Colonization of poultry with Campylobacter at the farm level is complex, poorly understood, and critically linked to contamination of poultry products, which is known to constitute a leading risk factor for human campylobacteriosis. Here, we investigated the use of a paired-farm design under standard production conditions and in the absence of experimental inoculations to assess potential impacts of farm and host genetics on prevalence, antimicrobial resistance and genotypes of Campylobacter in commercial turkeys of two different breeds. Data suggest impacts of farm proximity to other commercial turkey farms on the onset of colonization, genotypes, and antimicrobial resistance profiles of Campylobacter colonizing the birds. Furthermore, the significant association of a specific multidrug-resistant Campylobacter jejuni strain with turkeys of one breed suggests colonization partnerships at the Campylobacter strain-turkey breed level. The study design avoids potential pitfalls associated with experimental inoculations, providing novel insights into the dynamics of turkey colonization with Campylobacter in actual farm ecosystems.
KEYWORDS: Campylobacter, antimicrobial resistance, colonization, genotype, turkey
ABSTRACT
Campylobacter is a leading foodborne pathogen, and poultry products are major vehicles for human disease. However, determinants impacting Campylobacter colonization in poultry remain poorly understood, especially with turkeys. Here, we used a paired-farm design to concurrently investigate Campylobacter colonization and strain types in two turkey breeds (Hybrid and Nicholas) at two farms in eastern North Carolina. One farm (the Teaching Animal Unit [TAU]) was a university teaching unit at least 40 km from commercial turkey farms, while the other (SIB) was a commercial farm in an area with a high density of turkey farms. Day-old birds were obtained from the same breeder flock and hatchery and placed at TAU and SIB on the same day. Birds were marked to identify turkey breed and then commingled on each farm. TAU birds became colonized 1 week later than SIB and had lower initial Campylobacter levels in the cecum. Interestingly, Campylobacter genotypes and antimicrobial resistance profiles differed markedly between the farms. Most TAU isolates were resistant only to tetracycline, whereas multidrug-resistant isolates predominated at SIB. Multilocus sequence typing revealed that no Campylobacter genotypes were shared between TAU and SIB. A bovine-associated genotype (sequence type 1068 [ST1068]) predominated in Campylobacter coli from TAU, while SIB isolates had genotypes commonly encountered in commercial turkey production in the region. One multidrug-resistant Campylobacter jejuni strain (ST1839) showed significant association with one of the two turkey breeds. The findings highlight the need to further characterize the impact of farm-specific factors and host genetics on antimicrobial resistance and genotypes of C. jejuni and C. coli that colonize turkeys.
IMPORTANCE Colonization of poultry with Campylobacter at the farm level is complex, poorly understood, and critically linked to contamination of poultry products, which is known to constitute a leading risk factor for human campylobacteriosis. Here, we investigated the use of a paired-farm design under standard production conditions and in the absence of experimental inoculations to assess potential impacts of farm and host genetics on prevalence, antimicrobial resistance and genotypes of Campylobacter in commercial turkeys of two different breeds. Data suggest impacts of farm proximity to other commercial turkey farms on the onset of colonization, genotypes, and antimicrobial resistance profiles of Campylobacter colonizing the birds. Furthermore, the significant association of a specific multidrug-resistant Campylobacter jejuni strain with turkeys of one breed suggests colonization partnerships at the Campylobacter strain-turkey breed level. The study design avoids potential pitfalls associated with experimental inoculations, providing novel insights into the dynamics of turkey colonization with Campylobacter in actual farm ecosystems.
INTRODUCTION
Campylobacter is a leading zoonotic foodborne pathogen, with an estimated 0.8 million cases of human disease annually in the United States alone (1). Contaminated poultry products are considered a major vehicle for human campylobacteriosis (2–4). In the United States and other industrialized nations, Campylobacter jejuni is implicated in the majority (approximately 85%) of human campylobacteriosis cases, with Campylobacter coli accounting for most of the remaining cases (1, 5).
Factors that contribute to the colonization of poultry flocks with Campylobacter remain poorly understood and have been primarily investigated with broilers. In addition to impacts stemming from the environment and management practices in the poultry production ecosystem (6–13), several studies also point to the importance of host genetics in colonization of the birds with Campylobacter (14–20). Markedly less information is available for turkeys, in spite of the fact that turkey flocks are frequently colonized with C. jejuni and C. coli, including strains with multiple antimicrobial resistance attributes (8, 9, 21–28).
In an earlier study, we investigated Campylobacter colonization in two different turkey flocks that were grown concurrently and derived from the same breeder flock. One flock was associated with a university teaching unit, while the other was at a farm operated under the control of a commercial turkey company in the same region (29). It was found that birds on the industry-operated farm became colonized with Campylobacter spp., mostly C. coli, within 2 weeks and remained colonized until processing, while Campylobacter could not be detected in the ceca of the birds from the university farm. The findings suggested a pronounced impact of farm-related factors on Campylobacter colonization and failed to provide evidence for vertical transmission of Campylobacter in turkeys (29).
The objective of the current study was to further assess the usefulness of a paired-farm system as a model to analyze the dynamics of Campylobacter colonization in turkeys. In addition, we wished to determine the potential impact of host genetics in colonization by including birds of two different breeds extensively employed in commercial turkey production.
RESULTS AND DISCUSSION
The onset of Campylobacter colonization differed between the farms.
