Abstract
Background
The World Health Organization recommends universal drug susceptibility testing for Mycobacterium tuberculosis complex to guide treatment decisions and improve outcomes. We assessed whether DNA sequencing can accurately predict antibiotic susceptibility profiles for first-line anti-tuberculosis drugs.
Methods
Whole-genome sequences and associated phenotypes to isoniazid, rifampicin, ethambutol and pyrazinamide were obtained for isolates from 16 countries across six continents. For each isolate, mutations associated with drug-resistance and drug-susceptibility were identified across nine genes, and individual phenotypes were predicted unless mutations of unknown association were also present. To identify how whole-genome sequencing might direct first-line drug therapy, complete susceptibility profiles were predicted. These were predicted to be pan-susceptible if predicted susceptible to isoniazid and to other drugs, or contained mutations of unknown association in genes affecting these other drugs. We simulated how negative predictive value changed with drug-resistance prevalence.
Results
10,209 isolates were analysed. The greatest proportion of phenotypes were predicted for rifampicin (9,660/10,130; (95.4%)) and the lowest for ethambutol (8,794/9,794; (89.8%)). Isoniazid, rifampicin, ethambutol and pyrazinamide resistance was correctly predicted with 97.1%, 97.5% 94.6% and 91.3% sensitivity, and susceptibility with 99.0%, 98.8%, 93.6% and 96.8% specificity, respectively. 5,250 (89.5%) drug profiles were correctly predicted for 5,865/7,516 (78.0%) isolates with complete phenotypic profiles. Among these, 3,952/4,037 (97.9%) predictions of pan-susceptibility were correct. The negative predictive value for 97.5% of simulated drug profiles exceeded 95% where the prevalence of drug-resistance was below 47.0%.
Conclusions
Phenotypic testing for first-line drugs can be phased down in favour of DNA sequencing to guide anti- tuberculosis drug therapy.
Mycobacterium tuberculosis killed more people than any other pathogen in 2016, when over 10 million active cases were estimated, and 1.7 million patients died.1 In 2014, the World Health Organization (WHO) set a target to ‘END TB’ by 2035, acknowledging that success depends on the development of better preventative, diagnostic and therapeutic interventions. The global emergence of antimicrobial resistance poses a major challenge. Despite a call for universal access to drug susceptibility testing to direct individualised therapies, high costs and skills shortages mean it is unavailable in many countries with greatest need. Consequently, only 22% of an estimated 600,000 patients requiring treatment for multidrug-resistant tuberculosis were diagnosed and treated in 2016,1 facilitating the onward transmission of multidrug-resistant strains.2
The Xpert MTB/RIF (Cepheid, Sunnyvale, California, USA) assay has partially eased the global diagnostic need. It uses polymerase chain reaction technology to identify both M. tuberculosis complex and mutations in the rpoB gene (predictive of multidrug resistance) directly from clinical samples.3 However, as it targets only a few potential resistance-conferring mutations, antimicrobial susceptibility cannot be reliably inferred from a negative result.4 To direct individualised therapies, a diagnostic assay is needed to determine which drugs to give, in addition to which to avoid.
Advances in whole-genome sequencing mean it is now the most promising solution to the need for universal drug susceptibility testing. It is faster, more scalable, and likely to become cheaper than phenotypic testing.5 As the number of genomic sites whole-genome sequencing covers are virtually unrestricted, it should be possible to infer M. tuberculosis antimicrobial susceptibility from the absence of resistance-conferring mutations.6 Here we assess how well this performs for first-line anti- tuberculosis drugs, considering WHO target product profiles for new molecular assays,7 and whether whole-genome sequencing can be used to accurately direct anti-tuberculosis therapy.
Methods
Sample selection
Collections of M. tuberculosis complex isolates unenriched for resistance and largely sequenced prospectively for routine diagnostic reasons, or for disease surveillance, were included from Germany, Italy, the Netherlands and the UK. Collections enriched for antimicrobial resistance, were included from across six continents (Table1, Supplement S1). Analyses of both the unenriched and complete collection were planned.
Table 1. Number of isolates by country and drug resistance profile.
| Country of sample origin | Time period of isolation | Enriched for resistance | Susceptible to all 4 drugs | Susceptible to 3 drugs, with missing pyrazinamide result | Isoniazid resistant, rifampicin susceptible | Isoniazid susceptible, rifampicin resistant | Isoniazid resistant, rifampicin resistant | Other pattern | Total |
|---|---|---|---|---|---|---|---|---|---|
| Australia | 2006-2016 | Yes | 0 | 0 | 4 | 0 | 38 | 0 | 42 |
| Belgium | 2007-2015 | Yes | 121 | 0 | 2 | 0 | 97 | 14 | 234 |
| Canada | 2003-2014 | Yes | 11 | 1,118 | 164 | 14 | 24 | 12 | 1343 |
| China | 2009-2012 | Yes | 0 | 44 | 0 | 0 | 236 | 0 | 280 |
| Germany | 1998-2015 | No | 248 | 0 | 9 | 1 | 13 | 2 | 273 |
| Italy | 2008-2016 | Yes and No* | 82 | 1 | 9 | 0 | 132 | 2 | 226 |
| Netherlands | 1993-2016 | No | 420 | 42 | 24 | 1 | 149 | 31 | 667 |
| Pakistan | 2014-2015 | Yes | 47 | 5 | 11 | 6 | 345 | 1 | 415 |
| Peru | 1997-2009 | Yes | 24 | 12 | 49 | 18 | 199 | 13 | 315 |
| Russia | 2008-2010 | Yes | 282 | 0 | 116 | 15 | 407 | 22 | 842 |
| Serbia | 2008-2014 | Yes | 0 | 0 | 0 | 0 | 105 | 0 | 105 |
| South Africa | 2012-2014 | Yes | 593 | 11 | 37 | 69 | 151 | 130 | 991 |
| Spain | 2013-2015 | Yes | 45 | 3 | 5 | 2 | 8 | 1 | 64 |
| Swaziland | 2009-2010 | Yes | 2 | 130 | 14 | 4 | 116 | 7 | 273 |
| Thailand | 1998-2013 | Yes | 0 | 53 | 7 | 4 | 188 | 0 | 252 |
| UK | 2009-2017 | Yes and No* | 3,036 | 82 | 167 | 6 | 442 | 154 | 3,887 |
| Total | 4911 | 1501 | 618 | 140 | 2650 | 389 | 10209 |
More than one collection was derived from Italy and the UK, some enriched and some not enriched for resistance. See supplement for details.
