Suppression of bacterial growth in raw milk under cold storage is crucial for the quality control of commercially supplied milk. The use of lytic phages as low-cost microbicides is an attractive prospect. Due to strict host specificities, phages must be isolated from the raw milk where the host bacteria are growing. We first isolated the P. lactis bacterial strain and then the phage infecting that strain. Partial phage genomic analysis showed that this is a newly isolated phage, different from any previously reported. This study reports a lytic phage for P. lactis, and we have presented evidence here that this phage reduced viable bacterial cell counts not only in rich medium but also in skim and whole milk. As a result, we have concluded that the phage reported in this study would be useful in milk processing.
KEYWORDS: bacteriophage, Pseudomonas lactis, biocontrol
ABSTRACT
The control of bacterial growth during milk processing is crucial for the quality maintenance of commercial milk and milk products. During a period of cold storage prior to heat treatments, some psychrotrophic bacteria grow and produce extracellular heat-resistant lipases and proteases that cause product defects. The use of lytic bacteriophages (phages) that infect and kill bacteria could be a useful tool for suppressing bacterial growth during this cold storage phase. In this study, we isolated a Pseudomonas lactis strain and a phage from raw cow's milk. Quantitative characterization of the phage was used to elucidate whether this phage was active under low temperatures and neutral pH and whether it was inactivated during pasteurization. Phage titer determination was possible under conditions ranging from pH 4 to 9 and from 3°C to 25°C; the phage was inactivated under pasteurization conditions at 63°C for 30 min. Furthermore, we showed that this phage reduced viable bacterial cell counts in both skim and whole milk. The results of this study represent the potential uses of phages for controlling psychrotrophic bacterial growth in raw cow's milk during cold storage.
IMPORTANCE Suppression of bacterial growth in raw milk under cold storage is crucial for the quality control of commercially supplied milk. The use of lytic phages as low-cost microbicides is an attractive prospect. Due to strict host specificities, phages must be isolated from the raw milk where the host bacteria are growing. We first isolated the P. lactis bacterial strain and then the phage infecting that strain. Partial phage genomic analysis showed that this is a newly isolated phage, different from any previously reported. This study reports a lytic phage for P. lactis, and we have presented evidence here that this phage reduced viable bacterial cell counts not only in rich medium but also in skim and whole milk. As a result, we have concluded that the phage reported in this study would be useful in milk processing.
INTRODUCTION
Raw cow's milk contains a variety of nutrients, such as proteins, lipids, carbohydrates, vitamins, minerals, and more (1). In addition to its abundance of nutrients, its neutral pH and high water content provide a suitable habitat for bacteria. In fact, many kinds of bacteria, including species of the genera Acinetobacter, Chryseobacterium, Corynebacterium, Enterococcus, Lactobacillus, Lactococcus, Staphylococcus, Streptococcus, Pseudomonas, and Microbacterium, have been found in fresh raw milk (2). Recently, von Neubeck et al. comprehensively studied the raw milk microbiota using 2,906 isolates from 20 different raw cow's milk samples, and concluded that Pseudomonas, Lactococcus, and Acinetobacter were the most dominant genera (3). Furthermore, it was shown that Pseudomonas lactis sp. nov. is one of the most dominant species, along with Pseudomonas proteolytica and Pseudomonas lundensis (3, 4). Therefore, control of bacterial growth throughout dairy processing, particularly the control of Pseudomonas growth, is crucial for maintaining the quality of the milk.
Following milking, raw cow's milk is stored in bulk farm milk tanks, and then transported to the dairy plant directly or via collection stations with milk tanker trucks. The temperature of milk is maintained below 4°C during this period of cold storage and transportation for 3 to 5 days, keeping mesophilic bacteria to a minimum; however, psychrotrophic bacteria, including Pseudomonas species, have been reported to dominate the microbiota of milk after this refrigeration period (3, 5, 6). It is well known that Pseudomonas species are responsible for causing the spoilage of raw milk under refrigerated conditions. Recently, von Neubeck et al. revealed that of all their isolates identified as Pseudomonas species exhibited either a combination of lipolytic and proteolytic activities or proteolytic activities alone (3). Although the bacteria are killed by pasteurization or ultrahigh-temperature sterilization, these lytic enzymes can survive in the sterilized milk, remaining in the manufactured milk and causing deterioration of the market milk (7–10). Therefore, inhibiting bacterial growth, especially psychrotrophic bacterial growth, while maintaining raw milk at lower temperatures is crucial for the quality control of commercial milk products. However, we currently have few effective methods for suppressing bacterial growth, other than cooling the milk and washing the equipment used for milking on the farm and storage in the dairy plant.
Recently, the use of lytic phages that infect and kill bacteria by cell lysis has been brought back to the spotlight as a potential microbicidal treatment, due to the emergence of multidrug-resistant bacteria (11, 12). Lytic phages kill bacteria by different mechanisms than antibiotics (13). They infect bacterial cells and then multiply over 10-fold in a very short period of time, with a goal of lysing the host bacterial cells to release their progenies, which then repeat the infection and lysis cycle until the host population is severely reduced (14). Among the phages that infect Pseudomonas species, those that infect Pseudomonas aeruginosa and Pseudomonas syringae were isolated and characterized, whereas only some phages that infect Pseudomonas fluorescens were isolated and characterized. Phages that infect P. fluorescens were isolated from sewage treatment plants and wastewater of dairy industries (15, 16), and some were able to infect a variety of P. fluorescens strains isolated from dairy industries (15). However, phages from raw cow's milk, including liquid milk and milk clarifier sludge (sludge), and phages that infect P. lactis have not been isolated and characterized previously.
In the current study, we isolated P. lactis from raw milk and isolated from sludge a phage that infects the P. lactis strain; sludge is the sediment fraction that remains after raw milk is passed through a centrifugal clarifier. Electron microscopic analysis showed that the phage belonged to the family Podoviridae. Partial phage genome analysis showed that this phage is different from others previously reported. Thermal growth profiling indicated that this unique phage would have infectivity at the raw milk storage temperature of 3°C. In this paper, we also have provided evidence that the phage was able to kill the P. lactis bacterial strain in the milk, allowing us to conclude that this newly isolated phage would have the potential to control the growth of psychrotrophic bacteria during raw milk storage under low temperatures.
RESULTS
Isolation of Gram-negative bacteria from raw cow's milk.
