. Some of the emissions from these prescribed burns were collected by maneuvering an aerostat-lofted sampler (sail) into the in situ burn plumes from a ship-mounted winch as described by Aurell et al. [2010]. The resulting oil-sail PM (OSPM) was analyzed minimally for concentrations of polychlorinated dibenzodioxins and dibenzofurans (dioxins) [Aurell and Gullett, 2010; Gullett et al., 2016], as well as for the extent of the dioxin dispersion to result in levels of exposure sufficient to produce human health effects [Schaum et al., 2010].
These limited monitoring efforts to assess concentrations of selected pollutants did not provide measures of cumulative pollutant burden to the environment or toxicity to natural resources. This lack of knowledge has limited the ability of environmental health agencies to assess the potential risks to ecosystem components from burning oil associated with the DWH spill. Although additional studies have been published recently on the ecological and human health effects of the DWH oil spill [D’Andrea and Reddy, 2013; Liu et al., 2016; Paruk et al., 2016; Peres et al., 2016; Putman et al., 2015; Savitz and Engel, 2010; Stieglitz et al., 2016; Zengel et al., 2016], a review 5 years after the DWH event [Joye, 2015] indicated that many of the potential or actual environmental effects were still unclear.
One of the many unresolved issues is the effect that the emissions might have had on the lungs of marine mammals in the vicinity of the burning oil, many of which died during the event [Lane et al., 2015]. Relative to land-dwelling mammals, cetaceans are thought to be more susceptible to inhaled pollutants because of their distinct respiratory anatomy and physiology, specifically their lack of nasal turbinates and their deep inhalation with a lower frequency of air exchanges [Venn-Watson et al., 2013]. Necropsy of dolphins within the DWH spill region showed adrenal gland and lung disease consistent with exposure via ingestion, inhalation, or aspiration to petroleum hydrocarbons such as PAHs [Schwacke et al., 2014a,b; Venn-Watson et al., 2015].
Relevant to the potential of the oil-burn emissions to induce oxidative stress in the dolphins, Jaligama et al. [2015] found that the OSPM collected during the in situ burns during the DWH event induced a variety of oxidative effects in vitro and in vivo. These included the production of superoxide radicals and reactive oxygen species in vitro, as well as systemic oxidative stress (increased serum 8-isoprostane) and airway inflammation (increased macrophage and eosinophil numbers in bronchoalveolar fluid) in mice [Jaligama et al., 2015]. Exposure to the electrophiles and secondary reactive species of oxygen and nitrogen from combustion emissions has been shown to result in activation of signaling pathways that lead to increased production of inflammatory proteins and damage to tissues. For example, a variety of combustion emissions, such as those from diesel exhaust and cigarette smoke, have electrophilic compounds that can induce inflammation, damage to DNA, mutations, and cancer [IARC, 2004, 2014].
Sheppard et al. (1983) found that a dimethyl sulfoxide (DMSO) extract of samples of the smoke plume from a controlled burn of Prudhoe Bay, Alaska, crude oil that had fallen on the surrounding ice and collected were mutagenic in strain TA98 of Salmonella. This is the only study on the mutagenicity of emissions from burning oil, and it did not employ standard procedures to collect and extract the emissions, nor were measurements made to permit the calculation of a mutagenicity emission factor.
To further explore the effects of PM from the emissions of burning oil, we burned oil that we floated on sea water in metal pans to simulate the oil burns that occurred during the DWH event, and we collected and analyzed the emissions for 16 PAHs and a variety of dioxins, which we report in a companion paper by Gullett et al. [2017]. Here we examined an organic extract of the PM from those simulated oil burns for its ability to induce mutagenicity and oxidative damage. We assessed the effect of the extract on extracellular flux of mitochondrial respiration in cultured human airway epithelial cells (BEAS-2B), as well as on expression of the heme oxygenase (HMOX1) gene. We have shown previously that HMOX1 expression, a marker of oxidative stress in mammalian cells, is induced by a wide variety of air pollutants [Cheng et al., 2012; Silbajoris et al., 2009; Wages et al., 2014].
In addition, we evaluated the mutagenicity of the extract in a variety of strains in the Salmonella mutagenicity assay, including strain TA104, which detects oxidant-dependent mutagenesis. We compared and discussed the HMOX1 results and calculated mutagenicity emission factors relative to those of other combustion emissions, providing mechanistic insights into the potential toxic effects of the emissions from the open burning of oil to natural resources in the area affected by the DWH incident.
MATERIALS AND METHODS
Extraction of PM from Burning Oil
Details of the generation, collection, and characterization of the PM2.5 of the crude oil burned for this study are described by Gullett et al. [2017] and summarized here. We burned Bayou Choctaw Sweet crude oil obtained from the Strategic Petroleum Reserve, Dept. of Energy, Bayou Choctaw Site, 60825B Highway 1148, Plaquemine, LA. This oil is a blend from multiple wells and is a close approximation to the oil released during the DWH disaster. To simulate oil burning at sea, we floated the oil on the surface of a sea-water-filled pan, ignited the oil, and collected the particulate material with an aerodynamic size ≤2.5 µm (PM2.5) onto 47-mm Teflon® filters in sampling equipment suspended from a construction crane over the plume. We conducted these burns multiple times during several days at the Army Research Laboratory (ARL) Airbase Range 6, Aberdeen Proving Ground, Aberdeen, MD.
We prepared a dichloromethane (DCM) extract of 2 blank filters as well as from 2 exposed filters, one each from burns 1 and 2 conducted on February 1, 2016. We extracted the blank filters twice with sonication with 150 ml of DCM per filter for 20 min. We extracted the exposed filters 3 times with sonication with 500, 400, and then 250 ml of DCM for 20 min each to assure we extracted as much of the extractable organic material (EOM) as possible.
We combined the extracts from the blank and separately combined extracts from exposed filters; filtered the extracts through two 1-µm Zefluor® filters; evaporated them to 10 ml; and then filtered four 2.5-ml portions through four Anotop® 0.2-µm filters. We combined the respective filtrates, evaporated them to 2.5 ml, and filtered them through a 0.02-µm Anotop® filter; we brought the filtrate for the blank and exposed samples each to a volume of 10 ml gravimetrically and determined the EOM gravimentrically.
We analyzed the DCM extract for the concentrations of 26 PAHs, and the results are reported in Table S9 in Gullett et al. [2017]. To prepare the DCM extract for bioassay, we used the % EOM to determine how much DCM extract to solvent-exchange into dimethyl sulfoxide (DMSO) to give a concentrate at 10 mg EOM/ml DMSO as described [Mutlu et al. 2016].
Extracellular Flux Analysis of Mitochondrial Respiration
We seeded BEAS-2B cells in a XF24 (Seahorse Biosciences) plate at 30,000 cells/well two days prior to assay and starved the cells of growth factors overnight. We used the Seahorse XF Cell Mito Stress Test Kit with a Seahorse XFe24 extracellular flux analyzer per manufacturer’s instructions (Seahorse Bioscience, Billerica, MA) to analyze the exposed cells. Briefly, we changed the medium to Cell Mito Assay Media (XF Base Medium, 10-mM glucose, 1-mM sodium pyruvate, 2-mM L-glutamine) and placed the cells in a CO2-free incubator at 37°C for 60 min. The oxygen-consumption rate was monitored by the instrument at 8-min intervals. After a 24-min baseline collection, we injected 0–21 µg/ml of EOM into the medium, followed by the sequential addition of the complex V inhibitor oligomycin (1 µM), the protonophore carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 1.25 µM), and the complex I and III inhibitors rotenone (0.5 µM) and antimycin A (0.5 µM). We used 1,2-naphthoquinone (1,2-NQ) at 25 µM as a positive control in the assay. In some experiments we exposed cells to 0–21 µg/ml EOM for 24 h in keratinocyte basal medium (KBM) at 37oC with 5% CO2 prior to the assay.
