The nitrification process is an important step in the nitrogen (N) cycle, affecting N availability and N losses to the wider environment. Ammonia oxidation, which is the first and rate-limiting step of nitrification, was widely accepted to be mainly regulated by AOA in acidic soils. However, in this study, nitrification activity was correlated with the abundance of AOB rather than that of AOA in acidic Ultisols. Nitrosospira cluster 8a, a phylotype of AOB which preferred warm temperatures, and low soil pH played a predominant role in the nitrification process in the test Ultisols. Our results also showed that long-term application of lime or pig manure rather than plant residues altered the community structure of AOA and AOB. Taken together, our findings contribute new knowledge to the understanding of the nitrification process and ammonia oxidizers in subtropical acidic Ultisol under long-term inorganic and organic fertilization.
KEYWORDS: nitrification, AOA, AOB, Nitrosospira, long-term fertilization
ABSTRACT
Long-term effects of inorganic and organic fertilization on nitrification activity (NA) and the abundances and community structures of ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) were investigated in an acidic Ultisol. Seven treatments applied annually for 27 years comprised no fertilization (control), inorganic NPK fertilizer (N), inorganic NPK fertilizer plus lime (CaCO3) (NL), inorganic NPK fertilizer plus peanut straw (NPS), inorganic NPK fertilizer plus rice straw (NRS), inorganic NPK fertilizer plus radish (NR), and inorganic NPK fertilizer plus pig manure (NPM). In nonfertilized soil, the abundance of AOA was 1 order of magnitude higher than that of AOB. Fertilization reduced the abundance of AOA but increased that of AOB, especially in the NL treatment. The AOA communities in the control and the N treatments were dominated by the Nitrososphaera and B1 clades but shifted to clade A in the NL and NPM treatments. Nitrosospira cluster 8a was found to be the most dominant AOB in all treatments. NA was primarily regulated by soil properties, especially soil pH, and the interaction with AOB abundance explained up to 73% of the variance in NA. When NL soils with neutral pH were excluded from the analysis, AOB abundance, especially the relative abundance of Nitrosospira cluster 8a, was positively associated with NA. In contrast, there was no association between AOA abundance and NA. Overall, our data suggest that Nitrosospira cluster 8a of AOB played an important role in the nitrification process in acidic soil following long-term inorganic and organic fertilization.
IMPORTANCE The nitrification process is an important step in the nitrogen (N) cycle, affecting N availability and N losses to the wider environment. Ammonia oxidation, which is the first and rate-limiting step of nitrification, was widely accepted to be mainly regulated by AOA in acidic soils. However, in this study, nitrification activity was correlated with the abundance of AOB rather than that of AOA in acidic Ultisols. Nitrosospira cluster 8a, a phylotype of AOB which preferred warm temperatures, and low soil pH played a predominant role in the nitrification process in the test Ultisols. Our results also showed that long-term application of lime or pig manure rather than plant residues altered the community structure of AOA and AOB. Taken together, our findings contribute new knowledge to the understanding of the nitrification process and ammonia oxidizers in subtropical acidic Ultisol under long-term inorganic and organic fertilization.
INTRODUCTION
In order to increase crop yields to meet the food demands of an increasing population, increasing amounts of synthetic fertilizers, including nitrogen (N), have been applied to agroecosystems and have had adverse environmental consequences (1). The process of nitrification is a critical step in the N cycle that affects plant uptake of soil inorganic N, nitrate (NO3−) leaching, and nitrous oxide (N2O) emissions (2). It was previously thought that ammonia (NH3) oxidation, which is the first and rate-limiting step of nitrification, was mediated by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) (3, 4) in the soil. However, recently, complete ammonia oxidizers (comammox) were also discovered; they can complete nitrification in one step on their own (5).
The population sizes of AOA in a range of agricultural soils have been found to be larger than those of AOB, especially in acidic soils (6–8). Generally, AOA showed stronger resistance to low soil pH and higher nitrification activity (NA) in acidic soils (7), whereas in neutral and alkaline agricultural soils, nitrification is dominated by AOB (6, 9). Numerous studies have shown that nitrification activity in acidic soils is positively correlated with AOA abundance (7, 10), and changes of AOA communities could significantly affect nitrification activity (7, 11, 12). However, recent studies of acidic upland soils have found that AOB are associated with potential nitrification rates (13) and act as metabolically active ammonia oxidizers (14–16). Such evidence could indicate that the assumption of AOA dominating nitrification in acidic soils may not hold true for all soil types and that the role of AOB in nitrification in acidic soils should be reevaluated.
The community composition of AOA and AOB plays a key role in the regulation of nitrification activity (7, 11). For example, AOA clade B was found to be actively linked to NH3 oxidation in Arctic soils, while clade A showed little or no nitrification activity (17). Using 13CO2-DNA stable isotope probing, Wang et al. (18) found that the clades Nitrososphaera and Nitrosospira cluster 3 were the most active AOA and AOB phylotypes, respectively, in an acidic Ultisol. The community structures of both AOA and AOB were strongly shaped by soil variables, especially soil pH and substrate (NH3) availability (6, 19, 20). AOA clade B and the AOB Nitrosospira cluster 2 have been found to be well adapted to low-pH soils, while the AOB Nitrosomonas preferred neutral or alkaline soils (17, 21). Other soil properties, such as temperature (22), salinity (23), organic carbon (C) concentration (24), and moisture content (25), have also been found to influence the abundance and composition of soil ammonia-oxidizing communities.
Long-term fertilizer application has been shown to affect AOA and AOB abundances and community structures in different soil ecosystems (26, 27). For example, two long-term studies reported that the application of inorganic fertilizer enhanced AOB abundance in grassland soils after 44 years of fertilization (26) and in calcareous agricultural soils after 31 years of fertilization (28), whereas the application of organic fertilizer promoted the abundance of AOA. In contrast, Peng et al. (29) found that although AOA and AOB community compositions were altered by 35 years of fertilizer application to a salt marsh, abundances remained stable.
Despite the widespread distribution of Ultisols and their importance in biogeochemical processes (30), studies of the effects of long-term fertilizer application on AOA and AOB community compositions and roles in nitrification in subtropical Ultisols are limited (21). Therefore, we evaluated the long-term effects of the application of inorganic and organic fertilizer to a subtropical acidic Ultisol on the abundances and community compositions of AOA and AOB and nitrification to test the hypotheses that nitrification in Ultisols is primarily regulated by AOA and that long-term fertilizer application increases nitrification activities of AOA and AOB and elevates the relative importance of AOB.
RESULTS
Soil nitrification activity.
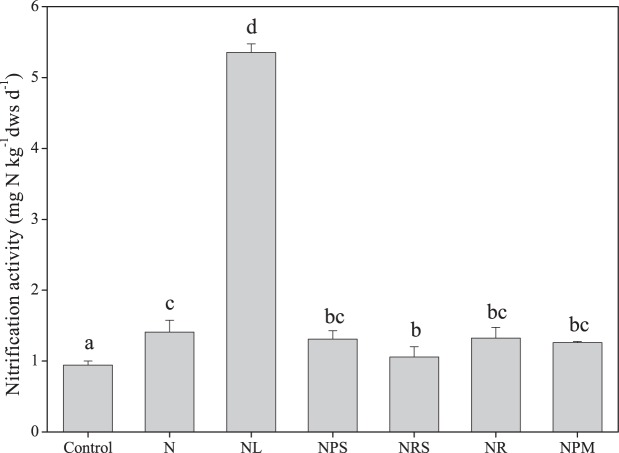
Long-term fertilization increased nitrification activity from 0.94 mg N kg−1 dry weight of soil (dws) day−1 in the control treatment to 1.06 to 5.35 mg N kg−1 dws day−1 in the fertilized treatments (Fig. 1). Nitrification activity was highest in the inorganic NPK fertilizer plus lime (CaCO3) (NL) treatment, which was significantly higher than those in the other treatments (P < 0.05). The application of organic materials in addition to NPK (N) did not increase nitrification activity (Fig. 1). Nitrification activity was correlated with soil pH that ranged from 4.96 to 6.59 (R = 0.942; P < 0.001) (Table 1; see also Table S1 in the supplemental material).
FIG 1.
Soil nitrification activity in the different treatments. Vertical bars represent standard errors of the means (SEMs) (n = 3). Different letters denote significant differences between treatments (P < 0.05).
TABLE 1.
