The function of thiaminase I has remained a long-standing, unsolved mystery. The enzyme is only known to be produced by a small subset of microorganisms, although thiaminase I activity has been associated with numerous plants and animals, and is implicated in thiamine deficiencies brought on by consumption of organisms containing this enzyme. Genomic and biochemical analyses have shed light on potential roles for the enzyme. Using the genetically amenable thiaminase I-producing soil bacterium Burkholderia thailandensis, we were able to demonstrate that thiaminase I helps salvage precursors from thiamine derivatives in the environment and degrades thiamine to its precursors, which are preferentially used by B. thailandensis auxotrophs. Our study establishes a biological role for this perplexing enzyme and provides insight into the complicated nature of thiamine metabolism. It also establishes B. thailandensis as a robust model system for studying thiamine metabolism.
KEYWORDS: Burkholderia thailandensis, HMP, thiC, thiG, thiaminase I, thiamine, thiazole
ABSTRACT
Thiamine is essential to life, as it serves as a cofactor for enzymes involved in critical carbon transformations. Many bacteria can synthesize thiamine, while thiamine auxotrophs must obtain it or its precursors from the environment. Thiaminases degrade thiamine by catalyzing the base-exchange substitution of thiazole with a nucleophile, and thiaminase I specifically has been implicated in thiamine deficiency syndromes in animals. The biological role of this secreted enzyme has been a long-standing mystery. We used the thiaminase I-producing soil bacterium Burkholderia thailandensis as a model to ascertain its function. First, we generated thiamine auxotrophs, which are still able to use exogenous precursors (thiazole and hydroxymethyl pyrimidine), to synthesize thiamine. We found that thiaminase I extended the survival of these strains, when grown in defined media where thiamine was serially diluted out, compared to isogenic strains that could not produce thiaminase I. Thiamine auxotrophs grew better on thiamine precursors than thiamine itself, suggesting thiaminase I functions to convert thiamine to useful precursors. Furthermore, our findings demonstrate that thiaminase I cleaves phosphorylated thiamine and toxic analogs, which releases precursors that can then be used for thiamine synthesis. This study establishes a biological role for this perplexing enzyme and provides additional insight into the complicated nature of thiamine metabolism and how individual bacteria may manipulate the availability of a vital nutrient in the environment.
IMPORTANCE The function of thiaminase I has remained a long-standing, unsolved mystery. The enzyme is only known to be produced by a small subset of microorganisms, although thiaminase I activity has been associated with numerous plants and animals, and is implicated in thiamine deficiencies brought on by consumption of organisms containing this enzyme. Genomic and biochemical analyses have shed light on potential roles for the enzyme. Using the genetically amenable thiaminase I-producing soil bacterium Burkholderia thailandensis, we were able to demonstrate that thiaminase I helps salvage precursors from thiamine derivatives in the environment and degrades thiamine to its precursors, which are preferentially used by B. thailandensis auxotrophs. Our study establishes a biological role for this perplexing enzyme and provides insight into the complicated nature of thiamine metabolism. It also establishes B. thailandensis as a robust model system for studying thiamine metabolism.
INTRODUCTION
Thiamine (vitamin B1) is an essential vitamin necessary for nearly all cellular life (1). Its diphosphorylated form, thiamine pyrophosphate (TPP), serves as a cofactor for enzymes that perform critical carbon transformations necessary for energy metabolism and the biosynthesis of precursors for cellular macromolecules (2). Thiamine is composed of pyrimidine and thiazole (THZ) moieties that are synthesized separately and then combined (3) (Fig. 1A). Generally, the pyrimidine moiety is derived from the purine intermediate 5-aminoimidazole ribotide, which is converted to hydroxymethyl pyrimidine phosphate (HMP-P) by the HMP synthase, ThiC. HMP-P is then phosphorylated to HMP-PP by the kinase ThiD. The THZ moiety is synthesized from glycolysis products, the sulfur carrier protein ThiS, and glycine or tyrosine in a multistep process where thiazole synthase (ThiG) ultimately forms THZ-P carboxylate. Thiamine phosphate synthase (ThiE) combines THZ-P and HMP-PP to form thiamine monophosphate (TMP), and typically ThiL phosphorylates TMP to produce the active cofactor TPP.
FIG 1.
Predicted thiamine biosynthesis in B. thailandensis and thiaminase I mode of action. (A) Schematic overview of the predicted thiamine biosynthesis pathway in B. thailandensis based on its genomic content. Enzymes in green text synthesize the thiazole moiety, while those in blue text synthesize the pyrimidine moiety. Those in red combine the moieties to form the active cofactor. DXP, 1-deoxy-d-xylulose 5-phosphate; G3P is glyceraldehyde 3-phosphate. The other compounds are defined in the introduction. (B) Schematic of the mode of action of thiaminase I. “X” represents a variety of organic nucleophiles that covalently attached to the HMP moiety, substituting for the THZ moiety.
Although the ability to synthesize thiamine is widespread in bacteria, many species are thiamine auxotrophs. Listeria monocytogenes abrogates the need to synthesize thiamine through uptake via the transporter ThiT (4). Members of the Enterobacteriaceae use the ThiBPQ system, which not only imports thiamine but also its phosphorylated forms (5). Imported thiamine can be diphosphorylated to TPP via thiamine pyrophosphokinase or stepwise as described above (6). Some bacteria take up thiamine precursors for growth (7, 8). Described HMP transporters include the YkoCDEF system (9, 10), CytX (11, 12), and HmpT (9). The import of THZ is less well defined in bacteria, but a THZ-specific transporter, ThiW, has been described (9, 12).
Bacteria also produce thiaminases, enzymes that degrade thiamine to its constituents by catalyzing the base exchange of thiazole with a nucleophile (Fig. 1B). Thiaminases are categorized as thiaminase II and thiaminase I. Although they perform a similar biochemical reaction, the two groups of proteins lack sequence similarity. Thiaminase II (known as TenA) uses water as a nucleophile, and this enzyme is widely distributed in bacteria and archaea (13, 14), with homologs also found in fungi (15) and plants (16). These intracellular enzymes recycle base-degraded thiamine, which comprises an intact HMP conjugated to a formylamino group. In Bacillus halodurans, base-degraded thiamine is imported and then deformylated to aminopyrimidine (17). TenA catalyzes the substitution of the amino group from aminopyrimidine with a hydroxyl from water, effectively restoring HMP, which can be subsequently recycled and used in thiamine synthesis.
Thiaminase I enzymes are less widely distributed, occurring in a number of phylogenetically unrelated bacteria, including some human pathogens and their close relatives (2). It has also been described in the amoeba Naegleria gruberi, where it is fused to transketolase (18). The bacterial enzymes are secreted through the general secretory pathway (19) and use a variety of organic nucleophiles, but not water, for the base-exchange reaction (20–22). Further, the enzyme has been shown to perform the base-exchange reaction on certain thiamine analogs with intact HMP moieties (23). The crystal structures of thiaminase I from both Paenibacillus thiaminolyticus (24) and Clostridium botulinum C143S (25) have been solved, revealing the active site and the likely mechanism by which the base-exchange reaction is catalyzed. Structural analysis further reveals that thiaminase I shares a common ancestor with TbpA (ThiB), the thiamine binding protein used for thiamine transport in Escherichia coli and other Enterobacteriaceae (26). Counterintuitively, and for unknown reasons, thiaminase I activity in culture is repressed by high levels of thiamine (27, 28). An analysis of the thiaminase I gene of C. botulinum A ATCC 19397 revealed that it is located in an operon with the biosynthesis genes for the HMP analog bacimethrin, which when combined with THZ, forms the antivitamin 2′-methoxythiamine (29). Despite being located in this operon, the role of thiaminase I, if any, in the synthesis of bacimethrin is unknown (29). In Paenibacillus species, thiaminase I is found in a conserved operon, including thiamine salvage and synthesis genes thiM, thiD, and thiE and possibly genes for bacimethrin synthesis (30). Despite these biochemical, mechanistic, and genomic analyses, the biological function of thiaminase I remains a great mystery in our understanding of thiamine metabolism.
Thiaminase I has been linked to thiamine deficiencies and death in animals. Despite only being known to be produced by microorganisms, thiaminase I activity has been found in plant and animal tissues (20, 31), though it is unclear whether these enzymes are produced endogenously or by host microbiota (32). Thiaminase I induced deficiencies affect economically important domesticated animals (33–37), as well as wild populations of a variety of species (2). Thiamine deficiency can also arise in humans that consume raw diets high in thiaminase I activity (2). The first characterized thiaminase I-producing bacteria were isolated from the feces of a thiamine deficient patient (20). It is increasingly apparent that thiamine and its precursors serve as drivers of ecological interactions between microorganisms (2, 8, 38); however, the role of thiaminase I in this complex interplay is not yet understood.
