The bacterial plant pathogen Xylella fastidiosa causes incurable diseases in multiple hosts, including grape, citrus, and blueberry. Historically restricted to the Americas, it was recently found to cause epidemics in olives in Italy and to infect other hosts in Europe and Asia. In this study, we report a short protocol to create deletion and complemented mutants using fusion PCR and natural transformation. We also determined the distinct function of two pilin paralogs, the main structural component of TFP involved in twitching motility, which allows this bacterium to move inside the xylem vessels against the flow. One of the paralogs is needed for twitching movement, whereas the other does not have an effect on motility but influences the number and position of TFP. Since twitching motility is fundamental for the virulence of this xylem-limited bacterium, this study contributes to the understanding of the regulation of virulence by this pathogen.
KEYWORDS: Xylella fastidiosa, fastidious prokaryote, hyperpiliation, mutagenesis, natural transformation systems, paralog, pilA, pilin, twitching
ABSTRACT
Twitching motility is one of the major virulence factors of the plant-pathogenic bacterium Xylella fastidiosa, and it is mediated by type IV pili (TFP) that are present at one of the cell poles. Genome analysis of X. fastidiosa showed the presence of at least four paralogs of the gene pilA, which encodes the TFP major pilin subunit. However, whether all of these paralogs have a functional role in TFP structure and function is unknown. Here, using a short and reliable protocol based on overlap extension PCR and natural transformation, deletion mutants of two pilA paralogs (pilA1 PD1924 and pilA2 PD1926) were generated in two X. fastidiosa subsp. fastidiosa strains, WM1-1 and TemeculaL, followed by assessment of twitching motility and biofilm formation. Deletion of pilA2 caused loss of twitching motility, whereas deletion of pilA1 did not influence twitching motility but caused hyperpiliation and extended distribution of TFP along the sides of the cell. Loss of twitching motility due to pilA2 deletion was restored when a wild-type copy of the pilA2 gene was added at a neutral site in the genome of mutants in both wild-type backgrounds. This study demonstrates that PCR templates generated by overlap extension PCR can be successfully used to rapidly generate gene knockouts and perform genetic complementation in X. fastidiosa, and that twitching motility in X. fastidiosa is controlled by regulating the transcription of the major pilin subunit, pilA2.
IMPORTANCE The bacterial plant pathogen Xylella fastidiosa causes incurable diseases in multiple hosts, including grape, citrus, and blueberry. Historically restricted to the Americas, it was recently found to cause epidemics in olives in Italy and to infect other hosts in Europe and Asia. In this study, we report a short protocol to create deletion and complemented mutants using fusion PCR and natural transformation. We also determined the distinct function of two pilin paralogs, the main structural component of TFP involved in twitching motility, which allows this bacterium to move inside the xylem vessels against the flow. One of the paralogs is needed for twitching movement, whereas the other does not have an effect on motility but influences the number and position of TFP. Since twitching motility is fundamental for the virulence of this xylem-limited bacterium, this study contributes to the understanding of the regulation of virulence by this pathogen.
INTRODUCTION
Xylella fastidiosa is a Gram-negative, xylem-limited, insect-vectored, plant-pathogenic bacterium causing incurable diseases and substantial economic losses to the production of grapevines, citrus, coffee, plum, and almond (1, 2). Pierce's disease (PD) of grapevines in the United States and citrus variegated chlorosis (CVC) in Brazil, two of the most impactful diseases caused by X. fastidiosa, are major limiting factors in the production of these crops in those countries (2). Xylem sap-feeding insect vectors, including sharpshooters and spittlebugs, transmit and inoculate the bacterial cells into xylem vessels, where the cells attach to the surface, form biofilms, and move along and across the xylem vessels (3–5). The severity of disease symptoms depends on the extent of xylem vessel occlusions produced by bacterial aggregates (3). Therefore, cell aggregation and systemic colonization of the xylem vessels are considered the major determinants of pathogenicity in X. fastidiosa.
X. fastidiosa cells are nonflagellated but possess two types of pili at one of their cell poles. Short type I pili (0.4 to 1.0 μm) are involved in cell attachment and biofilm formation, and long type IV pili (TFP) (1.0 to 5.8 μm) are involved in twitching motility (6–8). Twitching motility involves flagellum-independent translocation of cells on a moist surface, facilitated by extension, tethering, and retraction of TFP (9). It was demonstrated in X. fastidiosa that a mutant lacking type I pili (fimA mutant) was biofilm deficient but twitching enhanced, whereas the opposite phenotype was true for mutants lacking TFP (6–8). Moreover, mutants that were twitching enhanced traveled further upstream from the point of inoculation in grapevines (8). In addition, X. fastidiosa wild-type (WT) strains that lacked or exhibited reduced twitching motility (10) had less severe disease symptoms in the model plant tobacco and in the natural host blueberry (11, 12). A polygalacturonase gene mutant that is compromised in lateral movement due to its inability to degrade xylem pit membranes was avirulent in grapevines (4) and was found to be twitching deficient in another study (13). Furthermore, seven transposon mutants, including a mutant of hemagglutinin that mediates cell-cell aggregation, showed hypervirulent phenotypes and faster movement than that of wild-type strains in grapevines (14). Previous studies demonstrated the role of cell-cell communication system involving a diffusible signaling factor (DSF) in regulating host colonization, movement, and biofilm formation in X. fastidiosa (15). Interestingly, it was shown that a mutant that was unable to produce DSF was hypervirulent to grapevines (15) and had increased expression of several TFP genes compared to that of the wild-type strain (16). Together these observations suggest that motility of cells is one of the important virulence factors of X. fastidiosa, although this has not been directly demonstrated for this bacterium. Nevertheless, involvement of TFP and twitching motility in host colonization and virulence was demonstrated for other Gram-negative bacteria, including human pathogens, such as Pseudomonas aeruginosa (17), Neisseria meningitidis (18), Neisseria gonorrhoeae (19), and plant pathogens such as Ralstonia solanacearum (20), Acidovorax citrulli (21), Xanthomonas citri (22), and Pseudomonas syringae (23, 24). TFP were also shown to contribute to other bacterial functions like biofilm formation, electron transfer (9, 25), and natural DNA transformation (26), in addition to twitching motility.
TFP are flexible, filamentous structures mainly composed of polymers of a single pilin protein subunit that is encoded by pilA gene(s); however, multiple protein components are involved during TFP biogenesis and function. For example, in P. aeruginosa, where twitching motility and TFP have been characterized in detail, approximately 40 genes are required for the function of TFP, including genes for the pilin subunit, pilin assembly and retraction, and regulatory genes (9). Genomes of X. fastidiosa contain numerous genes (27, 28) that share signature motifs and properties of well-described pilus genes. The role in X. fastidiosa of a few of these genes in TFP production and twitching motility was demonstrated in previous studies (6–8, 29, 30). At least six open reading frames (ORFs) that encode characteristic pilin subunits (pilA homologs) were predicted in the genome of CVC strain 9a5c and one of the ORFs was found to be regulated by RpoN, the alternative sigma 54 (σ54) factor (31). At least four homologs of the pilA ORFs are also present in the grapevine strain Temecula1 (28). Protein products of two of these homologs, designated PilA1 (PD1924) and PilA2 (PD 1926) in previous studies (16, 32), share functional features of the TFP pilin subunit (9). Whole-transcriptome analysis by transcriptome sequencing (RNA-Seq) showed that both of these pilA paralogs are expressed in Temecula1, although the expression of pilA2 was greater than that of pilA1 (32). However, the exact role of these pilA paralogs in X. fastidiosa remains poorly understood, and whether both or only one of them has a major functional role in TFP formation and twitching motility is unknown. A clear understanding of this issue could help researchers in recognizing the regulation of TFP formation and twitching motility in X. fastidiosa.
