Abstract
Specialization onto different host plants has been hypothesized to be a major driver of diversification in insects, and traits controlling olfaction have been shown to play a fundamental role in host preferences. A diverse set of olfactory genes control olfactory traits in insects, and it remains unclear whether specialization onto different hosts is likely to involve a nonrandom subset of these genes. Here, we test the role of olfactory genes in a novel case of specialization in Drosophila orena. We report the first population‐level sample of D. orena on the West African island of Bioko, since its initial collection in Cameroon in 1975, and use field experiments and behavioral assays to show that D. orena has evolved a strong preference for waterberry (Syzygium staudtii). We then show that a nonrandom subset of genes controlling olfaction‐–those controlling odorant‐binding and chemosensory proteins–‐have an enriched signature of positive selection relative to the rest of the D. orena genome. By comparing signatures of positive selection on olfactory genes between D. orena and its sister species, D. erecta we show that odorant‐binding and chemosensory have evidence of positive selection in both species; however, overlap in the specific genes with evidence of selection in these two classes is not greater than expected by chance. Finally, we use quantitative complementation tests to confirm a role for seven olfactory loci in D. orena’s preference for waterberry fruit. Together, our results suggest that D. orena and D. erecta have specialized onto different host plants through convergent evolution at the level of olfactory gene family, but not at specific olfactory genes.
Keywords: Adaptation, behavioral genetics, chemosensory, host preference, olfaction, speciation
Impact Summary.
Ecological specialization is a widespread evolutionary outcome. In insects, for example, specialization can have important economic consequences and can result in the evolution of species that damage crops or transmit disease. A major goal in evolutionary biology is therefore to understand the genes and evolutionary processes that underlie independent bouts of specialization. Here, we describe a novel case of behavioral specialization in the fruit fly Drosophila orena. We show that D. orena has evolved a strong preference for the host plant Syzygium staudtii and that genes acting at the periphery of the olfactory system (those encoding odorant‐binding and chemosensory proteins) show an enriched signature of positive selection and are associated with this preference. We also find a shared signature of positive selection acting on genes encoding odorant‐binding and chemosensory protein in D. orena's sister species, D. erecta, a species specialized to use fruits of Pandanus trees. Our study highlights how species can employ a diverse, but non‐random, subset of genes involved in olfaction during specialization into different environments.
Introduction
Local adaptation is a fundamental evolutionary process that can drive phenotypic and genetic diversification and ultimately can be responsible for the origin of new traits and species (Darwin 1859; Schluter 2000; Rundle and Nosil 2005; Nosil 2012; Shafer and Wolf 2013). One conspicuous example of trait diversification is ecological specialization, the process whereby related species evolve to utilize different subsets of the total niche space available to them. Many groups of insects that rely on plants for food or breeding sites display such a pattern and have evolved to specialize on a small fraction of the total plant species available to them (Ehrlich and Raven 1964; Futuyma and Moreno 1988; Jaenike 1990; Nosil 2002). Given that insects are the most diverse group of animals on Earth, understanding the evolutionary processes underlying their diversification is central to our understanding of biodiversity (Jaenike 1990).
Host specialization in insects can involve the evolution of physiological, life history, and behavioral traits (Thompson 1988; Craig et al. 1989; Thompson and Pellmyr 1991; Gripenberg et al. 2010). Evolution of physiological traits can increase an organism's fitness in an environment that was previously suboptimal (e.g., Drosophila sechellia: Jones 2005; Dworkin and Jones 2009; Huang and Erezyilmaz 2015) and can include changes in life‐history strategies such as dispersal and reproductive potential (Southwood et al. 1974; Resetarits 1996; Denno et al. 2008). The evolution of behavioral traits can result in individuals preferentially seeking out certain environments, and in plant‐associated insects, preferences tend to be controlled by traits used to detect chemical cues generated by their preferred host plants (Tilmon 2008). Understanding how these traits evolve during host specialization is important for understanding potential trade‐offs encountered during specialization into a given ecological niche (Thompson 1988; Shoval et al. 2012; Anderson et al. 2013; Schick et al. 2015).
Chemoreception in insects is controlled by a diverse set of proteins that includes odorant‐binding proteins (OBPs), chemosensory proteins (CSPs), olfactory receptors (ORs), and gustatory receptors (GRs) (Hallem et al. 2006; Sánchez‐Gracia et al. 2009). OBPs and CSPs are small soluble proteins expressed in the sensory organs of insects that bind to distinct hydrophobic odorants and pheromones, facilitate their transport to ORs or GRs, and initiate signal transduction in sensory neurons (Vogt et al. 1991; Xu et al. 2005; Hallem et al. 2006; Sánchez‐Gracia et al. 2009). Here, we collectively refer to OBPs, CSPs, ORs, and GRs as “olfactory proteins” and their underlying genes as “olfactory genes.”
Olfactory genes or the sensory neurons in which they are expressed have been implicated in a number of cases of host specialization in insects (e.g., fruit flies: Matsuo et al. 2007; McBride 2007; Linz et al. 2013; Ramasamy et al. 2016; mosquitoes: McBride et al. 2014; aphids: Smadja et al. 2012; Duvaux et al. 2015; Eyres et al. 2016; apple maggot flies: Tait et al. 2016). Genetic variation at individual olfactory genes has also been shown to affect olfaction (Xu et al. 2005; Matsuo et al. 2007; Macharia et al. 2016), and olfactory genes are commonly thought to evolve through positive natural selection (McBride and Arguello 2007; Whiteman and Pierce 2008; Sánchez‐Gracia et al. 2009; Lavagnino et al. 2012). These findings suggest that specialization into different host environments likely involve evolution at olfactory genes. However, whether certain classes of olfactory genes may be more evolutionary labile than others, and therefore more likely to underlie different bouts of specialization, remains unknown.
A more general, and outstanding question in evolutionary biology is where phenotypic convergence occurs in terms of the hierarchy of genetic control. For example, phenotypic convergence can occur due to the same mutations, different mutations in the same gene, or different genes found in a common gene network (Manceau et al. 2010; Rosenblum et al. 2014). Identifying the genetic basis of convergence, and where it lies in the hierarchy of genetic control is important because it can shed light on constraints or biases in evolution. For example, repeated bouts of adaptation that involve the same gene (e.g., melanocortin‐1 receptor [Mc1r] in lizards [Rosenblum et al. 2010], birds [Uy et al. 2016], and mice [Steiner et al. 2007]) suggest that evolution is either constrained to use that locus, or the mutational spectrum at that locus is biased to generate adaptive phenotypes. At the other extreme, traits that are controlled by many genes may be less constrained to evolve using the same gene, especially in scenarios where there is redundancy in gene function (Yeaman 2015).
The Drosophila melanogaster species subgroup is well suited to study the ecological and genetic basis of host specialization. This group contains nine described species that have evolved over the last 10–15 million years to exploit a wide diversity of ecological niches (Lachaise et al. 1988; Tamura et al. 2004; David et al. 2007). Five of the species within this subgroup (D. melanogaster, D. simulans, D. mauritiana, D. yakuba, and D. teissieri) are considered dietary generalists, while three species (D. sechellia, D. santomea, and D. erecta) are thought to be dietary specialists. Of the specialist species, Drosophila sechellia specializes on the toxic fruits of Morinda citrifolia (Rubiaceae) (R'Kha et al. 1991), D. santomea is found in association with Ficus chlamydocarpa fernandesiana (Lachaise et al. 1988; Cariou et al. 2001), and D. erecta is seasonally abundant in regions with fruiting Pandanus spp. (Pandanaceae) (Rio et al. 1983; David et al. 2007). The ninth species from the clade, D. orena, was originally collected in 1975 in the highlands (2100 m) of the High Valley of Bafut N'Guemba, Cameroon (Tsacas and David 1978); however, the biology of this species has remained mostly unknown due to an inability to identify extant populations (Cariou 1987; David et al. 2007).