Sequencing
Isolates were sequenced on Illumina platforms and reads processed by the Public Health England bioinformatics pipeline at Genomics England,8 as described.6 Reads were mapped to the pan- susceptible M. tuberculosis reference genome (Genbank NC_000962.2) using Stampy (v.1.0.17)9, with repetitive regions masked. SAMtools mpileup10 (v.0.1.18) made variant-calls based on a minimum depth of 5X and at least one read on each strand. Mixed-calls were assigned where minority alleles composed >10% of read depth. Insertions and deletions were determined using Cortex (v.1.0.5.21).11
Drug susceptibility testing and prediction
Phenotypic drug susceptibility testing was performed locally using MGIT 960 (Becton Dickinson, New Jersey, USA), 7H10 or Löwenstein-Jensen agar, or by microscopic-observation drug- susceptibility (MODS), with method-specific critical concentrations for isoniazid (MGIT 0.1-0.2μg/mL; Agar 0.2μg/mL; MODS 0.4μg/mL), rifampicin (MGIT 1.0μg/mL; 40μg/mL Agar), ethambutol (MGIT 5.0μg/mL; Agar 0.2μg/mL), and pyrazinamide (100μg/mL). Not all laboratories routinely tested all agents (S1). Genotypic predictions were based on mutations in, or upstream of, genes associated with resistance to isoniazid (ahpC, inhA, fabG1, katG), rifampicin (rpoB), ethambutol (embA, embB, embC), and pyrazinamide (pncA).6 A knowledgebase of mutations predicting antimicrobial resistance, or not, was informed by (i) the molecular targets of WHO-recommended line-probe assays (MTBDRplus, MTBDRsl v1.0, HAIN Lifesciences, Germany), (ii) a systematic literature review,12 (iii) the CDC, Atlanta, USA, panel and (iv) two recent studies, with no isolates in common with this study (S2),6,13 of which one became available after this study commenced.13
Isolates containing resistance-mutations were predicted phenotypically resistant, whereas isolates containing only wild-type sequence, phylogenetic mutations,6 or mutations considered consistent with susceptibility, were predicted susceptible. Predictions were withheld for isolates containing mutations affecting target genes but of unknown association, or where no nucleotide-call could be determined at a resistance-associated site. In these circumstances, the genotype was reported ‘unknown’ or ‘failed’, respectively. Using phenotypic results as a gold-standard, sensitivity, specificity, negative and positive predictive value were calculated for the correct assignment of susceptibility or resistance. Primary analyses excluded phenotypes without a prediction.
Laboratory error was assumed where three or more phenotypes were discordant with an isolate’s genotype, or where susceptible phenotypes were recorded despite the presence of high-level resistance katG S315T mutations for isoniazid, or rpoB S450L mutations for rifampicin.14 Such isolates were excluded from further analysis.
Analysis was performed using STATA (Texas, USA, v13.1). No institutional review board approval was required except in Thailand, it was granted through Mahidol University (Si029/2557).
The study was first designed by TMW,TEAP,DWC, with subsequent contributions from others (supplement). Data were gathered at participating centres. Initial analysis was performed by TMW,TEAP,ASW,ZI,MH,SL,DW,PF,PM with later input from others (supplement). TMW wrote the first draft. TMW vouches for the analysis and had full access to the data; all authors agreed to publication.
Results
10,290 isolates were available for the study. 81 (0.8%) were excluded due to likely laboratory error. 10,209 isolates remained, for which full first-line phenotypic profiles were available for 7,516 (73.6%), and partial profiles for the remainder. 4,911 (48.1%) isolates were phenotypically susceptible to all drugs (Table 1).
For each isolate, the complete sequence of nine genes and their promoter regions was interrogated to make genotypic predictions of each available phenotypic result. Predictions could be made for 8,405/8,976 (93.6%) resistant and 26,879/28,746 (93.5%) susceptible phenotypes. The remainder contained uncharacterised mutations, or missing key nucleotide calls. For isoniazid and rifampicin, ethambutol and pyrazinamide, sensitivity (proportion of resistant phenotypes predicted resistant) was 97.1%, 97.5%, 94.6% and 91.3%, and specificity (proportion of susceptible phenotypes predicted susceptible) was 99.0%, 98.8%, 93.6% and 96.8%, respectively. By comparison, an in-silico prediction of the results that would have been obtained from WHO-recommended molecular assays (Xpert MTB/RIF, MTBDRplus, MTBDRsl v1.0) had a significantly lower sensitivity than whole-genome sequencing for isoniazid, rifampicin and ethambutol (p<0.001), but greater specificity for isoniazid and ethambutol (p<0.001) (Table 2a,b).
Table 2. Prediction of individual drug phenotypes.
| Resistant phenotype, n (%) | Susceptible phenotype, n (%) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | S | U | F | Total | R | S | U | F | Total | Sensitivity of predictions, %(95% CI) | Specificity of predictions, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) | Sensitivity (all*), % | Specificity (all*), % | No genotypic prediction made, % | Resistance prevalence (all), % | ||
| (a) All isolates | |||||||||||||||||||