HMOX1 Expression in Human Airway Epithelial Cells
We cultured human lung BEAS-2B cells to 80% confluence in KGM, which is KBM plus growth factors, as described [Cheng et al., 2012] and deprived the cells of growth factors overnight prior to exposure to 0–20 µg/mL EOM in KBM for 4 h at 37°C, 5% CO2, 100% humidity. We quantified relative gene expression of HMOX1 by real-time PCR using the ABI® Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) as described by Cheng et al. [2012] using 20-µM 1,2-NQ as a positive control. Briefly, RNA was quantified using a Nanodrop ND-1000 (ThermoFisher, Wilmington DE). Complementary DNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) with 1000 ng of purified total RNA. Transcript abundance was then quantified by TaqMan qPCR with cycling conditions 95°C at 3:00 min, 95°C at 0:15 min, 60°C at 0:45 min x 40 using a CFX96 Touch real time PCR apparatus (Bio-Rad). Gene expression was determined by calculating the fold change between O3 and clean air treatments, which were calculated with respect to actual PCR reaction efficiency (calculated for each primer/probe set on each PCR plate) and normalized to the corresponding fold change in ACTB transcript using the method of Pfaffl (2001).
Oligonucleotide primer pairs and fluorescent probes for HMOX1 were as follows: HMOX-1 Forward 5’ GACATTTTAGGGAGCTGGA, Reverse 5’ TCCCAGAAGGTTCCAGAAAGC 3’, Probe 5’ CTGATGTTGCCCACCAGGCTATTGC 3’; β-Actin Forward 5’ CTCAATCTCGCTCTCGCTCT 3’, Reverse 5’ CTCGAGCCATAAAAGGCAAC 3’, Probe 5’ AAATGCTGCACTGTGCGGCG 3’. We designed the oligonucleotides using a primer-design program (Primer Express, Applied Biosystems) obtained from Integrated DNA Technologies (Coralville, IA). We normalized the data to β-ACTIN mRNA levels [Cheng et al., 2012; Silbajoris et al., 2009; Wages et al., 2014], which were not found to change in response to treatment with the oil-burn extract.
Mutagenicity Assays
We evaluated the extract in the Salmonella mutagenicity assay using the plate-incorporation method [Maron and Ames 1983] at 10, 25, 50, 100, 125, 250, and 500 µg EOM/plate in the presence and absence of metabolic activation (S9 mix) made from liver S9 (Moltox, Boone, NC) prepared from Aroclor-induced Sprague-Dawley rats. Due to sample availability, we evaluated the extract at 1 plate/dose in at least 2 independent experiments in 6 strains that permitted us to infer the induction of various types of mutations or what the primary class of compound was that induced the mutations as described below. We incubated the plates at 37oC for 3 days, after which we counted the colonies with an automatic colony counter (AccuCount 1000, Manassas, VA). We defined a positive mutagenic response as a reproducible, dose-related response with a twofold or greater increase in revertants (rev) relative to the DMSO control plates.
As described by Mutlu et al. [2013], TA98 [hisD3052 chl-1008 (bio uvrB gal) rfa-1004 pKM101+, Fels-1+, Fels-2+ Gifsy-1+ Gifsy-2+] detects frameshift mutagens. In the absence of S9, TA98 has some specificity for nitroarenes, and in the presence of S9 it can detect some PAHs. TA100 [hisG46 chl-1005 (bio uvrB gal) rfa-1004 pKM101+, Fels-1+, Fels-2+ Gifsy-1+ Gifsy-2+] detects base-substitution mutagens at GC sites, and in the presence of S9 detects PAHs. Strain genotypes are detailed by Porwollik et al. [2001]. TA104 detects base-substitution mutagens at both GC and AT sites, and because it can detect mutagens at AT sites, it has some specificity for oxidative mutagens. This type of mutagen is of special interest in the evaluation of oil-burn emissions. YG1041 is a derivative of TA98 that overexpresses nitroreductase and o-acetyltransferase, permitting it to detect frameshift mutagens that are nitroarenes in the absence of S9 or aromatic amines in the presence of S9. YG1024 is a derivative of TA98 that overexpresses o-acetyltransferase only, and thus, in the presence of S9, it is especially sensitive to aromatic amines that are frameshift mutagens. YG1042 is a derivative of TA100 that overexpresses nitroreductase and o-acetyltransferase, permitting it to detect base-substitution mutagens such as nitroarenes in the absence of S9 or aromatic amines in the presence of S9.
The negative control (DMSO at 100 µl/plate) and positive controls (described below) were included with each experiment; typically, 3 plates of each type of control were made for each experiment. In the presence of S9 the positive controls were 2-aminoanthracene at 0.5 µg/plate for all strains. In the absence of S9, the positive controls were 2-nitrofluorene at 3 µg/plate for TA98 and its derivatives; sodium azide at 3 µg/plate for TA100, and methylglyoxal at 200 µg/plate for TA104.
Calculation of Mutagenicity Emission Factors
Using the GLM procedure in SAS/State v.9.4 software, we performed linear regression analyses over the linear portions of the dose-response curves to determine the mutagenic potencies (rev/µg EOM) ± standard error (SE). The linear portion was defined by the line with the highest r2 value relative to that produced by inclusion of the lower doses. Thus, in some cases, the highest dose or two highest doses were not included in the regression analysis because their inclusion would have lowered the r2 value. To test the differences between slopes of +S9 and −S9 for each strain, we included a regression model for each that assumed a common intercept but estimated separate slopes for +S9 and −S9 and performed a t-test in this model to test for any differences between the two slopes. Because we performed 6 tests (6 strains), we calculated a Bonferroni adjustment for multiple comparison to the P-values.
By multiplying the rev/µg by the %EOM, we expressed the data as rev/µg PM2.5, which we then expressed as rev/mg PM2.5 and as rev/g PM2.5. We calculated the mutagenicity emission factors (rev/megajoulethermal, MJth, and rev/kg fuel) using values determined by Gullett et al. [2017]. Briefly, the mass of the particles equals the mass of the PM2.5; thus, we multiplied the rev/g PM2.5 by 1.38 g PM2.5/MJth ± 0.08 (determined by Gullett et al. [2016a]) to give rev/MJth. Likewise, we multiplied the rev/g PM2.5 by 58 g PM2.5/kg fuel ± 3.5 (determined by Gullett et al. [2017]) to give rev/kg fuel. The mutagenicity emission factor rev/MJth in TA98 +S9 was then compared to those for other combustion emissions, such as diesel exhaust, coal, wood, etc., which are reported in Mutlu et al. [2016].
RESULTS
Percent EOM
There was no detectable EOM from the blank filters (data not shown), and the PM2.5 extracted from the exposed filters had a % EOM value of 7.0%, which we used in calculations for the preparation of the DMSO concentrate and for converting rev/µg EOM to rev/µg PM2.5.
