Abstract
Pancreatic ductal adenocarcinoma (PDAC) has a poor prognosis with frequent post-surgical local recurrence. The combination of adjuvant chemotherapy with radiotherapy is under consideration to achieve a prolonged progression-free survival (PFS). To date, few studies have determined the proteome profiles associated with response to adjuvant chemoradiation. We herein analyzed the proteomes of primary PDAC tumors subjected to additive chemoradiation after surgical resection and achieving short PFS (median 6 months) versus prolonged PFS (median 28 months). Proteomic analysis revealed the overexpression of Aldehyde Dehydrogenase 1 Family Member A1 (ALDH1A1) and Monoamine Oxidase A (MAOA) in the short PFS cohort, which were corroborated by immunohistochemistry. In vitro, specific inhibition of ALDH1A1 by A37 in combination with gemcitabine, radiation, and chemoradiation lowered cell viability and augmented cell death in MiaPaCa-2 and Panc 05.04 cells. ALDH1A1 silencing in both cell lines dampened cell proliferation, cell metabolism, and colony formation. In MiaPaCa-2 cells, ALDH1A1 silencing sensitized cells towards treatment with gemcitabine, radiation or chemoradiation. In Panc 05.04, increased cell death was observed upon gemcitabine treatment only. These findings are in line with previous studies that have suggested a role of ALDH1A1 chemoradiation resistance, e.g., in esophageal cancer. In summary, we present one of the first proteome studies to investigate the responsiveness of PDAC to chemoradiation and provide further evidence for a role of ALDH1A1 in therapy resistance.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is projected to be the second leading cause of cancer related deaths in the western world by the year 2030 [1], with reported 5-year survival rates as low as 8% [2], [3]. For most patients, curative treatment options do not exist at the time of diagnosis. Surgery is considered the best treatment option, but is restricted to localized pancreatic cancer without evidence of distant metastasis [4], [5], [6]. About 20% of patients are eligible for surgical resection at initial diagnosis. Recently, the ESPAC-4 trial reported 5-year survival rates of 15% to 30% in patients with margin negative resection (R0) and adjuvant chemotherapy with either gemcitabine alone or gemcitabine in combination with capecitabine [4]. Local recurrence of the tumor occurs in up to 86% of surgical cases despite adjuvant chemotherapy [3], [4], [5], [7], [8], [9], [10]. More efficient adjuvant therapy regimens are being actively sought. One of the concepts under investigation is chemo-radiation therapy (CRT). The clinical benefit of CRT (e.g., as compared to chemotherapy alone) is a topic of ongoing research and debate [11], [12]. However, it should be noted that the standard mode of treatment in pancreatic cancer following surgery is additive and/or adjuvant chemotherapy.
Several recently published studies have successfully highlighted molecular PDAC subtyping and stratification with a focus on response to chemotherapy [11], [12], [13], [14], [15], [16], [17]. In addition to establishing biological marker profiles of potential chemotherapy responders, these studies also yielded an improved understanding of the molecular basis of therapy resistance. However, this type of research has not yet covered resistance to CRT in PDAC unlike the case of other tumor entities [18], [19].
The availability of formalin fixed paraffin embedded (FFPE) tissues provides a retrospective source of proteomic information, especially in conjunction with clinicopathological annotation and follow-up data [20], [21]. Coupled with label-free shotgun proteomics and immunohistochemistry, these FFPE tissues have been widely used to identify biomarkers and drug targets in different disease settings [22], [23], [24], [25]. Several recent reports highlight a limited correlation between mRNA levels and corresponding protein levels, thus arguing in favor of proteomic approaches [26], [27]. In PDAC, drug combination therapies have been experimented to improve treatment efficacy. A good example is the use of erlonitib, an epidermal growth factor receptor (EGFR) inhibitor, together with gemcitabine to target advanced PDAC, which has improved survival in unresectable cases [28], [29]. Therefore, the identification of genes/proteins that drive therapy resistance is essential as it creates a foundation for novel, efficient co-treatment strategies.
In the present study, we employed quantitative proteomics to distinguish the proteome signature of short and prolonged disease-free survival in surgically resected PDAC tumors subjected to additive chemoradiation. The identification of this proteome signature is important as it lays a basis for the future development of effective co-treatment regimens. We applied the acid labile surfactant-based protein retrieval on FFPE tissues, liquid chromatography–tandem mass spectrometry (LC–MS/MS) and label free proteomics. We identified aldehyde dehydrogenase 1 family member A1 (ALDH1A1) as one of the proteins up-regulated in the short PFS subcohort. We further show that using a specific ALDH1A1 inhibitor, we sensitized PDAC cells to gemcitabine, radiation, and chemoradiation therapy in vitro. In line with further studies on other solid tumor entities, our study emphasizes a role of ALDH1A1 in contributing resistance to chemoradiation treatment.
Materials and Methods
Ethics Statement
The clinical proteomic study of human PDAC tissue was approved by the local Ethics Committee of the University of Lübeck / University Hospital of Schleswig-Holstein, Lübeck (AZ 15–368, January 2016).
Patient Cohort
This study included primary tumor tissue from 12 patients with primary diagnosis of localized (non-metastatic) PDAC between 1999 and 2014 who underwent chemoradiation at University Hospital of Schleswig-Holstein, Campus Lübeck. Only cases with sufficient follow-up information and availability of tissue were included. Details on the patient cohort are provided in the results section.
Tissue Collection, Fixation and Microdissection
Tissue specimens were harvested at the time of pancreatic surgery. Tissue fixation in formalin and paraffin embedding was conducted according to established protocols. Hematoxylin - Eosin (HE) - stained sections of the 12 samples were microscopically inspected by experienced pathologists to confirm diagnosis and to mark appropriate tumor areas for microdissection as described previously [30].
Sample Preparation for LC-MS/MS-analysis and Data Acquisition
An acid-labile surfactant protocol for heat antigen retrieval and direct trypsinization was employed as described previously [30], [31]. The microscopically dissected tumor regions were scrapped into 1.5 ml reaction tubes. 100 μl of an aqueous buffer containing 0.1% Rapigest (Waters), 1 mM dithiothreitol (DTT, AppliChem) and 1 M Hepes pH 8.0 (AppliChem) was added into each tube. Antigen retrieval and protein extraction were performed by incubating the samples at 95°C and 750 rpm for 1 hour. Afterwards, the pH of each sample was controlled to be in the range of 7.0–8.0. The samples were trypsinized at 37°C, 18 h using sequencing grade trypsin (Worthington), with 2 μg trypsin added per 1 mm3 tissue volume. Undigested tissue debris was removed by centrifugation (19,000 g for 15 minutes).
The supernatants were transferred to new 1.5 ml reaction tubes followed estimation of the protein concentration by bicinchoninic acid (BCA) assay (Thermo Scientific). Cysteine resides in the digested samples were reduced and alkylated using 10 mM DTT for 30 minutes at 37°C, followed by 30 mM iodoacetamide (Sigma-Aldrich) for 15 minutes at 37°C, and quenching of excess iodoacetamide with 10 mM DTT for 15 minutes at 37°C. Prior to Rapigest decomposition, guanidinium chloride (AppliChem) was added to a final concentration of 3 M before acidification by 0.2 M hydrochloric acid and incubation at 37°C for 30 minutes. The samples were then centrifuged (19,000 g for 15 minutes) and the supernatants were transferred to 1.5 ml reaction tubes. 12.5 μg of each sample were desalted using self-assembled C18 Stage tips (Empore, St. Paul, MN). Protein concentrations were determined using the BCA assay and 2 μg peptides per sample were vacuum dried using a centrifugal concentrator (Eppendorf) and stored at −80°C prior to LC–MS/MS analysis.
LC–MS/MS for Proteome Analysis
For LC–MS/MS, samples were analyzed by an Orbitrap Q-Exactive Plus (Thermo Scientific) mass spectrometer coupled to an Easy nano-LC 1000 (Thermo Scientific) with a flow rate of 300 nl/min. Buffer A was 0.1% (v/v) formic acid and buffer B was 0.1% (v/v) formic acid in 80% acetonitrile. For reverse phase chromatography, a gradient of increasing acetonitrile proportion was applied in combination with a C18 separating column (Acclaim PepMap, 2 μm particle size, 100 Å pore size, 150 mm length, and inner diameter 50 μm). The mass spectrometer operated in data dependent mode with a maximum of 10 MS/MS scans following each MS1 scan.
Data Analysis
Data analysis was performed using MaxQuant (v1.5.2.8) [21]. For peptide identification, the Andromeda search engine and the reviewed human database without isoforms downloaded from Uniprot on 25th July, 2016 (20,193 entries) were used. A decoy database was generated using the revert function. The precursor mass tolerance for the first search was 20 ppm, 4.5 ppm for the main search and 20 ppm for the fragment mass tolerance. Peptide-spectrum matching included a fixed modification of carbamidomethyl cysteine and variable modifications of methionine oxidation and N-terminal acetylation. Tryptic cleavage specificity was set with a maximum of two missed cleavages and minimum peptide length set at seven amino acids. For protein identification, a minimum of one peptide was set. The false discovery rate (FDR) at both the peptide and protein level was set to 0.01.
Label free quantitation was performed using the MaxLFQ algorithm with at least one peptide per protein considered. The MaxQuant output was further processed using Perseus (v1.5.0.0). Reverse decoy sequences and potential contaminant entries were removed and the LFQ intensities transformed to log 2 values. Statistical analysis was performed in R (v3.3.1) using linear models for microarray data (LIMMA) with a P value cut-off set at 0.01 to identify significantly regulated proteins. The choice for LIMMA was based on the small sample size as well as correcting for the multiple testing problem in this case study. For classification of interacting proteins/protein groups, STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) [32] was used on proteins with a p-value cut-off of 0.05.
Immunohistochemistry
Immunohistochemical corroboration of ALDH1A1 and MAOA was performed as described earlier [30], [33] using specific antibodies mouse anti-human ALDH1A1 (R&D, MAB5869) and rabbit anti-human MAOA (ProteinTech, 10,539-1AP). Briefly, 2 μm tissue sections were deparaffinized and subjected to heat-induced antigen retrieval. Tissue sections were then stained using the following steps: incubation in H2O2 for 5 minutes, with primary antibodies for 1 hour, with mouse/rabbit linker (15 minutes), with horseradish peroxidase and secondary antibody for 20 minutes and final incubation with 3, 3′-diaminobenzidine for 10 minutes. Sections were then counterstained in hematoxylin for a minute; with xylene used as permanent mounting medium. We evaluated the intensity of immunohistochemical staining using a well-established pathological scoring system with 0 = negative, 1 = weak, 2 = moderate, and 3 = strong [34]. For all samples, we only considered those tumor areas that corresponded to HE-stained templates that underwent proteomic analysis.
Cell Culture
MiaPaCa-2, HPAF-II and Panc 05.04 cell lines were purchased from the American Type Culture Collection (ATCC). MiaPaCa-2 and HPAF-II were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, Panc 05.04 was cultured in RPMI medium containing 15% fetal calf serum supplemented with 0.1% insulin. Cell lines were incubated at 37°C in humidified air, containing 5% CO2.