The two farms were concurrently monitored for Campylobacter at weekly intervals over the entire production period, i.e., weeks 1 to 9. Both flocks became colonized with Campylobacter, but the timing and initial levels in the cecum were different. Campylobacter was detected 1 week later at TAU (week 3) than at SIB (week 2) (Table 1). During the first 2 weeks of colonization, levels of Campylobacter (CFU/g cecal content) were also lower at TAU than at SIB. However, by week 5, cecal levels were similar between the two farms and remained similar for the remainder of the study (Table 1). During the first 2 weeks of TAU colonization (weeks 3 and 4), Campylobacter levels were lower in turkeys of breed Hybrid, but levels were similar between the two breeds thereafter (Table 1). We were unable to detect a similar difference between the two breeds during the first weeks of colonization in SIB (Table 1). The data suggest that potential breed-associated differences in Campylobacter levels in the cecum are transient and may be best detectable when total levels are relatively low, as was the case in the first 2 weeks of colonization of TAU (Table 1).
TABLE 1.
Campylobacter colonization at SIB and TAU
| Week(s) |
Campylobacter colonization (CFU/g cecal content) at: |
|||
|---|---|---|---|---|
| SIB |
TAU |
|||
| Hybrid | Nicholas | Hybrid | Nicholas | |
| 1 | NDa | ND | ND | ND |
| 2 | 7.20 × 105 | 6.92 × 105 | ND | ND |
| 3 | 2.80 × 107 | 4.00 × 106 | 6.00 × 103 | 3.00 × 105 |
| 4 | 5.60 × 106 | 3.00 × 107 | 2.00 × 105 | 8.40 × 105 |
| 5–9 | 2.75 × 107 | 5.56 × 107 | 8.94 × 106 | 2.37 × 107 |
ND, not detected.
Campylobacter antimicrobial resistance profiles and genotypes differed markedly between the two farms.
We analyzed a total of 110 Campylobacter isolates, including 48 from TAU and 62 from SIB, with an average of 4 isolates per positive cecal sample (range, 1 to 6 isolates/positive sample). C. coli predominated at both farms, both in brooders (87.5 and 93.8% of the Campylobacter isolates in SIB and TAU, respectively) and in grow-out birds (83.9 and 93.8% in SIB and TAU, respectively). All remaining Campylobacter isolates were C. jejuni.
Even though the same Campylobacter species (C. coli) predominated at both farms, the antimicrobial resistance (AMR) profiles of the isolates differed significantly (P < 0.0001). TAU isolates were primarily C. coli resistant only to tetracycline (designated CC: T), while a variety of other AMR profiles were noted among isolates from SIB (Fig. 1). Interestingly, the few C. jejuni isolates encountered at TAU had the same AMR phenotype as the TAU C. coli isolates, i.e., resistance only to tetracycline (designated CJ: T). In contrast, all tested C. jejuni isolates from SIB were multidrug resistant, being susceptible only to erythromycin (AMR profile TSKQG) (Fig. 1). The only species/AMR profile combination encountered in both SIB and TAU was C. coli with resistance to tetracycline and the quinolones nalidixic acid and ciprofloxacin (AMR profile TQ) (Fig. 1). C. coli TQ was encountered just once at TAU (week 4), but from multiple time points at SIB (Fig. 1 and Table 2). However, as discussed below, the C. coli TQ isolates in TAU and SIB had unrelated genotypes.
FIG 1.
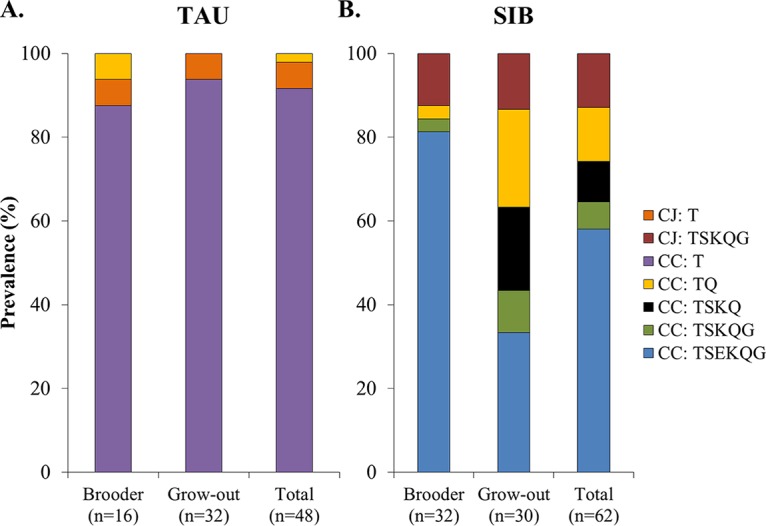
Campylobacter species and AMR profiles at the two farms. Isolates were from cecal contents of birds from (A) TAU and (B) SIB. Species and AMR profile determinations were as described in Materials and Methods. Findings are shown for brooders (from onset of colonization in week 2 or 3 to week 5), grow-out birds (weeks 6 to 9), and total duration of the study (from onset of colonization to week 9) for each farm. CJ and CC in the color code inset indicate C. jejuni and C. coli, respectively. AMR profile acronyms indicate resistance to tetracycline (T), streptomycin (S), kanamycin (K), erythromycin (E), the quinolones nalidixic acid and ciprofloxacin (Q), and gentamicin (G); combination of letters indicates specific AMR profile, e.g., T, resistance only to tetracycline; TQ, resistance to tetracycline and quinolones; TSEKQG, resistance to all tested antimicrobials listed above; TSKQG, resistance to all tested antimicrobials listed above except for erythromycin.