We isolated 40 Gram-negative bacterial strains from raw cow's milk obtained from the dairy plant of Aomori Prefecture in Japan. By examining the susceptibility to phage infection, described below, we selected one susceptible bacterium for further examination and designated it as YT4 (Table 1). We identified its genus and species based on 16S rRNA sequence comparisons and multilocus sequence analysis (MLSA), respectively. First, we amplified the 16S rRNA gene by PCR using the primers listed in Table 2 and performed direct sequencing to determine the 16S rRNA sequence of YT4. Second, we conducted 16S rRNA sequence comparisons with those of previously published bacterial sequences using the similarity rank program of the Ribosomal Database Project (RDP; Michigan State University, East Lansing) (17). This analysis showed that the closest cultivable relatives were 99.9% similar to the type strains of Pseudomonas lurida, Pseudomonas poae, Pseudomonas costantinii, and Pseudomonas simiae, confirming the genus of the isolate as Pseudomonas, but we were not able to accurately identify the species using only 16S rRNA sequence comparisons. We conducted MLSA using combined 16S rRNA, gyrB, rpoD, and rpoB sequences, according to Mulet et al. (18). A phylogenetic tree was constructed using the sequences and available genomes of the type strains from the Pseudomonas fluorescens group, which are presented in Table S1 in the supplemental material. This phylogenetic tree showed that our isolated bacterium, YT4, was included in the P. fluorescens subgroup and was closely related to P. lactis (Fig. S1). The similarity of the four concatenated genes (16S rRNA, gyrB, rpoD, and rpoB) was 98.3% between YT4 and P. lactis DSM 29167T. As Mulet et al. proposed a threshold of 97% similarity for the species in the genus Pseudomonas (18, 19), we identified the YT4 strain as P. lactis (4).
TABLE 1.
Bacterial strains and bacteriophage used in this study
| Strain or phage | Sourcea |
|---|---|
| Pseudomonas | |
| Pseudomonas lactis YT4 | This study |
| Pseudomonas lactis DSM 29167T | DSMZ |
| Pseudomonas paralactis DSM 29164T | DSMZ |
| Pseudomonas fluorescens NBRC 14160T | NBRC |
| Pseudomonas putida NBRC 14164T | NBRC |
| Pseudomonas syringae NBRC 3310 | NBRC |
| Pseudomonas alcaligenes NBRC 14159T | NBRC |
| Bacteriophage | |
| HU1 | This study |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Germany); NBRC, Biological Resource Center, National Institute of Technology and Evaluation (Japan).
TABLE 2.
Primers used to identify YT4
| Target gene | Primer name | Primer sequence (5′–3′)a | Fragment length (bp)b | Source or reference |
|---|---|---|---|---|
| 16S rRNA | 27F | AGAGTTTGATCCTGGCTCAG | 1,392 | 41 |
| 1401R | ACGGGCGGTGTGTAC | |||
| gyrB | PfgyrBF | TGCACGGYGTRGGYGT | 939 | This study |
| PfgyrBR | CMGCRGAGTCACCTTCCA | |||
| rpoD | PsEG30F | ATYGAAATCGCCAARCG | 763 | 43 |
| PfrpoD 804R | CCTCRCCGATCGACATG | This study | ||
| rpoB | LAPS5 | TGGCCGAGAACCAGTTCCGCGT | 1,230 | 44 |
| LAPS27 | CGGCTTCGTCCAGCTTGTTCAG |
For sequencing, the same primers were used, except that 341F (5′-CCTACGGGAGGCAGCAG-3′) (42) was also used for the 16S rRNA gene in addition to 27F and 1401R. M, A+C; R, A+G; Y, C+T; K, G+T.
Values for the Pseudomonas fluorescens type strain listed in Table S1.
Isolation of the phage from sludge and determination of host range.
To isolate a phage from the dairy plant of Aomori prefecture in Japan, we obtained supernatant from sludge collected in April 2015 and enriched phages by coincubating with a mixture of Gram-negative bacteria (strains YT1 to YT40) isolated in this study for potential hosts, as described in the Materials and Methods section. We conducted spot tests using these enriched phage suspensions and host strains and found that the phage suspension enriched with strains YT1 to YT10 made plaques on the strain YT4 (Fig. 1). We purified the phage and designated it as HU1.
FIG 1.
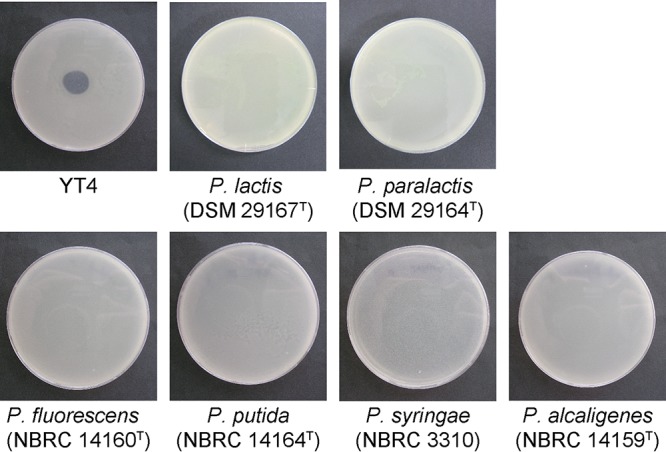
The spectrum of lytic activity by phage HU1 against Pseudomonas strains. Plaque formation by phage HU1 is shown against the P. lactis YT4 strain and the reference strains of P. lactis (DSM 29167T), P. paralactis (DSM 29164T), P. fluorescens (NBRC 14160T), P. putida (NBRC 14164T), P. syringae (NBRC 3310), and Pseudomonas alcaligenes (NBRC 14159T).
To determine the host range of this phage, we conducted spots test against seven bacteria listed in Table 1, including YT4 and the six reference strains P. lactis, Pseudomonas paralactis, P. fluorescens, Pseudomonas putida, Pseudomonas syringae, and Pseudomonas alcaligenes. The HU1 phage formed a plaque only on the YT4 strain and not on the other strains, including the P. lactis type strain, DSM 29167T. These results provided evidence that the phage existed in the cow's milk clarifier sludge and that the phage could only infect the P. lactis YT4 strain that was isolated from the raw milk in this study.
Morphological and genetic characterization of the phage.