Soil physicochemical variables following application of long-term inorganic and organic fertilizersa
| Treatment | Mean value ± SEM |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pH (1:5 soil/H2O ratio) | SOC concn (g C kg−1) | DOC concn (mg C kg−1) | TN concn (g N kg−1) | NH4+ concn (mg N kg−1) | NO3− concn (mg N kg−1) | DON concn (mg N kg−1) | AP concn (mg P kg−1) | AK concn (mg K kg−1) | DCo (10−6 m2 s−1) | |
| Control | 4.96 ± 0.02a | 5.69 ± 0.10a | 4.27 ± 0.37a | 0.59 ± 0.01a | 0.54 ± 0.17ab | 12.50 ± 0.39c | 9.22 ± 0.37c | 5.25 ± 0.54a | 84.7 ± 10.38a | 12.58 ± 0.39a |
| N | 5.20 ± 0.01b | 6.44 ± 0.26b | 4.38 ± 0.23a | 0.70 ± 0.01b | 0.66 ± 0.04b | 8.60 ± 0.77ab | 5.56 ± 0.24ab | 15.0 ± 3.12bc | 177.6 ± 23.00b | 10.71 ± 0.60b |
| NL | 6.59 ± 0.04d | 6.32 ± 0.08b | 5.33 ± 0.34a | 0.72 ± 0.02b | 0.56 ± 0.19ab | 5.21 ± 0.81a | 4.28 ± 0.75a | 16.4 ± 1.82c | 203.1 ± 5.62c | 9.95 ± 0.19b |
| NPS | 5.13 ± 0.03b | 6.62 ± 0.06bc | 4.35 ± 0.20a | 0.78 ± 0.02c | 0.69 ± 0.10b | 8.71 ± 0.60ab | 5.38 ± 0.57ab | 10.3 ± 0.95b | 178.0 ± 2.42b | 9.96 ± 0.85b |
| NRS | 5.16 ± 0.02b | 7.13 ± 0.28c | 5.53 ± 0.60a | 0.82 ± 0.02cd | 0.41 ± 0.13ab | 8.66 ± 0.08ab | 8.90 ± 2.88bc | 10.8 ± 0.52b | 206.7 ± 4.16c | 8.20 ± 0.55c |
| NR | 5.17 ± 0.02b | 6.76 ± 0.29bc | 5.63 ± 0.85a | 0.82 ± 0.01cd | 0.25 ± 0.07a | 11.6 ± 2.24bc | 5.93 ± 0.46abc | 11.6 ± 0.72bc | 204.8 ± 0.99c | 9.70 ± 0.66bc |
| NPM | 5.58 ± 0.02c | 8.21 ± 0.20d | 12.3 ± 1.80b | 0.85 ± 0.02d | 0.49 ± 0.03ab | 9.56 ± 0.50bc | 5.89 ± 0.45abc | 157 ± 8.85d | 170.8 ± 3.86b | 2.81 ± 0.35d |
n = 3. Values within the same row followed by different letters indicate significant differences at a P value of <0.05. SOC, soil organic carbon; DOC, dissolved organic carbon; TN, soil total nitrogen; DON, dissolved organic nitrogen; AP, available phosphorus; AK, available potassium; DCo, effective diffusion coefficient of oxygen.
Abundances of AOA and AOB.
The AOA and AOB amoA gene abundances ranged from 4.08 × 107 to 6.15 × 107 copies g−1 dws and from 4.94 × 106 to 17.15 × 106 copies g−1 dws, respectively (Fig. 2). Fertilizer application reduced the AOA amoA gene abundance in the NL, inorganic NPK fertilizer plus rice straw (NRS), and inorganic NPK fertilizer plus pig manure (NPM) treatments (P < 0.05), and all fertilizer treatments increased the AOB amoA gene abundance (P < 0.05). The abundance of amoA genes for AOA was positively correlated with soil NO3−-N (P < 0.01) and negatively associated with soil pH and available potassium (AK) (P < 0.01). In contrast, the abundance of amoA genes for AOB was negatively correlated with soil NO3−-N and dissolved organic nitrogen (DON) (P < 0.05) and positively associated with soil pH and AK (P < 0.01).
FIG 2.
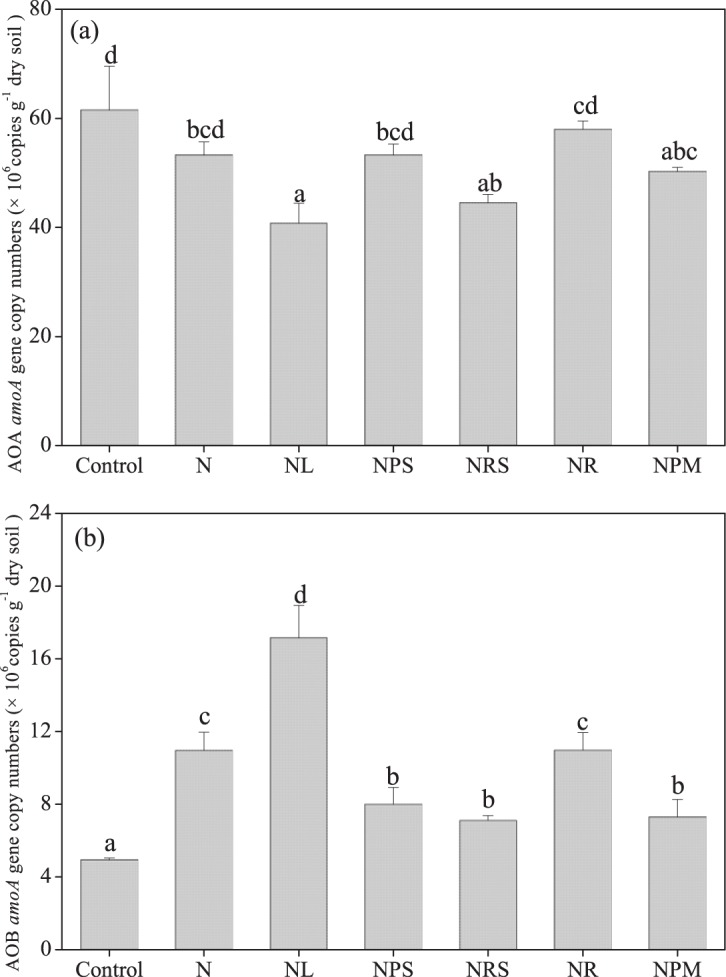
Abundances of amoA genes of ammonia-oxidizing archaea (AOA) (a) and ammonia-oxidizing bacteria (AOB) (b) in the treatments. Vertical bars represent SEMs (n = 3). Different letters denote significant differences between treatments (P < 0.05).
Community diversity and composition of AOA and AOB.
After quality filtering, totals of 998,950 and 1,226,551 high-quality sequences were obtained for AOA and AOB, with minima of 28,389 and 30,073 sequences per sample, respectively. There were differences in AOA and AOB community diversity among the treatments (P < 0.05) (see Fig. S1 in the supplemental material), where AOA diversity was lowest in the NL and NPM treatments and highest in the N, inorganic NPK fertilizer plus peanut straw (NPS), NRS, and inorganic NPK fertilizer plus radish (NR) treatments. In contrast, AOB community diversity was highest in the NL and NPM treatments (Fig. S1).