The objective of this study was to characterize the function of the two pilin paralogs, pilA1 (PD1924) and pilA2 (PD1926), in two Xylella fastidiosa subsp. fastidiosa strains, WM1-1 and TemeculaL. We found that while pilA1 was not necessary for twitching motility (unlike pilA2), mutation on this gene generated hyperpiliated cells. For this study, we designed a new protocol for mutagenesis that simplifies and expedites the protocols used before, which relied on either electroporation (30, 33) or natural competence (34, 35) to introduce plasmids for homologous recombination. The new protocol for gene deletion and complementation used in this study utilizes overlap extension PCR and natural genetic transformation of X. fastidiosa. Results show that this method is reliable and can be used to rapidly generate mutants in X. fastidiosa.
RESULTS
Sequence analysis of pilA paralogs and their promoter.
Based on gene expression analysis, a pilin paralog (XF2542) was predicted to be the major pilin of Xylella fastidiosa subsp. pauca strain 9a5c (31). A BLAST search using the 9a5c sequence of this paralog as query against the Temecula1 genome showed PD1926 (pilA2) as the closest homologue, with 98% coverage and 87% amino acid sequence identity, while a second paralog PD1924 (pilA1) had 97% coverage and 65% identity. On pairwise alignment, the two paralogs of Temecula1 (PD1924 and PD1926) were 68% identical by nucleotide sequence and 62% identical by amino acid sequence. They are not part of any operons (32), and they are separated by a hypothetical protein (PD1925) that is transcribed in a different strand. However, both contain all the characteristic features of a pilin subunit, including a leader peptide, cleavage site, N-terminal hydrophobic region, and pilin-cytoplasmic, transmembrane, and extracellular domains (data not shown). On comparing regulatory regions in the promoter based on the regions predicted by the previous study on X. fastidiosa subsp. pauca 9a5c (31), putative binding sites of the alternative sigma factor σ54 were identified in the promoter of PD1926 in both Temecula1 and WM1-1 (see Fig. S1 in the supplemental material). Furthermore, sequences of the coding regions and promoters of these pilin genes (pilA1 and pilA2) were identical in WM1-1, TemeculaL, and the reference strain Temecula1 (data not shown). Therefore, DNA sequence analyses suggest that the pilA paralog encoded by PD1926 (pilA2) is the functional pilin in X. fastidiosa subsp. fastidiosa strains WM1-1 and TemeculaL, and its expression is predicted to be under σ54 regulation.
Generation of mutants by overlap PCR and natural transformation.
Transformants were successfully generated from all overlap PCR-generated homologous templates that targeted two pilin genes, as well as two additional X. fastidiosa genomic regions, in the strains WM1-1 and TemeculaL (Table 1). The number of transformants ranged from 6.2 ± 3.4 to 103.6 ± 9.5 per 100 μl of recipient cell culture, depending on the construct and the strain used (Table 1). The estimated transformation frequency (ratio of transformant CFU to total viable CFU) with the protocol used here is ∼10−4 to 10−6 per recipient cell, based on our previous characterization (10, 13) of the same strains using plasmid DNA and heat-killed donor cells. The mutagenesis protocol was not optimized, since it worked satisfactorily (i.e., at least a few recombinant colonies were recovered on every experiment), but we observed that some important steps to consider in the success of this method for X. fastidiosa are as follows: (i) overlapping regions must be at least 21 bp between the two fragments, as regions shorter than this did not result in successful fusion of the PCR products (data not shown), and (ii) the growth stage of the cells, as well as the growth medium, influences the rate of natural transformation, as demonstrated in our previous studies (10, 13). We therefore suggest performance of natural transformation at the logarithmic growth phase and use of Pierce's disease 3 (PD3) (36) or PD2 medium in solid agar condition (36). Periwinkle wilt (PW) medium (37) and X. fastidiosa medium (XFM) (38) could be used without bovine serum albumin (BSA) supplementation. The basic steps of the protocol are summarized in Fig. S2 in the supplemental material.
TABLE 1.
PCR transformants and twitching phenotypes of mutants generated with various PCR templates
| Overlap PCR region (construct)a | Recipient strain/mutant | Mutation designation | No. of transformants (CFU ± SE)b | Mutant phenotypec |
|---|---|---|---|---|
| pilA1 (1924) | WM1-1 | pilA1W | 53.4 ± 15.9 | Twitching ~ to WT |
| TemeculaL | pilA1T | 35.6 ± 20.7 | Twitching ~ to WT | |
| pilA2 (1926) | WM1-1 | pilA2W | 27.6 ± 4.5 | Twitching deficient |
| TemeculaL | pilA2T | 6.2 ± 3.4 | Twitching deficient | |
| pilA1 WM1-1 | doubleW | 19 ± 5 | Twitching deficient | |
| pilA1 TemeculaL | doubleT | 103.6 ± 9.5 | Twitching deficient | |
| pilA2 complementation | pilA2 WM1-1 | pilA2CW | 20 ± 3.7 | Twitching ~ to WT |
| pilA2 TemeculaL | pilA2CT | 8.5 ± 3.5 | Twitching ~ to WT | |
| Upstream pilQ (1691) | WM1-1 | UpilQW | 51 ± 24.8 | Twitching ~ to WT |
| mrcA (1695) | WM1-1 | mrcAW | 57 ± 29.2 | Twitching deficient |
Name indicates target gene used to design homologous regions. All constructs include an antibiotic resistance cassette used for selection (see Materials and Methods).
Values indicate the mean number of transformant cells ± standard error (SE), obtained from a 0.1-ml cell suspension from three replications.
Twitching ~ to WT, twitching similar to the wild-type value.
Growth curve and biofilm formation of pilA mutants.
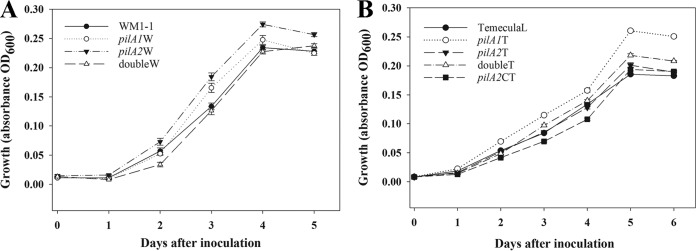
Growth curves (Fig. 1) show that growth of strains was similar for the mutants and wild-type strains. Slight growth increases were observed in the stationary phase for the pilA2W mutant (Fig. 1A) and pilA1T mutant (Fig. 1B). But these differences are probably due to differences in surface attachment of the mutant cells to the microtiter plate. Biofilm formation increased in both pilA mutants compared to that of the wild-type strains (Fig. 2). Similar tendencies were observed in biofilm formation with both TemeculaL and WM1-1 backgrounds, where the increase was significant (P = 0.0004 [pilA1W], P < 0.001 [pilA2W, pilA1T, and pilA2T]) (Fig. 2). Both double mutants had significantly (P < 0.0001) higher biofilm formation than that of wild-type strains, and higher than that of the single pilA mutants (Fig. 2). Complementation of pilA2 (pilA2C) in TemeculaL reduced biofilm formation to the level of the wild type (P > 0.05) (Fig. 2).
FIG 1.
Growth curves of Xylella fastidiosa strains. Growth curves of wild-type X. fastidiosa WM 1-1 (A) and TemeculaL (B) strains with their corresponding mutants in pilA1 and pilA2, double mutants, and complemented pilA2 were assessed by measuring optical density at 600 nm (OD600) every day for 5 to 6 days. Experiments were repeated independently three times. At least 6 wells per plate (and 1 to 3 plates per experiment) were used for each strain. pilA2CW was not used in these experiments.
FIG 2.
