The rapid emergence of infectious diseases and multiresistant pathogens has increased the necessity for new bioactive compounds; thus, novel strategies have to be developed to find them. Actinomycetes isolated in symbiosis with insects have attracted attention in recent years as producers of metabolites with important bioactivities. Sipanmycins are glycosylated macrolactams produced by Streptomyces sp. CS149, isolated from leaf-cutting ants, and show potent cytotoxic activity. Here, we characterize the sip cluster and propose a biosynthetic pathway for sipanmycins. As far as we know, it is the first time that the cooperation between two different glycosyltransferases is demonstrated to be strictly necessary for the incorporation of the same sugar. Also, a third protein with homology to P450 monooxygenases, SipO2, is shown to be essential in the second glycosylation step, forming a complex with the glycosyltransferase pair SipS9-SipS14.
KEYWORDS: Streptomyces, actinomycetes, antibiotics, biosynthesis gene cluster, glycosylation, glycosyltransferases, sugars
ABSTRACT
Macrolactams comprise a family of natural compounds with important bioactivities, such as antibiotic, antifungal, and antiproliferative activities. Sipanmycins A and B are two novel members of this family, with two sugar moieties attached to the aglycon. In the related macrolactam vicenistatin, the sugar moiety has been proven to be essential for cytotoxicity. In this work, the gene cluster responsible for the biosynthesis of sipanmycins (sip cluster) in Streptomyces sp. strain CS149 is described and the steps involved in the glycosylation of the final compounds unraveled. Also, the cooperation of two different glycosyltransferases in each glycosylation step is demonstrated. Additionally, the essential role of SipO2 as an auxiliary protein in the incorporation of the second deoxy sugar is addressed. In light of the results obtained by the generation of mutant strains and in silico characterization of the sip cluster, a biosynthetic pathway for sipanmycins and the two deoxy sugars attached is proposed. Finally, the importance of the hydroxyl group at C-10 of the macrolactam ring and the sugar moieties for cytotoxicity and antibiotic activity of sipanmycins is shown.
IMPORTANCE The rapid emergence of infectious diseases and multiresistant pathogens has increased the necessity for new bioactive compounds; thus, novel strategies have to be developed to find them. Actinomycetes isolated in symbiosis with insects have attracted attention in recent years as producers of metabolites with important bioactivities. Sipanmycins are glycosylated macrolactams produced by Streptomyces sp. CS149, isolated from leaf-cutting ants, and show potent cytotoxic activity. Here, we characterize the sip cluster and propose a biosynthetic pathway for sipanmycins. As far as we know, it is the first time that the cooperation between two different glycosyltransferases is demonstrated to be strictly necessary for the incorporation of the same sugar. Also, a third protein with homology to P450 monooxygenases, SipO2, is shown to be essential in the second glycosylation step, forming a complex with the glycosyltransferase pair SipS9-SipS14.
INTRODUCTION
Searching for novel bioactive natural products is a great challenge for researchers in order to find novel antibiotics to combat emergent infectious diseases and multiresistant pathogens or novel chemotherapeutic drugs in the field of cancer. Microorganisms produce a large number of natural products, with actinomycetes being responsible for the production of approximately two-thirds of all natural bioactive products known so far (1). However, due to the low hit rate for novel compounds obtained using classical screening approaches and the frequent rediscovery of already-known compounds (2), novel strategies must be developed. Some of these approaches involve the isolation of bioactive compound microbial producers from unexplored environments, paying special attention to microorganisms associated with marine or terrestrial macroorganisms, such as sponges, plants, ants, termites, and wasps (3–6), but also to mammals (7), including human beings (8). Furthermore, the development of DNA sequencing technologies and the increasing number of genomes sequenced facilitate the use of other strategies, such as transcriptome analysis by RNA sequencing (9), mining of microbial genomes or metagenomes in searching specific gene homologs (10), or new biosynthetic gene clusters (BGCs) (11), and the activation of low-expressed or silent clusters (12).
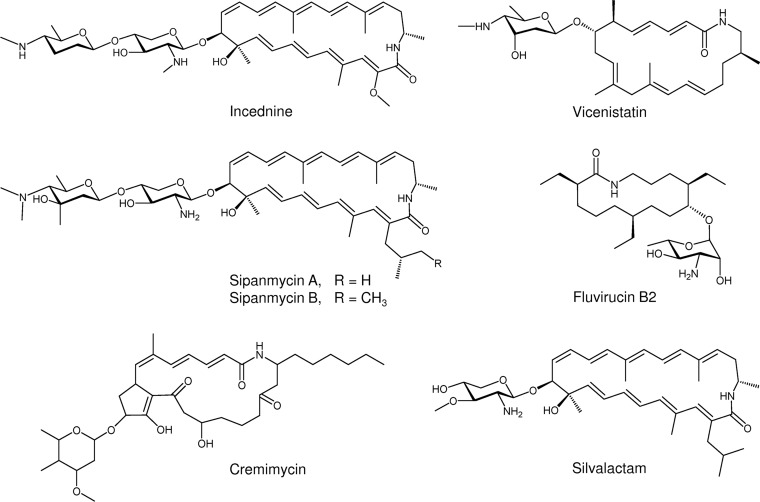
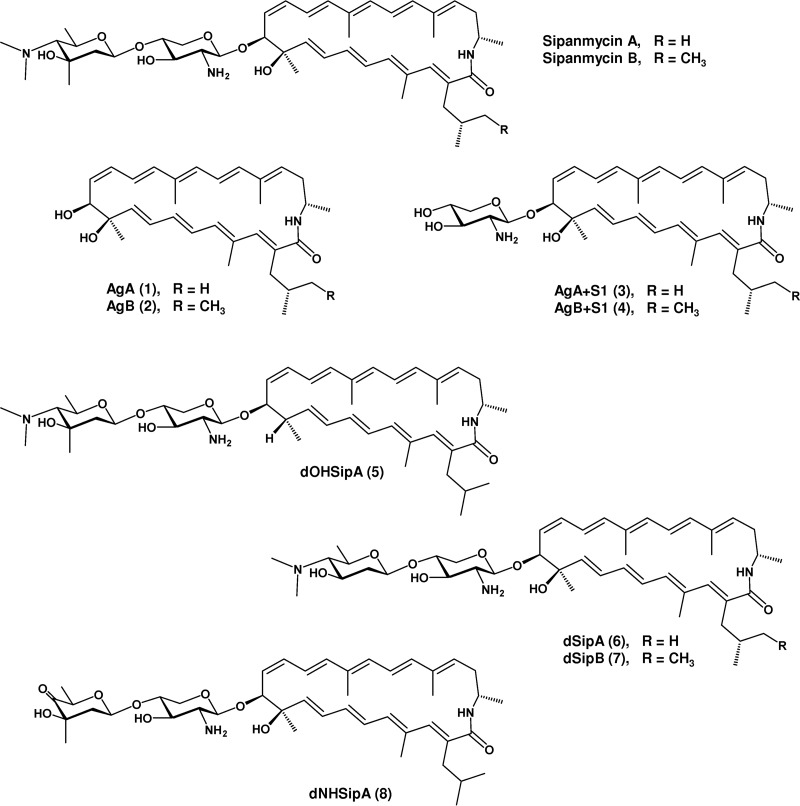
We have recently applied a targeted screening in the search for the presence of glycosylated bioactive natural products in a collection of Streptomyces strains isolated from leaf-cutting ants (13). By combining PCR-based screening, mass spectrometry (MS) dereplication, and generation of mutants, we identified two novel macrolactams, sipanmycins A and B (Fig. 1). Macrolactams are a growing family of macrocyclic polyketides that have attracted attention since they show potent bioactivities, such as antibacterial activity (rifamycin) (14), antifungal and antiviral activities (fluvirucins) (15), antiproliferative activity (vicenistatin and leinamycin) (16, 17), and even to overcome the resistance to chemotherapeutic drugs in certain tumor cell lines caused by overexpression of the antiapoptotic oncoprotein Bcl-xL (incednine) (18). Interestingly, many of the macrolactam family members have at least one deoxy sugar moiety attached to the macrocyclic ring, as in the case of sipanmycins, vicenistatin, cremimycin, fluvirucin B2, incednine, and silvalactam (Fig. 1). It has been demonstrated that this deoxy sugar is essential for the bioactivity of vicenistatin (19). Thus, the study of the glycosylation steps during glycosylated macrolactam biosynthesis could be a great opportunity to obtain different derivatives with improved biological properties.
FIG 1.
Structures of glycosylated macrolactams.
Sipanmycins A and B belong to the family of 24-membered macrolactams (Fig. 1) that include incednine (18) and silvalactam (20). The macrolactam rings of sipanmycin A and silvalactam are identical, but they differ in the number of amino sugars attached. In contrast, sipanmycins and incednine possess very similar macrolactam rings only differing in the side-chain substituent at C-2:an isobutyl chain or 2-methylbutyl (in sipanmycins A and B, respectively) and a methoxy group (in incednine). Both sipanmycins and incednine contain a disaccharide attached to the ring but with different amino sugars.
We report herein the identification of the gene cluster for sipanmycin biosynthesis (sip cluster) and the functional characterization of genes involved in deoxy sugar biosynthesis. Furthermore, we prove the complex glycosylation steps during sipanmycin biosynthesis that include the unexpected cooperation of two different glycosyltransferases for the attachment of each deoxy sugar, and the need for a P450-like helper protein in the incorporation of the second deoxy sugar. We also describe the isolation and structural elucidation of several novel macrolactam derivatives and test their antibiotic activity and cytotoxicity against several tumor cell lines.
RESULTS
The sipanmycin gene cluster, identification and characterization.
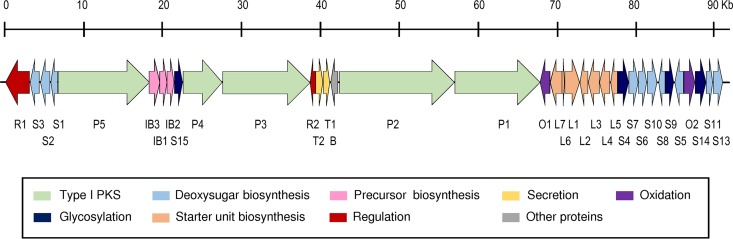
For the identification of the sip cluster in Streptomyces sp. strain CS149, the genome of this strain was sequenced and searched for the presence of putative BGCs using the bioinformatic tool antibiotics and Secondary Metabolite Analysis Shell (antiSMASH version 4) (21). Thirty-one putative BGCs were identified. They included three clusters containing nonribosomal peptide synthetases (NRPSs), three clusters containing polyketide synthases (PKSs; one type I and two type III), seven clusters comprising hybrid PKS-NRPS, and five clusters coding for enzymes involved in the biosynthesis of terpenes. In addition, antiSMASH analysis revealed the presence of other putative BGCs related to the biosynthesis of two lantipeptides, two butyrolactones, two siderophores, one oligosaccharide, one lasso peptide, one bacteriocin, one thiopeptide, ectoine, and melanin (see Table S1 in the supplemental material). Cluster 20, classified as belonging to the PKS type I-oligosaccharide class, appeared as a clear candidate to encode the sip cluster since it contained all genes coding for the different enzymatic functions that are necessary for the biosynthesis of sipanmycins (Table 1). In addition, amino acid sequence analysis of this cluster showed similarity to proteins from a Streptomyces sp. strain ML694-90F31 cluster involved in incednine (idn) biosynthesis (22) (Table 1). Furthermore, three genes of the cluster involved in amino sugar biosynthesis (sipS6, sipS7, and sipS10) contained DNA sequences that were previously used to generate sipanmycin-nonproducing mutants in this strain (13). In addition, a cluster very similar to the sip cluster is also present in Streptomyces sp. strain Tü 6075, with percentages of protein identity higher than 70% (Table 1). Comparison of the genetic organization between cluster 20 in CS149 and the idn cluster allowed the putative limits of the cluster to be defined. The sip cluster comprises approximately 93 kb and 36 open reading frames (ORFs) (Fig. 2 and Table 1). Genes coding for structural enzymes in the sip cluster can be divided into four different groups depending on which part of the final molecule they synthesize: (i) β-amino acid starter unit, (ii) polyketide chain, (iii) branched-chain extender unit, and (iv) deoxy sugar moieties.