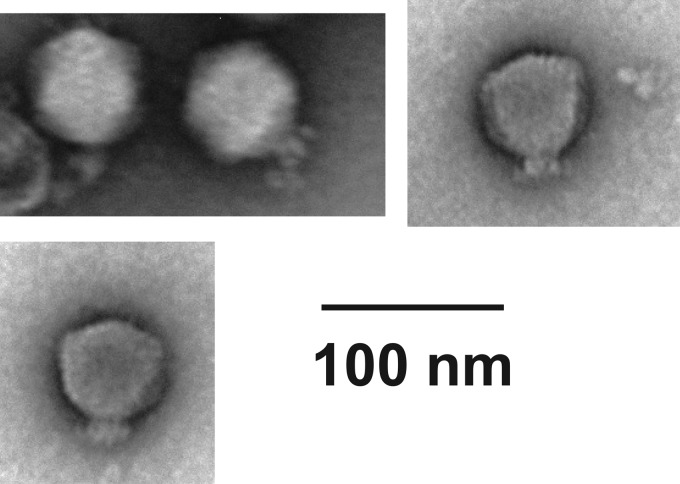
To identify the phage, we conducted transmission electron microscopy (TEM) and genomic analysis for HU1. The morphological analysis using TEM showed that HU1 had a hexagonal head and short tail (Fig. 2). Therefore, the phage belonged to the family Podoviridae.
FIG 2.
Morphology of phage HU1. Transmission electron micrograph of negatively stained phage HU1 observed at a magnification of 50,000×. Bar, 100 nm.
To characterize the genome, we extracted genetic material and digested it with restriction enzymes (Fig. 3). The genome was sensitive to restriction endonucleases and resistant to RNase treatment (data not shown). When we conducted endonuclease digestion, XbaI did not digest the phage genome, but SpeI, NdeI, and XhoI digested it successfully. By calculating the genome size based on the standard curves obtained from 3 DNA molecular weight markers, the sum of the fragments digested with NdeI and XhoI resulted in genome sizes of approximately 47 kbp and 49 kbp, respectively; therefore, HU1 has double-stranded DNA with approximately a 48-kbp genome size. To determine the sequence similarity with previously published bacterial sequences, we partially determined the sequences by cloning approximately 4,000-base-long fragments into the pBR322 vector. BLASTN analysis showed that no sequences with more than 11% similarity have been observed.
FIG 3.
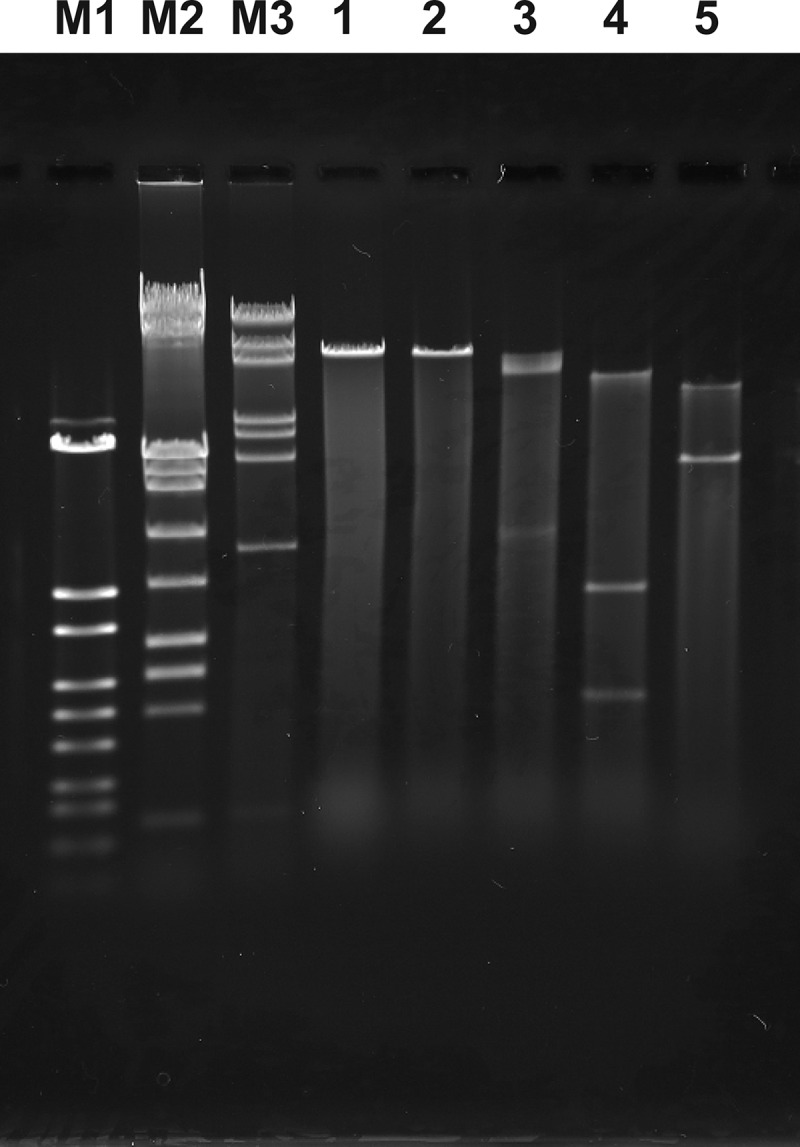
Restriction endonuclease digestion analysis of the phage HU1 genome. Lanes M1, M2, and M3, molecular weight marker of λ-StyI digestion, 8GT, and 7GT, respectively; lane 1, undigested; lane 2, XbaI; lane 3, SpeI; lane 4, NdeI; and lane 5, XhoI.
Characterization of stability of phage.
To control the spoilage of raw milk by bacteria during low-temperature storage before pasteurization or sterilization, it is necessary for this phage to be active at a neutral pH and at low temperatures. In addition, the phage must be deactivated during pasteurization (63 to 65°C for 30 min), along with the bacteria. To confirm that the phage HU1 meets the above criteria, we determined the thermostability, pH stability, and plaque-forming ability at low temperatures.