Oxidative Stress Assays
As shown in Figure 1, acute exposure of BEAS-2B cells to 0–21 µg/ml of EOM did not result in an appreciable difference in mitochondrial respiration. Mitochondrial stress-test parameters reflected by oxygen-consumption rates attributable to baseline mitochondrial respiration, proton leak, maximal mitochondrial respiration, ATP production, spare mitochondrial capacity, and non-mitochondrial respiration were not affected by exposure to the EOM compared to control cells that received DMSO alone (Fig. 1). Similarly, exposure to the EOM for 24 h also did not induce changes in oxygen-consumption rate indices in BEAS-2B cells (data not shown).
Figure 1.
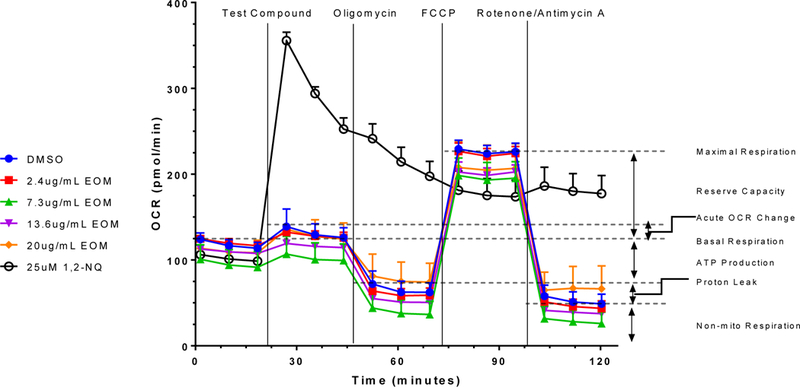
Exposure to the EOM from the oil-burn PM2.5 did not affect mitochondrial respiration in BEAS-2B cells. The oxygen-consumption rate (OCR) was monitored in an extracellular flux analyzer (Seahorse Bioscience Cell Mito Stress Test) for a baseline period, followed by an acute injection of test compound consisting of DMSO, 0.27–7.3 µg/ml EOM, or 25-µM 1,2-NQ (a positive control for non-mitochondrial redox cycling), oligomycin (1 µM), FCCP (1.25 µM), and rotenone/antimycin A (0.5 µM each), as indicated. Segments of the oxygen-consumption rate curve corresponding to indices of mitochondrial and non-mitochondrial functions are marked by dashed lines. Data shown are mean +/− SEM of samples tested in quadruplicate and are representative of three independent experiments.
In contrast, exposure of BEAS-2B cells to the extract produced a dose-dependent increase in HMOX1 expression, normalized to levels of β-ACTIN, relative to the DMSO-treated cells that was statistically significant at an exposure concentration of 20 µg EOM/ml for 4 h (Fig. 2). The positive control, 1,2-NQ, which is an organic electrophile that is associated commonly with ambient PM, induced a robust increase in HMOX1 expression (Fig. 2).
Figure 2.
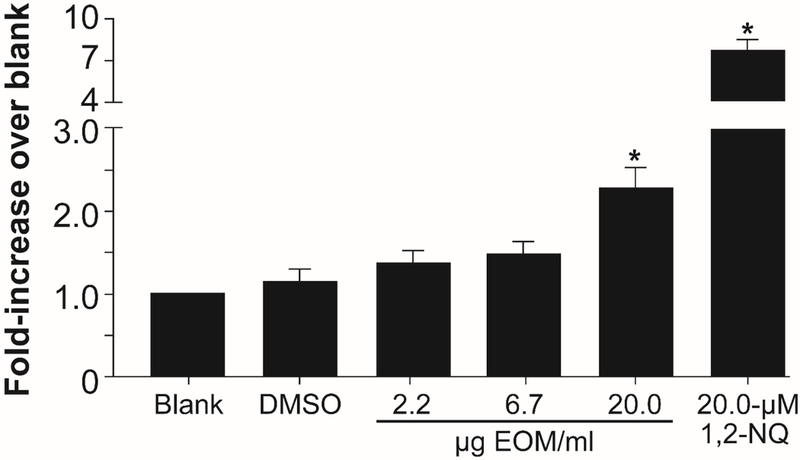
HMOX1 by RT-PCR relative to β-ACTIN mRNA expression after exposure of BEAS-2B cells for 4 h to oil-burn extract. Data are expressed as the fold-increase over control, with 20-µM 1,2-NQ as a positive control; mean ± standard error,* denotes P < 0.05, n = 6.
Mutagenic Potencies and Mutagenicity Emission Factors
Extracts of the filter blanks were not mutagenic (Table I), but the oil-burn extract was mutagenic in all 6 strains (Table I). The mutagenic potencies expressed as rev/µg EOM (Table II) and rev/mg PM2.5 (Table III) showed that for all strains except YG1041, the extract was more mutagenic in the presence of S9 than in the absence of S9 (P < 0.05); in YG1041 there was no difference in the mutagenic potency with or without S9 (P > 0.05). Conversion of these mutagenic potency values to mutagenicity emission factors by the calculations described in the Materials and Methods resulted in rev/MJth (Table IV) and rev/kg fuel (Table V). Again, S9 enhanced the mutagenicity emission factors of the extract in all strains (P < 0.05) except in strain YG1041, where there was no difference with or without S9 (P > 0.05).
Table I.