TABLE 1.
Sipanmycin biosynthesis gene cluster in Streptomyces sp. CS149
| Protein | Proposed function | Protein encoded in Streptomyces sp. ML694-90F3, % identity/similarity | Proteins by Streptomyces sp. Tü 6075 accession no., % identity/similarity |
|---|---|---|---|
| SipR1 | Transcriptional regulator (LuxR family) | IdnR1, 54/67 | WP_075268275, 97/98 |
| SipS3 | Nucleoside-diphosphate sugar epimerase | IdnS3, 76/84 | WP_075263857, 99/99 |
| SipS2 | UDP-glucose 6-dehydrogenase | IdnS2, 72/80 | WP_075263856, 99/99 |
| SipS1 | N-Acetylglucosaminyl deacetylase | IdnS1, 65/78 | WP_075263855, 99/99 |
| SipP5 | Type I PKS | IdnP5, 64/74 | WP_075263854, 98/98 |
| SipIB3 | Crotonyl-CoA carboxylase/reductase | WP_075263853, 99/99 | |
| SipIB1 | 3-Oxoacyl-ACP synthase | WP_075263852, 99/99 | |
| SipIB2 | 3-Hydroxybutyryl-CoA dehydrogenase | WP_075268274, 99/98 | |
| SipS15 | Glycosyltransferase | IdnS15, 40/50 | WP_075263851, 96/97 |
| SipP4 | Type I PKS | IdnP4, 62/70 | WP_075263850, 97/97 |
| SipP3 | Type I PKS | IdnP3, 65/73 | WP_075263849, 95/96 |
| SipR2 | Transcriptional regulator (TetR family) | IdnR2, 58/73 | WP_075263848, 99/100 |
| SipT2 | ABC transporter | IdnT2, 73/83 | WP_075263847, 99/99 |
| SipT1 | ABC transporter | IdnT1, 68/83 | |
| SipB | Thioesterase | IdnB, 58/72 | WP_075263846, 98/98 |
| SipP2 | Type I PKS | IdnP2, 64/72 | WP_079181603, 70/75 |
| SipP1 | Type I PKS | IdnP1, 60/69 | WP_079181603, 89/90 |
| SipO1 | Cytochrome P450 | IdnO1, 82/89 | WP_075263845, 99/99 |
| SipL7 | AMP-dependent synthetase | IdnL7, 64/73 | WP_075263844, 94/95 |
| SipL6 | Acyl carrier protein | IdnL6, 58/70 | WP_075263843, 96/97 |
| SipL1 | AMP-dependent synthetase | IdnL1, 70/81 | WP_075263842, 98/98 |
| SipL2 | Malonyl-CoA-ACP transacylase | IdnL2, 69/77 | WP_079181433, 94/94 |
| SipL3 | Aminotransferase | IdnL3, 74/80 | WP_075268271, 98/98 |
| SipL4 | Lysine 2,3-aminomutase | IdnL4, 75/86 | WP_075263841, 99/99 |
| SipL5 | l-Proline amide hydrolase | IdnL5, 75/84 | WP_075268270, 99/99 |
| SipS4 | Glycosyltransferase | IdnS4, 77/83 | WP_079181602, 99/99 |
| SipS7 | Glucose-1-phosphate thymidylyltransferase | IdnS7, 63/77 | WP_075263839, 99/99 |
| SipS6 | dTDP-glucose 4,6-dehydratase | IdnS6, 66/74 | WP_075263838, 97/98 |
| SipS10 | NDP-hexose 2,3-dehydratase | IdnS10, 51/61 | WP_075263837, 98/98 |
| SipS8 | N-Methyltransferase | IdnS8, 58/71 | |
| SipS9 | Glycosyltransferase | IdnS9, 47/57 | WP_075263836, 98/98 |
| SipS5 | C-Methyltransferase | WP_075263835, 99/99 | |
| SipO2 | P450-derived glycosyltransferase activator | IdnO2, 47/57 | WP_075263834, 95/95 |
| SipS14 | Glycosyltransferase | IdnS14, 60/76 | WP_075263833, 99/99 |
| SipS11 | NDP-hexose-3-ketoreductase | IdnS11, 68/75 | WP_079181432, 96/98 |
| SipS13 | dTDP-4-dehydro-6-deoxyglucose aminotransferase | IdnS13, 74/83 | WP_075263832, 99/98 |
FIG 2.
Sipanmycin biosynthesis gene cluster. Gene names of the sipanmycin (sip) cluster are indicated beneath its corresponding ORF representation.
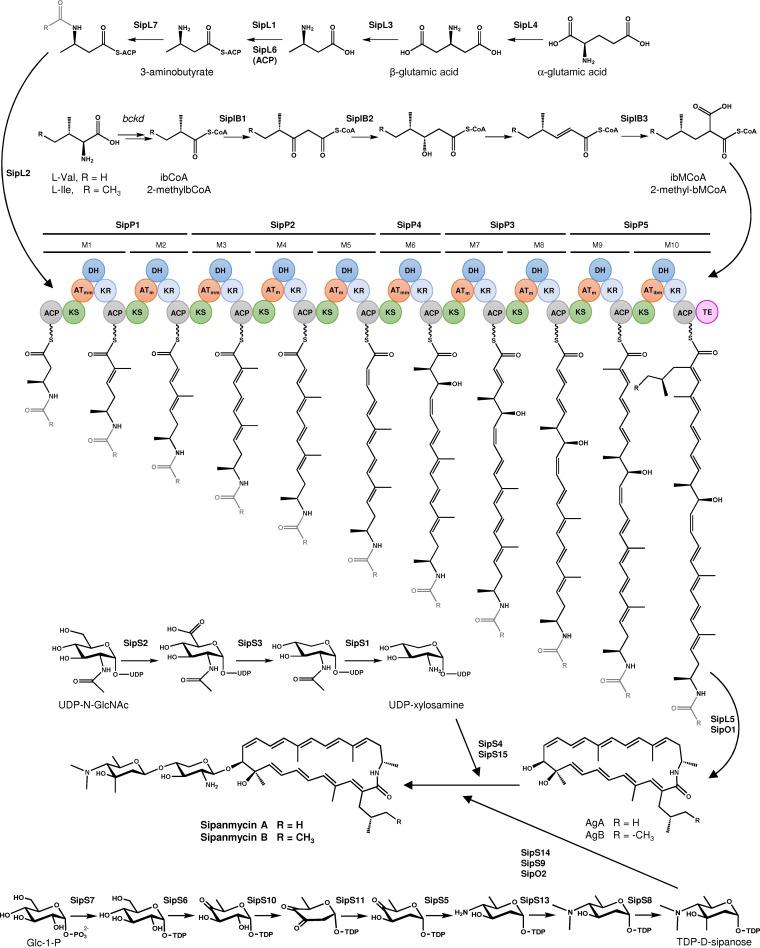
The chemical structure of sipanmycins suggests that their biosynthesis should start with the incorporation of a β-amino acid to the PKS assembly line. In silico analysis of the sip cluster showed the presence of seven genes (sipL1 to sipL7) putatively involved in the biosynthesis of this starter unit which are conserved in other BGCs involved in the biosynthesis of related macrolactams, such as incednine, vicenistatin, ML-449, cremimycin, and hitachimycin (22–26). Five genes coding for type I PKSs (sipP1 to sipP5) were found within the sip cluster, and their gene products showed high similarity to orthologs from idn cluster (ranging from 69 to 74%; Table 1). The fact that the chemical structure of the sipanmycin A aglycon is identical to the incednine one (except for the last unit introduced by the PKS), together with the same distribution of the 10 modules within PKS genes in sip and idn clusters, led us to assume that the order of PKS reactions is the same as that proposed for incednine biosynthesis, starting at SipP1 and ending at SipP5 (Fig. 3). A comparison of the predicted substrate specificities of AT domains revealed that the only difference between them in sipanmycins and incednine is AT10 (see Fig. S1 in the supplemental material at https://figshare.com/s/49de24d11120b7b114ba), which recognizes methoxymalonyl-acyl-carrier protein (methoxymalonyl-ACP) in incednine biosynthesis or, based on the chemical structure, isobutylmalonyl-CoA (ibMCoA) or 2-(2-methylbutyl)-malonyl-CoA (2-methylbMCoA) in sipanmycin A/B biosynthesis, respectively.
FIG 3.
Proposed biosynthesis pathway for sipanmycins.
As mentioned above, the last unit introduced by module 10 of SipP5 may be ibMCoA or 2-methylbMCoA. Bioinformatic analysis of the sip cluster revealed the presence of three genes putatively involved in the biosynthesis of this branched-chain extender unit, sipIB1 to sipIB3, which showed a high degree of similarity to divS, divT, and divR, respectively, involved in the biosynthesis of divergolides and germicidins in the mangrove endophyte Streptomyces sp. strain HKI0576 (27). For clarity and brevity, aglyca from sipanmycin A and sipanmycin B are referred to here as AgA and AgB, respectively.
The genes responsible for the biosynthesis of the two amino sugars attached to the sipanmycin aglyca are grouped into two separate subclusters within the sip cluster. The first one consisted of three genes (sipS1 to sipS3) that might be involved in the biosynthesis of UDP-xylosamine (Fig. 3).
The second subcluster (sip6, sip7, sip8, sip10, sip11, and sip13) might be involved in the biosynthesis of a N,N-dimethyl derivative of a monosaccharide which formally corresponds to a 3,5-diepimer of lemonose (the amino sugar contained in lemonomycin) (28, 29) and which we propose to trivially name d-sipanose (Fig. 3). For the transfer of the amino sugars to the aglyca, four glycosyltransferase (GT) genes are present in the cluster (sipS4, sipS9, sipS14, and sipS15) that are described below (Table 1 and Fig. 2).
