Abstract
It has been suggested that hepatic stellate cells (HSCs) could be used in the regulation of liver microcirculation and portal hypertension. The effects of tensile strain on the microRNA (miRNA) profile of HSCs are largely unknown. In this study, we aimed to explore the changes of miRNA expression in tensile strain-treated HSCs. The purity and activation of HSCs were determined by immunofluorescence staining with antibody against desmin and a-SMA, respectively. miRNA profile analysis was performed on HSCs with and without tensile strain treatment (n=3) using microarray analysis. We identified 6 significantly differentially expressed miRNAs (DEMs), including 1 downregulated (rno-miR-125b-2-3p) and 5 upregulated (rno-miR-1224, rho-miR-188-5p, rho-miR-211-3p, rho-miR-3584-5p and rho-miR-466b-5p), which were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) experiments. Further analysis of the DEMs revealed that many important biological processes and signal pathways were triggered in tensile strain-treated HSCs. These include the signal transduction mechanisms associated with protein binding, apoptosis, proliferation, and the FoxO and Wnt signaling pathways. In conclusion, this study presents the specific DEMs in tensile strain-treated HSCs. Our study provide novel miRNA-based information that may enhance our understanding of the pathophysiological processes leading to portal hypertension.
Keywords: microRNAs, hepatic stellate cells, portal hypertension
Introduction
Portal hypertension, defined as an increased pressure gradient between portal vein and hepatic veins, is the result of increased vascular resistance and portal blood flow (1,2). The causes of portal hypertension include pre-hepatic (e.g., arteriovenous fistula, portal or splenic vein thrombosis), intra-hepatic (e.g., sinusoidal obstruction syndrome, schistosomiasis and cirrhosis) and post-hepatic causes (e.g., inferior vena cava obstruction and Budd-Chiari syndrome), among which cirrhosis is the most common cause of portal hypertension (1,2). Hepatic stellate cells (HSCs) located in the perisinusoidal space of the liver accounts for 5–8% of all liver cells (3). HSCs have been considered as the main mediator of liver fibrosis (4–6) for years. Recent studies have reported that activated HSCs can contract or relax in response to various vasoactive substances [e.g., nitric oxide (7), carbon monoxide (8) and endothelin (9)] and suggested the involvement of HSCs in the regulation of liver microcirculation and portal hypertension.
MicroRNAs (miRNAs) are 21-to-23-nucleotide-long, non-coding RNA molecules. miRNAs bind to the 3′-untranslated region of target genes and negatively regulate their expression by mRNA cleavage or translational repression. Thus, miRNAs play an central role in the regulation of various cell functions such as cell proliferation, differentiation, and apoptosis (10). Some miRNAs, e.g., miR-19b (11), miR-29 (12), miR-126 (13), miR-132 (14), miR-150 (15) and miR-194 (15), have been found involved in HSC transdifferentiation and downregulated in fibrotic livers. Some studies have also been performed to investigate the association of miRNAs and portal hypertension. For example, miR-666 and miR-708 could suppress the expression of a water channel aquaporin-1, which promotes fibrosis and portal hypertension (16). Circulating miRNA-122 levels are found inversely correlated with portal pressure of patients with HIV/hepatitis C virus (HCV) coinfection (17). However, it is not known whether miRNAs play a role in the mechanical response of HSCs to portal hypertension.
In this study, tensile strain was applied to HSCs to mimic the mechanical force caused by portal hypertension. Microarray was used for miRNA profile analysis in HSCs with or without tensile strain. We characterized 6 significant differentially expressed miRNAs (DEMs) in response to tensile strain. The target genes of DEMs were predicted, and subjected to Gene Ontology (GO) and pathway analysis. This study may provide comprehensive information on the molecular mechanisms of portal hypertension.
Materials and methods
Isolation, culture and identification of primary HSCs
The animal experiments were approved by the Animal Care and Use Committee of the Second Military Medical University (Shanghai, China). Male Sprague-Dawley rats (10–14 weeks), weighing 400–600 g were purchased from Shanghai Experimental Animal Center (Shanghai, China). The rats were housed in a temperature controlled room (21 ± 2°C) on a 12:12-h light/dark cycle (lights on at 06:00). All rats had free access to water and food. HSCs were isolated from rat liver by in situ perfusion with 0.025% type IV collagenase and density gradient centrifugation with 12% Nycodenz (both from Sigma-Aldrich: Merck KGaA, St. Louis, MO, USA). The isolated cells (1×105 cells/cm2) were plated in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C in 5% CO2/95% air. The medium was replaced every other day. HSCs were quiescent within 2 days and completely activated at day 14. Cell survival was determined by trypan blue staining. The purity of HSCs was identified by immunofluorescence staining with anti-desmin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and anti-α-SMA (Abcam, Cambridge, MA, USA).
Application of tensile strain to HSCs and RNA extraction
Activated HSCs were plated onto collagen-coated BioFlex amino-culture plates (Flexcell International, McKeesport, PA, USA) and grown in DMEM without FBS for 24 h. HSCs were then subjected to a 10% tensile strain, 0.5 Hz for 24 h by using Flexercell FX-2000 system (Flexcell International), as previously described (18).