Mutagenicity of EOM of Oil-Burn Emissions
| Dose | Rev/platea | |||||||
|---|---|---|---|---|---|---|---|---|
| (µg EOM/ | −S9 | +S9 | ||||||
| Strain | plate) | Exp 1 | Exp 2 | Exp 3 | Exp 1 | Exp 2 | Exp 3 | |
| TA100 | 0 | 127 | 121 | 130 | 120 | |||
| 1 | 108 | 130 | ||||||
| 5 | 112 | 158 | ||||||
| 10 | 168 | 149 | 258 | 269 | ||||
| 25 | 195 | 175 | 383 | 416 | ||||
| 50 | 229 | 182 | 599 | 529 | ||||
| 100 | 245 | 264 | 809b | 707b | ||||
| NaN3 | 722 | 732 | ||||||
| 2AA | 871 | 895 | ||||||
| Blank | 129 | 109 | ||||||
| YG1041 | 0 | 87 | 89 | 81 | 106 | |||
| 1 | 77 | 70 | ||||||
| 5 | 104 | 107 | ||||||
| 10 | 142 | 168 | 147 | 170 | ||||
| 25 | 202 | 265 | 200 | 248 | ||||
| 50 | 380 | 391 | 346 | 377 | ||||
| 100 | 535 | 616 | 643 | 633 | ||||
| 2NF | 1648 | 1547 | ||||||
| 2AA | 1237 | 1786 | ||||||
| Blank | 100 | 118 | ||||||
| TA98 | 0 | 43 | 56 | 49 | 64 | |||
| 10 | 44 | 60 | 86 | 92 | ||||
| 25 | 70 | 82 | 99 | 102 | ||||
| 50 | 106 | 95 | 161 | 152 | ||||
| 100 | 115b | 119b | 256 | 250 | ||||
| 2NF | 372 | 305 | ||||||
| 2AA | 297 | 594 | ||||||
| Blank | 38 | 45 | 59 | 75 | ||||
| TA104 | 0 | 225 | 210 | 199 | 296 | 273 | 279 | |
| 10 | 243 | 383 | ||||||
| 25 | 278 | 268 | 240 | 453 | 494 | 513 | ||
| 50 | 295 | 292 | 287 | 535 | 602 | 568 | ||
| 100 | 322 | 348 | 345 | 588b | 635b | 716b | ||
| 252.5 | 458 | 454 | 906b | 856b | ||||
| MG | 1338 | 1563 | 1153 | |||||
| 2AA | 497 | 655 | 565 | |||||
| Blank | 221 | 253 | 285 | 326 | ||||
| YG1024 | 0 | 30 | 33 | 46 | 48 | |||
| 10 | 99 | 50 | 108 | 94 | ||||
| 25 | 120 | 90 | 222 | 128 | ||||
| 50 | 177 | 132 | 345 | 215 | ||||
| 100 | 199b | 181b | 360b | 323b | ||||
| 2NF | 2116 | 2065 | ||||||
| 2AA | 1769 | 2226 | ||||||
| Blank | 23 | 33 | 48 | 40 | ||||
| YG1042 | 0 | 108 | 108 | 112 | 123 | |||
| 10 | 138 | 133 | 253 | 215 | ||||
| 25 | 188 | 155 | 362 | 312 | ||||
| 50 | 167 | 181 | 554 | 414 | ||||
| 100 | 232 | 191 | 711b | 599b | ||||
| NaN3 | 977 | 956 | ||||||
| 2AA | 621 | 830 | ||||||
| Blank | 115 | 109 | 123 | 115 | ||||
Data for DMSO (0) controls and the positive controls are the average ± SE of 3 plates each per experiment for a total of 6 plates. Data for the method blanks are for 100 µl/plate and are the average of 2 plates (1 each from 2 independent experiments) except for TA100 and YG1041, which are based on 1 plate from only 1 experiment. Data for the EOM are single plates/dose for each experiment. The positive control data for each experiment represent the average of 3 plates. The positive controls were 2-nitrofluorene (2NF) at 3 µg/plate, 2-aminoanthracene (2AA) at 0.5 µg/plate, sodium azide (NaN3) at 3 µg/plate, and methylglyoxal (MG) at 200 µg/plate. The mean ± SE rev/plate of the DMSO controls for all experiments combined for −/+ S9 were 124 ± 3/125 ± 5 for TA100, 88 ± 1/94 ± 13 for YG1041, 50 ± 7/57 ± 8 for TA98, 211 ± 8/283 ± 7 for TA104, 32 ± 2/47 ± 1 for YG1024, and 108 ± 0/118 ± 6 for YG1042. The mean ± SE rev/plate of the method blanks for all experiments combined for −/+ S9 were 129/109 for TA100, 100/118 for YG1041, 42 ± 4/67 ± 8 for TA98, 237 ± 16/306 ± 21 for TA104, 28 ± 5/44 ± 4 for YG1024, and 112 ± 3/119 ± 4 for YG1042. The mean ± SE rev/plate for the positive controls for all experiments combined for −/+ S9 were 727 ± 5 NaN3/883 ± 12 2AA for TA100, 1598 ± 51 2NF/1512 ± 275 2AA for YG1041, 339 ± 34 2NF/446 ± 149 2AA for TA98, 1155 ± 302 MG/768 ± 198 2AA for TA104, 2091 ± 26 2NF/1998 ± 229 2AA for YG1024, and 967 ± 11 NaN3/726 ± 105 2AA for YG1042.
These data were not used in the linear regressions because they were outside of the linear portion of the dose-response curves.
Table II.
Mutagenic Potency of EOM of Oil-Burn Emissions
| Rev/µg EOM ± SEa | |||
|---|---|---|---|
| Strain | −S9 | +S9 | P-valueb |
| TA100 | 1.3 ± 0.2 | 8.9 ± 0.6 | <0.0001 |
| TA98 | 1.1 ± 0.2 | 1.9 ± 0.1 | <0.0001 |
| TA104 | 0.9 ± 0.1 | 5.6 ± 0.6 | <0.0001 |
| YG1024 | 2.4 ± 0.4 | 4.6 ± 0.9 | 0.0076 |
| YG1041 | 5.0 ± 0.3 | 5.5 ± 0.2 | 0.7629 |
| YG1042 | 0.9 ± 0.2 | 7.1 ± 0.9 | <0.0001 |
Values are the slopes of the linear regressions of the dose-response curves from data in Table I.
Comparison of –S9 versus +S9 based on Student’s t-test.
Table III.
Mutagenic potency of PM2.5 of Oil-Burn Emissions
| Rev/mg PM2.5 ± SEa | |||
|---|---|---|---|
| Strain | −S9 | +S9 | P-valueb |
| TA100 | 93 ± 12 | 620 ± 44 | <0.0001 |
| TA98 | 76 ± 11 | 135 ± 5 | <0.0001 |
| TA104 | 65 ± 4 | 394 ± 44 | <0.0001 |
| YG1024 | 164 ± 30 | 322 ± 63 | 0.0076 |
| YG1041 | 349 ± 19 | 385 ± 11 | 0.7629 |
| YG1042 | 64 ± 12 | 496 ± 65 | <0.0001 |
Values were calculated by multiplying the values in Table II by 1000 and then by the % EOM, which was 7.0%.
Comparison of –S9 versus +S9 based on Student’s t-test.
Table IV.
Mutagenicity Emission Factors per MJth of Oil-Burn Emissions
| Rev x 105/MJth ± SEa | |||
|---|---|---|---|
| Strain | −S9 | +S9 | P-valueb |
| TA100 | 1.3 ± 0.2 | 8.6 ± 1.1 | <0.0001 |
| TA98 | 1.0 ± 0.2 | 1.9 ± 0.2 | <0.0001 |
| TA104 | 0.9 ± 0.1 | 5.4 ± 0.9 | <0.0001 |
| YG1024 | 2.2 ± 0.5 | 4.5 ± 1.1 | 0.0076 |
| YG1041 | 4.8 ± 0.5 | 5.3 ± 0.5 | 0.7629 |
| YG1042 | 0.9 ± 0.2 | 6.8 ± 1.3 | <0.0001 |
Values were calculated by multiplying the values in Table III by 1000 to give rev/g PM2.5, then the resulting values were multiplied by 1.38 g PM2.5/MJth ± 0.08 to give rev/MJth.
Comparison of –S9 versus +S9 based on Student’s t-test.
Table V.
Mutagenicity Emission Factors per kg fuel of Oil-Burn Emissions
| Rev x 105/kg fuel ± SEa | |||
|---|---|---|---|
| Strain | −S9 | +S9 | P-valueb |
| TA100 | 53.9 ± 1.0 | 359.4 ± 47.0 | <0.0001 |
| TA98 | 44.0 ± 9.1 | 78.1 ± 7.6 | <0.0001 |
| TA104 | 37.7 ± 4.7 | 228.8 ± 39.2 | <0.0001 |
| YG1024 | 95.2 ± 23.2 | 188.0 ± 47.0 | 0.0076 |
| YG1041 | 202.4 ± 23.3 | 223.0 ± 19.7 | 0.7629 |
| YG1042 | 37.4 ± 9.5 | 287.6 ± 55.3 | <0.0001 |
Values were calculated by multiplying the values in Table III by 1000 to give rev/g PM2.5, and then the resulting values were multiplied by 58 g PM2.5/kg fuel ± 3.5 to give rev/kg fuel.
Comparison of –S9 versus +S9 based on Student’s t-test.