Quantitative Real Time PCR (qPCR)
RNA expression levels of genes of interest (ALDH1A1 and MAOA) were quantified using qPCR, essentially performed as described previously [35]. Briefly, RNA was isolated using the RNAeasy system (Qiagen), 2 μg each of total RNA was reverse transcribed using the iScript cDNA synthesis system (Bio-Rad) and 10 ng of each sample was analyzed in technical triplicates. Relative mRNA expression of ALDH1A1 and MAOA was normalized to β-actin mRNA as a control housekeeping gene. Relative mRNA abundance was calculated by the ddCt method. The following primer pairs were used, ALDH1A1 forward: 5′-ATCAAAGAAGCTGCCGGGAA-3′, ALDH1A1 reverse: 5′-TCTTAGCCCGCTCAACACTC-3′, MAOA forward: 5′-GCATTTCAGGACTATCTGCTGC-3′, MAOA reverse: 5′-GGTCCCACATAAGCTCCACC-3′, Actin forward AGCACTGTGTTGGCGTACAG, and Actin reverse CTCTTCCAGCCTTCCTTCCT. The cycling conditions were as follows: one cycle of 72°C for 1 min; 40 cycles of 95°C for 15 s, 56°C for 30 s, 72°C for 30 s; and one cycle of 72°C for 5 min.
Western Blot Analysis
Immunoblotting was performed using specific primary antibodies mouse anti-human ALDH1A1 (R&D, MAB5869, 1:500) and rabbit anti-human MAOA (ProteinTech, 10,539-1AP, 1:500). Briefly, total protein extraction was conducted as described previously [35], [36]. Protein concentration was determined using the bicinchoninic acid (BCA) assay. 50 μg of protein lysates were loaded on to 10% SDS-polyacrylamide gels followed by transfer onto PVDF membranes using the semidry blot system (Bio-Rad). The membranes were blocked with 5% milk in PBS-Tween, incubated overnight at 4°C with primary antibodies followed by 1 hour incubation with respective secondary antibodies at room temperature. The membranes were then developed using the West Pico Chemiluminescent substrate (Pierce) and peroxidase activity detected using the Lumilmager device (Roche Applied Science).
Inhibitor Sensitivity Assay
The sensitivity of MiaPaCa-2, HPAF-II and Panc 05.04 cells to ALDH1A1 and MAOA inhibitors, A37 (Tocris), DEAB (Tocris) and clorgyline (Sigma-Aldrich) was quantified using the MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) cell survival assay (Sigma). Briefly, cells were seeded at a density of 10,000 cells/well in 24-well plates and cultured at 37°C. After 24 hours, the inhibitors were added in increasing concentration and cells cultured for further 72 hours. Afterwards, 50 μl of the solubilized MTT salt was added to each well containing 500 μl of the cell culture medium and incubated for 3 hours. The purple crystals were then dissolved in acid isopropanol and incubated for a further 10 minutes. The samples were then transferred to a 96-well plate and absorbance read at 570 nm with a reference wavelength of 630 nm in a microplate reader. The dose response curve of three independent experiments was calculated relative to the DMSO treated controls.
MAOA and ALDH Activity Assays
MAOA activity assay (Sigma-Aldrich- MAK136) and ALDH activity assay (Stem Cell Technologies- #01700) were conducted according to the manufacturers' protocols.
Cell Sensitivity to 5-fluorouracil, Gemcitabine and/or Radiation
The in vitro sensitivity of these cells to 5-fluorouracil, gemcitabine and radiation was determined using the MTT assay or annexin V/PI staining. To quantify cell viability, 10,000 cells/well were seeded in 24-well plates and sub-cultured at 37°C. If applicable, after 24 hours, cells were treated with either ALDH1A1 or MAOA inhibitors (EC20 values depending on cell line – see Supplementary Figure 3, A–C) for 48 hours at 37°C. Next, fresh medium supplemented with gemcitabine (EC50 value- see Supplementary Figure 3E) was added to the cells and incubated for a further 72 hours. For radiation assays, cells were subjected to 10 Gy radiation dose from a Cesium-137 source (Cis Bio International) and incubated for a further 72 hours. For chemoradiation, cells were treated with both gemcitabine (EC50 values- see Supplementary Figure 3E) and 10 Gy radiation. As a control, we used cells treated with gemcitabine only, 10 Gy radiation only or a combination of both. Cell viability was then quantified using the MTT assay as previously described. For apoptosis assay, 50,000 cells/well were seeded in a 6 well plate and sub-cultured at 37°C. The treatment regimen described above was followed and cells harvested for apoptosis quantification by flow cytometry. Cells were trypsinized and washed once in annexin V binding buffer (Thermo Fischer) followed by staining using Annexin-V Alexa 488/PI (Thermo Fischer) for 15 minutes in the dark. The percentage of Annexin-V positive cells was measured using FACSCalibur (BD Bioscience).
For 3D experiments, MiaPaCa-2 cells were seeded in conical agarose microwell array (CAMA) to form non-spheroidal cell aggregates as previously described [37]. Briefly, 1000 cells were seeded into each microwell of these CAMA plates, cultured for 48 hours and treated with increasing concentrations (10 μM, 20 μM, and 40 μM) of ALDH1A1 inhibitor A37. After 3 hours, the cell aggregates were irradiated (8.5 Gy), followed by replenishing the inhibitor after 24 hours. The cell aggregates were left to grow for 14 days. The volume changes of the cell aggregates were quantified by scanning using a high-resolution flatbed scanner (CanoScan 9000F Mark II, Canon Inc.) as previously described [37].
ALDH1A1 Expression Silencing
The ALDH1A1 expression silencing by small hairpin RNA (shRNA) was generated using the MISSION shRNA lentivirus system (Sigma-Aldrich). Three different shRNA constructs were used: shRNA-ctrl (non-targeting shRNA, SHC002), shRNA-1 (TRCN0000026415), shRNA-2 (TRCN0000026498). Viral transduction and selection of stable transfectants was performed as previously described [36].
BrdU (Proliferation) and Metabolic Assays
Cell proliferation was quantified using the BrdU assay (Roche). Briefly, 100 μl of culture medium containing 2 x 103 cells were added to a 96-well plate and cultured for 48 hrs. 10 μl/well of BrdU labeling solution was added followed by 24 hours incubation. The cells were then fixed at room temperature followed by addition of 100 μl/well anti-BrdU-POD solution and incubated for 90 minutes. The cells were washed thrice and developed using 100 μl/well substrate solution. Sample absorbance in each well was measured at 370 nm with background measurement at 492 nm. For metabolic rate assays, 20,000 cells were seeded per well and cultured for 3 days. 50 μl of MTT salt was added to each well and incubated for 3 hours. The salt was solubilized using 100 μl of acid isopropanol and absorbance read at 570 nm.
Colony Formation Assays
To investigate the effect of ALDH1A1 on colony formation, 250 cells were added to each well of a 6-well plate. The cells were incubated for 12–14 days at 37°C with medium change every 3 days. The cells were washed twice with PBS and stained with 0.1% crystal violet for 30 minutes. The number of colonies per well was counted (≥50 cells) under the microscope and colony efficiency formation determined.
Cell Death Assays
The effect of ALDH1A1 expression silencing on sensitivity to gemcitabine, radiation and chemoradiation was probed as described above. Briefly, 50,000 cells were seeded in each well of a 6-well plate and cultured for 24 hours. Cells were then treated with gemcitabine (EC50 values), 10 Gy radiation or a combination of both gemcitabine and radiation. After 72 hours, cell death was quantified using annexin V/PI staining.
Reactive Oxygen Species Levels and DNA Damage Quantitation
Reactive oxygen species (ROS) levels were quantified using CellRox® Green Reagent (C10444, Life Technologies) according to manufacturer's instructions. Briefly, cells were irradiated at 10 Gy and incubated for 2 and 24 hours. The CellRox® reagent was added to the cultures to a final concentration of 5 μM and incubated for 30 minutes at 37°C. Non-irradiated cells were used as a control. The cells were washed thrice in PBS and resuspended in FACs buffer (1% FCS in PBS, 5 mM EDTA) and ROS levels quantified by flow cytometry.
Radiation generated ROS induces DNA damage that leads to H2A.X phosphorylation (Ser139). This phosphorylation was quantified by flow cytometry using pH2A.X (Ser139) mouse mAb Alexa488 conjugate (#20304, 1:50 Cell Signaling Technology). Briefly, irradiated cells were trypsinized at 0.5 and 2 hours after irradiation and washed in PBS solution. Methanol free formaldehyde was added to a final concentration of 4% and fixed at room temperature for 15 minutes. The cells were washed in cold PBS, permeabilized using ice-cold 100% methanol to a final concentration of 90% methanol and incubated for 30 minutes on ice. This was followed by washing in PBS, resuspension in 100 μl antibody solution (0.5 g BSA in 100 ml PBS) and incubation for 1 hour in the dark at room temperature. Cell were washed in PBS, resuspended in the antibody solution and analyzed by flow cytometry. Non-irradiated cells were used as a positive control.
Analysis of Intracellular Metabolites by LC-ESI-MS/MS
Cultured MiaPaCa-2 and Panc 05.04 cells were isolated by trypsinization, washed with PBS, and frozen at -80°C as dry pellets until further analysis. Cell pellets were thawed and quickly resuspended with 200 μL PBS. A 40 μL aliquot of the resuspended cells was mixed with an equal volume of an aminothiol preserving solution made up of 0.1 M Borate buffer pH 8.6, 0.1 M L-Serine, 10 mM iodoacetamide and 50 μM methylglutathione (GSMe, internal standard). Reaction of free reduced thiols with iodoacetamide limits artificial formation of mixed disulfides and spontaneous oxidation of reduced thiols during sample preparation. Formation of a borate-Ser complex inhibits the enzyme γ-glutamyltranspeptidase responsible for the degradation of glutathione. Lysis was performed by three cycles of freeze-thawing by alternating incubations in dry-ice and room temperature. An aliquot of 10 μl of lysate was separated and stored at -80°C for further measurement of concentration of proteins with the Bicinchoninic assay (Pierce, Thermofisher). For each sample, 20 μL lysate was mixed with 20 μL of H2O (basal pool of free oxidized thiols) or with 20 μL 0.5 M DTT (basal pool of mixed disulfides, both free and bound to proteins, converted to their free reduced forms upon reduction by DTT). The samples were incubated on ice 5 minutes, and 0.1 mL 0.1% formic acid in MeOH was added to precipitate proteins. The extracts were cleared by centrifugation at 13,000 rpm for 15 minutes at RT. An aliquot of 60 μl of cleared supernatant was transferred into high performance liquid chromatography (HPLC) vials and 4 μl of each sample was injected into the liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS QTrap 6500+, Sciex). Aminothiols were separated on a Sunfire C8 3.5 μm, 2.1 × 50 mm column (Waters) under isocratic conditions of 95% solvent A (0.1% formic acid in water) and 5% solvent B (0.1% formic acid in MeOH), at a flow rate of 0.75 mL/min, over 5 minutes. The concentration of aminothiols was determined with respect to a calibration curve created with commercial standards (homocysteine (Hcy), homocystine (Hcy-SS), cysteine (Cys), cystine (Cys-SS), reduced glutathione (GSH), oxidized glutathione (GSGG), cystathionine (Cysta), methionine (Met) and methionine sulfoxide (MSO), and the addition of GSMe as an internal standard. Values of intracellular metabolites were normalized to total protein concentration.
Statistical Analysis
For all experiments, statistical analysis was done for at least three independent experiments employing the unpaired Student's t test using GraphPad Prism 6.0 software (GraphPad Software) with P < .05 considered significant.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [38] via the PRIDE partner repository with the dataset identifier PXD009254 (Reviewer account details: username: reviewer44874@ebi.ac.uk, password: DwMsRVKM).
