Abstract
The present study provides a retrospective overview of the cohort of phenylketonuria (PKU) patients in Estonia. Based on the available data, the patients clearly cluster into two distinct groups: the patients with late diagnosis and start of therapy (N = 46), who were born before 1993 when the national newborn screening programme was launched, and the screened babies (N = 48) getting their diagnoses at least in a couple of weeks after birth.
Altogether 153 independent phenylalanine hydroxylase (PAH) alleles from 92 patients were analysed in the study, wherein 80% of them were carrying the p.Arg408Trp variation, making the relative frequency of this particular variation one of the highest known. Additionally, 15 other different variations in the PAH gene were identified, each with very low incidence, providing ground for phenotypic variability and potential response to BH4 therapy. Genealogical analysis revealed some “hotspots” of the origin of the p.Arg408Trp variation, with especially high density in South-East Estonia. According to our data, the incidence of PKU in Estonia is estimated as 1 in 6,700 newborns.
Electronic supplementary material
The online version of this article (10.1007/8904_2017_61) contains supplementary material, which is available to authorized users.
Keywords: Hyperphenylalaninaemia, PAH variation spectrum, Phenylalanine hydroxylase, Phenylketonuria, Variations in Estonian population
Introduction
Hyperphenylalaninaemia (HPA) is a condition usually caused by the inability of the organism to metabolise phenylalanine (Phe). Most of the HPA cases occur due to variations in the phenylalanine hydroxylase (PAH, EC1.14.16.1) gene leading to recessively inherited metabolic disease – phenylketonuria (PKU, OMIM#261600). If untreated, high levels of Phe lead to severe intellectual disability, while early restriction of dietary protein together with the intake of Phe-free substituted protein allows undisturbed mental and physical development (Blau et al. 2010). During the last decades, also tetrahydrobiopterin (BH4) has been introduced to improve the life quality of PKU patients, or substitute dietary treatment in certain cases (Cunningham et al. 2012).
The prevalence of distinct variations in the PAH gene varies highly between populations (Zschocke 2003). While in ethnically close Finland PKU is an extremely rare condition (Guldberg et al. 1995), Estonian neighbours Latvia (Pronina et al. 2003) and Lithuania (Kasnauskiene et al. 2003) exhibit similar occurrence and variation structure, also more distant East-European countries like Poland (Bik-Multanowski et al. 2013) and Czech Republic (Reblova et al. 2013); however, oversea neighbours Sweden, Denmark and Norway exhibit much more heterogeneous spectra (Eisensmith et al. 1992; Bayat et al. 2016; Ohlsson et al. 2016).
Previously, an overview about the spectrum of PKU allele variations in Estonia has been reported in 1996 in a cohort of 34 PKU patients (Lillevali et al. 1996). Now, we are able to present information about additional 60 PKU patients. The cohort includes new well-documented cases from 1996 to 2016, as well as cases from the period 1974–1996 not included in the previous study.
The aim of the present study is to provide an updated overview of all patients with a HPA diagnosis born and/or living in Estonia since 1974, to update the spectrum of allele variations among persons with HPA, and to gain insight into geographical distribution of the most prevalent variation p.Arg408Trp. We also compare the spectrum of PAH gene variations among distinct ethnic groups in Estonia and present corrected data on PKU incidence in Estonia.
Material and Methods
Patient Group
To create the Estonian database about available HPA cases, case histories of the patients born or living in Estonia from 1974 to 2016 were selected and analysed. Data fields with the following aspects were filled: last name as an identifier, year of birth, age and Phe level at diagnosis, method of diagnosis (fluorometrical, tandem mass spectrometry, or Fölling test), highest known Phe concentration value, PAH genotype, remarks about medical condition, data about the start and continuation/discontinuation of the treatment, and genealogical data of the patient, including ethnicity of the parents and grandparents of the proband. Additionally, data about general medical/social status of the person were included.
The whole cohort was divided into two subgroups according to the year of birth: 1974–1992 and 1993–2016, i.e. before and after the initiation of national newborn screening programme for PKU in Estonia.
Genealogical Survey
Parents of the PKU patients were requested to fill a questionnaire for genealogical search. It included fields about the names, maiden names, birth dates and birthplaces of the parents and grandparents of the patients, who had at least one grandparent of Estonian ethnic origin.