Expressing the mutagenicity emission factor as rev/MJth (Table IV), the extract ranked generally as follows in the strains in the presence of S9 based on Student’s t-tests: TA98 < YG1024 ≈ YG1041 ≈ TA104 ≈ YG1042 < TA100. We note that the emission factor in YG1042 was not significantly different than in TA100 (P = 0.11); however, the emission factor in YG1042 was not significantly different from those in the other strains except in TA98. The high mutagenicity emission factor of the extract in the base-substitution strain TA100 +S9 indicated that PAHs played a significant role in the mutagenicity of the extract. This was confirmed by the high mutagenicity emission factor in the base-substitution strain YG1042 +S9, which in addition to PAHs, also responds to aromatic amines and nitroarenes.
In the absence of S9, the mutagenicity emission factor (rev/MJth) of the extract ranked generally as follows: TA104 ≈ TA98 ≈ YG1042 ≈ TA100 < YG1024 < YG1041. These results showed that in the absence of S9, the extract had the highest mutagenicity emission factor in the frameshift strains YG1024 and YG1041. This indicated an important role for nitroarenes. Nonetheless, the extract produced the highest mutagenicity emission values in TA100 +S9, and secondarily in YG1024 –S9 and YG1041 –S9, indicating that PAHs were the predominant class of mutagen and that nitroarenes were a secondary class of mutagens in the oil-burn extract (Tables IV and V).
DISCUSSION
Oxidative Damage
Our finding that the organic extract of the PM2.5 from burning oil induced HMOX1 mRNA in human lung cells provides evidence that the emissions from the burning oil during the DWH event may have played a role in the damage found in the lungs and adrenal glands of dead dolphins whose deaths were associated with the DWH event [Venn-Watson et al., 2015]. Heme oxygenase-1, the enzyme that catalyzes the degradation of heme into biliverdin and carbon monoxide, has been ascribed a variety of protective antioxidant functions in the adaptive response to physiologic and xenobiotic oxidative stress [Calay et al., 2014; Li et al., 2016; Loboda et al., 2016; Nussler et al., 2010; Teng et al., 2013].
HMOX1 expression is regulated tightly at the transcriptional level by the Kelch-like ECH-associated protein 1/nuclear erythroid 2-related factor 2/antioxidant response element (KEAP1/Nrf2/ARE) pathway, which is induced by cellular exposure to a broad variety of electrophilic and oxidative stressors [Buendia et al., 2016; Lambros et al., 2016; Loboda et al., 2016]. We have shown previously that exposure of human airway epithelial cells to environmental electrophiles found in ambient air induces the expression of HMOX1 through an oxidant dependent mechanism [Cheng et al., 2012; Silbajoris et al., 2009; Wages et al., 2014].
The HMOX1 expression findings are consistent with an oxidative effect of exposure to the extract of the oil-burn PM on human airway epithelial cells. We have recently reported that environmental electrophiles, including 1,2-naphthoquinone, induce increased ROS production and oxidative modification of regulatory proteins in BEAS-2B cells [Cheng et al., 2012; Wages et al., 2015]. In order to determine the possible contribution of mitochondrial dysregulation in the oxidative effects of the extract of the PM from burning oil, we examined the effect of the extract on mitochondrial function in BEAS-2B cells using extracellular flux analysis. We found that the oxygen-consumption rate was not different from controls following exposure of the cells to EOM for up to 24 h. This suggests that the concentration of electrophilic organic compounds present in the extract was below that required to effect a change in mitochondrial respiration, which would imply a non-mitochondrial source of any reactive oxygen species involved in the mechanism leading to HMOX1 expression.
Our results with the PM extract from the simulated burning of oil are consistent with those of Jaligama et al. [2015] with the OSPM collected during the in situ burns during the DWH event. They showed that the OSPM induced oxidative stress and pulmonary inflammation both systemically and locally as evidenced by increased numbers of macrophages and eosinophils in the lungs of mice. The OSPM also enhanced the allergic asthma response in a mouse model of asthma based on an increase in the number of T-helper 2 cells, resulting in increased peri-bronchiolar inflammation and airway mucus production in the lungs of mice. Similarly, Liu et al. [2016] observed changes in gene expression in cells exposed to crude oil that are similar to those found among asthmatics.
Oxidative stress can account for some portion of these observations, consistent with our finding of the ability of PM from burning oil to induce expression of HMOX1, which is a marker of oxidative stress in mammalian lung cells. Our chemical analysis of the emissions identified various dioxins [Gullett et al., 2017], which also could contribute to oxidative damage. Nonetheless, from a mechanistic standpoint, the induction of HMOX1 expression implicates an oxidant mode of action for the adverse health effects of inhaled PM from burning oil. This would be consistent with the basis for the lesions in the lungs and adrenal glands of dead dolphins associated with the DWH event [Schwacke et al., 2014a,b; Venn-Watson et al., 2015].
Mutagenicity
The extract of the emissions from the open burning of oil was most mutagenic in strain TA100 +S9 whether expressed as a mutagenic potency (rev/µg EOM or rev/mg PM2.5) or as a mutagenicity emission factor (rev/MJth or rev/kg fuel). This implied that PAHs were the predominant class of mutagen, which was consistent with our chemical analysis showing that the total concentration of the 16 EPA Priority PAHs in these emissions was quite high, accounting for 1 g/kg of oil consumed [Gullett et al., 2017]. Consistent with this was the high mutagenicity of the extract in the two other base-substitution strains in the presence of S9 (TA104 and YG1042); PAHs induce primarily base-substitution mutations [DeMarini et al., 1995]. In contrast, the extract was most mutagenic in the frameshift strains in the absence of S9, indicating a role for nitroarenes. Although Sheppard et al. (1983) found that the emissions from burning Prudhoe Bay crude oil were mutagenic in TA98, we cannot compare their results with ours due to the differences in collection and extraction of the samples between the two studies.
Although TA104 permits recovery of mutants due to mutations at either GC or AT sites, the extract was consistently less mutagenic in TA104 than in TA100. Because of the availability of AT sites for mutagenesis in TA104, this strain can respond to oxidative mutagens, which typically cause DNA damage at AT sites [Levin et al., 1982]. However, the lower amount of mutagenesis in this strain relative to TA100, which contains only GC sites for mutant recovery, indicates that oxidative mutagenesis, if induced, was minor relative to that induced by PAHs, which produces mutations primarily at GC sites [DeMarini et al., 1995].
Although we did not conduct chemical analyses of the emissions for aromatic amines and nitroarenes [Gullett et al., 2017], the strong mutagenesis of the extract in YG1042 +S9 and YG1024 +S9 implies a role for aromatic amines, and the strong response in YG1041 –S9 and YG1024 –S9 indicates a role for nitroarenes. These results are consistent with those from other combustion emissions whose mutagenicity is also associated with PAHs, nitroarenes, and aromatic amines, such as diesel exhaust [IARC, 2010; Mutlu et al., 2013, 2015], woodsmoke [Mutlu et al., 2016], and air pollution due to combustion emissions [IARC, 2016].
To evaluate the mutagenicity of the oil-burn extract relative to other combustion emissions, we compared the mutagenicity emission factor of the extract to that of other combustion emissions in TA98 +S9 because mutagenicity emission values for some of the emissions are available only in TA98+S9 (Table VI). We have shown previously that the mutagenicity emission factors for a variety of combustion emissions rank similarly across 16 strain/S9 combinations of Salmonella; thus comparisons among a single strain/S9 combination result in a reliable comparative analysis among a set of emissions [Mutlu et al., 2015].