Results
General Approach and Overview of Patient Cohort
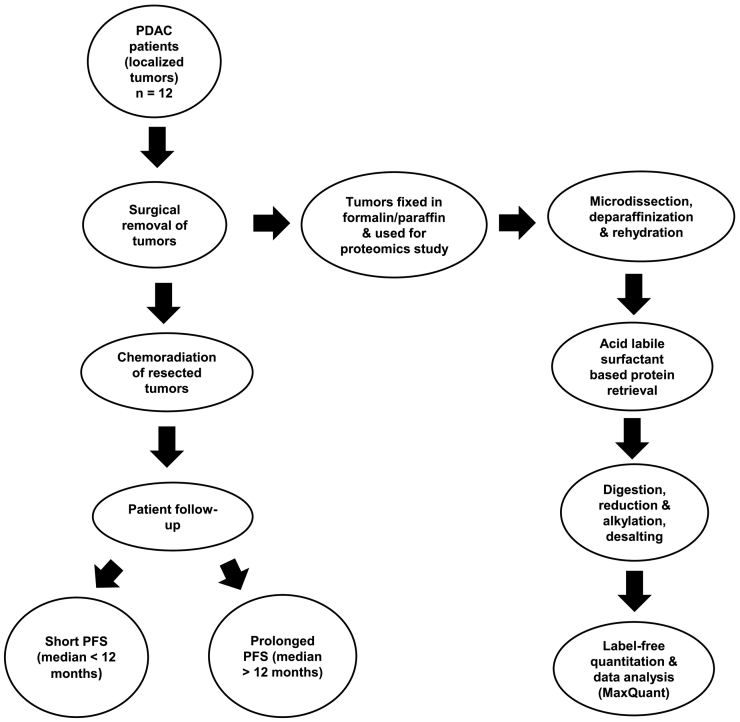
We aimed to investigate the proteome biology of localized PDAC subjected to chemoradiation and displaying short PFS (PFS< 12 months, n = 6) versus prolonged PFS (PFS >12 months, n = 6). In this case, PFS determination was based on macroscopic local recurrence or metastasis. The set-up is summarized in Figure 1. Patients with primary diagnosis of localized, non-metastatic PDAC underwent surgical exploration and resection at the University Hospital of Schleswig-Holstein, Campus Lübeck. Due to microscopically margin-positive resection (R1) or local irresectability upon exploration (n = 1, included due to prolonged PFS), surgery was followed by chemoradiation with 5-fluorouracil as radio-sensitizer between 1999 and 2014. Specimens of the resected primary tumor were preserved as FFPE samples. A detailed histopathological analysis of the patients involved in the study is shown in Table 1. We employed FFPE tissue samples since these represent the standard tissue for histopathological diagnosis and evaluation of human malignancies. Proteomic analysis of FFPE tissues using “direct trypsinization with acid-labile surfactants” demonstrated robustness in protein identification and quantification [30], [31].
Figure 1.
Schematic representation of the experimental design used in this proteomics study.
Table 1.
Histopathological Characteristics of the Patients Used in this Proteomics Study
| ID | age | sex | Tumor location | T | N | M | Grade | Margin status | Radiation dose | Chemo | FUP_OS | FUP_PFS | FUP_local recurrence | FUP_recurrence type | Tumor % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1 | 59 | F | head | T4 | N1 | M0 | G3 | R2 | 50.4 Gy | 5 FU | 12 | 6 | 6 | local | >80 |
| E2 | 61 | M | head | T3 | N1 | M0 | G2 | R2 | 50.4 Gy | 5 FU | 12 | 10 | 10 | distant | >80 |
| E3 | 65 | M | head | T3 | N1 | M0 | G2 | R1 | 50.4 Gy | no | 7 | 5 | 5 | local | >80 |
| E4 | 39 | M | head | T3 | N1 | M0 | G3 | R1 | 45 Gy | 5 FU | 5 | 5 | 5 | distant | >80 |
| E5 | 66 | F | head | T4 | N1 | M0 | G2 | R1 | 55.8 Gy | 5 FU | 5 | 5 | 5 | distant | >80 |
| E6 | 72 | F | head | T1 | N1 | M0 | G2 | R1 | 45 Gy | 5 FU | 2 | 2 | 2 | distant | >70 |
| L1 | 63 | F | tail | T4 | N0 | M0 | G2 | NA | 40 Gy | Gemzar+5FU | 33 | 33 | 33 | distant | >80 |
| L2 | 56 | M | head | T4 | N0 | M0 | G3 | R2 | 45 Gy | 5 FU | 14 | 13 | 13 | both | >80 |
| L3 | 63 | M | tail | T3 | N1 | M0 | G3 | R1 | 45 Gy | 5 FU | 27 | 27 | 27 | local | >80 |
| L4 | 66 | M | head | T3 | N1 | M0 | G2 | R1 | 50.4 Gy | 5 FU | 27 | 25 | 25 | distant | >80 |
| L5 | 64 | M | tail | T2 | N1 | M0 | G2 | R2 | 50.4 Gy | 5 FU | 29 | 29 | 29 | both | >80 |
| L6 | 65 | M | head | T3 | N1 | M0 | G3 | R0 | 45 Gy | 5 FU | 65 | 65 | 65 | distant | >80 |
Key: All these tumors were histologically classified as PDAC, FUP: follow up, OS (Overall survival), PFS (Progression-free survival), and local recurrence were all in months, NA: Not assessable, 5FU: 5-Fluorouracil, Chemo: Chemotherapy, Tumor %: Percent of FFPE tissue classified as tumor and used for proteomics.
Protein Identification and Relative Quantitation
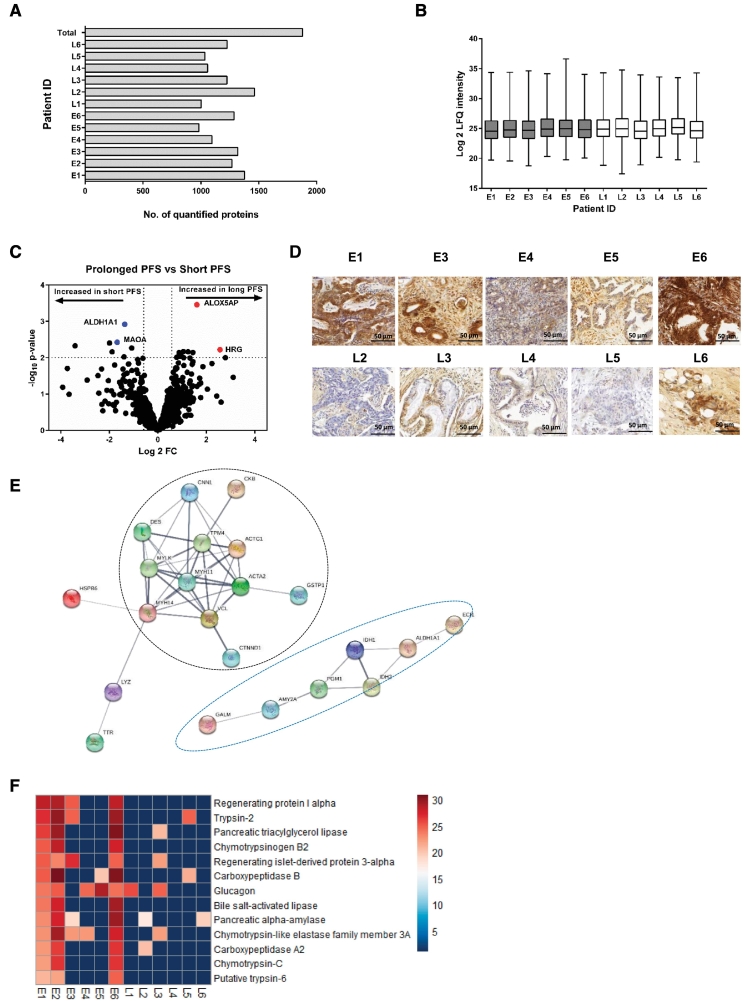
A total of 1878 proteins were identified and quantified across the 12 samples (Figure 2A and Supplementary Table 1). This number is in the range of several other FFPE-based proteomic studies [30], [31], [33]. Limited proteome overlap between biological and technical replicates continues to be a characteristic limitation of explorative shotgun proteomics [26], [27]. Although each case allowed for the identification and quantification of a comparable number of proteins (Figure 2A), the number of proteins that were consistently observed in all 12 samples remained below 600 (Supplementary Figure 1A). Each case also yielded a comparable distribution of label-free quantified protein intensities (Figure 2B). 1347 proteins were quantified in at least four samples (Supplementary Table 2). This “core” proteome was employed to determine proteomic differences with regard to short or prolonged PFS. We employed partial least squares discriminant analysis (PLS-DA) as an initial step to probe whether PDAC tumor tissues of short or prolonged PFS display distinguishable proteome profiles. Supervised PLS-DA separated short and prolonged PFS, hence supporting our aim to use quantitative proteomics for their unbiased distinction (Supplementary Figure 1C).
Figure 2.
Proteome profiling of PDAC patients. (A) Bar graph showing the number of proteins quantified in each patient sample. A total of 1878 proteins were quantified in the 12 patient samples. (B) The distribution and mean log 2 LFQ intensities (horizontal line) of quantified proteins in each patient. (C) Volcano plot showing the altered proteins in the PDAC patient cohort. Moderated t-test p-values were plotted against the mean protein fold changes per group. Data points in blue show the proteins overexpressed in short PFS group and those in red show the proteins overexpressed in prolonged PFS cohort. (D) Immunohistochemical corroboration of ALDH1A using representative tissues of the short PFS and prolonged PFS groups. The immunohistochemical staining shows clear differences in the expression of ALDH1A1 with more expression observed in the short PFS group than in the prolonged PFS group. Representative images were taken at x400 magnification. Sufficient material for immunohistochemical staining was available for only 10 of the 12 samples. (E) STRING analysis of proteins overexpressed in the early recurrence cohort based on P < .05 from LIMMA analysis show an enrichment in actin cytoskeleton proteins, oxidoreductases, and dehydrogenases. Connections show confidence (line thickness indicating the overall strength of data supports) and are based on categories “Co-expression”, “Co-occurrence”, “Databases”, “Experiments”, “Gene Fusion”, “Neighborhood”, and “Textmining” at a medium confidence interval of 0.4. MAOA does not appear in this figure because it is part of the 14 other proteins/nodes that were not connected to any other protein following STRING analysis. For the purpose of this analysis, we show connected proteins/nodes only that define the two distinct protein groups. (F) Heat map comparison of proteins expressed in at least 50% of patients in one group (short PFS) but absent in the other group. This comparison reveals a predominant exocrine-like fingerprint in 3 out of 6 patients in the short PFS cohort (E1, E2, and E6). The scale represents the log 2 values of LFQ intensities of the quantified proteins.
Further, statistical analysis was performed using linear models for microarray data (LIMMA) in-built in R (v3.3.1) that has been successfully employed for similar experiments [30], [31], [33] and which is particularly powerful with regard to multiple testing correction and prevention of false-positive discoveries in the analysis of omics-style data. As a cutoff to distinguish significantly regulated proteins, we chose a LIMMA moderated p-value of <0.01. Seven proteins were up-regulated in the short-PFS subcohort while 11 proteins were up-regulated in prolonged PFS subcohort as depicted by the volcano plot (Figure 2C, Table 2). Among the proteins of interest identified as up-regulated in the short-PFS subcohort is aldehyde dehydrogenase 1 family member A1 (ALDH1A1) and monoamine oxidase A (MAOA). Quantitation of ALDH1A1 and MAOA was corroborated by immunohistochemistry on adjacent tissue specimens of same patients as shown in Figure 2D and Supplementary Figure 1D respectively. Sufficient material for immunohistochemical staining was available for only 10 of the 12 samples.