Only D. sechellia and D. erecta have been studied with respect to the genetic or physiological basis of behavioral preferences for their respective hosts. Matsuo et al. (2007) showed that genetic variation at Obps affect D. sechellia’s preference for volatile chemicals produced by M. citrifolia and McBride and Arguello (2007) showed that both D. sechellia and D. erecta are losing Or and Gr genes more rapidly than related generalist species. Physiological work has also shown that D. erecta’s olfactory sensory neurons show increased excitation to volatiles produced by Pandanus fruits (Linz et al. 2013).
Here, we study the phenotypic and genetic basis of host specialization in the least known species of the melanogaster subgroup: D. orena. We use both laboratory and field experiments to show that a population of D. orena on the island of Bioko, West Africa displays a behavioral preference for Syzygium staudtii (waterberry) over other potential hosts. We then use both comparative genomic and classical genetic approaches to test the role of olfactory genes in D. orena’s preference for waterberry. Our results suggest that D. orena has evolved a strong preference for waterberry through positive selection on Obps and Csps. Signatures of positive evolution at olfactory loci in D. orena’s sister species, D. erecta, mirror the enrichment we observe in D. orena and suggest that host specialization in these two species is predisposed to involve an evolutionarily labile subset of olfactory genes that operate at the periphery of the olfactory system (Obps and Csps). Interestingly, the number of Obps and Csps that share a signature of positive selection in both D. orena and D. erecta is not greater than random expectations, indicating that convergence may occur at the level of gene family, but is not constrained to use the same locus or loci.
Methods
SAMPLE SITES ON THE ISLAND OF BIOKO, WEST AFRICA
We sampled Drosophila at five locations on the island of Bioko (Table S1). We set up five trapping stations, each consisting of one trap baited with lightly yeasted waterberry fruits, one with banana, and one with mango, at each location. We hung each trap from tree branches at an approximate height of 1 m, and collected all species present in the trap after 48 hours using an aspirator. We anesthetized each individual with triethylamine (FlyNap, Carolina Biological Supply Company) and identified and counted individuals from each species of the melanogaster subgroup under a light microscope. Species other than D. orena were immediately preserved in 70% ethanol and D. orena were placed in vials containing cornmeal in groups of 15–20 until later experiments.
PREFERENCE FOR WATERBERRY AS A HOST
In situ estimates of host preference
We assessed host preference in D. orena and four other species of the melanogaster subgroup by testing whether each species displayed variation in the host fruit it was collected from across our five sampling stations on Bioko (Pearson's χ2 tests, chisq.test function in R).
We also tested host preference using an “eclosion” experiment, which identified the species of flies that eclosed from waterberry, Parinari, and fig fruits. These were the three most abundant fruits on the forest floor in regions where we collected D. orena on the east side of Mount Biao (1100–2020 m above sea level). We collected and placed fruit in 259 mL glass bottles supplemented with a pupation substrate (Kimwipes, Kimberly Clark, Roswell, GA), for a total of 15 bottles of each type of fruit. Larvae and pupae were allowed to develop within the bottles and upon emergence we identified and counted adults belonging to the melanogaster subgroup. We restricted our counts to male flies because species‐specific male traits are easy to unambiguously identify relative to female traits in living individuals in the field (Markow and O'Grady 2006; Orgogozo and Stern 2009). We tested whether eclosion rates for each species varied across the three substrates with Pearson's χ2 tests.
Lab‐measured preference for waterberry fruits
We next tested the host preference of D. orena, D. melanogaster, D. simulans, D. teissieri, and D. yakuba using food‐choice behavioral assays. We collected adult flies from the trapping stations described above and maintained them in species and sex‐specific vials containing cornmeal food for 4 days prior to behavioral assays to allow them to recover from the anesthesia. In total, we assayed 210 D. orena, 512 D. melanogaster, 410 D. simulans, 422 D. teissieri, and 398 D. yakuba. Prior to behavioral assays, we placed an average of 26 flies of the same species and sex (range: 21–31) into empty vials with a source of water (i.e., hydrated cellulose acetate plugs, Genesee Scientific) and starved them for 12 hours overnight. The following morning, we connected the vials containing flies to vials containing cornmeal fly food on one side and waterberry fruit on the other (see Turissini et al. 2017 for additional details). We let flies choose a side for three hours and scored the number of flies in each vial (either waterberry, cornmeal, or center) at the end of the assay. The proportion of flies in the waterberry vial relative to the cornmeal vial was used as an estimate of food preference.
We tested whether D. orena preferred waterberry more than the other four species by fitting a generalized linear model (GLM; glm function in the R library “stats”) with binomially distributed error that modeled the proportion of individuals in the waterberry vial as a function of species, sex, and the interaction between species and sex. We also analyzed the ratio of individuals choosing waterberry over those choosing cornmeal, for each species, using exact binomial tests (EBTs; binom.test function in R) with the null expectation that a fly was equally likely to choose waterberry or cornmeal. These two tests allowed us to ask whether D. orena has a stronger preference for waterberry compared to other closely related species and if they have a general preference for waterberry over cornmeal, respectively.
PERFORMANCE ON WATERBERRY ACROSS THE MELANOGASTER SUBGROUP
In addition to preference, we estimated performance of D. orena, D. yakuba, D. teissieri, D. melanogaster, and D. simulans when raised in each of six different host environments: mango, fig, banana, cornmeal, waterberry, and instant Drosophila medium (Carolina Biological, Burlington, NC). Females (N = 600 of each species) were individually mated to conspecific males and pooled into groups of 10 individuals (N = 60 groups per species). We randomly assigned 10 groups of each species to each of the six host environments. After 10 days of laying eggs we removed the females from the vials and added a Kimwipe (Kimberly Clark) dampened with 0.5% propionic acid (to prevent fungal growth) as a pupation substrate. We counted the number of adult flies that eclosed from each replicate vial over the following 3 weeks (i.e., until no more flies emerged) as a measure of performance. This approach integrates performance across the propensity of females to lay eggs on a given substrate and the ability of those eggs to hatch, larvae to develop and pupate, and pupae to successfully eclose into adult flies.
To determine whether performance varied across host environments we first fitted a GLM testing for an interaction between species and host environment. We assessed significance of this interaction with a likelihood ratio test that compared the fit of the full model (fixed effects: species, host environment, and the interaction between species and host environment) to one lacking the interaction term. This analysis showed a highly significant interaction between species and host environment (see Results) on composite performance; therefore, we also looked at variation in performance across host environments for each species independently by fitting a GLM where the number of eclosing adults was the response and host environment was the fixed effect. We then compared performance between environments using Tukey's post‐hoc contrasts. Because we were specifically interested in performance when raised on waterberry fruits, we only report pairwise contrasts between waterberry and each of the other host environments. GLMs were fitted assuming Poisson‐distributed error.
THE ROLE OF OLFACTORY GENES IN HOST SPECIALIZATION
Because our phenotypic results indicate that D. orena has evolved a strong preference for waterberry fruits (see Results), we used both comparative genomic and classical genetic approaches to determine the role of genes associated with olfaction in this behavioral specialization.
Evolutionary rates of olfactory genes
We resequenced the genome of a single female D. orena and mapped the reads to the D. erecta reference genome to a mean per‐site coverage of 26.16X (D. erecta is D. orena’s sister species; see SI for details). We computed the ratio of synonymous (Ks) to nonsynonymous (Ka) substitutions (ω) in both the D. orena and D. erecta lineages using codeml from the PAML 4.8 package (Yang 2007). We computed ω for 13,605 D. erecta genes using a three‐species alignment of D. orena, D. erecta, and D. yakuba and polarized substitutions as being derived in either the D. orena or D. erecta lineage, whenever possible, using D. yakuba as the outgroup (D. yakuba is the sister species of D. orena and D. erecta; see SI for information regarding the D. yakuba sequence we used in our analysis). We categorized each gene as being subject to positive selection if ω was greater than or equal to 1 (Li et al. 1985; Yang and Nielsen 2000). Sites where both D. orena and D. erecta were derived (i.e., different alleles for D. orena, D. erecta, and D. yakuba) were not used in the lineage‐specific ratios because we could not infer the ancestral state.