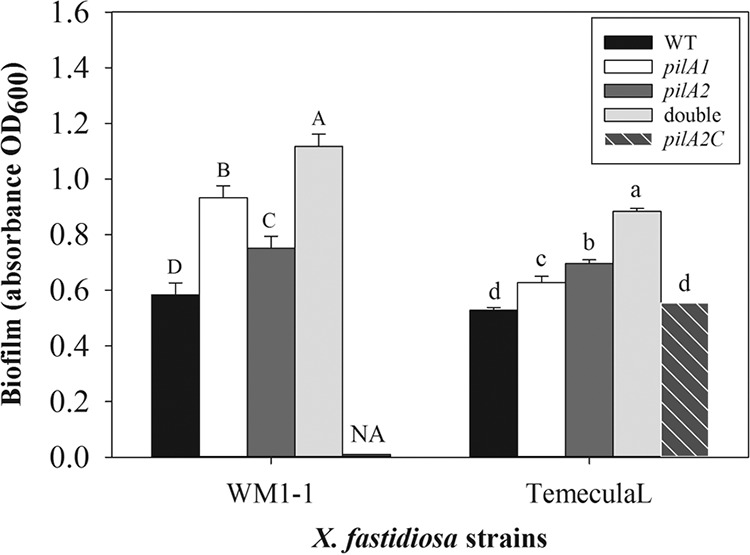
Biofilm formation of Xylella fastidiosa wild-type strains and mutants, as measured by crystal violet staining. Staining of biofilm was performed at the end of the growth curve experiment presented in Fig. 2. Experiments were repeated independently three times. At least 6 wells per plate (and 1 to 3 plates per experiment) were used for each strain. Data were analyzed in SAS 9.3 with PROC GLIMMIX. Error bars indicate standard error of the mean. Different uppercase (WM1-1 and its mutants) or lowercase (TemeculaL and its mutants) letters within each group indicate significant differences at 5% significance level. NA, nonavailable data.
Twitching motility and pilus formation by wild type and mutants.
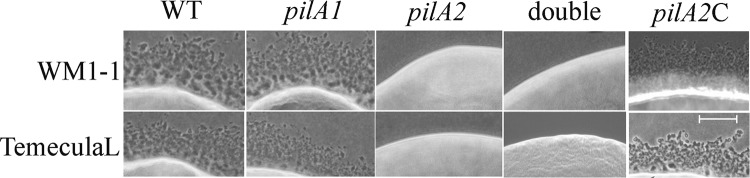
Formation of colony fringe was observed to assess twitching motility, as shown in Fig. 3. Both wild-type and pilA1 mutants of WM1-1 and TemeculaL displayed twitching motility, whereas the pilA2 mutants were twitching deficient for both strains. As expected, double mutants of both wild-type strains were also twitching deficient (Fig. 3). Similar patterns of twitching were observed in media supplemented with 50% xylem sap (vol/vol) from an X. fastidiosa-susceptible grapevine variety, Chardonnay (data not shown).
FIG 3.
Twitching motility of wild-type, pilA1 and pilA2 mutants, and complemented pilA2 mutant colonies. Twitching motility was observed for at least 10 independent transformants, with similar results obtained in all cases. For selected transformants for each background, observations were made at different time points. Bar (lower right panel), 100 μm.
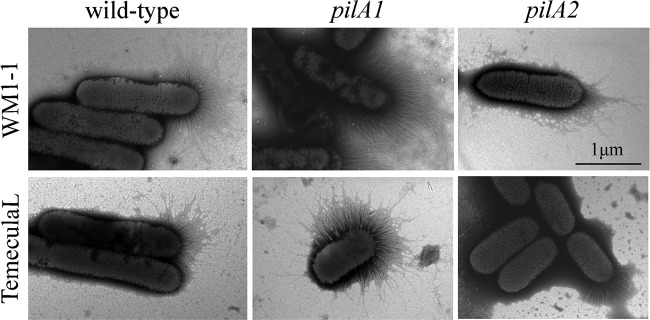
To evaluate the effect of these mutations on pilus formation, wild-type and mutant cells were observed using transmission electron microscopy (TEM), as shown in Fig. 4. Pili were observed on one of the polar ends of wild-type X. fastidiosa cells. Compared with those of wild-type strains, pili of pilA1 mutants were more abundant and not limited to the cell pole, as they were also found on the sides of the cells (Fig. 4). However, unlike in wild-type and pilA1 mutant strains, long pili were not observed on pilA2 mutants (Fig. 4). Average cell lengths of the wild type (WT) and mutants were compared, and no significant difference was observed (data not shown).
FIG 4.
Transmission electron microscopy of pilus formation by X. fastidiosa wild-type and mutant strains. Wild-type and mutant cells were negatively stained by phosphotungstic acid. Images were captured at 10,000× magnification. Bar, 1 μm. Note the hyperpiliation and TFP distribution in both pilA1 mutants.
Complementation of pilA2 restored twitching motility.
pilA2 mutants that were deficient in twitching motility were complemented using the wild-type copy of the pilA2 ORF and its native promoter and terminator at the neutral site 1 (NS1) of X. fastidiosa TemeculaL and WM1-1 genomes, using the method shown in Fig. S2B. Both TemeculaL and WM1-1 mutants could be successfully complemented with the overlap fusion PCR product of the upstream-pilA2-downstream region PCR template. Several transformants (Table 1) were generated, and five were restreaked onto new PW plates supplemented with both antibiotics (kanamycin and chloramphenicol). Two of these colonies for each wild-type background were confirmed with PCR and sequencing for the insertion of the pilA2 fragment in the neutral site. Twitching motility of five transformant colonies selected for restreaking showed that complementation restored the twitching motility lost by pilA2 deletion in both wild-type backgrounds (Fig. 3).
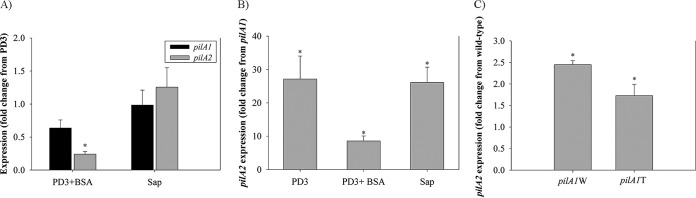
Expression of pilA2 was higher than that of pilA1, and supplementation of BSA in PD3 reduced pilA2 expression.
Absence, and therefore lack of expression of the respective mutated gene (pilA1 or pilA2), was confirmed for mutants in both wild-type backgrounds, as shown in Fig. S3 in the supplemental material, with pilA-specific primers. Expression of the two pilA paralogs in TemeculaL was compared among three different media using reverse transcription-quantitative PCR (qRT-PCR), and results are shown in Fig. 5A. Expression of pilA1 was not changed by growth in PD3 supplemented with BSA (PD3+BSA) or grapevine variety Chardonnay sap (Sap). However, expression of pilA2 was significantly reduced (P = 0.006) by addition of BSA to PD3. Expression of pilA2 was significantly higher than that of pilA1 in all media tested (Fig. 5B), suggesting that cells synthesize much more PilA2 protein than PilA1, which again supports pilA2 being the major pilin subunit. Expression of pilA2 was quantified in pilA1 mutants of both WM1-1 and TemeculaL backgrounds (Fig. 5C), and in both cases an overexpression of pilA2 was observed compared to expression in the respective wild-type strains, possibly explaining the greater number of TFP in pilA1 mutants.
FIG 5.
Expression analysis of the two pilA paralogs of TemeculaL and WM1-1 strains. For panels A and B, only TemeculaL and its mutants were used. (A) Results are expressed as fold change of pilA gene expression in cells cultured in PD3 medium supplemented with 3% BSA (PD3+BSA) and 100% grapevine variety Chardonnay sap (Sap) relative to expression in PD3. (B) Fold change in expression of pilA2 relative to that of pilA1 for the same cultures under three different media. (C) Expression of pilA2 in pilA1 mutants in both wild-type backgrounds (WM1-1 and TemeculaL) in PD3 media. Gene expressions was normalized by using nuoA as the endogenous control. An asterisk indicates significant difference in fold change, as compared by one-sample t test (fold change ≠ 1; P < 0.05). Error bars indicate standard errors of mean in fold change from three independent replicates of one experiment.