TABLE 2.
AMR profiles and genotypes of C. jejuni and C. coli from TAU and SIB at different time points
| Farm | Species | AMR profilea | Time (weeks)b | STc |
|---|---|---|---|---|
| TAU | C. coli | T | 3, 4, 5, 6, 7, 8, 9 | 1068 |
| C. coli | TQ | 4 | 1068 | |
| C. jejuni | T | 5, 6, 8 | 1698 | |
| SIB | C. coli | TSEKQG | 2, 3 | 1604 |
| 5, 7 | 1149 | |||
| 8 | 9064 | |||
| C. coli | TQ | 5 | 1161 | |
| 8, 9 | 8094 | |||
| C. coli | TSKQ | 8 | 889 | |
| 9 | 8094 | |||
| C. coli | TSKQG | 5, 6, 7 | NDd | |
| C. jejuni | TSKQG | 2, 5, 6, 7 | 1839 |
Abbreviations are as indicated in the legend to Fig. 1.
Boldface indicates weeks from which isolates were genotyped by MLST.
Boldface indicates a novel ST.
ND, not determined because isolates could not be resuscitated.
A total of 24 isolates, representing different species/AMR profile combinations at each farm, were analyzed by multilocus sequence typing (MLST). The findings revealed the absence of common Campylobacter strains shared between the two farms (Fig. 2). In fact, the sole AMR profile (C. coli TQ) shared between the two farms corresponded to unrelated sequence types. C. coli TQ from TAU had sequence type 1068 (ST1068), which was clearly distinct from C. coli TQ isolates from SIB, which had either ST1161 or ST8094 (Fig. 2). ST1068 differed markedly from ST1161 (all seven alleles) and ST8094 (six alleles), indicating that C. coli TQ from TAU and SIB represent unrelated strains. C. coli T isolates which predominated at TAU and were obtained at multiple time points between weeks 3 to 9 had ST1068, which was also noted as mentioned above in the sole C. coli TQ strain from TAU (Table 2 and Fig. 2). ST1068 was not detected among any of the C. coli SIB isolates, which instead exhibited six different STs, two of which (ST8094 and ST9064) were novel STs found in this study (Table 2 and Fig. 2). None of the C. coli STs encountered in SIB were detected in TAU (Fig. 2).
FIG 2.
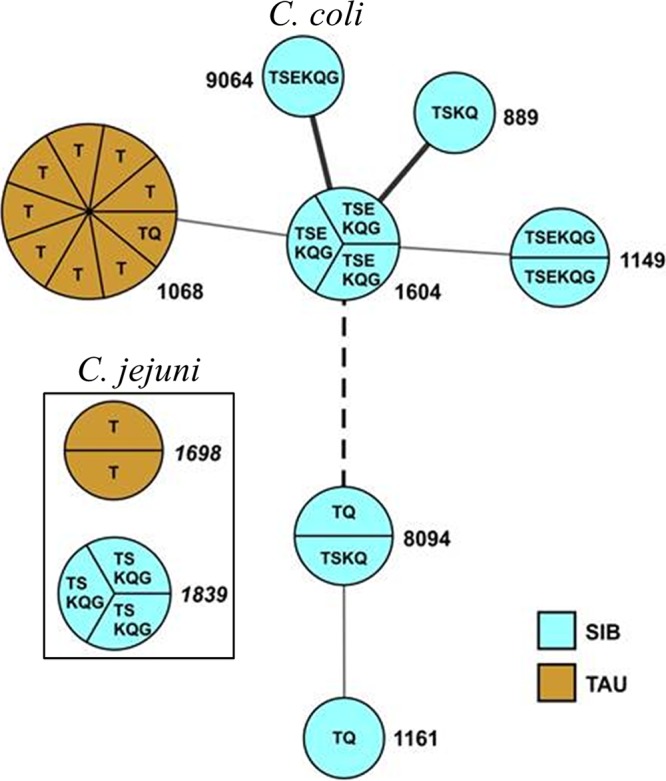
Minimum spanning tree of MLST-based STs in Campylobacter isolates from TAU and SIB. TAU and SIB are indicated in brown and blue, respectively. STs encountered in C. jejuni are italicized and separated from the rest of the STs within a black rectangle, while C. coli STs are in normal font. Thick solid line, 2-allele difference; thin solid line, 3-allele difference; dashed line, 5-allele difference. STs not linked with any lines (ST1698 and ST1839) shared no alleles with any of the other shown STs (i.e., 7-allele difference). Sizes of circles correspond to numbers of isolates with the corresponding ST, and circle segments corresponding to each isolate are indicated for STs encountered in more than one isolate. Letters inside the circles indicate the antimicrobial resistance profile, as described in the legend to Fig. 1.