Due to its occurrence with genes involved in thiamine salvage and biosynthesis in some genomes, it has been hypothesized that thiaminase I plays a role in thiamine salvage (26, 39), although this hypothesis has not been tested. Here we use B. thailandensis, a thiaminase I-producing soil bacterium first isolated from a rice field in Thailand, to explore the contribution of thiaminase I activity to thiamine metabolism. Since B. thailandensis is capable of synthesizing thiamine, we generated thiamine auxotrophic mutants incapable of synthesizing either HMP or THZ and examined the impact of thiaminase I production on growth in defined media. We found that thiaminase I, either produced by the cell or added to the medium, provided a growth advantage to thiamine auxotrophs. Furthermore, we found that thiamine auxotrophs grew better on exogenous HMP and THZ than when they were provided thiamine at an equimolar concentration. Thiaminase I also allowed for growth of thiamine auxotrophs on the phosphorylated forms of thiamine and allowed growth on certain thiamine analogs, suggesting a role not only in the salvage of thiamine precursors but also in detoxifying and recycling components of thiamine analogs. This study ascribes a biological function to the elusive enzyme. We propose to name the thiaminase I gene locus (BTH_II1306) thiA due to its current inconsistent nomenclature and its apparent role in thiamine salvage.
RESULTS
Thiaminase I confers a growth advantage to thiamine auxotrophs.
To test if thiaminase I plays a role in thiamine salvage or biosynthesis, we generated thiamine auxotroph mutants in the wild-type strain B. thailandensis E264 and in BT10432, an isogenic strain with a transposon inserted into thiA (see Fig. S1 in the supplemental material for the genomic context of disrupted genes). Both strains are capable of synthesizing thiamine. When assayed for ThiA activity under growth conditions that normally produce substantial thiaminase I activity in strain E264, no thiaminase activity was detected in BT10432 (not shown). The eight strains—E264, E264-thiC, E264-thiG, E264-thiCthiG, BT10432, BT10432-thiC, BT10432-thiG, and BT10432-thiCthiG—were serially transferred every 24 h, first from tryptic soy broth (TSB) to defined medium (DM4) containing 1 mM thiamine and then to thiamine-free DM4 until they would no longer grow. The optical density at 600 nm (OD600) values of cultures at 24 h were compared via an analysis of variance (ANOVA; the experimental design depicted in Fig. 2).
FIG 2.
Experimental overview of the serial transfer experiments. All strains were first grown in TSB; then, after 24 h, they were transferred to DM4 with 1 mM thiamine. Cultures were then serially transferred to fresh DM4 every 24 h. In the illustration, the shading of the liquid medium in tubes indicates growth, while the clear tube indicates no growth. At each transfer, cultures were inoculated to a starting OD600 of 0.025. Supernatants were taken from the tubes for the rescue experiments, as indicated by the red arrow, during the final transfer in which growth was observed. The addition of rescuing factors (thiaminase I, thiamine or its precursors or analog) is indicated with a green arrow. Growths of strains were compared using ANOVAs on the OD600 at 24 h.
We observed a decline in bacterial growth with serial transfers to a progressively diminished thiamine medium and noted that viability at each transfer was dependent on the presence of intact thiA and different subsets of thiamine biosynthesis genes. After the first transfer into thiamine-free DM4, a growth defect was observed in the BT10432-thiC and BT10432-thiCthiG mutants, since both strains had a significantly lower OD600 at 24 h (Fig. 3A). This suggests that a defect in thiazole synthesis, via ThiG, was tolerated better than a defect in the ability to make HMP.
FIG 3.
Survival of B. thailandensis strains in thiamine-free DM4. These data illustrate the decline of thiamine auxotrophs that cannot make thiaminase I. (A) Indicates the growth of the eight strains in the first transfer into thiamine-free DM4. Strains incapable of making ThiA and defective in synthesizing HMP or HMP and THZ were the first to show a significant reduction in growth (P < 0.0001). (B) After the second transfer to thiamine-free medium, the strain defective in both ThiA and THZ production dropped out. The growth curves are displayed as the average of six replicate cultures with standard error bars at each time point. In the key, strains with the same superscript designations indicate no significant difference (for example, AB is not significantly different from A or B). For all of the pairwise comparisons described here, P < 0.0001.
In the next transfer, none of the BT10432 thiamine auxotrophs were able to grow whereas all the E264 thiamine auxotrophs were able to grow (Fig. 3B). The E264-thiC and E264-thiCthiG strains took longer to reach an OD600 of 1.0 in this transfer compared to the other growing strains. This demonstrated that ThiA provided the E264 strains with a growth advantage compared to isogenic auxotrophs without an intact thiA gene. Consistent with observations in the BT10432 background, in the next serial transfer, the E264-thiC and E264-thiCthiG mutants were unable to grow, while the E264-thiG mutant was able to grow (see Fig. S2A in the supplemental material), and eventually did not grow in the next serial transfer (see Fig. S2B in the supplemental material). To ensure that thiaminase I was produced by the E264 thiamine auxotrophs during the transfers, supernatant was assayed for thiaminase I activity (see Fig. S3 in the supplemental material). All four strains in the E264 background produced thiaminase I in DM4, while the BT10432 control did not. Peak thiaminase I activity was detected at 18 h in transfer 1 in DM4, while peak activity was detected at 12 h in transfers 2 and 3 (see Fig. S3 in the supplemental material). Taken together, these results demonstrate that thiaminase I provided thiamine auxotrophic B. thailandensis strains a growth advantage, and these mutants were more sensitive to the loss of the ability to synthesize HMP than to the loss of the THZ synthesis pathway.
Thiaminase I is the rescuing factor allowing for growth.
Our first set of experiments indicated that ThiA was somehow allowing for growth of thiamine auxotrophs in DM4. To further explore this possibility, we purified ThiACb (BcmE), the thiaminase I from C. botulinum (25) and added it to DM4 at the terminal transfer step of the serial transfers for all auxotrophic strains. ThiACb rescued growth for all of the BT10432 strains (Fig. 4A). This suggests that ThiACb quenched the specific auxotrophies by generating the required thiamine precursors. When ThiACb was added to the E264 auxotrophs, it did not rescue growth in the thiC or the thiC thiG mutant cultures (Fig. 4B). Although there was some growth of E264-thiG, this was not at the same level as the BT10432 strains supplied with ThiACb (Fig. 4A). This lack of growth in the E264 strains is likely due to the presence of ThiA already acting on and transforming the media in the previous transfers, leaving little substrate for ThiACb to act on.
FIG 4.
Thiaminase I rescues of thiamine auxotrophs. (A) ThiACb rescued the growth of the BT10432 thiamine auxotrophs. (B) ThiACb addition did not substantially improve the growth of the E264 thiamine auxotrophs. The growth curves displayed are the average of three replicate cultures with standard error bars at each time point. In each case, growth (OD600) at 24 h was compared and significant differences are designated by different superscripts next to the names in the key (P < 0.0001 for all pairwise comparisons in these experiments).
We examined whether the growth of the BT10432-thiC auxotroph could be rescued with the addition of conditioned media containing B. thailandensis ThiA. To do this, we collected supernatant from BT10432-thiC and E264-thiC cultures at the penultimate transfer before each strain could no longer grow (see Fig. 2 for the experimental setup). The supernatant was filter sterilized and combined with fresh thiamine-free DM4 in a 1:1 mixture. We used this as the medium for the terminal transfer of BT10432-thiC and for transfer 2 into DM4 of E264-thiC. In both cases, the uninoculated 1:1 mixture exhibited no growth (data not shown). The addition of BT10432-thiC conditioned medium did not rescue growth of BT10432-thiC. When added to E264-thiC, this culture had a significantly higher OD600 at 24 h (see Fig. S4 in the supplemental material). This could possibly be a consequence of the cultures having a higher thiamine content due to their reduced growth during this transfer (Fig. 3A). The lack of growth of the BT10432-thiC strain indicated that cell lysis, which would release thiamine or thiamine precursors into the culture supernatant during growth, was not enough to support growth of the auxotrophs. When BT10432-thiC was grown in the E264-thiC conditioned medium mixture, growth was rescued, though the OD600 was significantly lower than for the E264-thiC strains (see Fig. S4 in the supplemental material). This finding indicated that the presence of ThiA, or its transformation of the medium components, was sufficient to rescue the growth of the BT10432-thiC auxotroph.
Thiamine auxotrophs lacking thiaminase I preferentially grow on thiamine precursors.