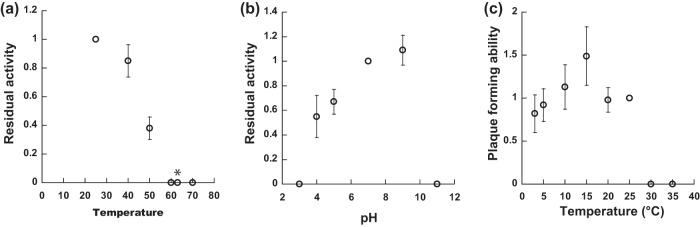
First, to elucidate the thermostability, we exposed HU1 to 25, 40, 50, 60, 63, and 70°C for 1 h (only 30 min at 63°C) and measured the titer. The remaining activities of HU1 at 40 and 50°C were approximately 80 and 40%, respectively, and no plaque was observed above 60°C (Fig. 4a). Second, to elucidate the pH stability, we exposed this phage to pHs of 3, 4, 5, 7, 9, and 11 for 1 h and measured the titer. The residual activity decreased below pH 7 and above pH 9 (Fig. 4b). At pHs of 4 to 5, the residual activity was 55% and 67%, respectively, and no plaque was observed at pHs of 3 or 11. Third, we measured plaque-forming ability at 3, 5, 10, 15, 20, 25, 30, and 35°C. We observed plaque-forming abilities from 3 to 25°C, but not at 30 and 35°C (Fig. 4c). Although this phage showed the greatest plaque-forming ability around 15°C, this phage could amplify at 3°C. These results indicated that HU1 is sufficiently active in raw milk under low-temperature storage conditions in which the temperature is about 3°C and the pH is around 7. HU1 is also sufficiently inactivated under pasteurization conditions.
FIG 4.
Characterization of phage HU1. (a) Thermal stability of HU1. Each residual activity was normalized with that of 25°C, as its value was 1.0. The values of 60, 63, and 70°C were below the detection limit (10−5). The asterisk represents the thermostability of HU1 at 63°C when incubation was conducted for only 30 min, corresponding to the conditions of pasteurization. (b) pH stability of HU1. Each residual activity was normalized with that of pH = 7, as the value was 1.0. The values of pH = 3 and pH = 11 are below the detection limit (10−3). (c) Plaque-forming abilities of HU1 at each temperature, normalized with that at 25°C, as the value was 1.0. The values at 30 and 35°C were below the detection limit (10−5). Data are expressed as average values ± SD (n = 3).
One-step growth experiment.
To investigate the latent period (i.e., the duration between phage infection and progeny release) and the burst size (i.e., the number of progeny phages per infected cell), we conducted one-step growth experiments at 25°C. HU1 was added during the logarithmic growth phase of YT4. Thirty minutes after infection, we used a 10−4 dilution for the solution to avoid the infection of uninfected YT4 by progeny phages. We monitored the infection center after dilution and calculated the latent period and the burst size by fitting a 4-parameter logistic curve (Fig. 5). The latent period was 105 ± 5 min and the burst size was 80 ± 16 progeny phages/infected cell at 25°C.
FIG 5.
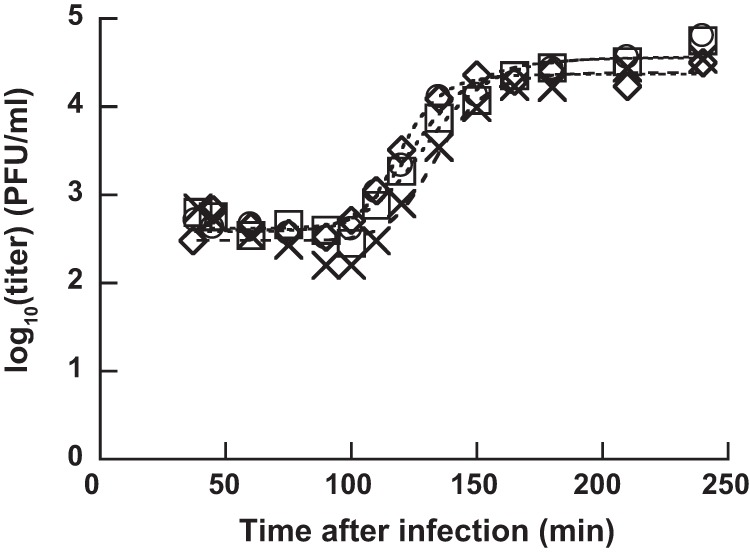
One-step growth curve of phage HU1 on P. lactis YT4. Four independent experiments were carried out at 25°C. Square, circle, diamond, and cross indicate independent replicates. The dashed curve indicates the four-parameter logistic curve fitted to a nonlinear regression curve.
Suppression of Pseudomonas growth in milk.
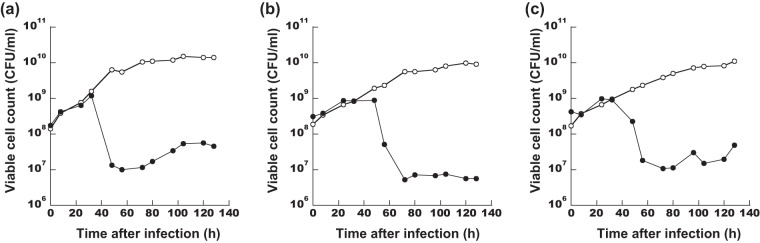
To investigate whether the phage suppressed P. lactis growth in milk under cold storage conditions (∼3°C), we monitored the decrease in viable cell numbers by adding HU1 into calcium-magnesium-LB (CM-LB) broth, skim milk, and whole milk (Fig. 6). Without phage infection, P. lactis YT4 cell numbers increased up to approximately 1 × 1010 CFU/ml in all treatments for CM-LB broth, skim milk, and whole milk. In contrast, although YT4 cell numbers continued to increase until ∼32 or 48 h after the addition of phage HU1, cell numbers gradually began to decrease in all treatments. In CM-LB broth, skim milk, and whole milk, viable cell numbers treated with phage decreased by 1,000-fold compared to those without phage treatment, and viable cell numbers remained low until 128 h (about 5 days) after infection (Fig. 6a, b, and c). These results clearly revealed that the HU1 phage was active under cold storage conditions, killing P. lactis not only in the CM-LB broth but also in the whole milk treatment.
FIG 6.
Decrease of the P. lactis population in the milk due to phage HU1 infection. Viable cell numbers for the P. lactis YT4 strain were monitored with the phage HU1 infection (closed circles) or without the phage infection (open circles) in CM-LB broth (a), skim milk (b), and whole milk (c). For closed-circle data, phages were added at time zero. Data are expressed as average values of CFU on 2 to 6 agar plates.