Prevalence Estimation
Children born during the period from 1993 to 01.09.2016 were taken under observation for estimation of the prevalence of PAH-dependent HPA-s in Estonia. Population data of all live births from 1993 to 2015 was obtained from national agency Statistics Estonia (http://www.stat.ee). The number of screened newborns between 01.01.2016 and 01.09.2016 was added according to the data recorded in the screening laboratory of the Department of Clinical Genetics, United Laboratories of Tartu University Hospital. The data about all Estonian HPA patients were collected at the Department of Clinical Genetics, United Laboratories of Tartu University Hospital.
Mutation Analysis
Mutation analysis of the PAH gene of the probands as well as their parents, when available, was performed as described previously (Lillevali et al. 1996) or/and PCR and automated dideoxy sequencing with ABI 3130XL capillary sequencer (Applied Biosystems) of all PAH gene NM_000277.1 exons (1–13) and exon–intron boundaries. First, the presence of the prevalent p.Arg408Trp variation was checked, if missing, all PAH gene exons were sequenced completely and MLPA analysis was performed using commercially available kit SALSA®MLPA® Probemix P055-PAH (MRC-Holland).
Statistical Analysis
Statistical analysis of the genealogical data was performed with SAS software (SAS®9.2 Analytics, SAS Institute Inc.). Data were collected about the birthplaces of the grandparents of PKU patients’ parental linage carrying the p.Arg408Trp allele of Estonian ethnicity p.Arg408Trp and determined with the fidelity of county. The analysed dataset consisted of 52 multivariate independent observations corresponding to 52 observed PKU patients. The pool of known localisations contained 162 birthplaces. The number of possible carrier grandparents in each of the 15 counties and the pre-World-War-II Petseri County was normalised to the population number of Estonian ethnicity of each county. Details of the analysis are provided in Supplementary Data 1. Confidence limits to the results were obtained by bootstrap method (Efron 1981).
Results
Description of the Patient Cohort
As a result of the study, a database of 95 records was created. These clustered into two groups according to the period of diagnosis. The first group comprises of 46 patients born between 1974 and 1992, before initiating the national PKU screening programme. These patients can be characterised by delay in diagnosis, mostly at the age between 6 months and 2 years, usually these children were taken under observation after abnormal symptoms had already occurred. This cohort involves also two patients diagnosed at the age of 9 and 11 years. Two late-diagnosed adult PKU patients have escaped intellectual disability. No genetic material and further medical data are available from two emigrated probands. All available genotypes of the patients in the late-diagnosed cohort consisted of homo- or compound heterozygotes for PAH alleles with minimal residual PAH activity. Nineteen patients continue low-Phe diet in their adulthood. One patient born in 1991 initially diagnosed PKU exhibited no thriving despite low-Phe diet and was subjected to BH4 loading test, unveiling dihydropteridine reductase (DHPR) deficiency.
The second group included 48 children with PKU born since 1993, after the introduction of the screening programme (Ounap et al. 1998). These patients are characterised by early diagnosis, constant observation and medical recordings, and continuous dietary treatment. Their age at diagnosis ranges from 10 to 35 days, with an average value of 24 days. Gradual tendency of earlier diagnosis together with logistical improvement of the screening programme occurred: while the average age of diagnosis in 1993–2000 was 33 days, it decreased to 20 days in 2001–2010, and further to 17 days in 2011–2016. This cohort includes two siblings with mild HPA diagnosed in Belgium, and one foetus prenatally diagnosed and terminated. Clear majority (39) of these patients are prescribed low-Phe diet and keep it continuously. Four patients receive BH4 supplement to reduce the dietary restrictions, having p.[Arg408Trp];[Leu48Ser], p.[Arg408Trp];[Arg261Gln] and p.[Arg408Trp];[Glu390Gly] genotypes accompanied with milder decrease of PAH activity.