Total RNA was extracted from HSCs with or without tensile strain treatment by using miRVana™ RNA Isolation kit (Thermo Fisher Scientific, Inc., Rockford, IL, USA) following the manufacturer's instructions. The extracted RNA was then quantified by using NanoDrop spectrophotometer (NanoDrop Technologies: Thermo Fisher Scientific, Inc., Wilmington, DE, USA) at 260 nm.
miRNA expression analysis using miRNA array
miRNA profiling was undertaken with Agilent Rat 8×15k miRNA microarray (Agilent Technologies, Inc., Santa Clara, CA, USA) containing 765 rat miRNAs annotated in miRBase version 21.0 according to the manufacturer's instructions. In brief, 100 ng total RNA was dephosphorylated, denaturated and labeled with cyanine 3-pCp. Subsequently, samples were purified, dried with a vacuum concentrator and hybridized to the microarrays in Agilent SureHyb Chamber base at 55°C for 20 h. After washing, the slides were scanned with a scanner (Agilent Technologies, Inc.). The raw data were exported by Agilent Feature Extraction (FE) software. The threshold for significant DEMs was set as fold-change ≥2.0 and P<0.05.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
To validate the microarray data, RT-qPCR was performed. The extracted RNA was reverse transcribed using M-MLV Reverse Transcriptase (Thermo Fisher Scientific, Inc.) with a special stem-loop primer (RT primer). Subsequently, real-time PCR was performed with SYBR-Green PCR kit (Thermo Fisher Scientific, Inc.) on ABI PRISM 7500 Real-Time PCR system (Applied Biosystems: Thermo Fisher Scientific, Inc., Foster City, CA, USA) according to manufacturer's instructions. The sequences of RT and PCR primers are listed in Table I. The expression levels of miRNAs were normalized to U6 expression. Statistical differences were determined using Student's t-test with a threshold of P<0.05.
Table I.
Primer sequences for RT-qPCR.
| Systematic name | RT primers |
|---|---|
| rno-miR-188-5p | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCTCCAC-3′ |
| rno-miR-125b-2-3p | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGTCCCA-3′ |
| rno-miR-211-3p | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCCCTTT-3′ |
| Systematic name | PCR primers |
| rno-miR-188-5p | 5′-ACACTCCAGCTGGGCATCCCTTGCATGGT-3′ |
| rno-miR-125b-2-3p | 5′-ACACTCCAGCTGGGACAAGTCAGGCTCTTG-3′ |
| rno-miR-211-3p | 5′-ACACTCCAGCTGGGGGCAAGGACAGCAA-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′ |
| Universal reverse | 5′-TGGTGTCGTGGAGTCG-3′ |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Target prediction of the DEMs
We used miRNAorg (http://www.microrna.org) and TargetScan (http://www.targetscan.org/) (19) to search for mRNA targets of the DEMs. The mRNA targets that were predicted by two programs were considered.
GO and pathway analysis of predicted mRNA targets
GO and pathway analysis was performed on predicted mRNA targets of the DEM using DAVID bioinformatics resources (20) and the KEGG database (21–23), respectively. Specific biological process categories and pathways were enriched. The threshold of significance was defined by P-value (<0.05) and Benjamini-Hochberg false discovery rate (FDR-bh, <0.25).
Statistical analysis
SPSS20.0 software (SPSS Chicago, Il, USA). SPSS 19.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. Students t-test was used for comparing the variables in control and treated groups.
Results
Identification of primary HSCs
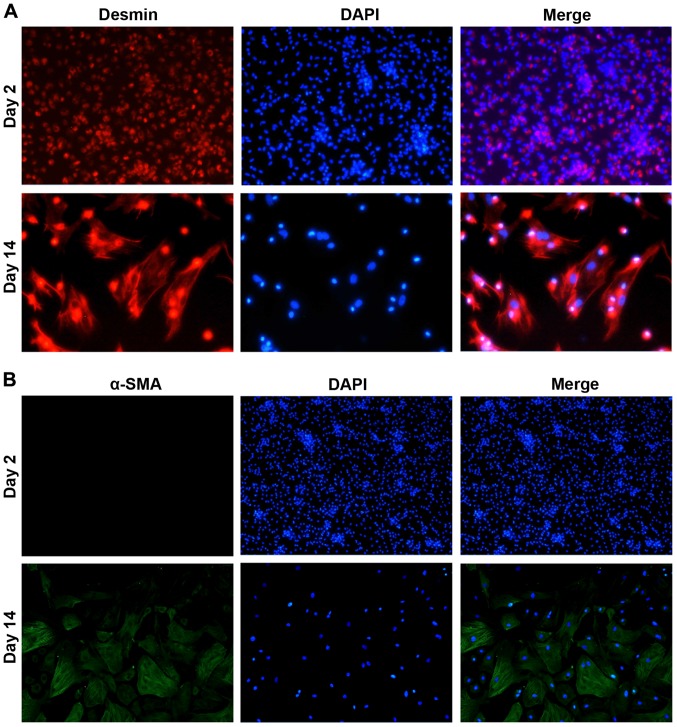
HSCs were isolated from rat liver by in situ collagenase digestion and density gradient centrifugation. The purity of HSCs was determined by immunofluorescence staining. As shown in Fig. 1, desmin, an HSC marker (24), was expressed in most cells at both day 2 and 4 after plating, suggesting the purity of HSCs. The signal of α-SMA, a marker of activated HSC (25), was not detected at day 2 and became obvious at day 14, indicating that the isolated HSCs were quiescent within 2 days and completely activated at day 14.