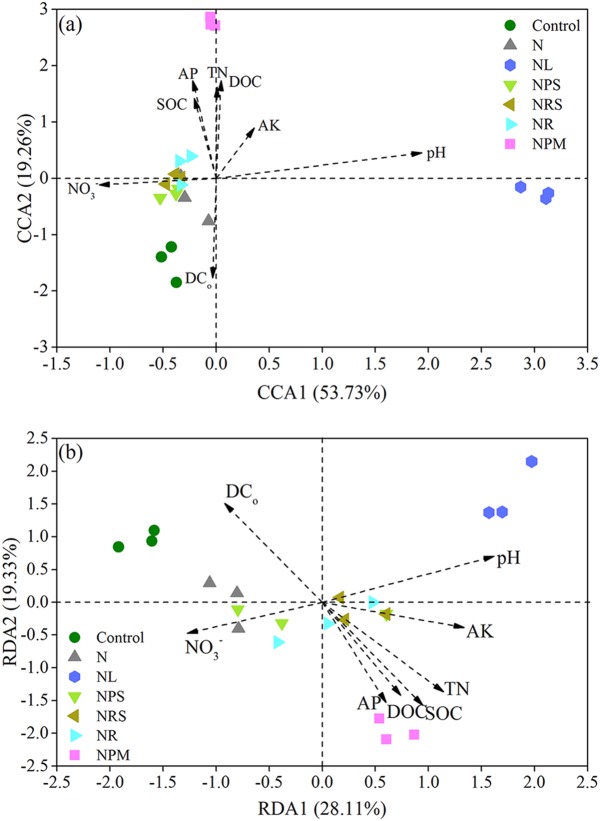
Canonical correspondence analysis (CCA) and linear redundancy analysis (RDA) showed that both AOA and AOB communities formed four clusters: control, NL, NPM, and a composite of the other treatments (N, NPS, NRS, and NR) (Fig. 3). AOA and AOB communities in the NL treatment were related to higher soil pH, while the communities in the NPM treatment were associated with soil organic carbon (SOC), dissolved organic carbon (DOC), available phosphorus (AP), and soil total nitrogen (TN). The multivariate regression tree (MRT) analysis of dominant operational taxonomic units (OTUs) (relative abundance of >0.1%) showed that the community composition of AOA and AOB was primarily shaped by soil pH (Fig. S2). Mantel tests confirmed that soil pH was the primary influencing factor in AOA and AOB communities (P = 0.001) (Table 2).
FIG 3.
Canonical correlation analysis (CCA) of AOA (a) and redundancy analysis (RDA) of AOB (b) communities in the treatments. The positions and lengths of the arrows indicate the directions and strengths, respectively, of the effects of soil variables on the communities.
TABLE 2.
Mantel test correlations between community structure of ammonia-oxidizing archaea or ammonia-oxidizing bacteria and soil physicochemical propertiesa
| Soil variable | AOA |
AOB |
||
|---|---|---|---|---|
| R | P | R | P | |
| Soil pH | 0.896 | 0.001 | 0.901 | 0.001 |
| SOC | 0.261 | 0.036 | 0.111 | 0.196 |
| DOC | 0.208 | 0.092 | −0.002 | 0.374 |
| TN | 0.200 | 0.044 | 0.185 | 0.054 |
| DON | 0.110 | 0.209 | 0.058 | 0.250 |
| NH4+-N | 0.060 | 0.280 | 0.023 | 0.379 |
| NO3−-N | 0.406 | 0.002 | 0.435 | 0.001 |
| AP | 0.289 | 0.042 | 0.018 | 0.393 |
| AK | 0.147 | 0.140 | 0.098 | 0.228 |
| DCo | 0.278 | 0.046 | 0.026 | 0.386 |
Values in boldface type indicate significant effects (P < 0.05).
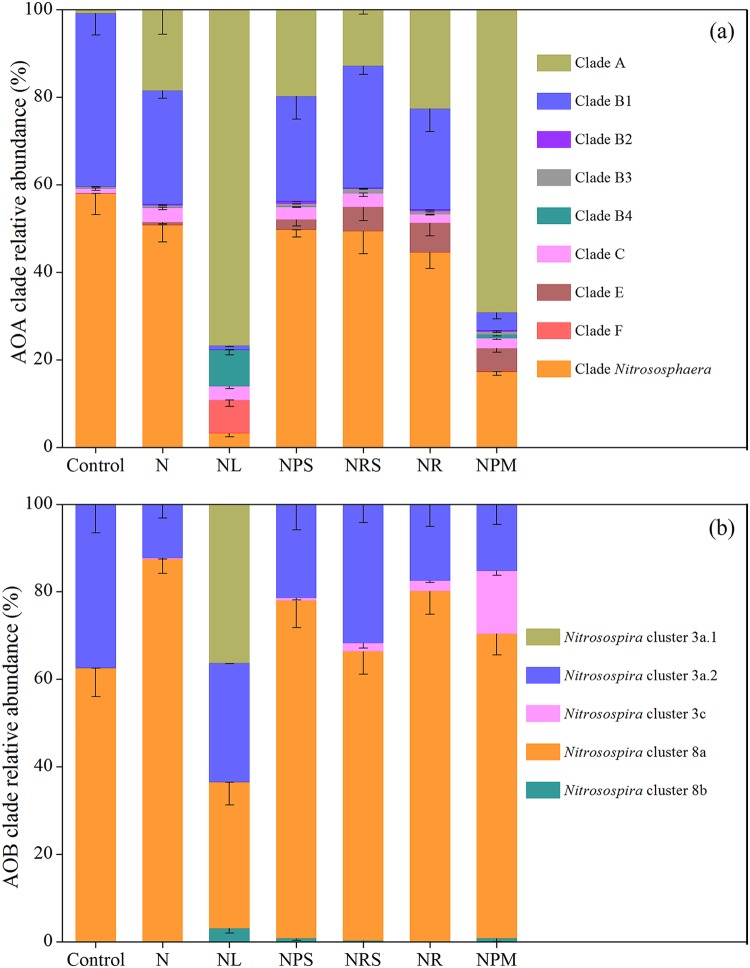
Sequences of dominant AOA and AOB amoA OTUs (relative abundance of >0.1%) were taxonomically classified into different phylotypes across the treatments (Fig. 4). Phylogenetic analysis of AOA communities showed that the dominant amoA OTU sequences were exclusively from the Nitrososphaera cluster (group I.1b lineage) and were distributed across nine robust phylogenetic clades (Fig. 4a and Fig. S3). In untreated soil (control), the Nitrososphaera and B1 clades represented 57.94% and 39.61% of AOA amoA sequences, respectively, and long-term fertilization reduced these proportions to 3.21 to 50.76% and 0.90 to 27.97%, respectively (P < 0.05), with the lowest proportions being recorded for the NL (3.21% and 0.90%, respectively) and NPM (17.28% and 4.12%, respectively) treatments. In contrast, the application of liming (NL) and pig manure (NPM) increased the proportions of clade A from 0.81% in the control soil to 76.80% and 69.23%, respectively (P < 0.001). The Nitrososphaera and B1 clades were negatively correlated with soil pH (P < 0.001), whereas clade A was positively associated (Table S2).
FIG 4.
Community compositions of AOA (a) and AOB (b) based on amoA gene sequences in the treatments. Error bars represent SEMs (n = 3).
The dominant detected AOB amoA OTUs were exclusively affiliated with the Nitrosospira-like genus and grouped into Nitrosospira clusters 3a.1, 3a.2, 3c, 8a, and 8b; no AOB sequence was affiliated with the Nitrosomonas genus (Fig. 4b and Fig. S4). Nitrosospira clusters 8a and 3a.2 dominated the unfertilized control soil, accounting for 62.56% and 37.43%, respectively. Long-term fertilization decreased the relative abundance of Nitrosospira cluster 3a.2 to 11.29 to 31.56% (P < 0.05), and the relative abundance of Nitrosospira cluster 3a.1 increased from 0.01% in the control soil to 36.33% in the NL soil (P < 0.001) with an increase in soil pH. The relative abundance of Nitrosospira cluster 8a increased from 62.56% in the control treatment to 66.13 to 87.44% in fertilization treatments, except for NL, in which it decreased to 33.35%, and was negatively correlated with soil pH (Table S3). The relative abundance of Nitrosospira cluster 3c was highest in the NPM treatment (P < 0.001) and was positively correlated with SOC, DOC, AP, and TN but negatively associated with DCo (effective diffusion coefficient of oxygen) (P < 0.001).
Factors affecting nitrification activity.
Variance partitioning was used to evaluate the relative contributions of different groups of variables to the variation of nitrification activity. The soil physicochemical properties and the abundances of AOA and AOB were separated into three groups of explanatory variables and used to explain each fraction of nitrification activity. Soil properties and their interaction with the AOB amoA gene abundance were found to explain up to 73% of the variance in nitrification activity (Fig. 5), and nitrification activity was positively associated with AOB amoA gene abundance (P < 0.001) (Fig. 6) and negatively associated with AOA amoA gene abundance (P = 0.005) (Fig. 6). When the NL treatment with neutral soil pH (6.59) was excluded from the analysis, nitrification activity continued to be positively associated with AOB amoA gene abundance (P = 0.007) (see Fig. S5 in the supplemental material), but there was a lack of an association with AOA amoA gene abundance; among the AOB communities, there was a positive association with Nitrosospira cluster 8a (Fig. 7).
FIG 5.