| Isoniazid | 3067 | 90 | 93 | 44 | 3294 | 65 | 6313 | 215 | 117 | 6710 | 97.1 (96.5-97.7) | 99.0 (98.7-99.2) | 97.9 (97.4-98.4) | 98.6 (98.3-98.9) | 93.1 | 94.1 | 4.7 | 32.9 | |
| Rifampicin | 2743 | 69 | 7 | 84 | 2903 | 85 | 6763 | 232 | 147 | 7227 | 97.5 (96.9-98.1) | 98.8 (98.5-99.0) | 97.0 (96.3-97.6) | 99.0 (98.7-99.2) | 94.5 | 93.6 | 4.6 | 28.7 | |
| Ethambutol | 1410 | 81 | 94 | 55 | 1640 | 468 | 6835 | 781 | 70 | 8154 | 94.6 (93.3-95.7) | 93.6 (93.0-94.1) | 75.1 (73.0-77.0) | 98.8 (98.5-99.1) | 86.0 | 83.8 | 10.2 | 16.7 | |
| Pyrazinamide | 863 | 82 | 117 | 77 | 1139 | 204 | 6146 | 197 | 108 | 6655 | 91.3 (89.3-93.0) | 96.8 (96.3-97.2) | 80.9 (78.4-83.2) | 98.7 (98.4-99.0) | 75.8 92.4 | 6.4 | 14.6 | ||
| (b) In silico prediction of performance of MTB/RIF Xpert and HAIN MTBDRplus/MTBDRsl line-probe assays for all isolates | |||||||||||||||||||
| Isoniazid | 2886 | 355 | 53 | 3294 | 27 | 6675 | 8 | 6710 | 89.0 (87.9-90.1)† | 99.6 (99.4-99.7)† | 99.1 (98.7-99.4)† | 95.0 (94.4-95.5)† | 0.6 32.9 | ||||||
| Rifampicin | 2669 | 143 | 91 | 2903 | 129 | 6826 | 272 | 7227 | 94.9 (94.0-95.7)†98.1 | (97.8-98.4)‡ | 95.4 (94.5-96.1)‡ | 97.9 (97.6-98.3)† | 3.6 | 28.7 | |||||
| Ethambutol | 961 | 641 | 38 | 1640 | 241 | 7895 | 18 | 8154 | 60.0 (57.5-62.4)† | 97.0 (96.6-97.4)† | 80.0 (77.6-82.2)‡ | 92.5 (91.9-93.0)† | 0.6 | 16.7 | |||||
| Pyrazinamide | |||||||||||||||||||
| (c) Collections from Germany, Italy, the Netherlands and the UK, unenriched for resistance | |||||||||||||||||||
| Isoniazid | 314 | 8 | 9 | 4 | 335 | 15 | 3770 | 104 | 90 | 3979 | 97.5 (95.2-98.9) | 99.6 (99.3-99.8)† | 95.4 (92.6-97.4)‡ | 99.8 (99.6-99.9)† | 93.7 | 94.7 | 4.8 | 7.8 | |
| Rifampicin | 126 | 0 | 0 | 9 | 135 | 31 | 3958 | 103 | 116 | 4208 | 100.0 (97.1-100.0) | 99.2 | (98.9-99.5)§ | 80.3 (73.2-86.2)† | 100.0 (99.9-100.0)† | 93.3 | 94.1 | 5.2 | 3.1 |
| Ethambutol | 72 | 1 | 0 | 0 | 73 | 47 | 3711 | 458 | 36 | 4252 | 98.6 (92.6-100.0) | 98.7 (98.3-99.1)† | 60.5 (51.1-69.3)† | 100.0 (99.8-100.0)† | 98.6 | 87.3 | 11.4 | 1.7 | |
| Pyrazinamide | 109 | 6 | 4 | 6 | 125 | 30 | 4003 | 14 | 58 | 4105 | 94.8 (89.0-98.1) | 99.3 (98.9-99.5)† | 78.4 (70.6-84.9) | 99.9 (99.7-99.9)† | 87.2 | 97.5 | 1.9 | 3.0 | |
| (d) In silico prediction of performance of MTB/RIF Xpert and HAIN MTBDRplus/MTBDRsl line-probe assays for collections unenriched for resistance | |||||||||||||||||||
| Isoniazid | 295 | 36 | 4 | 335 | 10 | 3965 | 4 | 3979 | 89.1 (85.3-92.3)† | 99.7 (99.5-99.9) | 96.7 (94.1-98.4) | 99.1 (98.8-99.4)† | 0.2 | ||||||
| Rifampicin | 114 | 11 | 10 | 135 | 22 | 3957 | 229 | 4208 | 91.2 (84.8-95.6)† | 99.4 (99.2-99.7) | 83.8 (76.5-89.6) | 99.7 (99.5-99.9)† | 5.5 | ||||||
| Ethambutol | 57 | 16 | 0 | 73 | 29 | 4220 | 3 | 4252 | 78.1 (66.9-86.9)† | 99.3 (99.0-99.5)§ | 66.3 (55.3-76.1) | 99.6 (99.4-99.8)† | 0.1 | ||||||
| Pyrazinamide | |||||||||||||||||||
PPV = Positive Predictive Value; NPV = Negative Predictive Value; R=resistant; S=susceptible; U=mutation of unknown association present; F=genotypic prediction failed due to missing data around a genomic resistance locus; All % results based on R/S genotypic predictions only, excluding U and F except where * for which denominator includes R, S, U and F. †p≤0.001 , ‡p≤0.01, and §p≤0.05 comparing sensitivity, specificity, NPV and PPV for each drug for (b) and (c) against (a), and comparing (d) against (c); p>0.05 for all results not marked †, ‡ or §. In silico predictions of resistance for Xpert and HAIN assays were based on the presence of non-wild type sequence within the genomic regions interrogated by these assays. 'F' was reported in the presence of minority alleles at relevant sites, just as for WGS predictions.
The negative predictive value (proportion of concordant susceptible predictions) was over 98.5% for all four drugs. Although dependent on prevalence, this also varied with isolates’ background phenotypic profiles. For example, at 20% prevalence of pyrazinamide resistance, the expected negative predictive value for pyrazinamide was 93.6% and 99.0% for isolates susceptible and resistant to the other three drugs, respectively (Table 3, S3).
| Phenotypic profiles | R | S | U | F | Total | R | S | U | F | Total | Prevalence of resistance among each of the listed drug profiles, % | Sensitivity, % | Specificity, % | PPV, % | NPV,% | Expected NPV at given prevalence of resistance based on simulations, % (95% CI)* | Calculated NPV at 20% prevalence of resistance, % (see table S3) | Calculated NPV at 40% prevalence of resistance, % (see table S3) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isoniazid | -SSS | 391 | 30 | 18 | 12 | 451 | 21 | 4,653 | 133 | 104 | 4,911 | 8.4 | 93 | 100 | 95 | 99.4 | 99.3-100 | 98.2 | 95.4 |
| -RSS | 459 | 21 | 20 | 6 | 506 | 7 | 85 | 5 | 1 | 98 | 83.8 | 96 | 92 | 98 | 80.2 | 83.5-100 | 98.8 | 96.9 | |
| -RRS | 424 | 3 | 13 | 4 | 444 | 2 | 2 | 2 | 0 | 6 | 98.7 | 99 | 50 | 100 | 40.0 | 73.7-85.6 | 99.6 | 99.1 | |
| -SRS | 24 | 4 | 1 | 0 | 29 | 0 | 10 | 1 | 0 | 11 | 72.5 | 86 | 100 | 100 | 71.4 | 90.5-95.6 | 96.6 | 91.3 | |
| -SSR | 24 | 1 | 2 | 1 | 28 | 0 | 95 | 6 | 3 | 104 | 21.2 | 96 | 100 | 100 | 99.0 | 98.5-99.7 | 99 | 97.4 | |
| -RRR | 662 | 3 | 11 | 4 | 680 | 0 | 0 | 0 | 0 | 0 | 100.0 | 100 | . | 100 | 0.0 | 73.7-85.6 | n/a | n/a | |