Figure 1.
Identification of primary cultured HSCs. Isolated HSCs were stained with (A) anti-desmin (red fluorescence) or (B) anti-α-SMA (green fluorescence) at day 2 and 14. Cell nucleus was stained with DAPI (blue fluorescence). Magnification, ×200. HSCs, hepatic stellate cells.
DEMs in response to tensile strain
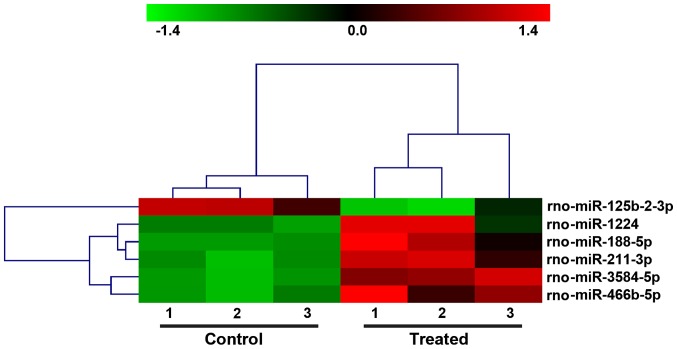
To investigate the effects of mechanical force on the miRNA profile of HSCs, tensile strain was applied to the activated HSCs, and RNA was extracted and subjected to microarray analysis. A total of 765 miRNAs was examined. With a threshold of fold-change ≥2.0 and P<0.05, we identified 6 significant DEMs, including 1 downregulated (rno-miR-125b-2-3p) and 5 upregulated (rno-miR-1224, rho-miR-188-5p, rho-miR-211-3p, rho-miR-3584-5p and rho-miR-466b-5p) (Fig. 2 and Table II) in tensile strain-treated HSCs compared with control HSCs.
Figure 2.
The DEMs differentiated the HSCs with (treated) and without tensile strain (control). The hierarchical clustering analysis is shown. DEMs, differentially expressed miRNAs; HSCs, hepatic stellate cells.
Table II.
Differentially expressed miRNAs.
| Systematic name | FC (abs) | P-value | Active sequence | miRBase accession no. |
|---|---|---|---|---|
| Upregulated | ||||
| rno-miR-1224 | 2.7449 | 0.0451 | CTCCACCTCCC | MIMAT0012827 |
| rno-miR-188-5p | 2.5143 | 0.0129 | CCCTCCACCAT | MIMAT0005301 |
| rno-miR-211-3p | 5.3486 | 0.0055 | CCCCCTTTGCTGTC | MIMAT0017157 |
| rno-miR-3584-5p | 4.5451 | 0.0003 | GCCTCCTGGACTCC | MIMAT0017875 |
| rno-miR-466b-5p | 5.6303 | 0.0071 | CATGGACATACACACACAC | MIMAT0005278 |
| Downregulated | ||||
| rno-miR-125b-2-3p | −15.5835 | 0.0131 | AGGTCCCAAGAGCCT | MIMAT0026467 |
miRNAs, microRNAs.
Validation of miRNAs microarray with RT-qPCR
To validate the microarray results, RT-qPCR was performed in tensile strain-treated HSCs and control HSCs for rno-miR-125b-2-3p, rho-miR-188-5p and rho-miR-211-3p. As shown in Fig. 3, the change trend of the 3 miRNAs detected by RT-qPCR was the same as the results of miRNA microarray.
Figure 3.
