Abstract
Giardia duodenalis and Cryptosporidium spp. are common human and animal pathogens. They have increasingly been reported in dairy calves in recent years; however, multilocus genotyping information for G. duodenalis and Cryptosporidium infecting pre-weaned dairy calves in southwestern China is limited. In the present study, the prevalence of G. duodenalis and Cryptosporidium spp. in pre-weaned dairy calves in central Sichuan province was determined and the pathogens were analyzed molecularly. Of 278 fecal samples from pre-weaned dairy calves, 26 (9.4%) were positive for G. duodenalis and 40 (14.4%) were positive for Cryptosporidium spp. Cryptosporidium bovis (n = 28), Cryptosporidium ryanae (n = 5) and Cryptosporidium parvum (n = 7) were detected. All seven C. parvum isolates were successfully subtyped based on the gp60 gene sequence, and only IIdA15G1 was detected. Multilocus sequence typing of G. duodenalis based on beta-giardin (bg), triose phosphate isomerase (tpi) and glutamate dehydrogenase (gdh) genes revealed 19 different assemblage E multilocus genotypes (two known and 17 unpublished genotypes). Based on eBURST analysis, a high degree of genetic diversity within assemblage E was observed in pre-weaned dairy calves in Sichuan province. To the best of our knowledge, this is the first study using multilocus sequence typing and eBURST analysis to characterize G. duodenalis in pre-weaned dairy calves in southwestern China.
Keywords: Giardia duodenalis, Cryptosporidium, Multilocus genotyping, Pre-weaned dairy calves, Sichuan province
Abstract
Giardia duodenalis et Cryptosporidium spp. sont des pathogènes humains et animaux communs. Ils ont été signalés de plus en plus chez les veaux laitiers au cours des dernières années; cependant, l’information de génotypage multilocus pour G. duodenalis et Cryptosporidium infectant les veaux laitiers pré-sevrés dans le sud-ouest de la Chine est limitée. Dans la présente étude, la prévalence de G. duodenalis et de Cryptosporidium spp. chez les veaux laitiers pré-sevrés dans la province centrale du Sichuan a été déterminée et les pathogènes ont été analysés moléculairement. Dans 278 échantillons fécaux de veaux laitiers pré-sevrés, 26 (9.4 %) étaient positifs pour G. duodenalis et 40 (14.4 %) étaient positifs pour Cryptosporidium spp. Cryptosporidium bovis (n = 28), Cryptosporidium ryanae (n = 5) et Cryptosporidium parvum (n = 7) ont été détectés. Les sept isolats de C. parvum ont été sous-typés avec succès sur la base de la séquence du gène gp60 et seul IIdA15G1 a été détecté. Le typage multilocus de G. duodenalis basé sur les gènes de béta-giardine (bg), triose phosphate isomérase (tpi) et glutamate déshydrogénase (gdh) a révélé 19 génotypes différents d’assemblage multilocus E (deux génotypes connus et 17 non-publiés). D’après l’analyse eBURST, on a observé un degré élevé de diversité génétique au sein de l’assemblage E chez les veaux laitiers pré-sevrés de la province du Sichuan. À notre connaissance, il s’agit de la première étude utilisant le typage de séquence multilocus et l’analyse eBURST pour caractériser G. duodenalis chez des veaux laitiers pré-sevrés dans le sud-ouest de la Chine.
Introduction
Protists of the genera Giardia and Cryptosporidium infect a wide range of animals as well as humans [3, 12, 19]. Typically, the infection is acquired following the ingestion of highly resilient, infective stages (oocysts or cysts) via the fecal-oral route [3, 4]. Disease is commonly associated with clinical signs including diarrhea, dehydration, fever, inappetence and anorexia. Infections are often self-limiting in immune-competent individuals [2, 31], but can be chronic and severe in infants, elderly people, and immune-compromised individuals [9, 16].