| -RSR | 217 | 3 | 5 | 5 | 230 | 0 | 3 | 0 | 0 | 3 | 98.7 | 99 | 100 | 100 | 50.0 | 73.7-85.6 | 99.7 | 99.1 | |
| -SRR | 13 | 0 | 0 | 2 | 15 | 0 | 0 | 0 | 0 | 0 | 100.0 | 100 | . | 100 | . | 73.7-85.6 | n/a | n/a | |
| Rifampicin | S-SS | 74 | 16 | 0 | 8 | 98 | 30 | 4,632 | 126 | 123 | 4,911 | 2.0 | 82 | 99 | 71 | 99.7 | 99.3-100 | 95.7 | 89.3 |
| S-RS | 6 | 0 | 0 | 0 | 6 | 1 | 9 | 1 | 0 | 11 | 35.3 | 100 | 90 | 86 | 100.0 | 97.8-99.5 | 100 | 100 | |
| S-SR | 1 | 2 | 0 | 0 | 3 | 0 | 100 | 3 | 1 | 104 | 2.8 | 33 | 100 | 100 | 98.0 | 99.3-100 | 85.7 | 69.2 | |
| S-RR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | . | . | . | . | . | . | n/a | n/a | |
| R-SS | 464 | 20 | 1 | 21 | 506 | 18 | 424 | 3 | 6 | 451 | 52.9 | 96 | 96 | 96 | 95.5 | 95.8-98.6 | 98.9 | 97.2 | |
| R-RS | 424 | 7 | 2 | 11 | 444 | 4 | 25 | 0 | 0 | 29 | 93.9 | 98 | 86 | 99 | 78.1 | 76.2-86.6 | 99.5 | 98.8 | |
| R-SR | 218 | 4 | 0 | 8 | 230 | 7 | 20 | 0 | 1 | 28 | 89.1 | 98 | 74 | 97 | 83.3 | 77.9-87.9 | 99.4 | 98.4 | |
| R-RR | 665 | 2 | 0 | 13 | 680 | 10 | 3 | 0 | 2 | 15 | 97.8 | 100 | 23 | 99 | 60.0 | 76.2-86.6 | 99.7 | 99.1 | |
| Ethambutol | SS-S | 1 | 9 | 1 | 0 | 11 | 4 | 4,399 | 472 | 36 | 4,911 | 0.2 | 10 | 100 | 20 | 99.8 | 98.8-99.9 | 81.6 | 62.5 |
| RS-S | 21 | 5 | 3 | 0 | 29 | 31 | 376 | 40 | 4 | 451 | 6.0 | 81 | 92 | 40 | 98.7 | 98.8-99.9 | 95.1 | 87.8 | |
| SR-S | 4 | 2 | 0 | 0 | 6 | 1 | 93 | 3 | 1 | 98 | 5.8 | 67 | 99 | 80 | 97.9 | 98.8-99.9 | 92.2 | 81.7 | |
| RR-S | 375 | 20 | 30 | 19 | 444 | 203 | 241 | 48 | 14 | 506 | 46.7 | 95 | 54 | 65 | 92.3 | 93.4-96.7 | 97.7 | 94.1 | |
| SS-R | 0 | 0 | 0 | 0 | 0 | 1 | 81 | 22 | 0 | 104 | 0.0 | . | 99 | 0 | 100.0 | 98.8-99.9 | n/a | n/a | |
| RS-R | 12 | 2 | 1 | 0 | 15 | 7 | 20 | 1 | 0 | 28 | 34.9 | 86 | 74 | 63 | 90.9 | 95.7-98.1 | 95.4 | 88.6 | |
| SR-R | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0.0 | . | 100 | . | 100.0 | 98.8-99.9 | n/a | n/a | |
| RR-R | 625 | 9 | 26 | 20 | 680 | 150 | 50 | 25 | 5 | 230 | 74.7 | 99 | 25 | 81 | 84.7 | 82.0-88.2 | 98.6 | 96.4 | |
| Pyrazinamide | SSS- | 74 | 28 | 0 | 2 | 104 | 12 | 4,826 | 13 | 60 | 4,911 | 2.1 | 73 | 100 | 86 | 99.4 | 98.6-99.6 | 93.6 | 84.5 |
| RSS- | 13 | 8 | 4 | 3 | 28 | 5 | 431 | 2 | 13 | 451 | 5.8 | 62 | 99 | 72 | 98.2 | 98.6-99.6 | 91.2 | 79.6 | |
| RRS- | 166 | 25 | 22 | 17 | 230 | 49 | 374 | 68 | 15 | 506 | 31.3 | 87 | 88 | 77 | 93.7 | 95.5-97.7 | 96.4 | 91 | |
| SRS- | 0 | 3 | 0 | 0 | 3 | 0 | 97 | 0 | 1 | 98 | 3.0 | 0 | 100 | . | 97.0 | 98.6-99.6 | 80 | 60 | |
| RRR- | 532 | 15 | 83 | 50 | 680 | 107 | 216 | 105 | 16 | 444 | 60.5 | 97 | 67 | 83 | 93.5 | 87.3-91.0 | 99 | 97.3 | |
| SRR- | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 6 | 0.0 | . | 100 | . | 100.0 | 98.6-99.6 | n/a | n/a | |
| RSR- | 10 | 2 | 1 | 2 | 15 | 0 | 28 | 0 | 1 | 29 | 34.1 | 83 | 100 | 100 | 93.3 | 95.0-97.3 | 96 | 90 | |
| SSR- | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 11 | 0.0 | . | 100 | . | 100.0 | 98.6-99.6 | n/a | n/a | |
Phenotypic profiles are listed in the following order: Isoniazid, Rifampicin, Ethambutol, Pyrazinamide. '-' under 'Phenotypic profiles' marks the drug phenotype being assessed. PPV = Positive Predictive Value; NPV = Negative Predictive Value; R=resistant; S=susceptible; U=mutation of unknown association present; F=genotypic prediction failed due to missing data around a genomic resistance locus; All % results based on R/S genotypic predictions only, excluding U and F. Expected NPV was calculated as follows: specificity x (1-prevlence) / (specificity x (1-prevlence)+(1-sensitivity) x prevalence). * indicates that for prevalence <10% or >90%, simulated values are given for 10% and 90% respectively as simulations were not performed below or above these values.
As some collections included clustered isolates, the analysis was repeated after randomly selecting one representative among genomically indistinguishable isolates, and again from isolates within five single nucleotide polymorphisms of another. No significant change in sensitivity or specificity was observed for any drugs (p>0.1, S4).
To reflect the emerging practice of routinely sequencing isolates for clinical care, the analysis was repeated for the subset of 4,397 isolates from German, Italian, Dutch and UK collections that were not enriched for resistance. Among these isolates, 335 (7.6%) were isoniazid resistant and 125 (2.8%) multidrug-resistant. For each drug, specificity and negative predictive values increased, whilst positive predictive values (the proportion of concordant resistant predictions) decreased relative to the overall results. There was no significant change in sensitivity (Table 2c).
Predicting complete phenotypic profiles
For DNA sequencing to help individualise therapy, a minimum requirement is that all first-line antimicrobial phenotypes are predicted. Phenotypic profiles were thus predicted for 7,516 isolates with phenotypic data available for all first-line drugs (S1&6). ‘Unknown’ or ‘failed’ was reported for at least one drug for 1,651 (22.0%) profiles. 5,865 (78.0%) were predicted completely, of which 5,250 (89.5%) were predicted correctly (S5). Among the 5,865 profiles, 4,007 were phenotypically pan-susceptible, of which 3952 (98.6%) were predicted correctly (Table 4).