Table 2.
List of Differentially Regulated Proteins as Determined by LIMMA Analysis With P < .01.
| Uniprot ID | Short PFS (Early) | Long PFS (Late) | Log 2 FC Late/Early | P | Protein Name |
|---|---|---|---|---|---|
| P12277 | 5 | 5 | −3.41 | .0047 | Creatine kinase B-type |
| O14558 | 6 | 5 | −1.99 | .0039 | Heat shock protein beta-6 |
| P98088 | 4 | 3 | −1.89 | .0069 | Mucin-5 AC |
| P21397 | 6 | 6 | −1.68 | .0038 | Monoamine oxidase type A |
| P36871 | 5 | 5 | −1.4 | .0094 | Phosphoglucomutase-1 |
| P00352 | 6 | 6 | −1.37 | .0012 | Aldehyde dehydrogenase family 1 member A1 |
| Q9P0M6 | 5 | 4 | −1.08 | .0055 | Core histone macro-H2A.2 |
| P38606 | 6 | 6 | 0.78 | .0098 | V-type proton ATPase catalytic subunit A |
| P61160 | 6 | 6 | 0.87 | .0073 | Actin-related protein 2 |
| P11413 | 6 | 6 | 0.94 | .0092 | Glucose-6-phosphate 1-dehydrogenase |
| P60953 | 6 | 6 | 0.98 | .0073 | Cell division control protein 42 homolog |
| O60506 | 5 | 4 | 1.05 | .0068 | Heterogeneous nuclear ribonucleoprotein Q |
| Q16658 | 6 | 6 | 1.06 | .0099 | Fascin |
| O60234 | 6 | 5 | 1.25 | .0069 | Glia maturation factor gamma |
| P16144 | 2 | 4 | 1.45 | .0072 | Integrin beta-4 |
| P20292 | 5 | 6 | 1.61 | .0004 | Arachidonate 5-lipoxygenase-activating protein |
| P04196 | 4 | 2 | 2.57 | .0061 | Histidine-rich glycoprotein |
| Q96SQ9 | 3 | 2 | 2.78 | .0099 | Cytochrome P450 2S1 |
Key: 7 proteins including ALDH1A1 and MAOA were up-regulated in the short PFS cohort while 11 proteins were up-regulated in the prolonged PFS cohort.
Increased Expression of Actin Cytoskeleton/Cell Adhesion Proteins, Oxidoreductases and Dehydrogenases in the short-PFS Subcohort
Altered protein clusters and interactions in the two patient cohorts were determined using a lower P (P < .05; two-sided Student t test). Using this criterion, 77 proteins were found to be differentially altered between the two patient groups. Proteins were then classified as up-regulated in either early or late recurrence on the basis of their fold change values. We used STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) to visualize the connections and interactions among the proteins up-regulated in each cohort / to probe for enriched proteome motives [32]. With a focus on the connected proteins only following STRING analysis, two distinct protein groups emerge as up-regulated in the short PFS cohort: (1) actin cytoskeleton/cell adhesion proteins and (2) oxidoreductases and dehydrogenases (Figure 2E). Four pathways (according to KEGG nomenclature) were enriched in the early recurrence cohort including metabolic pathways, glutathione metabolism, tricarboxylic acid (TCA) cycle, and glycolysis/gluconeogenesis (Table 3). This data suggests globally elevated metabolic activity in the early recurrence subcohort. Interestingly, elevated metabolic activity was also a proteome motif of metastasizing prostate cancer (PCa) compared to non-metastasizing (PCa) [30].
Table 3.
List of KEGG Pathways Enriched in the Short PFS Cohort.
| KEGG pathways | ||
|---|---|---|
| Pathway description | Gene count | FDR |
| Metabolic pathways | 9 | 0.0074 |
| Glutathione metabolism | 3 | 0.0074 |
| Drug metabolism-Cyt P450 | 3 | 0.0074 |
| Glycolysis/gluconeogenesis | 3 | 0.0074 |
The short-PFS Subcohort Exhibits an Exocrine-Like Proteome Fingerprint
Upon manual inspection of the differential proteome data, we noticed that the early recurrence subcohort is characterized by the abundant expression of several proteins that are associated with exocrine pancreas function. Examples include trypsin-2, carboxypeptidase B, regenerating protein I alpha, and pancreatic alpha-amylase among others. This proteomic fingerprint was observed for at least three of the six short PFS cases but only for one case of the prolonged PFS subcohort (Figure 2F, Table 4). The significance of this observation is limited due to the small sample size of our cohort. However, an exocrine-like PDAC subtype has been reported in two independent studies [16], [17]. In comparison to the classical PDAC subtype, exocrine-like PDAC is characterized by shortened overall survival and elevated resistance to small molecule drugs such as paclitaxel, erlotinib, and dasatinib [13], [17]. The specific response to adjuvant chemoradiation has however not been assessed in the aforementioned studies.
Table 4.
List of Proteins Contributing Towards the Exocrine-Like Fingerprint of the Short PFS Cohort.
| Uniprot ID | Short PFS | Long PFS | Protein name |
|---|---|---|---|
| P05451 | 4 | 0 | Regenerating protein I alpha |
| P07478 | 4 | 1 | Trypsin-2 |
| P16233 | 3 | 1 | Pancreatic triacylglycerol lipase |
| Q6GPI1 | 3 | 0 | Chymotrypsinogen B2 |
| Q06141 | 4 | 1 | Regenerating islet-derived protein 3-alpha |
| P15086 | 4 | 1 | Carboxypeptidase B |
| P01275 | 3 | 2 | Glucagon |
| P19835 | 4 | 0 | Bile salt-activated lipase |
| P04746 | 5 | 2 | Pancreatic alpha-amylase |
| P09093 | 3 | 1 | Chymotrypsin-like elastase family member 3A |
| P48052 | 3 | 1 | Carboxypeptidase A2 |
| Q99895 | 3 | 0 | Chymotrypsin-C |
| Q8NHM4 | 3 | 0 | Putative trypsin-6 |
Key: The values represent the number of samples in which a given protein was quantified in each subcohort (n = 6 for each subcohort).
We further analyzed the presence of mutated protein and peptide sequence as previously described [31]. In the short PFS subcohort we observed an average of 24.7 mutated sequences per patient. This number was slightly lower for the prolonged PFS subcohort (20.8 mutated sequences per patient); however this difference failed to be significant (P = .43; two-sided Student t test, Supplementary Figure 2, A and B).
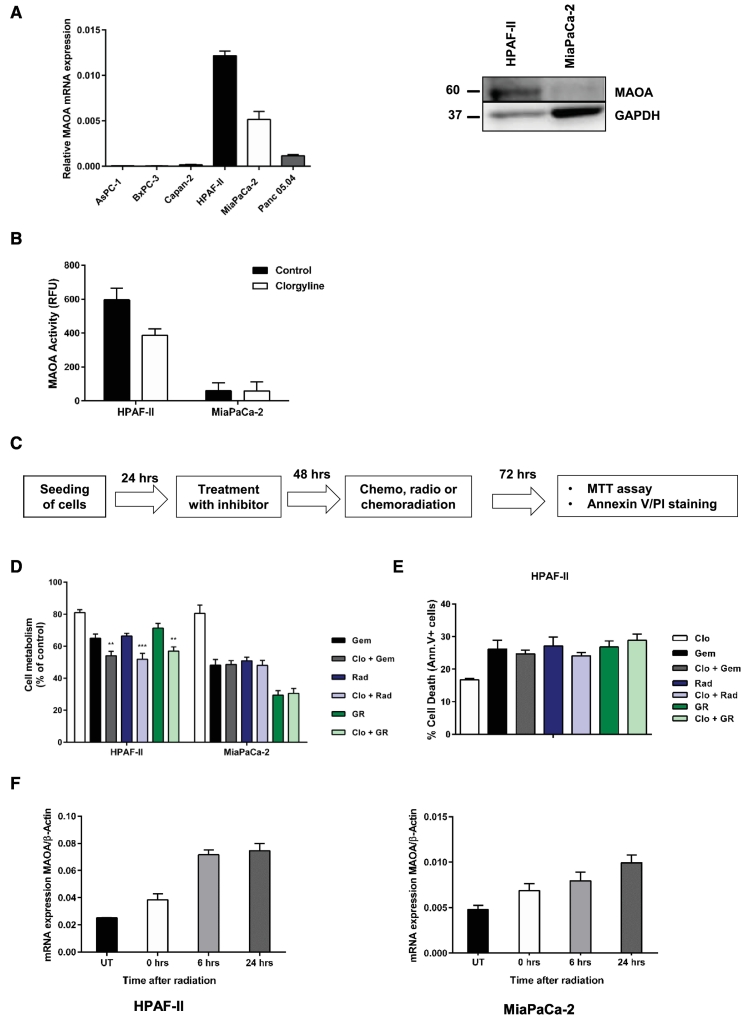
MAOA inhibition has a limited sensitizing effect on chemo- or radiation treatment in vitro
We investigated the role of MAOA inhibition, overexpressed in the short-PFS subcohort, in sensitizing PDAC cells to gemcitabine, radiation or chemoradiation. As an initial step, we established dose–response curves and EC50 values for 5-fluorouracil (5-FU), gemcitabine, and radiation (Supplementary Figure 3, D–F). Given the elevated EC50 value of MiaPaCa-2 and Panc 05.04 cells to 5-FU, we decided to use gemcitabine for further in vitro assays due to its comparably lower EC50 profile. We probed MAOA mRNA levels in a total of six established PDAC cell lines (AsPC-1, BxPC-3, Capan-2, HPAF-II, MiaPaCa-2, and Panc 05.04). Of these, only HPFAF-II and MiaPaCa-2 showed substantial MAOA levels and were chosen for further studies. Substantial MAOA activity was detected in HPAF-II that was inhibited upon administration of clorgyline (MAOA selective inhibitor) unlike in MiaPaCa-2 cells (Figure 3B). Clorgyline is an irreversible and specific inhibitor of MAOA with limited inhibitory effect on MAOB, and has been extensively used in research to study MAOA biology [39]. The EC20 values of MAOA inhibitor clorgyline in HPAF-II and MiaPaCa-2 were determined using the MTT assay (Supplementary Figure 3A). Pre-treatment of both cell lines with clorgyline for 48 hours followed by gemcitabine, radiation or combined chemoradiation (EC50 values) lowered cell metabolism in HPAF-II cells only (P < .05; two-sided Student t test, Figure 3D). No effect was observed in MiaPaCa-2 cells. Since a reduction in cell metabolism does not necessarily imply cytotoxicity, we further investigated whether pretreatment of HPAF-II cells with clorgyline followed by gemcitabine, radiation or chemoradiation augmented cell death. No effect on cell death, quantified by annexin V/PI staining, was observed in HPAF-II cells despite the reduced cell metabolism (Figure 3E). Interestingly, while we observed that MAOA inhibition has a limited sensitizing effect on treatment, the mRNA levels increased in a time-dependent manner upon radiation (Figure 3F).
Figure 3.