Ruminants are recognized as a significant reservoir of Giardia and Cryptosporidium taxa that infect animals and humans [19, 21]. Current data indicate that of the eight assemblages within Giardia duodenalis, assemblages A and E and the Cryptosporidium species C. parvum, C. andersoni, C. ryanae, and C. bovis predominate in cattle worldwide [4, 20].
Unlike in other countries (e.g. Australia, Sudan, Japan and India) [6, 14, 15, 24], where C. parvum is known to be the predominant species in pre-weaned calves, this does not appear to be the case everywhere in China. Some studies have shown that C. parvum is a major species in pre-weaned calves in some regions, whereas C. bovis is a major species in other regions [5, 25, 28].
According to the National Bureau of Statistics of the People’s Republic of China, in 2016, the total population of dairy cattle in Sichuan Province was 176 thousand heads. However, no information about G. duodenalis and Cryptosporidium infection of pre-weaned dairy calves was previously available in Sichuan Province. We undertook a molecular epidemiological study to obtain a preliminary snap-shot of the prevalence of G. duodenalis assemblages and Cryptosporidium genotypes in pre-weaned calves in Sichuan province, China.
Materials and methods
Sample collection
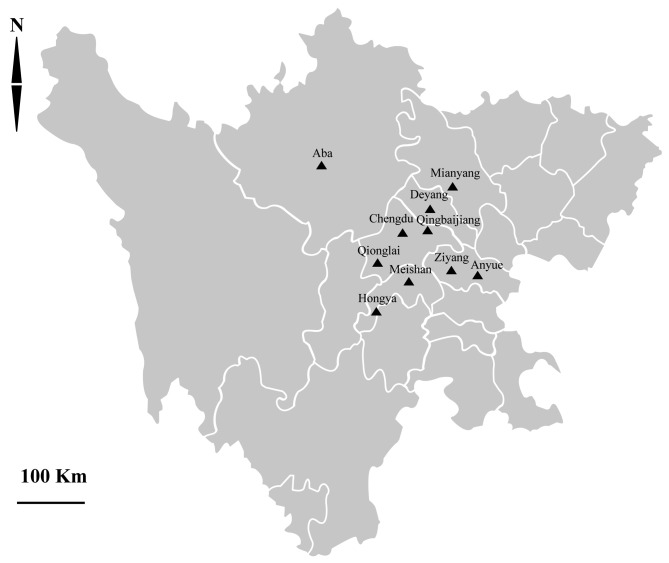
We collected 278 rectal fecal samples from pre-weaned dairy calves (<1 month of age) from 10 farms with a history of bovine diarrhea in 10 regions in Sichuan province, southwestern China, between June 2016 and March 2017. Collection sites included: Chengdu (104°06′E, 30°57′N), Hongya (103°37′E, 29°91′N), Aba (102°22′E, 31°90′N), Meishan (103°84′E, 30°08′N), Mianyang (104°67′E, 31°47′N), Ziyang (104°62′E, 30°13′N), Anyue (105°33′E, 30°10′N), Qionglai (103°46′E, 30°41′N), Qingbaijiang (104°25′E, 30°88′N), and Deyang (104°39′E, 31°13′N). The 10 farms are distributed in central Sichuan Province (Fig. 1). The city-level map was provided by the National Geomatics Centre of China (National Geomatics Centre of China, Beijing, China, http://ngcc.sbsm.gov.cn/).
Figure 1.
Distribution of sampling sites in Sichuan province in this study.
Of the 10 farms, six (Chengdu, Hongya, Aba, Mianyang, Ziyang, and Qionglai) are intensive feeding farms, while the other four are free-ranging. For intensive farms, there were approximately 1000–2500 cattle per farm, with more than 100 pre-weaned dairy calves, and the fecal samples were randomly collected from about 20% in each farm. For free-ranging farms, there were approximately 100–120 cattle per farm and the herd sizes of pre-weaned dairy calves were less than 50; in this case we collected fecal samples from all of the pre-weaned dairy calves at each farm (Table S1). In intensive farms, calves were bred in different calf stalls, with one hour outdoor time after eating and excretion in the morning and afternoon, respectively. Calves shared one yard during the outdoor time in intensive farms. In free-ranging farms, calves were kept in a field with a half cover and were raised together. The farms we selected had solely cattle, and no other animals.