Table 4. Genotypic drug profile predictions of pan-susceptibility.
| Prediction | Genotypic drug profile | Number predicted to have drug profile | Number predicted to have drug profile that are phenotypic ally pansusceptible (%) | Sensitivity % | Specificity % | PPV % | NPV % | Predictions made % | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inh | Rif | Emb | Pza | ||||||||
| (a) Predicted pan-susceptible | S | S | S | S | 4,037 | 3952 (97.9) | |||||
| (b) Predicted pansusceptible after inferring that 'U' mutations are consistent with susceptibility in this context | S | S | S | U | 11 | 11 (100) | |||||
| S | S | U | S | 410 | 399 (97.3) | ||||||
| S | S | U | U | 2 | 2 (100) | ||||||
| S | U | S | S | 93 | 88 (94.6) | ||||||
| S | U | U | S | 29 | 29 (100) | ||||||
| Total | 4,582 | 4481 (97.8) | |||||||||
| (c) Predicted to have some phenotypic resistance | R | S | R or S | 397 | 18 (4.5) | ||||||
| S | At least one R, no U or F | 158 | 36 (22.8) | ||||||||
| R | R | R or S | 1273 | 1 (0.1) | |||||||
| Total | 1828 | 55 (3.0) | |||||||||
| 95.4 | 98.6 | 97.0 | 97.9 | 78.0 | |||||||
| 94.6 | 98.8 | 97.0 | 97.8 | 85.1 | |||||||
| No prediction made (drug profile prediction incomplete) | U | S or U | 150 | 126 (84.0) | |||||||
| At least one F, no R | 280 | 240 (85.7) | |||||||||
| At least one R and U, no F | 499 | 6 (1.2) | |||||||||
| At least one R and F, no U | 159 | 3 (1.9) | |||||||||
| At least one R, U, and F | 18 | 0 (0.0) | |||||||||
| Total | 1106 | 375 (33.9) | |||||||||
As the proportion of incompletely predicted profiles was substantial (22.0%), we assessed whether pan-susceptibility could be accurately predicted for some of these isolates anyway. Because isoniazid susceptibility predicts susceptibility to other first-line drugs,15 we maximised confidence in isoniazid predictions by conditioning predictions on the absence of ‘unknown’ mutations in isoniazid- related genes. ‘Unknown’ mutations relevant to other drugs were permitted. Doing this, pan- susceptibility was correctly predicted for 4,481/4,582 (97.8%) isolates, including 545/1,651 (33.0%) previously incompletely predicted profiles (Table 4). Among the collections unenriched for resistance, 3439/3450 (99.7%) profiles were thereby correctly predicted pan-susceptible (S7).
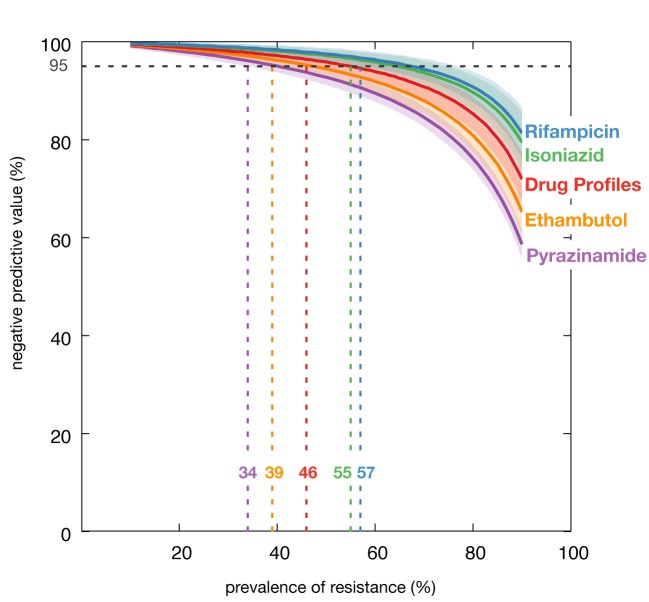
To simulate how this approach would perform in settings with differing burdens of antimicrobial resistance, we assessed the decline in negative predictive value with increasing prevalence of resistance to individual drugs, and with prevalence of any resistance within drug profiles. We randomly sub-sampled 1,000 isolates to represent every 1% increment in antimicrobial-resistance prevalence between 10%-90%, repeating this 1,000 times for each drug and for complete drug profiles. Negative predictive value declined further for ethambutol and pyrazinamide than for complete drug profiles, but declined least for isoniazid and rifampicin. Below 47.0% prevalence of resistance to any drug, the simulated negative predictive value remained above 95% for 97.5% of drug profiles (Figure 1).
Figure 1. Simulated negative predictive values for individual drugs and complete drug profiles.
Negative predictive vales shown for individual drugs and complete drug profiles, according to simulated prevalence of resistance to each drug, or within each drug profile (‘any resistance’). For each percentage prevalence between 10% and 90%, 1,000 isolates were randomly selected, 1,000 times. Lines indicate the median with shaded areas showing the 95% confidence intervals.
Discrepancy analyses
In Australia, eleven ethambutol susceptible isolates containing embB mutations were re- phenotyped. Three repeat assays failed, but seven of the remaining eight yielded, now consistent, resistant phenotypes. In Peru, 10 of 16 repeated assays remained phenotypically susceptible by MODS despite fabG1 C-15T or G-17T mutations. In isolates from the Netherlands, six resistant phenotypes predicted susceptible were identified as clerical errors, and three susceptible phenotypes predicted resistant tested phenotypically resistant by alternative phenotypic assays (S8). Although additional re- phenotyping was not possible, we conducted a ‘per mutation’ analysis to further assess discrepancies.
Of the 322 resistant phenotypes predicted susceptible, 290 (90.1%) had no mutations affecting targeted genes, and 32 (9.9%) had one or more of 15 mutations per isolate, each previously characterised as consistent with antimicrobial susceptibility. Supporting this, across all isolates in which these 15 mutations occurred as the sole mutation, they correctly predicted isoniazid susceptibility in 286/293 (97.6%) isolates and ethambutol susceptibility in 95/119 (79.8%) isolates. The one mutation relevant to pyrazinamide was seen in two isolates, both of which were phenotypically resistant. None of these mutations were relevant to rifampicin (S9).
Among 822 susceptible phenotypes predicted resistant, 145 different resistance-conferring mutations were found. Of these, 142 (97.9%) featured as the only resistance-conferring mutation in at least one isolate in the dataset, allowing assessment of individual predictive performance. They correctly predicted resistance to isoniazid in 308/371 (83.0%) isolates, rifampicin in 548/627 (87.4%) isolates, ethambutol in 1280/1743 (73.4%) isolates, and pyrazinamide in 459/663 (69.2%) isolates (S9). 14 of 17 (82.3%) mutations leading to rifampicin resistance predictions in phenotypically susceptible isolates were in the genetic region targeted by Xpert MTB/RIF and MTBDRplus.
Laboratory sample mislabelling probably also contributed discrepant results. This was estimated for each collection from the proportion of isolates excluded because of katG S315T or rpoB S450L mutations and susceptible phenotypes, the collection’s discrepancy rate, and the prevalence of antimicrobial resistance (S10). Overall, about 43% of isoniazid, and 12% of rifampicin discrepancies were thereby attributable to mislabelling.