Similarly to findings with C. coli, genotypes of C. jejuni were unrelated between TAU and SIB; that is, C. jejuni isolates from TAU had ST1698, while SIB isolates had ST1839, with the two STs differing at all 7 alleles (Fig. 2). An earlier analysis of isolates from commercial turkeys in the same region (eastern North Carolina) indicated that C. jejuni isolates of ST1698 were uncommon, being encountered only twice among 80 isolates, but exhibited the same AMR profile (resistance only to tetracycline) as the TAU isolates (22). In contrast, C. jejuni ST1839 has been frequently encountered in commercial turkey flocks in this region (22, 30).
SIB isolates exhibited notable strain compositional shifts between brooder and grow-out.
No major strain composition shifts between brooders and grow-out birds were noted in TAU, where C. coli T was predominant in brooders (14/16, 87.5%) and remained predominant in grow-out birds (30/32, 93.8%) (Fig. 1). However, a significant difference (P = 0.0004) in strain distribution between total brooder and grow-out bird isolates was observed. This difference was most apparent in SIB, where four strain profiles detected in brooders were also detected in grow-out birds, but at different proportions (Fig. 1). In addition, C. coli isolates with a new multidrug resistance profile (resistance to tetracycline, streptomycin, kanamycin, and the quinolones ciprofloxacin and nalidixic acid, i.e., AMR profile TSKQ) were noted only in grow-out birds (Fig. 1). Especially noteworthy was the shift in the incidence of C. coli resistant to all tested antibiotics, i.e., AMR profile TSEKQG. These isolates predominated in brooders (26/32, 81.3%), but were less common (10/30, 33.3%) among grow-out isolates (Fig. 1). Their prevalence decreased as birds aged, being encountered among 9 of the 16 total Campylobacter isolates (56.3%) from weeks 6 and 7, only once among 6 isolates at week 8, and not at all among the 8 isolates at week 9.
Competition by new strains colonizing the older birds, such as C. coli TSKQ, may have contributed to the observed decreases in C. coli TSEKQG as birds aged. In addition, fitness of different Campylobacter strains may change in response to age-related changes in the cecal microbiota (31), and this may result in displacement of previously common strains such as C. coli TSEKQG. The potential impacts of gut microbial community shifts on Campylobacter strain dynamics remain to be elucidated.
Evidence for strain-dependent impact of host genetics.
Strain (Campylobacter species/AMR profile) distributions among all isolates were significantly different between breeds (P = 0.0059). The most apparent difference was observed with C. jejuni TSKQG (ST1839). This strain was detected in 8/52 (15%) of the total Nicholas isolates but was not detected among any of the 58 isolates from Hybrid birds (P = 0.0018). More specifically, this strain accounted for 4/16 (25%) and 4/14 (28.6%) of isolates from Nicholas brooders and grow-out birds, respectively, i.e., 26.7% of the total Nicholas-derived isolates from SIB, but it was not detected among any of the 32 isolates from Hybrid birds at the same farm (SIB) (P = 0.0017). C. jejuni TSKQG (ST1839 and related STs) is a widely disseminated C. jejuni clone in turkey farms in eastern North Carolina (22, 30), possibly reflecting high colonization fitness in certain turkey breeds. Certain other strains from SIB, e.g., C. coli with AMR profile TSEKQG, also showed breed-associated differences (Fig. 3).
FIG 3.
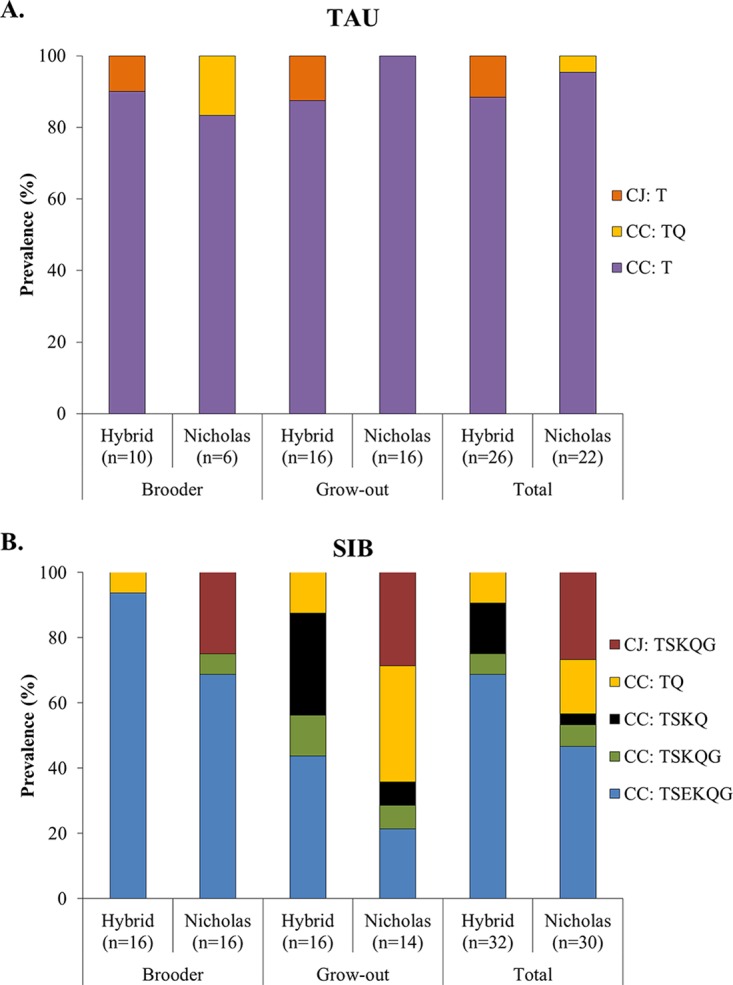
Campylobacter species and AMR profiles in turkeys of breeds Hybrid and Nicholas at (A) TAU and (B) SIB. Species and AMR determinations of isolates obtained from cecal contents were as described in Materials and Methods. Findings are shown for brooders (from onset of colonization in week 2 or 3 to week 5), grow-out birds (weeks 6 to 9), and total duration of the study (from onset of colonization to week 9) for each breed. Species and AMR profile acronyms in the color code inset are as in the legend to Fig. 1.
