Abstract
Transcriptional coactivator PPAR γ coactivator (PGC)-1α and its splice variant N-terminal (NT)-PGC-1α mediate transcriptional regulation of brown adipose tissue (BAT) thermogenesis in response to changes in ambient temperature. PGC-1α is dispensable for cold-induced BAT thermogenesis as long as NT-PGC-1α is present. However, the functional significance of NT-PGC-1α in BAT has not been determined. In the present study, we generated NT-PGC-1α−/− mice to investigate the effect of NT-PGC-1α deficiency on adaptive BAT thermogenesis. At thermoneutrality, NT-PGC-1α−/− mice exhibited abnormal BAT phenotype with increased accumulation of large lipid droplets concomitant with marked downregulation of FA oxidation (FAO)-related genes. Consistent with transcriptional changes, mitochondrial FAO was significantly diminished in NT-PGC-1α−/− BAT. This alteration, in turn, enhanced glucose utilization within the NT-PGC-1α−/− BAT mitochondria. In line with this, NT-PGC-1α−/− mice had higher reliance on carbohydrates. In response to cold or β3-adrenergic receptor agonist, NT-PGC-1α−/− mice transiently exhibited lower thermogenesis but reached similar thermogenic capacities as their WT littermates. Collectively, these findings demonstrate that NT-PGC-1α is an important contributor to the maintenance of FAO capacity in BAT at thermoneutrality and provide deeper insights into the relative contributions of PGC-1α and NT-PGC-1α to temperature-regulated BAT remodeling.
Keywords: thermogenesis, beta-3 adrenergic receptor, fatty acid, N-terminal peroxisome proliferator-activated receptor gamma coactivator 1-alpha
Brown adipose tissue (BAT) is specialized in dissipating energy in the form of heat. This process, known as nonshivering thermogenesis, requires an abundant fuel supply, a high number of mitochondria, and high levels of uncoupling protein 1 (UCP1), a BAT-specific transport protein located in the inner mitochondrial membrane (IMM) (1). UCP1 uncouples fuel-driven proton gradient from ATP synthesis and promotes proton leak across the IMM, thus resulting in heat production (2, 3). BAT not only allows rodents to maintain body temperature at temperatures below the thermoneutral zone, but also plays a role in whole-body energy balance (4, 5). Adult humans also have functional BAT depots that contribute to metabolic homeostasis (6, 7).
BAT undergoes cellular remodeling in response to changes in ambient temperature. Cold-induced stimulation of β3-adrenergic receptor (β3-AR) on brown adipocyte cells leads to a marked increase in UCP1 expression/activity, mitochondrial biogenesis, intracellular lipolysis, mitochondrial FA oxidation (FAO), and de novo lipogenesis (8–10). FA metabolism plays an essential role in maximizing UCP-mediated heat production. Lipolysis-derived FAs are directed to the mitochondria, where they activate UCP1 (11–14). FAs also undergo β-oxidation to generate acetyl-coA, which enters the tricarboxylic acid cycle and is oxidized to generate NADH and FADH2. The NADH and FADH2 are then used by the electron transport system to produce a proton gradient required for UCP1-mediated heat production (1). Prolonged cold exposure simultaneously increases FA synthesis and storage in BAT that contributes to replenish triacylglycerol (TAG) in lipid droplets, thus allowing continuous FA supply to be matched with increased rates of FAO (8–10, 15, 16). At thermoneutral temperature (28–32°C), where heat-producing demands are minimal, the aforementioned BAT thermogenic program is inactivated (1).
Temperature-sensitive transcriptional coactivator PPAR γ coactivator 1-α (PGC-1α) is a master regulator of adaptive BAT thermogenesis (17, 18). PGC-1α is upregulated by cold/β3-AR stimulation and activates a number of transcription factors, leading to increased expression of UCP1 and many mitochondrial genes involved in mitochondrial biogenesis and oxidative metabolism (17, 18). Previously, we showed that alternative splicing of the PGC-1α gene produces a shorter but functional isoform of PGC-1α [N-terminal (NT)-PGC-1α] in rodents and humans (19). Mice lacking both PGC-1α and NT-PGC-1α are unable to produce heat from BAT due to impaired induction of UCP1 and mitochondrial gene expression in response to cold (20). In contrast, mice selectively deficient in full-length PGC-1α can maintain normal BAT function and activate cold-induced thermogenesis because NT-PGC-1α upregulates UCP1 and many mitochondrial genes involved in FA transport, β-oxidation, the TCA cycle, and electron transport in the absence of PGC-1α (19, 21–23).
The present study aimed at elucidating the functional significance of NT-PGC-1α in adaptive BAT thermogenesis using newly generated NT-PGC-1α−/− mice. By assessing the thermogenic response to warm (28°C) or cold (4°C)/β3-AR agonist, we here demonstrate that NT-PGC-1α is dispensable for mitochondrial biogenesis and respiration, but specifically important for mitochondrial FAO in quiescent BAT and in fully activated BAT where the maximum FA-oxidizing capacity is needed. Taken together with our previous studies, our findings underscore the relative contributions of PGC-1α and NT-PGC-1α to temperature-regulated BAT remodeling.
MATERIALS AND METHODS
Animal studies
The animal protocol for the study was approved by the Institutional Animal Care and Use Committee at the Pennington Biomedical Research Center, and the procedures were carried out in accordance with the approved guidelines. Mice were housed at a room temperature of 22°C under a 12 h light/12 h dark cycle with ad libitum feeding (standard chow 5001, LabDiet, St. Louis, MO). For cohort 1, 4 week old female mice were kept at 22°C or housed at near thermoneutrality (28°C) for 3 weeks. For cohort 2, female mice housed at 28°C for 3 weeks were singly housed and exposed to 4°C. Core rectal temperature was measured at baseline and every 1 h over the 5 h period of cold exposure. For cohort 3, male mice housed at 28°C were weighed, and their body composition was measured using a Bruker Mouse Minispec (Bruker Optics, Billerica, MA). Mice were then placed in indirect calorimetry chambers (Columbus Instruments, Columbus, OH) and monitored for VO2 and VCO2 for 4 h prior to and after CL316243 injection (1 mg/kg body weight). For the cohort 4, male mice housed at 28°C were determined for body weight and composition and placed in indirect calorimetry chambers. After 3 days in the chambers, mice were intraperitoneally injected with CL316243 (1 mg/kg body weight/day) for 4 days and continuously monitored for VO2 and VCO2. Locomotor activity was measured while the mice were in the chambers. Energy expenditure (EE) was calculated as VO2 × [3.815 + (1.232 × RQ)] × 4.187 and expressed as kilojoules per hour. After removing from indirect calorimetry chambers, mice were injected with CL316243 for an additional 3 days. At the end of experiments (cohorts 1–4), BAT and white adipose tissue (WAT) were collected in the fed state.
Generation of NT-PGC-1α−/− mice
Gene targeting and chimeric mouse production were performed by the Transgenic Core at Pennington Biomedical Research Center. DNA derived from a C57BL6 genomic BAC library clone (RP24-399J12) was used to generate a targeting vector containing fused exons 6 and 7 and the loxP-neo-loxP. Targeted albino B6 embryonic stem cells carrying the mutant allele were injected into C57BL6 blastocytes, and chimeric mice were mated to C57BL6 mice to obtain heterozygous offspring containing a germline mutant allele. These heterozygotes were subsequently mated to Zp3-Cre transgenic mice (Jackson Laboratory catalog no. 003651) that express Cre recombinase under the control of ZP3 promoter, which is activated in growing oocytes (24). The Cre recombinase-mediated deletion of the neo cassette was confirmed by PCR. Heterozygous NT-PGC-1α+/− mice were mated to obtain homozygous NT-PGC-1α−/− mice. Genotyping was performed by PCR using ear punch DNA.
