Abstract
Variation in social behavior is common yet our knowledge of the mechanisms underpinning its evolution is limited. The fire ant Solenopsis invicta provides a textbook example of a Mendelian element controlling social organization: alternate alleles of a genetic element first identified as encoding an odorant binding protein (OBP) named Gp‐9 determine whether a colony accepts one or multiple queens. The potential roles of such a protein in perceiving olfactory cues and evidence of positive selection on its amino acid sequence made it an appealing candidate gene. However, we recently showed that recombination is suppressed between Gp‐9 and hundreds of other genes as part of a >19 Mb supergene‐like region carried by a pair of social chromosomes. This finding raises the need to reassess the potential role of Gp‐9. We identify 23 OBPs in the fire ant genome assembly, including nine located in the region of suppressed recombination with Gp‐9. For six of these, the alleles carried by the two variants of the supergene‐like region differ in protein‐coding sequence and thus likely in function, with Gp‐9 showing the strongest evidence of positive selection. We identify an additional OBP specific to the Sb variant of the region. Finally, we find that 14 OBPs are differentially expressed between single‐ and multiple‐queen colonies. These results are consistent with multiple OBPs playing a role in determining social structure.
Keywords: Green beard, olfaction, pheromone, social behavior, social chromosome, Solenopsis invicta, supergene
Impact Summary.
The invasive red fire ant provides a unique opportunity to investigate how changes in social behavior can evolve. In this species, two distinct forms of social organization coexist: colonies either have strictly one queen or up to dozens of reproductive queens.
Pioneering work in the early 2000s demonstrated that this social dimorphism has a genetic basis: one of the two alleles (versions) of the gene Gp‐9 is always and exclusively present in multiple‐queen colonies but never in single‐queen colonies. Gp‐9 encodes an odorant binding protein (OBP), a type of protein that can be involved in the production and in the perception of pheromones. The two alleles of Gp‐9 differ in amino acid sequence, and thus likely also in function. Furthermore, workers with the b allele of Gp‐9 behave spitefully toward queens that lack the b allele—in this sense the b allele appears to be a selfish gene with a so‐called “green beard” effect.
However, it was recently shown that the two alleles of Gp‐9 are part of a pair of “social chromosomes.” Like a pair of sex chromosomes, the two social chromosomes differ from each other, so that hundreds of genes in addition to Gp‐9 have alleles present exclusively in multiple‐queen colonies. This challenges the idea that Gp‐9 is pivotal in determining social organization.
Here, to determine whether there is still reason to believe that OBPs play roles in this system, we characterize all fire ant OBPs. We find that fire ants have 24 OBPs, 10 of which are in the social chromosome. Two of the social chromosome OBPs are exclusive to alternate social chromosome variants, and four OBPs in addition to Gp‐9 have differences in amino acid sequence between the variants. We also find differences in the activity levels of 14 OBP genes between single‐ and multiple‐queen colonies.
In sum, our study provides evidence that multiple OBPs may be responsible for differences between single‐ and multiple‐queen colonies. These results represent a significant step toward understanding the mechanisms by which the social chromosomes function.
Introduction
Variation in social behavior is common yet our knowledge of the mechanisms underpinning its evolution is limited (Robinson et al. 2005; Johnson and Linksvayer 2010). The fire ant Solenopsis invicta provides a rare, textbook example of variation in a fundamental social trait: some colonies have one queen, whereas others have up to dozens of queens. Queens that will form their own single‐queen colony typically disperse over greater distances and can effectively colonize newly available habitats. In contrast, multiple‐queen colonies can outcompete single‐queen colonies in saturated habitats and harsh environments, and can split by fission (Bourke and Heinze 1994; Ross and Keller 1995; Tschinkel 2006). Multiple additional traits differ between the two social forms, including in queen fecundity, colony size, worker size distribution, and worker aggressiveness (Ross and Keller 1995; DeHeer et al. 1999; Keller and Ross 1999; Goodisman et al. 2000; DeHeer 2002; Buechel et al. 2014; Huang et al. 2014).
A series of landmark studies (Ross 1997; Keller and Ross 1998; Ross and Keller 1998) demonstrated that the two social forms are under the control of a Mendelian element. This element was first identified in a screen of electrophoretic markers as a polymorphic protein coding gene, Gp‐9, with two alleles: Gp‐9B and Gp‐9b (Ross 1997). If a colony includes only Gp‐9 BB workers, they will accept a single Gp‐9 BB queen and execute any additional queens. In contrast, if more than ∼20% of the workers in a colony are Gp‐9 Bb heterozygotes, they will execute reproductively active Gp‐9 BB queens but accept dozens of Gp‐9 Bb queens (Ross 1997; Keller and Ross 1998, 1999; Ross and Keller 1998, 2002; DeHeer et al. 1999; Gotzek & Ross 2007). In contrast, Gp‐9 bb queens die before becoming reproductively active (Ross 1997; DeHeer et al. 1999; Keller and Ross 1999; Gotzek and Ross 2007; Trible and Ross 2016). The workers discriminate between queens of alternate genotypes based on olfactory cues (Keller and Ross 1998; Ross and Keller 1998, 2002), such as differences in the queens’ cuticular hydrocarbon profiles (Eliyahu et al. 2011; Trible and Ross 2016). Because workers carrying the Gp‐9b allele recognize whether queens also carry this allele and execute those that do not, this system represents a rare example of a “green beard gene” (Keller and Ross 1998), named after a theoretical model of a behavioral selfish genetic element (West and Gardner 2010).
In another landmark study, Krieger and Ross (2002) demonstrated that Gp‐9 encodes an odorant binding protein (OBP). OBPs are essential components of insect communication systems: they bind and transport pheromones and other semiochemicals, generally mediating their perception and sometimes their secretion (Pelosi et al. 2006, 2014; Leal 2013). Furthermore, tests of historical selection on Gp‐9 reveal a significant excess of nonsynonymous (amino acid replacing) substitutions relative to synonymous (silent) substitutions between the lineage of Gp‐9 b‐like alleles and Gp‐9 B‐like alleles in the fire ant and its relatives. This implies that directional or diversifying selection has driven the molecular evolution of Gp‐9, and is associated with differentiation between the two forms of social organization in these ants (Krieger and Ross 2002, 2005). Several models lay out the potential function of Gp‐9, generally involving differential production or perception of pheromones in queens as well as workers of alternate genotypes (Krieger 2004; Gotzek and Ross 2007, 2009).
However, recent genome‐wide analyses of the social dimorphism revealed that the association between genotype and form of social organization is not limited to Gp‐9 (Wang et al. 2013). Instead, genetic maps obtained using Restriction site Associated DNA (RAD) markers from crosses in seven families showed that this association extends over a large chromosomal region of suppressed recombination. The two variants of this region, respectively, marked by the Gp‐9B and Gp‐9b alleles are carried by a pair of “social chromosomes” named SB and Sb. The region is genetically differentiated over 10.8 Mb (55%) of the mapped assembly of the social chromosomes, although its total length could be 19.4–31.5 Mb given the estimated size of the nonassembled portion of the genome (Pracana et al. 2017). Based on the current NCBI gene set, this region contains at least 443 protein coding genes, including Gp‐9. The two chromosomes differ by at least one large inversion affecting a large portion of the region and an additional small (48 kb) inversion. The region of suppressed recombination can be described as a supergene, a locus containing multiple genes with tightly linked allelic combinations that control a complex polymorphic phenotype (Linksvayer et al. 2013; Schwander et al. 2014; Thompson and Jiggins 2014).