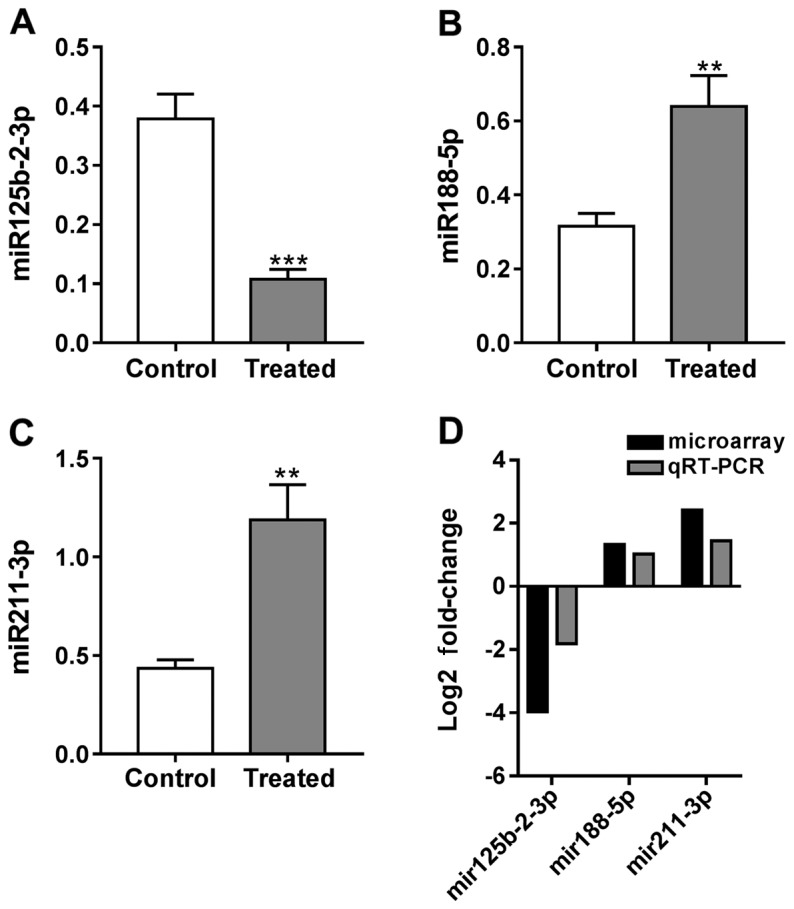
Expression of (A) rho-miR125b-2-3p, (B) rho-miR88-5p and (C) rho-mir211-3p by RT-qPCR analysis. (D) Comparison of microarray and RT-qPCR results of selected DEMs. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; DEMs, differentially expressed miRNAs. Compared with control, **P<0.01; Compared with control, ***P<0.001.
GO and pathway analysis
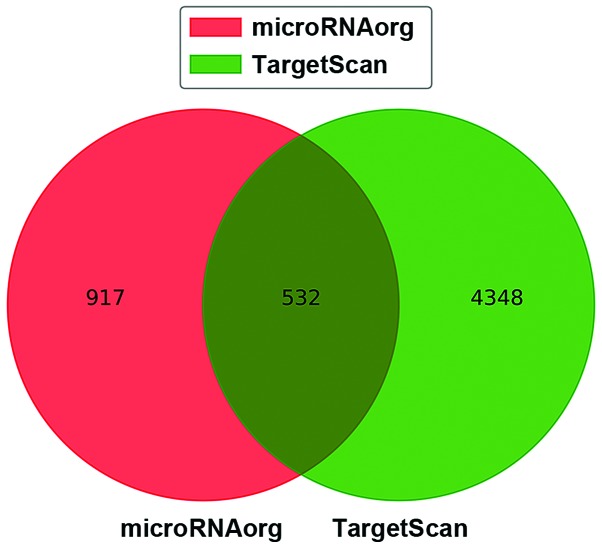
We then searched the potential target genes of the 6 DEMs by using two bioinformatic algorithms. There were 1,449 and 4,880 target genes predicted in miRNAorg and TargetScan, respectively (Fig. 4). To reduce the false positives, the target genes that were predicted in both algorihms were considered and are listed in Table III.
Figure 4.
Venn diagram of target genes predicted in miRNAorg and TargetScan.
Table III.
Target genes of DEMs.
| Systematic name | Target genes |
|---|---|
| rno-miR-1224 | Sorbs2, Il24, Pdpn, Dpp3, Kcnip2, Nupl1, Pkig, Fos, Pnkp, Maob, Fmod, Dnm1l, Ankrd1, Atf5, Ccdc47, Pde1b, Il11, Sbds, Anxa4, Tmem55a, Nsmf, Ugp2, Arpc1b, Ubac2, Nipsnap3b, B3gnt7, Abcc12, Kat7, Tmem43, Camkk1, Rhobtb2, Asrgl1, Creb3, Plin2, Sumo2, Srek1ip1, Tmem150a, Mina, Sipa1l3, Pld1, Dagla, Rgs12, Gas7, Arpc5, Capzb, Zhx1, Rfk, Ahrr, Gmcl1, Slc5a6, Arfgap2, Adgrl4, Tmem183a, Degs1, Hmgn3, Selk, Lias, Ppap2c, Gal, Cp, Cnot8, Gmpr, Tac4, Tspan3, Apln, Mapk14, Skap1, Becn1, Cd200, Vamp1, Cdc42bpa, Nudt3, Prss8, Gpha2, Smarca2, Narfl, Bcan, P2rx2, Rab15, Kpna1, Raf1, Synpr, Ndfip1, Clcn7, Gtf2i, Slc25a39, Gjb6, Smarcd2, Arl2, Pdap1, Myzap, Csf1r, Pyy, Prap1, Dync1i2, Klhl24, Syt15, Asah1, Acads, Prkag2, L1cam, Has2, Mks1, Polr1a, Parp16, Ssrp1, Ap5m1, Cacnb2, Copb1, Pole3, Wnt4, Phyhip, Psen1, Ubr7, Zc3h14, Atp6v1b2, Zmynd19, Haao, Stxbp1, Rnf135, Epn2, Fgf13, Il1rl1, Ddx21, Gulp1, Sfrp4, Ing3, Myod1, Skp1, Clip2, Trim9, Tpm3, Trim26, Sqstm1, Polm, Fgl2, Ptprr, Slc46a3, Map2k6, Fbln5, Cntn2, Klhl7, Sdccag3, Nup210, Cntn4, Lpar1, Prpf4b, Inppl1, Rnf39, Rnf185, Cdc25a, Slc30a3, Zfp422, Ik, Gtdc1, Cabs1, Calcoco1, Ucp2, Imp4, Ppil3, Tom1, Atg9a, Got2, Ppt2, Rab11a, Tor3a, Gnb3, Fut4, Chst15, Arhgap17, Rpp25, Tbrg1, Hnrnpu, Tmem106a, Clasp2, Ctsf, Insr, Tmem176b, Fkbp10, Csnk1g2, Idh3a, Stat5a, Plrg1, H3f3b, Gabarapl2, Polr3d, Amn1, Ccdc82, Uchl1, Kcnh3, Rhbg, Sdcbp2, Arfgap1, Dctn2, Arid4b, Zcchc12, Crat, Khdrbs2, Dhrs1, Tmem97, Igf2r, Dvl1, Syp, Cd276, Elavl2, Naa11, Tmem255a, Gga1, Sox10, Hyou1, Ybx3, Lrrc2, Atp6v1g2, Ralyl, Thrb |
| rno-miR-188-5p | Gjb2, Rnf146, Sp4, Tmem43, Tfb1m, Gpnmb, Elovl6, Bcl2l2, Ap5m1, Hhex, Khdrbs2, Impg2, Gimap8, Casp2, Sh2d4a, Jmjd8, Nek6, Prep, Ube2n, Efnb1, Chm, Ppp2ca, Msx1, Glra2, Crls1, Dffa, Snf8, Il13ra1, Mal2, Galnt11, Fignl1, Tomm70a, Cyb5b, Psmc3ip, Thtpa, Neurod1, Cnksr2, Glg1, Cnppd1, Nip7, Rassf1, Tspan12, Il22ra2, Arl8b, Yipf4, Emp1, Selt, Clec14a, Sumo2, Tmprss5, Tbrg1, Gpm6b, Snn, Fkrp, Lhx8, Ms4a2, Trub1, Mafb, Dynlt3, Smpd1, Extl3, Rassf5, Kcng3, Lpar1, Gnrhr, Trak2, Rras2, Cat, Pde4a, Slc22a3, Pts, Dcps, Rnf185, Sqstm1, P2rx7, Tspan11, Ptprr, Syt1, Gpc2, Smad3, Fut1, Ccdc50, Chrdl1, Thpo, Unc50, Hnf1a, Ube2i, Prps2, Pole3, Tmem50b, Klf6, Wdr45, Ing3, Zfp354a, Cdc42se1, Spef1, Rab9a, Oprm1, Cpt1a, Ccnd2, Armcx1, Tmem30a, Mgat1, Cbln2, Tlr3, Cnga2, Abcb6, Trip12, Syndig1, P4hb, Prkra, Galr1, Kitlg, Slc15a2, Ctps2, Tmem39a, Cnot4, Clmp, Ina, Fshb |
| rno-miR-466b-5p | Slc26a1, Hfe, Ccdc82, Slc25a22, Ak2, Adk, Net1, Oprk1, Rims1, Psma4, Kcnj3, Popdc2, Ret, Nupl1, Lypla1, Cnga2, Slc25a10, Slc6a3, Nxph1, Chrm5, Tagln2, Nlgn3, Gng10, Bicd2, Cabs1, Pmpcb, Foxa2, Dync1li1, Slco2a1, Fmod, Epha7, Chm, Polr3d, Nr3c1, Lppr4, Aqp4, Sp4, Fam110c, Ankrd34a, Metap2, Zhx1, Gjb4, Tmem38b, Tmx1, Adh7, Insig2, Kcnk10, Chst1, Asic4, Matr3, Capn6, Sharpin, Rbbp7, Flot2, Vsnl1, Rnd1, Gcnt2, Gpm6a, Nme8, Xkr5, Ifit2, Spry2, Il1rn, Slc35a3, Aspn, Hmgcs1, Sod3, Dek, Lztfl1, Kcnv1, Kpna4, Srr, Cacybp, Gabrb3, Coro6, Gldn, Rbl2, Has2, Pex3, Drd2, Dusp9, Tacr1, Chrnb2, Uggt1, Chn1, Fat2, Plcg1, Epcam, Cd53, Casp3, Flvcr2, Sorbs2, Slc18a2, Kcnq3, Xrcc4, Cyb5r3, Cldn1, C1galt1c1, Hsd17b12, Psmd1, Crebbp, Ntrk2, Cd38, Bcap29, Dclk1, Plscr3, Ctsd, Rassf5, Slc3a1, Ehd2, Cckbr, Armcx3, Bdkrb2, Chi3l1, Yipf6, Osgepl1, Asic2, Idi1, Nefl, Sirt6, Cyb561a3, Txn2, Slc2a13, Acsl4, Sstr1, Wscd1, Calcoco1, Tram1, P2rx1, Enpp5, Ppp1r1a, Khdrbs2, Fbxl12, Lpar1, Exoc8, Qprt, Npy5r, Rgs4, Stap1, Atad3a, Sema4f, Slc47a1, Gnb1, Ggact, Extl3, Rnls, Ube2v2, Fgl2, Gls2, Pot1, Comt, Nrg1, Arl8b, Cbwd1, Akap7, Abhd6, Cat, Bcl2, Crhbp, Chmp4c, Kat7, Acly, Brinp3, Prmt3, Bsnd, Cd81, Selt, Ctps2, Wars, Tcea1, Prim2, Btbd10, C2cd2, Gss, Strbp, Hrsp12, Tnfaip1, Opcml, Kpna1, Igfbp7, Cyp11a1, Hexim1, Mafk, Slc27a1, Foxg1, Rabl2a, Slc13a3, Prkag2, Bmp4, Rhoq, Elovl6, Atp2b1, Vps33b, Ssbp4, Nufip1, Adam7 |
DEMs, differentially expressed miRNAs.
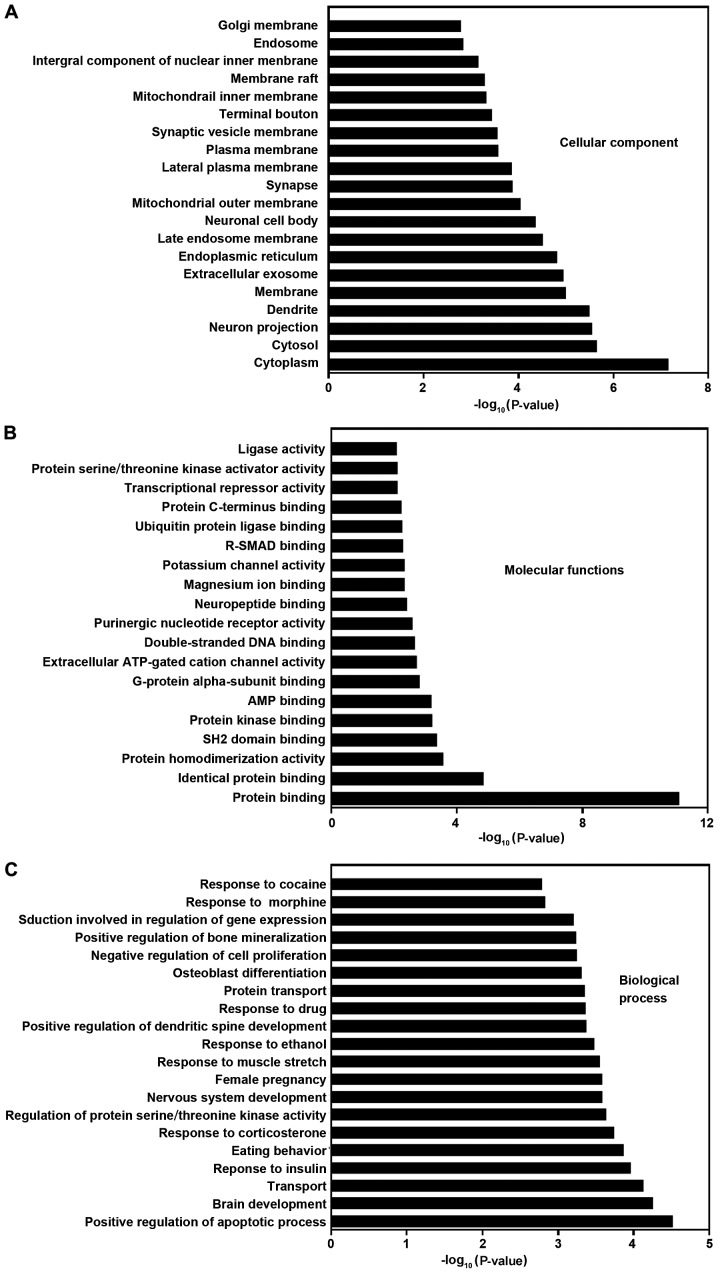
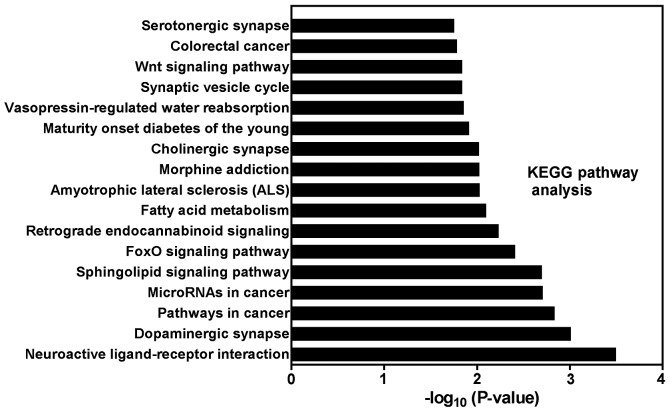
To understand the biological significance of DEMs, we conducted a functional annotation of the target genes with GO (Fig. 5) and pathway analysis (Fig. 6). By using a threshold of P<0.05 and FDR-bh<0.25, 259 GO terms were considered significant. Additionally, 64, 19 and 176 GO terms belong to cellular component, molecular function and biological process category, respectively (supplementary Tables available upon request). The target genes of DEMs were mainly enriched in the terms of membranes, protein binding, apoptosis, proliferation, response to muscle stretch and signal transduction. Through KEGG pathway analysis, we identified 17 significant KEGG pathways (Fig. 6) at a cut-off P<0.05 and FDR-bh<0.25, including diseases (pathways in cancer, colorectal cancer, miRNAs in cancer, amyotrophic lateral sclerosis, and maturity onset diabetes of the young) and signaling pathway (sphingolipid, FoxO and Wnt).