Quantitative real-time PCR analysis
Total RNA was reverse-transcribed for quantitative real-time PCR (qPCR) analysis as described previously (19, 23, 25). Relative expression levels of genes were determined after normalization to cyclophilin by the 2-ΔΔCt method. For quantitative analysis of mitochondrial biogenesis, mitochondrial and nuclear DNA were isolated, and their respective copy numbers were measured using qPCR with primers for ND1 and LPL as described previously (23).
FAO assay
Palmitate oxidation was measured as previously described (22, 26). In brief, freshly isolated adipose explants were homogenized in SET buffer containing 250 mM sucrose, 1 mM EDTA, and 10 mM Tris-HCl (pH 7.4). Tissue homogenates were incubated with reaction mixture containing 62.5 mM sucrose, 12.5 mM potassium phosphate, 100 mM potassium chloride, 1.25 mM magnesium chloride, 0.5 mM EDTA, 1.25 mM l-carnitine, 0.125 mM malic acid, 1.25 mM DTT, 62.5 µM NAD+, 2 mM ATP, 62.5 µM CoA, 10 mM Tris-HCl (pH 7.4), 200 µM palmitic acid coupled to BSA, and [1-14C]palmitate (0.65 µCi/ml). After 30 min of incubation at 37°C, 70% perchloric acid was added to stop the reaction. The 14CO2 produced during the incubation was trapped in 1 M sodium hydroxide, and the acidified portion of the incubation mixture was collected for liquid scintillation counting. Total palmitate oxidation was determined and normalized to total protein content in each sample.
Lipolysis assay
Interscapular BAT was freshly isolated, cut into small pieces (∼20 mg), and incubated at 37°C under agitation in Krebs–Henseleit buffer (KHB) containing 5 mM glucose and 2% BSA with or without isoproterenol (1 µM). After 6 h of incubation, an aliquot (25 µl) of the supernatants was incubated with 125 µl of glycerol assay buffer (pH 9.5) containing 1 mg/ml β-NAD, 1.8 mM ATP, GAPDH, and glycerokinase. Glycerol release was determined by measuring the absorbance at 320 nm. For inguinal and visceral adipose tissue, fresh fat explants were digested at 37°C with collagenase in KHB containing 5 mM glucose and 2% BSA, filtered through nylon mesh, and centrifuged. Isolated adipocytes were then incubated with or without isoproterenol (0.1 and 1 µM) at 37°C in KHB containing 5 mM glucose and 2% BSA. Glycerol was measured from infranatants after 2 h of incubation. Glycerol levels were normalized to total genomic DNA content in each sample.
Tissue O2 consumption rates
Oxygen consumption rates (OCRs) of BAT were measured as described previously (22). Briefly, fresh BAT explants were cut into small pieces (∼5 mg) and placed in a respirometric chamber of the OROBOROS Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria) containing the respiration buffer. Oxygen consumption was measured in the presence of malate/pyruvate, rotenone/succinate, or malate/palmitoyl carnitine, followed by injection of antimycin A. Mitochondrial respiration was determined by subtracting antimycin A-independent nonmitochondrial respiration and normalized by tissue weight.
Histological analysis
Tissue samples were fixed in 10% neutral-buffered formalin, paraffin embedded, and sectioned (5 µm) by the Cell Biology and Bioimaging Core at Pennington Biomedical Research Center. H&E-stained paraffin sections were scanned using a Hamamatsu NanoZoomer slide scanner (Hamamatsu, Japan).
TAG analysis
Tissue samples (40–60 mg) were homogenized in the Standard Diluent Assay Reagent provided by a Triglyceride Colorimetric Assay kit (Cayman Chemical, Ann Arbor, MI), and TAG concentrations were measured as described in the manufacturer’s instructions.
Western blot analysis and Immunoprecipitation
Whole-cell extracts were prepared from tissues by homogenization in RIPA buffer containing protease and phosphatase inhibitor cocktail (25) and subjected to Western blot analysis using following antibodies: anti-phospho-HSL (S563), anti-HSL, anti-ATGL (Cell Signaling), and anti-actin (Sigma). For immunoprecipitation, the whole-cell extracts were incubated with IgG or polyclonal anti-PGC-1α (19, 22) overnight at 4°C, followed by incubation with protein A agarose beads for 3 h at 4°C. After washings, immunoprecipitates were subjected to Western blot analysis using anti-PGC-1α antibody. Protein concentration was determined using Bio-Rad DC protein assay reagents according to the manufacturer’s instructions.
Statistical analysis
Data are presented as mean ± SEM. Student t-test or two-way ANOVA was used to compare the difference between groups using GraphPad Prism 6 software. Values of P < 0.05 were considered statistically significant.
RESULTS
Generation of NT-PGC-1α−/− mice
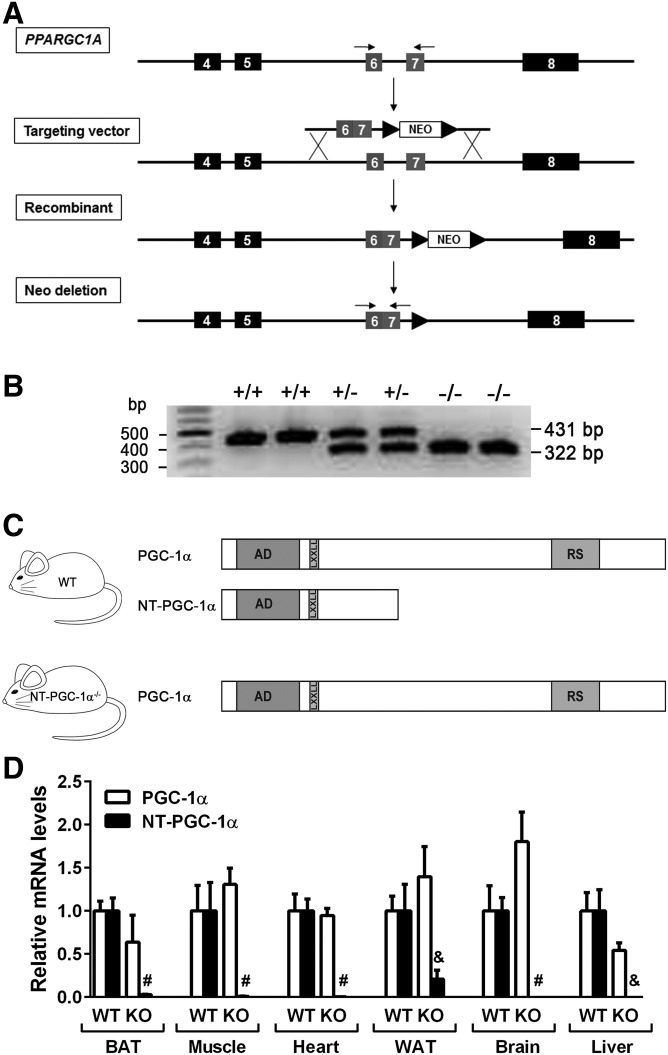
We previously reported that alternative 3′ splicing of the PPARGC1A gene produces an additional transcript encoding a shorter isoform of PGC-1α named NT-PGC-1α (19) and that NT-PGC-1α is sufficient to maintain normal BAT function and activate cold-induced thermogenesis in mice selectively deficient in full-length PGC-1α (21–23). To evaluate whether NT-PGC-1α is necessary for BAT function and adaptive thermogenesis, we generated mice with selective loss of NT-PGC-1α. A neomycin-based gene-targeting vector was generated to delete a 108 bp intron sequence between exons 6 and 7 of the murine PPARGC1A gene that contains an alternative 3′ splice site for NT-PGC-1α (19) (Fig. 1A). The insertion/recombination event resulted in a fused exon 6–7 without the intron. It is predicted that this fused exon 6–7 prevents generation of NT-PGC-1α as well as additional NT-PGC-1α-b (also known as PGC-1α4) and NT-PGC-1α-c isoforms produced from the alternative exon 1b (23, 27). The neo cassette was subsequently removed by crossing to the Zp3-Cre transgenic mouse that expresses Cre recombinase in growing oocytes (24). The resulting heterozygous NT-PGC-1α+/− mice were mated to produce homozygous NT-PGC-1α−/− mice. The mutant allele was confirmed using PCR analysis of genomic DNA isolated from NT-PGC-1α+/+, NT-PGC-1α+/−, and NT-PGC-1α−/− mice (Fig. 1B). The efficacy of the gene targeting was further evaluated by qPCR analysis with the previously verified isoform-specific primers (19, 23). Both PGC-1α and NT-PGC-1α transcripts were detected in BAT, muscle, heart, WAT, brain, and liver of WT littermates, but NT-PGC-1α transcripts were absent in NT-PGC-1α−/− mice (Fig. 1D), demonstrating that the mutant allele blocks an alternative splicing event producing the NT-PGC-1α transcript.