TAU birds of either breed were predominantly colonized by C. coli resistant only to tetracycline (Fig. 3). As discussed above, C. jejuni was uncommon in TAU, but when encountered, it was ST1698 and resistant only to tetracycline. It is worthy of note, however, that C. jejuni T isolates from TAU were only detected in Hybrid birds (Fig. 3).
Even though substantial evidence indicates the impact of host genetics for Campylobacter colonization of chickens (14–20), relevant reports have been lacking for turkeys. In addition, previous analyses typically employed experimental infections of chickens with a single Campylobacter strain and thus could not assess preference of different Campylobacter strains for different breeds of poultry. However, a study that tested a panel of C. jejuni strains with two chicken breeds identified potential breed- and strain-dependent differences in extraintestinal spread of C. jejuni to the liver (19). In chickens, resistance to C. jejuni colonization is a complex trait accompanied by multiple immunomodulatory responses (17, 18, 20, 32, 33), and it is reasonable to expect similar complexity in the case of turkeys. Future studies may unravel mechanisms underlying colonization dynamics at the strain level for both Campylobacter and its avian host.
Campylobacter strain types and AMR profiles may reflect proximity to other farms.
The finding that most isolates from TAU birds were C. coli with ST1068 was surprising, as this ST was previously detected frequently among C. coli from cattle and, to a lesser extent, from swine but not from poultry (34, 35). Even though Campylobacter strains may be able to transfer between poultry and cattle (13), the source of the C. coli ST1068 isolates predominating at TAU is currently unknown. On the other hand, all six C. coli STs detected in SIB were either identical or related to STs such as ST889 and ST1161 (Fig. 2) that were previously frequently detected among C. coli isolates from commercial turkey farms (34, 36). Such STs were uncommonly encountered among C. coli isolates from other animal hosts (34, 35), suggesting that they represent turkey-adapted strains.
Similar findings were obtained for C. jejuni strains. ST1698, detected at TAU, was only rarely detected before among C. jejuni isolates from commercial turkey farms in North Carolina (22, 30). In contrast, ST1839, which was detected in SIB, appears to belong to a turkey-associated clone frequently encountered among C. jejuni isolates from commercial turkeys in North Carolina (22, 30) (W. G. Miller and S. Kathariou, unpublished findings).
Such findings may reflect a common pool of Campylobacter strains shared among farms in a region dense in turkey farms, such as where SIB was located. Even though cattle and swine are also frequently produced in this turkey-dense region (23, 37) (H. Bolinger and S. Kathariou, unpublished), there is evidence for host preference among Campylobacter strains from different agricultural animal hosts, especially for C. coli (34, 35). Transmission of Campylobacter from one turkey farm to another may be facilitated by human and equipment traffic, flies, and other vectors (21). In addition, commercial farms in the vertically integrated turkey industry typically employ the same service staff to visit multiple farms, providing opportunities for the spread of Campylobacter strains from farm to farm (H. Bolinger, D. Carver, and S. Kathariou, unpublished).
One can speculate that in the absence of nearby commercial turkey farms, as was the case for TAU, birds can still become colonized by strains from other Campylobacter reservoirs in the vicinity. In the current study, the sources and transmission routes of Campylobacter into the TAU turkey farm remain unidentified. Nonetheless, the data suggest that, even though C. coli ST1068 has not before been encountered in commercial poultry and appears to be associated primarily with mammalian sources, especially cattle, it can still colonize turkeys. Under natural conditions, such colonization capacity can be best demonstrated when the birds are at a distance from other commercial turkey flocks, as was the case here, and thus do not have easy access to the C. jejuni or C. coli strains commonly associated with commercial turkey production.
Conclusions.
This paired-farm study indicated a lag in colonization at one farm (TAU) in comparison to the other (SIB). Even though both farms were in the same geographical region (eastern North Carolina), only SIB was in an area with a high density of commercial turkey farms; TAU was at a distance of >40 km from commercial turkey farms. The complete lack of overlap in Campylobacter genotypes between the two farms was of special interest. It was also noteworthy that multidrug-resistant strains predominated in SIB, whereas virtually all Campylobacter isolates from TAU were resistant only to tetracycline.
Differences in colonization onset and in Campylobacter strains between the two farms likely reflect differences in access and sources of Campylobacter. This was supported by the finding that SIB strains had genotypes commonly found in commercial turkey production from the region, in contrast to TAU birds, which were colonized by Campylobacter strains with genotypes that are absent or rarely encountered in commercial turkey flocks in the region.