Two gene products, SipO1 and SipO2, showed similarity to cytochrome P-450 monooxygenases (Table 1). A thorough analysis of their sequences revealed that SipO1 might act as a P450-monooxygenase catalyzing the C-10 hydroxylation of sipanmycin aglyca. In contrast, SipO2 lacks the Cys residue at the active site responsible for the coordination of the heme iron; thus, it could not act as a monooxygenase (Fig. S2, https://figshare.com/s/49de24d11120b7b114ba). These kinds of enzymes have been described as GT helper proteins with a chaperone-like function. These genes are normally located directly upstream of the corresponding GT gene, as is the case of sipO2. Several examples of GT auxiliary protein/GT systems have been described, including DesVIII/DesVII (methymycin/pikromycin) (30), TylM3/TylM2 (tylosin) (31), EryCII/EryCIII (erythromycin) (32), and AknT/AknS (aclacinomycin) (33). Thus, we proposed a similar role for SipO2 in sipanmycin biosynthesis (see below).
Apart from structural genes, two genes with a high degree of similarity to transcriptional regulators were found in the sip cluster (Table 1 and Fig. 2). Sequence analysis showed that sipR1 could code for a LuxR family transcriptional regulator (usually described as activators of secondary metabolite pathways) and sipR2 could code for a putative TetR family transcriptional repressor. In trans overexpression of sipR1 (149esipR1 strain) enhanced sipanmycin production during the first 3 days of cultivation in R5A medium (Fig. S3), supporting the role of SipR1 as an activator. After day 4, sipanmycin accumulation decreased in the wild-type and 149esipR1 strains, with this reduction being faster in 149esipR1, probably due to the higher depletion of precursor supply in this culture.
Finally, two putative ABC transporters, SipT1 and SipT2, would participate in sipanmycin export, and sipB may code for a type II thioesterase, often related to the removal of aberrant extender units loaded onto PKSs (34) (Table 1 and Fig. 2).
Involvement of four glycosyltransferases in the transfer of two amino sugars.
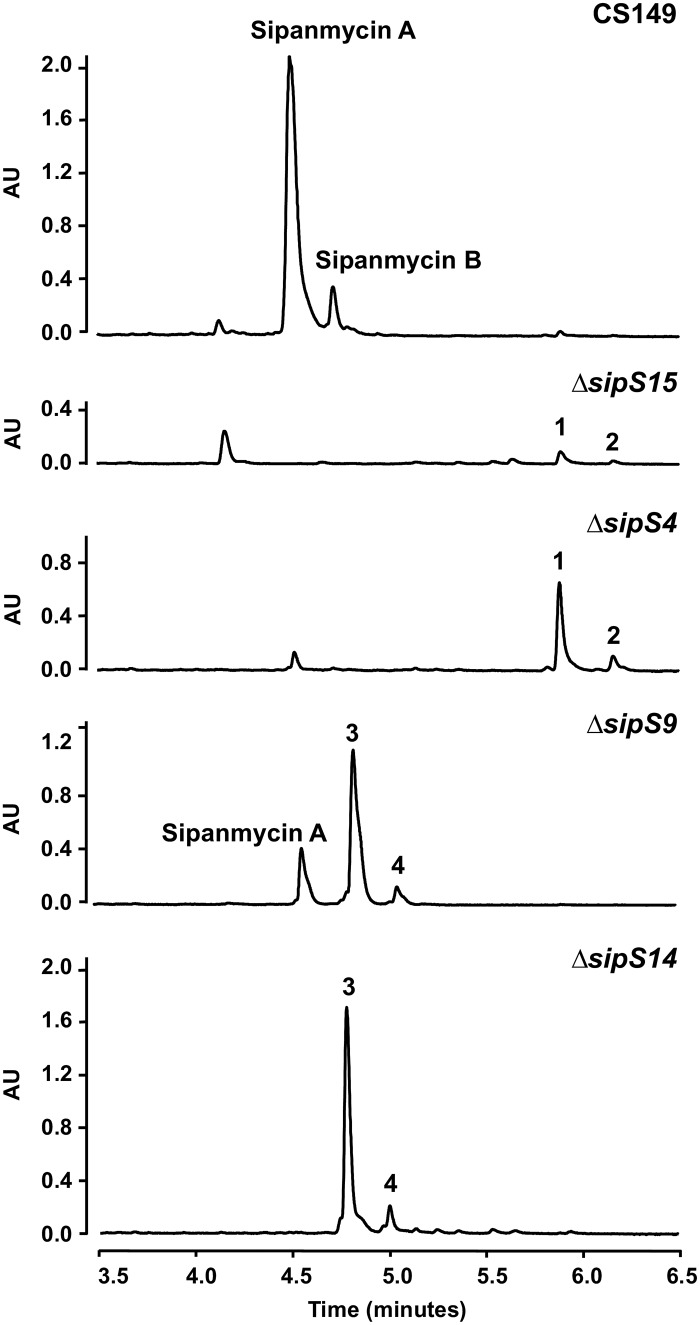
As mentioned above, sipanmycins contain two amino sugars in their structures, but four GT-encoding genes are present in the cluster (sipS4, sipS9, sipS14, and sipS15). To get further insight on the role and possible involvement of these GTs in amino sugar transfer, we individually inactivated the four genes by gene replacement through the insertion of the apramycin resistance cassette aac(3)-IV within each gene. The resultant strains were grown on R5A liquid medium and ethyl acetate extracts of cultures of each mutant analyzed by ultrahigh-performance liquid chromatography (UPLC). Inactivation of either sipS4 or sipS15 completely abolished the biosynthesis of sipanmycins, and instead, two new peaks were detected by UPLC (Fig. 4). These peaks showed the characteristic absorption spectrum of sipanmycins and ions in their mass spectra at m/z 492 [M+H]+ for compound 1 and m/z 506 [M+H]+ for compound 2, which matched the expected masses for AgA and AgB, respectively. The accumulation of compounds 1 and 2 was considerably higher in the ΔsipS4 mutant than in the ΔsipS15 mutant. On the other hand, the inactivation of sipS14 also blocked the production of sipanmycins A and B, while inactivation of sipS9 abolished the production of sipanmycin B, but trace amounts of sipanmycin A could be detected (Fig. 4). Both mutants showed the presence of two new peaks with the characteristic absorption spectrum of sipanmycins and ions at m/z 623 [M+H]+ for compound 3 and m/z 637 [M+H]+ for compound 4, both consistent with those expected for AgA and AgB, with just the first amino sugar of sipanmycins (d-xylosamine) attached, respectively (referred to here for brevity as AgA+S1 and AgB+S1, respectively) (Fig. 5). Compound 3 is nearly identical to silvalactam (20), only lacking methylation at 3′-OH in the amino sugar unit; thus, it can be named 3′-O-demethylsilvalactam. All four mutants recovered the capability to produce sipanmycins A and B upon complementation with the corresponding native gene expressed under the control of ermE*p in an integrative plasmid (Fig. S4), thus confirming that there were no polar effects upon amikacin resistance (Amr) cassette insertions. Major compounds (peaks 1 and 3) were purified by preparative high-performance liquid chromatography (HPLC) and their chemical structures confirmed by MS and nuclear magnetic resonance (NMR) (Fig. S5 to S16 and Table S2 for compound 1 and Fig. S17 to S24 and Table S3 for compound 3, https://figshare.com/s/49de24d11120b7b114ba).
FIG 4.
Effect of glycosyltransferase knockout experiments on sipanmycin biosynthesis. UPLC analysis of ethyl acetate extracts of mutants in the four GTs involved in the attachment of the two deoxy sugars to the sipanmycin aglycon. Peak 1, AgA; peak 2, AgB; peak 3, AgA+S1; peak 4, AgB+S1.
FIG 5.
Structures of sipanmycin derivatives obtained during this work.
The conclusion of these experiments is that the four GTs in the sip cluster are required for the transfer of the two amino sugars, with two GTs, SipS15 and SipS4, working together in the transfer of d-xylosamine, and the other two GTs, SipS9 and SipS14, working together in the transfer of d-sipanose.
Role of P450-like genes in sipanmycin biosynthesis.
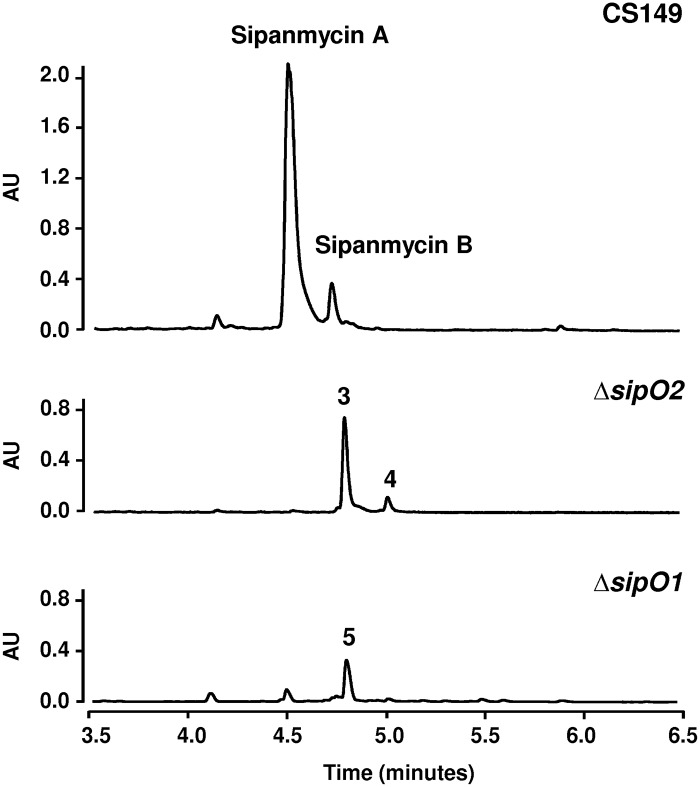
As mentioned above, two Sip proteins showed similarity to P450 monooxygenases, but only one seemed to be a real monooxygenase. To investigate the possible role of both proteins, we inactivated sipO1 and sipO2 by gene replacement. Analysis of cultures of the ΔsipO2 mutant revealed results similar to those obtained by the inactivation of GTs sipS9 and sipS14, an accumulation of AgA+S1 and AgB+S1 (compounds 3 and 4, respectively) (Fig. 6). These results suggest that SipO2 is an auxiliary protein of GTs SipS9 and SipS14 during the incorporation of the second amino sugar in sipanmycin biosynthesis. In cultures of the ΔsipO1 mutant, we could not detect sipanmycins but rather the accumulation of a new compound (compound 5) with the characteristic UV absorption spectrum of sipanmycins and a mass of m/z 778 [M+H]+ that matched that expected for sipanmycin A lacking the hydroxyl group at C-10 (10-deoxysipanmycin A [dOHSipA]; Fig. 6). This was confirmed after purification of the compound and elucidation of its chemical structure by high-resolution mass spectrometry (HRMS) and NMR (Fig. 5 and S25 to S35 and Table S4). Genetic complementation of each P450 monooxygenase-coding genes restored the wild-type phenotype in both mutant strains (Fig. S4).