Table VI.
Mutagenicity Emission Factors of Various Emissions in TA98+S9
| Emissiona | Rev/MJth ± SEb |
|---|---|
| Utility Natural Gas | 20 |
| Utility coal | 230 |
| Utility Wood | 1,000 |
| Residential Oil | 2,500 |
| 150-kw Diesel Engine | 2,760 |
| 4.3-kw Diesel Generator | 42,460 ± 8,000 |
| Forced-Draft Stove | 20,000 ± 4,399 |
| Natural-Draft Stove | 123,005 ± 20,663 |
| Oil Burn | 190,000 ± 20,000 |
| Three-Stone Fire | 242,739 ± 21,226 |
| Residential Wood | 250,000 |
| Open Burning of Agricultural Plastic | 250,000 |
| Open Burning of Tires | 2,000,000 |
Data for Oil Burn are from Table IV; data for Open Burning of Tires are from DeMarini et al. [1994]; data for Forced-Draft Stove, Natural-Draft Stove, and Three-Stone Fire are from Mutlu et al. [2016]; data for the two diesel sources are from Mutlu et al. [2015, 2016]; and the remainder are from DeMarini et al. [1992]. All of these data are plotted in Figure 3.
Comparisons among the 5 values with SEs are significantly different based on Student’s t-tests, P < 0.05.
Such a comparison is plotted in Figure 3, and where available, standard errors are shown in both Table VI and Figure 3. The mutagenicity emission factor of the extract of burning oil emissions was between that of wood burned in a natural-draft stove and a 3-stone fire, and was significantly different from both (P < 0.05). However, it was within an order of magnitude of those of a variety of other open-burning emissions, such as residential wood fireplaces and the open burning of agricultural plastic, which is itself a petroleum product [Table VI, Fig. 3]. As with the emissions from the burning oil, our chemical analyses and mutagenicity data in various strains of Salmonella showed that most of the mutagenicity of the emissions from open burning as well as from cookstoves was due largely to PAHs [DeMarini et al., 1992, 1994; Mutlu et al., 2015, 2016]. Our analysis placed the mutagenicity emission factor of the oil-burn emissions high among other emissions we have evaluated, and it was exceeded by an order of magnitude only by that of the emissions from the open burning of tires.
Figure 3.
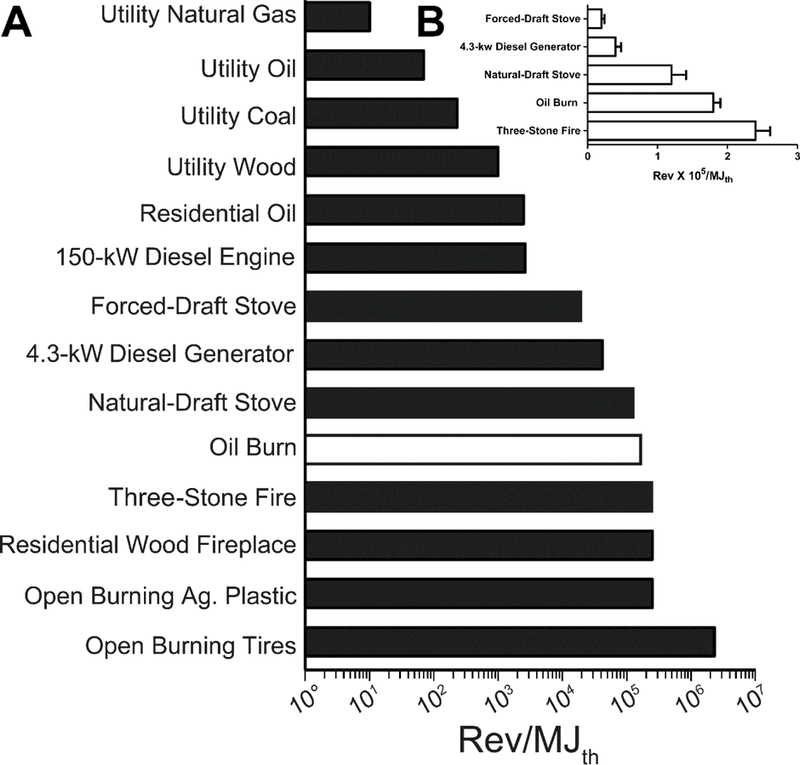
Mutagenicity emission factors in TA98 +S9 for a variety of emissions; data from Table VI. (A) Data plotted on a log scale; (B) subset of the data replotted on a linear scale showing standard error values; these mutagenicity emission factors were significantly different from one another (P <0.05).
Collectively, our data show that (a) the organics from the PM2.5 of the emissions from the open burning of oil were highly mutagenic, (b) PAHs played a predominant role in the mutagenesis, (c) the mutagenicity emission factor was within an order of magnitude of that of other types of open burning of either wood or plastic, and (d) the emissions induced oxidative damage to human lung cells in vitro. The adverse health effects of combustion-derived PM, especially to the lung, are well documented [IARC, 2004, 2014, 2016] and consistent with the adverse effects shown here of PM from combusted oil. The ability of the organics from the open burning oil to induce oxidative effects [Jaligama et al., 2015] and mutagenesis due to PAHs and other chemical classes also likely contributed to the various types of damage that occurred to natural resources within the DWH area [Paruk et al., 2016; Schwacke et al., 2014a,b; Venn-Watson et al., 2015; Zengel et al., 2016], the full impact of which has yet to be fully characterized [Joye, 2015].