DISCUSSION
In this study, we isolated a P. lactis strain, YT4, and a phage, HU1, from raw cow's milk. We showed that this phage had the ability to reduce the growth of YT4 in milk under cold storage conditions. Partial genome analysis showed that HU1 is a novel phage. P. lactis is also a recently described bacterial species belonging to the P. fluorescens subgroup (4), and this report describes a lytic phage acting on P. lactis.
Many Podoviridae phages known to infect the P. fluorescens subgroup, which is based on the phylogenic relationships represented by Mulet et al. (18), have been isolated from various environments; their whole genome sequences also have been reported (ΦGP100, SBW25Φ2, KNP, WRT, UNO-SLW1 to UNO-SLW4, φIBB-PF7A, Φ-S1, UFV-P2, etc.) (15, 16, 20–25). Their genome sizes range from approximately 39 to 50 kbp. We identified phage HU1 as a member of the family Podoviridae based on morphological characterizations using TEM; at 48 kbp, its genome size was comparable to those of others in the same family. Of those relatives, φIBB-PF7A and UFV-P2 were isolated from a sewage treatment plant from dairy industries/cow farms in Portugal and from wastewater from dairy industries in Brazil, respectively, for the purpose of controlling the growth of Pseudomonas fluorescens strains during milk processing (15, 16).
The lytic activities of the phages mentioned above were generally studied at 25 to 30°C. However, to apply phages as biocontrol agents of spoilage bacteria in raw milk and refrigerated foods, it is necessary to evaluate their activities under refrigerated conditions. Bacteriophages capable of lysing Pseudomonas strains at low temperatures (0 to 7°C) also have also been isolated, partially characterized, and named psychrophilic, psychrotrophic, or cold-active phages (26–33). Phage VSW-3 can lyse P. fluorescens and was isolated from wetlands, revealing that optimal activity at pH 7.0 decreased when the phage was exposed to temperatures exceeding 60°C, and it could propagate and generate plaques between 4 and 28°C (29). Phage VSW-3 was identified as a member of the family Podoviridae based on TEM results and complete genome sequence analysis (40,556 bp) (33).
After introducing the phage to suppress the growth of Pseudomonas in raw milk, it is necessary that the phage is inactivated by the pasteurization process used for market milk and other dairy products. For phage VSW-3, approximately 30 and 15% of initial activities remained after heat treatments at 60°C for 1 h and 70°C for 20 min, respectively (29). In contrast, the activity of phage HU1 decreased to a nondetectable level after heating at 63°C for 30 min (Fig. 4a). These data confirm a reduced need for concern for consumers and manufacturers regarding side effects due to residual activities.
The decrease in viable count of P. lactis YT4 cells in the whole milk following HU1 infection is shown in Fig. 6. The standards of bacterial limit in raw cow's milk are <1 × 105 to 3 × 105 CFU/ml in the United States (34), <1 × 105 CFU/ml in the European Union (35), and <4 × 106 cells/ml in Japan (36). Because we showed the decrease in viable count from 108 CFU/ml, further research using low-density YT4 cultures is required. However, in this study, we showed that HU1 has the ability to reduce the P. lactis YT4 strain in whole milk without being inhibited by the ingredients present in milk.
Although many studies have isolated phages from sewage derived from wastewater treatment plants or dairy plants, compost, or sediments from surface water, we obtained our phage from raw milk clarifier sludge. This confirms that raw milk clarifier sludge is also a useful source for isolating phages that can be used for various applications.
In conclusion, our study indicates that the psychrotrophic phage HU1 has the potential to improve the quality and safety of raw milk and dairy products, including those of market milk. This is because HU1 is active in a broad range of pHs (4 to 9) and temperatures (3 to 25°C) against P. lactis YT4, which is one of the predominant species that causes milk spoilage via production of lipolytic and proteolytic enzymes, has activity in milk, and is inactivated easily during the pasteurization process.
However, further research is necessary to characterize phage HU1 by complete genome sequencing and to evaluate its lysis activity against P. lactis strains other than YT4 and DSM 29167T, along with other major Pseudomonas species in raw cow's milk, including P. proteolytica and P. lundensis.
MATERIALS AND METHODS
Culture media.
Plate count agar (105463; Merck KGaA, Darmstadt, Germany) and LB broth (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl) were used to culture bacteria. Crystal violet tetrazolium (CVT) medium (2.5 g/liter yeast extract, 5 g/liter Trypticase peptone, 1 g/liter d-glucose, 1 mg/liter crystal violet, and 50 mg/liter 2,3,5-triphenyltetrazolium chloride) supplemented with 1.5% agar was used to isolate Gram-negative bacteria. CM-LB broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl, 0.4 g/liter CaCl2, 0.4 g/liter MgCl2·6H2O) and CM-LB broth supplemented with 0.6% agar (CM-LB soft agar) were used for multiplication of the phage.
Isolation and identification of bacteria.
We isolated 40 types of Gram-negative bacteria from raw cow's milk obtained from Hagiwara Dairy Product Industry Ltd. (Aomori, Japan). To isolate bacteria, we divided raw milk into two tubes, one stored at 4°C and the other stored at room temperature for 1 day. A portion of the milk was suspended and diluted with sterilized phosphate-buffered saline (PBS) (3.63 g/liter Na2HPO4·12H2O, 0.24 g/liter KH2PO4, 8 g/liter NaCl, and 0.2 g/liter KCl; pH 6.9) and cultured on CVT agar medium at room temperature (∼25°C) for 2 days. We picked up seven red-colored colonies from the 4°C sample and 33 red-colored colonies from the room temperature sample, all with different colony morphologies as distinguished visually. We designated these as YT1 to YT40.
Genomic DNA from the bacterial strain YT4 was isolated using a Quick Gene SP kit DNA tissue (SP-DT; Kurabo Industries Ltd., Osaka, Japan) according to the manufacturer's instructions. The 16S rRNA, rpoB, rpoD, and gyrB genes were amplified with the primers shown in Table 2 and with 2× Quick Taq HS Dye Mix (Toyobo, Osaka, Japan). The PCR products were purified and the sequences were analyzed by Fasmac Co., Ltd. (Kanagawa, Japan), using the primers shown in Table 2. To identify this strain, 1,276 bases were determined from the 16S rRNA gene, corresponding to positions 51 to 1326 in the nucleotide sequence (GenBank accession number D84013), and the 16S rRNA sequence was matched with previously published bacterial 16S rRNA sequences using the similarity rank program of the Ribosomal Database Project (RDP) (17). We also conducted multilocus sequence analysis (MLSA) using sequences corresponding to 3,410 bp of combined fragments from (i) the 16S rRNA gene (1,245 bp corresponding to positions 85 to 1,329 from GenBank accession number D84013), (ii) the gyrB gene (671 bp corresponding to positions 131 to 801 from GenBank accession number AB178888), (iii) the rpoD gene (642 bp corresponding to positions 79 to 720 from GenBank accession number AB039545), and (iv) the rpoB gene (852 bp corresponding to positions 8,016 to 8867 from GenBank accession number BDAA01000017) in the P. fluorescens type strain. The phylogenetic tree was constructed with these combined sequences and the available genomes of type strains from the P. fluorescens group shown in Table S1 using Molecular Evolutionary Genetics Analysis version 6 (MEGA6) (37).