PAH Variation Distribution
Genotype data for 92 PKU probands (of 94) was available. Two patients have emigrated, one has died and data about only one of her alleles exist. Mutation analysis of one proband (mild PKU phenotype) has been successful for only one allele. Altogether, we were able to include 182 alleles into the study, 19 of which being related. Accordingly, the known Estonian PAH allele pool contains 153 independent alleles. Among these, 123 harboured the p.Arg408Trp variation characteristic to East-European populations, constituting 80.4% of all PKU alleles and thus being one of the highest prevalence reported (Tighe et al. 2003). The full data about PAH variations are presented in Table 1.
Table 1.
PAH variations among PKU patients in Estonia (independent alleles)
| Variation in the PAH gene | Number of independent alleles | Percentage | Ethnic origin |
|---|---|---|---|
| p.Arg408Trp c.1222C>T | 123 | 80.4 | Therein 89 Estonian 31 Slavic 3 Mixed |
| p Leu48Ser c.143T>C | 5 | 3.3 | |
| p.(?)/IVS12+1G>A c.1315+1G>A | 4 | 2.6 | |
| p.Arg261Gln c.782G>A | 3 | 2 | |
| p.Arg252Trp c.754C>T | 2 | <1 | |
| p.Glu280Lys c.838G>A | 2 | <1 | |
| p.Pro281Leu c.842C>T | 2 | <1 | |
| p.Ile306Val c.916A>G | 2 | <1 | |
| p.Ile65Thr c.194T>C | 1 | <1 | |
| p.Arg158Gln c.473G>A | 1 | <1 | |
| p.Asp222* c.663_664delAG | 1 | <1 | Armenian |
| p.Ala300Ser c.898G>T | 1 | <1 | |
| p.Ser349Pro c.1045T>C | 1 | <1 | |
| p.Glu390Gly c.1169A>G | 1 | <1 | Georgian |
| p.Gln355_Tyr356insGlyLeuGln/IVS10-11G>A c.1066-11G>A | 1 | <1 | Slavic |
| p.Ala403Val c.1208C>T | 1 | <1 | Azerbaijan |
| Unknown (not p.Arg408Trp) | 2 | 1.3 | |
| Total | 153 | 100 |
Ethnical Structure of the Patient Cohort
The ethnical structure of Estonian PKU patients resembles that of the republic. Out of 94 patients, 63 (67%) were Estonians, 24 (26%) Slavic (Russian or Ukrainian) and seven of mixed origin, including Estonian, Latvian, Armenian and Azerbaijan. This structure is highly similar to general Estonian population, which comprises mostly of Estonians (68.8%) and people of East Slavic ethnicities (27.8%), according to Statistics Estonia (http://www.stat.ee) (01.01.2016).
Phenotypic Structure of Estonian HPA Patient Cohort
Vast majority of Estonian PKU patients (87%) exhibit the classical PKU phenotype with high Phe levels if untreated and minimal or zero PAH activity. This is in good correlation with the genotypic data, as the variations p.Arg408Trp, p.Arg158Gln, p.Pro281Leu, p.Arg252Trp, p.(?)/IVS12+1G>A (Okano et al. 1991; Danecka et al. 2015) retain negligible residual PAH enzymatic activity to the mutated protein. Only nine patients exhibit mild and/or BH4-responsive PKU harbouring p.Arg261Gln, p.Ala403Val, p.Ala300Ser, p.Glu390Gly beside the p.Arg408Trp in the second allele, whereas four patients exhibited good response to BH4 supplementation and are constantly on Kuvan® treatment now. Four patients do not require treatment. Genotype/phenotype correlation of Estonian HPA patients is presented in Table 2.
Table 2.