Our findings indicated that thiaminase I rescued growth by providing the auxotrophs with the necessary precursors to synthesize thiamine. We set out to confirm this by adding back the thiamine or its precursors to each auxotroph culture. Strikingly, the BT10432-thiC (Fig. 5A) and BT10432-thiCthiG (Fig. 5C) cultures reached significantly higher OD600 at 24 h when precursors were added to the media rather than when an equimolar amount of thiamine was added. In both cases, a longer delay in growth was observed when thiamine was added, indicating preferential use of precursors over intact thiamine. E264-thiC (Fig. 5B) reached a significantly higher OD600 at 24 h when growing on HMP compared to growth on thiamine. The E264-thiC strain provided with thiamine displayed a similar growth curve when HMP was provided, and the OD600 at 24 h was much higher in comparison to its BT10432-thiC counterpart, indicating that the conversion of thiamine to HMP was responsible for this. The preferential use of precursors was more pronounced in the BT10432-thiCthiG mutant (Fig. 5C). No statistically significant difference was found between growth of the E264-thiCthiG mutant on thiamine or both precursors after 24 h (Fig. 5D). The similar growth when precursors were added indicated that B. thailandensis preferentially uses the substrates of thiamine to grow, rather than thiamine itself, and ThiA provides this by degrading thiamine. The BT10432-thiG and E264-thiG mutant did not exhibit any significant difference when grown on thiamine or THZ (see Fig. S5 in the supplemental material). Although inconsistent with the HMP and HMP+THZ mutant rescues, it is consistent with our previous findings that the thiG mutants showed a weaker growth defect. Further, a thiochrome assay for thiamine was performed to ensure that the thiamine was not simply degrading in the culture medium and thus making precursors available. Throughout a 24-h incubation period, thiamine concentrations in sterile media remained at 200 nM, the initial thiamine concentration of the media.
FIG 5.
Growth of B. thailandensis strains on thiamine and its precursors. In the absence of thiaminase I, thiamine auxotrophs grew better on thiamine precursors than thiamine. (A) The growth curve showing BT10432-thiC reached a significantly higher OD600 at 24 h (P = 0.0001) with HMP in contrast to thiamine. (B) E264-thiC reached a significantly higher OD600 (P < 0.0001) at 24 h when rescued with HMP in contrast to thiamine. (C) BT10432-thiCthiG reached a significantly higher OD600 (P < 0.0001) when rescued with HMP+THZ in contrast to thiamine. (D) No significant difference was observed between growth on thiamine and HMP+THZ for E264-thiCthiG at 24 h. The growth curves displayed are the average of six replicate cultures for precursors and nine for thiamine additions, with standard error bars at each time point. In each case, growth at 24 h was compared, and the significant differences are designated by different superscripts next to the names in the key.
Phosphorylated thiamine compounds support growth only when thiaminase I is present.
In bacteria, compounds that need to be phosphorylated for activation are typically imported in their unphosphorylated form and then phosphorylated in the cytoplasm (26). However, in thiamine transport, TbpA and ThiT can bind phosphorylated forms of thiamine (26). It has also been observed that thiaminase I from P. thiaminolyticus is as active against the phosphorylated forms of thiamine as it is upon thiamine (21, 40). Based on these observations, we hypothesized that thiaminase I may function to free unphosphorylated HMP from phosphorylated forms of thiamine so the bacteria can more readily take up and use the precursor. Supporting our hypothesis, there was no substantial growth when either TMP or TPP was added to the BT10432-thiC or BT10432-thiCthiG mutants (Fig. 6A). However, either compound added at the terminal transfer supported the growth of both E264-thiC and E264-thiCthiG (Fig. 6B). There was no difference between growth on either compound or between the different thiamine auxotrophs. This suggests that B. thailandensis can use ThiA to free HMP from phosphorylated thiamine and has mechanisms available to use the various phosphorylated THZ products produced by ThiA. Consistent with other findings throughout this study, the BT10432-thiG mutant did not behave like the other BT10432 mutants, since it was able to grow slowly with TMP but did not grow substantially with TPP (see Fig. S6A in the supplemental material). In contrast, the E264-thiG cells were able to recycle the THZ-P (from TMP) and THZ-PP (from TPP) equally well (see Fig. S6B in the supplemental material), further supporting the notion that B. thailandensis can use phosphorylated THZ.
FIG 6.
Growth of B. thailandensis strains on TMP and TPP. Thiamine auxotrophs can grow when medium is supplemented with phosphorylated forms of thiamine, but only when the strains produce ThiA. (A) No substantial growth was observed for BT10432-thiC and BT10432-thiCthiG on TMP and TPP. (B) Substantial growth was observed for E264-thiC and E264-thiCthiG on TMP and TPP. When TMP was present E264-thiC had a significantly higher OD600 at 24 h than when TPP was added (P = 0.0044). The growth curves displayed are the averages of six replicate cultures, with standard error bars at each time point. In each case, growth at 24 h was compared, and significant differences are designated by different superscripts next to the names in the key.
Thiaminase I facilitates growth of auxotrophs provided with certain thiamine analogs.
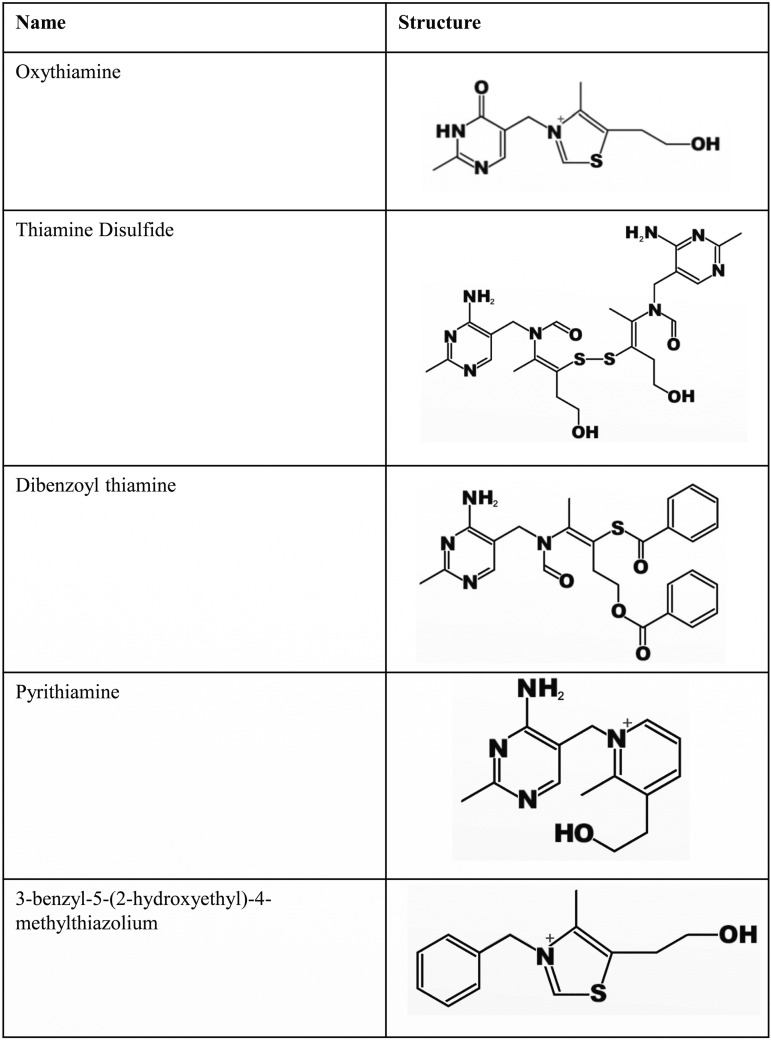
Previous work has shown that P. thiaminolyticus thiaminase I is active on thiamine analogs such as pyrithiamine, an antagonist to TPP-dependent enzymes and transporters (21, 23, 41). This led us to hypothesize that ThiA may function to release portions of thiamine analogs for salvage and thiamine synthesis. To test this, we added diverse thiamine analogs (Table 1) to DM4 at the terminal transfer step. These include compounds such as pyrithiamine and oxythiamine, which contain an intact HMP or THZ, respectively, as well as thiamine disulfide and dibenzoyl thiamine, which have an intact HMP with a bulky side chain that contains a ring-open form of THZ. We found that when pyrithiamine was added to the medium, E264-thiC reached a significantly higher OD600 at 24 h than the BT10432-thiC (Fig. 7A; see also Fig. S7A in the supplemental material); however, by 48 h, the BT10432 thiC mutant exceeded the growth of E264-thiC. This suggests that early in the growth of E264-thiC, ThiA was able to free HMP from pyrithiamine, giving the culture an initial boost, which did not occur in the BT10432-thiC culture. The samples were subcultured into fresh media either supplemented with pyrithiamine or without it (see Fig. S7A and B in the supplemental material) to determine whether there was an adapted ability to use it in the BT10432-thiC strain. When pyrithiamine was present, the initial OD600 of BT10432-thiC at 18 h was a bit higher than in the previous transfer, but it was not at the level of the E264-thiC strain, suggesting a lack of adaptation to pyrithiamine. When no pyrithiamine was present, the BT10432-thiC subculture did not grow, while the E264-thiC subculture did grow. From this we conclude that there was HMP contamination in the pyrithiamine. It allowed for initial growth of BT10432-thiC, but once the cells consumed the HMP, very little remained for transfer over in the next serial transfer, and additional growth was not supported. In contrast, the E264-thiC strain grew on the contaminating HMP but also converted the pyrithiamine to HMP, allowing it to grow in the subsequent serial transfer.