Of the 13,605 genes, 12,387 have a known homolog in D. melanogaster. We used our estimate of ω for these 12,387 genes to test whether genes annotated as Obps (46 of the 12,387 genes), Csps (18 genes), Grs (59 genes), or Ors (60 genes) (Graham and Davies 2002; Hekmat‐Scafe et al. 2002; Vieira et al. 2007) showed elevated rates of adaptive evolution across the D. orena and D. erecta dyad or along the D. orena or D. erecta branches. We tested whether the fraction of a given class of olfactory genes with ω ≥ 1 was greater than the genomic background with Fisher's exact tests (FETs; fisher.test function in R). We also compared the distribution of ω for each of the classes of olfactory genes to that of the remaining genes in our dataset using Wilcoxon rank sum tests (WRSTs; wilcox.test function in R).
Because we have estimates of ω for both D. orena and D. erecta, we tested whether any olfactory genes showed evidence of positive selection along both these lineages. For each class of olfactory gene, we determined whether the proportion of genes with ω ≥ 1 along both the D. orena and D. erecta lineage was enriched relative to random expectations. To test for convergent positive selection, we only considered genes with ω ≥ 1 as a result of different substitutions along the D. orena and D. erecta branches. We took this approach because we cannot rule out the possibility that a shared substitution evolved independently along both the D. orena and D. erecta branches or once along the branch leading from the common ancestor of the clade ((D. orena, D. erecta), D. yakuba) to the split between D. orena and D. erecta. We used randomization tests to assess whether the overlap in genes with ω ≥ 1 was greater than expected by chance. These tests compute the expected number of “convergent” genes of a given class relative to the observed number of genes in that class with evidence of positive selection along the D. orena and D. erecta branches. We generated 10,000 randomized data sets for each class of gene and calculated empirical P‐values as the proportion of the randomized samples where the number of convergent loci (i.e., loci with ω ≥ 1 in both lineages) was equal to or greater than the observed number between D. orena and D. erecta. Finally, we tested for a correlation (Spearman's) between estimates of ω along the D. orena and D. erecta branches for each class of olfactory gene (cor.test function in R).
In addition to our primary analysis of D. orena and D. erecta, we calculated ω for two additional species of the melanogaster subgroup: D. santomea and D. yakuba (see SI for details). This analysis allowed us to ask whether patterns of enrichment in ω we observe for olfactory genes in D. orena and D. erecta are shared in other specialist (D. santomea) or generalist (D. yakuba) species. If patterns of ω were shared for a given group of olfactory genes across all species, this would indicate that evolutionary rates we observe in D. orena are not related to host specialization, but are instead due to some intrinsic property of the gene family itself (e.g., if olfactory‐binding proteins evolve rapidly in all lineages). We tested whether the proportion of olfactory genes of a given family with ω ≥ 1 was enriched relative to genome‐wide expectations (i.e., compared to “nonolfactory” genes) for both D. santomea and D. yakuba using FETs.
Complementation tests
We took advantage of the fact that D. melanogaster females can be forced to hybridize with D. orena males to conduct complementation tests that isolate the effect of D. orena alleles on the behavioral preference for waterberry. (The rate of hybridization is ∼1/400 trials.) We focused on Obps because our analysis of ω identified the largest number of candidate loci for this class (Results). Consistent with previous food‐choice experiments in hybrid Drosophila (Turissini et al. 2017), D. melanogaster × D. orena F1 hybrid females are, in general, less able to locate food than their parents (FETs: F1s compared to D. melanogaster: odds ratio = 14.99, P < 1.0 × 10−11; F1s compared to D. orena: odds ratio = 7.53; P < 1.0 × 10−7). More importantly, the F1 hybrids that do locate food prefer cornmeal to waterberry (EBT: 26/30 chose cornmeal; P < 1.0 × 10−4; Fig. 4B) and do not differ from D. melanogaster in their preference (FET: odds ratio = 0.36; P = 0.0609). Drosophila orena preference alleles are, therefore, mostly recessive to D. melanogaster alleles, allowing us to test their effect in hemizygous hybrids that carry a deficiency along the chromosome inherited from D. melanogaster.
Figure 4.

Comparative genomics and deficiency mapping show that Obp and Csp alleles underlie D. orena’s preference for waterberry. (A) Estimates of Ka/Ks for genes of different types (see main text for descriptions of the different “types”). Obps and Csps (red points) are enriched for the proportion of loci with Ka/Ks ≥ 1 compared to Grs, Ors (light blue points), and the nonolfactory genes (“genome”; gray points) (FETs; all P < 0.05). The overall distributions of Ka/Ks values for Obps and Csps are also greater than those of Grs, Ors, and nonolfactory genes (Wilcoxon rank sum tests; all P < 0.0001). (B) Bars show the proportion of flies of a given genotype that choose waterberry versus cornmeal in food‐choice behavioral assays. The proportion of pure species and their F1 hybrids (three leftmost bars) indicate that D. orena preference alleles are recessive to D. melanogaster alleles. When recessive D. orena alleles present over a D. melanogaster deficiency (df/ore), hybrids prefer waterberry over cornmeal when compared to D. orena alleles found over D. melanogaster balancer chromosomes (Bal/ore). This pattern was not observed for seven genetic “controls” (“other” deficiencies) located adjacent to the Obp deficiencies. Error bars were computed using the “binconf” function of the Hmisc R library and represent 95% confidence intervals. See Table S6 for data.
We used seven D. melanogaster deficiency (df) stocks (i.e., lines containing chromosomal aberrations resulting in a deleted stretch of their genome) that spanned nine Obp loci with ω > 2 along the D. orena branch (Table S2) and have breakpoints that have been molecularly characterized. Five of these dfs span a single Obp locus and two span three Obps. Both dfs spanning more than one Obp covered three loci (Obp56e, Obp56g, and Obp56i; and Obp57a, Obp57b, and Obp57e); therefore, we can only assess the effect of these alleles jointly. The effect of Obp19b, Obp22a, and Obp83cd were assessed using three unique dfs and those of Obp22a and Obp83cd were each assessed using two independent but overlapping dfs. We also attempted to cross deficiency stocks spanning five additional Obps (Obp50a, Obp991, Obp99c, Obp99d, and Obp93a), but obtained no hybrid progeny. Each Obp df was paired with a df that spanned a region adjacent to the targeted Obp. This set of dfs acted as a genetic “control” to verify that not all df‐carrying hybrids were attracted to waterberry fruit. Each df spanned additional loci that are not predicted to affect the olfactory system (Table S3). All dfs were maintained over a Balancer chromosome (Bal; FM7 for X‐linked deficiencies, CyO for deficiencies on the second chromosome, and TM3, Sb for deficiencies on the third chromosome). We crossed females from D. melanogaster stocks (Bal/df) to D. orena males. The cross between a D. melanogaster Bal/df female and a D. orena male produces mel/ore F1 female hybrids with two genotypes: those carrying a single copy of the D. orena Obp allele over a melanogaster chromosome that is deficient with respect to this allele (df/ore) and those carrying a copy of the D. melanogaster Balancer chromosome and the D. orena Obp allele (Bal/ore). The complementation test we carried out is therefore similar to quantitative deficiency mapping (Anholt and Mackay 2004). We tested whether the flies expressing the D. orena allele (df/ore) were more attracted to waterberry fruit than their sisters that carried both D. melanogaster and D. orena alleles (Bal/ore). We obtained at least 50 hybrid females for each genotype and ran them through behavioral assays that tested their propensity to choose waterberry as a resource as described in “Lab‐measured preference for waterberry fruit.” For each df we compared the proportion of df/ore hybrids that chose waterberry fruit over cornmeal to the same proportion of Bal/ore hybrids using FETs.