ACKNOWLEDGMENTS
This project was part of the commitment made by the U.S. EPA as a trustee under the Natural Resource Damage Assessment (NRDA) and was funded by the National Pollution Funds Center of the U.S. Coast Guard. We thank the Aberdeen Test Center of the U.S. Army in Aberdeen, MD, and Kevin Powell, Dan Kogut, and colleagues who organized and enabled our access to the Center to conduct the oil burns. We also thank Roy Habbaz and Samuel Gauthe for providing oil from the Strategic Petroleum Reserve of the U.S. Department of Energy. We also thank the staff at the Office of Water of the U.S. EPA who headed the NRDA program, especially Catherine Aubee, Elizabeth Skane, Gale Bonanno, Jim Pendergast, Mary Kay Lynch, Tom Wall, Jim Bove, Bob Brown, and Marcia McCain. This article was reviewed by the National Health and Environmental Effects Research Laboratory, U.S. EPA, and approved for publication. Approval does not signify that the contents reflect the views of the agency nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
REFERENCES
- Allan SE, Smith BW, Anderson KA. 2012. Impact of the Deepwater Horizon oil spill on bioavailable polycyclic aromatic hydrocarbons in Gulf of Mexico coastal waters. Environ Sci Technol 46:2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurell J, Gullett BK. 2010. Aerostat Sampling of PCDD/PCDF Emissions from the Gulf Oil Spill In Situ Burns. Environ Sci Technol 44:9431–9437. [DOI] [PubMed] [Google Scholar]
- Buendia I, Michalska P, Navarro E, Gameiro I, Egea J, Leon R. 2016. Nrf2-ARE pathway: An emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol Therapeutics 157:84–104. [DOI] [PubMed] [Google Scholar]
- Calay D, Mason JC. 2014. The multifunctional role and therapeutic potential of HO-1 in the vascular endothelium. Antioxidants Redox Signal 20:1789–1809. [DOI] [PubMed] [Google Scholar]
- Cheng WY, Currier J, Bromberg PA, Silbajoris R, Simmons SO, Samet JM. 2012. Linking oxidative events to inflammatory and adaptive gene expression induced by exposure to an organic particulate matter component. Environ Health Perspect 120:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea MA, Reddy GK. 2013. Health consequences among subjects involved in Gulf oil spill clean-up activities. Am J Med 126:966–974. [DOI] [PubMed] [Google Scholar]
- de Gouw JA, Middlebrook AM, Warneke C, Ahmadov R, Atlas EL, Bahreini R, Blake DR, Brock CA, Brioude J, Fahey DW, Fehsenfeld FC, Holloway JS, Le Henaff M, Lueb RA, McKeen SA, Meagher JF, Murphy DM, Paris C, Parrish DD, Perring AE, Pollack IB, Ravishankara AR, Robinson AL, Ryerson TB, Schwarz JP, Spackman JR, Srinivasan A, Watts LA. 2011. Organic aerosol formation downwind from the Deepwater Horizon oil spill. Science 331:1295–1299. [DOI] [PubMed] [Google Scholar]
- DeMarini DM, Lemieux PM, Ryan JV, Brooks LR, Williams RW. 1994. Mutagenicity and chemical analysis of emissions from the open burning of scrap rubber tires. Environ Sci Technol 28:136–141. [DOI] [PubMed] [Google Scholar]
- DeMarini DM, Shelton ML, Levine JG. 1995. Mutation spectrum of cigarette smoke condensate in Salmonella: comparison to mutations in smoking-associated tumors. Carcinogenesis 16:2535–2542. [DOI] [PubMed] [Google Scholar]
- DeMarini DM, Williams RW, Perry E, Lemieux PM, Linak WP. 1992. Bioassay-directed chemical analysis of organic extracts of emissions from a laboratory-scale incinerator: combustion of surrogate compounds. Combust Sci Technol 85:437–453. [Google Scholar]
- Gullett BK, Aurell J, Holder A, Mitchell W, Greenwell D, Hays M, Conmy R, Tabor D, Preston W, George I, Abrahamson JP, Vander Wall R, Holder E. 2017. Characterization of emissions and residues from simulations of the Deepwater Horizon surface oil burns. In press. [DOI] [PMC free article] [PubMed]
- Gullett BK, Hays MD, Tabor D, Vander Wal R. 2016. Characterization of the particulate emissions from the BP Deepwater Horizon surface oil burns. Marine Pollut Bull 107:216–223. [DOI] [PubMed] [Google Scholar]
- IARC. 2004. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco Smoke and Involuntary Smoking Vol 83 Available at: http://monographs.iarc.fr/ENG/Monographs/vol83/mono83.pdf [Accessed 20 May 2016]. [PMC free article] [PubMed] [Google Scholar]
- IARC. 2014. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Diesel and Gasoline Engine Exhausts and Some Nitroarenes Vol 105 Available at: http://monographs.iarc.fr/ENG/Monographs/vol105/mono105.pdf [Accessed 20 May 2016]. [PMC free article] [PubMed] [Google Scholar]
- IARC. 2016. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Outdoor Air Pollution Vol 109 Available at: http://monographs.iarc.fr/ENG/Monographs/vol109/mono109.pdf [Accessed 20 January 2017.]. [PMC free article] [PubMed] [Google Scholar]
- Jaligama S, Chen Z, Saravia J, Yadav N, Lomnicki SM, Dugas TR, Cormier SA. 2016. Exposure to Deepwater Horizon crude oil burnoff particulate matter induces pulmonary inflammation and alters adaptive immune response. Environ Sci Technol 49:8769–8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joye SB. 2015. Deepwater Horizon, 5 years on. Science 349:592–593. [DOI] [PubMed] [Google Scholar]
- Lambros ML, Plafker SM. 2016. Oxidative Stress and the Nrf2 Anti-Oxidant Transcription Factor in Age-Related Macular Degeneration. Advances Exper Med Biol 854:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SM., Smith CR, Mitchell J, Balmer BC, Barry KP, McDonald T, Mori CS, Rosel PE, Rowles TK, Speakman TR, Townsend FI, Tumlin MC, Wells RS, Zolman ES, Schwacke LH. 2015. Reproductive outcome and survival of common bottlenose dolphins sampled in Barataria Bay, Louisiana, USA, following the Deepwater Horizon oil spill. Proc Royal Soc B-Biol Sci 282:20151944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Hollstein M, Christman MF, Schwiers EA, Ames BN. 1984. A new Salmonella tester strain (TA102) with A X T base pairs at the site of mutation detects oxidative mutagens. Proc Natl Acad Sci USA 79:7445–7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ye F, Li L, Chang W, Wu X, Chen J. 2016. The role of HO-1 in protection against lead-induced neurotoxicity. Neurotoxicology 52:1–11. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Roy-Engel AM, Baddoo MC, Flemington EK, Wang G, Wang H. 2016. The impact of oil spill to lung health—insights from an RNA-seq study of human airway epithelial cells. Gene 578:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Damulewicz M, Pyza E, Jozkowicz A, Dulak J. 2016. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell Mol Life Sci, in press. [DOI] [PMC free article] [PubMed]
- Mabile N, Allen A. 2010. Controlled burns: after-action report. Controlled Burn Group report Available at: http://www.mdl2179trialdocs.com/releases/release201501260800005/TREX-241730.pdf [Accessed 29 May 2016].
- Maron DM, Ames BN. 1983. Revised methods for the Salmonella mutagenicity test. Mutat Res 113:173–215. [DOI] [PubMed] [Google Scholar]
- Middlebrook AM., Murphy DM, Ahmadov R, Atlas EL, Bahreini R, Blake DR, Brioude J, de Gouw JA, Fehsenfeld FC, Frost GJ, Holloway JS, Lack DA, Langridge JM, Lueb RA, McKeen SA, Meagher JF, Meinardi S, Neuman JA, Nowak JB, Parrish DD, Peischl J, Perring AE, Pollack IB, Roberts JM, Ryerson TB, Schwarz JP, Spackman JR, Warneke C, Ravishankara AR. 2012. Air quality implications of the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109: 20280–20285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu E, Warren SH, Ebersviller SM, Kooter IM, Schmid JE, Dye JA, Linak WP, Gilmour MI, Jetter JJ, Higuchi M, DeMarini DM. 2016. Mutagenicity and pollutant emission factors of solid-fuel cookstoves: comparison with other combustion sources. Environ Health Perspect 124, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu E, Warren SH, Matthews PP, King C, Linak WP, Kooter IM, Schmid JE, Ross JA, Gilmour MI, DeMarini DM. 2013. Bioassay-directed fractionation and sub-fractionation for mutagenicity and chemical analysis of diesel exhaust particles. Environ Mol Mutagen 54:719–736. [DOI] [PubMed] [Google Scholar]
- Mutlu E, Warren SH, Matthews PP, King C, Walsh L, Kligerman AD, Schmid JE, Janek D, Kooter IM, Linak WP, Gilmour MI, DeMarini DM. 2015. Health effects of soy biodiesel emissions: mutagenicity emission factors. Inhal Toxicol 27:585–596. [DOI] [PubMed] [Google Scholar]
- National Commission on the BP Deepwater Horizon Oil Spill and Offshore Drilling. 2011. Deep Water: The Gulf Oil Disaster and the Future of Offshore Drilling. Report to the President Available at: http://cybercemetery.unt.edu/archive/oilspill/20121211005728/http://www.oilspillcommission.gov/sites/default/files/documents/DEEPWATER_ReporttothePresident_FINAL.pdf [Accessed 29 May 2016].