Allelic combinations and phenotype distribution of Estonian PKU patients
| Genotype (mutations in the PAH gene) | Number of patients | Frequency (%) | Phenotype |
|---|---|---|---|
| p.[Arg408Trp];[Arg408Trp] c.1222 [C>T];[C>T] | 58 | 62 | Classical PKU |
| p.[Arg408Trp];[Leu48Ser] c.[1222C>T];[143T>C] | 4 | 4.3 | 3 classical PKU/1 BH4-responsive PKU |
| p.[Arg408Trp];[(?)/IVS12+1G>A] c.[1222C>T];[1315+1G>A] | 3 | 3.2 | Classical PKU |
| p.[Arg408Trp];[Pro281Leu] c.[1222C>T];[842C>T] | 2 | 2.1 | Classical PKU |
| p.[Arg408Trp];[Arg261Gln] c.[1222C>T];[782G>A] | 4 | 4.3 | 2 classical PKU/2 BH4-responsive PKU |
| p.[Arg408Trp];[Glu390Gly] c.[1222C>T];[1169A>G] | 1 | 1.1 | BH4-responsive PKU |
| p.[Arg158Gln];[(?)/IVS12+1G>A] c.[473>A];[1315+1G>A] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[Gln355_Tyr356insGlyLeuGln/IVS10-11G>A] c.[1222C>T];[1066-11G>A] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[Arg252Trp] c.[1222C>T];[754C>T] | 4 | 4.3 | Classical PKU |
| p.[Arg408Trp];[Asp222*] c.[1222C>T];[c.663_664delAG] | 2 | 2.1 | Classical PKU |
| p.[Arg408Trp];[Ser349Pro] c.[1222C>T];[1045T>C] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[Ile306Val] c.[1222C>T];[916A>G] | 2 | 2.1 | Mild HPA |
| p.[Arg408Trp];[Glu280Lys] c.[1222C>T];[838G>A] | 2 | 2.1 | Classical PKU |
| p.[Leu48Ser];[Glu280Lys] c.[143T>C];[838G>A] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[Ile65Thr] c.[1222C>T];[194T>C] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[Ala300Ser] c.[1222C>T];[898G>T] | 2 | 2.1 | Mild HPA |
| p.[Arg408Trp];[Ala403Val] c.[1222C>T];[1208C>T] | 1 | 1.1 | Mild PKU |
| p.[Arg408Trp];[NA] c.[1222C>T];[NA] | 1 | 1.1 | Classical PKU |
| p.[Arg408Trp];[NA] c.[1222C>T];[NA] | 1 | 1.1 | Mild PKU |
| c.[NA];[NA] (no DNA) | 2 | 2.1 | Classical PKU |
| Total | 94 | 100 |
Possible Local Origins of P.Arg408Trp Allele Distribution
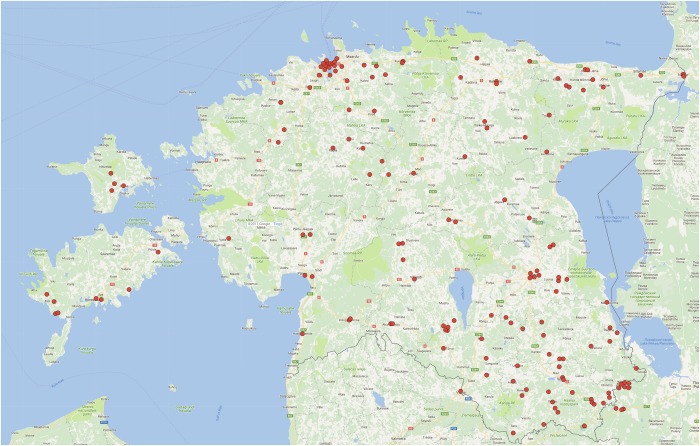
Statistical analysis of the birthplaces of the grandparents of PKU patients of Estonian ethnicity carrying the p.Arg408Trp variation revealed several counties of Estonia providing higher input of this allele into Estonian PAH variation pool. Considering the population density, the local ‘hotspot’ of p.Arg408Trp locates to three (and one former) counties of Southeastern Estonia, especially to former Petseri County, as well as Võru, Põlva and Valga Counties (3.5, 2.63, 2.04 and 2.01 carrier origins per 10,000 Estonians, respectively), while the number of p.Arg408Trp carrier origins per 10,000 Estonians for the whole country was 0.88. Relatively high input came from Estonian islands, Saaremaa and Hiiumaa, however, with wider confidence limits; a ‘hotspot’ was also found in Northeastern Ida-Viru County (2.1 carrier origins per 10,000), while Northern, Western mainland and Central Estonia had relatively few carriers in comparison with their population density (Supplementary Table 1).
The birthplaces of 160 grandparents of PKU patients were marked on a map with red dots and presented in Fig. 1. Each dot shows the origin of p.Arg408Trp with 50% probability.
Fig. 1.