Isolation of phage from raw milk clarifier sludge.
We isolated a phage from sludge that was obtained when passing raw cow's milk through a centrifugal clarifier in the Hagiwara Dairy Product Industry Ltd. plant on 7 April 2015. Samples of sludge in 1.8-ml aliquots were centrifuged at 9,057 × g and 4°C for 10 min, and the cream layer was removed with a sterilized cotton swab. The intermediate liquid layer was recovered as the phage suspension. To prepare hosts for phage enrichment, we cultured the YT1 to YT40 strains separately in LB or CM-LB broth at 25°C overnight. We mixed 10 strains by adding 10 μl of each culture into 1 ml of CM-LB broth, and then 50 μl of these bacterial mixtures plus 500 μl of phage suspension was added into 5 ml CM-LB broth and cultured overnight at 25°C with shaking. After centrifugation at 9,057 × g and 4°C for 10 min, we added 20 μl of chloroform to the 1-ml supernatant. The phage suspension was spotted on soft agar where each host bacterium from the phage enrichment procedure again was used as a host strain. To isolate the phage, a single plaque was sliced out and suspended in CM-LB broth with 2% chloroform. The isolation step was repeated again to confirm successful isolation. The phage was purified as follows. An overnight culture of YT4 was inoculated into CM-LB broth and incubated for 10 h at 25°C with agitation, and then the phage was added into the culture to obtain a final concentration of 1 × 107 PFU/ml. After the culture was incubated for another 24 h under the same conditions, supernatant was obtained by centrifugation at 3,260 × g for 10 min at 4°C. Phage particles were precipitated from the supernatant by adding polyethylene glycol 6000 (PEG 6000; final concentration of 10%) and NaCl (final concentration of 1 M) and then held on ice for 1 h, followed by centrifugation at 11,000 × g for 20 min at 4°C. The phage pellet was suspended in Tris-magnesium 1 (TM1) buffer (10 mM Tris-HCl and 0.1 mM MgCl2; pH = 7.5), and any impurities were removed by filtering successively using the 5-, 1.2-, and 0.45-μm-pore-sized syringe filters (Minisart; Sartorius, Gottingen, Germany). An equivalent volume of chloroform was added to the filtrate, and the aqueous phase was obtained after centrifugation at 3,000 × g for 15 min at 4°C. To precipitate the phage particles, the solution was split into two tubes, which were centrifuged at 100,000 × g for 3 h at 4°C. The pellets were suspended with Tris-magnesium 2 (TM2) buffer (10 mM Tris-HCl and 5 mM MgCl2; pH = 7.5), the suspensions of the two tubes were combined, and the impurities were removed by centrifugation at 18,000 × g for 10 min at 4°C. The supernatant was used as purified phage.
Morphological characterization of phage.
Aliquots of purified phage were spotted onto carbon-film-coated transmission electron microscopy grids and were negatively stained with 2% uranyl acetate. These were analyzed using an H-7600 transmission electron microscope (Hitachi, Tokyo, Japan). The electron microscopy analysis was conducted by the Hanaichi UltraStructure Research Institute (Aichi, Japan).
DNA analysis of phages.
The genome from the phage was isolated using a phage DNA isolation kit (Norgen Biotek Corporation, ON, Canada) according to the manufacturer's instructions. The purified nucleic acid was treated with XbaI, SpeI, NdeI, or XhoI. We analyzed these digests on a 0.3% agarose H (Nippon Gene, Tokyo, Japan) gel using an 80 × 100 mm gel set at 20 V for 7 h. The molecular weight was calculated using two standard curves of DNA ladder fragments, which were obtained using 3 markers, as follows: λ-StyI digestion marker, 7GT, and 8GT (Nippon Gene). The first curve was obtained by 10 fragments, ranging from 2.69 to 10.65 kbps, and the other curve was obtained by 11 fragments, ranging from 10.06 to 50.31 kbps. To search sequence similarities, fragments of approximately 4 kbp from digests with BglII were subcloned into the BamHI site of pBR322. Both ends of three different clones were sequenced using primer pBR322_f (5′-CGCAGTCAGGCACCGTGTATG-3′) or pBR322_r2 (5′-GCACCTGTCCTACGAGTTGC-3′). We carried out a BLAST search using the total 2,213-base-length sequences in the BLASTN program (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Determination of the lysis spectrum.
To determine the host specificity of the phage, we used the seven Pseudomonas strains listed in Table 1 as indicator strains. We cultured these strains overnight in LB broth at 25°C for YT4, at 28°C for P. lactis and P. paralactis, or at 30°C for the other strains, with shaking at 100 rpm. An aliquot of the overnight culture was added into the CM-LB soft agar, which was then allowed to solidify for 1 h. Then, 2 × 108 PFU of HU1 were spotted onto the agar as well. After culturing these at 25°C overnight, we observed the existence or nonexistence of plaques.
Effects of pH and temperature on phage stability and plaque formation.
To determine the pH stability of the phage, we added approximately 3 × 106 PFU of phage into 2.97 ml of Tris-sodium chloride-gelatin (TSG) buffer (pH = 3, 4, 5, 7, 9, and 11; 10 mM Tris-HCl, 150 mM NaCl, and 0.03% gelatin [pH = 7.5]) and incubated at 25°C for 1 h. To neutralize, 3.8 ml of suspended medium (SM) buffer (50 mM Tris-HCl, 100 mM NaCl, 8 mM MgSO4·7H2O, and 0.01% gelatin [pH = 7.5]) was added to the 200-μl phage solution. The neutralized phage solution was diluted with CM-LB broth and subjected to a plaque assay. The residual titer was calculated as x = Ni/NpH = 7, where Ni and NpH = 7 represent PFU exposed at each pH and at pH = 7, respectively.