DISCUSSION
X. fastidiosa possesses TFP (6, 8), which mediates twitching motility, at one of the cell poles. Involvement of twitching motility in X. fastidiosa virulence was suggested by indirect evidence, since mutants lacking TFP (due to mutations in pilB and pilQ genes) were deficient in twitching motility and had reduced colonization of grapevines (remaining close to the inoculation point) compared to that of the wild type (8). But since disease symptoms were not assessed in that study, a conclusion on the direct effect of twitching motility on virulence was not provided. The role in X. fastidiosa of some TFP-related genes, such as pilB, pilO, pilQ, pilR, and pilY1, in TFP biogenesis and twitching motility was demonstrated in previous studies (6–8, 29, 30). Moreover, environmental signals, such as calcium (30), pectin (39), and chitin (40), were shown to transcriptionally regulate twitching motility genes. Other studies demonstrated the involvement of chemotaxis-related genes (Pil-Chp) in regulating twitching motility and expression of TFP genes (29, 41). But there have been no studies in X. fastidiosa (besides the mentioned study in X. fastidiosa subsp. pauca; see reference 31) on the role of the major pilin(s) of TFP, a fundamental protein for this appendage.
In the present study, the role in twitching motility of two paralogs of type IV pilin (PD1924 and PD1926) was assessed. Despite X. fastidiosa genomes possessing several genes encoding pilin subunits (31) and at least two of these paralogs possessing similar structural features in the genomes of strains used in this study, the finding that only one of the pilA paralog (PilA2, PD1926) is essential for twitching motility was surprising. Of all the mutants generated, pilA2 and pilA1pilA2 double mutants lost twitching motility. Motility was restored in pilA2 mutants when a copy of pilA2 gene was reinserted at a different location in the genome. Interestingly, the pilA1 mutants showed similar twitching motility to that of the wild types, although biofilm formation was increased in these mutants, suggesting a different role of PilA1. By electron microscopy, we were able to detect the formation of profuse TFP, which were distributed on the sides of the pilA1 mutant cells, as opposed to the polar location in wild-type cells. Moreover, expression analysis indicated that higher production of pilA2 is detected in pilA1 mutants, which may explain the hyperpiliation observed in the pilA1 mutants and the consequent increase in biofilm formation, since TFP is known to be involved in this process (24). The molecular basis for TFP regulation of localization and the number of appendages formed has not been completely elucidated. In the case of other appendages, such as flagella, proteins FlhF and FlhG in various bacteria are known to be involved in assembly and regulation of number and localization of these structures (42). In Caulobacter crescentus, polar localization of pili was shown to involve a two-component signal transduction histidine-kinase, PleC (43) and PodJ (44); but mutations in those proteins render pilus-less cells, a different phenotype than the one observed here. In P. aeruginosa, the membrane-associated polar organelle coordinator (Poc) complex regulates localization of flagellum and TFP (45), and mutations in their components led to random localization of these structures and lack of twitching motility. These phenotypes are different from the ones observed here, namely hyperpiliation and extended localization without losing the ability of twitching motility. The putative role of pilA1 in regulating TFP localization needs to be further investigated.
Previous studies with X. fastidiosa (30, 46–48) have considered pilA1 paralog in their expression analysis to determine the influence of various environmental parameters on twitching motility, but our results suggest that pilA2 may be a better target to assess twitching in this bacterium. Expression analysis by RNA-Seq in our previous study (32) and by qRT-PCR in this current study showed that pilA2 is expressed at a significantly higher level than that of pilA1. Moreover, the closest homologue of pilA2 was demonstrated to be regulated differently by a σ54 factor in a X. fastidiosa subsp. pauca strain (31). Regulatory elements of σ54 in the pilA2 promoter region of X. fastidiosa subsp. fastidiosa strains WM1-1 and TemeculaL used in this study were found by sequence comparison with the promoter of the X. fastidiosa subsp. pauca strain. Previous studies in P. aeruginosa have demonstrated that mutants defective in σ54 protein production were nonmotile and unable to produce TFP (49) and flagella (50). Furthermore, expression of type IV pilin was demonstrated to be regulated by σ54 and a PilR/PilS two-component regulatory system in a variety of other bacterial species (51–54). In X. fastidiosa, a mutant of the pilR gene, the response regulator of the PilR/PilS system, was deficient in twitching (7). Therefore, the findings from previous studies that one of the pilA paralogs is differentially expressed in a RpoN mutant (31) and that the mutant of predicted σ54 activator is deficient in twitching (7), and the results of the present study that the pilin paralog suggested to be under σ54 regulation is twitching deficient, suggest that type IV pilin production in different X. fastidiosa strains may be regulated by σ54, but this needs to be further studied.
In addition to its involvement in the regulation of genes involved in motility, the primary role of σ54 in various bacteria was described to be in nitrogen assimilation under nitrogen-limited conditions (55–57). Interestingly, in previous studies (13, 58), supplementation of a nitrogen-rich compound, BSA, in the culture medium of X. fastidiosa significantly reduced twitching motility, and in the present study, BSA was shown to decrease the expression of the major pilin pilA2. Although it is not known if X. fastidiosa cells use BSA as a nitrogen source, we speculate that it could act as an environmental signal that regulates the activation of the σ54 factor, which then activates genes related to nitrogen assimilation and motility.
Although X. fastidiosa was the first plant-associated bacterium to have its genome sequenced (27), there is still limited understanding regarding its virulence factors. This is because, similarly to other bacteria, almost 40% of the proteins encoded by the genome do not have a known function assigned and are annotated as hypothetical proteins (28), and, moreover, X. fastidiosa has been a difficult pathogen for genetic manipulation studies (59, 60). Although transposon mutagenesis (4, 8) and homologous recombination protocols (15, 30, 33, 61) have been used, these protocols are time-consuming and inconsistent in efficiency. This is further complicated by the fastidious and slow-growing nature of the bacterium (62). In this study, we developed a rapid and simplified protocol of genetic manipulation using overlap extension PCR and natural transformation, since X. fastidiosa was shown to be capable of natural competence (10, 63). Using this method with careful planning, gene knockouts can be generated within a month from the start of the bacterial culture at −80°C, and this method does not require any plasmid vectors or steps such as restriction digestion, ligation, Escherichia coli transformation, and plasmid preparation. Only a few PCR steps are needed to generate an allelic exchange template that recombines with the recipient bacterium, inserting a marker in exchange for the target gene. Moreover, the same process was successfully used to generate complemented mutants of the knocked-out genes. We believe that this simple, rapid, and reliable technique will augment genetic and functional studies aimed at understanding the virulence mechanisms of X. fastidiosa. This method was used here to tag five genomic locations with a marker gene with successful transformation. Transformation of X. fastidiosa strains with linear DNA has been reported in a previous study (63), and overlap extension PCR fragments were introduced in other bacteria via electroporation (64, 65) and natural transformation (66). The protocol for X. fastidiosa presented here is a simplified and faster approach compared to the one mentioned above (66), and it should be useful for research in this fastidious prokaryote.
In our previous studies, we showed that twitching-deficient strains or mutants lost the ability of natural transformation (10, 13). In previous research by our group and others (10, 35), X. fastidiosa twitching-deficient mutants of pilO, pilQ, and pilM genes were not transformable, whereas another twitching-deficient mutant in pilB was transformable. In this study, however, we could naturally transform both twitching-deficient pilA2 mutants. Therefore, we speculate that although higher twitching motility increases the rate of natural transformation (10), a partial, rather than the complete, set of TFP structures may be sufficient for natural transformation of X. fastidiosa.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
X. fastidiosa subsp. fastidiosa strains WM1-1 (67) and TemeculaL (a variant of Temecula1) (28), isolated from infected vineyards in Georgia and California, respectively, were used in this study. Strains were cultured in PW (37) agar plates, modified by omitting phenol red and using 1.8 g · liter−1 of bovine serum albumin (BSA; Gibco Life Sciences Technology), for 1 week at 28°C from −80°C glycerol stock, restreaked onto new PW plates, and cultured for another week before use. PD3 medium (36) was used for culturing and suspending cells in liquid. Whenever needed, antibiotics kanamycin (Km) and chloramphenicol (Cm) were used at concentrations of 30 and 10 μg · ml−1, respectively.