Figure 5.
GO analysis based on miRNA-targeted genes. (A) Cellular component. (B) Molecular function. (C) Biological process. The histograms of significant pathways targeted by DEMs are shown. The vertical axis is the pathway category, and the horizontal axis represents -log (P-value) of the pathways. GO, Gene Ontology; miRNA, microRNA; DEMs, differentially expressed miRNAs.
Figure 6.
Pathway analysis based on miRNA-targeted genes. The histogram of significant pathways targeted by DEMs is shown. The vertical axis is the pathway category, and the horizontal axis represents -log (P-value) of the pathways. miRNA, microRNA; DEMs, differentially expressed miRNAs.
Discussion
HSCs have been suggested to be involved in the regulation of liver microcirculation and portal hypertension. Recently, evidence has accumulated supporting that miRNAs are associated with liver diseases (26). However, there are few studies on the mechanical response of HSCs to portal hypertension. In this microarray study, we identified differentially expressed miRNAs in HSCs in response to tensile strain. We found that 6 miRNAs (1 downregulated and 5 upregulated) significantly differentially expressed. DEMs included those that were previously associated with liver diseases, such as alcoholic liver disease [miR-1224 (27)], HBV/HCV infection [miR-211 (28)], and liver cancer [miR-125b-2-3p (29) and miR-188-5p (30)], and others that are novel in liver diseases (miR-3584-5p and miR-466b-5p). miR-3584-5p (31) and miR466b-5p (32) have been found involved in S. japonicum infection and lipopolysaccharide-induced inflammation, respectively.
Moreover, GO analysis was performed on the target mRNAs of DEMs. The results showed that the target mRNAs are involved in diverse pathophysiological processes including protein binding, cell apoptosis, cell proliferation and response to muscle stretch, which may be implicated in the repose to tensile strain and the pathogenesis of portal hypertension. To reveal miRNA regulation of pathways, KEGG pathway analysis was performed based on the predicted target mRNAs of DEMs. Notably, FoxO and Wnt pathways were identified as significant. Constitutively active FoxO1 negatively regulates the proliferation and transdifferentiation of HSCs in liver fibrosis (33). Wnt signaling promotes hepatic fibrosis by enhancing HSC activation and survival (34,35). These previous reports have supported the critical role of FoxO and Wnt pathways in the pathogenesis of liver fibrosis. Our results suggested the possible involvement of these two pathways in the mechanical response of HSCs and in the pathogenesis of portal hypertension, although there is still much to learn about the underlying mechanisms.