TABLE 1.
Thiamine analog structures
FIG 7.
Growth of B. thailandensis strains on thiamine analogs. Thiaminase I enhances the ability of thiamine auxotrophs to grow on some thiamine analogs. (A) In this comparison of growth on thiamine analogs in the thiC mutant background, E264-thiC was able to grow when pyrithiamine, thiamine disulfide, and dibenzoyl thiamine was added. Eventually, the BT10432-thiC mutant grew on pyrithiamine. The E264-thiC OD600s at 24 h were significantly higher than for all other cultures (P < 0.0001 for all pairwise comparisons), whereas BT10432-thiC reached the second highest OD600 when grown on pyrithiamine (P < 0.0001 for all pairwise comparisons), which eventually reached the highest OD600 by 48 h. E264-thiC growth on thiamine disulfide was significantly higher than all others (P < 0.001 for all pairwise comparisons). Despite not being significantly higher than the negative controls at 24 h, E264-thiC grew on dibenzoyl thiamine, while its BT10432 (thiA mutant) counterpart did not. The growth curves displayed are the averages of three replicate cultures, with standard error bars at each time point. In each case, growth at 24 h was compared, and significant differences are designated by different superscripts next to the names in the figure key. (B) Comparison of the growth of thiC thiG mutant strains on thiamine analogs. No growth was observed in either background when just pyrithiamine or oxythiamine was added; however, substantial growth occurred in the E264 background when both oxythiamine and pyrithiamine were supplied (P < 0.0001 for all pairwise comparisons). Slight growth occurred when both compounds were provided in the BT10432 thiaminase mutant background. (C) Comparison of the growth of the thiC thiG mutants in media supplemented with thiamine disulfide or dibenzoyl thiamine from fresh and older stocks. The older dibenzoyl thiamine supplemented medium resulted in E264-thiCthiG having a significantly higher OD600 at 24 h. E264-thiCthiG grown on the older thiamine disulfide stock had the second highest OD600 at 24 h. This demonstrated that older stocks were abiotically converting to thiamine, providing a growth advantage (P < 0.0001 for all pairwise comparisons).
In addition, the BT10432-thiC mutants were unable to grow substantially using thiamine disulfide and dibenzoyl thiamine; however, after 24 h the E264-thiC mutants were able to grow on both analogs (Fig. 7A; see also Fig. S7C and D in the supplemental material). This suggested that ThiA might be able to degrade thiamine disulfide and dibenzoyl thiamine, releasing HMP, which can then be used by the cell. This is in contrast to previous reports that described thiamine disulfide as recalcitrant to thiaminase I (41) and our own observations that thiamine disulfide cannot substitute for thiamine in our thiaminase colorimetric assay (42). It could be that the reaction rate is very slow and resulted in no detected turnover after an hour of incubation. A more likely interpretation is that thiamine disulfide was abiotically converted to thiamine (43) and then ThiA acted on the thiamine, causing a prolonged time for the culture to reach a high OD600. For dibenzoyl thiamine, the sulfur was also likely reduced abiotically in these cultures, leaving thiamine with a bulky benzoyl group still attached. Despite this, thiaminase I appears to be acting on the dibenzoyl thiamine derivative, as E264-thiC showed considerable growth (Fig. 7A). Serial transfers from media containing these derivatives with bulky side groups into fresh media containing them again, showed that E264-thiC grew better on both compounds in the second transfer (see Fig. S7D in the supplemental material). When E264-thiC was transferred to fresh media lacking the substrates, only the subculture from thiamine disulfide grew, while the dibenzoyl thiamine subculture did not. This suggests that ThiA acted upon thiamine disulfide more than dibenzoyl thiamine and that enough HMP was transferred over to support growth.
When examining the growth of the thiG mutants, we observed similar patterns to our previous experiments (see Fig. S8 in the supplemental material). In both the E264 and the BT10432 backgrounds, no significant difference in growth was observed when oxythiamine was added. This suggests that B. thailandensis has another mechanism for detoxifying this compound and restoring the THZ. Our thiaminase I assay detected no activity when oxythiamine was used as a substrate instead of thiamine, suggesting ThiA is not acting on it. No activity on oxythiamine was observed with P. thiaminolyticus thiaminase I as well (23). However, ThiA provided a growth benefit to the thiG mutants when thiamine disulfide or dibenzoyl thiamine was added, as the cultures reached a significantly higher OD600 at 24 h compared to the BT10432-thiG mutants (see Fig. S7 in the supplemental material).
Next, we added pyrithiamine and/or oxythiamine to DM4 at the terminal transfer in the BT10432 and E264 double mutants (Fig. 7B). Although little growth was observed at 24 h, the E264-thiCthiG mutant had significantly more growth when pyrithiamine and oxythiamine were both added. By 30 h, the OD600 nearly doubled, and the culture reached a high density by 48 h. In the BT10432 background, the OD600 only reached ∼0.5 at 48 h. This further demonstrated that ThiA was freeing HMP from pyrithiamine, while the THZ from oxythiamine was acquired through another mechanism. The observation of some growth of the BT10432-thiCthiG mutant supports our assertion that pyrithiamine was contaminated with HMP. To test if abiotic degradation of thiamine disulfide and dibenzoyl thiamine was occurring in the stock solutions, we rescued the thiC thiG mutants with both freshly made and older stocks of both compounds (Fig. 7C). BT10432-thiCthiG did not grow well when either older or fresh analogs were present, though there was slight growth in the cells rescued with the older dibenzoyl thiamine (Fig. 7C). The E264-thiCthiG strain was able to grow when either the fresh or older stocks of both compounds were used for supplementation. They reached a significantly higher OD600 at 24 h when supplemented with the older stocks (Fig. 7C). This suggests that abiotic conversion to thiamine was occurring in the stock solutions. Fig. S9 in the supplemental material summarizes the rescue data.
DISCUSSION
In this study, we used the genetic model Burkholderia thailandensis to demonstrate that the enigmatic enzyme thiaminase I plays a role in thiamine salvage. We found that thiamine auxotrophs with ThiA were able to survive more serial dilutions in defined thiamine-free medium than the same auxotrophs lacking thiaminase I. This survival advantage is likely due to the ability of ThiA to degrade thiamine to its precursors, which are preferentially used over intact thiamine by B. thailandensis thiamine auxotrophs. Further, thiaminase I allows B. thailandensis to salvage phosphorylated forms of thiamine, as well as some thiamine analogs. Our proposed model for the role of thiaminase I in thiamine salvage is shown in Fig. 8, although some details still need to be resolved. First, it is unclear whether the HMP with a covalently attached nucleophile, HMP-R, can be incorporated directly into to the thiamine biosynthesis pathway or if the R group needs to first be removed. If it is processed, it is unknown if this occurs extracellularly or intracellularly, and if it is imported, whether CytX or another transporter is responsible. In the case of THZ, it is unknown how THZ and its phosphorylated forms enter and are used by B. thailandensis, since no thiW homolog has been identified, but it is possible some THZ may enter through diffusion. Our experiments with thiamine analogs showed that the intact moieties could be recycled, while damaged or defective moieties could be avoided. In the case of the thiamine disulfide and dibenzoyl thiamine analogs, our results suggest that these compounds are abiotically converting to thiamine, or thiamine with a benzoyl group, and then acted upon by ThiA.
FIG 8.