Fig. 1.
Generation of NT-PGC-1α−/− mice. A: Schematic of the gene-targeting strategy. A region of the mouse PPARGC1A gene containing exons 4–8 is shown at the top. The targeting vector contains a fused exon 6–7 and a neomycin (neo) cassette flanked with loxP sites. Homologous recombination between the targeting vector and the PPARGC1A gene is indicated with solid lines. Insertion of the targeting construct into the PPARGC1A gene results in deletion of the 108 bp intron between exons 6 and 7. The neo cassette was removed by crossing to a Zp3-Cre transgenic mouse line. Two arrows above the exons 6 and 7 indicate a primer pair used for genotyping. B: PCR analysis of ear punch samples from NT-PGC-1α+/+, NT-PGC-1α+/−, and NT-PGC-1α−/− mice using a primer pair described in A. C: Schematic of WT and NT-PGC-1α−/− mice expressing PGC-1α/NT-PGC-1α and PGC-1α, respectively. AD, transcription activation domain; LXXLL, leucine-rich nuclear receptor binding motif; RS, arginine/serine-rich domain. D: qPCR analysis for detection of PGC-1α and NT-PGC-1α transcripts using the previously validated isoform-specific primers (19, 23) (n = 6–9). All data are presented as mean ± SEM. &P < 0.001; #P < 0.0001 determined by Student’s t-test.
NT-PGC-1α is required for basal expression of FAO-related genes in BAT of mice housed at thermoneutral temperature
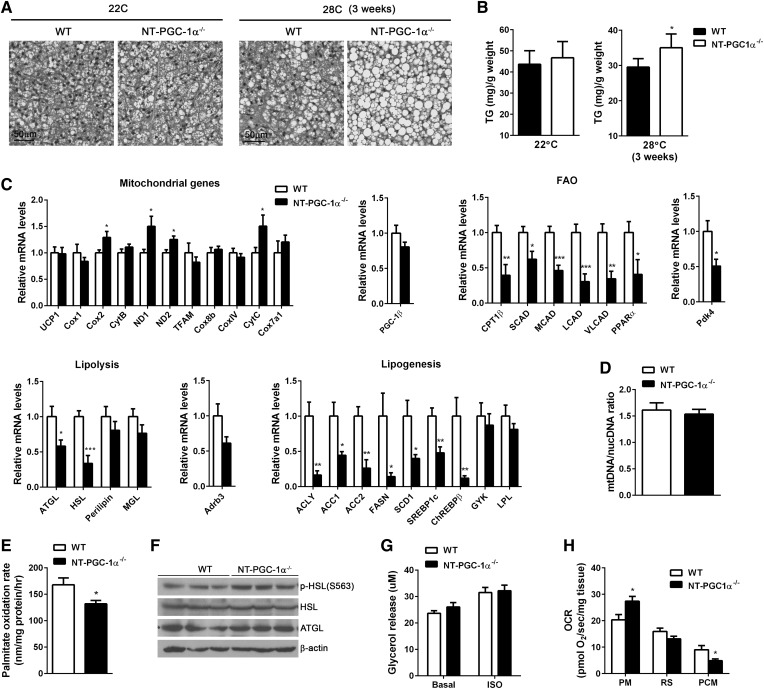
Pups lacking NT-PGC-1α were born at the expected Mendelian ratio at room temperature (22°C). After weaning, mice were housed at 22°C or near thermoneutral temperature (28°C). H&E-stained BAT sections at 22°C revealed normal histology without difference in triacyglycerol levels between WT and NT-PGC-1α−/− BAT (Fig. 2A, B). In contrast, NT-PGC-1α−/− mice housed at 28°C exhibited abnormal BAT phenotype with increased accumulation of large lipid droplets (Fig. 2A), which is a feature associated with impaired BAT function. The TAG content was also higher in NT-PGC-1α−/− BAT compared with WT BAT (Fig. 2B).
Fig. 2.
FAO is lower in BAT of NT-PGC-1α−/− mice housed at thermoneutrality. A: Representative H&E images of BAT from 7 week old WT and NT-PGC-1α−/− mice housed at 22°C (22C) or 28°C (28C) for 3 weeks (n = 4 or 5 per group). Scale bars, 50 µm. B: Triacyglycerol (TG) content of BAT from WT and NT-PGC-1α−/− mice (n = 8 per genotype). C: qPCR analysis of BAT gene expression in WT and NT-PGC-1α−/− mice housed at 28°C for 3 weeks (n = 8 per genotype). D: qPCR analysis of mitochondrial biogenesis (28°C). The relative ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nucDNA) was analyzed (n = 8 per genotype). E: The [14C] palmitate oxidation rates measured in fresh BAT homogenates in triplicate (n = 7 per genotype) (28°C). F: Western blot analysis of lipolytic enzymes in BAT. G: Basal and isoproterenol-stimulated lipolysis of BAT tissues (28°C). Glycerol release was measured in the absence and presence of isoproterenol (ISO; 1 µM) (n = 7 or 8 per genotype). H: OCRs of BAT explants in the presence of malate/pyruvate (PM), rotenone/succinate (RS), or malate/palmitoyl carnitine (PCM) (28°C). Measurements were performed in duplicate (n = 5 per genotype). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 determined by Student’s t-test.
Our previous microarray analyses showed that expression of NT-PGC-1α in PGC-1α−/− brown adipocytes upregulates many genes involved in thermogenesis, mitochondrial respiration, FA transport/oxidation, and FA synthesis (21). We thus assessed changes in gene expression associated with these biological pathways that might account for the observed phenotype of NT-PGC-1α-deficient BAT at 28°C. Surprisingly, transcript levels of mitochondrial UCP1 and electron transport chain (ETC) genes were comparable between WT and NT-PGC-1α−/− BAT (Fig. 2C). Consistent with gene expression results, there was no difference in mitochondrial number (Fig. 2D), indicating that NT-PGC-1α is not required for expression of mitochondrial genes involved in mitochondrial biogenesis, respiration, and uncoupling in BAT.