Fecal samples were collected from the rectum using disposable gloves, transferred into disposable plastic bags, and stored in 2.5% potassium dichromate at 4 °C.
DNA extraction
Before DNA extraction, feces were washed with distilled water to remove potassium dichromate. Genomic DNA was extracted from 250 mg (approximately) of individual samples using the Power Soil DNA isolation kit (MOBIO, USA), according to the manufacturer’s instructions, and frozen at −20 °C until use.
PCR amplification and sequencing
G. duodenalis was detected by nested PCR amplification of the bggene. The bg-positive samples were further characterized by amplifications of gdh and tpi. Genotyping of Cryptosporidium was based on amplification of the small subunit (SSU) rRNA gene by nested PCR and subsequent sequence analysis. All the C. parvum isolates were further characterized by amplification of the gp60 gene. The primers and amplification conditions in this study were described previously [1, 11, 23]. Positive and negative controls were included in each test. The secondary PCR products were visualized under UV light after electrophoresis on a 1% agarose gel mixed with Golden View.
All positive secondary PCR products were sent to Invitrogen (Shanghai, China) and sequenced in both directions. Sequences were aligned with reference sequences from GenBank using BLAST (http://blast.ncbi.nlm.nih.gov) and ClustalX.
A previous nomenclature system was used to name subtypes at each genetic locus [29, 30]. Specimens that were successfully subtyped at all three loci were included in multilocus genotyping of G. duodenalis. The genetic pedigree of the assemblage E multilocus genotypes (MLGs) was assessed by using eBURST 3.0 (http://eBURST.mlst.net).
Statistical analysis
The χ2 test was used to compare the infection rates of G. duodenalis and Cryptosporidium in different feeding patterns. Differences were considered significant at p < 0.05.
Results and discussion
G. duodenalis was detected in 9.4% of 278 pre-weaned dairy calves on 6 of 10 farms, with prevalences ranging from 7.7% to 46.4% (Table 1). Its prevalence shows substantial differences, ranging from 7.1% to 60.1% in other studies in China [7, 13, 18, 26, 29]. In this study, the overall infection rate in southwestern China was close to the prevalence in northwestern (9.7% [18]), northeastern (13.3% [13]) and north China (7.1% [7]), but much lower than the infection rates in central (17.6% [26]) and southeastern (60.1% [29]) China. Prior to the present study, these results were interpreted as related to differences in geographic distribution, environmental management and cultivation scale [7, 13, 18, 26, 29]. Cattle were kept in groups or in free stalls, which might promote the transmission of G. duodenalis infection among animals and lead to the high infection rates [26, 29]. Furthermore, we analyzed the infection rates between intensive feeding and free-ranging farms; there was no significant difference between the two breeding patterns (X 2 = 0.629, df = 1, p = 0.428).
Table 1.
Prevalence of Cryptosporidium and G. duodenalis in pre-weaned diary calves in Sichuan province.
| Region | No. tested |
Cryptosporidium
|
Cryptosporidium No. (%) of positive specimens | G. duodenalis | G. duodenalis infection rate | ||
|---|---|---|---|---|---|---|---|
| C. bovis | C. ryanae | C. parvum | |||||
| Chengdua | 39 | 2 | 1 | 3 (7.7%) | 3 | 7.7% | |
| Hongyaa | 24 | 1 | 1 (4.2%) | ||||
| Abaa | 20 | 7 | 7 (35.0%) | 2 | 10.0% | ||
| Meishanb | 20 | ||||||
| Mianyanga | 58 | 8 | 3 | 11 (19.0%) | |||
| Ziyanga | 26 | 8 | 8 (30.8%) | 2 | 7.7% | ||
| Anyueb | 22 | 2 | 2 (9.1%) | 3 | 13.6% | ||
| Qionglaia | 28 | 4 | 4 (14.3%) | 13 | 46.4% | ||
| Qingbaijiangb | 20 | 2 | 2 (10.0%) | 3 | 15.0% | ||
| Deyangb | 21 | 1 | 1 | 2 (9.5%) | |||
| Total | 278 | 28 | 5 | 7 | 40 (14.4%) | 26 | 9.4% |
Intensive farming;
free-ranging.