- Nussler AK, Hao L, Knobeloch D, Yao P, Nussler NC, Wang Z, Liu L, Ehnert S. 2010. Protective role of HO-1 for alcohol-dependent liver damage. Digestive Diseases 28:792–798. [DOI] [PubMed] [Google Scholar]
- Paruk JD, Adams EM, Uher-Koch H, Kovach KA, Long D IV, Perkins C, Schoch N, Evers DC. 2016. Polycyclic aromatic hydrocarbons in blood related to lower body mass in common loons. Sci Total Environ 565:360–368. [DOI] [PubMed] [Google Scholar]
- Peres LC, Trapido E, Rung AL, Harrington DJ, Oral E, Fang Z, Fontham E, Peters ES. 2016. The Deepwater Horizon oil spill and physical health among adult women in Southern Louisiana: the women and their children’s health (WaTCH) study. Environ Health Perspect 124:1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perring AE, Schwarz JP, Spackman JR, Bahreini R, de Gouw JA, Gao RS, Holloway JS, Lack DA, Langridge JM, Peischl J, Middlebrook AM, Ryerson TB, Warneke C, Watts LA, Fahey DW. 2011. Characteristics of black carbon aerosol from a surface oil burn during the Deepwater Horizon oil spill. Geophys Res Lett 38:L17809. [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT PCR. Nucleic Acids Res 29:(9)e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S, Wong RM-Y, Sims SH, Schaaper RM, DeMarini DM, Mcclelland M. 2001. The ∆uvrB mutations in the Ames strains of Salmonella span 47–199 genes. Mutat Res 483:1–11. [DOI] [PubMed] [Google Scholar]
- Putman NF, Abreu-Grobois FA, Iturbe-Darkistade I, Putman EM, Richards PM, Verley P. 2015. Deepwater Horizon oil spill impact on sea turtles could span the Atlantic. Biol Lett 11:20150596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz DA, Engel LS. 2010. Lessons for study of the health effects of oil spills. Ann Intern Med 153:540–541. [DOI] [PubMed] [Google Scholar]
- Schaum J, Cohen M, Perry S, Artz R, Draxler R, Frithsen JB, Heist D, Lorber M, Phillips L. 2010. Screening Level Assessment of Risks Due to Dioxin Emissions from Burning Oil from the BP Deepwater Horizon Gulf of Mexico Spill. Environ Sci Technol 44:9383–9389. [DOI] [PubMed] [Google Scholar]
- Schwacke LH, Smith CR, Townsend FI, Wells RS, Hart LB, Balmer BC, Collier TK, De Guise S, Fry MM, Guiillete LJ Jr, Lamb SV, Lane SM, McFee WE, Place NJ, Tumlin MC, Ylitalo GM, Zolman ES, Rowles TK. 2014a. Health of common bottlenose dolphins (Tursiops truncatus) in Barataria Bay, Louisiana, following the Deepwater Horizon Oil Spill. Environ Sci Technol 48:93–103. [DOI] [PubMed] [Google Scholar]
- Schwacke LH, Smith CR, Townsend FI, Wells RS, Hart LB, Balmer BC, Collier TK, De Guise S, Fry MM, Guiillete LJ Jr, Lamb SV, Lane SM, McFee WE, Place NJ, Tumlin MC, Ylitalo GM, Zolman ES, Rowles TK. 2014b. Response to comment on health of common bottlenose dolphins (Tursiops truncatus) in Barataria Bay, Louisiana, following the Deepwater Horizon Oil Spill. Environ Sci Technol 48:4209–4211. [DOI] [PubMed] [Google Scholar]
- Sheppard EP, Wells RA, Georghiou PE. 1983. The mutagenicity of a Prudhoe bay crude oil and its residues from an experimental in situ burn. Environ Res 30:427–441. [DOI] [PubMed] [Google Scholar]
- Silbajoris R, Huang JM, Cheng W-Y, Dailey L, Tal T, Jaspers I, Ghio AJ, Bromberg PA, Samet JM. 2009. Nanodiamond particles induce IL-8 expression through a transcript stabilization mechanism in human airway epithelial cells. Nanotoxicology 3:152–160. [Google Scholar]
- Stieglitz JD, Mager EM, Hoenig RH, Benetti DD, Grosell M. 2016. Impacts of Deepwater Horizon crude oil exposure on adult mahi-mahi (Coryphaena hippurus) swim performance. Environ Toxicol Chem, in press. [DOI] [PubMed]
- Stout SA, Payne JR. 2016. Chemical composition of floating and sunken in-situ burn residues from the Deepwater Horizon oil spill. Marine Pollut Bull, in press. [DOI] [PubMed]
- Teng YC, Tai YI, Lee YH, Lin AM. 2013. Role of HO-1 in the arsenite-induced neurotoxicity in primary cultured cortical neurons. Mol Neurobiol 48:281–287. [DOI] [PubMed] [Google Scholar]
- Tidwell LA, Allan SE, O’Connell SG, Hobbie KA, Smith BW, Anderson KA. 2015. Polycyclic aromatic hydrocarbon (PAH) and oxygenated PAH (OPAH) air-water exchange during the Deepwater Horizon oil spill. Environ Sci Technol 49:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Venn-Watson S, Colegrove KM, Litz J, Kinsel M, Terio K, Saliki J, Fire S, Carmichael R, Chevis C, Hatchett W, Pitchford J, Tumlin M, Field C, Smith S, Ewing R, Fauquier D, Lovewell G, Whitehead H, Rotstein D, McFee W, Fougeres E, Rowles T. 2015. Adrenal Gland and Lung Lesions in Gulf of Mexico Common Bottlenose Dolphins (Tursiops truncatus) Found Dead following the Deepwater Horizon Oil Spill. PLoS One 10(5):e0126538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venn-Watson S, Smith CR, Jensen ED, Rowles T. 2013. Assessing the potential health impacts of the 2003 and 2007 firestorms on bottlenose dolphins (Tursiops truncatus) in San Diego Bay. Inhal Toxicol 25:481–491. [DOI] [PubMed] [Google Scholar]
- Wages PA, Lavrich KS, Zhang Z, Cheng WY, Corteselli E, Gold A, Bromberg P, Simmons SO, Samet JM. 2015. Protein Sulfenylation: A Novel Readout of Environmental Oxidant Stress. Chem Res Toxicol 28:2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wages PA, Silbajoris R, Speen A, Brighton L, Henriquez A, Tong H, Bromberg PA, Simmons SO, Samet JM. 2014. Role of H2O2 in the oxidative effects of zinc exposure in human airway epithelial cells. Redox Bio 3:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel S, Montague CL, Pennings SC, Powers SP, Steinhoff M, Fricano G, Schlemme C, Zhang M, Oehring J, Nixon Z, Rouhani S, Michel J. 2016. Impact of the Deepwater Horizon oil spill on salt marsh periwinkles (Littoraria irrorata). Environ Sci Technol 50:643–652. [DOI] [PubMed] [Google Scholar]


