Abstract
Human cytomegalovirus (HCMV) has been detected in various types of tumors. We studied the prevalence of HCMV in ovarian cancer and its relation to clinical outcome. Paraffin-embedded tissues obtained prospectively from 45 patients with ovarian cancer and 30 patients with benign ovarian cystadenoma were analyzed for expression of HCMV immediate-early protein (IE) and HCMV tegument protein (pp65) by immunohistochemistry. Plasma was analyzed for HCMV serology. HCMV-IgG levels were higher in patients with ovarian cancer or benign cystadenoma than in age-matched controls (P = .002, P < .0001, respectively). HCMV IgM was detected in 12% of ovarian cancer patients and 3% of patients with benign tumors but was absent in controls. In patients with ovarian cancer, higher IgG levels were associated with better outcomes (P = .04). Extensive HCMV-IE protein expression was detected in 75% of ovarian cancers and 26% of benign tumors; pp65 was detected in 67% of ovarian cancers and 14% of benign tumors. A higher grade of HCMV infection was associated with higher stage of disease. Extensive HCMV-pp65 expression was associated with shorter median overall survival than focal expression (39 versus 42.5 months, P = .03). At study closure, 58% of ovarian cancer patients with focal pp65 expression were alive versus 27% of patients with extensive pp65 expression (P = .03). Thus, HCMV proteins are detected at different levels in ovarian tumors and benign cystadenomas. Ovarian cancer patients with focal HCMV-pp65 expression in their tumors and high IgG levels against HCMV lived longer, highlighting a need for in-depth studies of the oncomodulatory role of HCMV in ovarian cancer.
Introduction
Ovarian cancer is a major cause of morbidity and mortality in women worldwide [1]. Often diagnosed at an advanced stage, ovarian cancer has a 5-year survival rate of less than 50% [2], [3]. Standard care consists of cytoreductive surgery and platinum-based chemotherapy. Despite significant progress in treatments, the 5-year relative survival rate has improved marginally.
The etiology of ovarian cancer is not fully elucidated. Genetic susceptibility is believed to explain about 10–15% of these tumors [4]. Hormonal, infectious, and immunological factors have also been implicated in tumor development [5]. Although many key proteins and molecular pathways are potentially important in ovarian carcinogenesis, the early steps leading to malignancy are poorly understood [6]. Factors that may increase the risk of ovarian cancer are inherited gene mutations in breast cancer (BRCA) genes 1 and 2, mutations associated with Lynch syndrome, a family history of ovarian cancer, estrogen hormone replacement therapy, and the age at onset of menstruation and menopause [7]. The tumor micro-environment, including inflammation, may also affect tumor development and should be considered to understand the early steps of oncogenesis.
Human cytomegalovirus (HCMV) proteins and nucleic acid have been detected with optimized protocols in various types of cancers, including glioblastoma multiforme, neuroblastoma, medulloblastoma, and breast, prostate, and colon cancers [8], [9], [10], [11], [12], [13]. In a recent study, HCMV-Glycoprotein B (gB) DNA was detected in 50% of ovarian cancers. [14] HCMV is a member of the herpes virus family with a worldwide seroprevalence of 50–100%. The virus infects many cell types and can establish latency in myeloid progenitor cells or specifically in CD34+ cells [15], [16]. HCMV can be reactivated in blood monocytes by inflammation and production of cytokines that result in differentiation of monocytes into macrophages or dendritic cells, which can transmit the virus to other cell types. [15] During active infection, HCMV expresses immediate-early proteins (IE), which serve as transcription factors that help regulate the expression of both viral and host cellular genes. These proteins activate production of early and late structural viral proteins, including the viral tegument protein pp65, and several also trans-activate the expression of host and viral genes that are important for efficient viral replication [17]. In the final phase of infection, structural viral proteins are produced and assemble into a new virus particle.
HCMV is estimated to produce about 200 proteins, of which 50 are essential for viral replication. New data from ribosome profiling analysis suggest that the virus encodes over 750 unique RNAs that may encode viral proteins. Many of these proteins will affect cellular and immunological functions that are highly relevant to tumor development. Indeed, emerging research suggests that HCMV's oncomodulatory properties are important in carcinogenesis; HCMV proteins interfere with the retinoblastoma protein family (Rb) [18], cyclins, p53, Wnt, PI3K/Akt, NF-κB [18], [19], [20], [21], [22], [23], and STAT3 and modulate cellular functions though the effects on cellular differentiation, proliferation, and migration [24]. HCMV can block apoptosis and avoid immune surveillance, giving infected cells a survival advantage. Furthermore, HCMV infection alters expression of matrix metalloproteinases [25] and MMP2 and 9 have been shown to be strongly expressed in both stromal and epithelial tumor cells of serous invasive carcinomas [26].
Since HCMV is highly prevalent in breast cancers, which are morphologically similar to ovarian cancer, and mutations in BRCA are found in both types of tumors, we set out to study the prevalence and possible impact of HCMV infection on the survival rate of ovarian cancer patients in a prospective study.
Materials and Methods
Patient Characteristics and Treatment
Between February 2010 and July 2012, 45 consecutive patients with presumed epithelial ovarian cancer were enrolled in the study. All patients gave informed consent and underwent surgery at the Department of Obstetrics and Gynecology, Surgery at Karolinska University Hospital, the only referral center for gynecological malignancy in the Stockholm / Gotland region in Sweden. Clinical follow-up continued to June 1, 2015. Thereafter, clinical data were retrospectively collected into a database by a gynecology surgeon (AFR) (Table 1).
Table 1.