DNA extraction and knockout template construction.
For DNA extraction, cells cultured on PW agar plates were suspended in 400 μl of sterile Milli-Q water and saved at −20°C until use. DNA was extracted using Quick-DNA fungal/bacterial miniprep kit (Zymo Research) using the manufacturers' protocol. Bead beating for DNA extraction was performed for 5 min using a Mini-BeadBeater-96 cell disruptor (BioSpec Products).
Knockout templates targeting four genomic regions, according to the Temecula1 reference sequence, were constructed. The four regions were the pilA1 coding region (PD1924), the pilA2 coding region (PD1926), the upstream noncoding region of pilQ (PD1691), and the downstream region of pilM (PD1695) that disrupted the mrcA gene encoding penicillin-binding protein 1A. These templates were named construct 1924, construct 1926, construct 1691, and construct 1695, respectively. Constructs 1691 and 1924 contained a Km-resistant cassette, while the other two constructs, 1695 and 1926, contained Cm-resistant cassettes. For creating each knockout construct, approximately 0.7- to 1.2-kb upstream and downstream flanking regions of the targeted genomic region were PCR amplified using the corresponding UP_F/UP_R and Dn_F/Dn_R primer pairs, respectively (Table 2). For example, 1924_up_F and 1924_up_R were used to amplify the region upstream from PD1924 (pilA1). Cassettes encoding Km or Cm resistance were amplified using primer pairs Kan_F and Kan_R or Cm_F and Cm_R, respectively (Table 2). DNA from strain WM1-1 was used as the template to amplify upstream and downstream sequences of gene of interest, while plasmids pUC4K and pAX1.Cm were used as the templates for Km and Cm cassettes, respectively. Primers Up_R and Dn_F were 5′ extended with at least 21 bp of homology to the antibiotic cassette, thereby facilitating fusion of the upstream and downstream genomic fragments to the antibiotic resistance cassettes by overlap extension PCR. To accomplish this fusion, the three separately amplified fragments (upstream, antibiotic, and downstream) were purified from agarose gel and mixed in equal proportions in an overlap extension PCR using the end primers (UP_F and Dn_R). The fusion product, which contained the antibiotic resistance cassette flanked by genomic sequences upstream and downstream of the target, was purified from gel and stored at −20°C before use or was used as the template for amplification with the internal primer pair int_F/int_R (Table 2). All of the primers (Table 2) were designed with Primer3 Primer Design Tool in Geneious software (Biomatters). PCR was performed with a standard protocol, using an iProof High-Fidelity PCR kit (Bio-Rad) in an S1000 thermal cycler (Bio-Rad). PCR products were gel purified using either a 5PRIME gel extract minikit or Freeze 'N Squeeze DNA gel extraction spin columns (Bio-Rad).
TABLE 2.
PCR and qRT-PCR primers and probes used in this study
| Primer name | Sequence (5′–3′)a | Purpose or description | Amplicon size(s) (bp) | Source |
|---|---|---|---|---|
| 1924_up_F | GCGGCACCACGTATATCAATAAAA | Amplify upstream region of pilA1 (PD1924) | 981 | This study |
| 1924_up_R | GCAACACCTTCTTCACGAGGCAGACAGAGAATTCATGTGTGGGGGTTTA | |||
| 1924_dn_F | GAGATTTTGAGACACAACGTGGCTTATGAATACACACAGCAACACGATC | Amplify downstream region of pilA1 (PD1924) | 1,190 | This study |
| 1924_dn_R | TTGGAGAAGAGGCGTGTTAAAAAC | |||
| 1924_int_F | ATATACAGGGTGCTTGCTGATTGA | Amplify internal region of pilA1 construct | 2,952 | This study |
| 1924_int_R | GGATGGGTTTAGGGATGCTGATAA | |||
| 1924_out_F | AAACCACCACCGATAACAACAATC | Confirm pilA1 mutants (primers target outside the pilA1 construct) | 3,509, 2,884b | This study |
| 1924_out_R | AATAGCGTTGGTAAGAAATCCAGC | |||
| 1926_up_F | CATTTCACTTTGACTTCACCCGAA | Amplify upstream region of pilA2 (PD1926) | 1,032 | This study |
| 1926_up_R | TGCCCCGTATTCAGTGTCGCTGATTAATAGCGTTGGTAAGAAATCCAGC | |||
| 1926_dn_F | GCCTGGTGCTACGCCTGAATAAGTGAAACACGATTCATGGGTAAATGCTC | Amplify downstream region of pilA2 (PD1926) | 700 | This study |
| 1926_dn_R | TGAGCGTCAATTTTAGAGGATGGA | |||
| 1926_int_F | TCAGCAATACTCATACTGGCACTT | Amplify internal region of pilA2 construct | 2,773 | This study |
| 1926_int_R | AACGTGTGCTTGAATCTTCGAATT | |||
| 1926_out_F | ACAAGAGTGAGCCGTTACAACTAT | Confirm pilA2 mutants (primers target outside the pilA2 construct) | 3,004, 2,352b | This study |
| 1926_out_R | CTTTTCCAATGAGCAGTTATCGGG | |||
| Kan_F | GTCTGCCTCGTGAAG | Amplify kanamycin cassette | 1,204 | This study |
| Kan_R | AAGCCACGTTGTGT | |||
| Cm_F | AATCAGCGACACTGAATACGG | Amplify chloramphenicol cassette | 1,119 | This study |
| Cm_R | TCACTTATTCAGGCGTAGCAC | |||
| pilA2C_up_F | CGCGCCCGTTATTAATCGAA | Amplify upstream fragment of pilA2 complementation construct (contains upstream of NS1 region and kanamycin cassette) | 1,957 | This study |
| pilA2C_up_R2 | ATATTGAAGGGTGCAATACAAAGCATCTAGTCTCAACCATCATCGATGAA | |||
| pilA2C_dn_F2 | GTTTCAAGAGAGAGAGCGTTCAACACGATGCTGTTAACCATTGTCATC | Amplify downstream fragment of pilA2 complementation construct (this is same as NS1 downstream fragment) | 799 | This study |
| pilA2C_dn_R | TAACCTTGTCAGCGTAGATG | |||
| pilA2_F | ACAATTCATCGATGATGGTTGAGACTAGATGCTTTGTATTGCACCCTTCAAT | Amplify pilA2 coding region, 237 bp upstream, and 216 bp downstream to include promoter and terminator regions | 950 | This study |
| pilA2_R | GCCATTGATGACAATGGTTAACAGCATCGTTTCAAGAGAGAGAGCGTTCAAC | |||
| pilA_F | AAACACCGGACTTGCCAACATCAC | Primer and probe to amplify fragment of pilA1 | 140 | 30 |
| pilA_R | TGTTGCATGTCCACTGACCTCCAT | |||
| pilA_Pc | AAACCATCGCTTGGAATCGTAGCGTCGA | |||
| PD1926_F | CACTCCTAACGCTATTGGACTAC | Primer and probe to amplify fragment of pilA2 | 117 | 32 |
| PD1926_R | TTGACCTGACCATTACCAATCA | |||
| PD1926_Pc | TGGTGGACATCACAACTACTGGCG | |||
| nuoA_F | AGACGCACGGATGAAGTTCGATGT | Reference gene for qRT-PCR | 30 | |
| nuoA_R | ATTCCAGCGCTCCCTTCTTCCATA | |||
| nuoA_Pc | TTCATCGTGCCTTGGACTCAGGTGTT | |||
| 1691_up F | AGGCAACCTGACAGCGATAC | Amplify upstream region of 1691 construct | 541 | This study |
| 1691_up_R2 | GCAACACCTTCTTCACGAGGCAGACCGTTGATGTTCGAGAAGTGCG | |||
| 1691_Dn_F2 | GAGATTTTGAGACACAACGTGGCTTTGCATCAACCCCAAAGCTGA | Amplify downstream region of 1691 construct | 504 | This study |
| 1691_Dn R | AAGCGATCCAATGAAGGGCT | |||
| 1691_int_F | ACGGCCCTTCTCTAAGATTGC | Amplify internal region of 1691 construct | 2,152, 987b | This study |
| 1691_int_R | TGTGTGGTGCTTGCATATTCTG | |||
| 1695_up_F | CGATGGCCTGTTGATAGCGATA | Amplify upstream region of 1695 construct | 1,027 | This study |
| 1695_up_R | CCGTATTCAGTGTCGCTGATTGTCACCACGTTTGAGGAGTTTG | |||
| 1695_Dn_F | GTGCTACGCCTGAATAAGTGAAACGGGGTGAATGGACATTAG | Amplify downstream region of 1695 construct | 1,018 | This study |
| 1695_Dn_R | CAGATGGGGAGTGCTGCTTTA | |||
| 1695_int_F | AAAGACGAAATCCTGGAGCTGTAT | Amplify internal region of 1695 construct | 2,640, 1,546b | This study |
| 1695_int_R | TTGATACCAATTGGAAGACAACGC |
Underlining indicates the 5′ extended region of the primer that is homologous to one of the antibiotic resistance cassettes to facilitate fusion of chromosomal sequence to a selectable marker by overlap extension PCR.