We observed significantly reduced expression of genes associated with FAO, lipolysis, and FA synthesis. Several key genes encoding a FA transporter (CPT1β) and FAO enzymes such as short-, medium-, long- and very-long-chain acyl-CoA dehydrogenases (SCAD, MCAD, LCAD, and VLCAD) were markedly downregulated in NT-PGC-1α−/− BAT (Fig. 2C). Diminished FAO gene expression was also correlated with lower transcript levels of a FAO gene-regulating transcription factor PPARα. PPARα primarily activates genes involved in FA uptake and β-oxidation, rather than genes in mitochondrial biogenesis, TCA cycle, or respiration (28, 29). We previously found that PPARα is one of NT-PGC-1α target genes, and PPARα-mediated transcription is enhanced by NT-PGC-1α (19, 21–23). Taken together, these findings indicate that NT-PGC-1α-mediated expression and activation of PPARα is required to maintain FAO gene expression in the basal (i.e., thermoneutral) state. Consistent with transcriptional changes, NT-PGC-1α−/− BAT exhibited the diminished capacity to oxidize palmitic acids (Fig. 2E). In addition to FAO genes, transcript levels of ATGL and HSL, which are predominant TAG hydrolases in BAT (30), were lower in NT-PGC-1α−/− BAT (Fig. 2C). However, this reduction in mRNA levels was not associated with reduced protein levels (Fig. 2F), leading to no change in basal and isoproterenol-stimulated lipolysis (Fig. 2G). ADRB3 gene expression (Fig. 2C) and PKA-mediated phosphorylation of HSL at serine 563 were relatively similar (Fig. 2F), suggesting that β3-ΑR signaling is not altered in NT-PGC-1α−/− BAT. Moreover, NT-PGC-1α−/− BAT showed reduced expression of several lipogenic genes, including the previously identified NT-PGC-1α targets, ACC1 and ACC2 (21) (Fig. 2C). Despite the effect on ACC1 and ACC2, overall effects of NT-PGC-1α ablation on lipolytic and lipogenic gene expression are likely indirect. Reduced FAO might induce alteration in lipolytic and lipogenic gene expression by the previously observed cross-talk among FA oxidation, lipolysis, and FA synthesis (8–10, 15, 16). This altered gene expression profile in FA metabolism was not observed in either NT-PGC-1α−/− BAT collected at 22°C or cultured NT-PGC-1α−/− brown adipocytes (supplemental Fig. S1).
Measurement of mitochondrial respiration of BAT explants with different respiratory substrates further revealed that FA (palmitoyl carnitine)-responsive respiration was lower in NT-PGC-1α−/− BAT compared with WT BAT (Fig. 2H). However, when pyruvate, the end product of glycolysis, was provided as a respiratory substrate, mitochondrial respiration was higher in NT-PGC-1α−/− BAT, indicating a shift in substrate utilization toward glucose. FAO within the mitochondria increases acetyl CoA and NADH, which subsequently inhibit the pyruvate dehydrogenase (PDH) complex that mediates conversion of pyruvate to acetyl CoA (31). The resulting decrease in FAO might relieve the inhibition of the PDH complex in NT-PGC-1α−/− BAT, favoring pyruvate oxidation. In particular, PDK4 gene expression was significantly lower in NT-PGC-1α−/− BAT (Fig. 2C). PDK4 phosphorylates and inhibits the PDH complex as well (32). Together, decreases in acetyl CoA, NADH, and PDK4 might contribute to the enhanced capacity of NT-PGC-1α−/− BAT to oxidize pyruvate in the mitochondria.
Effects of NT-PGC-1α deficiency on WAT
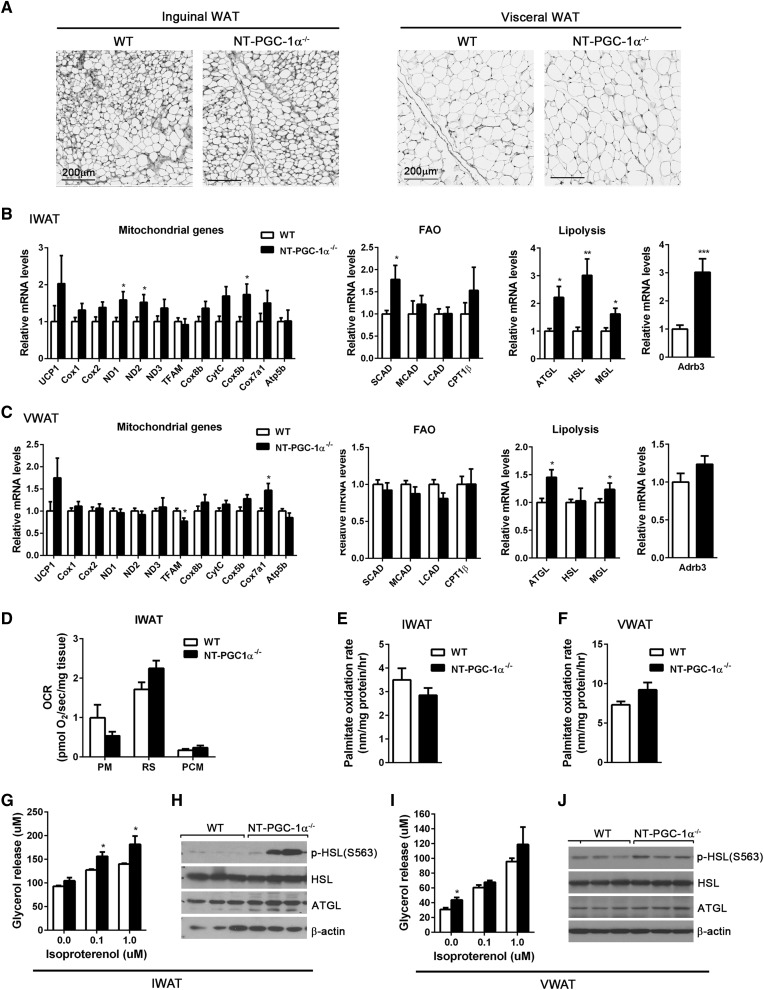
Histological analyses showed that inguinal and visceral (gonadal) fat pads from NT-PGC-1α−/− mice housed at 28°C had normal adipose tissue morphology (Fig. 3A). Given that NT-PGC-1α regulates a number of genes involved in thermogenesis, mitochondrial respiration, mitochondrial FA transport/oxidation, and FA synthesis in 3T3-L1 adipocytes (21), we assessed the effect of NT-PGC-1α deficiency on gene expression in inguinal and visceral (gonadal) WAT. Inguinal NT-PGC-1α−/− WAT exhibited the trend toward higher expression of mitochondrial electron transport and FAO genes (Fig. 3B). However, these changes were not sufficient to increase mitochondrial respiration (Fig. 3D) and FAO (Fig. 3E). Interestingly, transcript levels of ATGL, HSL, and MGL, as well as ADRB3, were higher in inguinal NT-PGC-1α−/− WAT (Fig. 3B), in conjunction with an increase in isopreterenol-stimulated lipolysis (Fig. 3G). Protein levels of ATGL and HSL were not increased but PKA-mediated phosphorylation of HSL at serine 563 was greatly increased in inguinal NT-PGC-1α−/− WAT (Fig. 3H). In contrast, NT-PGC-1α ablation had no discernable effects on gene expression and FA metabolism in visceral (gonadal) adipose tissue (Fig. 3C, F, I, J).