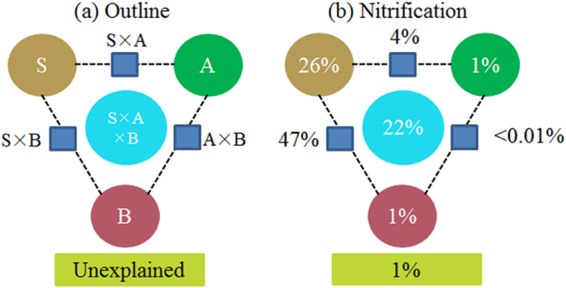
Percent variation in nitrification activity partitioned into soil physicochemical properties (S), abundance of AOA amoA genes (A), and abundance of AOB amoA genes (B). (a) Model scheme; (b) analysis.
FIG 6.
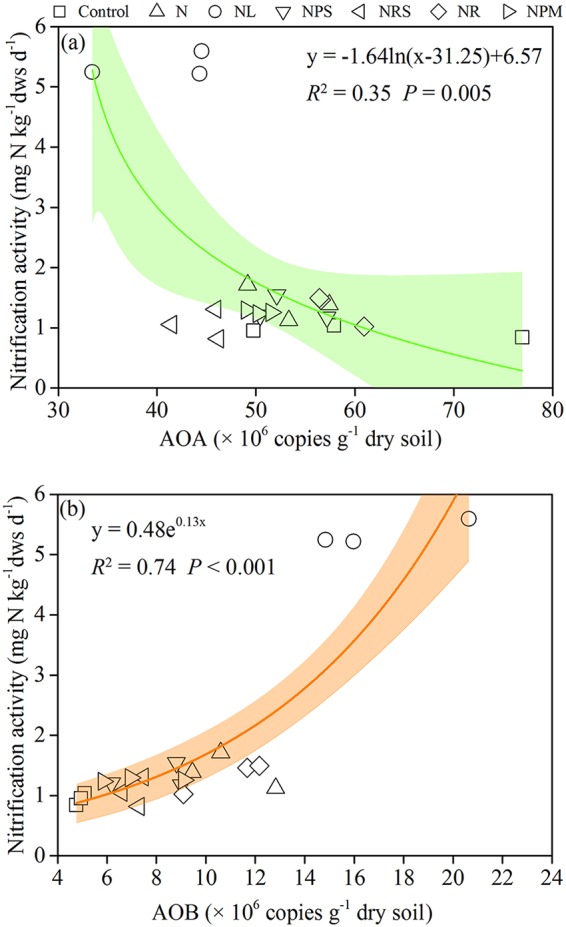
Association between nitrification activity and AOA (a) and AOB (b) amoA gene abundances in the treatments. The shaded area indicates the 95% confidence interval of the regression models.
FIG 7.
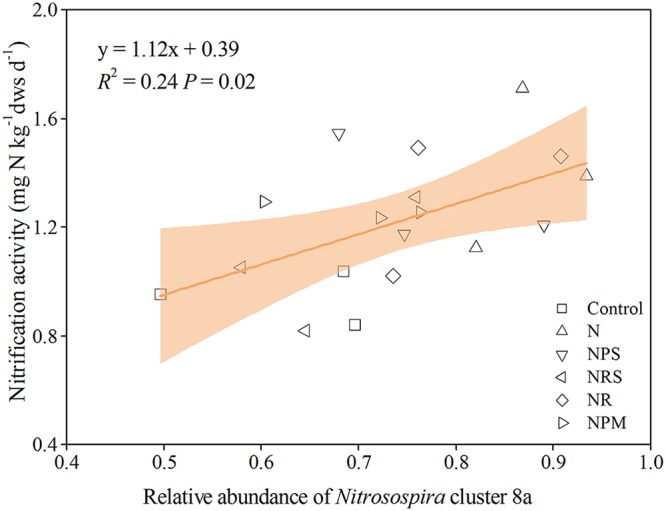
Association between nitrification activity and relative abundance of Nitrosospira cluster 8a in the treatments, excluding NL. The shaded area indicates the 95% confidence interval of the regression models.
DISCUSSION
Effect of fertilizer on AOA and AOB.
We found that the abundance of amoA genes for AOA was 1 order of magnitude higher than that for AOB in unfertilized soil, supporting previous reports of much higher abundances of AOA than their bacterial counterparts in acidic soils (6, 8, 18). The availability of ammonia (NH3) in acidic soils is generally low, due to the strong ionization of NH3 to NH4+, and subsequently decreases NH3 diffusion into microbial cells (31). However, genomic analysis suggested that AOA genes putatively encode pH homeostasis and high-affinity NH3 acquisition systems (32) and thus could play a crucial role in nitrification in acidic soils where pH and NH3 availability are particularly low (21). In contrast, AOB, which were also adapted to low soil pH (21), had a far lower substrate affinity than that of AOA (33), which suggests that AOA outcompeted AOB for substrates in the control soil. Our experiment showed that the long-term application of liming (NL) reduced AOA abundance, possibly because the increased soil pH reduced the competitive ability of AOA in soils (6, 34). In contrast, the abundance of AOB has been found to increase with soil pH (8, 34, 35). Intriguingly, the application of organic materials in our experiment had no apparent effect on the abundance of AOA compared to N, despite previous work showing benefits to AOA of mineralization of manure or plant residues in the absence of the addition of inorganic fertilizer (28, 36, 37). The reasons for this lack of an effect of organic fertilizer on AOA abundance may be due to the possible suppression of AOA growth in the treatments with N and N plus organic material due to high concentrations of NH4+ immediately after fertilization (38, 39), which overrode the possible stimulation effect induced by organic material amendment, or higher rates of turnover of the labile fraction of amended organic materials in our experimental soils due to higher temperature and precipitation than in previous studies (28, 36). The labile fraction of amended organic materials may have been quickly mineralized before soil ammonia decreased to a low level following fertilization; however, the remaining recalcitrant fraction of the organic material may not have stimulated AOA growth (37) under low levels of ammonia.
As has been reported in previous studies (6, 40), we found that pH was the primary driver for the shift of AOA community structure. Although the soil pH varied from 4.96 to 6.59, AOA sequences were within the Nitrososphaera cluster, which was recognized as a neutrophilic or alkaline, rather than acidic, archaeon (40). The Nitrososphaera cluster has been found to be metabolically active in acidic Ultisols using a DNA-SIP technique (18) and dominant in fertilized acidic soils (8). In this study, we found that dominant members of AOA were from the clades Nitrososphaera and B1 in the more acidic control soil and from clade A in the higher-pH NL and NPM soils, indicating the possibility of the occurrence of the Nitrososphaera cluster in neutrophilic or acidic soils, depending on its specific sublineages. Our results suggested that the phylogeny and characteristics of the Nitrososphaera cluster were more complicated than previously understood, so we suggest that phylogenetic analysis of AOA communities should focus on the sublineages of the Nitrososphaera cluster in soils, where they are known to be dominant ammonia oxidizers (8, 17).
Previous studies have suggested that NH4+ is the main factor affecting AOB composition (20); however, in our study, Mantel tests showed that soil pH rather than NH4+ shaped AOB communities. The weak effect of NH4+ on AOB community structure in this study may be due to the particularly low NH4+ content (<0.7 mg N kg−1) caused by intensive crop uptake and/or N losses in summer (41). We found that Nitrosospira was exclusively dominant in the AOB communities, probably because Nitrosospira generally outcompeted Nitrosomonas, which was previously reported for acidic soils (8, 21, 42). In contrast to previous studies that found that Nitrosospira cluster 2 dominated in acidic soils (21, 43), we found a dominance of Nitrosospira cluster 8a. Generally, Nitrosospira cluster 2 is restricted to cold temperate soils (44), whereas Nitrosospira cluster 8a, which is also referred to as clusters 10 and 11 (45, 46), tends to be dominant (relative abundance of >90%) in soils that experience long periods of warm temperatures (47), with optimum growth temperatures of 31°C to 33°C (48), and is active in extremely acidic soils (49). It is likely that low soil pH and, more importantly, long periods of warm temperatures at this subtropical study site selected for Nitrosospira cluster 8a rather than cluster 2. We found that the long-term application of lime increased the relative abundance of Nitrosospira cluster 3a.1, reflecting the known preference for neutral and alkaline soils (19, 21). Likewise, Pommerening-Röser and Koops (50) found that the optimum pH for Nitrosospira cluster 3a.1 was 6.0 under urea addition, and Zhang et al. (51) noted a preference for lower-altitude soils with higher temperatures in the Mount Everest region. It appears that high annual temperatures may have enhanced the suitability of the neutral soil pH of the NL treatment soil for Nitrosospira cluster 3a.1. In contrast, we found that the abundance of Nitrosospira cluster 3c increased with the application of pig manure, which also led to the high levels of SOC. Since cluster 3c has been found to be dominant in an upland Ultisol with an SOC content of >13.6 g C kg−1 (9) and a Mollisol with an SOC content of >22.8 g C kg−1 (52), we postulate that this cluster responded to the high SOC content.