A study of general patterns of divergence and diversity showed that Sb has two orders of magnitude lower diversity than SB and than the rest of the genome, and that there is high ratio of nonsynonymous to synonymous substitutions between SB and Sb (Pracana et al. 2017). These results suggest that the evolution of Sb has been shaped by Hill–Robertson effects (the effects of selection on linked loci) due to the rarity of recombination in Sb (Wang et al. 2013; Pracana et al. 2017). However, little work has been done to characterize the genes present in the supergene region and to identify the mechanisms by which SB and Sb control the phenotypic differences between single‐ and multiple‐queen colonies. Studies using cDNA microarrays representing 3673 genes demonstrated that the supergene region is enriched for genes that are differentially expressed between queens (Nipitwattanaphon et al. 2014) and workers (Wang et al. 2008, 2013) of the two colony types. This suggests that genes other than Gp‐9 could be responsible for the social dimorphism. Given that the determination of queen number requires the differential production and perception of semiochemicals by individuals of each genotype, it remains likely that OBPs play a part in determining the dimorphism.
Here, we determine to which extent OBPs have potentially functional divergence between social forms. For this, we identify all OBPs in the fire ant reference genome and map them to their genomic locations. Subsequently, we use population‐sequencing data to identify allelic differences between OBPs found on alternate variants of the social chromosome supergene. We also sequence an outgroup species, Solenopsis geminata, which allows us to determine which supergene variant carries the derived allele for each substitution. Finally, we compare gene expression profiles of all OBPs and gene coexpression modules between social forms. We show that there are nucleotide and amino acid sequence level differences between SB and Sb in the supergene OBPs, and that OBPs inside and outside the supergene are differentially expressed between single‐ and multiple‐queen colonies.
Methods
OBP DISCOVERY AND MANUAL GENE MODEL CURATION
The sequences of 18 fire ant OBP genes were previously reported, based on searches of Sanger‐sequenced Expressed Sequence Tag (EST) libraries (Table S1; Wang et al. 2007; Xu et al. 2009; Gotzek et al. 2011; Wurm et al. 2011). We used a curation approach similar to those previously used on other genes (Ingram et al. 2012; Corona et al. 2013; Kulmuni et al. 2013; Privman et al. 2013) to find the position of these OBP genes in the fire ant genome assembly (Wurm et al. 2011) and to discover previously unreported OBP genes. Our curation pipeline is described in detail in Supporting Information Methods. Briefly, we iteratively performed blastn and blastp (Camacho et al. 2009; Priyam et al. 2015) searches of the fire ant genome assembly (Wurm et al. 2011) using as queries the previously known fire ant OBP sequences as well as UniProt sequences that are part of the Pfam family “PBP_GOBP” (Finn et al. 2014; UniProt Consortium 2015). We manually curated the results of these searches by inspecting alignments of transcriptomic and genomic reads, which allowed us to infer intron–exon boundaries and coding sequences of these OBPs. We labeled the curated gene predictions that correspond to the previously known OBP genes (SiOBP1–17) according to the notation used by Gotzek et al. (2011) and we labeled newly discovered loci SiOBPZ1–Z7. We used a genetic map (Pracana et al. 2017) to assign OBPs to linkage groups. We generated a codon‐level alignment of the S. invicta OBPs using MAFFT‐linsi (version 6.903b; Katoh and Toh 2008) and PRANK (version 120626; Löytynoja and Goldman 2005), and built a phylogenetic tree using RaxML (version 8.2.9; Stamatakis 2006).
IDENTIFYING ALLELIC DIFFERENCES FOR OBPs CARRIED BY ALTERNATE VARIANTS OF THE SOCIAL CHROMOSOME
We used whole‐genome sequences from one SB and one Sb male from each of seven colonies that had been sequenced at low coverage (Illumina 2*100 bp paired‐end genome shotgun sequences; ∼6×–8× coverage) in 2012 (NCBI SRP017317; Wang et al. 2013). Each of these samples is a haploid male (ants have a haplodiploid sex determination system). We filtered the reads, aligned them to the reference genome using bowtie2 (version 2.1.0; Langmead and Salzberg 2012), and used samtools and bcftools (version 1.3.1 for both; Li et al. 2009) to call variants among the individuals (Supporting Information Methods).
We produced whole‐genome sequencing reads of the outgroup species S. geminata. We sequenced a pool of 10 workers (sampled in Thailand by Dr. Adam Devenish, University College London, United Kingdom) using Illumina HiSeq 4000 (×11 coverage; Supporting Information Methods). We called variants between the sample and the reference assembly (using freebayes version 1.0.2‐33‐gdbb6160; Garrison and Marth 2012) within the coding sequence of each OBP using freebayes (Supporting Information Methods). We classed the alleles in each SB‐Sb substitution as ancestral or derived based on the allele carried in the outgroup species. We estimated the rate of synonymous and nonsynonymous divergence (dS and dN, respectively) between SB and Sb using seqinR (version 3.0‐7; Charif and Lobry 2007).
DETECTION OF COPY NUMBER AND STRUCTURAL VARIATION IN OBPs
We visually inspected the alignments of the seven SB and the seven Sb haploid male samples against each OBP region. Deletions were identified as regions with no coverage and duplications were identified as regions where the coverage was higher than the background (Supporting Information Methods). Using the de novo assembler MIRA (version 4.0.2; Chevreux et al. 1999), we produced the sequence of the duplicate copy of SiOBP12, which we named SiOBPZ5 (approach detailed in Supporting Information Methods).
GENE EXPRESSION OF S. INVICTA OBPs IN PUBLICALLY AVAILABLE RNA SEQUENCING DATASETS
We analyzed all available RNA sequencing (RNA‐Seq) data from the NCBI SRA database for S. invicta (data from Wurm et al. 2011; Morandin et al. 2016 and PRJNA266847; details in Table S2). We determined the expression levels of S. invicta transcripts using the Kallisto count mode (version 0.43.0; Bray et al. 2016). Each sample was independently normalized using the DESeq2 method (version 1.14.1; Love et al. 2014). Additionally, we performed genome‐wide analysis of differential expression of data from Morandin et al. (2016), comparing three pools of queens from multiple‐queen colonies with two pools from single‐queen colonies, as well as two pools of workers from multiple‐queen colonies with three pools from single‐queen colonies. The pools of workers from multiple‐queen colonies contain a mix of individuals of both genotypes, whereas the pool of queens from multiple‐queen colonies has only SB/SB queens. We used a standard DESeq2 approach to identify expression differences between single‐ and multiple‐queen samples in queens and in workers. Additional details regarding these analyses are in Supporting Information Methods.
DIFFERENTIAL EXPRESSION OF GENE COEXPRESSION MODULES ACROSS SOCIAL FORMS
We created gene coexpression modules from two cDNA microarray datasets (Platform GPL6930, with 25,344 probes representing 3673 genes; Supporting Information Methods and Table S3; Wang et al. 2007), one with queen samples (GSE42062; Nipitwattanaphon et al. 2013), the other with worker samples (E‐GEOD‐11694; Wang et al. 2008). Both datasets included SB/SB and SB/Sb samples. We created modules for each set using weighted gene coexpression network analysis (WGCNA) (version 1.49; Langfelder and Horvath 2008). We used t‐tests to determine whether any module eigengene is correlated with genotype or social form. In queens, we compared SB/SB to SB/Sb samples because all samples originate in multiple‐queen colonies. In workers, we separated the effect of genotype from the effect of social form following the approach in Wang et al. (2008): we compared genotypes (SB/SB vs SB/Sb) using samples from multiple‐queen colonies, and we compared across social forms (single queen vs multiple queen) using SB/SB samples only.
EVIDENCE FOR SELECTION BASED ON NUCLEOTIDE DIVERSITY
Genomic regions that underwent recent selective sweeps are characterized by low nucleotide diversity (π) (Smith and Haigh 1974; Nei 1987; Nachman 2001). We used measurements of π along a sliding window of the genome, originally produced by Pracana et al. (2017), to identify selection pressure acting on S. invicta OBPs. Measurements of π were taken from nonoverlapping 10 kb windows (Supporting Information Methods).