Patient Characteristics
| Patients Characteristics | |
|---|---|
| Ovarian cancer (n = 45) | |
| Median age, years (range) | 66 (25–90) |
| Stage | |
| IA, IB, IC, IIA, IIB, IIC, IIIC, IV | n = 12 |
| IIIC-IV | n = 33 |
| Median BMI (range) | 25.6 (19–37.8) |
| Median CA125 level | |
| Initial | 436 U/ml |
| After treatment | 18 U/ml |
| Surgery | n = 45 |
| R0 | n = 25 (57%) |
| R1 | n = 12 (27%) |
| R2 | n = 8 (18%) |
| Neoadjuvant chemotherapy before surgery | |
| Yes | n = 15 (33%) |
| No | n = 30 (65%) |
| Adjuvant chemotherapy after surgery | |
| Yes | n = 41 (91%) |
| No | n = 4 (9%) |
| Dead at study closure | n = 20 (44%) |
| Alive at study closure | n = 25 (56%) |
| Benign tumors (n = 30) | |
| Median age, years (range) | 57 (31–82) |
| BMI (range) | 19–36 |
| Median initial CA125 (n = 28) | 34 U/ml |
BMI, body mass index.
The median age of the patients with ovarian carcinoma was 66 years (range 25–90 years). All underwent surgery during 2010–2012 by a gynecology surgeon; 30 patients had primary debulking surgery, and 15 had interval debulking surgery after neoadjuvant chemotherapy. Fifty-seven percent of the patients had a complete cytoreduction (R0, no visible tumor at the end of surgery), 27% had a R1 resection (1–2 cm2 of tumor left at the end of surgery) and 18% had a R2 resection (>2 cm2 of tumor left at the end of surgery) (Table 1). All patients received conventional adjuvant chemotherapy (carboplatin AUC5 and paclitaxel 175 mg/m2 intravenously every third week in total 6 cycles) according to the Swedish guidelines for standard of care of these patients (Table 1). At study closure, 22 (49%) of the patients were alive. Thirty-one women who had a benign ovarian cystadenoma served as controls; their median age was 57 years (range 31–82 years). In addition to tissue specimens, blood samples were also collected before surgery from 34 patients with ovarian cancer and 30 patients with benign tumors. All diagnoses were confirmed by a reference pathologist of gynecological cancer (JC) at the Department of Pathology, Karolinska University Hospital. Blood plasma samples from 31 healthy aged-matched women in a biobank at our hospital served as controls for HCMV serology. This study was approved by the Stockholm regional ethical committee and the regional ethical committee at the Karolinska Institutet (Dnr: 2008/628–31/2, Dnr: 01–420, Dnr: 2009/1412–31).
Immunohistochemistry
Paraffin-embedded tissue sections were analyzed by immunohistochemistry as described but with minor modifications as follows [10], [12]. The sections (4 μm) were deparaffinized in xylene (Sigma-Aldrich, Apoteket Farmaci) and Tissue Clear (Sigma-Aldrich) and rehydrated in an ethanol series (99.5%, 95%, 70%, and 50%; Apoteket Farmaci). For antigen retrieval, tissue sections were placed in Antigen Retrieval Citra Plus solution (Biosite), pH 7.6, in a 37 °C water bath overnight and treated with pepsin (BioSite) at 37 °C. Endogenous nonspecific binding of antibodies was blocked with 3% H2O2 (Histolab), avidin/biotin kit (Dako), FC receptor blocker (Innovex Biosciences), and Background Buster (Innovex Biosciences).
HCMV proteins were detected with monoclonal antibodies against HCMV-IE (IgG2a, Chemicon International) and HCMV-pp65 (IgG1, BioGenex). Antibodies against von Willebrand factor (IgG1, Dako) and keratin 20 (IgG2a, Chemicon) were used as controls. Biotin-labeled anti-mouse secondary antibody, horseradish peroxidase–labeled streptavidin, and chromogen diaminobenzidine (both from Innovex Biosciences) were used to detect positive signals. Few tissue sections were lost during pre-treatment for immunohistochemistry. The extent of HCMV infection was scored as negative (0% positive cells), focal (<50% positive cells), or extensive (≥50% positive cells), as judged from the estimated number of cells expressing HCMV proteins. All staining results were independently collected by researchers blinded to patient records, and the diagnoses were reviewed by a pathologist (JC) and a senior scientist (AR).
HCMV Serology
Plasma from fresh blood samples was collected by centrifugation at 1500 rpm for 10 minutes and stored at −80 °C. HCMV-IgG was detected with an established in-housed ELISA method as described (Cut off; OD: 0.2 = 1.7 U/ML) [27], [28], [29]. HCMV-IgM was detected with a kit for HCMV-IgM (Cut off; OD: 0.2, Dade Behring).
Polymerase Chain Reaction (PCR) and DNA Sequencing
Fresh frozen tissues specimens and matched PBMCs (peripheral blood mononuclear cells) [30] were randomly selected from 5 patients with ovarian cancer and 2 patients with benign ovarian cystadenoma (n = 14 samples). DNA samples were prepared with QIAamp DNA mini-kits (Qiagen, Valencia, CA), and analyzed by Taqman PCR for the HCMV major immediate-early (IE) gene [10], [31]. DNA from PCR products were sequenced with an ABI 3730 DNA analyzer. Sequence data were analyzed by BLAST searches.
Cell Culture
SKOV3 ovarian carcinoma cells were cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum and 2 mM penicillin and streptomycin (ThermoFisher Scientific). SKOV3 cells were maintained at 37 °C in 5% CO2 in a humidified chamber.
Western Blotting
For HCMV infection experiments, SKOV3 cells were infected with HCMV strain VR1814 at a multiplicity of infection of 5 for 5 days. Cells were lysed with RIPA buffer (150 mM sodium chloride, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0), and protein was quantified by BCA assay (Thermofisher Scientific). Proteins (25 μg) were separated on a NuPAGE 4–12% Bis Tris gel (Thermofisher Scientific), transferred to a PVDF membrane (Millipore), and detected with antibodies against HCMV-IE (Argene), MUC16/CA125 (ABCAM), matrix metalloproteinase (MMP) 2, MMP9, and smooth muscle cell alpha actin (Sigma Aldrich).