Thiamine metabolism in B. thailandensis E264. The illustration depicts thiamine metabolism in E264. Thiazole synthetic pathways are highlighted in green. Synthesis of the pyrimidine moiety and the CytX transporter for HMP uptake are represented in blue. The proteins that combine the moieties to make TMP and TPP are depicted in red. Extracellular thiaminase I, ThiA, is depicted as orange octagons acting on both thiamine and pyrithiamine. The “X” on thiamine represents the side chains thiamine, TMP, or TPP. The “R” on the HMP molecule represents the diversity of organic nucleophiles attached to it. Unclear or unknown pathways are represented as dashed lines. In our model, thiamine disulfide is abiotically converted to thiamine (represented by gray lines), which ThiA can act on. In the model, dibenzoyl thiamine loses the benzoyl group attached at the sulfur abiotically; the compound is then acted on by ThiA. It is unclear how the second benzoyl group is removed. We suspect the HMP-R is imported through an unknown transporter (white oval with a “?”), and processed intracellularly by an unknown enzyme (white triangle with a “?”) to HMP. THZ is likely imported by an unknown transporter (white oval with a “?”). For THZ molecules containing phosphates, they are likely removed extracellularly, and the THZ is then imported.
One confounding finding in our results was the growth of BT10432-thiC on pyrithiamine. Growth on this compound suggests multiple possibilities. (i) B. thailandensis is able to use pyrithiamine as a cofactor, which is highly unlikely since the thiC thiG mutants were not rescued by pyrithiamine addition. (ii) Pyrithiamine was slowly degraded abiotically in the medium, freeing HMP for import and salvage. This seems unlikely because serial transfers from medium containing pyrithiamine to pyrithiamine-free medium did not support BT10432-thiC growth. If pyrithiamine abiotically degraded, we would expect some HMP to be transferred over to allow for growth (see Fig. S7B in the supplemental material). In contrast, sufficient HMP to support growth was transferred only when thiaminase I was present, thus in E264-thiC cultures. Further, the use of fresh pyrithiamine stocks as opposed to older solutions did not diminish BT10432-thiC growth (compare the growth from Fig. 7A with that in Fig. S7A in the supplemental material). (iii) Pyrithiamine was contaminated with HMP. Based on the observation of Carini et al. (8) that thiamine and 4-amino-5-aminomethyl-2-methylpyrimidine stocks contained HMP, we consider that similar contamination in pyrithiamine is a likely explanation. ThiA still provided an initial advantage by acting on this compound, but a small amount of HMP in the pyrithiamine stocks may have been sufficient to eventually promote the growth of the BT10432-thiC mutant. Wild-type B. thailandensis E264 is a very capable thiamine producer and does not appear to encode any known thiamine transporter; therefore, it is unclear how thiamine could enter the cell. When exogenous ThiA is present, it can convert thiamine to an accessible form, rescuing growth. This may account for why added ThiACb rescues growth. At the terminal transfer, there is still likely some thiamine present that either cannot be accessed by the BT10432 mutants or is used slowly. The growth exhibited by the BT10432 strains when rescued with thiamine (Fig. 5) might also be due to precursor contamination.
The biological significance of the preference of B. thailandensis thiamine auxotrophs to grow on thiamine precursors rather than thiamine itself is unclear. However, this phenomenon has been observed in other bacteria. Members of the SAR11 clade are thiamine auxotrophs as they lack thiC, and in these bacteria, thiamine-limited growth in laboratory culture is not rescued by the addition of thiamine; rather, it only occurs when HMP is present (8). This might be an evolutionary consequence of their symbiosis with cyanobacteria that exude HMP and/or the higher concentration of HMP than thiamine near the deep chlorophyll maximum zone where SAR11 are found (8). Similarly, eukaryotic phytoplankton have been shown to more efficiently use pyrimidine precursors than thiamine for growth (44). In contrast, B. thailandensis has been isolated from rice field soil and water, where the concentrations of available thiamine, HMP, and THZ are unknown. Thiamine adsorbs to clay particles in soil, and the strength of adsorption is pH dependent (45, 46). Adsorption may reduce the bioavailability of thiamine, requiring soil microbes to evolve unique strategies for acquiring environmental thiamine. It is possible that by degrading thiamine to its precursors, these resources become more biologically available.
Aside from adsorbing to clay particles, thiamine is also labile in soil (17). Temperature, pH, and the presence of bisulfites and other inorganic bases, metal complexes, and UV radiation can all contribute to the abiotic destruction of thiamine (47). Jenkins et al. demonstrated that thiamine degradation occurs not only in alkaline soils but also in neutral soils (17). Consequently, abiotic thiamine degradation may influence the ecology of thiamine metabolism in multiple ways. First, thiaminase I may be active on multiple naturally occurring thiamine breakdown products present in its environment. This would free intact precursors, most likely the HMP, since THZ is less stable, essentially serving as an extracellular TenA. Differences in the specificities of ThiA and TenA (41) suggest that ThiA can accommodate a greater diversity of thiamine breakdown compounds, as the amino group of the pyrimidine moiety is important for thiaminase I, but unlike TenA the side chain of the thiazole moiety does not play an important role. Our results clearly demonstrate that HMP compounds produced by ThiA can be recycled despite the addition of a nucleophile to HMP, though the details are unclear.
The second likely influence of ThiA on the ecological context of B. thailandensis is that extracellular processing of thiamine and thiamine analogs might serve as a means for protecting the cell against incorporation of toxic analogs. ThiA could be used to prevent the import of compounds that would be phosphorylated and then compete with TPP for binding of TPP-dependent enzymes. Many organisms take preemptive measures to prevent the use of toxic metabolites and cofactors (48). Recently, a metabolic housecleaning function was described for a Nudix family hydrolase coded for in thiamine biosynthesis operons in bacteria, plants, and fungi (49). This intracellular enzyme removes the phosphate groups from oxythiamine and oxothiamine, preventing their incorporation into TPP-dependent proteins (49). In B. thailandensis, the gene for a Nudix family protein (see Fig. S1 in the supplemental material) is found in the ThiA operon, suggesting that it may serve this function. ThiA could be another preventative component and provide the added bonus of being able to recycle usable moieties from these toxic compounds. Our understanding of thiamine metabolism, particularly thiamine salvage and detoxification, is still limited, as there are new proteins that appear seemingly essential to this metabolism and yet have unidentified functions (50). Although our study supports a role for thiaminase I in thiamine salvage, further investigation will be required to clarify its roles further.
Converting thiamine to its precursors and using them instead of thiamine may provide an additional competitive advantage. ThiA activity reduces the already scarce amount of thiamine available in the environment, but it would increase the amount of precursors present. This may provide B. thailandensis with an advantage when competing against thiamine auxotrophs that can only import thiamine. Furthermore, by salvaging thiamine from environmental precursors B. thailandensis can bypass multiple steps in the thiamine synthesis pathway, thus reducing the energy input needed to make them. Overall, B. thailandensis E264 has an arsenal of factors to compete against other bacteria (51), and ThiA and may be another competition factor.
Although not known to be pathogenic to humans, B. thailandensis is able to infect mammals at high doses (51–53) and insect models (54, 55). Thus, thiaminase I may be another competition/pathogenicity factor that influences ecological interactions with animals. Since animals are unable to synthesize thiamine, the production of ThiA would convert any free thiamine into a form that B. thailandensis, but not its host, can use during infection. Further, HMP has been characterized as toxic to rats as it interferes with enzymes that require pyridoxal 5′-phosphate (56). Therefore, ThiA-mediated production of HMP may have a 3-fold effect: (i) it would supplement B. thailandensis growth, (ii) it would reduce host thiamine levels, and (iii) it would generate a compound potentially toxic to the host. Additional studies are needed to substantiate these hypotheses.
Our results shed light on a long-standing mystery in thiamine metabolism. The role of thiaminase I has remained elusive, and here we provide evidence supporting the role of this enzyme in thiamine salvage. Still, future studies are needed to develop a comprehensive understanding of the role of thiaminase I in thiamine metabolism by B. thailandensis and other organisms that produce this enzyme. Our study also raises caution with using thiaminase I as a tool to eliminate thiamine from media to evaluate the thiamine requirements of a bacterium (1). Specifically, the use of thiaminase I might confound results with bacteria that have the ability to take up and grow well on the thiamine precursors generated by the enzyme. Instead, we recommend using a defined medium to better understand the capacity of a microorganism to grow in a thiamine-free environment.
MATERIALS AND METHODS
Bacteria and yeast growth conditions.
Strains are listed in Table 2. The accession numbers for the B. thailandensis E264 genome are CP000085 for chromosome I and CP000086 for chromosome II. B. thailandensis strains were grown in TSB at 27°C, while E. coli strains were grown in Luria broth (LB) at 37°C. Saccharomyces cerevisiae was grown YPD medium (57) at 27°C. Chemically defined medium (DM4) for B. thailandensis was based on M9 medium (58); however, it was buffered with 0.1 M MES (pH 6.0) and contained 10 mM FeSO4, 9.5 mM NH4Cl, 0.276 mM K2SO4, 0.5 μM CaCl2, 0.525 mM MgCl2, 50 mM NaCl, 1.32 mM K2HPO4, 1% (vol/vol) vitamin supplement (ATCC MD-VS) excluding thiamine, 1% (vol/vol) trace mineral solution (ATCC MD-TMS), 0.1% (wt/vol) glucose, and 12.5% (vol/vol) of a mixture of all 20 amino acids, based on published concentrations (59). Due to the potential for thiamine adsorption to glassware (60), we took precautions to ensure no contamination of DM4 occurred using our published glassware washing protocol (61).