Results
THE FIRE ANT REFERENCE GENOME ASSEMBLY CONTAINS 23 PUTATIVE OBPs
We combined automatic and manual curation approaches incorporating genomic and gene expression data to identify the sequence, exon structure, and location of 23 putative OBP genes in the S. invicta reference genome. Seventeen of these matched fire ant OBP gene sequences that had been previously reported, although with differences in sequence or in their inferred location in linkage groups (Table S1 and Supporting Information Methods). The remaining seven putative OBP genes are novel to S. invicta (Table S4). Interestingly, the coverage depth of SiOBPZ6 is fourfold higher (95% confidence interval [3.66–4.78]; t‐test t df = 6 = 14.0, P < 10−5) than that of 1000 randomly selected genes, suggesting that there are four copies of this gene. There is little genetic variation among reads mapping to this gene across the 14 individuals in our dataset (4.2 Single Nucleotide Polymorphisms [SNPs] per 1000 bp). The alignment of whole‐genome sequencing reads of the outgroup species S. geminata to the S. invicta reference assembly shows that all OBPs are covered in this outgroup species. The coverage depth of SiOBPZ6 is threefold higher in S. geminata (95% confidence interval [2.78–3.16]; t‐test t df = 999 = 20.7, P < 10−15), suggesting that this species also carries multiple copies of this gene.
Nine of the 23 OBPs in the genome are adjacent to unrelated genes, the remainder are organized into gene clusters. There are three locations in the genome each containing a cluster of four OBPs (two in linkage group 16, one in linkage group 3) and one containing a cluster of two OBPs (in linkage group 6). Intriguingly, none of these clusters appear to be completely monophyletic (Fig. 1). For previously known OBPs, the topology of our phylogenetic tree agrees with previously published trees (Gotzek et al. 2011; Zhang et al. 2016), with the exception of the position of SiOBP15 (low bootstrap values in all trees) and SiOBP5.
Figure 1.
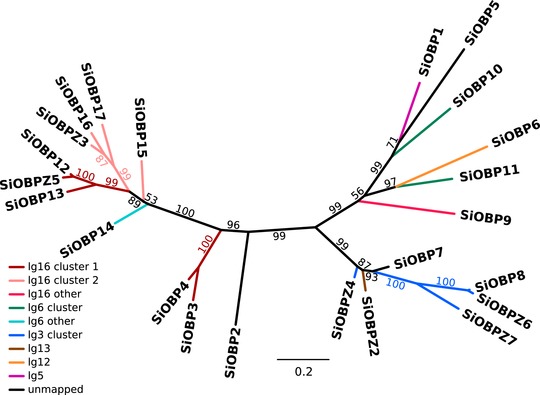
Phylogenetic tree based on a codon‐level alignment of revised gene predictions for previously described OBPs (SiOBP1–17) and novel OBPs (SiOBPZ2–Z6). Branches are colored by gene cluster and linkage group (lg). SiOBPZ1 was removed from this analysis because the high divergence of its sequence led to unreliable alignments and positioning in the phylogeny. All OBPs on linkage group 16 (lg16) are within the supergene‐like region of the social chromosomes (Fig. 2).
NONSYNONYMOUS DIFFERENTIATION BETWEEN SB And Sb IN OBPs
Eight of the OBPs are located in scaffolds of the SB fire ant genome assembly that map to the supergene region, with two clusters of four OBPs (Fig. 2). One of the clusters includes Gp‐9 (which was named SiOBP3 in Gotzek et al. 2011). A ninth gene, SiOBP9, is located in an unmapped scaffold that likely also belongs to the supergene region based on high levels of SB‐Sb differentiation (Fig. 2). To determine whether the supergene OBPs have allelic differences between SB and Sb, we used whole‐genome sequence data from seven SB males and seven Sb males.
Figure 2.
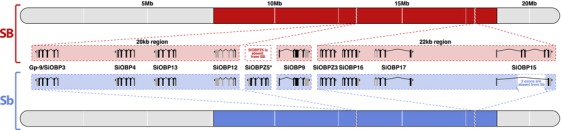
Relative positions on the social chromosome (i.e., linkage group 16) of 10 OBP loci, highlighting intron–exon structures and differences between the supergene region of Sb (blue) and SB (red). SiOBPZ5 is specific to Sb but we do not know its exact location; SiOBP15 is missing a 3‐exon region in Sb; SiOBP9 is in an unmapped scaffold that likely belongs to the supergene region based on high levels of SB‐Sb differentiation (Pracana et al. 2017).
These data confirmed the previous finding that Gp‐9/SiOBP3 has eight nonsynonymous and one synonymous fixed single nucleotide substitutions between SB and Sb in the North American study population (Krieger and Ross 2002). Of the other OBPs in the supergene region, SiOBP4 has three nonsynonymous and two synonymous substitutions. Two additional supergene OBPs have one fixed nonsynonymous substitution between SB and Sb (Table 1). Performing an analysis of the ratio of nonsynonymous to synonymous substitutions between alleles (dN/dS) was only possible for the two genes with the most divergent alleles: Gp‐9/SiOBP3 had the highest ratio of nonsynonymous to synonymous substitutions (dN/dS = 1.48), followed by SiOBP4 (dN/dS = 0.74).
Table 1.
OBP differentiation between SB and Sb: the number of sequence‐level differences between SB and Sb and differential OBP gene expression between multiple‐ and single‐queen colonies
| Significant differential expression between colonies types | |||||
|---|---|---|---|---|---|
| S. invicta OBP locus | Nonsynonymous differences | Synonymous differences | Total differences | In queens | In workers |
| SiOBP3 (Gp‐9) | 8 | 1 | 9 | Yes | No |
| SiOBP4 | 3 | 2 | 5 | Yes | Yes |
| SiOBP13 | 1 | 1 | 2 | Yes | Yes |
| SiOBP12 | Frameshift insertion in SB and duplication in Sb | Yes | No | ||
| SiOBPZ5 | Present exclusively in Sb | ||||
| SiOBPZ3 | 1 | 0 | 1 | No | No |
| SiOBP9 | 0 | 1 | 1 | No | No |
| SiOBP16 | 0 | 0 | 0 | Yes | No |
| SiOBP17 | 0 | 0 | 0 | Yes | Yes |
| SiOBP15 | ∼2600 bp deletion in Sb | No | No | ||
All differentially expressed genes between social forms were overexpressed in multiple‐queen colonies.
We analyzed the OBP sequences from an outgroup species, S. geminata, estimated to have diverged from S. invicta 3–3.5 million years ago (Moreau and Bell 2013; Ward et al. 2015), that is, before the divergence between SB and Sb in S. invicta (estimated 0.35–0.42 million years ago; Wang et al. 2013). These sequences allowed us to determine the ancestral allele in each substitution. Sb carried the derived allele in most of the positions with nonsynonymous substitutions between SB and Sb (seven out of eight in Gp‐9/SiOBP3 and all in SiOBP4 and SiOBPZ3; we could not derive the two SiOBP13 substitutions, as S. geminata read coverage was too low for this gene). This pattern is consistent with most nonsynonymous substitutions between SB and Sb having arisen in the lineage leading to Sb.