Cryptosporidium was detected in 14.4% of 278 fecal samples, on 9 out of 10 farms, with prevalences ranging from 4.2% to 35.0% (Table 1). The overall infection rate for Cryptosporidium is lower than the average prevalence of 19.5% reported previously in pre-weaned cattle in China [5], but similar to the infection rate reported in Xinjiang (15.6%) [17], and much higher than the rate in Hebei and Tianjin, China (1.0%) [7]. Prevalence of Cryptosporidium was significantly different (X 2 = 4.924, df = 1, p = 0.026) between intensive feeding and free-ranging farms in this study, which suggests that cultivation scale may lead to differences in infection rates with Cryptosporidium. Other studies also showed that geographic distribution and host health status may lead to the difference [5, 10, 17]. Three species of Cryptosporidium (28 C. bovis, 7 C. parvum [subtype IIdA15G1] and 5 C. ryanae) were identified in this study. Previous studies have shown that C. parvum is a major species in pre-weaned calves in Beijing [10], Xinjiang [17] and Ningxia [8], whereas C. bovis predominated in pre-weaned calves in this study, similar to reports from Henan [27] and Heilongjiang [32].
Giardia duodenalis in all 26 positive samples corresponded to assemblage E. G. duodenalis infection is relatively common in pre-weaned dairy calves. We further characterized the 26 G. duodenalis bg-positive samples at the tpi and gdh loci. Among these 26 samples, the tpi and gdh loci were successfully amplified and sequenced in 24 and 25 specimens, respectively. The bg, tpi and gdh loci all showed high levels of sequence polymorphism; seven subtypes were identified at each locus. Of the bg subtypes, E1 (MF671885), E8 (KY769093), E9 (KY769091), and E15 (KT698677) were known, and E13 (MF671880), E14 (MF671883), and E16 (MF671886) were unpublished. At the tpi locus, five known subtypes E1 (MF671900), E3 (KT922259), E9 (EF654690), E15 (KY432848) and E19 (KY769103) and two unpublished subtypes, E21 (MF671904) and E24 (MF671907), were found. The sequences from the gdh locus represented five known subtypes E1 (MF671891), E3 (KT369780), E8 (KT368785), E10 (KT698971), E13 (KY432838) and two unpublished subtypes, E19 (MF671896) and E20 (MF671899).
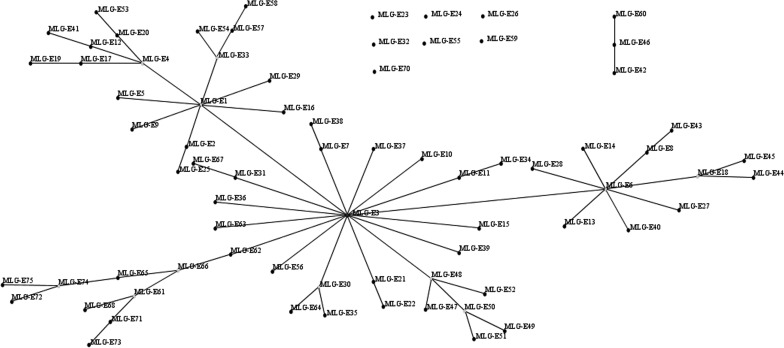
For G. duodenalis, multi-locus genotyping analysis suggested a high genetic diversity of assemblage E in pre-weaned dairy calves in this study. Based on the combination of bg, tpi and gdh loci, 19 MLGs of assemblage E were detected (Table 2). A high degree of nucleotide variation in assemblage E has been also detected in previous studies [18, 22, 26, 29]. Of the 19 MLGs, 17 were unpublished MLGs. The majority of MLGs were MLG-E3 and MLG-E13, which have also been detected in dairy calves in Shanghai [29]. To further analyze the evolutionary descent of the 19 assemblage E MLGs, we used eBURST analysis of the 19 assemblage E MLGs and 58 reference MLGs, which revealed two clonal complexes and seven singletons (Fig. 2). MLG-E3 is the primary founder of clonal complex 1, which is consistent with findings in a previous study in Shanghai [29]. The majority of MLGs (14/19) originated from MLG-E3. Furthermore, MLG-E60 is a variant of clonal complex 2, and MLG-E59 and MLG-E70 were singletons. The latter three MLG subtypes showed distant evolution from other assemblage E MLGs, which may indicate substantial differences in their evolutionary divergence [29].