Geographical distribution of the origins of the p.Arg408Trp variation in the phenylalanine hydroxylase (PAH) gene in Estonia. Birthplaces of the grandparents of PKU patients with the p.Arg408Trp variation in the PAH gene are shown with red dots. Note that each dot presents 50% probability of the grandparent being a carrier of the variation
Prevalence of PAH Deficiency
The number of live births in Estonia during 1993 to 01.09.2016 was 321,210. This number was divided with 48, the number of the second sub-cohort of HPA patients, and thus the prevalence of PAH-dependent HPA-s in Estonia was estimated as 1 in 6,700. Although some newborns miss from the screening programme due to lack of parental consent, the likelihood of missing a PKU patient from medical observation during last 24 years is negligible; therefore, total national statistics of live births was used. The two probands born in Belgium and one terminated prenatally diagnosed pregnancy were taken into account when determining the prevalence of PAH-deficient HPA-s.
Discussion
We present an updated cohort and data about the genotypic and phenotypic distribution of Estonian PKU patients. The relative frequency (80%) of the major p.Arg408Trp variation has remained among the highest across populations, with the most similar prevalence in our mainland neighbour Latvia, followed by Lithuania: 76% and 73.5%, respectively (Kasnauskiene et al. 2003; Pronina et al. 2003), gradually decreasing southward – 62% in Poland, 42% in the Czech Republic (Reblova et al. 2013), 23–27% in Germany (Aulehla-Scholz and Heilbronner 2003), and westward – 14–19% in Sweden (Ohlsson et al. 2016), being also very high (71%) in North-Western direction, the St. Petersburg region (Baranovskaya et al. 1996). The exceptionally high prevalence (84%) of this variation reported two decades ago could be considered a result of insufficient diagnostic capabilities in the past, leaving milder HPA cases out of the reach of paediatricians and clinical geneticists. However, the presence of only six PKU patients exhibiting mild HPA suggests that this could not have been prevalent. The distribution of this disease-causing allele highly resembles the total genetic structure of various European populations obtained by wide-screen analysis of neutral SNPs (Nelis et al. 2009).
Regional overviews about the diagnostic and management practices have been published lately (Gizewska et al. 2016). In 2012, national guidelines for treatment, diagnostics and management of PKU were approved in Estonia (Uudelepp et al. 2012). These are in good accordance with European guidelines (van Spronsen et al. 2017).
Analysis of the genealogical data and the birthplaces of possible carriers revealed the prevalence of p.Arg408Trp in relatively sparsely populated areas of Southern and Southeastern Estonia. It has been a subject of discussions that heterozygosity for PAH deficiency might possess selective advantage, as suggested by the incidence of PKU in distinct populations and with wide diversity in variation spectrum (Krawczak and Zschocke 2003; Zschocke 2003; Saugstad 2006). We speculate for a possible bottleneck effect or genetic drift, as the period of plagues in the seventeenth century and tremendous population loss due to the Great Northern War in the beginning of the eighteenth century may have led to the observed distribution of this particular allele.
Previous research on BH4-responsiveness has shown the need to study each patient individually, as the response may differ significantly due to particular molecular structure of the mutant PAH protein. Based on the genotypic structure of Estonian population it is not surprising that our cohort included a small number of BH4-sensitive patients, as is responsible for complete ablation of PAH enzymatic activity with no change if excess cofactor provided (Danecka et al. 2015). During the recent 6 years, every HPA newborn has undergone BH4 loading test, while the patients born earlier were not tested. Thus, three older patients with p.[Arg408Trp];[Leu48Ser] genotype are considered as classical PKU and one as BH4 responsive. Now four BH4-responsive patients have been discovered, providing them and their families better life-quality.
In conclusion, Estonia exhibits a notably homogenous pool of disease-causing PKU alleles with high prevalence of the classical severe form of PKU.
Electronics Supplementary Material
Details of statistical analysis (DOC 22 kb)
Relative impact of geographically distinct regions of Estonia (counties) to historical formation of Estonian pool of PAH alleles carrying the p.R408W mutation (DOC 42 kb)
Acknowledgements
The current study has been supported by grant PUT0355 from the Estonian Science Foundation.