COPY NUMBER AND STRUCTURAL DIFFERENTIATION BETWEEN SB AND Sb IN OBPs
We also found structural differences between SB and Sb affecting two OBPs. For the first, SiOBP15, we detected a ∼2600 bp deletion unique to Sb individuals (Fig. 2, Table 1). This deletion is derived (i.e., it is not present in the outgroup species, S. geminata) and causes the loss of three out of five coding exons (89 out of 139 amino acids), although it does not cause a frameshift. The second OBP with a major structural difference is SiOBP12. In Sb individuals, this gene is duplicated, forming the Sb‐specific SiOBPZ5 (Fig. 2, Table 1). This gene increases the total OBP count of S. invicta to 24. There are 18 fixed amino acid differences between SiOBPZ5 and the SB allele of SiOBP12 sequence (one deleted codon, 21 nonsynonymous and four synonymous nucleotide‐level fixed differences; four codons each contain two single‐nucleotide fixed differences; dN/dS = 2.67). Intriguingly, SiOBP12 has an early stop codon (TAG) at codon position 16 of 176 in all seven SB individuals and the reference genome. These individuals are also affected by six nonsynonymous SNPs and two polymorphic indels downstream of the early stop codon. Sb individuals have the CAG allele at position 16 of SiOBP12, but have a slightly later early stop codon at position 37 due to a frameshifting insertion of 17 bp at codon position 25 (nucleotide position 74). The outgroup species S. geminata has neither of the early stop codons. However, the very low S. geminata read coverage observed in the two terminal exons of this gene (median < 3; t df = 999 = −11.29, P < 10−27) could indicate a deletion in this species. SiOBP12 is thus nonfunctional in Sb and SB individuals, and putatively nonfunctional in the outgroup species. The Sb‐specific gene SiOBPZ5 appears to be functional as it has no early stop codons. None of the other OBPs showed differences in structure or in copy number between SB or Sb.
FOURTEEN OBPs ARE DIFFERENTIALLY EXPRESSED BETWEEN SOCIAL FORMS
We compared expression levels between single‐ and multiple‐queen colonies in workers and in queens (Fig. 3; Table 1) using RNA‐Seq data from Morandin et al. (2016). General expression patterns showed an enrichment in differentially expressed genes in the supergene region in queens (expected proportion = 0.022, observed proportion = 0.059, Chi2 df = 1 = 32.84, P = 10−8) but not in workers (expected proportion = 0.021, observed proportion = 0.024, Chi2 df = 1 = 0.05, P = 0.82).
Figure 3.
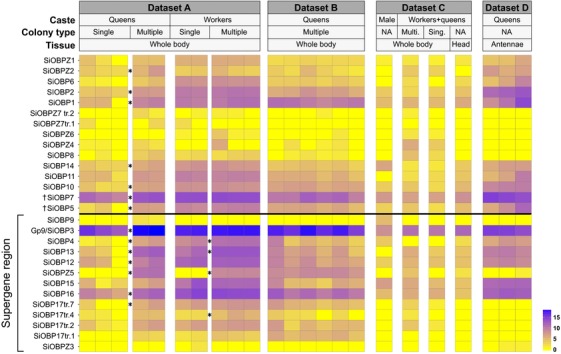
Expression patterns for all analyzed RNAseq datasets. Each tile represents the logarithm base 2 of DESeq normalized transcript counts. The rows with asterisks (*) correspond to those OBPs with significant differential expression between social forms within castes in dataset A (Morandin et al. 2016). Information about each dataset is available in Table S2 (A: PRJDB4088, B and C: PRJNA49629, D: PRJNA266847). † The exons of SiOBP5 and SiOBP7 are split across three unmapped scaffolds; we do not know whether these genes are within or outside the supergene region.
In queens, fourteen OBPs, including seven in the supergene region, were significantly differentially expressed between multiple‐queen and single‐queen colonies (DESeq2 Wald test; Benjamini–Hochberg adjusted P < 0.05). Consistent with this, the entire group of 24 fire ant OBPs showed significantly stronger P‐values for differential expression between queens from single‐ and multiple‐queen colonies than would be expected by chance (tested among 12,693 transcripts, two‐sided Kolmogorov–Smirnov test; P < 10−11; Fig. S1). Surprisingly, all of the OBPs that were differentially expressed between social forms in queens were more highly expressed in multiple‐queen colonies than in single‐queen colonies (14 significant OBPs in queens, binomial test 14 out of 14; null probability = 0.5; P < 10−4). In workers, only four OBPs (all in the supergene region) were significantly differentially expressed between social forms (DESeq2's Wald test Benjamini–Hochberg adjusted P < 0.05). All the differentially expressed OBPs in workers were also differentially expressed in queens. For one of these OBPs (SiOBP17), a different splice form was differentially expressed between colony types in queens than in workers (Fig. 3).
We additionally obtained qualitative gene expression profiles of all OBPs across 18 additional samples, in total representing seven different conditions of body part, social form, and caste (Table S2). We find generally consistent expression patterns for OBPs across all independent samples (Fig. 3). For instance, in every sample, Gp‐9/SiOBP3 was the most highly expressed of all OBPs, whereas SiOBPZ3 was only residually expressed (0.26 or fewer transcripts per million reads). Six OBPs had only residual expression in queen antennae and in heads, although most of these showed at least some expression in whole‐body samples. The expression of one of these genes, SiOBP9, appears to be limited to males.
GENE COEXPRESSION MODULES CORRELATED WITH SOCIAL FORM
We used WGCNA (Langfelder and Horvath 2008) to produce modules of coexpressed genes from a set of worker samples (Wang et al. 2008) and a set of queen samples (Nipitwattanaphon et al. 2013). Both datasets compare SB/SB and SB/Sb samples. The queen and worker datasets, respectively, clustered into 30 and 37 coexpression modules (Table S5). Most modules in one dataset share a significant number of probes with a module in the other dataset (30 out of 31 in queens and 35 out of 37 in workers; Fisher's exact test for the overlap of the pairs of modules across the datasets, Bonferroni corrected P > 0.05; Fig. S2). However, in most cases there was no one‐to‐one correspondence between datasets (17 out of 31 modules in queens and 19 out of 37 in workers have significant overlaps with more than one module). Eight of the OBPs discovered in the present study are represented in the microarray (Table S5). In the worker dataset, the module “worker_D” includes four of the OBPs (SiOBP3, SiOBP12, SiOBP13, and SiOBP16), accounting 25% of the 16 genes in the module. OBPs were present in nine other modules, although in all nine cases the OBP represented a very small proportion of the genes in the module (Table S5).
We tested whether there were gene coexpression modules with differential eigengene expression between genotypes or social forms. In queens, four modules had differential expression between genotypes (Table S7). In workers, one module had differential eigengene expression between genotypes, and one module had differential gene expression between social forms (Table S7). One of the modules that had differential expression between genotypes in queens (“queen_X”) corresponded with the module with differential expression between genotypes in workers (“worker_Z”). Only one of the modules with differential eigengene expression includes an OBP (SiOBP15 in “queen_D”). None of these modules were enriched for any GO term.
THREE OBPs ARE IN A REGION OF THE GENOME WITH CHARACTERISTICS OF A RECENT SELECTIVE SWEEP
We used measurements of π among SB individuals in nonoverlapping 10 kb windows from Pracana et al. (2017) to determine whether any OBPs are in regions of low π, characteristic of recent selective sweeps. Among windows overlapping OBPs, two neighboring windows had π within the lower quartile of the whole‐genome distribution (π < 0.0004; Fig. S3). These two windows overlap the loci SiOBPZ4, SiOBPZ7, and SiOBPZ6, which are within 19 kb of each other on linkage group 3. We did not perform an equivalent analysis on Sb individuals because the entire region of suppressed recombination has the signature of a recent sweep in Sb (Pracana et al. 2017).
Discussion
THE PUTATIVE ROLE OF OBPs IN DETERMINING SOCIAL DIMORPHISM
The description of Gp‐9 as a green beard gene (Keller and Ross 1998) and its subsequent characterization as an OBP (Krieger and Ross 2002) led to the proposal of different models of how this single gene can control the dimorphism in social organization (reviewed by Gotzek and Ross 2007). At their most basic level, these models propose that Gp‐9 controls the production of a green‐beard odor in queens and the differential perception of this odor by workers of alternate genotypes. However, it was also proposed that Gp‐9 additionally controls differential odor production in workers (Gotzek and Ross 2007), as well as a number of physiological and morphological traits in queens (Keller and Ross 1995; DeHeer et al. 1999; DeHeer 2002) and males (Lawson et al. 2012). The discovery that Gp‐9 is tightly linked to hundreds of other genes (Wang et al. 2013; Pracana et al. 2017)—including the nine additional OBPs we report here—suggests that the roles previously attributed to Gp‐9 could be split between multiple genes.