Table 2.
Multilocus sequence genotypes of G. duodenalis in pre-weaned dairy calves in Sichuan province.
| Isolate | Geographic source | Subtype |
MLG | ||
|---|---|---|---|---|---|
| bg | tpi | gdh | |||
| ABG3417 | A’ba | E9 | E15 | #E19/MF671896 | #MLGE72 |
| ABG3422 | E1 | E15 | #E19/MF671896 | #MLG E74 | |
| AYG6943 | Anyue | #E13/MF671880 | E1 | E10 | #MLG E70 |
| AYG6950 | #E14/MF671883 | E3 | E10 | #MLG E67 | |
| AYG6953 | E1 | E3 | E3 | #MLG E62 | |
| CDG16089 | Chengdu | E9 | E3 | E1 | #MLG E61 |
| CDG16090 | E8 | E9 | E10 | #MLG E60 | |
| CDG16100 | E9 | E19 | E1 | #MLG E68 | |
| QBJG13 | Qingbaijiang | #E14/MF671883 | E3 | E10 | #MLG E67 |
| QBJG17 | #E16MF671886 | E3 | E3 | #MLG E63 | |
| QBJG18 | E9 | E3 | E10 | MLG E3 | |
| QLG5065 | Qionglai | E1 | #E24MF671907 | #E19/MF671896 | #MLG E75 |
| QLG5066 | E9 | E3 | E10 | MLG E3 | |
| QLG5067 | E1 | E3 | E8 | #MLG E64 | |
| QLG5070 | E1 | E15 | E1 | #MLG E65 | |
| QLG5071 | E9 | E1 | #E19MF671896 | #MLG E73 | |
| QLG5073 | E1 | E10 | |||
| QLG5074 | E1 | E15 | E1 | #MLG E65 | |
| QLG5075 | #E13/MF671880 | E3 | E1 | MLG E13 | |
| QLG5076 | #E13/MF671880 | E3 | #E20/MF671899 | #MLG E59 | |
| QLG5083 | E1 | E3 | E1 | #MLG E66 | |
| QLG5091 | #E13/MF671880 | E3 | E1 | MLG E13 | |
| QLG5092 | #E13/MF671880 | E3 | E1 | MLG E13 | |
| QLG5093 | #E13 | ||||
| ZYG6863 | Ziyang | E9 | E1 | E1 | #MLGE71 |
| ZYG6844 | E15 | #E21/MF671904 | E13 | #MLG E69 | |
Unpublished subtypes and MLGs.
Figure 2.
eBURST networks for G. duodenalis assemblage E. Each MLG is represented by a dot. MLG-E3 is the primary founder, and the subgroup founders are MLG-E1, MLG-E4, MLG-E33, MLG-E66, MLG-E61, MLG-E74, MLG-E30, MLG-E48, MLG-E50, MLG-E6, and MLG-E18. The variants are connected by lines.
Conclusion
This is the first study to genotype G. duodenalis and Cryptosporidium in pre-weaned dairy calves in Sichuan province. C. bovis and G. duodenalis assemblage E are the dominant species in pre-weaned dairy calves in Sichuan, and high genetic diversity of assemblage E MLGs was observed.