RNA Isolation and Taqman PCR
Five days after infection, SKOV3 cells were lysed with RLT buffer supplemented with 5% β-mercaptoethanol (Sigma); total RNA was isolated with the RNeasy Mini kit (Qiagen), and cDNA was synthesized with SuperScript III First-Strand kit (Invitrogen, Life Technologies) and analyzed by quantitative TaqMan PCR. TaqMan Fast Universal PCR Master Mix (Life Technologies) was used with the following primers/probes: MUC16 (assay ID, Hs01065189_m1), MMP2 (assay ID, Hs01548727_m1), MMP9 (assay ID, Hs00234579_m1), human β2-microglobulin (B2M, assay ID, Hs00984230_m1), and HCMV custom made TaqMan assays (Applied Biosystems) for detection of HCMV IE and pp65. TaqMan assays were done as recommended by the manufacturer, using a 7900HT Fast Real-Time PCR system (Applied Biosystems) for 40 cycles with a final volume of 10 μl per reaction. The results were analyzed with SDS 2.4 software. The 2-ΔΔCt method was used to quantify expression relative to that of human β2-microglobulin (B2M).
Statistical Analysis
To detect a significant correlation between HCMV grade and overall survival (OS) at a power of 0.99, we calculated that a sample size of 42 patients was required. OS data are presented as Kaplan–Meier survival curves; patients who were alive at the time of the analysis (June 1, 2015) were censored. The results are presented as hazard ratios with 95% confidence intervals. Categorical data were analyzed with the chi-square test for trend. Differences in IgG levels were analyzed with the unpaired nonparametric Mann–Whitney U test. The correlation between HCMV-IgG and CA125 levels was examined with the Spearman correlation coefficient test. Values are expressed as medians; P < .05 was considered significant. Statistical analyses were done with Graph Pad Prism 6.
Results
Plasma HCMV Levels are High in Ovarian Cancer Patients
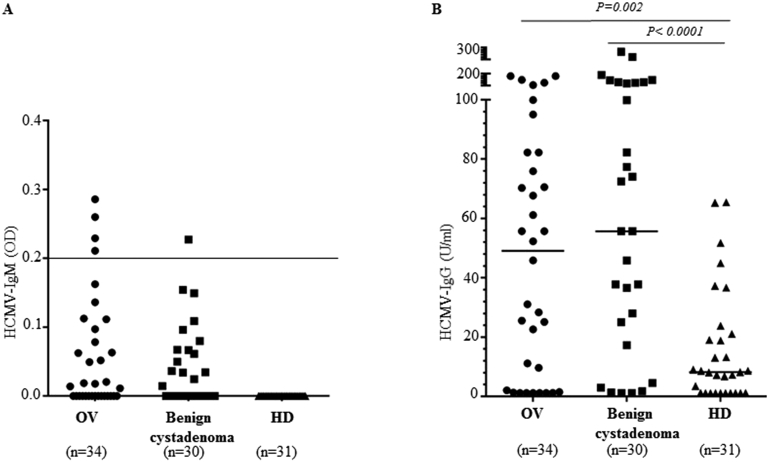
Plasma samples were available from 34 patients with ovarian cancer, 30 patients with benign cystadenomas, and 31 age-matched controls. HCMV IgM was detected in 12% of ovarian cancer patients, 3% of patients with cystadenomas, and 0% of healthy blood donors (Figure 1A); HCMV-IgG was detected in 77%, 90%, and 68% in their respective cohort. HCMV-IgG levels were higher in patients with ovarian cancer (P = .002) and cystadenomas (P < .0001) than in controls (Figure 1B).
Figure 1.
HCMV-specific antibodies are elevated in patients with ovarian cancer and benign cystadenoma. (A) HCMV-IgM was detected in 4 (12%) patients with ovarian cancer patients and in 1 (3%) patient with benign cystadenoma, but not in healthy age-matched controls. (B) HCMV-IgG levels were significantly higher in patients with ovarian cancer (P = .002) and benign cystadenoma (P < .0001) than in age-matched healthy female blood donors who served as controls.
High Prevalence of HCMV-IE and pp65 Protein in Ovarian Cancer and Benign Ovarian Cystadenomas
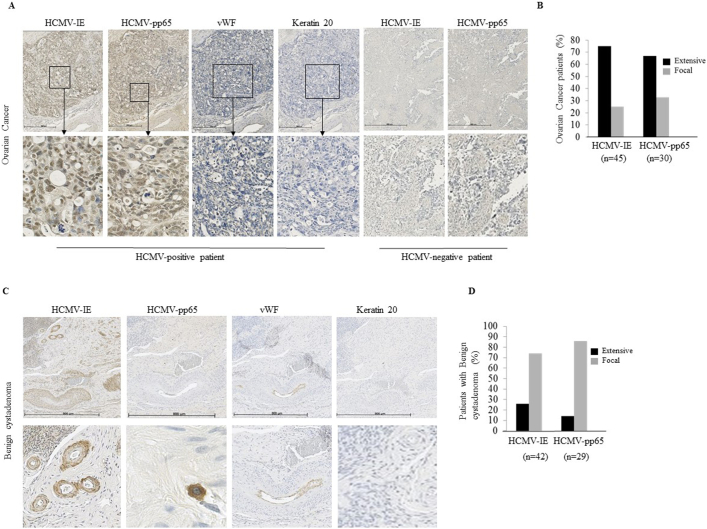
HCMV-IE protein was detected in tumor specimens from 89% of ovarian cancer patients and 93% of those with benign cystadenomas; protein expression was extensive in 75% and 67% in respective cohort. HCMV-pp65 was detected in 81% and 97%, respectively (Figure 2A, C), and the expression was extensive in 26% and 14%, respectively (Figure 2B-D, Table 2).
Figure 2.
HCMV protein expression in ovarian cancer and benign cytstadenomas. (A and B) HCMV proteins (IE, pp65) are frequently detected in ovarian cancer and (C and D) in benign cystadenoma. vWF, von Willebrand's factor.
Table 2.