FIG 6.
Effect of P450 cytochrome knockout experiments on sipanmycin biosynthesis. UPLC analysis of ethyl acetate extracts of mutants in sipO2 and sipO1 genes. Peak 3, AgA+S1; peak 4, AgB+S1; peak 5, dOHSipA.
Identification and role of aminotransferases and methyltransferases in amino sugar biosynthesis.
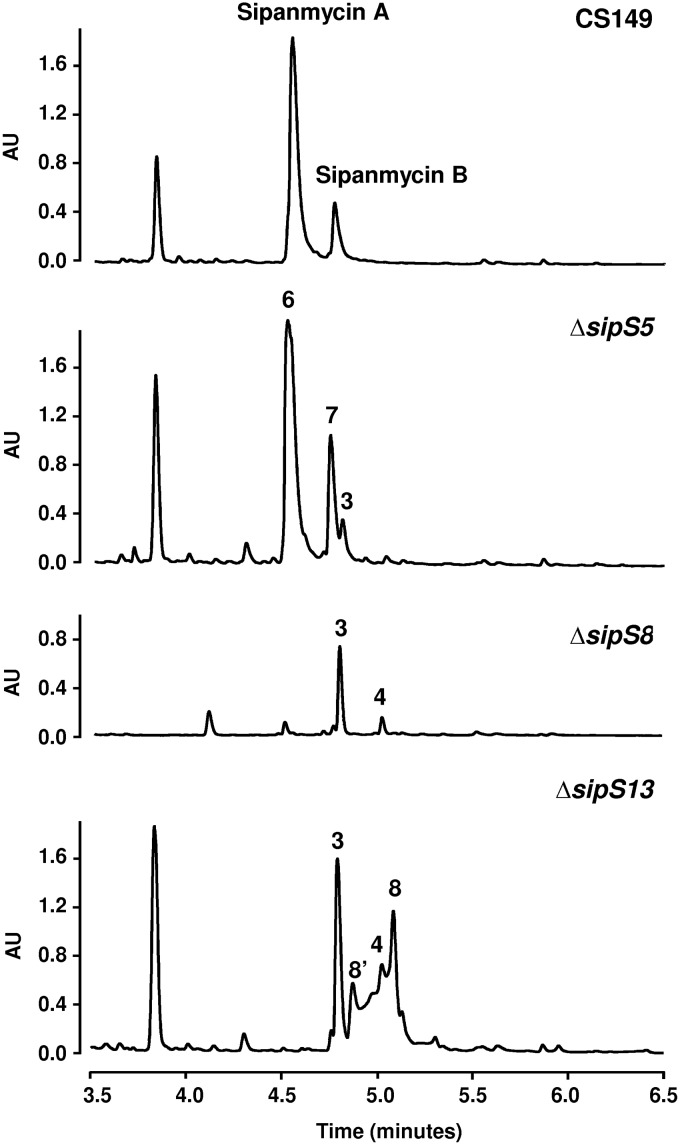
Biosynthesis of the second amino sugar requires the involvement of two methyltransferases and an aminotransferase. BLAST analysis of the sip cluster pointed to sipS8 and sipS5 as potential candidates as N- and C-methyltransferase genes, respectively, and to sipS13 as the aminotransferase gene. To clarify their role, we generated replacement mutants in each of these genes, analyzed the corresponding mutants, and isolated and determined the structures of the accumulated compounds. UPLC analysis of the ΔsipS5 mutant in comparison to the wild-type strain showed the absence of peaks corresponding to sipanmycins and the appearance of three new peaks (Fig. 7). Compounds in these peaks showed the characteristic UV absorption spectrum of sipanmycins and masses of m/z 781 [M+H]+ for compound 6, m/z 795 [M+H]+ for compound 7, and m/z 623 [M+H]+ for compound 3. Structural elucidation of compounds 6 and 7 identified them as 3″-demethyl-sipanmycins A and B (dSipA and dSipB), respectively, both lacking the methyl group at position C-3 in the second amino sugar, which in this case corresponds to N,N-dimethyl-d-pyrrolosamine, the enantiomer of the amino sugar found as l- form in the antitumor antibiotics lomaiviticin A and B (35) (Fig. 5 and S36 to S46 and Table S5 for compound 6 and Fig. S47 to S56 and Table S6 for compound 7). The third peak (compound 3) was the smallest one and corresponds to AgA+S1, confirming the role of SipS5 in the biosynthesis of the second amino sugar (Fig. 7).
FIG 7.
Effect of methyltransferases and aminotransferase knockout experiments on sipanmycin production. Chromatograms correspond to mutants in sipS5, sipS8, and sipS13 genes. Peak 3, AgA+S1; peak 4, AgB+S1; peak 6, dSipA; peak 7, dSipB; peak 8, dNHSipA; 8′, dNHSipA ketal derivative.
A comparative profile analysis of the ΔsipS8 mutant versus the wild-type strain showed the accumulation of two already-described compounds (compounds 3 and 4; Fig. 7), thus indicating that the inactivation of this N-methyltransferase gives rise to a sugar that is not recognized and transferred by the joint action of SipS14, SipS9, and SipO2.
Analysis of ethyl acetate extracts from the ΔsipS13 mutant showed the presence of a main peak (compound 3) and several small peaks absent in the wild-type strain that share absorption UV spectra with sipanmycins (Fig. 7). In a comparison of retention time and mass (m/z 637 [M+H]+), one of them was identified as compound 4, which is previously described in other mutants in this study. The other two peaks (labeled 8 and 8′ in Fig. 7) contained compounds that did not show clear pseudomolecular ion mass spectra, making a structural proposal impossible. After isolating compound 8, it was observed that its analytical chromatographic reanalysis surprisingly rendered two peaks (8 and 8′) rather than the expected single peak. This immediately pointed out a probable interconversion of both compounds under the acidic chromatographic conditions. Analysis of HRMS and NMR data allowed the identification of compound 8 (m/z 765 [M+H]+) as a sipanmycin A derivative with the dimethylated amino group at C-4 of the second amino sugar replaced by a ketone, rendering the monosaccharide 4-keto-β-d-olivomycose. Based on the structure of compound 8, it could be proposed that compound 8′ (m/z 783 [M+H]+) was an artifact that originated under acidic chromatographic conditions in which the ketone of compound 8 was hydrated rendering the monosaccharide 4-hydroxy-β-d-olivomycose. Compound 8 is proposed to be named 4″-deamino-4″-oxosipanmycin A (dNHSipA), and compound 8′ corresponded to its ketal derivative (Fig. 5 and S57 to S68 and Table S7). Taking into account the accumulation of compound 3 and the production phenotype of the ΔsipS13 mutant, it is probably the case that in the absence of amination at C-4, recognition of the second amino sugar by GTs SipS14 and SipS9 would be much less efficient.
The ΔsipS8, ΔsipS5, and ΔsipS13 mutants recovered the sipanmycin production after genetic complementation (Fig. S4).
In vitro cytotoxicity and antibiotic activity analyses of sipanmycins and derivatives.
Sipanmycins exerted potent cytotoxicity against several tumor cell lines (Table 2), with sipanmycin A being the one with strongest activity, showing 50% inhibitory concentrations (IC50s) ranging from 0.095 to 0.796 μM, depending on the cell line tested. This family of compounds neither exhibited antifungal activity against Candida albicans nor antibacterial activity against Escherichia coli. In contrast, they were strong antibiotics against Gram-positive bacteria Micrococcus luteus and Staphylococcus aureus (Table S8). The conclusions are that the deoxy sugars attached to the aglycon and the hydroxyl group at C-10 are essential for both the antiproliferative and antibiotic activities of this kind of compounds.
TABLE 2.
In vitro cytotoxicities of sipanmycins and derivatives
| Sipanmycin or derivative | Cytotoxicity (IC50 [μM]) by cell type (source) |
|||||
|---|---|---|---|---|---|---|
| 3T3 (fibroblasts) | A549 (lung) | HT29 (colon) | HL60 (leukemia) | CAPAN-1 (pancreas) | MDAMB231 (breast) | |
| Sipanmycin A | 0.302 | 0.117 | 0.189 | 0.796 | 0.178 | 0.095 |
| Sipanmycin B | 0.411 | 0.118 | 0.264 | 1.52 | 0.747 | 0.188 |
| AgA (compound 1) | ≫10 | ≫10 | ≫10 | 2.549 | ≫10 | ≫10 |
| AgB (compound 2) | ≫10 | >10 | >10 | 0.698 | ≫10 | >10 |
| AgA+S1 (compound 3) | 0.91 | 0.647 | 0.431 | 1.16 | 2.17 | 0.827 |
| dOHSipA (compound 5) | 1.614 | 1.75 | 0.873 | 1.089 | 2.68 | 1.2 |
| dSipA (compound 6) | 0.163 | 0.147 | 0.147 | 0.468 | 0.181 | 0.085 |
| dSipB (compound 7) | 0.293 | 0.211 | 0.213 | 1.129 | 0.493 | 0.241 |
DISCUSSION
It has been previously described that the deoxy sugar moieties attached to the aglyca from several bioactive compounds play a major role in their activities, usually participating in the interaction between the compound and its cellular target. Thus, we focused our attention on those genes involved in amino sugar biosynthesis and its transfer to the sipanmycin aglyca.
The sip cluster contains four genes encoding GTs, while only two amino sugars form part of the sipanmycin structure. There are reports in the literature of BGCs containing a lower number of GT genes with respect to the number of sugars attached to the aglycon (36–39), and it has been claimed that some GTs can act twice by incorporating the same sugar at different positions (38), incorporating different sugars (40, 41) or acting cooperatively to achieve dual O- and C-glycosyltransferase activities (39). However, reports on BGCs containing more GT genes than sugars in the molecule are scarce, with PM100117/PM100118 (42) and oleandomycin (43) being two examples of them. In the PM100117/PM100118 BGC, there is no clear role for a fourth GT in the biosynthesis pathway that otherwise involves the incorporation of three deoxy sugars (44). In the case of oleandomycin, which contains two deoxy sugars in its structure, the third GT acts as an inactivation mechanism (43). In addition to these examples, the BGC for incednine has also been reported to possess more GT genes than amino sugars in the molecule, and the authors proposed that two of the GT genes could be inactive since they lack internal protein sequences, which are conserved in active GTs, such as the essential histidine responsible for the deprotonation of the hydroxyl group of the glycosyl acceptor molecules (22). However, no experimental data were provided, supporting the lack of functionality of these GTs. In contrast, for the incorporation of the two amino sugars into the sipanmycin aglycon, we have demonstrated that the participation of the four GTs is required. They work together in pairs (Sip4 and Sip15 for the transfer of the first amino sugar and Sip9 and Sip14 for the second amino sugar). Apparently, none of them can replace the activity of any of the others, since independent inactivation of each of the four GT genes caused the disappearance of sipanmycins in culture broths (the exception is the ΔsipS9 mutant, in which the presence of a small amount of sipanmycin A still can be detected by LC-MS analysis).