Synopsis
The retrospective overview of Estonian phenylketonuria (PKU) patients revealed that the incidence of PKU in Estonia is 1 in 6,700 newborns with exceptionally high genetic homogeneity, as 80% of all PKU alleles carry the p.Arg408Trp variation typical for Eastern Europe; the genealogical part of the study disclosed certain regions of the country, where said variation was of higher prevalence compared to whole Estonia.
Author Contribution
All the listed authors have contributed to planning, conduct, and writing of the study; the statistical analysis was carried out by Dr. Tõnu Möls.
Guarantor
We declare Prof Katrin Õunap, MD, PhD as guarantor of this study.
Conflict of Interest
Hardo Lilleväli, Karit Reinson, Kai Muru, Kristi Simenson, Ülle Murumets, Tõnu Möls and Katrin Õunap declare that they have no conflict of interest. None of the authors received remunerations or honorariums of any manner or have any relationship that could inappropriately influence results. During the past 5 years the following authors have obtained reimbursement for attending a symposium/conference: K. Õunap (Biomarine, Sanofi, Shire); K. Reinson (Shire), K. Muru (Biomarine, Nutricia, Shire); K. Simenson (Shire).
Compliance with Ethical Standards
This study was approved by Research Ethics Committee of the University of Tartu (approval date 21.09.2015 number 251/T-6). The approval includes obligatory informed consent from all subjects whose personal or genealogical data has been used in the study.
Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Funding
The current study has been supported by grant PUT0355 from the Estonian Science Foundation.
Compliance with Ethics Guidelines
This study was approved by Research Ethics Committee of the University of Tartu (approval date 21.09.2015 number 251/T-6).
Contributor Information
Hardo Lilleväli, Email: hardo.lillevali@kliinikum.ee.
Collaborators: Matthias Baumgartner, Marc Patterson, Shamima Rahman, Verena Peters, Eva Morava, and Johannes Zschocke
References
- Aulehla-Scholz C, Heilbronner H. Mutational spectrum in German patients with phenylalanine hydroxylase deficiency. Hum Mutat. 2003;21:399–400. doi: 10.1002/humu.9116. [DOI] [PubMed] [Google Scholar]
- Baranovskaya S, Shevtsov S, Maksimova S, Kuzmin A, Schwartz E. The mutations and VNTRs in the phenylalanine hydroxylase gene of phenylketonuria in St Petersburg. J Inherit Metab Dis. 1996;19:705. doi: 10.1007/BF01799853. [DOI] [PubMed] [Google Scholar]
- Bayat A, Yasmeen S, Lund A, Nielsen JB, Moller LB. Mutational and phenotypical spectrum of phenylalanine hydroxylase deficiency in Denmark. Clin Genet. 2016;90:247–251. doi: 10.1111/cge.12692. [DOI] [PubMed] [Google Scholar]
- Bik-Multanowski M, Kaluzny L, Mozrzymas R, et al. Molecular genetics of PKU in Poland and potential impact of mutations on BH4 responsiveness. Acta Biochim Pol. 2013;60:613–616. [PubMed] [Google Scholar]
- Blau N, van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- Cunningham A, Bausell H, Brown M, et al. Recommendations for the use of sapropterin in phenylketonuria. Mol Genet Metab. 2012;106:269–276. doi: 10.1016/j.ymgme.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Danecka MK, Woidy M, Zschocke J, Feillet F, Muntau AC, Gersting SW. Mapping the functional landscape of frequent phenylalanine hydroxylase (PAH) genotypes promotes personalised medicine in phenylketonuria. J Med Genet. 2015;52:175–185. doi: 10.1136/jmedgenet-2014-102621. [DOI] [PubMed] [Google Scholar]
- Efron B. Nonparametric estimates of standard error: the jackknife, the bootstrap and other methods. Biometrika. 1981;68:589–599. doi: 10.1093/biomet/68.3.589. [DOI] [Google Scholar]
- Eisensmith RC, Okano Y, Dasovich M, et al. Multiple origins for phenylketonuria in Europe. Am J Hum Genet. 1992;51:1355–1365. [PMC free article] [PubMed] [Google Scholar]