The importance of farm location to the types of Campylobacter strains colonizing the birds could not be ascertained from a previous paired-study longitudinal analysis in the same region, where the birds at one of the farms (in the same facility as the current TAU) remained free of Campylobacter for the entire study, while birds at the other site became colonized (29). The current study design, furthermore, allowed us to assess potential impacts of turkey breed on the Campylobacter status of the birds at each farm. Even though birds of both tested breeds became colonized with Campylobacter at both farms, it was noteworthy that a specific C. jejuni strain encountered at the SIB farm showed significant association with one of the two turkey breeds. The findings suggest the potential for preferred Campylobacter genotype-turkey breed partnerships and warrant further studies.
Even though SIB was operated under conditions standard to the industry and was located in a turkey-dense region, analysis of additional farms from this and other regions would be valuable. It will be of special interest to determine whether the breed preference observed here can be observed in other turkey farms colonized with the same strains and whether Campylobacter strains predominating in other regions may also show preference for certain turkey breeds. The current study was ideally designed to assess the impact of turkey breed in a commercial setting, as birds of each breed were physically marked and then commingled on the same farm and at the same time. This type of study is logistically not easy to do, especially in a commercial setting. Even though experimental inoculations of turkeys with mixtures of specific strains can also be employed to assess potential breed-associated competitive advantages of specific strains, further investigations of flocks which are naturally colonized with Campylobacter during production will be especially useful, as they better reflect the complexities of the poultry production ecosystem.
In conclusion, the paired-farm study described here has provided considerable insight into the dynamics of Campylobacter colonization in turkeys, while also clearly illustrating the complexity of Campylobacter transmission and adaptations in poultry production. Further studies are needed to elucidate transmission of Campylobacter during agricultural production of turkeys and other poultry, leading to improved intervention strategies.
MATERIALS AND METHODS
Paired-farm design and flock management.
Birds were of two commercial turkey breeds, Hybrid and Nicholas, and originated from a single hatchery and breeder source. Day-old poults were randomly assigned and transferred on the same day to either the Teaching Animal Unit (TAU) farm, also employed in our previous study (29), at North Carolina State University, College of Veterinary Medicine (Raleigh, NC), or a commercial farm (sibling production, designated SIB) in a rural area with a high density of turkey farms. Both TAU and SIB were in eastern North Carolina. Turkey breeds Hybrid and Nicholas were physically marked (clipping of a claw on the left versus right leg) and then commingled and raised as a single flock at each farm. Stocking densities for SIB and TAU were 21.5 and 30.1 birds/m2, respectively. Totals of 2,000 birds and 8,000 birds were placed at TAU and SIB, respectively. SIB birds were in one house during the brooder stage (weeks 1 to 5) but were moved at 5 weeks to a different house at the same farm for the grow-out stage (weeks 6 to 9), while TAU birds remained in the same turkey house during the entire 9-week trial. Feed was the same for both TAU and SIB and included nitarsone at 170.1 g/ton to help prevent blackhead, caused by Histomonas meleagridis. General management and biosecurity practices followed industry standards and were comparable between TAU and SIB, with the exception of a hemorrhagic enteritis vaccine given only to SIB birds. No antibiotics or probiotics were administered at TAU or SIB at any time. Both flocks were monitored over a period of 9 weeks.
Sample collection and processing for Campylobacter spp.
TAU and SIB birds were monitored for Campylobacter at their respective farms at weekly intervals during weeks 1 to 9. Previous analyses of ceca from individual turkeys grown commercially in the same region indicated that, typically, 100% of the birds became colonized by week 3 and, in addition, the different birds tended to harbor the same Campylobacter strains (29, 38). Thus, in the current study analysis of Campylobacter utilized cecal samples pooled from multiple birds. Each week, 10 birds of each turkey breed were randomly chosen from each farm and euthanized following institutional animal welfare guidelines. The contents of one cecum from each bird were placed into sterile 50-ml conical tubes (Corning Inc., Corning, NY) and thoroughly mixed with a sterile glass rod to produce a composite cecal sample for each farm/breed combination. This led to 36 composite samples, 18 each from TAU and SIB, including nine composite samples of each breed from each farm. For Campylobacter isolation, 1 g from the interior of each composite sample was resuspended in 10 ml of Mueller-Hinton broth (MHB; Becton, Dickinson & Co., Sparks, MD). Appropriate dilutions were plated on modified charcoal-cefoperazone-deoxycholate agar (mCCDA; Oxoid Ltd., Hampshire, UK) and incubated microaerobically at 42°C for 48 h as described previously (29). For determination of the number of CFU/g cecal content, colonies with appearance typical of Campylobacter were enumerated from the mCCDA plates at the completion of the incubation period.
Characterization of Campylobacter spp.
Up to six individual colonies from positive samples were selected at random, purified, and preserved at −80°C, as described previously (29). Species designations (C. jejuni or C. coli) were determined via multiplex PCR as described previously (29). Susceptibility to a panel of antimicrobials, including tetracycline (T), streptomycin (S), erythromycin (E), kanamycin (K), nalidixic acid and ciprofloxacin (Q), and gentamicin (G), was determined as described previously (22). The pansensitive C. jejuni strain ATCC 33560 was included in each antimicrobial susceptibility test for quality assurance, as described previously (22). A subset of isolates representing distinct species and antimicrobial resistance profiles were characterized by multilocus sequence typing (MLST), as previously described (34). A minimum spanning tree (MST) displaying relationships between the MLST sequence types was constructed as previously described (34).