The key roles of OBPs in semiochemical perception (Leal 2013) and secretion (Li et al. 2008; Iovinella et al. 2011; Sun et al. 2012) lead to the prediction that such proteins are involved in determining the two colony types. Our results support this hypothesis, as we find divergence in protein coding sequence between SB and Sb in the OBPs in the supergene region, as well as differences in the regulation of OBP expression between single‐ and multiple‐queen colonies.
The differences in protein coding sequence affect seven of the ten OBPs in the supergene region, including Gp‐9/SiOBP3. The biggest differences are in SiOBPZ5, absent in SB, and in SiOBP15, which is missing three exons in Sb. Such differences could have a major effect on semiochemical communication. Additionally, among the four intact OBPs with nonsynonymous divergence between SB and Sb, both Gp‐9/SiOBP3 and SiOBP4 have dN/dS ratios indicative of adaptive differentiation between the alleles of these genes (Krieger and Ross 2002). This interpretation comes with some caution due to our relatively low sample size (14 individuals from an invasive population).
Additionally, 14 out of the 24 fire ant OBPs were differentially expressed between social forms in queens or in workers. Our analysis uncovers three potentially important aspects of the differential regulation of OBP expression in the two social forms. First, all of the differentially expressed OBPs are more highly expressed in multiple‐queen colonies than in single‐queen colonies, suggesting that multiple‐queen colony traits are associated with the activation of semiochemical communication pathways. Second, this activation seems to be stronger in queens, as more OBPs were differentially expressed between social forms in queens (14 OBPs) than in workers (four OBPs). This result reflects the more general pattern that the supergene region was enriched for differentially expressed genes between colony types in queens, but not in workers. The pools of workers from multiple‐queen colonies contain a mix of individuals of both genotypes (36% SB/SB and 64% SB/Sb workers expected; Buechel et al. 2014), which could mask differences between SB/SB workers from single‐queen colonies and SB/Sb workers from multiple‐queen colonies. Indeed, previous studies using cDNA microarray data and a different gene set suggest that the supergene region is enriched for differentially expressed genes in both queens (Nipitwattanaphon et al. 2013) and workers (Wang et al. 2013). Third, several of the queen‐specific differentially expressed OBPs are located outside the supergene, implying that they are regulated in trans by elements in the supergene. It is important to note that all three patterns could be affected by our use of samples from whole bodies, which is known to introduce several types of biases if the differences in expression are tissue specific (Johnson et al. 2013; Montgomery and Mank 2016). A particular issue is differences in allometry (i.e., relative body‐size proportions) between the individuals of different groups, for instance the larger gaster of queens in single‐queen colonies relative to queens in multiple‐queen colonies (Tschinkel 2006). These biases cannot be resolved by standard normalization methods, which are designed to normalize by entire library size rather than by the relative abundance of different transcripts (Dillies et al. 2013). Tissue‐specific gene expression profiling (Bastian et al. 2008; Robinson et al. 2013; Jasper et al. 2015) would be needed to control for such allometric differences.
Our results also support the idea that along with OBPs, other genes are likely involved in defining the social polymorphism of S. invicta. For instance, only one of the coexpression modules with significantly different eigengene expression contained an OBP. Furthermore, other genes inside and outside the supergene region were differentially expressed between social forms. Thus, a venue of further investigation would be to examine the potential roles of other genes, including genes from families known to be involved in communication, including chemosensory proteins (Kulmuni et al. 2013), desaturases (Helmkampf et al. 2015), fatty‐acid reductases (Lassance et al. 2010; Niehuis et al. 2013), and olfactory (Wurm et al. 2011), gustatory (Robertson et al. 2003; Zhou et al. 2012), and ionotropic receptors (Benton et al. 2009; Zhou et al. 2012). It is important to note that additional experimental work would be necessary to demonstrate whether OBPs or any of these proteins have a functional role. An interesting approach would be to measure the effect of artificially modifying the sequence or expression level of each gene to test their specific function (Gaj et al. 2013; Mohr et al. 2014).
GENERAL EVOLUTIONARY PATTERNS OF OBPs IN S. INVICTA
The evolution of the OBP gene family is generally thought to follow the birth‐and‐death model, where gene duplication is followed by either the pseudogenization or the rapid functional divergence of the duplicate gene (Nei and Rooney 2005; Vieira et al. 2007). The S. invicta OBPs are organized in clusters along the genome, as in other insect species (Xu et al. 2003; Foret and Maleszka 2006; Vieira et al. 2007). However, none of these clusters appear to be monophyletic (Fig. 1). This is consistent with the birth–death model, where the fast evolution of genes can mask their true phylogenetic relationship (Vieira et al. 2007; Gotzek et al. 2011; Vieira and Rozas 2011). Alternative explanations include translocations affecting the OBPs during or after duplication, or ectopic gene conversion across different clusters after duplication (Arguello and Connallon 2011). Another argument in support of the birth‐and‐death model is that we find evidence of expansions in OBP number. One example is the putative ant‐specific OBP expansion reported previously (the OBP cluster including SiOBP14 in Fig. 1; Gotzek et al. 2011). We found no one‐to‐one orthologous sequences for these genes in other ants or in other arthropods (the 11 genes in this group of OBPs have BLAST similarity to only three genes in the ant Monomorium pharaonis; phylogenetic group 1 in Table S7). A cluster with several novel genes identified in our study (the group including SiOBP7 and SiOBP8 in Fig. 1) follows a similar pattern (five OBPs have BLAST similarity to one M. pharaonis gene, two have BLAST similarity to one Pogonomyrmex barbatus gene; phylogenetic group 2 in Table S7). These groups of genes may have expanded in the lineage leading to S. invicta and S. geminata, although this conclusion would require the exhaustive identification of OBPs in the present study to be replicated for other ant species. An example of a putatively recent expansion is SiOBPZ6, which seems to be present in multiple copies both in S. invicta and in S. geminata. Lack of heterozygosity in the region suggests that the gene copies have been recently affected by ectopic gene conversion (Arguello and Connallon 2011). Furthermore, finding that the S. invicta SiOBPZ6 quadruplication is in a region that has a signature of a recent selective sweep makes it tempting to speculate that SiOBPZ6 is involved in a recent adaptive process (Kondrashov 2012)—for example, to the invasive range of this species (Ascunce et al. 2011).
Conclusion
Previous studies have focused on how the evolution of the social chromosomes has been affected by restricted recombination (Wang et al. 2013; Pracana et al. 2017), whereas the work presented here focuses on the putative mechanisms by which these chromosomes control social organization. In summary, our analyses provide a comprehensive overview of OBPs in the fire ant genome, describing patterns of differentiation and expression that are consistent with the predicted roles of OBPs in determining social organization in this species. Our study highlights the need for tissue‐specific expression profiles, as well as for broader taxonomic sampling to understand OBP evolution during the origin of the multiple‐queen colony organization. Finally, our work provides a starting point for future functional studies on the roles of OBPs in the social chromosome system.
We deposited the genomic reads of the Solenopsis geminata sample on NCBI SRA (SRX3045159). We manually produced gene models for 24 OBPs, which we deposited to NCBI. Additionally, all data is available at https://wurmlab.github.io/data.
Associate Editor: Z. Gompert
Supporting information
Figure S1. Density distribution of the p‐values for differential expression between social forms in queens for OBPs (in green) and all other protein‐coding genes (red). The p‐values for OBPs are strongly skewed towards 0. This result is based on the expression levels from the Morandin et al. (2016) dataset.
Figure S2. Correspondence between queen and worker modules. Numbers in the table indicate probe counts in the intersection of the corresponding modules. Coloring of the table encodes − log(p), with p being the Fisher's exact test p‐value for the overlap of the two modules. A module in one dataset would be preserved across both sets if it had a single corresponding module in the other dataset with a large number of probes in common.