Fig. 3.
Effects of NT-PGC-1α ablation on WAT. A: Representative H&E images of inguinal and visceral (gonadal) WAT from WT and NT-PGC-1α−/− mice adapted at 28°C for 3 weeks. Scale bars, 200 µm. B, C: qPCR analysis of gene expression in IWAT and visceral WAT (VWAT) from WT and NT-PGC-1α−/− mice (n = 8 per genotype). D: OCRs of IWAT explants in the presence of malate/pyruvate (PM), rotenone/succinate (RS), or malate/palmitoyl carnitine (PCM). Measurements were performed in duplicate (n = 5 per genotype). E, F: [14C] palmitate oxidation rates of fresh IWAT and VWAT homogenates (n = 7 per genotype). G, I: Basal and isoproterenol-stimulated lipolysis of primary adipocytes isolated from fresh IWAT and VWAT. Glycerol release was measured in the absence and presence of isoproterenol (0.1 and 1 µM) (n = 7 or 8 per genotype). H, J: Western blot analysis of lipolytic enzymes in IWAT and VWAT. All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 determined by Student’s t-test.
NT-PGC-1α−/− mice exhibit lower thermogenic response to cold
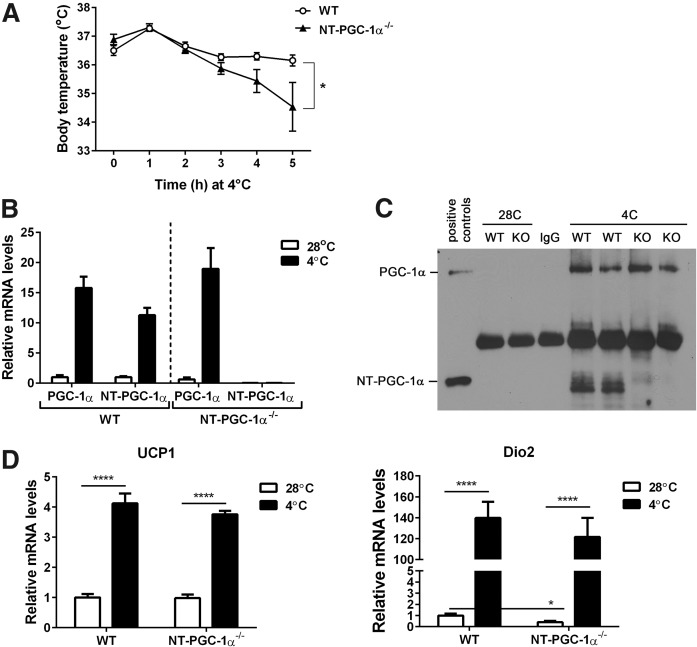
To determine whether NT-PGC-1α is required for cold-induced activation of BAT thermogenesis, WT and NT-PGC-1α−/− mice were exposed to cold (4°C) for 5 h while core body temperature was monitored. NT-PGC-1α−/− mice exhibited a larger drop (2.35°C) in body temperature than WT mice (0.34°C) after 5 h of cold exposure (Fig. 4A). qPCR and Western blot analyses confirmed no cold-dependent induction of NT-PGC-1α expression in BAT (Fig. 4B, C and supplemental Fig. S2). Moreover, NT-PGC-1α deficiency did not alter the mRNA and protein levels of PGC-1α in BAT (Fig. 4B, C and supplemental Fig. S2). Surprisingly, cold-induced expression of UCP1 and DIO2, which are indispensable for cold-induced BAT thermogenesis (3, 33), was comparable between WT and NT-PGC-1α−/− BAT (Fig. 4D), indicating that lower thermogenic response of NT-PGC-1α−/− mice is not due to impaired induction of UCP1 and DIO2 mRNA in BAT. This may be in part due to reduced rates of FAO in NT-PGC-1α−/− BAT.
Fig. 4.
NT-PGC-1α−/− mice have lower body temperature during acute cold exposure. A: Body temperature of 8 week old WT and NT-PGC-1α−/− mice (n = 7 or 8 per genotype) exposed to 4°C. Core body temperature was measured using a rectal thermometer every hour. Data are presented as mean ± SEM. *P < 0.05 determined by two-way ANOVA. B: qPCR analysis of PGC-1α and NT-PGC-1α in WT and NT-PGC-1α−/− BAT (n = 7 or 8 per group). C: Immunoprecipitation of PGC-1α and NT-PGC-1α proteins in BAT. 28C, 28°C; 4C, 4°C. D: qPCR analysis of UCP1 and DIO2 in WT and NT-PGC-1α−/− BAT (n = 7 or 8 per group).
NT-PGC-1α−/− mice exhibit normal thermogenic capacity during the prolonged β3-adrenergic stimulation but have higher reliance on carbohydrate utilization
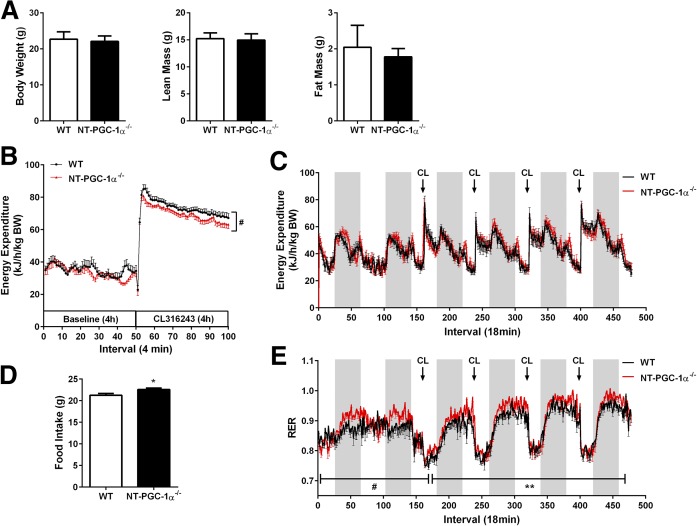
During acute cold exposure, mice can defend their body temperature by both BAT-mediated nonshivering thermogenesis and skeletal muscle-mediated shivering thermogenesis. To minimize the effect of muscle shivering, we activated BAT thermogenesis using a cold-mimetic β3-AR agonist, CL316243, under a thermoneutral zone and measured EE using indirect calorimetry. Given that β3-AR expression is limited to adipose tissue (34, 35), CL316243-induced increase in whole-body EE is attributable to the CL316243-induced activation of BAT thermogenesis. NT-PGC-1α−/− mice exhibited no difference in body weight and body composition (Fig. 5A). Baseline EE normalized by body weight was comparable between WT and NT-PGC-1α−/− mice (Fig. 5B). Administration of CL316243 rapidly increased EE in both WT and NT-PGC-1α−/− mice. However, NT-PGC-1α−/− mice exhibited a lower increase in EE compared with WT littermates during 4 h of β3-AR stimulation (Fig. 5B). A daily injection of CL316243 for 4 days further revealed that, despite the lower thermogenesis during the acute phase, NT-PGC-1α−/− mice were able to gradually increase CL316243-induced EE to levels comparable to WT littermates during the prolonged β3-AR stimulation (Fig. 5C).
Fig. 5.