Results
DISCOVERY OF D. orena ON THE ISLAND OF BIOKO, WEST AFRICA
We initially sampled a single D. orena female in 2009 from a hanging banana trap on Bioko (DRM). J.R. David inspected this individual and concluded it was D. orena based on morphology. The genital morphology of male D. orena is distinct from other species of the melanogaster subgroup: they have a large truncated phallus in the shape of a boomerang, basal phallic hooks, and no epandrial posterior lobes (Yassin and Orgogozo 2013). They also differ from their sister species D. erecta in that the latter has a hook shaped phallus curved dorsally. Female D. orena can be identified based on the presence of a sclerotinized vulval shield (Yassin and Orgogozo 2013). The D. orena that we report and analyze here were collected during an expedition to the island of Bioko in 2013. All D. orena we collected were sampled above 1200 m at cool and wet sites relative to the rest of the island (Figs. 1 and 2A; SI). No Pandanus spp. plants, the preferred host of D. erecta was found in Bioko, decreasing the possibility of our collections being D. erecta. We confirmed that the flies we sampled were not D. erecta by performing controlled crosses between the isofemale lines we established and the one D. orena line that was collected from Cameroon in 1975. All flies mated readily and produced fertile offspring (data not shown).
Figure 1.
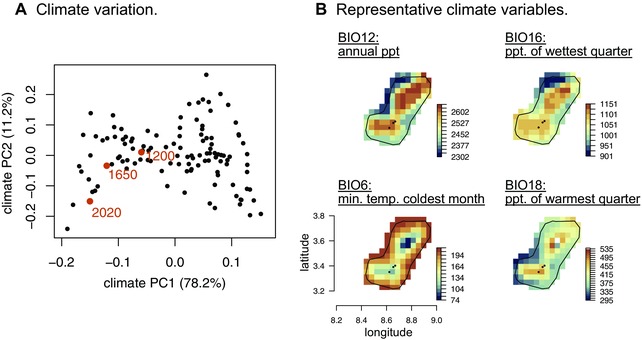
Drosophila orena’s climatic niche. (A) Climatic space across the island of Bioko and the position in this space for sites where we collected D. orena (red points; elevation in meters above sea level is shown beside these points). (B) Variation in four representative bioclimatic variables that loaded heavily on the PC axes shown in (A) across the island of Bioko (ppt. = precipitation; min. = minimum; temp. = temperature).
Figure 2.
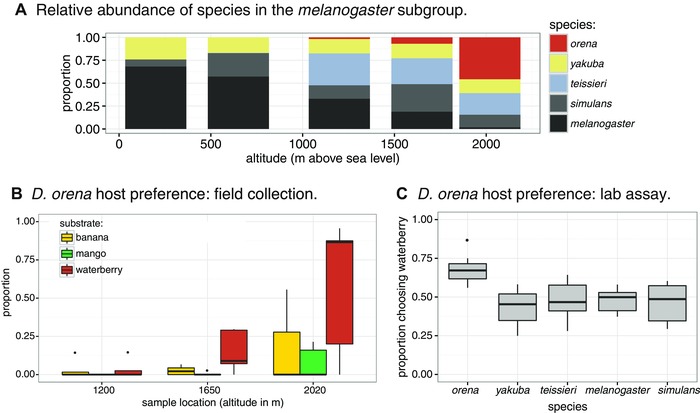
Drosophila orena is a high altitude specialist found on waterberry. (A) D. orena are only found at high elevations and are the primary species of the melanogaster species subgroup found at high elevations. The specific altitudes of the five sample locations are reported in Table S1. (B) At the high elevation sites, D. orena were preferentially collected from traps baited with waterberry. (C) Of the five species from the melanogaster subgroup we sampled on Bioko, D. orena was the only one showing a preference for waterberry fruit in food‐choice behavioral assays when given the choice between waterberry and a cornmeal substrate.
PREFERENCE FOR WATERBERRY AS A HOST
We first tested D. orena’s host preference in the field. Across the three locations where we collected D. orena (Figs. 1A, 2A and 2B), over 80% were collected over waterberry, while we tended to collect all other species of the melanogaster subgroup banana or mango (Table 1). This indicates that D. orena has a strong preference for waterberry. The number of male flies emerging from waterberry, Parinari, and fig fruit support this conclusion: Drosophila orena was the only species where the majority of individuals emerged from waterberry fruits (Table 2). At least one D. orena emerged from 9 of the 15 waterberry replicates and two of the 15 fig replicates, while none emerged from Parinari. Despite this strong bias to emerge from waterberry, it is possible that D. orena utilizes species of hosts we did not sample, and/or vary in their resource use throughout the year, as has been observed in D. erecta (Rio et al. 1983; David et al. 2007). While at least one individual of each of the other melanogaster subgroup species emerged from waterberry, the majority of individuals of D. yakuba, D. melanogaster, and D. simulans emerged from figs, while D. teissieri emerge mainly from Parinari fruits (Table 2).
Table 1.
Drosophila orena strongly prefer waterberry fruit over other suitable substrates
| Species | Waterberry | Banana | Mango | χ2 | df | P‐value |
|---|---|---|---|---|---|---|
| D. melanogaster | 618 | 614 | 643 | 0.39 | 2 | 0.823 |
| D. simulans | 314 | 334 | 290 | 1.56 | 2 | 0.468 |
| D. teissieri | 70 | 513 | 197 | 200.88 | 2 | 0.000 |
| D. yakuba | 186 | 299 | 306 | 18.78 | 2 | 8.0 × 10−5 |
| D. orena | 154 | 27 | 12 | 87.96 | 2 | 0.000 |
The number of flies caught over each type of fruit was pooled across sample locations and replicates (see main text for details; Table S4 for numbers grouped by the elevation collected from).
Table 2.
Species from the melanogaster subgroup vary in their frequency of host use
| Species | Waterberries | Parinari | Figs. | χ2 | P‐value |
|---|---|---|---|---|---|
| D. melanogaster | 5 | 12 | 39 | 34.5 | 3.17 × 10−8 |
| D. simulans | 4 | 10 | 16 | 7.2 | 2.73 × 10−2 |
| D. teissieri | 2 | 45 | 3 | 39.9 | 2.02 × 10−16 |
| D. yakuba | 10 | 1 | 22 | 20.2 | 4.15 × 10−5 |
| D. orena | 15 | 0 | 2 | 23.4 | 8.25 × 10−6 |
Species and number of individuals that eclosed from waterberries, Parinari, and figs that we collected on the island of Bioko.
Finally, in lab‐based choice assays, D. orena was more likely to move toward waterberry than any of the four other species (GLM: species term; deviance = 35.83; P = 3.13 × 10−7; Tukey contrast of D. orena versus each other species: all P < 0.05, except for with D. melanogaster, where P = 0.051; Fig. 2C). Drosophila orena was also the only species to show a general preference for waterberry fruit over cornmeal (EBT: flies attracted to waterberry fruit/total: D. orena = 144/210; P < 1 × 10−15; D. melanogaster = 250/512, P = 0.6269; D. simulans = 187/410; P = 0.08377; D. yakuba = 180/398, P = 0.06351; D. teissieri = 200/422; P = 0.3067; Fig. 2C). Together, sampling in the field, rates of emergence from field‐collected hosts, and lab‐based behavioral assays all indicate that D. orena has a strong preference for waterberry.