Contribution of AOA and AOB to nitrification activity.
We used the increase in the rate of the soil NO3− concentration to assess nitrification activity (18), and soil pH and substrate availability (usually NH3) have been suggested to be the two key factors affecting nitrification activity in acidic soils (21, 53). We found that the application of lime increased the abundance of AOB and substantially enhanced nitrification activity. While this increase in nitrification activity may be a result of reduced ionization of NH3 to NH4+ and a rise in the associated substrate availability under the increased-pH conditions and increased abundance of AOB, maximum rates of nitrification by AOB have been shown to be 20-fold higher than those by AOA (33).
Previous studies have suggested that nitrification in acidic soils is primarily affected by AOA (7, 10, 54). However, in contrast to previous findings that showed that nitrification rates in acidic soils were closely correlated with AOA abundance (21), we found a negative and a lack of association between AOA abundance and nitrification activity with all treatments and with all treatments except NL (neutral soil pH), respectively. This may be due to a dominance of AOA-mediated nitrification processes in previous studies of fertilizer addition that benefited AOA abundance as a result of reduced soil pH (6). For example, AOA played a key role in nitrification in 50-year-old tea orchard soil that received 450 kg N ha−1 year−1 and led to a reduction in the soil pH from 5.16 to 4.01 (7, 55). In that study, long-term fertilization slightly increased the soil pH due to the addition of P fertilizer as calcium magnesium phosphate (56), and we found that the dominant AOA clades shifted from the Nitrososphaera and B1 clades in the control and N treatments to clade A in the NL and NPM treatments. Alves et al. (17) noted that the Nitrososphaera and B1 clades were actively linked to NH3 oxidation, and in contrast, clade A elicited little or no nitrification activity. Our results suggest that AOA may have played a minor role in nitrification in the experimental Ultisol, and the contribution of AOA to nitrification activity may have been reduced in the NL and NPM soils.
The abundance of AOB in our study was more closely coupled with nitrification activity than AOA, differing from most previous findings for acidic soils (27, 57). Although similar correlations have been reported for some acidic soils (13, 58), Li et al. (21) argued that those studies used atypical acidic soils (pH >6), as were used in our study. However, when the NL soil (neutral pH) was excluded from the analysis, AOB abundance remained weakly correlated with nitrification activity, indicating that nitrification was primarily affected by AOB in soils under long-term N fertilization. It is likely that the increased substrate availability by long-term fertilization favored AOB, due to its much lower substrate affinity than that of AOA (33). He et al. (59) suggested that AOA may play more-important roles in ammonia oxidation than AOB from acid soils, due to the reduced availability of ammonia, and they highlighted that a wide range of AOB members could also contribute to nitrification in acidic environments and that more investigations were required. Using DNA-SIP, Wang et al. (18) found that AOA played a key role in nitrification under long-term fertilization in Ultisols; however, they also suggested that DNA-SIP relied on cell proliferation, and it is likely that AOB conducted ammonia oxidation without cell division and that RNA-SIP might be more helpful. Furthermore, the correlation of Nitrosospira cluster 8a with nitrification activity is weakly positive and at best suggests an association when NL treatment was excluded from the analysis, indicating that cluster 8a may play a key role in nitrification in acidic Ultisols. This study suggests that a specific phylotype of AOB has been associated with nitrification activity in acidic Ultisols, and it is possibly supported by Nitrosospira cluster 8a being the dominant AOB phylotype in the test soil and favored by warm temperatures and low soil pH in the study soils. Thus, Nitrosospira cluster 8a may play a key role in nitrification in tropical and subtropical acidic Ultisols. DNA/RNA-SIP experiments would be helpful to directly identify the active player in nitrification in further investigations.
Overall, our results show that the abundance of AOA was higher than that of AOB in the unfertilized control soil; however, long-term fertilization, especially the application of lime, reduced AOA abundance and increased AOB abundance. The dominant members of the AOA were the Nitrososphaera and B1 clades for all treatments, except for NL and NPM, in which clade A was dominant. In contrast, Nitrosospira cluster 8a was the dominant phylotype of AOB in the treatments, while the addition of lime and pig manure increased the relative abundances of Nitrosospira clusters 3a.1 and 3c, respectively. Nitrification activity was primarily regulated by the interaction of soil pH and AOB abundance in fertilized soils. When NL treatment was excluded from the analysis, nitrification activity was weakly positive and at best correlated with Nitrosospira cluster 8a. Our findings indicated that AOB rather than AOA played a key role in nitrification in the test Ultisols.
MATERIALS AND METHODS
Study site and soil sampling.
The field experiment was established in April 1988 at the Yingtan Red Soil Ecology Experimental Station, Chinese Academy of Sciences, Yujiang, China (28°15′20″N, 116°55′30″E), in a cropping system of continuous summer peanut followed by winter fallow. The area has a typical subtropical monsoon climate, with an annual average temperature of 17.6°C and precipitation of 1,795 mm. The soil, derived from quaternary red clay, is classified as a Typic Plinthudult (Ultisol) based on U.S. Department of Agriculture soil taxonomy, comprising 25.6% sand, 33.2% silt, and 41.2% clay.
Three replicates of seven treatments were arranged in a randomized block design in 34.6-m2 plots, where the treatments consisted of no fertilizer (control), inorganic NPK fertilizer (N), inorganic NPK fertilizer plus lime (CaCO3) (NL), inorganic NPK fertilizer plus peanut straw (NPS), inorganic NPK fertilizer plus rice straw (NRS), inorganic NPK fertilizer plus radish residues (NR), and inorganic NPK fertilizer plus pig manure (NPM). Annual application of NPK fertilizer in the N treatment comprised 120 kg N ha−1 as urea, 30 kg P ha−1 as calcium magnesium phosphate, and 90 kg K ha−1 as potassium chloride. In the NPS, NRS, NR, and NPM treatments, 30% of the inorganic N fertilizer was replaced by organic N. Pig manure was stockpiled on a concrete slab for 3 months before application. All fertilizer treatments received the same total N, P, and K; however, as the amounts of P and K in the organic material were generally smaller than the prescribed doses, the organic fertilizer was supplemented accordingly with calcium magnesium phosphate and potassium chloride, depending on the P and K contents of the organic materials. For NL, 1,500 kg ha−1 CaCO3 was applied annually. Prior to seed drilling, fertilizers were spread evenly onto the soil surface by hand and immediately incorporated into the top 20 cm of soil by plowing. Each year on 10 April, peanut (cv. Ganhua 5) was sown manually by placing two seeds per hole to give 20-cm plant spacing in rows 30 cm apart. Field management practices were the same for all treatments during the experimental period.
On 13 December 2015, 10 tillage layer (0 to 20 cm) soil samples were collected at random from each plot by using a 10-cm-diameter auger and mixed to form a single, composite sample. Samples were transported to the laboratory in a constant-temperature box containing ice, within 2 days. Visible stones and plant residues were carefully removed by using forceps, and soil was gently separated along natural breakpoints, passed through an 8-mm sieve, and then divided into three subsamples, which were air dried for physicochemical analysis, stored at 4°C to measure nitrification activity for no more than 3 days, and sieved to <2 mm and immediately stored at −80°C for subsequent DNA extraction.
Analysis of physicochemical properties.
Soil pH was measured by using a glass electrode with a 1:5 soil-to-water ratio. Concentrations of soil organic C (SOC) and total N (TN) were determined by using the wet oxidation redox titration and micro-Kjeldahl methods, respectively. Dissolved organic C (DOC) was extracted by incubating 10 g fresh soil (on an oven-dried basis) with 50 ml of deionized water for 30 min, followed by shaking (end-over-end) at 25°C, and the samples were then centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was filtered through a 0.45-μm membrane filter (Whatman, Clifton, NJ, USA), and the DOC content was quantified by using a Shimadzu C analyzer (TOC Vcph; Shimadzu, Kyoto, Japan). NH4+-N, NO3−-N, and dissolved total N (DTN) were extracted by using 2 M KCl; the content was measured by using a continuous-flow analyzer (San++; Skalar, Holland); and dissolved organic N (DON) was calculated as DTN − NH4+-N − NO3−-N. Soil available K (AK) was extracted by using 1 M ammonium acetate and measured by using flame photometry (catalog no. FP640; INASA, China). Soil available P (AP) was extracted by using 0.0125 M H2SO4 in 0.05 M HCl, and its concentration was determined by using the molybdenum blue method.