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotypea | Source or reference |
|---|---|---|
| Organism strains | ||
| E. coli | ||
| DH5α | Invitrogen | |
| BL21(DE3) | Overexpression strain | Invitrogen |
| B. thailandensis | ||
| E264 | Wild type | 67 |
| BT10432 | E264 thiA::ISlacZ-PrhaBo-Tp/FRT | 63 |
| BT10432-thiC | BT10432 ΔthiC::tetR | This study |
| BT10432-thiG | BT10432 ΔthiG::cat | This study |
| BT10432-thiCthiG | BT10432 ΔthiC::tetR ΔthiG::cat | This study |
| E264-thiC | E264 ΔthiC::tetR | This study |
| E264-thiG | E264 ΔthiG::cat | This study |
| E264-thiCthiG | E264 ΔthiC::tetR ΔthiG::cat | This study |
| S. cerevisiae | ||
| InvSc1 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | 62 |
| Plasmids | ||
| pMQ87 | CEN6 ARSH4 aaC1 lacZa oriT oriColE1; Gmr | 62 |
| pMQ87 thiC | pMQ87 ΔthiC::tetR | This study |
| pMQ87 thiG | pMQ87 ΔthiG::cat | This study |
| pET-28 bcmE | Overexpression vector containing bcmE(thiACb) | 25 |
Gmr, gentamicin resistance.
Mutant construction.
Plasmids containing thiC (BTH_I2844) and thiG (BTH_I3006) disruptions were generated using the S. cerevisiae recombineering method developed by Shanks et al. (57, 62). Briefly, the PCR products of the 5′ and 3′ ends of the thiC gene were amplified from E264 genomic DNA and recombined with the PCR product of a tetracycline resistance gene amplified from B. thailandensis BT02155 genomic DNA (63), partially deleting the thiC gene. These PCR products were recombined into the pMQ87 plasmid. The thiG disruption was created with the same approach, except using the thiG specific primers to generate the 5′- and 3′-end fragments and primers to amplify the chloramphenicol resistance gene (cat) from the pBeloBAC11 plasmid (64). All PCR primers are listed in Table 3.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′−3′)a |
|---|---|
| pMQ87-thiC 5′ F | AGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCTCTGTCCCCTGTTGAAAC |
| thiC 5′ F | TCTGTCCCCTGTTGAAAC |
| thiC 5′ R | CAGGCTTTCCAGATACTC |
| thiC 3′ F | GACTGGTGCAAGGAAGCG |
| thiC 3′ R | TTACGCTGCTGGGTGGTC |
| pMQ87-thiC 3′ R | AAACAGCTATGACCATGATTACGAATTCGAGCTCGTTACGCTGCTGGGTGGTC |
| thiC-tetR F | CGCGATCCGCGAGAACCAGCGCCGCGCCGAGTATCTGGAAAGGCTAACGCAGTCAGGCAC |
| tetR-thiC R | GTCGTAGCCGGGCGCGATGTCGGTCGTGAGCGGCCCGAGCGTTCCGTTAGCGAGGTGCCG |
| thiC-tetR construct confirmation F | GCGCAACATGGCATCCGG |
| thiC-tetR construct confirmation R | CGTCGGCCCATTCGAGGG |
| pMQ87-thiG 5′ F | AGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCAGCTGCACGGTCCGCAC |
| thiG 5′ F | AGCTGCACGGTCCGCAC |
| thiG 5′ R | GCGTGCTGCTGAACACG |
| thiG 3′ F | AGCGCGACCGTCACCATC |
| thiG 3′ R | GAACTGAATGCGCAATGC |
| pMQ87-thiG 3′ R | AAACAGCTATGACCATGATTACGAATTCGAGCTCGGAACTGAATGCGCAATGC |
| thiG-cat F | GAACGCGCGCGCCATGATCTCCGGATGCGTCGCCTGCGATACGGCGAAAATGAGACGTTG |
| cat-thiG R | GTCGCTGCAGTCGCTGTCCGATTCGATCGCGGCCGCGCGGCCAGGCGTAGCAACCAGGCG |
| thiG-cat construct confirmation F | GTTCAGGTTCGACAGGAC |
| thiG-cat construct confirmation R | ATGCGATGAACATGCGTG |
Underlining signifies the sections of the primers that overlap with pMQ87. Boldfacing indicates the sections of the antibiotic resistance gene primers that overlap with sections of the thiamine biosynthesis genes.
Generation of B. thailandensis mutants.
To introduce the thiC or thiG disruptions, we followed the natural competence protocol for B. thailandensis to generate competent E264, BT10432 and mutants (65). Briefly, PCR products of the disruption constructs amplified from their cognate pMQ87 plasmids were added to competent E264 and BT10432 cells to generate the single mutants. The E264-thiC transformants were selected on 50 μg/ml tetracycline and the BT10432-thiC transformants were selected on 50 μg/ml tetracycline and 100 μg/ml trimethoprim. The thiG disruption mutants (E264-thiG and BT10432-thiG) were generated the same way. The E264-thiG transformation culture was plated on LB with 256 μg/ml chloramphenicol, and the BT10432-thiG transformation cultures were plated on LB with 256 μg/ml chloramphenicol and 100 μg/ml trimethoprim. To generate the E264-thiCthiG disruption mutant, genomic DNA was isolated from E264-thiC and used to transform competent E264-thiG. For the BT10432-thiCthiG mutant, E264-thiCthiG was transformed with BT10432 genomic DNA. Cells were plated on LB containing the antibiotics specific for each mutation. Mutations were verified by PCR. Primers outside the construct region were used to amplify across the disruption. PCR products were compared to PCR products amplified from E264 genomic DNA. Those PCR products that differed in size and appeared to contain the disruption were confirmed via Sanger sequencing at the Cornell Institute of Biotechnology.
Serial transfer experiments.
For each experiment, B. thailandensis strains were plated from frozen stocks on TSA. After 3 days of growth at 30°C, individual colonies were grown in TSB overnight at 27°C with shaking at 200 rpm. The culture OD600 was read with an Ultrospec 2000 spectrophotometer (Pharmacia Biotech) and then subcultured in 3 ml of DM4 supplemented with 1 mM thiamine-HCl at a starting OD600 of 0.025. Growth was monitored at 18, 24, 30, and 48 h. At 24 h, each culture was subcultured into DM4 without any thiamine at the starting OD600 of 0.025. Serial dilutions were repeated until the auxotrophs could not grow. E264 and BT10432 were grown as positive controls for each experiment, depicted in Fig. 2. For the original serial transfer experiments, all 8 strains were used, n = 6 for each strain. For the serial transfers from pyrithiamine-containing media, cells were transferred at 24 h. For the serial transfers from thiamine disulfide- and dibenzoyl thiamine-containing media, samples were transferred at 30 h. OD600 at 24 h for each experimental grouping was statistically compared in JMP Pro 12.0.1 via an analysis of variance (ANOVA). If there were significant differences, pairwise comparisons were made using Student t test. P values were Bonferroni corrected based on the number of comparisons.
Supernatant rescue experiments.
Supernatant was collected after centrifugation of E264-thiC and BT10432-thiC cultures after 24 h of growth at the transfer before each strain cannot grow (Fig. 2). The supernatant was sterilized using a 0.2-μm-pore size filter. Then, 1.5 ml of each supernatant was mixed with 1.5 ml of fresh thiamine-free DM4. E264-thiC and BT10432-thiC were serially transferred as described in the serial transfer experiments. However, at the terminal transfer where BT10432-thiC could no longer grow (Fig. 2), it was subcultured in the 1:1 mixture E264-thiC-DM4 supernatant:DM4 and the 1:1 mixture BT10432-thiC-DM4 supernatant:DM4, while E264-thiC was grown in the mixture at the second transfer step.
Thiaminase I activity.
To assay thiaminase I activity in cultures grown in DM4, 1.5 ml samples of culture supernatant were collected at 12, 18, 24, and 30 h for each transfer. Large molecules were concentrated from the supernatant using Amicon Ultracel 30K centrifugal filters (Millipore). Thiaminase I assays were then conducted using 3 μl of the concentrated samples according to a published protocol (42), which is a spectrophotometric assay measuring the consumption of 4-nitrothiophenol (4-NTP). 4-NTP is a preferred nucleophile for thiaminase I and is consumed when exchanged with the THZ moiety in thiamine by thiaminase I. The rate of consumption of the compound and its associated colorimetric change delineates thiaminase I activity.