MAOA inhibition has limited sensitizing effect on chemo or radiation treatment. (A) Expression of MAOA mRNA in a panel of 6 PDAC cell lines and MAOA protein in HPAF-II and MiaPaCa-2 cells. (B) MAOA activity in HPAF-II and MiaPaCa-2 after treatment with 1 μM MAOA inhibitor clorgyline. Cell lysates were incubated with clorgyline for 1 hour in the dark at 37°C followed by fluorometric measurement of enzymatic activity at 37°C. (C) Schematics representation of the study design followed in these inhibitor sensitization experiments. (D) Inhibition of MAOA for 48 hours using clorgyline (EC20 values) followed by gemcitabine (1 μM), radiation (10 Gy), and chemoradiation for a further 72 hours reduced cell viability in HPAF-II cells. Cell viability was confirmed using MTT assay. (E) Inhibition of MAOA has no effect on cell death in HPAF-II cells. Cell death was confirmed using annexin V/PI staining. Data represents mean ± SD of three independent experiments. Statistical significance was determined using a two-sided Student t test with P < .01 (**) and P < .001 (***) against cells treated with gemcitabine, radiation or chemoradiation only. (F) Increased expression MAOA mRNA following radiation of HPAF-II and MiaPaCa-2 cells.
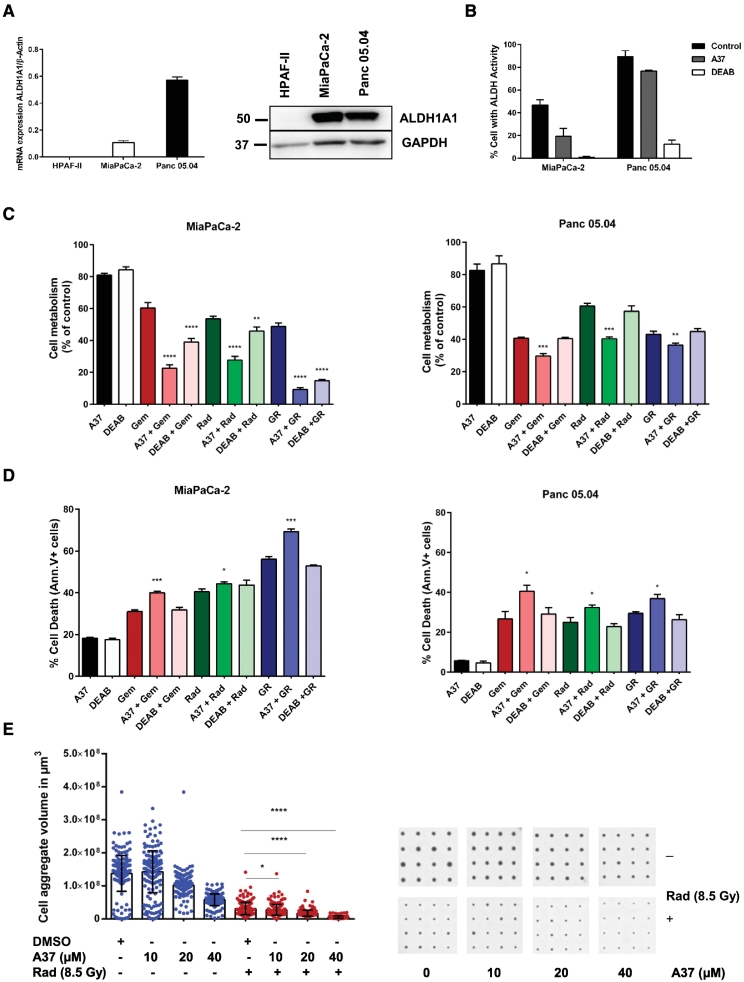
Inhibition of ALDH1A1 Activity Synergistically Sensitizes PDAC Cells to Chemo-, Radiation- or Chemoradiation Treatment in vitro
We further focused on ALDH1A1 to probe whether inhibition of the enzyme, which is overexpressed in the short-PFS cohort might sensitize PDAC cells for gemcitabine and/or radiation treatment in vitro. We detected expression of ALDH1A1 in the two PDAC cell lines MiaPaCa-2 and Panc 05.04 both on the mRNA and protein level (Figure 4A). In both cell lines, we detected substantial levels of ALDH1A1 activity, which was inhibited using the well-characterized ALDH1A1 inhibitors A37 and DEAB (Figure 4B). DEAB is a broad spectrum inhibitor of most of the ALDH family members including ALDH1A1. A37 is an ALDH1A1 specific inhibitor, a finding demonstrated by its lower normalized residual activity against ALDH1A1 than to other ALDH family of enzymes in the presence of 20 μM A37 [40]. The EC20 values of these inhibitors in MiaPaCa-2 and Panc 05.04 cells were determined using the MTT assay (Supplementary Figure 3, B and C). Pre-treatment of MiaPaCa-2 cells with both ALDH1A1 inhibitors followed by gemcitabine, radiation or combined chemoradiation (EC50 values) lowered cell metabolism significantly (P < .05; two-sided Student t test) (Figure 4C). For Panc 05.04 cells, decreased cell metabolism was observed using the A37 inhibitor only (Figure 4C). The role of ALDH1A1 in chemo- and radioresistance was assessed by quantifying the level of cell death in the presence of inhibitors. Cells were pretreated with inhibitors for 48 hours followed by gemcitabine, radiation or chemoradiation. The specific inhibition of ALDH1A1 activity using A37 augmented cell death in both cell lines (P < .05; two-sided Student t test) as determined by annexin V/PI staining, while we observed no effect with the broad spectrum inhibitor DEAB (Figure 4D). The co-efficient of drug interaction (CDI) upon treatment with A37 in combination with gemcitabine, radiation, and chemoradiation was calculated to determine whether the effect observed was synergistic or additive. We used the formula CDI = AB/(A × B), with A or B as the individual treatment, AB the combined treatment, and CDI value >1, =1, or <1 show that the drugs are antagonistic, additive or synergistic respectively [41]. The specific inhibition of ALDH1A1 with A37 followed by gemcitabine, radiation, or chemoradiation indicated a synergistic effect as the CDI value is less than 0.7 (Table 5).
Figure 4.
Inhibition of ALDH1A1 activity reduces cell viability and sensitizes PDAC cells to chemoradiation treatment. (A) Relative mRNA and protein expression of ALDH1A1 in PDAC cells HPAF-II, MiaPaCa-2, and Panc 05.04 cells. (B) Flow cytometry measurement of ALDH1A1 activity in MiaPaCa-2 and Panc 05.04 cells following treatment with ALDH1A1 inhibitors. DEAB treated cells served as negative controls and were used to set the FACS gate. (C) Inhibition of ALDH1A1 for 48 hours using A37 and DEAB (EC20 values) followed by gemcitabine (1 μM for MiaPaCa-2 and 0.25 μM for Panc 05.04), radiation (10 Gy), and chemoradiation for a further 72 hours reduced cell viability. Cell viability was confirmed using MTT assay. (D) Inhibition of ALDH1A1 for 48 hours using A37 and DEAB (EC20 values) followed by gemcitabine, radiation, and chemoradiation augmented cell death. Cell death was confirmed using annexin V/PI staining. For Panc 05.04, inhibition with DEAB had no effect on viability or cell death. Data represents mean ± SD of three independent experiments. Statistical significance was determined using a two-sided Student t test with P < .05 (*), P < .01 (**), P < .001 (***), and P < .0001 (****) against cells treated with gemcitabine, radiation or chemoradiation only. (E) Combined A37 and radiotherapy treatment of MiaPaCa-2 cell aggregates shows a reduction in volume of the cell aggregates at higher doses of the inhibitor. A scatter plot analysis with means ± SD (blue spots: inhibitor treatment only, red spots: A37 + radiation treatment). The bars represent mean of at least 175 replicates per condition. Statistical comparison was done between radiation treatment only vs radiation and A37 treatment using a two-sided Student t test with P < .0001 (****). A representative of each of the experimental conditions is shown in the corresponding CAMA scans.
Table 5.
The Co-Efficient of Drug Interactions (CDI) in MiaPaCa-2 and Panc 05.04 Upon Treatment With ALDH1A1 Specific Inhibitor (A37), in Combination With Gemcitabine, Radiation, and Chemoradiation.
| Co-efficient of drug interactions | |||||
|---|---|---|---|---|---|
| MiaPaCa-2 | |||||
| Treatment | A37 | A37 + Gem | A37 + Rad | A37 + GR | CDI |
| A37 | 18.22 ± 0.35 | ||||
| Gem | 30.94 ± 0.87 | 39.99 ± 0.76 | 0.07094 | ||
| Rad | 40.49 ± 1.27 | 44.27 ± 0.91 | 0.06001 | ||
| GR | 56.08 ± 1.19 | 69.22 ± 1.38 | 0.06774 | ||
| Panc 05.04 | |||||
| Treatment | A37 | A37 + Gem | A37 + Rad | A37 + GR | CDI |
| A37 | 05.68 ± 0.16 | ||||
| Gem | 26.69 ± 3.67 | 40.53 ± 3.01 | 0.26735 | ||
| Rad | 24.99 ± 2.34 | 32.37 ± 1.26 | 0.22805 | ||
| GR | 29.47 ± 0.77 | 36.86 ± 2.06 | 0.22021 | ||
| MiaPaCa-2 3D culture | |||||
| A37 (μM) | Vol. (μm3) | Rad (Gy) | Vol. (μm3) | CDI | |
| 0 | 137,776,090 | 8.5 | 31,224,455 | ||
| 10 | 142,544,081 | 8.5 | 27,296,987 | 6.133E-09 | |
| 20 | 101,913,975 | 8.5 | 17,333,234 | 5.449E-09 | |
| 40 | 58,002,988 | 8.5 | 5,128,585 | 2.837E-09 | |
Calculations
CDI = Survival AB/(Survival A x Survival B).
CDI <1, = 1 or >1 indicates that the drugs are synergistic, additive or antagonistic, respectively.
CDI <0.7 indicates that the drug is significantly synergistic.
Inhibition of ALDH1A1 in a 3D Assay Sensitizes MiaPaCa-2 Cells to Radiation Therapy
For 3D assays, only MiaPaCa-2 cells were used as Panc 05.04 did not form cell aggregates under conical agarose microwell array (CAMA) conditions. This method allows for monitoring of dose-dependent effects of combinatorial treatments using cell aggregate volumes as the read out [37]. The presence of A37 alone at higher concentrations (20 μM and 40 μM) lowers cell aggregate volume. In combination with radiation, a further decrease in cell aggregate volumes is observed (Figure 4E). These findings support a role of ALDH1A1 in both chemo- and radioresistance.