Amplicon size in the wild-type strain without the insertion of antibiotic resistance gene.
These probes were used for reverse transcription-quantitative PCR (qRT-PCR) and were labeled with 6-carboxyfluorescein (FAM) at the 5′ end and Black Hole 588 Quencher-1 (BHQ1) at the 3′ end.
Natural transformation of X. fastidiosa strains with PCR template and confirmation of mutants.
Transformation of X. fastidiosa cells was performed with the gel-purified or internal primer-amplified PCR templates, using a natural transformation protocol as previously described (10, 13). Briefly, recipient X. fastidiosa strain WM1-1 and TemeculaL cultures from PW plates were suspended in PD3 liquid medium and optical density at 600 nm (OD600) was adjusted to 0.25 (∼108 cells · ml−1). Ten μl of this suspension was spotted onto PD3 agar plates, 10 μl of PCR template was added on top of the spots, and the spots were dried before incubation at 28°C for 3 days. After 3 days, spots were suspended in 0.5 ml of PD3. Aliquots (100 μl) were spread plated onto PW plates containing necessary antibiotics for selection. After 2 weeks of incubation at 28°C, mutant CFU were enumerated and restreaked onto new antibiotic PW plates. Control spots of the strain cells without the addition of PCR template were also included for each experiment. Double mutants were generated by using pilA1 mutants as recipients with the gel purified PCR template of pilA2 deletion (construct 1926). Ten colonies per strain per construct were restreaked onto new antibiotic plates. After confirming that the colonies were resistant to the corresponding antibiotic, one colony per strain for each construct was selected for further analysis. Confirmation of gene deletion was performed by PCR and sequencing. PCR was performed with three sets of primer pairs, namely, the out pairs (out_F/out_R) that target the genomic region outside the recombination region of the construct; antibiotic primers (antb_F/antb_R); and respective gene primers (pilA1_F/pilA1_R and PD1926_F/PD1926_R). For each PCR, wild-type strains were included as controls (Fig. S3). Sanger sequencing of the mutant at the deleted genomic region was performed using primers targeting the outside region, internal region, and antibiotic region. Information on all of the primers is presented in Table 2.
Growth curve and biofilm formation.
Growth curve and biofilm formation of the mutant and wild-type strains were assessed in 96-well plates during 5 days, as previously described (10). Experiments were repeated three times independently, with one or three identical 96-well plates used per experiment. For each wild-type strain or mutant background at least six wells (repetitions) on each plate were used.
Twitching motility.
Twitching motility assessment of wild-type and mutant strains was performed in PD3 plates and PD3 plates supplemented with 50% Chardonnay sap, as previously described (10). Twitching motility was observed for at least 10 transformant colonies for each mutant. Selected colonies were restreaked, and repeated observation of these colonies for twitching motility was performed.
Complementation of pilA2 mutants.
Complementation of pilA2 mutants was performed with the PCR-amplified and gel-purified complementation template prepared from the fusion PCR of three fragments (upstream pilA2 complementation template, pilA2 template containing the pilA2 ORF its promoter and terminator region, and downstream pilA2 complementation template). The upstream pilA2 complementation construct was amplified from NS1::Km WM1-1 (10) using the primer pair pilA2C_up_F and pilA2C_up_R (Table 2). These primers amplify the upstream flanking region of NS1 and the Km cassette in one fragment. The 950-bp pilA2 fragment contained a 237-bp upstream promoter region, a pilA2 coding region, and a 216-bp downstream region. This fragment was amplified using primer pair pilA2_F and pilA2_R and using the wild-type WM1-1 DNA as the template. The downstream complementation product was also amplified from wild-type WM1-1 DNA using primers pilA2C_dn_F and pilA2C_dn_R and is the same as the downstream region of NS1. Primers pilA2C_up_R, pilA2F, pilA2R, and pilA2C_Dn_F had a 27-bp extension at their 5′ ends that was homologous to the adjoining fragment. The individual fragments were amplified, gel purified, and fused by extension PCR as described above. The PCR fragments were then used in a natural transformation assay with both the wild-type strains WM1-1 and TemeculaL and their pilA2 mutants.
RNA preparation and quantitative PCR.
Wild-type X. fastidiosa TemeculaL, WM1-1, or mutant cells cultured in PW plates were scraped and adjusted to an OD600 of 0.25 as previously mentioned. Three 15-ml plastic tubes containing 3 ml of media (PD3, PD3 + BSA [PD3 supplemented with 3 g · liter−1 BSA], and Sap [100% Vitis vinifera var. Chardonnay sap; see reference 13]) were supplemented with 120 μl of the OD600-adjusted TemeculaL strain. Tubes were incubated at 28°C with shaking at 150 rpm for 3 days. Cells were then precipitated by centrifugation of the tubes at 5,000 rpm for 5 min at 4°C. Pellet formed was suspended in 200 μl of DNA/RNA Shield (Zymo Research), and RNA was extracted using a Quick-RNA MiniPrep Plus kit (Zymo Research), using in-column DNase treatment following the manufacturers' protocol. RNA concentration was measured by a NanoDrop 2000 spectrophotometer (Thermo Scientific). cDNA was prepared using qScript cDNA SuperMix (Quantabio) with 170 ng of normalized RNA in 20 μl volume. Quantitative PCR was performed using 1 μl of cDNA in 20 μl volume containing 4 μl of 5× PerfeCTa multiplex quantitative PCR (qPCR) ToughMix (Quantabio), 0.4 μM forward and reverse primers, and 0.2 μM TaqMan probe (labeled with 5′ 6-carboxyfluorescein [FAM] and 3′ Black Hole 588 Quencher-1 [BHQ-1]). Reactions were performed using a CFX96 real-time system (Bio-Rad) with the following cycling parameters: 95°C for 1 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Fold change in expression of pilA1 and pilA2 was calculated by the threshold cycle (2−ΔΔCT) method (68) using expression of nuoA gene as the endogenous control (30). Primers and probes used for gene expression are included in Table 2. Three independent replications were used.
Electron microscopy.
Pili of wild-type and mutant strains were observed under a transmission electron microscope, following a protocol previously described (34). Two-day old cultures were scraped from PW agar plates and suspended in 200 μl of distilled water. Cell suspension (10 μl) was pipetted on Formvar-coated grid and subsequently stained with 50 μl of phosphotungstic acid. After 2 min, the excess liquid on the grid was removed by a filter paper, and then the grids were observed on a Zeiss EM 10 transmission electron microscope (Carl Zeiss, Jena, Germany). Two independent experiments were performed.
Statistical analysis.
Data from biofilm formation were analyzed in SAS 9.3 (SAS Institute, Inc., Cary, NC) using GLIMMIX procedure, and means were separated by the Tukey-Kramer method (P < 0.05). To compare gene expression data, differences in fold change were analyzed by one-sample t test (fold change ≠ 1, P < 0.05).