HCMV Serology Status and Expression of HCMV-IE and pp65 in Ovarian Cancer and Benign Ovarian Cyst Adenomas
| Type of tumor | HCMV IE |
HCMV PP65 |
HCMV IgG |
HCMV IgM | ||
|---|---|---|---|---|---|---|
| Focal/Negative n (%) |
Extensive n (%) |
Focal/Negative n (%) |
Extensive n (%) |
|||
| Ovarian cancer | ||||||
| Serous adenocarcinoma, | 5/31 (16) | 26/31 (84) | 19/28 (68) | 9/28 (32) | 20/23 (87) | 4/23 (17) |
| Other histiotypes, | 7/14 (50) | 8/14 (57) | 12/14 (86) | 2/14 (14) | 7/10 (70) | 0/11 (0) |
| Benign ovarian cyst adenoma, | 10/30 (50) | 20/30 (67) | 25/29 (86) | 4/29 (14) | 28/30 (93) | 1/29 (3) |
Focal (<50% positive cells), Extensive (≥50% positive cells).
To further confirm the presence of HCMV in ovarian cancer and benign ovarian cystadenoma, 14 DNA samples from PBMCs (n = 7) and matched frozen surgical tumor specimens of five patients with newly diagnosed ovarian cancer and two patients with benign ovarian cystadenoma were amplified by a PCR assay using primers specific for the HCMV IE gene [10], [31]. All samples except for one ovarian cancer tissue specimen were positive for HCMV IE (data not shown). DNA sequence analysis of the HCMV IE PCR products confirmed HCMV DNA sequences in all positive (n = 13) examined tissue samples, but the DNA sequences were distinctly different from wild type (clinical HCMV strains or laboratory virus strains), which excluded contamination.
Poor Survival Rate Among Ovarian Cancer Patients With Extensive HCMV-pp65 Expression in their Tumors
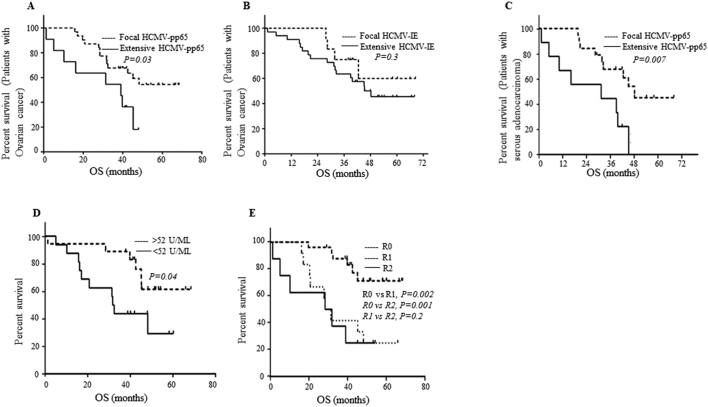
Next we analyzed the effects of HCMV on the median overall survival (OS) of ovarian cancer patients. At study closure, 58% of patients with focal expression of HCMV-pp65 in their tumors were alive versus 27% of those with extensive expression. Median OS was shorter in patients with extensive expression (defined as ≥50% positive cells) of HCMV-pp65 proteins than in those with focal expression (defined as <50% positive cells) (39 versus 42.5 months, P = .03) (Figure 3A). Similarly, 67% of patients with focal HCMV-IE protein expression were alive versus 48% with extensive expression; however, median OS did not differ between the two groups (42 versus 40 months, respectively; P = .3) (Figure 3B). At 5 years, all patients with extensive HCMV-pp65 expression were deceased, whereas 45% of those with focal expression were still alive. Three of five patients with focal expression of HCMV-IE were alive at 5 years.
Figure 3.
Poor survival of ovarian cancer patients with extensive HCMV-pp65 protein expression. Ovarian cancer patients who had focal HCMV-pp65 expression in their tumors had significantly longer median OS than those with extensive HCMV-pp65 expression (42.5 vs. 39 months, P = .03). (A) Furthermore, 58% and 27% of patients with focal and extensive HCMV-pp65, respectively, were alive at time of study closure. (A) At 5 years, all patients with extensive HCMV-pp65 were dead but 45% (5/11) of patients with focal pp65 in their tumor were still alive. (B) No significant difference in OS was observed in ovarian cancer patients with focal or extensive HCMV-IE expression in their tumors. (C) A significantly prolonged OS was observed in patients with serous ovarian cancer having focal HCMV-pp65 compared to those with extensive HCMV-pp65 (45 vs 31 months). (D) Ovarian cancer patients with higher IgG levels (Median; >52 U/ML) against HCMV had better survival (median OS of 32.8 months versus 45 months in patients with low levels of IgG). (E) Ovarian cancer patients with R0 (no visible tumor) has significantly longer OS compared to R1 (resection; 1–2 cm2 of tumor, median OS; 45 vs 30 months) and R2 (resection (>2 cm2 of tumor, median OS; 45 vs 30 months). There was no significant difference in OS between patients with R1 and R2 surgery (median OS: 30 months).
Among ovarian cancer patients, 31 had serous adenocarcinoma and 14 had other histotypes: carcinosarcoma (n = 3), endometriod adenocarcinoma (n = 4), mucinous adenocarcinoma (n = 6), and clear cell adenocarcinoma (n = 1). In patients with serous adenocarcinoma, extensive HCMV-IE expression was detected in 84% and pp65 in 32% (Table 2); focal HCMV-pp65 expression was associated with significantly longer OS (45 vs 31 months, P = .007) (Figure 3C). OS was also longer in patients with focal HCMV-IE expression than in those with extensive expression (39 vs 42 months), but the difference was not significant (P = .4). Median OS was longer in patients with higher HCMV IgG levels (32.8 versus 45 months) (Figure 3D). HCMV-IgG levels did not correlate with CA125 levels (P = .2).