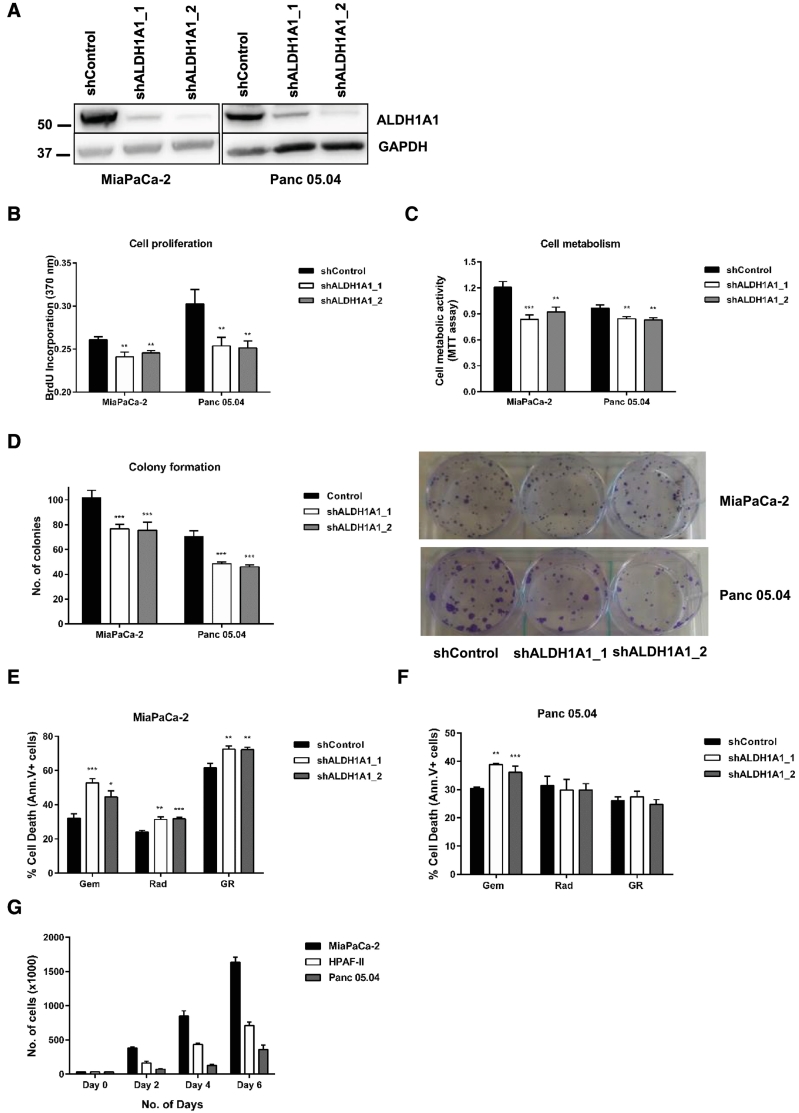
ALDH1A1 Expression Silencing Reduces Cell Growth, Proliferation and Clonogenic Capacity
To further investigate a putative role of ALDH1A1 in chemo/radiation resistance and/or recurrence, we silenced ALDH1A1 expression by shRNA in PDAC cells (Figure 5A). We investigated cell proliferation (BrdU incorporation), metabolic activity (MTT assay), and clonogenic capacity (Figure 5, B–D). All three cellular characteristics were attenuated by ALDH1A1 expression silencing.
Figure 5.
ALDH1A1 expression silencing lowers cell proliferation, colony formation and sensitizes cells to chemoradiation treatment. (A) ALDH1A1 expression silencing in MiaPaCa-2 and Panc 05.04 cells was confirmed using western blot analysis. ALDH1A1 knockdown reduced cell proliferation (B), lowered cell viability/metabolic activity (C), and attenuated the number of colonies formed by these cell lines (D). Cell proliferation was determined using the BrdU incorporation assay while metabolic rate was quantified using the MTT assay. Data represents mean ± SD of three independent experiments. Statistical significance was determined using a two-sided t-test with P < .01 (**) and P < .001 (***) against shControl cells. (E) ALDH1A1 expression silencing sensitizes MiaPaCa-2 cells to gemcitabine, radiation and chemoradiation. (F) For Panc 05.04 cells, ALDH1A1 expression silencing sensitizes the cells to gemcitabine only with no effect observed upon radiation or chemoradiation. Cell death was assayed using annexin V/PI staining. Data represents mean ± SD of three independent experiments. Statistical significance was determined using a two-sided t-test with P < .05 (*), P < .01 (**), P < .001 (***), and P < .0001 (****) against shControl cells. (G) Bar chart representing the proliferation rate of the three cell lines used for in vitro experiments. MiaPaCa-2 has the fastest proliferation rate while Panc 05.04 has the slowest rate of proliferation.
ALDH1A1 Expression Silencing Sensitizes PDAC Cells to Gemcitabine, Radiation, and Chemoradiation in vitro
Cells with ALDH1A1 expression silencing or corresponding controls were subjected to either gemcitabine, radiation or combined chemo-radiation (EC50 values) treatment in vitro for 72 hours. Cell death/cytotoxicity was quantified via annexin V/PI staining. Expression silencing of ALDH1A1 in MiaPaCa-2 cells led to increased cell death following treatment with gemcitabine, radiation, and combined chemoradiation (Figure 5E). For Panc 05.04 cells, increased cell death was observed under gemcitabine treatment only (Figure 5F). However, unlike for MiaPaCa-2 cells, combined chemoradiation did not lead to elevated cell death rates in Panc 05.04 cells as compared to gemcitabine or radiation alone. These results, at least for MiaPaCa-2 cells, complement the effects observed using the specific ALDH1A1 inhibitor (A37) suggesting specific rather than off-target effects of this inhibitor. The strong combinatorial effect of chemoradiation on MiaPaCa-2 cells can be attributed to their faster proliferation as compared to Panc 05.04 cells (Figure 5G). This possibly renders these cells sensitive to DNA damaging agents such as gemcitabine and radiation. We consider it likely that the mild effect observed in Panc 05.04 cells is associated with their slower proliferation, which would limit the impact of DNA damaging agents.
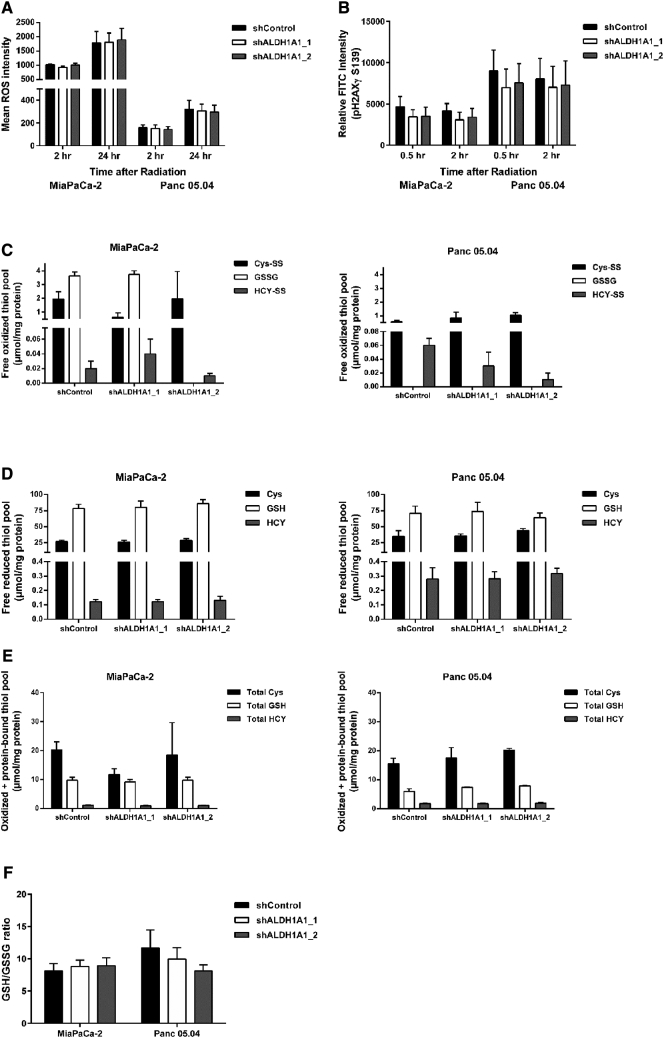
ALDH1A1 Expression Silencing has no Effect on ROS Levels, DNA Damage and Glutathione Metabolism
We hypothesized that ALDH1A1 drives chemoradiation resistance in PDAC cells, especially in MiaPaCa-2, by quenching ROS thereby limiting DNA damage and cell death. Therefore, ALDH1A1 expression silencing should lead to ROS accumulation with concomitantly enhanced DNA damage and cell death. Quantification of absolute ROS levels showed no difference between shControl and shALDH1A1 in both cell lines (Figure 6A). In addition, absolute quantification of pH2A.X (Ser139), a marker for DNA damage after irradiation, showed no differences in the phosphorylation levels of this histone marker upon ALDH1A1 expression silencing at 0.5 and 2 hours after irradiation (Figure 6B). These results suggest that the observed ALDH1A1 mediated CRT resistance is independent of ROS production and DNA damage. A link to DNA damage biology remains to be established.
Figure 6.
Analysis of cellular oxidative stress. (A) Quantification of absolute ROS levels at 2 hours and 24 hours post radiation showed no differences between shControl and shALDH1A1 in both cell lines. (B) DNA damage induced by radiation was assessed by quantifying absolute H2A.X phosphorylation (Ser139) levels with no significant differences observed in either cell line upon ALDH1A1 silencing. Analysis of glutathione metabolism intermediates revealed no changes in the levels of free oxidized thiol pool (C), free reduced thiol pool (D), oxidized plus protein-bound thiol pool (E), and the GSH/GSSG ratio (F) upon ALDH1A1 silencing in both cell lines.
Next we investigated whether silencing ALDH1A1 in PDAC cells affected the cellular oxidative stress levels through impaired oxidized and reduced thiol pools involved in glutathione metabolism. Using an LC-ESI-MS/MS system we analyzed different levels of both oxidized and reduced glutathione, cysteine and homocysteine pools in both MiaPaCa-2 and Panc 05.04 cell lines. Although there are strong cell type specific differences with regard to glutathione metabolism, expression silencing of ALDH1A1 did not significantly impact the reduced free thiol pool, oxidized free thiol pool, the oxidized plus protein-bound thiol pool, and the GSH/GSSG ratio (a measure of cellular oxidative stress) (Figure 6, C–F). These results show that expression silencing of ALDH1A1 has no impact on the cellular oxidative stress system. In summary, we probed a number of potential pathways via which ALDH1A1 may drive radiation resistance. These hypotheses were falsified, hence suggesting alternative mechanisms.
Discussion
Despite an aggressive treatment regimen involving radical surgical resection, tumor recurrence is virtually universal in PDAC patients. Local recurrence occurs in about 35–86% of patients following surgical resection with adjuvant therapy and contributes significantly to PDAC mortality [4], [5], [6], [7], [42]. Therefore, recurrence is a hallmark of therapy resistance leading to more aggressive tumors. Multiple studies have aimed at the molecular profiling and classification of PDAC [14], [15], [16], [43], [44], [45], [46]. Despite these efforts, molecular (e.g.. genome, transcriptome or proteome) profiles associated with short or prolonged PFS upon adjuvant chemoradiation have so far remained outside the focus of these studies. To our knowledge, we present one of the first studies aiming to distinguish the proteome biology of localized PDAC with short or prolonged PFS upon additive chemoradiation. Although adjuvant chemotherapy for PDAC following resection remains the most actively researched topic, a number of clinical studies are also focusing on adjuvant chemoradiation (as opposed to chemotherapy alone) in order to improve patient management [12], [47], [48]. The short PFS tumors presented increased expression of actin cytoskeleton and cell adhesion proteins together with an overexpression of oxidoreductases and dehydrogenases. Furthermore, four out of six patients in the short PFS group exhibited an exocrine-like phenotype characterized by the expression of tumor cell-derived digestive enzyme genes. Exocrine-like PDAC has been shown to be resistant to a number of existing chemotherapies contributing to its poor prognosis [13], [17], [49]. A combination of two or more of these factors could possibly contribute to chemoradiation resistance leading tumor recurrence.