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by Agriculture and Food Research Initiative competitive grant 2015-67014-23085 from the USDA National Institute of Food and Agriculture and by the HATCH Alabama Agricultural Experiment Station (AAES) program.
We thank Michael E. Miller, director of the AU Research and Instrumentation Facility, for his help with TEM, and Sy Traore for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01167-18.
REFERENCES
- 1.Chatterjee S, Almeida RP, Lindow S. 2008. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol 46:243–271. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins DL, Purcell AH. 2002. Xylella fastidiosa: cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis 86:1056–1066. doi: 10.1094/PDIS.2002.86.10.1056. [DOI] [PubMed] [Google Scholar]
- 3.Newman KL, Almeida RP, Purcell AH, Lindow SE. 2003. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microbiol 69:7319–7327. doi: 10.1128/AEM.69.12.7319-7327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roper MC, Greve LC, Warren JG, Labavitch JM, Kirkpatrick BC. 2007. Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol Plant Microbe Interact 20:411–419. doi: 10.1094/MPMI-20-4-0411. [DOI] [PubMed] [Google Scholar]
- 5.Saponari M, Loconsole G, Cornara D, Yokomi RK, De Stradis A, Boscia D, Bosco D, Martelli GP, Krugner R, Porcelli F. 2014. Infectivity and transmission of Xylella fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. J Econ Entomol 107:1316–1319. doi: 10.1603/EC14142. [DOI] [PubMed] [Google Scholar]
- 6.De La Fuente L, Burr TJ, Hoch HC. 2007. Mutations in type I and type IV pilus biosynthetic genes affect twitching motility rates in Xylella fastidiosa. J Bacteriol 189:7507–7510. doi: 10.1128/JB.00934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Hao G, Galvani CD, Meng Y, De La Fuente L, Hoch H, Burr TJ. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell–cell aggregation. Microbiology 153:719–726. doi: 10.1099/mic.0.2006/002311-0. [DOI] [PubMed] [Google Scholar]
- 8.Meng Y, Li Y, Galvani CD, Hao G, Turner JN, Burr TJ, Hoch HC. 2005. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol 187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS. 2002. Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 10.Kandel PP, Almeida RPP, Cobine PA, De La Fuente L. 2017. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol Plant Microbe Interact 30:589–600. doi: 10.1094/MPMI-02-17-0053-R. [DOI] [PubMed] [Google Scholar]
- 11.Oliver JE, Cobine PA, De La Fuente L. 2015. Xylella fastidiosa isolates from both subsp. multiplex and fastidiosa cause disease on southern highbush blueberry (Vaccinium sp.) under greenhouse conditions. Phytopathology 105:855–862. doi: 10.1094/PHYTO-11-14-0322-FI. [DOI] [PubMed] [Google Scholar]
- 12.Oliver JE, Sefick SA, Parker JK, Arnold T, Cobine PA, De La Fuente L. 2014. Ionome changes in Xylella fastidiosa-infected Nicotiana tabacum correlate with virulence and discriminate between subspecies of bacterial isolates. Mol Plant Microbe Interact 27:1048–1058. doi: 10.1094/MPMI-05-14-0151-R. [DOI] [PubMed] [Google Scholar]
- 13.Kandel PP, Lopez SM, Almeida RP, De La Fuente L. 2016. Natural competence of Xylella fastidiosa occurs at a high frequency inside microfluidic chambers mimicking the bacterium's natural habitats. Appl Environ Microbiol 82:5269–5277. doi: 10.1128/AEM.01412-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilhabert MR, Kirkpatrick BC. 2005. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol Plant Microbe Interact 18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 15.Newman KL, Almeida RP, Purcell AH, Lindow SE. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Narl Acad Sci U S A 101:1737–1742. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N, Li J-L, Lindow SE. 2012. RpfF-dependent regulon of Xylella fastidiosa. Phytopathology 102:1045–1053. doi: 10.1094/PHYTO-07-12-0146-R. [DOI] [PubMed] [Google Scholar]
- 17.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 18.Morand PC, Drab M, Rajalingam K, Nassif X, Meyer TF. 2009. Neisseria meningitidis differentially controls host cell motility through PilC1 and PilC2 components of type IV Pili. PLoS One 4:e6834. doi: 10.1371/journal.pone.0006834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfgang M, Park H-S, Hayes SF, Van Putten JP, Koomey M. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 95:14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Kang Y, Genin S, Schell MA, Denny TP. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215–3229. doi: 10.1099/00221287-147-12-3215. [DOI] [PubMed] [Google Scholar]
- 21.Bahar O, Goffer T, Burdman S. 2009. Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol Plant Microbe Interact 22:909–920. doi: 10.1094/MPMI-22-8-0909. [DOI] [PubMed] [Google Scholar]
- 22.Dunger G, Guzzo CR, Andrade MO, Jones JB, Farah CS. 2014. Xanthomonas citri subsp. citri type iv pilus is required for twitching motility, biofilm development, and adherence. Mol Plant Microbe Interact 27:1132–1147. doi: 10.1094/MPMI-06-14-0184-R. [DOI] [PubMed] [Google Scholar]
- 23.Taguchi F, Ichinose Y. 2011. Role of type IV pili in virulence of Pseudomonas syringae pv. tabaci 6605: correlation of motility, multidrug resistance, and HR-inducing activity on a nonhost plant. Mol Plant Microbe Interact 24:1001–1011. doi: 10.1094/MPMI-02-11-0026. [DOI] [PubMed] [Google Scholar]
- 24.Burdman S, Bahar O, Parker JK, De La Fuente L. 2011. Involvement of type IV pili in pathogenicity of plant pathogenic bacteria. Genes 2:706–735. doi: 10.3390/genes2040706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 26.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 27.Simpson AJG, Reinach F, Arruda P, Abreu F, Acencio M, Alvarenga R, Alves LC, Araya J, Baia G, Baptista C. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- 28.Van Sluys MA, de Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, Camargo LE, da Silva AC, Moon DH, Takita MA, Lemos EG, Machado MA, Ferro MI, da Silva FR, Goldman MH, Goldman GH, Lemos MV, El-Dorry H, Tsai SM, Carrer H, Carraro DM, de Oliveira RC, Nunes LR, Siqueira WJ, Coutinho LL, Kimura ET, Ferro ES, Harakava R, Kuramae EE, Marino CL, Giglioti E, Abreu IL, Alves LM, do Amaral AM, Baia GS, Blanco SR, Brito MS, Cannavan FS, Celestino AV, da Cunha AF, Fenille RC, Ferro JA, Formighieri EF, Kishi LT, Leoni SG, Oliveira AR, Rosa VE Jr, Sassaki FT, Sena JA, de Souza AA, Truffi D, Tsukumo F, Yanai GM, Zaros LG, Civerolo EL, Simpson AJ, Almeida NF Jr, Setubal JC, Kitajima JP. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J Bacteriol 185:1018–1026. doi: 10.1128/JB.185.3.1018-1026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cursino L, Galvani CD, Athinuwat D, Zaini PA, Li Y, De La Fuente L, Hoch HC, Burr TJ, Mowery P. 2011. Identification of an operon, Pil-Chp, that controls twitching motility and virulence in Xylella fastidiosa. Mol Plant Microbe Interact 24:1198–1206. doi: 10.1094/MPMI-10-10-0252. [DOI] [PubMed] [Google Scholar]
- 30.Cruz LF, Parker JK, Cobine PA, De La Fuente L. 2014. Calcium-enhanced twitching motility in Xylella fastidiosa is linked to a single PilY1 homolog. Appl Environ Microbiol 80:7176–7185. doi: 10.1128/AEM.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silva Neto JF, Koide T, Abe CM, Gomes SL, Marques MV. 2008. Role of σ54 in the regulation of genes involved in type I and type IV pili biogenesis in Xylella fastidiosa. Arch Microbiol 189:249–261. doi: 10.1007/s00203-007-0314-x. [DOI] [PubMed] [Google Scholar]