Interestingly, the joint cooperation of Sip9 and Sip14 is not sufficient for the incorporation of the second amino sugar since there is a requirement for the presence of a P450 helper protein encoded by sipO2. The in vitro activity of the GT EryCIII has been shown to be enhanced by the addition of its auxiliary P450-like EryCII (45), and it has been proposed that this auxiliary protein has the function of stabilizing the fold and quaternary structure of its partner GT (32). Thus, the catalytic complex of GT-auxiliary cytochrome P450 adopted a tetrameric complex formed by two homodimers, one of the GT and the other of the auxiliary P450. In the case of the sipanmycin GT pair SipS9/SipS14 in combination with auxiliary SipO2, this picture might be slightly different. Since both GTs are required for the attachment of the second amino sugar and strictly require the assistance of SipO2, the structure of the complex might involve a GT heterodimer (SipS9/SipS14) in combination with the auxiliary enzyme. Interestingly, an important structural difference between these two GTs exists, in that SipS9 (homolog of IndS9 and proposed by Takahishi et al. [22] as inactive) is 99 amino acids shorter than SipS14. Structural elucidation and domain-swapping experiments have shown that GTs contain two distinctive domains for substrate binding, an N-terminal domain recognizing the aglycon and a C-terminal domain involved in nucleotidyl-diphosphate (NDP)-deoxy sugar binding (46–48). The main differences between SipS9 and SipS14 are located at the N-terminal region where the aglycon recognition domain might reside. According to these differences, one member of the GT heterodimer (SipS14) could be in charge of recognizing the sipanmycin aglycon, while the other member (SipS9), or both in cooperation, could be involved in amino sugar binding. Another fact that should be taken into account is that in this N-terminal region resides a putative motif involved in GT-auxiliary protein interaction (H-X-R-X5-D-X5-R-X12–20-D-P-X3-W-L-X12–18-E-X4-G) described for EryCIII and SpnP GTs (49). This motif is present in SipS14 but absent in SipS9 (Fig. S68, https://figshare.com/s/49de24d11120b7b114ba), indicating that the auxiliary protein SipO2 could interact only with one of the partners of the GT pair, giving SipS9 a secondary role in this glycosylation step, as indicated by the residual sipanmycin A production in the ΔsipS9 mutant (Fig. 4).
A similar scenario may occur in the first glycosylation step, with the difference that the GT pair SipS15/SipS4 is not dependent on the auxiliary protein SipO2, as xylosamine is attached to AgA and AgB in the ΔsipO2 mutant (compounds 3 and 4, respectively). In a way similar to the GT pair SipS9/SipS14, the N-terminal region of SipS15 is shorter than the SipS4 one, lacking the aglycon recognition domain and the motif involved in the auxiliary protein interaction (Fig. S68). This last motif is present in SipS4 but with a lower level of conservation than in other GTs previously described, or even in SipS14, which could explain the independence from SipO2 of this GT pair.
Xylosamine, transferred from UDP-xylosamine, is the first amino sugar directly attached to the aglycon and present in both sipanmycins and incednine, but the second amino sugar is different. Three genes present in the sip cluster would be responsible for the aforementioned differences. The sip5 gene would participate in the C-methylation step at C-3 of this monosaccharide, as was confirmed by the isolation of dSipA and dSipB (compounds 6 and 7, respectively), both lacking this methyl group. Furthermore, a sipanmycin A derivative with a ketone in C-4″ of the second deoxy sugar, instead of a dimethylated amino group (compound 8), has been isolated from cultures of the ΔsipS13 mutant, indicating the aminotransferase activity of SipS13. The incorporation of the unmethylated amino sugar (in compounds 6 and 7) or neutral deoxy sugar (in compound 8) shows a certain degree of flexibility of the SipS9/SipS14/SipO2 complex. Such flexibility regarding the sugar donor has been reported for other GTs, such as ElmGT (for the biosynthesis of elloramycin) (50, 51) and UrdGT2 (for the biosynthesis of urdamycin) (52). In contrast, the lack of the dimethyl group at the amino group at C-4 restricts this flexibility, since a mutant in which the sip8 N-dimethyltransferase gene was inactivated accumulates biosynthetic intermediates in which only the first amino sugar was incorporated (compounds 3 and 4).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptomyces sp. CS149, a sipanmycin producer, has been previously reported (13). Medium A (MA) (53) was used for sporulation. For metabolite production, strains were added to flasks containing 30 ml tryptic soy broth (TSB) medium and grown at 30°C and 250 rpm. After 24 h, this seed culture was used to inoculate 50 ml of R5A medium (36) to a final optical density at 600 nm (OD600) of 0.2. For intergeneric conjugation, mannitol soy medium (MS) (54) was used. Escherichia coli strains were grown in 2×TY medium supplemented with the appropriate antibiotic. All bacterial strains used in this work are listed in Table 3. Culture media were supplemented with antibiotics when needed, as follows: apramycin, 100 μg/ml for E. coli and 25 μg/ml for Streptomyces; hygromycin, 200 μg/ml; kanamycin, 25 μg/ml; tetracycline, 10 μg/ml; chloramphenicol, 25 μg/ml; and/or nalidixic acid, 50 μg/ml.
TABLE 3.
Strains and plasmids used in this work
| Strain or plasmid | Genotype/characteristics | Use | Reference(s) or source |
|---|---|---|---|
| Strains | |||
| Escherichia coli | |||
| DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 ϕ80Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Cloning | 63 |
| ET12567/pUB307 | dam13::Tn9 dcm6 hsdM hsdR recF143 zjj201::Tn10 galK2 galT22 ara14 lacY1 xyl5 leuB6 thi1 tonA31 rpsL136 hisG4 tsx78 mtli glnV44 F−, carries plasmid pUB307 | Intergeneric conjugation | 64, 65 |
| Streptomyces | |||
| CS149 | Wild type | 1 | |
| ΔsipS15 mutant | ΔsipS15∷aac(3)-IV | This work | |
| ΔsipS4 mutant | ΔsipS4∷aac(3)-IV | This work | |
| ΔsipS9 mutant | ΔsipS9∷aac(3)-IV | This work | |
| ΔsipS14 mutant | ΔsipS14∷aac(3)-IV | This work | |
| ΔsipS8 mutant | ΔsipS8∷aac(3)-IV | This work | |
| ΔsipS5 mutant | ΔsipS5∷aac(3)-IV | This work | |
| ΔsipS13 mutant | ΔsipS13∷aac(3)-IV | This work | |
| ΔsipO1 mutant | ΔsipO1∷hyg | This work | |
| ΔsipO2 mutant | ΔsipO2∷hyg | This work | |
| 149esipR1 | ermE*p-sipR1 | This work | |
| Plasmids | |||
| pSETec | Integrating vector, aac(3)-IV oriTRK2 intΦC31 attPΦC31 ermE*p | Gene overexpression | 66 |
| pSETeTc | Integrating vector, aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 ermE*p | Gene overexpression | 66 |
| pLHyg | lacZ bla hyg | Source of hyg gene | 57 |
| pUK21 | lacZ km | Backbone for pUKHyg construction | 58 |
| pUKHyg | lacZ km hyg | Replacement cassette construction | This work |
| pUO9090 | aac(3)-IV km | Replacement cassette construction | 67 |
| pHZ1358 | bla tsr oriTRK2 | Knockout mutant generation | 68 |
| pUO149ΔsipS15 | Carries up- and downstream flanking regions of sipS15 at both sides of aac(3)-IV km | Source of sipS15 replacement cassette | This work |
| pUH149ΔsipS15 | Carries sipS15 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS15 replacement | This work |
| pSETT149esipS15 | Carries sipS15 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS15 complementation | This work |
| pUO149ΔsipS4 | Carries up- and downstream flanking regions of sipS4 at both sides of aac(3)-IV km | Source of sipS4 replacement cassette | This work |
| pUH149ΔsipS4 | Carries sipS4 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS4 replacement | This work |
| pSETT149esipS4 | Carries sipS4 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS4 complementation | This work |
| pUO149ΔsipS9 | Carries up- and downstream flanking regions of sipS9 at both sides of aac(3)-IV km | Source of sipS9 replacement cassette | This work |
| pUH149ΔsipS9 | Carries sipS9 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS9 replacement | This work |
| pSETT149esipS9 | Carries sipS9 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS9 complementation | This work |
| pUO149ΔsipS14 | Carries up- and downstream flanking regions of sipS14 at both sides of aac(3)-IV km | Source of sipS14 replacement cassette | This work |
| pUH149ΔsipS14 | Carries sipS14 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS14 replacement | This work |
| pSETT149esipS14 | Carries sipS14 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS14 complementation | This work |
| pUO149ΔsipS8 | Carries up- and downstream flanking regions of sipS8 at both sides of aac(3)-IV km | Source of sipS8 replacement cassette | This work |
| pUH149ΔsipS8 | Carries sipS8 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS8 replacement | This work |
| pSETT149esipS8 | Carries sipS8 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS8 complementation | This work |
| pUO149ΔsipS5 | Carries up- and downstream flanking regions of sipS5 at both sides of aac(3)-IV km | Source of sipS5 replacement cassette | This work |
| pUH149ΔsipS5 | Carries sipS5 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS5 replacement | This work |
| pSETT149esipS5 | Carries sipS5 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS5 complementation | This work |
| pUO149ΔsipS13 | Carries up- and downstream flanking regions of sipS13 at both sides of aac(3)-IV km | Source of sipS13 replacement cassette | This work |
| pUH149ΔsipS13 | Carries sipS13 replacement cassette, aac(3)-IV bla tsr oriTRK2 | sipS13 replacement | This work |
| pSETT149esipS13 | Carries sipS13 under control of ermE*p aac(3)-IV tsr oriTRK2 intΦC31 attPΦC31 | sipS13 complementation | This work |
| pUK149ΔsipO1 | Carries up- and downstream flanking regions of sipO1 at both sides of hyg km | Source of sipO1 replacement cassette | This work |
| pHH149ΔsipO1 | Carries sipO1 replacement cassette, hyg bla tsr oriTRK2 | sipO1 replacement | This work |
| pSET149esipO1 | Carries sipO1 under control of ermE*p aac(3)-IV oriTRK2 intΦC31 attPΦC31 | sipO1 complementation | This work |
| pUK149ΔsipO2 | Carries up- and downstream flanking regions of sipO2 at both sides of hyg km | Source of sipO2 replacement cassette | This work |
| pHH149ΔsipO2 | Carries sipO2 replacement cassette, hyg bla tsr oriTRK2 | sipO2 replacement | This work |
| pSET149esipO2 | Carries sipO2 under control of ermE*p aac(3)-IV oriTRK2 intΦC31 attPΦC31 | sipO2 complementation | This work |
| pSET149eLuxR | Carries sipR1 under control of ermE*p aac(3)-IV oriTRK2 intΦC31 attPΦC31 | sipR1 overexpression | This work |
DNA manipulation and vectors.