Statistical analysis.
Fisher's exact test was used to investigate relative frequencies of strain profiles across age groups, breeds, and farms. Pairwise comparisons of specific strains were also conducted using Fisher's exact tests. All calculations were conducted using the EXACT statement and FISHER option within SAS PROC FREQ (SAS Institute, Cary, NC).
ACKNOWLEDGMENTS
This study was partially supported by USDA-NIFA grant 2011-51110-31050.
We thank Michael Martin and John Barnes for their guidance and assistance in the study. We are grateful to the turkey industry collaborators for their unwavering support throughout the project. We thank all members of our laboratory for feedback and support.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Desai Ahuja S, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis 38:S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 3.Rosner BM, Schielke A, Didelot X, Kops F, Breidenbach J, Willrich N, Gölz G, Alter T, Stingl K, Josenhans C, Suerbaum S, Stark K. 2017. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci Rep 7:5139. doi: 10.1038/s41598-017-05227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald C. 2015. Campylobacter. Clin Lab Med 35:289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Agunos A, Waddell L, Léger D, Taboada E. 2014. A systematic review characterizing on-farm sources of Campylobacter spp. for broiler chickens. PLoS One 9:e104905. doi: 10.1371/journal.pone.0104905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newell DG, Elvers KT, Dopfer D, Hansson I, Jones P, James S, Gittins J, Stern NJ, Davies R, Connerton I, Pearson D, Salvat G, Allen VM. 2011. Biosecurity-based interventions and strategies to reduce Campylobacter spp. on poultry farms. Appl Environ Microbiol 77:8605–8614. doi: 10.1128/AEM.01090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arsenault J, Letellier A, Quessy S, Normand V, Boulianne M. 2007. Prevalence and risk factors for Salmonella spp. and Campylobacter spp. caecal colonization in broiler chicken and turkey flocks slaughtered in Quebec, Canada. Prev Vet Med 81:250–264. doi: 10.1016/j.prevetmed.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Luangtongkum T, Morishita TY, Ison AJ, Huang S, McDermott PF, Zhang Q. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl Environ Microbiol 72:3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bull SA, Allen VM, Domingue G, Jørgensen F, Frost JA, Ure R, Whyte R, Tinker D, Corry JEL, Gillard-King J, Humphrey TJ. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl Environ Microbiol 72:645–652. doi: 10.1128/AEM.72.1.645-652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerin MT, Martin W, Reiersen J, Berke O, McEwen SA, Bisaillon J-R, Lowman R. 2007. A farm-level study of risk factors associated with the colonization of broiler flocks with Campylobacter spp. in Iceland, 2001–2004. Acta Vet Scand 49:18. doi: 10.1186/1751-0147-49-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nather G, Alter T, Martin A, Ellerbroek L. 2009. Analysis of risk factors for Campylobacter species infection in broiler flocks. Poult Sci 88:1299–1305. doi: 10.3382/ps.2008-00389. [DOI] [PubMed] [Google Scholar]
- 13.Zweifel C, Scheu KD, Keel M, Renggli F, Stephan R. 2008. Occurrence and genotypes of Campylobacter in broiler flocks, other farm animals, and the environment during several rearing periods on selected poultry farms. Int J Food Microbiol 125:182–187. doi: 10.1016/j.ijfoodmicro.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 14.Stern NJ, Meinersmann RJ, Cox NA, Bailey JS, Blankenship LC. 1990. Influence of host lineage on cecal colonization by Campylobacter jejuni in chickens. Avian Dis 34:602–606. doi: 10.2307/1591251. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Swaggerty CL, Kogut MH, Chiang H, Wang Y, Genovese KJ, He H, Stern NJ, Pevzner IY, Zhou H. 2008. The paternal effect of Campylobacter jejuni colonization in ceca in broilers. Poult Sci 87:1742–1747. doi: 10.3382/ps.2008-00136. [DOI] [PubMed] [Google Scholar]
- 16.Boyd Y, Herbert EG, Marston KL, Jones MA, Barrow PA. 2005. Host genes affect intestinal colonisation of newly hatched chickens by Campylobacter jejuni. Immunogenetics 57:248–253. doi: 10.1007/s00251-005-0790-6. [DOI] [PubMed] [Google Scholar]
- 17.Connell S, Meade KG, Allan B, Lloyd AT, Kenny E, Cormican P, Morris DW, Bradley DG, O'Farrelly C. 2012. Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing. PLoS One 7:e40409. doi: 10.1371/journal.pone.0040409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connell S, Meade KG, Allan B, Lloyd AT, Downing T, O'Farrelly C, Bradley DG. 2013. Genome-wide association analysis of avian resistance to Campylobacter jejuni colonization identifies risk locus spanning the CDH13 gene. G3 (Bethesda) 3:881–890. doi: 10.1534/g3.113.