In summary, we have shown the differentially expressed miRNA in tensile strain-treated HSCs. The results of GO and pathway analysis further provided novel miRNA-based information that may enhance our understanding of the pathophysiological processes leading to portal hypertension and may contribute to improving outcome for patients with portal hypertension.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural Science Foundation of China (General Program) (11272342).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
SY and LZ designed the study. SY, XQ and XL performed the animal experiments. YZ and ZL performed RT-qPCR. SY, XQ, XL and LZ analyzed the RT-qPCR data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Care and Use Committee of the Second Military Medical University (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Berzigotti A, Seijo S, Reverter E, Bosch J. Assessing portal hypertension in liver diseases. Expert Rev Gastroenterol Hepatol. 2013;7:141–155. doi: 10.1586/egh.12.83. [DOI] [PubMed] [Google Scholar]
- 2.Bloom S, Kemp W, Lubel J. Portal hypertension: Pathophysiology, diagnosis and management. Intern Med J. 2015;45:16–26. doi: 10.1111/imj.12590. [DOI] [PubMed] [Google Scholar]
- 3.Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311–335. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- 4.Reeves HL, Friedman SL. Activation of hepatic stellate cells - a key issue in liver fibrosis. Front Biosci. 2002;7:d808–d826. doi: 10.2741/reeves. [DOI] [PubMed] [Google Scholar]
- 5.Gäbele E, Brenner DA, Rippe RA. Liver fibrosis: Signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci. 2003;8:d69–d77. doi: 10.2741/887. [DOI] [PubMed] [Google Scholar]
- 6.Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 7.Rockey DC, Chung JJ. Reduced nitric oxide production by endothelial cells in cirrhotic rat liver: Endothelial dysfunction in portal hypertension. Gastroenterology. 1998;114:344–351. doi: 10.1016/S0016-5085(98)70487-1. [DOI] [PubMed] [Google Scholar]
- 8.Suematsu M, Goda N, Sano T, Kashiwagi S, Egawa T, Shinoda Y, Ishimura Y. Carbon monoxide: An endogenous modulator of sinusoidal tone in the perfused rat liver. J Clin Invest. 1995;96:2431–2437. doi: 10.1172/JCI118300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallowfield JA, Hayden AL, Snowdon VK, Aucott RL, Stutchfield BM, Mole DJ, Pellicoro A, Gordon-Walker TT, Henke A, Schrader J, et al. Relaxin modulates human and rat hepatic myofibroblast function and ameliorates portal hypertension in vivo. Hepatology. 2014;59:1492–1504. doi: 10.1002/hep.26627. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Huang C, Zhang SP, Sun X, Long XR, Li J. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268–2272. doi: 10.1016/j.cellsig.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300–310. doi: 10.1002/hep.25613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 13.Guo CJ, Pan Q, Xiong H, Qiao YQ, Bian ZL, Zhong W, Sheng L, Li H, Shen L, Hua J, et al. Dynamic expression of miR-126* and its effects on proliferation and contraction of hepatic stellate cells. FEBS Lett. 2013;587:3792–3801. doi: 10.1016/j.febslet.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 14.Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138(705–714):714. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, Zern MA. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebert RC, Jagavelu K, Hendrickson HI, Vasdev MM, Arab JP, Splinter PL, Trussoni CE, Larusso NF, Shah VH. Aquaporin-1 promotes angiogenesis, fibrosis, and portal hypertension through mechanisms dependent on osmotically sensitive microRNAs. Am J Pathol. 2011;179:1851–1860. doi: 10.1016/j.ajpath.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen C, Reiberger T, Huang J, Eischeid H, Schierwagen R, Mandorfer M, Anadol E, Schwabl P, Schwarze-Zander C, Warnecke-Eberz U, et al. Circulating miRNA-122 levels are associated with hepatic necroinflammation and portal hypertension in HIV/HCV coinfection. PLoS One. 2015;10:e0116768. doi: 10.1371/journal.pone.0116768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi SH, Zhang Y, Tang D, Zhu L. Mechanical force and tensile strain activated hepatic stellate cells and inhibited retinol metabolism. Biotechnol Lett. 2015;37:1141–1152. doi: 10.1007/s10529-015-1785-5. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: A comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba S, Fujii H, Hirose T, Yasuchika K, Azuma H, Hoppo T, Naito M, Machimoto T, Ikai I. Commitment of bone marrow cells to hepatic stellate cells in mouse. J Hepatol. 2004;40:255–260. doi: 10.1016/j.jhep.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Di Sario A, Bendia E, Macarri G, Candelaresi C, Taffetani S, Marzioni M, Omenetti A, De Minicis S, Trozzi L, Benedetti A. The anti-fibrotic effect of pirfenidone in rat liver fibrosis is mediated by downregulation of procollagen alpha1(I), TIMP-1 and MMP-2. Dig Liver Dis. 2004;36:744–751. doi: 10.1016/j.dld.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 29.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–885. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Han H, Peng J, Hong Y, Zhang M, Han Y, Fu Z, Shi Y, Xu J, Tao J, Lin J. Comparison of the differential expression miRNAs in Wistar rats before and 10 days after S. japonicum infection. Parasit Vectors. 2013;6:120. doi: 10.1186/1756-3305-6-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh CH, Rau CS, Jeng JC, Chen YC, Lu TH, Wu CJ, Wu YC, Tzeng SL, Yang JC. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J Biomed Sci. 2012;19:69. doi: 10.1186/1423-0127-19-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adachi M, Osawa Y, Uchinami H, Kitamura T, Accili D, Brenner DA. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology. 2007;132:1434–1446. doi: 10.1053/j.gastro.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Jiang F, Parsons CJ, Stefanovic B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J Hepatol. 2006;45:401–409. doi: 10.1016/j.jhep.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Myung SJ, Yoon JH, Gwak GY, Kim W, Lee JH, Kim KM, Shin CS, Jang JJ, Lee SH, Lee SM, et al. Wnt signaling enhances the activation and survival of human hepatic stellate cells. FEBS Lett. 2007;581:2954–2958. doi: 10.1016/j.febslet.2007.05.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.