Six undisturbed soil cores collected by using a 100-cm3 cylinder were sampled from each plot and used to measure water retention curves with a ceramic pressure plate in a pressure chamber at equilibrium matrix potentials of −0.1, −0.2, −1, −3.5, −6, −10, −33, −50, −100, −200, −500, and −1,500 kPa. The effective diffusion coefficient of oxygen in the soil (DCo) (square meters per second) was calculated as (60), where N is soil porosity; Dao is the free diffusion coefficient of oxygen in air at 20°C (1.8 × 10−5 m2 s−1); KH is Henry's equilibrium constant at 20°C (0.03); Dwo is the free diffusion coefficient of oxygen in water at 20°C (2.2 × 10−9 m2 s−1); Qa and Qw are the proportions of soil porosity occupied by air and water, respectively (Qa + Qw = 1); and p is the power constant (3.4). Soil porosity (N) was calculated as N = 1 − ρ/ρ0, where ρ is soil bulk density (grams per cubic centimeter) and ρ0 is soil particle density (grams per cubic centimeter). The proportion of soil porosity occupied by water (Qw) was calculated as Qw = ρ × w/N, where w is the soil gravimetric moisture content (cubic centimeter per gram).
Soil physicochemical properties are presented in Table 1.
Nitrification activity.
Nitrification activity (NA) was determined according to a modified version of the method used by Ma et al. (61), where 20 g fresh soil (on an oven-dried basis) was placed into a 125-ml glass jar, to which 50 mg N kg−1 as (NH4)2SO4 was added, and soil moisture was adjusted to a 60% water-holding capacity with deionized water. The jars were incubated in darkness at 25°C, and 0, 1, 4, and 10 days after the addition of the substrate, three replicates for each treatment were destructively sampled to determine the nitrate concentration, where soil nitrate was extracted by using 2.0 M KCl and measured with a continuous-flow analyzer (San++; Skalar, Holland). NA was calculated as the linear accumulation of nitrate over the incubation period.
DNA extraction and real-time quantitative PCR.
Soil DNA was extracted from 0.5 g fresh soil (on an oven-dried basis) by using the Fast DNA spin kit for soil (MP Biomedicals, CA, USA) and subsequently purified by using a PowerClean DNA cleanup kit (Mobio, CA, USA) according to the manufacturer's protocols. The quality and concentration of the extracted DNA were measured by using gel electrophoresis (0.8% agarose) and a spectrophotometer (NanoDrop Technologies), and the DNA was stored at −20°C prior to additional analyses.
Real-time quantitative PCR (qPCR) to identify and quantify archaeal and bacterial amoA genes was done by using a CFX96 optical real-time detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA). A standard curve was generated by using plasmid DNA from one representative clone containing each target gene. Each reaction mixture (25 μl) comprised 12.5 μl SYBR qPCR master mix (Vazyme), 0.25 μl of each primer, and 1 μl of the DNA template containing approximately 1 to 10 ng of DNA. A negative control was always run with sterilized distilled water as the template instead of a DNA sample. A serial dilution of the DNA template was also used to determine whether the PCR had been inhibited during amplification. Details of gene-specific primers and thermal conditions are presented in Table 3. Amplifications resulted in single peaks, and efficiencies were 90.1 to 101.7%, with R2 values of between 0.992 and 0.999.
TABLE 3.
PCR primers and thermal cycling conditions used for gene quantification
| Gene | Primer | Sequence | Amplicon size (bp) | Thermal conditions | Reference |
|---|---|---|---|---|---|
| Archaeal amoA | Arch-amoAF | STAATGGTCTGGCTTAGACG | 635 | 95°C for 3 min and 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s + plate read; melt curve, 65.0°C–95.0°C; increment, 0.5°C, 5 s, + plate read | 75 |
| Arch-amoAR | GCGGCCATCCATCTGTATGT | ||||
| Bacterial amoA | amoA-1F | GGGGTTTCTACTGGTGGT | 491 | 95°C for 3 min and 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s + plate read; melt curve, 65.0°C–95.0°C; increment, 0.5°C, 5 s, + plate read | 63 |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC |
High-throughput sequencing and bioinformatics analysis.
We used primer sets Arch-amoA26F/Arch-amoA417R (62) and amoA-1F/amoA-2R (63) for AOA and AOB amoA gene amplification, respectively, where the PCR program and reaction composition of AOA and AOB were described previously by Park et al. (62) and Lin et al. (64), respectively. A unique sample-identifying barcode was added to the forward primer in PCR amplification, triplicate PCR amplifications were conducted and pooled for each sample, and PCR products were subsequently purified by using a Qiagen gel extraction kit. Sequencing libraries were generated by using the TruSeq Nano DNA LT library prep kit (Illumina, USA) according to the manufacturer's recommendations, and library quality was assessed on the Qubit 2.0 fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 systems. Finally, high-throughput sequencing of AOA and AOB amoA genes was carried out by using Illumina MiSeq, and 300-bp paired-end reads were generated.
Pairs of reads were merged in FLASH version 1.2.7 (65), where forward and reverse reads had overlapping base lengths of ≥10 bp and base mismatches were prevented. Sequence data were processed by using Quantitative Insights into Microbial Ecology (QIIME) v1.8.0 (66), and low-quality sequences that had quality scores of <20, contained ambiguous nucleotides, or did not match the primer or barcode were removed. Barcode and primer sequences were deleted after sample sequences were sorted according to the barcodes. Next, the sequences were compared with sequences in the reference database (67) by using the UCHIME algorithm to detect chimera sequences that were subsequently removed. The remaining high-quality sequences at 97% similarity were assigned to the same OTUs by using Uparse v7.0.1001 software (68), and the most abundant sequence from each OTU was selected as its representative sequence. The representative OTUs were taxonomically classified from the construction of neighbor-joining phylogenetic trees in MEGA 6 using representative sequences of the AOA or AOB amoA genes, together with taxonomically determined reference sequences from GenBank. We used nomenclature for AOA amoA clusters as defined by Alves et al. (17) and Zhang et al. (8) and for AOB amoA clusters as defined by Avrahami et al. (69) and He et al. (9). Heterogeneity in the number of sequences per sample was removed by rarefying sequences prior to calculation of Chao1 (70) and Shannon (71) diversity indices in QIIME.
Data analysis and statistics.
Variances in nitrification activity, amoA gene copy numbers, alpha diversity, and relative abundances of AOA and AOB clades were tested by using one-way analysis of variance (ANOVA) in SPSS 18.0 for Windows (SPSS Inc., Chicago, IL, USA). All data were checked for normality and homogeneity of variance (Levene's test) prior to testing for treatment differences; data were ln transformed to meet the assumptions of the ANOVA where necessary. Least-significant-differences (LSD) tests at a P value of 0.05 were used to test for differences among treatments. Associations between soil physicochemical variables, AOA or AOB amoA gene copy numbers, and nitrification activity were tested by using Pearson's correlation coefficient.
The following analyses were computed in R (version 3.4.1). A multivariate regression tree (MRT) was built to identify the most important abiotic factors for AOA and AOB community compositions using the mvpart package (72). Correlations between soil physicochemical variables and AOA or AOB community composition were calculated in the vegan package (73) by using a Mantel test. As best-fit models, unimodal canonical correspondence analysis (CCA) and linear redundancy analysis (RDA) were conducted for AOA and AOB communities, respectively, and only environmental variables with a VIF of <20 were selected for analysis. Variance partitioning of nitrification activity was analyzed by using the varpart function in the vegan package, as described previously by Domeignoz-Horta et al. (74).
Accession number(s).
Sequences have been deposited in the DNA Data Bank of Japan under accession no. DRA006528 and DRA006529.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Program of China (2016YFD0200309), the National Natural Science Foundation of China (41471077), and the Frontier Project from the Institute of Soil Science, Chinese Academy of Sciences (no. ISSASIP1607).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01031-18.