Supplementary Material
Acknowledgments
The authors thank Guangwen Yan for giving advice on sample collection. We sincerely thank Robin B Gasser and Tao Wang from the University of Melbourne, Australia for technical support and comments on the draft manuscript.
Cite this article as: Zhong Z, Dan J, Yan G, Tu R, Tian Y, Cao S, Shen L, Deng J, Yu S, Geng Y, Gu X, Wang Y, Liu H & Peng G. 2018. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 25, 45.
Statements
Ethics approval and consent to participate
This study was reviewed and approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University under permit number DYY-S20174604.
Consent for publication
Not applicable.
Availability of data and material
The datasets supporting the conclusions of this article are included within the article. Cryptosporidium and G. duodenalis sequences were deposited in the GenBank database under accession numbers MF671870–MF671908.
Competing interests
The authors declare that they have no conflict of interest.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0501009); the National Natural Science Foundation of China (31272620); and the Chengdu Giant Panda Breeding Research Foundation (CPF2015-4).
Author contributions
Experiments were conceived and designed by Z.J.Z and G.N.P. R.T, Y.N.T, S.Z.C and L.H.S collected samples. Experiments were performed by Z.J.Z, J.M.D, Y.W, X.B.G and J.L.D, and the data were analyzed by Z.J.Z, S.M.Y, H.F.L and Y.G. The manuscript was written by Z.J.Z, J.M.D and G.W.Y. All authors read and approved the final manuscript.
References
- 1. Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. 2008. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. International Jounal for Parasitology, 38(13), 1523–1531. [DOI] [PubMed] [Google Scholar]
- 2. Eckmann L. 2003. Mucosal defences against Giardia. Parasite Immunology, 25(5), 259–270. [DOI] [PubMed] [Google Scholar]
- 3. Fayer R. 2004. Cryptosporidium: a water-borne zoonotic parasite. Veterinary Parasitology, 126(1–2), 37–56. [DOI] [PubMed] [Google Scholar]
- 4. Feng Y, Xiao L. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clinical Microbiology Reviews, 24(1), 110–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gong C, Cao XF, Deng L, Li W, Huang XM, Lan JC, Xiao QC, Zhong ZJ, Feng F, Zhang Y, Wang WB, Guo P, Wu KJ, Peng GN. 2017. Epidemiology of Cryptosporidium infection in cattle in China: a review. Parasite, 24, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hingole AC, Gudewar JG, Pednekar RP, Gatne ML. 2017. Prevalence and molecular characterization of Cryptosporidium species in cattle and buffalo calves in Mumbai region of India. Journal of Parasitic Diseases, 41(1), 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu S, Liu Z, Yan F, Zhang Z, Zhang G, Zhang L, Jian F, Zhang S, Ning C, Wang R. 2017. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Veterinary Parasitology, 248, 68–73. [DOI] [PubMed] [Google Scholar]
- 8. Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, Shi K, Wang M, Zhang L. 2014. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Veterinary Research, 10, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hunter PR, Nichols G. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clinical Microbiology Reviews, 15(1), 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li F, Wang H, Zhang Z, Li J, Wang C, Zhao J, Hu S, Wang R, Zhang L, Wang M. 2016. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Veterinary Parasitology, 219, 61–65. [DOI] [PubMed] [Google Scholar]
- 11. Li J, Qi M, Chang Y, Wang R, Li T, Dong H, Zhang L. 2015. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou zoo, China. Journal of Eukaryotic Microbiology, 62(6), 833–839. [DOI] [PubMed] [Google Scholar]
- 12. Li J, Wang H, Wang R, Zhang L. 2017. Giardia duodenalisinfections in humans and other animals in China. Frontiers in Microbiology, 8, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu G, Su Y, Zhou M, Zhao J, Zhang T, Ahmad W, Lu H, Jiang N, Chen Q, Xiang M, Yin J. 2015. Prevalence and molecular characterization of Giardia duodenalis isolates from dairy cattle in northeast China. Experimental Parasitology, 154(3), 20–24. [DOI] [PubMed] [Google Scholar]