This study identified ALDH1A1 as one of the proteins up-regulated in the short PFS cohort out of the seven ALDH family members quantified via LC–MS/MS (Supplementary Table 4). ALDH1A1 has been implicated in cancer therapy resistance in various studies [50], [51], [52], [53], [54], [55], [56]. ALDH1A1 is a cytosolic enzyme and a member of the ALDH family of enzymes. It catalyzes the intracellular oxidation of different aldehydes, possesses antioxidant activity, is a key player in retinoic acid signaling, and used as a marker to identify various types of cancer stem cells [50], [52], [55], [56]. Expression studies for ALDH1A1 in cohorts of lung cancer and esophageal cancer have shown correlation between ALDH1A1 expression and shortened recurrence-free survival or chemoradiation resistance, respectively [57], [58], [59]. In both cases, data of The Cancer Genome Atlas (TCGA) suggests a lack of correlation between ALDH1A1 expression and overall survival. However, TCGA provides a rather global expression survey without a specific focus on recurrence-free survival or chemoradiation resistance. Additional analysis of non-TCGA data sets from the International Cancer Genome Consortium (ICGC), some of which are accessible via the PROGgeneV2 online resource, revealed no correlation between ALDH1A1 expression and overall survival of PDAC [60]. It has been suggested that neoadjuvant chemoradiation of PDAC enriches for a subpopulation of aldehyde dehydrogenase expressing cells that are resistant to this treatment [61]. There are diverse reports regarding the association between ALDH1A1 expression in pancreatic cancer and overall survival [56], [57], [58], [59]. Kim et al. demonstrated both in vitro and in vivo that high ALDH expressing human PDAC-derived cells were important in the mediating resistance towards different therapies [61]. Duong et al. reported that ALDH1A1 contributes to gemcitabine resistance in MiaPaCa-2 cells, a finding that corroborates our experimental results [62]. Further investigations on DNA damage levels, ROS levels, and cellular oxidative stress system revealed that ALDH1A1 silencing has no impact on these pathways (Figure 6, A–F). These results suggest that ALDH1A1 mediated chemoradiation resistance might be occurring through a pathway that is not clear at this point and warrants further investigation.
Conclusion
In summary we present an initial study towards an improved understanding of adjuvant therapy responsiveness (and lack thereof) in pancreatic cancer. Our results reemphasize an interest in the role of ALDH1A1 in pancreatic cancer biology. Since our small-scale study successfully identified proteome differences between the short and prolonged PFS subcohorts, we envisage the possibility that a similar large-scale study, also including further multimodal therapy regimens, might pave the way towards the establishment of tumor-specific proteome profiles that potentially identify patients that are likely to benefit from individualized therapy.
The following are the supplementary data related to this article.
List of all quantified proteins by LC-MS/MS
List of proteins quantified in at least 4 patient samples and used for LIMMA analysis
Additional histopathological characteristics of the patients used in this study
List of Aldehyde Dehydrogenase family members identified in this patient cohort by quantitative proteomics.
Supplementary Fig. 1: Proteome profiling of PDAC patients. (a) Bar chart representing protein groups interactions in 12 patients. The black nodes show the presence of a protein while the grey node shows the absence of the protein. Note that 553 proteins were quantified in all the 12 patients. Interactions with less than 10 proteins are not displayed in this figure. (b) Log 2 LFQ intensity expression of ALDH1A1 and MAOA in both the short PFS and prolonged PFS groups as determined by mass spectrometry analysis. Statistical significance was determined using a two-sided Student t test with p < 0.05 considered significant. (c) Partial least squares discriminant analysis (supervised clustering with z-score normalization) of the 12 samples display distinct proteome profiles between the short and prolonged PFS cohorts. E1-E6 are the short PFS patient samples while L1-L6 are the prolonged PFS patient samples. (d) Immunohistochemical corroboration of MAOA using representative tissues of the short and prolonged PFS groups. The immunohistochemical staining shows clear differences in the expression of MAOA with more expression observed in the short PFS than in the prolonged PFS group. Representative images were taken at x400 magnification. Sufficient material for immunohistochemical staining was available for only 10 of the 12 samples.
Supplementary Fig. 2: Mutation analysis. (a) Bar graph showing the average number of detected mutated peptides in each patient cohort. (b) Heat map showing mutated peptides together with the corresponding number of peptides detected in each tumor. Only mutated peptides observed in at least 6 tumor samples were considered.
Supplementary Fig. 3: Dose response curves. (a) Dose response curves of HPAF-II and MiaPaCa-2 to MAOA inhibitor clorgyline, (b and c) dose response curves of MiaPaCa-2 and Panc 05.04 to ALDH1A1 inhibitors A37 and DEAB respectively. (d, e, and f) Dose response curve of PDAC cell lines to 5-fluorouracil, gemcitabine, and radiation respectively. Data represents results from three independent experiments. EC20 and EC50 values were determined from these plots.
Declarations
Ethics Approval
The clinical proteomic study of human PDAC tissue was approved by the local Ethics Committee of the University of Lübeck / University Hospital of Schleswig-Holstein, Lübeck (AZ 15–368, January 2016).
Consent for Publication
All authors agree with the publication. This manuscript has not been published and is not under consideration for publication elsewhere.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflict of Interest
The authors declare no conflicts of interest.
Funding
We acknowledge support by Deutsche Forschungsgemeinschaft (GR 1748/6–1, SCHI 871/8–1, SCHI 871/9–1, SCHI 871/11–1, INST 39/900–1, and SFB850-Project Z1 (INST 39/766–3)), the Excellence Initiative of the German Federal and State Governments (EXC 294, BIOSS; GSC-4, Spemann Graduate School), and the German-Israel Foundation (Grant No. I-1444-201.2/2017). M.W. acknowledges support by the Mushett Family Foundation (Chester New Jersey).
Authors' Contribution
VOO performed the experiments and analyzed data, PB conceived the study and performed the IHC analysis, ART performed the 3D radiation experiment, MCF established the proteomics protocol and contributed to data analysis, CZ analyzed data, LH and SB performed and analyzed the metabolomics experiments, MLB performed LC–MS/MS measurements, CS, AG, LB, DR, TK, UFW conceived the study and assembled the patient cohort, and OS conceived the study and analyzed data. All authors contributed to writing of the manuscript.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Walston S, Salloum J, Grieco C, Wuthrick E, Diaz DA, Barney C, Manilchuk A, Schmidt C, Dillhoff M, Pawlik TM. Identifying Clinical Factors Which Predict for Early Failure Patterns Following Resection for Pancreatic Adenocarcinoma in Patients Who Received Adjuvant Chemotherapy Without Chemoradiation. Am J Clin Oncol. 2018 doi: 10.1097/COC.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM, Faluyi O, O'Reilly DA, Cunningham D, Wadsley J. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicenter, open-label, randomized, phase 3 trial. Lancet. 2017;389(10073):1011–1024. doi: 10.1016/S0140-6736(16)32409-6. [DOI] [PubMed] [Google Scholar]
- 5.Kyriazanos ID, Tsoukalos GG, Papageorgiou G, Verigos KE, Miliadis L, Stoidis CN. Local recurrence of pancreatic cancer after primary surgical intervention: How to deal with this devastating scenario? Surg Oncol. 2011;20(4):e133–e142. doi: 10.1016/j.suronc.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 6.La Torre M, Nigri G, lo Conte A, Mazzuca F, Tierno SM, Salaj A, Marchetti P, Ziparo V, Ramacciato G. Is a preoperative assessment of the early recurrence of pancreatic cancer possible after complete surgical resection? Gut Liver. 2014;8(1):102–108. doi: 10.5009/gnl.2014.8.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones OP, Melling JD, Ghaneh P. Adjuvant therapy in pancreatic cancer. World J Gastroenterol. 2014;20(40):14733–14746. doi: 10.3748/wjg.v20.i40.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami Y, Uemura K, Sudo T, Hashimoto Y, Kondo N, Nakagawa N, Sasaki H, Sueda T. Early initiation of adjuvant chemotherapy improves survival of patients with pancreatic carcinoma after surgical resection. Cancer Chemother Pharmacol. 2013;71(2):419–429. doi: 10.1007/s00280-012-2029-1. [DOI] [PubMed] [Google Scholar]
- 9.Ostapoff KT, Gabriel E, Attwood K, Kuvshinoff BW, Nurkin SJ, Hochwald SN. Does adjuvant therapy improve overall survival for stage IA/B pancreatic adenocarcinoma? HPB (Oxford) 2017;19(7):587–594. doi: 10.1016/j.hpb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda M, Endo I, Nakashima M, Minami Y, Takeda K, Matsuo K, Nagano Y, Tanaka K, Ichikawa Y, Togo S. Prognostic factors after resection of pancreatic cancer. World J Surg. 2009;33(1):104–110. doi: 10.1007/s00268-008-9807-2. [DOI] [PubMed] [Google Scholar]
- 11.Franke AJ, Rosati LM, Pawlik TM, Kumar R, Herman JM. The role of radiation therapy in pancreatic ductal adenocarcinoma in the neoadjuvant and adjuvant settings. Semin Oncol. 2015;42(1):144–162. doi: 10.1053/j.seminoncol.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Silvestris N, Brunetti O, Vasile E, Cellini F, Cataldo I, Pusceddu V, Cattaneo M, Partelli S, Scartozzi M, Aprile G. Multimodal treatment of resectable pancreatic ductal adenocarcinoma. Crit Rev Oncol Hematol. 2017;111:152–165. doi: 10.1016/j.critrevonc.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Noll EM, Eisen C, Stenzinger A, Espinet E, Muckenhuber A, Klein C, Vogel V, Klaus B, Nadler W, Rösli C. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22(3):278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muckenhuber A, Berger AK, Schlitter AM, Steiger K, Konukiewitz B, Trumpp A, Eils R, Werner J, Friess H, Esposito I. Pancreatic Ductal Adenocarcinoma Subtyping Using the Biomarkers Hepatocyte Nuclear Factor-1A and Cytokeratin-81 Correlates with Outcome and Treatment Response. Clin Cancer Res. 2018;24(2):351–359. doi: 10.1158/1078-0432.CCR-17-2180. [DOI] [PubMed] [Google Scholar]
- 15.Knudsen ES, Vail P, Balaji U, Ngo H, Botros IW, Makarov V, Riaz N, Balachandran V, Leach S, Thompson DM. Stratification of Pancreatic Ductal Adenocarcinoma: Combinatorial Genetic, Stromal, and Immunologic Markers. Clin Cancer Res. 2017;23(15):4429–4440. doi: 10.1158/1078-0432.CCR-17-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 17.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumann M, Krause M, Overgaard J, Debus J, Bentzen SM, Daartz J, Richter C, Zips D, Bortfeld T. Radiation oncology in the era of precision medicine. Nat Rev Cancer. 2016;16(4):234–249. doi: 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- 19.Berger AC, Garcia M, Jr., Hoffman JP, Regine WF, Abrams RA, Safran H, Konski A, Benson AB, 3rd, MacDonald J, Willett CG. Post resection CA 19-9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: a prospective validation by RTOG 9704. J Clin Oncol. 2008;26(36):5918–5922. doi: 10.1200/JCO.2008.18.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azimzadeh O, Barjaktarovic Z, Aubele M, Calzada-Wack J, Sarioglu H, Atkinson MJ, Tapio S. Formalin-Fixed Paraffin-Embedded (FFPE) Proteome Analysis Using Gel-Free and Gel-Based Proteomics. J Proteome Res. 2010;9(9):4710–4720. doi: 10.1021/pr1004168. [DOI] [PubMed] [Google Scholar]
- 21.Mason JT. Proteomic analysis of FFPE tissue: barriers to clinical impact. Expert Rev Proteomics. 2016;13(9):801–803. doi: 10.1080/14789450.2016.1221346. [DOI] [PubMed] [Google Scholar]