Discussion
This analysis of over 10,000 M. tuberculosis isolates collected from 16 countries across six continents, and representing all major lineages, demonstrates that whole-genome sequencing can now characterise susceptible first-line anti-tuberculosis drug profiles sufficiently accurately for clinical use. The importance of this is twofold: First, it demonstrates that the genomic approach can be used to tailor individual treatment regimens. Extended to all drugs, individualised therapies promise to improve cure rates over those achieved by semi-empiric regimens directed by more limited diagnostic tests.1 Second, it is now possible to reduce the phenotypic workload where routine whole-genome sequencing is performed.
The WHO’s target product profiles for new molecular assays for M. tuberculosis require over 90% and 95% sensitivity and specificity, respectively.7 Overall, both these targets were met for all drugs with the exception of specificity for ethambutol (93.6%). This is no surprise as phenotyping is an imperfect gold standard, in particular for isolates with embB mutations.6,13,16 For the collections unenriched for resistance, all drugs did however meet these targets, as did the predictions of pan- susceptibility in all collections. Only categorical agreement was assessed for complete drug profile predictions because of the number of permutations. These met the external quality assurance criteria (>80% concordance) for the European TB reference laboratory network.17
There are three reasons why pan-susceptibility predictions were particularly accurate. First, the knowledgebase included both resistance-associated genomic mutations, and mutations compatible with phenotypic susceptibility. Second, anti-tuberculosis drug susceptibility phenotypes are not independent of one another, allowing the use of isoniazid susceptibility to predict susceptibility to other drugs. Third, no predictions were attempted for isolates containing genomic variation of unknown association in genes affecting isoniazid. This maximised confidence in isoniazid predictions that were made. Consequently, the prediction of drug profiles performed better than the per-drug analysis for ethambutol and pyrazinamide, and although there was a slight corresponding decline in performance for isoniazid and rifampicin, simulations showed that the prevalence of resistance would have to exceed that seen in most of the worst affected countries in the world before these predictions no longer satisfied the WHO targets.1
Our findings showed substantial improvements over the in-silico predictions for the sensitivity of WHO-recommended PCR-based assays because whole-genome sequencing is able to identify many more mutations. These additional mutations were however simultaneously responsible for the losses in specificity, largely because of the number of mutations for which a minority of isolates did not manifest a resistant phenotype. A typical example of such is the rpoB I491F mutation which frequently gives a susceptible rifampicin result in liquid culture but has been linked to treatment failure.4,18,19
The broader discrepancy analysis highlighted the same phenomenon. Whilst the predictive performance of individual mutations, whether probed by WHO-recommended assays or not, was good, each mutation has an error rate, occasionally leading to an unexpected phenotype in a minority of isolates. This is most likely where a mutation elevates the minimum drug concentration required to inhibit bacterial growth to close to the concentration above which an isolate is considered resistant. Canonical ethambutol mutations are a classic example,20 but there are many others including the mutations missed by the MODS assay in Peru.16,21,22 Such phenomena are thus likely to explain the majority of isolates that were predicted resistant, yet were phenotypically susceptible. They are also the most likely reason why predicting pan-susceptible drug profiles was more accurate than predicting profiles apparently resistant to one or more drugs.
One study limitation is that the scale and cost of repeat sequencing and phenotyping of isolates meant that we could not definitively resolve most discrepancies. This was most concerning for phenotypically resistant isolates predicted susceptible. For these, possible explanations include phenotypic error, resistant minority bacterial populations undetected by sequencing, mechanisms of resistance linked to genes we did not interrogate, or laboratory labelling error.
More work remains to be done before predictions can be extended to second and third-line drugs, and to newer compounds. However, following external review, Public Health England has already decided to stop phenotyping isolates predicted pan-susceptible to first-line drugs (personal communication, Derrick Crook, Director, National Infection Service). Similar moves are expected in the Netherlands (Dick van Soolingen, Rijksinstituut voor Volksgezondheid en Milieu) and New York (Kimberlee Musser, Wadsworth Center, New York State Department of Health). For low and middle- income countries without easy access to phenotyping, there is now the prospect that emerging mobile sequencing platforms could be used to implement sequence-directed therapies, a potential solution to the call for universal susceptibility testing. Portable platform sequencing directly from spiked-samples has been achieved, although real-world systematic evaluation is still required.23
Should whole-genome sequencing perform as well for second and third-line drugs as for first- line, a clinical trial could be needed to assess the performance of individualised over standardized treatment regimens in countries with a high drug-resistant disease burden.24 Individualised therapies would be expected to reduce the amplification of resistance (to other drugs) in individual patients, side- effects, likelihood of onward transmission, and to exert a weaker selection pressure on strains at a population level, which is key where empiric regimens have been targeted on the basis of very narrow data on antimicrobial susceptibility.4 Welcome public health benefits could result from monitoring transmission using the very same sequences.2
The current investment in whole-genome sequencing in high-income countries is likely to help accelerate implementation in lower-income, higher-burden countries where the potential benefit is greatest.25 These data demonstrate how our understanding of the molecular determinants of resistance to first-line anti-tuberculosis drugs is now sufficiently good to start using DNA sequencing to guide therapy. Similar performance must now be replicated for the remaining drugs.
Supplementary Appendix
Acknowledgments
Authors contributing are (in alphabetical order): Caroline Allix-Béguec, Irena Arandjelovic, Patrick Beckert, Lijun Bi, Maryline Bonnet, Phelim Bradley, Andrea M Cabibbe, Irving Cancino-Muñoz, Mark J Caulfield, Angkana Chaiprasert, Daniela Cirillo, David Clifton, Iñaki Comas, Derrick W Crook, Maria Rosaria De Filippo, Han de Neeling, Roland Diel, Francis A Drobniewski, Kiatichai Faksri, Maha R Farhat, Joy Fleming, Philip Fowler, Tom A Fowler, Qian Gao, Jennifer Gardy, Deborah Gascoyne-Binzi, Ana Gibertoni Cruz, Ana Gil-Brusola, Tanya Golubchik, Ximena Gonzalo, Louis Grandjean, Jennifer L Guthrie, Guangxue He, Sarah Hoosdally, Martin Hunt, Zamin Iqbal, Nazir Ismail, James Johnston, Faisal Masood Khanzada, Chiea Chuen Khor, Thomas A Kohl, Clare Kong, Sam Lipworth, Qingyun Liu, Gugu Maphalala, Elena Martinez, Vanessa Mathys, Matthias Merker, Paolo Miotto, Nerges Mistry, David Moore, Megan Murray, Stefan Niemann, Rick Twee-Hee Ong, Tim E A Peto, James E Posey, Therdsak Prammananan, Alexander Pym, Camilla Rodrigues, Mabel Rodrigues, Timothy Rodwell, Gian Maria Rossolini, Elisabeth Sánchez Padilla, Marco Schito, Xin Shen, Jay Shendure, Vitali Sintchenko, Alex Sloutsky, E Grace Smith, Matthew Snyder, Karine Soetaert, Angela M Starks, Philip Supply, Prapat Suriyapol, Sabira Tahseen, Patrick Tang, Yik-Ying Teo, Thuong Nguyen Thuy Thuong, Guy Thwaites, Enrico Tortoli, Shaheed Vally Omar, Dick van Soolingen, A Sarah Walker, Timothy M Walker, Mark Wilcox, Daniel J Wilson, David Wyllie, Yang Yang, Hongtai Zhang, Yanlin Zhao, Baoli Zhu.