DNA manipulations were performed according to standard procedures for E. coli (55) and Streptomyces spp. (56). All amplifications were carried out with the Herculase II Fusion high-fidelity polymerase (Agilent Technologies), and PCR conditions were initial denaturation at 98°C for 2 min, 30 cycles of 98°C for 30 s, 58°C for 30 s, and 72°C for x s (1 kb/30 s), and a final extension at 72°C for 3 min. All plasmids used in this work are summarized in Table 3. pUKHyg was constructed by cloning the PstI-HindIII fragment containing the hygromycin resistance cassette from pLHyg (57) into pUK21 (58).
DNA sequencing and analysis.
The Streptomyces sp. CS149 chromosome was sequenced at the Department of Biochemistry, University of Cambridge (Cambridge, UK) using Illumina MiSeq sequencing technology. De novo assemblies were achieved using default parameters in the Newbler assembler software version 2.9. Annotation was performed using the PGAAP pipeline (https://submit.ncbi.nlm.nih.gov/subs/genome/) (59). Database searching and sequence analysis were carried out with the bioinformatic tool antiSMASH (21).
Plasmid construction for gene replacement.
The same steps were carried out to construct sipO1 and sipO2 gene replacement plasmids. Briefly, upstream and downstream flanking regions of the genes of interest were amplified by PCR using genomic DNA from CS149 as the template and oligonucleotides indicated in Table 4. These amplicons were gel purified and cloned into pUKHyg (using restriction enzyme sites added to the oligonucleotide sequences) at both sides of the hygromycin resistance gene. The resulting plasmids were digested with SpeI, and the gene replacement cassette was cloned into a compatible XbaI site of pHZ1358, resulting in the final conjugative plasmids used for knockout experiments. For the rest of the genes, the same strategy was applied, but flanking regions were cloned into pUO9090 (instead of pUKHyg) at both sides of the apramycin resistance gene. Plasmids were introduced into their respective strains by intergeneric conjugation. Mutant strains were confirmed by PCR using oligonucleotides listed in Table 5.
TABLE 4.
Oligonucleotides used for gene replacement in Streptomyces sp. CS149
| Name | Sequence (5′–3′) | Restriction site | Use |
|---|---|---|---|
| 149dGT47.5F | TATGAATTCCGAACTCGTCCTGCTCGT | EcoRI | sipS15 replacement (5′ flanking region) (1,067 bp) |
| 149dGT47.5R | TATAAGCTTACCAGGCTGTGCAGTCGT | HindIII | |
| 149dGT47.3F | TATGGATCCGGAACTGCGGGAGGAGAT | BamHI | sipS15 replacement (3′ flanking region) (1,069 bp) |
| 149dGT47.3R | TATTCTAGACGCTTGATGACCTTCACCTG | XbaI | |
| 149dGT66.5F | TATGAATTCTGAGCTCGGACAGCACATAG | EcoRI | sipS4 replacement (5′ flanking region) (1,357 bp) |
| 149dGT66.5R | TATAAGCTTCGGAAACGACATCAACATGA | HindIII | |
| 149dGT66.3F | TATCATATGCGATCTCGTCCCCGAACT | NdeI | sipS4 replacement (3′ flanking region) (1,124 bp) |
| 149dGT66.3R | TATTCTAGAGATCTGGCTGGTGTCGATG | XbaI | |
| 149dGT71.5F | TATGAATTCCCACCACGTCAACGTCCAG | EcoRI | sipS9 replacement (5′ flanking region) (1,101 bp) |
| 149dGT71.5R | TATAAGCTTGAGGACGGCACGGTCAGC | HindIII | |
| 149dGT71.3F | TATCATATGGAGGTCGTGCCGCTGCTG | NdeI | sipS9 replacement (3′ flanking region) (1,127 bp) |
| 149dGT71.3R | TATTCTAGACGATCTGTCGCTCCTGTACG | XbaI | |
| 149dGT74.5F | TATGAATTCATGCGAGGGAATAAGTGACG | EcoRI | sipS14 replacement (5′ flanking region) (1,408 bp) |
| 149dGT74.5R | TATAAGCTTGCGAAGACCGTGAACAGAAC | HindIII | |
| 149dGT74.3F | TATGGATCCGTGACCAAGGACAGCGAGAG | BamHI | sipS14 replacement (3′ flanking region) (1,118 bp) |
| 149dGT74.3R | TATTCTAGACCTGTCTGATCCGTTCCAGT | XbaI | |
| 149-DORF70.5F | TATGAATTCGAACATCCCGATCGTCTCC | EcoRI | sipS8 replacement (5′ flanking region) (1,420 bp) |
| 149-DORF70.5R | TATAAGCTTTCTGCTCATTCTCGATGACG | HindIII | |
| 149-DORF70.3F | TATCATATGGCGTCTGTGACCGGAGCA | NdeI | sipS8 replacement (3′ flanking region) (1,377 bp) |
| 149-DORF70.3R | TATTCTAGAGTGATGCGCACCGAGGAC | XbaI | |
| 149-DORF72.5F | TATGAATTCTCCCAACCGGTTTCTCAA | EcoRI | sipS5 replacement (5′ flanking region) (1,527 bp) |
| 149-DORF72.5R | TATAAGCTTCTCGGCATGCAGAAATGG | HindIII | |
| 149-DORF72.3F | TATCATATGAGGCTGTCCGAGGCGTAGT | NdeI | sipS5 replacement (3′ flanking region) (1,369 bp) |
| 149-DORF72.3R | TATTCTAGAGCGATCAGGTCGTGAATTTG | XbaI | |
| 149dAT76.5F | TATGAATTCAGTTGCAGCAGGAGATCCAC | EcoRI | sipS13 replacement (5′ flanking region) (1,125 bp) |
| 149dAT76.5R | TATAAGCTTCGAGGATGGCGAGTTCAG | HindIII | |
| 149dAT76.3F | TATGGATCCGGAGTGACGCTCGACCAG | BamHI | sipS13 replacement (3′ flanking region) (1,419 bp) |
| 149dAT76.3R | TATTCTAGATTCAACTCGATGCACGACAC | XbaI | |
| 149dP450_58.5F | TATTCTAGATCGAAGTTCCGCATGCTCAT | XbaI | sipO1 replacement (5′ flanking region) (2,028 bp) |
| 149dP450_58.5R | TATGGATCCCCTCCATGTCGAACGGGTAG | BamHI | |
| 149dP450_58.3F | TATAAGCTTATTCAGTGGAAGTTCGGCGT | HindII | sipO1 replacement (3′ flanking region) (4,050 bp) |
| 149dP450_58.3R | TATAGATCTGGTATGGGGCGTGGTTTGTA | BglII | |
| 149dP450_73.5F | TATTCTAGAGTCCGGTTCCTTCCCTACG | XbaI | sipO2 replacement (5′ flanking region) (2,022 bp) |
| 149dP450_73.5R | TATCATATGAGCCTCGGGTCGATCTGATA | NdeI | |
| 149dP450_73.3F | TATAAGCTTGCTGCTTGAGAAACCGGTTG | HindIII | sipO2 replacement (3′ flanking region) (2,203 bp) |
| 149dP450_73.3R | TATAGATCTCGATGATCTCTCGACGCTGA | BglII |
TABLE 5.
Oligonucleotides used for PCR confirmation of Streptomyces sp. CS149 mutants
| Name | Sequence (5′–3′) | Gene | Amplicon size (bp) in: |
|
|---|---|---|---|---|
| Wild-type strain | Mutant strain | |||
| comp149dGT47.5 | TACGCGGAGTTCAAGGAACC | sipS15 | 1,223 | 1,739 |
| comp149dGT47.3b | CCGTTCACGAGAGGTCTGTG | |||
| comp149dGT66.5 | CAGGGGTCGGACGGTATTTC | sipS4 | 1,297 | 2,004 |
| comp149dGT66.3 | AGCGGTTCCTCCTGGTAGAT | |||
| comp149dGT71.5 | TCTTCCGGCGTTGTACTGTC | sipS9 | 1,262 | 1,769 |
| comp149dGT71.3 | GAACATCCCGATCGTCTCCG | |||
| comp149dGT74.5 | GCTGCTTGAGAAACCGGTTG | sipS14 | 1,474 | 1,684 |
| comp149dGT74.3 | GGTCCGCCTTCTCCTTGTC | |||
| comp149dMT70.5 | GACAGTACAACGCCGGAAGA | sipS8 | 1,266 | 2,037 |
| comp149dMT70.3 | GTGCACCCCGGAGAACTAC | |||
| comp149dMT72.5 | TGGGTGCGTCACTTATTCCC | sipS5 | 1,655 | 1,922 |
| comp149dMT72.3 | CAGCTGATCCTCACGGACG | |||
| comp149dAT76.5 | TGACGGCACATCTGACCTTC | sipS13 | 1,983 | 2,410 |
| comp149dAT76.3 | CTGGAGTCGCTGAACTTCGT | |||
| comp149dP450_58.5 | CCGAAGCACTACCACCATCT | sipO1 | 1,738 | 2,141 |
| comp149dP450_58.3 | AGCAGTACGCCTCGGTGAT | |||
| comp149dP450_73.5 | ATGCGAGGGAATAAGTGACG | sipO2 | 1,408 | 1,691 |
| comp149dP450_73.3 | GCGAAGACCGTGAACAGAAC | |||
| ermE | TAGCTTGCGAGTGTCCGT | sipR1 | ||
| 149-LuxR.3 | TATGAATTCATCGGCTACGAAGTGCTCTG | |||
Plasmid construction for genetic complementation of CS149 mutant strains.
The complete ORF of each gene was amplified by PCR using the oligonucleotides listed in Table 6, and the whole sequences were verified by sequencing. Amplicons were gel purified, digested with restriction enzymes indicated in Table 6, and cloned into pSETec (sipO1 and sipO2) or pSETeTc (rest of genes); thus, each gene was constitutively expressed under the control of the ermE* promoter (ermE*p).
TABLE 6.