Thiochrome assay to test for deterioration of thiamine in uninoculated medium.
Thiamine was added to DM4 at a final concentration of 200 mM, and the production of thiochrome was assayed after incubations of 24 h, 12 h, 1 h, and 30 min. We followed the fluorescence assay procedure previously described (66). Thiochrome is the fluorescent, oxidized form of thiamine generated by the addition of potassium ferrocyanide and sodium hydroxide. Thiochrome only forms when thiamine is intact, and the amount of thiochrome formed is proportional to the amount of intact thiamine that was oxidized.
Overexpression and purification of thiaminase I.
The pET-28 plasmid containing His-tagged ThiACb, Clostridium botulinum thiaminase I (1), was transformed into E. coli BL21 as described previously (66). To extract the protein, we modified a published protocol (25). Briefly, USB PrepEase histidine-tagged high-yield purification resin (Affymetrix) was added to the clear cell lysate, followed by incubation overnight in an orbital shaker at 4°C. We used buffers recommended by the manufacturer. The slurry was poured into a column and washed with 50 ml of lysis buffer. The protein was eluted with 15 ml of elution buffer added in 5-ml increments. Fractions were tested for thiaminase activity (42). Samples were dialyzed overnight in 100 mM NaCl–50 mM potassium phosphate buffer (pH 7.2).
Rescue experiments.
Purified ThiACb, thiamine precursors, thiamine, phosphorylated thiamine, or thiamine analogs were added to bacterial cultures to evaluate conditions that could recover growth of thiamine auxotrophs. ThiACb was added to a concentration of 1 μg/ml (n = 3). HMP (n = 6), THZ (n = 6), a combination of both (n = 6), or thiamine (n = 9) was added to cultures at a concentrations of 50 mM each. The thiamine analogs and derivatives, TMP (n = 6), and TPP (n = 6), oxythiamine (n = 6), pyrithiamine (n = 6), 3-benzyl-5-(2-hydroxyethyl)-4-methylthiazolium chloride (n = 3), dibenzoyl thiamine (n = 6), and thiamine disulfide (n = 6) were added at 10 μg/ml. For the thiC thiG mutants, oxythiamine and pyrithiamine were added together at a concentration of 10 μg/ml for each compound.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jen Fownes for her work on testing defined media for growing Burkholderia strains. We are grateful to Peter Newell for supplying the yeast recombineering methods, materials, and advice. We thank Chunhao Li and Steve Ealick for providing the ThiACb overexpression construct. Finally, we are grateful to Francine Arroyo and Nicolas Buchon for helpful discussions and advice.
This study was supported by National Science Foundation grants MCB 1244378 and IOS 1354911.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01268-18.
REFERENCES
- 1.Zhang K, Bian J, Deng Y, Smith A, Nunez RE, Li MB, Pal U, Yu A-M, Qiu W, Ealick SE. 2016. Lyme disease spirochaete Borrelia burgdorferi does not require thiamin. Nat Microbiol 2:16213. doi: 10.1038/nmicrobiol.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraft CE, Angert ER. 2017. Competition for vitamin B1 (thiamin) structures numerous ecological interactions. Q Rev Biol 92:151–168. doi: 10.1086/692168. [DOI] [PubMed] [Google Scholar]
- 3.Jurgenson CT, Begley TP, Ealick SE. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schauer K, Stolz J, Scherer S, Fuchs TM. 2009. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol 191:2218–2227. doi: 10.1128/JB.01636-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb E, Claas K, Downs D. 1998. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem 273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 6.Melnick J, Lis E, Park J-H, Kinsland C, Mori H, Baba T, Perkins J, Schyns G, Vassieva O, Osterman A. 2004. Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J Bacteriol 186:3660–3662. doi: 10.1128/JB.186.11.3660-3662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karunakaran R, Ebert K, Harvey S, Leonard M, Ramachandran V, Poole P. 2006. Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J Bacteriol 188:6661–6668. doi: 10.1128/JB.00641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carini P, Campbell EO, Morré J, Sanudo-Wilhelmy SA, Thrash JC, Bennett SE, Temperton B, Begley T, Giovannoni SJ. 2014. Discovery of a SAR11 growth requirement for thiamin's pyrimidine precursor and its distribution in the Sargasso Sea. ISME J 8:1727–1738. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodionov DA, Hebbeln P, Eudes A, Ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devedjiev Y, Surendranath Y, Derewenda U, Gabrys A, Cooper DR, Zhang R-G, Lezondra L, Joachimiak A, Derewenda ZS. 2004. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J Mol Biol 343:395–406. doi: 10.1016/j.jmb.2004.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2002. Comparative genomics of thiamin biosynthesis in procaryotes: new genes and regulatory mechanisms. J Biol Chem 277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 12.Anderson LN, Koech PK, Plymale AE, Landorf EV, Konopka A, Collart FR, Lipton MS, Romine MF, Wright AT. 2015. Live cell discovery of microbial vitamin transport and enzyme-cofactor interactions. ACS Chem Biol 11:345–354. doi: 10.1021/acschembio.5b00918. [DOI] [PubMed] [Google Scholar]
- 13.Toms AV, Haas AL, Park J-H, Begley TP, Ealick SE. 2005. Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II. Biochemistry 44:2319–2329. doi: 10.1021/bi0478648. [DOI] [PubMed] [Google Scholar]
- 14.Benach J, Edstrom WC, Lee I, Das K, Cooper B, Xiao R, Liu J, Rost B, Acton TB, Montelione GT. 2005. The 2.35 Å structure of the TenA homolog from Pyrococcus furiosus supports an enzymatic function in thiamine metabolism. Acta Crystallogr D Biol Crystallogr 61:589–598. doi: 10.1107/S0907444905005147. [DOI] [PubMed] [Google Scholar]
- 15.Onozuka M, Konno H, Kawasaki Y, Akaji K, Nosaka K. 2007. Involvement of thiaminase II encoded by the THI20 gene in thiamin salvage of Saccharomyces cerevisiae. FEMS Yeast Res 8:266–275. doi: 10.1111/j.1567-1364.2007.00333.x. [DOI] [PubMed] [Google Scholar]
- 16.Zallot R, Yazdani M, Goyer A, Ziemak MJ, Guan J-C, McCarty DR, de Crécy-Lagard V, Gerdes S, Garrett TJ, Benach J. 2014. Salvage of the thiamin pyrimidine moiety by plant TenA proteins lacking an active-site cysteine. Biochem J 463:145–155. doi: 10.1042/BJ20140522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. 2007. A new thiamin salvage pathway. Nat Chem Biol 3:492–497. doi: 10.1038/nchembio.2007.13. [DOI] [PubMed] [Google Scholar]
- 18.Kreinbring CA, Remillard SP, Hubbard P, Brodkin HR, Leeper FJ, Hawksley D, Lai EY, Fulton C, Petsko GA, Ringe D. 2014. Structure of a eukaryotic thiaminase I. Proc Natl Acad Sci U S A 111:137–142. doi: 10.1073/pnas.1315882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costello CA, Kelleher NL, Abe M, McLafferty FW, Begley TP. 1996. Mechanistic studies on thiaminase I overexpression and identification of the active site nucleophile. J Biol Chem 271:3445–3452. doi: 10.1074/jbc.271.7.3445. [DOI] [PubMed] [Google Scholar]
- 20.Fujita A. 1954. Thiaminase. Adv Enzymol Relat Areas Mol Biol 15:389–421. [DOI] [PubMed] [Google Scholar]
- 21.Agee CC, Airth R. 1973. Reversible inactivation of thiaminase I of Bacillus thiaminolyticus by its primary substrate, thiamine. J Bacteriol 115:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebata J, Murata K. 1961. The purification of thiaminase I produced by Bacillus thiaminolyticus. J Vitaminol (Kyoto) 7:115–121. doi: 10.5925/jnsv1954.7.115. [DOI] [PubMed] [Google Scholar]
- 23.Douthit H, Airth R. 1966. Thiaminase I of Bacillus thiaminolyticus. Arch Biochem Biophys 113:331–337. doi: 10.1016/0003-9861(66)90194-9. [DOI] [PubMed] [Google Scholar]
- 24.Campobasso N, Costello CA, Kinsland C, Begley TP, Ealick SE. 1998. Crystal structure of thiaminase-I from Bacillus thiaminolyticus at 2.0 Å resolution. Biochemistry 37:15981–15989. doi: 10.1021/bi981673l. [DOI] [PubMed] [Google Scholar]