PRODUCTION OF OFFSPRING ON WATERBERRY ACROSS THE MELANOGASTER SUBGROUP
Specialist species of Drosophila commonly utilize a host that is either toxic or nutrient poor compared to other potential hosts (e.g., (Heed and Kircher 1965; Etges 1993; Jones 2005; Dworkin and Jones 2009; Linz et al. 2013). We assessed performance on waterberry for five species of the melanogaster subgroup. The interaction between host environment and species has a large effect on performance (LRT: χ2 = 557.31, P < 10−15; Fig. 3). When contrasting the production of offspring in different environments for each species separately, we find that D. orena is the only species that performs best when raised on waterberry (Table 3). Drosophila melanogaster, D. simulans, D. teissieri, and D. yakuba typically performed worse on waterberry than when raised on mango, cornmeal, fig, instant food, or banana (Table 3), indicating that waterberry was a suboptimal host for all these species.
Figure 3.
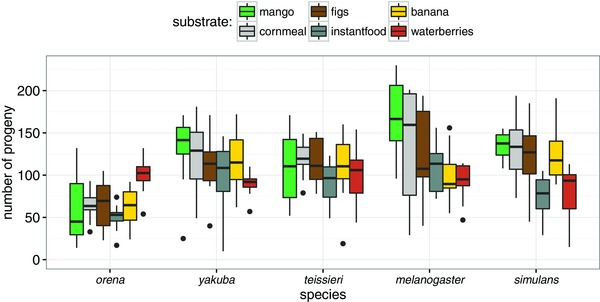
Performance on different dietary substrates. Of the five species in the melanogaster subgroup we sampled on Bioko, D. orena was the only species showing higher levels of performance on waterberry when compared to five different host environments (i.e., substrates).
Table 3.
Tukey's pairwise comparisons of composite performance when raised on waterberries versus five different food substrates
| Comparison | D. orena | D. yakuba | D. teiss. | D. melan. | D. simu. |
|---|---|---|---|---|---|
| Waterberry vs. mango | 9.885 | −8.8 | −1.911 | −14.682 | −11.79 |
| Waterberry vs. cornmeal | 8.885 | −7.388 | −4.355 | −9.219 | −11.202 |
| Waterberry vs. fig | 8.271 | −4.949 | −3.7 | −6.85 | −10.015 |
| Waterberry vs. instant food | 12.463 | −0.07146 | 1.809 | −3.333 | 1.222 |
| Waterberry vs. banana | 9.127 | −0.27412 | −2.061 | −2.067 | −9.88 |
bold text: waterberry > other at P < 0.05; italic text: waterberry < other at P < 0.01; plain text = no significant difference.
Values represent test statistics (Z statistics) for each substrate comparison for each of the five species we focus on in this manuscript.
THE ROLE OF OLFACTORY GENES IN HOST SPECIALIZATION
Evolutionary rates of olfactory genes
Given the strong preference of D. orena for waterberry, we hypothesized that olfactory genes would show a signature of adaptive (or at least accelerated) evolution since the split between D. orena and its sister species, D. erecta. We measured ω along both lineages for 46 Obps, 18 Csps, 59 Grs, and 60 Ors (Table S6) and found that Obps and Csps are more likely to show signatures of positive selection than nonolfactory genes in both D. orena and D. erecta: 26 (56.5%) and 21 (45.6%) Obps and 11 and 10 Csps have ω ≥ 1 along the D. orena and D. erecta branches, respectively, compared to 2958 (24.2%) and 2102 (17.2%) of 12,204 nonolfactory genes (Fig. 4A). When comparing between classes of olfactory genes, the proportion of loci with ω ≥ 1 does not differ between Obps and Csps in either species (FETs: D. orena: odds ratio = 0.77; P = 0.64, D. erecta: odds ratio = 0.65; P = 0.45) but is greater for both Obps and Csps when compared to Grs and Ors (FETs; Fig. 4A).
We next addressed the possibility of convergent positive selection within olfactory gene families. We found 12 Obps (26.1%), 8 Csps (44.4%), 7 Grs (11.9%), and 2 Ors (3.3%) versus 1257 of 12,204 nonolfactory genes (10.3%) with ω ≥ 1 in D. orena and D. erecta. The proportion of genes of each family with evidence of positive selection in D. orena and D. erecta was only enriched for Grs (randomization tests: expected median numbers [empirical P] for Obps: 12 [0.59]; Csps: 7 [0.41], Grs 2 [0.0003], and Ors 1 [0.42]). Estimates of ω for Obps and Csps are not correlated between D. orena and D. erecta (ρ = 0.28 and 0.20; P = 0.07 and 0.50, respectively) but they are for Grs and Ors (ρ = 0.62 and 0.46; P = 1.94 × 10−7 and 3.86 × 10−4, respectively). This result suggests that Obps and Csps have been more evolutionarily labile than Grs and Ors in D. orena and D. erecta. Our comparative analysis of ω shows that Obps and Csps display elevated rates of nonsynonymous substitution, suggesting that they have repeatedly been involved in host‐use specialization; however, the specific loci used in a bout of specialization do not tend to overlap more than expected by chance.
In the related generalist species D. yakuba, only Csps show an enrichment in ω compared to the genome‐wide expectation (3/6 vs 823/5611; FET: P = 0.045; Table S7); however, this result is based on only 6 Csp loci that have one or more derived synonymous substitution along the D. yakuba branch. For the related specialist species D. santomea, Csps (6/12; P = 0.012), Grs (13/28; P < 0.001), and Ors (14/35; P = 0.003) all have enriched estimates of ω compared to genome‐wide expectations (1185/6517), but Obps does not (0/10; P = 0.22) (Table S7). These results support the idea that specialist species have accelerated rates of evolution at olfactory genes, but the specific loci and locus family varies across independently evolved behavioral preferences.
Complementation tests
We tested whether df‐carrying hybrids (i.e., where the Obporena allele is hemizygous: df/ore) are more attracted to waterberry fruit than their Bal‐carrying sisters (i.e., where the orena allele is accompanied by a melanogaster allele: Bal/ore). For six out of the seven Obp‐spanning dfs we screened, this was the case (FETs; all P < 0.015; Fig. 4). F1 hybrids carrying the df that spanned the three Obps at band 57 (Obp57aore, Obp57bore, and Obp57eore) (i.e., Df(2R)BSC702/ore) were marginally more attracted to waterberry fruit than their Bal/ore siblings (FET: odds ratio = 0.25; P = 0.080). Despite this small difference between the two chromosomal types, the df/ore F1s showed a slight preference for waterberry fruit compared to the random expectation (EBT: 13/17 individuals chose waterberry; P = 0.049), suggesting that alleles at one (or more) of the three loci affect the preference for waterberry fruit. For seven “control” deficiencies, none of the df/ore F1s showed a preference for waterberry (Fig. 4; Table S8). These results confirm that D. orena Obp alleles that have evolved through strong positive selection (ω > 2) can causally affect levels of preference for waterberry fruits, however further validation of the Obps, Csps, Grs, and Ors with evidence of positive selection are required.
Discussion
Our results show that D. orena found on the island of Bioko have evolved a strong preference for waterberry fruits over other suitable substrates. This finding adds to the growing body of work that describes the natural history and evolution of the nine species within the melanogaster species subgroup (Lee and Watanabe 1987; David et al. 2007; Dworkin and Jones 2009; Linz et al. 2013; Yassin et al. 2016). Together, these studies suggest that host specialization within the melanogaster subgroup is not a rare phenomenon, with four of the nine species, and one population of the generalist species D. yakuba (Yassin et al. 2016), primarily being found on a single host species. Specialization may not be rare in Drosophila: upwards of 70% of Hawaiian Drosophila are estimated to be specialists (Heed 1971), other Drosophila have specialized on rotting cacti (Morales‐Hojas and Vieira 2012) or flowers (Brncic 1983), while others have evolved anatomical specializations for hard‐bodied fruits (e.g., Atallah et al. 2014). Despite these examples, a formal test of the frequency of specialization across Drosophila is, to our knowledge, yet to be conducted.