Effects of NT-PGC-1α deficiency on EE and RER during β3-AR stimulation. A: Body weight and body composition of 7 week old WT and NT-PGC-1α−/− mice housed at 28°C for 3 weeks (n = 8 per genotype). B: Acute thermogenic response of WT and NT-PGC-1α−/− mice to an β3-AR agonist CL316243. Mice (n = 7 or 8 per genotype) were measured for VO2 and VCO2 prior to and after 4 h stimulation with CL316243 at 30°C. EE was calculated as described in Materials and Methods and expressed per body weight (BW). Data are presented as mean ± SEM. #P < 0.05 determined by two-way ANOVA. C: Effect of prolonged β3-AR stimulation on EE in WT and NT-PGC-1α−/− mice at 30°C (n = 8 per genotype). D: Food intake of WT and NT-PGC-1α−/− mice during 6 days of indirect calorimetry (n = 8 per genotype). E: Effect of prolonged β3-AR stimulation on RER (VCO2/VO2) in WT and NT-PGC-1α−/− mice (n = 8 per genotype). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; #P < 0.0000001 determined by Student’s t-test.
Surprisingly, the respiratory exchange ratio (RER; VCO2/VO2), an indicator of what metabolic fuel is preferentially used to produce energy, was higher in NT-PGC-1α−/− mice compared with WT littermates, reflecting preferential use of carbohydrate energy sources (Fig. 5E). Increased utilization of carbohydrates was closely associated with an increase in chow intake (Fig. 5D). The higher RER can be explained in part by our finding that NT-PGC-1α−/− BAT shows a shift toward glucose utilization for mitochondrial respiration (Fig. 2F).
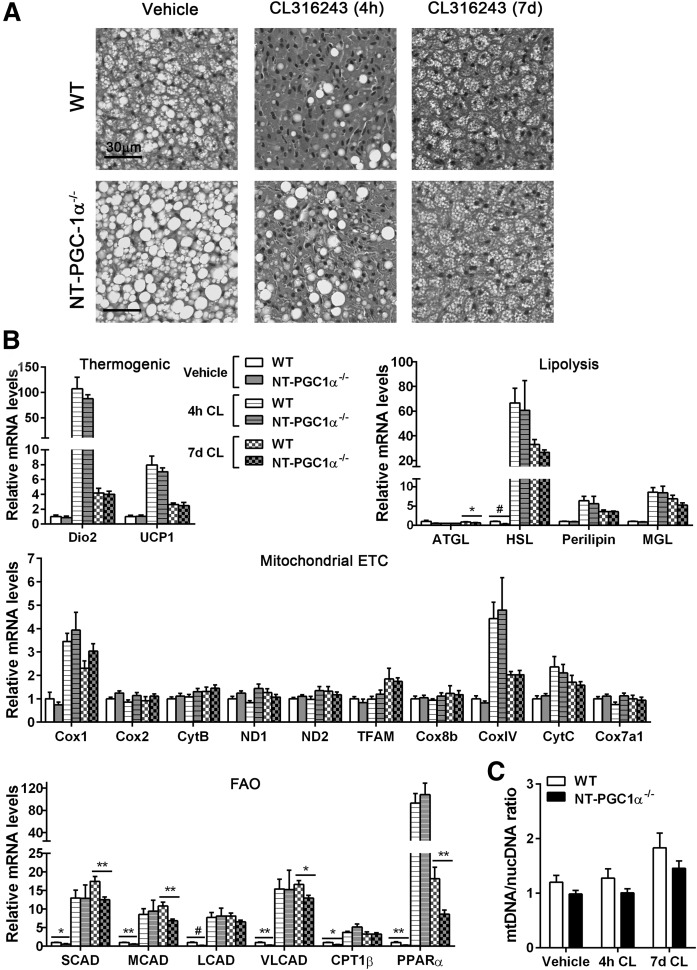
NT-PGC-1α is required for FAO gene expression during the prolonged activation of BAT by CL316243
Despite the diminished FAO at thermoneutrality, NT-PGC-1α−/− BAT underwent relatively normal remodeling in response to β3-AR stimulation (Fig. 6A). Large lipid droplets rapidly disappeared after 4 h of stimulation with CL316243. Prolonged stimulation of β3-AR increased brown adipocytes containing multilocular lipid droplets in both WT and NT-PGC-1α−/− BAT (Fig. 6A). Acute and prolonged stimulation with CL316243 induced similar changes in expression of thermogenic, lipolytic, and mitochondrial ETC genes in WT and NT-PGC-1α−/− BAT (Fig. 6B), indicating that NT-PGC-1α is not required to regulate these genes. It might be that PGC-1α is sufficient to mediate CL316243-induced expression of thermogenic, lipolytic, and mitochondrial ETC genes. In agreement with gene-expression data, CL316243-induced mitochondrial biogenesis was comparable between WT and NT-PGC-1α−/− BAT (Fig. 6C). Despite the lower basal expression of FAO genes in NT-PGC-1α−/− BAT, acute induction of FAO gene expression in response to β3-AR stimulation was comparable between WT and NT-PGC-1α−/− BAT (Fig. 6B). However, FAO gene transcript levels were significantly lower in NT-PGC-1α−/− BAT stimulated with CL316243 for 7 days (Fig. 6B). These results suggest that PGC-1α is sufficient for acute induction of FAO gene expression in response to β3-AR stimulation, but both PGC-1α and NT-PGC-1α are required for the maintenance of FAO gene expression during the prolonged BAT activation.
Fig. 6.
Effects of NT-PGC-1α ablation on CL316243-induced remodeling of BAT. A: Representative H&E images of BAT from WT and NT-PGC-1α−/− mice. Mice (n = 7 or 8 per group) housed at 28°C for 3 weeks were injected with vehicle or CL316243 for 4 h or 7 days. Scale bars, 30 µm. B: qPCR analysis of BAT gene expression (n = 7 or 8 per group). (C) qPCR analysis of mitochondrial biogenesis. The relative ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nucDNA) was analyzed (n = 7 or 8 per group). All data are presented as mean ± SEM. *P < 0.05; **P < 0.01; #P < 0.001 determined by Student’s t-test. CL, CL316243.
NT-PGC-1α deficiency had no effect on CL316243-induced browning of inguinal WAT (supplemental Fig. S3). WT and NT-PGC-1α−/− inguinal WAT (IWAT) exhibited similar transcriptional changes with upregulation of brown adipocyte-specific genes (DIO2, UCP1, and CIDEA), mitochondrial FAO and ETC genes, and lipolytic genes (supplemental Fig. S3).
DISCUSSION
BAT thermogenic activity is tightly regulated to maintain a constant core body temperature in spite of changes in ambient temperature. Transcriptional coactivator PGC-1α and its splice variant NT-PGC-1α mediate temperature-dependent regulation of adaptive thermogenesis by coordinating the transcriptional response to changes in ambient temperature. Mice lacking both PGC-1α and NT-PGC-1α are unable to produce heat in response to cold due to impaired induction of UCP1 and mitochondrial genes encoding the enzymes involved in mitochondrial FAO and electron transport in BAT (20). In contrast, mice selectively lacking full-length PGC-1α are cold-tolerant because NT-PGC-1α can mediate cold-induced activation of the aforementioned genes in BAT (23). The present study using NT-PGC-1α−/− mice further reveals that NT-PGC-1α plays a particularly important role in regulating FAO gene expression in response to changes in ambient temperature. NT-PGC-1α is required for the maintenance of FAO gene expression in quiescent and fully activated BAT, whereas it is dispensable for acute induction of FAO gene expression in response to cold/β3-AR stimulation.