- 22.Dowling P, Moran B, McAuley E, Meleady P, Henry M, Clynes M, McMenamin M, Leonard N, Monks M, Wynne B. Quantitative label-free mass spectrometry analysis of formalin-fixed, paraffin-embedded tissue representing the invasive cutaneous malignant melanoma proteome. Oncol Lett. 2016;12(5):3296–3304. doi: 10.3892/ol.2016.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gámez-Pozo A, Berges-Soria J, Arevalillo JM, Nanni P, López-Vacas R, Navarro H, Grossmann J, Castaneda CA, Main P, Díaz-Almirón M. Combined Label-Free Quantitative Proteomics and microRNA Expression Analysis of Breast Cancer Unravel Molecular Differences with Clinical Implications. Cancer Res. 2015;75(11):2243–2253. doi: 10.1158/0008-5472.CAN-14-1937. [DOI] [PubMed] [Google Scholar]
- 24.Negishi A, Masuda M, Ono M, Honda K, Shitashige M, Satow R, Sakuma T, Kuwabara H, Nakanishi Y, Kanai Y. Quantitative proteomics using formalin-fixed paraffin-embedded tissues of oral squamous cell carcinoma. Cancer Sci. 2009;100(9):1605–1611. doi: 10.1111/j.1349-7006.2009.01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quesada-Calvo F, Massot C, Bertrand V, Longuespée R, Blétard N, Somja J, Mazzucchelli G, Smargiasso N, Baiwir D, De Pauw-Gillet MC. OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages. Clin Proteomics. 2017;14:9–21. doi: 10.1186/s12014-017-9143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Beyer A, Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165(3):535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Fortelny N, Overall CM, Pavlidis P, Freue GVC. Can we predict protein from mRNA levels? Nature. 2017;547(7664):E19–E20. doi: 10.1038/nature22293. [DOI] [PubMed] [Google Scholar]
- 28.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA. Erlotinib plus Gemcitabine Compared With Gemcitabine Alone in Patients with Advanced Pancreatic Cancer: A Phase III Trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 29.Takano S, Togawa A, Yoshitomi H, Shida T, Kimura F, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Tomonaga T. Annexin II Overexpression Predicts Rapid Recurrence after Surgery in Pancreatic Cancer Patients Undergoing Gemcitabine-Adjuvant Chemotherapy. Ann Surg Oncol. 2008;15(11):3157–3168. doi: 10.1245/s10434-008-0061-5. [DOI] [PubMed] [Google Scholar]
- 30.Müller AK, Föll M, Heckelmann B, Kiefer S, Werner M, Schilling O, Biniossek ML, Jilg CA, Drendel V. Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases. Neoplasia. 2017;20(2):140–151. doi: 10.1016/j.neo.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Föll MC, Fahrner M, Gretzmeier C, Thoma K, Biniossek ML, Kiritsi D, Meiss F, Schilling O, Nyström A, Kern JS. Identification of tissue damage, extracellular matrix remodeling and bacterial challenge as common mechanisms associated with high-risk cutaneous squamous cell carcinomas. Matrix Biol. 2017;66:1–21. doi: 10.1016/j.matbio.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drendel V, Heckelmann B, Chen C-Y, Weisser J, Espadas G, Schell C, Sabido E, Werner M, Jilg CA, Schilling O. Proteome profiling of clear cell renal cell carcinoma in von Hippel-Lindau patients highlights upregulation of Xaa-Pro aminopeptidase-1, an anti-proliferative and anti-migratory exoprotease. Oncotarget. 2017;8(59):100066–100078. doi: 10.18632/oncotarget.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Diest PJ, van Dam P, Henzen-Logmans SC, Berns E, van der Burg ME, Green J, Vergote I. A scoring system for immunohistochemical staining: consensus report of the task force for basic research of the EORTC-GCCG. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Cooperative Group. J Clin Pathol. 1997;50(10):801–804. doi: 10.1136/jcp.50.10.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tholen S, Biniossek ML, Gessler AL, Müller S, Weisser J, Kizhakkedathu JN, Reinheckel T, Schilling O. Contribution of cathepsin L to secretome composition and cleavage pattern of mouse embryonic fibroblasts. Biol Chem. 2011;392(11):961–971. doi: 10.1515/BC.2011.162. [DOI] [PubMed] [Google Scholar]
- 36.Sigloch FC, Knopf JD, Weisser J, Gomez-Auli A, Biniossek ML, Petrera A, Schilling O. Proteomic analysis of silenced cathepsin B expression suggests non-proteolytic cathepsin B functionality. Biochim Biophys Acta. 2016;1863(11):2700–2709. doi: 10.1016/j.bbamcr.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Thomsen AR, Aldrian C, Bronsert P, Thomann Y, Nanko N, Melin N, Rücker G, Follo M, Grosu AL, Niedermann G. A deep conical agarose microwell array for adhesion independent three-dimensional cell culture and dynamic volume measurement. Lab Chip. 2017;18(1):179–189. doi: 10.1039/c7lc00832e. [DOI] [PubMed] [Google Scholar]
- 38.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillem K, Vouillac C, Azar MR, Parsons LH, Koob GF, Cador M, Stinus L. Monoamine oxidase A rather than monoamine oxidase B inhibition increases nicotine reinforcement in rats. Eur J Neurosci. 2006;24(12):3532–3540. doi: 10.1111/j.1460-9568.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 40.Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, Matei D. beta-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34(18):2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Lin Z, Zhang B, Guo L, Liu S, Li H, Zhang J, Ye Q. beta-elemene sensitizes hepatocellular carcinoma cells to oxaliplatin by preventing oxaliplatin-induced degradation of copper transporter 1. Sci Rep. 2016;6 doi: 10.1038/srep21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas RM, Truty MJ, Nogueras-Gonzalez GM, Fleming JB, Vauthey JN, Pisters PW, Lee JE, Rice DC, Hofstetter WL, Wolff RA. Selective Reoperation for Locally Recurrent or Metastatic Pancreatic Ductal Adenocarcinoma Following Primary Pancreatic Resection. J Gastrointest Surg. 2012;16(9):1696–1704. doi: 10.1007/s11605-012-1912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49(3):358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49(3):367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, Konukiewitz B, Öllinger R, Zwiebel M, Strong A. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–68. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohman KA, Liu J, Linehan DC, Tan MC, Tan BR, Fields RC, Strasberg SM, Hawkins WG. Interferon-Based Chemoradiation Followed by Gemcitabine for Resected Pancreatic Adenocarcinoma: Long-Term Follow-Up. HPB (Oxford) 2017;19(5):449–457. doi: 10.1016/j.hpb.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright JP, Schlegel C, Snyder RA, Du L, Shyr Y, Cardin DB, Merchant NB, Parikh AA. Selective benefit of adjuvant chemoradiation in resectable pancreatic cancer. J Clin Oncol. 2017;35(15_Suppl) 4122-4122. [Google Scholar]
- 49.Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, Hong R, Moffat J, Gao M. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci U S A. 2015;112(32):E4410–E4417. doi: 10.1073/pnas.1501605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cojoc M, Peitzsch C, Kurth I, Trautmann F, Kunz-Schughart LA, Telegeev GD, Stakhovsky EA, Walker JR, Simin K, Lyle S. Aldehyde Dehydrogenase Is Regulated by beta-Catenin/TCF and Promotes Radioresistance in Prostate Cancer Progenitor Cells. Cancer Res. 2015;75(7):1482–1494. doi: 10.1158/0008-5472.CAN-14-1924. [DOI] [PubMed] [Google Scholar]
- 51.Duong HQ, You KS, Oh S, Kwak SJ, Seong YS. Silencing of NRF2 Reduces the Expression of ALDH1A1 and ALDH3A1 and Sensitizes to 5-FU in Pancreatic Cancer Cells. Antioxidants. 2017;6(3):52–61. doi: 10.3390/antiox6030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schafer A, Teufel J, Ringel F, Bettstetter M, Hoepner I, Rasper M, Gempt J, Koeritzer J, Schmidt-Graf F, Meyer B. Aldehyde dehydrogenase 1A1-a new mediator of resistance to temozolomide in glioblastoma. Neuro-Oncology. 2012;14(12):1452–1464. doi: 10.1093/neuonc/nos270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hilton J. Role of aldehyde dehydrogenase in cyclophosphamide-resistant L1210 leukemia. Cancer Res. 1984;44(11):5156–5160. [PubMed] [Google Scholar]
- 54.Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75(10):1947–1950. [PubMed] [Google Scholar]
- 55.Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA, Palle K. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S-L, Liu S, Cui W, Shi Y, Liu Q, Duan JJ, Yu SC, Zhang X, Cui YH, Kung HF. Aldehyde dehydrogenase 1A1 circumscribes high invasive glioma cells and predicts poor prognosis. Am J Cancer Res. 2015;5(4):1471–1483. [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7(3):330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ajani JA, Wang X, Song S, Suzuki A, Taketa T, Sudo K, Wadhwa R, Hofstetter WL, Komaki R, Maru DM. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol Oncol. 2014;8(1):142–149. doi: 10.1016/j.molonc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7(10):11018–11032. doi: 10.18632/oncotarget.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goswami C.P., Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970–975. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim SK, Kim H, Lee DH, Kim TS, Kim T, Chung C, Koh GY, Kim H, Lim DS. Reversing the intractable nature of pancreatic cancer by selectively targeting ALDH-high, therapy-resistant cancer cells. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duong H-Q, Hwang JS, Kim HJ, Kang HJ, Seong Y-S, Bae I. Aldehyde dehydrogenase 1A1 confers intrinsic and acquired resistance to gemcitabine in human pancreatic adenocarcinoma MIAPaCa-2 cells. Int J Oncol. 2012;41(3):855–861. doi: 10.3892/ijo.2012.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all quantified proteins by LC-MS/MS
List of proteins quantified in at least 4 patient samples and used for LIMMA analysis
Additional histopathological characteristics of the patients used in this study
List of Aldehyde Dehydrogenase family members identified in this patient cohort by quantitative proteomics.
Supplementary Fig. 1: Proteome profiling of PDAC patients. (a) Bar chart representing protein groups interactions in 12 patients. The black nodes show the presence of a protein while the grey node shows the absence of the protein. Note that 553 proteins were quantified in all the 12 patients. Interactions with less than 10 proteins are not displayed in this figure. (b) Log 2 LFQ intensity expression of ALDH1A1 and MAOA in both the short PFS and prolonged PFS groups as determined by mass spectrometry analysis. Statistical significance was determined using a two-sided Student t test with p < 0.05 considered significant. (c) Partial least squares discriminant analysis (supervised clustering with z-score normalization) of the 12 samples display distinct proteome profiles between the short and prolonged PFS cohorts. E1-E6 are the short PFS patient samples while L1-L6 are the prolonged PFS patient samples. (d) Immunohistochemical corroboration of MAOA using representative tissues of the short and prolonged PFS groups. The immunohistochemical staining shows clear differences in the expression of MAOA with more expression observed in the short PFS than in the prolonged PFS group. Representative images were taken at x400 magnification. Sufficient material for immunohistochemical staining was available for only 10 of the 12 samples.
Supplementary Fig. 2: Mutation analysis. (a) Bar graph showing the average number of detected mutated peptides in each patient cohort. (b) Heat map showing mutated peptides together with the corresponding number of peptides detected in each tumor. Only mutated peptides observed in at least 6 tumor samples were considered.
Supplementary Fig. 3: Dose response curves. (a) Dose response curves of HPAF-II and MiaPaCa-2 to MAOA inhibitor clorgyline, (b and c) dose response curves of MiaPaCa-2 and Panc 05.04 to ALDH1A1 inhibitors A37 and DEAB respectively. (d, e, and f) Dose response curve of PDAC cell lines to 5-fluorouracil, gemcitabine, and radiation respectively. Data represents results from three independent experiments. EC20 and EC50 values were determined from these plots.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [38] via the PRIDE partner repository with the dataset identifier PXD009254 (Reviewer account details: username: reviewer44874@ebi.ac.uk, password: DwMsRVKM).
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.