Unlike some examples of host specialization in Drosophila (e.g., D. sechellia [Farine et al. 1996; Dworkin and Jones 2009] and D. pachea [Heed and Kircher 1965; Lang et al. 2012]), waterberry fruit is not toxic to any of the other four species of the melanogaster species subgroup found on Bioko (Fig. 3). However, all four species we assayed (other than D. orena) performed best on a substrate other than waterberry fruits (Fig. 3; Table 3). Drosophila orena, on the other hand, performs best on waterberry. One explanation for this would be that performance trade‐offs resulted in physiological specialization on waterberry before, during, or after D. orena evolved a behavioral preference for waterberry. An important caveat is that our measurement of performance does not allow us to differentiate between a female's propensity to lay eggs versus a decrease in egg hatchability or larval development. Given the numbers of offspring produced by all other species on the substrates we assayed, we suggest that the most likely explanation is that D. orena’s behavioral preference for waterberry extends to a female's propensity to lay eggs. Future work is needed to test this hypothesis and whether D. orena’s preference for waterberry has led to local adaptation and strict specialization.
THE GENETIC BASIS OF HOST PREFERENCE
The fact that D. orena and D. erecta are sister species that show behavioral preferences for different substrates allowed us to ask whether the same olfactory genes show evidence of positive selection in both species. We find two general patterns: (1) enrichment in ω suggest that Obps and Csps have been under positive selection in both lineages (Fig. 4A) and (2) unlike enrichment at the level of gene family, the overlap in the specific genes with evidence of positive selection is not greater than we would expect by chance. These findings indicate that the evolution of host preferences might be predisposed to involve Obps and Csps; however, the specific genes underlying a bout of specialization can differ. Interestingly, estimates of ω for olfactory genes along the generalist D. yakuba lineage do not show enrichment for positive selection relative to the rest of the genome, and in the specialist species D. santomea, Csps, Ors, and Grs are all enriched for estimates of positive selection. A related study found that Ors and Grs are evolving more rapidly (including being lost) in the specialist species D. sechellia and D. erecta than in related generalists (McBride and Arguello 2007); however these specialists also show elevated Ka/Ks across their genome. The analyses we conducted here indicate that Ors and Grs do not show enriched Ka/Ks relative to the rest of the genome. Others have suggested that genes acting at the periphery of the olfactory system (such as Obps and Csps) evolve more rapidly than those acting in more central positions, potentially due to pleiotropic effects being less constraining for “peripheral” genes (Lavagnino et al. 2012). This hypothesis could explain the lower and correlated estimates of ω we observe for Grs and Ors in D. orena and D. erecta (Fig. 4A).
Outside of protein coding substitutions, olfactory genes can evolve through changes in gene expression or copy number (i.e., duplication or loss). We did not test mechanisms other than protein coding substitutions along the D. orena lineage; however, the growing number of studies testing the genetic basis of olfactory preferences in insects suggest that these preferences can be controlled by a diverse genetic toolkit (e.g., Grs, Ors, and Obps in Acyrthosiphon pisum [Duvaux et al. 2015; Eyres et al. 2016]; an Or in Aedes aegypti [McBride et al. 2014]). Explicit tests of the relative role of protein‐changing substitution, changes in expression, or gene duplication are needed to help us better understand the dynamics of evolving host preferences.
While did not find statistical evidence for convergent positive selection within Obps and Csps, our analysis of ω in D. orena and D. erecta would not allow us to detect convergence due to changes in a small number (one or two) of amino acid residues or gene expression. Moreover, a lack of statistical convergence does not negate the possibility that individual loci are important for, and repeatedly used during, specialization onto different hosts. For example, Obp99a, Obp99c, and Obp99d, show signs of positive selection within D. orena and D. erecta, and mutations in different amino acid residues within these loci have been implicated in behavioral responses to acetophenone (Obp99a and Obp99d; Wang et al. 2010) and benzaldehyde in D. melanogaster (Wang et al. 2007). Two other Obps with signatures of positive selection in D. orena – Obp57d and Obp57e – have been implicated in D. sechellia’s preference for the fruit of Morinda citrifolia (Matsuo et al. 2007). Notably, the complementation tests we conducted show that hemizygous individuals jointly expressing Obp57aore, Obp57bore, and Obp57eore alleles have a slight preference for waterberry fruit (Fig. 4), suggesting that Obp57e is involved in host specialization in both D. sechellia and D. orena. We also find evidence for positive selection on Obp93a alleles in D. orena and D. erecta, a gene that is differentially expressed among population of D. mojavensis adapted to different species of cacti (Bono et al. 2011; Matzkin and Markow 2013). Finally, we find a signature of positive selection on Obp83a in D. orena, a locus that has previously been implicated in host seeking behavior in tsetse flies (Liu et al. 2012; Macharia et al. 2016). These individual examples could represent genes that are important for the evolution of different preference traits. It would be interesting to test this hypothesis by comparing these genes’ role in behavioral specialization across specialist and generalist species, spanning a large phylogenetic distance. These types of tests will add to our growing understanding of the genetic basis of adaptation, and the genetic toolkit deployed during specialization into different environments, a common evolutionary strategy.
Associate Editor: S. Wright
Handling Editor: J. Slate
Supporting information
Table S1. Location of sites sampled on the island of Bioko.
Table S2. Drosophila melanogaster deficiency stocks used for complementation mapping.
Table S3. See attached Excel spreadsheet for information on additional genes spanned by the deficiencies used in this study.
Table S4. Distribution of the five species of the melanogaster species group found on Bioko along an altitudinal gradient. Counts are aggregates from three different trap substrate (bananas, mangoes, and waterberries).
Table S5. Loadings for the principle components analysis carried out on ‘bioclime’ variables sampled across the island of Bioko.
Table S6. Ka/Ks estimates for olfactory genes in the D. orena and D. erecta linages.
Table S7. Ka/Ks estimates for olfactory genes in the D. yakuba and D. santomea linages.
Table S8. Complementation mapping in mel/ore hybrids.
Table‐S3
ACKNOWLEDGMENTS
We thank Barret Miles, Gabriel Ousmane, and the Bioko Biodiversity Protection Program for logistical support in the field and B. Cooper, C.H. Martin, the Associate Editor, and two anonymous reviewers for helpful comments.
AUTHOR CONTRIBUTIONS
A.A.C. and D.R.M. designed the study, P.J.M., J.R.D., and D.R.M. collected phenotypic data, A.A.C., D.A.T., A.S.C., and D.R.M. analyzed the data, A.A.C., A.S.C., and D.R.M. wrote the initial manuscript with all authors contributing to the final version.
DATA ACCESSIBILITY
Data are available online in supplementary tables, in Dryad (doi:https://doi.org/10.5061/dryad.0q9g6), and in NCBI's short‐read archive (BioProject: PRJNA380781). The authors declare no conflicts of interest.