Longer OS Among Ovarian Cancer Patients With Complete Cytoreductive Surgery
Among ovarian cancer patients, 25 (57%) underwent macroscopic radical debulking surgery (R0, no visible tumor), 12 (27%) had an R1 resection 1–2 cm2 of the tumor, and 8 (18%) had an R2 resection >2 cm2 of the tumor. As expected, median OS was longer in patients with R0 resection (45 months) than in those with R1 resection (30 months; P = .002, 95% CI 1.9–19.6) or R2 resection (30 months; P = .001, 95% CI 2.8–64). Median OS was the same in patients with R1 or R2 resection (P = .6) (Figure 3E).
Higher Tumor HCMV Activity is Associated With More Advanced Disease
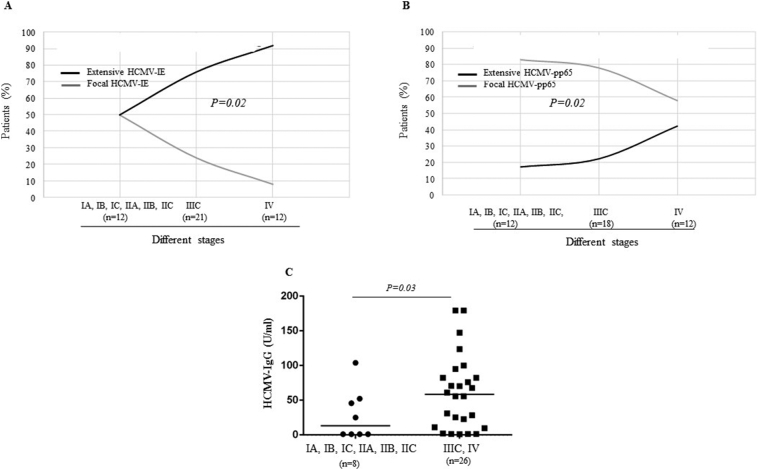
HCMV-IE expression was extensive in 50% of Stage IA–IIC tumors, 76% of Stage IIIC tumors, and 92% of Stage IV tumors; HCMV-pp65 expression was extensive in 17%, 22%, and 42%, respectively. Tumor stage correlated with extensive expression of either HCMV-IE or HCMV-pp65 (both P = .02) (Figure 4A-B). Stage IIIC and IV tumors had significantly higher levels of HCMV-IgG than tumors with earlier stages (IA, IB, IC, IIA, IIB, IIC) (P = .03) (Figure 4C).
Figure 4.
(A, B) Extensive HCMV-IE and HCMV-pp65 protein expression are associated with increased staging of ovarian cancer. (C) Levels of HCMV-IgG were higher in ovarian cancer patients with Stages IIIC and IV tumors than in those Stages IA, IB, IC, IIA, IIB, IIC, tumors.
HCMV-IE and HCMV-pp65 Levels are Not Associated With Increased Levels of CA125
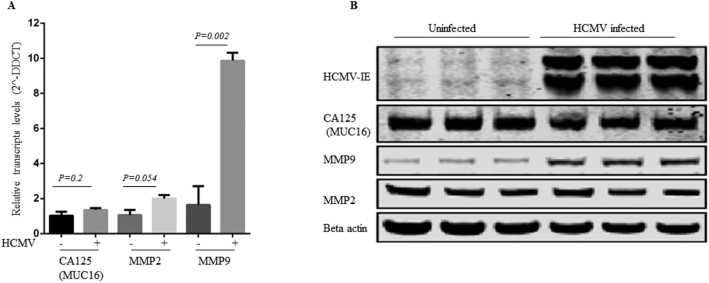
Serum CA125 was measured at diagnosis in 43 patients with ovarian cancer. Five were negative for CA125 but had extensive HCMV-IE expression in their tumors, and one had extensive expression of HCMV-pp65. The CA125 level (U/ml) was >35–100 in 7 patients, >100–1000 and> 1000–5000 in 13 each, and> 5000 in 5. Extensive HCMV-IE and pp65 expression was detected in 80% and 17%, respectively, of patients with CA125> 35–1000 U/ml and 61% and 35% of those with CA125> 1000 U/ml. From this, no association was found between CA125 and HCMV pp65/IE expression levels. To further confirm this, we examined CA125 expression level and MMP2 and MMP9 expression levels (known as potential markers of aggressive ovarian cancer with normal CA125 [32]) using in vitro model. HCMV infection did not affect CA125 (P = .2) or MMP2 (P = .054) on either mRNA or protein levels, but significantly increased the expression of MMP9 in both transcript and protein (P = .002) (Figure 5).
Figure 5.
HCMV infection does not affect the transcript and protein levels of CA125 (P = .2) and MMP2 (P = .054), but significantly increased MMP9 transcript and protein levels (P = .002).
Discussion
We prospectively studied the prevalence and clinical relevance of HCMV in patients with ovarian cancer. We found that HCMV proteins can be frequently detected at different levels in both ovarian cancer and benign ovarian cystadenomas. Median OS was shorter among ovarian cancer patients who had extensive expression of HCMV-pp65 in their tumors than in those with focal expression. HCMV IgG levels were significantly higher in ovarian cancer patients and patients with benign cystadenomas than in age-matched controls, and were highest in those with stage IIIC and IV tumors. However, the prognosis was better among ovarian cancer patients who had higher IgG levels. These findings suggest that HCMV affects both the tumor stage and survival of ovarian cancer, and that patients who mount a stronger immune response, as indicated by higher IgG antibody levels, have a better prognosis. Thus, HCMV may have an oncomodulatory effect that contributes to disease progression and spread of peritoneal carcinosis in ovarian cancer patients.
We detected HCMV-IE and pp65 protein expression in different cell types in ovarian cancer specimens. The viral proteins were detected in tumor cells, inflammatory cells, blood vessel walls, smooth muscle cells, and epithelial cells. In benign cystadenoma samples, however, HCMV-pp65 was detected mainly in inflammatory cells.