Oligonucleotides used for genetic complementation of Streptomyces sp. CS149 mutants
| Name | Sequence (5′–3′) | Restriction site | Gene expression (size [bp]) |
|---|---|---|---|
| 149eGT47.F | TATGGATCCCCGATGTACACCCGATTCCC | BamHI | sipS15 (1,060) |
| 149eGT47.R | TATGAATTCGAGGTCTGTGTCGGTCGTG | EcoRI | |
| 149eGT66.F | TATGGATCCCAGGGGTCGGACGGTATTTC | BamHI | sipS4 (1,511) |
| 149eGT66.R | TATGAATTCGAGGAAAACCAGGCGGACAT | EcoRI | |
| 149eGT71.F | TATTCTAGATCTTCCGGCGTTGTACTGTC | XbaI | sipS9 (1,138) |
| 149eGT71.R | TATGAATTCACGATGGTGTTCCCGCTG | EcoRI | |
| 149eGT74.F | TATGGATCCGCTGCTTGAGAAACCGGTTG | BamHI | sipS14 (1,469) |
| 149eGT74.R | TATGAATTCGCCTTCTCCTTGTCGCGG | EcoRI | |
| 149eMT70.F | TATGGATCCCTCACGCTTGGGATGATCGT | BamHI | sipS8 (750) |
| 149eMT70.R | TATGAATTCGTGCTCCGGTCACAGACG | EcoRI | |
| 149eMT72.F | TATTCTAGACTGGCGGGATTTCGTTTGGA | XbaI | sipS5 (1,272) |
| 149eMT72.R | TATGAATTCCTCGGACTACGCCTCGGA | EcoRI | |
| 149eAT76.F | TATGGATCCCCATGGGGCCTGAACTCG | BamHI | sipS13 (1,202) |
| 149eAT76.R | TATGAATTCCTTCCCGTCTCACGCCAG | EcoRI | |
| 149eP450_58.F | TATGGATCCCCACGATGAGCACGGAGAAG | BamHI | sipO1 (1,203) |
| 149eP450_58.R | TATGAATTCCCTACCAGGTGACACGCAG | EcoRI | |
| 149eP450_73.F | TATGGATCCGCCGGATCATGACATGACCA | BamHI | sipO2 (1,344) |
| 149eP450_73.R | TATGAATTCTCCCAACCGGTTTCTCAAGC | EcoRI |
Plasmid construction for sipR1 overexpression.
sipR1 was amplified by PCR using the oligonucleotides 149-LuxR.5 (5′-TATGGATCCCGGGGGTAGGTGTACATGAG-3′) and 149-LuxR.3 (5′-TATGAATTCATCGGCTACGAAGTGCTCTG-3′). The resulting 3,042-bp fragment was gel purified, digested with BamHI/EcoRI, and cloned into pSETec, leading to the final plasmid pSET149eLuxR.
Extraction, analysis, and isolation of metabolites by UPLC and HPLC-MS.
Whole cultures (1 ml) of selected strains were extracted with one volume of ethyl acetate at three different times and analyzed by UPLC, as described previously (13).
Compounds were isolated from two-liter cultures of CS149 mutant strains in R5A medium, as previously described (13), but using a different mobile phase during semipreparative HPLC with Milli-Q (MQ) water plus 0.05% trifluoroacetic acid-acetonitrile (TFA-can; ranging from 40 to 45% ACN, depending on the compound).
Structural elucidation of new compounds.
Compounds 1, 3, 5, 6, 7, and 8 (and the artifact 8′) were analyzed by liquid chromatography-diode array detector-electrospray ionization-time of flight (LC-DAD-ESI-TOF) to determine their UV-Vis (DAD) spectra and their molecular formula based on the experimental accurate masses and the corresponding isotopic distribution. HRMS-based dereplication against our in-house library (60) and the Dictionary of natural products version 26:2 (61) was carried out to ensure their novelty. The structural elucidation of these compounds was carried out by detailed analysis of one-dimensional (1D) and two-dimensional (2D) NMR spectra, further assisted by comparison with the spectroscopic data reported for incednine (18), silvalactam (20), and especially sipanmycins A and B (13). Relative configurations were determined by coupling constants and nuclear Overhauser effect (NOE) analyses, assisted by comparison with the NMR data of sipanmycins A and B (13).
LC-DAD-ESI-TOF and NMR analyses.
HRMS and UV-Vis spectra were obtained by LC-DAD-ESI-TOF analyses performed using an Agilent 1200RR HPLC equipped with a SB-C8 column (2.1 by 30 mm; Zorbax) coupled to a Bruker maXis spectrometer. The chromatographic and ionization conditions were identical to those previously employed for sipanmycins A and B (13).
NMR spectra were recorded in deuterated methanol (CD3OD) at 24°C on a Bruker Avance III-500 MHz (500 and 125 MHz for 1H and 13C NMR, respectively) equipped with a 1.7-mm TCI MicroCryoProbe, using the residual solvent signal as an internal reference.
In vitro cytotoxicity and antibiotic activity assays.
The cytotoxic activities of compounds were tested against the following human tumor cell lines: colon adenocarcinoma (HT29), non-small cell lung cancer (A549), breast adenocarcinoma (MDA-MB-231), promyelocytic leukemia (HL-60), and pancreatic cancer (CAPAN-1) cells. The mouse embryonic fibroblast cell line NIH/3T3 was used as a control to evaluate cytotoxicity against nonmalignant cells. Cells were previously grown for a week on Dulbecco's modified Eagle medium (DMEM)–10% fetal bovine serum (FBS) medium, aliquoted to 5,000 cells per well in 96-well plates using the cell counting kit CCK-8 (catalog no. 96992; Sigma-Aldrich), and grown for an extra 24 h. The compounds were dissolved in DMEM. After the incubation, 10 μl of compound (in diverse concentrations) was added to each well and incubated for another 48 h. Last, 10 μl of CCK-8 reagent (Sigma-Aldrich) was added, left to develop for 2 h in the incubator, and measured at 450 nm using a Bio-Tek ELx 800 enzyme-linked immunosorbent assay (ELISA) kit (BioTek).
Antibiotic activity tests (in MIC) were performed in 96-wells microtiter plates. Fresh cultures of each microorganism were used as seed cultures to inoculate the plates, with the appropriate compound concentration, to a final OD of 0.1 and total volume of 150 μl per well. Plates were incubated overnight at 37°C (30°C for C. albicans).
Accession number(s).
The whole-genome shotgun project of Streptomyces sp. CS149 has been deposited at DDBJ/ENA/GenBank under the accession number PVZY00000000. The version described in this paper is version PVZY01000000. The nucleotide sequence of the sip cluster was deposited in the European Nucleotide Archive under accession number LT986736 and at the Minimum Information about a Biosynthetic Gene Cluster (MIBiG) repository (62) under the accession number BGC0001452.
ACKNOWLEDGMENTS
This research was supported by grants of the Spanish Ministry of Economy and Competitiveness (MINECO) (grant BIO2015-64161-R to J.A.S.) and “Apoyo a grupos de excelencia” Principado de Asturias-FEDER (grant FC-15-GRUPIN14-014).
We thank Fundación Bancaria Cajastur for financial support to C.O.
REFERENCES
- 1.Genilloud O. 2017. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep 34:1203–1232. doi: 10.1039/C7NP00026J. [DOI] [PubMed] [Google Scholar]
- 2.Zhang MM, Qiao Y, Ang EL, Zhao H. 2017. Using natural products for drug discovery: the impact of the genomics era. Expert Opin Drug Discov 12:475–487. doi: 10.1080/17460441.2017.1303478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulsen M, Oh DC, Clardy J, Currie CR. 2011. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS One 6:e16763. doi: 10.1371/journal.pone.0016763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valliappan K, Sun W, Li Z. 2014. Marine actinobacteria associated with marine organisms and their potentials in producing pharmaceutical natural products. Appl Microbiol Biotechnol 98:7365–7377. doi: 10.1007/s00253-014-5954-6. [DOI] [PubMed] [Google Scholar]
- 5.Holmes NA, Innocent TM, Heine D, Bassam MA, Worsley SF, Trottmann F, Patrick EH, Yu DW, Murrell JC, Schiøtt M, Wilkinson B, Boomsma JJ, Hutchings MI. 2016. Genome analysis of two Pseudonocardia phylotypes associated with Acromyrmex leafcutter ants reveals their biosynthetic potential. Front Microbiol 7:2073. doi: 10.3389/fmicb.2016.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behie SW, Bonet B, Zacharia VM, McClung DJ, Traxler MF. 2017. Molecules to ecosystems: actinomycete natural products in situ. Front Microbiol 7:2149. doi: 10.3389/fmicb.2016.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motley JL, Stamps BW, Mitchell CA, Thompson AT, Cross J, You J, Powell DR, Stevenson BS, Cichewicz RH. 2017. Opportunistic sampling of roadkill as an entry point to accessing natural products assembled by bacteria associated with non-anthropoidal mammalian microbiomes. J Nat Prod 80:598–608. doi: 10.1021/acs.jnatprod.6b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brötz-Oesterhelt H, Grond S, Peschel A, Krismer B. 2016. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 9.Jones MB, Nierman WC, Shan Y, Frank BC, Spoering A, Ling L, Peoples A, Zullo A, Lewis K, Nelson KE. 2017. Reducing the bottleneck in discovery of novel antibiotics. Microb Ecol 73:658–667. doi: 10.1007/s00248-016-0889-3. [DOI] [PubMed] [Google Scholar]
- 10.Chang FY, Brady SF. 2013. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Proc Natl Acad Sci U S A 110:2478–2483. doi: 10.1073/pnas.1218073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HS. 2017. Phylogeny-guided (meta)genome mining approach for the targeted discovery of new microbial natural products. J Ind Microbiol Biotechnol 44:285–293. doi: 10.1007/s10295-016-1874-z. [DOI] [PubMed] [Google Scholar]
- 12.Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA. 2014. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malmierca MG, González-Montes L, Pérez-Victoria I, Sialer C, Braña AF, García Salcedo R, Martín J, Reyes F, Méndez C, Olano C, Salas JA. 2018. Searching for glycosylated natural products in actinomycetes and identification of novel macrolactams and angucyclines. Front Microbiol 9:39. doi: 10.3389/fmicb.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floss HG, Yu TW. 2005. Rifamicin: mode of action, resistance and biosynthesis. Chem Rev 105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 15.Hedge V, Patel M, Horan A, Gullo V, Marquez J, Gunnarsson I, Gentile F, Loebenberg D, King A, Puar M, Pramanik B. 1992. Macrolactams: a novel class of antifungal antibiotics produced by Actinomadura spp. SCC 1776 and SCC 1777. J Antibiot 45:624–632. doi: 10.7164/antibiotics.45.624. [DOI] [PubMed] [Google Scholar]
- 16.Shindo K, Kamishohara M, Odagawa A, Matsuoka M, Kawai H. 1993. Vicenistatin, a novel 20-membered macrocyclic lactam antitumor antibiotic. J Antibiot 46:1076–1081. doi: 10.7164/antibiotics.46.1076. [DOI] [PubMed] [Google Scholar]
- 17.Hara M, Asano K, Kawamoto I, Takiguchi T, Katsumata S, Takahashi K, Nakano H. 1989. Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J Antibiot 42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 18.Futamura Y, Sawa R, Umezawa Y, Igarashi M, Nakamura H, Hasegawa K, Yamasaki M, Tashiro E, Takahashi Y, Akamatsu Y, Imoto M. 2008. Discovery of incednine as a potent modulator of the anti-apoptotic function of Bcl-xL from microbial origin. J Am Chem Soc 130:1822–1823. doi: 10.1021/ja710124p. [DOI] [PubMed] [Google Scholar]
- 19.Matsushima Y, Nakayama T, Fujita M, Bhandari R, Eguchi T, Shindo K, Kakinuma K. 2001. Isolation and structure elucidation of vicenistatin M, and importance of the vicenisamine amino sugar for exerting cytotoxicity of vicenistatin. J Antibiot 54:211–219. doi: 10.7164/antibiotics.54.211. [DOI] [PubMed] [Google Scholar]