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey S, Lacharme-Lora L, Chaloner G, Gibbs K, Humphrey T, Williams N, Wigley P. 2015. Heterogeneity in the infection biology of Campylobacter jejuni isolates in three infection models reveals an invasive and virulent phenotype in a ST21 isolate from poultry. PLoS One 10:e0141182. doi: 10.1371/journal.pone.0141182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Psifidi A, Fife M, Howell J, Matika O, van Diemen PM, Kuo R, Smith J, Hocking PM, Salmon N, Jones MA, Hume DA, Banos G, Stevens MP, Kaiser P. 2016. The genomic architecture of resistance to Campylobacter jejuni intestinal colonisation in chickens. BMC Genomics 17:293. doi: 10.1186/s12864-016-2612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed MFEM, El-Adawy H, Hotzel H, Tomaso H, Neubauer H, Kemper N, Hartung J, Hafez HM. 2016. Prevalence, genotyping and risk factors of thermophilic Campylobacter spreading in organic turkey farms in Germany. Gut Pathog 8:28. doi: 10.1186/s13099-016-0108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu W, Siletzky RM, Wright S, Islam M, Kathariou S. 2009. Antimicrobial susceptibility profiles and strain type diversity of Campylobacter jejuni isolates from turkeys in eastern North Carolina. Appl Environ Microbiol 75:474–482. doi: 10.1128/AEM.02012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright SL, Carver DK, Siletzky RM, Romine S, Morrow WEM, Kathariou S. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J Food Prot 71:1791–1796. doi: 10.4315/0362-028X-71.9.1791. [DOI] [PubMed] [Google Scholar]
- 24.Lutgen EM, McEvoy JM, Sherwood JS, Logue CM. 2009. Antimicrobial resistance profiling and molecular subtyping of Campylobacter spp. from processed turkey. BMC Microbiol 9:203. doi: 10.1186/1471-2180-9-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Adawy H, Hotzel H, Düpre S, Tomaso H, Neubauer H, Hafez HM. 2012. Determination of antimicrobial sensitivities of Campylobacter jejuni isolated from commercial turkey farms in Germany. Avian Dis 56:685–692. doi: 10.1637/10135-031912-Reg.1. [DOI] [PubMed] [Google Scholar]
- 26.Giacomelli M, Andrighetto C, Lombardi A, Martini M, Piccirillo A. 2012. A longitudinal study on thermophilic Campylobacter spp. in commercial turkey flocks in northern Italy: occurrence and genetic diversity. Avian Dis 56:693–700. doi: 10.1637/10141-032312-Reg.1. [DOI] [PubMed] [Google Scholar]
- 27.Giacomelli M, Salata C, Martini M, Montesissa C, Piccirillo A. 2014. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb Drug Resist 20:181–188. doi: 10.1089/mdr.2013.0110. [DOI] [PubMed] [Google Scholar]
- 28.Kashoma IP, Kumar A, Sanad YM, Gebreyes W, Kazwala RR, Garabed R, Rajashekara G. 2014. Phenotypic and genotypic diversity of thermophilic Campylobacter spp. in commercial turkey flocks: a longitudinal study. Foodborne Pathog Dis 11:850–860. doi: 10.1089/fpd.2014.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith K, Reimers N, Barnes HJ, Lee BC, Siletzky R, Kathariou S. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J Food Prot 67:1463–1468. doi: 10.4315/0362-028X-67.7.1463. [DOI] [PubMed] [Google Scholar]
- 30.Qiu Y. 2013. Antimicrobial susceptibility and clonal population structure of multidrug resistant Campylobacter jejuni isolates from commercial turkeys in North Carolina. MSc thesis North Carolina State University, Raleigh, NC. [Google Scholar]
- 31.Danzeisen JL, Clayton JB, Huang H, Knights D, McComb B, Hayer SS, Johnson TJ. 2015. Temporal relationships exist between cecum, ileum, and litter bacterial microbiomes in a commercial turkey flock, and subtherapeutic penicillin treatment impacts ileum bacterial community establishment. Front Vet Sci 2:56. doi: 10.3389/fvets.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Swaggerty CL, Kogut MH, Chiang H-I, Wang Y, Genovese KJ, He H, Zhou H. 2010. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS One 5:e11827. doi: 10.1371/journal.pone.0011827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li XY, Swaggerty CL, Kogut MH, Chiang HI, Wang Y, Genovese KJ, He H, Pevzner IY, Zhou HJ. 2011. Caecal transcriptome analysis of colonized and non-colonized chickens within two genetic lines that differ in caecal colonization by Campylobacter jejuni. Anim Genet 42:491–500. doi: 10.1111/j.1365-2052.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Fedorka-Cray PJ, Mandrell RE. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- 35.Roux F, Sproston E, Rotariu O, Macrae M, Sheppard SK, Bessell P, Smith-Palmer A, Cowden J, Maiden MCJ, Forbes KJ, Strachan NJC. 2013. Elucidating the aetiology of human Campylobacter coli infections. PLoS One 8:e64504. doi: 10.1371/journal.pone.0064504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'lima CB, Miller WG, Mandrell RE, Wright SL, Siletzky RM, Carver DK, Kathariou S. 2007. Clonal population structure and specific genotypes of multidrug-resistant Campylobacter coli from turkeys. Appl Environ Microbiol 73:2156–2164. doi: 10.1128/AEM.02346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.North Carolina Department of Agriculture and Consumer Services (NCDACS). 2012. Census volume 1, chapter 2: county level data. North Carolina Department of Agriculture and Consumer Services (NCDACS), Raleigh, NC. [Google Scholar]
- 38.Kirchner MK. 2017. Response of the turkey gut microbiome, Campylobacter, and Salmonella to health-disease transitions and antimicrobial interventions. MSc thesis North Carolina State University, Raleigh, NC. [Google Scholar]