REFERENCES
- 1.Ju X, Xing G, Chen X, Zhang S, Zhang L, Liu X, Cui Z, Yin B, Christie P, Zhu Z. 2009. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc Natl Acad Sci U S A 106:3041–3046. doi: 10.1073/pnas.0813417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu H, Chen D, He J. 2015. Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol Rev 39:729–749. doi: 10.1093/femsre/fuv021. [DOI] [PubMed] [Google Scholar]
- 3.Schleper C, Nicol GW. 2010. Ammonia-oxidising archaea—physiology, ecology and evolution. Adv Microb Physiol 57:1–41. doi: 10.1016/B978-0-12-381045-8.00001-1. [DOI] [PubMed] [Google Scholar]
- 4.Norton JM. 2011. Diversity and environmental distribution of ammonia-oxidizing bacteria, p 39–55. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 5.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang LM, Hu HW, Shen JP, He JZ. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045. doi: 10.1038/ismej.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M, Alves RJE, Zhang D, Han L, He J, Zhang L. 2017. Time-dependent shifts in populations and activity of bacterial and archaeal ammonia oxidizers in response to liming in acidic soils. Soil Biol Biochem 112:77–89. doi: 10.1016/j.soilbio.2017.05.001. [DOI] [Google Scholar]
- 9.He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di H. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 10.Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L, Han W, Singh BK. 2011. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625. doi: 10.1128/AEM.00136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubry-Rangin C, Nicol GW, Prosser JI. 2010. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 12.Lu L, Han W, Zhang J, Wu Y, Wang B, Lin X, Zhu J, Cai Z, Jia Z. 2012. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J 6:1978–1984. doi: 10.1038/ismej.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Jin C, Sun B. 2014. Soil aggregate stratification of nematodes and ammonia oxidizers affects nitrification in an acid soil. Environ Microbiol 16:3083–3094. doi: 10.1111/1462-2920.12339. [DOI] [PubMed] [Google Scholar]
- 14.Robinson A, Di HJ, Cameron KC, Podolyan A, He J. 2014. The effect of soil pH and dicyandiamide (DCD) on N2O emissions and ammonia oxidiser abundance in a stimulated grazed pasture soil. J Soils Sediments 14:1434–1444. doi: 10.1007/s11368-014-0888-2. [DOI] [Google Scholar]
- 15.Dai S, Liu Q, Zhao J, Zhang J. 2018. Ecological niche differentiation of ammonia-oxidising archaea and bacteria in acidic soils due to land use change. Soil Res 56:71–79. [Google Scholar]
- 16.Huang X, Zhao J, Su J, Jia Z, Shi X, Wright AL, Zhu-Barker X, Jiang X. 2018. Neutrophilic bacteria are responsible for autotrophic ammonia oxidation in an acidic forest soil. Soil Biol Biochem 119:83–89. doi: 10.1016/j.soilbio.2018.01.016. [DOI] [Google Scholar]
- 17.Alves RJ, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T. 2013. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620–1631. doi: 10.1038/ismej.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B, Zheng Y, Huang R, Zhou X, Wang D, He Y, Jia Z. 2014. Active ammonia oxidizers in an acidic soil are phylogenetically closely related to neutrophilic archaeon. Appl Environ Microbiol 80:1684–1691. doi: 10.1128/AEM.03633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu H, Zhang L, Dai Y, Di H, He J. 2013. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J Soils Sediments 13:1439–1449. doi: 10.1007/s11368-013-0726-y. [DOI] [Google Scholar]
- 20.Guo J, Ling N, Chen H, Zhu C, Kong Y, Wang M, Shen Q, Guo S. 2017. Distinct drivers of activity, abundance, diversity and composition of ammonia-oxidizers: evidence from a long-term field experiment. Soil Biol Biochem 115:403–414. doi: 10.1016/j.soilbio.2017.09.007. [DOI] [Google Scholar]
- 21.Li Y, Chapman SJ, Nicol GW, Yao H. 2018. Nitrification and nitrifiers in acidic soils. Soil Biol Biochem 116:290–301. doi: 10.1016/j.soilbio.2017.10.023. [DOI] [Google Scholar]
- 22.Hu H, Macdonald CA, Trivedi P, Anderson IC, Zheng Y, Holmes B, Bodrossy L, Wang J, He J, Singh BK. 2016. Effects of climate warming and elevated CO2 on autotrophic nitrification and nitrifiers in dryland ecosystems. Soil Biol Biochem 92:1–15. doi: 10.1016/j.soilbio.2015.09.008. [DOI] [Google Scholar]
- 23.Zhang Y, Chen L, Dai T, Tian J, Wen D. 2015. The influence of salinity on the abundance, transcriptional activity, and diversity of AOA and AOB in an estuarine sediment: a microcosm study. Appl Microbiol Biotechnol 99:9825–9833. doi: 10.1007/s00253-015-6804-x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, Huang L, Deng Y, Wang S, Zhou Y, Liu L, Dong H. 2014. Latitudinal distribution of ammonia-oxidizing bacteria and archaea in the agricultural soils of eastern China. Appl Environ Microbiol 80:5593–5602. doi: 10.1128/AEM.01617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, Macdonald CA, Trivedi P, Holmes B, Bodrossy L, He J, Singh BK. 2015. Water addition regulates the metabolic activity of ammonia oxidizers responding to environmental perturbations in dry subhumid ecosystems. Environ Microbiol 17:444–461. doi: 10.1111/1462-2920.12481. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Fornara D, Wasson EA, Wang D, Ren G, Christie P, Jia Z. 2015. Effects of 44 years of chronic nitrogen fertilization on the soil nitrifying community of permanent grassland. Soil Biol Biochem 91:76–83. doi: 10.1016/j.soilbio.2015.08.031. [DOI] [Google Scholar]
- 27.Tao R, Wakelin SA, Liang Y, Chu G. 2017. Response of ammonia-oxidizing archaea and bacteria in calcareous soil to mineral and organic fertilizer application and their relative contribution to nitrification. Soil Biol Biochem 114:20–30. doi: 10.1016/j.soilbio.2017.06.027. [DOI] [Google Scholar]
- 28.Ai C, Liang G, Sun J, Wang X, He P, Zhou W. 2013. Different roles of rhizosphere effect and long-term fertilization in the activity and community structure of ammonia oxidizers in a calcareous fluvo-aquic soil. Soil Biol Biochem 57:30–42. doi: 10.1016/j.soilbio.2012.08.003. [DOI] [Google Scholar]
- 29.Peng X, Yando E, Hildebrand E, Dwyer C, Kearney A, Waciega A, Valiela I, Bernhard A. 2013. Differential responses of ammonia-oxidizing archaea and bacteria to long-term fertilization in a New England salt marsh. Front Microbiol 3:445. doi: 10.3389/fmicb.2012.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lal R. 2004. Soil carbon sequestration to mitigate climate change. Geoderma 123:1–22. doi: 10.1016/j.geoderma.2004.01.032. [DOI] [Google Scholar]
- 31.Frijlink MJ, Abee T, Laanbroek HJ, de Boer W, Konings WN. 1992. The bioenergetics of ammonia and hydroxylamine oxidation in Nitrosomonas europaea at acid and alkaline pH. Arch Microbiol 157:194–199. doi: 10.1007/BF00245290. [DOI] [Google Scholar]
- 32.Lehtovirta-Morley LE, Sayavedra-Soto LA, Gallois N, Schouten S, Stein LY, Prosser JI, Nicol GW. 2016. Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra.” Appl Environ Microbiol 82:2608–2619. doi: 10.1128/AEM.04031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang Y, Norton JM, Stark JM. 2017. Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol Biochem 113:161–172. doi: 10.1016/j.soilbio.2017.06.010. [DOI] [Google Scholar]
- 34.Xiao H, Schaefer DA, Yang X. 2017. pH drives ammonia oxidizing bacteria rather than archaea thereby stimulate nitrification under Ageratina adenophora colonization. Soil Biol Biochem 114:12–19. doi: 10.1016/j.soilbio.2017.06.024. [DOI] [Google Scholar]
- 35.Egan G, Zhou X, Wang D, Jia Z, Crawley M, Fornara DA. 2018. Long-term effects of grazing, liming and nutrient fertilization on the nitrifying community of grassland soils. Soil Biol Biochem 118:97–102. doi: 10.1016/j.soilbio.2017.12.005. [DOI] [Google Scholar]