HCMV-pp65 is an immunomodulatory protein. It affects expression of HLA-class II molecules and interferes with presentation of IE peptides in HLA class I molecules and thereby helps the virus to avoid recognition and killing of infected cells by T cells. [33], [34] HCMV-pp65 also accumulates and degrades HLA class II molecules in lysosomes of infected cells, thereby blocking antigen presentation to CD4+ lymphocytes [34]. HCMV-pp65 avoids immune recognition and killing of HCMV-infected cells by binding to the NKp30 activating receptor and interfering with the cross talk of dendritic cells and natural killer cells (NK) [33], [35]. Furthermore, HCMV-pp65 contributes to immunosuppression by down-regulating the interferon response [35], [36] and induces production of TGF-beta and viral interleukin-10 [36], [37], which are both immunosuppressive cytokines. Thus, HCMV-pp65 expression might help ovarian tumor cells avoid recognition and killing by the immune system and worsen patient outcome by mediating an immunosuppressive state in the tumor microenviroment.
On the other hand, inflammation in the tumor micro-environment is strongly linked to tumor development and is a hallmark of cancer [38]. Frequent reactivation of latent HCMV in tumor tissues by inflammation would exacerbate inflammation by increasing production of inflammatory factors that may contribute to tumor progression [39], [40]. HCMV induces COX-2 and 5-LO expression, production of prostaglandins and leukotrienes, and pro-inflammatory cytokines (IL-1, IL-6, IL-8 MCP-1, RANTES, MIP-1α, TNF-α, and IFN-γ) [41], [42], [43], [44]. The presence of HCMV proteins we observed in inflammatory cells in ovarian cancer specimens may reflect inflammation-induced reactivation of HCMV. Thus, a delicate balance is expected to occur between this virus and the immune system in tumors.
The interactions among inflammatory, stromal, and neoplastic cells in the tumor microenvironment can regulate tumorigenesis and tumor progression by promoting tumor growth, metastasis, and vascularization. Furthermore, tumor-associated macrophages—the major inflammatory cell type in tumors—have a high capacity to produce cytokines/chemokines, which may further drive HCMV reactivation and contribute to inflammation, immune evasion by the virus, and tumorigenesis [45]. HCMV promotes cellular proliferation by interfering with p53 and cyclins and by expressing anti-apoptotic viral proteins including UL36, UL38 and viral mitochondrion-localized inhibitor (vMIA) of apoptosis [46]. HCMV-US28 promotes angiogenesis by enhancing both the production of vascular endothelial growth factor and the expression of thrombospondin [47]. Four of 5 (80%) patients with extensive HCMV-IE and 3 (60%) with extensive pp65 expression in their tumors had high plasma levels of CA125 (≥5000 U/ml). However, HCMV infection did not by itself enhance CA125 in cells infected in vitro. The enhanced CA125 levels in HCMV-infected ovarian cancer patients may instead reflect increased cellular proliferation and inflammation stimulated by the viral infection; however, this was not evident in all patients. Furthermore, MMP9 expression was significantly increased in HCMV infected ovarian cancer cells as shown in our in vitro study. MMP9 and MMP2 have been linked to ovarian cancer in patients with low or absent CA125 levels [32], where high levels indicate more aggressive ovarian cancers. CA125 is involved in the epithelial to mesenchymal transition and may have a role in metastatic disease [32], [48], [49]. Thus, HCMV-induced expression of MMP9 may be linked to a more aggressive phenotype of ovarian cancer with higher metastatic potential and poor outcome.
HCMV-IgG levels were significantly higher in patients with benign cystadenomas and ovarian cancer than in age-matched controls, and were also higher in patients with Stage IIIC and IV tumors than in those with lower-stage tumors. HCMV-IgM was detected in 12% of patients with ovarian cancer and only one patient with benign cystadenoma, but not in any of the controls. These observations indicate a higher activity of HCMV in ovarian cancer patients, and higher HCMV-pp65 activity in ovarian tumors was indeed associated with lower survival rates. Thus, HCMV may have an oncomodulatory role in these tumors and should be further evaluated as a potential biomarker of more aggressive disease.
Previously we reported that OS was significantly longer in glioblastoma patients whose tumors had focal expression of HCMV-IE proteins (<50% positive cells) [10] than in those with more extensive infection. HCMV-pp65 was not examined, but survival did not correlate with expression of HCMV-late protein [10]. In the present study, HCMV-IE and pp65 were frequently detected in ovarian cancer tissue specimens, but only extensive expression of HCMV-pp65 was significantly associated with OS. Indeed, median OS was 14 months longer in patients with late-stage serous ovarian carcinoma who had focal expression of HCMV-pp65 in their tumor than in those with extensive expression (45 vs 31 months, P = .007). Evidently HCMV affects the clinical outcome of ovarian cancer and merits further study in a larger cohort of patients. Since higher activity of HCMV was associated with worse outcome, this virus may provide a new therapeutic target in ovarian cancer. As patients with enhanced immune reactivity to HCMV had a better prognosis, immunotherapy for HCMV might improve outcomes in patients with HCMV-positive ovarian cancers. Adoptive T-cell therapy [50] and pp65 mRNA dendritic cell vaccinations show promise for improved outcome in patients with HCMV-positive glioblastoma [51].
Ovarian cancer patients have poor OS despite advanced surgery and modern standard therapy. Our findings provide hope for alternative HCMV-directed therapies for ovarian cancer patients. Preliminary studies in HCMV-positive glioblastoma patients showed higher survival among patients who receive the anti-HCMV drug valganciclovir as an add-on to standard treatment [52]. Among glioblastoma patients who underwent a radical resection of their tumors and received early and continuous antiviral treatment, 90% were alive at 2 years versus 18% of patients who received only standard therapy [52]. Future studies are merited to evaluate antiviral treatment in patients with HCMV-positive tumors that have a poor prognosis.