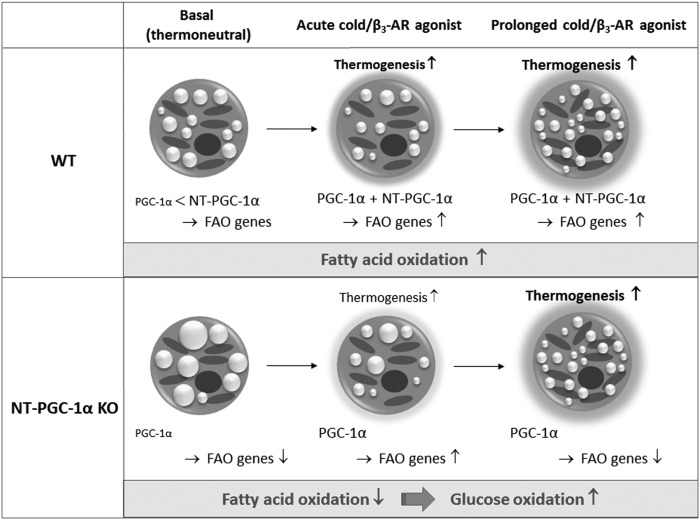
PGC-1α is a short-lived protein, and its stability and activity are highly elevated by posttranslational modifications induced by cold/β3-AR signaling (36). In contrast, NT-PGC-1α is relatively stable and undergoes a cold/β3-AR-induced increase in its nuclear localization (19, 25). We speculate that NT-PGC-1α primarily mediates FAO gene expression in the thermoneutral condition where PGC-1α stability and activity are low (Fig. 7). During the acute activation of BAT by cold/β3-AR agonist, cold/β3-AR-activated PGC-1α is likely sufficient to upregulate FAO genes without NT-PGC-1α. However, two isoforms are required for the maintenance of FAO gene expression in the prolonged cold/β3-AR-stimulated condition where maximal mitochondrial FAO is needed. Although we cannot exclude the possibility that this defect in FAO gene expression results from reduced levels of total PGC-1α proteins, our observation of this temperature-dependent effect of NT-PGC-1α ablation on FAO gene expression is more likely to support that the relative contribution of PGC-1α and NT-PGC-1α to FAO gene expression depends on ambient temperature. FAO plays an essential role in regulating UCP-mediated heat production in BAT. Therefore, regulation of FAO gene expression by two PGC-1α and NT-PGC-1α proteins, whose activities are differentially regulated by ambient temperature, may be a way for brown adipocytes to efficiently adapt their FA-oxidizing capacity to changes in ambient temperature.
Fig. 7.
Model illustrating the relative contribution of NT-PGC-1α to the FAO capacity in BAT. Our data suggest that NT-PGC-1α primarily mediates FAO gene expression in BAT in the thermoneutral condition where PGC-1α stability and activity are low. In response to cold/β3-AR agonist, cold/β3-AR-activated PGC-1α is sufficient to acutely upregulate FAO genes without NT-PGC-1α. However, two isoforms are required for the maintenance of FAO gene expression in the prolonged cold/β3-AR-stimulated condition where maximal mitochondrial FAO is needed. Diminished mitochondrial FAO in NT-PGC-1α−/− BAT induces enhanced oxidation of pyruvate in mitochondria.
Despite our previous findings that NT-PGC-1α upregulates many mitochondrial genes involved in thermogenesis and respiration in PGC-1α−/− brown adipocytes (21, 22, 37), NT-PGC-1α deficiency did not alter expression of thermogenic and electron-transport chain genes in BAT. Transcriptional coactivator PGC-1β, which is one of the PGC-1 family but not cold-inducible, primarily regulates basal mitochondrial biogenesis and respiration in BAT (20, 38), whereas PGC-1α and NT-PGC-1α play a relatively more important role in cold-induced increase in mitochondrial biogenesis and respiration (20, 38). We found that PGC-1β gene expression is similar between WT and NT-PGC-1α−/− BAT (Fig. 2C). Thus, PGC-1β might be sufficient to regulate basal expression of mitochondrial genes involved in biogenesis and respiration in the thermoneutral condition, and PGC-1α might be sufficient to mediate acute induction of mitochondrial gene expression in the cold/β3-AR-stimulated condition. Our finding that NT-PGC-1α deficiency specifically downregulates FAO gene expression can be in part attributable to the reduced expression of PPARα in NT-PGC-1α−/− BAT. PPARα predominantly activates genes involved in FA uptake and β-oxidation, rather than in mitochondrial biogenesis, TCA cycle, or respiration (28). We previously showed that NT-PGC-1α interacts with and coactivates PPARα (19) and that NT-PGC-1α not only upregulates PPARα gene expression but also enhances PPARα-mediated transcription (19, 21–23). Together, the lack of NT-PGC-1α-mediated expression and activation of PPARα might be responsible for the downregulated expression of FAO genes in NT-PGC-1α−/− BAT.
Consistent with gene-expression changes, NT-PGC-1α−/− BAT exhibited the diminished capacity to oxidize FAs for mitochondrial respiration. More interestingly, PGC-1α−/− BAT displayed higher mitochondrial respiration when pyruvate was used as a respiratory substrate, indicating a shift in fuel utilization toward glucose. We speculate that decreases in FA-derived acetyl CoA, NADH, and inhibitory PDK4 relieve inactivation of PDH, thus enhancing pyruvate oxidation in NT-PGC-1α−/− BAT mitochondria (Fig. 7). In agreement with this finding, NT-PGC-1α−/− mice exhibited a greater reliance on carbohydrates in association with an increase in food intake. Interestingly, we previously showed that mice selectively lacking full-length PGC-1α have a higher reliance on fat (22, 23). Mechanistically, NT-PGC-1α overexpression in full-length PGC-1α-deficient BAT led to enhanced FAO. Given that these previous studies were carried out at room temperature, further experiments at thermoneutrality will be needed to more accurately compare the phenotypes. Taken together, the opposing phenotypes in NT-PGC-1α loss-of-function versus overexpression models further provide strong evidence for the particularly important role of NT-PGC-1α in FAO in BAT.
Because PGC-1α and NT-PGC-1α are coexpressed and regulate many common target genes in BAT, their relative contributions to adaptive thermogenesis have been unclear. Our findings demonstrate that the relative contribution of PGC-1α and NT-PGC-1α to FAO gene expression depends on ambient temperature. Having two PGC-1α and NT-PGC-1α proteins whose activities are differentially regulated by ambient temperature may allow brown adipocytes to efficiently adapt their FA-oxidizing capacity to changes in ambient temperature.
Supplementary Material
Acknowledgments
The authors thank Dr. Tom Gettys (Pennington Biomedical Research Center) for kindly providing anti-PGC-1α antibodies and Ms. Cindi Tramonte for administrative support.