Author contributions: CAB, IA, PBe, LB, MB, AMC, AC, DMC, IC, MJC, RD, FAD, KF, MRF, JF, PF, TAF, QG, JGa, DGB, AGB, TG, XG, LG, JLG, GH, NI, JJ, CK, FMK, CCK, TAK, QL, GM, EM, ICM, VM, MMe, MMu, DM, HDN, SN, RTHO, TP, ESP, GMR, MR, TR, AS, VS, EGS, JS, KS, MSc, MSn, PhS, PrS, XS, PT, YYT, ST, ET, SVO, DvS, MW, HZ, YZ, and BZ contributed towards data acquisition (including whole-genome sequencing and phenotypic drug susceptibility testing); DMC, NI, DM, SN, CR, EGS, PhS, ST, DC, DWC, AGC, SH, PM, NM, TEAP, JEP, AP, AMS, TNTT, GT, ASW, TMW, YY, AMC, HDN, MRF, and PF contributed towards study design; DMC, DC, PM, TEAP, ASW, TMW, YY, AMC, PF, PBr, MRDF, MH, ZI, SL, DJW, and DW contributed towards data analysis; TEAP, ASW and TMW wrote the manuscript; all authors contributed feedback on the manuscript.
We would like to thank Stéphanie Duthoy, Carina Hahn, Alamdar Hussain, Yannick Laurent, Mathilde Mairey, Vanessa Mohr and Mahmood Qadir and for technical assistance.
The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England. Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the U.S. Public Health Service, or the CDC. The findings and conclusions expressed by authors contributing to this journal do not necessarily reflect the official opinion of the Centers for Disease Control and Prevention, or the authors’ affiliated institutions.
Institutional funding acknowledgements:
The UK element of this research was made possible in part by the 100,000 Genomes Project which is managed by Genomics England Limited (a wholly owned company of the Department of Health). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, the Medical Research Council and Public Health England have also funded research infrastructure. The 100,000 Genomes Project uses data and samples collected by the National Health Service as part of care and support of patients.
This work was also supported by Wellcome Trust/Newton Fund-MRC Collaborative Award [200205/Z/15/Z]; and Bill & Melinda Gates Foundation [OPP1133541] and by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC) and National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at University of Oxford in partnership with Public Health England (PHE). This work was also facilitated by the NIHR Biomedical Research Centre at Barts and by the NIHR Biomedical Research Centre at Imperial.
National Science and Technology Key Program of China (2014ZX10003002), National Basic Research program of China (2014CB744403). BCCDC Foundation for Population and Public Health in Canada.
Borstel has been supported by a grant from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the Patho-NGen- Trace project, and by the German Center for Infection Research (DZIF). Leibniz Science Campus Evolutionary Medicine of the Lung (EvoLUNG). Belgian Reference Centre for Tuberculosis & Mycobacteria from Bacterial Diseases Service is partially supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System. Genoscreen was supported by the French governmental Program ‘Investing for the Future’ (Equipex LIGAN platform) and by a grant from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the PathoNGenTrace project. EU FP& EUROGEN and PANNET grants.
Individual funding acknowledgements:
Angkana Chaiprasert (research grant Siriraj Research grant No. R015833003, Drug Resistant Tuberculosis Fund and Chalermprakiat grant, Faculty of Medicine Siriraj Hospital, Mahidol University). Iñaki Comas (MINECO research grant SAF2016-77346-R and the European Research Council (ERC) (638553-TB-ACCELERATE) (to IC)); Daniel Wilson and Zamin Iqbal are Sir Henry Dale Fellows, jointly funded by the Wellcome Trust and the Royal Society (grant nos. 101237/Z/13/Z and 102541/A/13/Z respectively)); Timothy Walker is an NIHR Academic Clinical Lecturer; Derrick Crook, Tim Peto and Mark Caulfield are NIHR Senior Investigators. Rick Twee-Hee Ong and Yik-Ying Teo (National University of Singapore Yong Loo Lin School of Medicine Aspiration Fund (NUHSRO/2014/069/AF- New Idea/04)). Francis A Drobniewski was supported by EU FP7 European Union Framework Programme 7 (Grant number 201483; TB-EUROGEN) and TB-PANNET (Grant FP7-223681).
Publisher's Disclaimer: This is an Author Final Manuscript, which is the version after external peer review and before publication in the Journal. The publisher’s version of record, which includes all New England Journal of Medicine editing and enhancements, is available at 10.1056/NEJMoa1800474..
Funding Statement
The UK element of this research was made possible in part by the 100,000 Genomes Project which is managed by Genomics England Limited (a wholly owned company of the Department of Health). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, the Medical Research Council and Public Health England have also funded research infrastructure. The 100,000 Genomes Project uses data and samples collected by the National Health Service as part of care and support of patients.
This work was also supported by Wellcome Trust/Newton Fund-MRC Collaborative Award [200205/Z/15/Z]; and Bill & Melinda Gates Foundation [OPP1133541] and by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC) and National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at University of Oxford in partnership with Public Health England (PHE). This work was also facilitated by the NIHR Biomedical Research Centre at Barts and by the NIHR Biomedical Research Centre at Imperial.
National Science and Technology Key Program of China (2014ZX10003002), National Basic Research program of China (2014CB744403). BCCDC Foundation for Population and Public Health in Canada.
Borstel has been supported by a grant from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the Patho-NGen- Trace project, and by the German Center for Infection Research (DZIF). Leibniz Science Campus Evolutionary Medicine of the Lung (EvoLUNG). Belgian Reference Centre for Tuberculosis & Mycobacteria from Bacterial Diseases Service is partially supported by the Belgian Ministry of Social Affairs through a fund within the Health Insurance System. Genoscreen was supported by the French governmental Program ‘Investing for the Future’ (Equipex LIGAN platform) and by a grant from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the PathoNGenTrace project. EU FP& EUROGEN and PANNET grants.