In conclusion, HCMV proteins are frequently detected at different levels in ovarian cancer and benign cystadenoma, and these patients have higher levels of HCMV-IgG and IgM than healthy controls. Higher activity of HCMV in ovarian cancer patients predicted poor outcome, while enhanced immune reactivity to HCMV was associated with a better prognosis. Further studies are needed to better understand the oncomodulatory and immunomodulatory roles of HCMV in ovarian cancer and to determine whether antiviral treatment and immunotherapies against HCMV in addition to standard therapies can improve the prognosis for these patients.
Conflicts of Interest
All authors have read the journal's authorship agreement and policy on disclosure of potential conflicts of interest. All authors have disclosed any financial or personal relationship with organizations that could potentially be perceived as influencing the described research (if there are none, a statement to this effect is included).
Acknowledgments
Conflicts of Interest: All authors have read the journal's policy on conflicts of interest and have none to declare. All authors have read the journal's authorship agreement and approved submission of the manuscript. The manuscript, has neither been published nor is currently under consideration for publication by any other journal. This study was supported by BILTEMA Foundation, Nexttobe, Stichting af Jochnicks Foundation, Sten A. Olssons Foundation for Research and Culture, Familjen Erling-Perssons Foundation, RATOS, independent grants from Hoffmann La Roche, Torsten and Ragnar Söderbergs Foundations (MF14/10), the Swedish Research Council (10350, K2014-99X-22627-01-4, K2013-57X-12615-16-5 and K2013-54X-20324-07-5) and Swedish Research Council Framework Grant in Infections and Antibiotics (K2014-99X-22627-01-4), the Swedish Cancer Foundation (5044-B05-01XAB), Dan och Jane Olssons Foundation, Swedish Cancer Foundation, Swedish Medical Research Council, Swedish Society for Medical Research (SLS), Goljes Memory Foundation, Magnus Bergvalls Foundation, Swedish Society for Medical Research (SSMF), Percy Falks Foundation, Karolinska Institutet Foundation, IngaBritt och Arne Lundbergs Foundation, and Tore Nilsons Foundation. The authors thank Berit Legerstam at the Women's Health Research Unit, Karolinska University Hospital, for data collection.
Footnotes
Declarations of interest: none.
Contributor Information
Afsar Rahbar, Email: afsar.rahbar@ki.se.
Cecilia Söderberg-Naucler, Email: cecilia.naucler@ki.se.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Coleman MP, Group CW Cancer survival: [corrected] the CONCORD-2 study - Authors' reply. Lancet. 2015;386:429–430. doi: 10.1016/S0140-6736(15)61443-X. [DOI] [PubMed] [Google Scholar]
- 3.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H, Ardanaz E. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE--5-a population-based study. Lancet Oncol. 2014;15:23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 4.Russo A, Calo V, Bruno L, Rizzo S, Bazan V, Di Fede G. Hereditary ovarian cancer. Crit Rev Oncol Hematol. 2009;69:28–44. doi: 10.1016/j.critrevonc.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Brown SB, Hankinson SE. Endogenous estrogens and the risk of breast, endometrial, and ovarian cancers. Steroids. 2015;99:8–10. doi: 10.1016/j.steroids.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Kurman RJ. Origin and molecular pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 2013;24(Suppl. 10):x16–x21. doi: 10.1093/annonc/mdt463. [DOI] [PubMed] [Google Scholar]
- 7.https://ocrfa.org/patients/about-ovarian-cancer/risk-factors/
- 8.Baryawno N, Rahbar A, Wolmer-Solberg N, Taher C, Odeberg J, Darabi A, Khan Z, Sveinbjornsson B, FuskevAg OM, Segerstrom L. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2011;121:4043–4055. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, Cobbs CS. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–1563. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 10.Rahbar A, Orrego A, Peredo I, Dzabic M, Wolmer-Solberg N, Straat K, Stragliotto G, Soderberg-Naucler C. Human cytomegalovirus infection levels in glioblastoma multiforme are of prognostic value for survival. J Clin Virol. 2013;57:36–42. doi: 10.1016/j.jcv.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- 12.Taher C, de Boniface J, Mohammad AA, Religa P, Hartman J, Yaiw KC, Frisell J, Rahbar A, Soderberg-Naucler C. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolmer-Solberg N, Baryawno N, Rahbar A, Fuchs D, Odeberg J, Taher C, Wilhelmi V, Milosevic J, Mohammad AA, Martinsson T. Frequent detection of human cytomegalovirus in neuroblastoma: a novel therapeutic target? Int J Cancer. 2013;133:2351–2361. doi: 10.1002/ijc.28265. [DOI] [PubMed] [Google Scholar]
- 14.Shanmughapriya S, Senthilkumar G, Vinodhini K, Das BC, Vasanthi N, Natarajaseenivasan K. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur J Clin Microbiol Infect Dis. 2012;31:2311–2317. doi: 10.1007/s10096-012-1570-5. [DOI] [PubMed] [Google Scholar]
- 15.Sinclair J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. J Clin Virol. 2008;41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Soderberg-Naucler C, Fish KN, Nelson JA. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 17.Depto AS, Stenberg RM. Regulated expression of the human cytomegalovirus pp65 gene: octamer sequence in the promoter is required for activation by viral gene products. J Virol. 1989;63:1232–1238. doi: 10.1128/jvi.63.3.1232-1238.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 19.Cinatl J, Jr., Vogel JU, Kotchetkov R, Wilhelm Doerr H. Oncomodulatory signals by regulatory proteins encoded by human cytomegalovirus: a novel role for viral infection in tumor progression. FEMS Microbiol Rev. 2004;28:59–77. doi: 10.1016/j.femsre.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J Virol. 2008;82:5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salvant BS, Fortunato EA, Spector DH. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song YJ, Stinski MF. Inhibition of cell division by the human cytomegalovirus IE86 protein: role of the p53 pathway or cyclin-dependent kinase 1/cyclin B1. J Virol. 2005;79:2597–2603. doi: 10.1128/JVI.79.4.2597-2603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yurochko AD, Kowalik TF, Huong SM, Huang ES. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobbs CS, Soroceanu L, Denham S, Zhang W, Kraus MH. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 2008;68:724–730. doi: 10.1158/0008-5472.CAN-07-2291. [DOI] [PubMed] [Google Scholar]