- Gizewska M, MacDonald A, Belanger-Quintana A, et al. Diagnostic and management practices for phenylketonuria in 19 countries of the South and Eastern European Region: survey results. Eur J Pediatr. 2016;175:261–272. doi: 10.1007/s00431-015-2622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldberg P, Henriksen KF, Sipila I, Guttler F, de la Chapelle A. Phenylketonuria in a low incidence population: molecular characterisation of mutations in Finland. J Med Genet. 1995;32:976–978. doi: 10.1136/jmg.32.12.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasnauskiene J, Giannattasio S, Lattanzio P, Cimbalistiene L, Kucinskas V. The molecular basis of phenylketonuria in Lithuania. Hum Mutat. 2003;21:398. doi: 10.1002/humu.9113. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Zschocke J. A role for overdominant selection in phenylketonuria? Evidence from molecular data. Hum Mutat. 2003;21:394–397. doi: 10.1002/humu.10205. [DOI] [PubMed] [Google Scholar]
- Lillevali H, Ounap K, Metspalu A. Phenylalanine hydroxylase gene mutation R408W is present on 84% of Estonian phenylketonuria chromosomes. Eur J Hum Genet. 1996;4:296–300. doi: 10.1159/000472217. [DOI] [PubMed] [Google Scholar]
- Nelis M, Esko T, Magi R, et al. Genetic structure of Europeans: a view from the north-east. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson A, Bruhn H, Nordenstrom A, Zetterstrom RH, Wedell A, von Dobeln U. The spectrum of PAH mutations and increase of milder forms of phenylketonuria in Sweden during 1965–2014. JIMD Rep. 2016;34:19–26. doi: 10.1007/8904_2016_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Yoshiyuki, Eisensmith Randy C., Güttler Flemming, Lichter-Konecki Uta, Konecki David S., Trefz Friedrich K., Dasovich Mary, Wang Tao, Henriksen Karen, Lou Hans, Woo Savio L.C. Molecular Basis of Phenotypic Heterogeneity in Phenylketonuria. New England Journal of Medicine. 1991;324(18):1232–1238. doi: 10.1056/NEJM199105023241802. [DOI] [PubMed] [Google Scholar]
- Ounap K, Lillevali H, Metspalu A, Lipping-Sitska M. Development of the phenylketonuria screening programme in Estonia. J Med Screen. 1998;5:22–23. doi: 10.1136/jms.5.1.22. [DOI] [PubMed] [Google Scholar]
- Pronina N, Giannattasio S, Lattanzio P, Lugovska R, Vevere P, Kornejeva A. The molecular basis of phenylketonuria in Latvia. Hum Mutat. 2003;21:398–399. doi: 10.1002/humu.9114. [DOI] [PubMed] [Google Scholar]
- Reblova K, Hruba Z, Prochazkova D, et al. Hyperphenylalaninemia in the Czech Republic: genotype-phenotype correlations and in silico analysis of novel missense mutations. Clin Chim Acta. 2013;419:1–10. doi: 10.1016/j.cca.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Saugstad LF. From genetics to epigenetics. Nutr Health. 2006;18:285–300. doi: 10.1177/026010600601800311. [DOI] [PubMed] [Google Scholar]
- Tighe O, Dunican D, O’Neill C, et al. Genetic diversity within the R408W phenylketonuria mutation lineages in Europe. Hum Mutat. 2003;21:387–393. doi: 10.1002/humu.10195. [DOI] [PubMed] [Google Scholar]
- Uudelepp ML, Joost K, Zordania R, Õunap K. Fenüülketonuuria Eesti ravijuhend (Treatment guidelines of phenylketonuria in Estonia; in Estonian) Eesti Arst. 2012;91:46–51. [Google Scholar]
- van Spronsen FJ, van Wegberg AM, Ahring K, et al. Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 2017;5(9):743–756. doi: 10.1016/S2213-8587(16)30320-5. [DOI] [PubMed] [Google Scholar]
- Zschocke J. Phenylketonuria mutations in Europe. Hum Mutat. 2003;21:345–356. doi: 10.1002/humu.10192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of statistical analysis (DOC 22 kb)
Relative impact of geographically distinct regions of Estonia (counties) to historical formation of Estonian pool of PAH alleles carrying the p.R408W mutation (DOC 42 kb)