- 20.Schultz D, Nachtigall J, Geisen U, Kalthoff H, Imhoff JF, Fiedler HP, Süssmuth RD. 2012. Silvalactam, a 24-membered macrolactam antibiotic produced by Streptomyces sp. Tü 6392. J Antibiot 65:369–372. doi: 10.1038/ja.2012.33. [DOI] [PubMed] [Google Scholar]
- 21.Blin K, Wolf T, Chevrette MG, Lu X, Schwale CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahishi M, Fumitaka K, Eguchi T. 2013. Identification of the incednine biosynthetic gene cluster: characterization of novel β-glutamate-β-decarboxylase IdnL3. J Antibiot 66:691–699. doi: 10.1038/ja.2013.76. [DOI] [PubMed] [Google Scholar]
- 23.Ogasawara Y, Katayama K, Minami A, Otsuka M, Eguchi T, Kakinuma K. 2004. Cloning, sequencing, and functional analysis of the biosynthetic gene cluster of macrolactam antibiotic vicenistatin in Streptomyces halstedii. Chem Biol 11:79–86. doi: 10.1016/j.chembiol.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Jørgensen H, Degnes KF, Dikiy A, Fjaervik E, Klinkenberg G, Zotchev SB. 2010. Insights into the evolution of macrolactam biosynthesis through cloning and comparative analysis of the biosynthetic gene cluster for a novel macrocyclic lactam, ML-449. Appl Environ Microbiol 76:283–293. doi: 10.1128/AEM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amagai K, Takaku R, Kudo F, Eguchi T. 2013. A unique amino transfer mechanism for constructing the β-amino fatty acid starter unit in the biosynthesis of the macrolactam antibiotic cremimycin. Chembiochem 14:1998–2006. doi: 10.1002/cbic.201300370. [DOI] [PubMed] [Google Scholar]
- 26.Kudo F, Kawamura K, Uchino A, Miyanaga A, Numakura M, Takayanagi R, Eguchi T. 2015. Genome mining of the hitachimycin biosynthetic gene cluster: involvement of a phenylalanine-2,3-aminomutase in biosynthesis. Chembiochem 16:909–914. doi: 10.1002/cbic.201500040. [DOI] [PubMed] [Google Scholar]
- 27.Xu Z, Ding L, Hertweck C. 2011. A branched extender unit shared between two orthogonal polyketide pathways in an endophyte. Angew Chem Int Ed Engl 50:4667–4670. doi: 10.1002/anie.201008265. [DOI] [PubMed] [Google Scholar]
- 28.He H, Shen B, Carter GT. 2000. Structural elucidation of lemonomycin, a potent antibiotic from Streptomyces candidus. Tetrahedron Lett 41:2067–2071. doi: 10.1016/S0040-4039(00)00116-7. [DOI] [Google Scholar]
- 29.Briegel AC, Cummings AK, Smith GR, Doroski MD, Boyko WJ, Piro NA, Kassel WS, Giuliano RM. 2015. Synthesis of lemonose derivatives: methyl 4-amino-3-O,4-N-carbonyl-2,4,6-trideoxy-3-C-methyl-α-l-lyxo-pyranoside and its phenyl thioglycoside. Carbohydr Res 409:63–68. doi: 10.1016/j.carres.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Hong JS, Park SJ, Parajuli N, Park SR, Koh HS, Jung WS, Choi CY, Yoon YJ. 2007. Functional analysis of desVIII homologues involved in glycosylation of macrolide antibiotics by interspecies complementation. Gene 386:123–130. doi: 10.1016/j.gene.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Melançon CE III, Takahashi H, Liu HW. 2004. Characterization of TylM3/TylM2 and MydC/MycB pairs required for efficient glycosyltransfer in macrolide antibiotic biosynthesis. J Am Chem Soc 126:16726–16727. doi: 10.1021/ja043900e. [DOI] [PubMed] [Google Scholar]
- 32.Moncrieffe MC, Fernandez MJ, Spiteller D, Matsumura H, Gay NJ, Luisi BF, Leadlay PF. 2012. Structure of the glycosyltransferase EryCIII in complex with its activating P450 homologue EryCII. J Mol Biol 415:92–101. doi: 10.1016/j.jmb.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu W, Leimkuhler C, Gatto GJ Jr, Kruger RG, Oberthür M, Kahne D, Walsh CT. 2005. AknT is an activating protein for the glycosyltransferase AknS in l-aminodeoxy sugar transfer to the aglycone of aclacinomycin A. Chem Biol 12:527–534. doi: 10.1016/j.chembiol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 34.Heathcote ML, Staunton J, Leadlay PF. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem Biol 8:207–220. doi: 10.1016/S1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 35.He H, Ding WD, Bernan VS, Richardson AD, Ireland CM, Greenstein M, Ellestad GA, Carter GT. 2001. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J Am Chem Soc 123:5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 36.Fernández E, Weissbach U, Sánchez Reillo C, Braña AF, Méndez C, Rohr J, Salas JA. 1998. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J Bacteriol 180:4929–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco G, Fernández E, Fernández MJ, Braña AF, Weissbach U, Künzel E, Rohr J, Méndez C, Salas JA. 2000. Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. Mol Gen Genet 262:991–1000. doi: 10.1007/PL00008667. [DOI] [PubMed] [Google Scholar]
- 38.Luzhetskyy A, Fedoryshyn M, Dürr C, Taguchi T, Novikov V, Bechthold A. 2005. Iteratively acting glycosyltransferases involved in the hexasaccharide biosynthesis of landomycin A. Chem Biol 12:725–729. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Salem SM, Weidenbach S, Rohr J. 2017. Two cooperative glycosyltransferases are responsible for the sugar diversity of saquayamycins isolated from Streptomyces sp. KY 40-1. ACS Chem Biol 12:2529–2534. doi: 10.1021/acschembio.7b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu W, Leimkuhler C, Oberthür M, Kahne D, Walsh CT. 2004. AknK is an l-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A. Biochemistry 43:4548–4558. doi: 10.1021/bi035945i. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Pahari P, Kharel MK, Chen J, Zhu H, Van Lanen SG, Rohr J. 2012. Cooperation of two bifunctional enzymes in the biosynthesis and attachment of deoxy sugars of the antitumor antibiotic mithramycin. Angew Chem Int Ed Engl 51:10638–10642. doi: 10.1002/anie.201205414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salcedo RG, Olano C, Gómez C, Fernández R, Braña AF, Méndez C, de la Calle F, Salas JA. 2016. Characterization and engineering of the biosynthesis gene cluster for antitumor macrolides PM100117 and PM100118 from a marine actinobacteria: generation of a novel improved derivative. Microb Cell Fact 15:44. doi: 10.1186/s12934-016-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quirós LM, Aguirrezabalaga I, Olano C, Méndez C, Salas JA. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol 28:1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 44.Salcedo RG, Olano C, Fernández R, Braña AF, Méndez C, de la Calle F, Salas JA. 2016. Elucidation of the glycosylation steps during biosynthesis of antitumor macrolides PM100117 and PM100118 and engineering for novel derivatives. Microb Cell Fact 15:187. doi: 10.1186/s12934-016-0591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. 2005. In vitro reconstitution of EryCIII activity for the preparation of unnatural macrolides. J Am Chem Soc 127:14128–14129. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. 2003. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci U S A 100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. 2003. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A 100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truman AW, Dias MVB, Wu S, Blundell TL, Huang FL, Spencer JB. 2009. Chimeric glycosyltransferases for the generation of hybrid glycopeptides. Chem Biol 16:676–685. doi: 10.1016/j.chembiol.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Isiorho EA, Jeon BS, Kim NH, Liu HW, Keatinge-Clay AT. 2014. Structural studies of the spinosyn forosaminyltransferase, SpnP. Biochemistry 53:4292–4301. doi: 10.1021/bi5003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco G, Patallo EP, Braña AF, Trefzer A, Bechthold A, Rohrm J, Méndez C, Salas JA. 2001. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem Biol 8:253–263. doi: 10.1016/S1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 51.Lombó F, Gibson M, Greenwell L, Braña AF, Rohr J, Salas JA, Méndez C. 2004. Engineering biosynthetic pathways for deoxy sugars: branched-chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem Biol 11:1709–1718. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 52.Hoffmeister D, Drager G, Ichinose K, Rohr J, Bechthold A. 2003. The C-glycosyltransferase UrdGT2 is unselective toward d- and l-configured nucleotide-bound rhodinoses. J Am Chem Soc 125:4678.4679. doi: 10.1021/ja029645k. [DOI] [PubMed] [Google Scholar]
- 53.Sánchez L, Braña AF. 1996. Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology 142:1209–1220. doi: 10.1099/13500872-142-5-1209. [DOI] [PubMed] [Google Scholar]
- 54.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. 1989. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol 31:272–277. doi: 10.1007/BF00258408. [DOI] [Google Scholar]
- 55.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 56.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 57.Olano C, Wilkinson B, Sánchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Braña AF, Martin CJ, Oliynyk M, Méndez C, Leadlay PF, Salas JA. 2004. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem Biol 11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Vieira J, Messing J. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189–194. doi: 10.1016/0378-1119(91)90365-I. [DOI] [PubMed] [Google Scholar]
- 59.Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O. 2008. Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS 12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pérez-Victoria I, Martín J, Reyes F. 2016. Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Med 82:857–871. doi: 10.1055/s-0042-101763. [DOI] [PubMed] [Google Scholar]
- 61.Buckingham J. 2017. Dictionary of natural products on DVD, version 26.2 CRC Press, London, United Kingdom. [Google Scholar]
- 62.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJ, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, et al. . 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 64.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 11:61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 65.Flett F, Mersinias V, Smith CP. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 66.Cano-Prieto C, García-Salcedo R, Sánchez-Hidalgo M, Braña AF, Fiedler HP, Méndez C, Salas JA, Olano C. 2015. Genome mining of Streptomyces sp. Tü 6176: characterization of the nataxazole biosynthesis pathway. Chembiochem 16:1461–1473. doi: 10.1002/cbic.201500153. [DOI] [PubMed] [Google Scholar]
- 67.Peláez AI, Ribas-Aparicio RM, Gomez A, Rodicio MR. 2001. Structural and functional characterization of the recR gene of Streptomyces. Mol Genet Genomics 265:663–672. doi: 10.1007/s004380100460. [DOI] [PubMed] [Google Scholar]
- 68.Sun Y, He X, Liang J, Zhou X, Deng Z. 2009. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl Microbiol Biotechnol 82:303–310. doi: 10.1007/s00253-008-1793-7. [DOI] [PubMed] [Google Scholar]