- 14. Matsuura Y, Matsubayashi M, Nukata S, Shibahara T, Ayukawa O, Kondo Y, Matsuo T, Uni S, Furuya M, Tani H, Tsuji N, Sasai K. 2017. Report of fatal mixed infection with Cryptosporidium parvum and Giardia intestinalis in neonatal calves. Acta Parasitologica, 62(1), 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nolan MJ, Jex AR, Mansell PD, Browning GF, Gasser RB. 2009. Genetic characterization of Cryptosporidium parvum from calves by mutation scanning and targeted sequencing – zoonotic implications. Electrophoresis, 30(15), 2640–2647. [DOI] [PubMed] [Google Scholar]
- 16. Petri WA Jr, Miller M, Binder HJ, Levine MM, Dillingham R, Guerrant RL. 2008. Enteric infections, diarrhea, and their impact on function and development. Journal of Clinical Investigation, 118(4), 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qi M, Wang H, Jing B, Wang D, Wang R, Zhang L. 2015. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Veterinary Parasitology, 212(3–4), 404–407. [DOI] [PubMed] [Google Scholar]
- 18. Qi M, Wang H, Jing B, Wang R, Jian F, Ning C, Zhang L. 2016. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, Northwestern China. Parasites & Vectors, 9, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryan U, Caccio SM. 2013. Zoonotic potential of Giardia. International Journal for Parasitology, 43, 943–956. [DOI] [PubMed] [Google Scholar]
- 20. Ryan U, Fayer R, Xiao L. 2014. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology, 141(13), 1667–1685. [DOI] [PubMed] [Google Scholar]
- 21. Ryan U, Zahedi A, Paparini A. 2016. Cryptosporidium in humans and animals – a one health approach to prophylaxis. Parasite Immunology, 38(9), 535–547. [DOI] [PubMed] [Google Scholar]
- 22. Santin M, Dargatz D, Fayer R. 2012. Prevalence of Giardia duodenalis assemblages in weaned cattle on cow-calf operations in the United States. Veterinary Parasitology, 183(3–4), 231–236. [DOI] [PubMed] [Google Scholar]
- 23. Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. Journal of Clinical Microbiology, 43(6), 2805–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taha S, Elmalik K, Bangoura B, Lendner M, Mossaad E, Daugschies A. 2017. Molecular characterization of bovine Cryptosporidium isolated from diarrheic calves in the Sudan. Parasitology Research, 116(11), 2971–2979. [DOI] [PubMed] [Google Scholar]
- 25. Tao W, Li Y, Yang H, Song M, Lu Y, Li W. 2018. Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from northeast China: public health concerns. Journal of Parasitology, 104(1), 10–17. [DOI] [PubMed] [Google Scholar]
- 26. Wang H, Zhao G, Chen G, Jian F, Zhang S, Feng C, Wang R, Zhu J, Dong H, Hua J, Wang M, Zhang L. 2014. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan, China. PLoS One, 9(6), e100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, Xiao L. 2011. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. Journal of Clinical Microbiology, 49(3), 1077–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang R, Zhao G, Gong Y, Zhang L. 2017. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Frontiers in Microbiology, 8, 1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Cai M, Jiang W, Wang Y, Jin Y, Li N, Guo Y, Feng Y, Xiao L. 2017. High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping. Parasitology Research, 116(8), 2101–2110. [DOI] [PubMed] [Google Scholar]
- 30. Wang X, Wang R, Ren G, Yu Z, Zhang L, Zhang S, Lu H, Peng X, Zhao G. 2016. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitology Research, 115(3), 1355–1361. [DOI] [PubMed] [Google Scholar]
- 31. Xiao L, Feng Y. 2008. Zoonotic cryptosporidiosis. Fems Immunology & Medical Microbiology, 52(3), 309–323. [DOI] [PubMed] [Google Scholar]
- 32. Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, Ling H, Liu A, Shen Y. 2013. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in northeastern China’s Heilongjiang province. PLoS One, 8(1), e54857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.