To analyze the thermostability of the phage, approximately 1.5 × 107 PFU were incubated at 25, 40, 50, 60, 70, and 80°C using an aluminum block bath (ALB-120; AGC Techno Glass Co., Ltd., Shizuoka, Japan) for 1 h; the 63°C sample was incubated for 30 min. The phages were diluted with CM-LB broth and the plaque assay was conducted. The residual titer was calculated as y = Nj/Nj = 25°C, where Nj and Nj = 25°C represent PFU exposed at each temperature and at 25°C, respectively.
To analyze the effects of culture temperature on plaque formation, plaque assays were conducted at 3, 5, 10, 15, 20, 25, 30, and 35°C. The plaque-forming abilities were calculated as y = Nk/Nk = 25°C, where Nk and Nk = 25°C represent PFU cultured at each temperature and at 25°C, respectively.
The titer assay was conducted using the standard double-layer agar method (38).
One-step growth experiment.
The one-step growth experiment was conducted according to the standard method (39). In brief, 150 μl of precultured YT4 in LB broth was transferred into 15 ml of CM-LB broth and incubated overnight with shaking at 25°C. HU1 was added until a PFU/CFU ratio of 0.1 to 0.3 was obtained, and the culture was incubated in a water bath at 25°C for 30 min. After 30 min, the culture was centrifuged at 12,000× g for 1 min, and the pellet was suspended with prewarmed CM-LB broth (15 ml) to a 10−4 dilution in the broth. An aliquot taken just after dilution was designated the 30-min sample. Next, aliquots were taken at 45, 60, 75, 90, 100, 110, 120, 135, 150, 165, 180, 210, and 240 min after infection, and the titer assay was conducted according to the method mentioned above. We conducted the one-step growth experiment four times independently. The log10-transformed data were fitted to a four-parameter sigmoid curve using ImageJ (40). Nonlinear regression analysis was performed to estimate the latent period and burst size.
Determination of activity in the milk.
We transferred 200 μl of an overnight culture of the YT4 strain into two tubes of 20 ml each CM-LB, skim milk (containing 0.1% milk fat), and whole milk with more than 3.5% milk fat. These were cultured at 3°C with 100 rpm shaking for 48 h. We added 6 × 107 PFU phage into one of the two tubes and added TM2 buffer into the second tube as a control; these were further cultured for 128 h at 3°C with agitation at 100 rpm. Aliquots of the culture were taken at 0, 8, 24, 32, 48, 56, 72, 80, 96, 104, 120, and 128 h after infection, and viable cell counts were measured on agar plates using an Autoplate 4000 spiral plater (Spiral Biotech, Norwood, MA), followed by incubation overnight at 25°C.
Accession number(s).
The sequences from the Pseudomonas lactis YT4 isolate determined in this study have been deposited at the DNA Data Bank Japan (DDBJ) under the accession numbers LC333862, LC333863, LC333864, and LC333865. Phage sequences determined in this study have been deposited in the DDBJ under the accession numbers LC333943, LC333944, and LC333945.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hagiwara Dairy Product Industry Ltd. for providing raw milk and clarifier sludge. We thank M. Senda for valuable technical advice and C. Nakata for technical support. Part of this study was done at the Gene Research Center of Hirosaki University.
A.K. and T.T. designed the research; C.T., K.Y., H.T., Y.I., A.K., and T.T. carried out the experiments; C.T., Y.I., A.K., and T.T. analyzed the data; and A.K. and T.T. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00111-18.
REFERENCES
- 1.Wong NP, Jenness R, Keeney M, Marth EM. 1988. Fundamentals of dairy chemistry, 3rd ed Van Nostrand Reihold Co, New York, NY. [Google Scholar]
- 2.Quigley L, O'Sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2013. The complex microbiota of raw milk. FEMS Microbiol Rev 37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 3.von Neubeck M, Baur C, Krewinkel M, Stoeckel M, Kranz B, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M. 2015. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int J Food Microbiol 211:57–65. doi: 10.1016/j.ijfoodmicro.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 4.von Neubeck M, Huptas C, Glück C, Krewinkel M, Stoeckel M, Stressler T, Fischer L, Hinrichs J, Scherer S, Wenning M. 2017. Pseudomonas lactis sp. nov. and Pseudomonas paralactis sp. nov., isolated from bovine raw milk. Int J Syst Evol Microbiol 67:1656–1664. doi: 10.1099/ijsem.0.001836. [DOI] [PubMed] [Google Scholar]
- 5.Raats D, Offek M, Minz D, Halpern M. 2011. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol 28:465–471. doi: 10.1016/j.fm.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Rasolofo EA, St-Gelais D, LaPointe G, Roy D. 2010. Molecular analysis of bacterial population structure and dynamics during cold storage of untreated and treated milk. Int J Food Microbiol 138:108–118. doi: 10.1016/j.ijfoodmicro.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Baur C, Krewinkel M, Kranz B, von Neubeck M, Wenning M, Scherer S, Stoeckel M, Hinrichs J, Stressler T, Fischer L. 2015. Quantification of the proteolytic and lipolytic activity of microorganisms isolated from raw milk. Int Dairy J 49:23–29. doi: 10.1016/j.idairyj.2015.04.005. [DOI] [Google Scholar]
- 8.Cousin MA. 1982. Presence and activity of psychrotrophic microorganisms in milk and dairy products: a review. J Food Prot 45:172–207. doi: 10.4315/0362-028X-45.2.172. [DOI] [PubMed] [Google Scholar]
- 9.Sørhaug T, Tepaniak L. 1997. Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends Food Sci Technol. 8:35–40. doi: 10.1016/S0924-2244(97)01006-6. [DOI] [Google Scholar]
- 10.Samaržija D, Zamberlin Š, Pogačić T. 2012. Psychrotrophic bacteria and milk and dairy products quality. Mljekarstvo 62:77–95. [Google Scholar]
- 11.Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiel K. 2004. Old dogma, new tricks—21st century phage therapy. Nat Biotechnol 22:31–36. doi: 10.1038/nbt0104-31. [DOI] [PubMed] [Google Scholar]
- 13.Young R, Wang IN. 2006. Phage lysis, p 104–125. In Calendar R. (ed), The bacteriophages. Oxford University Press, New York, NY. [Google Scholar]