Footnotes
Abbreviations:
- AR
- adrenergic receptor
- BAT
- brown adipose tissue
- EE
- energy expenditure
- ETC
- electron transport chain
- FAO
- FA oxidation
- IWAT
- inguinal white adipose tissue
- KHB
- Krebs-Henseleit buffer
- NT-PGC1α
- N-terminal PPAR γ coactivator 1-α
- PDH
- pyruvate dehydrogenase
- qPCR
- quantitative real-time PCR
- RER
- respiratory exchange ratio
- TAG
- triacylglycerol
- UCP1
- uncoupling protein 1
- VWAT
- visceral white adipose tissue
- WAT
- white adipose tissue
This work was supported by National Institutes of Health Grant NIH R01DK104748 (J.S.C.) and used Cell Biology and Bioimaging and Genomics Core facilities that are supported in part by National Institutes of Health Center of Biomedical Research Excellence Grant NIH8 1P30GM118430-01 and National Institutes of Health Nutrition Obesity Research Center Grant NIH P30-DK072476. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Cannon B., and Nedergaard J.. 2004. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84: 277–359. [DOI] [PubMed] [Google Scholar]
- 2.Nedergaard J., Golozoubova V., Matthias A., Asadi A., Jacobsson A., and Cannon B.. 2001. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim. Biophys. Acta. 1504: 82–106. [DOI] [PubMed] [Google Scholar]
- 3.Golozoubova V., Hohtola E., Matthias A., Jacobsson A., Cannon B., and Nedergaard J.. 2001. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 15: 2048–2050. [DOI] [PubMed] [Google Scholar]
- 4.Rothwell N. J., and Stock M. J.. 1979. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 281: 31–35. [DOI] [PubMed] [Google Scholar]
- 5.Himms-Hagen J. 1990. Brown adipose tissue thermogenesis: interdisciplinary studies. FASEB J. 4: 2890–2898. [PubMed] [Google Scholar]
- 6.Ouellet V., Labbe S. M., Blondin D. P., Phoenix S., Guerin B., Haman F., Turcotte E. E., Richard D., and Carpentier A. C.. 2012. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 122: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Lans A. A., Hoeks J., Brans B., Vijgen G. H., Visser M. G., Vosselman M. J., Hansen J., Jorgensen J. A., Wu J., Mottaghy F. M., et al. . 2013. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 123: 3395–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X. X., Lewin D. A., Forrest W., and Adams S. H.. 2002. Cold elicits the simultaneous induction of fatty acid synthesis and beta-oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 16: 155–168. [DOI] [PubMed] [Google Scholar]
- 9.Mottillo E. P., Balasubramanian P., Lee Y. H., Weng C., Kershaw E. E., and Granneman J. G.. 2014. Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. J. Lipid Res. 55: 2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Gurmaches J., Tang Y., Jespersen N. Z., Wallace M., Martinez Calejman C., Gujja S., Li H., Edwards Y. J. K., Wolfrum C., Metallo C. M., et al. . 2017. Brown fat AKT2 is a cold-induced kinase that stimulates ChREBP-mediated de novo lipogenesis to optimize fuel storage and thermogenesis. Cell Metab 27: 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., and Lass A.. 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 50: 3–21. [DOI] [PubMed] [Google Scholar]
- 12.Holm C., Fredrikson G., Cannon B., and Belfrage P.. 1987. Hormone-sensitive lipase in brown adipose tissue: identification and effect of cold exposure. Biosci. Rep. 7: 897–904. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D., Gong D., Londos C., Yamaguchi T., Holm C., et al. . 2009. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 284: 32116–32125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divakaruni A. S., Humphrey D. M., and Brand M. D.. 2012. Fatty acids change the conformation of uncoupling protein 1 (UCP1). J. Biol. Chem. 287: 36845–36853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack J. G., and Denton R. M.. 1977. Evidence that fatty acid synthesis in the interscapular brown adipose tissue of cold-adapted rats is increased in vivo by insulin by mechanisms involving parallel activation of pyruvate dehydrogenase and acetyl-coenzyme A carboxylase. Biochem. J. 166: 627–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trayhurn P. 1981. Fatty acid synthesis in mouse brown adipose tissue. The influence of environmental temperature on the proportion of whole-body fatty acid synthesis in brown adipose tissue and the liver. Biochim. Biophys. Acta. 664: 549–560. [DOI] [PubMed] [Google Scholar]
- 17.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., and Spiegelman B. M.. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 92: 829–839. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., et al. . 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 98: 115–124. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Huypens P., Adamson A. W., Chang J. S., Henagan T. M., Lenard N. R., Burk D., Klein J., Perwitz N., Shin J., et al. . 2009. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1{alpha}. J. Biol. Chem. 284: 32813–32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jager S., Vianna C. R., Reznick R. M., et al. . 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 119: 121–135. [DOI] [PubMed] [Google Scholar]
- 21.Kim J., Fernand V. E., Henagan T. M., Shin J., Huypens P., Newman S., Gettys T. W., and Chang J. S.. 2016. Regulation of brown and white adipocyte transcriptome by the transcriptional coactivator NT-PGC-1alpha. PLoS One. 11: e0159990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun H. J., Joshi Y., Patil Y., Noland R. C., and Chang J. S.. 2014. NT-PGC-1alpha activation attenuates high-fat diet-induced obesity by enhancing brown fat thermogenesis and adipose tissue oxidative metabolism. Diabetes. 63: 3615–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang J. S., Fernand V., Zhang Y., Shin J., Jun H. J., Joshi Y., and Gettys T. W.. 2012. NT-PGC-1alpha protein is sufficient to link beta3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. J. Biol. Chem. 287: 9100–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandoski M., Wassarman K. M., and Martin G. R.. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7: 148–151. [DOI] [PubMed] [Google Scholar]
- 25.Chang J. S., Huypens P., Zhang Y., Black C., Kralli A., and Gettys T. W.. 2010. Regulation of NT-PGC-1alpha subcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. J. Biol. Chem. 285: 18039–18050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huynh F. K., Green M. F., Koves T. R., and Hirschey M. D.. 2014. Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 542: 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruas J. L., White J. P., Rao R. R., Kleiner S., Brannan K. T., Harrison B. C., Greene N. P., Wu J., Estall J. L., Irving B. A., et al. . 2012. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 151: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finck B. N., Lehman J. J., Leone T. C., Welch M. J., Bennett M. J., Kovacs A., Han X., Gross R. W., Kozak R., Lopaschuk G. D., et al. . 2002. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 109: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leone T. C., Weinheimer C. J., and Kelly D. P.. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA. 96: 7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zechner R., Zimmermann R., Eichmann T. O., Kohlwein S. D., Haemmerle G., Lass A., and Madeo F.. 2012. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batenburg J. J., and Olson M. S.. 1976. Regulation of pyruvate dehydrogenase by fatty acid in isolated rat liver mitochondria. J. Biol. Chem. 251: 1364–1370. [PubMed] [Google Scholar]
- 32.Cadoudal T., Distel E., Durant S., Fouque F., Blouin J. M., Collinet M., Bortoli S., Forest C., and Benelli C.. 2008. Pyruvate dehydrogenase kinase 4: regulation by thiazolidinediones and implication in glyceroneogenesis in adipose tissue. Diabetes. 57: 2272–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jesus L. A., Carvalho S. D., Ribeiro M. O., Schneider M., Kim S. W., Harney J. W., Larsen P. R., and Bianco A. C.. 2001. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 108: 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muzzin P., Revelli J. P., Kuhne F., Gocayne J. D., McCombie W. R., Venter J. C., Giacobino J. P., and Fraser C. M.. 1991. An adipose tissue-specific beta-adrenergic receptor. Molecular cloning and down-regulation in obesity. J. Biol. Chem. 266: 24053–24058. [PubMed] [Google Scholar]
- 35.Granneman J. G., Lahners K. N., and Chaudhry A.. 1991. Molecular cloning and expression of the rat beta 3-adrenergic receptor. Mol. Pharmacol. 40: 895–899. [PubMed] [Google Scholar]
- 36.Fernandez-Marcos P. J., and Auwerx J.. 2011. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93: 884S–890S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang J. S., and Ha K.. 2017. An unexpected role for the transcriptional coactivator isoform NT-PGC-1alpha in the regulation of mitochondrial respiration in brown adipocytes. J. Biol. Chem. 292: 9958–9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., and Spiegelman B. M.. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3: 333–341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.