Contributor Information
Aaron A. Comeault, Email: aacomeault@gmail.com
Daniel R. Matute, dmatute@email.unc.edu
LITERATURE CITED
- Anderson, J. T. , Lee C.‐R., and Mitchell‐Olds T.. 2013. Strong selection genome‐wide enhances fitness trade‐offs across environments and episodes of selection. Evolution 63:16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt, R. R. H. , and Mackay T. F. C.. 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 5:838–849. [DOI] [PubMed] [Google Scholar]
- Atallah, J. , Teixeira L., Salazar R., Zaragoza G., and Kopp A.. 2014. The making of a pest: the evolution of a fruit‐penetrating ovipositor in Drosophila suzukii and related species. P R Soc. B 281:20132840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, J. M. , Matzkin L. M., Kelleher E. S., and Markow T. A.. 2011. Postmating transcriptional changes in reproductive tracts of con‐ and heterospecifically mated Drosophila mojavensis females. Proc. Natl. Acad. Sci. USA 108:7878–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brncic, D. 1983. Ecology of flower‐breeding Drosophila, vol. 3b Pp. 333–382 in Ashburner M., Carson H. L., Thompson J. N. J., eds. The genetics and biology of Drosophila. Academic Press, New York. [Google Scholar]
- Cariou, M. L. 1987. Biochemical phylogeny of the eight species in the Drosophila melanogaster subgroup, including D. sechellia and D. orena . Genet. Res. 50:181–185. [DOI] [PubMed] [Google Scholar]
- Cariou, M. L. , Silvain J. F., Daubin V., Da Lage J. L., and Lachaise D.. 2001. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol. Ecol. 10:649–660. [DOI] [PubMed] [Google Scholar]
- Craig, T. P. , Itami J. K., and Price P. W.. 1989. A strong relationship between oviposition preference and larval performance in a shoot‐ galling sawfly. Ecology 70:1691–1699. [Google Scholar]
- Darwin, C. 1859. On the origin of the species. John Murray, London, UK. [Google Scholar]
- David, J. R. , Lemeunier F., Tsacas L., and Yassin A.. 2007. The historical discovery of the nine species in the Drosophila melanogaster species subgroup. Genetics 177:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denno, R. F. , Peterson M. A., Weaver M. R., and Hawthorne D. J.. 2008. Life‐history evolution in native and introduced populations Pp. 296–310 in Tilmon K. J., ed. The evolutionary biology of herbivorous insects: Specialization, speciation, and radiation. California Univ. Press, Berkeley, CA. [Google Scholar]
- Duvaux, L. , Geissmann Q., Gharbi K., Zhou J‐J., Farrari J., Smadja C. M., and Butlin R.. 2015. Dynamics of copy number variation in host races of the pea aphid. Mol. Biol. Evol. 32:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin, I. , and Jones C. D.. 2009. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia . Genetics 181:721–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, P. R. , and Raven P. H.. 1964. Butterflies and plants: a study of coevolution. Evolution 18:586–608. [Google Scholar]
- Etges, W. J. 1993. Genetics of host‐cactus response and life‐history evolution among ancestral and derived populations of cactophilic Drosophila mojavensis . Evolution 47:750–767. [DOI] [PubMed] [Google Scholar]
- Eyres, I. , Duvaux L., Gharbi K., Tucker R., Hopkins D., Simon J‐C., Ferrari J., Smadja C. M., and Butlin R. K.. 2016. Targeted re‐sequencing confirms the importance of chemosensory genes in aphid host race differentiation. Mol. Ecol. 26:43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farine, J.‐P. , Legal L., Moreteauf B., and Quere J. L. Le.. 1996. Volatile components of ripe fruits of Morinda citrifolia and their effects on Drosophila . Phytochemistry 41:433–438. [Google Scholar]
- Futuyma, D. J. , and Moreno G.. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19:207–233. [Google Scholar]
- Graham, L. A. , and Davies P. L.. 2002. The odorant‐binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene 292:43–55. [DOI] [PubMed] [Google Scholar]
- Gripenberg, S. , Mayhew P. J., Parnell M., and Roslin T.. 2010. A meta‐analysis of preference‐performance relationships in phytophagous insects. Ecol. Lett. 13:383–393. [DOI] [PubMed] [Google Scholar]
- Hallem, E. A. , Dahanukar A., and Carlson J. R.. 2006. Insect odor and taste receptors. Annu. Rev. Entomol. 51:113–135. [DOI] [PubMed] [Google Scholar]
- Heed, W. B. 1971. Host plant specificity and speciation in Hawaiian Drosophila . TAXON 20:115–121. [Google Scholar]
- Heed, W. B. , and Kircher H. W.. 1965. Unique sterol in the ecology and nutrition of Drosophila pachea . Science 149:758 LP–761. [DOI] [PubMed] [Google Scholar]
- Hekmat‐Scafe, D. S. , Scafe C. R., McKinney A. J., and Tanouye M. A.. 2002. Genome‐wide analysis of the odorant‐binding protein gene family in Drosophila melanogaster . Genome Res. 12:1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , and Erezyilmaz D.. 2015. The genetics of resistance to morinda fruit toxin during the postembryonic stages in Drosophila sechellia . G3 5:1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J. 1990. Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 21:243–273. [Google Scholar]
- Jones, C. D. 2005. The genetics of adaptation in Drosophila sechellia . Genetica 123:137–145. [DOI] [PubMed] [Google Scholar]
- Lachaise, D. , Cariou M.‐L., David J. R., Lemeunier F., Tsacas F., and Ashburner M.. 1988. Historical biogeography of the Drosophila melanogaster species subgroup Pp. 159–225 in Hecht M. K., Wallace B., Prance G. T., eds. Evolutionary biology. Springer US, Boston, MA. [Google Scholar]
- Lang, M. , Murat S., Clark A. G., Gouppil G., Blais C., Matzkin L. M., Guittard É., Yoshiyama‐Yanagawa T., Kataoka H., Niwa R., Lafont R., et al 2012. Mutations in the neverland gene turned Drosophila pachea into an obligate specialist species. Science 337:1658 LP–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavagnino, N. , Serra F., Arbiza L., Dopazo H., and Hasson E.. 2012. Evolutionary genomics of genes involved in olfactory behavior in the Drosophila melanogaster species group. Evol. Bioinform. 8:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. H. , and Watanabe T. K.. 1987. Evolutionary genetics of the Drosophila melanogaster subgroup I. Phylogenetic relationships based on matings, hybrids and proteins. Jap. J. Genet. 62:225–239. [Google Scholar]
- Li, W.‐H. , Wu C.‐I., Luo C.‐C.. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2:150–174. [DOI] [PubMed] [Google Scholar]
- Linz, J. , Baschwitz A., Strutz A., Dweck H. K. W., Sachse S., Hansson B. S., and Stensmyr M. C.. 2013. Host plant‐driven sensory specialization in Drosophila erecta . P R Soc. B 280:20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , He X., Lehane S., Lehane M., Hertz‐Fowler C., Berriman M., Field L. M., and Zhou J.‐J.. 2012. Expression of chemosensory proteins in the tsetse fly Glossina morsitans morsitans is related to female host‐seeking behaviour. Insect Mol. Biol. 21:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macharia, R. , Mireji P., Murungi E., Murilla G., Christoffels A., Aksoy S., and Masiga D.. 2016. Genome‐wide comparative analysis of chemosensory gene families in five tsetse fly species. PLoS Negl. Trop. Dis. 10:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau, M. , Domingues V. S., Linnen C. R., Rosenblum E. B., and Hoekstra H. E.. 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Phil. Trans. R Soc. B 365:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T. , Sugaya S., Yasukawa J., Aigaki T., and Fuyama Y.. 2007. Odorant‐binding proteins OBP57d and OBP57e affect taste perception and host‐plant preference in Drosophila sechellia . PLoS Biol 5:0985–0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin, L. M. , and Markow T. A.. 2013. Transcriptional Differentiation Across the Four Subspecies of Drosophila mojavensis Pp. 119–135 in Michalak P., ed. Speciation: Natural processes, genetics and biodiversity. Nova Science Publishers, Inc, New York. [Google Scholar]