Figure S3. Nucleotide distribution (π, measured from SB individuals in Pracana et al. 2017) of 10kb windows of the assembled genome that overlap coding sequences. Vertical bars represent π of windows overlapping OBPs; orange bars representing those overlapping supergene OBPs.
Table S1. Summary of correspondences between identifiers of sequences produced in this project and previously published sequences, including the number of sequence differences between the two groups.
Table S2. Accession numbers of the gene expression data used. “Project” and “SRA” columns indicate NCBI identifiers. The descriptions provided and the sequencing method used are based on metadata available on NCBI and in the manuscripts. Two samples (marked with an asterisk) were discarded because of very low coverage after aligning the reads to the S. invicta genome.
Table S3. Table showing the results of the alignment of ESTs from the S.invicta microarray (Platform GPL6930) to its reference genome. The columns “Scaffold”, “Start” and “End” indicate the coordinates of the EST in the reference. “Probe” refers to probe to which each EST belongs and “Gene” refers to the gene intersecting that particular region of the S.invicta genome. ESTs tagged with an asterisk (*) map to more than one location in the genome.
Table S4. Details of closest BLASTP hit to NCBI “nr” database for each newly produced S. invicta OBP sequence.
Table S5. Number of genes represented in each co‐expression module; OBPs represented in each module. Each gene may be represented by multiple probes. Probes in queen_E_1 are not assinged to a module.
Table S6. Gene co‐expression modules with module eigene differential expression between genotypes in queens (SB/SB versus SB/Sb), between genotypes in workers from multiple‐queen colonies (SB/SB versus SB/Sb), and betwen social forms in SB/SB workers (single‐queen colony versus multiple‐queen colony). Differential expression was tested with t‐tests within each comparison within each dataset, with p‐values corrected for multiple testing using Bonferroni correction.
Table S7. Putative OBP orthologs in other species. First, we ran a tblastn search of all S. invicta OBPs against all non‐S. invicta arthropod sequences, accepting hits where e‐value < 10−3. We then ran a blastx search of these hits against the S. invicta gene predictions (including our newly curated OBP set). We report the hits with the lowest e‐value of the blastx search. We repeated this analysis by searching non‐ant arthropods (not Formicidae). The coloured cells represent cases where the same non‐S. invicta sequence aligns to multiple S. Invicta OBPs.
AUTHOR CONTRIBUTIONS
YW, RP, and IL conceived and designed the study; IL and RP performed the majority of genome‐level analyses of OBPs; ES sequenced S. geminata; AP performed automated gene prediction; CMR analyzed gene expression; YW, RP, and IL drafted the manuscript and all authors contributed to later versions of the manuscript.
ACKNOWLEDGMENTS
We thank K. G. Ross, R. A. Nichols, C. Eizaguirre, L. Henry, E. Favreau, T. Colgan, two anonymous reviewers, the editor and the associate editor for advice and comments on the manuscript, and QMUL's SBCS Evolution group for support and stimulating discussion. We thank A. Devenish for supplying Solenopsis geminata samples. This work was supported by the Biotechnology and Biological Sciences Research Council (grant BB/K004204/1), the Natural Environment Research Council (grant NE/L00626X/1), NERC EOS Cloud, the Deutscher Akademischer Austauschdienst (DAAD) Postdoc‐Programm (570704 83), Marie Curie Actions (PIEF‐GA‐2013‐623713), and QMUL Research‐IT and MidPlus computational facilities (The Engineering and Physical Sciences Research Council grant EP/K000128/1).
DATA ARCHIVING
This analysis relies on the following data:
Illumina sequences from 15 fire ant males: NCBI SAMN00014755 and SRP017317.
Fire ant reference genome assembly: GCA_000188075.1.
LITERATURE CITED
- Arguello, J. R. , and Connallon T.. 2011. Gene duplication and ectopic gene conversion in Drosophila . Genes 2:131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascunce, M. S. , Yang C.‐C., Oakey J., Calcaterra L., Wu W.‐J., Shih C.‐J., Goudet J., Ross K. G., and Shoemaker D.. 2011. Global invasion history of the fire ant Solenopsis invicta . Science 331:1066–1068. [DOI] [PubMed] [Google Scholar]
- Bastian, F. , Parmentier G., Roux J., Moretti S., Laudet V., and Robinson‐Rechavi M.. 2008. Bgee: integrating and comparing heterogeneous transcriptome data among species Pp. 124–131 in Bairoch A., Cohen‐Boulakia S., and Froidevaux C., eds. Data integration in the life sciences, Lecture notes in computer science. Springer, Berlin, Heidelberg. [Google Scholar]
- Benton, R. , Vannice K. S., Gomez‐Diaz C., and Vosshall L. B.. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila . Cell 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke, A. F. G. , and Heinze J.. 1994. The ecology of communal breeding: the case of multiple‐queen Leptothoracine ants. Proc. R. Soc. B 345:359–372. [Google Scholar]
- Bray, N. L. , Pimentel H., Melsted P., and Pachter L.. 2016. Near‐optimal probabilistic RNA‐seq quantification. Nat. Biotechnol. 34:525–527. [DOI] [PubMed] [Google Scholar]
- Buechel, S. D. , Wurm Y., and Keller L.. 2014. Social chromosome variants differentially affect queen determination and the survival of workers in the fire ant Solenopsis invicta . Mol. Ecol. 23:5117–5127. [DOI] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T. L.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif, D. , and Lobry J. R.. 2007. SeqinR 1.0‐2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis Pp. 207–232 in Bastolla U., Porto M., Roman H. E., and Vendruscolo M., eds. Structural approaches to sequence evolution, Biological and medical physics, biomedical engineering. Springer, Berlin, Heidelberg. [Google Scholar]
- Chevreux, B. , Wetter T., and Suhai S.. 1999. Genome sequence assembly using trace signals and additional sequence information Pp. 45–56 in Computer science and biology: Proceedings of the German Conference on Bioinformatics GCB’99, Heidelberg,. [Google Scholar]
- Corona, M. , Libbrecht R., Wurm Y., Riba‐Grognuz O., Studer R. A., and Keller L.. 2013. Vitellogenin underwent subfunctionalization to acquire caste and behavioral specific expression in the harvester ant Pogonomyrmex barbatus . PLoS Genet. 9:e1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHeer, C. J. 2002. A comparison of the colony‐founding potential of queens from single‐ and multiple‐queen colonies of the fire ant Solenopsis invicta . Anim. Behav. 64:655–661. [Google Scholar]
- DeHeer, C. J. , Goodisman M. A. D., and Ross K. G.. 1999. Queen dispersal strategies in the multiple‐queen form of the fire ant Solenopsis invicta . Am. Nat. 153:660–675. [Google Scholar]
- Dillies, M.‐A. , Rau A., Aubert J., Hennequet‐Antier C., Jeanmougin M., Servant N., et al. 2013. A comprehensive evaluation of normalization methods for Illumina high‐throughput RNA sequencing data analysis. Brief. Bioinform. 14:671–683. [DOI] [PubMed] [Google Scholar]
- Eliyahu, D. , Ross K. G., Haight K. L., Keller L., and Liebig J.. 2011. Venom alkaloid and cuticular hydrocarbon profiles are associated with social organization, queen fertility status, and queen genotype in the fire ant Solenopsis invicta . J. Chem. Ecol. 37:1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret, S. , and Maleszka R.. 2006. Function and evolution of a gene family encoding odorant binding‐like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, T. , Gersbach C. A., and Barbas C. F. III. 2013. ZFN, TALEN, and CRISPR/Cas‐based methods for genome engineering. Trends Biotechnol. 31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E. , and Marth G.. 2012. Haplotype‐based variant detection from short‐read sequencing. arXiv:1207.3907 [q‐bio.GN]. [Google Scholar]
- Goodisman, M. A. D. , DeHeer C. J., and Ross K. G.. 2000. Unusual behavior of polygyne fire ant queens on nuptial flights. J. Insect Behav. 13:455–468. [Google Scholar]