- 36.Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, Sharma S, Wilke BM, Matthies M, Smalla K, Munch JC, Amelung W, Kaupenjohann M, Schloter M, Schleper C. 2009. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- 37.Wessén E, Nyberg K, Jansson JK, Hallin S. 2010. Responses of bacterial and archaeal ammonia oxidizers to soil organic and fertilizer amendments under long-term management. Appl Soil Ecol 45:193–200. doi: 10.1016/j.apsoil.2010.04.003. [DOI] [Google Scholar]
- 38.Wu Y, Xiang Y, Wang J, Zhong J, He J, Wu QL. 2010. Heterogeneity of archaeal and bacterial ammonia-oxidizing communities in Lake Taihu, China. Environ Microbiol Rep 2:569–576. doi: 10.1111/j.1758-2229.2010.00146.x. [DOI] [PubMed] [Google Scholar]
- 39.Hou J, Song C, Cao X, Zhou Y. 2013. Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res 47:2285–2296. doi: 10.1016/j.watres.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 40.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 108:21206–21211. doi: 10.1073/pnas.1109000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao W, Hong H, Zhang Y, Chen N, Zeng Y, Wang W. 2006. Anthropogenic nitrogen sources and exports in a village-scale catchment in southeast China. Environ Geochem Health 28:45–51. doi: 10.1007/s10653-005-9010-4. [DOI] [PubMed] [Google Scholar]
- 42.Song H, Che Z, Cao W, Huang T, Wang J, Dong Z. 2016. Changing roles of ammonia-oxidizing bacteria and archaea in a continuously acidifying soil caused by over-fertilization with nitrogen. Environ Sci Pollut Res Int 23:11964–11974. doi: 10.1007/s11356-016-6396-8. [DOI] [PubMed] [Google Scholar]
- 43.Nugroho RA, Röling WF, van Straalen NM, Verhoef HA. 2009. Changes in nitrification and bacterial community structure upon cross-inoculation of Scots pine forest soils with different initial nitrification rates. Soil Biol Biochem 41:243–250. doi: 10.1016/j.soilbio.2008.10.020. [DOI] [Google Scholar]
- 44.Avrahami S, Conrad R. 2005. Cold-temperate climate: a factor for selection of ammonia oxidizers in upland soil? Can J Microbiol 51:709–714. doi: 10.1139/w05-045. [DOI] [PubMed] [Google Scholar]
- 45.Avrahami S, Conrad R. 2003. Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl Environ Microbiol 69:6152–6164. doi: 10.1128/AEM.69.10.6152-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avrahami S, Liesack W, Conrad R. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol 5:691–705. doi: 10.1046/j.1462-2920.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- 47.Avrahami S, Bohannan BJM. 2007. Response of Nitrosospira sp. strain AF-like ammonia oxidizers to changes in temperature, soil moisture content, and fertilizer concentration. Appl Environ Microbiol 73:1166–1173. doi: 10.1128/AEM.01803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Q, Bakken LR. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol 30:171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 49.Cytryn E, Levkovitch I, Negreanu Y, Dowd S, Frenk S, Silber A. 2012. Impact of short-term acidification on nitrification and nitrifying bacterial community dynamics in soilless cultivation media. Appl Environ Microbiol 78:6576–6582. doi: 10.1128/AEM.01545-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pommerening-Röser A, Koops HP. 2005. Environmental pH as an important factor for the distribution of urease positive ammonia-oxidizing bacteria. Microbiol Res 160:27–35. doi: 10.1016/j.micres.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Wang M, Prosser JI, Zheng Y, He J. 2009. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol Ecol 70:208–217. doi: 10.1111/j.1574-6941.2009.00775.x. [DOI] [PubMed] [Google Scholar]
- 52.Xue C, Zhang X, Zhu C, Zhao J, Zhu P, Peng C, Ling N, Shen Q. 2016. Quantitative and compositional responses of ammonia-oxidizing archaea and bacteria to long-term field fertilization. Sci Rep 6:28981. doi: 10.1038/srep28981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao H, Campbell CD, Chapman SJ, Freitag TE, Nicol GW, Singh BK. 2013. Multi-factorial drivers of ammonia oxidizer communities: evidence from a national soil survey. Environ Microbiol 15:2545–2556. doi: 10.1111/1462-2920.12141. [DOI] [PubMed] [Google Scholar]
- 54.Hu H, Xu Z, He J. 2014. Ammonia-oxidizing archaea play a predominant role in acid soil nitrification. Adv Agron 125:261–302. doi: 10.1016/B978-0-12-800137-0.00006-6. [DOI] [Google Scholar]
- 55.Xue D, Yao H, Huang C. 2006. Microbial biomass, N mineralization and nitrification, enzyme activities, and microbial community diversity in tea orchard soils. Plant Soil 288:319–331. doi: 10.1007/s11104-006-9123-2. [DOI] [Google Scholar]
- 56.Liu W, Wang Q, Wang B, Wang X, Franks AE, Teng Y, Li Z, Luo Y. 2015. Changes in the abundance and structure of bacterial communities under long-term fertilization treatments in a peanut monocropping system. Plant Soil 395:415–427. doi: 10.1007/s11104-015-2569-3. [DOI] [Google Scholar]
- 57.Wakelin S, Williams E, O'Sullivan CA, Cameron KC, Di HJ, Cave V, O'Callaghan M. 2014. Predicting the efficacy of the nitrification inhibitor dicyandiamide in pastoral soils. Plant Soil 381:35–43. doi: 10.1007/s11104-014-2107-8. [DOI] [Google Scholar]
- 58.Di H, Cameron K, Shen JP, Winefield C, O'Callaghan M, Bowatte S, He J. 2009. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624. doi: 10.1038/ngeo613. [DOI] [Google Scholar]
- 59.He JZ, Hu HW, Zhang LM. 2012. Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154. doi: 10.1016/j.soilbio.2012.06.006. [DOI] [Google Scholar]
- 60.Aachib M, Mbonimpa M, Aubertin M. 2004. Measurement and prediction of the oxygen diffusion coefficient in unsaturated media, with applications to soil covers. Water Air Soil Pollut 156:163–193. doi: 10.1023/B:WATE.0000036803.84061.e5. [DOI] [Google Scholar]
- 61.Ma W, Jiang S, Assemien F, Qin M, Ma B, Xie Z, Liu Y, Feng H, Du G, Ma X, Le Roux X. 2016. Response of microbial functional groups involved in soil N cycle to N, P and NP fertilization in Tibetan alpine meadows. Soil Biol Biochem 101:195–206. doi: 10.1016/j.soilbio.2016.07.023. [DOI] [Google Scholar]
- 62.Park SJ, Park BJ, Rhee SK. 2008. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615. doi: 10.1007/s00792-008-0165-7. [DOI] [PubMed] [Google Scholar]
- 63.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y, Ding W, Liu D, He T, Yoo G, Yuan J, Chen Z, Fan J. 2017. Wheat straw-derived biochar amendment stimulated N2O emissions from rice paddy soils by regulating the amoA genes of ammonia-oxidizing bacteria. Soil Biol Biochem 113:89–98. doi: 10.1016/j.soilbio.2017.06.001. [DOI] [Google Scholar]
- 65.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 69.Avrahami S, Conrad R, Braker G. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl Environ Microbiol 68:5685–5692. doi: 10.1128/AEM.68.11.5685-5692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chao A, Bunge J. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539. doi: 10.1111/j.0006-341X.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 71.Shannon CE, Weaver W. 1949. The mathematical theory of information. University of Illinois Press, Urbana, IL. [Google Scholar]
- 72.De'Ath G. 2002. Multivariate regression trees: a new technique for modeling species-environment relationships. Ecology 83:1105–1117. doi: 10.1890/0012-9658(2002)083[1105:MRTANT]2.0.CO;2. [DOI] [Google Scholar]
- 73.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2008. The vegan package version 2.2-1 Community ecology package. https://cran.r-project.org/web/packages/vegan.
- 74.Domeignoz-Horta LA, Philippot L, Peyrard C, Bru D, Breuil MC, Bizouard F, Justes E, Mary B, Leonard J, Spor A. 2017. Peaks of in situ N2O emissions are influenced by N2O-producing and reducing microbial communities across arable soils. Glob Chang Biol 24:360–370. doi: 10.1111/gcb.13853. [DOI] [PubMed] [Google Scholar]
- 75.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.