- 14.Guttman B, Raya R, Kutter E. 2005. Basic phage biology, p 29–66. In Kutter E, Sulakvelidze A (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 15.Sillankorva S, Neubauer P, Azeredo J. 2008. Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens. BMC Biotechnol 8:80. doi: 10.1186/1472-6750-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eller MR, Vidigal PMP, Salgado RL, Alves MP, Dias RS, da Silva CC, de Carvalho AF, Kropinski A, de Paula SO. 2014. UFV-P2 as a member of the Luz24likevirus genus: a new overview on comparative functional genome analyses of the LUZ24-like phages. BMC Genomics 15:7. doi: 10.1186/1471-2164-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulet M, Lalucat J, García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 19.Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E. 2012. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol 35:455–464. doi: 10.1016/j.syapm.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Keel C, Ucurum Z, Michaux P, Adrian M, Haas D. 2002. Deleterious impact of a virulent bacteriophage on survival and biocontrol activity of Pseudomonas fluorescens strain CHAO in natural soil. Mol Plant Microbe Interact 15:567–576. doi: 10.1094/MPMI.2002.15.6.567. [DOI] [PubMed] [Google Scholar]
- 21.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, Quail M, Smith F, Walker D, Libberton B, Fenton A, Hall N, Brockhurst MA. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowicki G, Walkowiak-Nowicka K, Zemleduch-Barylska A, Mleczko A, Frąckowiak P, Nowaczyk N, Kozdrowska E, Barylski J. 2017. Complete genome sequences of two novel autographiviruses infecting a bacterium from the Pseudomonas fluorescens group. Arch Virol 162:2907–2911. doi: 10.1007/s00705-017-3419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu G, Luhr J, Stoecklein A, Warner P, Tapprich W. 2017. Complete genome sequences of Pseudomonas fluorescens bacteriophages isolated from freshwater samples in Omaha, Nebraska. Genome Announc 5:e01501-16. doi: 10.1128/genomeA.01501-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sillankorva S, Kropinski AM, Azeredo J. 2012. Genome sequence of the broad-host-range Pseudomonas phage Φ-S1. J Virol 86:10239. doi: 10.1128/JVI.01605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelln RA, Warren RAJ. 1971. Isolation and properties of a bacteriophage lytic for a wide range of pseudomonads. Can J Microbiol 17:677–382. doi: 10.1139/m71-109. [DOI] [PubMed] [Google Scholar]
- 26.Greer GG. 1982. Psychrotrophic bacteriophages for beef spoilage pseudomonads. J Food Prot 45:1318–1325. doi: 10.4315/0362-028X-45.14.1318. [DOI] [PubMed] [Google Scholar]
- 27.Olsen RH, Metcalf ES, Todd JK. 1968. Characteristics of bacteriophages attacking psychrophilic and mesophilic pseudomonads. J Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel TR, Jackman DM. 1986. Susceptibility of psychrotrophic pseudomonads of milk origin to psychrotrophic bacteriophages. Appl Environ Microbiol 51:446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin K, Ji X, Zhang C, Ding Y, Kuang A, Zhang S, Zhang Q, Lin L, Wei Y. 2017. Isolation and characterization of wetland VSW-3, a novel lytic cold-active bacteriophage of Pseudomonas fluorescens. Can J Microbiol 63:110–118. doi: 10.1139/cjm-2016-0368. [DOI] [PubMed] [Google Scholar]
- 30.Sajben-Nagy E, Maróti G, Kredics L, Horváth B, Párducz A, Vágvölgyi C, Manczinger L. 2012. Isolation of new Pseudomonas tolaasii bacteriophages and genomic investigation of the lytic phage BF7. FEMS Microbiol Lett 332:162–169. doi: 10.1111/j.1574-6968.2012.02592.x. [DOI] [PubMed] [Google Scholar]
- 31.Whitman PA, Marshall RT. 1971. Isolation of psychrophilic bacteriophage-host systems from refrigerated food products. Appl Microbiol 22:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitman PA, Marshall RT. 1971. Characterization of two psychrophilic Pseudomonas bacteriophages isolated from ground beef. Appl Microbiol 22:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Zhang Z, Li J, Qin K, Wei Y, Zhang Q, Lin L, Ji X. 2017. Complete genome sequence of the lytic cold-active Pseudomonas fluorescens bacteriophage VSW-3 from Napahai plateau wetland. Virus Genes 53: 146–150. doi: 10.1007/s11262-016-1403-1. [DOI] [PubMed] [Google Scholar]
- 34.U.S. Food and Drug Administration. 2015. Grade “A” pasteurized milk ordinance—2015 revision. U.S. Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/Milk/UCM513508.pdf. [Google Scholar]
- 35.European Parliament and Council of the European Union. 2004. Regulation (EC) No 853/2004 of the European Parliament and of the council of 29 April 2004 laying down specific hygiene rules for on the hygiene of foodstuffs. Off J Eur Union L139:55 https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32004R0853&from=EN. [Google Scholar]
- 36.Ministry of Health and Welfare, Japan. 1951. Ordinance on milk and milk products concerning compositional standards, etc. (Ministry of Health and Welfare Ordinance No 52). Ministry of Health and Welfare, Tokyo, Japan: http://www.mhlw.go.jp/english/topics/foodsafety/dl/t-1.pdf. [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay, p 69–76. In Clokie MR, Kropinski AM (ed), Bacteriophages: methods and protocols: isolation, characterization, and interactions, vol 1 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 39.Carlson K. 2005. Working with bacteriophages: common techniques and methodological approaches, p 437–494. In Kutter E, Sulakvelidze A (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL. [Google Scholar]
- 40.Rasband WS. 1997–2016. ImageJ. U.S. National Institutes of Health, Bethesda, MD: http://rsb.info.nih.gov/ij/. [Google Scholar]
- 41.Hagi T, Sasaki K, Aso H, Nomura M. 2013. Adhesive properties of predominant bacteria in raw cow's milk to bovine mammary gland epithelial cells. Folia Microbiol (Praha) 58:515–522. doi: 10.1007/s12223-013-0240-z. [DOI] [PubMed] [Google Scholar]
- 42.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulet M, Bennasar A, Lalucat J, García-Valdés E. 2009. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol Cell Probes 23:140–147. doi: 10.1016/j.mcp.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Ait Tayeb L, Ageron E, Grimont F, Grimont PA. 2005. Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res Microbiol 156:763–773. doi: 10.1016/j.resmic.2005.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.