- 25.Straat K, de Klark R, Gredmark-Russ S, Eriksson P, Soderberg-Naucler C. Infection with human cytomegalovirus alters the MMP-9/TIMP-1 balance in human macrophages. J Virol. 2009;83:830–835. doi: 10.1128/JVI.01363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Alem L, Curry TE., Jr. Ovarian cancer: involvement of the matrix metalloproteinases. Reproduction. 2015;150:R55–R64. doi: 10.1530/REP-14-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grillner L. Screening of blood donors for cytomegalovirus (CMV) antibodies: an evaluation of different tests. J Virol Methods. 1987;17:133–139. doi: 10.1016/0166-0934(87)90076-0. [DOI] [PubMed] [Google Scholar]
- 28.Rahbar AR, Sundqvist VA, Wirgart BZ, Grillner L, Soderberg-Naucler C. Recognition of cytomegalovirus clinical isolate antigens by sera from cytomegalovirus-negative blood donors. Transfusion. 2004;44:1059–1066. doi: 10.1111/j.1537-2995.2004.03292.x. [DOI] [PubMed] [Google Scholar]
- 29.Warnke C, Ramanujam R, Plavina T, Bergstrom T, Goelz S, Subramanyam M, Kockum I, Rahbar A, Kieseier BC, Holmen C. Changes to anti-JCV antibody levels in a Swedish national MS cohort. J Neurol Neurosurg Psychiatry. 2013;84:1199–1205. doi: 10.1136/jnnp-2012-304332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsson S, Soderberg-Naucler C, Wang FZ, Moller E. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion. 1998;38:271–278. doi: 10.1046/j.1537-2995.1998.38398222871.x. [DOI] [PubMed] [Google Scholar]
- 31.Soderberg C, Larsson S, Bergstedt-Lindqvist S, Moller E. Definition of a subset of human peripheral blood mononuclear cells that are permissive to human cytomegalovirus infection. J Virol. 1993;67:3166–3175. doi: 10.1128/jvi.67.6.3166-3175.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coticchia CM, Curatolo AS, Zurakowski D, Yang J, Daniels KE, Matulonis UA, Moses MA. Urinary MMP-2 and MMP-9 predict the presence of ovarian cancer in women with normal CA125 levels. Gynecol Oncol. 2011;123:295–300. doi: 10.1016/j.ygyno.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 33.Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, Gazit R, Gonen-Gross T, Hanna J, Nahari E. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 34.Odeberg J, Plachter B, Branden L, Soderberg-Naucler C. Human cytomegalovirus protein pp65 mediates accumulation of HLA-DR in lysosomes and destruction of the HLA-DR alpha-chain. Blood. 2003;101:4870–4877. doi: 10.1182/blood-2002-05-1504. [DOI] [PubMed] [Google Scholar]
- 35.Arnon TI, Markel G, Mandelboim O. Tumor and viral recognition by natural killer cells receptors. Semin Cancer Biol. 2006;16:348–358. doi: 10.1016/j.semcancer.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A. 2003;100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Cobbs CS. Evolving evidence implicates cytomegalovirus as a promoter of malignant glioma pathogenesis. Herpesviridae. 2011;2:10. doi: 10.1186/2042-4280-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soderberg-Naucler C, Johnsen JI. Cytomegalovirus in human brain tumors: Role in pathogenesis and potential treatment options. World J Exp Med. 2015;5:1–10. doi: 10.5493/wjem.v5.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmen KA, Singh J, Luukkonen BG, Lopper M, Bittner A, Miller NE, Jackson MR, Compton T, Fruh K. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc Natl Acad Sci U S A. 2001;98:7140–7145. doi: 10.1073/pnas.121177598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer. 2016;2 doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maussang D, Verzijl D, van Walsum M, Leurs R, Holl J, Pleskoff O, Michel D, van Dongen GA, Smit MJ. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc Natl Acad Sci U S A. 2006;103:13068–13073. doi: 10.1073/pnas.0604433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comamala M, Pinard M, Theriault C, Matte I, Albert A, Boivin M, Beaudin J, Piche A, Rancourt C. Downregulation of cell surface CA125/MUC16 induces epithelial-to-mesenchymal transition and restores EGFR signalling in NIH:OVCAR3 ovarian carcinoma cells. Br J Cancer. 2011;104:989–999. doi: 10.1038/bjc.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Hu XX, Yang XZ, Wang Q, Cheng H, Wang SM, Hu YL, Yang ZJ, Li L. Combined detection of serum matrix metalloproteinase 9, acetyl heparinase and cathepsin L in diagnosis of ovarian cancer. Chin J Cancer Res. 2012;24:67–71. doi: 10.1007/s11670-012-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuessler A, Smith C, Beagley L, Boyle GM, Rehan S, Matthews K, Jones L, Crough T, Dasari V, Klein K. Autologous T-cell therapy for cytomegalovirus as a consolidative treatment for recurrent glioblastoma. Cancer Res. 2014;74:3466–3476. doi: 10.1158/0008-5472.CAN-14-0296. [DOI] [PubMed] [Google Scholar]
- 51.Batich KA, Reap EA, Archer GE, Sanchez-Perez L, Nair SK, Schmittling RJ, Norberg P, Xie W, Herndon JE, II, Healy P. Long-term Survival in Glioblastoma with Cytomegalovirus pp65-Targeted Vaccination. Clin Cancer Res. 2017;23:1898–1909. doi: 10.1158/1078-0432.CCR-16-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderberg-Naucler C, Rahbar A, Stragliotto G. Survival in patients with glioblastoma receiving valganciclovir. N Engl J Med. 2013;369:985–986. doi: 10.1056/NEJMc1302145. [DOI] [PubMed] [Google Scholar]