- 32.Parker JK, Chen H, McCarty SE, Liu LY, De La Fuente L. 2016. Calcium transcriptionally regulates the biofilm machinery of Xylella fastidiosa to promote continued biofilm development in batch cultures. Environ Microbiol 18:1620–1634. doi: 10.1111/1462-2920.13242. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto A, Young GM, Igo MM. 2009. Chromosome-based genetic complementation system for Xylella fastidiosa. Appl Environ Microbiol 75:1679–1687. doi: 10.1128/AEM.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Kandel PP, Cruz LF, Cobine PA, De La Fuente L. 2017. The major outer membrane protein MopB is required for twitching movement and affects biofilm formation and virulence in two Xylella fastidiosa strains. Mol Plant Microbe Interact 30:896–905. doi: 10.1094/MPMI-07-17-0161-R. [DOI] [PubMed] [Google Scholar]
- 35.Kung SH, Almeida RP. 2014. Biological and genetic factors regulating natural competence in a bacterial plant pathogen. Microbiology 160:37–46. doi: 10.1099/mic.0.070581-0. [DOI] [PubMed] [Google Scholar]
- 36.Davis MJ, French WJ, Schaad NW. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol 6:309–314. doi: 10.1007/BF01566883. [DOI] [Google Scholar]
- 37.Davis M, Purcell A, Thomson S. 1980. Isolation media for the Pierce's disease bacterium. Phytopathology 70:425–429. doi: 10.1094/Phyto-70-425. [DOI] [Google Scholar]
- 38.Killiny N, Almeida RPP. 2009. Host structural carbohydrate induces vector transmission of a bacterial plant pathogen. Proc Natl Acad Sci U S A 106:22416–22420. doi: 10.1073/pnas.0908562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Killiny N, Almeida RP. 2009. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl Environ Microbiol 75:521–528. doi: 10.1128/AEM.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Killiny N, Prado SS, Almeida RP. 2010. Chitin utilization by the insect-transmitted bacterium Xylella fastidiosa. Appl Environ Microbiol 76:6134–6140. doi: 10.1128/AEM.01036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao L, Athinuwat D, Johnson K, Cursino L, Burr T, Mowery P. 2017. Xylella fastidiosa pil-chp operon is involved in regulating key structural genes of both type I and IV pili. VITIS 56:55–62. [Google Scholar]
- 42.Kazmierczak BI, Hendrixson DR. 2013. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol 88:655–663. doi: 10.1111/mmi.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viollier PH, Sternheim N, Shapiro L. 2002. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J 21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viollier PH, Sternheim N, Shapiro L. 2002. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A 99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowles KN, Moser TS, Siryaporn A, Nyakudarika N, Dixon W, Turner JJ, Gitai Z. 2013. The putative Poc complex controls two distinct Pseudomonas aeruginosa polar motility mechanisms. Mol Microbiol 90:923–938. doi: 10.1111/mmi.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Souza AA, Takita MA, Coletta-Filho HD, Caldana C, Goldman GH, Yanai GM, Muto NH, de Oliveira RC, Nunes LR, Machado MA. 2003. Analysis of gene expression in two growth states of Xylella fastidiosa and its relationship with pathogenicity. Mol Plant Microbe Interact 16:867–875. doi: 10.1094/MPMI.2003.16.10.867. [DOI] [PubMed] [Google Scholar]
- 47.de Souza AA, Takita MA, Pereira EO, Coletta-Filho HD, Machado MA. 2005. Expression of pathogenicity-related genes of Xylella fastidiosa in vitro and in planta. Curr Microbiol 50:223–228. doi: 10.1007/s00284-004-4447-8. [DOI] [PubMed] [Google Scholar]
- 48.Caserta R, Takita M, Targon M, Rosselli-Murai L, De Souza A, Peroni L, Stach-Machado D, Andrade A, Labate C, Kitajima E. 2010. Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation. Appl Environ Microbiol 76:4250–4259. doi: 10.1128/AEM.02114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishimoto KS, Lory S. 1989. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A 86:1954–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Totten PA, Lara JC, Lory S. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol 172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kehl-Fie TE, Porsch EA, Miller SE, StGeme JW. 2009. Expression of Kingella kingae type IV pili is regulated by σ54, PilS, and PilR. J Bacteriol 191:4976–4986. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker D, Kennan RM, Myers GS, Paulsen IT, Songer JG, Rood JI. 2006. Regulation of type IV fimbrial biogenesis in Dichelobacter nodosus. J Bacteriol 188:4801–4811. doi: 10.1128/JB.00255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ray SK, Kumar R, Peeters N, Boucher C, Genin S. 2015. rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum. Front Microbiol 6:229. doi: 10.3389/fmicb.2015.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bretl DJ, Müller S, Ladd KM, Atkinson SN, Kirby JR. 2016. Type IV-pili dependent motility is co-regulated by PilSR and PilS2R2 two-component systems via distinct pathways in Myxococcus xanthus. Mol Microbiol 102:37–53. doi: 10.1111/mmi.13445. [DOI] [PubMed] [Google Scholar]
- 55.Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. 2011. Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang N, Buck M. 2015. A perspective on the enhancer dependent bacterial RNA polymerase. Biomolecules 5:1012–1019. doi: 10.3390/biom5021012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao K, Liu M, Burgess RR. 2010. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res 38:1273–1283. doi: 10.1093/nar/gkp1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galvani CD, Li Y, Burr TJ, Hoch HC. 2007. Twitching motility among pathogenic Xylella fastidiosa isolates and the influence of bovine serum albumin on twitching-dependent colony fringe morphology. FEMS Microbiol Lett 268:202–208. [DOI] [PubMed] [Google Scholar]
- 59.da Silva Neto JF, Koide T, Gomes SL, Marques MV. 2002. Site-directed gene disruption in Xylella fastidiosa. FEMS Microbiol Lett 210:105–110. [DOI] [PubMed] [Google Scholar]
- 60.Monteiro PcB, Teixeira DC, Palma RR, Garnier M, Bové J-M, Renaudin J. 2001. Stable transformation of the Xylella fastidiosa citrus variegated chlorosis strain with oriC plasmids. Appl Environ Microbiol 67:2263–2269. doi: 10.1128/AEM.67.5.2263-2269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Navarrete F, De La Fuente L. 2015. Zinc detoxification is required for full virulence and modification of the host leaf ionome by Xylella fastidiosa. Mol Plant Microbe Interact 28:497–507. doi: 10.1094/MPMI-07-14-0221-R. [DOI] [PubMed] [Google Scholar]
- 62.Wells JM, Raju BC, Hung HY, Weisburg WG, Mandelco-Paul L, Brenner DJ. 1987. Xylella fastidiosa gen. nov., sp. nov.: Gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int J Syst Bacteriol 37:136–143. doi: 10.1099/00207713-37-2-136. [DOI] [Google Scholar]
- 63.Kung SH, Almeida RP. 2011. Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 77:5278–5284. doi: 10.1128/AEM.00730-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomaa AE, Deng Z, Yang Z, Shang L, Zhan Y, Lu W, Lin M, Yan Y. 2017. High-frequency targeted mutagenesis in Pseudomonas stutzeri using a vector-free allele-exchange protocol. J Microbiol Biotechnol 27:335–341. doi: 10.4014/jmb.1608.08019. [DOI] [PubMed] [Google Scholar]
- 65.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodríguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol 10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang S, Zou ZZ, Kreth J, Merritt J. 2017. Recombineering in Streptococcus mutans using direct repeat-mediated cloning-independent markerless mutagenesis (DR-CIMM). Front Cell Infect Microbiol 7:202. doi: 10.3389/fcimb.2017.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker JK, Havird JC, De La Fuente L. 2012. Differentiation of Xylella fastidiosa strains via multilocus sequence analysis of environmentally mediated genes (MLSA-E). Appl Environ Microbiol 78:1385–1396. doi: 10.1128/AEM.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.