- McBride, C. S. 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia . Proc. Nat. Acad. Sci. USA 104:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C. S. , and Arguello J. R.. 2007. Five drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177:1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C. S. , Baier F., Omondi A. B., Spitzer S. A., Lutomiah J., Sang R., Ignell R., and Vosshall L. B.. 2014. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales‐Hojas, R. , and Vieira J.. 2012. Phylogenetic patterns of geographical and ecological diversification in the subgenus Drosophila . PLoS One 7(11):e49552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil, P. 2002. Transition rates between specialization and generalization in phytophagous insects. Evolution 56:1701–1706. [DOI] [PubMed] [Google Scholar]
- Nosil, P. 2012. Ecological speciation. Oxford Univ. Press, New York. [Google Scholar]
- R'Kha, S. , Capy P., and David J. R.. 1991. Host‐plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc. Natl. Acad. Sci. USA 88:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy, S. , Ometto L., Crava C. M., Revadi S., Kaur R., Horner D., Pisani D., Dekker T., Anfora G. and Rota‐Stabelli O.. 2016. The evolution of olfactory gene families in Drosophila and the genomic basis of chemical‐ecological adaptation in Drosophila suzukii . Genome Biol. Evol. 8:2297–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resetarits, W. J. J. 1996. Oviposition site choice and life history evolution. Am. Zool. 215:205–215. [Google Scholar]
- Rio, B. , Couturier G., Lemeunier F., and Lachaise D.. 1983. Evolution d'une specialisation saisonniere chez Drosophila erecta (Dipt., Drosophilidae). Annals de la Societe entomologique de France (N.S.) 19:235–248. [Google Scholar]
- Rosenblum, E. B. , Parent C. E., and Brandt E. E.. 2014. The molecular basis of phenotypic convergence. Annu. R Ecol. Evol. Syst. 45:203–226. [Google Scholar]
- Rosenblum, E. B. , Römpler H., Schöneberg T., and Hoekstra H. E.. 2010. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl. Acad. Sci. USA 107:2113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle, H. D. , and Nosil P.. 2005. Ecological speciation. Ecol. Lett. 8:336–352. [Google Scholar]
- Sánchez‐Gracia, A. , Vieira F. G., and Rozas J.. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103:208–216. [DOI] [PubMed] [Google Scholar]
- Schick, A. , Bailey S. F., and Kassen R.. 2015. Evolution of fitness trade‐offs in locally adapted populations of Pseudomonas fluorescens . Am. Nat. 186:S48–S59. [DOI] [PubMed] [Google Scholar]
- Schluter, D. 2000. The ecology of adaptive radiation. Oxford Univ. Press, Oxford. [Google Scholar]
- Shafer, A. B. A. , and Wolf J. B. W.. 2013. Widespread evidence for incipient ecological speciation: a meta‐analysis of isolation‐by‐ecology. Ecol. Lett. 16:940–950. [DOI] [PubMed] [Google Scholar]
- Shoval, O. , Sheftel H., Shinar G., Hart Y., Ramote O., Mayo A., Dekel E., Kavanagh K., and Alon U.. 2012. Evolutionary trade‐offs, pareto optimality, and the geometry of phenotype space. Science 336:1157–1160. [DOI] [PubMed] [Google Scholar]
- Smadja, C. M. , Canbäck B., Vitalis R., Gautier M., Ferrari J., Zhou J.-J., and Butlin R. K.. 2012. Large‐scale candidate gene scan reveals the role of chemoreceptor genes in host plant specialization and speciation in the pea aphid. Evolution 66:2723–2738. [DOI] [PubMed] [Google Scholar]
- Southwood, T. R. E. , May R. M., Hassell M. P., and Conway G. R.. 1974. Ecological strategies and population parameters. Am. Nat. 108:791–804. [Google Scholar]
- Steiner, C. C. , Weber J. N., and Hoekstra H. E.. 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 313:1880–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait, C. , Batra S., Ramaswamy S. S., Feder J. L., and Olsson S. B.. 2016. Sensory specificity and speciation: a potential neuronal pathway for host fruit odour discrimination in Rhagoletis pomonella . P R Soc. B 283:20162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Subramanian S., and Kumar S.. 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21:36–44. [DOI] [PubMed] [Google Scholar]
- Thompson, J. N. 1988. Evolutionary ecology of the relationship between oviposition preference and performance of off spring in phytophagons insects. Entomol. Exp. Appl. 47:3–14. [Google Scholar]
- Thompson, J. N. , and Pellmyr O.. 1991. Evolution of oviposition behavior and host preference in Lepidoptera. Annu. Rev. Entom. 36:65–89. [Google Scholar]
- Tilmon, K. J. 2008. in Tilmon K. J., ed. The evolutionary biology of herbivorous insects: Specialization, speciation, and radiation. California Univ. Press, Berkeley, CA. [Google Scholar]
- Tsacas, L. , and David J.. 1978. Une septieme espece appartenant au sous‐groupe Drosophila melanogaster Meigen: Drosophila orena spec. nov. du Cameroun (Diptera: Drosophilidae)/Seventh species belonging to the sub‐group Drosophila melanogaster Meigen: Drosophila orena spec. nov. of Cam. Beitrage zur Entomologie 28:179–182. [Google Scholar]
- Turissini, D. A. , Comeault A. A., Liu G., Lee Y. C. G., and Matute D. R.. 2017. The ability of Drosophila hybrids to locate food declines with parental divergence. Evolution 4:960‐973. [DOI] [PubMed] [Google Scholar]
- Uy, J. A. C. , Cooper E. A., Cutie S., Concannon M. R., Poelstra J. W., Moyle R. G., and Filardi C. E.. 2016. Mutations in different pigmentation genes are associated with parallel melanism in island flycatchers. P R Soc B. 283:20160731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, F. G. , Sánchez‐Gracia A., and Rozas J.. 2007. Comparative genomic analysis of the odorant‐binding protein family in 12 Drosophila genomes: purifying selection and birth‐and‐death evolution. Genome Biol. 8:R235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, R. G. , Prestwich G. D., and Lerner M. R.. 1991. Odorant‐binding‐protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 22:74–84. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Lyman R. F., Mackay T. F. C., and Anholt R. R. H.. 2010. Natural variation in odorant recognition among odorant‐binding proteins in Drosophila melanogaster . Genetics 184:759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Lyman R. F., Shabalina S. A., Mackay T. F. C., and Anholt R. R. H.. 2007. Association of polymorphisms in odorant‐binding protein genes with variation in olfactory response to benzaldehyde in Drosophila . Genetics 177:1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman, N. K. , and Pierce N. E.. 2008. Delicious poison: genetics of Drosophila host plant preference. Trend Ecol. Evol. 23:473–478. [DOI] [PubMed] [Google Scholar]
- Xu, P. , Atkinson R., Jones D. N. M., and Smith D. P.. 2005. Drosophila OBP LUSH is required for activity of pheromone‐sensitive neurons. Neuron 45:193–200. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , and Nielsen R.. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17:32–43. [DOI] [PubMed] [Google Scholar]
- Yassin, A. , Debat V., Bastide H., Gidaszewski N., David J. R., and Pool J. E.. 2016. Recurrent specialization on a toxic fruit in an island Drosophila population. Proc. Natl. Acad. Sci. USA 113:4771–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin, A. , and Orgogozo V.. 2013. Coevolution between male and female genitalia in the Drosophila melanogaster species subgroup. PLoS One 8(2):e57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman, S. 2015. Local adaptation by alleles of small effect. Am. Nat. 186:S1, S74-S89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Location of sites sampled on the island of Bioko.
Table S2. Drosophila melanogaster deficiency stocks used for complementation mapping.
Table S3. See attached Excel spreadsheet for information on additional genes spanned by the deficiencies used in this study.
Table S4. Distribution of the five species of the melanogaster species group found on Bioko along an altitudinal gradient. Counts are aggregates from three different trap substrate (bananas, mangoes, and waterberries).
Table S5. Loadings for the principle components analysis carried out on ‘bioclime’ variables sampled across the island of Bioko.
Table S6. Ka/Ks estimates for olfactory genes in the D. orena and D. erecta linages.
Table S7. Ka/Ks estimates for olfactory genes in the D. yakuba and D. santomea linages.
Table S8. Complementation mapping in mel/ore hybrids.
Table‐S3