- 25.Sikowitz MD, Shome B, Zhang Y, Begley TP, Ealick SE. 2013. Structure of a Clostridium botulinum C143S thiaminase I/thiamin complex reveals active site architecture. Biochemistry 52:7830–7839. doi: 10.1021/bi400841g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soriano EV, Rajashankar KR, Hanes JW, Bale S, Begley TP, Ealick SE. 2008. Structural similarities between thiamin-binding protein and thiaminase-I suggest a common ancestor. Biochemistry 47:1346–1357. doi: 10.1021/bi7018282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Airth R. 1967. Repression of thiaminase I in Bacillus thiaminolyticus. Biochem Biophys Res Commun 27:325–330. doi: 10.1016/S0006-291X(67)80101-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Wilkins JH, Airth R. 1968. Repression of thiaminase I by thiamine and related compounds in Bacillus thiaminolyticus. Can J Microbiol 14:1143–1147. doi: 10.1139/m68-191. [DOI] [PubMed] [Google Scholar]
- 29.Cooper LE, O'Leary SE, Begley TP. 2014. Biosynthesis of a thiamin antivitamin in Clostridium botulinum. Biochemistry 53:2215–2217. doi: 10.1021/bi500281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sannino D, Angert ER. 2017. Genomic insights into the thiamin metabolism of Paenibacillus thiaminolyticus NRRL B-4156 and P. apiarius NRRL B-23460. Stand Genomic Sci 12:59. doi: 10.1186/s40793-017-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honeyfield DC, Hinterkopf JP, Brown SB. 2002. Isolation of thiaminase-positive bacteria from alewife. Trans Am Fish Soc 131:171–175. doi:. [DOI] [Google Scholar]
- 32.Richter CA, Evans AN, Wright-Osment MK, Zajicek JL, Heppell SA, Riley SC, Krueger CC, Tillitt DE. 2012. Paenibacillus thiaminolyticus is not the cause of thiamine deficiency impeding lake trout (Salvelinus namaycush) recruitment in the Great Lakes. Can J Fish Aquat Sci 69:1056–1064. doi: 10.1139/f2012-043. [DOI] [Google Scholar]
- 33.Green R, Evans C. 1940. A deficiency disease of foxes. Science 92:154–155. doi: 10.1126/science.92.2381.154. [DOI] [PubMed] [Google Scholar]
- 34.Evans C, Carlosn W, Green R. 1942. The pathology of Chastek paralysis in foxes: a counterpart of Wernicke's hemorrhagic polioencephalitis of man. Am J Pathol 18:79. [PMC free article] [PubMed] [Google Scholar]
- 35.Yudkin WH. 1949. Thiaminase, the Chastek-paralysis factor. Physiol Rev 29:389–402. doi: 10.1152/physrev.1949.29.4.389. [DOI] [PubMed] [Google Scholar]
- 36.Linklater K, Dyson D, Morgan K. 1977. Faecal thiaminase in clinically normal sheep associated with outbreaks of polioencephalomalacia. Res Vet Sci 22:308–312. [PubMed] [Google Scholar]
- 37.Shreeve J, Edwin E. 1974. Thiaminase-producing strains of Cl. sporogenes associated with outbreaks of cerebrocortical necrosis. Vet Rec 94:330. doi: 10.1136/vr.94.15.330. [DOI] [PubMed] [Google Scholar]
- 38.Paerl RW, Bouget F-Y, Lozano J-C, Vergé V, Schatt P, Allen EE, Palenik B, Azam F. 2017. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J 11:753. doi: 10.1038/ismej.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bettendorff L. 2007. At the crossroad of thiamine degradation and biosynthesis. Nat Chem Biol 3:454–455. doi: 10.1038/nchembio0807-454. [DOI] [PubMed] [Google Scholar]
- 40.Erkens GB, Slotboom DJ. 2010. Biochemical characterization of ThiT from Lactococcus lactis: a thiamin transporter with picomolar substrate binding affinity. Biochemistry 49:3203–3212. doi: 10.1021/bi100154r. [DOI] [PubMed] [Google Scholar]
- 41.Murata K. 1982. Actions of two types of thiaminase on thiamin and its analogues. Ann N Y Acad Sci 378:146–156. doi: 10.1111/j.1749-6632.1982.tb31193.x. [DOI] [PubMed] [Google Scholar]
- 42.Kraft CE, Gordon ER, Angert ER. 2014. A rapid method for assaying thiaminase I activity in diverse biological samples. PLoS One 9:e92688. doi: 10.1371/journal.pone.0092688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin W, Chun K. 1987. Structure of thiamin disulfide dinitrate. Acta Crystallogr C 43:2123–2125. doi: 10.1107/S0108270187088784. [DOI] [Google Scholar]
- 44.Gutowska MA, Shome B, Sudek S, McRose DL, Hamilton M, Giovannoni SJ, Begley TP, Worden AZ. 2017. Globally important haptophyte algae use exogenous pyrimidine compounds more efficiently than thiamin. mBio 8:e01459-. doi: 10.1128/mBio.01459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidhalter U, Evéquoz M, Studer C, Oertli J, Kahr G. 1994. Adsorption of thiamin (vitamin B1) on soils and clays. Soil Sci Soc Am J 58:1829–1837. doi: 10.2136/sssaj1994.03615995005800060036x. [DOI] [Google Scholar]
- 46.Cain AH, Sullivan GR, Roberts JD. 1977. The protonation site of vitamin B1 as determined from natural-abundance nitrogen-15 nuclear magnetic resonance spectra. J Am Chem Soc 99:6423–6425. doi: 10.1021/ja00461a040. [DOI] [PubMed] [Google Scholar]
- 47.Dwivedi BK, Arnold RG. 1973. Chemistry of thiamine degradation on food products and model systems. Rev J Agric Food Chem 21:54–60. doi: 10.1021/jf60185a004. [DOI] [PubMed] [Google Scholar]
- 48.Linster CL, Van Schaftingen E, Hanson AD. 2013. Metabolite damage and its repair or pre-emption. Nat Chem Biol 9:72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 49.Goyer A, Hasnain G, Frelin O, Ralat MA, Gregory JF, Hanson AD. 2013. A cross-kingdom Nudix enzyme that pre-empts damage in thiamin metabolism. Biochem J 454:533–542. doi: 10.1042/BJ20130516. [DOI] [PubMed] [Google Scholar]
- 50.Pribat A, Blaby IK, Lara-Núñez A, Jeanguenin L, Fouquet R, Frelin O, Gregory JF, Philmus B, Begley TP, de Crécy-Lagard V. 2011. A 5-formyltetrahydrofolate cycloligase paralog from all domains of life: comparative genomic and experimental evidence for a cryptic role in thiamin metabolism. Funct Integr Genomics 11:467–478. doi: 10.1007/s10142-011-0224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeShazer D. 2007. Virulence of clinical and environmental isolates of Burkholderia oklahomensis and Burkholderia thailandensis in hamsters and mice. FEMS Microbiol Lett 277:64–69. doi: 10.1111/j.1574-6968.2007.00946.x. [DOI] [PubMed] [Google Scholar]
- 53.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun 76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilátová M, Dionne MS. 2012. Burkholderia thailandensis is virulent in Drosophila melanogaster. PLoS One 7:e49745. doi: 10.1371/journal.pone.0049745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher NA, Ribot WJ, Applefeld W, DeShazer D. 2012. The Madagascar hissing cockroach as a novel surrogate host for Burkholderia pseudomallei, B. mallei, and B thailandensis. BMC Microbiol 12:117. doi: 10.1186/1471-2180-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haughton BG, King H. 1958. Toxopyrimidine phosphate as an inhibitor of bacterial enzyme systems that require pyridoxal phosphate. Biochem J 70:660. doi: 10.1042/bj0700660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley Interscience, New York, NY. [Google Scholar]
- 60.Farrer K, Hollenberg W. 1953. Adsorption of thiamine on glassware. Analyst 78:730–731. [Google Scholar]
- 61.Sannino DR, Dobson AJ, Edwards K, Angert ER, Buchon N. 2018. The Drosophila melanogaster gut microbiota provisions thiamine to its host. mBio 9:e00155-. doi: 10.1128/mBio.00155-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. 2013. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio 4:e00604-13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim U-J, Birren BW, Slepak T, Mancino V, Boysen C, Kang H-L, Simon MI, Shizuya H. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 65.Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. 2008. Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Appl Environ Microbiol 74:2985–2989. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters JE. 2007. Gene transfer in Gram-negative bacteria, p 735–755. In Methods for general and molecular microbiology, 3rd ed American Society of Microbiology, Washington, DC. [Google Scholar]
- 67.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48(Pt 1):317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.