Angkana Chaiprasert (research grant Siriraj Research grant No. R015833003, Drug Resistant Tuberculosis Fund and Chalermprakiat grant, Faculty of Medicine Siriraj Hospital, Mahidol University). Iñaki Comas (MINECO research grant SAF2016-77346-R and the European Research Council (ERC) (638553-TB-ACCELERATE) (to IC)); Daniel Wilson and Zamin Iqbal are Sir Henry Dale Fellows, jointly funded by the Wellcome Trust and the Royal Society (grant nos. 101237/Z/13/Z and 102541/A/13/Z respectively)); Timothy Walker is an NIHR Academic Clinical Lecturer; Derrick Crook, Tim Peto and Mark Caulfield are NIHR Senior Investigators. Rick Twee-Hee Ong and Yik-Ying Teo (National University of Singapore Yong Loo Lin School of Medicine Aspiration Fund (NUHSRO/2014/069/AF- New Idea/04)). Francis A Drobniewski was supported by EU FP7 European Union Framework Programme 7 (Grant number 201483; TB-EUROGEN) and TB-PANNET (Grant FP7-223681).
Contributor Information
CRyPTIC consortium:
Caroline Allix-Béguec, Irena Arandjelovic, Lijun Bi, Patrick Beckert, Maryline Bonnet, Phelim Bradley, Andrea M Cabibbe, Irving Cancino-Muñoz, Mark J Caulfield, Angkana Chaiprasert, Daniela Cirillo, David Clifton, Iñaki Comas, Derrick W Crook, Maria Rosaria De Filippo, Han de Neeling, Roland Diel, Francis A Drobniewski, Kiatichai Faksri, Maha R Farhat, Joy Fleming, Philip Fowler, Tom A Fowler, Qian Gao, Jennifer Gardy, Deborah Gascoyne-Binzi, Ana Gibertoni Cruz, Ana Gil-Brusola, Tanya Golubchik, Ximena Gonzalo, Louis Grandjean, Guangxue He, Jennifer L Guthrie, Sarah Hoosdally, Martin Hunt, Zamin Iqbal, Nazir Ismail, James Johnston, Faisal Masood Khanzada, Chiea Chuen Khor, Thomas A Kohl, Clare Kong, Sam Lipworth, Qingyun Liu, Gugu Maphalala, Elena Martinez, Vanessa Mathys, Matthias Merker, Paolo Miotto, Nerges Mistry, David Moore, Megan Murray, Stefan Niemann, Rick Twee-Hee Ong, Tim E A Peto, James E Posey, Therdsak Prammananan, Alexander Pym, Camilla Rodrigues, Mabel Rodrigues, Timothy Rodwell, Gian Maria Rossolini, Elisabeth Sánchez Padilla, Marco Schito, Marco Schito, Xin Shen, Jay Shendure, Vitali Sintchenko, Alex Sloutsky, E Grace Smith, Matthew Snyder, Karine Soetaert, Angela M Starks, Philip Supply, Prapat Suriyapol, Sabira Tahseen, Patrick Tang, Yik-Ying Teo, Thuong Nguyen Thuy Thuong, Guy Thwaites, Enrico Tortoli, Shaheed Vally Omar, Dick van Soolingen, A Sarah Walker, Timothy M Walker, Mark Wilcox, Daniel J Wilson, David Wyllie, Yang Yang, Hongtai Zhang, Yanlin Zhao, and Baoli Zhu
References
- 1.World Health Organization. Global Tuberculosis Report 2017. 2017;:1–262. [Google Scholar]
- 2.Shah NS, Auld SC, Brust JCM, et al. Transmission of Extensively Drug-Resistant Tuberculosis in South Africa. N Engl J Med 2017;376(3):243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehme CC, Nicol MP, Nabeta P, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 2011;377(9776):1495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Padilla E, Merker M, Beckert P, et al. Detection of Drug-Resistant Tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med 2015;372(12):1181–2. [DOI] [PubMed] [Google Scholar]
- 5.Pankhurst LJ, Del Ojo Elias C, Votintseva AA, et al. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. The Lancet Respiratory Medicine 2016;4(1):49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker TM, Kohl TA, Omar SV, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. 2014:1–98. [Google Scholar]
- 8.The 100,000 Genomes Project Protocol v3, Genomics England. doi:10.6084/m9.figshare.4530893.v2. 2017. [Google Scholar]
- 9.Gerton Lunter MG. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Research 2011;21(6):936–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 2012;44(2):226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miotto P, Tessema B, Tagliani E, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. 2017; 50: 1701354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadon AN, Maharaj K, Adamson JH, et al. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Comms 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casali N, Nikolayevskyy V, Balabanova Y, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 2014;46(3):279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manson AL, Cohen KA, Abeel T, et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet 2017;49(3):395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schön T, Miotto P, Köser CU, Viveiros M, Boettger E, Cambau E. Mycobacterium tuberculosis drug resistance testing: challenges, recent developments and perspectives. Clinical Microbiology and Infection 2016:1–33. [DOI] [PubMed] [Google Scholar]
- 17.Nikolayevskyy V, Hillemann D, Richter E, et al. External Quality Assessment for Tuberculosis Diagnosis and Drug Resistance in the European Union: A Five Year Multicentre Implementation Study. PLoS ONE 2016;11(4):e0152926–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigouts L, Gumusboga M, de Rijk WB, et al. Rifampin Resistance Missed in Automated Liquid Culture System for Mycobacterium tuberculosis Isolates with Specific rpoB Mutations. Journal of Clinical Microbiology 2013;51(8):2641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.André E, Goeminne L, Colmant A, Beckert P, Niemann S, Delmee M. Novel rapid PCR for the detection of Ile491Phe rpoB mutation of Mycobacterium tuberculosis, a rifampicin-resistance- conferring mutation undetected by commercial assays. Clinical Microbiology and Infection 2017;23(4):267.e5–267.e7. [DOI] [PubMed] [Google Scholar]
- 20.Sreevatsan S, Stockbauer KE, Pan X et al. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother 1997;41(8):1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miotto P, Cabibbe AM, Feuerriegel S, et al. Mycobacterium tuberculosis Pyrazinamide Resistance Determinants: a Multicenter Study. mBio 2014; 5(5):e01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coronel J, Roper M, Mitchell S, et al. MODS accreditation process for regional reference laboratories in Peru: validation by GenoType® MTBDRplus. Int J Tuberc Lung Dis 2010;14(11):1475–80. [PMC free article] [PubMed] [Google Scholar]
- 23.Votintseva AA, Bradley P, Pankhurst L, et al. Same-Day Diagnostic and Surveillance Data for Tuberculosis via Whole-Genome Sequencing of Direct Respiratory Samples. Journal of Clinical Microbiology 2017;55(5):1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyckendorf J, Andres S, Köser CU, et al. What is resistance? Impact of phenotypic versus molecular drug resistance testing on multi- and extensively drug-resistant tuberculosis therapy. Antimicrob Agents Chemother 2017:AAC.01550–17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben J Marais, Walker TM, Cirillo DM, et al. Comment: Aiming for zero tuberculosis transmission in low-burden countries. The Lancet Respiratory 2017:1–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.