- Gotzek, D. , Robertson H. M., Wurm Y., and Shoemaker D.. 2011. Odorant binding proteins of the red imported fire ant, Solenopsis invicta: an example of the problems facing the analysis of widely divergent proteins. PLoS One 6:e16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotzek, D. , and Ross K. G.. 2007. Genetic regulation of colony social organization in fire ants: an integrative overview. Q. Rev. Biol. 82:201–226. [DOI] [PubMed] [Google Scholar]
- Gotzek, D. , and Ross K. G. 2009. Current status of a model system: the gene Gp‐9 and its association with social organization in fire ants. PLoS One 4:e7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmkampf, M. , Cash E., and Gadau J.. 2015. Evolution of the insect desaturase gene family with an emphasis on social Hymenoptera. Mol. Biol. Evol. 32:456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.‐C. , Yu‐Ching H., and Wang J.. 2014. Did the fire ant supergene evolve selfishly or socially? Bioessays 36:200–208. [DOI] [PubMed] [Google Scholar]
- Ingram, K. K. , Kutowoi A., Wurm Y., Shoemaker D., Meier R., and Bloch G.. 2012. The molecular clockwork of the fire ant Solenopsis invicta . PLoS One 7:e45715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovinella, I. , Dani F. R., Niccolini A., Sagona S., Michelucci E., Gazzano A., et al. 2011. Differential expression of odorant‐binding proteins in the mandibular glands of the honey bee according to caste and age. J. Proteome Res. 10:3439–3449. [DOI] [PubMed] [Google Scholar]
- Jasper, W. C. , Linksvayer T. A., Atallah J., Friedman D., Chiu J. C., and Johnson B. R.. 2015. Large‐scale coding sequence change underlies the evolution of postdevelopmental novelty in honey bees. Mol. Biol. Evol. 32:334–346. [DOI] [PubMed] [Google Scholar]
- Johnson, B. R. , Atallah J., and Plachetzki D. C.. 2013. The importance of tissue specificity for RNA‐seq: highlighting the errors of composite structure extractions. BMC Genomics 14:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B. R. , and Linksvayer T. A.. 2010. Deconstructing the superorganism: social physiology, groundplans, and sociogenomics. Q. Rev. Biol. 85:57–79. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , and Toh H.. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298. [DOI] [PubMed] [Google Scholar]
- Keller, L. , and Ross K. G.. 1995. Gene by environment interaction: effects of a single gene and social environment on reproductive phenotypes of fire ant queens. Funct. Ecol. 9:667–676. [Google Scholar]
- Keller, L. , and Ross K. G. 1998. Selfish genes: a green beard in the red fire ant. Nature 394:573–575. [Google Scholar]
- Keller, L. , and Ross K. G. 1999. Major gene effects on phenotype and fitness: the relative roles of Pgm‐3 and Gp‐9 in introduced populations of the fire ant Solenopsis invicta . J. Evol. Biol. 12:672–680. [Google Scholar]
- Kondrashov, F. A. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B 279:5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, M. J. B. 2004. To b or not to b: a pheromone‐binding protein regulates colony social organization in fire ants. Bioessays 27:91–99. [DOI] [PubMed] [Google Scholar]
- Krieger, M. J. B. , and Ross K. G.. 2002. Identification of a major gene regulating complex social behavior. Science 295:328–332. [DOI] [PubMed] [Google Scholar]
- Krieger, M. J. B. , and Ross K. G. 2005. Molecular evolutionary analyses of the odorant‐binding protein gene Gp‐9 in fire ants and other Solenopsis species. Mol. Biol. Evol. 22:2090–2103. [DOI] [PubMed] [Google Scholar]
- Kulmuni, J. , Wurm Y., and Pamilo P.. 2013. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant‐specific duplicates. Heredity 110:538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , and Horvath S.. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , and Salzberg S. L.. 2012. Fast gapped‐read alignment with Bowtie 2. Nat. Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassance, J.‐M. , Groot A. T., Liénard M. A., Antony B., Borgwardt C., Andersson F., Hedenström E., Heckel D. G., and Löfstedt C.. 2010. Allelic variation in a fatty‐acyl reductase gene causes divergence in moth sex pheromones. Nature 466:486–489. [DOI] [PubMed] [Google Scholar]
- Lawson, L. P. , Vander Meer R. K., and Shoemaker D.. 2012. Male reproductive fitness and queen polyandry are linked to variation in the supergene Gp‐9 in the fire ant Solenopsis invicta . Proc. R. Soc. B 279:3217–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal, W. S. 2013. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58:373–391. [DOI] [PubMed] [Google Scholar]
- Li, H. , Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Picimbon J.‐F., Ji S., Kan Y., Chuanling Q., Zhou J.‐J. et al. 2008. Multiple functions of an odorant‐binding protein in the mosquito Aedes aegypti . Biochem. Biophys. Res. Commun. 372:464–468. [DOI] [PubMed] [Google Scholar]
- Linksvayer, T. A. , Busch J. W., and Smith C. R.. 2013. Social supergenes of superorganisms: do supergenes play important roles in social evolution? Bioessays 35:683–689. [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Wolfgang H., and Simon A.. 2014. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja, A. , and Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S. E. , Smith J. A., Shamu C. E., Neumüller R. A., and Perrimon N.. 2014. RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 15:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, S. H. , and Mank J. E.. 2016. Inferring regulatory change from gene expression: the confounding effects of tissue scaling. Mol. Ecol. 25:5114–5128. [DOI] [PubMed] [Google Scholar]
- Morandin, C. , Tin M. M. Y., Abril S., Gómez C., Pontieri L., Schiøtt M., Schiøtt M., Sundström L., Tsuji K., Pedersen J. S., et al. 2016. Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome Biol. 17:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, C. S. , and Bell C. D.. 2013. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution 67:2240–2257. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W. 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17:481–485. [DOI] [PubMed] [Google Scholar]
- Nei, M. 1987. Molecular evolutionary genetics. Columbia Univ. Press, New York. [Google Scholar]
- Nei, M. , and Rooney A. P.. 2005. Concerted and birth‐and‐death evolution of multigene families. Annu. Rev. Genet. 39:121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis, O. , Buellesbach J., Gibson J. D., Pothmann D., Hanner C., Mutti N. S., Judson A. K., Gadau J., Ruther J., and Schmitt T.. 2013. Behavioural and genetic analyses of Nasonia shed light on the evolution of sex pheromones. Nature 494:345–348. [DOI] [PubMed] [Google Scholar]
- Nipitwattanaphon, M. , Wang J., Dijkstra M. B., and Keller L.. 2013. A simple genetic basis for complex social behaviour mediates widespread gene expression differences. Mol. Ecol. 22:3797–3813. [DOI] [PubMed] [Google Scholar]
- Nipitwattanaphon, M. , Wang J., Ross K. G., Riba‐Grognuz O., Wurm Y., Khurewathanakul C., et al. 2014. Effects of ploidy and sex‐locus genotype on gene expression patterns in the fire ant Solenopsis invicta . Proc. R. Soc. B. 281:20141776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P. , Iovinella I., Felicioli A., and Dani F. R. 2014. Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P. , Zhou J.‐J., Ban L. P., and Calvello M.. 2006. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63:1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracana, R. , Priyam A., Levantis I., Nichols R., and Wurm Y.. 2017. The fire ant social chromosome supergene variant Sb shows low diversity but high divergence from SB. Mol. Ecol. 26:2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privman, E. , Wurm Y., and Keller L.. 2013. Duplication and concerted evolution in a master sex determiner under balancing selection. Proc. R. Soc. B 280:20122968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyam, A. , Woodcroft B. J., Rai V., Munagala A., Moghul I., Ter F., et al. 2015. Sequenceserver: a modern graphical user interface for custom BLAST databases. bioRxiv 033142. 10.1101/033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M. , Warr C. G., and Carlson J. R.. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster . Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, G. E. , Grozinger C. M., and Whitfield C. W.. 2005. Sociogenomics: social life in molecular terms. Nat. Rev. Genet. 6:257–270. [DOI] [PubMed] [Google Scholar]
- Robinson, S. W. , Herzyk P., Dow J. A. T., and Leader D. P.. 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster . Nucleic Acids Res. 41:D744–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K. G. 1997. Multilocus evolution in fire ants: effects of selection, gene flow and recombination. Genetics 145:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K. G. , and Keller L.. 1995. Ecology and evolution of social organization: insights from fire ants and other highly eusocial insects. Annu. Rev. Ecol. Syst. 26:631–656. [Google Scholar]
- Ross, K. G. , and Keller L. 1998. Genetic control of social organization in an ant. Proc. Natl. Acad. Sci. USA 95:14232–14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K. G. , and Keller L. 2002. Experimental conversion of colony social organization by manipulation of worker genotype composition in fire ants (Solenopsis invicta). Behav. Ecol. Sociobiol. 51:287–295. [Google Scholar]
- Stamatakis, A. 2006. RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Schwander, T. , Libbrecht R., and Keller L.. 2014. Supergenes and complex phenotypes. Curr. Biol. 24:R288–R294. [DOI] [PubMed] [Google Scholar]
- Smith, J. M. , and Haigh J.. 1974. The hitch‐hiking effect of a favourable gene. Genet. Res. 23:23–35. [PubMed] [Google Scholar]
- Sun, Y.‐L. , Huang L.‐Q., Pelosi P., and Wang C.‐Z.. 2012. Expression in antennae and reproductive organs suggests a dual role of an odorant‐binding protein in two sibling Helicoverpa species. PLoS One 7:e30040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, M. J. , and Jiggins C. D.. 2014. Supergenes and their role in evolution. Heredity 113:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible, W. , and Ross K. G.. 2016. Chemical communication of queen supergene status in an ant. J. Evol. Biol. 29:502–513. [DOI] [PubMed] [Google Scholar]
- Tschinkel, W. R. 2006. The fire ants. The Belknap Press of Harvard Univ. Press, Cambridge, Massachusetts, London, England. [Google Scholar]
- UniProt Consortium . 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, F. G. , and Rozas J.. 2011. Comparative genomics of the odorant‐binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, F. G. , Sánchez‐Gracia A., and Rozas J.. 2007. Comparative genomic analysis of the odorant‐binding protein family in 12 Drosophila genomes: purifying selection and birth‐and‐death evolution. Genome Biol. 8:R235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Jemielity S., Uva P., Wurm Y., Gräff J., and Keller L.. 2007. An annotated cDNA library and microarray for large‐scale gene‐expression studies in the ant Solenopsis invicta . Genome Biol. 8:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ross K. G., and Keller L.. 2008. Genome‐wide expression patterns and the genetic architecture of a fundamental social trait. PLoS Genet. 4:e1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wurm Y., Nipitwattanaphon M., Riba‐Grognuz O., Huang Y.‐C., Shoemaker D., and Keller L. 2013. A Y‐like social chromosome causes alternative colony organization in fire ants. Nature 493:664–668. [DOI] [PubMed] [Google Scholar]
- Ward, P. S. , Brady S. G., Fisher B. L., and Schultz T. R.. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst. Entomol. 40:61–81. [Google Scholar]
- West, S. A. , and Gardner A.. 2010. Altruism, spite, and greenbeards. Science 327:1341–1344. [DOI] [PubMed] [Google Scholar]
- Wurm, Y. , Wang J., Riba‐Grognuz O., Corona M., Nygaard S., Hunt B. G., et al. 2011. The genome of the fire ant Solenopsis invicta . Proc. Natl. Acad. Sci. USA 108:5679–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. X. , Zwiebel L. J., and Smith D. P.. 2003. Identification of a distinct family of genes encoding atypical odorant‐binding proteins in the malaria vector mosquito, Anopheles gambiae . Insect Mol. Biol. 12:549–560. [DOI] [PubMed] [Google Scholar]
- Xu, Y.‐L. , He P., Zhang L., Fang S.‐Q., Dong S.‐L., Zhang Y.‐J., and Li F.. 2009. Large‐scale identification of odorant‐binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics 10:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Wanchoo A., Ortiz‐Urquiza A., Xia Y., and Keyhani N. O.. 2016. Tissue, developmental, and caste‐specific expression of odorant binding proteins in a eusocial insect, the red imported fire ant, Solenopsis invicta . Sci. Rep. 6:35452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Slone J. D., Rokas A., Berger S. L., Liebig J., Ray A., Reinberg D., and Zwiebel L. J., 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex‐specific signatures of odor coding. PLoS Genet. 8:e1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Density distribution of the p‐values for differential expression between social forms in queens for OBPs (in green) and all other protein‐coding genes (red). The p‐values for OBPs are strongly skewed towards 0. This result is based on the expression levels from the Morandin et al. (2016) dataset.
Figure S2. Correspondence between queen and worker modules. Numbers in the table indicate probe counts in the intersection of the corresponding modules. Coloring of the table encodes − log(p), with p being the Fisher's exact test p‐value for the overlap of the two modules. A module in one dataset would be preserved across both sets if it had a single corresponding module in the other dataset with a large number of probes in common.
Figure S3. Nucleotide distribution (π, measured from SB individuals in Pracana et al. 2017) of 10kb windows of the assembled genome that overlap coding sequences. Vertical bars represent π of windows overlapping OBPs; orange bars representing those overlapping supergene OBPs.
Table S1. Summary of correspondences between identifiers of sequences produced in this project and previously published sequences, including the number of sequence differences between the two groups.
Table S2. Accession numbers of the gene expression data used. “Project” and “SRA” columns indicate NCBI identifiers. The descriptions provided and the sequencing method used are based on metadata available on NCBI and in the manuscripts. Two samples (marked with an asterisk) were discarded because of very low coverage after aligning the reads to the S. invicta genome.
Table S3. Table showing the results of the alignment of ESTs from the S.invicta microarray (Platform GPL6930) to its reference genome. The columns “Scaffold”, “Start” and “End” indicate the coordinates of the EST in the reference. “Probe” refers to probe to which each EST belongs and “Gene” refers to the gene intersecting that particular region of the S.invicta genome. ESTs tagged with an asterisk (*) map to more than one location in the genome.
Table S4. Details of closest BLASTP hit to NCBI “nr” database for each newly produced S. invicta OBP sequence.
Table S5. Number of genes represented in each co‐expression module; OBPs represented in each module. Each gene may be represented by multiple probes. Probes in queen_E_1 are not assinged to a module.
Table S6. Gene co‐expression modules with module eigene differential expression between genotypes in queens (SB/SB versus SB/Sb), between genotypes in workers from multiple‐queen colonies (SB/SB versus SB/Sb), and betwen social forms in SB/SB workers (single‐queen colony versus multiple‐queen colony). Differential expression was tested with t‐tests within each comparison within each dataset, with p‐values corrected for multiple testing using Bonferroni correction.
Table S7. Putative OBP orthologs in other species. First, we ran a tblastn search of all S. invicta OBPs against all non‐S. invicta arthropod sequences, accepting hits where e‐value < 10−3. We then ran a blastx search of these hits against the S. invicta gene predictions (including our newly curated OBP set). We report the hits with the lowest e‐value of the blastx search. We repeated this analysis by searching non‐ant arthropods (not Formicidae). The coloured cells represent cases where the same non‐S. invicta sequence aligns to multiple S. Invicta OBPs.


